Abstract
Sorting-related receptor with A-type repeats (SORLA) is an intracellular sorting receptor that directs cargo proteins, such as kinases, phosphatases, and signaling receptors, to their correct location within the cell. The activity of SORLA assures proper function of cells and tissues, and receptor dysfunction is the underlying cause of common human malignancies, including Alzheimer’s disease, atherosclerosis, and obesity. Here, we discuss the molecular mechanisms that govern sorting of SORLA and its cargo in multiple cell types, and why genetic defects in this receptor results in devastating diseases.
Keywords: VPS10P domain receptors, Protein sorting, Retromer, GGA, Alzheimer’s disease, Obesity
Introduction
Sorting of proteins to their destined location in subcellular compartments is essential for proper cell function, and faulty protein sorting will result in cellular dysfunction and disease. Protein sorting is essential for all cell types, but particularly challenging in neurons in which cell compartments of axons and dendrites may be as far away as 1 m from the soma of motor neurons. Within cells, the Golgi is the central hub that sorts the bulk of proteins. Protein sorting proceeds in the trans-most cisterna of this organelle called the trans-Golgi network (TGN) that consists of an elaborate web of branching tubular membrane domains. From the TGN, proteins may be targeted to the apical or basolateral plasma membranes, to the endosomal/lysosomal system, or to specialized secretory granules for activity-dependent release. Directed protein trafficking is mediated by sorting receptors, transmembrane proteins that interact with cytosolic adaptors at the Golgi membranes to guide their protein cargo to and from the TGN (reviewed in [1]).
In recent years, one group of sorting receptors received particular attention because of their causal involvement in human diseases, such as Alzheimer’s and Huntington’s disease, psychiatric disorders, but also atherosclerosis, dyslipidemia, and diabetes. These sorting receptors are called VPS10P domain receptors. Initially characterized in neurons in the brain, VPS10P domain receptors now emerge as key regulators of intracellular protein sorting not only in the nervous system but also in many other tissues as well [2]. Here, we focus on the sorting-related receptor with A-type repeats (SORLA), a VPS10P domain receptor that is paradigmatic for the mode of action of this class of sorting receptors. We describe the molecular interactions of SORLA with adaptor complexes that control traffic between TGN, plasma membrane, and endosomes. We discuss the functional significance of this trafficking pathway for proper routing of enzymes, growth factors, and signaling receptors, and why SORLA dysfunction may result in devastating pathologies, including neurodegeneration, impaired renal ion homeostasis, and obesity.
Cell biology of SORLA sorting
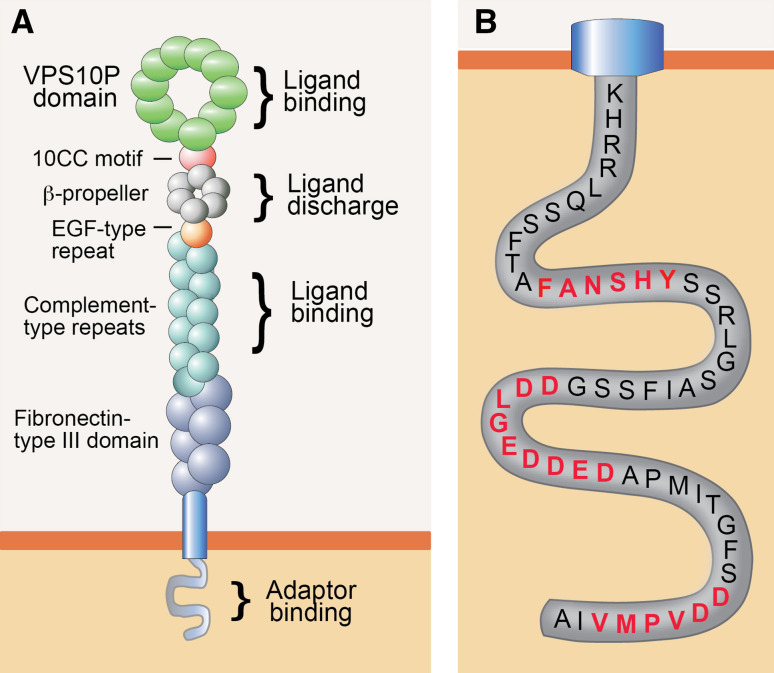
SORLA (also known as LR11 or SORL1) was initially uncovered in the search for receptors that share structural similarity to the low-density lipoprotein (LDL) receptor, the main endocytic receptor for uptake of lipid-loaded lipoproteins into vertebrate cells. These studies identified a 250-kDa type 1 transmembrane protein in brain [3] and liver [4] that contained complement-type repeats and a β-propeller, structural elements in the LDL receptor required for binding and for pH-dependent release of ligands, respectively (Fig. 1a). The ability of SORLA to internalize lipoproteins seemingly supported the notion of a novel species of lipoprotein receptor [5, 6]. However, this assumption was questioned by the presence of additional structural elements in SORLA not found in the LDL receptor, namely a VPS10P domain and six fibronectin-type III domains (Fig. 1a). The VPS10P domain was noteworthy as it was identified earlier in an intracellular sorting protein in yeast called the vacuolar protein sorting 10 proteins (VPS10P) [7]. VPS10P directs newly synthesized peptidases from the TGN to the vacuole (the yeast lysosome) where they act in proteolytic breakdown of internalized proteins. A similar function for SORLA in intracellular protein sorting in mammalian cell types was supported by the fact that the bulk of the receptor molecules was present in the Golgi rather than at the cell surface, a finding that argued against a role as endocytic receptor [6, 8].
Fig. 1.
Structural organization of SORLA. a Organization of the SORLA polypeptide is shown, indicating the main structural elements and their documented functions. The VPS10P domain and the cluster of complement-type repeats serve as major ligand-binding sites in the luminal receptor domain. The β-propeller interacts with the molecular chaperone MESD to facilitate folding of the receptor polypeptide, and it may be involved in pH-dependent release of bound ligands in acidic endosomal compartments. b Amino-acid sequence of the cytoplasmic receptor tail highlighting three main binding motifs for cytosolic adaptors, termed FANSHY, the acidic motif (DDLGEDDED), and the GGA-binding site (DDVPMV). GGA, Golgi-localizing, γ-adaptin ear homology domain, ARF-interacting protein; MESD, mesodermal development deletion interval; VPS10P, vacuolar protein sorting 10 proteins. a adapted from [43]
By now, an extensive body of work has substantiated the relevance of SORLA as an intracellular sorting receptor that shuttles between TGN, cell surface, and endosomes in neurons and multiple other cell types. SORLA is synthesized as a pro-receptor containing a 53 amino-acid pro-peptide at the ultimate amino terminus. This pro-peptide is believed to block the binding site for ligands in the VPS10P domain, a major site for interaction with peptide ligands [9, 10]. Removal of the pro-peptide by convertases in the TGN activates the ligand-binding capability of the receptor [6]. This activation step may be required to prevent premature binding of ligands to nascent receptor molecules in the biosynthetic pathway of the cell. Apart from the VPS10P domain, the cluster of complement-type repeats in SORLA constitutes another site for ligand recognition [11, 12]. Binding of ligands to the VPS10P domain or the complement-type repeats is lost at low pH (<5.5) [13], suggesting ligand interaction to partake in the secretory pathway and at the cell surface, but to be disrupted in the acidic milieu of late endosomes. The significance of additional structural elements in the extracellular domain for receptor functions is less clear. Based on analogy to other proteins, the fibronectin-type III domain may be involved in protein–protein interactions [14], while the β-propeller may facilitate pH-dependent release of ligands in endocytic compartments [15].
