Abstract
Breast cancer cells frequently home to the bone marrow, where they may enter a dormant state before forming a bone metastasis. Several members of the interleukin-6 (IL-6) cytokine family are implicated in breast cancer bone colonization, but the role for the IL-6 cytokine leukemia inhibitory factor (LIF) in this process is unknown. We tested the hypothesis that LIF provides a pro-dormancy signal to breast cancer cells in the bone. In breast cancer patients, LIF receptor (LIFR) levels are lower with bone metastases and are significantly and inversely correlated with patient outcome and hypoxia gene activity. Hypoxia also reduces the LIFR:STAT3:SOCS3 signaling pathway in breast cancer cells. Loss of the LIFR or STAT3 enables otherwise dormant breast cancer cells to down-regulate dormancy, quiescence, and cancer stem cell-associated genes, and to proliferate in and specifically colonize the bone, suggesting LIFR:STAT3 signaling confers a dormancy phenotype in breast cancer cells disseminated to bone.
Breast cancer cells disseminated to the bone marrow possess the ability to remain in a dormant state for years prior to emerging as a clinically detectable bone metastasis1. The mechanisms enabling tumor cells to emerge from dormancy are poorly understood, but there is increasing evidence that tumor-stromal interactions, and the osteoblast2, 3, perivascular4 and perisinusoidal5 niche are critical mediators of tumor cell dormancy and bone colonization. Hypoxia, or very low oxygen tensions, has also been implicated in modulating tumor dormancy6, but the role for hypoxia in tumor cell dormancy in the bone has not been investigated7.
Several members of the interleukin-6 (IL-6) family of cytokines, such as IL-6 and oncostatin M (OSM), have been demonstrated to promote breast cancer colonization of the bone marrow8, 9. The leukemia inhibitory factor (LIF) receptor (LIFR), whose ligand LIF also belongs to the IL-6 family of cytokines, was recently identified as a breast tumor suppressor and lung metastasis suppressor10, 11. Previous correlations between LIF and LIFR expression in breast cancer cell lines capable of colonizing the bone12 suggest that the LIF signaling pathway may play a key role in tumor establishment in bone.
Results
LIFR is down-regulated in patients with bone metastases
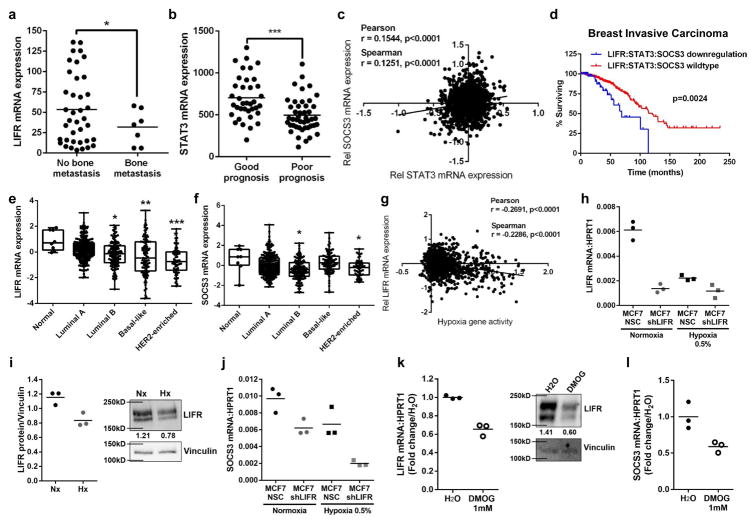
We first investigated LIFR expression in primary tumors of breast cancer patients who were predicted to have a poor prognosis13, and found that LIFR mRNA levels were significantly lower in those patients with bone metastases (Fig. 1a). In this same patient dataset14, signal transducer and activator 3 (STAT3) mRNA levels were significantly lower in breast cancer patients with a poor prognosis compared to those with a good prognosis (Fig. 1b). STAT3 is a mediator of downstream LIF:LIFR signaling and can repress or activate target genes, including suppressor of cytokine signaling 3 (SOCS3), which is activated by LIF and can negatively regulate STAT315. In patients with invasive breast carcinoma, STAT3 mRNA levels positively correlated with SOCS3 mRNA levels (Fig. 1c), suggesting this signaling axis may be important in patient outcome. Indeed, patients with mRNA down-regulation of LIFR:STAT3:SOCS3 genes had significantly reduced overall survival (Fig. 1d, Supplementary Fig. 1a–c), and there was a significant co-occurrence of alterations (amplification, homozygous deletion, mutation, or mRNA expression changes) within the LIFR and STAT3 genes, as well as STAT3 and SOCS3 (Supplementary Fig. 1d). LIFR and SOCS3 mRNA levels were significantly lower in breast cancer patients with the luminal B subtype, which is the tumor type that most frequently metastasizes to bone16, as well as in basal-like (LIFR only), and HER2-enriched tumor types, which are more aggressive subtypes (Fig. 1e,f), suggesting that LIFR and SOCS3 are down-regulated in patients most likely to develop bone metastases.
Figure 1. LIFR:STAT3 signaling is down-regulated in patients with bone metastases and repressed by hypoxia.
(a) LIFR mRNA levels (Minn et al. dataset) in breast tumors from patients with a poor prognosis based on Van’t Veer signature (n=38 patients with no bone metastasis, n=7 patients with bone metastasis). Student’s unpaired t-test with Welch’s Correction. (b) STAT3 mRNA levels (Minn et al. dataset) in breast tumors (n=37 good prognosis, n=45 poor prognosis). Student’s unpaired t-test. (c) Correlation of STAT3 and SOCS3 mRNA levels in The Cancer Genome Atlas (TCGA) provisional dataset, n=999 patients. Pearson and Spearman Correlation. (d) Analysis of TCGA invasive breast carcinoma (Nature 2012 dataset) patient samples for LIFR, STAT3, and SOCS3 mRNA down-regulation (n=682 patients with no mRNA down-regulation, n=55 patients with mRNA down-regulation). Logrank test. (e,f) LIFR and SOCS3 mRNA levels in TCGA invasive breast carcinoma (Nature 2012 dataset) samples stratified by PAM50 breast cancer subtype (Normal n=8, Luminal A n=230, Luminal B n=125, Basal-like n=98, HER2-enriched n=58 patients). Kruskal-Wallis test. Minima=25% percentile, maxima=75% percentile, center=median. (g) Correlation of Li et al. hypoxia gene signature with LIFR mRNA levels in TCGA invasive breast carcinoma patient dataset, as in (c). Pearson and Spearman Correlation. n=998 patients. (h) LIFR mRNA levels following 24hrs in normoxia or hypoxia. 3 technical replicates from a single experiment representative of 2 independent experiments. (i) Quantification of 3 Western blots, representative LIFR blot after 24hrs in normoxia or hypoxia. 3 technical replicates from a single experiment. (j) SOCS3 mRNA levels following 24hr culture in normoxia or hypoxia. 3 technical replicates from a single experiment representative of 2 independent experiments. (k) LIFR mRNA levels in MCF7 cells following 16hr treatment with PHD inhibitor DMOG (1mM) and LIFR Western blot representative of 3 technical replicates from a single experiment. PCR: 3 technical replicates from a single experiment. (l) SOCS3 mRNA levels in MCF7 cells following 16hr treatment with DMOG (1mM). 3 technical replicates from a single experiment. Source data for 1h–l in Supplementary Table 1 and unprocessed blots in Supplementary Fig. 9. Graphs represent mean/group and error bars=SEM. *p<0.05, **p<0.01, and ***p<0.001.
Hypoxia represses the LIFR in breast cancer cells and is negatively correlated with LIFR in patients
Since alterations in the LIFR result in significantly worse patient survival and it has been demonstrated that hypoxia in breast tumors is linked to poor patient survival17–20, we hypothesized that hypoxia may down-regulate the LIFR in breast cancer cells and signal cells to exit dormancy. Analysis of invasive breast carcinoma patients from The Cancer Genome Atlas (TCGA) revealed that as hypoxia gene activity increases21, LIFR mRNA levels are significantly reduced (Fig. 1g), indicating that hypoxia may be a mechanism by which breast cancer cells down-regulate the LIFR. Hypoxia has been previously identified as a regulator of the LIFR in embryonic stem cells22. To investigate this relationship, MCF7 human breast cancer cells were cultured in hypoxic conditions (0.5% O2) and LIFR mRNA and protein levels, and SOCS3 mRNA levels, were reduced (Fig. 1h–j; Supplementary Fig. 2a, shRNA #3), indicating that hypoxia down-regulates the entire LIFR:STAT3:SOCS3 signaling pathway. PGK1 mRNA levels were used as a positive control for activation of hypoxia-inducible signaling (Supplementary Fig. 2b). The breast cancer cell line SUM159, with known low metastatic potential for the lungs, showed a reduction in LIFR mRNA levels similar to MCF7 cells following culture in hypoxia (Supplementary Fig. 2c). A similar effect on LIFR and SOCS3 mRNA levels was observed when MCF7 cells were treated with dimethyloxaloylglycine (DMOG), a pharmacological activator of HIF signaling (Fig. 1k,l).
Hypoxic regulation of the LIFR is HIF-independent
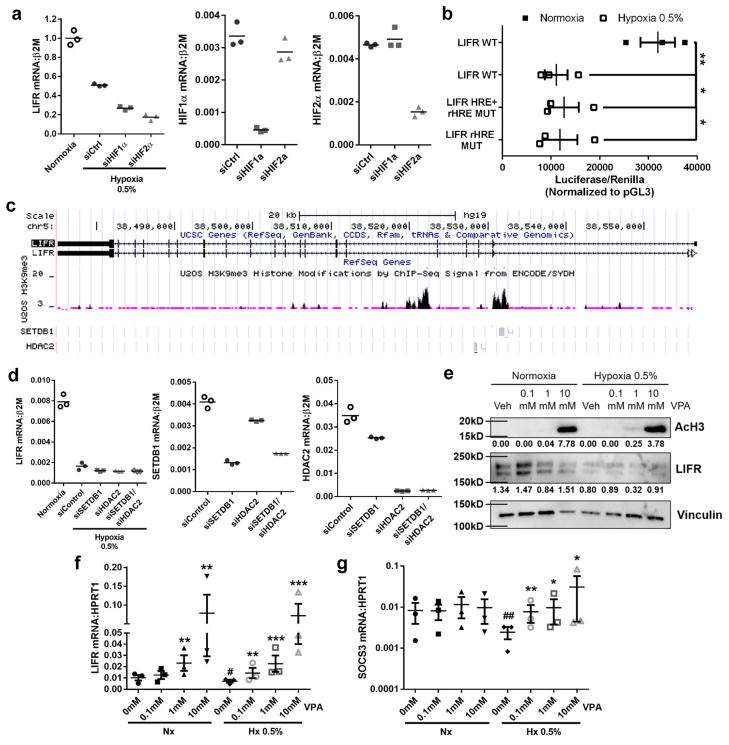
Although hypoxia and DMOG were able to induce LIFR down-regulation, we found that neither HIF1α nor HIF2α were needed for hypoxic repression of the LIFR (Fig. 2a). We also identified six hypoxia responsive elements (HREs), and one reverse HRE23, in the proximal promoter of the LIFR. Mutation of activating HREs and the reverse HRE, or the reverse HRE alone, indicated that these regions of the promoter are not required for hypoxic repression of the LIFR (Fig. 2b). These data suggest a HIF-independent, but DMOG-inducible mechanism for hypoxic repression of LIFR transcription.
