Abstract
The transmembrane protein Smoothened (Smo) is activated in response to the extracellular protein signal, Hedgehog (Hh), and transmits this state of pathway activity into the cell. Previous studies in Drosophila have correlated pathway activation with Smo accumulation and increased phosphorylation. Using immunopurification and mass spectrometry, we identify here 26 serine/threonine residues within the Smo C-terminal cytoplasmic tail that are phosphorylated in Hh-stimulated cells. By systematically substituting alanine or glutamic acid to block or simulate phosphorylation, we provide evidence for a functional role of collective phosphorylation of a subset of phosphoresidues in pathway activation. This role is indicated by the ability of altered Smo proteins to produce changes in transcription of Hh-responsive genes in vivo and in cultured cells. These altered Smo proteins also affect biochemical indicators of pathway activity, such as Smo accumulation and phosphorylation of other pathway components. The prevalence and arrangement of phosphoresidues within the Smo cytoplasmic tail at recognition sites for cAMP-dependent protein kinase and casein kinase 1 suggest a role for these kinases in Smo phosphorylation, and such a role is supported by the effects of manipulating kinase activities in cultured cells. Our studies confirm and extend previous studies showing a positive effect for cAMP-dependent protein kinase and uncover a positive role for casein kinase 1α in Hh pathway activation.
The Hedgehog (Hh) family of secreted protein signals specifies cell differentiation and proliferation in a diverse array of patterning events in embryos ranging from insects to mammals. Hh signaling also plays a postembryonic role in homeostatic tissue maintenance and repair, and continuous pathway activity plays a pathological role in the growth of a group of endodermally derived cancer types that together account for approximately one-fourth of human cancer deaths (reviewed in refs. 1–3).
Hh protein binding to the transporter-like receptor Patched (Ptc) (4–6) releases an inhibitory effect of Ptc on Smoothened (Smo) (7), a seven-pass transmembrane protein (8, 9). In Drosophila, activated Smo accumulates at the plasma membrane (10–12) and signals pathway activation by recruiting a complex of intracellular pathway components, including the kinesin-like molecule Costal-2 (Cos2) (13, 14), the putative serine/threonine protein kinase Fused (Fu) (15), and Cubitus interruptus (Ci) (16, 17), a zinc finger transcription factor that is the major transcriptional effector of the pathway (18–20). In the absence of Hh stimulation, Cos2 tethers Ci in the cytoplasm (21–23) and promotes its cleavage to a repressor form, CiR (14, 24, 25), whereas the Suppressor of Fused [Su(fu)] protein (26, 27) associates with and negatively regulates the fraction of full-length Ci that is not associated with the Cos2/Fu/Ci complex (28, 29), at least in part by influencing its subcellular localization (23, 30, 31). Upon Hh-induced pathway activation, the Cos2/Fu/Ci complex is recruited to the membrane through Cos2 association with the Smo cytoplasmic tail (29, 32–34), and Su(fu) is inactivated through the activity of Fu (26, 31, 35). These events lead to decreased Ci cleavage and loss of CiR repressor, accumulation and nuclear translocation of Ci, and expression of Hh pathway transcriptional targets (reviewed in ref. 2).
Hh pathway activation is marked by decreased phosphorylation of Ci (21, 25) and increased phosphorylation of Smo, Cos2, Fu, and Su(fu), which can be observed within 10–15 min of stimulation (10, 13, 14, 29, 36). Phosphorylation of Ci maintains pathway quiescence by promoting cleavage of Ci to CiR, and at least three kinases, cAMP-dependent protein kinase (PKA) (37–40), casein kinase 1α (CK1α) (41, 42), and glycogen synthase kinase 3β (GSK3β) (41, 43), likely exert their negative effects on pathway activity through Ci phosphorylation. Curiously, genetic studies demonstrate that at least one of these kinases, PKA, also exerts a positive effect on pathway activity. Thus, although loss of PKA was initially linked to ectopic Ci accumulation and expression of Hh pathway target genes, the degree of Hh pathway activation is limited, and PKA is required for full activity in both embryos (44) and wing imaginal discs (45). Furthermore, ectopic PKA activity in embryos is sufficient to induce expression of Hh pathway target genes in a manner that requires Smo (44). Although PKA activity in vertebrates has been reported to reduce response to Sonic hedgehog (Shh) signaling (46–49), it is possible that PKA has dual roles, as noted in Drosophila, and that the negative actions of PKA prevail in the experiments reported. Beyond the genetic studies with PKA, a positive role for phosphorylation in triggering pathway activity in Drosophila is further suggested by the preferential interaction of the Cos2/Fu/Ci complex with phosphorylated Smo (29).
The observations that the positive role of PKA requires Smo and that the Cos2/Fu/Ci complex preferentially binds phosphorylated Smo together suggest that Smo phosphorylation upon Hh stimulation may play a modulatory role in pathway activity in Drosophila. Consistent with this possibility, the 481-residue cytoplasmic tail of Smo contains several consensus PKA sites (8) and several CK1 sites that would be primed by phosphorylation of these PKA sites (see below). Here we report the existence of 26 phosphorylated serine/threonine residues, including residues within three or four PKA sites, seven or eight CK1 sites, and one possible GSK3 site, in endogenous Smo isolated from Hh N-terminal signaling domain (HhN)-stimulated Drosophila S2 cells. Through manipulation of PKA and CK1α activities in cultured cells, we have observed that these kinases promote Smo phosphorylation and accumulation. Our data confirm and extend previous studies showing a positive effect for PKA on Hh signal transduction and uncover a positive role for CK1α. Our analyses of Smo variants that either prevent or simulate phosphorylation suggest a functional role for collective phosphorylation of specific subsets of Smo residues in stabilization and accumulation of Smo, in stimulation of phosphorylation of components of the intracellular signaling complex, and in induction of Hh target gene transcription.
Materials and Methods
Immunoaffinity Purification of Smo. Preparative scale-affinity purifications of endogenous Smo using anti-Smo 20C6 and anti-Fu 26F11 mAb affinity matrices were performed as described (29) (for details, see Supporting Text, which is published as supporting information on the PNAS web site).
Phosphopeptide Analysis. Immunoaffinity-purified protein mixtures were digested with trypsin or chymotrypsin. Immobilized metal affinity chromatography was performed according to Ficarro et al. (50), with minor modifications. Phosphopeptides were analyzed by using nanoflow HPLC/microelectrospray ionization tandem MS on a ThermoFinnigan (San Jose, CA) LCQ Deca XP Plus ion trap mass spectrometer. (For details, see Supporting Text.)
Smo Variants and Other Expression Constructs. Smo variants used for reporter assays were created by PCR-based mutagenesis and subcloning into a pAcSV Smo plasmid containing the actin 5C promoter and lacking the endogenous 5′ and 3′ UTRs (29).
