Abstract
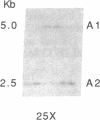
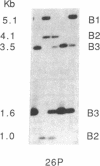
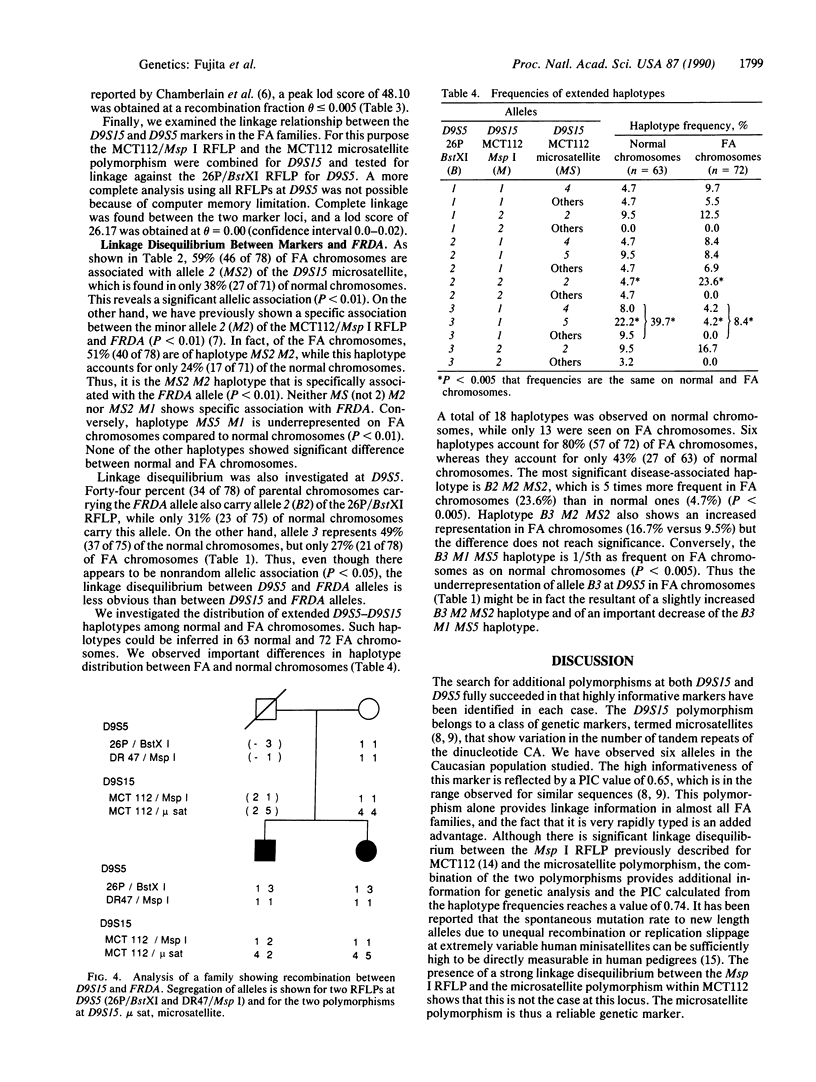
The gene for Friedreich ataxia (FA), a severe recessive neurodegenerative disease, has previously been shown to be tightly linked to the polymorphic markers D9S15 and D9S5 on human chromosome 9. In addition, the observation of linkage disequilibrium suggested that D9S15 is within 1 centimorgan (cM) of the disease locus, FRDA. Although D9S5 did not show recombination with FRDA, its localization was less precise (0-5 cM) due to its lower informativeness. We have now identified additional polymorphisms at both marker loci. Two cosmids spanning 50 kilobases around D9S5 were isolated, and a probe derived from one of them detects an informative three-allele polymorphism. We have found a highly polymorphic microsatellite sequence at D9S15 which is rapidly typed by the DNA polymerase chain reaction. The polymorphism information contents at the D9S5 and D9S15 loci have been increased from 0.14 to 0.60 and from 0.33 to 0.74, respectively. With the additional polymorphisms the lod (log10 odds ratio) score for the D9S15-FRDA linkage is now 48.10 at recombination fraction theta = 0.005 and for D9S5-FRDA, the lod score is 27.87 at theta = 0.00. We have identified a recombinant between D9S15 and FRDA. However, due to the family structure, it will be of limited usefulness for more precise localization of FRDA. The linkage disequilibrium previously observed between D9S15 and FRDA is strengthened by analysis of the haplotypes using the microsatellite polymorphism, while weaker but significant disequilibrium is found between D9S5 and FRDA. Extended haplotypes that encompass D9S5 and D9S15 show a strikingly different distribution between chromosomes that carry the FA mutation and normal chromosomes. This suggests that both marker loci are less than 1 cM from the FRDA gene and that a small number of mutations account for the majority of FA cases in the French population studied. D9S5 and D9S15 are thus excellent start points to isolate the disease gene.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barbeau A., Roy M., Sadibelouiz M., Wilensky M. A. Recessive ataxia in Acadians and "Cajuns". Can J Neurol Sci. 1984 Nov;11(4 Suppl):526–533. doi: 10.1017/s0317167100034995. [DOI] [PubMed] [Google Scholar]
- Barbeau A. The Quebec Cooperative Study of Friedreich's Ataxia: 1974-1984--10 years of research. Can J Neurol Sci. 1984 Nov;11(4 Suppl):646–660. doi: 10.1017/s0317167100035228. [DOI] [PubMed] [Google Scholar]
- Carlson M., Nakamura Y., Krapcho K., Fujimoto E., O'Connell P., Leppert M., Lathrop G. M., Lalouel J. M., White R. Isolation and mapping of a polymorphic DNA sequence pMCT112 on chromosome 9q (D9S15). Nucleic Acids Res. 1987 Dec 23;15(24):10614–10614. doi: 10.1093/nar/15.24.10614-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain S., Shaw J., Rowland A., Wallis J., South S., Nakamura Y., von Gabain A., Farrall M., Williamson R. Mapping of mutation causing Friedreich's ataxia to human chromosome 9. Nature. 1988 Jul 21;334(6179):248–250. doi: 10.1038/334248a0. [DOI] [PubMed] [Google Scholar]
