Abstract
Familial adenomatous polyposis (FAP) is an autosomal-dominantly inherited form of colorectal cancer (CRC) caused by mutation in the adenomatous polyposis coli (APC) gene. Our ability to exhaustively screen for APC mutations identify microsatellite-stable and APC-mutation negative familial CRC patients, enabling us to search for novel genes. We performed genome-wide scan on two affected siblings of one family and 88 ethnicity- and gender-matched healthy controls to identify deletions shared by the siblings. Combined loss of heterozygosity, copy number and allelic-specific copy number analysis uncovered 5 shared deletions. Long-range polymerase chain reaction (PCR) confirmed chromosome 19q13 deletion, which was subsequently found in one other family. The 32 kb deleted region harbors the CYP2A7 gene and was enriched with enhancer, repressor and insulator sites. The wildtype allele was lost in the polyps of the proband. Further, real-time RT-PCR assays showed that expressions of MIA and MIA-RAB4B located 35 kb upstream of the deletion, were up-regulated in the polyps compared to the matched mucosa of the proband. MIA-RAB4B, the read-through long non-coding RNA (lncRNA), RAB4B, PIM2 and TAOK1 share common binding site of a microRNA, miR-24, in their 3’UTRs. PIM2 and TAOK1, two target oncogenes of miR-24, were co-ordinately up-regulated with MIA-RAB4B in the polyps, suggesting that MIA-RAB4B could function as competitive endogenous RNA to titrate miR-24 away from its other targets. The data suggest that the 19.13 deletion disrupted chromatin boundary, leading to altered expression of several genes and lncRNA, could contribute to colorectal cancer via novel genetic and epigenetic mechanisms.
Introduction
Colorectal cancer (CRC) is one of the most frequent cancers and a leading cause of cancer mortality in the developed world. There are two main autosomal-dominantly inherited CRC, familial adenomatous polyposis (FAP) and hereditary non-polyposis colorectal cancer (HNPCC). The former is caused mainly by germline mutation in the adenomatous polyposis coli (APC) gene [1–3] and the latter by mismatch repair genes (MMR) characterized by microsatellite instability [4–6]. Our ability to exhaustively screen for these genes have enabled us to identify familial CRC patients who are microsatellite-stable and APC-mutation negative, implying that the underlying defect is in other genes although the clinical manifestation and mode of inheritance is similar to FAP [7, 8].
We have previously identified a copy number variant (CNV) region that regulates PPM1L, a novel serine-threonine phosphatase, in APC-mutation negative familial CRC patients with aggressive polyposis via genome-wide scan [9]. A recent comprehensive survey has also reported that microsatellite stable, non-polyposis familial Caucasian patients had increased CNV burden indicating further that structural variation could play significant role in familial colorectal tumorigenesis [10]. In this study, we embarked on the search of novel genes in one of the APC-mutation negative families with attenuated polyposis in which other possible tumour suppressors had been ruled out. Whole genome genotyping and CNV analysis identified a 32 kb genomic deletion at chromosome 19q13 shared by both affected siblings of the family that was not found in 88 ethnicity- (Chinese) and gender- (male) matched healthy controls.
We showed that the expression of CYP2A7 located within the deletion was down- regulated. We further showed that MIA, and its run-through lncRNA, MIA-RAB4B, located 35 kb upstream of the deletion, were specifically up-regulated in the polyps compared to matched colonic mucosa of the proband but was correspondingly down regulated in that of APC mutation-positive FAP patients. Moreover, MIA-RAB4B could possibly function as a competitive endogenous RNA (ceRNA) to titrate away micro-RNA 24, resulting in the up-regulation of its other target oncogenes, PIM2 and TAOK1.
The results imply that the 32 kb genomic deletion at chromosome 19q13 remove regulatory elements and reposition several upstream oncogenes, leading to their activation thus contributing to tumorigenesis.
Materials and methods
Patient samples and phenotypic classification
Polyposis patients were registered with the Singapore Polyposis Registry. The diagnostic criteria for attenuated FAP were the presence of 20 or more adenomas throughout the colon and rectum as documented in the patient’s histopathological records. All probands were diagnosed at colonoscopy or colectomy. The proband was interviewed and the detailed pedigree of the family and clinical history of proband and other affected members documented. Clinicopathological data were retrieved from histopathology reports and patient medical records of Singapore General Hospital (SGH). Peripheral lymphocyte and tissue specimens were collected with written informed consent. This study was approved by SingHealth Centralized Institutional Review Board B.
Exclusion of APC, MutYH, BMPR1A, PTEN, p53, POLD1, POLE and NTHL1 germline mutations
APC germline mutation was excluded by a combination of the protein truncation test, multiplex ligation-dependent probe amplification (MLPA) and differential expression assay as previously described [7]. MutYH, BMPR1A, PTEN and p53 germline mutations were excluded by amplification of exonic fragments followed by Sangers’ sequencing on ABI 3100 automated sequencer. Exclusion of POLD1, POLE and NTHL1 were performed on the mutation hot spots c.1421T>C, c.1433G>A (POLD1), c.1270C>G (POLE), and c.268C>T (NTHL1) respectively. The sequences of the primer pairs used are available upon request.