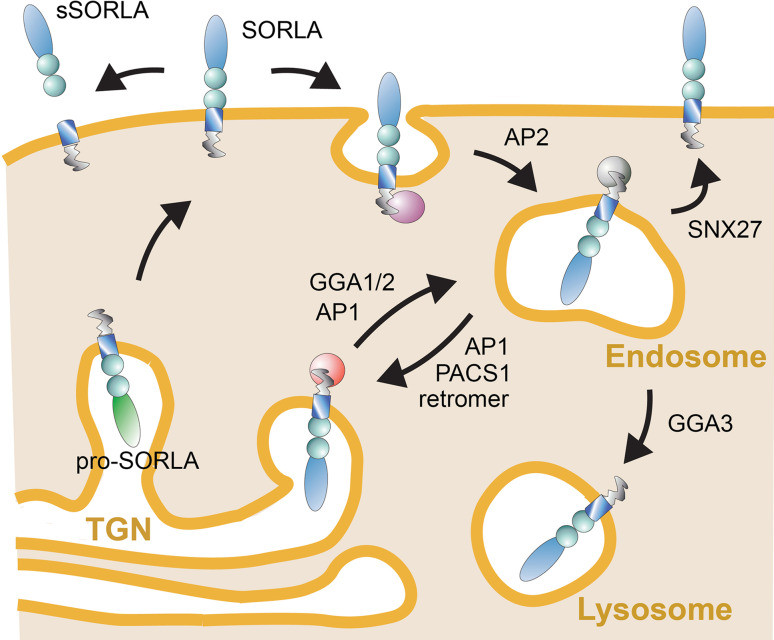
As well as by the ability to recognize distinct ligands through its luminal domain, the function of SORLA in protein sorting is determined by information encoded in its short cytoplasmic tail. This tail domain encodes recognition motifs for cytosolic adaptors that direct the complex trafficking path of SORLA between cell surface and various intracellular compartments (Fig. 1b). Typically, newly synthesized SORLA molecules follow the constitute secretory pathway from the endoplasmic reticulum through the Golgi to the cell surface, a default route for transmembrane proteins that does not require distinct sorting motifs (Fig. 2). At the cell surface, some SORLA molecules are subject to proteolytic shedding releasing the soluble ectodomain of the receptor, termed soluble (s) SORLA [16, 17]. However, most SORLA molecules at the cell surface remain intact and undergo clathrin-dependent endocytosis guided by the clathrin adaptor protein 2 (AP2) that interacts with an acidic motif D2190DLGEDDED in the receptor tail [8]. Internalized receptors move to the early endosomes from where most receptors will sort to the TGN to continuously shuttle between TGN and endosomes thereafter. Retrograde movement of SORLA from endosomes to the TGN is guided by phosphofurin acidic cluster sorting protein 1 (PACS1) that also binds to the acidic cluster [18], and by the multimeric adaptor complex retromer that binds to the F2172ANSHY tail motif [19, 20]. Anterograde sorting of SORLA from the TGN to endosomes is mediated by the monomeric clathrin adaptors GGA1 and GGA2 (Golgi-localizing, γ-adaptin ear homology domain, and ARF-interacting proteins) that interact with the D2207DVPMVIA element in the SORLA tail [18, 21, 22]. Finally, binding of the adaptor protein AP1 to the acidic tail motif may aid in anterograde as well as retrograde sorting of SORLA [8]. The shuttling of protein cargo between TGN and endosomes likely constitutes the major trafficking route taken by SORLA in neurons. However, some studies also report the ability of SORLA to move ligands from endosomes to the cell surface guided by the sorting nexin family member (SNX) 27 [23], or from endosomes to lysosomes, potentially sorted by GGA3 [24, 25]. The complex trafficking path for SORLA has mainly been elucidated in established cell lines. However, recent studies in mouse models expressing mutant SORLA variants lacking individual adaptor binding sites have substantiated this model in the brain by documenting impaired anterograde sorting in receptor mutants lacking the GGA-binding site [25] and impaired retrograde sorting in mutants unable to interact with PACS1 [26] or retromer [25].
Fig. 2.
Intracellular trafficking path for SORLA. Nascent SORLA is an inactive pro-receptor (pro-SORLA) that is activated by proteolytic removal of an amino-terminal pro-peptide in the TGN, resulting in transfer of the active receptor (SORLA) through the constitutive secretory pathway to the cell surface. Some receptor molecules at the cell surface are subjected to ectodomain shedding, resulting in release of the extracellular receptor domain. Ectodomain shedding disrupts the ability of SORLA to act as a sorting receptor, but may serve to produce a soluble receptor fragment termed soluble (s)SORLA that acts as a signaling molecule. Still, most SORLA molecules at the cell surface remain intact and undergo clathrin-dependent endocytosis facilitated by the clathrin adaptor protein (AP) 2. The bulk of internalized receptors move from endosomes back to the TGN to continuously shuttle between TGN and endosomal compartments thereafter. Adaptors GGA1 and GGA2 guide anterograde movement of SORLA from the TGN to endosomes, whereas PACS1 and the retromer complex facilitate retrograde sorting from endosomes back to the Golgi. AP1 may be involved in bi-directional sorting. As alternative routes, SORLA may sort from endosomes to the cell surface (aided by adaptor SNX27) or to lysosomes (aided by GGA3). Figure adapted from [43]. AP, adaptor protein; GGA, Golgi-localizing, γ-adaptin ear homology domain, ARF-interacting protein; PACS1, phosphofurin acidic cluster sorting protein 1; SNX27, sorting nexin family member 27
SORLA controls amyloidogenic processes in the brain
Given the predominant expression of SORLA in the brain and its complex trafficking path in neurons, major efforts have been focused on identifying the protein cargo sorted by this receptor and its relevance for brain (patho)physiology. These studies have highlighted an important role for SORLA in control of amyloidogenic processes in the brain and as a major risk factor for Alzheimer’s disease (AD).
