Abstract
Chromatin remodelers are ATP-dependent enzymes that are critical for reorganizing and repositioning nucleosomes in concert with many basic cellular processes. For the Chromodomain Helicase DNA Binding Protein 1 (Chd1) remodeler, nucleosome sliding has been shown to depend on DNA flanking the nucleosome, transcription factor binding at the nucleosome edge, and the presence of the histone H2A/H2B dimer on the entry side. Here we report that Chd1 is also sensitive to the sequence of DNA within the nucleosome, and slides nucleosomes made with the 601 Widom positioning sequence asymmetrically. Kinetic and equilibrium experiments show that poly(dA:dT) tracts perturb remodeling reactions if within one and a half helical turns of superhelix location 2 (SHL2), where the Chd1 ATPase engages nucleosomal DNA. These sequence-dependent effects do not rely on the Chd1 DNA-binding domain and are not due to differences in nucleosome affinity. Using site-specific cross-linking, we show that internal poly(dA:dT) tracts do not block engagement of the ATPase motor with SHL2, yet they promote multiple translational positions of DNA with respect to both Chd1 and the histone core. We speculate that Chd1 senses the sequence-dependent response of DNA as the remodeler ATPase perturbs the duplex at SHL2. These results suggest that the sequence sensitivity of histones and remodelers occurs at unique segments of DNA on the nucleosome, and therefore can work together or in opposition to determine nucleosome positions throughout the genome.
Keywords: Chromatin remodeling, nucleosome sliding, Snf2 ATPase, poly(dA:dT) tracts, Widom 601 positioning sequence
Introduction
A core feature of eukaryotic genomes is the extensive packaging of DNA into nucleosomes. Many nucleosomes are specifically positioned throughout the genome by chromatin remodelers, which dictate remarkably predictable placement of nucleosomes in gene promoters and can pack intragenic nucleosomes into evenly spaced arrays 1. Chromatin remodelers can be categorized into distinct families, which exhibit unique biochemical properties and appear specialized for particular biological settings 2. These remodelers are uniquely influenced by a variety of inputs including transcription factor binding, DNA linker length, and DNA sequence, often competing and collaborating with each other 3–7. A key question is therefore how different remodelers select preferred nucleosome substrates, and what nucleosome features either activate or inhibit remodeler actions.
In recent years, there has been a growing appreciation that DNA sequence directly impacts chromatin remodeling. In one study, seven different remodelers were found to reposition a starting pool of mononucleosomes into different products, with each remodeler showing characteristic patterns of nucleosome repositioning 8. For the ISWI-type remodeler ACF, the authors found that the remodeling pattern was strongly influenced by a particular sequence of DNA within the nucleosome. A separate study on ACF, however, failed to observe evidence of sequence-directed sliding 9. Although the reason for this discrepancy is unclear, one possibility is that remodeling is biased by the sequence context of a particular DNA segment on the nucleosome. One example where sequence context appears paramount is for the INO80 remodeler. In a genome-wide study of nucleosome positioning in Saccharomyces cerevisiae, INO80 was discovered to be responsible for the precise positioning of +1 nucleosomes, and could recapitulate in vivo nucleosome positions through DNA sequence alone 6. Although further work will be needed to tease out the relationship between DNA sequence and INO80 action, nucleosome positioning by INO80 correlated with a predicted greater twist of DNA over the dyad of the +1 nucleosome, suggesting a mechanism of responding to DNA shape on the nucleosome. Sequence-directed remodeling also guides action of the SWI/SNF-type remodeler RSC. In S. cerevisiae, RSC plays a primary role in removing and/or shifting nucleosomes away from nucleosome-free regions (NFRs) in gene promoters 10–12. NFRs in yeast promoters were shown to be commonly associated with poly(dA:dT) tracts 13, 14, which, given their unique structural characteristics of a narrow minor groove, bifurcated hydrogen bond base pairing, and increased twist 15–18, gave rise to the idea that nucleosomes would intrinsically disfavor assembly on DNA containing poly(dA:dT) tracts 19, 20. However, poly(dA:dT) tracts were found to wrap into nucleosomes with surprisingly little or no energetic penalty 21–23, and nucleosomes possessing these tracts exhibited rather canonical conformations of DNA 24, 25. Poly(dA:dT) tracts were instead found to exert their effects by directing RSC, resulting in preferential shifting of nucleosomes off of poly(dA:dT) tracts 6, 26.
In this study, we investigate the effects of DNA sequence on the Chd1 chromatin remodeler. Chd1 is known for preferentially shifting mononucleosomes toward the center of short DNA fragments, which is believed to underlie its ability to evenly distribute nucleosomes in arrays 27–29. To date, directional sliding by Chd1 has been shown to arise from responding to three distinct nucleosome characteristics: the length and availability of DNA flanking the nucleosome 28, 30, the presence of a transcription factor bound at the nucleosome edge 4, and the absence of the histone H2A/H2B dimer on the entry side of hexasomes 31. Our work here reveals that Chd1 activity is also impacted by the DNA sequence adjacent to SHL2. As we demonstrate using internal poly(dA:dT) tracts, responding to sequence elements within nucleosomal DNA was not solely due to binding defects and did not require the Chd1 DNA-binding domain. While the Chd1 ATPase was not blocked from engaging DNA at SHL2, cross-linking experiments suggested that internal poly(dA:dT) tracts promoted multiple translational positions of nucleosomal DNA. We speculate that the influence of DNA sequence on remodeling reflects a sensitivity of Chd1 to DNA energetics at intermediate steps in the catalytic cycle. The location of sequence sensitivity appears to be distinct from regions where histones most tightly grip DNA within the nucleosome. On a genome-wide scale, this independence would allow thermodynamically preferred positions to either be reinforced or overridden by chromatin remodelers such as Chd1.
Results
The Chd1 remodeler shows a directional preference in sliding nucleosomes made with the Widom 601 sequence
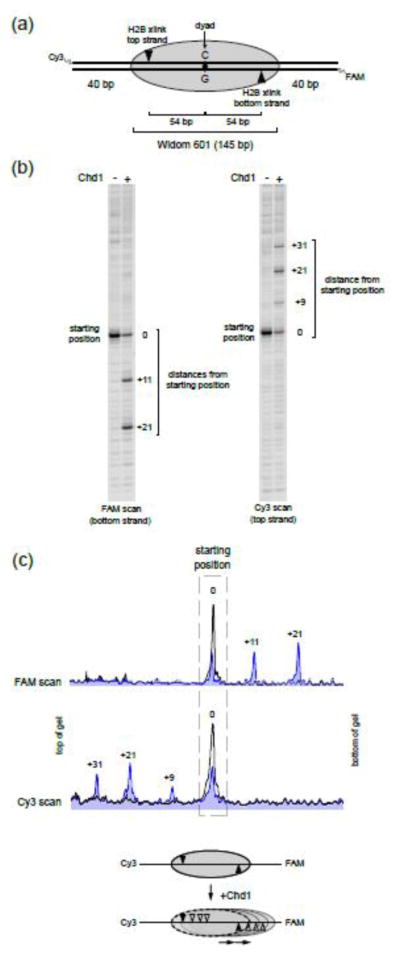
Chd1 has been shown to preferentially slide histone octamers of end-positioned mono-nucleosomes toward the center of short DNA fragments 28, 30. Since Chd1 acts throughout the genome and its DNA-binding domain appears to lack sequence-specificity 32, we expected that nucleosome sliding with equal lengths of DNA flanking each side of the nucleosome would produce a symmetric distribution of the histone octamer about the center of the DNA fragment. Using the Widom 601 sequence to initially position nucleosomes, we unexpectedly found that Chd1 sliding was biased in one direction (Fig. 1). Nucleosome positions were determined using a cross-linking technique called histone mapping 33. With this technique, an introduced cysteine (H2B-S53C) that is adjacent to nucleosomal DNA is labeled with the photoactivatable cross-linker 4-azidophenacyl bromide (APB). The formation of an APB adduct on DNA bases allows for generating abasic sites, resulting in cleavage of the DNA backbone and thus revealing the locations of cross-linked sites at base pair (bp) resolution 34. The Widom 601 positioning sequence is asymmetric, with a G:C base pair on the dyad. Here we refer to the orientation of the Widom 601 with the dyad C base on the top strand, and G on the bottom strand (Fig. 1a). DNA sequence influences the efficiency of histone-DNA cross-linking and may therefore bias interpretation of histone mapping experiments. For histone mapping with H2B(S53C), however, each copy of H2B cross-links to only one DNA strand ~19 bp from the nucleosome edge 33, thereby allowing the two sides of the nucleosome to be independently monitored. For each side, nucleosome sliding reactions with Chd1 produced cross-linking patterns with ~10–11 bp periodicity, as expected for the strong rotational dominance of the Widom 601. As shown with double-labeled DNA, both sides of the nucleosome reported a similar and marked bias for the histone octamer shifting toward the right side of the 601 sequence (Fig. 1b,c).
Figure 1. The Chd1 remodeler distributes Widom 601 nucleosomes asymmetrically.