Myc-tagged Smo variants were generated by PCR amplification and subcloning into pAcSV of a restriction fragment containing a 6×Myc insertion just C-terminal to Smo residue 34 from pUAST Myc-Smo (32) to generate pAcSV Myc-Smo. Altered Smo sequences from pAcSV Smo constructs were then cloned into this pAcSV Myc-Smo backbone.
GFP-tagged Smo expression constructs in pUAST were generated by subcloning C-terminal coding sequences of pAcSV Smo variants into pUAST GFP-Smo (32), thus removing the endogenous 3′ UTR. Expression constructs for Myc-Cos2, Myc-His-Fu, V5-Su(fu), Hh, CK1α, CK1α K49R, PKA mC*, and GAL4 are described in Supporting Text.
Cell Culture, RNA Interference, and ptc-Luciferase Reporter Assays. Conditions for cl-8, S2R+, and HhN S2 stable cell culture were as described (29). Hh-responsive ptc-Luciferase reporter assays were performed as described (29). For smo rescue assays, endogenous smo transcript was depleted by transfection of cells with dsRNA targeting the 5′ UTR, whereas smo alleles were overexpressed from cotransfected plasmids lacking the endogenous 5′ UTR. For overexpression assays, control (yfp) dsRNA and pAct5C GAL4 were transfected alongside smo expression constructs. Transfection with plasmid expressing GFP (no Smo) was used as a negative control. Luciferase activity was normalized to constitutively expressed Renilla luciferase control plasmid.
Characterization of Intracellular Hh Pathway Components. S2R+ cells were cotransfected with 0.05–0.1 μg each of pAcSV or pAct5C constructs expressing control protein (GFP) or Myc-Smo forms, Myc-Cos2, MycHis-Fu, V5-Su(fu), mC* (PKA), CK1α, and/or CK1α K49R. Cells were either cotransfected with 0.1 μg of pAct5C GAL4/UAS-hh (42) or treated at 20–30 h after transfection with HhN-conditioned medium. Approximately 24 h after HhN addition or 48 h after transfection with pAct5C GAL4/UAS-hh, cell lysates were harvested and analyzed by SDS/PAGE and Western blotting by using anti-Myc rabbit polyclonal Ab (A-14; Santa Cruz Biotechnology), anti-V5 mouse mAb (Invitrogen), anti-tubulin mouse mAb E7 (Developmental Studies Hybridoma Bank, Iowa City, IA; contributed by M. Klymkowsky, University of Colorado, Boulder), and anti-lamin rabbit polyclonal Ab R836 (gift from P. Fisher, Stony Brook University, Stony Brook, NY).
Fly Stocks and Production of Transgenic Lines. GAL4 driver lines used were prd-GAL4, ptc-GAL4, and 71B (51). pUAST-derived plasmids (with the mini white gene) containing GFP-smo coding sequences were injected into w1118 fly embryos by Genetic Services, Inc. (Sudbury, MA). Several independent transgenic lines for each construct were recovered. w1118 flies were used for wild-type controls.
Analysis of Fly Embryos and Wings. Immunostaining of fly embryos and mounting of adult wings were performed according to standard protocols. Two independent transgenic fly lines were assayed for each construct. Abs were diluted as follows: 1 μg/ml anti-Wingless (Wg) [mouse monoclonal 4D4; gift from S. Cohen, European Molecular Biology Laboratory, Heidelberg, and contributed to Developmental Studies Hybridoma Bank (DSHB)], 10 μg/ml anti-Engrailed (mouse monoclonal 4D9; DSHB, contributed by C. Goodman, University of California, Berkeley), 1 μg/ml anti-GFP (rabbit polyclonal; Molecular Probes), and 1:400 dilution of Alexa Fluor 488 goat anti-rabbit IgG or Alexa Fluor 546 goat anti-mouse IgG (Molecular Probes). Embryos were examined by confocal microscopy.
Results
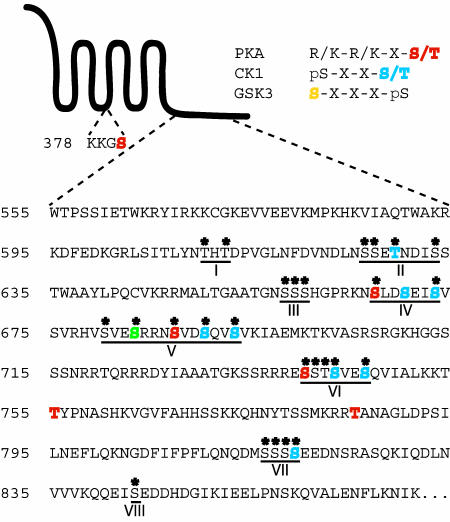
Extensive Phosphorylation of the Smo Cytoplasmic Tail in Hh-Stimulated Cells. To identify Drosophila Smo residues that are phosphorylated under conditions of Hh stimulation, we used mouse mAb directed against the Smo extracellular N-terminal domain or against the Smo-associated protein Fu for preparative scale immunoprecipitation of endogenous Smo from Drosophila embryonic S2 cells stably expressing HhN. These immunoprecipitates were digested with trypsin, which cleaves after lysine or arginine residues, or with chymotrypsin, which cleaves after aromatic or large hydrophobic residues, to increase the diversity of peptides present for further analysis. Phosphopeptides were enriched by using immobilized metal affinity chromatography, based on the affinity of phosphopeptides for Fe3+ (52, 53), and phosphorylated residues were physically mapped by using nanoflow reverse-phase HPLC/microelectrospray ionization tandem MS analysis. Twenty-six distinct phosphopeptides were identified (Table 1), encompassing eight distinct regions (I-VIII) of the Smo cytoplasmic tail that undergo phosphorylation on a total of 26 serine/threonine residues (Fig. 1). Three of the 26 identified phosphoresidues fall within consensus PKA recognition motifs in regions IV, V, and VI, and an additional five phosphoresidues within regions V and VI could be accounted for by PKA priming followed by a cascade of CK1 and GSK3 phosphorylation events (54). Three additional phosphorylation events within regions II, IV, and VII could be primed for CK1 recognition by non-PKA-targeted phosphoresidues. Phosphorylation was not detected on three predicted PKA sites nor on one predicted CK1 site in region IV.