Central to the pathology of AD is the amyloid precursor protein (APP), a type-1 transmembrane protein expressed in many cell types, including neurons. In a naturally occurring process, APP is broken down into various proteolytic fragments, including the amyloid-β peptides (Aβ), peptides of 37–43 amino-acid length that encompass part of the transmembrane, and extracellular domains of APP. Amyloid-β peptides, notably Aβ42, are considered main culprits in neurodegenerative processes as they exhibit a tendency to aggregate to neurotoxic oligomers and senile plaques, pathological features causative of neuronal dysfunction and cell loss in AD patients (reviewed in [27]). Amyloidogenic processing requires endocytosis of APP molecules from the cell surface and delivery to endosomes whereby proteolytic breakdown to Aβ occurs [28–30]. As it turns out, SORLA acts as a sorting receptor for APP that shuttles internalized precursor molecules from endosomes back to the TGN to decrease production of Aβ [18, 31, 32]. Binding of APP proceeds through the cluster of complement-type repeats in SORLA that forms a 1:1 stoichiometric complex with the luminal domain of APP [11, 12, 33]. Overexpression of SORLA in cells reduces Aβ formation [18, 31, 32, 34], while loss of expression accelerates Aβ production and senile plaque deposition [35, 36], documenting a protective function for SORLA in AD progression. The interaction of SORLA and APP is blocked by signaling through β-adrenergic receptors via a yet unknown mechanism, resulting in impaired Golgi retrieval and in increased endosomal accumulation of APP [37]. In line with the presumed sorting path of the receptor, prevention of APP processing depends on the ability of SORLA to move retrogradely from endosomes to the TGN and is lost in receptor mutants that cannot interact with retromer [25] or PACS1 [26]. As well as by sorting APP, SORLA has also been shown to reduce the amyloidogenic burden by sorting of newly produced Aβ peptides to lysosomes for catabolism [13]. This activity depends on the interaction of Aβ with a recognition site in the VPS10P domain of SORLA [10] and on the ability of the receptor to interact with GGAs to move in an anterograde fashion from the TGN to endocytic compartments [25].
Taken together, the ability of SORLA to sort APP and Aβ likely represents major mechanisms, whereby this receptor reduces the amyloidogenic burden and delays progression of neurodegeneration. This hypothesis received strong support from genetic studies in AD patients that identified gene variants in SORL1, the gene encoding SORLA, as being associated with the risk of the sporadic form of AD [34, 38, 39]. Some of these sequence variants have been shown to impair efficiency of SORL1 transcription [34, 40, 41] or translation [42], in line with low levels of SORLA being disease promoting in patients and mouse models (reviewed in [43]). Furthermore, a missense mutation in SORL1, that disrupts its ability to bind Aβ, has been identified in a family with the autosomal dominant form of AD [44].
SORLA in neurotrophin signaling
Protein cargo sorted by SORLA in neurons is not restricted to APP and its processing products, but also encompasses a number of neurotrophin receptors, cell surface proteins that transmit trophic signals to support growth and survival of neurons. Specifically, SORLA acts as a sorting factor for the tropomyosin-related kinase receptor (TrkB), the receptor for brain-derived neurotrophic factor (BDNF). SORLA facilitates trafficking of TrkB between synaptic membranes and the cell soma, a step critical for BDNF signal transduction into cells [45]. Loss of SORLA results in impaired neuritic transport of TrkB and in a blunted response to BDNF [45]. Intriguingly, SORLA is also a downstream target of BDNF with receptor gene transcription being induced almost 10-fold by BDNF signaling in neurons [41, 46]. These data suggest a positive feedback loop, whereby BDNF enhances trophic signaling through induction of SORL1, the gene encoding the sorting receptor for TrkB.
Another trophic pathway modulated by SORLA acts through glial cell-line-derived neurotrophic factor (GDNF) that promotes survival of distinct populations of central and peripheral neurons, such as midbrain dopaminergic neurons and spinal motor neurons. SORLA interacts with GDNF to increase its regulated secretion from cells [47]. In addition, SORLA interacts with GFRα1, the co-receptor for GDNF [48]. SORLA facilitates internalization of GFRα1/GDNF complexes from the plasma membrane, resulting in lysosomal catabolism of GDNF but cell surface recycling of GFRα1. This sorting route provides an efficient pathway for clearance of GDNF from the extracellular space and counteracts consequences of excessive GDNF signaling, such as hyperactivity and reduced anxiety (as seen in mice lacking SORLA) [48]. Finally, SORLA also impacts signaling through a heterodimeric neurotrophic cytokine called cardiotrophin-like cytokine:cytokine-like factor 1 (CLC:CLF-1) [49]. Specifically, SORLA interacts with the CLF-1 moiety to facilitate internalization of the cytokine in complex with the ciliary neurotrophic factor receptor (CNTFR) α. SORLA-dependent endocytosis is required for neurotrophic signaling through CLC:CLF-1, but it also downregulates signal reception by directing ligand and receptor to lysosomal degradation [49].
The cell biology of SORLA-dependent sorting of neurotrophins and their receptors, and the implication of cytosolic adaptors in this process, still awaits further investigation. However, the ability of this receptor to impact pathways both for trophic support but also of amyloidogenic insult to neurons makes this sorting pathway an important target in control of neurodegenerative processes in patients.
SORLA in renal ion homeostasis
While major attention has been focused on the relevance of SORLA for protein sorting in neurons, other studies have uncovered important roles for this protein in non-neuronal cell types, as well. Thus, SORLA is abundantly expressed in the thick ascending limb (TAL) of Henle’s loop, a distal segment of the renal nephron responsible for water and ion homeostasis [50, 51]. Lack of SORLA expression in epithelial cells of the TAL results in failure to properly reabsorb sodium and chloride, a defect attributed to the inability of these cells to activate the major sodium transporter in the distal nephron Na–K-Cl cotransporter 2 (NKCC2) [51]. As it turns out, SORLA controls the phospho-regulation of NKCC2 by interacting with both the Ste-20-related proline-alanine-rich kinase (SPAK) [51] and the calcineurin phosphatase [52] that carry out phosphorylation and dephosphorylation of NKCC2, respectively. These findings suggest SORLA-mediated sorting of kinases and phosphatases as a regulatory process in modulation of renal ion balance.
SORLA in vascular cell migration and atherosclerosis
Atherosclerosis, or thickening of the artery wall, is a major risk factor for cardiovascular morbidity and mortality, including myocardial infarction and stroke. Atherosclerosis is caused by excessive accumulation of lipids in macrophages in the vessel wall (foam cells) and by the proliferation of intimal smooth muscle cells. Jointly, these processes contribute to the formation of fibrous plaques that may obstruct the vessel lumen. Interestingly, Sorl1 has been mapped as a pro-atherogenic locus in mice [53]. The relevance of SORLA for atherosclerotic processes was further supported by correlating circulating levels of the shedded ectodomain sSORLA (indicative of full-length receptor levels in tissues) with intima-media thickness in subjects with coronary artery disease [54] or acute coronary syndrome [55]. Currently, there are two main hypotheses how SORLA impacts atherosclerotic plaque formation. One model suggests a role for SORLA in control of plasma triacylglyceride levels through regulation of lipolysis. Triacylglyceride-rich lipoproteins are highly pro-atherogenic particles. Their turnover is determined by hydrolysis of triacylglycerides to free fatty acids through lipoprotein lipase (LPL) in the circulation. SORLA traffics newly synthesized LPL molecules from the TGN to lysosomes, reducing the amount of the enzyme being secreted by cultured cells [56]. In addition, SORLA mediates the endocytosis of apoA-V, an activator of LPL [57, 58]. Modulation of LPL activity through clearance of apoA-V is supported by the loss of SORLA binding in an apoA-V variant found in individuals with severe hypertriglyceridemia [59]. Potentially, either through control of LPL or apoA-V levels, SORLA may inhibit lipolysis and raise the levels of pro-atherogenic lipoprotein particles in the circulation.
