Abstract
Neurofibromatosis 2 (NF2) is a tumor suppressor, although the molecular mechanism accounting for this effect remains unknown. Here, we show that merlin exerts its activity by inhibiting phosphatidylinositol 3-kinase (PI3-kinase), through binding to PIKE-L. Wild-type merlin, but not patient-derived mutant (L64P), binds PIKE-L and inhibits PI3-kinase activity. This suppression of PI3-kinase activity results from merlin disrupting the binding of PIKE-L to PI3-kinase. In addition, merlin suppression of PI3-kinase activity as well as schwannoma cell growth is abrogated by a single PIKE-L point mutation (P187L) that cannot bind merlin but can still activate PI3-kinase. Knocking down PIKE-L with RNA interference abolishes merlin's tumor-suppressive activity. Our data support the hypothesis that PIKE-L is an important mediator of merlin growth suppression.
Neurofibromatosis 2 (NF2) is a dominantly inherited disorder characterized by bilateral occurrence of vestibular schwannomas and other brain tumors, including meningiomas and ependymomas (1). The NF2 tumor suppressor gene encodes an intracellular membrane-associated protein, called merlin or schwannomin, which belongs to the band 4.1 family of cytoskeleton-associated proteins (2, 3). Inactivation of the NF2 gene and consequent lack of gene expression are the primary cause of this disease, although the molecular mechanism accounting for the tumor-suppressive activity remains unknown (4).
Merlin, like other ERM (ezrin/radix/moesin) proteins, is highly enriched in micovilli and filopodia as well as the ruffling edges of motile cells (5, 6). Overexpression of merlin results in dramatic changes in the actin cytoskeleton and impairs cell attachment and motility (7). Moreover, it can effectively suppress the growth of rat schwannoma cells, both in vitro and in vivo (4, 8). Knocking down merlin leads to alterations in actin cytoskeleton-mediated events and increases cell proliferation (9). merlin exists in “open” (inactive) and “closed” (growth-suppressive) conformations dictated by the ability of merlin to form an intramolecular association between the N and C termini of the protein (10–13). The full-length merlin I containing exon 17 exists in a closed conformation, whereas merlin II with exon 16 or disease-oriented mutants display open conformation. merlin cycles between open and closed conformations in vivo that differentially determine whether it forms heterooligomers with ERM proteins or other binding targets to transduce its growth-regulatory signal (14). Numerous merlin-binding partners have been identified, but none of these molecules provides substantial clues as to the tumor-suppressive activity of merlin.
PIKE [phosphatidylinositol 3-kinase (PI3-kinase) enhancer] is a brain-specific GTPase that binds to PI3-kinase and stimulates its lipid kinase activity (15). It exists in three isoforms, PIKE-S (short form), PIKE-L (long form), and PIKE-A, as the result of alternative splicing (PIKE-L and -S) or a differential transcription initiation site (PIKE-A). PIKE-S originally was identified in a yeast two-hybrid screen searching for binding partners of protein 4.1N, a neuronal member of the band 4.1 superfamily. Nerve growth factor treatment leads to PIKE-S activation by triggering the nuclear translocation of phospholipase C γ1, which acts as a physiological guanine-nucleotide-exchange factor for PIKE through its SH3 domain (16). Nerve growth factor treatment also elicits translocation of membrane-associated 4.1N to the nucleus, where it binds to PIKE-S. We showed previously that the PIKE-S/PI3-kinase signaling pathway is negatively regulated by protein 4.1N (15). PIKE-L occurs in both the cytoplasm and the nucleus. Recently, we showed that it forms a complex with Homer 1 and couples PI3-kinase to the metabotropic glutamate receptor, preventing neuronal apoptosis (17).
Here, we report that merlin specifically binds to PIKE-L and abolishes its stimulatory effect on PI3-kinase by blocking the association between PIKE-L and PI3-kinase. Patient-derived mutant L64P merlin does not interact with PIKE-L and has no effect on PI3-kinase activity. Moreover, P187L mutation on PIKE-L disrupts its interaction with merlin, leading to its failure to inhibit PI3-kinase.
Materials and Methods
Plasmids and Reagents. GST-tagged merlin, merlin N-terminal domain (NTD; residues 1–332), and merlin C-terminal domain (residues 342–595) in pGex vector were kindly provided by Vijaya Ramesh (Massachusetts General Hospital, Harvard Medical School, Boston). Mouse monoclonal anti-hemagglutinin (HA)-horseradish peroxidase, anti-Myc-horseradish peroxidase, anti-Flag, and anti-GST antibodies were from Sigma. Mouse monoclonal anti-Ser-473, anti-Thr-308, and anti-Akt antibodies were from Cell Signaling Technology (Beverly, MA). Rabbit polyclonal anti-NF2, anti-p85, and anti-p110 antibodies were from Santa Cruz Biotechnology. Protein A/G-conjugated agarose beads were from Calbiochem. Glutathione sepharose 4B was supplied by Amersham Pharmacia. Adenovirus expressing short hairpin (sh)-PIKE RNA was supplied by Welgene (Worcester, MA). All chemicals not listed above were purchased from Sigma.
Coimmunoprecipitation and in Vitro Binding and PI3-Kinase Assays.The experimental procedures for coimmunoprecipitation and in vitro binding and in vitro PI3-kinase assays are described in ref. 18.
