Abstract
Background
Differences in response to cancer treatments have been observed among racially and ethnically diverse gastric cancer patient populations. In the era of targeted therapy, mutation profiling of cancer is a crucial aspect of making therapeutic decisions. Mapping driver gene mutations for the gastric cancer patient population as a whole has significant potential to advance precision therapy.
Methods
Gastric cancer patient cases with sequencing data (total n=473) were obtained from The Cancer Genome Atlas (TCGA; n=295), Moffitt Cancer Center Total Cancer Care™ (TCC; n=33), and three published studies (n=145). Relevant somatic mutation frequency data were obtained from cBioPortal, TCC database and in-house analysis tool, and relevant publication
Results
We have found somatic mutation rates of several driver genes significantly vary between gastric cancer patients of Asian and Caucasian descent, with substantial variation across different geographic regions. Non-parametric statistical tests were performed to examine significant differences in protein-altering somatic mutations between Asian and Caucasian gastric cancer patient groups. Frequencies of somatic mutations of 5 genes were APC(Asian: Caucasian 6.06% vs. 14.40%, p=0.0076) ARIDIA(20.7% vs. 32.1%, p=0.01) KMT2A(4.04% vs. 12.35%, p=0.003) PIK3CA(9.6% vs. 18.52%, p=0.01) PTEN(2.52% vs. 9.05%, p=0.008), showing significant differences between Asian and Caucasian gastric cancer patients.
Conclusions
Our study has found significant differences in protein-altering somatic mutation frequencies in diverse geographic populations. In particular, we found that the somatic patterns may offer better insight and important opportunities for both targeted drug development and precision therapeutic strategies between Asian and Caucasian gastric cancer patients.
1. Introduction
Nearly one million new cases of gastric cancer (GC) are estimated to occur each year, making it the fifth most common malignancy in the world. (GLOBOCAN 2012)(1). There is substantial geographic variation in the incidence of gastric cancer, with the highest rates in East Asian and the lowest in North American populations(2). Certain environmental factors such as H. pylori infection, dietary factors, and smoking patterns may contribute to these disparities (3–5). The study of the genetic basis of gastric cancer, including host genetic susceptibility, has improved understanding of the pathogenesis of this disease and has highlighted the role of infection and chronic inflammation in gastric cancer.
Precision therapy research has opened up opportunities for cancer treatments, commercialization, and personal understanding on highly heterogeneous human tumors. Advances in high-throughput cancer genome sequencing and profiling technologies are rapidly transforming the development and approval of targeted agents, paving the way for a more precise therapeutic selection for individual patients. Multiple signaling pathways have been identified as key drivers of cancer patient outcome and therapeutic response through genetic and epigenetic aberrations, allowing for the expansion of gastric cancer classification from epidemiology to molecular disease biology.
Given the power and throughput of next generation sequencing, the application of this technology is driving the development of new therapies by targeting the most frequently occurring molecular abnormalities. However, much of our knowledge of somatic mutations have been obtained from tumors of Western Caucasian patient populations, while the greatest burden of gastric cancer is among Eastern Asian patient populations. This raises the question of whether gastric cancers from Asian- and Caucasian-descended patients exhibit different somatic genomic alterations, leading to an inadvertent disparity in development and application of new therapies.
2. Materials and Methods
2.1 Integrating multi-country sequencing somatic mutation data from TCGA
Mutation data of Hong Kong cases (N=100) (6) and Japanese cases (N=30) (7) were identified in genes relevant to gastric cancer (Table 1) using cBioPortal. Singapore (N=15) cases were obtained from the published study (8).
Table 1.
Similarities and differences between Asian and Caucasian gastric cancer patients. Shown are the mutation rates for 11 genes of interests to in each ethnicity.
