Abstract
Prolonged (8 weeks) oral administration of clofazimine results in a profound pharmacodynamic response- bioaccumulation in macrophages (including Kupffer cells) as intracellular crystal-like drug inclusions (CLDIs) with an associated increase in interleukin-1 receptor antagonist production. Notably, CLDI formation in Kupffer cells concomitantly occurs with the formation of macrophage-centric granulomas. Accordingly, we sought to understand the impact of these events on host metabolism using 1H-nuclear magnetic resonance metabolomics. Mice received a clofazimine - or vehicle-enriched (sham) diet for at least 8 weeks. At two weeks, the antimicrobial activity of clofazimine was evident by changes in urine metabolites. From 2 to 8 weeks, there was a striking change in metabolite levels indicative of a reorientation of host energy metabolism paralleling the onset of CLDI and granuloma formation. This was evidenced by a progressive reduction in urine levels of metabolites involved in one-carbon metabolism with corresponding increases in whole blood, and changes in metabolites associated with lipid, nucleotide and amino acid metabolism, and glycolysis. Although clofazimine-fed mice ate more, they gained less weight than control mice. Together, these results indicate that macrophage sequestration of clofazimine as CLDIs and granuloma formation is accompanied by a profound metabolic disruption in energy homeostasis and one-carbon metabolism.
Keywords: intracellular drug crystals, granulomas, metabolomics, microbiome, pharmacodynamics, pharmacometabolomics, nuclear magnetic resonance
INTRODUCTION
Drug bioaccumulation and the formation of insoluble drug complexes in humans is a pharmacological phenomenon that is traditionally perceived as an unwanted side effect and these drugs are typically excluded from the drug development pipeline. One exception to this is the red-pigmented antibiotic clofazimine (CFZ), which is included in the World Health Organization’s List of Essential Medicines and has been used as part of the standard treatment of leprosy since the 1960s.1,2 In patients, CFZ accumulates to very high levels in tissues3–6 resulting in visible changes in the pigmentation of skin and other organs, as well as the formation of intracellular crystal-like drug inclusions (CLDIs) – a biocrystalline form of CFZ inside macrophages.3,7–12 In spite of its bioaccumulation, several clinical trials have established CFZ as a potentially useful therapeutic agent for treating a variety of chronic inflammatory and infectious diseases.13–21 In a similar manner, therapeutic doses of CFZ administered to mice results in red pigmentation of the skin, with the soluble form of CFZ accumulating in adipose tissue, that is evident within two weeks.22,23 This is followed by the appearance of red CLDIs that exclusively form in resident tissue macrophages.23–25
The predominant site of CLDI formation is the liver where most CLDIs are sequestered in massive quantities inside macrophages (Kupffer cells) that aggregate to form granulomas.22,26 These CLDI-containing granulomas can modulate the immune response of mice by boosting the production of hepatic interleukin-1 receptor antagonist (IL-1RA), while suppressing inflammasome activation to provide protection from lethal injury.22,26 The immunomodulatory properties of CLDIs coincide with the onset of macrophage granuloma formation, and therefore, it is reasonable to expect a metabolic consequence considering the relationship between metabolism and immunomodulation.27–29 Moreover, the identification of host metabolic changes could enhance the mechanistic understanding of the macrophage-mediated drug sequestration response.30
To this end, the aim of the work presented here was to employ metabolomics of urine and whole blood (WB) samples and targeted assays of select analytes to better understand the metabolic consequences of CFZ bioaccumulation and macrophage-mediated drug sequestration. Quantitative proton (1H)-1-dimensional (1D) nuclear magnetic resonance (NMR) metabolomics generates metabolite data which can be used to identify metabolic changes associated with phenotype.31–33 Metabolites are the end-products of gene and protein activity and represent the organism’s physiological condition.33,34 This principle has been illustrated by a number of pharmacometabolomics studies34–38 for which metabolomics has served as a sensitive indicator of phenotype and has informed biological mechanisms. Here, we show that CFZ administration resulted in metabolic reprogramming that is linked to its pharmacodynamics and provides insight into the mechanisms involved in its sequestration by macrophages.
MATERIALS and METHODS
Animals
Male mice (4 week old, C57Bl6) were purchased from the Jackson Laboratory (Bar Harbor, ME) and acclimatized for 1 week in a specific-pathogen-free animal facility. All animal care was provided by the University of Michigan's Unit for Laboratory Animal Medicine (ULAM), and the experimental protocol was submitted to and approved by the University of Michigan’s Institutional Committee on Use and Care of Animals. Clofazimine (C8895; Sigma-Aldrich, St. Louis, MO) was dissolved in sesame oil (Shirakiku, Japan) to achieve a concentration of 3 mg/ml, which was mixed with Powdered Lab Diet 5001 (PMI International, Inc., St. Louis, MO; http://www.labdiet.com/cs/groups/lolweb/@labdiet/documents/web_content/mdrf/mdi4/~edisp/ducm04_028021.pdf) to produce a 0.03% drug to powdered feed mix, as previously described.22 A corresponding amount of sesame oil was mixed with chow for sham treatment (control). Mice had access to food and water ad libitum with a 12 h artificial light and 12 h dark cycle.
Microscopy
CFZ- and sham-treated mice were euthanized by exsanguination while deeply anesthetized with an intraperitoneal injection of ketamine and xylazine (100 and 10 mg/kg, respectively), and the whole mouse was perfused blood-free by an infusion of ice-cold DPBS into the heart. For fluorescence immunohistochemistry, the liver was removed en bloc and frozen in Tissue-Plus OCT compound (Fisher HealthCare, Houston, TX). Tissue blocks were sectioned (6 μm) using a Leica 3050S cryostat, fixed in paraformaldehyde (4% for 10 min), blocked with 1% bovine serum albumin (BSA, Sigma, St. Louis, MO), 5% goat serum (Sigma), and 2.5 mM glycine (Sigma) in DPBS (Life Technologies, Carlsbad, CA) for 2 h. The samples were then incubated with anti-F4/80 antibody (1 μg/ml in 1% BSA; Cat. No. ab6640 Abcam, UK) overnight (4°C) followed by Alexa Fluor 488-conjugated secondary antibody (5 μg/ml in 1% BSA; 1h at RT; Cell Signaling Technology, Danvers, MA) and Hoechst 33342 (1 μg/ml, Life Technologies) was used for nuclear detection. Sections were mounted on glass slides with ProlongGold (Life Technologies). For hematoxylin and eosin (H&E) staining, the liver was formalin-fixed and paraffin-embedded and stained by the Pathology Core for Animal Research (PCAR) in the ULAM at the University of Michigan. Brightfield and fluorescence (DAPI and FITC) images were acquired using a Nikon Eclipse Ti (Japan) inverted microscope equipped with a Nikon Digital Sight DS-Fi2 camera (Japan) for brightfield and Photometrics Coolsnap Myo camera (Tucson, AZ) for fluorescence.
Quantification of clofazimine in mouse liver
At predetermined time points, mice were sacrificed as described above. Each liver was harvested, weighed, washed in cold DPBS, minced with a scalpel and homogenized to a suspension in DPBS. This solution was centrifuged (500 x g for 10 min) with the addition of 9 M H2SO4. The absorbance of the supernatants was measured at λ=540 nm (A540) and 750 nm (A750) using a Synergy II plate reader (Biotek, Winooski, VT USA). Corrected absorbance was determined as (A540 – A750) and CFZ content was calculated using calibrated standards in 9 M H2SO4 and the value was corrected for organ weight.39
Metabolic studies
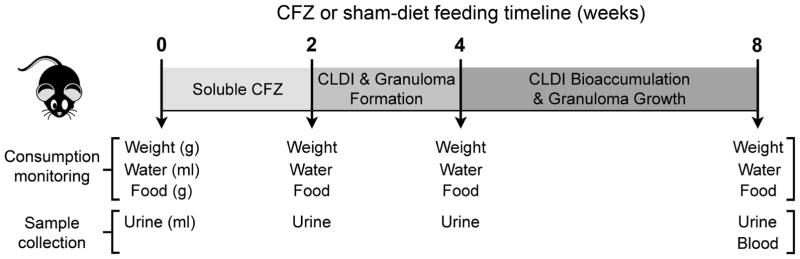
For the metabolic studies, mice (4 mice/cage) were placed in a Tecniplast Metabolic Cage (TECNIPLAST S.p.A. Via I Maggio, 6 - 21020 BUGUGGIATE (VA) Italy) prior to and at 2, 4 and 8 weeks after CFZ or sham treatment. These specialized cages allow for the separate collection of urine and feces. In advance of the study, the urine collection containers were coated with 10% NaN3 to help retard bacterial growth.40 The containers were air-dried in a biosafety cabinet before being attached to the cage. The water bottle and the food dispenser were filled and the volume and weight, respectively, were recorded before assembly to the cage. In advance of the metabolic studies, each mouse was ear tagged so that the same mice were assigned to the same metabolic cage at each time point. Metabolic studies were consistently conducted over 18 h (3:00pm to 9:00am) during which food (regular chow) and water were available ad libitum. At the conclusion of each 18 h study, the volume of water (mL) and the amount of food consumed (g) were determined (Fig. 1).
Figure 1.
Timeline of clofazimine (CFZ) or sham-diet administration and time points at which mouse weight, water and food consumption were measured, and urine and blood samples were collected for 1H-NMR metabolomics.
Upon completion of the 8-week study, mice were anesthetized with an intraperitoneal dose of ketamine and xylazine (100 and 10 mg/kg, respectively). Blood samples were collected by cardiac puncture and immediately transferred into a microfuge tube containing sodium heparin. They were then quickly placed in an ice-water bath for no more than 10 min after which they were stored (−80°C) until the time of assay (Fig. 1).
Urine sample processing
At the conclusion of each 18 h metabolic study, urine volume was measured and each sample was transferred to a 15 mL conical tube and centrifuged (2000 x g for 10 min at 4°C) to precipitate sediment. 40,41 The supernatants were carefully transferred to new 15 mL conical tubes so as not to disturb the pellet. The initial pH was recorded and the volume of each urine sample was measured again. An internal standard (Chenomx IS-2; deuterated 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS-d6) and sodium azide in D2O, pH 7.0) equivalent to 10% of the sample volume was added, the sample was gently mixed by pipetting it up and down several times and then aliquots (500μL) were transferred into screw-top cryotubes. Each tube was sealed with parafilm and they were stored (−80°C) until the time of assay. At the time of assay, samples were thawed at room temperature. The pH of each sample was measured and corrected to 6.5–7.5 with the addition of NaOD or DCl, as applicable, and transferred to an NMR tube (528-PP-7, 5mm; Wilmad, Vineland, NJ, USA) for assay.
Whole blood sample processing
At the time of assay WB samples were thawed on ice and subjected to a 1:1 methanol (MeOH)/chloroform (CHCl3)/water extraction to remove macromolecules as previously described.35,42 The aqueous fractions of each sample were dried by lyophilization and resuspended in 500uL D2O with DSS-d6 internal standard Chenomx IS-2 (Chenomx) of known concentration. As with urine samples, pH was adjusted and samples were transferred to NMR tubes for assay.
