Abstract
In mouse spermatogenesis, differentiating germ line cells initiate expression of specific genes at subsequent developmental steps. The Calmegin (Clgn) gene is first expressed in meiotic prophase, in primary spermatocytes, and encodes a protein that acts as a chaperone. To identify testis-specific transcription factors that control expression of the Clgn gene in spermatogenesis, we performed a yeast one-hybrid screening with a Clgn promoter sequence as bait DNA. This screening resulted in the identification of mouse Tcfl5 as a candidate Clgn promoter-binding protein. Tcfl5 is a member of the basic helix–loop–helix (bHLH) family of transcription factors, and mouse Tcfl5 shows 83% amino acid sequence identity with human TCFL5. Gel-shift and yeast one-hybrid experiments showed that Tcfl5 interacts with a non-canonical CACGCG site that is present in the Clgn promoter. By using northern blot, RT–PCR and in situ hybridization, mouse Tcfl5 mRNA was detected only in testis, with the highest expression level in primary spermatocytes and round spermatids. The highest level of Tcfl5 protein was found in primary spermatocytes at the diplotene stage of meiotic prophase, where the protein colocalizes with transcriptionally active chromatin.
INTRODUCTION
Development of the highly unique molecular and cellular properties of mammalian spermatozoa requires well-controlled expression of a variety of genes, many of which are expressed exclusively in spermatogenesis. The availability of the complete sequence of the mouse genome, followed by the analysis of genome-wide gene expression patterns, facilitates the identification of novel mouse testis-specific genes. Microarray analysis identified 1652 testis-specific transcripts, and it was estimated that more than 2300 genes are specifically expressed in meiotic and post-meiotic male germ cells (1). Many of these genes encode proteins that are functionally unique to the developing germ cells, or protein isoforms, which can take over functions of proteins encoded by genes that are silenced during spermatogenesis [reviewed in (2–4)].
The developmental series of subsequent steps in spermatogenesis starts with the mitotic proliferation and maintenance of spermatogonial stem cells, and the entry of differentiated spermatogonia into meiotic prophase. Progression of primary spermatocytes through meiotic prophase takes around 10 days in the mouse, and is followed by the first meiotic division resulting in the formation of haploid secondary spermatocytes, and then by the second meiotic division, which gives rise to round spermatids. Without further cell division, round spermatids engage in a long and complex differentiation process, to become spermatozoa ready to be released from supporting Sertoli cells (5,6).
For many of the testis-specific genes that are expressed in differentiating male germ line cells, a developmental time-point of first expression during spermatogenesis can be indicated. In meiotic prophase, the initiation of expression can occur when the cells progress through the early prophase steps, from preleptotene to early pachytene, but also at later prophase steps, in mid-to-late pachytene spermatocytes. Several genes expressed in meiotic prophase may encode proteins that play some role in post-meiotic differentiation of spermatids and in sperm function, rather than in meiotic prophase. An example of a gene first expressed in mid-to-late meiotic prophase, is the gene encoding Calmegin (Clgn). In spermatogenesis, the Calmegin protein (Clgn) is thought to act as a chaperone for proteins in transition from endoplasmic reticulum to the spermatid surface (7).
The initiation of expression of the Clgn gene in mid-to-late meiotic prophase (8,9) coincides with that of the genes encoding proacrosin (10), Tcp10-bt (11), testis-specific histone H2B (12), testis-specific histone H1t (13) and Zfp35 (14). Initiation of expression of different genes at the same developmental step in spermatogenesis might be controlled by first expression of a transcription factor that activates transcriptional complexes for those genes. One example of such a key transcription activator is the well-studied CREMτ, or activator CREM protein isoform (15). CREMτ has been implicated in the activation of testis-specific gene promoters that contain a cyclic AMP response element (CRE), post-meiotically in spermatids (16,17). Several other transcription factors that play an exclusive role in spermatogenesis have been identified, but an assigned role in the regulation of specific gene promoters has been described for only a very few factors [reviewed in (18,19)]. Known testis-specific transcription factors belong to several classes, including zinc finger proteins (20), homeodomain proteins (21) and basic helix–loop–helix (bHLH) proteins (22). In addition, testis-specific homologues of the general transcription factors TFIIA, TFIID, elongin and TBP have been identified (23–26).
Promoters of several testis-specific genes have been functionally analysed, including analysis of transcriptional activity in transgenic mouse models. It was observed that relatively short promoter sequences can be sufficient to drive first expression of a reporter gene at the correct spermatogenic developmental step, corresponding to initiation of expression of the respective endogenous gene. For example, testis-specific expression of the gene encoding the C-subunit of lactate dehydrogenase LDH-C4, was shown to be driven by a genomic DNA sequence containing 100 bp immediately upstream of the transcription start site (27). In addition, endogenous expression of the round spermatid-specific gene mSP-10 is reliably mimicked in transgenic mice bearing a 294 bp 5′-flanking region of the mSP-10 promoter fused to the green fluorescent protein reporter gene (28). Even smaller promoter regions that can control spermatogenic gene expression have been identified. For example, DNase footprinting identified a 14 bp sequence in the promoter of the testis-specific mouse β4-galactosyltransferase-I gene, and the expression from this promoter was lost in transgenic mice carrying a mutation in this 14 bp region (29).
Despite all knowledge about transcription factors and regulatory promoter elements in spermatogenesis, little is known about which transcription factor regulates which testis-specific promoter(s). In order to identify transcription factors that can take part in transcriptional regulation of known genes in spermatogenesis, we have used the yeast one-hybrid system (30) with a testis-specific promoter element as bait. The one-hybrid system is an in vivo genetic assay that can be used for the identification of genes encoding transcription factors that bind to a small cis-acting regulatory element.
In transgenic mice, a 330 bp sequence, containing a 152 bp putative promoter upstream of the Clgn transcription initiation site, is sufficient for testis-specific expression, and testis-specific proteins bind this promoter region in band shift assays (31). Based on this information, we selected the Clgn gene for the present experiments, and we have used the 152 bp putative promoter sequence, which was suggested to contain the cis-acting element regulating testis-specific transcription (31), as a bait in the one-hybrid screen. We identified Tcfl5 as a testis-specific transcription factor, which interacts with the promoter of the Clgn gene in meiotic prophase, in primary spermatocytes.
MATERIALS AND METHODS
Yeast one-hybrid screening
Reporter constructs were created by cloning the 152 bp sequence from the putative Clgn promoter (31) in the EcoRI–XbaI sites of pHISi-1 vector (Clontech, Palo Alto, CA) and in the EcoRI–KpnI sites of pLacZi vector (Clontech). The Clgn promoter was obtained by PCR on genomic DNA isolated from the liver of an adult male FVB mouse. Reporter strains were constructed by subsequent integration of the linearized reporter constructs in the genome of yeast strain YM4271 according to the manufacturer's instructions (Clontech). The resulting strain, CalHis/CalLacZ, was transformed with the mouse testis Matchmaker cDNA library (Clontech) and plated on the Synthetic DropOut medium lacking histidine and leucine containing 30 mM 3-amino-triazole (3-AT). Plates were incubated at 30°C for 4–6 days. Positive clones were named One Hybrid Calmegin 1–14 (OHC1–14). [His+] positive clones were assayed for blue colouring by colony-lift filter X-Gal staining (Yeast Protocols Handbook; Clontech). From [His+, LacZ+] double positives, the library plasmids were isolated using a yeast plasmid isolation kit (Bio101, Vista, CA), and these plasmids were amplified in Escherichia coli strain DH5α. To confirm the one-hybrid interaction, isolated plasmids were re-introduced into the reporter strains using the one-step yeast transformation protocol (32). Reporter constructs containing consensus or variant E-box binding sites were created by cloning trimers of these sequences in the EcoRI–XbaI sites of pHISi-1 vector and in the EcoRI–KpnI sites of pLacZi vector. Annealing complementary oligomers with added restriction site sequences yielded trimers of these binding sites. The used oligomers are as follows:
- Group A bHLH canonical E-box
- 5′-AATTCcagctgcagctgcagctgT-3′
- 5′-CTAGAcagctgcagctgcagctgG-3′
- 5′-AATTCcagctgcagctgcagctgGGTAC-3′
- 5′-CcagctgcagctgcagctgG-3′
- Group B bHLH canonical E-box
- 5′-AATTCcacgtgcacgtgcacgtgT-3′
- 5′-CTAGAcacgtgcacgtgcacgtgG-3′
- 5′-AATTCcacgtgcacgtgcacgtgGGTAC-3′
- 5′-CcacgtgcacgtgcacgtgG-3′
- Hairy-related non-canonical E-box
- 5′-AATTCcacgcgcacgcgcacgcgT-3′
- 5′-CTAGAcgcgtgcgcgtgcgcgtgG-3′
- 5′-AATTCcacgcgcacgcgcacgcgGGTAC-3′
- 5′-CcgcgtgcgcgtgcgcgtgG-3′
Reporter strains were constructed by subsequent integration of the linearized reporter constructs in the genome of yeast strain YM4271 according to the manufacturer's instructions. To test protein–DNA interaction, the resulting strains were transformed with clone OHC1 and empty pACT2 vector using the one-step yeast transformation protocol (32). Transformants were spotted on plates lacking histidine and leucine containing 7.5 mM 3-AT. After 6 days at 30°C, the spots were assayed for blue colouring by colony-lift filter X-Gal staining. For quantification of the one-hybrid interactions, overnight cultures of transformants in medium lacking leucine were assayed by the liquid culture assay using o-nitrophenyl β-d-galactopyranoside (ONPG) as substrate (Yeast Protocols Handbook; Clontech). Statistical differences in the number of β-galactosidase units per strain were calculated using the multiple comparison Tuckey test, with 95% confidence interval.