Table 1. Identified phosphopeptides of endogenous Smo from HhN-stimulated cells.
| Phosphopeptide* | Region |
|---|---|
| α-Smo, chymotrypsin | |
| NTHTDPVGL | I |
| NFDVNDLNSSETNDISSTW | II |
| NFDVNDLNSSETNDISSTW | II |
| NFDVNDLNSSETNDISSTW | II |
| NFDVNDLNSSETNDISSTW | II |
| DVNDLNSSETNDISSTW | II |
| DVNDLNSSETNDISSTW | II |
| IAAATGKSSRRRESSTSVESQVIAL | VI |
| LQNQDMSSSSEEDNSRASQKIQDL | VII |
| α-Smo, trypsin | |
| MALTGAATGNSSSHGPR | III |
| RNSVDSQVSVK | V |
| RNSVDSQVSVKIAEMK | V |
| HVSVESRRNSVDSQVSVK | V |
| RRESSTSVESQVIALK | VI |
| α-Fu, chymotrypsin | |
| NFDVNDLNSSETNDISSTW† | II |
| NFDVNDLNSSETNDISSTW† | II |
| NFDVNDLNSSETNDISSTW† | II |
| α-Fu, trypsin | |
| KNSLDSEISVSVR | IV |
| RNSVDSQVSVK | V |
| RNSVDSQVSVK | V |
| RNSVDSQVSVK | V |
| HVSVESR | V |
| RESSTSVESQVIALK† | VI |
| RESSTSVESQVIALK† | VI |
| RESSTSVESQVIALK† | VI |
| QQEISEDDHDGIK | VIII |
Phosphorylated residues are underlined.
The phosphopeptide(s) isolated represent one or a combination of these three possible species.
Fig. 1.
Predicted and detected phosphorylation in Smo. A schematic diagram of Smo is shown, alongside specific residues within intracellular loop two and a portion of the C-terminal cytoplasmic tail. Identified phosphoserine/threonine residues in endogenous Smo purified from HhN-stimulated Drosophila S2 cells are marked with asterisks. Phosphopeptides were isolated from eight distinct regions, as designated (I–VIII). Residues within the consensus kinase recognition motifs shown for PKA (red) and CK1 (blue) are highlighted. One residue that may be phosphorylated by either CK1 or GSK3, depending on the priming phosphoresidue, is shown in green. Designations of CK1 and GSK3 target residues assume that all possible priming phosphorylation has occurred.
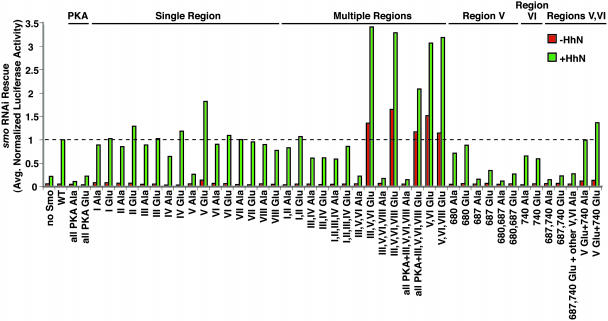
Smo Activation Linked to Collective Phosphorylation of Multiple Residues. We used the identity of these phosphoresidues as a basis for investigating the role of phosphorylation in Smo activity by altering phosphorylated serine and threonine residues to Ala to prevent phosphorylation and to Glu to simulate constitutive phosphorylation (Table 2). Our initial assays (Fig. 2) measured the activity of Smo variants expressed in cells in which endogenous Smo activity was depleted by RNA interference (RNAi) directed against 5′ UTR sequences absent in the altered Smo expression constructs (29). We note that RNAi targeting alone abolished response to HhN signaling, and that responsiveness to HhN was rescued by introduction of wild-type Smo (Fig. 2).
Table 2. Phosphorylation site mutations in Smo.
| Allele name | Mutated region(s)* | Region |
|---|---|---|
| All PKA Ala | KKGAYFH | PKA |
| RKNALDS | ||
| RRNAVDS | ||
| RREASTS | ||
| KKTAYPN | ||
| KRRAANA | ||
| I Ala | LYNAHADPV | I |
| II Ala | DLNAAEANDIASTW | II |
| III Ala | TGNAAAHGP | III |
| IV Ala | RKNALDAEIAVSV | IV |
| V Ala | RHVAVEARRNAVDAQVAVKI | V |
| VI Ala | RREAAAAVEAQVI | VI |
| VII Ala | QDMAAAAEED | VII |
| VIII Ala | QEIAEDD | VIII |
| 680 Ala | RHVAVESRRNSVDSQVSVKI | V |
| 687 Ala | RHVSVESRRNAVDSQVSVKI | V |
| 680,687 Ala | RHVAVESRRNAVDSQVSVKI | V |
| 740 Ala | RREASTSVESQVI | VI |
| 687,740 Ala | RHVSVESRRNAVDSQVSVKI | V |
| RREASTSVESQVI | VI | |
| 687,740 Glu + other V,VI Ala | RHVAVEARRNEVDAQVAVKI | V |
| RREEAAAVEAQVI | VI | |
| V Glu+740 Ala | RHVEVEERRNEVDEQVEVKI | V |
| RREASTSVESQVI | VI | |
| V Glu+740 Glu | RHVEVEERRNEVDEQVEVKI | V |
| RREESTSVESQVI | VI |
Shown in most cases are the Ala variants created to mutate a given region or individual phosphoresidue to alanine. The corresponding Glu variants (not shown) change the underlined residues to glutamic acid instead. The following alleles are simply combinations of the alleles listed above, encoding variants with phospho-residues in two or more regions mutated all to Ala or all to Glu: I,II Ala; I,II Glu; III,IV Ala; III,IV Glu; I,II,III,IV Ala; I,II,III,IV Glu; III,V,VI Ala; III,V,VI Glu; III,V,VI,VIII Ala; III,V,VI,VIII Glu; all PKA+ III,V,VI,VIII Ala; all PKA+III,V,VI,VIII Glu; V,VI Glu; V,VI,VIII Glu.
Mutated residues are underlined.
Fig. 2.
Mutation of Smo phosphorylation sites modulates Hh-responsive reporter expression. Hh pathway responsiveness was monitored by using the ptc-Luc reporter after transfection with dsRNA targeting the smo 5′ UTR to deplete endogenous smo transcript and plasmids to express mutant smo alleles. See Table 2 for details on the smo alleles shown. Shown is the normalized reporter activity produced by each variant relative to that of wild-type Smo in the presence of HhN (“+HhN”) (set to a value of 1; dotted line). Each variant was tested in a minimum of two independent assays, and the results were averaged.
As a first test of the potential importance of Smo phosphorylation, we altered all six predicted PKA recognition sites, at least three of which are phosphorylated in vivo. We found that substitution of these six residues with either Ala or Glu caused a loss of Smo responsiveness. It is worth noting that substitution by Glu likely disrupts the ability to prime further phosphorylation by kinases like CK1 or GSK3. Alteration of PKA sites to Glu therefore simulates phosphorylation of PKA target residues, potentially permitting some phosphorylation-dependent interactions with other proteins but disrupting priming for further phosphorylation by other kinases.
Further analysis focused on alterations of all phosphoresidues within one or more of the eight regions identified (Fig. 1 and Table 1). For region IV, a predicted CK1 site situated between the identified PKA and probable CK1 sites was included as part of the cluster. Alteration of all phosphoresidues within single regions (up to five missense mutations per allele), either to Ala or to Glu, produced only modest effects for seven of the eight regions. In contrast, alteration of region V phosphoresidues to Ala produced a loss of signaling activity, whereas mutation of the same residues to Glu led to an ∼2-fold increase in reporter expression both in the absence and presence of HhN stimulation.
