Abstract
In animal cells, stable RNA silencing can be achieved by vector-based small interfering RNA (siRNA) expression system, in which Pol III RNA gene promoters are used to drive the expression of short hairpin RNA, however, this has not been demonstrated in plants. Whether Pol III RNA gene promoter is capable of driving siRNA expression in plants is unknown. Here, we report that RNA silencing was achieved in plants through stable expression of short hairpin RNA, which was driven by Pol III RNA gene promoters. Using glucuronidase (GUS) transformed tobacco as a model system, the results demonstrated that 21 nt RNA duplexes, targeting at different sites of GUS gene, were stably expressed under the control of either human H1 or Arabidopsis 7SL RNA gene promoter, and GUS gene was silenced in 80% of siRNA transgenics. The severity of silencing was correlated with the abundance of siRNA expression but independent of the target sites and uridine residue structures in siRNA hairpin transcripts. Thus, the specific expression of siRNA provides a new system for the study of siRNA silencing pathways and functional genomics in plants. Moreover, the effectiveness of the human H1 promoter in a plant background suggested a conserved mechanism underlying Pol III complex functionality.
INTRODUCTION
Small interfering RNAs (siRNAs) of ∼21 nt have been reported to play a crucial role in RNA silencing, a term referring to post-transcriptional gene silencing in plants (1–3), quelling in fungi (4) and RNA interference (RNAi) in animals (5–7). The mechanism of siRNA biogenesis and function [for reviews see (8–15)] are thought to be highly conserved in almost all the eukaryotes including plants and animals, in which siRNAs are produced from double-stranded RNA (dsRNA) by an RNase III termed Dicer in animal cells or DCL (Dicer-like) in plants, and then incorporated into a RNA-induced silencing complex (RISC), in which siRNAs play a guiding role in sequence-specific cleavage of target mRNAs. Moreover, in some organisms, such as Caenorhabditis elegans, Drosophila and plants, the siRNA signal is found to spread along the mRNA target, which results in the production of secondary siRNAs and the induction of transitive RNA silencing (16–21).
However, clear differences in siRNA-mediated RNA silencing have also been found between plants and animals cells. In an example, siRNAs produced in Drosophila embryos (22) and in mammalian cells (23) show only a ∼21 nt class, while siRNAs in plants and in fungi fall into two distinct classes: a short (∼21 nt) and a long (∼24 nt) size class (19,21,24–28). In another example, only 5′ direction spreading exists in nematodes (16), while both 5′ and 3′ direction spreading is present in plants (18–21). These observations suggest the likelihood that different mechanisms of siRNA-mediated RNA silencing occur in plants and animal cells.
On the other hand, the siRNA-mediated RNA silencing has been employed to develop new technology for the study of function genomics in various organisms [for reviews see (8–15,29)]. In plants, a common technique for inducing RNA silencing is the use of transgenes driven by a constitutive 35S or other plant promoters to express a dsRNA structure in the length of ∼200–1000 bp [for reviews see (9)]. The expressed long dsRNA is then thought to be cleaved via the mediation of endonucleolytic enzyme DCL into siRNAs, which lead to the silencing of its target mRNA (7,30).
However, in most mammalian cells, the introduction of RNA silencing by expressing a long dsRNA structure has not been very successful due to the global shut down of protein synthesis intrigued by dsRNA (>30 bp) that activates the interferon (IFN)-related pathways (31–33). To circumvent this cytotoxic non-specificity, synthetic siRNAs of 21–22 nt RNA duplexes have been successfully used for inducing strong and specific RNA silencing (7). However, such an RNA silencing is transient and generally fails to yield a stable phenotype for further characterization. This limitation in mammalian cells was alleviated with the development of Pol III RNA gene promoter-based systems for stable expression of short hairpin RNAs in vivo (34–40). Whether Pol III RNA gene promoter is capable of driving siRNA expression in plants is unknown.
To address this question and to develop a useful system for the study of siRNA signal pathways and functional genomics in plants, we designed vector-based siRNA expression systems under the control of a human H1 or a plant 7SL RNA gene promoter. Then, we tested these systems in glucuronidase (GUS) transformed tobacco. The results demonstrated that target GUS gene was silenced in 80% of siRNA transgenics. The severity of silencing was correlated with the abundance of siRNA expression but was independent of the target sites and uridine residue structures in siRNA hairpin.