(a) Diagram of a centered nucleosome on the Widom 601 positioning sequence. The orientation of the 601 sequence is defined with a dyad having a cytosine (C) on the top strand. As indicated, each H2B(S53C) cross-link occurs only on one strand.
(b) Chd1 shifts centered nucleosomes preferentially to the right side. Shown are two scans from the same remodeling reaction, which reveal the locations of H2B(S53C) cross-links for each DNA strand. For these reactions, 150 nM nucleosome was incubated plus or minus 50 nM Chd1 and 2 mM ATP for 64 min. This gel is representative of four independent experiments.
(c) Intensity scans of the gels shown in (b). The black trace represents the cross-linking distribution before remodeling, and blue trace is 64 min after addition of ATP and Chd1. Diagram below summarizes direction of octamer movement by Chd1.
Poly(dA:dT) tracts in the vicinity of SHL2 alter equilibrium nucleosome positions resulting from Chd1 remodeling
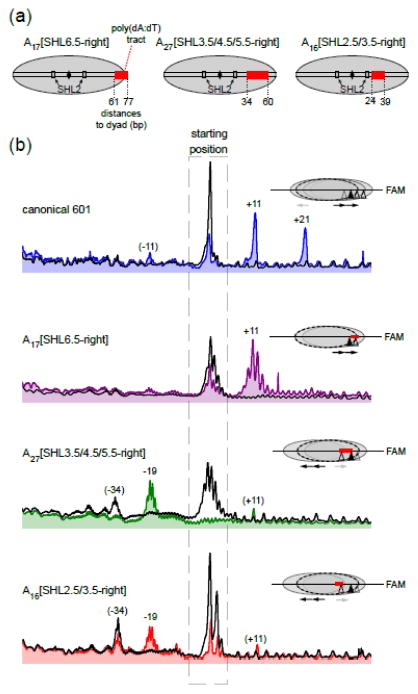
To investigate the role of DNA sequence in sliding directionality, we introduced poly(dA:dT) tracts into the 601 positioning sequence. Poly(dA:dT) tracts were substituted at three regions on the right side of the 601, spanning from the edge of the nucleosome to a more interior position extending up to 24 bp from the nucleosome dyad. We use the notation of superhelix locations (SHLs), defined with the first nucleosome crystal structure 35, to describe the primary location of poly(dA:dT) tracts. SHLs identify where the major groove faces the histone core, with the dyad defined as SHL0 (zero). Each additional helical turn of DNA away from the dyad corresponds with increasing SHL numbering, with the intervening segments where the minor groove faces the histone core referred to with the traditional 0.5n SHL numbering (SHL0.5, SHL1.5, etc). Rather than positive or negative SHL numbering as is sometimes used, we instead refer to each SHL position as being to the right or left of the dyad, according to the orientation where the top strand has a cytosine at the 601 dyad. For these studies, the tracts were organized such that the poly(A)-containing strand was always 5 with respect to the nucleosome dyad. In addition to tract length, the 601 constructs are referred to by the histone contact sites encompassed by each tract: A17[SHL6.5-right], A27[SHL3.5/4.5/5.5-right], and A16[SHL2.5/3.5-right] (Fig. 2a). The tracts of A17[SHL6.5-right] and A16[SHL2.5/3.5-right] are at similar distances from the dyad as studied by Anderson and Widom 36, though on the other side of the 601 sequence.
Figure 2. The presence of internal poly(dA:dT) tracts bias nucleosome sliding in the opposite direction.
(a) Schematic illustrating the locations of poly(dA:dT) tracts used in this study. Numbers indicate distances (bp) to dyad. Superhelix location 2 (SHL2), where Chd1 and several other chromatin remodelers have been shown to act on the nucleosome, is also highlighted.
(b) Cross-linking distributions of 40-N-40 nucleosomes before and after sliding by Chd1, monitored with FAM (bottom strand) scans. Black traces show starting nucleosome positions prior to remodeling, and colored traces show distributions 32 min after addition of 2 mM ATP and 300 nM Chd1. The positions of peaks that are weak but reproducibly show up in different experiments are given in parentheses. Scans are oriented with the bottom of the gel on the right. These scans are representative of three or more experiments.
See Supplementary Figure S1 for the gel and Supplementary Figure S2 for scans of the other DNA strand.
After incubation with Chd1 and ATP, each nucleosome containing a poly(dA:dT) insertion produced a cross-linking pattern distinct from canonical 601. For the centered A17[SHL6.5-right] nucleosome, the FAM scan, which reports on the right side of the nucleosome, only showed an 11 bp shift to the right, in contrast to the 11 and 21 bp pattern observed with canonical 601 (Fig. 2b and S1). However, rather than distinct positioning of the nucleosome, this difference may reflect a change in cross-linking efficiency, as a rightward shift of the histone octamer would put the cross-linking position of H2B on the poly(dA:dT) tract itself. Indeed, the Cy5 scan, reporting on the left side of the nucleosome, showed a pattern similar to the canonical 601 (Fig. S2). Thus, the equilibrium distribution of the histone octamer did not appear significantly different for the 40-N-40 A17[SHL6.5-right] nucleosome compared with the canonical 601 nucleosome.
In contrast to A17[SHL6.5-right], 40-N-40 nucleosomes made with both A27[SHL3.5/4.5/5.5-right] and A16[SHL2.5/3.5-right] were remodeled to different equilibrium positions than the canonical 601 sequence, favoring repositioning in the opposite direction (Fig. 2b). For A27[SHL3.5/4.5/5.5-right], in addition to generating a small population 11 bp to the right, Chd1 shifted the histone octamer to the left by 19 bp. A similar, though weaker pattern, was also observed with the Cy5 label, consistent with a 19 bp shift to the left side of A27[SHL3.5/4.5/5.5-right] 40-N-40 (Fig. S2). For A16[SHL2.5/3.5-right], Chd1 sliding yielded a similar distribution as A27[SHL3.5/4.5/5.5-right], with a dominant histone octamer position 19 bp to the left (Fig. 2b and S2). For both A16[SHL2.5/3.5-right] and A27[SHL3.5/4.5/5.5-right], a peak at 34 bp on the left side of the 601 sequence was present in the starting material, prior to addition of Chd1. This pre-shifted species, which appears to be favored by these two internal poly(dA:dT) tracts, was largely maintained in A16[SHL2.5/3.5-right] after sliding by Chd1, perhaps further indicating a preference for sliding the histone octamer to the left.
Previous work with Chd1 showed that directional nucleosome sliding relies on the DNA-binding domain 30, 37. Deletion of the Chd1 DNA-binding domain greatly diminishes both sliding activity and the preference of Chd1 to slide nucleosomes away from DNA ends 30, 38. We therefore tested a Chd1 construct lacking the DNA-binding domain (residues 118–1014, referred to as Chd1[ΔDBD]) to determine whether the directional sliding in response to internal poly(dA:dT) tracts required the DNA-binding domain. Compared to reactions carried out with Chd1 possessing the DBD, repositioning reactions with Chd1[ΔDBD] also showed a marked difference between canonical 601 and A27[SHL3.5/4.5/5.5-right] nucleosomes, with the poly(dA:dT) tract corresponding with a prominent +19 bp shift to the left (Fig. 3 and S3). While all reactions with Chd1[ΔDBD] displayed increased retention of the starting material, consistent with weaker activity, a strong sliding defect was observed for 40-N-40 A16[SHL2.5/3.5-right] nucleosomes, which were not detectably shifted by Chd1[ΔDBD]. These results confirm that internally positioned poly(dA:dT) tracts can interfere with nucleosome sliding by Chd1, and reveal that this sequence bias does not require the Chd1 DNA-binding domain.
Figure 3. Internal poly(dA:dT) tracts influence nucleosome sliding by Chd1 independently of the DNA-binding domain.
Cross-linking distributions of 40-N-40 nucleosomes before and after addition of Chd1[ΔDBD]. Black traces show starting nucleosome positions prior to remodeling, and colored traces show distributions 30 min after addition of 2mM ATP and 300 nM Chd1[ΔDBD]. Scans are oriented with the bottom of the gel on the right. These scans are representative of three or more experiments. Cy5 (top strand) scans are shown in Supplementary Figure S3.