Figure 2. Hypoxic regulation of LIFR:STAT3 signaling is independent of HIF1α or HIF2α.
(a) LIFR mRNA levels in MCF7 cells in normoxia or hypoxia for 24hrs and transfected with HIF1α or HIF2α siRNA. HIF1α and HIF2α mRNA knockdown levels shown as controls. 3 technical replicates from a single experiment representative of 3 independent experiments. (b) LIFR promoter activity of wildtype promoter (WT) or promoter with mutations in hypoxia responsive elements + reverse hypoxia element (LIFR HRE+rHRE mut) or reverse hypoxia element mutated only (LIFR rHRE mut) following culture for 48hrs in normoxia or hypoxia. Student’s unpaired t-test. n=3 biological replicates, one each from 3 independent experiments. (c) UCSC genome browser tracks for LIFR gene variants 1 and 2, H3K9me3 histone modifications by ChIP-Seq analysis from ENCODE database, and SETDB1 and HDAC2 binding sites on the LIFR gene. (d) LIFR mRNA levels in MCF7 cells in 24hrs normoxia or hypoxia and transfected with SETDB1 or HDAC2 siRNA. SETDB1 and HDAC2 mRNA knockdown levels shown as controls. 3 technical replicates from a single experiment representative of 3 independent experiments. (e) Western blot for acetylated histone H3 (AcH3) and LIFR protein levels in MCF7 cells treated with vehicle (Veh), 0.1mM, 1mM, or 10mM valproic acid (VPA), a histone deacetylase (HDAC) inhibitor, for 24hrs with actin loading control. Representative of 2 independent biological replicates. (f) LIFR mRNA levels after 24hrs treatment with 0, 0.1mM, 1mM, or 10mM VPA in normoxia or hypoxia. n=3 biological replicates, each being an average from 3 independent experiments. Student’s unpaired t-test. (g) SOCS3 mRNA levels after 24hrs treatment with 0, 0.1mM, 1mM, or 10mM VPA in normoxia or hypoxia. Student’s unpaired t-test. n=3 biological replicates, each being an average from 3 independent experiments. Source data for 2a,d available in Supplementary Table 1 and unprocessed blots in Supplementary Fig. 9. Graphs represent the mean/group and error bars represent standard error of the mean (SEM). *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001. #p<0.05 versus 0mM Nx.
It has previously been reported that DMOG can promote methylation on histone H3, and thus induce gene repression through stimulation of histone methyltransferases and inhibition of histone demethylases24. Interestingly, DMOG can promote repression of some genes while still activating HIF target genes24, suggesting that DMOG could inhibit LIFR through HIF-independent effects. Since we found that the histone methyltransferase SETDB1 and histone deacetylase HDAC2 both bind to the LIFR and make modifications on histone H3 on lysine 9 (Fig. 2c), and SETDB1 is hypoxia-inducible25, we evaluated these enzymes in the hypoxic repression of the LIFR in breast cancer cells. Knockdown of either gene alone or in combination was not sufficient to rescue down-regulation of the LIFR in hypoxia (Fig. 2d). However, it is well documented that HDACs and histone methyltransferases possess compensatory functions26. Therefore we used the pan-HDAC inhibitor valproic acid (VPA)27, which enhanced histone H3 acetylation in MCF7 cells in vitro (Fig. 2e), and significantly increased LIFR and SOCS3 mRNA levels in hypoxia (Fig. 2f,g), suggesting HDAC inhibition enhances LIFR:STAT3:SOCS3 signaling.
We also investigated whether the LIFR was methylated at the DNA level in patients with breast carcinoma, and whether increased LIFR methylation may relate to poor patient outcome. Analysis of the invasive breast carcinoma cohort from TCGA revealed that LIFR and STAT3 mRNA levels significantly decrease as LIFR DNA methylation increases (Supplementary Fig. 2d,e), suggesting that methylation, or repression, of the LIFR gene may lead to repression of STAT3 in patients. LIFR mRNA levels are significantly lower in breast cancer patients who have recurred/progressed (Supplementary Fig. 2f), and most importantly, LIFR DNA methylation is significantly increased in breast cancer patients who have recurred/progressed (Supplementary Fig. 2g), suggesting that LIFR DNA methylation is directly related to a negative patient outcome. Since up to 70% of breast cancer patients who succumb to disease present with bone metastases upon autopsy7, we hypothesize that many of those patients who recurred/progressed will also harbor bone metastases.
LIF inhibits breast cancer cell growth in tumor cells known to lie dormant in bone
Since cells of the osteoblast lineage and bone marrow stromal cells produce LIF28–32, we hypothesized that LIF may act as an inhibitory signal to breast cancer cells known to home to the bone marrow. MCF7 cells have been shown to home to the bone marrow following intracardiac33 and intrailiac inoculation3, where they induce limited bone destruction, thus acting as dormant tumor cells in the bone marrow, similar to their dormant phenotype in the lungs following intravenous inoculation34. MCF7 cells were growth inhibited in response to recombinant LIF (Supplementary Fig. 3a), while bone metastatic variants of MDA-MB-231 (MDA-MB-231b35, 36) and 4T1 cells (4T1BM237) were growth resistant to recombinant LIF in vitro (Supplementary Fig. 3b,c). Likewise, low doses of LIF induced SOCS3 expression in MCF7 (Supplementary Fig. 3d), but not MDA-MB-231b cells (Supplementary Fig. 3e), indicating that cells like the MCF7s that home to the bone and essentially lie dormant are sensitive to LIF.
Breast cancer cells with low metastatic potential maintain a functional LIFR
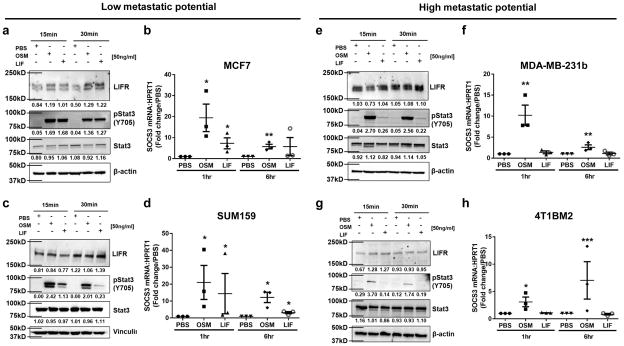
Human breast and mouse mammary carcinoma cell lines with low metastatic potential, MCF7, SUM159, PyMT, and D2.0R mouse mammary carcinoma cells, all showed abundant expression of the LIFR at the protein level, and rapidly phosphorylated STAT3 and induced SOCS3 transcription in response to recombinant oncostatin M (OSM) or recombinant LIF (Fig. 3a–d, Supplementary Fig. 4a–d), indicating that both signaling pathways are functional. Conversely, the MDA-MB-231b, 4T1BM2 and D2A1 mouse mammary carcinoma cells were unresponsive to recombinant LIF, but maintained a STAT3:SOCS3 response to recombinant OSM (Fig. 3e–h, Supplementary Fig. 4e,f), suggesting loss of the LIFR:STAT3 signaling pathway may confer loss of dormancy.
Figure 3. The LIFR:STAT3 signaling pathway is intact in breast cancer cells with low metastatic potential and not high metastatic potential.
(a,c,e,g) Western blot for LIFR, pSTAT3 (Y705), total Stat3, and β-actin (loading control) after 15 or 30 minute treatment with PBS (vehicle control), recombinant OSM (50ng/ml) or recombinant LIF (50ng/ml) in (a,c) MCF7 and SUM159 cells with low metastatic potential and (e,g) MDA-MB-231b and 4T1BM2 cells with high metastatic potential. Blots represent 3 independent biological replicates. (b,d,f,h) SOCS3 mRNA levels after 1 or 6 hour treatment with PBS, recombinant OSM (50ng/ml) or recombinant LIF (50ng/ml). b,f: Student’s unpaired t-test. d,h: Mann-Whitney test. n=3 biological replicates, each being an average from 3 independent experiments. Unprocessed blots in Supplementary Fig. 9. Graphs represent the mean/group and error bars represent standard error of the mean (SEM). *p<0.05 and **p<0.01.
We also compared the 4T1 and MDA-MB-231 parental cell lines to their bone metastatic variants (4T1BM2 and MDA-MB-231b cell lines). In 4T1 parental cells, LIFR was semi-functional in terms of inducing Stat3 phosphorylation in response to LIF (Supplemental Fig. 4g), but not Socs3 mRNA induction (Supplemental Fig. 4h). In MDA-MB-231 parental cells multiple glycosylated forms of the LIFR were expressed (Supplementary Fig. 4i), but Stat3 and Socs3 signaling were not activated downstream (Supplementary Fig. 4i,j). It is important to note that 4T1 and MDA-MB-231 parental lines will readily grow in and colonize the bone, and therefore may possess other mechanisms or mutations that can promote bone invasion independent of the effects of the LIFR.
Loss of the LIFR enhances invasion and down-regulates dormancy, quiescence and cancer stem cell-associated genes
In order to determine the impact of LIFR expression on MCF7 tumor cell behavior, we analyzed MCF7 control (MCF7 non-silencing control; MCF7NSC) and MCF7 cells in which LIFR was functionally knocked down (MCF7shLIFR #3; Supplementary Fig. 2a, Supplementary Fig. 5a,b). LIFR knockdown did not alter proliferation in vitro or in the mammary fat pad in vivo (Supplementary Fig. 5c,d), but increased MCF7 migration and invasion in 3D cultures (Supplementary Fig. 5e,f), consistent with the phenotype of LIFR knockdown in SUM159 cells10. These data suggest that loss of LIFR signaling disrupts autocrine LIF signaling to downstream targets and promotes tumor cell migration and invasion.
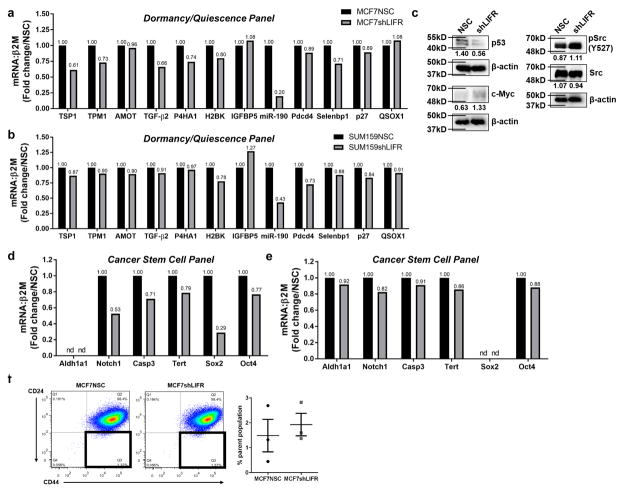
We next identified key dormancy and quiescence genes validated across tumor models (Supplementary Fig. 6a)38–48, and tested their expression in our LIFR knockdown model. Loss of LIFR signaling in MCF7 cells resulted in lower mRNA levels in 6 out of 12 genes associated with a dormancy/quiescence phenotype, including thrombospondin-1 (TSP1)38, 44, tropomyosin-1 (TPM1)38, 44, transforming growth factor-β2 (TGF-β2)39, 44, prolyl 4 hydroxylase α-1 (P4HA1)38, 44, miRNA-190 (miR-190)43, and Selenbp145 (Fig. 4a), suggesting that intact LIFR signaling regulates genes involved in dormancy. A similar, but less dramatic reduction in dormancy/quiescence genes was detected in SUM159shLIFR cells (Fig. 4b; Supplementary Fig. 6b). LIFR knockdown in MCF7 cells also resulted in lower p5338, 41, 46 protein levels, and greater c-Myc42, 45 and pSrc34, 40 (Y527) protein levels (Fig. 4c), consistent with an exit from dormancy40–42.