In testing whether phosphorylation of multiple regions might act cooperatively, we found that combined Glu substitutions in regions III, V, and VI produced a dramatic and synergistic increase in reporter expression. This increase was observed even in the absence of HhN (to a level similar to that for HhN-stimulated cells with wild-type Smo), and reporter expression increased even further with HhN stimulation. Additional Glu substitution of residues in region VIII or of VIII plus the remaining predicted PKA sites produced no further increase in reporter activity. Further analysis revealed that Glu substitution of residues in regions V and VI was sufficient for this increase, which, notably, was greater than the additive effect of the two regions alone. We thus conclude that collective simulated phosphorylation of multiple phosphoresidues can contribute to Smo activity, with phosphoresidues in region V producing the greatest effect.
The III,V,VI Ala variant displayed a complete loss of Smo activity, similar to Ala substitution of region V alone (see above). Alteration of individual phosphoresidues within regions V and VI focused initially on PKA target sites, including the N-terminal-most phosphoresidue (Ser-680) in region V, which could be a low consensus PKA recognition site (55). These alterations revealed a hierarchy of importance for these phosphoresidues, with alteration of residue 687 producing a nearly complete loss of Smo response and of residues 680 and 740 producing far less dramatic effects. Interestingly, either Glu or Ala substitution for each of these residues produced an equivalent reduction in Smo function, suggesting that the major effect of these alterations likely is due to a loss of priming for subsequent phosphorylation by CK1 or GSK3 (see Fig. 1). We also found that combined alteration to Glu of residues 687 and 740, either with or without Ala substitution of the remaining residues in regions V and VI, was insufficient to produce Smo activation. This supports the importance of collective and sequential phosphorylation, because loss of full phosphorylation due to Ala substitution of residues potentially targeted by priming-dependent kinases is equivalent to loss of priming due to substitution of critical priming phosphoresidues themselves.
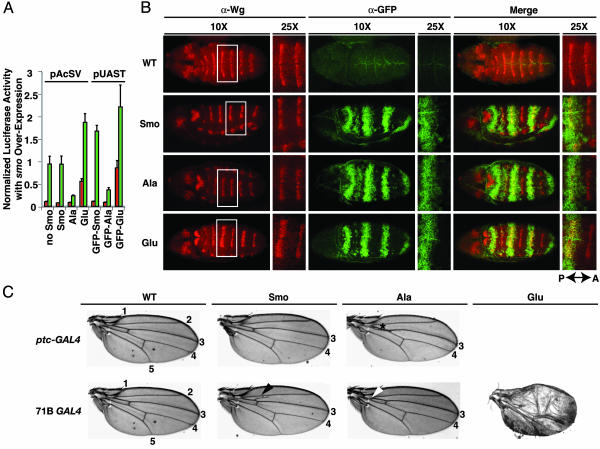
Transdominant Activities of Smo Phosphoresidue Variants in Vivo. For further analysis of the role of phosphorylation in modulation of Smo activity, we focused on III,V,VI Ala and III,V,VI Glu, hereafter referred to as the Smo-Ala and Smo-Glu variants, respectively. The Smo-Ala variant not only failed to complement loss of endogenous Smo (see above), but also showed transdominant negative activity when overexpressed in the presence of endogenous Smo in our cultured cell assay system; overexpression of the Glu variant in contrast showed transdominant positive effects, because it increased reporter activity above the normal level afforded by endogenous Smo, with or without Hh induction (Fig. 3A). These transdominant effects facilitated an analysis of Smo variant function in Drosophila embryos and wing imaginal discs using the GAL4/UAS system (51). We first tested the activities of the GFP-tagged Smo-Ala and Smo-Glu variants, as well as wild-type Smo, in our cultured cell reporter assay and found that their relative activities were similar to those of untagged Smo and its variants, although the actual levels were different, presumably due to differences in GAL4-driven protein expression levels (Fig. 3A).
Fig. 3.
Smo phosphorylation site variants show transdominant effects in vitro and in vivo. (A) Overexpressed untagged and GFP-tagged Smo variants cause transdominant effects on reporter activity. Either pAcSV- or pUAST-based plasmids expressing the Smo variants shown were transfected in the presence of endogenous Smo. The results of a representative ptc-Luc reporter assay are shown. (B) Overexpression of GFP-tagged Smo-Glu in embryos leads to expansion of Wg expression. Immunofluorescence using anti-GFP (green) and anti-Wg (red) Abs is shown for dorsal views at ×10 and ×25 of embryos at extended germ-band stage that are either wild-type (WT; w1118) or expressing UAS GFP-tagged wild-type (Smo), III,V,VI Ala (Ala), or III,V,VI Glu (Glu) forms of Smo using the prd-GAL4 driver. P ↔ A, orientation of the posterior/anterior axis. (C) Overexpression of Smo variants leads to wing patterning alterations. Wings collected from adult flies that are either wild-type (WT) or heterozygous for the ptc-GAL4 (Upper) or 71B GAL4 (Lower) driver and expressing GFP-Smo variants as in B are shown. Longitudinal veins 1–5 are labeled. Asterisk, proximal L3 and L4 fusion for the GFP-Smo-Ala variant overexpressed with ptc-GAL4; arrowheads, ectopic veination near the proximal end of L3 when GFP-Smo is overexpressed (black) and proximal L3 and L4 fusion when GFP-Smo-Ala is overexpressed (white) using 71B GAL4. Flies expressing GFP-Smo-Glu die as pupae with ptc-GAL4 and as pharate adults with 71B GAL4. A severely defective wing from an escaper from the GFP-Smo-Glu 71B GAL4 line is shown (Lower Right).
Embryonic expression of Smo and its variants was produced by using a prd-GAL4 driver, and appropriate expression of GFP-tagged proteins was noted in alternate segments of transgenic but not control embryos analyzed at the extended germ-band stage (Fig. 3B). Wild-type GFP-Smo expressed in this manner had little effect on expression of Wg, a target for Hh pathway signaling (56) (Fig. 3B). In embryos expressing the GFP-Smo-Glu variant, the Wg stripe in contrast was expanded to the anterior limit of GFP-Smo-Glu expression but retained its normal posterior boundary within the zone of variant protein expression (Fig. 3B), presumably because Engrailed continues to repress Ci expression in the posterior of the segment (57) (Fig. 5, which is published as supporting information on the PNAS web site). This gain of function of the GFP-Smo-Glu variant in vivo is consistent with its activity in our cell-based transcriptional reporter assay (Figs. 2 and 3A). Contrary to expectation from the transdominant negative effect of the GFP-Smo-Ala variant in our reporter assay (Fig. 3A), overexpression of the GFP-Smo-Ala variant did not alter Wg expression in embryos (Fig. 3B), possibly because the level of variant protein expression driven by prd-GAL4 was insufficient to interfere with the function of endogenous Smo.
