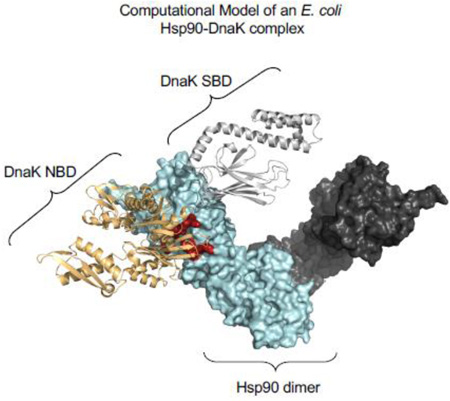
Abstract
Hsp90 is a widely conserved and ubiquitous molecular chaperone that participates in ATP-dependent protein remodeling in both eukaryotes and prokaryotes. It functions in conjunction with Hsp70 and the Hsp70 cochaperones, an Hsp40 (J-protein) and a nucleotide exchange factor. In E. coli the functional collaboration between Hsp90Ec and Hsp70, DnaK, requires that the two chaperones directly interact. We used molecular docking to model the interaction of Hsp90Ec and DnaK. The top-ranked docked model predicted that a region in the nucleotide-binding domain of DnaK interacted with a region in the middle domain of Hsp90Ec. We then made substitution mutants in DnaK residues suggested by the model to interact with Hsp90Ec. Eleven of the twelve mutants tested were defective or partially defective in their ability to interact with Hsp90Ec in vivo in a bacterial two-hybrid assay and in vitro in a Bio-Layer Interferometry assay. These DnaK mutants were also defective in their ability to function collaboratively in protein remodeling with Hsp90Ec, but retained the ability to act with DnaK cochaperones. Taken together these results suggest that a specific region in the nucleotide-binding domain of DnaK is involved in the interaction with Hsp90Ec and this interaction is functionally important. Moreover, the region of DnaK that we found to be necessary for Hsp90Ec binding includes residues that are also involved in J-protein binding, suggesting a functional interplay between DnaK, DnaK cochaperones and Hsp90Ec.
Keywords: Hsp40, CbpA, HtpG, molecular chaperone, protein remodeling
Graphical abstract
Introduction
The 90-kDa heat shock protein (Hsp90) is a highly ubiquitous and evolutionarily conserved molecular chaperone [1–5]. It is essential in eukaryotes, where it is involved in the folding, stability and activation of more than 200 client proteins including many transcription factors, steroid hormone receptors and protein kinases [4, 6, 7]. In addition, Hsp90 stabilizes and activates oncoproteins and therefore is a potential target for drug discovery for the treatment of cancer [8]. Escherichia coli Hsp90, referred to as Hsp90Ec and encoded by htpG, is an abundant protein and is further induced upon heat shock and other stress conditions [9]. Strains lacking Hsp90Ec exhibit modest phenotypes, including slow growth at elevated temperature [9], accumulation of aggregated proteins at high temperature [10], loss of adaptive immunity conferred by the CRISPR system [11], decreased ability to form biofilms at elevated temperature [12] and decreased ability to swarm [13]. Overexpression of Hsp90Ec causes defects in cell division that results in filamentous cells as well as SDS sensitivity [14].
Hsp90 is a homodimer with each monomer consisting of three domains: an N-terminal domain that possesses an ATP binding site [15, 16]; a middle domain containing residues that participate in binding client proteins [7, 14, 15, 17]; and a C-terminal domain that is essential for dimerization and is also involved in client binding [3, 14]. ATP binding and hydrolysis by Hsp90 triggers large conformational changes that are necessary for the cycle of client binding, remodeling and release [4, 15, 16, 18–21]. The Hsp90 dimer exists in a predominantly open V-shaped conformation in the absence of nucleotide with the protomers interacting via the C-terminal domain [21]. ATP binding triggers closing of the ATP lid on the ATP-binding site, followed by dimerization of the two N-terminal domains [4, 18, 21, 22]. ATP hydrolysis and ADP release leads to the dissociation of the N-domains [4, 18, 21] and Hsp90 returns to the open conformation [18–20, 22]. Cochaperones and client protein binding bias the Hsp90 chaperone cycle and stabilize or destabilize various conformations of Hsp90 [1, 3, 4].
Hsp90 functions with the Hsp70 chaperone system in protein activation and remodeling [1, 5, 23]. Eukaryotic Hsp70 and its prokaryotic homolog, DnaK, are highly conserved proteins [24–27]. Hsp70/DnaK is comprised of an N-terminal nucleotide binding domain (NBD) and a C-terminal substrate-binding domain (SBD) that are connected by a flexible linker [25]. It collaborates with two cochaperones, an Hsp40 (J-domain protein) and a nucleotide exchange factor (NEF). The Hsp40 protein stimulates ATP hydrolysis by Hsp70/DnaK and presents substrate to Hsp70/DnaK while the NEF stimulates nucleotide exchange by Hsp70/DnaK [24, 27, 28].
In addition to collaborating with the Hsp70 chaperone system, eukaryotic Hsp90 also functions with numerous cochaperones, including Hop/Sti1, Aha1/Hch1, p23/Sba1, Cdc37 and Sgt1 [1, 15, 16]. Cochaperones regulate Hsp90 in various ways, such as imparting client protein specificity, modulating ATPase activity or stabilizing specific Hsp90 conformations [1, 4]. Moreover, Hop/Sti1 stabilizes the interaction between eukaryotic Hsp90 and Hsp70 by simultaneously interacting with tetratricopeptide repeat domains at the extreme C-terminus of each chaperone. In contrast to eukaryotes, bacterial Hsp90 functions with the DnaK chaperone system independently of other Hsp90 cochaperones.
Protein reactivation by bacterial Hsp90 in vitro is simpler in its requirements than its eukaryotic homolog, requiring only DnaK and a J-domain protein [23, 29]. GrpE, the bacterial NEF stimulates reactivation, but is not essential [23]. ATP hydrolysis and client binding by both chaperones is essential for reactivation [30]. Moreover, bacterial Hsp90 physically interacts with DnaK in vivo and in vitro [29, 30] through a region identified on the M-domain of Hsp90Ec [30].
In the work presented here, we examined the collaboration between Hsp90Ec and DnaK. We used molecular modeling to identify a region on DnaK that interacts with the middle domain of Hsp90Ec. We then constructed DnaK mutants in some of the residues suggested by the docked model and tested the mutants for defects in interaction and functional collaboration with Hsp90Ec. We found that the region of DnaK involved in the physical and functional interaction with Hsp90Ec comprises residues in the DnaK NBD. This region of DnaK overlaps the region where DnaJ binds, suggesting a mechanism where Hsp90Ec directly interacts with a client bound DnaK-DnaJ complex to displace DnaJ and promote the transfer of substrate to Hsp90Ec.
Results
Identification of residues in DnaK potentially involved in protein interactions with Hsp90Ec
We previously showed that E. coli DnaK collaborates with Hsp90Ec in protein reactivation and that the two chaperones physically interact through a region in the middle domain of Hsp90Ec [23, 30]. To elucidate the region of DnaK essential for binding Hsp90Ec, we used molecular docking to predict potential interactions between the two chaperones. Four combinations of Hsp90Ec and DnaK molecules available in the protein data bank were tested [21, 31, 32]: 1) ADP-bound DnaK with apo Hsp90Ec, 2) ADP-bound DnaK with ADP-bound Hsp90Ec, 3) ATP-bound DnaK with apo Hsp90Ec and 4) ATP-bound DnaK with ADP-bound Hsp90Ec (see Materials and Methods). Without imposing restraints, the proteins were docked using ZDOCK [33, 34] and the top model for each combination was selected based on the lowest energy using ZRANK [35]. Three of the four models (2, 3 and 4, above) could be eliminated since they predicted interacting regions that were inconsistent with earlier work [30] or with results shown in Supplemental Information (Supplemental Results and Supplemental Fig. S1-S5 and Supplemental Tables S2-S4).
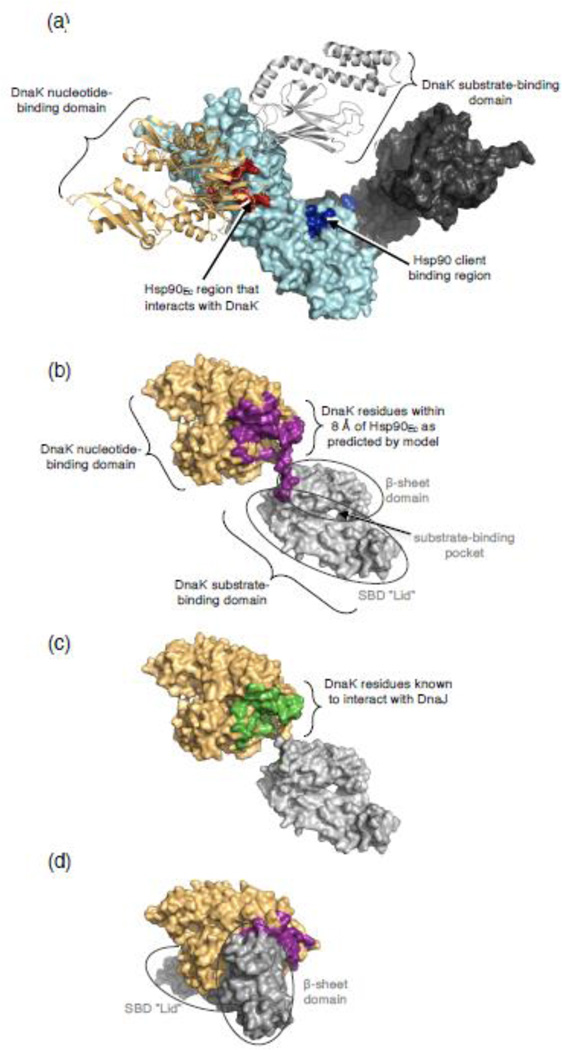
The model of the complex of ADP-bound DnaK and apo Hsp90Ec predicted that DnaK interacted exclusively with residues on the Hsp90Ec middle domain, many of which had been shown experimentally to be involved in binding DnaK [30] (Fig. 1a and Table 1). To gain additional evidence for this model, we constructed two Hsp90Ec mutants with substitutions in residues predicted by the model to be important in interaction with DnaK (Supplemental Fig. S6a). Hsp90Ec(Q302A, R303A) was defective in functional and physical interaction with DnaK in vitro; Hsp90Ec(D329A, L330A, P331A) was slightly defective (Supplemental Fig. S6b and S6c).