Immunofluorescent Staining of Schwannoma Cells. Induced and uninduced cells were fixed with cold (–20°C) methanol for 5 min and then rehydrated by PBS for 1 min. Nonspecific sites were blocked by incubating with 200 μl of 1% BSA in PBS at 37°C for 15 min. PIKE-L was stained with mouse anti-PIKE antibody (1:300 dilution in PBS containing 1% BSA) and incubated (200 μl), and merlin was stained with rabbit polyclonal anti-NF2 antibody (1:250 dilution). The secondary antibodies are Texas red-labeled goat anti-rabbit and FITC-conjugated goat antimouse antibodies, respectively. The staining was performed as described in ref. 19.
Subcellular Fractionation from Schwannoma Cells. For membrane extracts, cells were lysed by mechanical disruption in cold hypotonic buffer (10 mM Hepes, pH 7.4/1 mM EDTA/protease inhibitors). The nuclei were pelleted by centrifugation at 750 × g for 10 min. Further centrifugation of the resulting supernatant at 1 × 105 × g for 1 h led to recovery of the cytosolic fraction (C). The pellet was extracted with a membrane extraction buffer (50 mM Tris, pH 7.4/1% Triton X-100/150 mM NaCl/1 mM EDTA/1 mM Na3VO4/protease inhibitors) and centrifuged at 1 × 105 × g for 1 h. This supernatant corresponded to the Triton X-100 soluble membrane extract (S). The final pellet was extracted with modified RIPA buffer (pH 7.5) and centrifuged at 1 × 105 × g for 5 min; this supernatant corresponded to the Triton X-100 insoluble fraction (I).
Infection of Schwannoma Cells with Adenovirus. Adenovirusexpressing WT dominant-negative (K413AS414N) or P187L point mutant PIKE-L was prepared as described in ref. 20. The virus was purified by CsCl banding with 1011 to 1012 plaque-forming units, introduced into doxycycline-induced or uninduced schwannoma cells, and cultured overnight. The GFP was monitored with an immunofluorescence microscope.
Assay with 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyl Tetrazolium Bromide (MTT). The same number of rat RT4-D6P2T schwannoma cells were induced with doxycycline for 1 day and infected with adenovirus expressing WT PIKE-L, dominant-negative PIKE-L-KS, or PIKE-L-P187L. Cells were incubated 48 h after infection with 0.5 mg of MTT per ml of fresh medium at 37°C for 1 h. The formazan products were dissolved in DMSO and quantified by measurement of the absorbance at 562 nm, which represents the number of proliferating cells.
Statistical Analysis. The results were expressed as means ± SEM calculated from the specified numbers of determination. Student's t test was used to compare individual data with control value.
Results
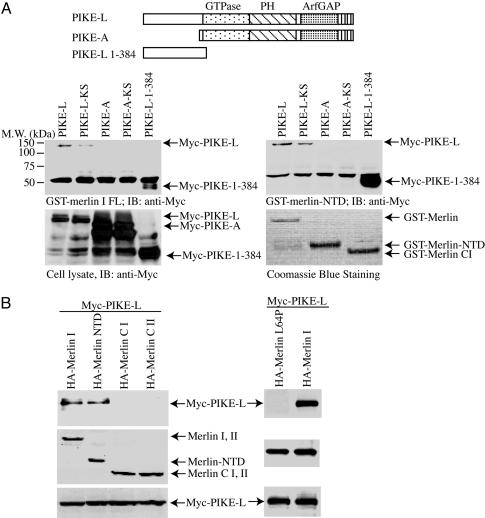
merlin Binds to PIKE-L. The homology between protein 4.1N and merlin suggests that merlin might have similar inhibitory effects on PI3-kinase activity. To explore this possibility, we conducted affinity chromatography interaction experiments to investigate the association between PIKE and full-length merlin and detected a robust interaction with PIKE-L. This interaction maps to the N terminus of PIKE-L (amino acid residues 1–384), consistent with the failure of the PIKE-A isoform, which lacks the N terminus of PIKE-L (21), to bind merlin in vitro (Fig. 1A Left Lower). Protein 4.1N binding to PIKE-L involves residues in the C terminus of protein 4.1N; thus, we examined the ability of the merlin NTD and C-terminal domain fragments to bind PIKE-L. We showed that merlin binding to PIKE-L requires residues in the FERM-containing NTD in vitro (Fig. 1A Right Lower). In contrast, we observed no binding with the C-terminal domain of merlin (Fig. 1B). We have demonstrated previously that the association between PIKE-S and PI3-kinase is GTP-dependent and can be abolished by mutations of K413 and S414 in the PIKE-L GTPase domain. The dominant inhibitory PIKE-L-KS mutant (K413AS414N) has been shown to prevent PIKE-L activation of PI3-kinase by binding to, but not activating, PI3-kinase (15, 17). We showed that the PIKE-L-KS mutant exhibits moderately reduced binding to merlin, suggesting that PIKE-L GTPase activity might be important in regulating the association between PIKE-L and merlin (Fig. 1A Left Lower).
Fig. 1.