MATERIALS AND METHODS
Constructs
In order to express short hairpin RNA in plant cells, the binary vector system that is amenable to Agrobacterium-mediated transformation was applied. Two promoters, one from human H1 RNA and the other from Arabidopsis 7SL RNA gene, were used to drive siRNA expression. pGPH1 and pGPSL were named for the constructs corresponding to the promoters, respectively. For each promoter, two or three siRNA constructs were made in order to compare the effects of siRNA sites on target mRNA and the uridine residue structures in siRNA hairpin.
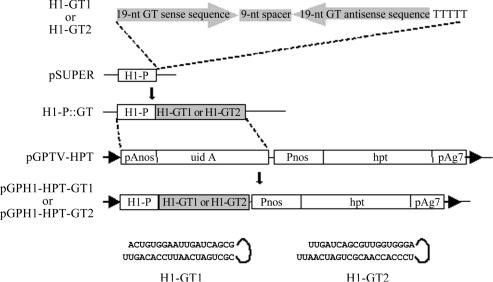
Two H1 promoter constructs were designed, pGPH1-GT1 and pGPH1-GT2. For pGPH1-GT1, a forward oligo GT1F: 5′-GATCCCCACTGTGGAATTGATCAGCGTTCAAGAGACGCTGATCAATTCCACAGTTTTTTGGAAA-3′ and a complementary oligo GT1R: 5′-AGCTTTTCCAAAAAACTGTGGAATTGATCAGCGTCTCTTGAACGCTGATCAATTCCACAGTGGG were synthesized, and another pair of oligos (forward oligo GT2F 5′-GATCCCCTTGATCAGCGTTGGTGGGATTCAAGAGATCCCACCAACGCTGATCAATTTTTGGAAA-3′ and complementary oligo GT2R 5′-AGCTTTTCCAAAAATTGATCAGCGTTGGTGGGATCTCTTGAATCCCACCAACGCTGATCAAGGG-3′) were designed for pGPH1-GT2 construct. The oligo contained sense and antisense 19 nt target sequences separated by a 9 nt spacer (in boldface). GT1 targeted at the sequence of 80–98 downstream from ATG in GUS-coding region and GT2 at the sequence of 89–107. The forward and complementary oligos were annealed and then cloned into pSUPER vector (34) at downstream of the H1 promoter (H1-P). After confirmation of sequence accuracy, the H1-P::GT expression cassette was then excised by double digestion with EcoRI and HindIII and cloned into binary vector pGPTV-HPT (41) by replacing pAnos-uidA fragment. Thus, the yielded siRNA expression vectors, pGPH1-HPT-GT1 and pGPH1-HPT-GT2, were amenable to Agrobacterium transformation system under selection of hygromycine (Figure 1).
Figure 1.
Preparation of human H1 RNA gene promoter-based siRNA expression constructs. The 19 nt GUS gene specific sequence (GT1 or GT2) separated by a 9 nt spacer from the reverse complement of the same sequence followed by a termination signal of five thymidines (H1-GT1 or H1-GT2) was cloned into pSUPER (34) downstream of the H1 promoter (H1-P). The H1-P::GT expression construct harboring H1-GT1 or H1-GT2 was then excised and cloned into the binary vector pGPTV-HPT (41). The resulting vector, pGPH1-HPT-GT1 or pGPH1-HPT-GT2, which contained a hygromycin phosphotransferase (hpt) selectable marker gene under the control of a nopaline synthase promoter (Pnos)-transcription terminator (pAg7, agropine synthase polyadenylation signal sequence) pair, was then mobilized into A.tumefaciens C58 for transforming tobacco. The predicted secondary siRNA structures of H1-GT1 and H1-GT2 are depicted.