An alternative model suggests a more direct role for SORLA in atherosclerotic processes in the vessel wall. It is based on the ability of SORLA to stimulate proliferation and migration of intimal smooth muscle cells (SMC) and monocytes, processes that accelerated intimal thickening and atherosclerotic plaque formation [60–63]. Potentially, the stimulation of SMC migration by SORLA works through modulation of cell surface expression of the urokinase receptor (uPAR) [64, 65]. The uPAR is a glycosylphosphatidyl inositol-anchored receptor for urokinase, a protease that activates plasminogen to plasmin, which, in turn, breaks down the extracellular matrix. Binding of urokinase to uPAR on the surface of cells increases their proteolytic potential and facilitates migration. The ability to regulate surface exposure of uPAR is seen for full-length SORLA but also for sSORLA, suggesting both cell autonomous and non-autonomous modes of action [65, 66].
SORLA is a risk factor for obesity
Genome-wide association studies not only confirmed the relevance of SORL1 as a genetic risk factor for sporadic AD [38] but also revealed a surprising association of this locus with metabolic traits (e.g., obesity and waist circumference) in humans and mouse models [67, 68]. In addition, loss of SORLA expression in mice with targeted Sorl1 disruption is protected from diet-induced obesity, suggesting a so far unknown function for this receptor in metabolic regulation [69, 70]. Recent studies in transgenic mouse models shed light on potential modes of receptor action, albeit proposing distinct roles for SORLA and sSORLA in this context. In a study by Whittle and colleagues, sSORLA was shown to impair thermogenesis in mice by binding to bone morphogenetic protein (BMP) receptors and inhibiting BMP/TGF β signaling in adipocytes [69]. Thermogenesis is the process of heat production from metabolic fuel and a driving force for consumption of body lipid stores by brown adipose tissue. Mice genetically deficient for SORLA are protected from diet-induced obesity because of enhanced thermogenesis in adipose tissue, providing an explanatory model for the association of SORL1 with obesity in the human population [69].
An alternative model to explain the role of SORLA in energy homeostasis entails intracellular sorting of the insulin receptor (IR) [70]. One of the actions of insulin signaling in adipocytes is the downregulation of lipolysis. This mechanism reduces energy production from breakdown of lipid stores in a state of sufficient energy supply from carbohydrates. Cellular signal transduction proceeds through binding of insulin to the IR on the surface of target cells and subsequent endocytosis of receptor and hormone complexes. Internalization serves two purposes. First, it delivers receptor-ligand complexes to endosomes, a prerequisite for signal transduction. Second, it moves receptor-ligand complexes to lysosomal compartments for catabolism, a mean to downregulate signal reception. In a process reminiscent of APP sorting in neurons, SORLA interacts with internalized IR molecules in endosomes and shuttles them back to the TGN. Retrograde trafficking reduces lysosomal catabolism and increases the fraction of IR molecules recycled back to the cell surface. SORLA-dependent recycling sensitizes adipocytes for insulin signal reception and enhances the impact of insulin on blockade of lipolysis. Consequently, overexpression of SORLA in adipose tissue of mice inhibits lipolysis and promotes the fat mass gain, while loss of the receptor expression increases lipolysis rate and protects animals from obesity and secondary metabolic complications [70].
In obese human subjects, the levels of SORLA in adipose tissue [70] and those of sSORLA in the circulation [69] positively correlate with the body mass index. Although the exact mode of action in adipose tissues as humoral factor or as sorting receptor still awaits further clarification, all current data support the significance of SORL1 as genetic risk factor of obesity in the human population.
Outlook: SORLA dysfunction as an explanatory model for comorbidities?
Studies on seemingly unrelated disease entities have converged on SORLA as a sorting receptor for multiple ligands in organs, such diverse as the brain, the kidney, or adipose tissue. These observations raise the intriguing possibility that SORLA (dys)function may explain some of the comorbidities commonly seen in the human population as exemplified for AD and type II diabetes (T2D). T2D, a disease characterized by lack of responsiveness of cells to insulin (insulin resistance), is one of the major risk factors for sporadic AD [71]. As well as in peripheral tissues, such as muscle, liver, and fat, insulin signaling is also widespread in neurons in the brain, where it modulates central control of metabolism but also behavior and memory [72–74]. In AD patients, brain insulin signaling is impaired, partially due to reduced levels of the hormone [75] and abnormal intracellular sequestration of the IR in neurons caused by Aβ [76, 77]. Conversely, alerted levels of insulin signaling in a diabetic state may impact Aβ metabolism by changing rates of production and clearance as suggested by studies in vitro and in rodent models [78, 79]. Currently, the mechanistic link between brain insulin resistance and amyloidogenic processes is a matter of intense debate and the reader is referred to excellent recent reviews in this subject ([80]). A role for SORLA in insulin signaling in neurons has not been documented yet. However, low levels of the receptor are likely to result in decreased neuronal sensitivity to the hormone as shown for adipocytes previously [70]. Although quite speculative at present, low levels of receptor expression in carriers of SORL1 risk alleles may thus cause both central insulin resistance and increased amyloidogenic burden, and prove to be an explanatory model for the link between neurodegenerative and metabolic diseases that warrant further exploration.
Acknowledgements
We are indebted to Ina-Maria Rudolph for help with the illustrations. Work in the authors’ laboratory was partially funding by the European Research Council (BeyOND No. 335692), the Helmholtz Association (iCEMED, International Research Groups), and the Berlin Institute of Health (Collaborative Research Group 11220008).