Microsatellite instability assay
Microsatellite instability assay was performed on matched mucosa and tumour samples using the MSI Analysis System from Promega (Madison, WI). Briefly, 1-2ng DNA from matched mucosa and tumour tissues was used in each multiplex PCR reaction according to manufacturer’s protocol [11]. The fluorescence labeled PCR products were separated on ABI PRISM 310 Genetic Analyzer and analyzed using the ABI Gene Scan Analysis software. A microsatellite-stable tumour (MSS) is a tumour with identical profile as matched mucosa in all 5 mononucleotide markers.
Genome-wide genotyping and copy number analysis
Genomic DNA was extracted using the high salt method [12]. Whole-genome scan was performed with Affymetrix GeneChip Human Mapping SNP Array 6.0 consisting 906, 600 SNPs and 946,000 copy number probes as per the manufacturer’s instructions (Affymetrix, Santa Clara, CA). Arrays were scanned with the Affymetrix 3000-7G scanner and analysed with the Genotyping Console v3. Only arrays with call rate 97% or more were accepted for further data processing [13].
The CEL (cell intensity) files from the genome-wide scan were imported into Partek Genomics Suite for bioinformatics analysis. The CEL files were subjected to principal component and copy number analysis. The copy number tool made use of the Hidden Markov Model (HMM). Unpaired copy number analysis were performed on CEL files from the lymphocytic DNAs of the affected siblings and that of the 88 ethnicity- and gender-matched controls as reference baseline to identify deletions common to both siblings but not found in the reference. Three different algorithms, loss of heterozygosity (LOH), copy number and allelic-specific copy number analysis were performed using default settings of the software. Only deletions that were called by all three algorithms were followed-up. In addition, the CEL files from the two polyp DNAs of the proband were compared to that of the matched lymphocytic DNA as reference (paired copy number analysis) to determine whether the second allele was deleted.
Data mining the confirmed deleted region
The flanking region surrounding the genomic deletion was imported into the Repeat Masker program (http://www.repeatmasker.org/) to search for repetitive sequences. The 1Mb region encompassing the deletion was further interrogated with the NCBI (http://www.ncbi.nlm.nih.bov/), Ensembl (http://www.ensembl.org/) and the UCSC (http://genome.ucsc.edu/) databases with emphasis on the chromatin modification tracks under ENCODE to identify regulatory elements.
Long range polymerase chain reaction
The reactions were performed with the Expand long Range, dNTPack (Roche). The 25-μl reaction volume contained 1X expand long range buffer with 12.5mM MgCl2, 500μM PCR nucleotide mix, 0.3μM of each primer, 1% DMSO, 3.5U enzyme mix and 250 ng template DNA. The internal control is a 6kb BMPR1A amplicon at chromosome 10q23. The amplification reaction consisted of denaturing at 92°C for 2 min, followed by 10 cycles of 92°C for 10s, 63°C for 15s and 68°C for 8 min, 25 cycles of 92°C for 10s, 63°C for 15s and 68°C for 8 min plus an extra 20s elongation per cycle, and final elongation at 68°C for 7 min as previously performed [14].
Real-time RT-PCR assay and primers
cDNA was synthesized from total RNA using ABI High-Capacity cDNA Reverse Transcription kit according to the protocols of the manufacturer (http://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_042557.pdf). The cDNA generated was used as template in real-time PCR reactions with ABI SYBR-Green PCR master mix on MicroAmp® Optical 384-well reaction plate and ran on ABI 7900HT. All reactions were performed in quadruplicate. The primers for the amplifications were available upon request. Validation experiments verified that the efficiencies of amplification of the target genes and beta-actin, the internal reference, were approximately equal. Hence, relative quantification for each gene (2-ΔΔCt) in the polyps compared to the matched mucosa was determined using the comparative Ct method [15].
Results
Identification of APC-mutation negative familial CRC patients
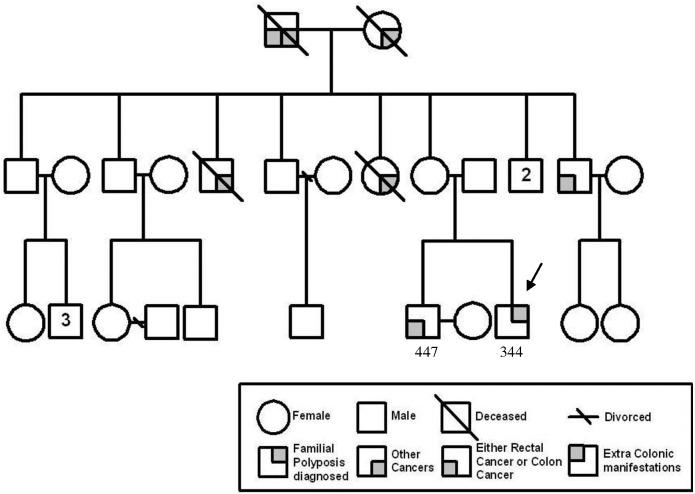
We screened for APC germline mutation in FAP patients by a combination of techniques that enabled us to achieve a high APC mutation detection rate of over 90% in the families registered in the Singapore Polyposis Registry [7]. Consequently, we were able to identify a sub-class of familial CRC patients clinically diagnosed as FAP or attenuated-FAP but were APC-mutation negative. One of these families is represented in Fig 1. The proband, 344, was diagnosed as an attenuated FAP patient with 25 adenomatous polyps, precursors to carcinomas, at age 23. Subsequently, the elder brother, 447, presented with multiple mixed (adenomatous as well as juvenile and hyperplastic) polyps and cecal mucinous adenocarcinoma at age 35 and underwent right hemicolectomy. No FAP-associated extracolonic manifestation was recorded.
