Abstract
We developed an allele-specific assay for transposase-accessible chromatin with high-throughput sequencing (ATAC–seq) to genotype and profile active regulatory DNA across the genome. Using a mouse hybrid F1 system, we found that monoallelic DNA accessibility across autosomes was pervasive, developmentally programmed and composed of several patterns. Genetically determined accessibility was enriched at distal enhancers, but random monoallelically accessible (RAMA) elements were enriched at promoters and may act as gatekeepers of monoallelic mRNA expression. Allelic choice at RAMA elements was stable across cell generations and bookmarked through mitosis. RAMA elements in neural progenitor cells were biallelically accessible in embryonic stem cells but premarked with bivalent histone modifications; one allele was silenced during differentiation. Quantitative analysis indicated that allelic choice at the majority of RAMA elements is consistent with a stochastic process; however, up to 30% of RAMA elements may deviate from the expected pattern, suggesting a regulated or counting mechanism.
Mammalian cells have two copies of every autosomal gene that are typically turned on or off together in the same nucleus. The mechanisms by which cells break this symmetry in some cases and express a gene from only one of the two alleles of the diploid genome are not yet fully understood. Classic monoallelic genes include X-chromosome-linked genes, olfactory receptor genes and developmentally imprinted genes1. There are no changes in DNA content between the two alleles in these classes of genes, and thus they are differentially regulated at the epigenetic level. This regulation involves noncoding RNAs, DNA methylation, histone modifications and heterochromatin formation2–4. Recently, a new class of random monoallelically expressed (RME) genes has been identified. These genes are expressed from one allele or the other in a clone-specific manner that is independent of parent of origin and underlying DNA sequence5–9. In neural progenitor cells (NPCs), many RME genes are candidate genes for neurodegenerative disorders, and clonal heterogeneity in their expression may contribute to variable disease severity and age of onset8,10. Allele choice in RME genes is stable in culture, but little is known about how it is established and epigenetically remembered4. There is some evidence that a subset of RME genes are asynchronously replicated and differentially methylated on the two alleles5,11. The epi-genetic mechanism by which the cell can break symmetry randomly in development and express one gene monoallelically among a sea of biallelically expressed genes is of great interest12,13.
We used ATAC–seq to define the DNA sequences related to monoallelic epigenetic memory, a method for profiling DNA accessibility with a small number of cells on a rapid timescale14. ATAC–seq can be used to comprehensively identify active regulatory elements, transcription factor binding sites and nucleosome position across the genome. However, standard ATAC–seq and other genomic analyses mask the effects of heterozygous mutations and regulatory changes. To interrogate the effects of an acquired mutation or genotype on regulatory changes in the clinic, it is important to be able to resolve individual haplotypes in accessibility data. Here we describe the optimization of allele-specific ATAC–seq. We used a tractable mouse hybrid system in which millions of fully phased SNPs can be interrogated in ATAC–seq reads15. Using this method, we identified the landscape of monoallelically accessible regulatory elements in embryonic stem cells (ESCs) and NPCs. We identified a new class of RAMA elements and characterized their distinctive genomic distribution, capacity for epigenetic memory and developmental ontogeny.
RESULTS
Optimization of allele-specific ATAC–seq
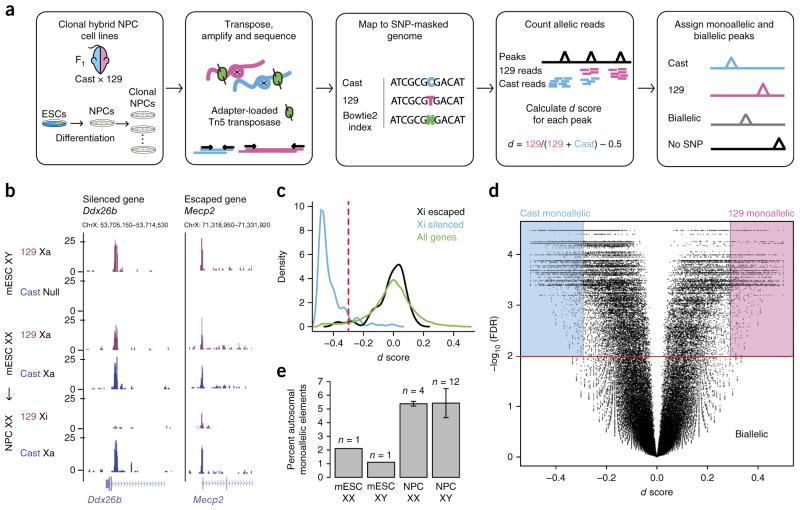
To identify allele-specific regulatory elements in the mouse genome, we performed ATAC–seq in highly polymorphic F1 hybrid mouse ESCs and ESC-derived clonal NPCs (Fig. 1a). An NPC clone was derived from a single colony, which was picked under a microscope. These cell lines, derived from a 129S1 (here referred to as 129) × Castaneous (Cast) cross, contain ~23 million SNPs (1 SNP for every ~110 bp)5. This SNP density is approximately tenfold the SNP density in human cells and thus provides high resolution to interrogate allelic chromatin regulation. We performed ATAC–seq in male and female ESCs (two lines) and NPCs (16 clones) and developed an allele-specific ATAC–seq analysis pipeline. For each clone, we sequenced two replicates to an average of 50 million usable reads and then merged them after verifying their reproducibility (Supplementary Fig. 1a and Supplementary Table 1). We mapped sequencing reads to a ‘SNP-masked’ genome index in which we replaced each SNP site with ‘N’ to eliminate reference bias. We then assigned each ‘N’-overlapping read (~55% of total reads) to its genome of origin on the basis of SNP identity (Fig. 1a). To confirm that our mapping strategy was highly accurate, we simulated reads from the 129 and Cast alleles and found that only 0.05% of reads mapped to the wrong allele16. Furthermore, for all monoallelic sites identified in NPCs, there was no allelic bias in ESCs, indicating that our allelic assignment was specific to this cell type and not a systematic bias in allelic analysis.
Figure 1.
Allele-specific ATAC–seq used to discover monoallelically accessible regulatory elements across the genome in mouse cells. (a) Experimental and analytic scheme. Cast and 129 mice were crossed, and F1 hybrid ESCs were isolated. ESCs were differentiated into NPCs and subcloned. ATAC–seq was performed on clonal cell lines. Sequencing reads were assigned to the 129 and Cast genomes. The d score of allelic imbalance was calculated for each ATAC–seq peak using SNP-informative reads. (b) Two examples of allele-specific ATAC–seq tracks on the X chromosome including the Ddx26b locus, which was silenced in differentiated cells, and the Mecp2 locus, which escaped silencing. Xa, active X chromosome; Xi, inactive X chromosome. (c) Distribution of d scores for ATAC–seq peaks at the promoters of silenced and escaped genes on the X chromosome as well as for all genes. The red dashed line corresponds to the cutoff of d score = −0.3 used to distinguish escaped from silenced elements. (d) Volcano plot showing d score versus –log10 (FDR) for all peaks in NPC clone XX1 across the genome. Background colors indicate how peaks are assigned on the basis of d score and FDR. (e) Percentage of total autosomal ATAC–seq peaks that are monoallelic in ESCs and NPCs derived from females and males. Error bars show s.d. across the number of clones indicated.
Identifying monoallelic regulatory elements
To identify regulatory elements that are differentially regulated on the two alleles, we developed a method for assigning allelic ATAC–seq peaks. First, we called high-confidence ATAC–seq peaks using combined reads from all NPC samples with MACS2 (ref. 17). We then counted the number of reads from the 129 and Cast alleles in each ATAC–seq peak and calculated a score of allelic bias, the d score (Fig. 1a)7.
The d score has a range of −0.5 to +0.5: +0.5 means all the reads are from the 129 allele and −0.5 means all the reads are from the Cast allele; 0 means the reads are equally distributed between the 129 and Cast alleles. In addition to the d score, we computed a P value for the d score using a permutation-based method to evaluate the significance of the deviation from biallelic accessibility (Online Methods and Supplementary Fig. 1b).
To determine the d-score cutoff to consider a peak monoallelic, we used the X chromosome in female differentiated and undifferentiated cells. In female ESCs, both X chromosomes are active and the d score for most peaks should be ~0. In an NPC clone in which the 129 X chromosome has been inactivated and the majority of genes should be monoallelically expressed, we found that the d score for most X-chromosome peaks was <−0.3 (Fig. 1b,c). Therefore, we used this d score as the threshold for assigning monoallelic and biallelic peaks on the autosomes (Fig. 1d).
We considered peaks with ≥10 allele-informative reads to be assignable with high confidence. The number of ATAC–seq peaks that could be assigned allelically increased with sequencing depth and plateaued at ~90% of total peaks (Supplementary Fig. 1c–e). To remove potential false positives, we further filtered out all sites of somatic DNA copy number variants (CNVs) by assessing chromosomal blocks of ATAC–seq signal variation for all cell lines studied (Online Methods).