| Gene | Asian (Alteration rates) | Caucasian (Alteration rates) | P-value |
|---|---|---|---|
| ACVR2A | 4.0% | 6.2% | 0.06 |
| APC | 6.0% | 14.4% | 0.007* |
| ARID1A | 20.0% | 32.1% | 0.01* |
| CDH1 | 13.1% | 10.7% | 0.52 |
| CTNNB1 | 5.5% | 8.2% | 0.37 |
| GLI3 | 11.1% | 13.2% | 0.61 |
| KMT2A | 4.0% | 12.4% | 0.003* |
| PIK3CA | 9.6% | 18.5% | 0.012* |
| PTEN | 2.5% | 9.1% | 0.008* |
| SMAD4 | 6.5% | 7.8% | 0.75 |
| TP53 | 51.5% | 44.4% | 0.17 |
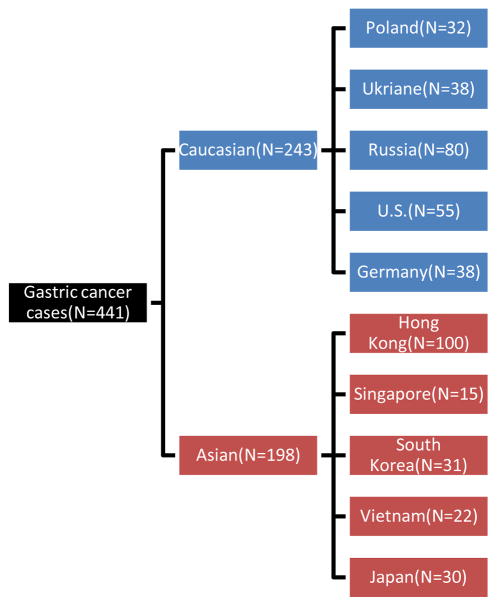
TCGA stomach adenocarcinoma (STAD) cases (N=295) were obtained with relevant clinical and somatic mutation (Level 2) data from the TCGA web portal (tcga-data.nci.nih.gov;June, 2014). These TCGA cases were further filtered with 1000 Genomes and re-annotated as described below. Mutation rates were calculated among patient groups defined by country of origin. TCGA identifier was used as a linker to clinical and genomic data. Finally, Germany (N=38), Poland (N=32), Russia (N=80), South Korea (N=31), Ukraine (N=38), U.S (N=22) and Vietnam (N=22) gastric cancer samples were obtained with mutation frequencies across relevant genes (Figure 1) (supplementary table 1).
Fig. 1.
Gastric cancer cases from diverse countries. *U.S. data are from TCGA U.S. and TCC cohort.
2.2 Patients and tissues, DNA extraction and quantification at Moffitt Cancer Center
Gastric cancer samples were identified at H. Lee Moffitt Cancer Center as part of a large population based study acquiring nearly 20,000 snap frozen, clinically characterized cancer specimens(9, 10). Gastric adenocarcinoma and signet-ring cell carcinoma (Supplementary Table 3) were all included.
Primary and metastatic samples from 33 gastric cancer patients (self-identified race = “white”) were available for analysis. In all cases, tissue and clinical data were collected on patients under institutional review board approval as part of the Total Cancer Care (TCC) project(9). Approval from Moffitt Cancer Center IRB was obtained to analyze clinical data for this study (Supplementary Table 2) from patients who consented to the TCC protocol and whose tumors were profiled with targeted sequencing.
All tumors were collected from curative survival resections and snap frozen in liquid nitrogen within 15–20 min of extirpation. Tumors then underwent a macro dissection quality control process to ensure >80% tumor was present in the specimen that underwent sequence analysis. DNA was then extracted from 33 gastric cancer specimens, followed by targeted sequencing using a custom designed Agilent Sure Select Capture, Agilent Technologies, Inc., Santa Clara, CA. 1,321 cancer-associated genes were selected by a joint committee (Merck Co., Inc. & Moffitt Cancer Center) for hybrid capture and sequencing. Capture probes for the 1,321 genes were based on the Agilent 50 MB Sure Select capture.
2.3 Targeted exome sequencing workflow and analysis at Moffitt Cancer Center
Tumor samples from Total Cancer Care were subjected to genomic capture (performed by BGI, Shenzhen using Sure Select custom designs targeting 1,321 genes, Agilent Technologies, Inc., Santa Clara, CA) and massively parallel sequencing (performed by BGI, Shenzhen using GAIIx, Illumina, Inc., San Diego, CA). Sequences were aligned to the hs37d5 human reference with the Burrows-Wheeler Aligner (BWA)(11). Insertion/deletion realignment, quality score recalibration, and variant identification were performed with the Genome Analysis ToolKit (GATK) (12). Sequence variants were annotated with ANNOVAR(13). Additional contextual information was incorporated, including allele frequency from the 1000 Genomes Project and the NHLBI Exome Sequence Project, in silico functional impact predictions, and observations from the Collection of Somatic Mutations in Cancer (COSMIC). Somatic mutations were enriched by filtering out variants observed in the 1000 Genomes Project and observed at >5% in an internal normal sample dataset (supplementary table 2).