Fig 1. Pedigree of APC mutation-negative familial CRC family 1.
Arrow indicates the proband.
Microsatellite instability assay performed indicated that the tumour is microsatellite- stable. We considered the possibility that mutation in other tumour suppressors could have caused the clinical phenotype. We searched for germline mutation in MutYH, BMPR1A, PTEN, p53, POLD1, POLE and NTHL1 genes by direct exonic Sanger’s sequencing and found the proband to be mutation-negative in these genes. It is thus likely that the disease-causing gene lies elsewhere although the clinical phenotype mimics that of FAP patients.
Prediction of 5 deletions shared by the two affected siblings
We embarked on genome wide scan of the lymphocytic and polyp DNA of the two affected siblings, 344 and 447, in this family (Fig 1) and 88 ethnicity- and gender-matched healthy controls using the Affymetrix SNP 6 arrays.
We imported the CEL files into third-party software, Partek Genomics Suite, and searched for germline deletions that were shared by these two brothers but were not found in the 88 healthy controls. The deletions were called only if they were identified by all three algorithms applied, loss of heterozygosity, copy number and allelic-specific copy number analysis. Partek Genomics Suite predicted 5 genomic deletions in four different chromosomes (Table 1).
Table 1. Five predicted germline deletions shared by the two affected siblings.
| Chr | Start* | Stop* | No. of Markers | Total size (bp) | Associated genes |
|---|---|---|---|---|---|
| 1q32.2 | 210076338 | 210085268 | 3 | 8931 | None |
| 3q28 | 189362217 | 189367813 | 11 | 5597 | TP63 |
| 11q11 | 55376390 | 55453292 | 55 | 76903 | OR4P4, OR4S2, OR4C6, OR4V1P, OR4P1P |
| 19q13.2 | 41360272 | 41392154 | 32 | 31883 | CYP2A7 |
| 19q13.31 | 43700184 | 43761448 | 23 | 61265 | PSG4, CEACAMP10, LOC284344, PSG9 |
* Physical positions based on GRCh37/hg19
Validation of the genomic deletion at chromosome 19q13
We ranked the 5 predicted deleted regions by the candidate genes residing within the deletions (Table 1). Other than the two highest ranked chromosomes 3q28 and 19q13.2 regions, the other three regions harbor either no candidate gene (chromosome 1q32.2) or genes unlikely to be implicated in polyposis or cancer such as olfactory receptor (chromosome 11q11) or pregnancy-specific genes (chromosome 19q13.31). We decided to validate the first two highest ranked regions at chromosomes 3q28 and 19q13 with a predicted deletion size of approximately 6 kb and 32 kb respectively by a secondary long-range PCR assay. Primers approximately 3 kb from the predicted deletion sites at 3q28 amplified a 10 kb TP63 fragment in the lymphocytic DNAs of 273 (the ethnicity- and gender-matched healthy control), 344 and 447 (affected siblings) as well as in two different polyp DNAs of 344 with approximately equal intensity after normalization to the internal control BMPR1A (S1 Fig). Hence, the predicted 6 kb deletion at 3q28 (encompassing TP63) was not validated.
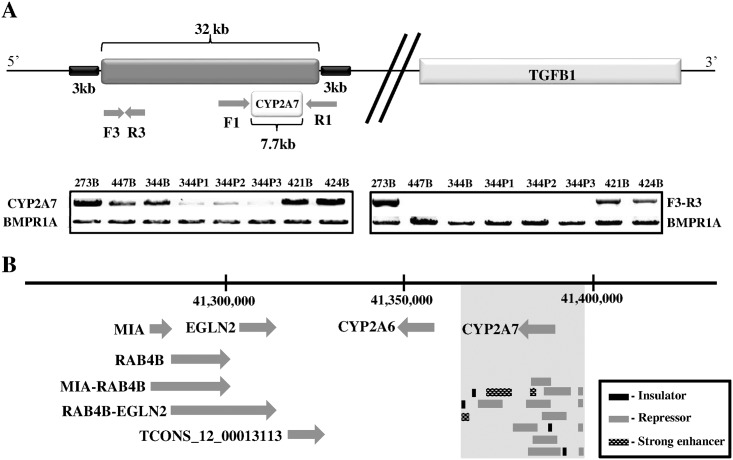
We then interrogated the predicted deleted region at 19q13 with a pair of long-range PCR primers (F1 and R1) which amplified a 7.7 kb fragment encompassing the CYP2A7 gene (Fig 2A). The amplified fragments from the lymphocytic DNA of the two affected siblings, 344 and 447, were only about half the intensity of 273, 421 and 424 after normalization to the internal control, indicating that the template DNA was probably halved. 421 and 424 were the APC mutation-positive controls. The fragment intensity was further reduced in the DNA isolated from the three polyps of 344 suggesting that the deletion could now be on both chromosomes. Copy number plot generated from the paired analysis between 344 and the polyps P1 and P2 showed that indeed a similar region of 28.5 kb is deleted in the polyps (S2 Fig, S1 Table). We designed another set of PCR primers (F3-R3) spanning the intergenic region within the deletion and showed further that the expected 2.5 kb fragment was clearly amplified in 273, 421 and 424 only (Fig 2A).
Fig 2.