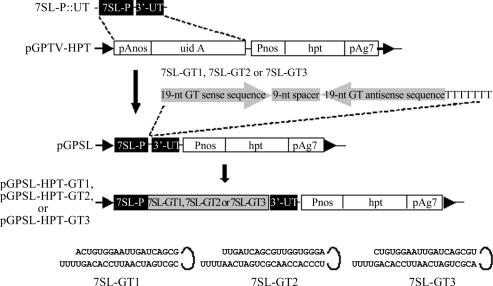
To clone At7SL4 promoter (7SL-P), Arabidopsis thaliana (Columbia ecotype) genomic DNA was PCR-amplified using a forward primer (SLpF 5′-GGAATTCTGCGTTTGAAGAAGAGTGTTTGA-3′) and a reverse primer (SLpR 5′-GCCCGGGAAGATCGGTTCGTGTAATATAT-3′). To facilitate subsequent cloning, a restriction site (EcoRI in forward primer and SmaI in reverse primer are underlined) was included at 5′ end. The PCR product was cloned into a pCR2.1-TOPO system (Invitrogen) and the accuracy of the promoter was subsequently confirmed (AY525344). The 3′-untranscribed region (3′-UT) of At7SL4 gene was also cloned by PCR amplification from A.thaliana (Columbia ecotype). Using forward primer SLtF (5′-GTCTAGATTTTGATTTTGTTTTCCAAAACTTTCTACG-3′, an XbaI site underlined at 5′ end) and reverse primer SLtR (5′-GAAGCTTGGTGTTGATCACAACGATACA-3′, a HindIII site underlined at 5′ end), 3′-UT fragment was amplified by PCR and cloned into pCR2.1-TOPO system. After the sequence was confirmed, the 3′-UT fragment was assembled with 7SL-P to form siRNA expression module, 7SL-P::UT. This module structure was then cloned into pGPTV-HPT (41) to replace uidA-pAnos fragment and resulted in a plasmid named pGPSL (Figure 2).
Figure 2.
Preparation of plant 7SL RNA gene promoter-based siRNA expression constructs. A promoter fragment (7SL-P, 289 bp) containing USE and TATA elements (47) and a 3′-UT region (267 bp) of Arabidopsis At7SL4 (AY525344) gene were cloned and ligated in pUC19, from which the 7SL-P::UT construct was excised and cloned into the pGPTV-HPT vector (41) to replace the pAnos-uidA fragment. The resulting vector, pGPSL, contained an hpt selectable marker gene under the control of a nopaline synthase promoter (Pnos)-transcription terminator (pAg7, agropine synthase polyadenylation signal sequence) pair. GUS gene-specific 7SL-GT1, 7SL-GT2 or 7SL-GT3 sequence module, which contained a termination signal of seven thymidines, for the generation of the corresponding hairpin, siRNA was inserted into pGPSL between 7SL-P and 3′-UT. The resulting binary vectors were named pGPSL-HPT-GT1, pGPSL-HPT-GT2 and pGPSL-HPT-GT3, respectively. The binary vector was then mobilized into A.tumefaciens C58 for transforming tobacco. The predicted secondary siRNA structures of 7SL-GT1, 7SL-GT2 and 7SL-GT3 are depicted.
Following the similar design of H1 promoter constructs, three 7SL promoter vectors, pGPSL-HPT-GT1, pGPSL-HPT-GT2 and pGPSL-HPT-GT3, were constructed. Three pairs of oligos, GT1 (GPSL1aF, 5′-TACACTGTGGAATTGATCAGCGTTCAGATGACGCTGATCAATTCCACAGTTTTTTTT and GPSL1aR, 5′-CTAGAAAAAAAACTGTGGAATTGATCAGCGTCATCTGAACGCTGATCAATTCCACAGTGTA), GT2 (GPSL2F, 5′-TACTTGATCAGCGTTGGTGGGATTCAGATGATCCCACCAACGCTGATCAATTTTTTT and GPSL2R, 5′-CTAGAAAAAAATTGATCAGCGTTGGTGGGATCATCTGAATCCCACCAACGCTGATCAAGTA), and GT3 (GPSL1F, 5′-TACCTGTGGAATTGATCAGCGTTTCAGATGAACGCTGATCAATTCCACAGTTTTTTT and GPSL1R 5′-CTAGAAAAAAACTGTGGAATTGATCAGCGTTCATCTGAAACGCTGATCAATTCCACAGGTA) were synthesized for the construction. These siRNA structures targeted at different sites of GUS mRNA, respectively, at 80–98, 89–107 and 81–99 sequence downstream of GUS ATG. As described above, the siRNA duplex was inserted into pGPSL between 7SL-P and 3′-UT fragment (Figure 2).
Agrobacterium-mediated transformation and GUS activity assay
The pGPH1 and pGPSL series plasmids were mobilized individually into Agrobacterium tumefaciens strain C58 by freeze-thaw method (42). Leaf disc transformation of tobacco (Nicotiana tabacum cv. Havana) and histochemical characterization of GUS activity was conducted as described previously (43). For GUS enzyme activity assay, ∼100 mg leaf tissues were ground in 800 ml GUS extraction buffer (50 mM phosphate buffer, pH 7.4, 10 mM DTT, 1 mM Na2-EDTA, 0.1% sodium lauryl sarcosine and 0.1% Triton-X 100) by FastPrep FP120 (Thermo Savant). The GUS activity was analyzed according to Jefferson et al. (44). The fluorescence was detected by a TD-700 Fluorometer (Turner Designs). The protein concentration was determined using Protein Assay Dye Reagent Concentrate (Bio-Rad) by DU 800 Spectrophotometer (Beckman Coulter).