Three classes of monoallelic elements
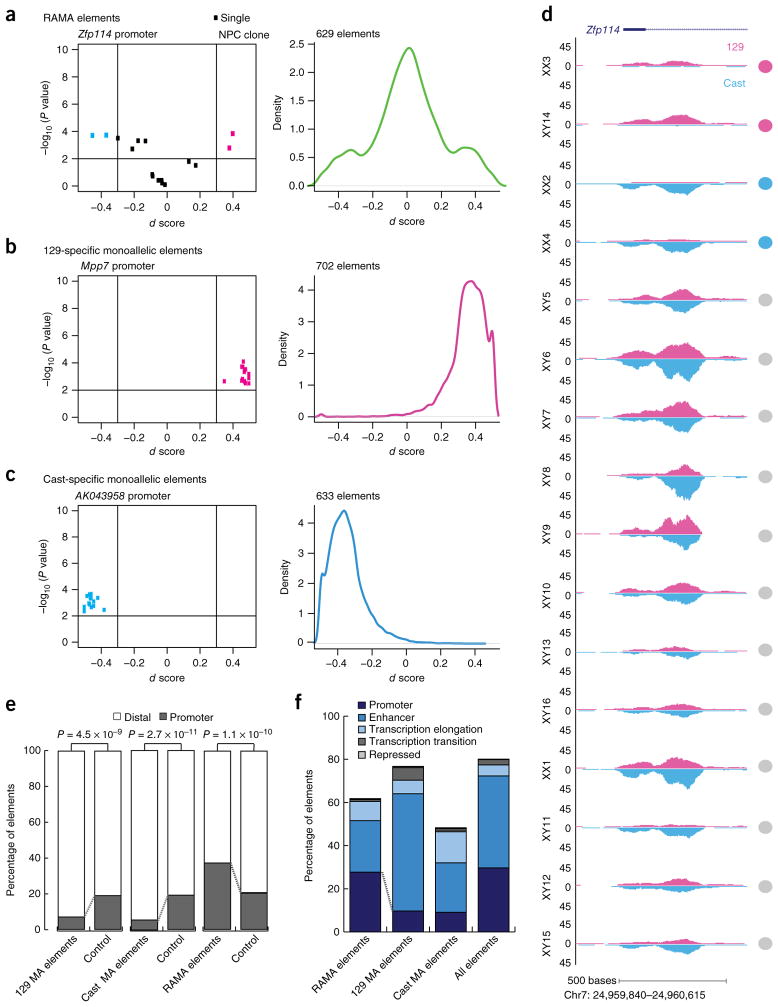
In each NPC clone, we identified between 2,800 and 4,500 monoal-lelic sites of DNA accessibility, comprising ~5% of all ATAC–seq peaks (Fig. 1e and Supplementary Fig. 1f). We classified all monoallelically accessible elements (1,964 elements) into 129-specific (702 elements), Cast-specific (633 elements) and RAMA elements (629 elements) (Fig. 2a–c). We defined RAMA elements as those that were monoallelic in at least two clones with at least one being 129 monoallelic and one being Cast monoallelic (Fig. 2a,d). We show one example in which the promoter of the RME gene Zfp114 had a RAMA pattern, with monoallelic accessibility in four NPC clones and biallelic accessibility in the remaining 12 (Fig. 2d). The sex of the clone did not affect allelic choice at RAMA elements (Fig. 2d). 129-specific and Cast-specific elements, arising owing to parent- or genotype-specific regulation, were monoallelic from the same allele in at least 50% of clones and biallelic in the other clones (Fig. 2b,c and Supplementary Fig. 2a,b). We filtered out known imprinted genes, as imprinting is eroded in ESC culture and thus is not faithfully maintained in our ESC-derived NPCs18,19. Overall, our results indicated pervasive monoallelic DNA accessibility occurring in three distinctive patterns.
Figure 2.
Distinct classes of monoallelic regulatory elements with different genomic locations. (a) Left, d score versus –log10 (P value) for the RAMA element at the Zfp114 promoter. Each point represents data for an NPC clone. Color corresponds to whether the element is biallelic (black) or monoallelic (pink or blue) in each clone. Right, density plot showing the distribution of d scores for all RAMA elements (n = 629). (b) Data as in a for 129-specific monoallelic elements: d score versus –log10 (P value) for the accessible element at the Mpp7 promoter (left) and d scores for 702 129-specific monoallelic elements (right). (c) Data as in a for Cast-specific monoallelic elements: d score versus –log10 (P value) for the accessible element at the 3′ end of the AK043958 gene (left) and d scores for 633 Cast-specific monoallelic elements (right). (d) Example allele-specific ATAC–seq tracks for 16 NPC clones at the RAMA element in the promoter of Zfp114. In each plot, 129 reads are shown on top (pink) and Cast reads are shown below (blue). Circles correspond to how each element was called in that line (gray, biallelic; pink, monoallelic 129; blue, monoalellic Cast). (e) Proportion of 129-specific, Cast-specific and RAMA elements located in distal elements (>2 kb from a TSS) and promoter elements (<2 kb from a TSS) (RefSeq). For each set of elements, control sets were size matched and matched for peak enrichment. MA, monoallelic. (f) ChromHMM annotation for RAMA, 129-specific and Cast-specific elements. Public ChIP–seq data and ChromHMM were used to annotate elements (Online Methods).
RAMA elements are enriched at promoters
We compared the three classes of monoallelic elements, using transcription start site (TSS) annotation and ChIP–seq data from NPCs to determine the genomic location and features of our identified monoallelic regulatory elements. Although 129- or Cast-specific elements were significantly enriched at distal elements (>2 kb from a TSS; P < 1 × 10−9 for each), RAMA elements were significantly enriched at promoters (<2 kb from a TSS; P = 1.1 × 10−10; Fig. 2e and Supplementary Fig. 2c). ChromHMM analysis confirmed that RAMA elements had the highest proportion of promoters, marked by trimethylation of histone H3 at lysine 4 (H3K4me3) and RNA polymerase II (Pol II) occupancy. In contrast, 129- and Cast-specific accessible elements had enhancers as the largest constituent class and were depleted of promoters (Fig. 2f and Supplementary Fig. 2d,e). Thus, genetic bias in DNA accessibility tended to occur at enhancers, whereas RAMA tended to occur at promoters. We found no significant enrichment for specific transcription factor binding sites at RAMA elements, indicating that there was no single transcription factor or family of transcription factors regulating these elements as a class.
The promoter bias of RAMA elements suggested that they might be tightly linked to monoallelic transcription. In contrast, the enhancer bias in genotype-specific monoallelic elements reflected the looser conservation of genomic sequence at distal elements and the use of distinct enhancers in evolutionarily divergent strains. Furthermore, this strain specificity of enhancers was reflected in the fact that 129-specific elements were more enriched for all states, including enhancers, as a result of its similarity to the reference strain in which most ChIP–seq experiments have been performed (Fig. 2f).
RAMA element choice is stable across cell generations and bookmarked in mitosis
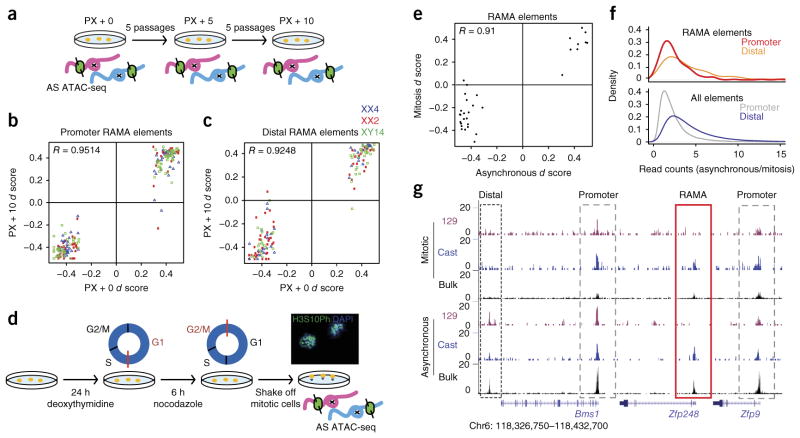
We focused on RAMA elements because they are a new class of regulatory DNA. Clonal monoallelic gene expression can arise as a result of stable silencing of one allele or transient monoallelic states coordinated within a clonal population4. To test whether RAMA elements are epigenetically stable over time, we performed ATAC–seq at five and ten additional passages after the first ATAC–seq experiment (Fig. 3a). We found that RAMA elements, 129-specific and Cast-specific elements maintained the same allelic bias across all passages (R = 0.95 and 0.92 for promoters and distal elements, respectively; Fig. 3b,c and Supplementary Fig. 3a–f). This is consistent with stability over multiple passages of a limited number of RME genes based on allele-specific RT–PCR analysis in ref. 5. We compared the slight changes in ATAC–seq d score at some monoallelic elements with the variability in d score owing to technical variation across three replicates of one clone at the same passage. The changes in d scores for RAMA elements across passages were very small and on the same scale as technical variation between replicates at the same passage (Supplementary Fig. 3d,e).
Figure 3.
RAMA elements are stable through mitosis and over many passages in the NPC state. (a) Experimental setup for allele-specific (AS) ATAC–seq across passages. Three NPC lines (XX4, XX2 and XY14) were cultured for five and ten additional passages (PX + 5 and PX + 10, respectively) after the initial ATAC–seq experiment (PX + 0). (b) ATAC–seq d scores at passage 0 versus passage 10 for promoter RAMA elements in NPC clones XX2, XX4 and XY14. (c) ATAC–seq d scores at passage 0 versus passage 10 for distal RAMA elements in NPC clones XX2, XX4 and XY14. (d) Experimental setup for mitotic ATAC–seq. NPC clone XX1 was blocked in M phase, and mitotic cells were then collected by shaking. The image shows mitotic NPCs stained with DAPI for mitotic chromosomes and antibody to H3S10ph, a marker of prometaphase cells (40× magnification). (e) ATAC–seq d scores for RAMA elements in clone XX1 in mitotic versus asynchronous cells. Monoallelic RAMA elements were included if there were ≥20 allele-informative reads under the peak in both mitotic and asynchronous ATAC–seq data. (f) Ratio of ATAC–seq read peaks in asynchronous/mitotic cells for promoter elements (<2 kb from a TSS) and distal elements (>2 kb from a TSS). RAMA elements are shown at the top, and all accessible elements are shown at the bottom. (g) Example locus showing ATAC–seq signal in mitotic and asynchronous NPCs. Allelic ATAC–seq signal is shown in magenta and blue, and non-allelic ATAC–seq signal is shown in black (bulk). Dashed black boxes indicate distal regulatory elements, dashed gray boxes indicate promoters and the red box indicates the RAMA element at the TSS of the RME gene Zfp248.
The mechanism by which monoallelic DNA accessibility can be transmitted through the cell cycle is of great interest. Most transcription factors are believed to dissociate from chromatin during mitosis, although some are believed to ‘bookmark’ DNA to preserve binding sites and facilitate reactivation of gene expression after cytokine-sis20. Hi-C studies have shown that hierarchical chromatin structure is erased during the cell cycle, although some specific accessibility patterns seem to be maintained21,22. We asked whether monoallelic DNA accessibility is bookmarked through mitosis. We isolated mitotic NPCs and performed ATAC–seq. DAPI and phosphorylation of his-tone H3 at Ser10 (H3S10ph) staining showed that 94% of cells were arrested in prometaphase (Fig. 3d). We found that, in mitosis, there were fewer ATAC–seq peaks overall, and many peaks were reduced but not entirely lost (Supplementary Fig. 3g). Promoter-proximal peaks were more highly preserved during mitosis than distal regulatory elements, supporting a model in which active genes are accessible through the cell cycle but lose contacts with other distal elements (Fig. 3f)21,22. We found that RAMA elements retained allele-specific accessibility during mitosis, and the d score at RAMA elements in the asynchronous cell population was highly correlated with d scores from mitotic NPCs (R = 0.75; Fig. 3e,f and Supplementary Fig. 3g). In Figure 3g, we show a locus in which the DNA accessibility of distal regulatory elements was not preserved during mitosis, whereas accessibility at promoters was maintained as well as the RAMA element choice. Thus, RAMA elements were stable and faithfully bookmarked throughout the cell cycle, as evidenced by differential mitotic accessibility on the two alleles.