2.4 Statistical analysis comparing Asian and Caucasian gastric cancer patient populations
We selected gastric cancer patients from Hong Kong, Vietnam, South Korea, Japan as Asian group (n=198), while gastric cancer patients from US, Germany, Poland, Russia, Ukraine and TCC were included in the Caucasian group (n=243). Somatic mutation frequency was calculated as GC patients bearing mutation for each gene divided by total number in each ethnic group (198 for Asian and 243 for Caucasian). We used exact binominal test to examine statistically significant differences in somatic mutation frequencies for 11 genes (supplementary table 4). Statistical analysis were performed using open source statistical software R (v3.1.0), and world maps were also drawn using R packages “rworldmap” and “rgdal”.
3. Results
Assessment of WES/WGS studies in Asian patients identified 11 genes that were consistently mutated in GC. Somatic mutation frequencies of five of those genes APC, ARID1A, KMT2A, PIK3CA, and PTEN, were significantly different between the Asian and Caucasian geographic populations (Table 1) (Supplementary figures). The mean frequency was higher in Caucasians compared to Asian patient populations for five genes. More detailed information for these genes and their somatic mutations is described below.
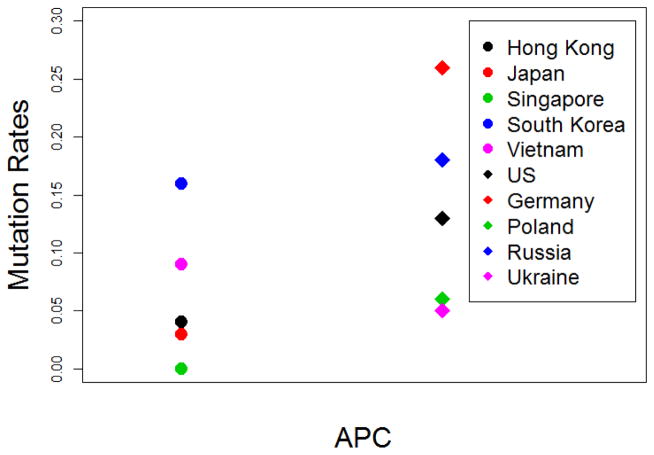
The mean frequency of APC alteration was more than twice as high in Caucasian than Asian patients (Figure 2), with the highest frequency in German patients and lowest in patients from Singapore. Within patients from Asian regions, the frequency was the highest in South Korea (16.1%) and lowest in Singapore (0%), whereas in Europe the frequency was the highest in Germany (26.3%) and lowest in Ukraine (5.3%).
Fig. 2.
APC mutation rates across diverse geographic area. Circles represent Asian countries while diamonds represent Caucasian countries.
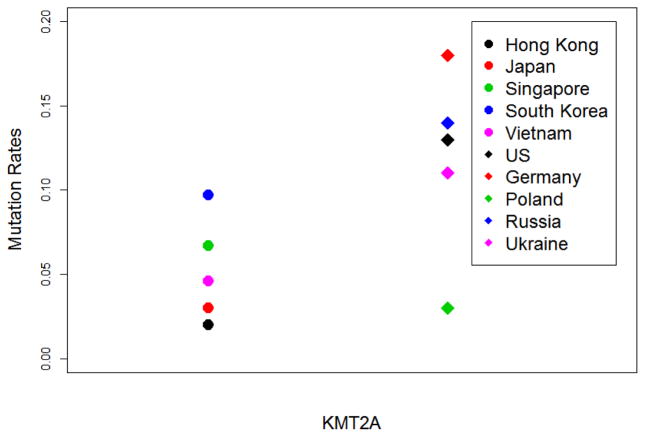
19 of 198 (9.6%) Asian gastric cancer patients had PIK3CA mutations, compared to 45 of 243 (18.5%) in Caucasian population (p=0.01). Amongst Asian countries, South Korea GC patients harbored the highest alteration frequency of APC, ARID1A, GLI3, PIK3CA and KMT2A (Figure 3). Further, PIK3CA, GLI3 and ARID1A mutation frequency are higher than any accessed cohorts. Chinese (Hong Kong) has the highest mutation rates of ACVR2A (8%) and PTEN (4%) amongst Asian countries, compared to the Asian means of 4% for ACVR2A and 2.5% for PTEN. Interestingly, other than China (Hong Kong), no other Asian GC patients harbored ACVR2A alterations. Although PTEN alterations were relatively frequent in Chinese patients compared to other Asian countries, it was altered less often than Caucasian patients (9.1%). Contrasting with the relatively high frequency of ACVR2A and PTEN, Chinese GC patients harbored the lowest frequency of KMT2A (2%) and PIK3CA (2%) alterations.