1H-NMR metabolomics
The 1-dimensional (D)-1H-NMR spectrum of each urine sample and the aqueous fraction of each WB sample was acquired at the University of Michigan’s Biochemical NMR Core Laboratory on a Varian (now Agilent Inc., Santa Clara, CA, USA) 11.74 Tesla (500 MHz) NMR spectrometer with a VNMRS console operated by host software VNMRJ 4.0, and equipped with a 5mm Agilent “One-probe.” NMR spectra were recorded using 32 scans of the first increment of a 1H,1H-NOESY (commonly referred to as a 1D-NOESY or METNOESY) pulse sequence.40 The NMR pulse sequence was as follows: a 1s recovery delay, a 990ms saturation pulse of 80 Hz (γB1) induced field strength empirically centered on the water resonance, two calibrated 90° pulses, a mixing time (t mix) of 100ms, a final 90° pulse, and an acquisition period of 4 seconds. Optimal excitation pulse widths were obtained by utilizing an array of pulse lengths to determine the 360° pulse null for water, and dividing by four to obtain the 90° optimum. This strategy avoids off resonance and radiation damping issues.40 Spectra were acquired at a room temperature of 295.45 ± 0.3 K.
The resulting NMR spectra of urine and WB were analyzed using Chenomx NMR Suite 8.0 (Chenomx, Inc., Edmonton, Alberta, Canada; chenomx.com). The Processor module was used to phase shift and baseline correct each spectrum.40,43 Compounds were then identified and quantified in the Profiler module of the software, which accounts for the pH of the sample and the concentration of the IS and quantifies metabolite concentration relative to the IS. Metabolite identity was confirmed using the Chenomx Compound Library, which contains 312 compounds and in some cases, a 2-dimensional spectrum was acquired (see supporting information, Fig. S2A–F). Spectral processing, compound identification and quantification of the urine and WB samples were each completed by users who were blinded to the samples’ treatment allocation.
Targeted high-performance liquid chromatography (HPLC) of urine analytes
CFZ has been reported to influence antioxidant capacity 44 and the production of reactive oxygen species.45 To assay for the concentrations of some of these redox analytes (cysteine, cystine, glutathione and glutathione disulfide), samples were derivatized with 2,3-dinitrofluorobenzene followed by HPLC analysis as previously described.46,47 This assay also simultaneously detects glutamate and aspartate. These metabolites are not routinely detected by our NMR assay of urine.48 The limit of detection of the assay is 2–3μM for cystine and glutathione disulfide and 3–4μM for glutamate, aspartate, glutathione, and cysteine; the coefficient of variation of the assay is < 5%.
Data analysis
To convert the urine metabolite and analyte data from μM to μmole, concentration values were multiplied by 0.001 and the resulting value by the measured volume of each sample. The entire NMR urine metabolite data set (μmole) was auto scaled and the WB metabolite data were range scaled and log transformed to achieve a normal distribution of the data.49 For parametric statistical analysis of the quantified data, the mean normalized value of each urine metabolite at each time point for sham and CFZ-treated mice were compared by repeated measures ANOVA followed by Sidak’s multiple comparison test.50 The mean normalized concentration of each WB metabolite of CFZ and sham-treated mice were compared by an unpaired Student’s t-test with Welch’s correction for different variances when applicable. Each resulting p value was corrected for false discovery by calculating a corresponding false discovery rate (FDR) using the method of Storey, et al.51 Urine metabolites at each time point (pre-treatment, and 2, 4, and 8 weeks) and WB metabolites were ranked by ascending FDR and those with an FDR ≤ 15% were considered as potentially discriminating52 for CFZ administration and were used in subsequent bioinformatics analysis (see below). Targeted urine analyte data were compared using a Mann-Whitney test. As an assessment of renal function, the rate of creatinine excretion was calculated by dividing the urine creatinine concentration (μmole/mL) by 18h (the length of sample collection). This and other measured parameters such as urine volume, body weight and food consumption were also compared between CFZ- and sham-treated mice at each time point by ANOVA followed by Sidak’s multiple comparison test. Statistical analyses were done and associated graphs were constructed using GraphPad Prism version 6g for Mac (GraphPad software, La Jolla, CA., www.graphpad.com) and Sigmaplot version 9 (Systat Software, San Jose, CA). Figures were constructed using Adobe Illustrator or Photoshop (CS6, Adobe Systems, San Jose, CA). Data, unless otherwise stated, are presented as the median with the interquartile range (IQR).
Bioinformatics analysis
To assess the relationship between the metabolites that differentiated CFZ and sham-treated mice, the 8-week urine and WB metabolites with associated Kyoto Encyclopedia of Genes and Genomes (KEGG) ID and FDR ≤ 15% were used to generate compound networks using Metscape53,54 a plugin for Cytoscape (http://www.cytoscape.org/). Metscape is a freely available online tool that permits metabolic network analysis and data visualization (http://metscape.ncibi.org/). Metscape contains nearly 2,700 compounds, 870 enzymes, 1,400 genes, and 3,000 metabolic reactions as defined in the Edinburgh human metabolic network and uses the KEGG ID as the primary compound identifier.
RESULTS
Prolonged (8 week) CFZ administration results in CFZ accumulation, increased macrophage number and granuloma formation in the liver
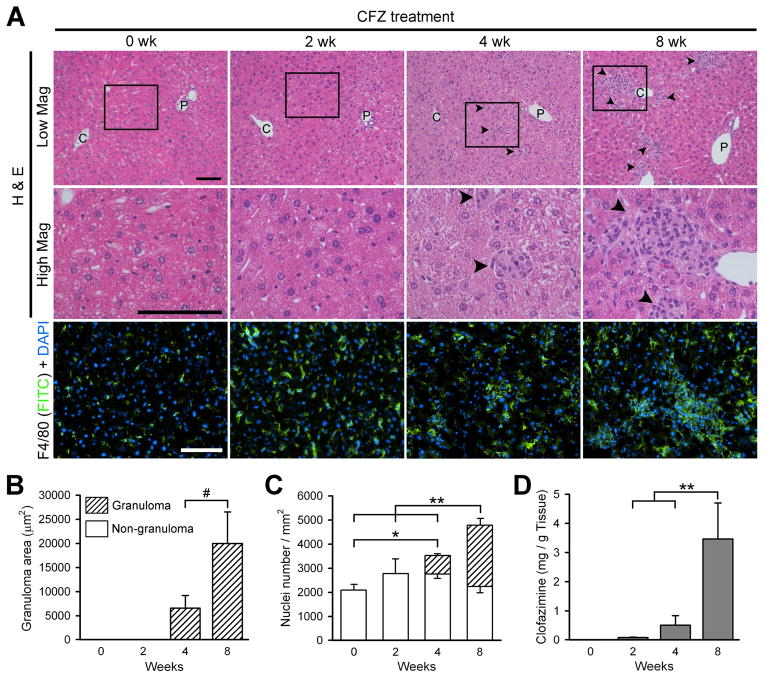
Histological analyses of liver H&E stained sections show that after two weeks of CFZ treatment, hepatic granulomas are not evident (Fig. 2A, H&E), since CFZ exists in the soluble form.26 However, between 2 and 4 weeks, CFZ crystalizes in the liver22,26 and by 4 weeks, small granulomas (Mean±S.D.: 6596±2609 μm2) are apparent (Fig. 2A and B). Over the next four weeks, granuloma area increased by 3-fold (Fig. 2B), and many are formed around the central vein (Fig. 2A). Nuclear counting of F4/80 and DAPI stained liver sections shows a gradual increase in F4/80 positive cells over the 8-week treatment period (Fig. 2A and C). By 8 weeks, liver CFZ concentration reached its peak22,26, which was accompanied by a 2.4-fold increase in total cell number (per field) (Fig. 2C). The latter represents a dramatic increase in macrophage numbers, which are concentrated in granulomas (Fig. 2A F4/80+nucleus and C). These changes coincided with a progressive increase in the liver CFZ concentration (Fig. 2D) which is consistent with previous studies in mice.22,26
Figure 2.
Prolonged (8 week) administration leads to the bioaccumulation of clofazimine (CFZ) in the liver and increases macrophage influx leading to granuloma formation. (A) Representative bright field (H & E, low and high magnification) and fluorescence immunohistochemistry (FITC for F4/80 and DAPI for nucleus) images of murine liver sections from untreated mice control mice and mice treated with CFZ for 2, 4 and 8 weeks. The square box in low magnification image (H&E) is represented in the high magnification image. Arrowheads represent granulomas. C, central vein; P, portal vein. Scale bar = 100 μm. (B) Granuloma area measurements from liver H & E bright field images. Data are the mean ± S.D. of 40 granulomas from two mice/timepoint. (C) Quantification of nuclei (DAPI) number per liver area (mm2) from DAPI images. Data are the mean ± S.D of 3 images/time point. One-way ANOVA was used to compare nuclei number between time points and p ≤ 0.05 was considered statistically significant. *p ≤ 0.05; **p ≤ 0.01; # ≤ 0.001
CLDI formation and macrophage-mediated sequestration of CLDIs is accompanied by a change in the urinary excretion of various key metabolites
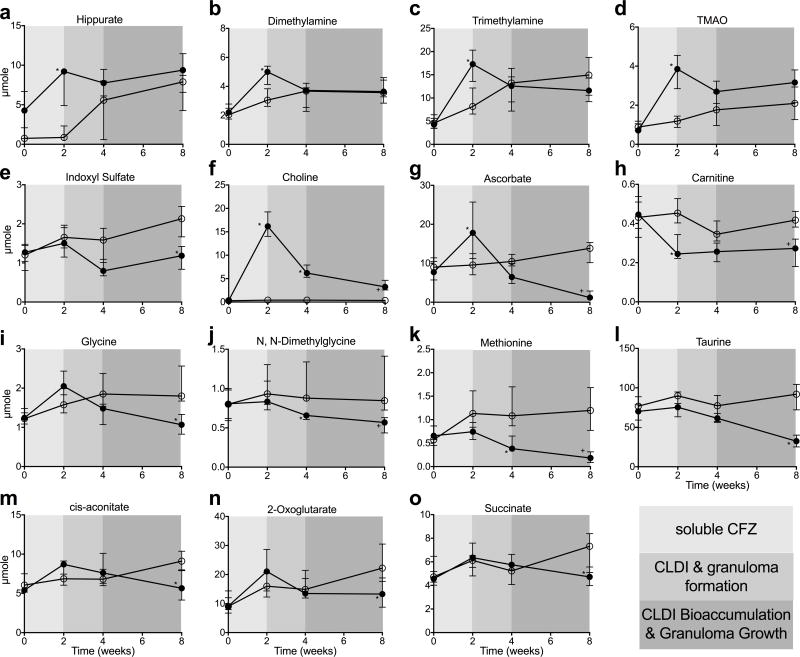
A total of 34 metabolites (including creatinine and urea) were identified and quantified in urine samples (see data file in supplemental material). There were no statistical differences in the amount of any of the detected urine metabolites between mice assigned to sham or CFZ chow at time 0. During the first 2 weeks of oral CFZ treatment, before any CLDIs are detectable and during which, in vivo, CFZ is in the soluble form, levels of metabolites that are typically associated with antibiotic mediated changes in the metabolome and/or alterations in the gut microbiome55–61 of preclinical models increased compared to control mice. These include hippurate, dimethylamine (DMA), trimethylamine (TMA), and trimethylamine N-oxide (TMAO) (Fig. 3A–D). However, indoxyl sulfate, which is also linked to gut microbiota disruption62, did not increase at 2 weeks, but rather progressively decreased at 4 and 8 weeks (Fig. 3E). Choline, an essential nutrient that is known to be altered by antibiotic administration58, also increased at 2 weeks (Fig. 3F). But after 2 weeks, concomitant with CFZ redistribution from fat to macrophages22, CLDI formation in macrophages and granuloma growth in the liver22, the urine excretion of choline progressively declined, but still maintained higher levels than control mice. Ascorbate excretion also increased at 2 weeks in CFZ-treated mice, but progressively declined afterwards such that by 8 weeks it was lower in CFZ-treated than in control mice (Fig.3G). Carnitine (Fig. 3H) was the only urine metabolite that was decreased by CFZ treatment at 2 weeks and remained lower compared with sham treatment. Seven other urine metabolites were impacted (FDR ≤ 15%) by CFZ treatment: glycine (Fig. 3I), N, N-dimethylglycine (DMG; Fig. 3J), methionine (Fig. 3K), taurine (Fig. 3L), and the citric acid cycle intermediates, cis-aconitate (Fig. 3M), 2-oxoglutarate (Fig. 3N), and succinate (Fig. 3O. Levels of these metabolites progressively declined and were significantly lower than in sham-treated mice at 8 weeks. Urine creatinine was also lower in CFZ-treated mice at 8 weeks (median+IQR: 8.2+2.0 vs. 13.3+2.2 μmole; p=0.014, FDR=5.2%) but this did not translate to a difference in the rate of creatinine excretion (see below). Representative NMR spectra of urine are shown in Fig. 4A and B; information about metabolite identification can be found in the supporting information.