Fig. 1.
Regions of interaction on DnaK and Hsp90Ec. (a) Docked model of the apo structure of Hsp90Ec [21] and ADP-bound DnaK [31] as determined using ZDOCK and ZRANK and described in Materials and Methods. Hsp90Ec is shown as a surface rendering with one protomer in dark gray and one protomer in light cyan. The DnaK interacting region of Hsp90Ec [30] is shown in red while the client binding region is in blue [14]. DnaK in the ADP-bound conformation is shown as a ribbon model with the NBD in light orange and the SBD in light gray. (b) DnaK in the ADP-bound conformation [31] showing residues (purple) on DnaK within 8 Å of Hsp90Ec as predicted by the docked model in (a). In (b-d) DnaK is shown as a surface rendering with the NBD in light orange and the SBD in light gray. (c) DnaK in the ADP-bound conformation [31] showing residues (green) experimentally identified as interacting with DnaJ [38–40]. (d) DnaK in the ATP-bound conformation [32] showing residues (purple) on DnaK within 8 Å of Hsp90Ec as predicted by the docked model in (a). In the ATP-bound conformation, only some of the DnaK residues within 8 Å of Hsp90Ec in the model are surface exposed. Images in (a-d) were made using PyMOL (Schrodinger, LLC; www.pymol.org).
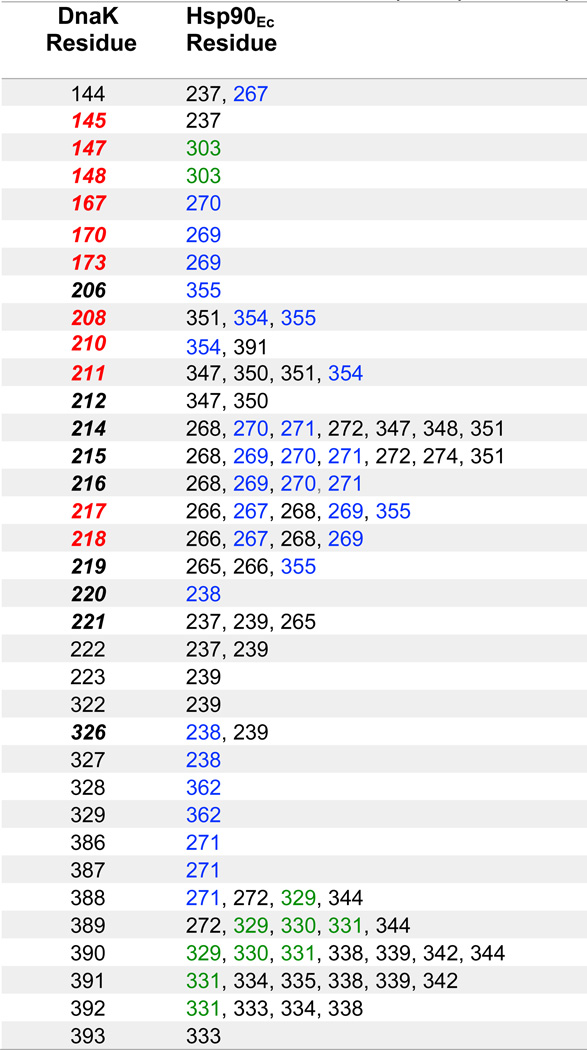
Table 1.
DnaK residues within 8 Å of Hsp90Ec residues in the model of the ADP-bound DnaK-apo Hsp90Ec complex.
Red indicates DnaK residues shown in this study to be important for interaction with Hsp90Ec. Blue indicates Hsp90Ec residues previously shown to be important for the interaction with DnaK [30]. Green indicates Hsp90Ec residues identified in this study to be important for interaction with DnaK. Black indicates residues not tested in this study, except for DnaK residue 214, which when mutated was similar to DnaK wild-type (Fig. 2). Bold, italics indicates DnaK residues interacting with DnaJ as identified from previous studies [38–40].
To further test our model, we used the PRISM webserver [36, 37] to predict the interface between DnaK in the ADP-bound conformation and apo Hsp90Ec by structural matching. The model we obtained was very similar to our top docking result obtained with ZDOCK and ZRANK (Supplemental Fig. S7a and S7b and Supplemental Table S1).
The model of ADP-bound DnaK and apo Hsp90Ec predicted that residues in the DnaK nucleotide-binding domain (NBD) interacted with Hsp90Ec (Fig. 1a and 1b, Table 1 and Supplemental Table S1). Interestingly, of the 35 DnaK residues predicted from this docked model to interact with Hsp90Ec, 20 were previously identified as important for interacting with DnaJ [38–40], suggesting that Hsp90Ec and DnaJ share a common binding interface on DnaK (Fig. 1c, Table 1). Additionally, 26 of the 35 predicted residues are also involved in the association between the DnaK NBD and substrate-binding domain (SBD) that occurs when DnaK is in the ATP conformation [32, 41] (Fig. 1d). The NBD and SBD of DnaK are not in contact with one another when DnaK is in the ADP-bound conformation [31], indicating that Hsp90Ec likely interacts with the ADP-bound conformation of DnaK. Importantly, the docked model of ADP-bound DnaK and apo Hsp90Ec, would allow a client protein bound to the SBD of DnaK to readily interact with the client-binding site of Hsp90Ec, making this model mechanistically plausible (Fig. 1a).
Mutations in the potential Hsp90Ec binding region of DnaK cause defective interaction with Hsp90Ec in vivo
We wanted to explore the validity of the docked model of the apo Hsp90Ec structure with DnaK in the ADP-bound form. To do this we constructed site-directed amino acid substitutions in surface exposed residues of DnaK in and near the residues suggested by the model to interact with Hsp90Ec and screened the mutants in a bacterial two-hybrid assay (Fig. 2a). Some substitutions were previously described mutants, including DnaKY145A,N147A,D148A [40], DnaKR167H [39], DnaKN170A,T173A [39] and DnaKE217A,V218A [40], while other substitutions were in residues that had previously been substituted with alternate amino acids, including DnaKD208R, DnaKE209C and DnaKV210R [38]. The remaining mutants were alanine substitutions or charge changes, including DnaKR84E, DnaKD211R, DnaKE213Q,K214A, DnaKD224K,T225A and DnaKS234A,I237A,N238A.
Fig. 2.
Identification of DnaK amino acid residues involved in Hsp90Ec interaction in vivo. (a) Model of E. coli DnaK in the ADP-bound conformation [31] with the mutated residues used in this study shown as CPK models. The NBD is colored light orange and the SBD is gray. DnaK was rendered using PyMOL. (b, c) Interaction between DnaK wild-type or mutant and Hsp90Ec in a bacterial two-hybrid system in vivo, as described in Materials and Methods. Interaction was measured by monitoring β-galactosidase activity on MacConkey indicator plates (b) and in liquid assays (c). In (b), a representative plate from three independent experiments is shown with colonies labeled 1 through 16. For colonies 2 through 13, all DnaK substitution mutants, named by substituted residues, have been constructed in T25-DnaK and are present in reactions with the T18-Hsp90Ec construct. In (c), β-galactosidase activity is shown as mean ± SEM (n = 3) and are also presented in Supplemental Table S5.
We had previously observed that DnaK wild-type and Hsp90Ec interact in the bacterial two-hybrid assay [23, 30]. For this assay one domain of the Bordetella pertussis adenylate cyclase protein, T18, was fused to Hsp90Ec and the other domain, T25, was fused to DnaK wild-type or to a DnaK mutant. If the two fusion proteins interact when they are coexpressed in an E. coli cya-strain, cyclic AMP is synthesized and the cAMP reporter gene, β-galactosidase, is expressed [42]. As we saw previously, the coexpression of T25-DnaK wild-type and T18-Hsp90Ec wild-type resulted in colonies that appeared red on indicator plates and expressed ~12-fold higher levels of β-galactosidase compared to coexpression of T25-DnaK wild-type and T18-vector [30] (Fig. 2b and 2c).