Fig. 3.
KMT2A mutation rates across diverse geographic area. Circles represent Asian countries while diamonds represent Caucasian countries.
Japanese GC patients harbored the lowest CTNNB1, GLI3, PTEN, SMAD4, and TP53 alterations (3%, 3%, 0%, 3% and 37% respectively) among Asian countries, compared to the mean values of 5.6%, 11.1%, 2.5%, 6.6% and 51.5% respectively, across the Asian region. Singapore GC patients’ alterations in ACVR2A, APC, CDH1 and PTEN were absent. Alterations in ARID1A were lowest among Asian countries (13.3% vs. Asian mean 21.7%). On the other hand, CTNNB1 alteration rates in Singapore GC patients were the highest among Asian GC patients (13.3% vs. Asian mean 5.6%).
Significant heterogeneity was seen across the Caucasian cohorts. However, consistent patterns could not be detected. German patients had the highest frequency of APC and KMT2A alterations, but lowest for CDH1. Polish GC patients harbored the highest CDH1 alteration rates while the lowest KMT2A and SMAD4 alteration rates; Ukrainian GC patients have the highest SMAD4, PIK3CA, GLI3 and lowest TP53, APC alteration rates. Caucasian patient population has larger variation of KMT2A alteration compared Asian group, ranging 3.1% to 18.4%, compared to 2% to 9.7% in Asian population. The mean KMT2A mutational frequency was three times higher in Caucasian compared to Asian GC patients (p=0.003) (Figure 3).
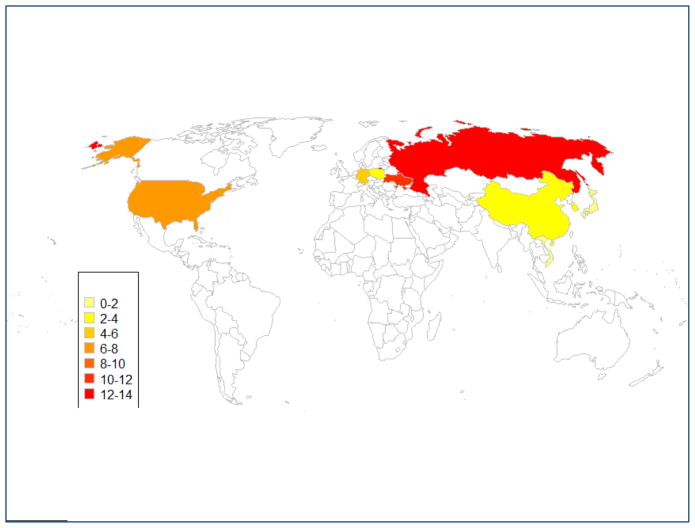
Dramatic differences were seen in PTEN gene, the mutation rates were 13.8% and 13.6% for Russia and American GC patients, while PTEN alterations were absent in Vietnamese, Singaporean and Japanese GC patients. PTEN mutation rates are higher among Caucasian gastric cancer patients compared to Asian cases. (p=0.008) (Figure 4).
Fig. 4.
PTEN mutation rates shown on the world map. Numbers on the left side of the figure indicate PTEN alteration rates.
4. Discussion
Understanding geographic differences in somatic mutation frequency is more than an academic interest and is important for rational global introduction of targeted cancer drugs due to the highly heterogeneous therapeutic responses in their worldwide applications. For example, from recent Phase II and III trials suggested that epidermal growth factor receptor (EGFR) inhibitors were more effective for Asian cancer patients than patients in other ethnicities, a characteristic that parallels a much higher incidence of EGFR-activating mutations among Asian patient populations (14–17). Consequently, clinical studies focusing on EGFR-targeted therapies have rapidly advanced based on the fact that Asian patients harboring the EGFR somatic mutation account for 30–40% of total patients, significantly higher than that of patients from European descents (18–21).