RME genes have randomly monoallelic promoters but not enhancers
RME genes had been identified by RNA–seq in seven NPC clones5 for which we have ATAC–seq data5. We asked whether RME genes in these clones have nearby RAMA elements reflecting their transcriptional state and marking allele-specific regulatory elements that may control RME. For this analysis, we considered only RME genes for which the promoter is accessible and contains informative SNPs (149 genes) and RAMA elements (87 elements) for which the adjacent transcript is expressed and contains informative SNPs.
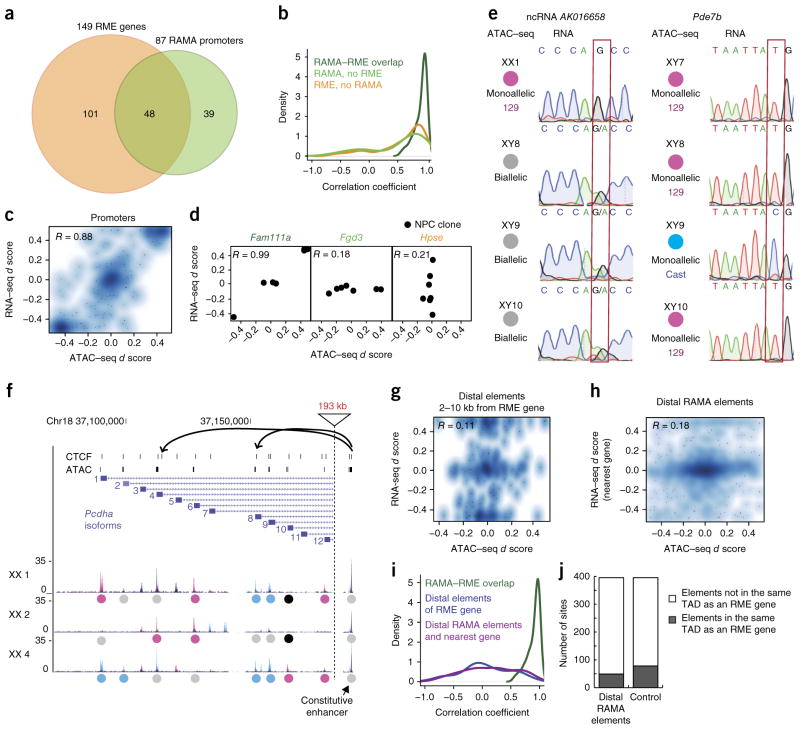
Forty-eight of 149 RME genes had RAMA elements at their promoters, corresponding to 48 of the 87 RAMA promoters (Fig. 4a). The overall correlation of d score for expression and d score for accessibility was 0.88 across all RME genes in all clones (Fig. 4b,c). Some RME genes, such as Fam111a, had accessibility d scores that were very highly correlated with expression d score, whereas others, such as Fgd3 and Hpse, did not (Fig. 4d). RT–PCR and Sanger sequencing in independently derived NPC lines for which RNA–seq data were not available confirmed this strong promoter RAMA–RME correlation at the RME genes Pde7b and Bag3 (Fig. 4e and Supplementary Fig. 4c). RME genes that did not have RAMA promoters tended to be weakly expressed, and RAMA elements that did not have adjacent RME genes tended to have low ATAC–seq peak enrichment scores (Supplementary Fig. 4a,b). In addition, there were 16 RAMA elements that were not adjacent to RME genes called by Gendrel et al. but that had been called as RME genes in another study of RME in NPCs7. Collectively, these results indicate that in many cases the promoter of an RME gene is only accessible to transcriptional machinery on the expressed allele, resulting in stable expression of only this allele. In other, more rare, cases such as the Hpse locus, an RME gene may have a biallelically accessible promoter, but other epigenetic or post-transcriptional regulation is in place to maintain RME (Fig. 4d).
Figure 4.
RME genes regulated at local promoter-proximal RAMA elements. (a) Overlap between RME genes from RNA–seq (ref. 5) and RAMA elements from ATAC–seq. Seven clones for which we had both RNA–seq and ATAC–seq data were used. Included are genes containing a SNP in the transcript as well as the promoter ATAC–seq peak. (b) Density plot of the correlation coefficient for promoter ATAC–seq d score and RNA–seq d score for RME genes with RAMA elements at the promoter (dark green), RME genes with no RAMA element at the promoter (orange), and promoter-proximal RAMA elements with no adjacent RME gene (light green). (c) Smooth scatterplot showing RNA–seq versus ATAC–seq d scores for RME–RAMA pairs. For all smooth scatterplots, the density of points is represented by color. Points are superimposed in low-density regions. (d) RNA–seq versus promoter ATAC–seq d scores for genes with RME–RAMA overlap (Fam111a), RME genes with no RAMA element at the promoter (Fgd3), and RAMA elements with no adjacent RME gene (Hpse). Points correspond to NPC clones for which RNA–seq and ATAC–seq data were available. (e) RT–PCR and Sanger sequencing for the noncoding RNA AK016658 (left) and Pde7b (right). Circles show the allelic status of the promoter element. The allele-informative SNP is highlighted in a red box. (f) Example of the Pcdha (protocadherin-α) locus in which there are multiple alternative isoforms independently selected on each allele. Top, CTCF sites and ATAC–seq peaks. Bottom, tracks for NPC clones showing 129 reads (pink) and Cast reads (blue). Colored circles indicate whether the peak is monoallelic 129 (pink), monoallelic Cast (blue), biallelic (gray) or unassignable (black). The constitutive, biallelic enhancer region is shown to the right. (g) Smooth scatterplot showing RNA–seq versus ATAC–seq d scores for RME genes and peaks 2–10 kb from the promoter. (h) As in g, for promoter-distal RAMA elements and their nearest genes. (i) Density plot showing the correlation coefficient for RNA–seq and ATAC–seq d scores for RME genes with RAMA promoters (green), RME genes and elements 2–10 kb from the promoter (blue), and distal RAMA elements with the nearest gene (purple). (j) Number of enhancer RAMA elements located in a TAD with or without an RME gene. Control is a set of size- and enrichment-matched elements.
Of the 39 promoter RAMA elements that were not located at described RME genes, some were located at the promoters of genes with multiple isoforms that are difficult to distinguish by RNA–seq. An example of this is the protocadherin-α cluster in which there are 12 alternative exons with highly repetitive sequences. In a given clone, one or more isoforms is expressed on each allele independently. The promoters of the chosen alleles form contacts with a constitutive enhancer ~200 kb downstream of the locus that is biallelically accessible23,24. Allele-specific combinatorial isoform choice is difficult to distinguish by RNA–seq but was identified by allele-specific ATAC–seq (Fig. 4f).
We next asked whether RME genes have distal RAMA elements nearby that might act as monoallelic enhancer switches. We were surprised to find that there was no correlation between RME gene d score and the d score of the non-promoter ATAC–seq peaks between 2 kb and 10 kb upstream or downstream (R = 0.11; Fig. 4g and Supplementary Fig. 4f). A well-studied enhancer–promoter pair is the Arc gene and its enhancer located 7 kb upstream, which loops over to contact the promoter in a neuronal-activity-dependent manner25. Although Arc is RME and its promoter is RAMA, the upstream enhancer was biallelic in all clones (Supplementary Fig. 4j). All of this evidence suggests a model in which the enhancer landscape near many RME genes is permissive for expression of both alleles but monoallelic accessibility of the promoter may serve as the gatekeeper for monoallelic gene expression.
Conversely, we tested whether the distal RAMA elements (>2 kb from a TSS) that we identified by ATAC–seq might regulate previously identified RME genes or new monoallelic transcripts. Using Hi-C data from these same cells16, we found that distal RAMA elements were not located in the same topologically associating domains (TADs) as RME genes more than expected by chance (Fig. 4j). We found that the d score at promoter-distal RAMA elements and the d score for expression at the nearest gene was not well correlated (R = 0.18). There were only a few sites for which the distal RAMA element might contribute to allelic bias in transcription at the nearest gene (Fig. 4h,i and Supplementary Fig. 4d,e,g,h). These distal RAMA elements may mark the promoters of currently unannotated transcripts or non-polyadenylated transcripts (Supplementary Fig. 4d,e). For the noncoding RNA AK016658, we confirmed that the allelic expression matched promoter ATAC–seq status (Fig. 4e).
RAMA elements are developmentally programmed but premarked in embryonic stem cells
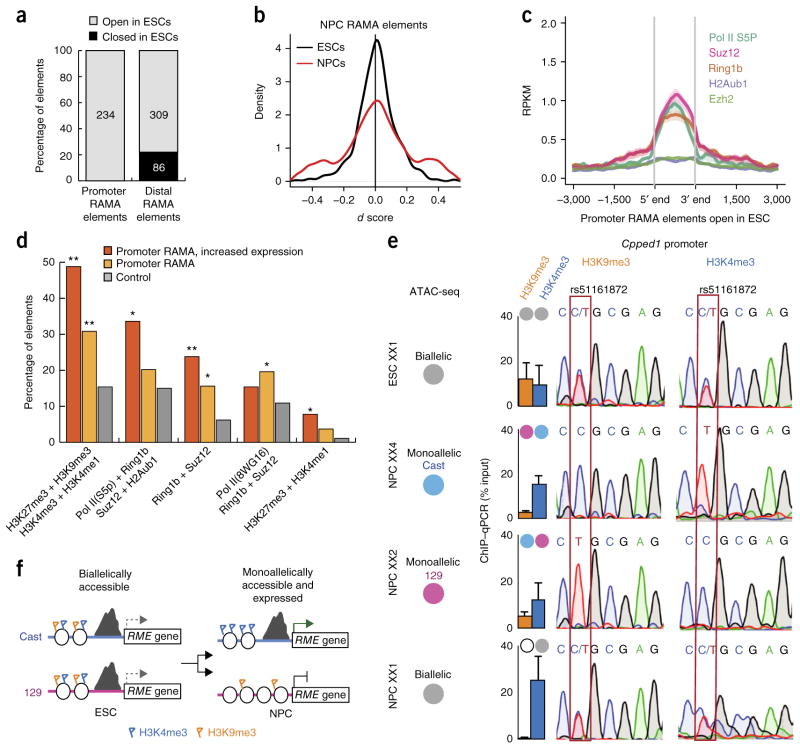
To investigate whether RAMA elements are prepatterned during development or arise de novo during neural differentiation, we performed ATAC–seq in ESCs that we used to generate the clonal NPCs. We found that the number of monoallelic ATAC–seq peaks in ESCs was one-fourth the number in NPCs (Fig. 1e and Supplementary Fig. 1f). The majority of RAMA elements in NPCs were accessible in ESCs (86%) (Fig. 5a), but only 2–4% were monoallelic in ESCs (as compared with 21% monoallelic in each NPC clone), indicating that their monoallelic accessibility is NPC specific (Fig. 5b).