We also assayed the effects of Smo variant expression on wing development using two drivers, 71B GAL4, which produces GAL4 throughout the prospective wing blade in the imaginal disk, and ptc-GAL4, which produces GAL4 throughout the anterior compartment under ptc promoter control, with highest levels in cells adjacent to the anterior/posterior compartment boundary. The wild-type wing veins L3 and L4 run parallel along the proximal/distal axis of the wing, flanking the anterior/posterior compartment boundary (Fig. 3C). Whereas expression of GFP-Smo using the ptc-GAL4 driver did not alter the pattern of the adult wing, ptc-GAL4-driven expression of the GFP-Smo-Ala variant caused proximal fusion of longitudinal veins L3 and L4 (Fig. 3C), a phenotype typical of decreased Hh pathway activity. Flies expressing the GFP-Smo-Glu variant using ptc-GAL4 died in the early pupal stage, probably due to widespread disturbances of pathway activity, and wing tissues could not be examined. The 71B GAL4 driver produces modest expression of transgenes throughout the wing blade at 25°C (51), and GFP-Smo expression under 71B control caused abnormal outgrowth of minor side veins at the anterior proximal end of vein L3 (Fig. 3C), indicative of ectopic pathway activation. In contrast, expression of GFP-Smo-Ala under 71B control caused proximal fusions of L3 and L4 near the base of the wing blade (Fig. 3C), a phenotype similar to mild pathway suppression by expression of ptc using the same driver (37). Expression of GFP-Smo-Glu driven by 71B GAL4 caused the death of most progeny as pharate adults, although a few escapers with overgrown and deformed wings emerged and died soon after eclosion (Fig. 3C). Overall, these observations validate the results from our cultured cell signaling assays and suggest that phosphorylation of the Smo cytoplasmic tail can modulate Smo activity in vivo.
Stability, Phosphorylation, and Biochemical Activity of Transdominant Smo Variants. Hh stimulation produces a Cos2-dependent accumulation of phosphorylated Smo protein on the cell surface, as well as Smo-dependent phosphorylation of Cos2, Fu, and Su(fu) (10, 11, 29). Given the transdominant positive and negative activities of Smo-Glu and Smo-Ala on wing patterning and on target gene expression in vivo and in cultured cell assays, we also tested the effects of these phosphosite alterations on Smo phosphorylation, accumulation, and surface localization, and on regulation of downstream phosphorylation events. Expression constructs for epitope-tagged Smo, Cos2, Fu, and Su(fu) were cotransfected to distinguish exogenous from endogenous protein and in an attempt to maintain the stoichiometry and interactions of intracellular pathway components that may be necessary for Hh response. Cos2, for example, is required for Hh-induced Smo stabilization (29). We noted that, although the mobility shifts associated with phosphorylation of endogenous Cos2, Fu, and Su(fu) depend largely upon Hh stimulation (13, 14, 29, 36), some degree of tonic phosphorylation of the epitope-tagged Fu and Su(fu) expressed in our experiments resulted simply from cotransfection of wild-type Myc-Smo in the absence of Hh (Fig. 4A, lane 5); this effect is consistent with the observation that some pathway activity in flies is produced simply by overexpression of GFP-Smo (Fig. 3C).
Fig. 4.
Phosphorylation in modulation of Smo stability and signaling activity. (A) Smo variants show different levels of Smo accumulation and phosphorylation of pathway components. S2R+ cells were transfected with expression constructs for wild-type Myc-Smo (Smo); Myc-Smo-III,V,VI Ala (Ala); Myc-Smo-III,V,VI Glu (Glu); Myc-Cos2; MycHis-Fu; V5-Su(fu); and/or pAct5C GAL4/UAS-hh (Hh). Approximately 48 h after transfection, lysates were analyzed by Western blotting with anti-Myc, anti-V5, or anti-lamin Ab. The lamin signal serves as a loading control. Arrowheads, dephosphorylated or minimally phosphorylated forms; bars, heavily phosphorylated species. (B) Levels of PKA and CK1α activity influence accumulation and phosphorylation of Smo. S2R+ cells were transfected with expression constructs for GFP (-); wild-type Myc-Smo (Smo); a constitutively active mouse PKA catalytic subunit (mC*); and/or wild-type or kinase-inactive (K/R) CK1α, as shown. Cells were treated with control (-) or HhN conditioned (+) medium for ∼24 h before lysis and analysis of lysates by Western blotting with anti-Myc or anti-tubulin Ab. Tubulin signal serves as a loading control. Arrowhead, minimally phosphorylated Smo; bar, heavily phosphorylated Smo.
Cotransfection with a plasmid expressing Hh nevertheless increased the degree of phosphorylation of epitope-tagged Fu and Su(fu), as well as the phosphorylation of Myc-Smo itself (Fig. 4A, lanes 5 and 6). In contrast, cells cotransfected with Smo variants had altered levels of phosphorylation of epitope-tagged Fu and Su(fu), with generally higher phosphorylation in the presence of Myc-Smo-Glu and lower in the presence of Myc-Smo-Ala (Fig. 4A, lanes 9, 10, 13, and 14). Similar changes in Cos2 phosphorylation also have been observed in other experiments (data not shown). Further, the level of phosphorylation of each component did not change in the presence of Hh. Perhaps the most striking effect of Smo alterations in these cotransfection experiments, however, was the dramatic accumulation of Myc-Smo-Glu even in the absence of Hh, as compared with Myc-Smo or Myc-Smo-Ala. This accumulation/stabilization was most evident upon coexpression of Cos2/Fu/Su(fu) and favored slower-migrating forms of Myc-Smo-Glu (Fig. 4A, lanes 9 and 10). This dramatic responsiveness to coexpression with Cos2/Fu/Su(fu) of Myc-Smo-Glu, which simulates partial phosphosite phosphorylation, is consistent with previous observations of a preferential interaction of the Cos2 complex with phosphorylated Smo (29). We also used surface biotinylation to examine localization at the cell surface for these Smo variants and found no clear difference in the proportion of total Smo protein that was localized to the plasma membrane (Fig. 6, which is published as supporting information on the PNAS web site). Overall, our results indicate that the phosphorylation status of Smo appears to modulate its stability and its propensity to be further phosphorylated, as well as its activity in promoting phosphorylation of Cos2, Fu, and Su(fu).
PKA and CK1α in Phosphorylation of Smo. Given the prevalence and arrangement of PKA and CK1 target sites in the Smo cytoplasmic tail, we investigated the effects of gain of PKA or CK1α activity by coexpressing Myc-Smo with a constitutively active mouse PKA catalytic subunit (mC*) (58) or with Drosophila CK1α. We noted increased accumulation and decreased mobility of Myc-Smo upon cotransfection with either of these kinases (Fig. 4B, compare lanes 5–8 with lanes 3 and 4). Combined expression of both kinases further increased accumulation/stabilization of Smo, as well as the proportion of more slowly migrating Smo protein (Fig. 4B, lanes 11 and 12).