Poly(dA:dT) tracts do not block Chd1 remodeling
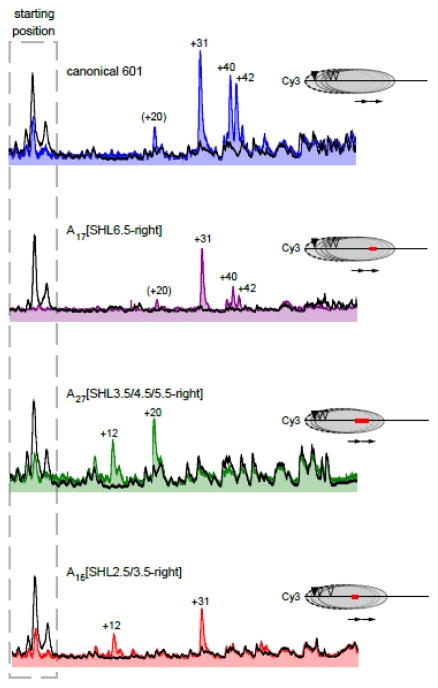
Like SWI/SNF- and ISWI-type remodelers, Chd1 shifts nucleosomes by translocating DNA at the internal SHL2 position on the nucleosome 30, 39–41. Due to two-fold symmetry of the histone core, an SHL2 position is located on either side of the dyad, and the side where the remodeler acts determines the direction the histone core will be shifted. The observation that Chd1 shifts nucleosomes away from poly(dA:dT) tracts indicates that the remodeler acts at the SHL2 on the opposite side of the nucleosome. The closest edges of A27[SHL3.5/4.5/5.5-right] and A16[SHL2.5/3.5-right] are ~15 and ~5 bp from the right SHL2, respectively, and action at this SHL2 would pull the poly(dA:dT) tracts toward the dyad, placing this sequence directly against the Chd1 ATPase motor. One potential explanation for why poly(dA:dT) tracts were not shifted onto SHL2 for 40-N-40 nucleosomes is that the unique structure of the poly(dA:dT) tract cannot easily be accommodated by the active site of Chd1, blocking DNA translocation of the remodeler at SHL2.
To explore this possibility, we generated 0-N-80 nucleosomes that should only allow movement of the histone octamer toward the right side, onto the tracts. With the canonical 601 nucleosomes, Chd1 shifted the histone octamer 20 bp, 31 bp, and 40/42 bp onto the 80 bp flanking DNA on the right side. As expected, the A17[SHL6.5-right] nucleosomes also populated these positions at equilibrium (Fig. 4 and S4). For A16[SHL2.5/3.5-right] and A27[SHL3.5/4.5/5.5-right] nucleosomes, Chd1 sliding also was able to move the histone octamer to the right, but with altered equilibrium distributions from that observed with 601. For A27[SHL3.5/4.5/5.5-right], histones primarily occupied positions 12 bp and 20 bp from the starting location, which would position the edge of the poly(dA:dT) tract around SHL1.5, such that the tract would extend over the SHL2 position where the Chd1 ATPase acts. For A16[SHL2.5/3.5-right], histones were shifted up to 31 bp, corresponding with translocation of the entire poly(dA:dT) tract past SHL2. These results indicate that, with a lack of flanking DNA on the opposing side, Chd1 can shift nucleosomes further on top of poly(dA:dT) tracts, and that these tracts therefore are not absolute barriers for remodeler action at SHL2.
Figure 4. Chd1 can pull internal poly(dA:dT) tracts further onto the nucleosome.
Cross-linking distributions of 0-N-80 nucleosomes before and after sliding by Chd1, using Cy3 (top strand) scans. Black traces show starting nucleosome positions prior to remodeling, and colored traces show distributions 16 min (for A17[SHL6.5-right]) or 64 min (all others) after addition of 2 mM ATP and 50 nM Chd1. The right side of each scan corresponds to the top of the gel. These scans are representative of three or more experiments. FAM (bottom strand) scans are shown in Supplementary Figure S4.
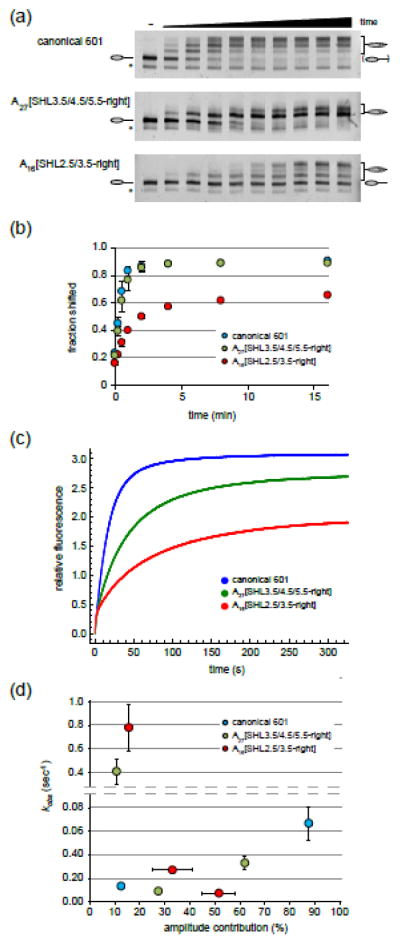
A poly(dA:dT) tract adjacent to SHL2 can slow down nucleosome sliding by Chd1
Although Chd1 can shift poly(dA:dT) tracts through SHL2, preferred nucleosome sliding in the opposite direction might be expected if poly(dA:dT) slowed down remodeling when close to SHL2 where the ATPase motor acts. To directly test this, we first analyzed nucleosome sliding reactions by native gels. End-positioned nucleosomes migrate more quickly through native gels than more centrally-positioned nucleosomes, allowing different nucleosome species to be followed over time. Consistent with histone mapping experiments, nucleosomes possessing internal poly(dA:dT) tracts were shifted by Chd1 to different equilibrium positions (Fig. 5a). As shown in Figure 5b, movement away from the end-positioned A27[SHL3.5/4.5/5.5-right] nucleosome occurred at a similar rate to that of the canonical 601 indicating that initial movement away from the end-position for this construct was not inhibited under these conditions. In contrast, nucleosome sliding of A16[SHL2.5/3.5-right] was significantly slower, and additionally failed to redistribute a large fraction of nucleosomes away from the starting position. This is consistent with the retention of a significant amount of starting material for the centered A16[SHL2.5/3.5-right] nucleosome as monitored by Cy5 (Fig. S2). For these reactions, we assume that the significant level of end-positioned nucleosomes is not due to a large immobile fraction, but instead results from nucleosomes dynamically shifted back toward the starting position. In a previous study using Lac repressor bound at the nucleosome edge, we observed a similar behavior of Chd1 sliding nucleosomes back to their starting position 4. In that study, we showed that Lac repressor acts as a barrier when bound to DNA being pulled onto the nucleosome, effectively slowing down the rate of sliding and thus favoring sliding in the opposite direction. We believe that the A16[SHL2.5/3.5-right] tract may be having a similar barrier effect, favoring action of Chd1 on the other side of the nucleosome and thus significantly repopulating the starting position of the nucleosome. These experiments were performed with sub-saturating concentrations of remodeler, and so one possibility was that the slower sliding of A16[SHL2.5/3.5-right] was due to a decreased binding affinity of Chd1. Alternatively, slower sliding by Chd1 could indicate that the poly(dA:dT) tract had a catalytic effect on the remodeling process.
Figure 5. Internal poly(dA:dT) tracts affect the rate of nucleosome sliding by Chd1.
(a) Shown are nucleosome sliding reactions, analyzed on native acrylamide gels. End-positioned 0-N-80 nucleosomes (150 nM) were incubated with Chd1 (50 nM) for 0, 0.25, 0.5, 1, 2, 4, 8, 16, 32, and 64 min. Nucleosomes were visualized using a DNA FAM label. Asterisks denote hexasomes, which were not shifted. Gels are representative of three independent experiments.
(b) Quantification of native gel nucleosome sliding experiments, such as those shown in (a). Each point is the average of three experiments, and error bars (sometimes obscured by the symbols) give the standard deviations.
(c) Chd1 nucleosome sliding reactions monitored with Cy3B-Dabcyl static quenching. Stopped flow experiments were carried out using 0-N-80 nucleosomes (10 nM) and saturating (600 nM) Chd1 in the presence of 25 μM ATP. Each trace, which is the average of 3–6 technical replicates, is representative of four or more independent experiments.
(d) Analysis of stopped flow nucleosome sliding rates. Observed rates from double (canonical 601) and triple exponential fits (poly(dA:dT) tract variants) are given along the y-axis, with the corresponding amplitudes along the x-axis. The reported values are the averages from three or more independent experiments, with standard deviations shown with error bars that are sometimes obscured by symbols.
To determine if the slower rate for A16[SHL2.5/3.5-right] was due to a catalytic or a binding effect, we monitored the kinetics of sliding for 0-N-80 nucleosomes with saturating Chd1 using a fluorescence-quenching assay 31. In this assay, Dabcyl was attached to the short DNA end of an 0-N-80 nucleosome, which placed it close enough to quench a Cy3B fluorophore attached to histone H2A at T120C. Nucleosome movement onto the 80 bp flanking DNA shifts the short DNA end away from the nucleosome core, decreasing static quenching of Cy3B as observed by recovery of fluorescence over time. Due to initial challenges in recording early time points with high Chd1 concentrations, the reactions were slowed down by lowering ATP concentration to 25 μM, which has been standard practice for kinetic analysis of other remodelers 5, 9, 42. Titration of Chd1 under limiting ATP showed that 600 nM remodeler yielded maximum rates (Fig. S5). With saturating remodeler, the progress curves for 0-N-80 nucleosomes with canonical 601, A27[SHL3.5/4.5/5.5-right], and A16[SHL2.5/3.5-right] were all distinct (Fig. 5C), indicating that differences in nucleosome binding alone could not explain behavior of the poly(dA:dT) tracts.