Gel blot analysis of GUS gene expression
An aliquot of 5 μg of total RNA isolated with TRIzol® Reagent (Invitrogen) was used in each lane of the northern blots. RNA gel electrophoresis, blotting and hybridization were performed as described previously (43). 32P-labeled probe was prepared from the entire GUS-coding sequence. Hybridization was performed at 65°C.
Gel blot analysis of siRNA expression
Northern hybridization of small RNA was performed according to Hutvágner et al. (45) with modifications. Total RNA (25 μg) isolated with TRIzol® Reagent (Invitrogen) was denatured for 10 min at 65–70°C, separated in 12% polyacrylamide/8 M urea gel (Amersham) in Protean II apparatus (BioRad), and electro-blotted onto Hybond-N+ membrane (Amersham) by using a Trans-Blot SD Semi-Dry Electrophoretic Transfer Cell (BioRad). After UV cross-linking and air drying, blots were prehybridized in 5× SSC, 5× Denhardt's solution and 0.5% SDS at 50°C for 2–3 h, and hybridized with a randomly primed 32P-labeled probe from the entire GUS-coding sequence at 50°C for 16 h. The membranes were washed once in 2× SSC at 50°C for 5 min and 3–4 times in 2× SSC and 1% SDS at 50°C for 20 min. Signals were visualized by autoradiography on X-ray at −80°C.
RESULTS AND DISCUSSION
Human H1 RNA gene promoter-based siRNA expression system
We first produced, via Agrobacterium-mediated transformation and kanamycin selection, transgenic tobacco plants expressing a GUS reporter gene under the control of a CaMV 35S promoter and a Nos terminator. A line exhibiting a strong GUS activity was selected, named GUS-line, for testing the siRNA expression vectors designed for silencing the GUS gene expression. Based on the standard design rules (46), two 19 nt sequences (designated GT1 and GT2) targeting at two distinct sites in GUS mRNA (nt 80–98 for GT1 and nt 89–107 for GT2) were selected for constructing the expression vectors. The siRNA expression cassette, H1-GT1 or H1-GT2 (Figure 1), consisting of the sense and the antisense 19 nt sequences linked through a 9 base spacer, was under the control of a human H1 RNA gene promoter (34). The siRNA expression construct (pGPH1-HPT-GT1 or pGPH1-HPT-GT2) containing a hygromycin phosphotransferase (hpt) marker gene (Figure 1) was transferred into the GUS-line via Agrobacterium and the transgenic plants were produced under hygromycin selection.
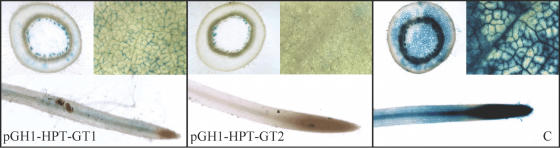
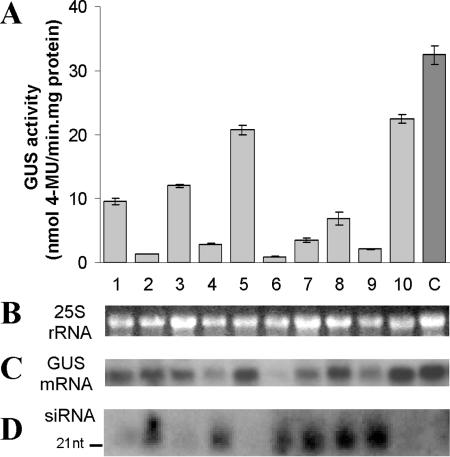
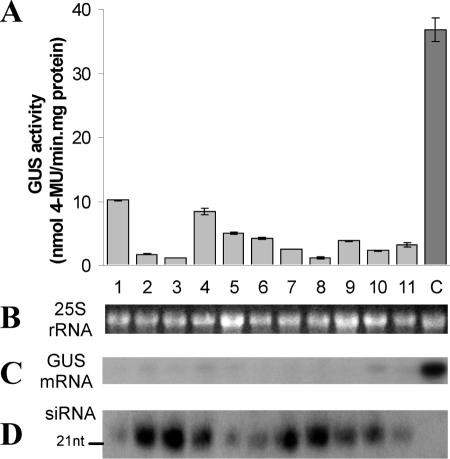
A total of 23 transgenic plants were produced from the pGPH1-HPT-GT1 construct and 19 from pGPH1-HPT-GT2. These siRNA transgenics and the GUS-line control plants were characterized when ∼1 month old. GUS histological analyses showed that the leaf, stem and root of a majority of the pGPH1-HPT-GT1 and pGPH1-HPT-GT2 transgenics had either reduced or had no GUS staining (Figure 3). Quantitative assay of GUS protein activity in leaves of siRNA transgenics and GUS-line control demonstrated that 74% of the pGPH1-HPT-GT1 transgenics had a GUS activity reduction, ranging from 12 to 94%, and 84% of the pGPH1-HPT-GT2 transgenics exhibited 31–97% GUS activity reduction. The reduction in GUS activity (Figure 4A) corresponded with the diminished GUS mRNA level (Figure 4C).