When plasmids expressing T25-DnaK mutant proteins were coexpressed with T18-Hsp90Ec in an E. coli cya-strain, we observed that 11 of the 12 colonies appeared white or pale pink on indicator plates and had lower levels of β-galactosidase activity than cells coexpressing T25-DnaK wild-type and T18-Hsp90Ec, suggesting that the DnaK mutant fusion proteins were defective in interaction with Hsp90Ec (Figure 2b and 2c). These mutants included DnaKY145A,N147A,D148A, DnaKR167H, DnaKN170A,T173A, DnaKD208R, DnaKV210R, DnaKE217A,V218A, DnaKS234A,I237A,N238A, DnaKE209C, DnaKD224K,T225A, DnaKD211R and DnaKR84E. One mutant, DnaKE213Q,K214A, produced red colonies on indicator plates and had β-galactosidase activity similar to T25-DnaK wild-type (Fig. 2b and 2c) and was therefore not studied further. Control experiments showed that all of the mutant fusion proteins were expressed at levels similar to the T25-DnaK wild-type protein (Supplemental Fig. S8). Thus, all but one of the DnaK substitution mutants constructed in residues that were predicted by the model to interact with Hsp90Ec or were in nearby surface exposed residues were defective in interaction in vivo. These results suggest the DnaK mutants may be defective in direct interaction with Hsp90Ec. However, in vivo the interaction may be affected by other cellular components [42].
DnaK mutant proteins defective in Hsp90Ec interaction in vivo are defective in direct complex formation with Hsp90Ec in vitro
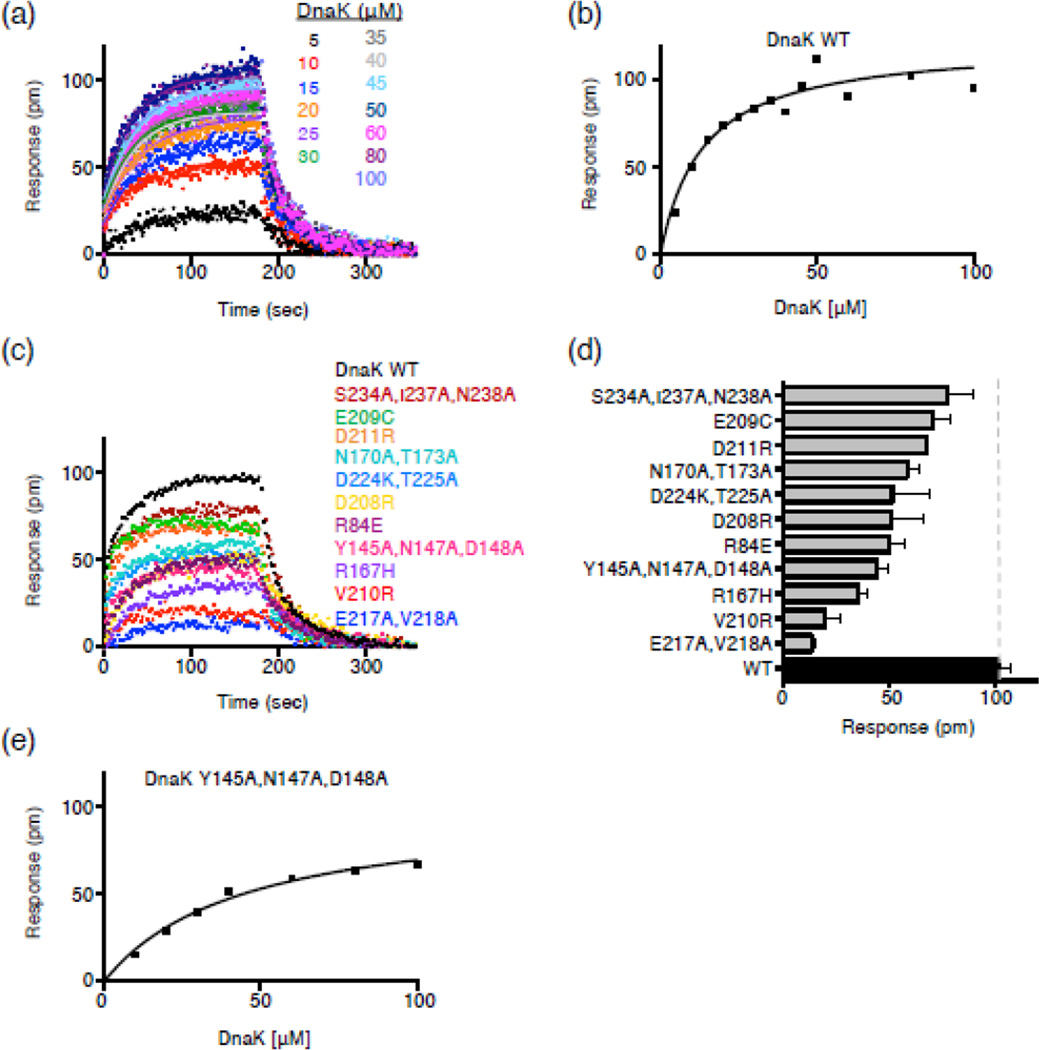
We next tested if the DnaK mutants that were defective in the two-hybrid screen were defective in direct protein-protein interaction in vitro. We first cloned, purified and characterized the mutant proteins (Supplemental Results and Supplemental Fig. S9 and S10). DnaK and Hsp90Ec proteins have been shown to form a binary complex in vitro in both the presence and absence of hydrolyzable ATP, although the interaction is very weak [29, 30]. We tested our DnaK mutant proteins for direct interaction with Hsp90Ec by Bio-Layer Interferometry (BLI). Hsp90Ec(E584C) was specifically labeled with biotin and immobilized on a streptavidin-coated biosensor (see Material and Methods). Binding was monitored using buffer conditions that allowed an optimal signal to noise ratio (see Material and Methods). We first monitored the binding of DnaK wild-type and Hsp90Ec (Fig. 3a and 3b). Since the binding curves at high DnaK concentrations were complex, we used a steady-state analysis and calculated the Kd to be ~13 µM under the conditions used in this assay (Fig. 3a and 3b). The observed Kd is consistent with the weak interaction seen previously between Hsp90Ec and DnaK wild-type [23, 30] as well as between Synechococcus elongates Hsp90 and DnaK [29] and mitochondrial Hsp90 (TRAP) and Hsp70 (Mortalin) [43].
Fig. 3.
DnaK NBD substitution mutants are defective in direct interaction with Hsp90Ec in vitro. (a) Bio-Layer Interferometry (BLI) was used to monitor the kinetics of association and dissociation between biotinylated Hsp90Ec and DnaK wild-type as described in Materials and Methods. Representative curves are shown for numerous concentrations of DnaK as indicated. The single-exponential fit of the association step was used to obtain the response value (pm) for the plateau plotted in (b). (b) Steady-state analysis of the DnaK wild-type-Hsp90Ec interaction. The response value (pm) for the plateau obtained in (a) is plotted vs. each DnaK concentration and fit as described in Materials and Methods. The Kd and Bmax for the Hsp90Ec interaction with DnaK wild-type are 13.4 ± 3.3 µM and 0.12 ± 0.01 nm, respectively. (c) Curves showing association and dissociation of 50 µM DnaK wild-type (black) or mutant (colored) and biotinylated Hsp90Ec using BLI. (d) Average plateau response value for the interaction between DnaK wild-type or mutant and biotinylated Hsp90Ec. Data are plotted as mean ± SEM (n=2 or more) and are also presented in Supplemental Table S5. (e) Steady-state analysis of the DnaKY145A,N147A,D148A-Hsp90Ec interaction as described in (b). The Kd and Bmax for the Hsp90Ec interaction with DnaKY145A,N147A,D148A are 44.4 ± 7.5 µM and 0.10 ± 0.01 nm, respectively.
We then used BLI to monitor the direct interaction between the DnaK mutant proteins and Hsp90Ec, at a concentration of DnaK (50 µM) that was saturating for the interaction of DnaK wild-type with Hsp90Ec (Fig. 3c and 3d). The results indicated that all of the DnaK mutants were defective to varying degrees in interaction with Hsp90Ec. The most defective were DnaKE217A,V218A, DnaKV210R and DnaKR167H (Fig. 3c and 3d). Others, including DnaKY145A,N147A,D148A, DnaKR84E, DnaKD208R, DnaKD224K,T225A, DnaKN170A,T173A, DnaKD211R and DnaKE209C were partially defective in Hsp90Ec binding (Fig. 3c and 3d). One mutant, DnaKY145A,N147A,D148A, was further evaluated and found to have ~3-fold lower affinity for Hsp90Ec than wild-type DnaK; the Kd was ~44 µM (Fig. 3e). DnaKS234A,I237A,N238A was the least defective mutant protein tested (Fig. 3c and 3d). These results show that the DnaK residues identified by molecular docking are important for the direct interaction with Hsp90Ec.