Figure 5.
RAMA elements are accessible and marked by active and repressive marks in the ESC state. (a) Percentage of NPC RAMA elements that are open and closed in ESCs. (b) Distribution of d scores for NPC RAMA elements in NPCs (red) and ESCs (black). (c) Average plot showing the enrichment for ChIP–seq signal of chromatin-modifying enzymes in ESCs at promoter-proximal NPC RAMA elements open in ESCs (n = 35). (d) Colocalization of chromatin-modifying enzymes and histone modifications by ChIP–seq in ESCs at all promoter NPC RAMA elements (orange) and promoter NPC RAMA elements at the promoters of genes with increased expression from the ESC to NPC state. P values (Fisher’s test) indicate enrichment: *P < 0.05, **P < 0.01. (e) ChIP–qPCR and Sanger sequencing for H3K4me3 and H3K9me3 at the Cpped1 promoter in ESC XX1 and NPC clones derived from this cell line (XX1, XX2 and XX4). Gray circles indicate biallelic ATAC–seq signal in a given clone, and blue and pink circles indicate monoallelic ATAC–seq signal. The allele-informative SNP is highlighted in a red box. (f) Model for how biallelic promoters in the ESC state give rise to RAMA elements in NPCs.
We asked whether NPC RAMA elements are marked to become RAMA in the earlier ESC state by histone modifications. We found that 47% of promoter-proximal NPC RAMA elements were marked by both active and repressive histone modifications (H3K4me3 or monomethylation of histone H3 at lysine 4 (H3K4me1) plus trimeth-ylation of histone H3 at lysine 27 (H3K27me3) or lysine 9 (H3K9me3)) and DNA-binding factors (Pol II Ser2 phosphorylation plus Suz12 or Ring1b) in ESCs, more often than non-RAMA accessible elements (Fig. 5c,d and Supplementary Fig. 5a–h)26 RAMA elements at promoters of genes whose expression increased from the ESC to NPC state were the most highly co-marked with active and repressive modifications (Fig. 5d). We confirmed this for some loci using ChIP–qPCR for the repressive mark H3K9me3 and the active mark H3K4me3 in female ESCs and NPCs derived from the same line. ESCs, which have biallelic accessibility at the Cpped1 promoter, were marked by both H3K9me3 and H3K4me3 at this locus. H3K4me3 was retained as ESCs differentiated into NPCs. Notably, H3K9me3 was lost in NPCs in which the promoter was biallelically accessible, whereas H3K9me3 was retained but reduced in other NPCs in which the promoter was monoallelically accessible. Sanger sequencing of ChIP DNA showed that both alleles were marked by both H3K4me3 and H3K9me3 in ESCs, but in NPCs the active and repressed alleles were marked only by H3K4me3 or H3K9me3, respectively (Fig. 5e and Supplementary Fig. 5i). These results suggest that both alleles are poised for activation or repression in the ESC state, and each has some probability of being activated or repressed as the cells exit the pluripotent state (Fig. 5f). These marks could be present on the same nucleosome or could reflect a mixture of nucleosome states in the ESC population27,28. This mechanism of shutting down no or one allele upon differentiation is similar to X-chromosome inactivation but distinct from other monoallelic systems like olfactory receptor choice, where it is the low probability of activation that assures only a single allele is expressed29.
A subset of RAMA elements are established by a non-stochastic mechanism
RAMA and RME may arise owing to stochastic binding of chromatin remodelers and trans factors at low-probability sites in the genome or alternatively arise from specific biological mechanisms for generating diversity such as allelic exclusion or counting4,11. These mechanisms exist in other monoallelic gene expression programs and include pre-marking of alleles by histone modifications or asynchronous replication, or feedback that ensures only a single allele is activated30–33.
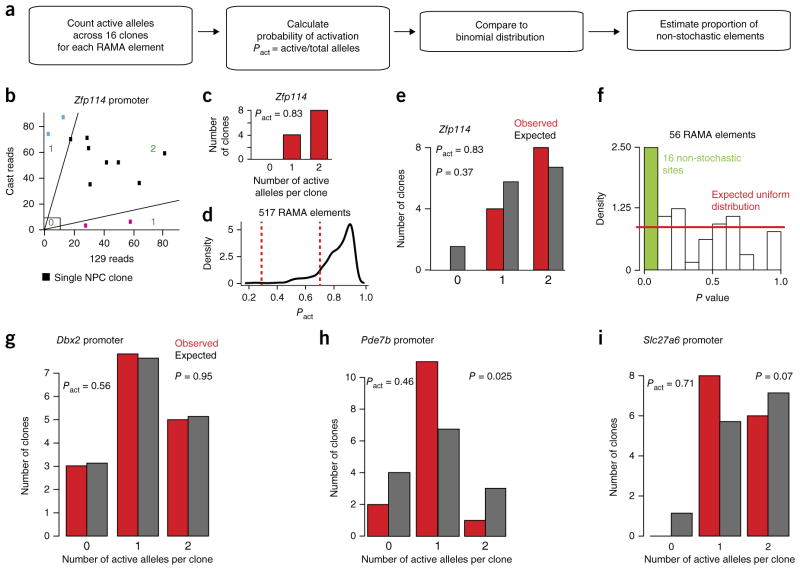
To address whether the RAMA/RME pattern, where one or two alleles can be expressed at each element, is stochastically established, we explored whether the two alleles are independently regulated. In the case that the alleles are independently opened or closed, the distribution of inaccessible, monoallelically accessible and biallelically accessible elements across clones would follow a binomial distribution based on the probability of activation (Pact = number of active alleles in 16 clones/number of alleles present) (Fig. 6a,b). In the case that the distribution is not approximately binomial, there could be a specific counting mechanism or selection mechanism in place after establishment.
Figure 6.
Establishment of some RAMA elements cannot be explained by a stochastic model. (a) Description of the method for testing whether the distribution of 0, 1 or 2 accessible alleles in ATAC–seq data across 16 NPC clones deviates from the binomial. (b) Spectrum of active alleles at the Zfp114 promoter locus. Blue and pink clones are monoallelic and considered ‘1’. Black clones are biallelic and were either accessible or closed on both alleles (‘2’ or ‘0’). (c) The number of 0, 1 and 2 alleles across 16 clones at the Zfp114 promoter. (d) Distribution of Pact for RAMA elements. Elements with Pact between 0.3 and 0.7 (between dashed red lines) were considered for analysis in f. (e) Comparison of the distribution of 0, 1 or 2 accessible alleles at the Zfp114 promoter locus to the binomial based on Pact = 0.83. (f) Estimate of the percentage of RAMA elements whose distribution of active alleles deviates significantly from the expected binomial distribution based on random activation. The plot shows the distribution of P values for 56 RAMA elements with probabilities of activation between 0.3 and 0.7. The red line corresponds to the expected uniform distribution, and the green bar indicates elements whose P values are significant. (g) Example of a RAMA element at the Dbx2 promoter showing a random distribution of 0, 1 and 2 alleles. (h) The RAMA element in the Pde7b promoter had a higher number of clones with 1 active allele then expected. (i) The RAMA element in the Slc27a6 promoter had a lower number of clones with 0 active alleles than expected.
To test whether RAMA elements become accessible independently on each allele, we tested whether the number of biallelically closed (0), monoallelically open (1) and biallelically open (2) alleles follows the binomial distribution. We calculated the expected number of 0, 1 and 2 states from Pact and then used the observed distribution to calculate a P value for the deviation from the expected (Fig. 6a–e and Supplementary Fig. 6b). We focused on elements for which 0.3 < Pact < 0.7 because we were sufficiently powered in this range (n = 16 clones) and because these elements are particularly interesting owing to their high probability of being monoallelically accessible (Fig. 6d and Supplementary Fig. 6a). First, we found that individual sites had very distinct distributions of 0, 1 and 2 accessible alleles. We found that 16 individual RAMA elements had distributions that rejected the binomial distribution with P < 0.1 (13 with P value < 0.05). Because we had low power at individual loci, we used the distribution of P values across RAMA elements to estimate that around 29% deviated from the binomial distribution, indicating that there may be a non-stochastic mechanism underlying the establishment of their RAMA pattern or selection of clones thereafter (Fig. 6f)34. We show three RAMA elements as examples: the Dbx2 promoter whose distribution did not differ from the binomial, the Pde7b promoter, which had more monoallelic clones than expected, and the Slc27a6 promoter, which had a ‘non-zero’ pattern (Fig. 6g–i and Supplementary Fig. 6c–e). Our analysis suggests that the majority of RAMA elements are consistent with a stochastic, independent allelic choice, whereas the minority have feedback or selection mechanisms in place. Furthermore, the distinct distribution at each RAMA site indicated that these loci are not regulated by a common mechanism.
DISCUSSION
Here we developed allele-specific ATAC–seq to relate DNA sequence variation and element accessibility. Our allele-specific ATAC–seq analysis framework can be easily adapted to other animal models and to patient genomes in the context of human disease. The latter has many clinical applications, as the regulome is highly dynamic and the effects of environmental triggers and medical treatments on heterozygous variants can be monitored on a clinically relevant time scale.
We applied allelic ATAC–seq to a highly polymorphic mouse hybrid F1 system, in which we identified over 1,800 monoallelic DNA regulatory elements across autosomes that showed as much allelic bias as genes subject to X-chromosome inactivation. Genetically determined monoallelically accessible elements tended to occur at enhancers, whereas RAMA elements—capable of monoallelic accessibility on either allele—tended to be located at promoters. These results highlight a new important functional distinction between enhancers and promoters and raise the possibility that epigenetic changes resulting in RME tend to be associated with the immediate environment of a gene’s promoter rather than its long-range regulatory landscape, at least once the changes have been established. Further, we found that the memory of allelic choice at RAMA elements was transmitted through cell generations and through the cell cycle when chromatin was highly compacted.
The mechanisms of establishment and function of RME are largely a black box. We found that RME genes in NPCs tended to have monoallelic promoter elements that may drive their monoallelic expression. To our surprise, nearby distal enhancer elements tended to be biallelic, indicating that they are permissive and not restrictive for monoallelic gene expression. It is intriguing that this is reminiscent of genes that escape silencing on the inactive X chromosome, the promoters of which are the only accessible elements within a sea of heterochromatin, indicating that the promoter region alone may be sufficient for gene control in these contexts16. This highly local unit of gene regulation at RME genes is interesting, as it may allow for allelic heterogeneity in expression of specific genes without dictating allelic states of nearby essential genes. This suggests a model in which transcription factors can bind to nearby enhancer elements but that they only have a functional relationship on the allele where the promoter is accessible, leading to productive transcription. This gatekeeper model indicates that the promoter itself is the locus control element in the context of RME.