(A) Schematic of the 32 kb genomic deletion at chromosome 19q13. The gel pictures represent the long range PCR products amplified with primer pairs F1-R1 and F3-R3. The internal control is a 6 kb BMPR1A fragment from chromosome 10q23. 273, male healthy control; 344, proband; 447, another affected sibling; 421 and 424; APC mutation-positive FAP patients; B, blood; P, polyp. (B) The genes and lncRNAs in the 35 kb region upstream of the deletion. The deletion is shown as vertical grey block, with relevant regulatory element tracks extracted from the UCSC browser.
We screened the probands of another eleven APC mutation-negative families using the same set of primers (F1-R1) and found this deletion in one other Chinese family.
In silico analysis of the 1 Mb region encompassing the deletion
Interrogating flanking regions of the deletion with the repeat masker program revealed multiple identical repetitive sequences of the MIR and Alu families (S3 Fig). Since the similarity scores of the Alu families of elements flanking the breakpoints are higher than the MIR’s, the genomic deletion is likely to be mediated by a homologous recombination event involving the Alu elements, as previously reported in a Singapore FAP family [16].
It is interesting to note that the only gene within the genomic deletion at 19q13 is CYP2A7, a member of a nicotine-metabolizing family of enzymes that was not previously considered as a typical tumour suppressor [17]. Hence, we speculated that the 32 kb deletion may harbour regulatory elements that could enhance or repress neighbouring genes as previously reported [9]. In silico analysis of the deleted region with the UCSC Browser tracts revealed a chromatin three-dimensional architecture highly inundated with many enhancer, insulator and repressor sites (Fig 2b). Insulators divide the chromosome into topologically associating domains (TAD) marked by the CTCF proteins [18]. Hence, the deletion could remove a TAD and alter the regulation of candidate tumour suppressors or oncogenes in the vicinity [19, 20].
We scanned a 1 Mb region encompassing the genomic deletion and identified several candidate genes. TGFβ1, located about 400kb downstream of the deletion (Fig 2A), is a ligand in the TGFβ signalling pathway that is known to play important role in colorectal tumourigenesis [21]. MIA, RAB4B and EGLN2 are three genes located in close proximity and about 35 kb upstream of the deletion (Fig 2B). MIA and EGLN2 were previously implicated in tumourigenic processes such as cellular proliferation and invasion [22, 23]. RAB4B is a small GTPase in the RAS signalling pathway, the dysregulation of which led to cancers in many organs [24]. Two related read-through transcripts that were predicted to be long non-coding RNA (lnc-RNA), MIA-RAB4B and RAB4B-EGLN2, interspersed these three genes. In addition, another lnc-RNA in the vicinity, lncRNA_TCONS_|2_00013113, was predicted by the Encode “transcript of uncertain coding potential (TUCP)” algorithm (Fig 2B).
Expression of CYP2A7 and TGFβ1
Several lines of evidence have indicated that disruption of chromatin boundaries by deletion, leading to heterochromatin spreading and enhancer or silencer blocking, are stage- and tissue-specific [18, 25]. We therefore investigated the expressions of these candidate genes in the polyps compared to that of the matched mucosa in affected sibling 344. This was then contrasted to that of 421, an APC mutation-positive FAP patient with no deletion at 19q13.
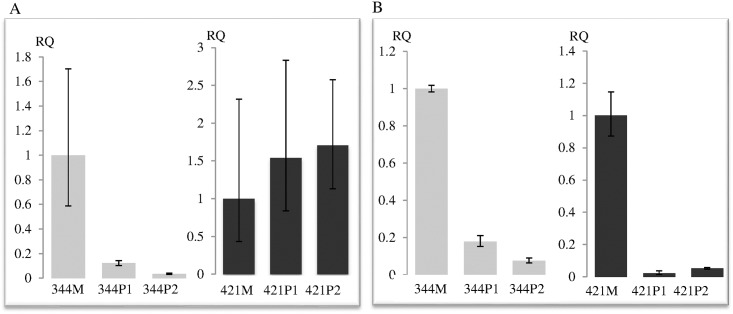
We interrogated first the expression of CYP2A7 and TGFβ1 by real-time RT-PCR assay. The expression of CYP2A7 was diminished ten-fold or more in the two polyps of 344 compared to the matched mucosa (Fig 3A). On the contrary, the expressions of CYP2A7 in the polyps of 421 were slightly elevated compared to that of the matched mucosa. The expression of TGFβ1, however, was similarly down-regulated in the polyps of both 344 and 421 (Fig 3B).
Fig 3.
Relative Quantitation (RQ) of (A) CYP2A7 and (B) TGFβ1 transcripts in the polyps normalized to matched mucosa of 344 and 421 respectively. Vertical bar denotes maximum and minimum RQ of each specimen. M, mucosa; P, polyp.
Expression of MIA, lncRNAs and EGLN2
We investigated next the expressions of the candidate genes 5’ to the genomic deletion, MIA, EGLN2, and their related read-through lncRNAs, MIA-RAB4B and RAB4B-EGLN2. It was not possible to interrogate the expression of RAB4B as it was not possible to design primers unique to this transcript alone.