Identification of cDNAs
The fusions of the yeast Gal4AD and testis cDNAs were cloned in vector pACT2 (Clontech). Using a primer specific for the C-terminal part of the Gal4AD the sequence of the cDNA inserts was determined. Plasmid DNA of the positive one-hybrid clones was isolated using the Plasmid Maxi Kit (Qiagen, Valencia, CA). Sequences of the cDNA inserts were determined by BaseClear Labservices (Leiden, The Netherlands). To determine the identity of the sequenced cDNA inserts, searches were performed against publicly available databases NCBI Blast (http://www.ncbi.nlm.nih.gov/BLAST/), TIGR (http://www.tigr.org/), MouseBlast (http://mouseblast.informatics.jax.org/) and Ensembl (www.ensembl.org/). Clone OHC1 and OHC13 contained the full-length cDNA of the Tcfl5 gene. The complete coding sequence of this gene was determined by sequencing overlapping fragments of these inserts.
Northern blotting
A mouse Tcfl5 cDNA probe was generated by PvuII digestion on positive one-hybrid clone OHC1, yielding a 840 bp internal fragment of the Tcfl5 cDNA. A mouse tissue northern blot with poly(A)+ RNA from 15 tissues isolated from ICR mice was obtained from RNway Laboratories (Seoul, Korea). Pachytene spermatocytes and round spermatids were isolated from FVB mice using testicular cell separation by sedimentation at unit gravity (33). Total RNA from whole testis of 4- to 35-day-old FVB mice, and from isolated and separated spermatocytes and spermatids, was obtained by LiCl–urea extraction (34). RNA samples (20 μg) were separated on a 1% (w/v) agarose, 2% (v/v) formaldehyde gel, and then transferred and UV cross-linked onto Hybond N+ membrane (Amersham Pharmacia Biotech, Uppsala, Sweden).
Membranes were pre-hybridized for 1 h at 65°C in 25 ml hybridization buffer (5× SSPE, 5× Denhardt's solution and 0.5% SDS), and then randomly 32P-labelled Tcfl5 cDNA probe was added for overnight hybridization at 65°C. After hybridization, membranes were washed twice with 2× SSPE, 0.1% SDS for 10 min at room temperature, once with 1× SSPE, 0.1% SDS for 15 min at 65°C, and once with 0.1× SSPE, 0.1% SDS for 10 min at 65°C. Membranes were subjected to autoradiography.
RT–PCR
A mouse total RNA panel containing RNA from 15 tissues was obtained from Clontech. Total RNA from testis of 3–24-day-old C57bl6 mice was isolated by TRIzol RNA extraction (Life Technologies, Carlsbad, CA). An aliquot of 1 μg of total RNA was reverse transcribed using AMV reverse transcriptase (Promega, Madison, WI) in accordance with the manufacturer's protocol. For all samples, 2 μg of total RNA was used as an input in a cDNA synthesis reaction, and 1 μg oligo(dT)15 primer (Life Technologies), 1 μg pd(N)6 (Amersham Pharmacia Biotech) and 3 M betaine (Sigma–Aldrich, St Louis, MO) was added. The mixture was heated at 80°C for 10 min and quickly chilled on ice for 2 min. cDNA was synthesized in a reaction containing 50 mM Tris–HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 0.5 mM dNTPs and 400 U Superscript II RNAse H− reverse transcriptase (Life Technologies). After 1 h incubation at 37°C, the cDNA was ready for use in the PCR reaction. A second cDNA reaction without Superscript II RNase H− reverse transcriptase was performed as a control for genomic DNA contamination. For PCR amplification, gene-specific primers were synthesized (Invitrogen, Carlsbad, CA).
Tcfl5 (accession no. AY234363): 5′-CTGCCAAGCACCAGGATATTG-3′ and 5′-TTGTTGCTTTATCTGTCTCC-3′; Clgn (accession no. D14117): 5′-AGAGGGCTGGTTCTGAAGTC-3′ and 5′-TCACCAGTGTCTGCTAGCAG-3′; and Actb (accession no. M12481): 5′-GCGGACTGTTACTGAGCTG-3′ and 5′-GAAGCAATGCTGTCACCTTC-3′. The primer set for Tcfl5 spans two introns, and the primer set for Clgn spans one intron. The Tcfl5 primers yield cDNA and genomic PCR products of 455 and 2323 bp, respectively. The Clgn primers yield cDNA and genomic PCR products of 166 and 7906 bp, respectively. The Actb primer set does not span an intron, and on both cDNA and genomic DNA these primers yield a product of 448 bp. The RT–PCR products were analysed on 1% agarose gels.
Western blotting
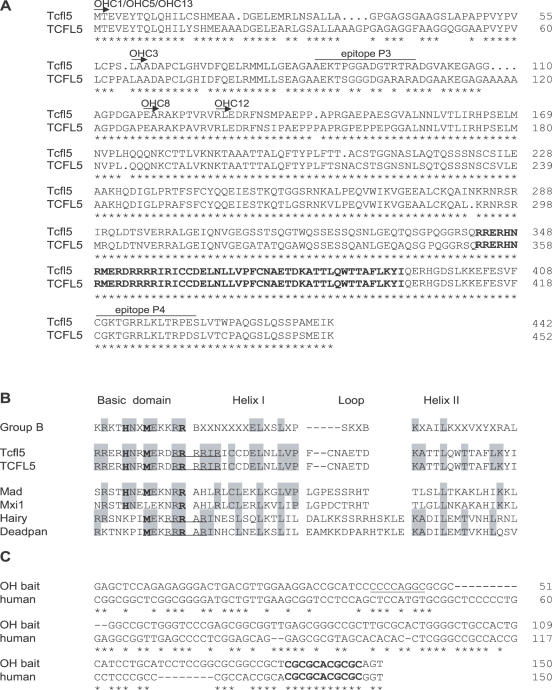
From the deduced amino acid sequence of Tcfl5, two peptides were selected to generate rabbit polyclonal antibodies (Eurogentec, Seraing, Belgium). The peptides chosen (Figure 1A) were as follows: P3 (AEKTPGGADGTRTRA, amino acids 86–98) and P4 (CGKTGRRLKLTRPES, amino acids 409–422).
Figure 1.
Alignment of bHLH proteins Tcfl5 and TCFL5. (A) Alignment of mouse Tcfl5 (accession no. AY234363) and human TCFL5 (accession no. AB012124). Arrows and clone names indicate the part of the protein that is encoded by the six positive one-hybrid clones. Both proteins show high-sequence conservation (83% identity), the bHLH domain (in boldface) at the C-terminus is 100% conserved. Asterisks indicate identical amino acid residues. The epitopes that were selected to generate antibodies against Tcfl5 are indicated (P3 and P4). (B) Alignment of Tcfl5, TCFL5 and other bHLH protein family members. Group B depicts the consensus bHLH amino acid sequence for this bHLH protein family (38). Amino acid residues at positions 5, 8 and 13 (in boldface) determine the family of bHLH proteins. Tcfl5 and TCFL5 follow the group B consensus at these positions. Mad and Mxi1 are the closest family members of Tcfl5 and TCFL5. Hairy and deadpan show similarity in arginine (R) content (underlined) with Tcfl5. (C) Alignment of the one-hybrid bait sequence and the putative human Calmegin promoter. The AP-2 site that is present in the mouse calmegin promoter is underlined and the 11 bp conserved element containing the non-canonical E-box CACGCG is indicated in boldface. Asterisks indicate identical nucleotides.