The ontogeny of random monoallelic gene expression is of great interest, as it lends clues to the establishment and function of variegated gene expression programs. The developmental specificity of RAMA for NPCs is especially interesting for brain development because heterogeneity in gene expression may yield unique combinations of proteins in neurons to create great diversity. RME genes are also enriched for gene sets associated with Alzheimer’s disease and schizophrenia10, further motivating their understanding. The biallelic accessibility of RAMA elements in ESCs suggests that it is stochastic silencing and closing of chromatin (as opposed to activation) during differentiation that leads to monoallelic expression. This is reminiscent of X-chromosome inactivation where one of two X chromosomes is silenced, but is the exact opposite of other forms of monoallelic expression such as olfactory receptor choice29. Furthermore, the observation that these accessible promoter regions were marked both by repressive and active marks in the ESC state suggests that each allele may be poised and easily tipped toward activation or repression upon receiving differentiation signals, thus leading to a RAMA pattern. The diploid cell has evolved to have two copies of every gene, buffering it against deleterious single-hit mutations. The discovery of RAMA elements, which defy this safeguard system, likely has some advantage at the organ level and sets the stage for further study. In the future, single-cell methods that combine DNA accessibility and RNA measurements may greatly increase throughput and shed light on these fascinating mechanisms.
URLs
Allele-specific ATAC–seq analysis code, https://github.com/jinxu9/AlleleSpecificATACseq; RefSeq genes, http://hgdownload.soe.ucsc.edu/goldenPath/mm9/database/refGene.txt.gz; imprinted genes, http://www.mousebook.org/imprinting-gene-list and http://geneim-print.com/site/genes-by-species.Mus+musculus; ChromHMM results, https://github.com/jinxu9/mESC_histone_chromHMM, https://github.com/jinxu9/mESC_TF_chromHMM and https://github.com/jinxu9/mNPC_epi_anno; Picard, https://github.com/broadinstitute/picard; MACS2, https://github.com/taoliu/MACS; EpiStemNet data, http://epistemnet.bioinfo.cnio.es/download/bam_files.
METHODS
Methods and any associated references are available in the online version of the paper.
ONLINE METHODS
Cell culture
Mouse ESCs were cultured in serum (Fisher Scientific, SH30071.03) and medium containing LIF (Millipore, ESG1107) on 0.2% gelatin-coated plates. NPCs were cultured in N2B27 medium (DMEM/F12 (Invitrogen, 11320-033), Neurobasal (Gibco, 21103-049), NDiff Neuro-2 Medium Supplement (Millipore, SCM012), B27 Supplement (Gibco, 17504-044)) supplemented with EGF and FGF (10 ng/ml, each) (315-09 and 100-18B, Peprotech). Cells were passaged every other day with Accutase (SCR005, Millipore) and seeded on 0.2% gelatin-coated plates. NPC differentiation from ESCs was performed as previously described35. Briefly, ESCs were plated on gelatin-coated plates in N2B27 medium for 7 d. On day 7, cells were dissociated with Accutase and cultured in suspension in N2B27 medium with FGF and EGF (10 ng/ml, each). On day 10, embryoid bodies were plated onto 0.2% gelatin-coated plates and allowed to grow for three passages before single cells were subcloned. For passage analysis, NPC clones XX2, XX4 and XY14 were grown for an additional five and ten passages after the initial ATAC–seq experiment.
ATAC–seq
ATAC–seq library preparation was performed exactly as described14. Briefly, ESCs and NPCs were dissociated using Accutase (SCR005, Millipore). 50,000 cells per replicate (two replicates per clone) were incubated with 0.1% NP-40 to isolate nuclei. Nuclei were then transposed for 30 min at 37 °C with adaptor-loaded Nextera Tn5 (Illumina, Fc-121-1030). Transposed fragments were directly PCR amplified and sequenced on an Illumina NextSeq 500 or HiSeq 4000 to generate 2× 75-bp paired-end reads.
ATAC–seq library quality control
Libraries were sequenced to an average depth of 42 million reads. The sequencing depth for each library and each clone is listed in Supplementary Table 1. The raw reads were first trimmed using cutadapt (version 1.6) (ref. 36) to remove adaptor sequence at the 3′ end. The trimmed reads were aligned to a modified reference genome (mm9) using Bowtie2 (v2.2.3) (ref. 37) using the ‘--very-sensitive’ parameter. Paired-end reads that aligned to the genome with mapping quality ≥10 were kept as usable reads (reads aligned to the mitochondrial genome were removed). PCR duplicates were removed using Picard (see URLs). Reproducibility between technical duplicates was estimated, and these agreed well (data not shown). Unique usable reads from technical duplicates and different batches of ATAC–seq were merged together for each clone. The fragment length distribution and TSS enrichment score for each clone is listed in Supplementary Table 2. The TSS enrichment score was defined as the ratio of the summit in a 4-kb window around the TSSs of all RefSeq genes from the UCSC Genome Browser to the background. Empirically, a TSS enrichment score >6 was required for a successful ATAC–seq library. All libraries passed that criterion.
Open/active chromatin region identification in NPCs
Unique usable reads from all NPC clones were further pooled together for global peak calling. Open chromatin regions (peak regions) were called using MACS2 (ref. 17), with the following parameters ‘-q 0.001 -n NPC_all_nomodel_shift50 --nomodel --shift −50 --extsize 100 --keep-dup all’. Peaks with an enrichment score less than 5 or within the mm9 blacklist region were filtered out. A total of 78,922 peak regions were identified in NPCs and used for the following allelic analysis.
Allele-specific alignment
SNP sites between 129S1/SvImJ (129) and Cast/EiJ (Cast) strains were collected from the dbSNP (v132) database. To make an unbiased mapping reference for the 129 and Cast alleles, SNP sites that were shared by 129 and Cast, but different from the reference genome, were replaced by the common 129/Cast SNP. SNP sites that differed in 129 and Cast were replaced by ‘N’ in the reference genome, and the position and genotype were recorded separately. This modified genome was used as the reference for Bowtie2. After alignment, all reads that mapped to an ‘N’ position were separated into 129- and Cast-specific reads according to their genotype. Reads containing non-concordant SNPs were rare and were discarded. 36–49% of usable reads in each clone contained a SNP and were considered allelically informative (Supplementary Table 3). The overall alignment and allele-specific alignment files were further converted into bigWigs using BEDtools38, which were normalized and can be visualized in the UCSC Genome Browser.
Evaluation of allelic reproducibility
To evaluate the reproducibility of the allelic accessibility measured by ATAC–seq, the correlation coefficients from technical replicates, biological replicates (same clone, different passage) and different clones were compared. The expectation was that the technical replicates should be highly correlated and should be the highest, comparing to the correlation between biological replicates and different clone comparison. Three NPC lines (XX2, XX4 and XY14) with biological replicates were used to test the reproducibility. Allelic reads were counted for each open chromatin peak in NPCs. The Pearson’s correlation coefficient was calculated for each comparison. The distribution of R values of chromosome 5 in each group is shown in Supplementary Figure 1a. Apparently, the technical replicates gave the highest R values globally, which validated the reproducibility of our allelic ATAC–seq measurement. After we confirmed the reproducibility, the sequences from technical replicates were merged for monoallelic open chromatin region identification.
Identification of monoallelic open chromatin regions
To assign monoal-lelically and biallelically accessible peaks, allelic reads mapping to 129 and Cast were counted for each peak in each clone and a d score was calculated as a measure of the strength of allelic imbalance7. Peaks with ≥10 allelic reads were considered as allelically informative peaks. For a given peak, the d score was calculated as the ratio of 129 reads to the total number of reads minus 1/2. The d score takes a value between –0.5 and 0.5, where negative values correspond to a Cast bias and positive values correspond to a 129 bias. A d score of 0 reflects equal accessibility on the two alleles.
To evaluate the statistical significance of any deviation from biallelic accessibility, a P value based on a permutation method was designed and applied to each peak as follows
We randomly sampled reads from the input file and assigned the sampled reads to the maternal or paternal allele on the basis of the binomial distribution. Znull was calculated in the same way as Zobs but using sampling reads. This step was repeated N times
where C129 is the number of allelic reads from the 129 allele and Ctotal is the total number of allelic reads.
This permutation scheme was followed for each chromosome separately, and a Benjamini–Hochberg FDR control method was applied to adjust for multiple testing. The permutation scheme is more stringent on the autosomes but more sensitive on the X chromosome when compared to the binomial test (Supplementary Fig. 1b).
An empirical threshold for monoallelic accessibility was determined by using the promoter elements of X-linked genes. We showed previously that there is a strong correlation between allelic activation of the promoter on the X chromosome with allelic expression16.
The same methods and threshold were applied to the autosomes in each clone to identify genome-wide monoallelic accessibility. Briefly, peaks with at least ten allelically informative reads, a |d score| ≥0.3 and Padj <0.01 were identified as monoallelic peaks.
To investigate the dependence of monoallelic peak identification on sequencing depth, the clone with the highest sequencing depth (NPC XY14) was used to make a systematic estimation. Usable reads were downsampled from 5% to 90%, and monoallelic peaks were counted at each sequencing depth. The results showed that the number of monoallelic peaks was saturated at above 40 million usable reads (Supplementary Fig. 1c). Fourteen of 18 lines were sequenced to a depth of more than 40 million reads.
Copy number variation detection for NPC clones using ATAC–seq data
To avoid calling false positive monoallelic peaks caused by aneuploidy or copy number variation, we estimated CNVs from ATAC–seq data. The principle is that sequencing from closed chromatin regions (background) should randomly distribute along the whole genome, and increases or decreases in this background should reflect CNVs. To call CNVs, we used total usable reads from ATAC–seq data in background regions. Peak regions were first called using MACS2 with loss criteria for each clone. Reads within the peak regions (extended by 500 bp on either side) were excluded. The background reads were used to estimate the average coverage of each 100-kb window and were tested for statistical derivation from diploidly using FreeC (v7.2) (ref. 39). Because we cannot detect copy-neutral aneuploidy in this manner, we used the d score to control for this. Specifically, chromosomes 12 and 1q were detected as copy-neutral aneuploidy regions. CNV regions (losses >100 kb and gains >500 kb) and amplified or lost chromosomes were defined on a clone-by-clone basis from our analyses. For a subset of clones, whole-genome sequencing data were used to tune CNV calling parameters (data not shown). The number of autosomal monoallelic peaks identified for each clone following CNV removal is listed in Supplementary Table 4.