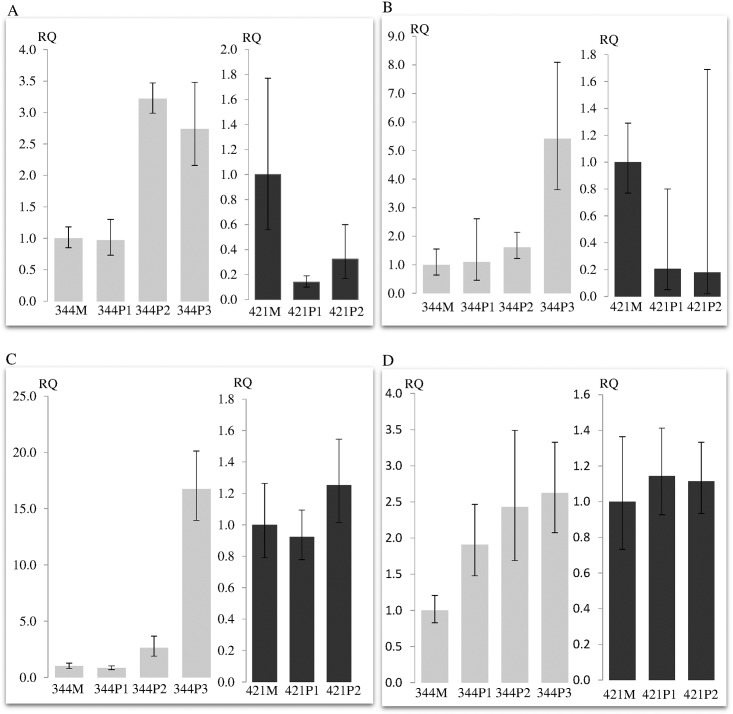
Although the expressions of MIA and MIA-RAB4B were consistently down-regulated in the polyps of 421 compared to the matched mucosa, the expressions of these genes were one to five fold up-regulated in the polyps of 344 (Fig 4A and 4B). The minimum and maximum relative quantitation (RQ-min and RQ-max) for MIA-RAB4B was large in the polyps of 421 as the expression of this read-through lncRNA is very low and thus cannot be accurately measured. The expression of EGLN2 and the second read-through lncRNA, RAB4B-EGLN2, remain in approximately the same level in the polyps of both 344 and 421 compared to their respective mucosa. Furthermore, the expression of another lncRNA in close proximity, lncRNA_TCONS_|2_00013113 (Fig 2B) remained at barely detectable level in the polyps of both 344 and 421 (data not shown).
Fig 4.
Relative quantitation of (A) MIA (B) MIA-RAB4B (C) PIM2 and (D) TAOK1 transcripts in the polyps normalized to matched mucosa of 344 and 421 respectively. MIA-RAB4B is the read-through lncRNAs of MIA and RAB4B. PIM2 and TAOK1 are two other target oncogenes of miR-24.
Expression of a subset of miR-24 regulated genes
In silico analysis with NCBI database indicated that MIA-RAB4B and RAB4B share the same 3’ untranslated region (UTR) including the seed recognition sites for miR-24. This suggests that RAB4B expression could be affected by the up-regualtion of MIA-RAB4B via miRNA titration [26]. The expression of RAB4B, however, cannot be investigated independently of MIA-RAB4B and RAB4B-EGLN2 (Fig 2B). We therefore searched for other miR-24-regulated, putative MIA-RAB4B ceRNA targets. Interrogation using miRNA target prediction softwares identified PIM2, TAOK1, and MAPK7 as other oncogeneic targets of miR-24. These genes were identified by all three predictive softwares, TargetScan, MiRanda and PicTar.
Interestingly, the expressions of PIM2 and TAOK1 were increased up to fifteen and two and half folds respectively in the polyps of 344 compared to the matched mucosa whilst the expressions of these genes in the polyps of 421 remained constant (Fig 4C and 4D). There was, however, no difference in the expression of MAPK7 in the polyps of 344 and 421 compared to their matched mucosa (data not shown).
Discussion
In this study, we report the identification of a 32 kb chromosome 19q13 deletion that specifically disrupts the expressions of CYP2A7, MIA and MIA-RAB4B lncRNA in an APC mutation-negative patient compared to two different APC mutation-positive patients. The overall workflow of this study is summarized in Supporting S4 Fig. It is postulated that this germline deletion could mimic the loss of APC and contribute to the initiation of the FAP-like phenotype in this APC mutation-negative microsatellite stable family 1.
Germline mutations in other known polyposis and cancer causing tumor suppressors such as MUTYH, BMPR1A, PTEN, TP53, POLD1, POLE and NTHL1 were also excluded. The polyps in proband 344 and affected sibling 447 of family 1 are adenomas with just a few mixed polyps documented in 447; hence it is unlikely that the SMAD4 gene (responsible for juvenile polyposis) is disease-causing. Chromosome 15q13.3 (harbouring GREM1) region was not amplified in both affected siblings 344 and 447 compared to the 88 ethnicity- and gender- matched controls (Table 1). Recent studies have not found GALNT12 at chromosome 9q22-33 to be a highly penetrant gene in Mendelian syndromes [27, 28]. We have also not identified this locus as a shared deleted region in family 1 which has autosomal dominant inheritance (Table 1).
Copy number analysis by three different algorithms of the dataset created from genome-wide scan of the two affected siblings of family 1 (Fig 1) and 88 ethnicity- and gender-matched healthy controls revealed 5 shared deletions in the siblings (Table 1). The deletion at chromosome 19q13 (but not the deletion at 3q28) was validated by long-range PCR (Fig 2A, S1 Fig). The data reemphasize that bioinformatics analysis has to be corroborated by independent secondary assay.