Compared to nucleosomes containing the canonical 601, introduction of a poly(dA:dT) tract close to SHL2 slowed down sliding by Chd1. Progress curves for the canonical Widom 601 were best fit with a double exponential rise, with the slower rate (0.013 ± .002s−1) contributing only 12 ± 1% of the amplitude compared to 88 ± 1% for the faster rate (0.067 ± .014s−1; Fig. 5D). Unlike the canonical 601, fitting showed that progress curves for A27[SHL3.5/4.5/5.5-right] and A16[SHL2.5/3.5-right] better matched a triple exponential rise. Interestingly, although the reactions were slower to complete overall, one of the three rates for both A27[SHL3.5/4.5/5.5-right] and A16[SHL2.5/3.5-right] was notably faster than canonical 601 (Fig. 5D). These faster rates had relatively small contributions to the overall amplitude (11 ± 1% and 16 ± 2% for A27[SHL3.5/4.5/5.5-right] and A16[SHL2.5/3.5-right], respectively), and instead the slower rates were dominant: 0.033 ± .005 s−1 of A27[SHL3.5/4.5/5.5-right] contributed 62 ± 4%, and 0.007 ± .0005 s−1 of A16[SHL2.5/3.5-right] contributed 52 ± 7%. The increased number of terms required to fit the progress curves suggests that the presence of the poly(dA:dT) tracts affected one or more rate-limiting steps in the nucleosome sliding reaction. The slower overall sliding suggests that poly(dA:dT) tracts near SHL2 pose an energetic barrier to Chd1 action. However, with the appearance of a faster rate, it also seems likely that one or more steps of the sliding reaction may be accelerated by an internal poly(dA:dT) tract. Although the complexity of the sliding reaction precludes a detailed interpretation of these kinetic data at present, the notably altered progress curves indicate that Chd1 is sensitive to the nature of DNA sequence adjacent to the location where the ATPase motor engages with the nucleosome.
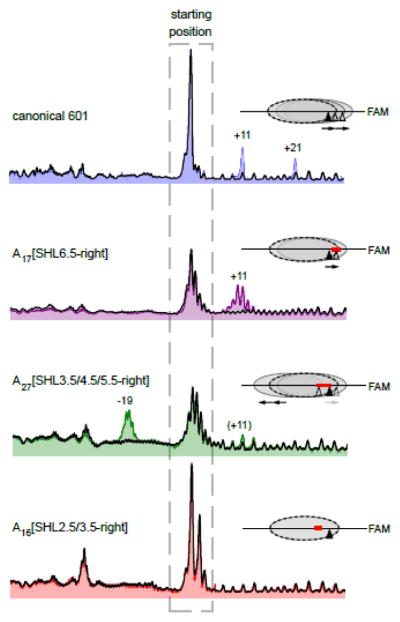
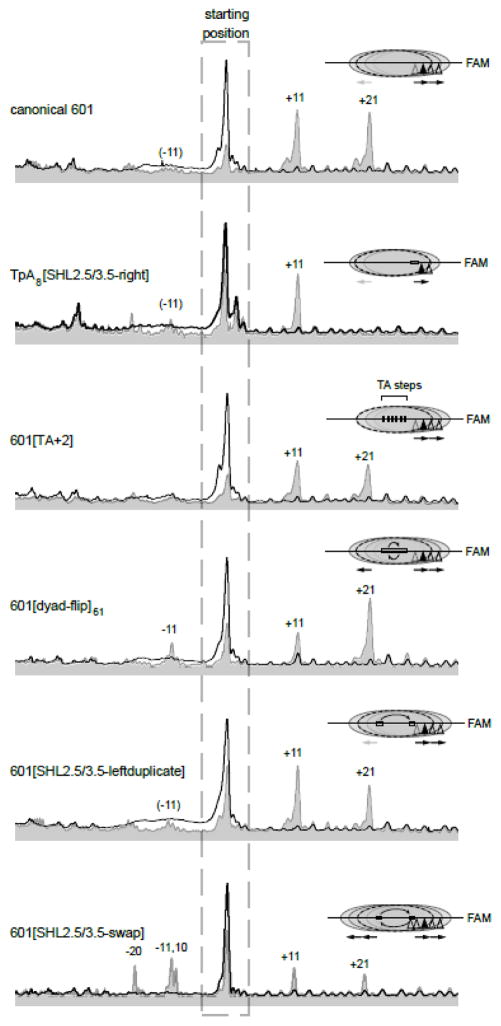
Given the expected rigidity of the poly(dA:dT) tract, we reasoned that a flexible sequence, which should be more easily accommodated on the nucleosome, would likely not interfere with sliding by Chd1. We therefore generated a 40-N-40 nucleosome containing a 16 nt tract of repeating TpA dinucleotide steps at the same location as A16[SHL2.5/3.5-right], referred to as (TpA)8[SHL2.5/3.5-right] (Fig. 6). Unexpectedly, we found that although the insertion of this flexible sequence did allow sliding in the same direction as canonical 601, the distribution of shifted species was altered. While canonical 601 had two shifted species, at 11bp and 21bp, (TpA)8[SHL2.5/3.5-right] only had one shifted species at 11bp (Fig. 7). This shifted species correlates with placement of the (TpA)8 tract directly on top of SHL2. Therefore, although it favored repositioning away from the starting position, the lack of a further shift suggests that having a highly flexible sequence at SHL2 may disfavor sliding by Chd1.
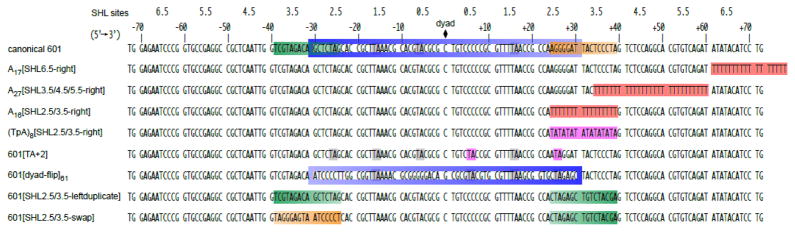
Figure 6. Sequence variants of the Widom 601 used in this study.
Shown are all of the Widom 601 variants described in this study, aligned and numbered based on the the central dyad. Coloring highlights regions of the 601 that were altered in these variants. See text for details.
Figure 7. A DNA segment of the Widom 601 spanning SHL2.5 to SHL3.5 impacts redistribution of nucleosomes by Chd1.
Shown are intensity scans from histone mapping reactions carried out with Chd1 (300 nM Chd1 for (TpA)8[SHL2.5/3.5-right], 50 nM Chd1 for all others), 150 nM nucleosomes (40-N-40), and 2 mM ATP. Black traces represent the nucleosome positions before sliding, and the gray traces show H2B(S53C) cross-linking distributions after 64 min (601[SHL2.5/3.5-swap]) or 32 min (all others). Nucleosome positions were mapped using the FAM-labeled DNA (bottom strand), oriented with the right side of each scan corresponding to the bottom of the gel. Each experiment is representative of two or more independent reactions. Sequence variants are described in Figure 6.
The DNA sequence spanning SHL2.5 to SHL3.5 strongly impacts redistribution of nucleosomes by Chd1
While the insertion of poly(dA:dT) tracts influenced the direction of Chd1 sliding, it is possible that asymmetric sliding of the canonical 601 arose from other regions of the nucleosome. The 601 is notably asymmetric: one side has a higher affinity for histones than the other, and the strength of histone-DNA are unevenly distributed, with strongest contacts near the dyad 31, 43, 44. The central portion of 601 is also asymmetric with respect to periodic TA steps, with one side of the dyad having a TA step at four consecutive turns of DNA where the minor groove faces the histone core, whereas the other side possesses just a single TA step (Fig. 6) 44. These TA steps were shown to dramatically impact DNA flexibility and DNA unwrapping under force 45. To see the extent that the pattern of TA steps influenced asymmetric sliding, we generated a 601 variant with TA steps at seven of the eight minor groove positions, such that TA steps were present at all minor groove contacts for the H3/H4 tetramer (called 601[TA+2]; Fig. 6). Nucleosome sliding reactions showed no discernable difference of 601[TA+2] from canonical 601 nucleosomes (Fig. 7), indicating that the pattern of TA steps is not responsible for the pattern of repositioned nucleosomes. To determine whether some other aspect of the 601 sequence that binds to the H3/H4 tetramer may dictate the asymmetric sliding by Chd1, we also tested a 601 variant where the middle 61 bp segment about the dyad was flipped (601[dyad-flip]61; Fig. 6). Sliding reactions with 40-N-40 601[dyad-flip]61 nucleosomes revealed an additional species 11 bp to the left, but still favored the 11 bp and 21 bp positions on the right side (Fig. 7). Thus, although this reversal of the central 601 segment produced some sliding in the opposite direction, the overall pattern was not reversed. We therefore conclude that while the central H3/H4-binding region of 601 can influence sliding, it is not the dominant element guiding the asymmetric repositioning by Chd1.
