Abstract
The p27 rs34330 (-79C/T) polymorphism has been widely studied for human cancer susceptibility. The current findings, however, still remained controversial. Therefore, we performed the meta-analysis to provide a more accurate result. Eligible studies were identified from PubMed database up to June 2015. The association of p27 rs34330 polymorphism and cancer susceptibility was estimated with odds ratios and corresponding 95% confidence intervals. The meta-analysis was performed with Stata 12. A total of ten studies with 11,214 cases and more than 8,776 controls were included in the meta-analysis (including breast, lung, thyroid, endometrial, and hepatocellular cancer). In pooled analysis, p27 gene rs34330 polymorphism significantly increased the cancer susceptibility. Subgroup analysis indicated that the elevated risk was observed under all the genetic models for Asians and under three genetic models for Caucasians. Results of sensitivity analysis were similar to the overall results. The results suggested that the p27 rs34330 polymorphism increased the cancer susceptibility, especially in Asians. Further well-designed and large sample size studies are warranted to verify the conclusion.
Cancer is a leading cause of death and a major public health problem in both economically developed and developing countries. The occurrence of cancer is elevating because of the growth and aging of global population and environmental factors, especially in less developed countries, in which approximately 82% of the world’s population resides. Based on GLBOCAN estimates, about 14.1 million new cancer cases and 8.2 million deaths occurred in 2012 worldwide1. Many risk factors, such as lifestyle behaviors2,3,4 and genetic factors5,6,7,8,9, have been identified. However, cancer prevention is still a challenging project. Therefore, it is urgent to identify other risk factors for preventing cancers.
The p27/Kip1 (p27) gene (also known as CDKN1B) is located on chromosome 12p13 and encodes cyclin-dependent kinase inhibitors (CDKIs) implicated in the negative regulation of the cell cycle10,11. Cell cycle arrest allows cells to repair DNA damage and replication errors. Therefore, the loss of cell cycle control may contribute to the development of malignancies12,13. Certain polymorphisms including rs2066827 (109T/G) and rs34330 (-79 C/T) of p27 gene have been identified as associated cancer susceptibility. In 2012, a meta-analysis had been performed to estimate the association between p27 gene rs2066827 polymorphism and cancer susceptibility14. The rs34330 polymorphism of p27 gene has also been widely studied for human cancer susceptibility. The existing evidence, however, still remains controversial and has not yet been investigated using meta-analytic methods. Therefore, we aimed in this study to investigate the association of p27 gene rs34330 polymorphism with cancer susceptibility.
Results
Study characteristics
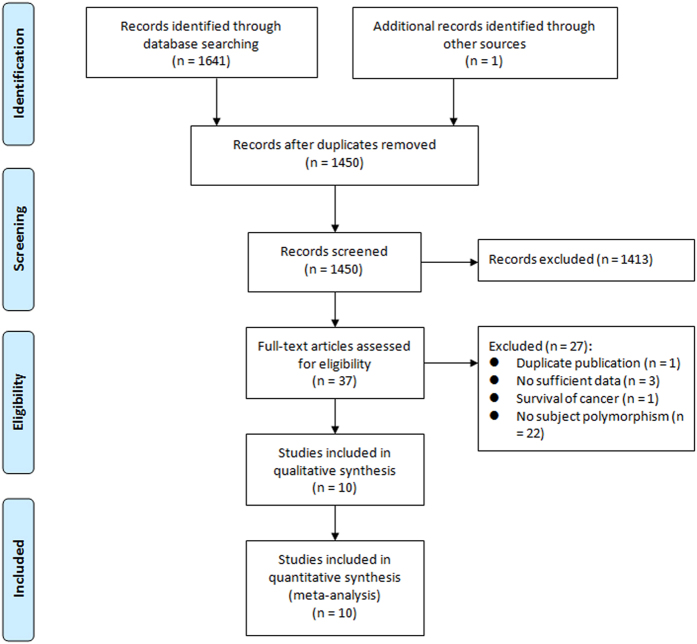
Figure 1 summarizes the detailed process of study selection. Based on the search strategy, 1,641 records were retrieved. In this meta-analysis, ten studies15,16,17,18,19,20,21,22,23,24 involving 11,214 cases and more than 8,776 controls were identified from the electronic databases according to the inclusion criteria. Characteristics of the identified studies are presented in Table 1. These case-controls studies were published between 2006 and 2014. Of them, three studies were conducted in China, two in the US, one in the UK, one in Australia, one in Turkey, one in Spain, and one in Brazil. Six types of malignant diseases were involved, shown as follows: breast cancer, lung cancer, bladder cancer, thyroid cancer, endometrial cancer, and hepatocellular cancer. The sample size in these studies ranged from 143 to 9,030 individuals. The genotype distributions of cases and controls were presented in seven studies15,16,17,18,20,23,24. Other three studies19,21,22 only reported the ORs with 95% CIs in more than two of genetic models, such as homozygous model, heterozygous model, recessive model, or allele model; of these three studies, one study19 reported ORs and 95% CIs in two different populations, which was treated as two distinct reports in the combined analysis. All studies were consistent with HWE in controls except one study that did not provide the genotype distribution of controls or report any information for HWE.