Figure 3.
Histological staining of GUS protein activity in tobacco plants harboring human H1 RNA gene promoter-based siRNA expression vectors. GUS staining of stem cross-section, leaf and root from 1-month-old siRNA-transgenic (pGPH1-HPT-GT1 and pGPH1-HPT-GT2) and GUS-expressing control (C) tobacco plants.
Figure 4.
Analysis of human H1 promoter-mediated siRNA silencing of GUS gene expression in transgenic tobacco. (A) GUS protein activity in leaves of the control plants (C) and 10 pGPH1-HPT-GT2 transgenic lines. Mean values were calculated from three independent measurements per line. (B) Loading control for gel blot analysis showing 25S rRNA transcript levels. (C) The same gel blot as in (B) was used to characterize the GUS mRNA level with a GUS cDNA probe. (D) Gel blot detection of small RNAs of ∼21 nt, as indicated, using a GUS cDNA probe. RNA was isolated from a portion of the leaves used for GUS protein activity assay in (A).
We then examined whether the siRNA transgenics produced GUS-specific small RNAs, the necessary molecules for the siRNA-mediated gene silencing. Total RNA was isolated from leaves of pGPH1-HPT-GT1 and pGPH1-HPT-GT2 transgenic and GUS-line control plants and gel blot analysis of small RNAs using GUS gene sequence-specific probes was performed as described by Hutvágner et al. (45). As shown in Figure 4D, a GUS-specific small RNA hybridizing band of ∼21 nt in size was present in the transgenic lines having reduced GUS mRNA and protein activity, but was absent from the GUS-line control. Furthermore, the abundance of the specific small RNA was inversely correlated to that of GUS mRNA (Figure 4C and D), indicating that the siRNA-guided degradation of the target mRNA was involved. That GT1 and GT2 expression vectors having siRNA sequences targeting at two GUS mRNA sites exerting a similar gene silencing efficiency is also consistent with the proposed siRNA mechanism (7,34,36), in which gene silencing efficiency is independent of the target sites.
According to the design of siRNA expression vector, the transcript of siRNA expression cassette was predicted to form an inverted hairpin RNA structure containing one (for H1-GT1) or two (for H1-GT2) 3′ overhanging uridines (Figure 1). Such a structure of 3′ overhanging uridines is reported to be necessary for an siRNA-guided mRNA cleavage (7). Our results showed that gene silencing efficiency might be independent of the number of 3′ overhanging uridine (U) residues as testified with 1 U in GT1 versus 2 in GT2. Overall, the results demonstrated an effective gene silencing system for stably inducing targeted RNA silencing through the expression of gene-specific siRNA transgenes. The effectiveness of the human H1 promoter in a plant background indicated that plant Pol III complex is capable of initiating transcription through the recognition of the mammalian promoter sequences, suggesting a conservative mechanism likely underlying Pol III RNA processing in plants and mammalian cells.
Plant 7SL RNA gene promoter-based siRNA expression system
We next tested a plant promoter-based system. A DNA-dependent RNA polymerase III 7SL RNA gene promoter from A.thaliana was chosen for this purpose because the transcription of small 7SL RNA genes is controlled exclusively by their upstream external regulatory sequence elements (USE and TATA) (47) and terminates at a run of five to seven thymidines. Therefore, their promoters were expected to direct the expression of siRNA duplexes that would contain 3′ overhanging uridines, affording the needed structure for an siRNA-guided mRNA cleavage (7).