PIKE-L interacts with merlin. (A) The FERM domain of merlin binds to the N terminus of PIKE-L in vitro. (Top) Illustration of the PIKE constructs used. GST-merlin and GST-FERM domain of merlin bind to PIKE-L but not PIKE-A. (Middle) Purified GST-merlin and GST-merlin-NTD were incubated with HEK293 cell lysates transfected with various myc-tagged PIKE constructs. After 3-h incubation at 4°C, the associated proteins were analyzed by Western blotting with anti-myc antibody. PIKE-L and PIKE-L N terminus (amino acid residues1–384) robustly binds to both merlin and merlin-NTD. The dominant-negative PIKE-L-KS displayed reduced binding activity to merlin. (Left Bottom) Protein expression of transfected constructs was confirmed by myc immunoblotting. (Right Bottom) Levels of GST-merlin recombinant proteins were verified by Coomassie blue staining. (B) PIKE-L interacts with merlin in vivo. Various HA-merlin constructs and myc-PIKE-L were cotransfected into HEK293 cells. PIKE-L was immunoprecipitated with anti-myc antibody, and bound proteins were visualized by Western blot with anti-HA antibody. Both full-length merlin and merlin NTD interacted with PIKE-L (Left Top, lanes 1 and 2). Patient-derived L64P mutant did not bind to PIKE-L (Right Top). Similar levels of all HA-merlin and myc-PIKE constructs were expressed in all experiments (Middle and Bottom).
To demonstrate the interaction between merlin and PIKE-L in vivo, we performed coimmunoprecipitation binding assays in HEK293 cells. In these experiments, both full-length merlin and the merlin NTD strongly bound to PIKE-L. In addition, merlin containing the patient-derived missense mutation (L64P) within the NTD, previously shown to lack growth-suppressor properties (8, 12), did not bind to PIKE-L (Fig. 1B Right).
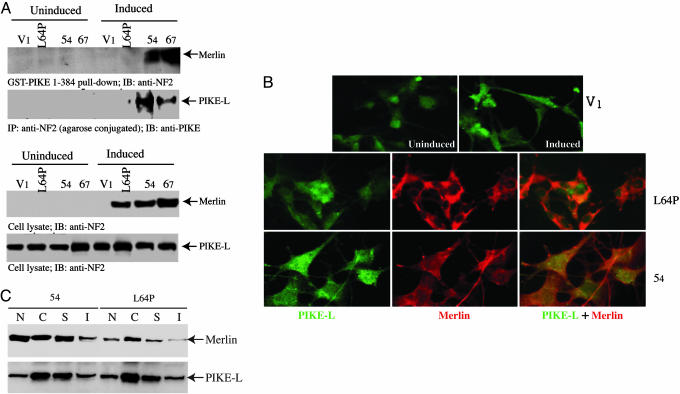
To determine whether the interaction between merlin and PIKE-L occurs in schwannoma cells, we used the RT4-D6P2T rat schwannoma cell line with inducible expression of either WT or L64P mutant merlin. Incubation of RT4 cell lysates with GST-PIKE-1–384 confirmed this interaction with WT, but not L64P mutant, merlin (Fig. 2A). We observed identical results by coimmunoprecipitation with agarose-conjugated anti-NF2 antibody in vivo (Fig. 2A). The induction of merlin in L64P, 54, and 67 cells was verified (Fig. 2A). An equal amount of PIKE-L was confirmed in both induced and uninduced cells (Fig. 2A).
Fig. 2.
PIKE-L interacts with WT merlin in schwannoma cells. (A) Rat RT4-D6P2T schwannoma cells stably transfected with empty vector (V1), merlin mutant L64P, or WT merlin (54 and 67, two clones) were either uninduced or induced with doxycycline for 1 day. One milligram of cell lysate was incubated with purified GST-PIKE-L-1–384. After 3-h incubation at 4°C, the associated proteins were analyzed by Western blotting with anti-NF2. (Top Upper) WT merlin, but not merlin mutant L64P, selectively binds to the N terminus of PIKE-L. (Top Lower) Coimmunoprecipitation with anti-NF2 reveals that PIKE-L specifically associates with WT merlin but not L64P mutant. (Bottom) The induced merlin was verified with anti-NF2 antibody. (B) PIKE-L colocalizes with WT merlin in schwannoma cells. Uninduced and induced schwannoma cells were stained with mouse anti-PIKE antibody and rabbit polyclonal anti-merlin antibody. (Top) PIKE-L resides in both the cytoplasm and the nucleus. (Middle and Bottom) Merlin L64P exclusively distributes in the cytoplasm, whereas WT merlin occurs in both compartments, colocalizing with PIKE-L in 54 cell line. (C) Subcellular fractionation of 54 and L64P cells. The cytosolic (C), membrane soluble (S), membrane insoluble (I), and nuclear (N) fractions were prepared. Immunoblotting analysis was performed with anti-PIKE and anti-merlin antibodies. (Upper) Both WT and mutant merlin display similar distribution patterns in C, S, and I fractions; by contrast, a very faint amount of L64P was observed in the nucleus compared to counterpart in the WT cells. (Lower) PIKE-L reveals identical subcellular distribution.
To investigate whether PIKE-L and merlin colocalize in intact cells, we conducted immunofluorescent staining with schwannoma cells. Before merlin induction, PIKE-L occurs in both the cytoplasm and the nucleus in V1 control cells (Fig. 2B Top). Similar subcellular distribution occurs in L64P, 54, and 67 cells (data not shown). The induced merlin L64P exclusively resides in the cytoplasm, whereas demonstrable PIKE-L locates in the nucleus (Fig. 2B Middle). By contrast, WT merlin (54 cell line) colocalizes with PIKE-L in the whole cell (Fig. 2B Bottom). To further evaluate these two proteins' colocalization, we performed subcellular fractionation with induced L64P and 54 cell lines. Both WT merlin and L64P display similar distribution in cytosolic (C), membrane soluble (S), and insoluble (I) fractions. However, compared with robust nuclear distribution of WT merlin in 54 cells, a faint amount of mutant merlin disperses in the nuclear fraction of L64P cells (Fig. 2C Upper), fitting with immunohistochemistry staining results. The modest level of L64P in the nuclear fraction might be due to unbroken cells. By contrast, the same distribution pattern of PIKE-L was detected in both cells (Fig. 2C Lower). Collectively, these results demonstrate that merlin and PIKE-L interact in vitro and in vivo and that this association requires residues in the N terminus of PIKE-L and the FERM (NTD) of merlin.