References
- 1.Guo Y, Sirkis DW, Schekman R. Protein sorting at the trans-Golgi network. Annu Rev Cell Dev Biol. 2014;30:169–206. doi: 10.1146/annurev-cellbio-100913-013012. [DOI] [PubMed] [Google Scholar]
- 2.Willnow TE, Petersen CM, Nykjaer A. VPS10P-domain receptors—regulators of neuronal viability and function. Nat Rev Neurosci. 2008;9(12):899–909. doi: 10.1038/nrg2454. [DOI] [PubMed] [Google Scholar]
- 3.Jacobsen L, Madsen P, Moestrup SK, Lund AH, Tommerup N, Nykjaer A, Sottrup-Jensen L, Gliemann J, Petersen CM. Molecular characterization of a novel human hybrid-type receptor that binds the alpha2-macroglobulin receptor-associated protein. J Biol Chem. 1996;271(49):31379–31383. doi: 10.1074/jbc.271.49.31379. [DOI] [PubMed] [Google Scholar]
- 4.Yamazaki H, Bujo H, Kusunoki J, Seimiya K, Kanaki T, Morisaki N, Schneider WJ, Saito Y. Elements of neural adhesion molecules and a yeast vacuolar protein sorting receptor are present in a novel mammalian low density lipoprotein receptor family member. J Biol Chem. 1996;271(40):24761–24768. doi: 10.1074/jbc.271.40.24761. [DOI] [PubMed] [Google Scholar]
- 5.Taira K, Bujo H, Hirayama S, Yamazaki H, Kanaki T, Takahashi K, Ishii I, Miida T, Schneider WJ, Saito Y. LR11, a mosaic LDL receptor family member, mediates the uptake of ApoE-rich lipoproteins in vitro. Arterioscler Thromb Vasc Biol. 2001;21(9):1501–1506. doi: 10.1161/hq0901.094500. [DOI] [PubMed] [Google Scholar]
- 6.Jacobsen L, Madsen P, Jacobsen C, Nielsen MS, Gliemann J, Petersen CM. Activation and functional characterization of the mosaic receptor SorLA/LR11. J Biol Chem. 2001;276(25):22788–22796. doi: 10.1074/jbc.M100857200. [DOI] [PubMed] [Google Scholar]
- 7.Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell. 1994;77(4):579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen MS, Gustafsen C, Madsen P, Nyengaard JR, Hermey G, Bakke O, Mari M, Schu P, Pohlmann R, Dennes A, Petersen CM. Sorting by the cytoplasmic domain of the amyloid precursor protein binding receptor SorLA. Mol Cell Biol. 2007;27(19):6842–6851. doi: 10.1128/MCB.00815-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quistgaard EM, Madsen P, Groftehauge MK, Nissen P, Petersen CM, Thirup SS. Ligands bind to Sortilin in the tunnel of a ten-bladed beta-propeller domain. Nat Struct Mol Biol. 2009;16(1):96–98. doi: 10.1038/nsmb.1543. [DOI] [PubMed] [Google Scholar]
- 10.Kitago Y, Nagae M, Nakata Z, Yagi-Utsumi M, Takagi-Niidome S, Mihara E, Nogi T, Kato K, Takagi J. Structural basis for amyloidogenic peptide recognition by sorLA. Nat Struct Mol Biol. 2015;22(3):199–206. doi: 10.1038/nsmb.2954. [DOI] [PubMed] [Google Scholar]
- 11.Andersen OM, Schmidt V, Spoelgen R, Gliemann J, Behlke J, Galatis D, McKinstry WJ, Parker MW, Masters CL, Hyman BT, Cappai R, Willnow TE. Molecular Dissection of the Interaction between Amyloid Precursor Protein and Its Neuronal Trafficking Receptor SorLA/LR11. Biochemistry. 2006;45(8):2618–2628. doi: 10.1021/bi052120v. [DOI] [PubMed] [Google Scholar]
- 12.Mehmedbasic A, Christensen SK, Nilsson J, Ruetschi U, Gustafsen C, Poulsen AS, Rasmussen RW, Fjorback AN, Larson G, Andersen OM. SorLA complement-type repeat domains protect the amyloid precursor protein against processing. J Biol Chem. 2015;290(6):3359–3376. doi: 10.1074/jbc.M114.619940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caglayan S, Takagi-Niidome S, Liao F, Carlo AS, Schmidt V, Burgert T, Kitago Y, Fuchtbauer EM, Fuchtbauer A, Holtzman DM, Takagi J, Willnow TE (2014) Lysosomal sorting of amyloid-beta by the SORLA receptor is impaired by a familial Alzheimer’s disease mutation. Sci Transl Med 6 (223):223ra220. doi:10.1126/scitranslmed.3007747 [DOI] [PubMed]
- 14.Campbell ID, Spitzfaden C. Building proteins with fibronectin type III modules. Structure. 1994;2(5):333–337. doi: 10.1016/S0969-2126(00)00034-4. [DOI] [PubMed] [Google Scholar]
- 15.Rudenko G, Henry L, Henderson K, Ichtchenko K, Brown MS, Goldstein JL, Deisenhofer J. Structure of the LDL receptor extracellular domain at endosomal pH. Science. 2002;298(5602):2353–2358. doi: 10.1126/science.1078124. [DOI] [PubMed] [Google Scholar]
- 16.Hampe W, Riedel IB, Lintzel J, Bader CO, Franke I, Schaller HC. Ectodomain shedding, translocation and synthesis of SorLA are stimulated by its ligand head activator. J Cell Sci. 2000;113(Pt 24):4475–4485. doi: 10.1242/jcs.113.24.4475. [DOI] [PubMed] [Google Scholar]
- 17.Hermey G, Sjogaard SS, Petersen CM, Nykjaer A, Gliemann J. Tumour necrosis factor alpha-converting enzyme mediates ectodomain shedding of Vps10p-domain receptor family members. Biochem J. 2006;395(2):285–293. doi: 10.1042/BJ20051364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt V, Sporbert A, Rohe M, Reimer T, Rehm A, Andersen OM, Willnow TE. SorLA/LR11 regulates processing of amyloid precursor protein via interaction with adaptors GGA and PACS-1. J Biol Chem. 2007;282(45):32956–32964. doi: 10.1074/jbc.M705073200. [DOI] [PubMed] [Google Scholar]
- 19.Seaman MN. Identification of a novel conserved sorting motif required for retromer-mediated endosome-to-TGN retrieval. J Cell Sci. 2007;120(Pt 14):2378–2389. doi: 10.1242/jcs.009654. [DOI] [PubMed] [Google Scholar]
- 20.Fjorback AW, Seaman M, Gustafsen C, Mehmedbasic A, Gokool S, Wu C, Militz D, Schmidt V, Madsen P, Nyengaard JR, Willnow TE, Christensen EI, Mobley WB, Nykjaer A, Andersen OM. Retromer Binds the FANSHY Sorting Motif in SorLA to Regulate Amyloid Precursor Protein Sorting and Processing. J Neurosci. 2012;32(4):1467–1480. doi: 10.1523/JNEUROSCI.2272-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobsen L, Madsen P, Nielsen MS, Geraerts WP, Gliemann J, Smit AB, Petersen CM. The sorLA cytoplasmic domain interacts with GGA1 and -2 and defines minimum requirements for GGA binding. FEBS Lett. 2002;511(1–3):155–158. doi: 10.1016/S0014-5793(01)03299-9. [DOI] [PubMed] [Google Scholar]
- 22.Herskowitz JH, Offe K, Deshpande A, Kahn RA, Levey AI, Lah JJ. GGA1-mediated endocytic traffic of LR11/SorLA alters APP intracellular distribution and amyloid-beta production. Mol Biol Cell. 2012;23(14):2645–2657. doi: 10.1091/mbc.E12-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang TY, Zhao Y, Li X, Wang X, Tseng IC, Thompson R, Tu S, Willnow TE, Zhang YW, Xu H. SNX27 and SORLA interact to reduce amyloidogenic subcellular distribution and processing of amyloid precursor protein. J Neurosci. 2016;36(30):7996–8011. doi: 10.1523/JNEUROSCI.0206-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puertollano R, Bonifacino JS. Interactions of GGA3 with the ubiquitin sorting machinery. Nat Cell Biol. 2004;6(3):244–251. doi: 10.1038/ncb1106. [DOI] [PubMed] [Google Scholar]