Figure 4. Inhibition of LIFR signaling alters dormancy, quiescence and cancer stem cell-associated genes.
(a) mRNA levels of genes associated with dormancy and quiescence in MCF7NSC and MCF7shLIFR cells. 3 technical replicates from a single experiment. (b) mRNA levels of genes associated with dormancy and quiescence in SUM159NSC and SUM159shLIFR cells. 3 technical replicates from a single experiment. (c) Western blot for signaling pathways associated with tumor dormancy (p53, Myc, Src). β-actin is loading control for each blot. (d) mRNA levels of genes associated with a cancer stem cell phenotype in MCF7NSC and MCF7shLIFR cells. 3 technical replicates from a single experiment. (e) mRNA levels of genes associated with a cancer stem cell phenotype in SUM159NSC and SUM159shLIFR cells. 3 technical replicates from a single experiment. (f) Representative FACS plot and graph of % parent population from n=3 biological replicates, each being an average from 3 independent experiments. Q3 = CD44High/CD24Low cancer stem cell population. Student’s unpaired t-test. Source data for 4a,b,d,e available in Supplementary Table 1 and unprocessed blots in Supplementary Fig. 9. Mean indicated on graph. Graphs represent the mean/group and error bars represent standard error of the mean (SEM). *p<0.05, **p<0.01, and ****p<0.0001.
Dormant or quiescent cancer cells may adopt a cancer stem cell phenotype49, and thus we evaluated a number of cancer stem cell-associated genes45, 50. We found a dramatic down-regulation of Notch1, Caspase-3 [Casp-3 (Sca-1)], Tert, and Sox2 in MCF7shLIFR cells (Fig. 4d), and all genes were mildly reduced in SUM159shLIFR cells (Fig. 4e), suggesting that LIFR signaling may maintain both a dormant and cancer stem cell phenotype. However, LIFR deletion did not alter the percentage of MCF7 cells exhibiting the cancer stem cell CD44Hi/CD24Lo marker phenotype51 (Fig. 4f).
We also confirmed that LIFR effects on patient outcome and tumor dormancy were not mediated via downstream PI3K:mTOR52 or MAPK53 signaling cascades (Supplementary Fig. 7a–h).
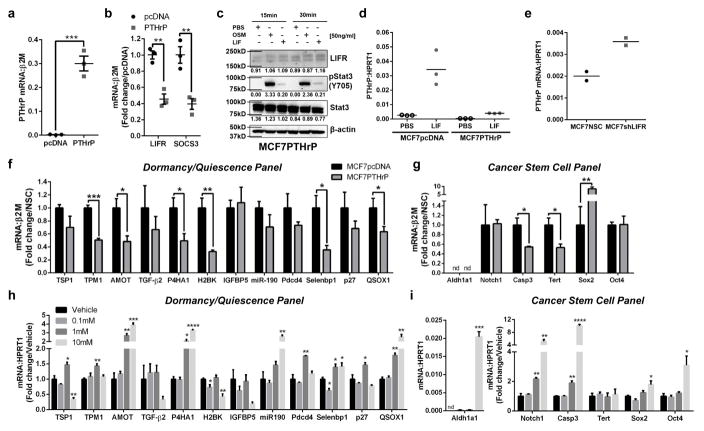
PTHrP over-expression ablates LIFR signaling in MCF7 cells
Parathyroid hormone-related protein (PTHrP) is a secreted factor required for breast tumor-induced bone destruction35. Studies indicate that direct inhibition of PTHrP shortly after tumor inoculation is beneficial in blocking tumor-induced osteolysis35, 54. When MCF7 cells are forced to overexpress PTHrP (Fig. 5a), these cells induce extensive bone destruction, and MCF7 cells no longer reside in a dormant state33. We found that both LIFR and SOCS3 mRNA levels and LIFR protein were significantly repressed in MCF7 cells overexpressing PTHrP (Fig. 5b,c). LIF stimulation of STAT3 phosphorylation (Fig. 5c) and SOCS3 induction (Fig. 5d) was dampened in MCF7PTHrP over-expressing cells, indicating PTHrP negatively regulates LIFR:STAT3:SOCS3. Interestingly, MCF7shLIFR cells expressed 2-fold higher levels of PTHrP (Fig. 5e), suggesting a potential regulatory loop between LIFR and PTHrP signaling. Similar to LIFR knockdown in MCF7 cells, we observed significant down-regulation of dormancy, quiescence, and cancer stem cell-associated genes in MCF7 cells over-expressing PTHrP (Fig. 5f,g). Thus, repression of the LIFR, either directly or perhaps through PTHrP overexpression, impacts expression of dormancy-associated genes.
Figure 5. Elevated PTHrP signaling blocks LIFR signaling and reduces dormancy, quiescence and cancer stem cell-associated genes.
(a) Parathyroid hormone-related protein (PTHrP) mRNA levels in MCF7 cells overexpressing PTHrP (MCF7PTHrP). Student’s unpaired t-test. n=3 biological replicates, one each from 3 independent experiments. (b) LIFR and SOCS3 mRNA levels in MCF7PTHrP over-expressing cells. Student’s unpaired t-test. n=3 biological replicates, one each from 3 independent experiments. (c) Western blot for LIFR, pSTAT3 (Y705), total Stat3, and β-actin (loading control) in MCF7PTHrP over-expressing whole cell lysates. (d) SOCS3 mRNA levels in MCF7pcDNA and MCF7PTHrP over-expressing cells after 1 hour treatment with PBS (vehicle control) or recombinant LIF (50ng/ml). 3 technical replicates from a single experiment representative of 2 independent experiments. (e) PTHrP mRNA levels (1–139aa) inn MCF7NSC and MCF7shLIFR cells. 2 biological replicates, each being an average from 2 independent experiments. (f,g) mRNA levels of genes associated with (f) dormancy and quiescence, and (g) cancer stem cells in MCF7pcDNA (control) and MCF7PTHrP over-expressing cells. Mann-Whitney test. n=3 biological replicates, one each from 3 independent experiments. (h,i) mRNA levels of genes associated with (h) dormancy and quiescence, and (i) cancer stem cells in MCF7 cells treated for 24 hours with 0, 0.1mM, 1mM, or 10mM valproic acid (VPA). Multiple t-test with Holm-Sidak post-test. n=3 biological replicates, one each from 3 independent experiments. Source data for 5d,e available in Supplementary Table 1 and unprocessed blots in Supplementary Fig. 9. Graphs represent the mean/group and error bars represent standard error of the mean (SEM). *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001.
Valproic acid stimulates dormancy and cancer stem cell-associated genes
Since valproic acid increases LIFR mRNA levels in MCF7 cells (Fig. 2f), we hypothesized that valproic acid may enhance expression of genes associated with dormancy, quiescence and a cancer stem cell phenotype. Indeed, valproic acid significantly stimulated 9 out of 12 dormancy and quiescence-associated genes, and 5 out of 6 cancer stem cell-associated genes (Fig. 5h,i). These data suggest that LIFR expression has a corresponding effect on dormancy and cancer stem cell-associated genes.
Loss of LIFR in MCF7 cells induces osteolytic bone destruction
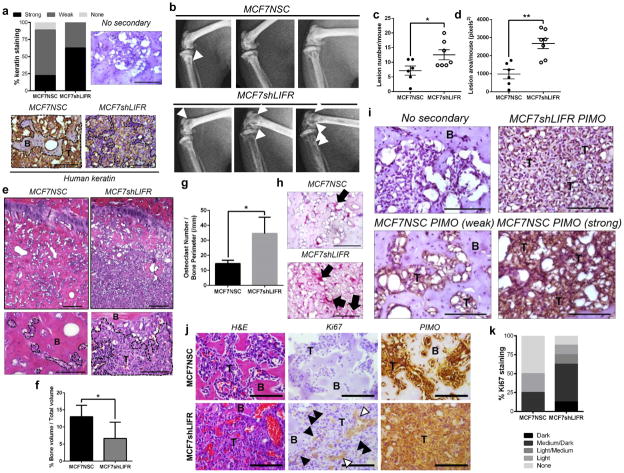
Since loss of LIFR signaling resulted in a decrease in genes associated with dormancy, we investigated whether deletion of LIFR increases MCF7 colonization and growth in the bone marrow following intracardiac inoculation. MCF7 cells in the bone marrow were identified in 6 of 7 MCF7NSC and 7 of 7 MCF7shLIFR tumor-bearing mouse tibiae by human keratin staining (Fig. 6a). As expected, MCF7NSC cells did disseminate to the bone, and we observed limited bone destruction. In contrast, MCF7shLIFR cells disseminated to the bone and induced significant bone destruction as indicated by lesion area and lesion number upon x-ray analysis (Fig. 6b–d). Histological analysis also revealed significantly lower bone volume in MCF7shLIFR tumor-bearing mice (Fig. 6e,f) which may be attributable to the significantly greater number of osteoclasts lining the bone surface in MCF7shLIFR tumor-bearing bones (Fig. 6g,h).
Figure 6. LIFR knockdown results in greater bone destruction via increased osteoclastogenesis and proliferation in vivo.
(a) Percent of animals with strong, weak or no human keratin staining in bone marrow of mouse tibiae (7 mice/group). Representative human keratin staining in the bone marrow for MCF7NSC and MCF7shLIFR tumor-bearing mouse. Brown = human keratin positive. Black dashed line outlines tumor burden. B=bone, T=tumor. Scale bar=100μm. (b) Radiographic images of mouse tibiae and femora 10 weeks after MCF7NSC (n=6 mice) or MCF7shLIFR (n=7 mice) intracardiac tumor cell inoculation. White arrowheads indicate osteolytic lesions. (c) Total lesion number/mouse and (d) total lesion area/mouse in MCF7NSC (n=6 mice) and MCF7shLIFR (n=7 mice) tumor-bearing bones. Student’s unpaired t-test. (e) Representative H&E images of MCF7NSC (6 mice) and MCF7shLIFR (7 mice) tumor-bearing tibiae. B=bone, T=tumor. Histological analysis and (f) quantification of bone volume/total volume (%BV/TV) in MCF7NSC and MCF7shLIFR tumor-bearing tibiae (n=6 mice/group). Mann-Whitney test. Scale bar (top)=200μm. Scale bar (bottom)=100μm. (g) Quantification of osteoclast number/bone perimeter (OcN/BPm) in millimeters (n=7 mice/group). Student’s unpaired t-test. (h) Representative images of TRAP-positive osteoclasts in MCF7NSC (n=6 mice) and MCF7shLIFR (n=7 mice) tumor-bearing bones. Scale bar=100μm. (i) Immunohistochemistry for pimonidazole (hypoxia probe) in MCF7shLIFR (representative of 2 mice) and MCF7NSC (representative of 4 mice) tumor-bearing mice. B=bone, T=tumor. Upper left image=no secondary control. Upper right image representative of weak pimonidazole staining where MCF7shLIFR tumor cells have infiltrated the bone. Lower right image representative of strong pimonidazole staining with extensive tumor infiltration in MCF7NSC mouse and lower left image representative of weak pimonidazole staining in a pocket with little tumor infiltration in the same MCF7NSC tumor-bearing limb. Scale bar=100μm. (j) Histology/immunostaining for H&E, Ki67 (negative controls Supplementary Figure 8a), and PIMO in MCF7NSC and MCF7shLIFR tumor-bearing tibiae (8 mice/group). White arrows indicate blood vessels; black arrows indicate Ki67-positive tumor cells. T=tumor, B=bone. Scale bar=100μm. (k) Quantification of Ki67 percent staining (8 mice/group). Source data for 6c,d,f,g available in Supplementary Table 1. Graphs represent mean/group and error bars=SEM. *p<0.05 and **p<0.01.
