Abstract
The generation of human induced pluripotent stem cell (hiPSC)-derived cardiomyocytes has been of utmost interest for the study of cardiac development, cardiac disease modeling, and evaluation of cardiotoxic effects of novel candidate drugs. Several protocols have been developed to guide human stem cells toward the cardiogenic path. Pioneering work used serum to promote cardiogenesis; however, low cardiogenic throughputs, lack of chemical definition, and batch-to-batch variability of serum lots constituted a considerable impediment to the implementation of those protocols to large-scale cell biology. Further work focused on the manipulation of pathways that mouse genetics indicated to be fundamental in cardiac development to promote cardiac differentiation in stem cells. Although extremely elegant, those serum-free protocols involved the use of human recombinant cytokines that tend to be quite costly and which can also be variable between lots. The latest generation of cardiogenic protocols aimed for a more cost-effective and reproducible definition of the conditions driving cardiac differentiation, using small molecules to manipulate cardiogenic pathways overriding the need for cytokines. This chapter details methods based on currently available cardiac differentiation protocols for the generation and characterization of robust numbers of hiPSC-derived cardiomyocytes under chemically defined conditions.
Keywords: Human induced pluripotent stem cells, Cardiomyocyte, Cardiac differentiation, Cardiac assays, Small molecule, Wnt signaling pathway
1 Introduction
Since their establishment, human induced pluripotent stem cells (hiPSCs) have been considered a new gold standard for modeling human genetic-based diseases. Similar to human embryonic stem cells, hiPSCs have the potential to differentiate into virtually any cell type in the human body, offering enticing possibilities to study organ- and system-specific disorders, including diseases affecting the heart (1). Additionally, hiPSC-derived cardiomyocytes constitute a robust platform for the assessment of cardiotoxicity, a major contributor to the failure of potential novel drugs in later stages of clinical trials (2). Several genetic diseases bearing cardiac phenotypes have been modeled with hiPSCs including LEOPARD syndrome (3), long QT syndrome (4–7), Timothy syndrome (8), catecholaminergic polymorphic ventricular tachycardia (9–12), familial dilated (13) and hypertrophic cardiomyopathies (14, 15), arrhythmogenic right ventricular cardiomyopathy (16–18), as well as an overlapping syndrome of a cardiac Na+ channel disease (19). As the field has evolved, several differentiation protocols have been developed for the differentiation of hiPSCs toward the cardiac lineages (1). Currently, the most popular protocols rely on small-molecule-mediated temporal modulation of the Wnt pathway (20, 21). Here we present an adapted version of a small-molecule-based protocol, which has been used successfully across many independent hiPSC control and diseased lines.
2 Materials
| 2.1 Commercially Available Reagents. | See Table 1 |
| 2.2 Preparation of Extracellular Matrices (ECMs) | Add the corresponding volume of diluted ECMs to coat different tissue culture plates according to Table 2. |
| 2.2.1 Growth Factor-Reduced Matrigel | Thaw a 10 ml vial of growth factor-reduced matrigel on ice for 1 hour (h) or overnight at 4 °C. Add 10 ml of cold DMEM/F12 medium and mix quickly and thoroughly. Quickly prepare 600 μl aliquots, to be stored at −80 °C. |
| 2.2.2 Diluted Growth Factor-Reduced Matrigel | Thaw a 600 μl aliquot of growth factor-reduced matrigel on ice for 1 h or overnight at 4 °C. Add 50 ml of cold DMEM/F12 medium to a 50 ml falcon tube. Dilute the matrigel aliquot into DMEM/ F12 and quickly mix thoroughly by inverting the falcon tube several times. Cell culture plates are subsequently incubated with matrigel solution to coat surfaces. Coated plates may be stored at 37 °C for up to 5 days prior to cell seeding. |
| 2.2.3 Laminin | A 1 ml vial of laminin is thawed on ice and 50 μl aliquots are prepared for storage at −80 °C. For laminin coating, a 50 μl laminin aliquot is thawed on ice and diluted into a 5 ml solution of PBS+, mixed thoroughly and incubated on tissue culture dish surfaces overnight at 37 °C. Coated dishes may be stored at 4 °C for up to 7 days prior to cell seeding. |