Pro-187 →Leu Mutation in PIKE-L Disrupts Its Interaction with merlin. To identify the region of PIKE-L required for merlin binding, we examined the binding of various truncations of PIKE-L to merlin in vitro, by using equal amounts of GST-PIKE truncation constructs and merlin (Fig. 5A, which is published as supporting information on the PNAS web site). In vitro binding assay revealed that amino acid residues 180–225 of PIKE-L appear to mediate binding to merlin (Fig. 5B Lower). The expression of purified recombinant GST proteins was verified (Fig. 5B Upper). Previously, we demonstrated that PIKE-L binds to Homer, an adaptor protein known to link metabotropic glutamate receptors to multiple intracellular targets including the inositol-1,4,5-trisphosphate receptor. Binding of Homer to PIKE-L mapped to a similar region in PIKE-L, and the PIKE-L P187L mutation disrupted the interaction between PIKE-L and Homer 1c (17). In this study, we found that this same amino acid substitution also abolished the binding of full-length PIKE-L to merlin (Fig. 5C, lane 2). Therefore, P187L mutation in PIKE-L abrogates its interactions with merlin.
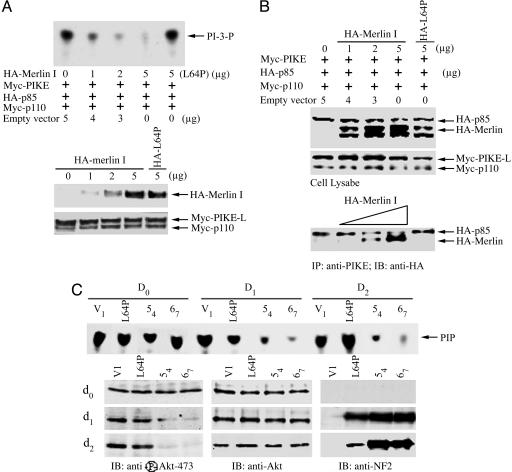
merlin Competes with PI3-Kinase for Binding to PIKE-L. Studies in our laboratory have shown that overexpression of protein 4.1N abolishes the ability of PIKE-S to stimulate PI3-kinase (15). To determine whether merlin similarly could inhibit PIKE activation of PI3-kinase, we cotransfected increasing amounts of merlin with PIKE-L and PI3-kinase into HEK293 cells. Cotransfected merlin significantly reduced PIKE-L-induced activation of PI3-kinase activity in a dose-dependent manner; by contrast, merlin L64P failed to inhibit PI3-kinase activity at the same concentration (Fig. 3A). To evaluate whether merlin competes with PI3-kinase for binding to PIKE-L, we transfected increasing amounts of merlin along with p110, p85, and PIKE-L into HEK293 cells (Fig. 5B). Binding of p85 to PIKE-L was dramatically reduced in proportion to the level of merlin expression. However, merlin (L64P) was unable to block PI3-kinase for binding to PIKE-L (Fig. 3B), suggesting that merlin competes with PI3-kinase for binding to PIKE-L, thus inhibiting PIKE-L activation of PI-3 kinase. In this model, if merlin impairs PI3-kinase binding to PIKE-L, merlin expression should result in reduced PI3-kinase activity in vivo. As predicted, we observed markedly reduced PI3-kinase activity in RT4-D6P2T rat schwannoma cells upon induction of WT merlin expression. High levels of PI3-kinase activity were observed in all cells before merlin induction. After induction of WT merlin expression, PI3-kinase activity was diminished substantially in both the WT merlin-inducible 54 and 67 cell lines. In contrast, PI3-kinase activity was not altered in RT4 cells induced to express mutant L64P merlin (Fig. 3C), consistent with the failure of this mutant to bind to PIKE-L. The inhibition of PI3-kinase activity in these cells is further demonstrated by significant reductions in the phosphorylation status of Akt, a downstream effector of PI3-kinase (Fig. 3C). As seen before, no effect of mutant L64P merlin on Akt activation was observed. Therefore, merlin inhibits PI3-kinase activity by competing with PI3-kinase for binding to PIKE-L.