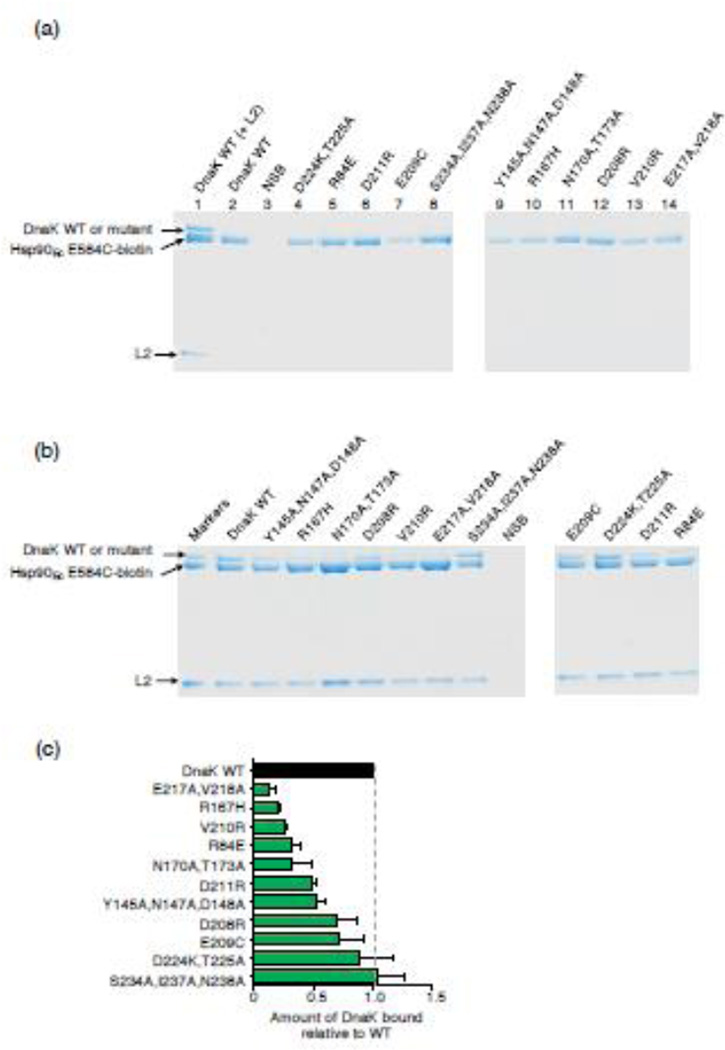
Based on our previous observation that a model client protein, ribosomal protein L2, stabilizes the DnaK-Hsp90Ec interaction [30], we next tested if L2 similarly stabilizes the physical interaction between the DnaK mutant proteins and Hsp90Ec. We used an in vitro protein-protein interaction assay (pull down assay) in which DnaK wild-type or mutant protein was incubated with biotin labeled Hsp90Ec in the presence of L2. Biotinylated Hsp90Ec and associated proteins were then captured on neutravidin agarose beads and analyzed by SDS-PAGE. As shown previously [30], in the absence of L2 the interaction between Hsp90Ec and DnaK wild-type was not detectable by Coomassie staining of the gels (Fig. 4a, compare lanes 1 and 2), although the weak interaction could be detected by Western blot analysis [30]. Similarly, interaction between Hsp90Ec and the DnaK mutant proteins in the absence of L2 was not observed by Coomassie staining (Fig. 4a, lanes 4-14). In the presence of L2, the three DnaK mutants that were most defective in binary interaction with Hsp90Ec (Fig. 3c and 3d), including DnaKE217A,V218A, DnaKR167H and DnaKV210R, were most defective in interaction with Hsp90Ec in the pull-down assay (Fig. 4b and 4c). Other mutants were partially defective, including DnaKR84E, DnaKN170A,T173A, DnaKD211R, DnaKY145A,N147A,D148A, DnaKD208R and DnaKE209C (Fig. 4b and 4c). DnaKD224K,T225A, which was partially defective in binary interaction with Hsp90Ec, bound Hsp90Ec in the presence of L2 to a similar extent as DnaK wild-type (Fig. 4b and 4c). This observation suggests that L2 rescues the defective Hsp90Ec-DnaKD224K,T225A interaction. The mutant that was least defective in the BLI assay, DnaKS234A,I237A,N238A, bound Hsp90Ec in the presence of L2 similarly to DnaK wild-type (Fig. 4b and 4c). In control experiments, all of the DnaK mutants were able to bind L2 like DnaK wild-type, showing that the differences in complex formation in the presence of L2 were not due to defects in client binding by the DnaK mutant proteins (Supplemental Fig. S10c). Together these results indicate that many of the DnaK mutant proteins are defective in direct physical interactions with Hsp90Ec and show decreased ability to interact with Hsp90Ec in the presence of L2.
Fig. 4.
DnaK mutants exhibit defective interaction with Hsp90Ec in the presence of L2. Hsp90Ec-biotin was incubated with DnaK wild-type or mutant, without or with L2. Hsp90Ec-biotin associated proteins were monitored using a pull-down assay and analyzed by Coomassie blue staining following SDS-PAGE as described in Materials and Methods. (a) In control experiments, Hsp90Ec-biotin was incubated with DnaK wild-type with L2 (lane 1) or without L2 (lane 2) or with DnaK mutant proteins without L2 (lanes 4-14). DnaK wild-type is seen in association with Hsp90Ec-biotin in the presence of L2 (lane 1), but not in the absence of L2 by this analysis (lane 2); however, the weak association between Hsp90Ec and DnaK is seen by Western blot analysis [30]. NSB indicates non-specific binding of DnaK wild-type in the absence of Hsp90Ec-biotin. (b) Interaction between Hsp90Ec-biotin and DnaK wild-type or mutant in the presence of L2 was determined as in a. Pure proteins are shown in the first lane as markers. NSB indicates non-specific binding of DnaK wild-type and L2 in the absence of Hsp90Ec-biotin. The gels shown are representative of at least three independent experiments. (c) Quantification of DnaK wild-type or mutant associated with Hsp90Ec-biotin in the presence of L2. The results were normalized to Hsp90Ec-biotin and the ratio of DnaK mutant to wild-type was plotted. Data from three or more replicates are presented as mean ± SEM and are also presented in Supplemental Table S5. The gray dashed line indicates DnaK wild-type binding and is meant to aid the eye.
DnaK mutants are defective in functioning synergistically with Hsp90Ec
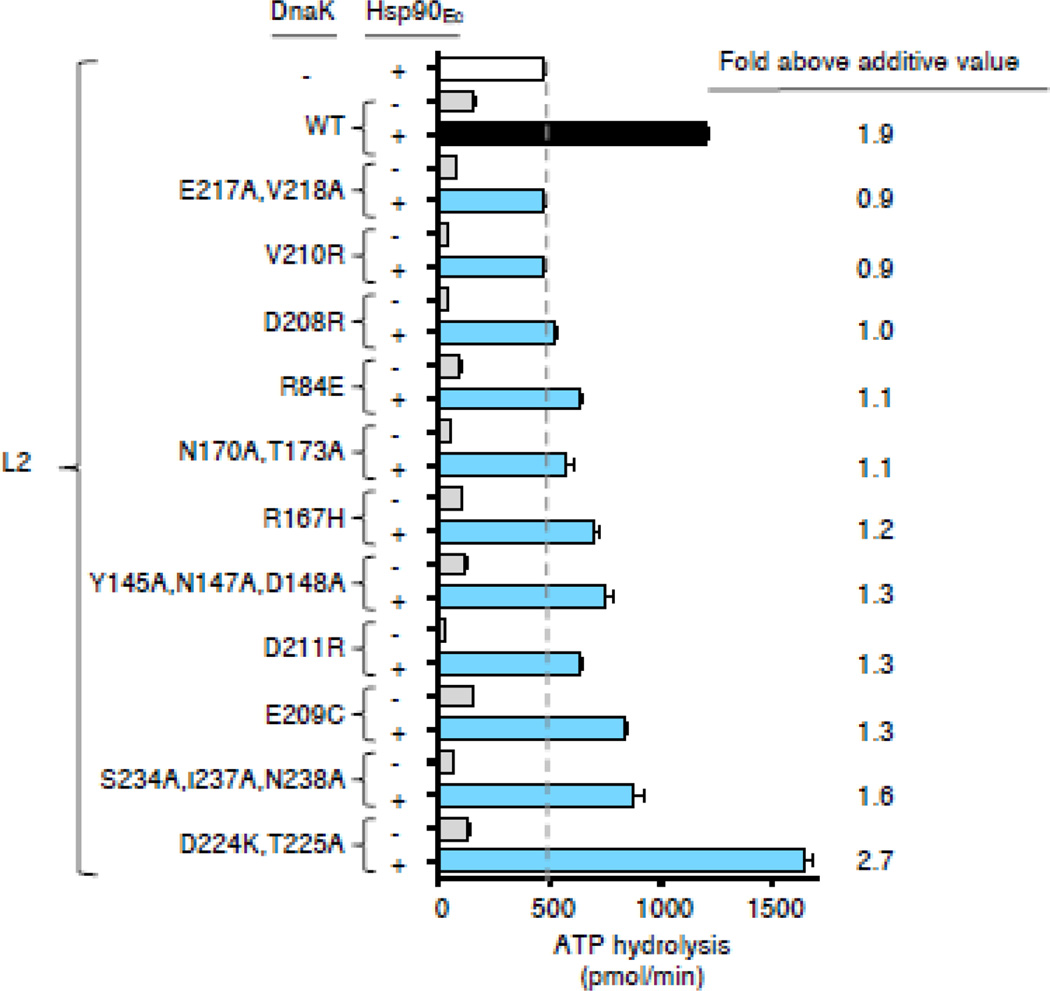
We next wanted to determine whether DnaK mutants that are defective in their ability to physically interact with Hsp90Ec are also defective in collaborating synergistically with Hsp90Ec in functional assays. It has previously been shown that ATP hydrolysis is stimulated ~2-fold above additive by the combined action of Hsp90Ec and DnaK in the presence of ribosomal protein L2, demonstrating a functional collaboration between the two chaperones [30] (Fig. 5). This synergy requires client binding by DnaK and Hsp90Ec as well as ATP hydrolysis by both chaperones [30]. Although all of our DnaK mutant proteins bound L2 similarly to wild-type (Supplemental Fig. S10c) and hydrolyzed ATP (Supplemental Fig. S10d), we observed that several of the DnaK mutants showed no synergy in ATP hydrolysis in combination with Hsp90Ec and L2, including DnaKE217A,V218A DnaKV210R and DnaKD208R (Fig. 5). Others stimulated ATP hydrolysis 1.1 to 1.3-fold above additive compared to the 1.9-fold stimulation seen with wild-type DnaK, including DnaKR84E, DnaKN170A,T173A, DnaKR167H, DnaKY145A,N147A,D148A, DnaKD211R and DnaKE209C (Fig. 5). DnaKS234A,I237A,N238A was slightly defective in stimulating ATP hydrolysis, consistent with its slight defect in the BLI assay. In combination with Hsp90Ec and L2, DnaKD224K,T225A stimulated ATP hydrolysis 2.7-fold above additive (Fig. 5). The stabilization of the Hsp90Ec-DnaKD224K,T225A interaction by L2 (Fig. 4b and 4c) may explain the increased synergy in ATP hydrolysis. Taken together, the results indicate that the DnaK mutants that are defective or partially defective in physical interaction with Hsp90Ec are defective or partially defective in functional interaction as well, with the exception of DnaKD224K,T225A.