In addition, coexpression of CK1α K/R, a catalytically inactive form of CK1α [K49R mutation (59)], inhibited phosphorylation and decreased accumulation of Myc-Smo (Fig. 4B, lanes 9 and 10). This result suggests that catalytically inactive CK1α K/R may function as a negative transdominant by retaining interactions with other pathway components sufficient to compete with endogenous CK1α and prevent its action. We also noted that CK1α K/R reduced phosphorylation of other pathway components (data not shown), consistent with CK1α-mediated phosphorylation as a modulator of Smo signaling activity.
Discussion
We demonstrate here that the Smo cytoplasmic tail is phosphorylated on at least 26 serine/threonine residues in HhN-stimulated cells, a degree of phosphorylation that to our knowledge is unprecedented among membrane-associated signaling proteins. By altering Smo phosphoresidues and manipulating the activity of candidate kinases, we have provided evidence for positive pathway effects for PKA and CK1α in phosphorylation and activation of Smo and have deconvoluted these positive effects from previously established inhibitory effects of PKA and CK1α on pathway activity via Ci. Overall, our results show that collective phosphorylation of multiple Smo residues can modulate Smo activity and its capacity to transduce the Hh signal.
Kinase Specificity in Smo Phosphorylation. Our mapping of in vivo phosphorylation sites in Smo revealed phosphorylation of residues potentially recognized by kinases such as PKA, CK1, and GSK3. In addition, our analysis revealed phosphorylation of residues apparently targeted by kinases of unknown specificity, as well as lack of phosphorylation on some predicted PKA and CK1 sites. Three apparent PKA sites on which phosphorylation was not detected are predicted to produce singly phosphorylated peptides upon proteolytic digestion, which may be underrepresented due to the phosphopeptide enrichment protocol used and/or the frequency and accessibility of nearby trypsin or chymotrypsin cleavage sites. Alternatively, the activity of PKA or CK1 may be selective for particular sites due to kinase specificity, Smo conformation, or Smo association with other proteins. Isolated phosphoresidues also might be subject physiologically to more efficient dephosphorylation than phosphoresidues that are capable of priming further phosphorylation nearby. Consistent with the latter possibility, 11 of the 12 phosphopeptides we isolated that include a potential PKA site contain phosphorylation on the PKA site plus additional sites nearby (Table 1).
PKA and CK1α in Phosphorylation of Hh Pathway Components at Multiple Levels. A negative pathway role for PKA and CK1 activity likely via phosphorylation of Ci was previously established (39, 41, 42). Here, we have shown that PKA and CK1α activity is associated with Smo phosphorylation and accumulation, and that these indicators of pathway activity are triggered more decisively by combined manipulation of both kinases, reminiscent of the enhanced Ci cleavage produced by combined overexpression of CK1 and PKA (41). This additive effect of the two kinases could be due to cooperative action, either because PKA primes CK1 target sites or because both kinases function as part of a larger protein complex.
Increased Smo phosphorylation and accumulation in response to kinase overexpression could result from direct action on Smo or from kinase targeting of other proteins that then influence Smo phosphorylation and accumulation. Although indirect action through other proteins cannot be excluded, at least part of the effect seems likely to result from direct kinase activity on Smo, because we observe phosphorylation of residues that constitute potential PKA and CK1 recognition sites, and simulated phosphorylation of some of these sites (along with a few sites presumably targeted by other kinases) collectively suffices to trigger positive biochemical indicators of pathway activity and target gene activation in vivo and in cultured cell assays. Our results further show that expression of a kinase-inactive form (K49R) of CK1α is sufficient to inhibit HhN-induced Smo accumulation and phosphorylation (Fig. 4) and phosphorylation of Fu and Su(fu) (data not shown), presumably by interfering with the activity of endogenous CK1α. Similar effects on Fu and Su(fu) are produced by RNA interference depletion of endogenous CK1α (data not shown). These results together demonstrate a significant positive role for CK1α in Hh pathway component phosphorylation and, potentially, in modulating signaling activity.
We should like to note that, whereas our results demonstrate a positive role for PKA activity in Smo accumulation, it has been reported that inhibition of PKA activity throughout Drosophila embryos by ectopic expression of a cAMP-insensitive regulatory subunit leads to widespread stabilization of epitope-tagged Smo from a transgene (11). This apparent increase in accumulation of Smo may be explained and resolved by the use of the same epitope to mark Smo and the ectopically expressed PKA regulatory subunit (ref. 60; data not shown).
The Role of Smo Phosphorylation in Hh Signal Transduction. How might Smo phosphorylation influence Hh pathway activation? Several recent studies have shown that recruitment of Cos2 into a complex with the Smo cytoplasmic tail (cytotail) is necessary for Hh signal transduction (29, 32–34), and that Cos2 appears preferentially to bind phosphorylated Smo (29). Smo phosphoresidues themselves, however, are unlikely to be part of an essential recognition surface for interaction with Cos2, because the known Cos2 interaction domains in the Smo cytotail do not overlap with regions V and VI, the regions containing phosphoresidues that when altered to simulate constitutive phosphorylation suffice to significantly increase signaling activity in the absence of HhN stimulation. In addition, Cos2 and Smo fusion proteins interact in vitro and presumably in the absence of phosphorylation. Thus, for example, a Cos2 fusion protein can bind a bacterially produced Smo fragment consisting of residues 557–686 (29), which contains phosphorylation regions I, II, III, IV, and part of V (Fig. 1). In addition, a myristoylated Smo cytotail fragment deleted for residues 625–818, which retains only phosphorylation regions I and VIII (Fig. 1), coimmunoprecipitates Cos2 in extracts from transfected S2 cells (32).
Given this dissociation between phosphorylation and the physical determinants required for Cos2 binding, we favor an alternative model in which phosphorylation within the Smo cytotail indirectly affects Cos2 binding by inducing a conformational shift that promotes interaction of Cos2 and perhaps other components with Smo, thereby suppressing negative and stimulating positive effects of Cos2 on pathway activity (22, 23, 29). Although further studies are needed to test this model of Smo activity modulation through a conformational shift triggered by collective phosphorylation, precedent for such a mechanism comes from the transcription factor NFAT1, which in its inactive state is phosphorylated on at least 21 residues. This protein becomes active after dephosphorylation of at least 13 of these sites, leading to a conformational shift that exposes a nuclear localization signal and masks a nuclear export signal (61).
Sequence alignments show that most of the phosphoresidues in Drosophila Smo are not conserved in mammalian Smo, particularly not in the critical regions V and VI, despite clear conservation of sequences N terminal to these regions. Thus, if such a phosphorylation-dependent mechanism for activation or stabilization of the Smo active state exists in mammals, it may use distinct kinases and recognition sequences. Alternatively, it is possible that such a mechanism has been uniquely preserved or is an innovation within the insect lineage, and that it functions to provide distinct latencies of pathway activation or inactivation. Curiously, some of the phosphoresidues whose alteration produced no evident effect on pathway activity are found in regions of sequence conservation. Phosphorylation of these residues may play a role that is masked by phosphorylation of other residues, so it may be interesting to explore the role of these more conserved phosphoresidues in the context of a Smo protein in which the phospho-switch we have uncovered is disabled. Future studies may also address the question of whether additional residues are phosphorylated primarily in the unstimulated state, and whether phosphorylation of these residues also modulates Smo activity.