It has been observed thus far that ethnic factors are likely to contribute to the disappointing results in advanced gastric cancer, so these factors should be carefully taken into account when conducing global clinical trials across different ethnic populations (22–25). Stratified analysis by race/ethnicity may need to be performed in such global studies in order to maximize benefits of alternative precision therapeutic modalities. For instance, the phase III GRANITE-1, first global gastric antitumor trial with everolimus (NCT00879333), did not show a significant survival benefit from everolimus beyond BSC (best supportive care) for the worldwide cohort of advanced gastric cancer patients whose disease relapsed after one or two lines of systemic chemotherapy(22). However, when ethnic subgroups were examined, a 15% reduced risk of death was seen for Caucasian patients (U.S., Canada, Israel, Russia, Mexico, Colombia, Brazil, Venezuela, Australia and New Zealand) compared to patients in Asia (China, Hong Kong, Japan, Korea, Taiwan and Thailand) (22). Di Nicolantonio and colleagues have demonstrated that PIK3CA mutations can sensitize cancer cells to everolimus (26). Interestingly, our study demonstrated PIK3CA mutation rates is almost twice as high in Caucasian gastric cancer patients as they are Asian cases (18.5% vs. 9.6%, p= 0.012). This suggests a mechanistic basis behind the differential response to everolimus in Caucasian gastric cancer patients. Even if Asian patients with PIK3CA mutations might also have been more likely to respond to the drug, the effect could have been masked in the overall analysis by a low mutation frequency among Asian patients. Initial investigations on biomarkers in the PI3K/Akt/mTOR pathway are ongoing and its results are being eagerly awaited (22).
Unfortunately, in-depth understanding of the molecular underpinnings of gastric cancer is currently lagging compared to many other cancers of similar incidence and morbidity. Thanks to recent advance in NGS technology, common somatic alterations (PTEN, PIK3CA, and TP53) are rapidly being discovered, some of which are being pursued clinically. For instance, a multination Phase I study (US, Germany, Japan, South Korea, Switzerland and Taiwan) of PIK3CA inhibitor BYL719 in combination with the HSP90 inhibitor AUY922 in advanced gastric cancer patients carrying either a molecular alteration of PIK3CA or an amplification of HER2 (NCT01613950). Also, a Phase II trial of AZD5363 (Akt inhibitor) plus paclitaxel/AZD2014 (mTOR inhibitor) plus paclitaxel in biomarker negative (PIK3CA/MEK/RAS/TP53/MET) advanced gastric adenocarcinoma patients as second-line chemotherapy are currently enrolling patients (NCT02449655). A single-arm phase II study of AZD1775 (Wee1 G2 checkpoint serine/threonine protein kinase inhibitor) in combination with paclitaxel in patients with advanced gastric adenocarcinoma harboring TP53 mutation as a second-line chemotherapy (NCT02448329) is also currently recruiting participants. These studies should yield greater understanding of the translational impact of mutated biological pathways, in addition to the anticipated therapeutic benefits.
Many novel somatic gene targets (CDH1, ARID1A, KMT2A, ACVR2A, CTNNB1, GLI3, SMAD4, and APC) have also been identified to be of great interest (6–8). Our study demonstrated that Caucasian gastric cancer patients have higher mutation rates of APC, ARID1A, KMT2A, PIK3CA, and PTEN genes than Asian patients. The mechanism responsible for the high frequency of these mutations in Caucasian patients will be a subject of great interest. In the near future considerably larger Asian clinical trials will need to understand ethnic differences in these biomarkers and their somatic mutation frequencies. If frequencies of such biomarkers are lower in Asian populations (as we have shown for mutation rates in five genes), more patients may need to be screened for eligibility to complete sufficiently powered biomarker-driven trials.
Global gastric cancer drug development is largely performed in Western countries whilst considerable somatic based prognostic indicator models of gastric cancer were identified and developed in Asian countries. Tan et al. identified two major intrinsic genomic subtypes as a statistically significant covariate which is associated with survival time following adjuvant, 5-fluorouracil-based therapy (27). Cho et al. (28) identified and validated robust prognostic markers (CTNNB1, EXOSC3, TOP2A, LBA1, LZTR1 and CCL5) in Korean gastric cancer patients and developed a prognostic risk score that can be easily translated into clinic. Lei Z et al. identified three subtypes of gastric adenocarcinoma: proliferative, metabolic, and mesenchymal based on gene expression patterns (29). Results from detailed molecular and/or pathological GC studies from Asian populations are promising, but it remains questionable whether they can be fully extrapolated to Caucasian populations. Our study has shown that there can be significant biomarker frequency differences across populations, suggesting that the potential clinical impact of a biomarker may also vary across populations. Although personalized research into these differences has just begun, molecular models should be carefully considered when transforming into clinic.