| 2.3 Cell Culture Medium | hiPSCs are maintained in Essential 8 (E8) medium (22). Prepare stock solutions and make aliquots as indicated in Table 3. See Note 1. |
| 2.3.1 hiPSC Culture Medium | Prepare a 500 ml bottle of E8 medium by adding supplements according to Table 4. See Note 2. |
| 2.3.2 Differentiation Initiation Medium: RPMI Medium with B27 Supplement Without Insulin (RB−) | Take a 500 ml bottle of RPMI and add one 10 ml vial of B27 without insulin. Add one 5 ml aliquot of P/S. Mix well. If the medium will be used within 1–10 days, then the bottle can be kept at 4 °C and 50 ml aliquots can be generated to be warmed when needed. Otherwise, 50 ml aliquots should be prepared for storage at −20 °C. |
| 2.3.3 Differentiation Medium: RPMI Medium with B27 Containing Insulin (RB+) | Take a 500 ml bottle of RPMI and add one 10 ml vial of B27 with insulin. Add one 5 ml aliquot of P/S. Mix well. If the medium will be used within 1–10 days, then the bottle can be kept at 4 °C and 50 ml aliquots can be generated to be warmed when needed. Otherwise, 50 ml aliquots should be prepared for storage at −20 °C. |
| 2.3.4 Maturation Medium: DMEM/M199 Medium with B27 Containing Insulin (D/199B+) | Take a fresh bottle of DMEM and remove 140 ml of the medium. Add 125 ml of M199. Add one 10 ml vial of B27 with insulin. Add one 5 ml aliquot of P/S. Mix well. If the medium will be used within 1–10 days, then the bottle can be kept at 4 °C and 50 ml aliquots can be generated to be warmed when needed. Otherwise, 50 ml aliquots should be prepared for storage at −20 °C. |
| 2.4 Small Molecules | For a 10 mM (2,000×) solution, re-suspend 10 mg vial in 3.122 ml of DMSO. Mix well, vortex, and make 25–50 μl aliquots, to be stored at −20 °C. Aliquots may be reused up to three times. |
| 2.4.1 Rock Inhibitor/ Y27632: Rocki | |
| 2.4.2 CHIR99021: CH | For a 12 mM (2,000×) solution, re-suspend 25 mg vial in 4.152 ml of DMSO. Mix well, vortex, and make 25–50 μl aliquots, to be stored at −20 °C. Aliquots may be reused up to three times. |
| 2.4.3 IWP-2 | For a 5 mM (1,000×) solution, re-suspend 10 mg vial in 4.286 ml of pre-warmed DMSO. Mix well, vortex, and make 25–50 μl aliquots, to be stored at −20 °C. Aliquots may be reused up to three times. |
| 2.5 Dissociation Reagents | Add 500 μl of 0.5 M EDTA to a bottle of PBS−. |
| 2.5.1 hiPSC Dissociation Solution: 0.5 mM EDTA | |
| 2.5.2 Collagenase Type II Solution | Cardiomyocytes are dissociated in a solution containing 200 units/ ml of HBSS−. Estimate the required volume of collagenase II solution, and calculate and weigh the mass necessary to reach the target concentration in units/ml. Dissolve into HBSS− and filter sterilize. The solution can be kept at 4 °C for up to 1 week. |
| 2.5.3 Cardiomyocyte Dissociation Solution | Prepare cardiomyocyte dissociation solution immediately before use. For 1 ml cardiomyocyte dissociation solution, prepare the stock solutions in tissue culture-grade water as described in Table 5. |
| 2.6 Buffers | In order to prepare 50 ml of immunofluorescence permeabilization buffer, dilute 100 μl of Triton-X in 50 ml of PBS+, for a final concentration of 0.2 % Triton-X. |
| 2.6.1 Immunofluorescence Permeabilization Buffer | |
| 2.6.2 Immunofluorescence Dilution Buffer | In order to prepare 50 ml of immunofluorescence buffer, prepare the stock solutions listed in Table 6 and add the appropriate volumes for the final desired concentrations. |
| 2.6.3 Immunofluorescence Blocking Buffer | In order to prepare 10 ml of immunofluorescence blocking buffer, take 9.5 ml of immunofluorescence dilution buffer and add 500 μl of donkey serum for a final concentration of 5 %. |
| 2.6.4 FACS Dilution Buffer | BD Perm/Wash buffer is sold as a 10× concentrate that must be diluted in PBS− before use. For 10 ml of 1× ready-to-use BD Perm/Wash buffer, add 1 ml of 10× concentrate BD Perm/Wash buffer to 9 ml of PBS−. |
| 2.6.5 FACS Blocking Buffer | For 10 ml of FACS blocking buffer, take 9.5 ml of 1× ready-to-use BD Perm/Wash buffer and add 500 μl of donkey serum for a final concentration of 5 %. |
Table 1.