Figure 3.
Temporal changes in the amount of urine metabolites that occurred throughout the
soluble phase (0 - 2 wks), crystal-like drug inclusion (CLDI) and granuloma
formation phase (2 – 4 wks), and CLDI bioaccumulation and granuloma
growth phases (4 – 8 wks) of clofazimine (CFZ;  ) distribution compared with sham-treated
control (
) distribution compared with sham-treated
control (  ). Levels of several
metabolites associated with antibiotic-induced changes in the metabolome
occurred during the first two weeks of CFZ treatment. (A) Hippurate
(KEGG ID: C01586;*p=0.002; FDR=1.2%); and the
by-products of choline metabolism (B) Dimethylamine (KEGG ID:
C00543;*p=0.018; FDR=8.2%); (C)
Trimethylamine (KEGG ID: C00565;*p=0.004, FDR: 2.1%) and
(D) Trimethylamine N-oxide (TMAO, KEGG ID:
C01104;*p=0.0001; FDR=0.3%) all increased in the
urine during the first two weeks of CFZ treatment but normalized afterwards.
Differentiating (FDR ≤ 15%) urine metabolites of CFZ at 8 weeks
are: (E) Indoxyl sulfate (KEGG ID: NA), a metabolite of the colonic
degradation of the amino acid, tryptophan, and also associated with the
microbiome continuously declined in CFZ-treated mice (*p=0.0001,
FDR=0.3%); (F) Choline (KEGG ID: C00114), an
essential nutrient that is required for one carbon metabolism, which initially
increased during the soluble phase then precipitously declined over the
remaining four weeks of the study (*p<0.0001,
FDR=0.3%; +p=0.002, FDR=1.1%);
(G) Ascorbate (KEGG ID: C00072), an anti-oxidant which is also
required for de novo synthesis of carnitine, declined during
the CLDI formation phase (*p=0.0007,
FDR=0.6%;+p<0.0001, FDR=0.3%);
(H) Carnitine (KEGG ID: C00318), which is required for the
transport of long-chain fatty acids into the mitochondria, was lower in
CFZ-treated mice (*p=0.0008, FDR=0.6%;
+p=0.002, FDR=1.1%); (I) glycine
(KEGG ID: C00037), an amino acid which is important in one carbon metabolism,
declined during CLDI and granuloma formation (*p<0.0001,
FDR=0.3%); (J) N, N-dimethylglycine (KEGG ID:
C01026), a derivative of glycine and a byproduct of choline metabolism
(*p=0.02, FDR=5.2%); (K) Methionine
(KEGG ID: C00073), a proteinogenic amino acid (*p<0.0001,
FDR=0.3%); and (L) Taurine (KEGG ID: C00245), an
endogenous sulfonic acid that is a major constituent of bile
(*p<0.0001, FDR=0.3%), all progressively declined
with CFZ treatment. Intermediate metabolites of the citric acid cycle, (M)
cis-aconitate (KEGG ID: C00417; *p=0.2, FDR=6%),
(N) 2-oxoglutarate (KEGG ID: C00026; *p=0.03,
FDR=8.8%), and (O) succinate (KEGG ID: C00042;
*p=0.02, FDR=5.2%) were reduced in CFZ-treated
mice at 8 weeks. Data are the median (IQR) of 6 urine samples/group.
). Levels of several
metabolites associated with antibiotic-induced changes in the metabolome
occurred during the first two weeks of CFZ treatment. (A) Hippurate
(KEGG ID: C01586;*p=0.002; FDR=1.2%); and the
by-products of choline metabolism (B) Dimethylamine (KEGG ID:
C00543;*p=0.018; FDR=8.2%); (C)
Trimethylamine (KEGG ID: C00565;*p=0.004, FDR: 2.1%) and
(D) Trimethylamine N-oxide (TMAO, KEGG ID:
C01104;*p=0.0001; FDR=0.3%) all increased in the
urine during the first two weeks of CFZ treatment but normalized afterwards.
Differentiating (FDR ≤ 15%) urine metabolites of CFZ at 8 weeks
are: (E) Indoxyl sulfate (KEGG ID: NA), a metabolite of the colonic
degradation of the amino acid, tryptophan, and also associated with the
microbiome continuously declined in CFZ-treated mice (*p=0.0001,
FDR=0.3%); (F) Choline (KEGG ID: C00114), an
essential nutrient that is required for one carbon metabolism, which initially
increased during the soluble phase then precipitously declined over the
remaining four weeks of the study (*p<0.0001,
FDR=0.3%; +p=0.002, FDR=1.1%);
(G) Ascorbate (KEGG ID: C00072), an anti-oxidant which is also
required for de novo synthesis of carnitine, declined during
the CLDI formation phase (*p=0.0007,
FDR=0.6%;+p<0.0001, FDR=0.3%);
(H) Carnitine (KEGG ID: C00318), which is required for the
transport of long-chain fatty acids into the mitochondria, was lower in
CFZ-treated mice (*p=0.0008, FDR=0.6%;
+p=0.002, FDR=1.1%); (I) glycine
(KEGG ID: C00037), an amino acid which is important in one carbon metabolism,
declined during CLDI and granuloma formation (*p<0.0001,
FDR=0.3%); (J) N, N-dimethylglycine (KEGG ID:
C01026), a derivative of glycine and a byproduct of choline metabolism
(*p=0.02, FDR=5.2%); (K) Methionine
(KEGG ID: C00073), a proteinogenic amino acid (*p<0.0001,
FDR=0.3%); and (L) Taurine (KEGG ID: C00245), an
endogenous sulfonic acid that is a major constituent of bile
(*p<0.0001, FDR=0.3%), all progressively declined
with CFZ treatment. Intermediate metabolites of the citric acid cycle, (M)
cis-aconitate (KEGG ID: C00417; *p=0.2, FDR=6%),
(N) 2-oxoglutarate (KEGG ID: C00026; *p=0.03,
FDR=8.8%), and (O) succinate (KEGG ID: C00042;
*p=0.02, FDR=5.2%) were reduced in CFZ-treated
mice at 8 weeks. Data are the median (IQR) of 6 urine samples/group.
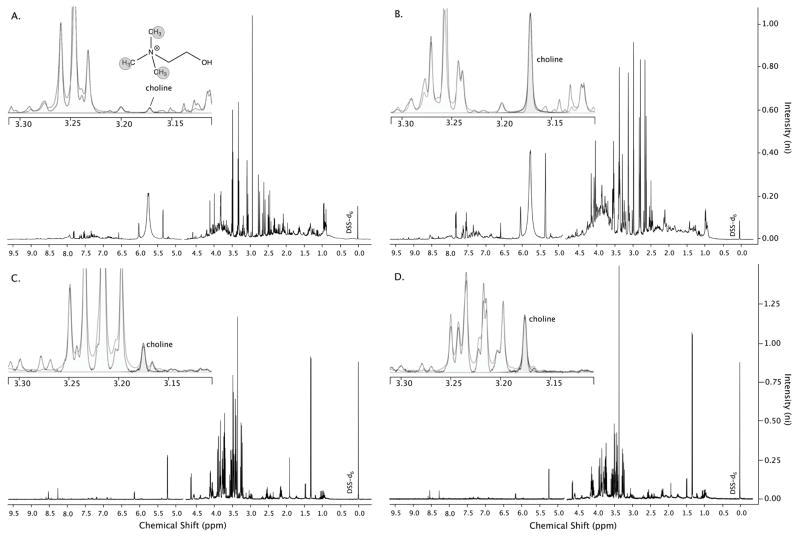
Figure 4.
Representative 1D-1H-NMR spectra of 8-week urine samples from mice that were (A) sham-treated and (B) clofazimine (CFZ)-treated highlighting (insets) the spectral region via Chenomx software where the methyl hydrogens of choline are detected. The peak height of choline (and other metabolites) relative to the internal standard DSS-d6 is representative of its concentration and is greater in the spectrum of the sample from CFZ-treated mice (B). Representative NMR spectra of 8-week whole blood samples from a (C) sham-treated and a (D) CFZ-treated mouse also show a higher level of choline in the sample from the CFZ-treated animal. DSS-d6 = 4,4- dimethyl-4-silapentane-1-sulfonic acid which is the internal standard added to each sample.