The chosen peptides are located 310 amino acids apart from each other in the protein, they are not located in the bHLH domain, and do not occur in other proteins according to the TIGR, NCBI and MouseBlast databases. Protein extracts were obtained by dounce disruption of testis or germ cells in 1 ml of 0.25 M sucrose, 1 mM EDTA, with one total protease inhibitor tablet (Roche, Basel, Switzerland). The lysate was sonicated for 30 s on ice and centrifuged at 13 000 r.p.m. at 4°C. Supernatants were aliquoted at protein concentrations of 5–10 mg/ml. Proteins (10 μg) were separated on 10% SDS–PAGE gels and transferred onto nitrocellulose filters (Schleicher and Schuell, Dassel, Germany). To detect the Tcfl5 protein, blots were incubated with 1:500 dilutions of polyclonal antibodies P3 or P4 and subsequently with 1:3000 goat-anti-rabbit second antibody coupled to horseradish peroxidase (HRP) (Sigma–Aldrich). Immobilized immunoglobins were visualized using chemoluminescence (Du Pont/NEN, Bad Homburg, Germany).
Immunohistochemistry
Testes were isolated from 28-day-old FVB mice and fixed overnight in neutral buffered formaline [4 g/l NaH2PO4·H2 8 g/l Na2HPO4·H2O and 1% (v/v) formaldehyde]. Fixed tissues were embedded in paraffin, according to the standard procedures. The 8 μm sections were dried overnight on 3-aminopropyltriethoxysilane-coated slides at 37°C. For immunological detection of Tcfl5, slides were deparaffinized and rehydrated. Sections were heated in a microwave oven for 20 min at 700 W in 0.01 M citric acid. Before incubation with antibody, slides were washed three times for 10 min in phosphate-buffered saline (PBS) and non-specific sites were blocked with 0.5% (w/v) BSA and 0.5% (w/v) milk powder in PBS. Slides were incubated with 1:100 diluted Tcfl5 antibody P3 in 10% (w/v) BSA in PBS overnight at room temperature. After washing in PBS (three times for 10 min), slides were incubated with 1:200 diluted biotinylated goat-anti-rabbit secondary antibody (DAKO, Carpinteria, CA) in 2% normal goat serum (Sigma–Aldrich) in PBS for 1 h at room temperature. Finally, slides were washed in PBS and subjected to signal detection. To detect the specific Tcfl5 signal, 1 μl streptavidin and 1 μl biotinylated HRP were diluted in 200 μl PBS (StreptABComplex; DAKO). Sections were incubated in 100 μl of this complex for 30 min at room temperature. After washing in PBS, sections were counterstained with DAB/Metal concentrate (1:10 in stable peroxide substrate buffer; Pierce, Rockford, IL) and haematoxylin.
Spread nuclei preparations of spermatocytes from 5-week-old FVB mice were prepared as described by Peters et al. (35). Immunocytochemistry was carried out with polyclonal Tcfl5 antibody P3 and monoclonal anti-SCP3 (a gift from C.Heyting, Wageningen University, The Netherlands). After blocking of non-specific sites in 0.5% (w/v) BSA and 0.5% (w/v) milk powder in PBS, slides were incubated with 1:100 diluted Tcfl5 antibody P4 and 1:1 diluted anti-SCP3 in 10% (w/v) BSA in PBS overnight in a humid chamber. Slides were washed in PBS (three times for 10 min), blocked in 10% normal goat serum in blocking buffer [supernatant of 5% (w/v) milk powder in PBS centrifuged at 14 000 r.p.m. for 10 min], and incubated with 1:800 FITC-labelled goat-anti-rabbit (Sigma–Aldrich) and 1:200 TRITC-labelled goat-anti-mouse antibodies (Sigma–Aldrich) in 10% normal goat serum in blocking buffer. After washing in PBS, slides were embedded in Vectashield containing DAPI to counterstain the DNA (Vector Laboratories, Burlingame, CA).
Images were observed using a fluorescent microscope (Axioplan 2; Carl Zeiss, Jena, Germany) equipped with a digital camera (Coolsnap-Pro; Photometrics, Waterloo, Canada). Digital images were processed using Adobe Photoshop software (Adobe Systems). The intensity of the blue and green signal in digital images of four diplotene spread nuclei was quantified using ImageJ (Wayne Randal, NIH, USA). Threshold was set to detect the upper 10% of signal, representing the visible Tcfl5 signal and intense DAPI-stained heterochromatin as observed through the microscope. Colocalization of signals was determined using the Advanced Image Microscopy software, version 3.2 (Carl Zeiss).
RNA in situ hybridization
Postnatal testes were isolated from 8-, 15- and 30-day-old FVB mice and fixed in 4% (v/v) paraformaldehyde in PBS. Fixed tissues were embedded in paraffin, according to the standard procedures. Sections were prepared and hybridized with [35S]UTP RNA probes (36). Sense and antisense Tcfl5 probes were generated by T7 transcription of BglII linearized pSG5 (Stratagene, La Jolla, CA) subclones of the original one-hybrid clone OHC1 cDNA insert. Sections were photographed using a fluorescent microscope (Axioplan 2; Carl Zeiss) equipped with a digital camera (Coolsnap-Pro).
Gel-mobility shift assay
Tcfl5 protein was synthesized using the TNT Quick Coupled Transcription/Translation System (Promega) in accordance with the manufacturer's instructions. End-labelled probes were obtained by kinase labelling of the group A, group B and non-canonical bHLH E-box oligomers. In short, 1 pmol of the sense oligomer was incubated with [γ-32P]ATP (Amersham) and T4 polynucleotide kinase (Roche) at 37°C for 30 min. Unincorporated label was removed by Sephadex G-50 gel filtration and the anti-sense oligomer was added to the eluate. After precipitation, sense and anti-sense oligomers were dissolved in 20 μl of 100 mM KCl, 10 mM Tris–HCl (pH 7.5) and 1 mM MgCl2. By submerging this solution in a 90°C water bath followed by cooling to room temperature, the oligomers were annealed. The annealed oligomers were diluted to 250 μl with 50 mM NaCl, 10 mM Tris–HCl (pH 7.5) and 1 mM MgCl2.
In binding reactions, 4 μl of labelled oligomer and 5 μl of in vitro translated Tcfl5 protein were incubated at 30°C for 30 min with 1 μg poly(dAdT) in a 15 μl vol containing 10% (v/v) glycerol, 12 mM HEPES (pH 7.9), 60 mM KCl and 0.5 mM EDTA. In supershift experiments, 1 μl of P4-αTcfl5 antibody was added to the binding reaction. Reaction mixtures were loaded onto a 5% non-denaturing polyacrylamide precast TBE gels (Bio-Rad Laboratories, Hercules, CA), and electrophoresed. Gels were dried and subjected to autoradiography.
RESULTS
One-hybrid screening
The 152 bp sequence from the Clgn promoter (31) was used as bait DNA to screen a mouse testis cDNA library for proteins that bind the Clgn promoter. Cloning a 152 bp PCR fragment in front of the yeast reporter genes HIS3 and LacZ created the reporter constructs CalHis and CalLacZ. After integration of both reporters in the genome of yeast strain YM4271, reporter strain CalHis/LacZ was transformed with the mouse testis Matchmaker cDNA library. From library screening on plates lacking histidine and leucine containing 30 mM 3-AT, we obtained 14 HIS+ positive clones (OHC1-14) from 200 000 transformants. Seven of these clones showed blue colouring in the β-Gal assay. To exclude the possibility of false positive interactions, plasmids were isolated from all positive clones, and after amplification in E.coli the plasmids were re-introduced into the CalHis/LacZ reporter strain. Transformants were re-tested for HIS3 activation and blue colouring. Six clones were again HIS+LacZ+, indicating that these six clones are true positives containing testis cDNA that encodes a protein that binds the Calmegin promoter.
Identification of Tcfl5
Using a primer specific for the pACT2 vector, the 5′ part of the selected cDNA sequences was determined. The sequences were used to screen public databases, such as NCBI Blast, MouseBlast, TIGR Gene Indices and Ensembl, to identify the corresponding genes. All six clones from the one-hybrid screening with the Clgn promoter contained cDNA or part of the cDNA representing the mouse homologue of the human gene TransCription Factor-Like 5 (TCFL5) (37). This human gene has been originally identified by differential display analysis of PCR products from mRNAs isolated from nine different human tissues, as a testis-specific cDNA encoding a 452 amino acid protein that contains a bHLH motif. Immunohistochemical staining located the gene product exclusively in cell nuclei of primary spermatocytes at the pachytene stage. It was suggested that TCFL5 may function as a transcription factor in spermatogenesis through binding to a specific DNA sequence (37).