Commercially available reagents
| Description | Manufacturer | Catalog number |
|---|---|---|
| Cell growth matrices | ||
| Growth factor-reduced matrigel | Corning | 354230 |
| Laminin | Life Technologies | 23017-015 |
|
| ||
| Cell culture media | ||
| DMEM/F12 with glutamine and HEPES | Life Technologies | 11330-032 |
| RPMI 1640 with L-glutamine | Life Technologies | 11875093 |
| DMEM with L-glutamine and without sodium pyruvate | Corning | 10-017 |
| M199 with L-glutamine | Corning | 10-060 |
|
| ||
| Medium supplements | ||
| L-Ascorbic acid 2-phosphate | Sigma-Aldrich | A8960 |
| Insulin | Life Technologies | 12585-014 |
| Transferrin | Sigma-Aldrich | T3705 |
| Sodium selenite | Sigma-Aldrich | S5261 |
| B-27 with insulin | Life Technologies | 17504044 |
| B-27 without insulin | Life Technologies | A1895601 |
| Penicillin/streptomycin antibiotics (P/S) | Corning | 30-002 |
|
| ||
| Cytokines | ||
| FGF2 | PeproTech | 100-18B |
| TGFβ | PeproTech | 100-21 |
|
| ||
| Buffers | ||
| PBS without Ca2+ and Mg2+ (PBS−) | Corning | 21-031 |
| PBS with Ca2+ and Mg2+ (PBS+) | Corning | 21-030 |
| HBSS without Ca2+ and Mg2+ (HBSS−) | Corning | 21-022 |
| 7.5 % sodium bicarbonate | Life Technologies | 25080-094 |
| 0.5 M EDTA | Life Technologies | 15575-020 |
| BD Perm/Wash buffer | BD Biosciences | 554723 |
|
| ||
| Cell dissociation reagents | ||
| Accutase | Innovative Cell Technologies | AT 104 |
| Collagenase type II | Worthington | LS004174 |
| Taurine | Sigma-Aldrich | T8691-100G |
| EGTA | Sigma-Aldrich | E-4378-25G |
| 25 % BSA | Life Technologies | A10008-01 |
|
| ||
| Small molecules | ||
| Y27632/Rock inhibitor (Rocki) | Selleckchem | S1049 |
| CHIR99021 (CH) | Selleckchem | S1263 |
| IWP-2 | Cayman Chemical | 13951 |
|
| ||
| Antibodies and staining reagents | ||
| Donkey serum | Jackson ImmunoResearch | 017-000-121 |
| Mouse anti-sarcomeric alpha actinin | Sigma-Aldrich | A7811 |
| Rabbit anti-NKX2.5 | Santa Cruz | sc-14033 |
| DyLight-488-conjugated donkey anti-mouse | Jackson ImmunoResearch | 715-485-150 |
| DyLight-549-conjugated donkey anti-rabbit | Jackson ImmunoResearch | 711-505-152 |
| R-phycoerythrin-conjugated donkey anti-mouse | Jackson ImmunoResearch | 715-116-150 |
| PerCP-conjugated donkey anti-rabbit | Jackson ImmunoResearch | 711-126-152 |
| Hoechst 33342 nuclear DNA stain | Life Technologies | H1399 |
Table 2.