Eight weeks of CFZ treatment induced changes in whole blood energy metabolites
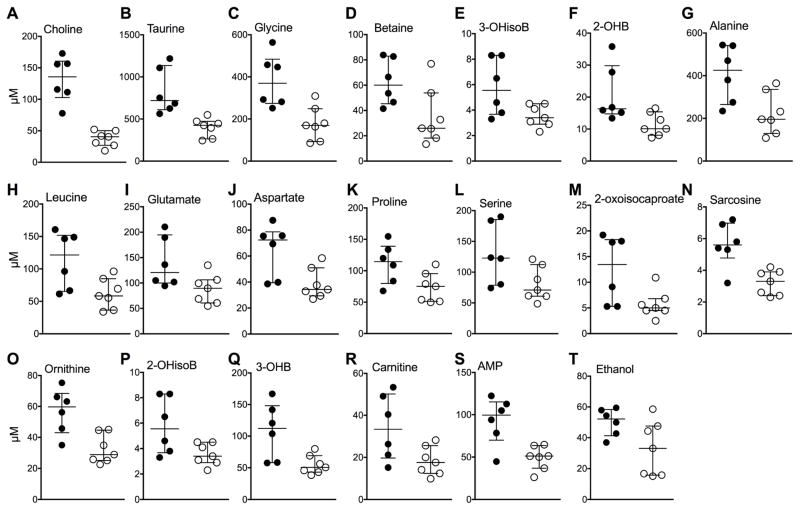
A total of 56 metabolites were detected in WB samples that were available from 8 week CFZ (n=6) and sham-treated mice (n=7), respectively (see data file in supplemental material). Reflecting the declining amounts of urine metabolites, most blood metabolite concentrations, including choline, taurine and glycine (Fig. 5A–C)63, were increased in CFZ-treated mice compared to sham-treated mice. Accompanying this change, was an increase in betaine (Fig. 5D), a methyl donor that is produced by the oxidation of choline.63 Overall, in WB, 20 of the 56 detected metabolites were different (FDR ≤ 15%) in CFZ- vs. sham-treated mice at 8 weeks (Fig. 5A–T). The CFZ-induced reorientation in energy metabolism was apparent in the lactate:glucose ratio which was higher in CFZ-treated mice (median+IQR: 0.94 + 0.69 vs. 0.32 ± 0.28, p = 0.034 by Mann-Whitney test) suggestive of increased glycolytic activity.64 However, whole blood glucose and lactate levels were not different between mice following 8-weeks of CFZ or sham treatment (see supplemental data file). The detected intermediates of amino acid metabolism such as 3-hydroxyisobutyrate (3-OHisoB) (Fig. 5E), an intermediate in valine metabolism65; 2-hydroxybutyrate (2-OHB) (Fig. 5F), which is derived from α-ketobutyrate produced by amino acid catabolism66; and the amino acids: alanine, leucine, glutamate, aspartate, proline, and serine (Fig. 5G–L); and 2-oxoisocaproate, a short-chain keto acid (Fig. 5M) were all increased by CFZ administration. In WB, sarcosine (Fig. 5N), an intermediate in choline and glycine metabolism, was increased by CFZ treatment; as was ornithine (Fig. 5O), a by-product of the urea cycle; 2-hydroxyisobutyrate (2-OHisoB) (Fig. 5P), which has been reported as a metabolite of the gut microbiota67,68; 3-hydroxybutyrate (3-OHB), a ketone body (Fig. 5Q); carnitine (Fig. 5R), which can be synthesized from lysine and methionine and is required for the transport of long-chain fatty acids into the mitochondria; the nucleotide adenosine monophosphate (AMP) (Fig. 5S); and ethanol (Fig. 5T), which is most likely formed by intestinal bacteria.69–71 Representative NMR spectra of WB are shown in Fig. 4C and D. Additional information about metabolite identification can be found in the supporting information and Fig. S1 and S2. There were 15 metabolites common to WB and urine samples. Of these metabolites, four were changed (FDR ≤ 15%) by CFZ treatment in both biofluids at 8 weeks (Fig. S3A–D); of the remaining 11, methionine was lower in the urine of CFZ-treated mice but was unchanged in WB (Fig. S3E). Betaine was increased by CFZ treatment in WB but was unchanged in urine (Fig. S3F). Notably, CFZ administration did not result in a significant change in the amount of targeted analytes, including redox analytes, in the 8-week urine samples (Fig. 6A–F). Three of these compounds were also detected in WB and of these glutamate (Fig. 5I) and aspartate (Fig. 5J) were increased by CFZ treatment, but glutathione was unchanged.
Figure 5.
Whole blood (WB) metabolites altered (FDR ≤ 15%) by 8 weeks of
clofazimine (CFZ;  ) treatment
compared with sham (
) treatment
compared with sham (  ) treatment.
These included, in order of ascending FDR, the essential nutrient,
(A) choline (KEGG ID: C00114; p=0.00001,
FDR=0.6%); (B) taurine (KEGG ID: C00245;
p=0.001, FDR=4%); (C) glycine (KEGG ID:
C00037; p=0.005, FDR=4.4%); (D) betaine
(KEGG ID:C00719; p=0.026, FDR=11%); (E)
3-hydroxyisobutyrate (3-OHisoB; KEGG ID: C06001; p=0.011,
FDR=8%); (F) 2-hydroxybutyrate (2-OHB; KEGG ID:
C05984; p=0.014, FDR=8.5%); (G) alanine
(KEGG ID: C00041; p=0.017, FDR=8.7%); (H)
the branched chain amino acid, leucine (KEGG ID: C00123; p=0.024,
FDR=11%); (I) glutamate (KEGG ID: C00025;
p=0.03, FDR=11%); (J) aspartate (KEGG ID:
C00049; p=0.017, FDR=8.7%); (K) proline,
(KEGG ID: C00148; p=0.03, FDR=11%); (L)
serine (KEGG ID: C00065; p=0.046; FDR=14%);
(M) 2-oxoisocaproate, also known as ketoleucine (KEGG ID:
C00233; p=0.03, FDR=11%); (N) sarcosine
(KEGG ID: C00213; p=0.003, FDR=5.3%), an intermediate in
choline and glycine metabolism; (O) ornithine, a product of the
urea cycle (KEGG ID: C00077; p=0.004, FDR=4.4%);
(P) 2-hydroxyisobutyrate (2-OHisoB; KEGG ID: NA;
p=0.030, FDR=11%), (Q) the ketone body,
3-hydroxybutyrate (3-OHB; KEGG ID: C01089; p=0.009,
FDR=7.4%); (R) carnitine, which is required for
long-chain fatty acid transport into the mitochondria, (KEGG ID: C00318
p=0.03, FDR= 11%); (S) adenosine
monophosphate (AMP; KEGG ID: 00020; p=0.007, FDR=6.7%);
and (T) ethanol (KEGG ID: C00469; p=0.051,
FDR=14%). The crossbars represent the medians and the error
bars, the interquartile ranges, of 6 and 7 WB samples from CFZ- and sham-treated
mice, respectively.
) treatment.
These included, in order of ascending FDR, the essential nutrient,
(A) choline (KEGG ID: C00114; p=0.00001,
FDR=0.6%); (B) taurine (KEGG ID: C00245;
p=0.001, FDR=4%); (C) glycine (KEGG ID:
C00037; p=0.005, FDR=4.4%); (D) betaine
(KEGG ID:C00719; p=0.026, FDR=11%); (E)
3-hydroxyisobutyrate (3-OHisoB; KEGG ID: C06001; p=0.011,
FDR=8%); (F) 2-hydroxybutyrate (2-OHB; KEGG ID:
C05984; p=0.014, FDR=8.5%); (G) alanine
(KEGG ID: C00041; p=0.017, FDR=8.7%); (H)
the branched chain amino acid, leucine (KEGG ID: C00123; p=0.024,
FDR=11%); (I) glutamate (KEGG ID: C00025;
p=0.03, FDR=11%); (J) aspartate (KEGG ID:
C00049; p=0.017, FDR=8.7%); (K) proline,
(KEGG ID: C00148; p=0.03, FDR=11%); (L)
serine (KEGG ID: C00065; p=0.046; FDR=14%);
(M) 2-oxoisocaproate, also known as ketoleucine (KEGG ID:
C00233; p=0.03, FDR=11%); (N) sarcosine
(KEGG ID: C00213; p=0.003, FDR=5.3%), an intermediate in
choline and glycine metabolism; (O) ornithine, a product of the
urea cycle (KEGG ID: C00077; p=0.004, FDR=4.4%);
(P) 2-hydroxyisobutyrate (2-OHisoB; KEGG ID: NA;
p=0.030, FDR=11%), (Q) the ketone body,
3-hydroxybutyrate (3-OHB; KEGG ID: C01089; p=0.009,
FDR=7.4%); (R) carnitine, which is required for
long-chain fatty acid transport into the mitochondria, (KEGG ID: C00318
p=0.03, FDR= 11%); (S) adenosine
monophosphate (AMP; KEGG ID: 00020; p=0.007, FDR=6.7%);
and (T) ethanol (KEGG ID: C00469; p=0.051,
FDR=14%). The crossbars represent the medians and the error
bars, the interquartile ranges, of 6 and 7 WB samples from CFZ- and sham-treated
mice, respectively.
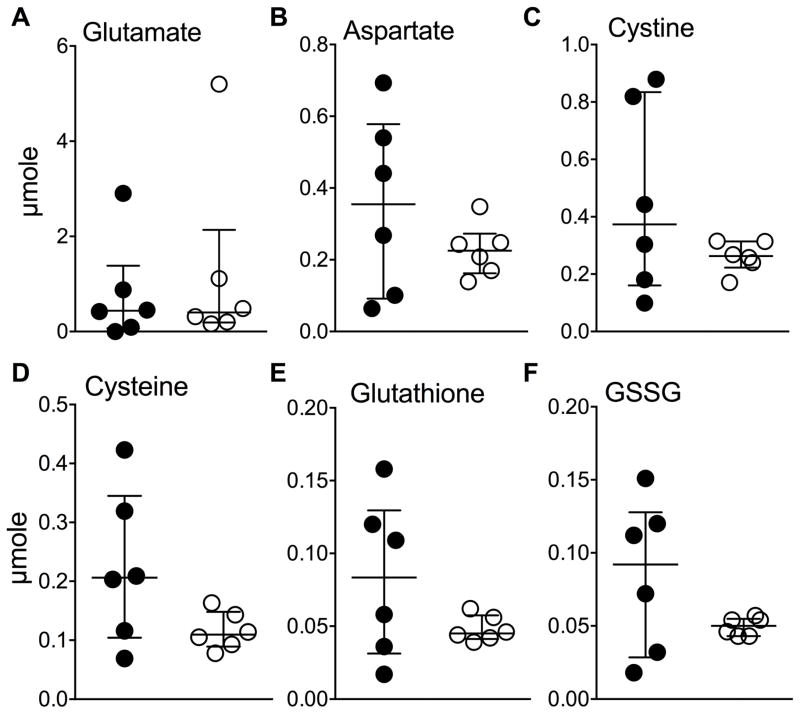
Figure 6.