Four A.thaliana 7SL genes have been cloned, including At7SL4 (AY525344) that we isolated. From At7SL4 gene, a 289 bp promoter fragment (7SL-P) containing USE and TATA elements and a 267 bp 3′-UT segment were cloned and fused into pGPTV-HPT (41) to assemble the siRNA expression cassette, pGPSL (Figure 2). In addition to GT1 and GT2 sequences, 19 nt GUS mRNA sequences, named GT3 targeting at nt 81–99 of the GUS-coding region, were also selected for constructing the siRNA expression constructs. These three constructs, pGPSL-HPT-GT1, pGPSL-HPT-GT2 and pGPSL-HPT-GT3 (Figure 2), were then mobilized into A.tumefaciens C58 cells individually for transforming the GUS-line. After hygromycin selection, a total of 89 plants were regenerated from these three expression constructs. The same analysis schemes as described in H1 promoter-based siRNA expression system were applied. The results demonstrated that 83% of these transgenic plants exhibited a reduction in GUS enzyme activity, ranging from 20 to 99%. No apparent difference in overall GUS activity reduction efficiency was observed among these three expression constructs. The GUS activity reduction corresponded with the diminished GUS mRNA level (Figure 5A and C) and with the appearance/abundance of the GUS-specific small RNAs (Figure 5D).
Figure 5.
Analysis of plant 7SL promoter-mediated siRNA silencing of GUS gene expression in transgenic tobacco. (A) GUS protein activity in leaves of the control plants (C) and 11 pGPSL-HPT-GT2 transgenic lines. Mean values were calculated from three independent measurements per line. (B) RNA loading control. (C) Same gel blot used in (B) above was used to characterize the GUS mRNA level with a GUS cDNA probe. (D) Gel blot detection of small RNAs of ∼21 nt, as indicated, using a GUS cDNA probe. RNA was isolated from a portion of the leaves used for GUS protein activity assay in (A).
Thus, the present study demonstrated that both the human H1 and the Arabidopsis 7SL RNA gene promoters were able to drive the expression of specific short hairpin RNA that subsequently resulted in the target gene silencing in a highly efficient manner in plants. Recent results showed that no secondary siRNA was produced from endogenous gene in rice (48), suggesting that the systems developed in this report could be used for specific silencing of the genes whose sequences are similar to each other, although this type of specificity needs to be further examined precisely. Moreover, the application of the systems would also promise high potential in the analysis of various aspects of plant siRNA signal pathways, such as the distinct functions of short (∼21 nt) and long (∼24 nt) sizes of siRNA, which were found to exist in plants (19,21,24–28) and the biogenesis of secondary siRNA (16–21). Furthermore, the successful development of small RNA expression system under the control of Pol III RNA promoter in plants would shed light on the research of small RNA function, such as the function of plant microRNAs (miRNAs). miRNAs are a large class of small non-coding RNAs and are believed to play crucial roles in regulatory pathways of many plant developmental processes (49–53). However, the function of most of miRNAs is yet to be characterized (51,53). In this context, the described siRNA systems could be a timely tool.
Acknowledgments
ACKNOWLEDGEMENT
This work was supported by a grant from the US Department of Energy Division of Energy Biosciences DE-FG02-03ER15442.