Pockets of tumor infiltration co-localize with strong staining for a hypoxia marker
MCF7NSC tumor infiltration coincided with strong staining for the hypoxia probe pimonidazole in the bone marrow, while areas where MCF7NSC cells appeared to lie dormant in the bone without inducing bone destruction stained weakly for pimonidazole within the same limb (Fig. 6i, lower left and lower right). In contrast, MCF7shLIFR tumors infiltrated the bone regardless of pimonidazole staining intensity (Fig 6i, upper right). These data are consistent with a mechanism by which hypoxia may promote outgrowth of indolent breast cancer cells disseminated to the bone marrow.
Loss of LIFR promotes MCF7 proliferation within the bone marrow
We found that the majority of MCF7NSC cells did not stain for Ki67 (a proliferative marker) while MCF7shLIFR cells stained for Ki67 within the bone marrow (Fig. 6j,k; Supplementary Fig. 8a). A small percentage of MCF7 tumor cells slowly invaded the bone over time and stained positive for Ki67. This is consistent with the gradual increase in osteolysis detected by radiography within MCF7 control tumor-bearing mice over the course of 10 weeks. However, the numbers of these cells still remained less than the Ki67 positive MCF7shLIFR tumor cells in the bone marrow (Fig. 6j,k). For the most part, MCF7 control cells appeared quiescent in vivo. These data support the idea of intrinsic differences in these cells after dissemination to the bone marrow.
We also stained for pimonidazole and found that MCF7NSC cells, although for the most part Ki67-negative, can be found in pimo-positive regions (Fig. 6i,j). Interestingly, there was pronounced pimonidazole staining along the tumor-bone interface in both MCF7NSC and MCF7shLIFR tumor-bearing limbs (Supplementary Fig. 8b), suggesting this region may be where the tumor cells are most likely to become invasive.
Many factors that have been found to regulate dormancy act in a tissue-specific manner4, 39, 55, 56. We confirmed that MCF7shLIFR cells colonize the lung at a similar rate to MCF7NSC cells (Supplementary Fig. 8c,d). This corroborates patient data derived from Minn et al.14, which shows no significant difference in LIFR mRNA levels in patients with a poor prognosis who developed lung metastases versus those with no lung metastases (Supplementary Fig. 8e), and is in contrast to previously published data showing a dramatic role for LIFR in suppressing breast cancer lung colonization 10. Since the previously published study used ER negative human breast cancer cells, and the MCF7 cells in our study are ER positive, we propose the difference in colonization may be due to ER expression.
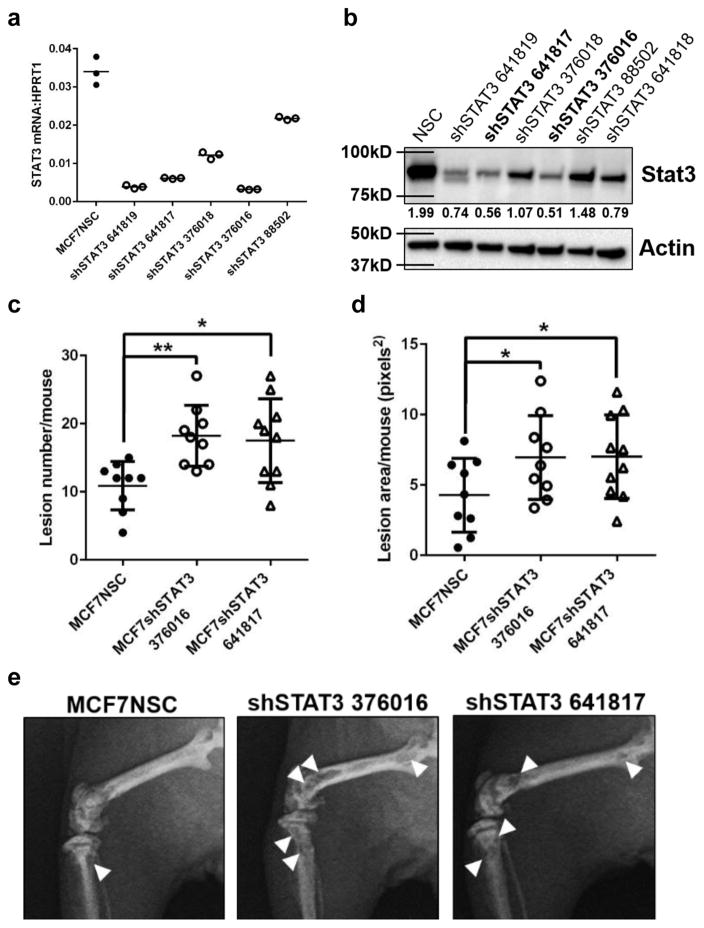
Loss of STAT3 signaling in breast cancer cells mimics loss of LIFR in vivo
To determine if LIFR effects on tumor cell dormancy in bone are mediated through STAT3 signaling, we inoculated MCF7 cells that had stable STAT3 knockdown (Fig. 7a,b; clones 641817 and 376016) by intracardiac injection. Loss of STAT3 in MCF7 cells induced significantly greater bone destruction in vivo with both clones (Fig. 7c–e), similar to MCF7 cells lacking the LIFR. These data indicate that STAT3 acting downstream of the LIFR may be an important mediator of tumor-induced osteolysis and MCF7 exit from dormancy.
Figure 7. Loss of Stat3 signaling results in greater bone destruction in vivo.
(a) STAT3 mRNA levels in MCF7 cells infected with STAT3 lentiviral clones tested for knockdown efficiency. 3 technical replicates from a single experiment. (b) Western blot for Stat3 protein levels in MCF7 cells infected with STAT3 lentiviral clones tested for knockdown efficiency. (c) Lesion number (Total number from both tibiae and femora from each mouse) and (d) lesion area (Total area from both tibiae and femora from each mouse) in the tibiae and femora of MCF7NSC (n=9 mice) or MCF7shSTAT3 tumor-bearing mice (shSTAT3 clone 376016 n =9 mice, shSTAT3 clone 641817 n=10 mice). Student’s unpaired t-test. (e) Representative images of osteolytic bone destruction in MCF7NSC (n=9 mice), MCF7shSTAT3 376016 (n=9 mice), and MCF7shSTAT3 641817 (n=10 mice) tumor-bearing mice. Source data for 7a,c,d available in Supplementary Table 1. Graphs represent the mean/group and error bars represent standard error of the mean (SEM). *p<0.05 and **p<0.01.
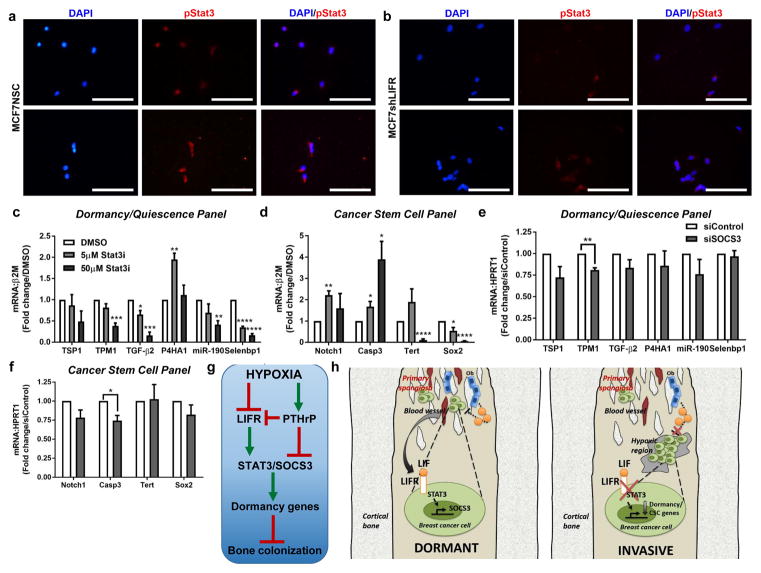
Inhibiting Stat3 signaling down-regulates dormancy-associated genes
Since STAT3 is a key mediator of LIFR signaling, we investigated STAT3 phosphorylation in MCF7shLIFR tumor cells by immunocytochemistry and found nuclear staining was reduced (Fig. 8a,b). We next inhibited STAT3/SOCS3 by treatment with a small molecule STAT3 inhibitor (ML116, which we termed Stat3i, Supplementary Fig. 8f–h) or siRNA against SOCS3. MCF7 cells exhibited a Stat3i dose-dependent reduction in 5 out of 6 of the dormancy genes that were down-regulated with LIFR knockdown (Fig. 8c), and 2 out of 4 of the cancer stem cell genes that were down-regulated with LIFR knockdown (Fig. 8d). Stat3i treatment also significantly down-regulated mRNA levels of all 6 of the remaining dormancy genes that were not regulated via LIFR signaling (Supplementary Fig. 8i). A similar, but modest effect on dormancy and cancer stem cell-associated genes was observed with siRNA against SOCS3 in MCF7 cells (Fig. 8e,f; Supplementary Fig. 8j) and SUM159 cells (Supplementary Fig. 8k,l).
Figure 8. Targeting STAT3 or SOCS3 mimics effects of loss of LIFR signaling on dormancy genes.
(a,b) Immunocytochemistry for basal pStat3 (Y705) protein levels in (a) MCF7NSC (2 biological replicates) and (b) MCF7shLIFR cells (2 biological replicates). Images representative of 3 independent replicates/group. Scale bar = 100μm. (c,d) mRNA levels of dormancy/quiescence and cancer stem cell-associated genes in MCF7 cells following 24 hour treatment with 5μM or 50μM of the small molecule Stat3 inhibitor ML116 (Stat3i). Multiple t-tests with Holm-Sidak method. n=3 biological replicates, one each from 3 independent experiments. (e,f) mRNA levels of dormancy/quiescence and cancer stem cell-associated genes in MCF7 cells transfected with siRNA against SOCS3 for 48 hours. Multiple t-tests with Holm-Sidak method. n=3 biological replicates, one each from 3 independent experiments. (g) Flow chart indicating hypoxia differentially regulates LIFR and PTHrP, which signal via STAT3 and SOCS3 to regulate dormancy-associated genes and thus influence bone colonization. (h) Working model for LIFR:STAT3 signaling in disseminated breast cancer cells transitioning from a dormant to invasive phenotype in strongly hypoxic regions of the bone marrow. Graphs represent the mean/group and error bars represent standard error of the mean (SEM).*p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001.