Three clones obtained in the present one-hybrid screening (OHC1, OHC5 and OHC13) contain the complete cDNA, and the other three clones miss sequences encoding the N-terminal part of the protein but still contain the sequence that encodes the DNA-binding bHLH domain of the protein (Figure 1A).
Through sequencing clones OHC1 and OHC13, we obtained the complete sequence of mouse Tcfl5 cDNA. Comparison of this sequence with different databases (TIGR, Ensembl and NCBI BLAST) was done to check and correct the sequence. This sequence was deposited in GenBank at http://www.ncbi.nlm.nih.gov/GenBank, and is available under accession no. AY234363. The mouse Tcfl5 cDNA is 2018 bp, containing an open reading frame of 1326 bp that encodes a protein of 442 amino acid residues. Mouse Tcfl5 and human TCFL5 proteins show a high degree of sequence conservation (83% overall amino acid identity), whereas the bHLH domain located at the C-terminus is 100% identical (Figure 1A).
Database searches with the bHLH domain of Tcfl5 identified Mad and Mxi1 as the closest bHLH protein family members, showing 30% amino acid sequence identity to Tcfl5. Outside the bHLH domain of Tcfl5, no similarity was found to other bHLH family members or other proteins. Extensive analysis of the protein sequence did not identify any other functional domain.
According to the classification described by Atchley and Fitch (38), there are five groups of bHLH proteins (A–E), and the amino acids at positions 5, 8 and 13 accurately determine the groups. In Tcfl5 protein, the histidine (H), methionine (M) and arginine (R) at these positions follow the consensus of group B bHLH proteins (Figure 1B). However, Tcfl5 protein seems to be an atypical group B protein, since Tcfl5 contains a basic domain with a high number of arginine (R) residues (Figure 1B). The R residues at positions 12, 13, 14, 15 and 17 compose a basic domain that is more extended as compared with other mammalian group B bHLH proteins. The Drosophila group B proteins hairy and deadpan also contain an extended basic domain, with R residues at positions 11, 12, 13 and 15 (39,40).
Tissue-specific expression of Tcfl5
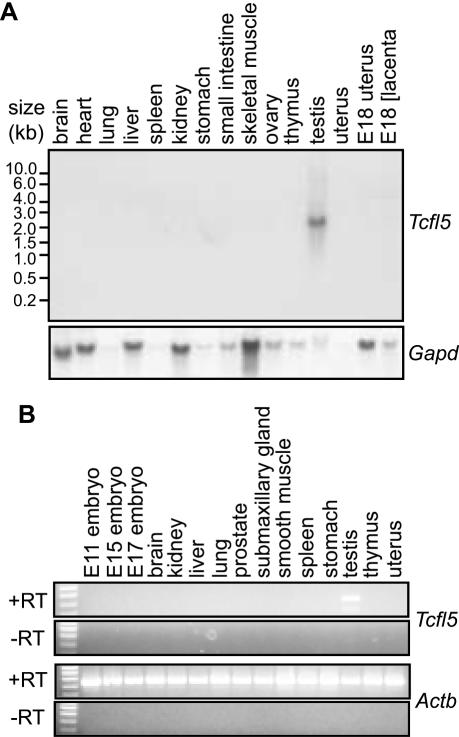
Using a northern blot containing poly(A)+ RNA from 15 mouse tissue samples, we detected a Tcfl5 mRNA species specifically in testis, whereas no detectable signal was obtained for the other tissues (Figure 2A). The length of the observed transcript, ∼2.4 kb, is similar to the single TCFL5 mRNA detected by Maruyama et al. (37). Testis-specific expression of the Tcfl5 gene was confirmed by RT–PCR analysis on a mouse total RNA panel (Figure 2B).
Figure 2.
Testis-specific expression of Tcfl5 mRNA. (A) Northern-blot analysis of poly(A)+ mRNA from 15 mouse tissues shows a specific signal in testis, while no signal was detected in any of the other tissues. Loading of the different lanes was checked by hybridization with a mouse glyceraldehyde-3-phosphate dehydrogenase (Gapd) cDNA probe. (B) RT–PCR analysis on total RNA panel from 15 tissues. The 455 bp Tcfl5 PCR product is only detected in testis. Primers specific for the ubiquitously expressed beta actin gene (Actb) were used to show that equal amounts of material were used. The 448 bp Actb PCR product is not detected in the control experiment without reverse transcriptase (−RT), ruling out contamination with genomic DNA.
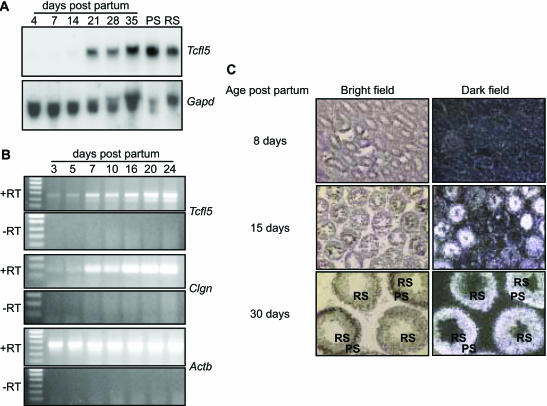
To determine at which developmental time point Tcfl5 expression is initiated, RNA samples were isolated from testis of mice of different ages, from postnatal day 4 to day 35. In newborn mice, the spermatogenic epithelium in the testis tubules consists of Sertoli cells and spermatogonia. In the first 3–5 days of postnatal testis development, the spermatogonia engage in mitotic proliferation and differentiation, from undifferentiated spermatogonia to type B spermatogonia, which undergo the final mitotic division to become preleptotene spermatocytes that go through the meiotic prophase: leptotene, zygotene, pachytene and diplotene. Early and late pachytene spermatocytes are populating the mouse testis around day 14–21 of postnatal development and concomitantly the expression level of the Tcfl5 transcript markedly increased (Figure 3A). Upon completion of the meiotic divisions, increasing numbers of haploid round and elongating spermatids appear around day 21–35 and the testicular Tcfl5 mRNA level remains high, which is explained by continued presence of Tcfl5 mRNA also in the post-meiotic cells. Tcfl5 mRNA is found in both isolated meiotic spermatocytes and in post-meiotic round spermatids (Figure 3A). It is to be noted that spermatogenesis is a continuous process, which is reinitiated from undifferentiated spermatogonia throughout adult life, so that the 35-day-old testis contains all spermatogenic cell types (5).
Figure 3.
Expression of Tcfl5 mRNA during spermatogenesis. (A) Northern-blot analysis of total RNA isolated from testis of mice of different ages (4-, 7-, 14-, 21-, 28- and 35-day-old mice) and purified spermatocytes (PS) and spermatids (RS). Only a faint signal is detected in day 4, 7 and 14 testis and the signal increases significantly between day 14 and day 21. Spermatocytes show a slightly higher expression level of Tcfl5 mRNA than spermatids. Loading of the different lanes was checked by hybridization with a Gapd cDNA. (B) Using RT–PCR, testicular RNAs from mice aged 3–24 days were analysed for Tcfl5 and Clgn expression. Both, the 455 bp Tcfl5 and the 166 bp Clgn PCR product intensified in later stages of spermatogenesis. Primers specific for the ubiquitously expressed Actb gene were used to show that equal amounts of material were used. The 448 bp Actb PCR product is not detected in the control experiment without reverse transcriptase (−RT), ruling out contamination with genomic DNA. (C) In situ hybridization on postnatal mouse testes from 8-, 15- and 30-day-old mice with an antisense Tcfl5 RNA probe shows that Tcfl5 mRNA expression starts in the spermatocytes. Little Tcfl5 mRNA signal is detected in the 8-day-old mouse testis. In the 15-day-old mouse testis, many tubule cross-sections show Tcfl5 mRNA signal, while other tubules show no signal. The cells that show the Tcfl5 mRNA signal are spermatocytes. Expression of Tcfl5 mRNA continues in spermatids as is shown in the 30-day-old mouse testis, where both spermatocytes and spermatids show the Tcfl5 mRNA signal. PS, pachytene spermatocytes; RS, round spermatids. With the sense probe no signal is visible, indicating the specificity of the antisense probe (data not shown).
RT–PCR on total RNA from testis from 3-, 5-, 7-, 10-, 16-, 20- and 24-day-old mice showed an increase in Tcfl5 mRNA signal from postnatal day 3 onwards (Figure 3B). The RT–PCR signal for Clgn mRNA also increases in testis after postnatal day 3 and the patterns of Clgn mRNA and Tcfl5 mRNA expression overlap (Figure 3B). Both Tcfl5 and Clgn RT–PCR signals show highest intensity in 20- to 24-day-old mouse testis (Figure 3B).