Fig. 5.
DnaK mutants defective in Hsp90Ec interaction are defective in synergistic stimulation of ATP hydrolysis with Hsp90Ec and a client protein, L2. ATP hydrolysis by DnaK wild-type or mutant in the presence of L2 was determined in the absence and presence of Hsp90Ec as described in Materials and Methods. The fold above additive is calculated by dividing the rate of ATP hydrolysis by Hsp90Ec and DnaK in the presence of L2 by the sum of the rate for Hsp90Ec in the presence of L2 and the rate for DnaK in the presence of L2. Data from three or more replicates are presented as mean ± SEM and are also presented in Supplemental Table S5. The dashed line indicates the rate of ATP hydrolysis by Hsp90Ec with L2 and is meant to aid the eye.
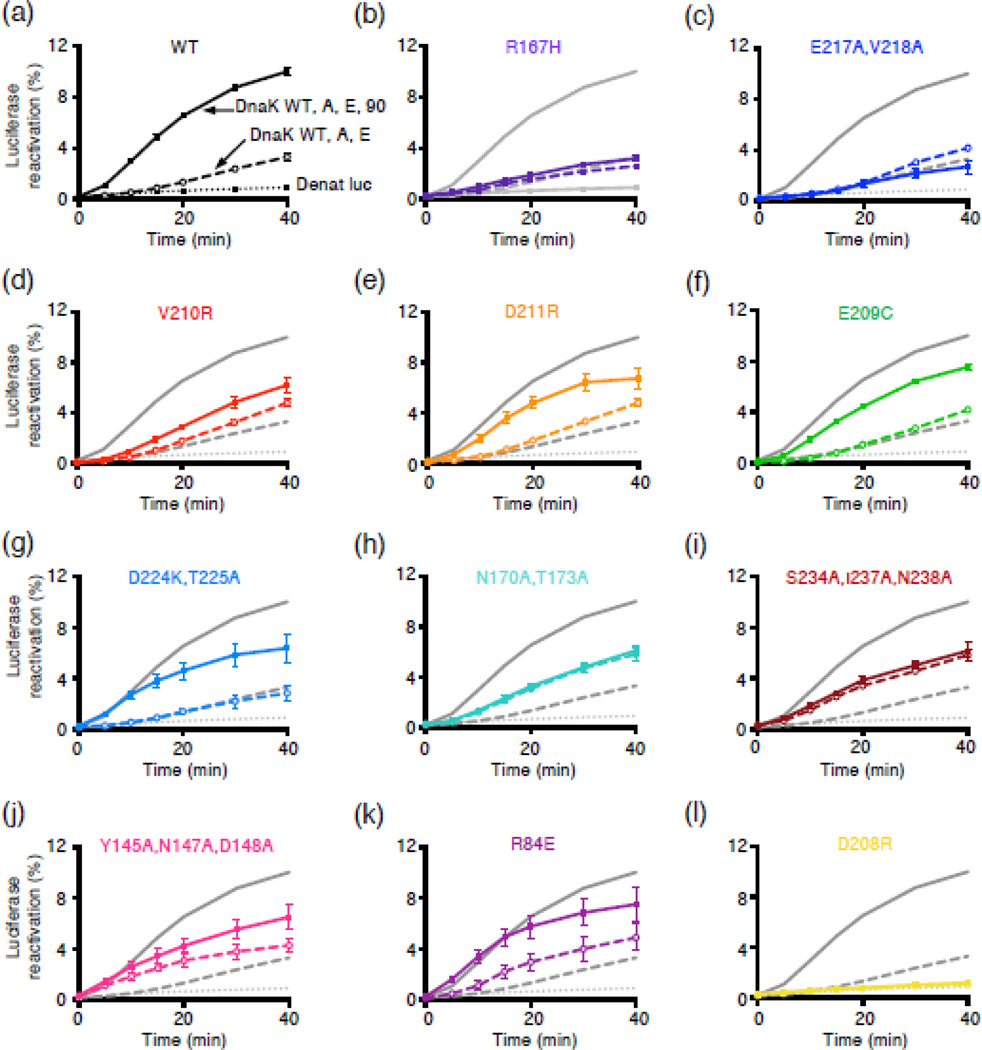
We next tested if the DnaK mutants were also defective in functional collaboration with Hsp90Ec in client protein remodeling. We assessed remodeling ability by monitoring reactivation of heat-inactivated luciferase by the combination of the DnaK chaperone system and Hsp90Ec. In this assay, the chaperone concentrations were optimized in order to observe the largest dependence on Hsp90Ec. Under these conditions, DnaK and two cochaperones, CbpA (a DnaJ homolog in E. coli) and GrpE (a nucleotide exchange factor), catalyze luciferase reactivation at a slow rate and Hsp90Ec stimulates the rate ~6-fold [30] (Fig. 6a). The percentage of luciferase reactivated is low; however, only ~20% of the total heat-inactivated luciferase is soluble and available for reactivation [23]. When we tested our DnaK mutant proteins, all were defective or partially defective in functioning synergistically with Hsp90Ec (Fig. 6). DnaKR167H, DnaKE217A,V218A and DnaKV210R, reactivated luciferase at rates similar to DnaK wild-type in the absence of Hsp90Ec, but were not stimulated or only slightly stimulated by Hsp90Ec (Fig. 6b-6d). These three DnaK mutants were also the most defective in binary complex formation with Hsp90Ec (Fig. 3) and in complex formation with Hsp90Ec and L2 (Fig. 4). The remainder of the mutants with the exception of DnaKD208R were also defective or partially defective in synergistic reactivation of luciferase with Hsp90Ec. However, in the absence of Hsp90Ec several of these mutants were similar to DnaK wild-type in the ability to reactivate luciferase with CbpA and GrpE, including DnaKD211R, DnaKE209C and DnaKD224K,T225A (Fig. 6e-6g), while others, including DnaKN170A,T173A, DnaKS234A,I237A,N238A, DnaKY145A,N147A,D148A and DnaKR84E, were more active than DnaK wild-type (Fig. 6h-6k). The reason for the increased activity of these mutants with CbpA and GrpE is not understood. One mutant, DnaKD208R, was defective in the absence and presence of Hsp90Ec (Fig. 6l), consistent with previous results showing that this mutant is defective in J-domain stimulated ATPase activity [38]. Altogether, the results indicate that many of the DnaK mutants are defective or partially defective in their ability to function collaboratively with Hsp90Ec in protein reactivation. They suggest that the functional defects of the DnaK mutants are a consequence of defects in the physical interaction between the mutants and Hsp90Ec.
Fig. 6.
DnaK NBD mutants defective in Hsp90Ec interaction are defective in functional collaboration with Hsp90Ec in luciferase reactivation. Heat-inactivated luciferase was reactivated as described in Materials and Methods using DnaK wild-type or mutant, CbpA, GrpE and Hsp90Ec as indicated. (a-l) Luciferase reactivation by DnaK wild-type (black/gray) or mutant (color), as indicated in panels a-l in combination with CbpA and GrpE (open symbols and dashed lines) or CbpA, GrpE and Hsp90Ec (filled symbols and solid lines) as a function of time. Dotted lines indicate luciferase alone control. In a-l, data from three or more replicates are presented as mean ± SEM. For some points, the symbols obscure the error bars.
In summary, we have identified a region of DnaK that is required for the interaction of DnaK with Hsp90Ec. The DnaK mutants define a site located in the nucleotide-binding domain of DnaK that is essential for Hsp90Ec interaction. This region overlaps the DnaK site known to bind DnaJ, suggesting an interplay between DnaK, DnaJ and Hsp90Ec during protein remodeling.
Discussion
In this work, we identified a region in the nucleotide-binding domain of DnaK that is important for the physical and functional interaction with a region in the middle domain of Hsp90Ec. Direct interactions between eukaryotic Hsp90 and Hsp70 have been observed previously. A recent study showed a direct interaction between the mitochondrial Hsp90 and Hsp70 homologs, TRAP1 and Mortalin [43]. Like bacteria, mitochondria lack Hop/Sti1, a cochaperone known to interact simultaneously with both Hsp90 and Hsp70 and facilitate the Hsp90-Hsp70 interaction [1, 44]. Therefore, the mechanism of action of TRAP1 may be more similar to that of bacterial Hsp90 than eukaryotic cytoplasmic Hsp90. However, in studies using crude lysates from rabbit reticulocytes or using purified cytoplasmic Hsp90 and Hsp70 from Neurospora crassa [45–47], direct interactions were also observed. In both systems, Hop/Sti1 increased the amount of Hsp90 and Hsp70 in the complex [45–47]. Thus, the functional significance of the observed direct interaction between cytoplasmic chaperones is not clear, and current models suggest that eukaryotic Hsp90 and Hsp70 need not contact one another directly, but rather Hop/Sti1 forms a bridge between the two chaperones [3, 4, 15, 16, 44].