Notably, the 32 kb deletion encompassed the CYP2A7 gene only (Fig 2B). Both copies of CYP2A7 gene appeared to be lost in the polyps of the proband, 344, by bioinformatics analysis as well as long-range PCR experiments (Fig 2A and S2 Fig). The loss of the wildtype allele is consistent with this deletion functioning as a tumor suppressor. Thus, it was not surprising that CYP2A7 expression in the polyps compared to the matched mucosa of 344 was very much down-regulated while the expression in the polyps of 421 (the APC mutation-positive patient with no deletion at 19q13) were similar to the matched mucosa (Fig 3A).
No miRNA gene resides in this 32 kb region. However, the region is inundated with several strong enhancers, repressors and insulators (Fig 2B). Thus, the deletion is predicted to disrupt chromatin architecture and consequently alter the regulation of neighbouring genes [19, 20]. We showed that this cis-regulation was specific as only the expressions of MIA and MIARAB4B, located 35 kb upstream of the deletion, were up-regulated in the polyps of 344 but consistently down-regulated in the polyps of 421 (Fig 4A and 4B). The expression of TGFβ1, on the other hand, was similarly down-regulated in the polyps of both 344 and 421, suggesting that the deletion did not affect TGFβ1 regulation in the colonic mucosa and that regulating element(s) for TGFβ1 expression resides outside of the deletion and was not affected by the chromatin repositioning (Fig 3B).
Melanoma inhibitory activity (MIA) has been shown to increase the invasiveness of cancer cells not only in melanomas but also in solid tumors such as pancreatic and gastric carcinomas [29]. Hence, MIA up regulation in the polyps of 344 is likely to lead to increased invasiveness in CRC (Fig 4A). Notably, the read-through lncRNA, MIA-RAB4B, was also up-regulated (Fig 4B). A recent study has proposed a novel mechanism whereby the high mobility group AT-hook 2 (HMG2A) transcript promotes non-small cell lung cancer progression by competing with transforming growth factor β receptor III (Tgfbr3) for let-7 binding [26]. Since MIA-RAB4B and RAB4B share the same seed recognition site of miR-24, we wondered whether a similar mechanism could operate in this instance i.e. increased expression of MIA-RAB4B could compete away the binding of miR-24 to RAB4B, thus reducing its degradation. This derepression could conceivably be extended to other targets of miR-24. We investigated the expressions of other target oncogenes of miR-24 as the expression of RAB4B cannot be studied directly (since no RAB4B unique primers can be designed). The expressions of PIM2 and TAOK1 were up-regulated in the polyps of 344 but not in 421 (Fig 4C and 4D). The expression of MAPK7 in the polyps of both 344 and 421, however, were similarly down-regulated compared to their matched mucosa. Thus, in contrast to MAPK7, the co-ordinated overexpression of MIA-RAB4B, PIM2 and TAOK1 suggests that the latter two oncogenes could be critical targets of MIA-RAB4B ceRNA. PIM2 is a serine-threonine kinase that was found to be overexpressed in various solid tumours and involved in anti-apoptotic, metabolic and cell cycle regulation [30–32]. TAOK1 is a member of sterile 20 (STE20)-like kinases previously implicated in microtubule organisation and play active role in cytoskeletal stability and mitotic progression [33, 34]. The overexpression of PIM2 and TAOK1 oncogenes in 344 but not 421 polyps implies that the dysregulation of these genes and consequently their participatory pathways could possibly mimic the dysregulation due to loss of APC.
The 32 kb genomic deletion at chromosome 19q13 disrupts chromatin organization and alters the expressions of several genes in two out of twelve (16.7%) of the APC mutation-negative familial CRC families screened. In healthy individuals, the frequency of copy number loss in this region from the combined Database of Genomic Variants (DGV) (http://dgv.tcag.ca/dgv/app/home) entries is 1.68% (n = 7620 individuals). Thus, this copy number loss is significantly (Fisher’s Exact Test, p = 0.017) enriched in a subset of familial CRC patients previously screened to be mutation-negative in other possible disease-causing genes such as APC and BMPR1A.
The main limitation of the study is the lack of samples from other members of family 1 to show co-segregation of the disease with the 32 kb genomic deletion. Furthermore, no polyps were available from the second affected sibling 447 for the expression studies. Nevertheless, the expressions of the various genes studied in the polyps of another APC mutation-positive patient, 424, closely resembles that of 421 (S5 Fig), indicating that the expressions of these genes were consistent for patients with different APC germline mutations and distinct from that of 344 polyps. Patient 421 has a novel APC codon 1309 deletion while patient 424 has whole APC gene deletion. Furthermore, there was a second somatic deletion in the wildtype allele consistent with this locus being important for polyp development. Thus, although it is conceivable that the causative gene may still remain to be discovered, the 32 kb genomic deletion could contribute to the FAP-like phenotype in these APC mutation-negative patients via both genetic and epigenetic mechanisms.
Supporting information
The internal control is a 6 kb BMPR1A fragment from chromosome 10q23. 273, male healthy control; 344, proband; 447, another affected sibling; 421, APC mutation-positive FAP patient; B, blood; P, polyp.
(TIF)
(TIF)
Upper panel depicts the repetitive elements flanking the 32 kb deletion. Lower panel shows the homology of the 13.6 kb regions 5’ and 3’ to the deletion. The red region [880bp] is 98% identical; green a [1890bp] and green b [3398bp] regions are 97% and 98% identical; yellow region [7427bp] is 95% identical and grey region [1153bp] is 98% identical to each other.