Targeted measurement of urine analytes by an HPLC assay were unchanged by
clofazimine (CFZ). CFZ (  ) compared
with sham-treatment (
) compared
with sham-treatment (  ) did not
change urine levels of (A) glutamate (KEGG ID:
C00217;p=0.7) or (B) aspartate (KEGG ID:
C00049;p=0.5). Both glutamate and aspartate are proteinogenic amino
acids that were increased by CFZ in the blood (see Fig. 5I and 5J). Neither urine (C) cystine (KEGG ID:
C00491;p=0.5) nor (D) cysteine (KEGG ID:
C00097;p=0.13) were changed by CFZ treatment. Also, urine
(E) glutathione (KEGG ID: C00051; GSH; p=0.5) the most
abundant anti-oxidant in mammals and (F) glutathione disulfide
(KEGG ID: C00127; GSSG; p=0.4) were unchanged by CFZ. Notably, blood
glutathione was also unchanged by CFZ (see supporting material, data file).
The crossbars represent the medians and the error bars, the interquartile
ranges, of 6 urine samples/group. Urine analyte p values are
those that resulted from a Mann-Whitney test of the μmole data.
) did not
change urine levels of (A) glutamate (KEGG ID:
C00217;p=0.7) or (B) aspartate (KEGG ID:
C00049;p=0.5). Both glutamate and aspartate are proteinogenic amino
acids that were increased by CFZ in the blood (see Fig. 5I and 5J). Neither urine (C) cystine (KEGG ID:
C00491;p=0.5) nor (D) cysteine (KEGG ID:
C00097;p=0.13) were changed by CFZ treatment. Also, urine
(E) glutathione (KEGG ID: C00051; GSH; p=0.5) the most
abundant anti-oxidant in mammals and (F) glutathione disulfide
(KEGG ID: C00127; GSSG; p=0.4) were unchanged by CFZ. Notably, blood
glutathione was also unchanged by CFZ (see supporting material, data file).
The crossbars represent the medians and the error bars, the interquartile
ranges, of 6 urine samples/group. Urine analyte p values are
those that resulted from a Mann-Whitney test of the μmole data.
CFZ treatment impacts body weight
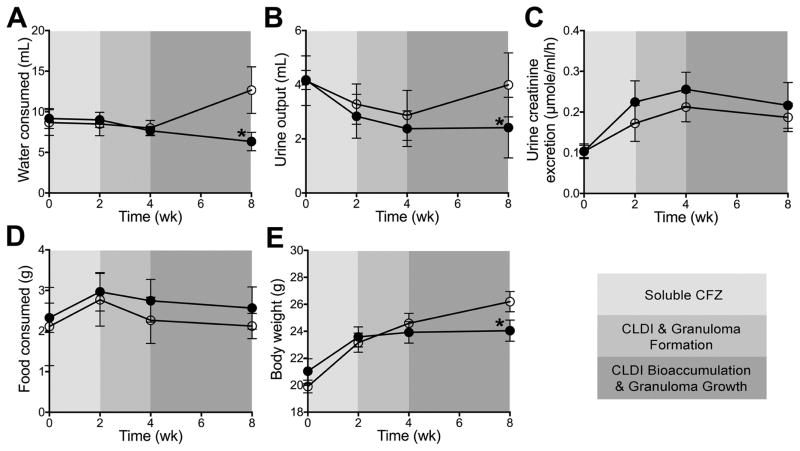
CFZ-induced changes in the urine and WB 1H-NMR detected metabolomes cannot be explained by variations in water intake, urine output, or renal function. The metabolic cage data show that water intake and urine output (Fig. 7A and B) did not differ between CFZ- and sham-treated mice until 8-weeks at which point, sham-treated mice consumed more water which was associated with an concurrent increase in urine output. Importantly, renal function, as assessed by the calculated urine creatinine excretion, was not different between the two groups during any phase of CFZ treatment (Fig. 7C). CFZ-treatment did not influence the amount of food consumed at each time point (Fig. 6D) but the mean (+SD) total amount of food during the study period was slightly higher in CFZ-treated mice (2.65+0.49 g vs. 2.32+0.65 g; p=0.050 by unpaired Student’s t-test). CFZ-treated mice produced more feces (by weight, 1.3+0.23 vs. 0.93+0.23 g, p<0.0001) than sham-treated mice but the consistency of feces between the two groups was not notably different. Over the course of the 8-week study, CFZ-treated mice gained less weight than sham-treated mice (Fig. 7E).
Figure 7.
Data acquired from 18h metabolic cage studies conducted at baseline
(pre-treatment) and then at each phase of clofazimine (CFZ;  ) distribution included (A) the
amount of water (mL) consumed (*p<0.0001) and (B) urine
(mL) output (*p=0.009) when compared to sham-treated (
) distribution included (A) the
amount of water (mL) consumed (*p<0.0001) and (B) urine
(mL) output (*p=0.009) when compared to sham-treated (
 ) mice. Using the amount
(μmole) of urine creatinine (KEGG ID: C00791) measured by 1H-
NMR and the corresponding urine volume output (mL), urine creatinine excretion
was calculated (C) and was not different between CFZ and
sham-treated mice. Food consumed at each time point (D) did not
differ between the two groups but the total amount (g) of food consumed was
higher in CFZ-treated mice (see text) and CFZ-treated mice did not gain as much
body weight (E) as sham-treated mice (*p=0.0002)
over the 8-week study. Data are the mean (+SD) of 6 samples per group
except for body weight which are 23–24 mice per group.
) mice. Using the amount
(μmole) of urine creatinine (KEGG ID: C00791) measured by 1H-
NMR and the corresponding urine volume output (mL), urine creatinine excretion
was calculated (C) and was not different between CFZ and
sham-treated mice. Food consumed at each time point (D) did not
differ between the two groups but the total amount (g) of food consumed was
higher in CFZ-treated mice (see text) and CFZ-treated mice did not gain as much
body weight (E) as sham-treated mice (*p=0.0002)
over the 8-week study. Data are the mean (+SD) of 6 samples per group
except for body weight which are 23–24 mice per group.
Bioinformatics analysis of CFZ-induced changes in urine and whole blood 1H-NMR metabolomics
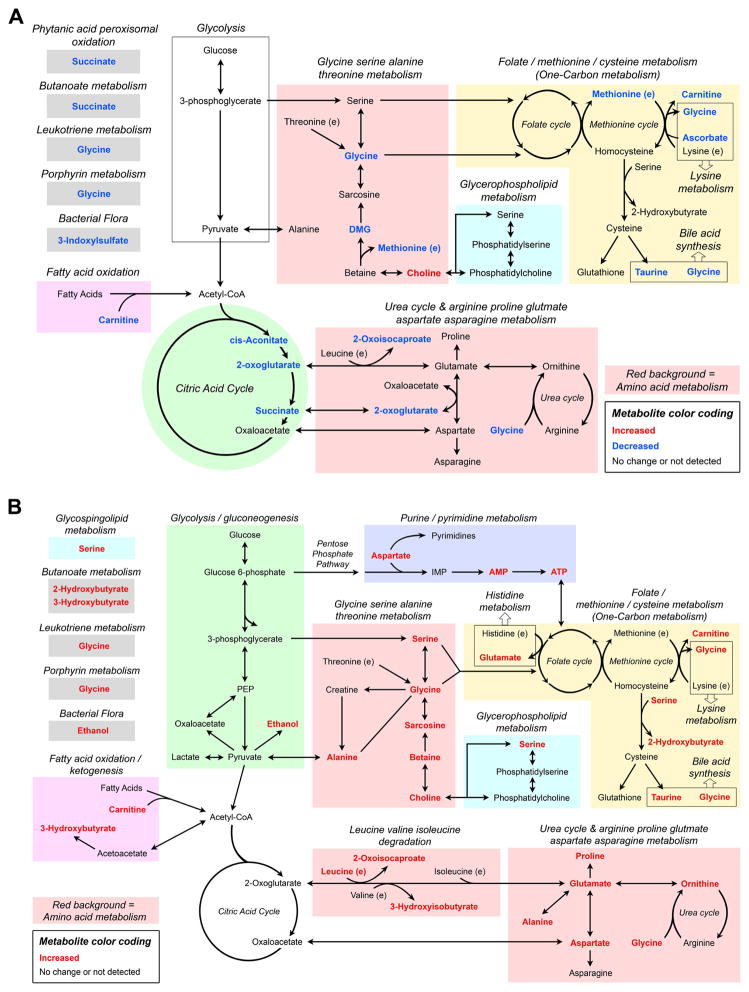
Bioinformatics is an important aid in the interpretation of metabolic data.53,72 Pathway analysis of the 12 differentiating (FDR ≤ 15%) urine metabolites of CFZ treatment (see Fig. 3) using Metscape shows that 12 metabolic pathways were changed at 8 weeks (Fig. S4). In WB, the 20 differentiating (FDR ≤ 15%) metabolites of CFZ treatment (see Fig. 5) mapped into 16 Metscape pathways (Fig. S5). There were 10 metabolic pathways that were common to both urine and blood, which include: 1) folate metabolism; 2) methionine and cysteine metabolism; 3) glycine, serine, alanine, threonine metabolism; 4) urea cycle and arginine, proline, glutamate, aspartate asparagine metabolism; 5) lysine metabolism; 6) glycerophospholipid metabolism; 7) bile acid synthesis; 8) butanoate metabolism; 9) leukotriene metabolism, and 10) porphyrin metabolism. This analysis shows that 8 weeks of CFZ administration leads to a large shift in energy metabolism that is related to the production of building blocks required for cellular biosynthetic processes by macrophages as CFZ is sequestered and bioaccumulates.
DISCUSSION
In this study, temporal changes in the amount of several urine metabolites as well as a profound shift in the concentration of a number of WB metabolites after 8 weeks of CFZ treatment in mice demonstrate a distinct metabolic change that accompanies a macrophage-mediated CFZ sequestration and detoxification response. This finding is consistent with the recent recognition that metabolic reprogramming is a prerequisite for immunological responses to diverse stimuli in macrophages.29 In our model, the signature pharmacodynamic response to CFZ administration is the formation of CLDIs in resident tissue macrophages22,24,25, which results in increased IL1RA production as we have previously reported.26,39 The formation of CLDIs occurs over time, such that after the first two weeks of drug administration most of the drug is present in the soluble form which circulates in the blood and is associated with adipose tissue.22 Over the next two weeks, CFZ bioaccumulation intensifies in tissue macrophages, particularly those of the liver, during which CLDIs begin to form and granuloma formation becomes apparent.22,23 From 4 to 8 weeks, drug-sequestering CLDIs intensively aggregate as granulomas grow. In these distinct phases of CFZ bioaccumulation and re-distribution there is a significant shift in the 1H-NMR-detected urine energy metabolome. These changes occurred concurrent with a modest increase in food consumption and a decrease in body weight, which suggests that prolonged (8-week) CFZ administration to mice results in a profound disruption in global energy metabolism that manifests in the urine and WB metabolomes. Specifically, we found disruptions in: 1) one carbon metabolism, as demonstrated by changes in urine levels of choline, methionine and glycine, while WB levels of the same metabolites increased along with glutamate, serine and 2-OHB; 2) amino acid metabolism, as shown by changes in levels of amino acids and amino acid intermediates such as 3-OHisoB and 2-oxoisocaproate; 3) carnitine utilization; and 4) glycolysis, as evidenced by a CFZ-induced increase in the lactate:glucose ratio in WB (Fig. 8A and B). Collectively, these findings indicate activation of biosynthetic pathways utilized by macrophages to form CLDI and CLDI-induced granulomas.