Our finding that many residues in the DnaK NBD that are involved in Hsp90Ec interaction are also involved in binding DnaJ/CbpA [38–40, 48], suggests the possibility of a functional interplay of DnaK with Hsp90Ec and DnaJ/CbpA. One hypothesis that invokes a DnaK binding site shared by DnaJ/CbpA and Hsp90Ec is that Hsp90Ec participates in displacing DnaJ/CbpA from DnaK once DnaJ/CbpA has promoted ATP hydrolysis by DnaK. This is consistent with our previous studies that showed substoichiometric concentrations of CbpA (~1:100, CbpA:Hsp90Ec) facilitated formation of the DnaK-Hsp90Ec-L2 ternary complex while CbpA was not detected in the complex [30]. Previous studies have also suggested a similar role for Hsp40, where Hsp40 stimulated formation of Hsp90-Hop-Hsp70 complexes but was not seen in the final ternary complex [49–51]. Together, these results suggest that the overlapping binding site on DnaK should lead to competition between DnaJ/CbpA and Hsp90Ec. However, in the assays used above, we have not seen competition, suggesting the reaction pathway may be more complex.
In E. coli, the concentrations of molecular chaperones and cochaperones varies under different conditions (i.e. cellular stress or starvation) and growth phase. At 30 °C in log phase growth, DnaK is highly abundant with a concentration ~5-fold higher than Hsp90Ec or GrpE, ~25-fold higher than DnaJ and >25-fold higher than CbpA [52, 53]. However, CbpA expression increases as cells enter stationary phase and DnaK may only be ~2-fold higher in concentration than CbpA in late stationary phase [54]. This suggests that the interplay between DnaK and DnaJ/CbpA or Hsp90Ec likely varies depending on the growth phase or stress condition.
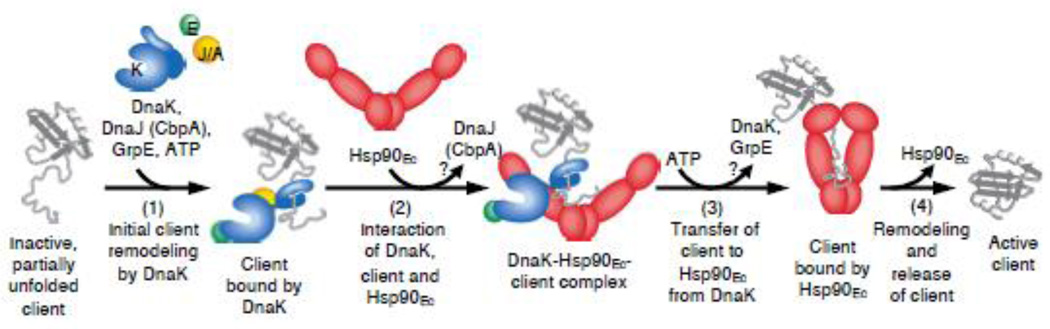
Our current working model for the mechanism of protein remodeling by Hsp90Ec and DnaK is speculative, but consistent with the current knowledge of the bacterial chaperones (Fig. 7). First, the client protein is bound by DnaK, in a reaction requiring ATP hydrolysis and facilitated by DnaJ/CbpA and GrpE (Fig. 7, step 1). This is in keeping with our previous work showing the DnaK chaperone system functions prior to Hsp90Ec in protein remodeling [23]. Then, client-bound DnaK recruits Hsp90Ec, likely through a direct interaction between the DnaK NBD and the middle domain of Hsp90Ec. Based on our molecular docking results, a client protein bound to the SBD of DnaK would be poised for interaction with the client binding site of Hsp90Ec (Fig. 7, step 2). The binding of Hsp90Ec to DnaK may displace DnaJ/CbpA, since our results suggest DnaJ/CbpA and Hsp90Ec bind overlapping regions on DnaK. Next, ATP binding and hydrolysis by Hsp90Ec leads to conformational changes in Hsp90Ec that promote client transfer from DnaK, stabilize client binding by Hsp90Ec and may release DnaK (Fig. 7, step 3). Then, nucleotide release from Hsp90Ec likely causes the release of an active client (Fig. 7, step 4). However, in cases where the client does not attain its active conformation it may enter another cycle of remodeling. Taken together, these studies are providing insight into the interactions and collaboration between Hsp90 and Hsp70.
Fig. 7.
Working model for the mechanism of action of the DnaK system in collaboration with Hsp90Ec. First, the client protein is bound by DnaK for initial ATP-dependent remodeling, a process that requires DnaJ/CbpA and GrpE (step 1). Next, DnaK, through a direct interaction of the DnaK NBD with the Hsp90Ec middle domain, recruits Hsp90Ec to the client, which further stabilizes the interaction between DnaK and Hsp90Ec (step 2). DnaJ/CbpA may be released at this time. Binding and hydrolysis of ATP by Hsp90Ec triggers conformational changes in the chaperone that lead to client transfer and stabilization of client binding to Hsp90Ec (step 3). DnaK and GrpE may be released at this step. Hsp90Ec promotes further client remodeling and releases the active native client (step 4). Client proteins that do not attain the active conformation may reenter the chaperone cycle.
Materials and Methods
Plasmids and Strains
Substitution mutations of Hsp90Ec and DnaK were made with the QuikChange Lightning mutagenesis kit (Agilent) using pET-HtpG [23], pRE-DnaK [55], pET-DnaK [56] or pT25-DnaK [23]. All mutations were verified by DNA sequencing.
Proteins
Hsp90Ec wild-type and mutants [23], DnaK wild-type and mutants [55], CbpA [57], GrpE [55] and His-tagged L2 [58] were isolated as described. All proteins were >95% pure as determined by SDS-PAGE. All DnaK substitution mutants exhibited similar physical properties as the wild-type, including partial proteolysis patterns with and without ATP (Supplemental Fig. S9a) and CD spectra (Supplemental Fig. S9b). All exhibited some DnaK functional activities, including: reactivation of GFP with DnaJ and GrpE (Supplemental Fig. S10a) and further stimulation of GFP reactivation by ClpB (Supplemental Fig. S10b), client binding (L2) (Supplemental Fig. S10c) and basal ATPase activity (Supplemental Fig. S10d). The Hsp90Ec E584C-biotin mutant was similar to Hsp90Ec wild-type in luciferase reactivation activity (Supplemental Fig. S11a). Luciferase and luciferin were from Promega. Concentrations given are for Hsp90Ec, CbpA and GrpE dimers and DnaK, L2 and luciferase monomers. Hsp90Ec E584C was labeled using a 20-fold excess of Maleimide-PEG11-Biotin (Thermo, Life Technologies) as recommended by the manufacturer. CbpA was labeled using a 1.5-fold excess of NHS-PEG4-Biotin (Thermo, Life Technologies). Excess biotin reagent was removed by extensive dialysis.
Luciferase reactivation
Luciferase reactivation was performed as previously described [23, 30]. 40 nM heat-denatured luciferase was incubated at 24 °C in reaction mixtures (75 µl) containing 25 mM Hepes, pH 7.5, 50 mM KCl, 0.1 mM EDTA, 2 mM DTT, 10 mM MgCl2, 50 µg/ml bovine serum albumin (BSA), 3 mM ATP, an ATP regenerating system (25 mM creatine phosphate, 6 µg creatine kinase), 0.95 µM DnaK wild-type or mutant, 0.15 µM CbpA, 0.05 µM GrpE and 0.5 µM Hsp90Ec. Aliquots were removed at the indicated times and light output was measured using a Tecan Infinite M200Pro in luminescence mode with an integration time of 1000 ms. Reactivation was determined compared to a non-denatured luciferase control. To test for potential defects in binding between the DnaK mutants and CbpA that could lead to defects in luciferase reactivation, we performed pull-down assays with our DnaK mutants using biotinylated-CbpA (Supplemental Fig. S11b).
ATPase activity
Steady state ATP hydrolysis was measured at 37 °C in 25 mM Hepes, pH 7.5, 50 mM KCl, 5 mM DTT, 5 mM MgCl2 and 2 mM ATP using a pyruvate kinase/lactate dehydrogenase enzyme-coupled assay as described [18] and using 1 µM Hsp90Ec, 1 µM L2 and 1 µM DnaK wild-type or mutant.