- Chamberlain S., Shaw J., Wallis J., Rowland A., Chow L., Farrall M., Keats B., Richter A., Roy M., Melancon S. Genetic homogeneity at the Friedreich ataxia locus on chromosome 9. Am J Hum Genet. 1989 Apr;44(4):518–521. [PMC free article] [PubMed] [Google Scholar]
- Dean G., Chamberlain S., Middleton L. Friedreich's ataxia in Kathikas-Arodhes, Cyprus. Lancet. 1988 Mar 12;1(8585):587–587. doi: 10.1016/s0140-6736(88)91378-5. [DOI] [PubMed] [Google Scholar]
- Fujita R., Agid Y., Trouillas P., Seck A., Tommasi-Davenas C., Driesel A. J., Olek K., Grzeschik K. H., Nakamura Y., Mandel J. L. Confirmation of linkage of Friedreich ataxia to chromosome 9 and identification of a new closely linked marker. Genomics. 1989 Jan;4(1):110–111. doi: 10.1016/0888-7543(89)90323-6. [DOI] [PubMed] [Google Scholar]
- Geoffroy G., Barbeau A., Breton G., Lemieux B., Aube M., Leger C., Bouchard J. P. Clinical description and roentgenologic evaluation of patients with Friedreich's ataxia. Can J Neurol Sci. 1976 Nov;3(4):279–286. doi: 10.1017/s0317167100025464. [DOI] [PubMed] [Google Scholar]
- Hanauer A., Chery M., Fujita R., Driesel A. J., Gilgenkrantz S., Mandel J. L. The Friedreich ataxia gene is assigned to chromosome 9q13-q21 by mapping of tightly linked markers and shows linkage disequilibrium with D9S15. Am J Hum Genet. 1990 Jan;46(1):133–137. [PMC free article] [PubMed] [Google Scholar]
- Harding A. E. Friedreich's ataxia: a clinical and genetic study of 90 families with an analysis of early diagnostic criteria and intrafamilial clustering of clinical features. Brain. 1981 Sep;104(3):589–620. doi: 10.1093/brain/104.3.589. [DOI] [PubMed] [Google Scholar]
- Heilig R., Lemaire C., Mandel J. L. A 230kb cosmid walk in the Duchenne muscular dystrophy gene: detection of a conserved sequence and of a possible deletion prone region. Nucleic Acids Res. 1987 Nov 25;15(22):9129–9142. doi: 10.1093/nar/15.22.9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu L. C., Yoshida A., Mohandas T. Chromosomal assignment of the genes for human aldehyde dehydrogenase-1 and aldehyde dehydrogenase-2. Am J Hum Genet. 1986 May;38(5):641–648. [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A. J., Royle N. J., Wilson V., Wong Z. Spontaneous mutation rates to new length alleles at tandem-repetitive hypervariable loci in human DNA. Nature. 1988 Mar 17;332(6161):278–281. doi: 10.1038/332278a0. [DOI] [PubMed] [Google Scholar]
- Kerem B., Rommens J. M., Buchanan J. A., Markiewicz D., Cox T. K., Chakravarti A., Buchwald M., Tsui L. C. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989 Sep 8;245(4922):1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- Lathrop G. M., Lalouel J. M., Julier C., Ott J. Multilocus linkage analysis in humans: detection of linkage and estimation of recombination. Am J Hum Genet. 1985 May;37(3):482–498. [PMC free article] [PubMed] [Google Scholar]
- Litt M., Luty J. A. A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am J Hum Genet. 1989 Mar;44(3):397–401. [PMC free article] [PubMed] [Google Scholar]
- Murray J. C., Shiang R., Carlock L. R., Smith M., Buetow K. H. Rapid RFLP screening procedure identifies new polymorphisms at albumin and alcohol dehydrogenase loci. Hum Genet. 1987 Jul;76(3):274–277. doi: 10.1007/BF00283622. [DOI] [PubMed] [Google Scholar]