The traditional approach of one-size fits all clinical trials and attempting to find a single optimal therapy to apply across gastric cancer has likely contributed to a slow progress on treating this disease with novel targeted drugs. The study from TCGA has enlightened understanding of this highly heterogeneous disease by cataloging genomic characteristics across a spectrum of gastric cancer patients from large international groups(30). Our analyses utilizing TCGA and other databases have found discordance of somatic mutations in specific clinically relevant genes between the two ethnic groups. Differences were also observed among countries with similar ethnic populations, suggesting that it also be important to consider local-regional differences as well as national/ethnic diversities.
There are several limitations in our current study. First, differences in mutation rates between different ethnic groups may also be due to different depths or other technologic aspects of sequencing methods. Also, although TCGA used similar sequencing and analysis methods for each sample, sample sizes and representative countries of Asian patient populations were limited. Despite these limitations, comparisons within TCGA showed the same trends as our overall analysis, which, at least partially confirmed the observations and analysis results obtained in our global comparison. It remains important for future large molecular cancer studies to include diverse patient populations in proportions that allow conclusions to be drawn. We believe that our findings help to understand the multifaceted nature of gastric cancer, underscore the importance of molecular-guided personalized medicine, and provide practical implications for further study and future personalized gastric cancer treatments.
Supplementary Material
Figure 1a APC mutation rates shown on the worldmap
Numbers on the left side of the figure indicate APC alteration rates.
Figure 1b ARID1A mutation rates shown on the worldmap
Numbers on the left side of the figure indicate ARID1A alteration rates
Figure 1c KMT2A mutation rates shown on the worldmap
Numbers on the left side of the figure indicate KMT2A alteration rates
Figure 1d PIK3CA mutation rates shown on the worldmap
Numbers on the left side of the figure indicate PIK3CA alteration rates
Figure 2a ARID1A mutation rates across diverse geographic area
Circles represent Asian countries while diamonds represent Caucasian countries
Figure 2b PIK3CA mutation rates across diverse geographic area
Circles represent Asian countries while diamonds represent Caucasian countries
Figure 2c PTEN mutation rates across diverse geographic area
Circles represent Asian countries while diamonds represent Caucasian countries
Supplementary Table 1 Multi-country sequencing somatic mutation data from TCGA gastric cancer study
Supplementary Table 2 Moffitt cancer center IRB approved somatic mutation data for gastric cancer samples
Supplementary Table 3 Moffitt cancer center gastric cancer patients’ clinicopathological subtype identification
Supplementary Table 4 Multi-country somatic mutation frequency statistical test for 11 genes
Key points.
The aim of the study was to find substantial ethnic and geographic variation of somatic mutations among Asian and Caucasian gastric cancer patients.
Our findings suggest that gastric cancer from Asian- and Caucasian-descended patients exhibit different somatic genomic alterations and help to understand multifaceted nature of gastric cancer, underlying the importance of molecular-guided precision medicine and provide practical clinical implications for further study in gastric cancer treatments.
Acknowledgments
The authors acknowledge the contributions of the investigators in the TCGA and TCC study for recruitment of patients and sample acquisition. Total Cancer Care® is enabled, in part, by the generous support of the DeBartolo Family, and we thank the many patients who so graciously provided data and tissue to the Total Cancer Care Consortium. Our study also received valuable assistance from the Cancer Informatics and Collaborative Data Services Core Facilities at the H. Lee Moffitt Cancer Center & Research Institute, an NCI designated Comprehensive Cancer Center, supported under NIH grant P30-CA76292.
Funding: Feifei Jia is supported by China Scholarship Council. This work was supported in part by Moffitt Cancer Center CCSG funding (P30-CA76292).
Footnotes
Compliance with Ethical Standards
Conflict of interest: The authors (FJ, JT, TK, JL YL & HM) declare that they have no competing interests.
Ethnical approval and informed consent: All patients declared informed consent and the studies were approved by Moffitt Cancer Center.