Bio-Layer Interferometry (BLI) Assay
BLI was used to monitor the interaction between Hsp90Ec and DnaK using a FortéBio (Menlo Park, CA) Octet RED96 instrument and streptavidin (SA) biosensors at 30 °C. Each assay step was 200 µL containing BLI buffer (20 mM Tris, pH 7.5, 25 mM NaCl, 0.01% Triton X-100 (vol/vol), 0.02% Tween-20 (vol/vol), 1 mg/ml BSA, 1 mM ATP, and 10 mM MgCl2). Hsp90Ec E584C-biotin was loaded on the biosensors to a BLI response signal of 1 nm. The biosensors were blocked in BLI buffer containing 10 µg/ml biocytin (Sigma) for 1 min, and a baseline was established in BLI buffer alone. Association of DnaK wild-type or mutant (5 to 100 µM) to the Hsp90-biotin bound sensor tip in BLI buffer was monitored over time. Lastly, dissociation was monitored in BLI buffer alone. For each experiment, nonspecific binding was monitored using a reference biosensor subjected to each of the above steps in the absence of biotinylated Hsp90Ec and non-specific binding signal was subtracted from the corresponding experiment. For steady state analysis of kinetic association data, the association curve at each DnaK concentration was fit using a single exponential equation without constraints in Prism (GraphPad Software, La Jolla California USA, www.graphpad.com) and the plateau value determined from the fit was plotted versus the concentration of DnaK. The resulting binding curve was analyzed using a one-site specific binding model in Prism to determine the Kd and Bmax values.
Protein-protein interaction assay
Interaction of Hsp90Ec with DnaK in the presence of L2 was measured using a pull down assay. Hsp90Ec E584C-biotin (1.5 µM) was incubated for 5 minutes at 23 °C in reaction mixtures (50 µL) containing GPD buffer (20 mM Tris-HCl, pH 7.5, 75 mM KCl, 10% glycerol (vol/vol), 0.01% Triton X-100 (vol/vol), 2 mM DTT) with 2 µM DnaK wild-type or mutant, and 2.3 µM L2. Neutravidin agarose (40 µL 1:1 slurry) (Thermo, Pierce) was then added and incubated 5 min at 23 °C with mixing. The reactions were diluted with 0.4 mL GPD buffer, centrifuged 1 min at 1000 × g and the recovered agarose beads were washed twice with 0.4 mL GPD buffer. Bound proteins were eluted with buffer containing 2 M NaCl and analyzed by Coomassie blue staining following SDS-PAGE. Where indicated, protein band intensities from replicate gels were quantified using ImageJ (http://imagej.nih.gov/ij). The results were normalized to Hsp90Ec E584C-biotin and the ratio of DnaK mutant relative to DnaK wild-type was calculated and plotted.
Bacterial two-hybrid assay
Bacterial two-hybrid assays were performed as previously described [23, 42].
Modeling DnaK-Hsp90Ec interaction
Starting with the ADP-bound conformation of DnaK, PDB ID 2KHO [31], or separately, the ATP-bound form of DnaK, PDB ID 4BQ9 [32], the CHARMM molecular modeling program [59] was used to build in missing atoms as described previously [60]. Similarly, the dimeric structure of ADP-bound Hsp90Ec was constructed from biological assembly 1 of PDB ID 2IOP [21] using CHARMM to build in missing atoms. The dimeric structure of apo Hsp90Ec had several missing regions at positions 1-15, 98-114, 493-501 and 544-565 [21]. Therefore, we constructed a full length monomer using the I-TASSER structure prediction software [61–63] by threading the sequence of E. coli Hsp90 along the apo conformation of Hsp90Ec (PDB ID 2IOQ, chain A). Two copies of the resulting full-length monomer were then oriented as to minimize the RMSD with each monomer of the dimer to create a dimer of apo Hsp90Ec.
Hsp90Ec-DnaK complexes were generated using ZDOCK [33, 34] (version 2.3), which employs rigid body docking and utilizes a scoring function based on pairwise shape complementarity, desolvation, and electrostatic energies. Docking complexes were created without restraining the interaction between DnaK and Hsp90Ec. The top 2000 docked complexes from ZDOCK were then reranked using ZRANK [35]; a detailed scoring function that takes into account desolvation energy and both attractive and repulsive van der Waals and electrostatic energies that are calculated using the CHARMM19 polar hydrogen force-field [64]. The docked complex with the best score was selected for each combination: 1) ADP-bound DnaK with apo Hsp90Ec, 2) ADP-bound DnaK with ADP-bound Hsp90Ec, 3) ATP-bound DnaK with apo Hsp90Ec and 4) ATP-bound DnaK with ADP-bound Hsp90Ec.
Contacts were computed by calculating distances from the center of geometry of residue pairs within an 8 Å distance measured from alpha carbons. This larger distance cutoff value was used to define a general region of DnaK that may participate in binding Hsp90Ec, which aided in the selection of residues for DnaK substitution mutants.
Supplementary Material
Research Highlights.
Molecular docking predicts Hsp90Ec interacts with the ADP conformation of DnaK
The predicted Hsp90Ec interacting region is in the DnaK nucleotide-binding domain
Many predicted Hsp90Ec interacting residues on DnaK are involved in DnaJ binding
Substitutions in predicted DnaK residues cause defects in interaction with Hsp90Ec
The DnaK mutants are defective in collaboration with Hsp90Ec in protein remodeling
Acknowledgments
We thank Grzegorz Piszczek (NHLBI Biophysical Core Facility) and Jonathan McMurry for helpful comments and suggestions with BLI experiments and analysis. We also thank George Stan for valuable discussions regarding molecular docking. This research was supported by the Intramural Research Program of the NIH, NCI, Center for Cancer Research.
Abbreviations
- NBD
nucleotide-binding domain
- SBD
substrate-binding domain
- NEF
nucleotide exchange factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions- A. N. K., S. M. D., J. R. H., E. D., O. G., and S. W. designed the experiments. A. N. K., S. M. D., J. R. H., and E. D. performed the experiments. All authors were involved in data interpretation and discussion. A. N. K., S. M. D and S. W. wrote the manuscript with contributions from all other authors. The authors declare no competing financial interests.
References
- 1.Johnson JL. Evolution and function of diverse Hsp90 homologs and cochaperone proteins. Biochim. Biophys. Acta. - Molecular Cell Research. 2012;1823:607–613. doi: 10.1016/j.bbamcr.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 2.Mayer MP. Gymnastics of molecular chaperones. Mol. Cell. 2010;39:321–331. doi: 10.1016/j.molcel.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 3.Röhl A, Rohrberg J, Buchner J. The chaperone Hsp90: Changing partners for demanding clients. Trends Biochem. Sci. 2013;38:253–262. doi: 10.1016/j.tibs.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Soroka J, Buchner J. The Hsp90 chaperone machinery: Conformational dynamics and regulation by co-chaperones. Biochim. Biophys. Acta. 2012;1823:624–635. doi: 10.1016/j.bbamcr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Zuehlke A, Johnson JL. Hsp90 and co-chaperones twist the functions of diverse client proteins. Biopolymers. 2010;93:211–217. doi: 10.1002/bip.21292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karagöz GE, Rüdiger SGD. Hsp90 interaction with clients. Trends Biochem. Sci. 2015;40:117–125. doi: 10.1016/j.tibs.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat. Rev. Cancer. 2010;10:537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bardwell JC, Craig EA. Ancient heat shock gene is dispensable. J. Bacteriol. 1988;170:2977–2983. doi: 10.1128/jb.170.7.2977-2983.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas JG, Baneyx F. ClpB and HtpG facilitate de novo protein folding in stressed Escherichia coli cells. Mol. Microbiol. 2000;36:1360–1370. doi: 10.1046/j.1365-2958.2000.01951.x. [DOI] [PubMed] [Google Scholar]
- 11.Yosef I, Goren MG, Kiro R, Edgar R, Qimron U. High-temperature protein G is essential for activity of the Escherichia coli clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system. Proc. Natl. Acad. Sci. U.S.A. 2011;108:20136–20141. doi: 10.1073/pnas.1113519108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grudniak AM, Markowska K, Wolska KI. Interactions of Escherichia coli molecular chaperone HtpG with DnaA replication initiator DNA. Cell Stress Chaperones. 2015;20:951–957. doi: 10.1007/s12192-015-0623-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Press MO, Li H, Creanza N, Kramer G, Queitsch C, Sourjik V, et al. Genome-scale Co-evolutionary Inference Identifies Functions and Clients of Bacterial Hsp90. PLoS Genet. 2013;9:e1003631. doi: 10.1371/journal.pgen.1003631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genest O, Reidy M, Street TO, Hoskins JR, Camberg JL, Agard DA, et al. Uncovering a Region of Heat Shock Protein 90 Important for Client Binding in E. coli and Chaperone Function in Yeast. Mol. Cell. 2013;49:464–473. doi: 10.1016/j.molcel.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prodromou C. The 'active life' of Hsp90 complexes. Biochim. Biophys Acta. 2012;1823:614–623. doi: 10.1016/j.bbamcr.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prodromou C, Morgan RML. "Tuning" the ATPase Activity of Hsp90. Adv. Biochem. Health D. 2016;14:469–490. [Google Scholar]