To see if we could pinpoint a region of the 601 that might explain the asymmetric sliding by Chd1 we looked to the regions replaced by the poly(dA:dT) tracts. Since A16[SHL2.5/3.5-right] had a significant impact on sliding, we focused on the same region of the 601 (24 to 39 bp from the dyad) on both sides of the dyad. We reasoned that the canonical 601 could have either a more barrier-like sequence on the left side, a remodeler-stimulating region on the right side, or a combination of the two. To test the idea of an intrinsic barrier on the left side, we generated a 601 variant with the left 24–39 bp segment duplicated on the right side (601[SHL2.5/3.5-leftduplicate]; Fig. 6). If this sequence were a barrier, we would expect to see blunted and more symmetric repositioning patterns. However, the equilibrium sliding positions of 601[SHL2.5/3.5-leftduplicate] were unchanged from canonical 601, arguing against the left segment acting as a barrier. To see if the right segment was stimulating, we swapped the 24–39 bp segment on the two sides of the 601 (601[SHL2.5/3.5-swap]; Fig. 6). Strikingly, this sequence swap dramatically altered the pattern of nucleosome repositioning, yielding a much more symmetric distribution (Fig. 7). We therefore conclude that, whereas sliding activity can be affected at multiple positions around the nucleosome, Chd1 was most strongly and positively influenced by the sequence on the right side of the 601 spanning SHL2.5 to SHL3.5.
Poly(dA:dT) tracts on the nucleosome increase positional variability of neighboring DNA segments
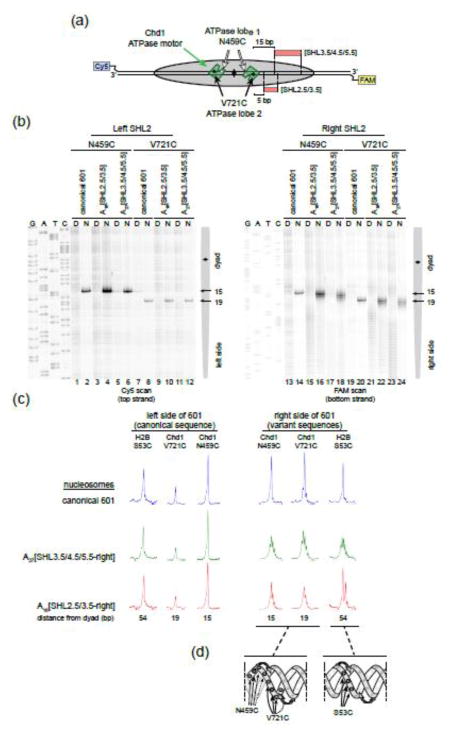
We hypothesized that the poly(dA:dT) tracts may be altering remodeler activity by changing how the ATPase motor of Chd1 engages with DNA. To pursue this idea, we probed remodeler-DNA interactions using site-specific cross-linking. We recently found that both lobes of the ATPase motor engage specifically with DNA at SHL2, visualized by APB-labeling of single cysteine substitutions on lobe 1 (N459C) or lobe 2 (V721C) 46. As expected, in the presence of the ATP mimic ADP BeF3, strong cross-links were observed 15 bp (N459C) and 19 bp (V721C) from the nucleosome dyad on both sides of the canonical 601 (Fig. 8a,b). For 40-N-40 nucleosomes containing A27[SHL3.5/4.5/5.5-right] and A16[SHL2.5/3.5-right] tracts, these cross-links remained strong at both SHL2 sites. These results indicate that in an ATP-bound state, the poly(dA:dT) tracts do not prevent the ATPase motor of Chd1 from robustly engaging with nucleosomal DNA.
Figure 8. Poly(dA:dT) tracts on the nucleosome increase positional variability of neighboring DNA segments.
(a) Schematic showing where two ATPase cross-links map onto nucleosomal DNA relative to poly(dA:dT) sequences.
(b) The Chd1 ATPase motor cross-links on both sides of the nucleosome in the presence and absence of poly(dA:dT) sequences. Shown are representative cross-linking reactions carried out with 40-N-40 nucleosomes (150 nM) and single cysteine variants of Chd1 (300 nM) prelabeled with APB. Nucleosomes (N) and control naked DNA samples (D) were cross-linked in parallel reactions.
(c) Analysis of cross-linking reactions shown in (b) as well as H2B mapping shown in Figure 2, highlighting the increased number of strong cross-linking positions corresponding with the presence of adjacent poly(dA:dT) tracts.
(d) Model of DNA reptation, where a screw-like motion of the DNA duplex allows for cross-linking to multiple neighboring bases.
While cross-linking indicated that the poly(dA:dT) tracts did not prevent the ATPase motor from engaging with SHL2, the poly(dA:dT) tracts made the patterns more diffuse. In fact, on the side of the nucleosome with the poly(dA:dT) tracts, the cross-linking from N459C and V721C on Chd1 was similar to that of H2B-S53C (Fig. 8c). Intriguingly, these diffuse patterns occurred despite the cross-linking positions occurring outside the poly(dA:dT) tracts: for A16[SHL2.5/3.5-right], one edge of the poly(dA:dT) tract was 4 and 8 nt from Chd1 V721C and N459C, respectively, whereas the other edge was 13 nt from H2B-S53C. While A27[SHL3.5/4.5/5.5-right] overlapped with H2B-S53C, it was more distant from the Chd1 ATPase motor, with the closest edge being 14–18 nt from cross-linking residues.
Taken together, we believe that these patterns reflect reptating motions of the DNA duplex on the histone core (Fig. 8d). When free in solution, poly(dA:dT) tracts have a narrow minor groove as well as a spine of hydration giving them a unique structure that decreases their overall DNA flexibility 15–18. When wrapped around the histone core, however, DNA is geometrically constrained, resulting in a narrowing of major and minor grooves facing inward toward the histones and compensatory widening where grooves face outward. The unique structural properties of poly(dA:dT) would therefore not be uniformly accommodated on the nucleosome, and we postulate that our cross-linking captures multiple discrete positions of DNA on the histone core due to unfavorable bending energetics of the poly(dA:dT) tracts. Strikingly, the sharpness of cross-links at the opposite SHL2 are unaffected by the poly(dA:dT) substitutions, indicating that perturbations in DNA placement do not extend to the other side of the nucleosome. Given the poorer nucleosome sliding, we postulate that changes in DNA geometry and/or energetics due to the poly(dA:dT) tracts is responsible for the reduced activity of Chd1.
Discussion
In this report we demonstrate an intrinsic sensitivity of the Chd1 remodeler to the sequence of nucleosomal DNA. Poly(dA:dT) tracts and other sequence changes to the Widom 601 had the greatest impact on Chd1 activity when located adjacent to the internal SHL2 site on the nucleosome. Given that SHL2 is the site of DNA translocation by Chd1 and other remodelers 30, 39–41, these findings suggest that DNA sequence may directly influence the Chd1 ATPase motor. Consistent with a direct impact on the ATPase motor, we observed sequence-dependent repositioning both in the presence and absence of the Chd1 DNA-binding domain (Fig. 2, 3). Interference with nucleosome sliding was most pronounced with a poly(dA:dT) tract 24 bp from the dyad and thus bordering SHL2, which overlaps with where Snf2-specific insertions of the Chd1 ATPase motor interact with DNA 46.
Due to the symmetry of the nucleosome, two SHL2 sites lie on opposite sides of the nucleosome, and unidirectional sliding at each site allows Chd1 to shift the nucleosome in either direction. When Chd1 activity at one SHL2 site is much poorer compared to the other, the histone octamer is shifted away from this site due to preferential action at the more active SHL2, thus producing a barrier-like effect. The relative activities of the remodeler at each SHL2 therefore determine the distributions of nucleosomes along DNA. An obvious complication in interpreting how DNA sequence affects particular distributions of nucleosomes is that DNA positioning changes with each movement of the octamer, giving unique nucleosome substrates after every shift. Therefore, sequences that do not affect Chd1 activity at one nucleosome position may exert greater influence as they migrate around the histone octamer. A further complication is that sequence context is also likely important. While our experiments have shown that the DNA sequence adjacent to SHL2 impacts Chd1 activity, we note that other sequences inserted at this location will likely have different effects depending on the nature of neighboring DNA segments. In addition, as our experiments were exclusively carried out with variants of the 601 positioning sequence, we cannot rule out the possibility that other regions of nucleosomal DNA could have a greater impact on Chd1 action in other sequence contexts.