In situ hybridization analysis showed that Tcfl5 signal becomes detectable in 15-day-old mouse testis, in tubules that contain pachytene spermatocytes (Figure 3C). The number of tubules that contain Tcfl5 mRNA is higher in 30-day-old mouse testis and Tcfl5 mRNA was detected in both spermatocytes and spermatids (Figure 3C).
Taken together, the present results indicate that Tcfl5 expression initiates in late spermatogonia or early prophase spermatocytes and the mRNA level increases in mid-to-late pachytene spermatocytes and is maintained in round spermatids.
Tcfl5 protein expression
To study Tcfl5 protein expression, we generated antibodies against two Tcfl5 peptides. One peptide was located between amino acids 86 and 98, the other between amino acids 409 and 422 (Figure 1A). Both antibodies yielded similar results. Western blotting using total testis lysate showed a faint signal at 55 kDa, but the signal intensity increased when a lysate from isolated spermatocytes was used (Figure 4A, left panel). Competition experiments with the immunizing peptide P3 showed that the observed band is specific for Tcfl5 (Figure 4A, right panel). As described above, northern-blot analysis and in situ hybridization showed that Tcfl5 mRNA is present in spermatids. However, with western blotting, we did not detect Tcfl5 protein in cell preparations containing round spermatids (Figure 4A, left panel).
Figure 4.
Tcfl5 protein expression in mouse testis. (A) Left panel: western-blot analysis using P3-αTcfl5 antibody shows that a specific 55 kDa protein is recognized in total testis and spermatocyte protein extracts. PS, pachytene spermatocytes; RS, round spermatids; and Te, total testis. Right panel: competition experiment with immunizing peptide P3. Western blots of spermatocyte protein extracts were incubated with P3-αTcfl5 in the absence (−) or presence (+) of excess amount of peptide P3. (B) Immunohistochemical staining of Tcfl5 protein on testis sections. The upper panel shows the results obtained with P3-αTcfl5. The middle panel represents the results of the control competition experiment using the immunizing peptide P3. Tcfl5 is detected mainly in mid-to-late pachytene spermatocytes (ps), and this signal is lost in the competition experiment. The lower panel shows larger magnifications of P3-αTcfl5 immunostaining (left) and the control competition experiment (right). rs, round spermatids; es, elongating spermatids; m, metaphase spermatocytes; and roman numerals indicate the stage of the tubule cross-section. Size bar indicates 100 μm in the upper and middle panel, and 20 μm in the lower panel.
Immunohistochemistry on mouse testis sections showed that Tcfl5 protein is not present in all tubule cross-sections representing the different stages of the spermatogenic cycle (41). Stage I and stages II and III tubule cross-sections contain early prophase spermatocytes and post-meiotic round spermatids, but these stages were negative for Tcfl5 protein (data not shown). However, Tcfl5 protein is found at stages IV–XII, in mid-to-late pachytene spermatocytes (Figure 4B, showing cross-sections of different stages).
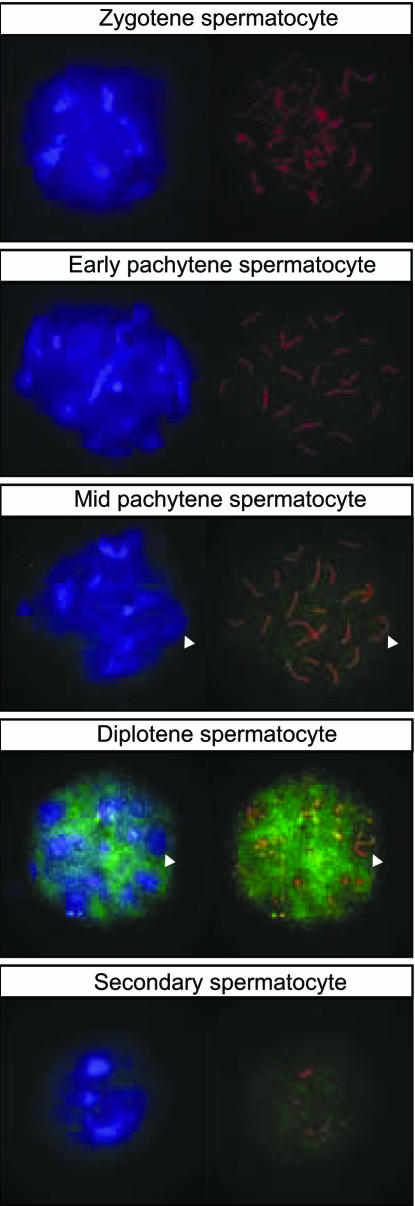
Nuclear localization of Tcfl5 was further investigated, using spread nuclei preparations of spermatocytes. To determine meiotic prophase stage in these spread nuclei, an antibody detecting the synaptonemal complex protein Sycp3 was used. Sycp3 is localized on lateral elements on the axes of paired chromosomes, and the visual appearance of these elements can be used to determine the progression of spermatocytes through meiotic prophase (42–44). This analysis showed presence of Tcfl5 protein in late pachytene primary spermatocytes, and also in secondary spermatocytes that arise from primary spermatocytes through the first meiotic division (Figure 5). Interestingly, and this was observed in particular for diplotene spermatocytes, which are near the end of meiotic prophase, less P3-αTcfl5 signal is observed in regions of the nucleus that show strong DAPI staining (Figure 5). This is also seen for the XY body, a heterochromatic nuclear region that contains the transcriptionally inactive X and Y chromosome (45). Quantitative analysis showed that ∼15% of Tcfl5 signal above threshold co-localizes with 13% of DAPI signal above threshold. Thus, Tcfl5 preferentially localizes to chromatin regions containing DAPI signal below the threshold, and these areas contain transcriptionally active DNA.
Figure 5.
Tcfl5 protein expression in nuclei of late spermatocytes. Using immunocytochemistry on spread nuclei of spermatocytes Tcfl5 protein is detected from mid-pachytene spermatocytes to secondary spermatocytes. Specificity of the antibody for Tcfl5 was verified by peptide competition assay (data not shown). Blue: DAPI DNA staining; green: P3-αTcfl5; red: αSycp3; and arrowheads indicate the XY body.
Tcfl5 preferentially binds the non-canonical sequence CACGCG
Searches in the Ensembl database with the 150 bp mouse Clgn promoter one-hybrid bait sequence, identified a putative promoter element for the human Calmegin gene (CLGN), which shows 58% sequence identity with the 150 bp mouse element. The mouse Clgn promoter contains a putative binding site for transcription factor AP-2 (31), but this site is not conserved in human (Figure 1C). All bHLH proteins recognize the canonical E-box CANNTG (46), and no such consensus site is present in the mouse one-hybrid bait sequence or the putative human CLGN promoter. However, within the homologous part of the mouse Clgn and human CLGN promoters a conserved element of 11 nt is present directly upstream of the transcription start site (Figure 1C). This element contains the non-canonical E-box sequence CACGCG, which might represent a putative binding site for Tcfl5, since the related Drosophila bHLH proteins hairy and deadpan both bind to this sequence (39,40).
DNA binding of Tcfl5 was studied using gel-mobility shift assays. In vitro translated Tcfl5 protein was found to bind both the group B bHLH canonical E-box site CACGTG and the hairy-related bHLH non-canonical E-box site CACGCG, but not the group A bHLH canonical E-box site CAGCTG (Figure 6A and B).
Figure 6.
Gel-mobility shift assay of Tcfl5 binding to bHLH E-box oligomers. (A) Reactions in lanes 1, 3 and 5 contain the in vitro translation product of the vector pGBKT7 (control), and the reactions in lanes 2, 4 and 6 contain the in vitro translated Tcfl5 protein. The Tcfl5 protein forms a complex with both the group B bHLH canonical E-box oligomer (BE) and the hairy-related bHLH non-canonical E-box oligomer (CE), whereas no retarded band is obtained with the group A bHLH canonical E-box oligomer (AE). (B) The retarded band obtained with Tcfl5 for oligomers BE and CE is supershifted by the addition of P4-αTcfl5, in lanes 3 and 6. Reactions in lanes 1 and 4 contain the in vitro translation product of the vector pGBKT7 (control).
Subsequently, we generated three different strains containing trimers of the E-box sites specific for group A, B and hairy-related bHLH proteins, fused to the HIS3 and lacZ reporter genes, named AEHis/LacZ, BEHis/LacZ and CEHis/LacZ, respectively (Table 1). Protein–DNA interactions in these strains were compared to protein–DNA interactions in strain CalHis/LacZ containing the 152 bp Clgn promoter (Table 1). All four strains were transformed with clone OHC1 expressing Tcfl5 and with the empty pACT2 vector.