- 25.Dumanis SB, Burgert T, Caglayan S, Fuchtbauer A, Fuchtbauer EM, Schmidt V, Willnow TE. Distinct functions for anterograde and retrograde sorting of SORLA in amyloidogenic processes in the brain. J Neurosci. 2015;35(37):12703–12713. doi: 10.1523/JNEUROSCI.0427-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burgert T, Schmidt V, Caglayan S, Lin F, Fuchtbauer A, Fuchtbauer EM, Nykjaer A, Carlo AS, Willnow TE. SORLA-dependent and -independent functions for PACS1 in control of amyloidogenic processes. Mol Cell Biol. 2013;33:4308–4320. doi: 10.1128/MCB.00628-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8(2):101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 28.Haass C, Koo EH, Mellon A, Hung AY, Selkoe DJ. Targeting of cell-surface beta-amyloid precursor protein to lysosomes: alternative processing into amyloid-bearing fragments. Nature. 1992;357(6378):500–503. doi: 10.1038/357500a0. [DOI] [PubMed] [Google Scholar]
- 29.Golde TE, Estus S, Younkin LH, Selkoe DJ, Younkin SG. Processing of the amyloid protein precursor to potentially amyloidogenic derivatives. Science. 1992;255(5045):728–730. doi: 10.1126/science.1738847. [DOI] [PubMed] [Google Scholar]
- 30.Rajendran L, Schneider A, Schlechtingen G, Weidlich S, Ries J, Braxmeier T, Schwille P, Schulz JB, Schroeder C, Simons M, Jennings G, Knolker HJ, Simons K. Efficient inhibition of the Alzheimer’s disease beta-secretase by membrane targeting. Science. 2008;320(5875):520–523. doi: 10.1126/science.1156609. [DOI] [PubMed] [Google Scholar]
- 31.Andersen OM, Reiche J, Schmidt V, Gotthardt M, Spoelgen R, Behlke J, von Arnim CA, Breiderhoff T, Jansen P, Wu X, Bales KR, Cappai R, Masters CL, Gliemann J, Mufson EJ, Hyman BT, Paul SM, Nykjaer A, Willnow TE. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci USA. 2005;102(38):13461–13466. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Offe K, Dodson SE, Shoemaker JT, Fritz JJ, Gearing M, Levey AI, Lah JJ. The lipoprotein receptor LR11 regulates amyloid beta production and amyloid precursor protein traffic in endosomal compartments. J Neurosci. 2006;26(5):1596–1603. doi: 10.1523/JNEUROSCI.4946-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt V, Baum K, Lao A, Rateitschak K, Schmitz Y, Teichmann A, Wiesner B, Petersen CM, Nykjaer A, Wolf J, Wolkenhauer O, Willnow TE. Quantitative modelling of amyloidogenic processing and its influence by SORLA in Alzheimer’s disease. EMBO J. 2012;31(1):187–200. doi: 10.1038/emboj.2011.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, Katayama T, Baldwin CT, Cheng R, Hasegawa H, Chen F, Shibata N, Lunetta KL, Pardossi-Piquard R, Bohm C, Wakutani Y, Cupples LA, Cuenco KT, Green RC, Pinessi L, Rainero I, Sorbi S, Bruni A, Duara R, Friedland RP, Inzelberg R, Hampe W, Bujo H, Song YQ, Andersen OM, Willnow TE, Graff-Radford N, Petersen RC, Dickson D, Der SD, Fraser PE, Schmitt-Ulms G, Younkin S, Mayeux R, Farrer LA, St George-Hyslop P. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39(2):168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohe M, Carlo AS, Breyhan H, Sporbert A, Militz D, Schmidt V, Wozny C, Harmeier A, Erdmann B, Bales KR, Wolf S, Kempermann G, Paul SM, Schmitz D, Bayer TA, Willnow TE, Andersen OM. Sortilin-related receptor with A-type repeats (SORLA) affects the amyloid precursor protein-dependent stimulation of ERK signaling and adult neurogenesis. J Biol Chem. 2008;283(21):14826–14834. doi: 10.1074/jbc.M710574200. [DOI] [PubMed] [Google Scholar]
- 36.Dodson SE, Andersen OM, Karmali V, Fritz JJ, Cheng D, Peng J, Levey AI, Willnow TE, Lah JJ. Loss of LR11/SORLA enhances early pathology in a mouse model of amyloidosis: evidence for a proximal role in Alzheimer’s disease. J Neurosci. 2008;28(48):12877–12886. doi: 10.1523/JNEUROSCI.4582-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y, Peng Y, Che P, Gannon M, Liu Y, Li L, Bu G, van Groen T, Jiao K, Wang Q. alpha(2A) adrenergic receptor promotes amyloidogenesis through disrupting APP-SorLA interaction. Proc Natl Acad Sci USA. 2014;111(48):17296–17301. doi: 10.1073/pnas.1409513111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thorton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Moron FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fievet N, Huentelman MW, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuiness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossu P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Deniz Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, European Alzheimer’s Disease I, Genetic, Environmental Risk in Alzheimer’s D, Alzheimer’s Disease Genetic C, Cohorts for H, Aging Research in Genomic E, Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannefelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH, Jr., Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltuenen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nothen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45(12):1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyashita A, Koike A, Jun G, Wang LS, Takahashi S, Matsubara E, Kawarabayashi T, Shoji M, Tomita N, Arai H, Asada T, Harigaya Y, Ikeda M, Amari M, Hanyu H, Higuchi S, Ikeuchi T, Nishizawa M, Suga M, Kawase Y, Akatsu H, Kosaka K, Yamamoto T, Imagawa M, Hamaguchi T, Yamada M, Morihara T, Takeda M, Takao T, Nakata K, Fujisawa Y, Sasaki K, Watanabe K, Nakashima K, Urakami K, Ooya T, Takahashi M, Yuzuriha T, Serikawa K, Yoshimoto S, Nakagawa R, Kim JW, Ki CS, Won HH, Na DL, Seo SW, Mook-Jung I, Alzheimer Disease Genetics C, St George-Hyslop P, Mayeux R, Haines JL, Pericak-Vance MA, Yoshida M, Nishida N, Tokunaga K, Yamamoto K, Tsuji S, Kanazawa I, Ihara Y, Schellenberg GD, Farrer LA, Kuwano R. SORL1 is genetically associated with late-onset Alzheimer’s disease in Japanese, Koreans and Caucasians. PloS One. 2013;8(4):e58618. doi: 10.1371/journal.pone.0058618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grear KE, Ling IF, Simpson JF, Furman JL, Simmons CR, Peterson SL, Schmitt FA, Markesbery WR, Liu Q, Crook JE, Younkin SG, Bu G, Estus S. Expression of SORL1 and a novel SORL1 splice variant in normal and Alzheimers disease brain. Mol Neurodegener. 2009;4:46. doi: 10.1186/1750-1326-4-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young JE, Boulanger-Weill J, Williams DA, Woodruff G, Buen F, Revilla AC, Herrera C, Israel MA, Yuan SH, Edland SD, Goldstein LS. Elucidating molecular phenotypes caused by the SORL1 Alzheimer’s disease genetic risk factor using human induced pluripotent stem cells. Cell Stem Cell. 2015;16(4):373–385. doi: 10.1016/j.stem.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caglayan S, Bauerfeind A, Schmidt V, Carlo AS, Prabakaran T, Hubner N, Willnow TE. Identification of Alzheimer Disease Risk Genotype That Predicts Efficiency of SORL1 Expression in the Brain. Arch Neurol. 2012;69(3):373–379. doi: 10.1001/archneurol.2011.788. [DOI] [PubMed] [Google Scholar]