In addition to revealing where Chd1 is sensitive to DNA on the nucleosome, our data suggest an unexpected adaptation of nucleosomes to DNA energetics. Interestingly, despite their unique structural properties when free in solution, poly(dA:dT) tracts sample a more classical B-form geometry at higher temperatures or with high dimethyl sulfoxide concentrations 47, 48, and it has been suggested that similar structural changes may also be stabilized through interactions with histones 22, 23, 48. While poly(dA:dT) tracts have been shown to adopt similar conformations to canonical nucleosomal DNA 24, 25, our cross-linking experiments suggest that poly(dA:dT) tracts are accompanied by bulging or stretching on the nucleosome. The nucleosome is well known for such structural heterogeneity, as the first crystal structure of a nucleosome revealed a different number of base pairs (72 vs 73) on either side of the dyad 35. In that structure, a bulge/stretch of one bp was observed at SHL2, and subsequent structures showed that a one bp bulge/stretch could also occur at SHL5 (ref. 49). For our cross-linking of the Chd1 ATPase motor to nucleosomal DNA, the canonical 601 sequence yielded single, sharp cross-links on one side of the dyad, whereas the poly(dA:dT) tract on the other side correlated with distributions of cross-links varying by 3–4 nt (Fig. 8c). If we assume that the distribution of DNA cross-links to the ATPase motor represents a screw-like reptation of the DNA duplex past the histone core (Fig. 8d), it would suggest that bulging/stretching of nucleosomal DNA must also occur within 15 bp on either side of the dyad. To our knowledge, such bulging/stretching has not previously been reported at SHL0 or SHL1 and suggests a remarkable ability of the nucleosome to buffer the length of DNA based on sequence or energetics. This idea directly supports the twist-diffusion mechanism for nucleosome sliding, where an ability of the histone core to accommodate two or more discrete lengths of DNA between minor groove contacts would allow the duplex to be effectively pumped around the histone octamer without disrupting nucleosomal wrapping of DNA 50. While this potential DNA buffering is supported by our findings, future studies will be required for further substantiating and defining the manner in which the nucleosome adapts to DNA energetics.
Given the low energetic cost of a single base pair bulge/stretch at SHL2, DNA sequence would be expected to influence the conformation of DNA in this region of the nucleosome, which is where Chd1 and other remodelers bind. Rippe et al. proposed that sequence-dependent activity of Chd1 was due to differences in nucleosome binding 8, yet our sliding experiments, carried out with saturated remodeler, argue instead that the poly(dA:dT) tracts altered catalytic turnover (Fig. 5). Moreover, the ATPase motor cross-linked to SHL2 similarly for the two internal poly(dA:dT) tracts (Fig. 8), yet sliding activity was worse for the A16[SHL2.5/3.5-right] tract, which was closer to the ATPase binding site. These results suggest that instead of influencing the initial encounter between Chd1 and the nucleosome, DNA sequence was sensed at an intermediate stage in the remodeling cycle. Although the details of how remodelers shift DNA past the histone core are presently unclear, a poly(dA:dT) tract adjacent to SHL2 would likely alter the energetics of DNA twisting or bending carried out by the ATPase motor. We speculate that Chd1 may therefore sense how DNA responds to structural perturbations by the ATPase motor, with sliding activity influenced by sequence-based properties of DNA.
Finally, our results highlight the notion that actions of chromatin remodelers do not necessarily reflect DNA-binding preferences of the histone core. Previous DNA unzipping experiments revealed a characteristic energy profile of histone-DNA contacts throughout the nucleosome, showing that contacts at SHL0.5 and SHL1.5 were the strongest 43. Here, by contrast, our data point to Chd1 being sensitive to the region between SHL2.5 and SHL3.5, sites that were dramatically weaker in DNA unzipping experiments 43. Consistent with a proposed “spring-loaded” mechanism, where distinct chromatin patterns are achieved through active (via remodelers) and passive repositioning (via histone-DNA contacts) 51, our work suggests that sequence-directed sliding by Chd1 is uncoupled from thermodynamic preferences of nucleosomes. As shown for RSC and INO80 remodelers, unique sequence signatures likely underlie many remodeler-specific differences 6, and it will be fascinating to discover how different remodeler types tailor their responses to the locations and characteristics of particular sequences on the nucleosome.
Material and Methods
Protein constructs and purification
His-tagged Saccharomyces cerevisiae Chd1 constructs were expressed and purified as described previously 38, 52, 53. The construct referred to as Chd1 throughout the text is truncated at both the N- and C-terminal regions and encompasses the conserved ATPase domain, DNA binding domain, and chromodomains (residues 118–1274). The Chd1[ΔDBD] construct (residues 118–1014) lacks the DNA binding domain. Site-specific cross-linking experiments utilized single-cysteine variants of Chd1, N459C and V721C in an otherwise cysteine-free background, as previously described 46. Xenopus laevis histones were expressed, purified, and reconstituted into octamers as described previously 54.
Nucleosome preparation
Poly(dA:dT) tracts were inserted into the 601 nucleosome positioning sequence using PCR mutagenesis. Using fluorescently-labeled primers (IDT), DNA fragments were amplified from the desired template via PCR, purified by MiniPrep Cell (BioRad) and then incorporated into nucleosomes as described previously 54. The nucleosome positioning sequence of the canonical Widom 601 and variants used in this study are given in Figure 6. The sequence of the central 145 bp along with the 40 bp flanking DNA used for 40-N-40 601 nucleosomes is given in Figure S6.
Nucleosome sliding and histone mapping
Histone mapping was carried out essentially as previously described 33, with some variations. Xenopus laevis histone octamers were generated with the variants H3(C110A), which removed the native cysteine, and H2B(S53C), which introduced a cysteine at the desired APB-labeling location. Nucleosomes were labeled with photo-reactive APB at room temperature for 2.5–3 hours. Sliding reactions were carried out at room temperature in 1X Slide buffer (20 mM Tris-HCl pH 7.5, 50 mM KCl, 5 mM MgCl2, 5% sucrose, 0.1 mg/mL BSA, 5 mM DTT) with 150 nM nucleosome and 2 mM ATP, using 300 nM or 50nM Chd1 as indicated in the figure legends. After quenching with EDTA, sliding reactions were UV irradiated and further processed to isolate DNA fragments as previously described. Samples were separated on 7.8M urea, 8% polyacrylamide gels (19:1 acrylamide:bisacrylamide) at 65 Watts and visualized on a GE Typhoon 9410 variable mode imager. Gel intensity scans were obtained using ImageJ.
Native gel sliding
Native gel sliding was carried out as described previously 53 with 0-N-80 nucleosomes labeled on the long end with the 6-FAM fluorophore. Briefly, reactions were carried out in 1X slide buffer (20 mM Hepes-KOH pH 7.6, 50 mM KCl, 5 mM MgCl2, 0.1mg/mL bovine serum albumin, 1 mM DTT, 5% sucrose) with 50 nM Chd1 and 150 nM nucleosome at room temperature. Reaction was initiated with 2.5 mM ATP and samples were removed and quenched with 1X slide with 5 mM EDTA and 125 ng/μL salmon sperm DNA at the designated timepoints. Samples were then separated on 6% acrylamide native gels and visualized using a GE Typhoon 9410 variable mode imager. Gel quantification was performed with ImageJ.
Kinetic measurements of nucleosome sliding
Histone H2A point-substitution T120C was labeled with maleimide Cy3B and refolded with H3(C110A) and wild type H4 and H2B into octamers. End-positioned 0-N-80 nucleosomes were generated by reconstituting the Cy3B-labeled octamer with DNA containing the quencher Dabcyl at the zero-end of the nucleosome and the fluorophore 6-FAM on the 80-end. We utilized the fluorophore Cy3B, which is incapable of undergoing Protein Induced Fluorescence Enhancement (PIFE) 55 to ensure that changes in fluorescence would be solely attributed to the movement of the DNA with respect to the octamer and insensitive to Chd1 binding. Kinetic traces were measured using either a Horiba Fluorolog-3 or an Applied Photophysics SX20 Stopped Flow Spectrophotometer. To determine Chd1 saturation levels, 10 nM nucleosome was incubated at 25°C with increasing concentrations of Chd1 (25 nM, 200 nM, 600 nM, 800 nM) in 1X Slide buffer (20 mM HEPES-KOH pH 7.5, 5 mM MgCl2, 0.1 mM EDTA, 5% sucrose (w/v), 1 mM DTT, 0.02% Nonidet P-40, 0.1 mg/mL BSA). A negative control baseline was taken before the addition of ATP. The reaction was initiated by the addition of ATP to a final concentration of 25 μM. The increase in fluorescence was monitored with an excitation wavelength of 510 nm and an emission wavelength of 565 nm.
For stopped flow experiments, two syringes were prepared, one containing a solution of 1200 nM Chd1 and 20 nM nucleosome in 1X Slide buffer and the other containing 50 μM ATP in 1X Slide buffer. Sliding reactions were initiated by rapidly mixing together 100 μL from each syringe such that the final concentrations were 600 nM Chd1, 10 nM nucleosome, and 25 μM ATP. Repositioning of the octamer containing Cy3B-labeled H2B away from Dabcyl on the short (zero) DNA end reduced quenching, and was therefore observed as an increase in Cy3B fluorescence over time. For each set of syringes, the progress curves from 3–6 injections (technical replicates) were averaged together. Progress curves were fit in Mathematica using the NonLinearModelFit function for a double exponential (y = a1*(1 − Exp[−k1*x]) + a2*(1 − Exp[−k2*x]) + c) or triple exponential function (y = a1*(1 − Exp[−k1*x]) + a2*(1 − Exp[−k2*x]) + a3*(1 − Exp[−k3*x]) + c), where an were amplitudes (fractions of total fluorescence range), kn were rates (sec−1), and c was a constant.