Supplementary Material
Acknowledgments
We thank J. Jiang (University of Texas Southwestern Medical Center, Dallas) for pUAST constructs and the prd-GAL4 and 71B GAL4 fly lines and for sharing data before publication; D. Kalderon (Columbia University, New York) for the mC* clone and for sharing data beforepublication; D. Montell (Johns Hopkins University School of Medicine, Baltimore) for the ptc-GAL4 fly line; S. Cohen for anti-Wg Ab; P. Fisher for anti-lamin Ab; R. Mann (Johns Hopkins University School of Medicine) for assistance with analysis of Smo phosphopeptide MS data and for affinity-purified anti-Wg Ab; S. Oh (Johns Hopkins University School of Medicine) for pAcSV V5-Su(fu); Y. Ma (Johns Hopkins University School of Medicine) for pAct5C GAL4. We also thank J. Nathans and J. Corden (Johns Hopkins University School of Medicine) for comments on the manuscript and J. G. N. Garcia (Johns Hopkins University School of Medicine) for the use of equipment. This work was supported by a National Research Service Award Kirschstein Postdoctoral Fellowship (to E.H.W.), a Life Sciences Research Foundation Fellowship (to L.L.), and a grant from the National Institutes of Health. P.A.B. is an Investigator of the Howard Hughes Medical Institute.
Author contributions: C.Z., E.H.W., Y.G., L.L., and P.A.B. designed research; C.Z., E.H.W., Y.G., and L.L. performed research; C.Z., E.H.W., Y.G., and L.L. contributed new reagents/analytic tools; C.Z., E.H.W., Y.G., L.L., and P.A.B. analyzed data; and C.Z., E.H.W., Y.G., L.L., and P.A.B. wrote the paper.
Abbreviations: Hh, Hedgehog; HhN, Hh N-terminal signaling domain; Smo, Smoothened; Fu, Fused; Ci, Cubitus interruptus; CiR, repressor form of Ci; Su(fu), Suppressor of Fused; PKA, cAMP-dependent protein kinase; CK1α, casein kinase 1α; GSK3β, glycogen synthase kinase 3β; Wg, Wingless; Cos2, Costal-2.
See accompanying Biography on page 17897.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 30, 2002.
References
- 1.Ingham, P. W. & McMahon, A. P. (2001) Genes Dev. 15, 3059–3087. [DOI] [PubMed] [Google Scholar]
- 2.Lum, L. & Beachy, P. A. (2004) Science 304, 1755–1759. [DOI] [PubMed] [Google Scholar]
- 3.Beachy, P. A., Karhadkar, S. S. & Berman, D. M. (2004) Nature 432, 324–331. [DOI] [PubMed] [Google Scholar]
- 4.Marigo, V., Davey, R. A., Zuo, Y., Cunningham, J. M. & Tabin, C. J. (1996) Nature 384, 176–179. [DOI] [PubMed] [Google Scholar]
- 5.Stone, D. M., Hynes, M., Armanini, M., Swanson, T. A., Gu, Q., Johnson, R. L., Scott, M. P., Pennica, D., Goddard, A., Phillips, H., et al. (1996) Nature 384, 129–134. [DOI] [PubMed] [Google Scholar]
- 6.Fuse, N., Maiti, T., Wang, B., Porter, J. A., Hall, T. M., Leahy, D. J. & Beachy, P. A. (1999) Proc. Natl. Acad. Sci. USA 96, 10992–10999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taipale, J., Cooper, M. K., Maiti, T. & Beachy, P. A. (2002) Nature 418, 892–896. [DOI] [PubMed] [Google Scholar]
- 8.Alcedo, J., Ayzenzon, M., Von Ohlen, T., Noll, M. & Hooper, J. E. (1996) Cell 86, 221–232. [DOI] [PubMed] [Google Scholar]
- 9.van den Heuvel, M. & Ingham, P. W. (1996) Nature 382, 547–551. [DOI] [PubMed] [Google Scholar]
- 10.Denef, N., Neubuser, D., Perez, L. & Cohen, S. M. (2000) Cell 102, 521–531. [DOI] [PubMed] [Google Scholar]
- 11.Alcedo, J., Zou, Y. & Noll, M. (2000) Mol. Cell 6, 457–465. [DOI] [PubMed] [Google Scholar]
- 12.Zhu, A. J., Zheng, L., Suyama, K. & Scott, M. P. (2003) Genes Dev. 17, 1240–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robbins, D. J., Nybakken, K. E., Kobayashi, R., Sisson, J. C., Bishop, J. M. & Therond, P. P. (1997) Cell 90, 225–234. [DOI] [PubMed] [Google Scholar]
- 14.Sisson, J. C., Ho, K. S., Suyama, K. & Scott, M. P. (1997) Cell 90, 235–245. [DOI] [PubMed] [Google Scholar]
- 15.Preat, T., Therond, P., Lamour-Isnard, C., Limbourg-Bouchon, B., Tricoire, H., Erk, I., Mariol, M. C. & Busson, D. (1990) Nature 347, 87–89. [DOI] [PubMed] [Google Scholar]
- 16.Orenic, T. V., Slusarski, D. C., Kroll, K. L. & Holmgren, R. A. (1990) Genes Dev. 4, 1053–1067. [DOI] [PubMed] [Google Scholar]
- 17.Eaton, S. & Kornberg, T. B. (1990) Genes Dev. 4, 1068–1077. [DOI] [PubMed] [Google Scholar]
- 18.Dominguez, M., Brunner, M., Hafen, E. & Basler, K. (1996) Science 272, 1621–1625. [DOI] [PubMed] [Google Scholar]
- 19.Alexandre, C., Jacinto, A. & Ingham, P. W. (1996) Genes Dev. 10, 2003–2013. [DOI] [PubMed] [Google Scholar]
- 20.Von Ohlen, T., Lessing, D., Nusse, R. & Hooper, J. E. (1997) Proc. Natl. Acad. Sci. USA 94, 2404–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen, C. H., von Kessler, D. P., Park, W., Wang, B., Ma, Y. & Beachy, P. A. (1999) Cell 98, 305–316. [DOI] [PubMed] [Google Scholar]
- 22.Wang, Q. T. & Holmgren, R. A. (2000) Development (Cambridge, U.K.) 127, 3131–3139. [DOI] [PubMed] [Google Scholar]
- 23.Wang, G., Amanai, K., Wang, B. & Jiang, J. (2000) Genes Dev. 14, 2893–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aza-Blanc, P., Ramirez-Weber, F. A., Laget, M. P., Schwartz, C. & Kornberg, T. B. (1997) Cell 89, 1043–1053. [DOI] [PubMed] [Google Scholar]