- 43.Andersen OM, Rudolph IM, Willnow TE. Risk factor SORL1: from genetic association to functional validation in Alzheimer’s disease. Acta Neuropathol. 2016 doi: 10.1007/s00401-016-1615-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pottier C, Hannequin D, Coutant S, Rovelet-Lecrux A, Wallon D, Rousseau S, Legallic S, Paquet C, Bombois S, Pariente J, Thomas-Anterion C, Michon A, Croisile B, Etcharry-Bouyx F, Berr C, Dartigues JF, Amouyel P, Dauchel H, Boutoleau-Bretonniere C, Thauvin C, Frebourg T, Lambert JC, Campion D, Collaborators PG. High frequency of potentially pathogenic SORL1 mutations in autosomal dominant early-onset Alzheimer disease. Mol Psychiatry. 2012;17(9):875–879. doi: 10.1038/mp.2012.15. [DOI] [PubMed] [Google Scholar]
- 45.Rohe M, Hartl D, Fjorback AN, Klose J, Willnow TE. SORLA-Mediated Trafficking of TrkB Enhances the Response of Neurons to BDNF. PLoS One. 2013;8(8):e72164. doi: 10.1371/journal.pone.0072164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohe M, Synowitz M, Glass R, Paul SM, Nykjaer A, Willnow TE. Brain-derived neurotrophic factor reduces amyloidogenic processing through control of SORLA gene expression. J Neurosci. 2009;29(49):15472–15478. doi: 10.1523/JNEUROSCI.3960-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geng Z, Xu FY, Huang SH, Chen ZY. Sorting protein-related receptor SorLA controls regulated secretion of glial cell line-derived neurotrophic factor. J Biol Chem. 2011;286(48):41871–41882. doi: 10.1074/jbc.M111.246413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glerup S, Lume M, Olsen D, Nyengaard JR, Vaegter CB, Gustafsen C, Christensen EI, Kjolby M, Hay-Schmidt A, Bender D, Madsen P, Saarma M, Nykjaer A, Petersen CM. SorLA controls neurotrophic activity by sorting of GDNF and its receptors GFRalpha1 and RET. Cell reports. 2013;3(1):186–199. doi: 10.1016/j.celrep.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 49.Larsen JV, Kristensen AM, Pallesen LT, Bauer J, Vaegter CB, Nielsen MS, Madsen P, Petersen CM. Cytokine-Like Factor 1, an Essential Facilitator of Cardiotrophin-Like Cytokine: Ciliary Neurotrophic Factor Receptor alpha Signaling and sorLA-Mediated Turnover. Mol Cell Biol. 2016;36(8):1272–1286. doi: 10.1128/MCB.00917-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riedel IB, Hermans-Borgmeyer I, Hubner CA. SorLA, a member of the LDL receptor family, is expressed in the collecting duct of the murine kidney. Histochem Cell Biol. 2002;118(3):183–191. doi: 10.1007/s00418-002-0445-8. [DOI] [PubMed] [Google Scholar]
- 51.Reiche J, Theilig F, Rafiqi FH, Carlo AS, Militz D, Mutig K, Todiras M, Christensen EI, Ellison DH, Bader M, Nykjaer A, Bachmann S, Alessi D, Willnow TE. SORLA/SORL1 functionally interacts with SPAK to control renal activation of Na(+)-K(+)-Cl(−) cotransporter 2. Mol Cell Biol. 2010;30(12):3027–3037. doi: 10.1128/MCB.01560-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borschewski A, Himmerkus N, Boldt C, Blankenstein KI, McCormick JA, Lazelle R, Willnow TE, Jankowski V, Plain A, Bleich M, Ellison DH, Bachmann S, Mutig K. Calcineurin and Sorting-Related Receptor with A-Type Repeats Interact to Regulate the Renal Na(+)-K(+)-2Cl(−) Cotransporter. J Am Soc Nephrol. 2016;27(1):107–119. doi: 10.1681/ASN.2014070728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang SS, Shi W, Wang X, Velky L, Greenlee S, Wang MT, Drake TA, Lusis AJ. Mapping, genetic isolation, and characterization of genetic loci that determine resistance to atherosclerosis in C3H mice. Arterioscler Thromb Vasc Biol. 2007;27(12):2671–2676. doi: 10.1161/ATVBAHA.107.148106. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi M, Bujo H, Jiang M, Noike H, Saito Y, Shirai K. Enhanced circulating soluble LR11 in patients with coronary organic stenosis. Atherosclerosis. 2010;210(2):581–584. doi: 10.1016/j.atherosclerosis.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 55.Ogita M, Miyauchi K, Dohi T, Tsuboi S, Miyazaki T, Yokoyama T, Yokoyama K, Shimada K, Kurata T, Jiang M, Bujo H, Daida H. Increased circulating soluble LR11 in patients with acute coronary syndrome. Clin Chim Acta. 2013;415:191–194. doi: 10.1016/j.cca.2012.10.047. [DOI] [PubMed] [Google Scholar]
- 56.Klinger SC, Glerup S, Raarup MK, Mari MC, Nyegaard M, Koster G, Prabakaran T, Nilsson SK, Kjaergaard MM, Bakke O, Nykjaer A, Olivecrona G, Petersen CM, Nielsen MS. SorLA regulates the activity of lipoprotein lipase by intracellular trafficking. J Cell Sci. 2011;124(Pt 7):1095–1105. doi: 10.1242/jcs.072538. [DOI] [PubMed] [Google Scholar]
- 57.Nilsson SK, Lookene A, Beckstead JA, Gliemann J, Ryan RO, Olivecrona G. Apolipoprotein A-V interaction with members of the low density lipoprotein receptor gene family. Biochemistry. 2007;46(12):3896–3904. doi: 10.1021/bi7000533. [DOI] [PubMed] [Google Scholar]
- 58.Nilsson SK, Christensen S, Raarup MK, Ryan RO, Nielsen MS, Olivecrona G. Endocytosis of apolipoprotein A-V by members of the low density lipoprotein receptor and the VPS10p domain receptor families. J Biol Chem. 2008;283(38):25920–25927. doi: 10.1074/jbc.M802721200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mendoza-Barbera E, Julve J, Nilsson SK, Lookene A, Martin-Campos JM, Roig R, Lechuga-Sancho AM, Sloan JH, Fuentes-Prior P, Blanco-Vaca F. Structural and functional analysis of APOA5 mutations identified in patients with severe hypertriglyceridemia. J Lipid Res. 2013;54(3):649–661. doi: 10.1194/jlr.M031195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanaki T, Bujo H, Hirayama S, Ishii I, Morisaki N, Schneider WJ, Saito Y. Expression of LR11, a mosaic LDL receptor family member, is markedly increased in atherosclerotic lesions. Arterioscler Thromb Vasc Biol. 1999;19(11):2687–2695. doi: 10.1161/01.ATV.19.11.2687. [DOI] [PubMed] [Google Scholar]