Classification of monoallelically accessible elements
Allelic information for peak regions in all NPC clones was merged as a matrix. Peaks in CNV regions were excluded on a clone-by-clone basis. Additionally, peaks located within 2 kb of known imprinted loci were filtered out using the list of imprinted genes from the combination of MouseBook and Geneimprint. After filtering CNVs and imprinted loci, peaks with at least ten allelically informative clones were further classified into RAMA, 129-specific monoallelically accessible (129 MA) and Cast-specific monoallelically accessible (Cast MA) elements (Supplementary Tables 5–7). Randomly monoallelically accessible elements were those for which at least one clone was 129 monoallelically accessible and at least one clone was Cast monoallelically accessible. The 129- and Cast-specific elements were those in which more than 50% of clones were monoal-lelically accessible from the same allele and zero clones were monoallelically accessible from the other allele.
Estimation of mappability for strain-specific monoallelically accessible elements
100 million 75-bp paired-end reads were simulated from the 129 and Cast genomes, respectively. The simulated reads were merged as the silicon sequencing from F1 mice and were subsequently mapped and counted using our allelic ATAC–seq pipeline. According to the simulation results, only two of the 129-specific elements showed allelic bias and were removed in the further analysis.
Annotation of accessible elements in NPCs
ChIP–seq data for histone modifications and transcription factors in mouse NPCs were collected from previously published data. The full list of marker and accession numbers is provided in Supplementary Table 8. Briefly, the raw data were downloaded from the Sequence Read Archive (SRA) database and then converted into fastq files. The fastq files were mapped to the mouse genome (mm9) using Bowtie2 (v2.2.3) with default parameters. Duplicates were removed using SAMtools (version: 0.1.19) (ref. 40) and then converted into the bed format required by chromHMM (version 1.10) (ref. 41). Parameters for chromHMM were optimized with 500-bp bin size and 16 states. The classification and state features are shown in Supplementary Figure 2e.
To test and compare the enrichment of a specific set of peaks, a control set was randomly simulated using the distribution of the enrichment scores from the tested set of peaks. The same number of peaks was simulated for the control set. The enrichment test was done by Fisher’s exact test (two-tailed). The results from chromHMM can be accessed on GitHub (see URLs).
Comparison across passages
Allelic reads for random monoallelic elements identified in all clones were counted in three clones (NPC XX2, NPC XX4 and NPC XY14) that had ATAC–seq data from passages PX + 0, PX + 5 and PX + 10 (where PX is the passage of the original ATAC–seq experiment). Informative monoallelic elements (|d score| ≥ 0.3 and allelic reads ≥ 10) were compared across different passages. Correlation coefficient was calculated by adding the three clones by Pearson’s correlation. To evaluate the consistency of monoallelic assignment across passages, the same analyses were performed for a set of three technical replicates. The maximum difference in d score among passages was compared to the maximum difference in d score among technical triplicates.
Correlation with RNA–seq data
Expression data including RPKM and allelic ratios from RNA–seq data were downloaded from a previous study5. The allelic ratio from RNA–seq data was converted into a d score, as described previously. ATAC–seq data including peak intensity and d score were extracted from the seven NPC clones for which RNA–seq data were available. ATAC–seq peaks, located within 2 kb around the TSS of a specific gene, were assigned as a promoter–transcript pair. Only promoter–transcript pairs with allelic expression ratios as well as allelically informative peaks were kept to estimate the proportion of RAMA–RME pairs. The promoter–transcript pairs were classified into three classes on the basis of whether they were called or not called as randomly monoallelic with the applied threshold. Correlation coefficient was calculated for all pairs across seven clones. Correlation between distal regulatory elements and transcripts was compared in two ways: (i) regulatory elements located 2–10 kb from the TSS of an RME gene were selected and tested and (ii) the nearest gene for a distal RAMA element were selected and tested. All the correlation coefficients were calculated by Pearson’s correlation using R (v3.2.2).
Colocalization within topologically associating domains
TADs called from Hi-C data in NPCs were used to test the colocalization of RMEs and distal RAMA elements16. TADs containing distal RAMA elements were extracted, and the proportion of these TADs that contained RME genes was then calculated. The same analysis was performed for randomly selected controls for enrichment comparison.
Quantifying the number of active alleles
It is easy to distinguish monoalleli-cally (1) and biallelically (2) accessible elements using the d score and P value. However, it is difficult to distinguish 0 from 2 active alleles because the d score is always around 0 for both cases. To resolve this problem, we estimated the baseline background signal for each element on the silenced allele of a monoallelic clone. We used this background read count to define a lack of accessibility.
Then, using normalized allelic counts from both alleles and TSS enrichment score we separated elements into those having 0, 1 and 2 accessible alleles on the basis of the following rules: (i) if the element has been called as a monoal-lelic peak (using the d-score method), count as 1 active allele, (ii) if neither of the two alleles is higher than the baseline, count as 0 active alleles, (iii) when both alleles are higher than the baseline and if |d score| < 0.3, count as 2 active alleles, and (iv) else, if |d score| ≥ 0.3, count as 1 active allele. Pact was estimated as the number of active alleles deviated by the total number of alleles.
To look at the global Pact across all RAMA sites, we filtered out the less confident peaks. Basically, a linear regression between allelic counts and enrichment score was applied for all RAMA sites. If the allelic count was not correlated with the enrichment score as expected, it indicated that the allelic reads might be located at the boundary of the peak region, instead of in the center. In this situation, the number of active alleles will not be accurately estimated. The top ~20% of RAMA sites with the highest deviation from the regression were removed in the following comparison to the stochastic model.
Stochastic model test
We tested whether the establishment of RAMA sites can be explained by a stochastic model in which the spectrum of active alleles in each clone should be the same as the spectrum from a binomial distribution with the same probability of activation.
The observed spectrum was calculated by counting the number of active alleles in each clone for each peak. Pact was then estimated by dividing the number of active alleles by the total number of alleles. Then, the expected spectrum was drawn from X~B(n, Pact), where n = 2, X = (0, 1, 2). The observed spectrum was compared to the expected spectrum, and the difference was tested using the likelihood-ratio test, which is similar to the χ2 test but allows zero observed values42.
With the distribution of P values by likelihood-ratio test of RAMA sites (those with Pact from 0.3 to 0.7), we estimated the proportion of elements that are truly null with the assumption that null P values are uniformly distributed34. This is quantified with
where m is the total number of tested elements when setting Λ = 0.5. Then, π1 = 1 – π0, which gives the proportion of elements that are truly alternative features.
Evaluating the accessibility of NPC RAMA elements in ESCs
To evaluate the accessibility of NPC RAMA elements in ESCs, we counted the total number of reads within the RAMA regions defined in NPC lines. Then, we normalized the number of reads in each region for each line with sequencing depth. The minimum enrichment score for peak calling in NPC lines was 5. Therefore, if there was more than 20% reads count in ESCs as compared to NPCs, the region was defined as an accessible region in ESCs.
Annotation with histone modifications in ESCs and bivalent region identification
ChIP–seq data for histone modifications and transcription factors in mouse ESCs were collected from a previous collection (EpiStemNet data; see URLs)43. The bam files were downloaded and converted into the bed format required by chromHMM41. The full list of marks and accession numbers is provided in Supplementary Table 9. A Control set was selected as previously described and following the same processing as the set of RAMA elements. Lineage-specific genes were defined as those with a twofold increase in NPCs as compared to ESCs at the expression level. The results from chromHMM can be accessed from GitHub. The enrichment of RAMA elements in bivalent or repressive regions was tested by Fisher’s exact test (two-tailed), comparing to the randomly selected control sets.
ChIP–seq signal for chromatin-modifying enzymes in ESCs at promoter-proximal NPC RAMA elements open in ESCs was plotted using ngsplot44.
Isolation of mitotic cells for ATAC–seq
NPC clone XX2 was plated at low density and treated for 24 h with deoxythymidine (dT; 2 mM). Following dT treatment, cells recovered in fresh medium for 3 h and were then treated for 6 h with nocodazole (40 μg/ml). Mitotic cells were shaken off the plate and collected in the medium. ATAC–seq was performed on 50,000 cells per replicate. Mitotic cells were stained with antibody (1:500 dilution) to H3S10ph (Cell Signaling, 9706S) and DAPI (Vector Laboratories, H-1200) to verify mitotic state.
Chromatin immunoprecipitation
Cells were fixed in 1% formaldehyde for 10 min at room temperature, and reactions were subsequently quenched with 0.125 M glycine. Cells were then snap frozen and stored at −80 °C. Cells were then lysed (50 mM HEPES-KOH, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100) for 10 min at 4 °C. Nuclei were lysed (100 mM Tris pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA) for 10 min at room temperature. Chromatin was resuspended in sonication buffer (10 mM Tris pH 8.0, 1 mM EDTA, 0.1% SDS) and sonicated using a Covaris Ultrasonicator to an average length of 220 bp. For H3K9me3 ChIP, chromatin from 5 million cells was incubated with 5 μg of anti-H3K9me3 antibody (abcam, AB8898) overnight at 4 °C. Antibody-bound chromatin was incubated with protein G Dynabeads (Invitrogen, 10004D) for 4 h at 4 °C and eluted in Tris buffer (10 mM Tris pH 8.0, 10 mM EDTA, 1% SDS). Cross-links were reversed by incubation overnight at 65 °C followed by treatment with 0.2 mg/ml proteinase K (Life Technologies, AM2548) and 0.2 mg/ml RNase A (Qiagen). DNA was purified using Qiagen MinElute columns (Qiagen, 28006).
For Sanger sequencing, SNP-containing regions were amplified using the primers listed in Supplementary Table 10, and amplicons were sequenced by ElimBio using the forward primer. qPCR primers used for ChIP are listed in Supplementary Table 10.
RT–PCR and Sanger sequencing
Whole-cell RNA was reverse transcribed using SuperScript III (Thermo Fisher, 18080051). cDNA was amplified using the primers listed in Supplementary Table 10 and sent for Sanger sequencing.
Supplementary Material
Supplementary Figure 1. Evaluation of allelically informative accessible regions and monoallelic sites.