Discussion
We conclude from our analysis of LIFR expression across all breast cancer cell lines that breast cancer cells with low metastatic potential, either to the lung (MCF7, D2.0R, PyMT) or to the bone (MCF7) express a functional LIFR and are LIF responsive. In contrast, breast cancer cells that aggressively colonize the lung (D2A1) or bone (MDA-MB-231b, 4T1BM2) lack a functional LIFR and do not respond to LIF in vitro. Interestingly, the 4T1BM2 and D2A1 cells expressed very low levels of the LIFR (Fig. 3g, Supplementary Fig. 4e), while MDA-MB-231b cells expressed high levels of the LIFR (Fig. 3e), although all three cell lines are highly metastatic. We suspect that the LIFR is inactive in MDA-MB-231b tumor cells, since they do not respond to LIF in vitro. Together these data suggest that breast cancer cells with high metastatic potential, which readily home to and colonize the bone, are unable to respond to paracrine LIF produced in the bone by marrow cells and the osteoblast niche.
Our findings indicate that LIFR is important for maintaining tumor cells in a dormant state. In support of this hypothesis, it has previously been reported that LIF and LIFR mRNA levels are elevated in dormant versus proliferative squamous cell carcinoma cells (supplemental expression data)57 and that LIFR levels are elevated in cells in a G0 (non-proliferative) state (supplemental expression data)45. STAT3 has also been identified as a dormancy-associated gene in estrogen receptor positive breast cancer cells, which are reported to have higher dormancy signature scores than ER negative breast cancer cells38.
LIFR signaling has been found to negatively regulate the stem cell pool58, and our data in tumor cells are in contrast to these findings. The physiological role for LIF is to maintain a stem-like state59, and thus our data are consistent with a role for LIFR signaling in maintaining a less differentiated cell phenotype. It has also been shown that cells with high metastatic potential (e.g. MDA-MB-231 cells) express higher levels of LIF60, which may enable these cells to secrete paracrine LIF within the bone marrow to drive LIFR signaling in other cell types and prevent differentiation. It is possible that all of the effects of LIF:LIFR signaling in MDA-MB-231 cells are mediated via AKT/mTOR signaling as previously reported60. However, we did not focus on this signaling pathway since it was not implicated in dormancy regulation. Importantly, while over-expression of LIF did result in significantly reduced relapse-free survival60, greater than 75% of patients over-expressing LIF in their tumors remained disease-free by 10 years out, compared to 0% overall survival in patients with LIFR downregulation at this same time point (Fig. 1d). This suggests that the loss of a functional LIFR may be critical for patient outcome and fits with the less dramatic effects of LIFR/AKT/mTOR signaling on overall survival that we found in patient data. Hypoxia has also been shown to induce LIF in colorectal carcinoma cell lines61, but the status of the LIFR is not clear in those cell lines, and thus what effect elevated LIF may have on autocrine signaling is unknown. It is possible that the HIF2α-driven up-regulation of LIF in those cells may also activate an alternative pathway to STAT3 such as AKT/mTOR. Our data suggests that activation of alternative pathways downstream of the LIFR may have a dramatically different impact on patient survival.
We observed an increase in pimonidazole staining along the tumor-bone interface, suggesting this may be where tumor cells are most likely to become invasive. Alternatively, the osteogenic niche may promote the outgrowth of early disseminated tumor cells in the bone marrow3 and support the homing of disseminated proliferative human breast cancer cells5. Furthermore, the presence of tumor cells in this niche may be driving down oxygen tensions due to localized increased oxygen consumption. The occasional Ki67-positive cells found in MCF7NSC samples may have encountered the hypoxic microenvironment and have shut down their dormancy machinery to become proliferative; however, our current technologies do not enable us to determine whether hypoxia is driving that process through LIFR.
Our data also indicate that STAT3 may be a key mediator of the LIFR-induced dormancy phenotype. However, STAT3 may play a diminished role in the LIFR-mediated effects on cancer stem cell gene expression, since many of those genes were unaltered or stimulated by STAT3 inhibition. Interestingly, treatment with the small molecule inhibitor Stat3i also significantly down-regulated mRNA levels of all 6 of the remaining dormancy genes that were not regulated by LIFR signaling (Supplementary Fig. 8i), suggesting that STAT3 may be a major mediator of tumor dormancy and that these effects are not restricted to a LIFR-mediated mechanism. While STAT3 inhibitors are generally used as anti-cancer agents62, our data suggest that loss of STAT3 signaling stimulates tumor cells to exit dormancy. Thus STAT3 inhibition in patients with disseminated tumor cells may be detrimental to patient outcome and may in part explain why STAT3 inhibitors have failed to produce a positive outcome in clinical trials for breast cancer62.
In summary, LIFR:STAT3:SOCS3 signaling in disseminated breast cancer cells confers a dormancy phenotype through maintenance of STAT3 signaling in response to LIF. In regions of extremely low oxygen tensions within the bone marrow, tumor cells become invasive through down-regulation of STAT3 and possibly SOCS3 signaling. These data provide a mechanism by which tumor cells may be directed to spontaneously exit dormancy by the bone microenvironment and indicate that stimulating the LIFR:STAT3:SOCS3 signaling pathway in breast cancer cells may prevent the outgrowth of indolent tumor cells disseminated to the marrow.
Methods
Cell culture and reagents
Cells
Human MCF7 and MDA-MB-231 breast cancer cells and 4T1 mouse mammary carcinoma cells were obtained from ATCC and cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin (P/S). Human SUM159 breast cancer cells were gifted by the Rutgers Cancer Institute of New Jersey and were cultured in Ham’s F12 medium supplemented with 5% FBS, 5μg/ml insulin, and 1μg/ml hydrocortisone. Mouse mammary carcinoma cell lines D2A1 and D2.0R were gifted by Jeffrey Green at the National Cancer Institute. PyMT-derived tumor cells were established in Robin Anderson’s laboratory. Human MDA-MB-231b bone metastatic cells were established from the original bone clone made by the Mundy laboratory, and passaged in bone periodically to maintain bone metastatic phenotype35, 36. 4T1BM2 bone metastatic mouse mammary carcinoma cells37 were gifted by Dr. Normand Pouliot at the Peter MacCallum Cancer Centre. MCF7pcDNA and MCF7PTHrP overexpressing cells33 were established in Jack Martin’s laboratory at St. Vincent’s Institute of Medical Research and were gifted by Dr. Theresa Guise at Indiana University. All cell lines, except SUM159 human breast cancer cells, were cultured in DMEM supplemented with 10% FBS and P/S. No cell lines used in this study were found in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI Biosample. MCF7 cells were re-authenticated by ATCC; none of the remaining cell lines have been re-authenticated. The MCF7 cells were tested for mycoplasma contamination; none of the remaining cell lines have been tested for mycoplasma contamination.
shRNA and siRNA
GIPZ lentiviral LIFR- and STAT3-targeting human shRNAs were purchased from Open Biosystems (catalog numbers in parentheses). LIFR shRNA #1=V2LHS_133981 (RHS4430-200216162), shRNA #2=V2LHS_133982 (RHS4430-200175346), shRNA #3=V3LHS_347493 (RHS4430-200297260). STAT3 shRNA = V2LHS_88502 (RHS4430-200162926), V3LHS_376016 (RHS4430-200250657), V3LHS_376018 (RHS4430-200248312), V3LHS_641817 (RHS4430-200225068), V3LHS_641818 (RHS4430-200227569), V3LHS_641819 (RHS4430-200223545). GIPZ vectors were transfected into 293T cells using Lipofectamine Plus (Life Technologies) to generate lentivirus, which was then transferred onto MCF7 or SUM159 cells. Breast cancer cells were selected for 6 days in 1μg/ml puromycin and evaluated for knockdown at the protein and mRNA level. ON-TARGETplus SMARTpool siRNAs for HIF1A (L-004018-00), EPAS1 (HIF2A) (M-004814-01), SETDB1 (L-020070-00), HDAC2 (L-003495-02), and SOCS3 (L-004299-00) were purchased from Dharmacon/GE Healthcare (catalog numbers in parentheses) along with ON-TARGETplus SMARTpool siControl (D-001810-10-20) and transfected into MCF7 or SUM159 cells using DharmaFECT 1 transfection reagent (Dharmacon/GE Healthcare).
Stat3 small molecule inhibitor
The STAT3 inhibitor probe referred to as Stat3i in the text was provided by William Roush at The Scripps Research Institute (Scripps Florida) and its Scripps probe ID is ML116. The inhibitor is a thienopyrimidine scaffold: 4-(4-benzylpiperidin-1-yl)thieno[2,3-d]pyrimidine and the PubChem ID is 2100018. The probe was identified in a high throughput screen for STAT3 inhibitors and was selected for its specificity for STAT3 over STAT1 and NFκB. Details of the screen can be found at the Scripps Research Institute website (http://mlpcn.florida.scripps.edu/index.php/probes/probe-reports.html).
To determine whether ML116 (Stat3i) may be a biologically active inhibitor, we used receptor-based virtual screening and molecular docking to determine whether Stat3i will specifically bind and inhibit the activity of STAT3. We obtained the crystalized 3D structure of a STAT3 monomer from the Protein Data Bank database and we were able to confirm docking and molecular interactions between STAT3 and ML116, with ML116 virtually docking adjacent to the DNA binding groove of the STAT3 monomer with a docking score of −7.3 (considered favorable binding affinity), suggesting disruption of STAT3 and DNA binding.
Recombinant proteins
Recombinant human LIF (R&D Systems), human oncostatin M (R&D Systems), mouse LIF (Miltenyi Biotec), mouse oncostatin M (R&D Systems), and human TGF-β1 (R&D Systems) were reconstituted in PBS + 0.1% bovine serum albumin (BSA) at 10–25μg/ml and aliquoted for storage at −80°C. For all experiments, mouse recombinant proteins were used on mouse cell lines, and human recombinant proteins were used on human cell lines, with the exception of human TGF-β1, which was used on all cell lines since TGF-β1 maintains approximately 99% sequence homology between human and mouse species63. Prior to cytokine treatment cells were serum starved in 2% FBS overnight and cytokine treatment was made up in media containing 2% FBS.
Hypoxia
For hypoxia experiments, cells were seeded at 1 ×105 cells/well in normoxia, allowed to settle overnight, and placed into a hypoxia (0.5% O2) chamber (Invivo2 Hypoxia Workstation 400) the following day for 24–48 hours, as indicated in the figure legends. Cells were harvested in situ for RNA with TRIzol (Life Technologies) or for protein in RIPA lysis buffer supplemented with protease and phosphatase inhibitors (Roche). For siRNA transfected cells, transfection was performed in normoxia and cells incubated overnight prior to being transferred to the hypoxia chamber.
DMOG treatment
Cells were seeded at 2.5 × 105 cells/well and incubated overnight. Cells were treated with 1mM DMOG or an equal volume of vehicle (water) for 16 hours in full serum conditions and harvested for RNA with TRIzol (Life Technologies) or for protein in RIPA lysis buffer supplemented with protease and phosphatase inhibitors (Roche).
Valproic acid (VPA) treatment
Cells were seeded at 4 × 105 cells/well and valproic acid (VPA, Sigma-Aldrich) treatment was performed in DMEM+10% FBS. Addition of VPA did not alter the pH of the media, even at the highest dose of 10mM (tested with pH strips). For experiments in which tumor cells were transfected with the LIFR WT promoter prior to treatment, VPA was added to the transfection media. For experiments in which mRNA levels were evaluated following culture in normoxia or hypoxia ± VPA treatment, VPA was added to the cells in normoxia and cells were moved into hypoxia one hour later.