Authors’ Contributions
Conception and design: Howard L. McLeod, Jae K. Lee, Jamie K. Teer, Yi-jing He
Development of methodology: Feifei Jia, Jae K. Lee, Jamie K. Teer, Todd C. Knepper
Acquisition of data: Feifei Jia, Jamie K. Teer, Todd C. Knepper
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): Feifei Jia, Jamie K. Teer, Jae K. Lee, Howard L. McLeod
Writing, review, and/or revision of the manuscript: Feifei Jia, Howard L. McLeod, Jamie K. Teer, Jae K. Lee, Hong-hao Zhou, Todd C. Knepper, Yi-jing He
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): Feifei Jia, Yi-jing He, Todd C. Knepper
Study supervision: Howard L. McLeod, Jae K. Lee, Jamie K. Teer, Hong-hao Zhou, Yi-Jing He
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer Journal international du cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Kamangar F, Dores GM, Anderson WF. Patterns of Cancer Incidence, Mortality, and Prevalence Across Five Continents: Defining Priorities to Reduce Cancer Disparities in Different Geographic Regions of the World. Journal of Clinical Oncology. 2006;24:2137–50. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 3.Kamangar F, Qiao YL, Blaser MJ, Sun XD, Katki H, Fan JH, et al. Helicobacter pylori and oesophageal and gastric cancers in a prospective study in China. Br J Cancer. 2006;96:172–6. doi: 10.1038/sj.bjc.6603517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertuccio P, Praud D, Chatenoud L, Lucenteforte E, Bosetti C, Pelucchi C, et al. Dietary glycemic load and gastric cancer risk in Italy. Br J Cancer. 2009;100:558–61. doi: 10.1038/sj.bjc.6604894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abnet CC, Freedman ND, Kamangar F, Leitzmann MF, Hollenbeck AR, Schatzkin A. Non-steroidal anti-inflammatory drugs and risk of gastric and oesophageal adenocarcinomas: results from a cohort study and a meta-analysis. Br J Cancer. 2009;100:551–7. doi: 10.1038/sj.bjc.6604880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nature genetics. 2014;46:573–82. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 7.Kakiuchi M, Nishizawa T, Ueda H, Gotoh K, Tanaka A, Hayashi A, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nature genetics. 2014;46:583–7. doi: 10.1038/ng.2984. [DOI] [PubMed] [Google Scholar]
- 8.Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nature genetics. 2012;44:570–4. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 9.Fenstermacher DA, Wenham RM, Rollison DE, Dalton WS. Implementing personalized medicine in a cancer center. Cancer journal. 2011;17:528–36. doi: 10.1097/PPO.0b013e318238216e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeatman TJ, Mule J, Dalton WS, Sullivan D. On the eve of personalized medicine in oncology. Cancer research. 2008;68:7250–2. doi: 10.1158/0008-5472.CAN-08-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. 2009;25:14. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukuoka M, Yano S, Giaccone G, Tamura T, Nakagawa K, Douillard JY, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL 1 Trial) [corrected] Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2003;21:2237–46. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 15.Thatcher N, Chang A, Parikh P, Rodrigues Pereira J, Ciuleanu T, von Pawel J, et al. Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: results from a randomised, placebo-controlled, multicentre study (Iressa Survival Evaluation in Lung Cancer) Lancet. 2005;366:1527–37. doi: 10.1016/S0140-6736(05)67625-8. [DOI] [PubMed] [Google Scholar]
- 16.Perez-Soler R, Chachoua A, Hammond LA, Rowinsky EK, Huberman M, Karp D, et al. Determinants of tumor response and survival with erlotinib in patients with non--small-cell lung cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2004;22:3238–47. doi: 10.1200/JCO.2004.11.057. [DOI] [PubMed] [Google Scholar]
- 17.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, et al. Erlotinib in previously treated non-small-cell lung cancer. The New England journal of medicine. 2005;353:123–32. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, Ma S, Song X, Han B, Cheng Y, Huang C, et al. Gefitinib versus placebo as maintenance therapy in patients with locally advanced or metastatic non-small-cell lung cancer (INFORM; C-TONG 0804): a multicentre, double-blind randomised phase 3 trial. The Lancet Oncology. 2012;13:466–75. doi: 10.1016/S1470-2045(12)70117-1. [DOI] [PubMed] [Google Scholar]