Fig. 3.
merlin blocks the stimulatory effect of PIKE-L on PI3-kinase. (A) TLC was used to assay PI3-kinase activity. HEK293 cells were transfected with the indicated expression constructs. PI3-kinase was immunoprecipitated by anti-p110 antibody and assayed for in vitro lipid kinase activity. (Top) Increasing amount of merlin progressively diminished PI3-kinase activity, but L64P failed to do so. (Middle) Gradually increasing levels of HA-merlin was expressed in transfected cells. (Bottom) Equal amount of Myc-p110 and Myc-PIKE-L was confirmed by anti-myc immunoblotting. (B) merlin competes with PI3-kinase for binding to PIKE-L. (Bottom) Myc-PIKE-L was immunoprecipitated by anti-PIKE antibody, and the coprecipitated HA-p85 and HA-merlin were visualized by Western blot with anti-HA antibody. (Middle) Equal amounts of Myc-p110 and PIKE-L were confirmed with anti-Myc antibody. (Top) Increased expression of merlin was confirmed. (C) Merlin mediates PI3-kinase in schwannoma cells. TLC was used to assay PI3-kinase activity. Rat RT4-D6P2T schwannoma cells stably transfected with empty vector, merlin mutant L64P, or WT merlin (54 or 67) were uninduced or induced with doxycycline for 0, 1, or 2 days. (First blot from top) PI3-kinase activity is abolished by induced WT merlin, but not merlin mutant L64P. Akt and its phosphorylation in the lysate of schwannoma cells were analyzed. (Left, lower three blots) WT, but not patient-derived L64P, merlin blocks Akt phosphorylation. (Center and Right, lower three blots) The expression of Akt and induced merlin was verified.
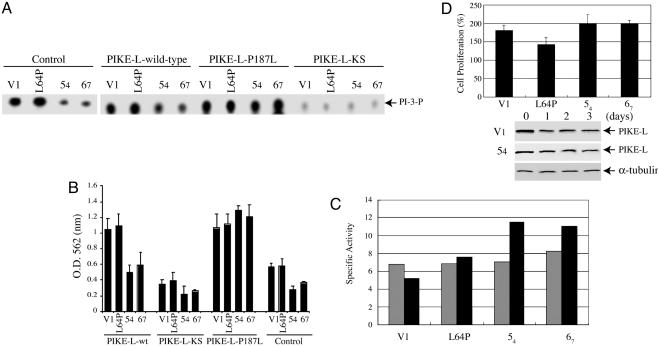
merlin Inhibits PI3-Kinase and Schwannoma Proliferation Through PIKE-L. We showed previously that PIKE-L containing the P187L mutation stimulates PI3-kinase as effectively as WT PIKE-L. In contrast, the dominant-negative PIKE-L-KS mutant prevents stimulation of PI3-kinase (17). To determine whether merlin suppresses PI3-kinase activity through PIKE-L, we infected RT4-D6P2T schwannoma cells with control adenovirus or adenovirus expressing WT PIKE-L, PIKE-L-KS, or PIKE-L-P187L and monitored PI3-kinase activity upon merlin induction. Compared with control (V1) or mutant L64P merlin-expressing cells, WT merlin-expressing cells (54 and 67) exhibited markedly reduced levels of PI3-kinase activity (Fig. 4A). Infection with WT PIKE-L resulted in an increase in PI3-kinase activity compared with control infected cells, presumably due to PIKE-L enhancement of PI3-kinase activity. This effect on PI3-kinase activity was reduced in the merlin-expressing 54 and 67 cell lines, but the magnitude of reduction was identical to that observed in control-infected cells. In striking contrast, we observed a reversal of the merlin-induced reduction in PI3-kinase activity in the 54 and 67 cell lines infected with PIKE-L-P187L, consistent with the fact that this mutant is incapable of binding to merlin but can still stimulate PI3-kinase activity. As previously shown for the dominant inhibitory PIKE-L-KS mutant, expression resulted in a dramatic decrease in PI3-kinase activity in all cell lines. These results suggest that merlin inhibition of PI3-kinase activity operates through PIKE-L binding.
Fig. 4.
merlin suppresses PI3-kinase activity through PIKE-L. (A) TLC was used to assay PI3-kinase activity in rat RT4-D6P2T schwannoma cells, which were infected with control adenovirus or adenovirus expressing WT PIKE-L, PIKE-L-KS, or PIKE-L-P187L. Infection with WT PIKE-L or PIKE-L-P187L resulted in an increase in PI3-kinase activity compared with control infected. This effect was suppressed in the merlin-expressing clones 54 and 67. PI3-kinase activity was unaffected in merlin clones infected with PIKE-L-P187L or merlin mutant L64P infected with either WT or P187L PIKE-L, presumably because of the inability of merlin or PIKE-L to cointeract. As expected, the dominant-negative mutant, PIKE-L-KS, inhibited PI3-kinase. (B) The number of proliferating rat RT4-D6P2T schwannoma cells was measured by MTT assay. Cellular proliferation paralleled PI3-kinase activity. (C) Caspase-3 activity assay. Empty vector (V1), merlin mutant L64P, and WT merlin (54 and 67) cells were induced for 3 days. Caspase-3 activity assay was conducted with 30 μg of protein from day 0 (gray bar) and 3 (black bar) samples. Induction of WT merlin substantially increases apoptosis compared with control or L64P mutant. (D) Knocking down PIKE-L abolishes the tumor-suppressive activity of merlin. The cells was induced and infected with adenovirus expressing sh-PIKE RNA. The expression of PIKE-L in both L64P and 54 cells was decreased substantially upon infection of PIKE RNA interference (blots). Surprisingly, knocking down PIKE-L robustly increases cell proliferation in 54 and 67 cells compared with control and L64P cells (graph). The results were expressed as means ± SEM calculated from five times of determination (P < 0.05).
To provide a functional link between merlin regulation of PI3-kinase and merlin growth suppression, we analyzed cell growth upon expression of PIKE-L in inducible merlin-expressing confluent RT4 cells in parallel experiments. In control adenovirusinfected cells, substantial cell proliferation occurs in V1 and L64P cells but not in 54 and 67 cells. Expression of PIKE-L escalates cell growth in four cell lines with smaller increases in 54 and 67 cells than in V1 and L64P cells. Consistent with the PI3-kinase regulation results, we observed a reversal of merlin growth suppression in the WT merlin-expressing RT4 cells upon the introduction of PIKE-L-P187L. Infection of dominant-negative PIKE-L-KS substantially inhibits all cell growth (Fig. 4B).