- 17.Southworth DR, Agard DA. Species-Dependent Ensembles of Conserved Conformational States Define the Hsp90 Chaperone ATPase Cycle. Mol. Cell. 2008;32:631–640. doi: 10.1016/j.molcel.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graf C, Stankiewicz M, Kramer G, Mayer MP. Spatially and kinetically resolved changes in the conformational dynamics of the Hsp90 chaperone machine. EMBO J. 2009;28:602–613. doi: 10.1038/emboj.2008.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krukenberg KA, Street TO, Lavery LA, Agard DA. Conformational dynamics of the molecular chaperone Hsp90. Q. Rev. Biophys. 2011;44:229–255. doi: 10.1017/S0033583510000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ratzke C, Mickler M, Hellenkamp B, Buchner J, Hugel T. Dynamics of heat shock protein 90 C-terminal dimerization is an important part of its conformational cycle. Proc. Natl. Acad. Sci. U.S.A. 2010;107:16101–16106. doi: 10.1073/pnas.1000916107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shiau AK, Harris SF, Southworth DR, Agard DA. Structural Analysis of E. coli hsp90 Reveals Dramatic Nucleotide-Dependent Conformational Rearrangements. Cell. 2006;127:329–340. doi: 10.1016/j.cell.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 22.Ali MMU, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, et al. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Genest O, Hoskins JR, Camberg JL, Doyle SM, Wickner S. Heat shock protein 90 from Escherichia coli collaborates with the DnaK chaperone system in client protein remodeling. Proc. Natl. Acad. Sci. U.S.A. 2011;108:8206–8211. doi: 10.1073/pnas.1104703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balchin D, Hayer-Hartl M, Hartl FU. In vivo aspects of protein folding and quality control. Science. 2016;353:aac4354. doi: 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- 25.Mayer MP. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem. Sci. 2013;38:507–514. doi: 10.1016/j.tibs.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Clerico EM, Tilitsky JM, Meng W, Gierasch LM. How Hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J. Mol. Biol. 2015;427:1575–1588. doi: 10.1016/j.jmb.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alderson Thomas R, Kim Jin H, Markley John L. Dynamical Structures of Hsp70 and Hsp70-Hsp40 Complexes. Structure. 2016;24:1014–1030. doi: 10.1016/j.str.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuiderweg ERP, Bertelsen EB, Rousaki A, Mayer MP, Gestwicki JE, Ahmad A. Allostery in the Hsp70 Chaperone Proteins. Top. Curr. Chem. 2013;328:99–153. doi: 10.1007/128_2012_323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamoto H, Fujita K, Ohtaki A, Watanabe S, Narumi S, Maruyama T, et al. Physical interaction between bacterial heat shock protein (Hsp) 90 and Hsp70 chaperones mediates their cooperative action to refold denatured proteins. J. Biol. Chem. 2014;289:6110–6119. doi: 10.1074/jbc.M113.524801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Genest O, Hoskins JR, Kravats AN, Doyle SM, Wickner S. Hsp70 and Hsp90 of E. coli Directly Interact for Collaboration in Protein Remodeling. J. Mol. Biol. 2015;427:3877–3889. doi: 10.1016/j.jmb.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ERP. Solution conformation of wild-type E. \coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc. Natl. Acad. Sci. U.S.A. 2009;106:8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kityk R, Kopp J, Sinning I, Mayer MP. Structure and Dynamics of the ATP-Bound Open Conformation of Hsp70 Chaperones. Mol. Cell. 2012;48:863–874. doi: 10.1016/j.molcel.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 33.Chen R, Li L, Weng Z. ZDOCK: An initial-stage protein-docking algorithm. Proteins. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 34.Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T, Weng Z. ZDOCK server: Interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30:1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pierce B, Weng Z. ZRANK: reranking protien docking predictions with an optimized energy function. Proteins. 2007;67:1078–1086. doi: 10.1002/prot.21373. [DOI] [PubMed] [Google Scholar]
- 36.Baspinar A, Cukuroglu E, Nussinov R, Keskin O, Gursoy A. PRISM: A web server and repository for prediction of protein-protein interactions and modeling their 3D complexes. Nuc. Acids Res. 2014;42:W285–W289. doi: 10.1093/nar/gku397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tuncbag N, Gursoy A, Nussinov R, Keskin O. Predicting protein-protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using PRISM. Nat. Protoc. 2011;6:1341–1354. doi: 10.1038/nprot.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahmad A, Bhattacharya A, McDonald RA, Cordes M, Ellington B, Bertelsen EB, et al. Heat shock protein 70 kDa chaperone/DnaJ cochaperone complex employs an unusual dynamic interface. Proc. Natl. Acad. Sci. U.S.A. 2011;108:18966–18971. doi: 10.1073/pnas.1111220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suh W-C, Burkholder WF, Lu CZ, Zhao X, Gottesman ME, Gross C. Interaction of the Hsp70 molecular chaperone , DnaK , with its cochaperone Dna. J. Proc. Natl. Acad. Sci. U.S.A. 1998;95:15223–15228. doi: 10.1073/pnas.95.26.15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gässler CS, Buchberger A, Laufen T, Mayer MP, Schröder H, Valencia A, et al. Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc. Natl. Acad. Sci. U.S.A. 1998;95:15229–15234. doi: 10.1073/pnas.95.26.15229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi R, Sarbeng EB, Liu Q, Le KQ, Xu X, Xu H, et al. Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat. Struct. Mol. Biol. 2013;20:900–907. doi: 10.1038/nsmb.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Battesti A, Bouveret E. The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods. 2012;58:325–334. doi: 10.1016/j.ymeth.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 43.Sung N, Lee J, Kim J-H, Chang C, Tsai FTF, Lee S. 2.4 Å resolution crystal structure of human TRAP1NM, the Hsp90 paralog in the mitochondrial matrix. Acta Crystallogr. D. Biol. Crystallogr. 2016;72:904–911. doi: 10.1107/S2059798316009906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, Buchner J. Structure, function and regulation of the hsp90 machinery. Biomedical J. 2013;36:106–117. doi: 10.4103/2319-4170.113230. [DOI] [PubMed] [Google Scholar]
- 45.Murphy PJM, Kanelakis KC, Galigniana MD, Morishima Y, Pratt WB. Stoichiometry, Abundance, and Functional Significance of the hsp90/hsp70-based Multiprotein Chaperone Machinery in Reticulocyte Lysate. J. Biol. Chem. 2001;276:30092–30098. doi: 10.1074/jbc.M103773200. [DOI] [PubMed] [Google Scholar]
- 46.Freitag DG, Ouimet PM, Girvitz TL, Kapoor M. Heat shock protein 80 of Neurospora crassa, a cytosolic molecular chaperone of the eukaryotic stress 90 family, interacts directly with heat shock protein 70. Biochemistry. 1997;36:10221–10229. doi: 10.1021/bi963030g. [DOI] [PubMed] [Google Scholar]
- 47.Britton ME, Kapoor M. The oligomeric state, complex formation, and chaperoning activity of hsp70 and hsp80 of Neurospora crassa. Biochem. Cell Biol. 2002;80:797–809. doi: 10.1139/o02-166. [DOI] [PubMed] [Google Scholar]
- 48.Mayer MP, Laufen T, Paal K, McCarty JS, Bukau B. Investigation of the interaction between DnaK and DnaJ by surface plasmon resonance spectroscopy. J. Mol. Biol. 1999;289:1131–1144. doi: 10.1006/jmbi.1999.2844. [DOI] [PubMed] [Google Scholar]
- 49.Alvira S, Cuéllar J, Röhl A, Yamamoto S, Itoh H, Alfonso C, et al. Structural characterization of the substrate transfer mechanism in Hsp70/Hsp90 folding machinery mediated by Hop. Nat. Commun. 2014;5:5484. doi: 10.1038/ncomms6484. [DOI] [PubMed] [Google Scholar]
- 50.Hernandez MP, Sullivan WP, Toft DO. The assembly and intermolecular properties of the hsp70-Hop-hsp90 molecular chaperone complex. J. Biol. Chem. 2002;277:38294–38304. doi: 10.1074/jbc.M206566200. [DOI] [PubMed] [Google Scholar]
- 51.Kirschke E, Goswami D, Southworth D, Griffin PR, Agard DA. Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell. 2014;157:1685–1697. doi: 10.1016/j.cell.2014.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mogk A, Tomoyasu T, Goloubinoff P, Rudiger S, Roder D, Langen H, et al. Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999;18:6934–6949. doi: 10.1093/emboj/18.24.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomoyasu T, Ogura T, Tatsuta T, Bukau B. Levels of DnaK and DnaJ provide tight control of heat shock gene expression and protein repair in Escherichia coli. Mol. Microbiol. 1998;30:567–581. doi: 10.1046/j.1365-2958.1998.01090.x. [DOI] [PubMed] [Google Scholar]
- 54.Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skowyra D, Wickner S. The interplay of the GrpE heat shock protein and Mg2+ in RepA monomerization by DnaJ and DnaK. J. Biol. Chem. 1993;268:25296–25301. [PubMed] [Google Scholar]
- 56.Miot M, Reidy M, Doyle SM, Hoskins JR, Johnston DM, Genest O, et al. Species-specific collaboration of heat shock proteins (Hsp) 70 and 100 in thermotolerance and protein disaggregation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6915–6920. doi: 10.1073/pnas.1102828108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ueguchi C, Kakeda M, Yamada H, Mizuno T. An analogue of the DnaJ molecular chaperone in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1994;91:1054–1058. doi: 10.1073/pnas.91.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Motojima-Miyazaki Y, Yoshida M, Motojima F. Ribosomal protein L2 associates with E. coli HtpG and activates its ATPase activity. Biochem. Biophys. Res. Commun. 2010;400:241–245. doi: 10.1016/j.bbrc.2010.08.047. [DOI] [PubMed] [Google Scholar]
- 59.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 60.Doyle SM, Shastry S, Kravats AN, Shih YH, Miot M, Hoskins JR, et al. Interplay between E. coli DnaK, ClpB and GrpE during protein disaggregation. J. Mol. Biol. 2015;427:312–327. doi: 10.1016/j.jmb.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat. Meth. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neria E, Fischer S, Karplus M. Simulation of activation free energies in molecular systems. J. Chem. Phys. 1996;105:1902–1921. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.