Chd1 cross-linking
Chd1 cross-linking was carried out as previously described 46. Briefly, in the dark APB was added to a 7.5 μM stock of a single-cysteine Chd1 variant to achieve a final concentration of 400 μM APB and 1% DMF. The labeling reaction was allowed to proceed for 2–3 hours at room temperature, then 300 nM labeled Chd1 variant was incubated with either nucleosome (150 nM) or naked DNA (150 nM) for 30 min in the presence of ADP-BeF3 (2 mM ADP, 15 mM NaF, 3 mM BeCl2, 6 mM MgCl2) and 1X Slide buffer (20 mM Tris-HCl pH 7.5; 50 mM KCl; 0.1 mg/mL BSA; 1 mM DTT; 5% sucrose; 5 mM MgCl2). Samples (50 μL) were transferred to a silanized coverslip and irradiated for 15 sec then subsequently quenched with 100 μL of 20 mM Tris-HCl pH 7.5, 50 mM KCl, 0.1 mg/mL BSA, 5 mM DTT, and 5 mM EDTA then further processed as described 33.
Supplementary Material
Supplementary Figure S1. Chd1 remodeling reactions rapidly reach equilibrium.
Remodeling reactions were carried out with 150 nM 40-N-40 nucleosomes and 300 nM Chd1, and the nucleosome positions were detected by histone mapping with H2B(S53C). These reactions were resolved on a urea denaturing gel, visualized with the FAM label, and are representative of similar experiments carried out more than five times.
Supplementary Figure S2. The presence of internal poly(dA:dT) tracts bias nucleosome sliding in the opposite direction.
Cross-linking distributions of 40-N-40 nucleosomes before and after sliding by Chd1, monitored with Cy5 (top strand) scans. Black traces show starting nucleosome positions prior to remodeling, and colored traces show distributions 32 min after addition of 2 mM ATP and 300 nM Chd1. The positions of peaks that are weak but reproducibly show up in different experiments are given in parentheses. Scans are oriented with the bottom of the gel on the right. These scans are representative of three or more experiments.
Supplementary Figure S3. Internal poly(dA:dT) tracts influence nucleosome sliding by Chd1 independently of the DNA-binding domain.
Cross-linking distributions of 40-N-40 nucleosomes before and after addition of Chd1[ΔDBD], monitored with Cy5 (top strand) scans. Black traces show starting nucleosome positions prior to remodeling, and colored traces show distributions 32 min after addition of 2 mM ATP and 300 nM Chd1[ΔDBD]. The positions of peaks that are weak but reproducibly show up in different experiments are given in parentheses. Scans are oriented with the bottom of the gel on the right. These scans are representative of three or more experiments.
Supplementary Figure S4. Chd1 can pull internal poly(dA:dT) tracts further onto the nucleosome.
Histone mapping reactions of 0-N-80 nucleosomes before and after sliding by Chd1, followed with the FAM (bottom strand) label. Black traces show starting nucleosome positions prior to remodeling, and colored traces show distributions 16 min (for A17[SHL6.5-right]) or 64 min (all others) after addition of 2 mM ATP and 50 nM Chd1 to 150 nM nucleosomes. These data are representative of three or more experiments.
Supplementary Figure S5. Rate of remodeling saturates at 600 nM Chd1.
Shown are progress curves of remodeling reactions followed using Cy3B-Dabcyl SQOF, with increasing fluorescence correlated to movement of the histone octamer away from the short DNA end of 0-N-80 nucleosomes. Reactions contained 10 nM nucleosome, 25 μM ATP, and the indicated concentrations of Chd1. These results are representative of three similar titrations.
Highlights.
How DNA sequence impacts chromatin remodeler action is poorly understood.
Nucleosome sliding by Chd1 is biased by the asymmetry of the Widom 601 sequence.
Poly(dA:dT) tracts adjacent to SHL2 introduce a rate-limiting step during remodeling.
Poly(dA:dT) tracts broaden the translational positioning of DNA on the histone core.
DNA sequence surrounding SHL2 influences Chd1 activity.
Acknowledgments
We thank Sarah Woodson for generously sharing equipment, Ilana Nodelman for providing Chd1(N459C) and Chd1(V721C) protein, and members of the Bowman lab for discussions and helpful comments. This work was supported by the National Institutes of Health (R01GM-084192).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 2015;16:178–189. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 2.Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li M, Hada A, Sen P, Olufemi L, Hall MA, Smith BY, Forth S, McKnight JN, Patel A, Bowman GD, Bartholomew B, Wang MD. Dynamic regulation of transcription factors by nucleosome remodeling. Elife. 2015;4:10. doi: 10.7554/eLife.06249. 7554/eLife.06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nodelman IM, Horvath KC, Levendosky RF, Winger J, Ren R, Patel A, Li M, Wang MD, Roberts E, Bowman GD. The Chd1 chromatin remodeler can sense both entry and exit sides of the nucleosome. Nucleic Acids Res. 2016;44:7580–7591. doi: 10.1093/nar/gkw406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang JG, Madrid TS, Sevastopoulos E, Narlikar GJ. The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat Struct Mol Biol. 2006;13:1078–1083. doi: 10.1038/nsmb1170. [DOI] [PubMed] [Google Scholar]
- 6.Krietenstein N, Wal M, Watanabe S, Park B, Peterson CL, Pugh BF, Korber P. Genomic Nucleosome Organization Reconstituted with Pure Proteins. Cell. 2016;167:709–721. e12. doi: 10.1016/j.cell.2016.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKnight JN, Tsukiyama T, Bowman GD. Sequence-targeted nucleosome sliding in vivo by a hybrid Chd1 chromatin remodeler. Genome Res. 2016;26:693–704. doi: 10.1101/gr.199919.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rippe K, Schrader A, Riede P, Strohner R, Lehmann E, Längst G. DNA sequence- and conformation-directed positioning of nucleosomes by chromatin-remodeling complexes. Proc Natl Acad Sci U S A. 2007;104:15635–15640. doi: 10.1073/pnas.0702430104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Partensky PD, Narlikar GJ. Chromatin remodelers act globally, sequence positions nucleosomes locally. J Mol Biol. 2009;391:12–25. doi: 10.1016/j.jmb.2009.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartley PD, Madhani HD. Mechanisms that specify promoter nucleosome location and identity. Cell. 2009;137:445–458. doi: 10.1016/j.cell.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parnell TJ, Huff JT, Cairns BR. RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. Embo j. 2008;27:100–110. doi: 10.1038/sj.emboj.7601946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, Gebbia M, Talukder S, Yang A, Mnaimneh S, Terterov D, Coburn D, Li Yeo A, Yeo ZX, Clarke ND, Lieb JD, Ansari AZ, Nislow C, Hughes TR. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 1985;82:8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 15.Fratini AV, Kopka ML, Drew HR, Dickerson RE. Reversible bending and helix geometry in a B-DNA dodecamer: CGCGAATTBrCGCG. J Biol Chem. 1982;257:14686–14707. [PubMed] [Google Scholar]
- 16.Dickerson RE, Drew HR. Structure of a B-DNA dodecamer. II Influence of base sequence on helix structure. J Mol Biol. 1981;149:761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- 17.Nelson HC, Finch JT, Luisi BF, Klug A. The structure of an oligo(dA). oligo(dT) tract and its biological implications. Nature. 1987;330:221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- 18.Coll M, Frederick CA, Wang AH, Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci U S A. 1987;84:8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. Embo j. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segal E, Widom J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr Opin Struct Biol. 2009;19:65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Getts RC, Behe MJ. Isolated oligopurine tracts do not significantly affect the binding of DNA to nucleosomes. Biochemistry. 1992;31:5380–5385. doi: 10.1021/bi00138a020. [DOI] [PubMed] [Google Scholar]
- 22.Puhl HL, Behe MJ. Poly(dA). poly(dT) forms very stable nucleosomes at higher temperatures. J Mol Biol. 1995;245:559–567. doi: 10.1006/jmbi.1994.0046. [DOI] [PubMed] [Google Scholar]
- 23.Mahloogi H, Behe MJ. Oligoadenosine tracts favor nucleosome formation. Biochem Biophys Res Commun. 1997;235:663–668. doi: 10.1006/bbrc.1997.6858. [DOI] [PubMed] [Google Scholar]
- 24.Bao Y, White CL, Luger K. Nucleosome core particles containing a poly(dA. dT) sequence element exhibit a locally distorted DNA structure. J Mol Biol. 2006;361:617–624. doi: 10.1016/j.jmb.2006.06.051. [DOI] [PubMed] [Google Scholar]
- 25.Hayes JJ, Clark DJ, Wolffe AP. Histone contributions to the structure of DNA in the nucleosome. Proc Natl Acad Sci U S A. 1991;88:6829–6833. doi: 10.1073/pnas.88.15.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorch Y, Maier-Davis B, Kornberg RD. Role of DNA sequence in chromatin remodeling and the formation of nucleosome-free regions. Genes Dev. 2014;28:2492–2497. doi: 10.1101/gad.250704.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lusser A, Urwin DL, Kadonaga JT. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat Struct Mol Biol. 2005;12:160–166. doi: 10.1038/nsmb884. [DOI] [PubMed] [Google Scholar]