Table 1. Strains constructed in this study to identify the Tcfl5 binding site.
| Strain | Promoter element |
|---|---|
| AEHis/LacZ | Trimer of CAGCTG |
| BEHis/LacZ | Trimer of CACGTG |
| CEHis/LacZ | Trimer of CACGCG |
| CalHis/LacZ | 152 bp Clgn promoter |
The one-hybrid interaction between Tcfl5 and these reporter constructs was investigated on selective medium and in colony lift X-Gal assays. Transformants were spotted on plates lacking histidine and leucine containing 7.5 mM 3-AT, which is the optimal 3-AT concentration for the E-box reporter strains. As summarized in Table 2, growth and blue colouring of reporter strains, BEHis/LacZ, CEHis/LacZ and CalHis/LacZ, indicate that Tcfl5 interacts with the promoter elements used in these strains. In contrast, absence of growth on selective medium of reporter strain AEHis/LacZ confirms that Tcfl5 is not able to interact with the canonical E-box site CAGCTG specific for group A bHLH proteins. Growth of CalHis/LacZ transformed with empty vector is due to the fact that the optimal 3-AT concentration for this reporter strain is 30 mM. Growth and blue colouring in strains CalHis/LacZ and CEHis/LacZ is stronger than in strain BEHis/LacZ, indicating that Tcfl5 is capable of binding CACGTG, but the CACGCG sequence specific for hairy-related bHLH proteins and present in the proximal Clgn promoter is the preferred binding site.
Table 2. Summary of results obtained with the transformed promoter reporter strains.
| Strain | Transformed witha | Growthb | Blue colouringc |
|---|---|---|---|
| AEHis/LacZ | Tcfl5 | − | − |
| Vector | − | − | |
| BEHis/LacZ | Tcfl5 | + | + |
| Vector | − | − | |
| CEHis/LacZ | Tcfl5 | ++ | ++ |
| Vector | − | − | |
| CalHis/LacZ | Tcfl5 | ++++ | ++++ |
| Vector | ++ | ++ |
aStrains were transformed with one-hybrid clone OHC1, expressing Tcfl5 and vector pACT2.
b‘+’ and ‘−’ indicate the different levels of growth after 6 days at 30°C on plates lacking leucine and histidine containing 7.5 mM 3-AT.
cThe difference in intensity of blue colouring in the colony-lift X-Gal assay.
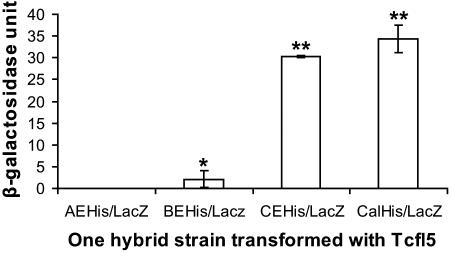
To quantify the one-hybrid interactions and to compare the relative strength of the protein–DNA interactions, we assayed liquid cultures for β-galactosidase. In this assay, Tcfl5 is able to activate the CELacZ and the CalLacZ reporters to comparable levels, but the level of activation of the group B E-box reporter is ∼15-fold lower (Figure 7). As expected, Tcfl5 cannot activate the group A E-box reporter. These results identify the Clgn promoter as a bona fide downstream target of Tcfl5.
Figure 7.
One-hybrid analysis of Tcfl5 binding site. Graphical representation of quantified one-hybrid interactions of Tcfl5 and the constructed promoter reporters. An aliquot of 1 U β-galactosidase is the amount that hydrolyses 1 μmol of ONPG to o-nitrophenol and d-galactose per minute per cell. The values represent the mean average of three independent measurements. Asteriks indicate a significant difference with reporter AEHis/LacZ (P < 0.01, Tuckey test). Double asteriks indicate that reporters CEHis/LacZ and CalHis/LacZ also are significantly different from BEHis/LacZ (P < 0.01, Tuckey test).
DISCUSSION
One-hybrid screening appears to be a powerful and useful tool to identify candidate transcription factors involved in the regulation of genes in spermatogenesis. Screening of a mouse testis cDNA library for proteins that bind the Clgn promoter resulted in the identification of a cDNA encoding the testis-specific bHLH protein Tcfl5, which is homologous to human TCFL5 (37). bHLH proteins are important gene regulatory factors, and contribute to control of many developmental pathways, such as neurogenesis, myogenesis, cell proliferation and differentiation, cell lineage determination, sex determination and other essential processes, in organisms ranging from yeast to mammals (38).
The present observations on Tcfl5 mRNA and Tcfl5 protein expression in mouse testis are in agreement with the report on TCFL5/TCFL5 expression in human testis (37). In the mouse testis, we found Tcfl5 mRNA in meiotic pachytene spermatocytes and in post-meiotic round spermatids, while the Tcfl5 protein is detected in spermatocytes, from late pachytene until the second meiotic division, but not in post-meiotic round spermatids. Tcfl5 protein expression overlaps with the onset of expression of the putative target gene Clgn in pachytene spermatocytes (9). However, a function of calmegin during meiotic prophase is not evident. In Clgn gene knockout mice, spermatogenesis is quite normal, but the Clgn deficient spermatozoa do not adhere to the zona pellucida, resulting in male infertility (7). There is impaired heterodimerization of fertilin α/β, which results in the absence of fertilin β from mature Clgn knockout sperm (47). Calmegin is thought to act as a chaperone for sperm surface proteins that mediate sperm–egg interaction, and expression of the Clgn gene in spermatocytes seems to be like a kind of foresight of the meiotic cells looking ahead to post-meiotic spermiogenesis and sperm function.
In this context, it may be of interest to note that the Factor In the Germline α gene (Figla in mouse and FIGLA in human) encodes a bHLH protein expressed in the female germ line in both mouse and human (48,49). In mice, Figla mRNA is first detected in oocytes at E13.5 and persists in adults. The Figla protein forms a heterodimer with bHLH protein E12, and regulates the expression of the oocyte-specific zona pellucida genes Zp1, Zp2 and Zp3 by binding to a conserved E-box upstream of the transcription start site (48,50). Therefore, both bHLH proteins Tcfl5 and Figla are expressed during meiotic prophase, of male and female germ cell development, respectively, whereas putative or known target genes are not involved in meiosis but function even as late as fertilization. Tcfl5 and Figla belong to different groups within the bHLH protein family, but still, the analogy in function between these two bHLH transcription factors suggests that similar mechanisms may be operative in the regulation of sex-specific aspects of male and female gametogenesis.
Numerous bHLH proteins have been identified and have been classified according to different criteria. Atchley et al. (51) defined five subgroups (A–E) based upon evolutionary conservation of several amino acid residues and DNA-binding properties. In addition, the presence or absence of a leucine zipper domain is taken into account. Later studies have defined additional conserved functional protein domains that are present in subfamilies of bHLH proteins. An example of such a domain is the orange domain adjacent to the bHLH domain in the so-called hairy-related group of bHLH proteins (52).
The basic domain of all bHLH proteins permits these proteins to bind to a consensus hexanucleotide that has been defined as the canonical E-box CANNTG. In addition, the HLH domain is important for homo- and/or heterodimerization (51). Following the criteria of Atchley and Fitch (38), we suggest that Tcfl5 is a member of the group B bHLH subfamily. However, the 152 bp putative Clgn promoter sequence that was used in the present one-hybrid screening does not contain the consensus group B hexanucleotide E-box sequence CACGTG. The present results show that Tcfl5 binds the non-canonical E-box CACGCG.
Conserved amino acid residues in the basic region determine binding site specificity of bHLH proteins. The highly conserved Glu (E) at position 9 contacts the 5′ CA nucleotides of the E-box consensus binding site (53). Studies with the group B bHLH protein Max pointed out that the conserved Arg (R) at position 13 is the key determinant for binding specificity of group B bHLH proteins. Analysis of protein–DNA co-crystals showed that R13 makes direct contact with the G nucleotide at position 4 in the group B binding site CACGTG (53). However, this sequence is not present in the Clgn promoter, and Tcfl5 preferentially binds the non-canonical CACGCG sequence. This sequence is also preferred by a subfamily of bHLH proteins, including Drosophila hairy and deadpan (39,40). These and other hairy-related bHLH proteins contain a conserved Pro (P) at position 6 of the basic region, but in Tcfl5 no Pro is found at this position. However, Tcfl5 contains an Arg at position 15 that is also found in hairy and deadpan (39,40). The presence of this Arg, and the overall high similarity of this part of the basic region of Tcfl5 to that of hairy-related bHLH proteins, suggests that this Arg might play an important role in determining the DNA-binding specificity of Tcfl5. Tcfl5 lacks the Orange domain that is typical for the hairy-related subfamily of bHLH group proteins (52), and it appears that Tcfl5 is an atypical member of the B group of bHLH proteins.