DDBJ/EMBL/GenBank accession no. AY525344
REFERENCES
- 1.Van der Krol A.R., Mur,L.A., Beld,M., Mol,J.N.M. and Stuitje,A.R. (1990) Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell, 2, 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Napoli C., Lemieux,C. and Jorgensen,R. (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell, 2, 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.English J.J., Mueller,E. and Baulcombe,D.C. (1996) Suppression of virus accumulation in transgenic plants exhibiting silencing of nuclear genes. Plant Cell, 8, 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cogoni C., Irelan,J.T., Schumacher,M., Schmidhauser,T.J., Selker,E.U. and Macino,G. (1996) Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA–DNA interactions or DNA methylation. EMBO J., 15, 3153–3163. [PMC free article] [PubMed] [Google Scholar]
- 5.Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 6.Tuschl T., Zamore,P.D., Lehmann,R., Bartel,D.P. and Sharp,P.A. (1999) Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev., 13, 3191–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 8.Dillin A. (2003) The specifics of small interfering RNA specificity. Proc. Natl Acad. Sci. USA, 100, 6289–6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waterhouse P.M. and Helliwell,C.A. (2003) Exploring plant genomes by RNA-induced gene silencing. Nature Rev. Genet., 4, 29–38. [DOI] [PubMed] [Google Scholar]
- 10.McManus M.T. and Sharp,P.A. (2002) Gene silencing in mammals by small interfering RNAs. Nature Rev. Genet., 3, 737–747. [DOI] [PubMed] [Google Scholar]
- 11.Ahlquist P. (2002) RNA-dependent RNA polymerases, viruses and RNA silencing. Science, 296, 1270–1273. [DOI] [PubMed] [Google Scholar]
- 12.Hannon G.J. (2002) RNA interference. Nature, 418, 244–251. [DOI] [PubMed] [Google Scholar]
- 13.Tuschl T. (2002) Expanding small RNA interference. Nat. Biotechnol., 20, 446–448. [DOI] [PubMed] [Google Scholar]
- 14.Voinnet O. (2002) RNA silencing: small RNAs as ubiquitous regulators of gene expression. Curr. Opin. Plant Biol., 5, 444–451. [DOI] [PubMed] [Google Scholar]
- 15.Zamore P.D. (2002) Ancient pathways programmed by small RNAs. Science, 296, 1265–1269. [DOI] [PubMed] [Google Scholar]
- 16.Sijen T., Fleenor,J., Simmer,F., Thijssen,K.L., Parrish,S., Timmons,L., Plasterk,R.H.A. and Fire,A. (2001) One the role of RNA amplification in dsRNA-triggered gene silencing. Cell, 107, 465–476. [DOI] [PubMed] [Google Scholar]
- 17.Lipardi C., Wei,Q. and Paterson,B.M. (2001) RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell, 107, 297–307. [DOI] [PubMed] [Google Scholar]
- 18.Voinnet O., Vain,P., Angell,S. and Baulcombe,D.C. (1998) Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell, 95, 177–187. [DOI] [PubMed] [Google Scholar]
- 19.Klahre U., Crete,P., Leuenberger,S.A., Iglesias,V.A. and Meins,F.,Jr (2002) High molecular weight RNAs and small interfering RNAs induce systemic posttranscriptional gene silencing in plants. Proc. Natl Acad. Sci. USA, 99, 11981–11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaistij F.E., Jones,L. and Baulcombe,D.C. (2002) Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell, 14, 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang G., Reinhart,B.J., Bartel,D.P. and Zamore,P.D. (2003) A biochemical framework for RNA silencing in plants. Genes Dev., 17, 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zamore P.D., Tuschl,T., Sharp,P.A. and Bartel,D.P. (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell, 101, 25–33. [DOI] [PubMed] [Google Scholar]
- 23.Billy E., Brondani,V., Zhang,H., Müller,U. and Filipowicz,W. (2001) Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl Acad. Sci. USA, 98, 14428–14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamilton A., Voinnet,O., Chappell,L. and Baulcombe,D. (2002) Two classes of short interfering RNA in RNA silencing. EMBO J., 21, 4671–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mallory A.C., Reinhart,B.J., Bartel,D., Vance,V.B. and Bowman,L.H. (2002) A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs tobacco. Proc. Natl Acad. Sci. USA, 99, 15228–15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Llave C., Kasschau,K.D., Rector,M.A. and Carrington,J.C. (2002) Endogenous and silencing-associated small RNAs in plants. Plant Cell, 14, 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papp I., Mette,M.F., Aufsatz,W., Daxinger,L., Schauer,S.E., Ray,A., van der Winder,J., Matzke,M. and Matzke,A.J.M. (2003) Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol., 132, 1382–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicolás F.E., Torres-Martínez,S., Ruiz-Vázquez,R.M. (2003) Two classes of small antisense RNAs in fungal RNA silencing triggered by non-integrative transgenes. EMBO J., 22, 3983–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall J. (2004) Opinion: Unravelling the general properties of siRNAs: strength in numbers and lessons from the past. Nature Rev. Genet., 5, 552–557. [DOI] [PubMed] [Google Scholar]