(TIF)
(TIF)
Expressions of the transcripts are similar for both 421 and 424 but distinct from that of 344. Vertical bar denotes maximum and minimum RQ of each specimen. M, mucosa; P, polyp.
(TIF)
(DOCX)
Acknowledgments
We thank Ms. Goh Wei Lin from the Department of Colorectal Surgery for technical assistance, and the Department of Clinical Research, SGH for use of core facilities.
Data Availability
The genomic deletion NC_000019.9:g.41360272_41392154del has been submitted to the ClinVar database (SCV000256863).
Funding Statement
This study was funded in part by the National Medical Research Council Singapore (NMRC/1193/2008 and NMRC/OFIRG/0004/2016). The rest of the funding was from the host institution, Singapore General Hospital. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66(3):589–600. [DOI] [PubMed] [Google Scholar]
- 2.Joslyn G, Carlson M, Thliveris A, Albertsen H, Gelbert L, Samowitz W, et al. Identification of deletion mutations and three new genes at the familial polyposis locus. Cell. 1991;66(3):601–13. [DOI] [PubMed] [Google Scholar]
- 3.Kinzler KW, Nilbert MC, Su LK, Vogelstein B, Bryan TM, Levy DB, et al. Identification of FAP locus genes from chromosome 5q21. Science. 1991;253(5020):661–5. [DOI] [PubMed] [Google Scholar]
- 4.Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368(6468):258–61. 10.1038/368258a0 [DOI] [PubMed] [Google Scholar]
- 5.Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75(6):1215–25. [DOI] [PubMed] [Google Scholar]
- 6.Papadopoulos N, Nicolaides NC, Wei YF, Ruben SM, Carter KC, Rosen CA, et al. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263(5153):1625–9. [DOI] [PubMed] [Google Scholar]
- 7.Cao X, Hong Y, Eu KW, Loi C, Cheah PY. Singapore familial adenomatous polyposis (FAP) patients with classical adenomatous polyposis but undetectable APC mutations have accelerated cancer progression. The American journal of gastroenterology. 2006;101(12):2810–7. 10.1111/j.1572-0241.2006.00842.x [DOI] [PubMed] [Google Scholar]
- 8.Yao J, Eu KW, Seow-Choen F, Vijayan V, Cheah PY. Microsatellite instability and aneuploidy rate in young colorectal-cancer patients do not differ significantly from those in older patients. Int J Cancer. 1999;80(5):667–70. [DOI] [PubMed] [Google Scholar]
- 9.Thean LF, Loi C, Ho KS, Koh PK, Eu KW, Cheah PY. Genome-wide scan identifies a copy number variable region at 3q26 that regulates PPM1L in APC mutation-negative familial colorectal cancer patients. Genes, chromosomes & cancer. 2010;49(2):99–106. [DOI] [PubMed] [Google Scholar]
- 10.Yang R, Chen B, Pfutze K, Buch S, Steinke V, Holinski-Feder E, et al. Genome-wide analysis associates familial colorectal cancer with increases in copy number variations and a rare structural variation at 12p12.3. Carcinogenesis. 2014;35(2):315–23. 10.1093/carcin/bgt344 [DOI] [PubMed] [Google Scholar]
- 11.Hong Y, Downey T, Eu KW, Koh PK, Cheah PY. A 'metastasis-prone' signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clinical & experimental metastasis. 2010;27(2):83–90. [DOI] [PubMed] [Google Scholar]
- 12.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thean LF, Li HH, Teo YY, Koh WP, Yuan JM, Teoh ML, et al. Association of Caucasian-identified variants with colorectal cancer risk in Singapore Chinese. PLoS One. 2012;7(8):e42407 10.1371/journal.pone.0042407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheah PY, Wong YH, Chau YP, Loi C, Lim KH, Lim JF, et al. Germline bone morphogenesis protein receptor 1A mutation causes colorectal tumorigenesis in hereditary mixed polyposis syndrome. The American journal of gastroenterology. 2009;104(12):3027–33. 10.1038/ajg.2009.542 [DOI] [PubMed] [Google Scholar]
- 15.Hong Y, Ho KS, Eu KW, Cheah PY. A susceptibility gene set for early onset colorectal cancer that integrates diverse signaling pathways: implication for tumorigenesis. Clin Cancer Res. 2007;13(4):1107–14. 10.1158/1078-0432.CCR-06-1633 [DOI] [PubMed] [Google Scholar]
- 16.Cao X, Eu KW, Seow-Choen F, Zhao Y, Cheah PY. Topoisomerase-I- and Alu-mediated genomic deletions of the APC gene in familial adenomatous polyposis. Hum Genet. 2001;108(5):436–42. [DOI] [PubMed] [Google Scholar]
- 17.Godoy W, Albano RM, Moraes EG, Pinho PR, Nunes RA, Saito EH, et al. CYP2A6/2A7 and CYP2E1 expression in human oesophageal mucosa: regional and inter-individual variation in expression and relevance to nitrosamine metabolism. Carcinogenesis. 2002;23(4):611–6. [DOI] [PubMed] [Google Scholar]
- 18.Heger P, Wiehe T. New tools in the box: an evolutionary synopsis of chromatin insulators. Trends in genetics: TIG. 2014;30(5):161–71. 10.1016/j.tig.2014.03.004 [DOI] [PubMed] [Google Scholar]