Database searches identified partial Tcfl5 expressed sequence tags (ESTs) from rat, bovine, pig, chicken and trout (data not shown). Most of these libraries were listed to be derived from reproductive tissue. Although the ESTs represent partial sequences, we found that the bHLH domain is almost completely conserved in all these species, and that the C-terminus is highly similar. Database searches also yielded information that the target gene Clgn is present and testis-specifically expressed in rat, bovine and pig (data not shown), and we would like to suggest an important role for Tcfl5 in the regulation of spermatogenesis in species ranging from fish and birds to mammals.
All bHLH proteins known to date function as homo- or heterodimers. In addition to Tcfl5, two other bHLH group family members have been found to be expressed in mouse spermatocytes. One is the hairy-related factor, Hey1, which interacts with a co-regulator Boip through its Orange domain (54). The second is a more distant family member, a germ cell-specific truncated variant of sterol regulatory element binding protein 2, which is detected in mouse spermatocytes and round spermatids (55). Future two-hybrid screens may identify these or other bHLH proteins as dimerization partners for Tcfl5. However, Tcfl5 may function as a homodimer. In the helix II domain, involved in dimerization, Tcfl5 is more similar to hairy and deadpan than to Mad and Mxi1 (Figure 1), and hairy binds DNA as a homodimer (39).
As discussed herein, timing of Tcfl5 expression in spermatogenesis would be in agreement with Tcfl5 acting as a transcriptional activator of the Clgn gene. However, it is to be noted that members of the hairy-related bHLH family act as transcriptional repressors, inhibiting transcription by binding to co-repressors, mediated by a sequence in the C-terminus (52). Tcfl5 does not contain such a co-repressor binding sequence, and we cannot predict whether Tcfl5 acts as transcriptional activator or repressor. The present finding that Tcfl5 protein is associated with transcriptionally active chromatin regions in spermatocytes would support a role as activator of transcription, possibly of several target genes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Christa Heyting (Wageningen University, The Netherlands) for the antibody targeting Sycp3, and Dr Mark Wijgerde for advice with the in situ hybridization. This work was supported by Organon N.V.
DDBJ/EMBL/GenBank accession no. AY234363
REFERENCES
- 1.Schultz N., Hamra,F.K. and Garbers,D.L. (2003) A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc. Natl Acad. Sci. USA, 100, 12201–12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baarends W.A. and Grootegoed,J.A. (1999) Molecular biology of male gametogenesis. In Fauser,B.C.J.M. (ed.), Molecular Biology in Reproductive Medicine. The Parthenon Publishing Group, NY, pp. 271–295. [Google Scholar]
- 3.Eddy E.M. (2002) Male germ cell gene expression. Recent Prog. Horm. Res., 57, 103–128. [DOI] [PubMed] [Google Scholar]
- 4.Wang P.J. (2004) X chromosomes, retrogenes and their role in male reproduction. Trends Endocrinol. Metab., 15, 79–83. [DOI] [PubMed] [Google Scholar]
- 5.Oakberg E.F. (1956) Duration of spermatogenesis in the mouse and timing of stages of the cycle of the seminiferous epithelium. Am. J. Anat., 99, 507–516. [DOI] [PubMed] [Google Scholar]
- 6.Nebel B.R., Amarose,A.P. and Hacket,E.M. (1961) Calendar of gametogenic development in the prepuberal male mouse. Science, 134, 832–833. [DOI] [PubMed] [Google Scholar]
- 7.Ikawa M., Wada,I., Kominami,K., Watanabe,D., Toshimori,K., Nishimune,Y. and Okabe,M. (1997) The putative chaperone calmegin is required for sperm fertility. Nature, 387, 607–611. [DOI] [PubMed] [Google Scholar]
- 8.Watanabe D., Sawada,K., Koshimizu,U., Kagawa,T. and Nishimune,Y. (1992) Characterization of male meiotic germ cell-specific antigen (Meg 1) by monoclonal antibody TRA 369 in mice. Mol. Reprod. Dev., 33, 307–312. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe D., Yamada,K., Nishina,Y., Tajima,Y., Koshimizu,U., Nagata,A. and Nishimune,Y. (1994) Molecular cloning of a novel Ca2+-binding protein (calmegin) specifically expressed during male meiotic germ cell development. J. Biol. Chem., 269, 7744–7749. [PubMed] [Google Scholar]
- 10.Nayernia K., Nieter,S., Kremling,H., Oberwinkler,H. and Engel,W. (1994) Functional and molecular characterization of the transcriptional regulatory region of the proacrosin gene. J. Biol. Chem., 269, 32181–32186. [PubMed] [Google Scholar]
- 11.Ewulonu U.K., Snyder,L., Silver,L.M. and Schimenti,J.C. (1996) Promoter mapping of the mouse Tcp-10bt gene in transgenic mice identifies essential male germ cell regulatory sequences. Mol. Reprod. Dev., 43, 290–297. [DOI] [PubMed] [Google Scholar]
- 12.Choi Y.C., Gu,W., Hecht,N.B., Feinberg,A.P. and Chae,C.B. (1996) Molecular cloning of mouse somatic and testis-specific H2B histone genes containing a methylated CpG island. DNA Cell Biol., 15, 495–504. [DOI] [PubMed] [Google Scholar]
- 13.Wolfe S.A., Mottram,P.J., vanWert,J.M. and Grimes,S.R. (1999) Localization of upstream elements involved in transcriptional regulation of the rat testis-specific histone H1t gene in somatic cells. Biol. Reprod., 61, 1005–1011. [DOI] [PubMed] [Google Scholar]
- 14.Cunliffe V., Koopman,P., McLaren,A. and Trowsdale,J. (1990) A mouse zinc finger gene which is transiently expressed during spermatogenesis. EMBO J., 9, 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foulkes N.S., Mellstrom,B., Benusiglio,E. and Sassone-Corsi,P. (1992) Developmental switch of CREM function during spermatogenesis: from antagonist to activator. Nature, 355, 80–84. [DOI] [PubMed] [Google Scholar]
- 16.Sun Z. and Means,A.R. (1995) An intron facilitates activation of the calspermin gene by the testis-specific transcription factor CREMτ. J. Biol. Chem., 270, 20962–20967. [DOI] [PubMed] [Google Scholar]
- 17.Blendy J.A., Kaestner,K.H., Weinbauer,G.F., Nieschlag,E. and Schutz,G. (1996) Severe impairment of spermatogenesis in mice lacking the CREM gene. Nature, 380, 162–165. [DOI] [PubMed] [Google Scholar]
- 18.Lele K. and Wolgemuth,D.J. (1998) The role of transcriptional control during spermatogenesis. J. Androl., 19, 639–649. [PubMed] [Google Scholar]
- 19.Sassone-Corsi P. (2002) Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science, 296, 2176–2178. [DOI] [PubMed] [Google Scholar]
- 20.Wu H., Zhang,S., Qiu,W., Zhang,G., Xia,Q., Xiao,C., Huang,X., Huang,M., Agen,P., Fan,T. et al. (2001) Isolation, characterization, and mapping of a novel human KRAB zinc finger protein encoding gene ZNF463. Biochem. Biophys. Acta, 1518, 190–193. [DOI] [PubMed] [Google Scholar]
- 21.Lai Y.L., Li,H., Chiang,H.S. and Hsieh-Li,H.M. (2002) Expression of a novel TGIF subclass homeobox gene, Tex1, in the spermatids of mouse testis during spermatogenesis. Mech. Dev., 113, 185–187. [DOI] [PubMed] [Google Scholar]
- 22.Hsu S.H., Shyu,H.W., Hsieh-Li,H.M. and Li,H. (2001) Spz1, a novel bHLH-Zip protein, is specifically expressed in testis. Mech. Dev., 100, 177–187. [DOI] [PubMed] [Google Scholar]
- 23.Aso T., Yamazaki,K., Amimoto,K., Kuroiwa,A., Higashi,H., Matsuda,Y., Kitajima,S. and Hatakeyama,M. (2000) Identification and characterization of Elongin A2, a new member of the Elongin family of transcription elongation factors, specifically expressed in the testis. J. Biol. Chem., 275, 6546–6552. [DOI] [PubMed] [Google Scholar]