Figure 8.
Schematic diagram of the overarching metabolic transformation induced in the 1H-NMR detected (A) urine and (B) whole blood (WB) metabolomes after 8-weeks of clofazimine (CFZ). The diagram was derived using the compound subnetworks generated by Metscape using the differentiating urine and WB metabolites (FDR < 15%; see Fig. S4 and S5 in supporting information) as well as the calculated CFZ-induced increase in lactate:glucose ratio. The grouping of metabolites illustrates the most profoundly reprogrammed metabolic pathways by macrophage-mediated CFZ sequestration and bioaccumulation in granulomas. Red font indicates increased amount/concentration in 8 week CFZ mice compared to control and blue font indicates no significant change or a decrease.
Increased one-carbon metabolism
Choline, along with the amino acids glycine, serine, and methionine is integral to one-carbon metabolism, which includes the folate, methionine and transsulfuration pathways, the outputs of which include nucleotides, proteins and lipids that are required for the construction of new cellular components.63 One carbon metabolism also maintains redox homeostasis and generates substrates for methylation reactions. Specifically, our data provide evidence that both physiologic and metabolic CFZ-induced disruptions in glycerophospholipid metabolism, as determined by pathway analysis, are related to one-carbon metabolism. First, we know that multi-lamellar membranes surround CLDIs that likely form secondary to changes in cellular membrane organization, leading to increased turnover of glycerophospholipids.24,39 This process resembles macroautophagy, the hallmark of which is the formation of multi-lamellar vesicles.73–76 Expression of monocyte chemoattractant protein-1 (CCL2), which is abundant in macrophages and is a known mediator of autophagy also increases during CLDI formation.22,77 Therefore, the progressive decline in urine choline between 4 and 8 weeks and the higher WB levels of choline in CFZ-treated mice could be explained by its utilization as macrophages sequester and compartmentalize increasing amounts of CFZ into CLDIs. In parallel, urine levels of glycine and methionine progressively declined in CFZ-treated mice and at 8 weeks, WB glycine and serine were elevated, providing additional evidence of the enhanced activation of one-carbon metabolism. Notably, CFZ did not alter urine concentrations of redox analytes, which suggests that the observed activation of one-carbon metabolism was not due to a loss of redox homeostasis.
Increased amino acid metabolism
In addition to the amino acids involved in one-carbon metabolism, our data also implicate a CFZ-induced reorientation in amino acid metabolism. The WB levels of leucine, glutamate, proline, ornithine, sarcosine, aspartate and alanine were increased by CFZ treatment at 8 weeks. This suggests that the drug-sequestration response of macrophages altered host amino acid metabolism. Moreover, increased levels of serine, glycine, sarcosine and aspartate in WB suggest the enhanced usage of 3-phosphoglycerate (an intermediate product of glycolysis) for serine de novo synthesis; oxaloacetate (a TCA cycle intermediate) for glycine, sarcosine and aspartate metabolism; and 2-oxoglutarate (a TCA cycle intermediate) for glutamate metabolism. The urine excretion of 2-oxoglutarate was lower in CFZ-treated mice, as well as other citric acid cycle intermediates, cis-aconitate and succinate. In addition, intermediate products of essential amino acid degradation, such as 2-oxoisocaproate and 3-OHB, which are produced from the degradation of leucine and valine, respectively, were elevated in the blood of CFZ-treated mice. Overall, the CFZ-induced increase in WB essential and non-essential amino acids, as well as intermediate products of amino acid metabolism implicate an increased requirement for amino acid driven biosynthetic processes such as protein and membrane synthesis that are likely required for the macrophage-mediated CFZ sequestration response.
Carnitine utilization
Carnitine, a short-chain keto-acid, that is required for the transport of long-chain fatty acids into the mitochondria78, declined in the urine of CFZ-treated mice while WB levels increased (Fig. 4R). In addition to the known relationship between carnitine and choline55, the de novo synthesis of carnitine requires methionine, lysine and ascorbate, which is the primary source of carnitine in mice. Urine methionine and ascorbate declined over time, but WB levels of lysine and methionine were unchanged by CFZ, which suggests that they were retained.79 Although CFZ treatment caused a significant decline in urine ascorbate, other urine redox analytes including glutathione did not change, suggesting that utilization of ascorbate as an anti-oxidant does not explain this change. As carnitine is essential for fatty acid oxidation, its retention could imply enhanced utilization of fat stores for energy production during CFZ-treatment, via β-oxidation and ketogenesis. This is evidenced by the elevated blood levels of the ketone body 3-OHB, and the lower body weight of CFZ-treated mice compared to control mice at 8 weeks. In aggregate, the CFZ-induced decrease in the urine excretion of methionine, ascorbate and carnitine; and the increase in WB carnitine levels suggest an increased requirement for carnitine that may be linked to increased fat utilization for energy.
Metabolic changes due to CFZ-induced alterations on the gut microbiome
We acknowledge that we cannot completely attribute the found metabolic disruptions to CLDI and granuloma formation. CFZ is an antibiotic and it is well known that antimicrobial disruption of the gut microbiome61,80 leads to changes in the metabolome.56–60 However, to the best of our knowledge, this is the first report of the pharmacometabolomics of CFZ and there is no evidence that prolonged (8-week) use of an anti-microbial in mice leads to the large and temporal change in energy metabolism that we report here.81 In our study, urine choline and choline-related metabolites, TMAO, TMA and DMA55,82 as well as hippurate, which has been attributed to the gut microbiota, were increased during the first two weeks of CFZ treatment (Fig. 1A–D), a finding consistent with other antibiotic metabolomics studies.57,58,60,82,83 However, after two weeks, the urine levels of choline begin to decline and TMAO, TMA and DMA levels returned to levels similar to sham-treated mice. During subsequent phases (> 2 weeks) of CFZ distribution, urine levels of indoxyl sulfate (Fig. 2E)58 progressively increased and WB ethanol was higher in CFZ treated mice than in sham mice (Fig. 5T).70
The observed CFZ-induced change in body weight and modest increase in total food consumption likely reflects the overall global shift in energy metabolism in these mice. Importantly, CFZ-induced metabolic changes cannot simply be explained by differences in the amount of food consumed or renal function between drug- and sham-treated animals. For example, even though CFZ-treated mice consumed an average of 0.3 g more chow, which translates to approximately an additional 675μg of choline over the course of the study than sham-treated mice, this neither explains the nearly 95% higher amount of urine choline nor the over 3-fold increase in the median choline concentration in WB. Renal function, as assessed by the calculated urine excretion rate of creatinine, was not different between CFZ- and sham-treated mice, indicating that the decreased concentrations of urinary metabolites are neither reflective of a general CFZ-induced change in renal function nor indicative of an inherent decline in the levels of metabolites circulating in the blood.
CONCLUSIONS
The combined, complementary NMR metabolomics analysis of WB and urine unveiled a previously unrecognized large and remarkable CFZ-induced change in host energy metabolism. These profound changes accompanied the drug’s sequestration and bioaccumulation response by macrophages and granuloma formation in the liver. These findings are consistent with activation of amino acid, nucleotide, glycophospholipid and one-carbon metabolism that occurs during biosynthetic processes indicative of protein synthesis, membrane biogenesis and cell proliferation. There is also a metabolic signature of anti-microbial activity. This study illustrates the informative capacity of NMR metabolomics84 and how the use of two biofluids can further understanding of the biological action of drugs.85 It also provides evidence of a profound shift in metabolism that can occur secondary to drug sequestration and bioaccumulation in macrophages that are connected to mechanisms of drug action and pharmacodynamics that were previously unrecognized.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of General Medical Sciences (NIGMS; R01GM078200 to GRR and R01GM111400 to KAS) and the University of Michigan’s M-Cubed initiative (http://mcubed.umich.edu). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIGMS or the National Institutes of Health. Portions of this work were presented at the 11th International Conference of the Metabolomics Society, June 29th–July 2nd, 2015 in San Francisco, CA USA.
Footnotes
Conflict of Interest Disclosures
Dr. Rosania is a consultant for Bristol-Myers Squibb. All other authors have nothing to disclose.