(a) Comparison of allelic read assignment for technical replicates, biological replicates and different clones. Pearson’s correlation coefficients were calculated from allelic counts for the 129 and Cast alleles. (b) Comparison of two statistical methods for evaluating allelic bias based on the binomial distribution and permutation method. Cumulative curves for P value (adjusted by the Benjamini– Hochberg procedure) from the X chromosome and chromosome 5 are shown in red and blue, respectively. The permutation method was compared to the exact binomial test. The right panel is a zoomed-out view of the left panel. The difference between the dark and light curves shows that the permutation method is more stringent on the autosomes, but more sensitive on the X chromosome. (c) Scatterplot showing the number of usable reads versus the number of allelically informative accessible regions. The number of allelically informative regions is close to stable when the number of usable sequencing reads is more than 40 million. The number of allelically informative accessible regions in ESCs is lower than for NPC clones with the same number of usable reads (red points). Blue points show downsampled data from a highly sequenced line (NPC XY14). (d) Scatterplot of TSS enrichment score versus the number of allelically informative accessible regions. There is no significant correlation between them (r = –0.186, P = 0.461). Red indicates the two ESC lines. (e) Simulation to determine the number of reads needed to identify monoallelic peaks in NPCs. 6–100 million usable reads were randomly sampled from a deeply sequenced NPC clone (NPC XY14). The number of monoallelically accessible regions dramatically increases up to 40 million reads where it plateaus. (f) Number of monoallelic peaks identified by ATAC–seq on the Cast X chromosome, 129 X chromosome, Cast autosomes and 129 autosomes in each cell line analyzed.
Supplementary Figure 2. Examples of genome-specific monoallelic regulatory elements and chromHMM annotation of accessible sites.
(a) Example of a 129-specific site located at the Mpp7 gene promoter on chromosome 18. 129 reads are shown in pink, and Cast reads are shown in blue. (b) Example of a Cast-specific site located at the AK043958 gene promoter on chromosome 4. 129 reads are shown in pink, and Cast reads are shown in blue. The peak to the left is biallelic, as indicated by pile-up of 129 (pink) and Cast (blue) reads, while the peak on the right is Cast specific. (c) Proportion of 129-specific, Cast-specific and RAMA elements located at distal elements (>2 kb from a TSS), promoter elements of expressed genes and promoter elements of unexpressed genes (<2 kb from a TSS). For each set of elements, control sets are size matched and matched for peak enrichment. (d) Chromatin states identified by chromHMM using available histone modification ChIP–seq data in NPCs. Enrichment for the histone modifications labeled on the x axis is represented in blue. Interpretations of each state are labeled on the right. (e) Allelically accessible regions (accessible regions for which ten clones have sufficient allelically informative information after removing CNV regions) show the same distribution of chromatin states as all accessible regions across the genome in NPCs.
Supplementary Figure 3. Genome-specific monoallelic sites and RAMA elements are stable across passages and during mitosis.
(a–c) Scatterplots showing the d score for Cast- and 129-specific monoallelically accessible regions in PX + 10 versus PX + 0 for NPC XX2, XX4 and XY14. (d) Distribution of changes in d score (max d score – min d score) across passages PX + 0, PX + 5, and PX + 10 for NPC clones XX2, XX4 and XY14. As a control, the changes in d score are compared to three replicates from a single clone at the same passage (NPC XX 1). (e) Distribution of changes in d score (max d score – min d score) across passages PX + 0, PX + 5, and PX + 10 for NPC clones XX2, XX4 and XY14 for genome-specific monoallelic elements. (f) Tracks showing an example RAMA element at the promoter of RME gene Fam149a across passages in NPC XX 4. 129 reads are shown in magenta, and Cast reads are shown in blue. (g) Scatterplot showing the number of reads under all ATAC–seq peaks in mitotic and asynchronous NPCs (XX 1). All peaks are shown in black, and RAMA elements are shown in red.
Supplementary Figure 4. RME genes are regulated at local promoter-proximal RAMA elements.
(a) Cumulative plot of the log2 (RPKM) values from RNA–seq data for RME genes with promoter RAMA elements (green) and RME genes with a non-RAMA element in the TSS region (yellow). (b) Cumulative plot showing the peak enrichment score for promoter RAMA elements located at RME genes (dark green) and non-RME genes (light green). (c) RT–PCR followed by Sanger sequencing for RME gene Bag3 in NPC lines XY7-10. Colored circles indicate the allelic status of the promoter ATAC–seq peak in that line. Red boxes outline the informative SNP at each locus. (d) Tracks showing allele-specific ATAC–seq at the noncoding RNA Vaultrc5, which is not detected by allelic RNA–seq due to poly(A) selection. 129 reads are shown in pink, and Cast reads are shown in blue. (e) Tracks showing allele-specific ATAC–seq at the noncoding RNA Mir34b, which is not detected by allelic RNA–seq due to poly(A) selection. 129 reads are shown in pink, and Cast reads are shown in blue. (f) Permutation of RNA–seq d score versus d score of all accessible elements within 2–10 kb of RME genes. Matched with Figure 4h, left. (g) Permutation of RNA–seq d score of nearest gene versus d score of distal RAMA elements. Matched with Figure 4h, left. (h) Density plot showing the correlation coefficient for distal RAMA elements and RNA–seq d score of the nearest gene compared to permutated values. (i) Density plot showing the correlation coefficient for ATAC–seq d score of accessible element and RNA–seq d score of the nearest gene for promoter and distal genetic MA separately. (j) Tracks showing allele-specific ATAC–seq at the Arc locus. The Arc promoter element is RAMA, while the upstream enhancer is biallelically accessible. 129 reads are shown in pink, and Cast reads are shown in blue.
Supplementary Figure 5. Chromatin signature of RAMA elements during differentiation from ESCs to NPCs.
(a) Histone modification signal from ESC ChIP–seq data at promoter RAMA elements that are accessible in ESCs. (b) Histone modification signal from NPC ChIP–seq data at promoter RAMA elements that are accessible in ESCs. (c) Histone modification signal from ESC ChIP–seq data at distal RAMA elements that are not accessible in ESCs. (d) Histone modification signal from NPC ChIP–seq data at distal RAMA elements that are not accessible in ESCs. (e) Histone modification signal from ESC ChIP–seq data at distal RAMA elements that are accessible in ESCs. (f) Histone modification signal from NPC ChIP–seq data at distal RAMA elements that are accessible in ESCs. (g) Chromatin states identified by chromHMM using histone modification ChIP–seq data from ESCs. Enrichment for histone modifications labeled on the x axis is represented in blue. Interpretations of each state are labeled on the right. (h) Chromatin states identified by chromHMM using Pol II and PRC complex ChIP–seq data from ESCs. Enrichment for transcription factors labeled on the x axis is represented in blue. Interpretations of each state are labeled on the right. (i) ChIP–qPCR for H3K4me3 and H3K9me3 at six RAMA promoters in XX1 ESCs.
Supplementary Figure 6. Test of the stochastic model of RAMA establishment in NPCs.
(a) Distribution of Pact (n accessible alleles/n total alleles) for promoter-distal (blue) and promoter-proximal (black) RAMA elements. (b) Distribution of P value by likelihood test for all RAMA elements. (c) Example RAMA site at the promoter of Mettl24 that has fewer monoallelic (1) clones than expected based on its Pact of 0.81. (d) Allelic ATAC–seq signal in 16 clones at the RAMA promoter element at Pde7b (related to Fig. 6h). Pink, 129 monoallelic; blue, Cast monoallelic; gray, biallelically accessible; black, not accessible; white, masked in CNV region. (d) Allelic ATAC–seq signal in 16 clones at the RAMA promoter element at Slc27a6 (related to Fig. 6i). Pink, 129 monoallelic; blue, Cast monoallelic; gray, biallelically accessible; black, not accessible; white, masked in CNV region.
Acknowledgments
We thank J. Dekker for sharing Hi-C data before publication and for feedback, and J. Pritchard, A. Urban and members of our laboratories for advice. This work was supported by the US National Institutes of Health (NIH; P50-HG007735 to H.Y.C. and W.J.G.). The Heard labratory is “Equipe labellisée par la Ligue Nationale Contre le Cancer” and is supported by a European Research Council (ERC) Advanced Investigator Award; Labex DEEP (ANR-11-LBX-0044) part of the IDEX Idex PSL (ANR-10-IDEX-0001-02 PSL).
Footnotes
Accession codes. Sequencing data are available on the Gene Expression Omnibus (GSE84646).