PI3K (BEZ235) and ERK1/2 (AZD6244) inhibitors
MCF7 cells were seeded at 4 × 105 cells/well in 6 well cluster dishes and treated the following day with either vehicle (water), 10nM, 100nM, or 1000nM BEZ235 or vehicle (DMSO), 0.1uM, 1uM, or 10uM AZD6244 for 24 hours in full serum conditions. Cells were harvested at 24 hours for RNA in TRIzol and processed for qPCR. For Western analysis, cells were seeded at 1 × 106 in 10cm dishes and treated the following day at the same concentrations as above and harvested for protein in RIPA buffer after 24 hours.
XTT assay
Cells were seeded at 2 × 104 cells/well in triplicate into 96 well plates and settled after 2 hours in culture. Plating media was removed and replaced with DMEM+2% FBS and either vehicle (PBS), recombinant LIF (50ng/ml) or recombinant TGF-β (5ng/ml). Transforming growth factor-β1 (TGF-β1) was used as a positive control, since breast cancer cells are growth inhibited by TGF-β1 early in tumor progression, which can also be detected in vitro64. Media and treatments were replenished daily while cells grew in culture. At end point cells were incubated in serum-phenol-red-free DMEM with PMS and XTT sodium salt for 2–4 hours until color change and measured for absorbance at 450nm using a Synergy H1-mono plate reader (Biotek).
Migration and invasion assays
Migration
Cells were seeded at 1 × 106 cells/well into 6 well plates and settled for 24 hours for migration (scratch) assays. Three vertical scratches were made through the cell layer using marker guidelines drawn on the bottom of the plate to ensure consistency in imaging time points. Brightfield images were captured at 0 and 24 hours after the scratch was made and wound closure was quantified as a percent of the initial wound area using Image Pro software.
Invasion/3D cultures
Cells were seeded at 12.5 × 103 cells/well into 48 well plates in quadruplicate in a 3:1 ratio with rat collagen type I (BD Biosciences) and allowed to polymerize at room temperature for 30 minutes. Plates were incubated at 37°C for 4 hours then 0.5ml media was added to each well. Media was changed every 1–2 days and brightfield images were acquired using an inverted microscope.
Immunostaining
Cells were seeded into 4-well chamber slides (Biotek) at 5 × 103 cells/chamber and incubated overnight. Cells were fixed in 10% formalin and permeabilized in 0.25% Triton-X in PBS. Cells were stained sequentially with a primary antibody against pStat3 Y705 (Cell Signaling, clone D3A7 XP, Catalog Number 9145, 1:50), secondary antibody biotinylated goat anti-rabbit, and streptdavadin-texas red, and counterstained with DAPI. Fluorescence images were acquired on a Leica CTR6000 microscope (Leica) and Hamamatsu digital camera (C11440) using LAS X software (Leica) and overlaid using ImageJ software.
Real-time PCR
Cells were harvested in TRIzol (Life Technologies), extracted, digested (TURBO DNA-free Kit, Life Technologies), and cDNA synthesized (200ng–1000ng RNA, iScript cDNA Synthesis Kit, Bio-Rad) per the manufacturer’s instructions. Real-time PCR was performed on a 7900HT Fast Real-time PCR System (Applied Biosystems) with the following cycling conditions: 2 minutes at 50°C, 10 minutes at 95°C, (15 seconds at 95°C, 1 minute at 60°C) x40 cycles followed by dissociation curve (15 seconds at 95°C, 1 minute at 60°C, 15 seconds at 95°C). Human primers for PTHrP65, QSOX166, PGK167, Pdcd4, and p27 coding sequence were previously published45. The following primers were designed using PrimerBlast (NCBI) against the human genome (Homo sapiens) and validated by dissociation: LIFR (F-GCGTACCGACTGACTGCATTG, R-CCAGAGGGTGCTTTCCAAGAA), SOCS3 (F-GCTCCAAGAGCGAGTACCAG, R-CTGTCGCGGATCAGAAAGGT), TSP1 (F-TCCCCATCCAAAGCGTCTTC, R-ACCACGTTGTTGTCAAGGGT), TPM1 (F-TCTCAGAAGGCCAAGTCCGA, R-CAAACTCAGCCCGAGTCTCA), AMOT (F-GGCATGCCACCCCAATCT, R-TTGTAGCAAGGGCAAGGACC), TGF-β2 (F-CAGCTTGTGCTCCAGACAGT, R-GCTCAATCCGTTGTTCAGGC), P4HA1 (F-GTACATGACCCTGAGACTGGA, R-GGGGTTCATACTGTCCTCCAA), H2BK (F-CAAGGCCGTCACCAAGTACA, R-GAAGGCAATTGTGCTTCTTTTGA), IGFBP5 (F-CCCAATTGTGACCGCAAAGG, R-CGTCAACGTACTCCATGCCT), miR-190 (F-GCAGGCCTCTGTGTGATATGT, R-GGCAAGACACTGTAGGAATATGT), Selenbp1 (F-AAGTGCGAACTGGCCTTTCT, R-CCCATCCAGCAGCACAAAAC), Aldh1α1 (F-CAAGATCCAGGGCCGTACAA, R-CAGTGCAGGCCCTATCTTCC), Notch1 (F-AGCCTCAACGGGTACAAGTG, R-CACACGTAGCCACTGGTCAT), Casp3 (F-GCGGTTGTAGAAGAGTTTCGTG, R-CTCACGGCCTGGGATTTCAA), Tert (F-CTTGCGGAAGACAGTGGTGA, R-GTCCGGGCATAGCTGGAGTA), Sox2 (F-ACCAGCGCATGGACAGTTAC, R-CCGTTCATGTAGGTCTGCGA), Oct4 (F-AGAAGCTGGAGCAAAACCCG, R-ACCTTCCCAAATAGAACCCCCA), Stat3 (F-CAATACCATTGACCTGCCGAT, R-GAGCGACTCAAACTGCCCT), Socs3, (F-GCTCCAAGAGCGAGTACCAG, R-CTGTCGCGGATCAGAAAGGT), β2M (F-GAGTATGCCTGCCGTGTGAA, R-TGCGGCATCTTCAAACCTCC), and HPRT1 (F-TTGCTTTCCTTGGTCAGGCA, R-ATCCAACACTTCGTGGGGTC). Mouse HMBS was designed using PrimerBlast against the mouse genome (Mus musculus) and validated by dissociation: HMBS (F-TCATGTCCGGTAACGGCG, R-CACTCGAATCACCCTCATCTTTG).
LIFR promoter construct and activity
The human LIFR sequence was identified using the UCSC Genome Browser December 2013 Assembly (most recent dataset available). The FASTA sequence for LIFR transcript variant 1 coding sequence was obtained and BLASTed (NCBI) against mRNA from NCBI LIFR variant 1, with 100% matching sequence. The transcription start site was identified and 2kb of the proximal promoter scanned for hypoxia responsive elements (HREs) and reverse HREs (rHRE) using consensus sequences CACGT, GCGTG, ACGTG, CGTG for HREs and TGCAC for rHREs. Six HREs and one rHRE were identified within 1.3kb upstream of the LIFR transcription start site (3 proximal to the transcription start site and 3 distal). There were no HREs identified between ~1.3–2kb upstream of the transcription start site. The consensus sequence ACGTG was the only HRE not found in the LIFR promoter. The following constructs were synthesized: LIFR wild type promoter construct, LIFR construct with all HREs and the rHRE mutated, and LIFR construct with the rHRE only mutated. All constructs were synthesized and cloned into the pGL3 basic vector by GenScript (Pascataway, NJ). Cells transfected with promoter constructs were harvested in 1X passive lysis buffer and assessed for activity after 48 hours in normoxic and hypoxic conditions (72 hours after transfection) using the Bright-Glo/Renilla luciferase assay system (Promega) per manufacturer’s instructions. Plates were read using a Synergy H1-mono plate reader (Biotek).
Western blotting
Cells grown in monolayer were rinsed with 1X PBS and harvested for protein in RIPA buffer supplemented with a protease and phosphatase inhibitor (Roche). Protein concentration was determined by BCA assay (Thermo Fisher) and 20μg protein was loaded onto an SDS-PAGE gel in reducing conditions using standard techniques. PVDF membranes were probed with antibodies against LIFR (Santa Cruz Biotechnology, C-19, Catalog Number sc-659, 1:1000), pSTAT3 Y705 (Cell Signaling, Catalog Number 9131, 1:1000), Stat3 (Cell Signaling, clone 124H6, Catalog Number 9139, 1:1000), p53 (Invitrogen, clone PAB1801, Catalog Number 13–4000, 1:2000), c-Myc (Cell Signaling, clone D84C12 XP, Catalog Number 5605, 1:1000), pSrc Y527 (Cell Signaling, Catalog Number 2105, 1:1000), Src (Cell Signaling, clone 36D10, Catalog Number 2109, 1:1000), pAkt Ser473 (Cell Signaling, Catalog Number 9271, 1:1000), Akt (Cell Signaling, Catalog Number 9272S, 1:1000), pErk1/2 Thr202/Tyr204 (Cell Signaling, Catalog Number 9101, 1:1000), Erk1/2 (Cell Signaling, Catalog Number 9102, 1:1000), β-actin (Sigma, clone AC-15, Catalog Number A5441, 1:5000), and vinculin (Millipore, Catalog Number AB6039, 1:10,000). All Western blots were quantified for adjusted relative density using ImageJ. Adjusted relative density values are listed below each lane. Briefly, blot images were converted to a histogram rendering for each lane and peaks were converted to the relative percentage for each blot. Proteins of interest were then normalized to the relative percentage of the loading control for the respective lane.
In silico analyses
TCGA patient data analysis
The Cancer Genome Atlas cBioPortal was accessed on September 24, 2014, February 2, 2015, February 19, 2015 and January 8, 2016 to determine overall patient survival and the tendency for co-occurrence of LIFR, STAT3, and SOCS3 alterations in the dataset for Breast Invasive Carcinoma (Nature 2012). LIFR, STAT3, and SOCS3 were entered as the query genes and overall survival and co-expression was provided through the cBioPortal user interface. Data were then downloaded and manually sorted for patients with mRNA down-regulation of LIFR, STAT3, and SOCS3. cBioPortal was accessed on March 25, 2016 for survival data for LIFR:PIK3CA:MTOR and LIFR:MAPK3:MAPK1 in the Breast Invasive Carcinoma dataset (Nature 2012) and data were downloaded and manually entered into Prism for survival curve analysis. CBioPortal was accessed on March 27, 2015 to determine whether the hypoxia gene signature from n=1104 patients with Breast Invasive Carcinoma (Provisional dataset) correlated with the LIFR mRNA expression. First, the hypoxia gene signature from Li et al.21 and LIFR were entered into the query gene field for cBioPortal downloads on the entry page. The gene expression for each gene across all 1104 patients was downloaded and saved in excel format. For each gene in the hypoxia gene signature, the gene expression provided by RNAseq was normalized to the median expression of that gene across all patients as previously published21. We next averaged the hypoxia gene activity score for all of the hypoxia genes in each individual patient. The average hypoxia gene score for each patient was the final output. For the LIFR gene expression analysis, gene expression levels of the LIFR were normalized for each patient to the median expression of the LIFR across all patients. A log conversion was applied to the normalized, averaged hypoxia gene expression and the LIFR expression and then plotted as a correlation graph in Prism software with a linear regression curve. SOCS3 and STAT3 mRNA levels were correlated for the same TCGA Provisional dataset using Prism software. TCGA cBioPortal was accessed on January 29, 2016 to determine LIFR and SOCS3 mRNA levels by PAM50 breast cancer subtype in the Breast Invasive Carcinoma dataset (Nature 2012). The raw data were downloaded from TCGA through the cBioPortal and manually graphed in Prism (Graphpad). TCGA cBioPortal was accessed on January 6, 2016 for DNA methylation data for the LIFR in disease free and recurred/progressed patients within the Breast Invasive Carcinoma dataset (Provisional). These data were downloaded from TCGA through cBioPortal and manually graphed in Prism as expression data and correlated with STAT3 and LIFR mRNA expression.