Tissue culture plate formats and coating volumes
| Tissue culture plate | ECM coating volume |
|---|---|
| 6 cm2 plate | 3 ml |
| 3.5 cm2 plate | 1.5 ml |
| 12-well plate | 0.5 ml/well |
| 96-well plate | 50 μl/well |
Table 3.
Stock solutions and aliquot volumes
| Stock solution | Desired concentration | MW | Quantity needed | Resuspension volume | Aliquot volume (μl) |
|---|---|---|---|---|---|
| L-Ascorbic acid 2-phosphate | 64 mg/ml | 289.54 | 5 g | 78.125 ml | 500 |
| Transferrin | 53.5 mg/ml | N/A | 1 g | 18.692 ml | 100 |
| Sodium selenite | 700 μg/ml | 172.94 | 35 mg | 50 ml | 10 |
| FGF2 | 100 μg/ml | N/A | 1 mg | 10 ml | 500 |
| TGFβ1 | 100 μg/ml | N/A | 100 μg | 1 ml | 10 |
Table 4.
Preparation of E8 medium
| Component | Stock solution | Take from stock | Final concentration |
|---|---|---|---|
| DMEM/F12 with glutamine and HEPES | 1× | 500 ml | 1× |
| Sodium bicarbonate | 75 mg/ml | 3.62 ml | 543 μg/ml |
| L-Ascorbic acid 2-phosphate | 64 mg/ml | 500 μl | 64 μg/ml |
| Insulin | 4 mg/ml | 2.5 μl | 20 μg/ml |
| Transferrin | 53.5 mg/ml | 100 μl | 10.7 μg/ml |
| Sodium selenite | 700 μg/ml | 10 μl | 14 ng/ml |
| FGF2 | 100 μg/ml | 500 μl | 100 ng/ml |
| TGFβ | 100 μg/ml | 10 μl | 2 ng/ml |
| Pen/strep | 100× | 5 ml | 1× |
Table 5.
Preparation of cardiomyocyte dissociation solution
| Component | Stock solution | Take from stock (μl) | Final concentration |
|---|---|---|---|
| Collagenase II solution | 200 units/ml | 880 | 200 units/ml |
| Taurine | 200 mM | 100 | 1 mM |
| EGTA | 0.2 mM | 10 | 0.1 mM |
| BSA | 250 mg/ml | 8 | 1 mg/ml |
Table 6.
Preparation of immunofluorescence dilution buffer
| Component | Stock solution | Take from stock | Final concentration |
|---|---|---|---|
| Milli-Q water | N/A | 42.13 ml | N/A |
| Tris base, pH 7.5 | 1 M | 1 ml | 20 mM |
| NaCl | 2 M | 3.87 ml | 155 mM |
| EGTA | 50 mM | 2 ml | 2 mM |
| MgCl2 | 100 mM | 1 ml | 100 mM |
3 Methods
| 3.1 hiPSC Culture | hiPSCs are grown as confluent monolayers on growth factor-reduced matrigel-coated 6 cm2 plates. Passaging is performed at a ratio of 1:4–1:10 every 4–6 days and achieved as outlined below:
|
|
| 3.2 hiPSC Seeding for Cardiac Differentiation | Cardiac differentiation is typically performed in a 12-well plate format, although other formats can be used with the appropriate adjustments to cell growth surfaces. Prior to seeding, coat a 12-well plate with diluted growth factor-reduced matrigel as described in Sections 2.2 and 2.2.1.