This article contains supplementary material available from the authors upon request or via the Internet at http://onlinelibrary.wiley.com/.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barry VC, Belton JG, Conalty ML, Denneny JM, Edward DW, O'Sullivan JF, Twomey D, Winder F. A new series of phenazines (rimino-compounds) with high antituberculosis activity. Nature. 1957;179(4568):1013–1015. doi: 10.1038/1791013a0. [DOI] [PubMed] [Google Scholar]
- 2.Cholo MC, Steel HC, Fourie PB, Germishuizen WA, Anderson R. Clofazimine: current status and future prospects. J Antimicrob Chemother. 2012;67(2):290–298. doi: 10.1093/jac/dkr444. [DOI] [PubMed] [Google Scholar]
- 3.Conalty ML, Barry VC, Jina A. The antileprosy agent B.663 (Clofazimine) and the reticuloendothelial system. Int J Lepr Other Mycobact Dis. 1971;39(2):479–492. [PubMed] [Google Scholar]
- 4.Atkinson AJ, Jr, Sheagren JN, Rubio JB, Knight V. Evaluation of B.663 in human leprosy. Int J Lepr Other Mycobact Dis. 1967;35(2):119–127. [PubMed] [Google Scholar]
- 5.Mansfield RE. Tissue concentrations of clofazimine (B663) in man. Am J Trop Med Hyg. 1974;23(6):1116–1119. doi: 10.4269/ajtmh.1974.23.1116. [DOI] [PubMed] [Google Scholar]
- 6.Venkatesan K, Deo N, Gupta UD. Tissue distribution and deposition of clofazimine in mice following oral administration with or without isoniazid. Arzneimittelforschung. 2007;57(7):472–474. doi: 10.1055/s-0031-1296634. [DOI] [PubMed] [Google Scholar]
- 7.Sukpanichnant S, Hargrove NS, Kachintorn U, Manatsathit S, Chanchairujira T, Siritanaratkul N, Akaraviputh T, Thakerngpol K. Clofazimine-induced crystal-storing histiocytosis producing chronic abdominal pain in a leprosy patient. Am J Surg Pathol. 2000;24(1):129–135. doi: 10.1097/00000478-200001000-00016. [DOI] [PubMed] [Google Scholar]
- 8.McDougall AC. Electron microscope studies of the antileprosy drug B663 (clofazimine; Lamprenel) Int J Lepr Other Mycobact Dis. 1974;42(1):1–12. [PubMed] [Google Scholar]
- 9.Harbeck RJ, Worthen GS, Lebo TD, Peloquin CA. Clofazimine crystals in the cytoplasm of pulmonary macrophages. Ann Pharmacother. 1999;33(2):250. doi: 10.1345/aph.18170. [DOI] [PubMed] [Google Scholar]
- 10.Lewis JT, Candelora JN, Hogan RB, Briggs FR, Abraham SC. Crystal-storing histiocytosis due to massive accumulation of charcot-leyden crystals: a unique association producing colonic polyposis in a 78-year-old woman with eosinophilic colitis. Am J Surg Pathol. 2007;31(3):481–485. doi: 10.1097/01.pas.0000213420.46127.9c. [DOI] [PubMed] [Google Scholar]
- 11.Jadhav MV, Sathe AG, Deore SS, Patil PG, Joshi NG. Tissue concentration, systemic distribution and toxicity of clofazimine--an autopsy study. Indian J Pathol Microbiol. 2004;47(2):281–283. [PubMed] [Google Scholar]
- 12.Desikan KV, Ramanujam K, Ramu G, Balakrishnan S. Autopsy findings in a case of lepromatous leprosy treated with clofazimine. Lepr Rev. 1975;46(3):181–189. doi: 10.5935/0305-7518.19750019. [DOI] [PubMed] [Google Scholar]
- 13.Mackey JP, Barnes J. Clofazimine in the treatment of discoid lupus erythematosus. Br J Dermatol. 1974;91(1):93–96. doi: 10.1111/j.1365-2133.1974.tb06723.x. [DOI] [PubMed] [Google Scholar]
- 14.Lo JS, Berg RE, Tomecki KJ. Treatment of discoid lupus erythematosus. Int J Dermatol. 1989;28(8):497–507. doi: 10.1111/j.1365-4362.1989.tb04599.x. [DOI] [PubMed] [Google Scholar]
- 15.Chuaprapaisilp T, Piamphongsant T. Treatment of pustular psoriasis with clofazimine. Br J Dermatol. 1978;99(3):303–305. doi: 10.1111/j.1365-2133.1978.tb02001.x. [DOI] [PubMed] [Google Scholar]
- 16.Podmore P, Burrows D. Clofazimine--an effective treatment for Melkersson-Rosenthal syndrome or Miescher's cheilitis. Clin Exp Dermatol. 1986;11(2):173–178. doi: 10.1111/j.1365-2230.1986.tb00443.x. [DOI] [PubMed] [Google Scholar]
- 17.Bezerra EL, Vilar MJ, da Trindade Neto PB, Sato EI. Double-blind, randomized, controlled clinical trial of clofazimine compared with chloroquine in patients with systemic lupus erythematosus. Arthritis Rheum. 2005;52(10):3073–3078. doi: 10.1002/art.21358. [DOI] [PubMed] [Google Scholar]
- 18.Venkateswarlu B, Venkataramana D, Rao AV, Prabhakar MC, Reddy BM. Role of rifampin and clofazimine ointments in the treatment of leprosy. Int J Lepr Other Mycobact Dis. 1992;60(2):269–270. [PubMed] [Google Scholar]
- 19.Tyagi S, Ammerman NC, Li SY, Adamson J, Converse PJ, Swanson RV, Almeida DV, Grosset JH. Clofazimine shortens the duration of the first-line treatment regimen for experimental chemotherapy of tuberculosis. Proc Natl Acad Sci U S A. 2015;112(3):869–874. doi: 10.1073/pnas.1416951112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lechartier B, Cole ST. Mode of Action of Clofazimine and Combination Therapy with Benzothiazinones against Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2015;59(8):4457–4463. doi: 10.1128/AAC.00395-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams K, Minkowski A, Amoabeng O, Peloquin CA, Taylor D, Andries K, Wallis RS, Mdluli KE, Nuermberger EL. Sterilizing activities of novel combinations lacking first- and second-line drugs in a murine model of tuberculosis. Antimicrob Agents Chemother. 2012;56(6):3114–3120. doi: 10.1128/AAC.00384-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baik J, Stringer KA, Mane G, Rosania GR. Multiscale distribution and bioaccumulation analysis of clofazimine reveals a massive immune system-mediated xenobiotic sequestration response. Antimicrob Agents Chemother. 2013;57(3):1218–1230. doi: 10.1128/AAC.01731-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swanson RV, Adamson J, Moodley C, Ngcobo B, Ammerman NC, Dorasamy A, Moodley S, Mgaga Z, Tapley A, Bester LA, Singh S, Grosset JH, Almeida DV. Pharmacokinetics and pharmacodynamics of clofazimine in the mouse model of tuberculosis. Antimicrob Agents Chemother. 2015 doi: 10.1128/AAC.00260-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baik J, Rosania GR. Molecular imaging of intracellular drug-membrane aggregate formation. Mol Pharm. 2011;8(5):1742–1749. doi: 10.1021/mp200101b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baik J, Rosania GR. Macrophages sequester clofazimine in an intracellular liquid crystal-like supramolecular organization. PLoS One. 2012;7(10):e47494. doi: 10.1371/journal.pone.0047494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoon GS, Keswani RK, Sud S, Rzeczycki PM, Murashov MD, Koehn TA, Standiford TJ, Stringer KA, Rosania GR. Clofazimine Biocrystal Accumulation in Macrophages Upregulates Interleukin 1 Receptor Antagonist Production To Induce a Systemic Anti-Inflammatory State. Antimicrob Agents Chemother. 2016;60(6):3470–3479. doi: 10.1128/AAC.00265-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly B, O'Neill LA. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell research. 2015;25(7):771–784. doi: 10.1038/cr.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kominsky DJ, Campbell EL, Colgan SP. Metabolic shifts in immunity and inflammation. J Immunol. 2010;184(8):4062–4068. doi: 10.4049/jimmunol.0903002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Neill LA, Pearce EJ. Immunometabolism governs dendritic cell and macrophage function. J Exp Med. 2016;213(1):15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cholo MC, Mothiba MT, Fourie F, Anderson R. Mechanisms of action and therapeutic efficacies of the lipophilic antimycobacterial agents clofazimine and bedaquiline. J Antimicrob Chemother. 2016 doi: 10.1093/jac/dkw426. [DOI] [PubMed] [Google Scholar]
- 31.Barding GA, Jr, Salditos R, Larive CK. Quantitative NMR for bioanalysis and metabolomics. Anal Bioanal Chem. 2012;404(4):1165–1179. doi: 10.1007/s00216-012-6188-z. [DOI] [PubMed] [Google Scholar]
- 32.Serkova NJ, Brown MS. Quantitative analysis in magnetic resonance spectroscopy: from metabolic profiling to in vivo biomarkers. Bioanalysis. 2012;4(3):321–341. doi: 10.4155/bio.11.320. [DOI] [PubMed] [Google Scholar]
- 33.Wishart DS. Quantitative metabolomics using NMR. Trac-Trend Anal Chem. 2008;27(3):228–237. [Google Scholar]
- 34.Serkova NJ, Standiford TJ, Stringer KA. The emerging field of quantitative blood metabolomics for biomarker discovery in critical illnesses. Am J Respir Crit Care Med. 2011;184(6):647–655. doi: 10.1164/rccm.201103-0474CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puskarich MA, Finkel MA, Karnovsky A, Jones AE, Trexel J, Harris BN, Stringer KA. Pharmacometabolomics of l-carnitine treatment response phenotypes in patients with septic shock. Ann Am Thorac Soc. 2015;12(1):46–56. doi: 10.1513/AnnalsATS.201409-415OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellero-Simatos S, Beitelshees AL, Lewis JP, Yerges-Armstrong LM, Georgiades A, Dane A, Harms AC, Strassburg K, Guled F, Hendriks MM, Horenstein RB, Shuldiner AR, Hankemeier T, Kaddurah-Daouk R. Oxylipid Profile of Low-Dose Aspirin Exposure: A Pharmacometabolomics Study. Journal of the American Heart Association. 2015;4(10) doi: 10.1161/JAHA.115.002203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaddurah-Daouk R, Weinshilboum R. Metabolomic Signatures for Drug Response Phenotypes: Pharmacometabolomics Enables Precision Medicine. Clinical pharmacology and therapeutics. 2015;98(1):71–75. doi: 10.1002/cpt.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Everett JR. Pharmacometabonomics in humans: a new tool for personalized medicine. Pharmacogenomics. 2015;16(7):737–754. doi: 10.2217/pgs.15.20. [DOI] [PubMed] [Google Scholar]
- 39.Yoon GS, Sud S, Keswani RK, Baik J, Standiford TJ, Stringer KA, Rosania GR. Phagocytosed Clofazimine Biocrystals Can Modulate Innate Immune Signaling by Inhibiting TNFalpha and Boosting IL-1RA Secretion. Mol Pharm. 2015;12(7):2517–2527. doi: 10.1021/acs.molpharmaceut.5b00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lacy P, McKay RT, Finkel M, Karnovsky A, Woehler S, Lewis MJ, Chang D, Stringer KA. Signal intensities derived from different NMR probes and parameters contribute to variations in quantification of metabolites. PLoS One. 2014;9(1):e85732. doi: 10.1371/journal.pone.0085732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emwas AH, Luchinat C, Turano P, Tenori L, Roy R, Salek RM, Ryan D, Merzaban JS, Kaddurah-Daouk R, Zeri AC, Nagana Gowda GA, Raftery D, Wang Y, Brennan L, Wishart DS. Standardizing the experimental conditions for using urine in NMR-based metabolomic studies with a particular focus on diagnostic studies: a review. Metabolomics. 2015;11(4):872–894. doi: 10.1007/s11306-014-0746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stringer KA, Younger JG, McHugh C, Yeomans L, Finkel MA, Puskarich MA, Jones AE, Trexel J, Karnovsky A. Whole Blood Reveals More Metabolic Detail of the Human Metabolome Than Serum as Measured by 1H-NMR Spectroscopy: Implications for Sepsis Metabolomics. Shock. 2015 doi: 10.1097/SHK.0000000000000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mercier P, Lewis MJ, Chang D, Baker D, Wishart DS. Towards automatic metabolomic profiling of high-resolution one-dimensional proton NMR spectra. Journal of biomolecular NMR. 2011;49(3–4):307–323. doi: 10.1007/s10858-011-9480-x. [DOI] [PubMed] [Google Scholar]