To explore whether increased cell proliferation after the introduction of PIKE-L is related to enhanced cell survival, we monitored the basal rate of cell survival upon induction of merlin. Caspase-3 activity assay demonstrated that expression of WT merlin triggers an ≈40–70% apoptotic activity increase in 67 and 54 cells at day 3 after induction vs. day 0, whereas no significant increase was observed in control (V1) or L64P cells (Fig. 4C). These observations are consistent with previous reports that merlin expression results in increased cell death (22, 23).
Induction of WT, but not mutant, merlin suppresses cell proliferation (Fig. 4B). If PIKE-L plays a critical role in mediating merlin's tumor-suppressive activity, then knocking down PIKE-L expression in schwannoma cells should compromise this effect. Accordingly, we prepared an adenovirus expressing sh-PIKE RNA to inhibit PIKE-L expression. As expected, PIKE-L was successfully decreased upon infection in both 54 and L64P cells; by contrast, α-tubulin was not affected (Fig. 4D). Strikingly, cell proliferation assay showed that both WT merlin cells present ≈200% growth at day 3 compared with day 0, whereas ≈175% and 145% increases were observed on control and L64P cells, respectively (Fig. 4D), indicating that WT merlin, instead of suppressing cell growth, provokes cell proliferation in the absence of PIKE-L. Collectively, these observations demonstrate that merlin growth suppression is mediated in part by binding to PIKE-L and its inhibitory effects on PI3-kinase activation.
Discussion
Our findings that PIKE-L mediates the tumor-suppressive activity of merlin through PI3-kinase provide a molecular mechanism that may account for the negative growth regulatory function of merlin. WT PIKE-L robustly binds to merlin. By contrast, GTPase mutated dominant-negative PIKE-L-KS faintly associates with merlin (Fig. 1), suggesting that the interaction between merlin and PIKE-L is mediated by the GTPase activity. Our previous study revealed that PIKE-L-KS binds to PI3-kinase but prevents its activation (17). WT PIKE-L-triggered PI3-kinase activity was decreased substantially in WT merlin-induced cells compared with L64P and control cells (Fig. 4A), indicating that PI3-kinase activity is regulated by merlin/PIKE-L interaction. However, expression of PIKE-L-KS in schwannoma cells potently inhibited PI3-kinase activity in all cells, regardless of WT or mutant merlin induction (Fig. 4A), suggesting that merlin does not interrupt the effect of PIKE-L-KS on PI3-kinase. This effect correlates with its crippled binding activity to merlin (Fig. 1).
The FERM domain of merlin binds to the N terminus of PIKE-L (Fig. 1). Both PIKE-L and -S isoforms share the same N-terminal region, suggesting that PIKE-S also might bind to merlin. As predicted, our in vitro binding assay with GST-PIKE-S revealed that these two proteins interact with each other. Moreover, this interaction also was observed in coimmunoprecipitation assays in transfected HEK293 cells (data not shown). However, PIKE-S was not detected in schwannoma cells, fitting with the previous finding that PIKE-S predominantly occurs in neuronal tissue (15). Given the high sequence conservation between merlin and ERM proteins in the FERM domain, it is possible that PIKE-L also might bind to other ERM proteins. merlin is directly phosphorylated on Ser-518 by members of the p21-activated kinase (PAK) family of kinases, including PAK1 and PAK2 (24, 25). Recently, we showed that a merlin mutant that mimics Ser-518 phosphorylation (S518D) cannot suppress cell growth or motility in RT4 rat schwannoma cells and results in dramatic changes in cell morphology and actin cytoskeleton organization (26). Consistently, S518D mutation attenuated the interaction between merlin and PIKE-L compared with WT and S518A merlin (data not shown).
To determine how merlin functions as a growth suppressor, several groups have used yeast two-hybrid cloning to identify novel merlin partners, including CD44 (27, 28), βII-spectrin (29), SCHIP-1 (30), hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) (31), NHE-RF (32), and β1-integrin (33). Among the merlin-associated proteins, CD44 and HRS are the potential candidates mediating the growth suppressive activity of merlin. At high cell density, merlin becomes hypophosphorylated and associates with the cytoplasmic tail of CD44 and inhibits cell growth in response to HA. At low cell density, merlin is phosphorylated, is growth permissive, and exists in a complex with ezrin, moesin, and CD44 (28). Hepatocyte growth factor is one of the most potent mitogens for Schwann cells and also promotes cell motility (34). HRS specifically interacts with the C-terminal domain of merlin. Merlin interacts with HRS in the unfolded, or open, conformation. However, merlin binding to HRS does not negatively regulate HRS growth suppressor activity (14).