- 30.Hammond S.M., Bernstein,E., Beach,D. and Hannon,G.J. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature, 404, 293–296. [DOI] [PubMed] [Google Scholar]
- 31.Paddison P.J., Caudy,A.A. and Hannon,G.J. (2002) Stable suppression of gene expression RNAi in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svovoda P., Stein,P. and Schultz,R.M. (2001) RNAi in mouse oocytes and preimplantation embryos: effectiveness of hairpin dsRNA. Biochem. Biophy. Res. Commun., 287, 1099–1104. [DOI] [PubMed] [Google Scholar]
- 33.Hunter T., Hunt,T., Jackson,R.J. and Robertson,H.D. (1975) The characteristics of inhibition of protein synthesis by double-stranded ribonucleic acid in reticulocyte lysates. J. Biol. Chem., 250, 409–417. [PubMed] [Google Scholar]
- 34.Brummelkamp T.R., Bernards,R. and Agami,R. (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science, 296, 550–553. [DOI] [PubMed] [Google Scholar]
- 35.Lee N.S., Dohjima,T., Bauer,G., Li,H., Li,M.-J., Ehsani,A., Salvaterra,P. and Rossi,J. (2002) Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol., 20, 500–505. [DOI] [PubMed] [Google Scholar]
- 36.Miyagishi M. and Taira,K. (2002) U6-promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol., 20, 497–500. [DOI] [PubMed] [Google Scholar]
- 37.Paddison P.J., Caudy,A.A., Bernstein,E., Hannon,G.J. and Conklin,D.S. (2002) Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev., 16, 948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paul C.P., Good,P.D., Winer,I. and Engelke,D.R. (2002) Effective expression of small interfering RNA in human cells. Nat. Biotechnol., 20, 505–508. [DOI] [PubMed] [Google Scholar]
- 39.Sui G., Soohoo,C., Affar,E.-B., Gay,F., Shi,Y., Forrester,W.C. and Shi,Y. (2002) A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 5515–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu J.-Y., DeRuiter,S.L. and Turner,D.L. (2002) RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 6047–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Becker D., Kemper,E., Schell,J. and Masterson,R. (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol., 20, 1195–1197. [DOI] [PubMed] [Google Scholar]
- 42.Chang S., Puryear,J. and Cairney,J. (1993) A simple method for isolating RNA from pine trees. Plant Mol. Biol. Rep., 11, 113–116. [Google Scholar]
- 43.Hu W.J., Kawaoka,A., Tsai,C.J., Lung,J., Osakabe,K., Ebinuma,H. and Chiang,V.L. (1998) Compartmentalized expression of two structurally and functionally distinct 4-coumarate: coA ligase genes in aspen (Populus tremuloides). Proc. Natl Acad. Sci. USA, 95, 5407–5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jefferson R.A., Kavanagh,T.A. and Bevan,M.W. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J., 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hutvágner G., Mlynárová,L. and Nap,J.-P. (2000) Detailed characterization of the posttranscriptional gene-silencing-related small RNA in a GUS gene-silenced tobacco. RNA, 6, 1445–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elbashir S.M., Harborth,J., Weber,K. and Tucchl,T. (2002) Analysis of gene function in somatic mammalian cells using small interfering DNAs. Methods, 26, 199–213. [DOI] [PubMed] [Google Scholar]
- 47.Yukawa Y., Matousek,J., Grimm,M., Vrba,L., Steger,G., Sugiura,M. and Beier,H. (2002) Plant 7SL RNA and tRNA(Tyr) genes with inserted antisense sequences are efficiently expressed in an in vitro transcription system from Nicotiana tabacum cells. Plant Mol. Biol., 50, 713–723. [DOI] [PubMed] [Google Scholar]
- 48.Miki D., Itoh,R., Moritoh,S. and Shimamoto,K. (2004) Gene-specific suppression of a multigene family in plants by RNA silencing and its epigenetic effects on the target genes. In: Report at the annual meeting of the American Society of Plant Biologists, Lake Buena Vista, FL. July 2004. [Google Scholar]
- 49.Bartel D.P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116, 281–297. [DOI] [PubMed] [Google Scholar]
- 50.Chen X. (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science, 303, 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bartel B. and Bartel,D.P. (2003) MicroRNAs: At the root of plant development. Plant Physiol., 132, 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palatnik J.F., Allen,E., Wu,X., Schommer,C., Schwab,R., Carrington,J.C. and Weigel,D. (2003) Control of leaf morphogenesis by microRNAs. Nature, 425, 257–263. [DOI] [PubMed] [Google Scholar]
- 53.Rhoades M.W., Reinhart,B.J., Lim,L.P., Burge,C.B., Bartel,B. and Bartel,D.P. (2002) Prediction of plant microRNA targets. Cell, 110, 513–520. [DOI] [PubMed] [Google Scholar]