- 25.Wang, Q. T. & Holmgren, R. A. (1999) Development (Cambridge, U.K.) 126, 5097–5106. [DOI] [PubMed] [Google Scholar]
- 26.Preat, T. (1992) Genetics 132, 725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pham, A., Therond, P., Alves, G., Tournier, F. B., Busson, D., Lamour-Isnard, C., Bouchon, B. L., Preat, T. & Tricoire, H. (1995) Genetics 140, 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monnier, V., Dussillol, F., Alves, G., Lamour-Isnard, C. & Plessis, A. (1998) Curr. Biol. 8, 583–586. [DOI] [PubMed] [Google Scholar]
- 29.Lum, L., Zhang, C., Oh, S., Mann, R. K., von Kessler, D. P., Taipale, J., Weis-Garcia, F., Gong, R., Wang, B. & Beachy, P. A. (2003) Mol. Cell 12, 1261–1274. [DOI] [PubMed] [Google Scholar]
- 30.Methot, N. & Basler, K. (2000) Development (Cambridge, U.K.) 127, 4001–4010. [DOI] [PubMed] [Google Scholar]
- 31.Lefers, M. A., Wang, Q. T. & Holmgren, R. A. (2001) Dev. Biol. 236, 411–420. [DOI] [PubMed] [Google Scholar]
- 32.Jia, J., Tong, C. & Jiang, J. (2003) Genes Dev. 17, 2709–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogden, S. K., Ascano, M., Jr., Stegman, M. A., Suber, L. M., Hooper, J. E. & Robbins, D. J. (2003) Curr. Biol. 13, 1998–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruel, L., Rodriguez, R., Gallet, A., Lavenant-Staccini, L. & Therond, P. P. (2003) Nat. Cell Biol. 5, 907–913. [DOI] [PubMed] [Google Scholar]
- 35.Ohlmeyer, J. T. & Kalderon, D. (1998) Nature 396, 749–753. [DOI] [PubMed] [Google Scholar]
- 36.Therond, P. P., Knight, J. D., Kornberg, T. B. & Bishop, J. M. (1996) Proc. Natl. Acad. Sci. USA 93, 4224–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson, R. L., Grenier, J. K. & Scott, M. P. (1995) Development (Cambridge, U.K.) 121, 4161–4170. [DOI] [PubMed] [Google Scholar]
- 38.Chen, Y., Gallaher, N., Goodman, R. H. & Smolik, S. M. (1998) Proc. Natl. Acad. Sci. USA 95, 2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price, M. A. & Kalderon, D. (1999) Development (Cambridge, U.K.) 126, 4331–4339. [DOI] [PubMed] [Google Scholar]
- 40.Wang, G., Wang, B. & Jiang, J. (1999) Genes Dev. 13, 2828–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price, M. A. & Kalderon, D. (2002) Cell 108, 823–835. [DOI] [PubMed] [Google Scholar]
- 42.Lum, L., Yao, S., Mozer, B., Rovescalli, A., Von Kessler, D., Nirenberg, M. & Beachy, P. A. (2003) Science 299, 2039–2045. [DOI] [PubMed] [Google Scholar]
- 43.Jia, J., Amanai, K., Wang, G., Tang, J., Wang, B. & Jiang, J. (2002) Nature 416, 548–552. [DOI] [PubMed] [Google Scholar]
- 44.Ohlmeyer, J. T. & Kalderon, D. (1997) Genes Dev. 11, 2250–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collier, L. S., Suyama, K., Anderson, J. H. & Scott, M. P. (2004) Genetics 167, 783–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan, C. M., Porter, J. A., Chiang, C., Chang, D. T., Beachy, P. A. & Tessier-Lavigne, M. (1995) Cell 81, 457–465. [DOI] [PubMed] [Google Scholar]
- 47.Hammerschmidt, M., Bitgood, M. J. & McMahon, A. P. (1996) Genes Dev. 10, 647–658. [DOI] [PubMed] [Google Scholar]
- 48.Concordet, J. P., Lewis, K. E., Moore, J. W., Goodrich, L. V., Johnson, R. L., Scott, M. P. & Ingham, P. W. (1996) Development (Cambridge, U.K.) 122, 2835–2846. [DOI] [PubMed] [Google Scholar]
- 49.Huang, Y., Roelink, H. & McKnight, G. S. (2002) J. Biol. Chem. 277, 19889–96. [DOI] [PubMed] [Google Scholar]
- 50.Ficarro, S. B., McCleland, M. L., Stukenberg, P. T., Burke, D. J., Ross, M. M., Shabanowitz, J., Hunt, D. F. & White, F. M. (2002) Nat. Biotechnol. 20, 301–305. [DOI] [PubMed] [Google Scholar]
- 51.Brand, A. H. & Perrimon, N. (1993) Development (Cambridge, U.K.) 118, 401–415. [DOI] [PubMed] [Google Scholar]
- 52.Muszynska, G., Andersson, L. & Porath, J. (1986) Biochemistry 25, 6850–6853. [DOI] [PubMed] [Google Scholar]
- 53.Andersson, L. & Porath, J. (1986) Anal. Biochem. 154, 250–254. [DOI] [PubMed] [Google Scholar]
- 54.Pearson, R. B. & Kemp, B. E. (1991) Methods Enzymol. 200, 62–81. [DOI] [PubMed] [Google Scholar]
- 55.Fujii, K., Zhu, G., Liu, Y., Hallam, J., Chen, L., Herrero, J. & Shaw, S. (2004) Proc. Natl. Acad. Sci. USA 101, 13744–13749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martizez Arias, A., Baker, N. E. & Ingham, P. W. (1988) Development (Cambridge, U.K.) 103, 157–170. [DOI] [PubMed] [Google Scholar]
- 57.Schwartz, C., Locke, J., Nishida, C. & Kornberg, T. B. (1995) Development (Cambridge, U.K.) 121, 1625–1635. [DOI] [PubMed] [Google Scholar]
- 58.Orellana, S. A. & McKnight, G. S. (1992) Proc. Natl. Acad. Sci. USA 89, 4726–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yanagawa, S., Matsuda, Y., Lee, J. S., Matsubayashi, H., Sese, S., Kadowaki, T. & Ishimoto, A. (2002) EMBO J. 21, 1733–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li, W., Ohlmeyer, J. T., Lane, M. E. & Kalderon, D. (1995) Cell 80, 553–562. [DOI] [PubMed] [Google Scholar]
- 61.Okamura, H., Aramburu, J., Garcia-Rodriguez, C., Viola, J. P., Raghavan, A., Tahiliani, M., Zhang, X., Qin, J., Hogan, P. G. & Rao, A. (2000) Mol. Cell 6, 539–550. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.