- 19.Phillips-Cremins JE, Corces VG. Chromatin insulators: linking genome organization to cellular function. Molecular cell. 2013;50(4):461–74. 10.1016/j.molcel.2013.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wala J, Beroukhim R. CANCER. The oncogene makes its escape. Science. 2016;351(6280):1398–9. 10.1126/science.aaf5542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu M, Trobridge P, Wang Y, Kanngurn S, Morris SM, Knoblaugh S, et al. Inactivation of TGF-beta signaling and loss of PTEN cooperate to induce colon cancer in vivo. Oncogene. 2014;33(12):1538–47. 10.1038/onc.2013.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aung PP, Oue N, Mitani Y, Nakayama H, Yoshida K, Noguchi T, et al. Systematic search for gastric cancer-specific genes based on SAGE data: melanoma inhibitory activity and matrix metalloproteinase-10 are novel prognostic factors in patients with gastric cancer. Oncogene. 2006;25(17):2546–57. 10.1038/sj.onc.1209279 [DOI] [PubMed] [Google Scholar]
- 23.Zhang Q, Gu J, Li L, Liu J, Luo B, Cheung HW, et al. Control of cyclin D1 and breast tumorigenesis by the EglN2 prolyl hydroxylase. Cancer cell. 2009;16(5):413–24. 10.1016/j.ccr.2009.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer cell. 2014;25(3):272–81. 10.1016/j.ccr.2014.02.017 [DOI] [PubMed] [Google Scholar]
- 25.Chetverina DA, Elizar'ev PV, Georgiev PG, Erokhin MM. [Interactions of 1A2 insulator with promoter of hsp 70 gene in Drosophila melanogaster]. Genetika. 2013;49(4):429–38. [DOI] [PubMed] [Google Scholar]
- 26.Kumar MS, Armenteros-Monterroso E, East P, Chakravorty P, Matthews N, Winslow MM, et al. HMGA2 functions as a competing endogenous RNA to promote lung cancer progression. Nature. 2014;505(7482):212–7. 10.1038/nature12785 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Segui N, Pineda M, Navarro M, Lazaro C, Brunet J, Infante M, et al. GALNT12 is not a major contributor of familial colorectal cancer type X. Human mutation. 2014;35(1):50–2. 10.1002/humu.22454 [DOI] [PubMed] [Google Scholar]
- 28.Tomlinson I. The Mendelian colorectal cancer syndromes. Annals of clinical biochemistry. 2015;52(Pt 6):690–2. 10.1177/0004563215597944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El Fitori J, Kleeff J, Giese NA, Guweidhi A, Bosserhoff AK, Buchler MW, et al. Melanoma Inhibitory Activity (MIA) increases the invasiveness of pancreatic cancer cells. Cancer cell international. 2005;5(1):3 10.1186/1475-2867-5-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu Z, Zhao X, Huang L, Zhang T, Yang F, Xie L, et al. Proviral insertion in murine lymphomas 2 (PIM2) oncogene phosphorylates pyruvate kinase M2 (PKM2) and promotes glycolysis in cancer cells. J Biol Chem. 2013;288(49):35406–16. 10.1074/jbc.M113.508226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aguirre E, Renner O, Narlik-Grassow M, Blanco-Aparicio C. Genetic Modeling of PIM Proteins in Cancer: Proviral Tagging and Cooperation with Oncogenes, Tumor Suppressor Genes, and Carcinogens. Frontiers in oncology. 2014;4:109 10.3389/fonc.2014.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narlik-Grassow M, Blanco-Aparicio C, Carnero A. The PIM family of serine/threonine kinases in cancer. Medicinal research reviews. 2014;34(1):136–59. 10.1002/med.21284 [DOI] [PubMed] [Google Scholar]
- 33.Tavares IA, Touma D, Lynham S, Troakes C, Schober M, Causevic M, et al. Prostate-derived sterile 20-like kinases (PSKs/TAOKs) phosphorylate tau protein and are activated in tangle-bearing neurons in Alzheimer disease. J Biol Chem. 2013;288(21):15418–29. 10.1074/jbc.M112.448183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zihni C, Mitsopoulos C, Tavares IA, Ridley AJ, Morris JD. Prostate-derived sterile 20-like kinase 2 (PSK2) regulates apoptotic morphology via C-Jun N-terminal kinase and Rho kinase-1. J Biol Chem. 2006;281(11):7317–23. 10.1074/jbc.M513769200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The internal control is a 6 kb BMPR1A fragment from chromosome 10q23. 273, male healthy control; 344, proband; 447, another affected sibling; 421, APC mutation-positive FAP patient; B, blood; P, polyp.
(TIF)
(TIF)
Upper panel depicts the repetitive elements flanking the 32 kb deletion. Lower panel shows the homology of the 13.6 kb regions 5’ and 3’ to the deletion. The red region [880bp] is 98% identical; green a [1890bp] and green b [3398bp] regions are 97% and 98% identical; yellow region [7427bp] is 95% identical and grey region [1153bp] is 98% identical to each other.
(TIF)
(TIF)
Expressions of the transcripts are similar for both 421 and 424 but distinct from that of 344. Vertical bar denotes maximum and minimum RQ of each specimen. M, mucosa; P, polyp.
(TIF)
(DOCX)
Data Availability Statement
The genomic deletion NC_000019.9:g.41360272_41392154del has been submitted to the ClinVar database (SCV000256863).