- 28.Stockdale C, Flaus A, Ferreira H, Owen-Hughes T. Analysis of Nucleosome Repositioning by Yeast ISWI and Chd1 Chromatin Remodeling Complexes. J Biol Chem. 2006;281:16279–16288. doi: 10.1074/jbc.M600682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pointner J, Persson J, Prasad P, Norman-Axelsson U, Stralfors A, Khorosjutina O, Krietenstein N, Peter Svensson J, Ekwall K, Korber P. CHD1 remodelers regulate nucleosome spacing in vitro and align nucleosomal arrays over gene coding regions in S. pombe. Embo j. 2012;31:4388–4403. doi: 10.1038/emboj.2012.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKnight JN, Jenkins KR, Nodelman IM, Escobar T, Bowman GD. Extranucleosomal DNA binding directs nucleosome sliding by Chd1. Mol Cell Biol. 2011;31:4746–4759. doi: 10.1128/MCB.05735-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levendosky RF, Sabantsev A, Deindl S, Bowman GD. The Chd1 chromatin remodeler shifts hexasomes unidirectionally. Elife. 2016;5:e21356. doi: 10.7554/eLife.21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma A, Jenkins KR, Héroux A, Bowman GD. Crystal structure of the chromodomain helicase DNA-binding protein 1 (Chd1) DNA-binding domain in complex with DNA. J Biol Chem. 2011;286:42099–42104. doi: 10.1074/jbc.C111.294462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kassabov SR, Bartholomew B. Site-directed histone-DNA contact mapping for analysis of nucleosome dynamics. Methods Enzymol. 2004;375:193–210. doi: 10.1016/s0076-6879(03)75013-7. [DOI] [PubMed] [Google Scholar]
- 34.Pendergrast PS, Chen Y, Ebright YW, Ebright RH. Determination of the orientation of a DNA binding motif in a protein-DNA complex by photocrosslinking. Proc Natl Acad Sci U S A. 1992;89:10287–10291. doi: 10.1073/pnas.89.21.10287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 36.Anderson JD, Widom J. Poly(dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites. Mol Cell Biol. 2001;21:3830–3839. doi: 10.1128/MCB.21.11.3830-3839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel A, Chakravarthy S, Morrone S, Nodelman IM, McKnight JN, Bowman GD. Decoupling nucleosome recognition from DNA binding dramatically alters the properties of the Chd1 chromatin remodeler. Nucleic Acids Res. 2013;41:1637–1648. doi: 10.1093/nar/gks1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel A, McKnight JN, Genzor P, Bowman GD. Identification of residues in Chromo-Helicase-DNA-Binding Protein 1 (Chd1) required for coupling ATP hydrolysis to nucleosome sliding. J Biol Chem. 2011;286:43984–43993. doi: 10.1074/jbc.M111.282970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwanbeck R, Xiao H, Wu C. Spatial contacts and nucleosome step movements induced by the NURF chromatin remodeling complex. J Biol Chem. 2004;279:39933–39941. doi: 10.1074/jbc.M406060200. [DOI] [PubMed] [Google Scholar]
- 40.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol. 2005;12:747–755. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- 41.Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
- 42.Blosser TR, Yang JG, Stone MD, Narlikar GJ, Zhuang X. Dynamics of nucleosome remodelling by individual ACF complexes. Nature. 2009;462:1022–1027. doi: 10.1038/nature08627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall MA, Shundrovsky A, Bai L, Fulbright RM, Lis JT, Wang MD. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat Struct Mol Biol. 2009;16:124–129. doi: 10.1038/nsmb.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chua EY, Vasudevan D, Davey GE, Wu B, Davey CA. The mechanics behind DNA sequence-dependent properties of the nucleosome. Nucleic Acids Res. 2012;40:6338–6352. doi: 10.1093/nar/gks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ngo TT, Zhang Q, Zhou R, Yodh JG, Ha T. Asymmetric unwrapping of nucleosomes under tension directed by DNA local flexibility. Cell. 2015;160:1135–1144. doi: 10.1016/j.cell.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nodelman IM, Bleichert F, Patel A, Ren R, Horvath KC, Berger JM, Bowman GD. Interdomain communication of the Chd1 chromatin remodeler across the DNA gyres of the nucleosome. Mol Cell. 2017;65:447–459. doi: 10.1016/j.molcel.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herrera JE, Chaires JB. A premelting conformational transition in poly(dA)-Poly(dT) coupled to daunomycin binding. Biochemistry. 1989;28:1993–2000. doi: 10.1021/bi00431a006. [DOI] [PubMed] [Google Scholar]
- 48.Premilat S, Albiser G. X-ray fibre diffraction study of an elevated temperature structure of poly(dA). poly(dT) J Mol Biol. 1997;274:64–71. doi: 10.1006/jmbi.1997.1378. [DOI] [PubMed] [Google Scholar]
- 49.Tan S, Davey CA. Nucleosome structural studies. Curr Opin Struct Biol. 2011;21:128–136. doi: 10.1016/j.sbi.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Holde K, Yager T. Models for chromatin remodeling: a critical comparison. Biochem Cell Biol. 2003;81:169–172. doi: 10.1139/o03-038. [DOI] [PubMed] [Google Scholar]
- 51.Sexton BS, Avey D, Druliner BR, Fincher JA, Vera DL, Grau DJ, Borowsky ML, Gupta S, Girimurugan SB, Chicken E, Zhang J, Noble WS, Zhu F, Kingston RE, Dennis JH. The spring-loaded genome: nucleosome redistributions are widespread, transient, and DNA-directed. Genome Res. 2014;24:251–259. doi: 10.1101/gr.160150.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hauk G, McKnight JN, Nodelman IM, Bowman GD. The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell. 2010;39:711–723. doi: 10.1016/j.molcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nodelman IM, Bowman GD. Nucleosome sliding by Chd1 does not require rigid coupling between DNA-binding and ATPase domains. EMBO Rep. 2013;14:1098–1103. doi: 10.1038/embor.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 55.Hwang H, Kim H, Myong S. Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc Natl Acad Sci U S A. 2011;108:7414–7418. doi: 10.1073/pnas.1017672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Chd1 remodeling reactions rapidly reach equilibrium.
Remodeling reactions were carried out with 150 nM 40-N-40 nucleosomes and 300 nM Chd1, and the nucleosome positions were detected by histone mapping with H2B(S53C). These reactions were resolved on a urea denaturing gel, visualized with the FAM label, and are representative of similar experiments carried out more than five times.
Supplementary Figure S2. The presence of internal poly(dA:dT) tracts bias nucleosome sliding in the opposite direction.
Cross-linking distributions of 40-N-40 nucleosomes before and after sliding by Chd1, monitored with Cy5 (top strand) scans. Black traces show starting nucleosome positions prior to remodeling, and colored traces show distributions 32 min after addition of 2 mM ATP and 300 nM Chd1. The positions of peaks that are weak but reproducibly show up in different experiments are given in parentheses. Scans are oriented with the bottom of the gel on the right. These scans are representative of three or more experiments.
Supplementary Figure S3. Internal poly(dA:dT) tracts influence nucleosome sliding by Chd1 independently of the DNA-binding domain.
Cross-linking distributions of 40-N-40 nucleosomes before and after addition of Chd1[ΔDBD], monitored with Cy5 (top strand) scans. Black traces show starting nucleosome positions prior to remodeling, and colored traces show distributions 32 min after addition of 2 mM ATP and 300 nM Chd1[ΔDBD]. The positions of peaks that are weak but reproducibly show up in different experiments are given in parentheses. Scans are oriented with the bottom of the gel on the right. These scans are representative of three or more experiments.
Supplementary Figure S4. Chd1 can pull internal poly(dA:dT) tracts further onto the nucleosome.
Histone mapping reactions of 0-N-80 nucleosomes before and after sliding by Chd1, followed with the FAM (bottom strand) label. Black traces show starting nucleosome positions prior to remodeling, and colored traces show distributions 16 min (for A17[SHL6.5-right]) or 64 min (all others) after addition of 2 mM ATP and 50 nM Chd1 to 150 nM nucleosomes. These data are representative of three or more experiments.
Supplementary Figure S5. Rate of remodeling saturates at 600 nM Chd1.
Shown are progress curves of remodeling reactions followed using Cy3B-Dabcyl SQOF, with increasing fluorescence correlated to movement of the histone octamer away from the short DNA end of 0-N-80 nucleosomes. Reactions contained 10 nM nucleosome, 25 μM ATP, and the indicated concentrations of Chd1. These results are representative of three similar titrations.