|
|
| 3.3 Cardiac Differentiation | The protocol described herein is an adaptation of a previously described small-molecule-based method (20, 23). The key differences are the following:
|
|
| 3.3.1 Day 0: Induction |
|
|
| 3.3.2 Day 2: Wash and Switch to RB− | After CH treatment, mild to considerable cell death is expected. Proceed with the protocol as outlined below:
|
|
| 3.3.3 Day 4: IWP-2 in Conditioned Medium |
|
|
| 3.3.4 Day 6: Medium Change |
See
Note 7.
|
|
| 3.3.5 Day 8: Medium Change |
|
|
| 3.3.6 Day 10: Medium Change (Switch to RB+) |
See
Note 8.
|
|
| 3.3.7 Day 11–30: Differentiation and Maintenance | Change medium every 2–3 days with RB+ until day 30. See Note 9. | |
| 3.3.8 Day 31–45+: Maturation | Switch medium to D/199B+:
|
|
| 3.4 Characterization of hiPSC-Derived Cardiomyocytes | Bona fide hiPSC-derived cardiomyocytes express appreciable levels of the contractile protein sarcomeric α-actinin and the transcription factor NKX2.5. Co-staining of cells with this combination of cardiac markers robustly identifies hiPSC-derived cardiomyocytes. | |
| 3.4.1 Cardiomyocyte Dissociation | Beating clusters or sheets of hiPSC-derived cardiomyocytes can be microdissected, dissociated, and replated for specific downstream applications. See
Note 11.
|
|
| 3.4.2 Flow Cytometry | Flow cytometry allows for robust, quantitative, and rapid estimation of cardiomyocyte throughputs in a given differentiation experiment. The staining protocol outlined below relies on co-staining of undifferentiated hiPSCs (negative control) and hiPSC-derived cardiomyocytes resulting from a differentiation experiment with sarcomeric α-actinin and NKX2.5. See
Note 13.
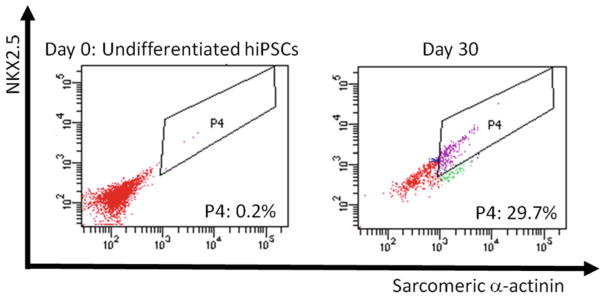
Expected Results Figure 1 exemplifies FACS analysis of NKX2.5 and sarcomeric α-actinin results from hiPSC before and after cardiac differentiation. Cellular debris and cell doublets were eliminated, as well as individual signals for both antibodies were analyzed through gates not shown here. Gate P4 was set after analysis of stained undifferentiated hiPSC and adjustment of the negative control signals to the first three decades of each axis. |
|
| 3.4.3 Immunofluorescence Microscopy | Cardiomyocytes may be dissociated and replated on laminin-coated dishes that are appropriate for cell imaging such as chamber slides, glass coverslips, and black-walled optic plates for immunofluorescence-based microscopy analysis.
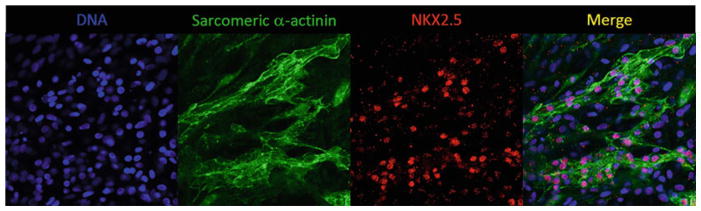
Expected Results Figure 2 exemplifies results obtained from immunofluorescence staining analysis of sarcomeric α-actinin and NKX2.5 in hiPSC following cardiac differentiation. Cells are considered positive for sarcomeric α-actinin when a clear striated pattern can be visualized. Cardiomyocytes are also defined by expression of NKX2.5, which is found confined to the cell nucleus. |
|
Fig. 1.