In addition to cell-growth regulation, merlin regulates actin cytoskeleton-mediated functions, such as spreading, motility, and attachment. Accordingly, merlin has been implicated in Rac/Cdc42 signaling (25, 35). Recently, merlin has been shown to inhibit directly the Rac/CDC42-dependent Ser/Thr kinase PAK1, which is essential for both Ras transformation and neurofibromatosis type 1 (36, 37). Moreover, merlin also has been suggested to inhibit Ras/mitogen-activated protein kinase cascade (38). Our findings that merlin specifically binds to PIKE-L and abrogates PIKE's effects on PI3-kinase provides further evidence that merlin acts as a tumor suppressor by antagonizing the PIKE/PI3-kinase pathway. The discovery that merlin regulates cell growth through PI3-kinase/Akt-mediated signaling pathways is intriguing in light of the established relationship between other FERM-containing proteins and apoptosis (39, 40). Although most previous studies of merlin function have focused on the ability of merlin to reduce cell proliferation, merlin expression also can result in increased cell death (22). Transduction of merlin into human schwannoma cells was found to decrease cell growth by inducing apoptosis (23). The PI3-kinase/Akt pathway plays an essential role in promoting cell survival in various cell types. In this fashion, merlin expression would result in decreased PIKE-induced PI3-kinase activity and decreased Akt activation, culminating in increased cell death. Thus, our studies, to our knowledge, provide the first mechanistic insights into how merlin might regulate cell growth by modulating PI3-kinase/Akt pathway.
Supplementary Material
Acknowledgments
We thank Drs. Helen Morrison and Peter Herrlich (Karlsruhe, Germany) for the WT merlin inducible RT4 schwannoma cell lines. This work was supported by Department of Defense New Investigator Award NF020013 (to K.Y.) and National Institutes of Health Grant R01-NS35848 (to D.H.G.).
Author contributions: K.Y. designed research; R.R., X.T., and K.Y. performed research; D.H.G. and K.Y. analyzed data; K.Y. contributed new reagents/analytic tools; and D.H.G. and K.Y. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ERM, ezrin/radix/moesin; HA, hemagglutinin; HRS, hepatocyte growth factor-regulated tyrosine kinase substrate; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide; NF2, neurofibromatosis 2; NTD, N-terminal domain; PI3-kinase, phosphatidylinositol 3-kinase.
References
- 1.Eldridge, R. (1981) Adv. Neurol. 29, 57–65. [PubMed] [Google Scholar]
- 2.Rouleau, G. A., Merel, P., Lutchman, M., Sanson, M., Zucman, J., Marineau, C., Hoang-Xuan, K., Demczuk, S., Desmaze, C., Plougastel, B., et al. (1993) Nature 363, 515–521. [DOI] [PubMed] [Google Scholar]
- 3.Trofatter, J. A., MacCollin, M. M., Rutter, J. L., Murrell, J. R., Duyao, M. P., Parry, D. M., Eldridge, R., Kley, N., Menon, A. G., Pulaski, K., et al. (1993) Cell 72, 791–800. [DOI] [PubMed] [Google Scholar]
- 4.Gutmann, D. H. (1997) Neurobiol. Dis. 3, 247–261. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez-Agosti, C., Xu, L., Pinney, D., Beauchamp, R., Hobbs, W., Gusella, J. & Ramesh, V. (1996) Oncogene 13, 1239–1247. [PubMed] [Google Scholar]
- 6.Scherer, S. S. & Gutmann, D. H. (1996) J. Neurosci. Res. 46, 595–605. [DOI] [PubMed] [Google Scholar]
- 7.Gutmann, D. H., Sherman, L., Seftor, L., Haipek, C., Hoang Lu, K. & Hendrix, M. (1999) Hum. Mol. Genet. 8, 267–275. [DOI] [PubMed] [Google Scholar]
- 8.Sherman, L., Xu, H. M., Geist, R. T., Saporito-Irwin, S., Howells, N., Ponta, H., Herrlich, P. & Gutmann, D. H. (1997) Oncogene 15, 2505–2509. [DOI] [PubMed] [Google Scholar]
- 9.Huynh, D. P. & Pulst, S. M. (1996) Oncogene 13, 73–84. [PubMed] [Google Scholar]
- 10.Gonzalez-Agosti, C., Wiederhold, T., Herndon, M. E., Gusella, J. & Ramesh, V. (1999) J. Biol. Chem. 274, 34438–34442. [DOI] [PubMed] [Google Scholar]
- 11.Gutmann, D. H., Geist, R. T., Xu, H., Kim, J. S. & Saporito-Irwin, S. (1998) Hum. Mol. Genet. 7, 335–345. [DOI] [PubMed] [Google Scholar]
- 12.Gutmann, D. H., Haipek, C. A. & Hoang Lu, K. (1999) J. Neurosci. Res. 58, 706–716. [PubMed] [Google Scholar]
- 13.Meng, J. J., Lowrie, D. J., Sun, H., Dorsey, E., Pelton, P. D., Bashour, A. M., Groden, J., Ratner, N. & Ip, W. (2000) J. Neurosci. Res. 62, 491–502. [DOI] [PubMed] [Google Scholar]