- 44.Schalcher TR, Borges RS, Coleman MD, Batista J, Junior, Salgado CG, Vieira JL, Romao PR, Oliveira FR, Monteiro MC. Clinical oxidative stress during leprosy multidrug therapy: impact of dapsone oxidation. PLoS One. 2014;9(1):e85712. doi: 10.1371/journal.pone.0085712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yano T, Kassovska-Bratinova S, Teh JS, Winkler J, Sullivan K, Isaacs A, Schechter NM, Rubin H. Reduction of clofazimine by mycobacterial type 2 NADH:quinone oxidoreductase: a pathway for the generation of bactericidal levels of reactive oxygen species. J Biol Chem. 2011;286(12):10276–10287. doi: 10.1074/jbc.M110.200501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vitvitsky V, Martinov M, Ataullakhanov F, Miller RA, Banerjee R. Sulfur-based redox alterations in long-lived Snell dwarf mice. Mech Ageing Dev. 2013;134(7–8):321–330. doi: 10.1016/j.mad.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garg SK, Yan V, Vitvitsky V, Banerjee R. In: Analysis of sulfur-containing metabolites involved in redox and methionine metabolism. Das DK, editor. New Rochelle, NY: Mary Ann Liebert; 2010. pp. 7–11. [Google Scholar]
- 48.Bouatra S, Aziat F, Mandal R, Guo AC, Wilson MR, Knox C, Bjorndahl TC, Krishnamurthy R, Saleem F, Liu P, Dame ZT, Poelzer J, Huynh J, Yallou FS, Psychogios N, Dong E, Bogumil R, Roehring C, Wishart DS. The human urine metabolome. PLoS One. 2013;8(9):e73076. doi: 10.1371/journal.pone.0073076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Curran-Everett D, Benos DJ. Guidelines for reporting statistics in journals published by the American Physiological Society. Physiol Genomics. 2004;18(3):249–251. doi: 10.1152/physiolgenomics.00155.2004. [DOI] [PubMed] [Google Scholar]
- 50.Sidak Z. Rectangular Confidence Regions for Means of Multivariate Normal Distributions. J Am Stat Assoc. 1967;62(318):626. [Google Scholar]
- 51.Storey JD. The positive false discovery rate: A Bayesian interpretation and the q-value. Ann Stat. 2003;31(6):2013–2035. [Google Scholar]
- 52.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karnovsky A, Weymouth T, Hull T, Tarcea VG, Scardoni G, Laudanna C, Sartor MA, Stringer KA, Jagadish HV, Burant C, Athey B, Omenn GS. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics. 2012;28(3):373–380. doi: 10.1093/bioinformatics/btr661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao J, Tarcea VG, Karnovsky A, Mirel BR, Weymouth TE, Beecher CW, Cavalcoli JD, Athey BD, Omenn GS, Burant CF, Jagadish HV. Metscape: a Cytoscape plug-in for visualizing and interpreting metabolomic data in the context of human metabolic networks. Bioinformatics. 2010;26(7):971–973. doi: 10.1093/bioinformatics/btq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WH, Bushman FD, Lusis AJ, Hazen SL. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19(5):576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Romick-Rosendale LE, Goodpaster AM, Hanwright PJ, Patel NB, Wheeler ET, Chona DL, Kennedy MA. NMR-based metabonomics analysis of mouse urine and fecal extracts following oral treatment with the broad-spectrum antibiotic enrofloxacin (Baytril) Magn Reson Chem. 2009;47(Suppl 1):S36–46. doi: 10.1002/mrc.2511. [DOI] [PubMed] [Google Scholar]
- 57.Sun J, Schnackenberg LK, Khare S, Yang X, Greenhaw J, Salminen W, Mendrick DL, Beger RD. Evaluating effects of penicillin treatment on the metabolome of rats. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;932:134–143. doi: 10.1016/j.jchromb.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 58.Swann JR, Tuohy KM, Lindfors P, Brown DT, Gibson GR, Wilson ID, Sidaway J, Nicholson JK, Holmes E. Variation in antibiotic-induced microbial recolonization impacts on the host metabolic phenotypes of rats. J Proteome Res. 2011;10(8):3590–3603. doi: 10.1021/pr200243t. [DOI] [PubMed] [Google Scholar]
- 59.Theriot CM, Koenigsknecht MJ, Carlson PE, Jr, Hatton GE, Nelson AM, Li B, Huffnagle GB, JZL, Young VB. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yap IK, Li JV, Saric J, Martin FP, Davies H, Wang Y, Wilson ID, Nicholson JK, Utzinger J, Marchesi JR, Holmes E. Metabonomic and microbiological analysis of the dynamic effect of vancomycin-induced gut microbiota modification in the mouse. J Proteome Res. 2008;7(9):3718–3728. doi: 10.1021/pr700864x. [DOI] [PubMed] [Google Scholar]
- 61.Zheng X, Xie G, Zhao A, Zhao L, Yao C, Chiu NH, Zhou Z, Bao Y, Jia W, Nicholson JK, Jia W. The footprints of gut microbial-mammalian co-metabolism. J Proteome Res. 2011;10(12):5512–5522. doi: 10.1021/pr2007945. [DOI] [PubMed] [Google Scholar]
- 62.Weber D, Oefner PJ, Hiergeist A, Koestler J, Gessner A, Weber M, Hahn J, Wolff D, Stammler F, Spang R, Herr W, Dettmer K, Holler E. Low urinary indoxyl sulfate levels early after transplantation reflect a disrupted microbiome and are associated with poor outcome. Blood. 2015;126(14):1723–1728. doi: 10.1182/blood-2015-04-638858. [DOI] [PubMed] [Google Scholar]
- 63.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13(8):572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Serkova NJ, Van Rheen Z, Tobias M, Pitzer JE, Wilkinson JE, Stringer KA. Utility of magnetic resonance imaging and nuclear magnetic resonance-based metabolomics for quantification of inflammatory lung injury. Am J Physiol Lung Cell Mol Physiol. 2008;295(1):L152–161. doi: 10.1152/ajplung.00515.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kedishvili NY, Popov KM, Jaskiewicz JA, Harris RA. Coordinated expression of valine catabolic enzymes during adipogenesis: analysis of activity, mRNA, protein levels, and metabolic consequences. Arch Biochem Biophys. 1994;315(2):317–322. doi: 10.1006/abbi.1994.1506. [DOI] [PubMed] [Google Scholar]
- 66.House JD, Hall BN, Brosnan JT. Threonine metabolism in isolated rat hepatocytes. Am J Physiol Endocrinol Metab. 2001;281(6):E1300–1307. doi: 10.1152/ajpendo.2001.281.6.E1300. [DOI] [PubMed] [Google Scholar]
- 67.Elliott P, Posma JM, Chan Q, Garcia-Perez I, Wijeyesekera A, Bictash M, Ebbels TM, Ueshima H, Zhao L, van Horn L, Daviglus M, Stamler J, Holmes E, Nicholson JK. Urinary metabolic signatures of human adiposity. Science translational medicine. 2015;7(285):285ra262. doi: 10.1126/scitranslmed.aaa5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Won EY, Yoon MK, Kim SW, Jung Y, Bae HW, Lee D, Park SG, Lee CH, Hwang GS, Chi SW. Gender-specific metabolomic profiling of obesity in leptin-deficient ob/ob mice by 1H NMR spectroscopy. PLoS One. 2013;8(10):e75998. doi: 10.1371/journal.pone.0075998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Engelke UF, Tangerman A, Willemsen MA, Moskau D, Loss S, Mudd SH, Wevers RA. Dimethyl sulfone in human cerebrospinal fluid and blood plasma confirmed by one-dimensional (1)H and two-dimensional (1)H-(13)C NMR. NMR Biomed. 2005;18(5):331–336. doi: 10.1002/nbm.966. [DOI] [PubMed] [Google Scholar]
- 70.Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, White MB, Monteith P, Noble NA, Unser AB, Daita K, Fisher AR, Sikaroodi M, Gillevet PM. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 2014;60(5):940–947. doi: 10.1016/j.jhep.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dorokhov YL, Shindyapina AV, Sheshukova EV, Komarova TV. Metabolic methanol: molecular pathways and physiological roles. Physiol Rev. 2015;95(2):603–644. doi: 10.1152/physrev.00034.2014. [DOI] [PubMed] [Google Scholar]
- 72.Xia J, Wishart DS. MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics. 2010;26(18):2342–2344. doi: 10.1093/bioinformatics/btq418. [DOI] [PubMed] [Google Scholar]
- 73.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13(10):722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell research. 2014;24(1):24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaur J, Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol. 2015;16(8):461–472. doi: 10.1038/nrm4024. [DOI] [PubMed] [Google Scholar]
- 76.Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12(9):831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 77.Roca H, Varsos ZS, Sud S, Craig MJ, Ying C, Pienta KJ. CCL2 and interleukin-6 promote survival of human CD11b+ peripheral blood mononuclear cells and induce M2-type macrophage polarization. J Biol Chem. 2009;284(49):34342–34354. doi: 10.1074/jbc.M109.042671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Indiveri C, Iacobazzi V, Tonazzi A, Giangregorio N, Infantino V, Convertini P, Console L, Palmieri F. The mitochondrial carnitine/acylcarnitine carrier: function, structure and physiopathology. Molecular aspects of medicine. 2011;32(4–6):223–233. doi: 10.1016/j.mam.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 79.Banhegyi G, Benedetti A, Margittai E, Marcolongo P, Fulceri R, Nemeth CE, Szarka A. Subcellular compartmentation of ascorbate and its variation in disease states. Biochim Biophys Acta. 2014;1843(9):1909–1916. doi: 10.1016/j.bbamcr.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 80.Yen S, McDonald JA, Schroeter K, Oliphant K, Sokolenko S, Blondeel EJ, Allen-Vercoe E, Aucoin MG. Metabolomic analysis of human fecal microbiota: a comparison of feces-derived communities and defined mixed communities. J Proteome Res. 2015;14(3):1472–1482. doi: 10.1021/pr5011247. [DOI] [PubMed] [Google Scholar]
- 81.Hwang TJ, Dotsenko S, Jafarov A, Weyer K, Falzon D, Lunte K, Nunn P, Jaramillo E, Keshavjee S, Wares DF. Safety and availability of clofazimine in the treatment of multidrug and extensively drug-resistant tuberculosis: analysis of published guidance and meta-analysis of cohort studies. BMJ Open. 2014;4(1):e004143. doi: 10.1136/bmjopen-2013-004143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bajaj JS, Cox IJ, Betrapally NS, Heuman DM, Schubert ML, Ratneswaran M, Hylemon PB, White MB, Daita K, Noble NA, Sikaroodi M, Williams R, Crossey MM, Taylor-Robinson SD, Gillevet PM. Systems biology analysis of omeprazole therapy in cirrhosis demonstrates significant shifts in gut microbiota composition and function. Am J Physiol Gastrointest Liver Physiol. 2014;307(10):G951–957. doi: 10.1152/ajpgi.00268.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lees HJ, Swann JR, Wilson ID, Nicholson JK, Holmes E. Hippurate: the natural history of a mammalian-microbial cometabolite. J Proteome Res. 2013;12(4):1527–1546. doi: 10.1021/pr300900b. [DOI] [PubMed] [Google Scholar]
- 84.Halouska S, Fenton RJ, Barletta RG, Powers R. Predicting the in vivo mechanism of action for drug leads using NMR metabolomics. ACS Chem Biol. 2012;7(1):166–171. doi: 10.1021/cb200348m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Do KT, Kastenmuller G, Mook-Kanamori DO, Yousri NA, Theis FJ, Suhre K, Krumsiek J. Network-based approach for analyzing intra- and interfluid metabolite associations in human blood, urine, and saliva. J Proteome Res. 2015;14(2):1183–1194. doi: 10.1021/pr501130a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.