Minn et al., Nature 2005 dataset
The supplementary microarray datasets nature03799-s11 and nature03799-s12 for breast cancer patient samples was downloaded from Minn et al.14 on April 17, 2015 to analyze LIFR expression in patients with a bone metastatic event. First, LIFR and STAT3 expression levels were gleaned from nature03799-211 for all patients in the dataset. For the LIFR analysis, patients were selected who were assigned a poor prognosis by van’t Veer signature13 in the nature03799-s12 dataset, since we felt these patients were the most likely to have recurred, and then stratified the data based on the occurrence of a bone metastatic event or lung metastatic event who had data for LIFR expression. For STAT3 analysis, all patients were stratified based on van’t Veer signature.
Animal studies and imaging
Animals
All animal studies were conducted with approval by the Institutional Animal Care and Use Committee (IACUC) at Stanford University. For animal studies, sample size was determined based on power calculations performed by a Radiation Oncology statistician, Rie von Eyben, at Stanford University. The experiments were not randomized. The investigator was blinded during the experiment for radiographical assessment and after the experiment for histological assessment.
For the shLIFR intracardiac study, the day prior to tumor cell inoculation, 4–6 week old female athymic nude mice were subcutaneously implanted with a slow-release 17β-estradiol pellet as previously described33. The following day mice were inoculated via intracardiac injection with 1 × 105 tumor cells as previously described36, and sacrificed at 5 weeks (MCF7NSC n=8 mice, MCF7shLIFR n=8 mice; and MCF7NSC n=9 mice, MCF7shSTAT3 cl376016 n=9 mice, MCF7shSTAT3 cl641817 n=10 mice) or 10 weeks (MCF7NSC and MCF7shLIFR, n=7 mice/group) post-tumor cell inoculation. For the mammary fat pad study, female athymic nude mice were inoculated with 5 × 105 MCF7NSC (n=4 mice) or MCF7shLIFR cells (n=3 mice) in 50μl volume of sterile PBS via mammary fat pad injection into the 4th and 9th inguinal gland as previously described36. Tumor volume was assessed by caliper measurement. For the tail vein study, mice were inoculated with 1 × 106 MCF7NSC or MCF7shLIFR cells (n=10 mice/group) in 100μl PBS and sacrificed after 6 weeks.
Radiography
Radiographic (x-ray) images of mice were obtained at weeks 5, 6, 7, 8, and 10 post-tumor cell inoculation as previously described36. X-ray images were obtained using a Progeny Dental apparatus (65kV, 4mA at 0.08 seconds) and mouse hind limbs were laid directly on the sensor. Images were quantified using Image Pro software (shLIFR study) or ImageJ software (shSTAT3 study; please note difference in pixel count between the two software programs resulting in different axes between the two graphs for lesion area). For x-ray analysis of the intracardiac study, one mouse from the MCF7NSC inoculated group was excluded from the final analysis as a significant outlier following a statistical Grubbs’ test with a significance level of alpha = 0.05.
Histology and immunohistochemistry
Hind limbs were dissected and fixed in 10% formalin for 72 hours and decalcified in EDTA (20% pH7.4) solution for 72 hours. Decalcified bones were embedded in paraffin and 5μm thick sections were collected for staining. Hematoxylin and eosin (H&E) staining was performed as previously described36 and the region of interest was defined beginning 200μm distal to the tibial growth plate, extending 900μm distal to the growth plate. ROIs were quantified for bone volume using Image-Pro software (Media Cybernetics). Immunohistochemistry (IHC) for human keratin (Cell Signaling, Pan-Keratin clone C11, Catalog Number 4545, 1:500), Ki67 (Thermo Scientific, SP6, Catalog Number RM-9106-S 0, 1:250), and Pimonidazole (PIMO, Hypoxyprobe-1 Omni Kit, Hypoxyprobe, Inc, Catalog Number HP3–100Kit, 1:100) was performed according to manufacturer’s instructions using citric acid antigen retrieval. TRAP staining was performed using the acid phosphatase, leukocyte (TRAP) kit (Sigma-Aldrich) per the manufacturer’s instructions using all dilutions as specified in the kit. Quantification of TRAP staining was performed in the same ROI as bone volume above using ImageJ software. Images were quantified at 40x magnification and results are reported using a conversion ratio of 810 pixels: 100 microns.
Statistics and reproducibility
For all studies n/group is as indicated in the figure legend, bar graphs or numbers on graph indicate mean of each group, and error bars indicate standard error of the mean. All graphs and statistical analyses were generated using Prism software (Graphpad). All in vitro and in vivo assays were analyzed for statistical significance using Student’s unpaired t-test, Mann-Whitney test, multiple t-tests with Holm-Sidak post-test or Welch’s correction or ANOVA with Sidak’s or Tukey’s post-test. Please see figure legends for detailed statistical analyses for each experiment. For in vitro assays, no statistical method was used to predetermine sample size. In vitro experiments were reproducible, with some variability between biological replicates and experiments. Animal studies were performed one time and included appropriate animal numbers to ensure statistical evaluation. For all analyses p<0.05 was considered statistically significant, and *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Data availability
Previously published microarray data-sets that we re-analyzed here to detect LIFR expression in breast cancer patients with a bone metastasis are available to download as an excel file in the supplemental data provided by Minn et al.14, which includes cross-referenced data for van’t Veer signature, as described above in the in silico section. Previously published human breast invasive carcinoma survival data-sets and RNA sequencing data-sets that we re-analyzed here were reported previously68,69 and are available from the TCGA Research Network (http://cancergenome.nih.gov/.) as described in detail above in the in silico section. Source data for Fig. 1(h–l), 2(a,d), 4(a,b,d,e), 5(d,e), 6(c,d,f,g), 7(a,c,d), and Supplementary Fig. 2b, 3(a–c), 5(b–e), 6b, and 8(c,d,h) have been provided as Supplementary Table 1. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
The authors wish to acknowledge Dr. Ahn Diep, Greg Nelson, and the Stanford Veterinary Service Center for technical assistance, and Drs. Natalie Sims and Jack Martin for thoughtful discussion of LIFR signaling. Cell sorting/flow cytometry analysis for this project was performed on instruments in the Stanford Shared FACS Facility. RWJ received fellowship support from the Stanford Cancer Institute. This work was funded by NIH grant K99CA194198 (RWJ). JAS was supported by VA grant 1I01BX001957 and NIH grant CA163499. AJG was supported by NIH grants CA67166, CA197713 and CA198291, the Silicon Valley Foundation, The Kimmelman Fund, and the Skippy Frank Foundation.
Footnotes
Contributions
RWJ directed the project, developed the hypothesis, performed experiments, analyzed data, and wrote the manuscript. ECF designed and developed the LIFR promoter construct, performed experiments and edited the manuscript. MMO, MV, TA, YM, ARM and JRJ performed experiments and edited the manuscript. JAS performed experiments, conceptualized and interpreted data. JYW conceptualized and interpreted data. AJG conceived the hypothesis, interpreted data and edited the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Sosa MS, Bragado P, Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nature reviews Cancer. 2014;14:611–622. doi: 10.1038/nrc3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Croucher PI, McDonald MM, Martin TJ. Bone metastasis: the importance of the neighbourhood. Nature reviews Cancer. 2016;16:373–386. doi: 10.1038/nrc.2016.44. [DOI] [PubMed] [Google Scholar]
- 3.Wang H, et al. The osteogenic niche promotes early-stage bone colonization of disseminated breast cancer cells. Cancer cell. 2015;27:193–210. doi: 10.1016/j.ccell.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghajar CM, et al. The perivascular niche regulates breast tumour dormancy. Nature cell biology. 2013;15:807–817. doi: 10.1038/ncb2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Price TT, et al. Dormant breast cancer micrometastases reside in specific bone marrow niches that regulate their transit to and from bone. Sci Transl Med. 2016;8:340ra373. doi: 10.1126/scitranslmed.aad4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bragado P, Sosa MS, Keely P, Condeelis J, Aguirre-Ghiso JA. Microenvironments dictating tumor cell dormancy. Recent Results Cancer Res. 2012;195:25–39. doi: 10.1007/978-3-642-28160-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson RW, Schipani E, Giaccia AJ. HIF targets in bone remodeling and metastatic disease. Pharmacol Ther. 2015;150:169–177. doi: 10.1016/j.pharmthera.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng Y, et al. Targeting IL-6 and RANKL signaling inhibits prostate cancer growth in bone. Clinical & experimental metastasis. 2014;31:921–933. doi: 10.1007/s10585-014-9680-3. [DOI] [PubMed] [Google Scholar]
- 9.Bolin C, et al. Oncostatin m promotes mammary tumor metastasis to bone and osteolytic bone degradation. Genes & cancer. 2012;3:117–130. doi: 10.1177/1947601912458284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen D, et al. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nature medicine. 2012;18:1511–1517. doi: 10.1038/nm.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iorns E, et al. Whole genome in vivo RNAi screening identifies the leukemia inhibitory factor receptor as a novel breast tumor suppressor. Breast cancer research and treatment. 2012;135:79–91. doi: 10.1007/s10549-012-2068-7. [DOI] [PubMed] [Google Scholar]
- 12.Sims NA, Johnson RW. Leukemia inhibitory factor: a paracrine mediator of bone metabolism. Growth factors. 2012;30:76–87. doi: 10.3109/08977194.2012.656760. [DOI] [PubMed] [Google Scholar]
- 13.van’t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 14.Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Starr R, et al. A family of cytokine-inducible inhibitors of signalling. Nature. 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 16.Kennecke H, et al. Metastatic behavior of breast cancer subtypes. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28:3271–3277. doi: 10.1200/JCO.2009.25.9820. [DOI] [PubMed] [Google Scholar]
- 17.Schindl M, et al. Overexpression of hypoxia-inducible factor 1alpha is associated with an unfavorable prognosis in lymph node-positive breast cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2002;8:1831–1837. [PubMed] [Google Scholar]
- 18.Bos R, et al. Levels of hypoxia-inducible factor-1alpha independently predict prognosis in patients with lymph node negative breast carcinoma. Cancer. 2003;97:1573–1581. doi: 10.1002/cncr.11246. [DOI] [PubMed] [Google Scholar]
- 19.Dales JP, et al. Overexpression of hypoxia-inducible factor HIF-1alpha predicts early relapse in breast cancer: retrospective study in a series of 745 patients. International journal of cancer Journal international du cancer. 2005;116:734–739. doi: 10.1002/ijc.20984. [DOI] [PubMed] [Google Scholar]
- 20.Generali D, et al. Hypoxia-inducible factor-1alpha expression predicts a poor response to primary chemoendocrine therapy and disease-free survival in primary human breast cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2006;12:4562–4568. doi: 10.1158/1078-0432.CCR-05-2690. [DOI] [PubMed] [Google Scholar]