An example of FACS analysis of sarcomeric α-actinin and NKX2.5 coexpression as an indicator of cardiomyocyte throughput. Gate P4 highlights double-positive cells in undifferentiated cells on day 0 and hiPSC-derived cardiomyocytes on day 30
Fig. 2.
An example of immunofluorescence analysis of sarcomeric α-actinin and NKX2.5 coexpression on day 30 of differentiation as an indicator of cardiomyocyte throughput
Acknowledgments
F.Z. is a current recipient of the American Heart Association Postdoctoral Fellowship. F.S. is supported by grants from the National Institutes of Health, American Heart Association and Saving tiny Heart Society grants.
Footnotes
The solvent for all stock solutions is tissue culture-grade water, except for TGFβ, for which 10 mM citric acid with pH = 3 should be used. When preparing the ascorbic acid stock solution, pre-warm, pre-warm tissue culture-grade water to 37 °C before slowly dissolving with constant agitation and vortexing. Crystals may take several hours to dissolve completely. Store all aliquots of stock solutions at −20 °C.
The cytokines in E8 medium are not stable at 37 °C for long periods of time. Therefore it is preferable to warm the medium to room temperature instead of 37 °C.
Since Accutase is an aggressive enzyme, cell dissociation should be monitored frequently as some cell lines will singularize and even detach at a shorter time. If it is noted that cells start to peel off, remove the enzyme immediately. If all cells become detached, add 3 ml of maintenance medium, pipette up and down to collect all cells, and transfer to a 15 ml Falcon tube. Add 5 μM of Rocki (2.5 μl of a 10 mM aliquot) and spin at 800 rpm for 5 min. Re-suspend cells in 4 ml of maintenance medium, add 5 μM of Rocki, and proceed to next step (4).
Seed 1 × 105–5 × 105 cells per well in matrigel-coated 12-well plates. Cell density should be optimized for each cell line, so confluence is reached within 2–5 days after plating. Ideally, cells should reach confluency within wells at the same approximate time frame that it took to reach confluency in 6 cm plates.
CH aliquots are prepared at 12 mM (2,000×), so for 24.5 ml of RB+, 12.25 μl of a CH aliquot is required. Also note that 6 μM CH for 48 h seems to be optimal for most cell lines; however, specific lines may have different requirements. It is recommended that a range of 6–14 μM is tested at treatment durations of 24 h (12–14 μM) and 48 h (6–8 μM).
IWP-2 aliquots are prepared at 5 mM (1,000×), so if a whole 12-well plate is used, add 25 μl of IWP to the 25 ml of combined medium prepared in step 3.
From this time point onward, cells may display a type of growth pattern that resembles epithelial-to-mesenchymal transition (EMT). As a result, instead of growing flat and spreading out horizontally, they may grow vertically and pile up in the form of foci in the well. This type of growth is indicative of successful mesoderm and precardiac mesoderm formation.
If medium is changed every 2 days, add 2 ml of medium per well. If medium is changed every 3 days, add 3 ml of medium per well.
At this time point, there should be sheets and/or clusters of beating cells that can be readily visualized by light microscopy. Beating is first observed between days 7 and 11 for most of cell lines.
If medium is changed every 2 days, add 2 ml of medium per well. If medium is changed every 3 days, add 3 ml of medium per well.
24 h before dissociation, tissue culture dishes should be coated with 10 μg/ml laminin in PBS+, overnight at 37 °C.
Dissociated cardiomyocytes may take up to 3–5 days to start beating again. Change medium every 2–3 days.
Cardiomyocytes can display considerable autofluorescence within the 488 nm wavelengths; thus, secondary antibodies with emissions within the yellow and far-red regions of the light spectrum are preferable.