- 19.Seto T, Kato T, Nishio M, Goto K, Atagi S, Hosomi Y, et al. Erlotinib alone or with bevacizumab as first-line therapy in patients with advanced non-squamous non-small-cell lung cancer harbouring EGFR mutations (JO25567): an open-label, randomised, multicentre, phase 2 study. The Lancet Oncology. 2014;15:1236–44. doi: 10.1016/S1470-2045(14)70381-X. [DOI] [PubMed] [Google Scholar]
- 20.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. The Lancet Oncology. 2010;11:121–8. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 21.Sekine I, Yamamoto N, Nishio K, Saijo N. Emerging ethnic differences in lung cancer therapy. Br J Cancer. 2008;99:1757–62. doi: 10.1038/sj.bjc.6604721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohtsu A, Ajani JA, Bai YX, Bang YJ, Chung HC, Pan HM, et al. Everolimus for previously treated advanced gastric cancer: results of the randomized, double-blind, phase III GRANITE-1 study. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2013;31:3935–43. doi: 10.1200/JCO.2012.48.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Satoh T, Xu RH, Chung HC, Sun GP, Doi T, Xu JM, et al. Lapatinib plus paclitaxel versus paclitaxel alone in the second-line treatment of HER2-amplified advanced gastric cancer in Asian populations: TyTAN--a randomized, phase III study. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2014;32:2039–49. doi: 10.1200/JCO.2013.53.6136. [DOI] [PubMed] [Google Scholar]
- 24.Lordick F, Kang YK, Chung HC, Salman P, Oh SC, Bodoky G, et al. Capecitabine and cisplatin with or without cetuximab for patients with previously untreated advanced gastric cancer (EXPAND): a randomised, open-label phase 3 trial. The Lancet Oncology. 2013;14:490–9. doi: 10.1016/S1470-2045(13)70102-5. [DOI] [PubMed] [Google Scholar]
- 25.Waddell T, Chau I, Cunningham D, Gonzalez D, Okines AF, Okines C, et al. Epirubicin, oxaliplatin, and capecitabine with or without panitumumab for patients with previously untreated advanced oesophagogastric cancer (REAL3): a randomised, open-label phase 3 trial. The Lancet Oncology. 2013;14:481–9. doi: 10.1016/S1470-2045(13)70096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Nicolantonio F, Arena S, Tabernero J, Grosso S, Molinari F, Macarulla T, et al. Deregulation of the PI3K and KRAS signaling pathways in human cancer cells determines their response to everolimus. The Journal of clinical investigation. 2010;120:2858–66. doi: 10.1172/JCI37539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan IB, Ivanova T, Lim KH, Ong CW, Deng N, Lee J, et al. Intrinsic subtypes of gastric cancer, based on gene expression pattern, predict survival and respond differently to chemotherapy. Gastroenterology. 2011;141:476–85. 85 e1–11. doi: 10.1053/j.gastro.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho JY, Lim JY, Cheong JH, Park YY, Yoon SL, Kim SM, et al. Gene expression signature-based prognostic risk score in gastric cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:1850–7. doi: 10.1158/1078-0432.CCR-10-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei Z, Tan IB, Das K, Deng N, Zouridis H, Pattison S, et al. Identification of molecular subtypes of gastric cancer with different responses to PI3-kinase inhibitors and 5-fluorouracil. Gastroenterology. 2013;145:554–65. doi: 10.1053/j.gastro.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 1a APC mutation rates shown on the worldmap
Numbers on the left side of the figure indicate APC alteration rates.
Figure 1b ARID1A mutation rates shown on the worldmap
Numbers on the left side of the figure indicate ARID1A alteration rates
Figure 1c KMT2A mutation rates shown on the worldmap
Numbers on the left side of the figure indicate KMT2A alteration rates
Figure 1d PIK3CA mutation rates shown on the worldmap
Numbers on the left side of the figure indicate PIK3CA alteration rates
Figure 2a ARID1A mutation rates across diverse geographic area
Circles represent Asian countries while diamonds represent Caucasian countries
Figure 2b PIK3CA mutation rates across diverse geographic area
Circles represent Asian countries while diamonds represent Caucasian countries
Figure 2c PTEN mutation rates across diverse geographic area
Circles represent Asian countries while diamonds represent Caucasian countries
Supplementary Table 1 Multi-country sequencing somatic mutation data from TCGA gastric cancer study
Supplementary Table 2 Moffitt cancer center IRB approved somatic mutation data for gastric cancer samples
Supplementary Table 3 Moffitt cancer center gastric cancer patients’ clinicopathological subtype identification
Supplementary Table 4 Multi-country somatic mutation frequency statistical test for 11 genes