- 24.Ozer J., Moore,P.A. and Lieberman,P.M. (2000) A testis-specific transcription factor IIA (TFIIAtau) stimulates TATA-binding protein–DNA binding and transcription activation. J. Biol. Chem., 275, 122–128. [DOI] [PubMed] [Google Scholar]
- 25.Zhang D., Penttila,T.L., Morris,P.L. and Roeder,R.G. (2001) Cell- and stage-specific high-level expression of TBP-related factor 2 (TRF2) during mouse spermatogenesis. Mech. Dev., 106, 203–205. [DOI] [PubMed] [Google Scholar]
- 26.Wang P.J. and Page,D.C. (2002) Functional substitution for TAF(II)250 by a retroposed homolog that is expressed in human spermatogenesis. Hum. Mol. Genet., 11, 2341–2346. [DOI] [PubMed] [Google Scholar]
- 27.Li S., Zhou,W., Doglio,L. and Goldberg,E. (1998) Transgenic mice demonstrate a testis-specific promoter for lactate dehydrogenase, LDHC. J. Biol. Chem., 273, 31191–31194. [DOI] [PubMed] [Google Scholar]
- 28.Reddi P.P., Flickinger,C.J. and Herr,J.C. (1999) Round spermatid-specific transcription of the mouse SP-10 gene is mediated by a 294-base pair proximal promoter. Biol. Reprod., 61, 1256–1266. [DOI] [PubMed] [Google Scholar]
- 29.Charron M., Shaper,N.L., Rajput,B. and Shaper,J.H. (1999) A novel 14-base-pair regulatory element is essential for in vivo expression of murine beta4-galactosyltransferase-I in late pachytene spermatocytes and round spermatids. Mol. Cell. Biol., 19, 5823–5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sieweke M. (2000) Detection of transcription factor partners with a yeast one hybrid screen. Methods Mol. Biol., 130, 59–77. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe D., Okabe,M., Hamajima,N., Morita,T., Nishina,Y. and Nishimune,Y. (1995) Characterization of the testis-specific gene ‘calmegin’ promoter sequence and its activity defined by transgenic mouse experiments. FEBS Lett., 368, 509–512. [DOI] [PubMed] [Google Scholar]
- 32.Chen D.C., Yang,B.C. and Kuo,T.T. (1992) One-step transformation of yeast in stationary phase. Curr. Genet., 21, 83–84. [DOI] [PubMed] [Google Scholar]
- 33.Grootegoed J.A. and den Boer,P.J. (1989) Energy metabolism of spermatids: a review. In Hamilton,D.W. and Waites,G.H.M. (eds), Cellular and Molecular Events in Spermiogenesis as Targets for Fertility Regulations. Cambridge University Press, Cambridge, pp. 193–215. [Google Scholar]
- 34.Auffray C. and Rougeon,F. (1980) Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur. J. Biochem., 107, 303–314. [DOI] [PubMed] [Google Scholar]
- 35.Peters A.H., Plug,A.W., van Vugt,M.J. and de Boer,P. (1997) A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res., 5, 66–68. [DOI] [PubMed] [Google Scholar]
- 36.Wilkinson D.G., Bailes,J.A., Champion,J.E. and McMahon,A.P. (1987) A molecular analysis of mouse development from 8 to 10 days post coitum detects changes only in embryonic globin expression. Development, 99, 493–500. [DOI] [PubMed] [Google Scholar]
- 37.Maruyama O., Nishimori,H., Katagiri,T., Miki,Y., Ueno,A. and Nakamura,Y. (1998) Cloning of TCFL5 encoding a novel human basic helix–loop–helix motif protein that is specifically expressed in primary spermatocytes at the pachytene stage. Cytogenet. Cell. Genet., 82, 41–45. [DOI] [PubMed] [Google Scholar]
- 38.Atchley W.R. and Fitch,W.M. (1997) A natural classification of the basic helix–loop–helix class of transcription factors. Proc. Natl Acad. Sci. USA, 94, 5172–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohsako S., Hyer,J., Panganiban,G., Oliver,I. and Caudy,M. (1994) Hairy function as a DNA-binding helix–loop–helix repressor of Drosophila sensory organ formation. Genes Dev., 8, 2743–2755. [DOI] [PubMed] [Google Scholar]
- 40.Robinson K.A. and Lopes,J.M. (2000) SURVEY AND SUMMARY: Saccharomyces cerevisiae basic helix–loop–helix proteins regulate diverse biological processes. Nucleic Acids Res., 28, 1499–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell L.D., Ettlin,R.A., Sinha Hikim,A.P. and Clegg,E.D. (1990) Histological and Histopathological Evaluation of the Testis, 1st edn. Cache River Press, Clearwater, Fl. [Google Scholar]
- 42.Heyting C. (1996) Synaptonemal complexes: structure and function. Curr. Opin. Cell Biol., 8, 389–396. [DOI] [PubMed] [Google Scholar]
- 43.Yuan L., Liu,J.G., Zhao,J., Brundell,E., Daneholt,B. and Hoog,C. (2000) The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol. Cell, 5, 73–83. [DOI] [PubMed] [Google Scholar]
- 44.Baarends W.M., Wassenaar,E., Hoogerbrugge,J.W., van Cappellen,G., Roest,H.P., Vreeburg,J., Ooms,M., Hoeijmakers,J.H. and Grootegoed,J.A. (2003) Loss of HR6B ubiquitin-conjugating activity results in damaged synaptonemal complex structure and increased crossing-over frequency during the male meiotic prophase. Mol. Cell. Biol., 23, 1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Handel M.A. (2004) The XY body: a specialized meiotic chromatin domain. Exp. Cell Res., 296, 57–63. [DOI] [PubMed] [Google Scholar]
- 46.Voronova A. and Baltimore,D. (1990) Mutations that disrupt DNA binding and dimer formation in the E47 helix–loop–helix protein map to distinct domains. Proc. Natl Acad. Sci. USA, 87, 4722–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ikawa M., Nakanishi,T., Yamada,S., Wada,I., Kominami,K., Tanaka,H., Nozaki,M., Nishimune,Y. and Okabe,M. (2001) Calmegin is required for fertilin alpha/beta heterodimerization and sperm fertility. Dev. Biol., 240, 254–261. [DOI] [PubMed] [Google Scholar]
- 48.Liang L., Soyal,S.M. and Dean,J. (1997) FIGα, a germ cell specific transcription factor involved in the coordinate expression of the zona pellucida genes. Development, 124, 4939–4947. [DOI] [PubMed] [Google Scholar]
- 49.Huntriss J., Gosden,R., Hinkins,M., Oliver,B., Miller,D., Rutherford,A.J. and Picton,H.M. (2002) Isolation, characterization and expression of the human Factor In the Germline alpha (FIGLA) gene in ovarian follicles and oocytes. Mol. Hum. Reprod., 8, 1087–1095. [DOI] [PubMed] [Google Scholar]
- 50.Soyal S.M., Amleh,A. and Dean,J. (2000) FIGα, a germ cell-specific transcription factor required for ovarian follicle formation. Development, 127, 4645–4654. [DOI] [PubMed] [Google Scholar]
- 51.Atchley W.R., Wollenberg,K.R., Fitch,W.M., Terhalle,W. and Dress,A.W. (2000) Correlations among amino acid sites in bHLH protein domains: an information theoretic analysis. Mol. Biol. Evol., 17, 164–178. [DOI] [PubMed] [Google Scholar]
- 52.Davis R.L. and Turner,D.L. (2001) Vertebrate hairy and enhancer of split related proteins: transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene, 20, 8342–8357. [DOI] [PubMed] [Google Scholar]
- 53.Ferre-D'Amare A.R., Prendergast,G.C., Ziff,E.B. and Burley,S.K. (1993) Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature, 363, 38–45. [DOI] [PubMed] [Google Scholar]
- 54.Van Wayenbergh R., Taelman,V., Pichon,B., Fischer,A., Kricha,S., Gessler,M., Christophe,D. and Bellefroid,E.J. (2003) Identification of BOIP, a novel cDNA highly expressed during spermatogenesis that encodes a protein interacting with the orange domain of the hairy-related transcription factor HRT1/Hey1 in Xenopus and mouse. Dev. Dyn., 228, 716–725. [DOI] [PubMed] [Google Scholar]
- 55.Wang H., Liu,F., Millette,C.F. and Kilpatrick,D.L. (2002) Expression of a novel, sterol-insensitive form of sterol regulatory element binding protein 2 (SREBP2) in male germ cells suggests important cell- and stage-specific functions for SREBP targets during spermatogenesis. Mol. Cell. Biol., 22, 8478–8490. [DOI] [PMC free article] [PubMed] [Google Scholar]