- 61.Ting HJ, Stice JP, Schaff UY, Hui DY, Rutledge JC, Knowlton AA, Passerini AG, Simon SI. Triglyceride-rich lipoproteins prime aortic endothelium for an enhanced inflammatory response to tumor necrosis factor-alpha. Circ Res. 2007;100(3):381–390. doi: 10.1161/01.RES.0000258023.76515.a3. [DOI] [PubMed] [Google Scholar]
- 62.Zhu Y, Bujo H, Yamazaki H, Hirayama S, Kanaki T, Takahashi K, Shibasaki M, Schneider WJ, Saito Y. Enhanced expression of the LDL receptor family member LR11 increases migration of smooth muscle cells in vitro. Circulation. 2002;105(15):1830–1836. doi: 10.1161/01.CIR.0000014413.91312.EF. [DOI] [PubMed] [Google Scholar]
- 63.McCarthy C, O’Gaora P, James WG, de Gaetano M, McClelland S, Fitzgerald DJ, Belton O. SorLA modulates atheroprotective properties of CLA by regulating monocyte migration. Atherosclerosis. 2010;213(2):400–407. doi: 10.1016/j.atherosclerosis.2010.09.025. [DOI] [PubMed] [Google Scholar]
- 64.Gliemann J, Hermey G, Nykjaer A, Petersen CM, Jacobsen C, Andreasen PA (2004) The mosaic receptor sorLA/LR11 binds components of the plasminogen activating system and PDGF-BB similarly to low density lipoprotein receptor-related protein (LRP1) but mediates slow internalization of bound ligand. Biochem J 381(Pt 1):203–212. doi:10.1042/BJ20040149 [DOI] [PMC free article] [PubMed]
- 65.Zhu Y, Bujo H, Yamazaki H, Ohwaki K, Jiang M, Hirayama S, Kanaki T, Shibasaki M, Takahashi K, Schneider WJ, Saito Y. LR11, an LDL receptor gene family member, is a novel regulator of smooth muscle cell migration. Circ Res. 2004;94(6):752–758. doi: 10.1161/01.RES.0000120862.79154.0F. [DOI] [PubMed] [Google Scholar]
- 66.Ohwaki K, Bujo H, Jiang M, Yamazaki H, Schneider WJ, Saito Y. A secreted soluble form of LR11, specifically expressed in intimal smooth muscle cells, accelerates formation of lipid-laden macrophages. Arterioscler Thromb Vasc Biol. 2007;27(5):1050–1056. doi: 10.1161/ATVBAHA.106.137091. [DOI] [PubMed] [Google Scholar]
- 67.Parks BW, Nam E, Org E, Kostem E, Norheim F, Hui ST, Pan C, Civelek M, Rau CD, Bennett BJ, Mehrabian M, Ursell LK, He A, Castellani LW, Zinker B, Kirby M, Drake TA, Drevon CA, Knight R, Gargalovic P, Kirchgessner T, Eskin E, Lusis AJ. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 2013;17(1):141–152. doi: 10.1016/j.cmet.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith EN, Chen W, Kahonen M, Kettunen J, Lehtimaki T, Peltonen L, Raitakari OT, Salem RM, Schork NJ, Shaw M, Srinivasan SR, Topol EJ, Viikari JS, Berenson GS, Murray SS. Longitudinal genome-wide association of cardiovascular disease risk factors in the Bogalusa heart study. PLoS Genet. 2010;6(9):e1001094. doi: 10.1371/journal.pgen.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Whittle AJ, Jiang M, Peirce V, Relat J, Virtue S, Ebinuma H, Fukamachi I, Yamaguchi T, Takahashi M, Murano T, Tatsuno I, Takeuchi M, Nakaseko C, Jin W, Jin Z, Campbell M, Schneider WJ, Vidal-Puig A, Bujo H. Soluble LR11/SorLA represses thermogenesis in adipose tissue and correlates with BMI in humans. Nat Commun. 2015;6:8951. doi: 10.1038/ncomms9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmidt V, Schulz N, Yan X, Schurmann A, Kempa S, Kern M, Bluher M, Poy MN, Olivecrona G, Willnow TE. SORLA facilitates insulin receptor signaling in adipocytes and exacerbates obesity. J Clin Invest. 2016;126(7):2706–2720. doi: 10.1172/JCI84708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sims-Robinson C, Kim B, Rosko A, Feldman EL. How does diabetes accelerate Alzheimer disease pathology? Nat Rev Neurol. 2010;6(10):551–559. doi: 10.1038/nrneurol.2010.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bruning JC, Gautam D, Burks DJ, Gillette J, Schubert M, Orban PC, Klein R, Krone W, Muller-Wieland D, Kahn CR. Role of brain insulin receptor in control of body weight and reproduction. Science. 2000;289(5487):2122–2125. doi: 10.1126/science.289.5487.2122. [DOI] [PubMed] [Google Scholar]
- 73.Koch L, Wunderlich FT, Seibler J, Konner AC, Hampel B, Irlenbusch S, Brabant G, Kahn CR, Schwenk F, Bruning JC. Central insulin action regulates peripheral glucose and fat metabolism in mice. J Clin Invest. 2008;118(6):2132–2147. doi: 10.1172/JCI31073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McNay EC, Ong CT, McCrimmon RJ, Cresswell J, Bogan JS, Sherwin RS. Hippocampal memory processes are modulated by insulin and high-fat-induced insulin resistance. Neurobiol Learn Mem. 2010;93(4):546–553. doi: 10.1016/j.nlm.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Steen E, Terry BM, Rivera EJ, Cannon JL, Neely TR, Tavares R, Xu XJ, Wands JR, de la Monte SM. Impaired insulin and insulin-like growth factor expression and signaling mechanisms in Alzheimer’s disease–is this type 3 diabetes? J Alzheimers Dis. 2005;7(1):63–80. doi: 10.3233/jad-2005-7107. [DOI] [PubMed] [Google Scholar]
- 76.Moloney AM, Griffin RJ, Timmons S, O’Connor R, Ravid R, O’Neill C. Defects in IGF-1 receptor, insulin receptor and IRS-1/2 in Alzheimer’s disease indicate possible resistance to IGF-1 and insulin signalling. Neurobiol Aging. 2010;31(2):224–243. doi: 10.1016/j.neurobiolaging.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 77.Zhao WQ, De Felice FG, Fernandez S, Chen H, Lambert MP, Quon MJ, Krafft GA, Klein WL. Amyloid beta oligomers induce impairment of neuronal insulin receptors. FASEB J. 2008;22(1):246–260. doi: 10.1096/fj.06-7703com. [DOI] [PubMed] [Google Scholar]
- 78.Shiiki T, Ohtsuki S, Kurihara A, Naganuma H, Nishimura K, Tachikawa M, Hosoya K, Terasaki T. Brain insulin impairs amyloid-beta(1-40) clearance from the brain. J Neurosci. 2004;24(43):9632–9637. doi: 10.1523/JNEUROSCI.2236-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gasparini L, Gouras GK, Wang R, Gross RS, Beal MF, Greengard P, Xu H. Stimulation of beta-amyloid precursor protein trafficking by insulin reduces intraneuronal beta-amyloid and requires mitogen-activated protein kinase signaling. J Neurosci. 2001;21(8):2561–2570. doi: 10.1523/JNEUROSCI.21-08-02561.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stanley M, Macauley SL, Holtzman DM. Changes in insulin and insulin signaling in Alzheimer’s disease: cause or consequence? J Exp Med. 2016;213(8):1375–1385. doi: 10.1084/jem.20160493. [DOI] [PMC free article] [PubMed] [Google Scholar]