References
- 1.Zanella F, Lyon RC, Sheikh F. Modeling heart disease in a dish: from somatic cells to disease-relevant cardiomyocytes. Trends Cardiovasc Med. 2014;24(1):32–44. doi: 10.1016/j.tcm.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma A, Wu JC, Wu SM. Induced pluripotent stem cell-derived cardiomyocytes for cardiovascular disease modeling and drug screening. Stem Cell Res Ther. 2013;4(6):150. doi: 10.1186/scrt380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carvajal-Vergara X, Sevilla A, D’Souza SL, Ang YS, Schaniel C, Lee DF, Yang L, Kaplan AD, Adler ED, Rozov R, Ge Y, Cohen N, Edelmann LJ, Chang B, Waghray A, Su J, Pardo S, Lichtenbelt KD, Tartaglia M, Gelb BD, Lemischka IR. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 2010;465(7299):808–812. doi: 10.1038/nature09005. nature09005 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, Feldman O, Gepstein A, Arbel G, Hammerman H, Boulos M, Gepstein L. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471(7337):225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 5.Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flugel L, Dorn T, Goedel A, Hohnke C, Hofmann F, Seyfarth M, Sinnecker D, Schomig A, Laugwitz KL. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363(15):1397–1409. doi: 10.1056/NEJMoa0908679. NEJMoa0908679 [pii] [DOI] [PubMed] [Google Scholar]
- 6.Lahti AL, Kujala VJ, Chapman H, Koivisto AP, Pekkanen-Mattila M, Kerkela E, Hyttinen J, Kontula K, Swan H, Conklin BR, Yamanaka S, Silvennoinen O, Aalto-Setala K. Model for long QT syndrome type 2 using human iPS cells demonstrates arrhythmogenic characteristics in cell culture. Dis Model Mech. 2012;5(2):220–230. doi: 10.1242/dmm.008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsa E, Rajamohan D, Dick E, Young L, Mellor I, Staniforth A, Denning C. Drug evaluation in cardiomyocytes derived from human induced pluripotent stem cells carrying a long QT syndrome type 2 mutation. Eur Heart J. 2011;32(8):952–962. doi: 10.1093/eurheartj/ehr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yazawa M, Hsueh B, Jia X, Pasca AM, Bernstein JA, Hallmayer J, Dolmetsch RE. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature. 2011;471(7337):230–234. doi: 10.1038/nature09855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itzhaki I, Maizels L, Huber I, Gepstein A, Arbel G, Caspi O, Miller L, Belhassen B, Nof E, Glikson M, Gepstein L. Modeling of catecholaminergic polymorphic ventricular tachycardia with patient-specific human-induced pluripotent stem cells. J Am Coll Cardiol. 2012;60(11):990–1000. doi: 10.1016/j.jacc.2012.02.066. [DOI] [PubMed] [Google Scholar]
- 10.Fatima A, Xu G, Shao K, Papadopoulos S, Lehmann M, Arnaiz-Cot JJ, Rosa AO, Nguemo F, Matzkies M, Dittmann S, Stone SL, Linke M, Zechner U, Beyer V, Hennies HC, Rosenkranz S, Klauke B, Parwani AS, Haverkamp W, Pfitzer G, Farr M, Cleemann L, Morad M, Milting H, Hescheler J, Saric T. In vitro modeling of ryanodine receptor 2 dysfunction using human induced pluripotent stem cells. Cell Physiol Biochem. 2011;28(4):579–592. doi: 10.1159/000335753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung CB, Moretti A, Mederos y Schnitzler M, Iop L, Storch U, Bellin M, Dorn T, Ruppenthal S, Pfeiffer S, Goedel A, Dirschinger RJ, Seyfarth M, Lam JT, Sinnecker D, Gudermann T, Lipp P, Laugwitz KL. Dantrolene rescues arrhythmogenic RYR2 defect in a patient-specific stem cell model of catecholaminergic polymorphic ventricular tachycardia. EMBO Mol Med. 2012;4(3):180–191. doi: 10.1002/emmm.201100194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kujala K, Paavola J, Lahti A, Larsson K, Pekkanen-Mattila M, Viitasalo M, Lahtinen AM, Toivonen L, Kontula K, Swan H, Laine M, Silvennoinen O, Aalto-Setala K. Cell model of catecholaminergic polymorphic ventricular tachycardia reveals early and delayed after depolarizations. PLoS One. 2012;7(9):e44660. doi: 10.1371/journal.pone.0044660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, Navarrete EG, Hu S, Wang L, Lee A, Pavlovic A, Lin S, Chen R, Hajjar RJ, Snyder MP, Dolmetsch RE, Butte MJ, Ashley EA, Longaker MT, Robbins RC, Wu JC. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med. 2012;4(130):130ra147. doi: 10.1126/scitranslmed.3003552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lan F, Lee AS, Liang P, Sanchez-Freire V, Nguyen PK, Wang L, Han L, Yen M, Wang Y, Sun N, Abilez OJ, Hu S, Ebert AD, Navarrete EG, Simmons CS, Wheeler M, Pruitt B, Lewis R, Yamaguchi Y, Ashley EA, Bers DM, Robbins RC, Longaker MT, Wu JC. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12(1):101–113. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han L, Li Y, Tchao J, Kaplan AD, Lin B, Li Y, Mich-Basso J, Lis A, Hassan N, London B, Bett GC, Tobita K, Rasmusson RL, Yang L. Study familial hypertrophic cardiomyopathy using patient-specific induced pluripotent stem cells. Cardiovasc Res. 2014;104:258. doi: 10.1093/cvr/cvu205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma D, Wei H, Lu J, Ho S, Zhang G, Sun X, Oh Y, Tan SH, Ng ML, Shim W, Wong P, Liew R. Generation of patient-specific induced pluripotent stem cell-derived cardiomyocytes as a cellular model of arrhythmogenic right ventricular cardiomyopathy. Eur Heart J. 2012;34:1122. doi: 10.1093/eurheartj/ehs226. [DOI] [PubMed] [Google Scholar]
- 17.Kim C, Wong J, Wen J, Wang S, Wang C, Spiering S, Kan NG, Forcales S, Puri PL, Leone TC, Marine JE, Calkins H, Kelly DP, Judge DP, Chen HS. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature. 2013;494:105. doi: 10.1038/nature11799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caspi O, Huber I, Gepstein A, Arbel G, Maizels L, Boulos M, Gepstein L. Modeling of arrhythmogenic right ventricular cardiomyopathy with human induced pluripotent stem cells. Circ Cardiovasc Genet. 2013;6(6):557–568. doi: 10.1161/CIRCGENETICS.113.000188. [DOI] [PubMed] [Google Scholar]
- 19.Davis RP, Casini S, van den Berg CW, Hoekstra M, Remme CA, Dambrot C, Salvatori D, Oostwaard DW, Wilde AA, Bezzina CR, Verkerk AO, Freund C, Mummery CL. Cardiomyocytes derived from pluripotent stem cells recapitulate electrophysiological characteristics of an overlap syndrome of cardiac sodium channel disease. Circulation. 2012;125(25):3079–3091. doi: 10.1161/CIRCULATIONAHA.111.066092. [DOI] [PubMed] [Google Scholar]
- 20.Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat Protoc. 2013;8(1):162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, Plews JR, Abilez OJ, Cui B, Gold JD, Wu JC. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11(8):855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, Wagner R, Lee GO, Antosiewicz-Bourget J, Teng JM, Thomson JA. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8(5):424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A. 2012;109(27):E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lian X, Zhang J, Zhu K, Kamp TJ, Palecek SP. Insulin inhibits cardiac mesoderm, not mesendoderm, formation during cardiac differentiation of human pluripotent stem cells and modulation of canonical Wnt signaling can rescue this inhibition. Stem Cells. 2013;31(3):447–457. doi: 10.1002/stem.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]