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
J.X., A.C.C., E.H. and H.Y.C. conceived the project. J.X. wrote the allele-specific ATAC–seq analysis pipeline and did most of the data analysis, and A.C.C. performed all experiments, generated ATAC–seq libraries and did some data analysis. A.-V.G., M.A. and E.H. provided cell lines and conceptual input. J.L. and R.T. helped design statistical analysis and permutations for the allele-specific ATAC–seq pipeline. W.J.G. provided insight and helped design experiments. H.Y.C. supervised the project and wrote the paper with J.X. and A.C.C.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Chess A. Random and non-random monoallelic expression. Neuropsychophar-macology. 2013;38:55–61. doi: 10.1038/npp.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- 3.Magklara A, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145:555–570. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chess A. Mechanisms and consequences of widespread random monoallelic expression. Nat Rev Genet. 2012;13:421–428. doi: 10.1038/nrg3239. [DOI] [PubMed] [Google Scholar]
- 5.Gendrel AV, et al. Developmental dynamics and disease potential of random monoallelic gene expression. Dev Cell. 2014;28:366–380. doi: 10.1016/j.devcel.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 6.Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 7.Eckersley-Maslin MA, et al. Random monoallelic gene expression increases upon embryonic stem cell differentiation. Dev Cell. 2014;28:351–365. doi: 10.1016/j.devcel.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeffries AR, et al. Stochastic choice of allelic expression in human neural stem cells. Stem Cells. 2012;30:1938–1947. doi: 10.1002/stem.1155. [DOI] [PubMed] [Google Scholar]
- 9.Reinius B, et al. Analysis of allelic expression patterns in clonal somatic cells by single-cell RNA–seq. Nat Genet. 2016;48:1430–1435. doi: 10.1038/ng.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeffries AR, et al. Random or stochastic monoallelic expressed genes are enriched for neurodevelopmental disorder candidate genes. PLoS One. 2013;8:e85093. doi: 10.1371/journal.pone.0085093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckersley-Maslin MA, Spector DL. Random monoallelic expression: regulating gene expression one allele at a time. Trends Genet. 2014;30:237–244. doi: 10.1016/j.tig.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schimenti J. Monoallelic gene expression in mice: who? When? How? Why? Genome Res. 2001;11:1799–1800. doi: 10.1101/gr.215601. [DOI] [PubMed] [Google Scholar]
- 13.Gendrel AV, Marion-Poll L, Katoh K, Heard E. Random monoallelic expression of genes on autosomes: parallels with X-chromosome inactivation. Semin Cell Dev Biol. 2016;56:100–110. doi: 10.1016/j.semcdb.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keane TM, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giorgetti L, et al. Structural organization of the inactive X chromosome in the mouse. Nature. 2016;535:575–579. doi: 10.1038/nature18589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frost J, et al. The effects of culture on genomic imprinting profiles in human embryonic and fetal mesenchymal stem cells. Epigenetics. 2011;6:52–62. doi: 10.4161/epi.6.1.13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Humpherys D, et al. Epigenetic instability in ES cells and cloned mice. Science. 2001;293:95–97. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- 20.Kadauke S, et al. Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell. 2012;150:725–737. doi: 10.1016/j.cell.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naumova N, et al. Organization of the mitotic chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsiung CCS, et al. Genome accessibility is widely preserved and locally modulated during mitosis. Genome Res. 2015;25:213–225. doi: 10.1101/gr.180646.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Y, et al. CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice. Proc Natl Acad Sci USA. 2012;109:21081–21086. doi: 10.1073/pnas.1219280110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tasic B, et al. Promoter choice determines splice site selection in protocadherin α and γ pre-mRNA splicing. Mol Cell. 2002;10:21–33. doi: 10.1016/s1097-2765(02)00578-6. [DOI] [PubMed] [Google Scholar]
- 25.Kawashima T, et al. Synaptic activity–responsive element in the Arc/Arg3.1 promoter essential for synapse-to-nucleus signaling in activated neurons. Proc Natl Acad Sci USA. 2009;106:316–321. doi: 10.1073/pnas.0806518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brookes E, et al. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell. 2012;10:157–170. doi: 10.1016/j.stem.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 28.Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyons DB, et al. Heterochromatin-mediated gene silencing facilitates the diversification of olfactory neurons. Cell Rep. 2014;9:884–892. doi: 10.1016/j.celrep.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farago M, et al. Clonal allelic predetermination of immunoglobulin-κ rearrangement. Nature. 2012;490:561–565. doi: 10.1038/nature11496. [DOI] [PubMed] [Google Scholar]
- 31.Mostoslavsky R, et al. Asynchronous replication and allelic exclusion in the immune system. Nature. 2001;414:221–225. doi: 10.1038/35102606. [DOI] [PubMed] [Google Scholar]
- 32.Ensminger AW, Chess A. Bidirectional promoters regulate the monoallelically expressed Ly49 NK receptors. Immunity. 2004;21:2–3. doi: 10.1016/j.immuni.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 33.Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 34.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conti L, et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mrxuqdo QHW, Iurp V, Wkurxjksxw K. Cutadapt removers adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 37.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boeva V, et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics. 2012;28:423–425. doi: 10.1093/bioinformatics/btr670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elston RC, Forthofer R. Testing for Hardy–Weinberg equilibrium in small samples. Soc Int Biometric. 2016;33:536–542. [Google Scholar]
- 43.Juan D, et al. Epigenomic co-localization and co-evolution reveal a key role for 5hmC as a communication hub in the chromatin network of ESCs. Cell Rep. 2016;14:1246–1257. doi: 10.1016/j.celrep.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 44.Shen L, Shao N, Liu X, Nestler E. ngs.plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Evaluation of allelically informative accessible regions and monoallelic sites.
(a) Comparison of allelic read assignment for technical replicates, biological replicates and different clones. Pearson’s correlation coefficients were calculated from allelic counts for the 129 and Cast alleles. (b) Comparison of two statistical methods for evaluating allelic bias based on the binomial distribution and permutation method. Cumulative curves for P value (adjusted by the Benjamini– Hochberg procedure) from the X chromosome and chromosome 5 are shown in red and blue, respectively. The permutation method was compared to the exact binomial test. The right panel is a zoomed-out view of the left panel. The difference between the dark and light curves shows that the permutation method is more stringent on the autosomes, but more sensitive on the X chromosome. (c) Scatterplot showing the number of usable reads versus the number of allelically informative accessible regions. The number of allelically informative regions is close to stable when the number of usable sequencing reads is more than 40 million. The number of allelically informative accessible regions in ESCs is lower than for NPC clones with the same number of usable reads (red points). Blue points show downsampled data from a highly sequenced line (NPC XY14). (d) Scatterplot of TSS enrichment score versus the number of allelically informative accessible regions. There is no significant correlation between them (r = –0.186, P = 0.461). Red indicates the two ESC lines. (e) Simulation to determine the number of reads needed to identify monoallelic peaks in NPCs. 6–100 million usable reads were randomly sampled from a deeply sequenced NPC clone (NPC XY14). The number of monoallelically accessible regions dramatically increases up to 40 million reads where it plateaus. (f) Number of monoallelic peaks identified by ATAC–seq on the Cast X chromosome, 129 X chromosome, Cast autosomes and 129 autosomes in each cell line analyzed.
Supplementary Figure 2. Examples of genome-specific monoallelic regulatory elements and chromHMM annotation of accessible sites.
(a) Example of a 129-specific site located at the Mpp7 gene promoter on chromosome 18. 129 reads are shown in pink, and Cast reads are shown in blue. (b) Example of a Cast-specific site located at the AK043958 gene promoter on chromosome 4. 129 reads are shown in pink, and Cast reads are shown in blue. The peak to the left is biallelic, as indicated by pile-up of 129 (pink) and Cast (blue) reads, while the peak on the right is Cast specific. (c) Proportion of 129-specific, Cast-specific and RAMA elements located at distal elements (>2 kb from a TSS), promoter elements of expressed genes and promoter elements of unexpressed genes (<2 kb from a TSS). For each set of elements, control sets are size matched and matched for peak enrichment. (d) Chromatin states identified by chromHMM using available histone modification ChIP–seq data in NPCs. Enrichment for the histone modifications labeled on the x axis is represented in blue. Interpretations of each state are labeled on the right. (e) Allelically accessible regions (accessible regions for which ten clones have sufficient allelically informative information after removing CNV regions) show the same distribution of chromatin states as all accessible regions across the genome in NPCs.
Supplementary Figure 3. Genome-specific monoallelic sites and RAMA elements are stable across passages and during mitosis.
(a–c) Scatterplots showing the d score for Cast- and 129-specific monoallelically accessible regions in PX + 10 versus PX + 0 for NPC XX2, XX4 and XY14. (d) Distribution of changes in d score (max d score – min d score) across passages PX + 0, PX + 5, and PX + 10 for NPC clones XX2, XX4 and XY14. As a control, the changes in d score are compared to three replicates from a single clone at the same passage (NPC XX 1). (e) Distribution of changes in d score (max d score – min d score) across passages PX + 0, PX + 5, and PX + 10 for NPC clones XX2, XX4 and XY14 for genome-specific monoallelic elements. (f) Tracks showing an example RAMA element at the promoter of RME gene Fam149a across passages in NPC XX 4. 129 reads are shown in magenta, and Cast reads are shown in blue. (g) Scatterplot showing the number of reads under all ATAC–seq peaks in mitotic and asynchronous NPCs (XX 1). All peaks are shown in black, and RAMA elements are shown in red.
Supplementary Figure 4. RME genes are regulated at local promoter-proximal RAMA elements.
(a) Cumulative plot of the log2 (RPKM) values from RNA–seq data for RME genes with promoter RAMA elements (green) and RME genes with a non-RAMA element in the TSS region (yellow). (b) Cumulative plot showing the peak enrichment score for promoter RAMA elements located at RME genes (dark green) and non-RME genes (light green). (c) RT–PCR followed by Sanger sequencing for RME gene Bag3 in NPC lines XY7-10. Colored circles indicate the allelic status of the promoter ATAC–seq peak in that line. Red boxes outline the informative SNP at each locus. (d) Tracks showing allele-specific ATAC–seq at the noncoding RNA Vaultrc5, which is not detected by allelic RNA–seq due to poly(A) selection. 129 reads are shown in pink, and Cast reads are shown in blue. (e) Tracks showing allele-specific ATAC–seq at the noncoding RNA Mir34b, which is not detected by allelic RNA–seq due to poly(A) selection. 129 reads are shown in pink, and Cast reads are shown in blue. (f) Permutation of RNA–seq d score versus d score of all accessible elements within 2–10 kb of RME genes. Matched with Figure 4h, left. (g) Permutation of RNA–seq d score of nearest gene versus d score of distal RAMA elements. Matched with Figure 4h, left. (h) Density plot showing the correlation coefficient for distal RAMA elements and RNA–seq d score of the nearest gene compared to permutated values. (i) Density plot showing the correlation coefficient for ATAC–seq d score of accessible element and RNA–seq d score of the nearest gene for promoter and distal genetic MA separately. (j) Tracks showing allele-specific ATAC–seq at the Arc locus. The Arc promoter element is RAMA, while the upstream enhancer is biallelically accessible. 129 reads are shown in pink, and Cast reads are shown in blue.
Supplementary Figure 5. Chromatin signature of RAMA elements during differentiation from ESCs to NPCs.
(a) Histone modification signal from ESC ChIP–seq data at promoter RAMA elements that are accessible in ESCs. (b) Histone modification signal from NPC ChIP–seq data at promoter RAMA elements that are accessible in ESCs. (c) Histone modification signal from ESC ChIP–seq data at distal RAMA elements that are not accessible in ESCs. (d) Histone modification signal from NPC ChIP–seq data at distal RAMA elements that are not accessible in ESCs. (e) Histone modification signal from ESC ChIP–seq data at distal RAMA elements that are accessible in ESCs. (f) Histone modification signal from NPC ChIP–seq data at distal RAMA elements that are accessible in ESCs. (g) Chromatin states identified by chromHMM using histone modification ChIP–seq data from ESCs. Enrichment for histone modifications labeled on the x axis is represented in blue. Interpretations of each state are labeled on the right. (h) Chromatin states identified by chromHMM using Pol II and PRC complex ChIP–seq data from ESCs. Enrichment for transcription factors labeled on the x axis is represented in blue. Interpretations of each state are labeled on the right. (i) ChIP–qPCR for H3K4me3 and H3K9me3 at six RAMA promoters in XX1 ESCs.
Supplementary Figure 6. Test of the stochastic model of RAMA establishment in NPCs.
(a) Distribution of Pact (n accessible alleles/n total alleles) for promoter-distal (blue) and promoter-proximal (black) RAMA elements. (b) Distribution of P value by likelihood test for all RAMA elements. (c) Example RAMA site at the promoter of Mettl24 that has fewer monoallelic (1) clones than expected based on its Pact of 0.81. (d) Allelic ATAC–seq signal in 16 clones at the RAMA promoter element at Pde7b (related to Fig. 6h). Pink, 129 monoallelic; blue, Cast monoallelic; gray, biallelically accessible; black, not accessible; white, masked in CNV region. (d) Allelic ATAC–seq signal in 16 clones at the RAMA promoter element at Slc27a6 (related to Fig. 6i). Pink, 129 monoallelic; blue, Cast monoallelic; gray, biallelically accessible; black, not accessible; white, masked in CNV region.