- 21.Li B, et al. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513:251–255. doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeong CH, et al. Hypoxia-inducible factor-1 alpha inhibits self-renewal of mouse embryonic stem cells in Vitro via negative regulation of the leukemia inhibitory factor-STAT3 pathway. The Journal of biological chemistry. 2007;282:13672–13679. doi: 10.1074/jbc.M700534200. [DOI] [PubMed] [Google Scholar]
- 23.Narravula S, Colgan SP. Hypoxia-inducible factor 1-mediated inhibition of peroxisome proliferator-activated receptor alpha expression during hypoxia. Journal of immunology. 2001;166:7543–7548. doi: 10.4049/jimmunol.166.12.7543. [DOI] [PubMed] [Google Scholar]
- 24.Tausendschon M, Dehne N, Brune B. Hypoxia causes epigenetic gene regulation in macrophages by attenuating Jumonji histone demethylase activity. Cytokine. 2011;53:256–262. doi: 10.1016/j.cyto.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Olcina MM, et al. H3K9me3 facilitates hypoxia-induced p53-dependent apoptosis through repression of APAK. Oncogene. 2015 doi: 10.1038/onc.2015.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelly RD, Cowley SM. The physiological roles of histone deacetylase (HDAC) 1 and 2: complex co-stars with multiple leading parts. Biochemical Society transactions. 2013;41:741–749. doi: 10.1042/BST20130010. [DOI] [PubMed] [Google Scholar]
- 27.Lee HJ, Kim KW. Suppression of HIF-1alpha by Valproic Acid Sustains Self-Renewal of Mouse Embryonic Stem Cells under Hypoxia In Vitro. Biomolecules & therapeutics. 2012;20:280–285. doi: 10.4062/biomolther.2012.20.3.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allan EH, et al. Osteoblasts display receptors for and responses to leukemia-inhibitory factor. Journal of cellular physiology. 1990;145:110–119. doi: 10.1002/jcp.1041450116. [DOI] [PubMed] [Google Scholar]
- 29.Ishimi Y, et al. Leukemia inhibitory factor/differentiation-stimulating factor (LIF/D-factor): regulation of its production and possible roles in bone metabolism. Journal of cellular physiology. 1992;152:71–78. doi: 10.1002/jcp.1041520110. [DOI] [PubMed] [Google Scholar]
- 30.Rathjen PD, et al. Developmentally programmed induction of differentiation inhibiting activity and the control of stem cell populations. Genes & development. 1990;4:2308–2318. doi: 10.1101/gad.4.12b.2308. [DOI] [PubMed] [Google Scholar]
- 31.Derigs HG, Boswell HS. LIF mRNA expression is transcriptionally regulated in murine bone marrow stromal cells. Leukemia. 1993;7:630–634. [PubMed] [Google Scholar]
- 32.Wetzler M, et al. Constitutive expression of leukemia inhibitory factor RNA by human bone marrow stromal cells and modulation by IL-1, TNF-alpha, and TGF-beta. Experimental hematology. 1991;19:347–351. [PubMed] [Google Scholar]
- 33.Thomas RJ, et al. Breast cancer cells interact with osteoblasts to support osteoclast formation. Endocrinology. 1999;140:4451–4458. doi: 10.1210/endo.140.10.7037. [DOI] [PubMed] [Google Scholar]
- 34.Barkan D, et al. Inhibition of metastatic outgrowth from single dormant tumor cells by targeting the cytoskeleton. Cancer research. 2008;68:6241–6250. doi: 10.1158/0008-5472.CAN-07-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guise TA, et al. Evidence for a causal role of parathyroid hormone-related protein in the pathogenesis of human breast cancer-mediated osteolysis. The Journal of clinical investigation. 1996;98:1544–1549. doi: 10.1172/JCI118947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson RW, et al. TGF-beta promotion of Gli2-induced expression of parathyroid hormone-related protein, an important osteolytic factor in bone metastasis, is independent of canonical Hedgehog signaling. Cancer research. 2011;71:822–831. doi: 10.1158/0008-5472.CAN-10-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kusuma N, et al. Integrin-dependent response to laminin-511 regulates breast tumor cell invasion and metastasis. International journal of cancer Journal international du cancer. 2012;130:555–566. doi: 10.1002/ijc.26018. [DOI] [PubMed] [Google Scholar]
- 38.Kim RS, et al. Dormancy signatures and metastasis in estrogen receptor positive and negative breast cancer. PloS one. 2012;7:e35569. doi: 10.1371/journal.pone.0035569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bragado P, et al. TGF-beta2 dictates disseminated tumour cell fate in target organs through TGF-beta-RIII and p38alpha/beta signalling. Nature cell biology. 2013;15:1351–1361. doi: 10.1038/ncb2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El Touny LH, et al. Combined SFK/MEK inhibition prevents metastatic outgrowth of dormant tumor cells. The Journal of clinical investigation. 2014;124:156–168. doi: 10.1172/JCI70259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holmgren L, Jackson G, Arbiser J. p53 induces angiogenesis-restricted dormancy in a mouse fibrosarcoma. Oncogene. 1998;17:819–824. doi: 10.1038/sj.onc.1201993. [DOI] [PubMed] [Google Scholar]
- 42.Shachaf CM, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 43.Almog N, et al. Transcriptional changes induced by the tumor dormancy-associated microRNA-190. Transcription. 2013;4:177–191. doi: 10.4161/trns.25558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Almog N, et al. Transcriptional switch of dormant tumors to fast-growing angiogenic phenotype. Cancer research. 2009;69:836–844. doi: 10.1158/0008-5472.CAN-08-2590. [DOI] [PubMed] [Google Scholar]
- 45.Oki T, et al. A novel cell-cycle-indicator, mVenus-p27K-, identifies quiescent cells and visualizes G0–G1 transition. Scientific reports. 2014;4:4012. doi: 10.1038/srep04012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adam AP, et al. Computational identification of a p38SAPK-regulated transcription factor network required for tumor cell quiescence. Cancer research. 2009;69:5664–5672. doi: 10.1158/0008-5472.CAN-08-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pernodet N, et al. High expression of QSOX1 reduces tumorogenesis, and is associated with a better outcome for breast cancer patients. Breast cancer research: BCR. 2012;14:R136. doi: 10.1186/bcr3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barkan D, et al. Metastatic growth from dormant cells induced by a col-I-enriched fibrotic environment. Cancer research. 2010;70:5706–5716. doi: 10.1158/0008-5472.CAN-09-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang N, et al. Mitotic quiescence, but not unique “stemness,” marks the phenotype of bone metastasis-initiating cells in prostate cancer. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2015 doi: 10.1096/fj.14-266379. [DOI] [PubMed] [Google Scholar]
- 50.Manrique I, et al. The inhibitor of differentiation isoform Id1b, generated by alternative splicing, maintains cell quiescence and confers self-renewal and cancer stem cell-like properties. Cancer letters. 2015;356:899–909. doi: 10.1016/j.canlet.2014.10.035. [DOI] [PubMed] [Google Scholar]
- 51.Chaffer CL, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu SC, et al. Leukemia inhibitory factor promotes nasopharyngeal carcinoma progression and radioresistance. The Journal of clinical investigation. 2013;123:5269–5283. doi: 10.1172/JCI63428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kritikou EA, et al. A dual, non-redundant, role for LIF as a regulator of development and STAT3-mediated cell death in mammary gland. Development. 2003;130:3459–3468. doi: 10.1242/dev.00578. [DOI] [PubMed] [Google Scholar]
- 54.Biswas S, et al. Anti-transforming growth factor ss antibody treatment rescues bone loss and prevents breast cancer metastasis to bone. PloS one. 2011;6:e27090. doi: 10.1371/journal.pone.0027090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao H, et al. The BMP inhibitor Coco reactivates breast cancer cells at lung metastatic sites. Cell. 2012;150:764–779. doi: 10.1016/j.cell.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kobayashi A, et al. Bone morphogenetic protein 7 in dormancy and metastasis of prostate cancer stem-like cells in bone. J Exp Med. 2011;208:2641–2655. doi: 10.1084/jem.20110840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sosa MS, et al. NR2F1 controls tumour cell dormancy via SOX9- and RARbeta-driven quiescence programmes. Nature communications. 2015;6:6170. doi: 10.1038/ncomms7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nandy SB, et al. MicroRNA-125a influences breast cancer stem cells by targeting leukemia inhibitory factor receptor which regulates the hippo signaling pathway. Oncotarget. 2015 doi: 10.18632/oncotarget.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tomida M, Yamamoto-Yamaguchi Y, Hozumi M. Characterization of a factor inducing differentiation of mouse myeloid leukemic cells purified from conditioned medium of mouse Ehrlich ascites tumor cells. FEBS letters. 1984;178:291–296. doi: 10.1016/0014-5793(84)80619-5. [DOI] [PubMed] [Google Scholar]
- 60.Li X, et al. LIF promotes tumorigenesis and metastasis of breast cancer through the AKT-mTOR pathway. Oncotarget. 2014;5:788–801. doi: 10.18632/oncotarget.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu L, et al. HIF-2alpha mediates hypoxia-induced LIF expression in human colorectal cancer cells. Oncotarget. 2015;6:4406–4417. doi: 10.18632/oncotarget.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wake MS, Watson CJ. STAT3 the oncogene - still eluding therapy? FEBS J. 2015;282:2600–2611. doi: 10.1111/febs.13285. [DOI] [PubMed] [Google Scholar]
- 63.Derynck R, Jarrett JA, Chen EY, Goeddel DV. The murine transforming growth factor-beta precursor. The Journal of biological chemistry. 1986;261:4377–4379. [PubMed] [Google Scholar]
- 64.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 65.Richard V, Luchin A, Brena RM, Plass C, Rosol TJ. Quantitative evaluation of alternative promoter usage and 3′ splice variants for parathyroid hormone-related protein by real-time reverse transcription-PCR. Clinical chemistry. 2003;49:1398–1402. doi: 10.1373/49.8.1398. [DOI] [PubMed] [Google Scholar]
- 66.Poillet L, et al. QSOX1 inhibits autophagic flux in breast cancer cells. PloS one. 2014;9:e86641. doi: 10.1371/journal.pone.0086641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Razorenova OV, et al. The apoptosis repressor with a CARD domain (ARC) gene is a direct hypoxia-inducible factor 1 target gene and promotes survival and proliferation of VHL-deficient renal cancer cells. Molecular and cellular biology. 2014;34:739–751. doi: 10.1128/MCB.00644-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Previously published microarray data-sets that we re-analyzed here to detect LIFR expression in breast cancer patients with a bone metastasis are available to download as an excel file in the supplemental data provided by Minn et al.14, which includes cross-referenced data for van’t Veer signature, as described above in the in silico section. Previously published human breast invasive carcinoma survival data-sets and RNA sequencing data-sets that we re-analyzed here were reported previously68,69 and are available from the TCGA Research Network (http://cancergenome.nih.gov/.) as described in detail above in the in silico section. Source data for Fig. 1(h–l), 2(a,d), 4(a,b,d,e), 5(d,e), 6(c,d,f,g), 7(a,c,d), and Supplementary Fig. 2b, 3(a–c), 5(b–e), 6b, and 8(c,d,h) have been provided as Supplementary Table 1. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.