- 14.Gutmann, D. H. (2001) Hum. Mol. Genet. 10, 747–755. [DOI] [PubMed] [Google Scholar]
- 15.Ye, K., Hurt, K. J., Wu, F. Y., Fang, M., Luo, H. R., Hong, J. J., Blackshaw, S., Ferris, C. D. & Snyder, S. H. (2000) Cell 103, 919–930. [DOI] [PubMed] [Google Scholar]
- 16.Ye, K., Aghdasi, B., Luo, H. R., Moriarity, J. L., Wu, F. Y., Hong, J. J., Hurt, K. J., Bae, S. S., Suh, P. G. & Snyder, S. H. (2002) Nature 415, 541–544. [DOI] [PubMed] [Google Scholar]
- 17.Rong, R., Ahn, J. Y., Huang, H., Nagata, E., Kalman, D., Kapp, J. A., Tu, J., Worley, P. F., Snyder, S. H. & Ye, K. (2003) Nat. Neurosci. 6, 1153–1161. [DOI] [PubMed] [Google Scholar]
- 18.Ye, K., Compton, D. A., Lai, M. M., Walensky, L. D. & Snyder, S. H. (1999) J. Neurosci. 19, 10747–10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye, K., Ke, Y., Keshava, N., Shanks, J., Kapp, J. A., Tekmal, R. R., Petros, J. & Joshi, H. C. (1998) Proc. Natl. Acad. Sci. USA 95, 1601–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, T. C., Zhou, S., da Costa, L. T., Yu, J., Kinzler, K. W. & Vogelstein, B. (1998) Proc. Natl. Acad. Sci. USA 95, 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahn, J. Y., Rong, R., Kroll, T. G., Van Meir, E. G., Snyder, S. H. & Ye, K. (2004) J. Biol. Chem. 279, 16441–16451. [DOI] [PubMed] [Google Scholar]
- 22.Shaw, R. J., McClatchey, A. I. & Jacks, T. (1998) J. Biol. Chem. 273, 7757–7764. [DOI] [PubMed] [Google Scholar]
- 23.Schulze, K. M., Hanemann, C. O., Muller, H. W. & Hanenberg, H. (2002) Hum. Mol. Genet. 11, 69–76. [DOI] [PubMed] [Google Scholar]
- 24.Kissil, J. L., Johnson, K. C., Eckman, M. S. & Jacks, T. (2002) J. Biol. Chem. 277, 10394–10399. [DOI] [PubMed] [Google Scholar]
- 25.Xiao, G. H., Beeser, A., Chernoff, J. & Testa, J. R. (2002) J. Biol. Chem. 277, 883–886. [DOI] [PubMed] [Google Scholar]
- 26.Surace, E. I., Haipek, C. A. & Gutmann, D. H. (2004) Oncogene 23, 580–587. [DOI] [PubMed] [Google Scholar]
- 27.Sainio, M., Zhao, F., Heiska, L., Turunen, O., den Bakker, M., Zwarthoff, E., Lutchman, M., Rouleau, G. A., Jaaskelainen, J., Vaheri, A. & Carpen, O. (1997) J. Cell. Sci. 110, 2249–2260. [DOI] [PubMed] [Google Scholar]
- 28.Morrison, H., Sherman, L. S., Legg, J., Banine, F., Isacke, C., Haipek, C. A., Gutmann, D. H., Ponta, H. & Herrlich, P. (2001) Genes Dev. 15, 968–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scoles, D. R., Huynh, D. P., Morcos, P. A., Coulsell, E. R., Robinson, N. G., Tamanoi, F. & Pulst, S. M. (1998) Nat. Genet. 18, 354–359. [DOI] [PubMed] [Google Scholar]
- 30.Goutebroze, L., Brault, E., Muchardt, C., Camonis, J. & Thomas, G. (2000) Mol. Cell. Biol. 20, 1699–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scoles, D. R., Huynh, D. P., Chen, M. S., Burke, S. P., Gutmann, D. H. & Pulst, S. M. (2000) Hum. Mol. Genet. 9, 1567–1574. [DOI] [PubMed] [Google Scholar]
- 32.Murthy, A., Gonzalez-Agosti, C., Cordero, E., Pinney, D., Candia, C., Solomon, F., Gusella, J. & Ramesh, V. (1998) J. Biol. Chem. 273, 1273–1276. [DOI] [PubMed] [Google Scholar]
- 33.Obremski, V. J., Hall, A. M. & Fernandez-Valle, C. (1998) J. Neurobiol. 37, 487–501. [PubMed] [Google Scholar]
- 34.Krasnoselsky, A., Massay, M. J., DeFrances, M. C., Michalopoulos, G., Zarnegar, R. & Ratner, N. (1994) J. Neurosci. 14, 7284–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw, R. J., Paez, J. G., Curto, M., Yaktine, A., Pruitt, W. M., Saotome, I., O'Bryan, J. P., Gupta, V., Ratner, N., Der, C. J., et al. (2001) Dev. Cell 1, 63–72. [DOI] [PubMed] [Google Scholar]
- 36.Kissil, J. L., Wilker, E. W., Johnson, K. C., Eckman, M. S., Yaffe, M. B. & Jacks, T. (2003) Mol. Cell 12, 841–849. [DOI] [PubMed] [Google Scholar]
- 37.Hirokawa, Y., Tikoo, A., Huynh, J., Utermark, T., Hanemann, C. O., Giovannini, M., Xiao, G. H., Testa, J. R., Wood, J. & Maruta, H. (2004) Cancer J. 10, 20–26. [DOI] [PubMed] [Google Scholar]
- 38.Lim, J. Y., Kim, H., Kim, Y. H., Kim, S. W., Huh, P. W., Lee, K. H., Jeun, S. S., Rha, H. K. & Kang, J. K. (2003) Biochem. Biophys. Res. Commun. 302, 238–245. [DOI] [PubMed] [Google Scholar]
- 39.Gautreau, A., Poullet, P., Louvard, D. & Arpin, M. (1999) Proc. Natl. Acad. Sci. USA 96, 7300–7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kondo, T., Takeuchi, K., Doi, Y., Yonemura, S., Nagata, S. & Tsukita, S. (1997) J. Cell Biol. 139, 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.