Figure 1. Flow diagram of the study identification and selection process.
Table 1. Characteristics of included studies in the meta-analysis.
| Reference | Country | Ethnicity | Cancer type | Sample size (case/control) | Control source | Genotyping method | HWE in controls |
|---|---|---|---|---|---|---|---|
| Ma15 | China | Asian | Breast cancer | 368/476 | Population based | PIRA-PCR | Yes |
| Wang16 | USA | Caucasian | Lung cancer | 1518/1518 | Hospital based | TaqMan | Yes |
| Driver17 | UK | Caucasian | Breast cancer | 4470/4560 | Population based | TaqMan | Yes |
| Ye18 | USA | Caucasian | Bladder cancer | 591/602 | Hospital based | PCR-RFLP | Yes |
| Spurdle19 | Australia | Caucasian | Breast cancer | 2359/NR | Hospital based | PCR | NA |
| Canbay20 | Turkey | Mixed | Breast cancer | 78/84 | Population based | PCR-RFLP | Yes |
| Landa21 | Spain | Caucasian | Thyroid cancer | 649/385 | Population based | qRT-PCR | Yes |
| Cai22 | China | Asian | Endometrial cancer | 1028/1003 | Population based | PCR | Yes |
| Liu23 | China | Asian | Hepatocellular cancer | 476/526 | Hospital based | MALDI-TOF | Yes |
| Barbieri24 | Brazil | Mixed | Thyroid cancer | 45/98 | Population based | TaqMan | Yes |
HWE = Hardy-Weinberg equilibrium; NA = not available.
Quantitative analysis
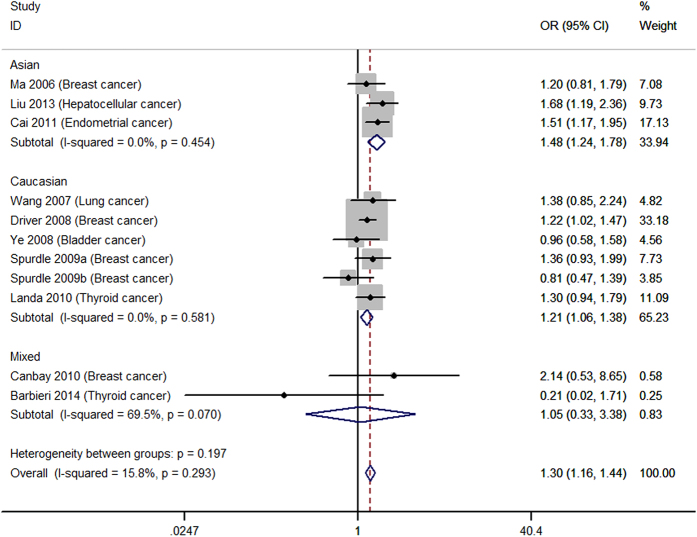
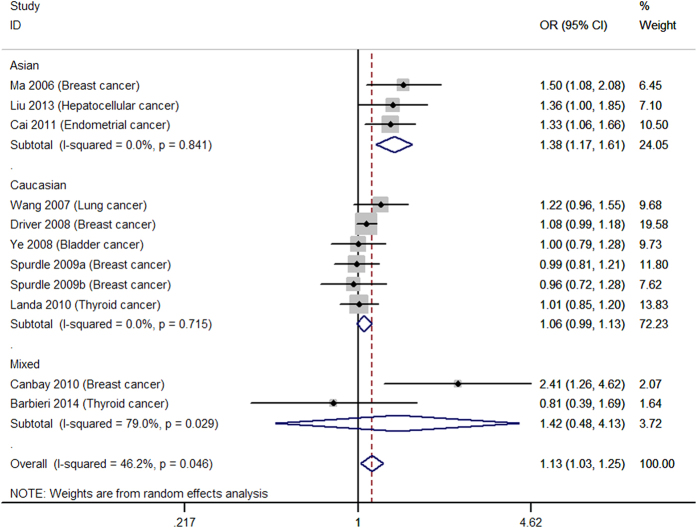
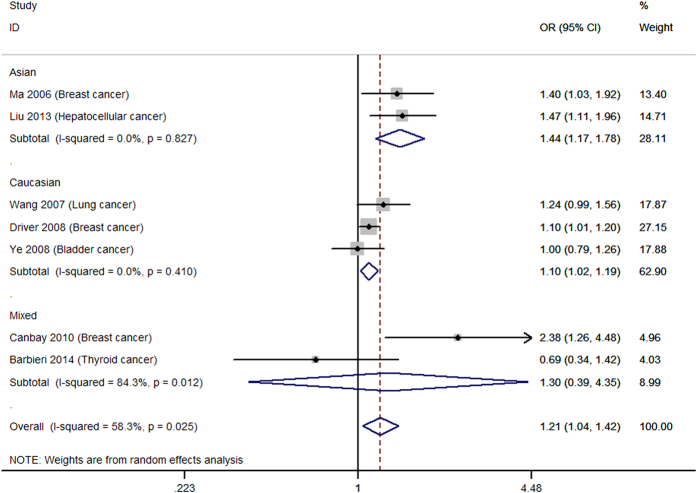
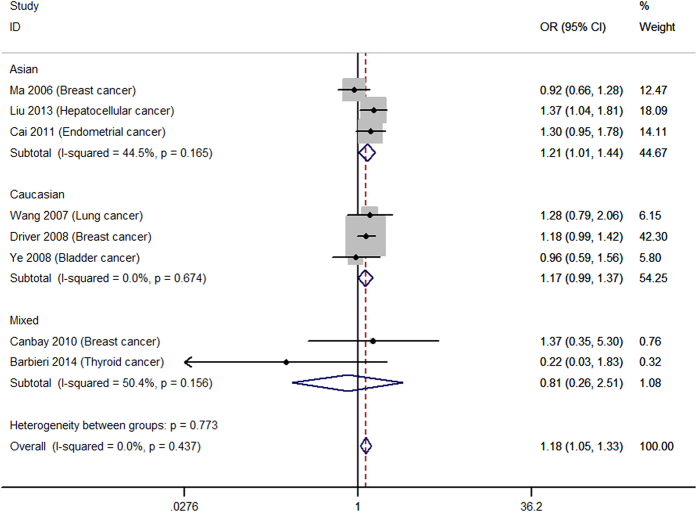
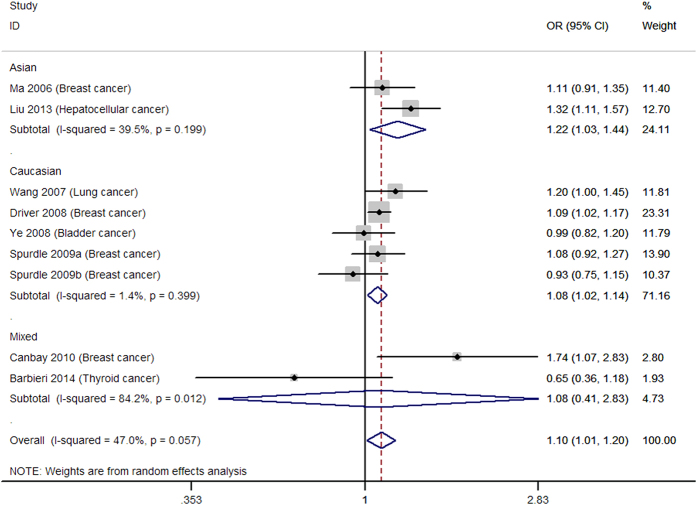
Table 2 shows the main results of summarized ORs and 95% CIs for all genetic models estimated in the present analysis of p27 rs34330 polymorphism and cancer susceptibility. Overall, significantly increased cancer susceptibility was observed in all the tested genetic models: homozygous model (TT vs. CC: OR = 1.30, 95% CI = 1.16–1.44, Fig. 2), heterogeneous model (CT vs. CC: OR = 1.13, 95% CI = 1.03–1.25, Fig. 3), dominant model (TT + CT vs. CC: OR = 1.21, 95% CI = 1.04–1.42, Fig. 4), recessive model (TT vs. CT + CC: OR = 1.18, 95% CI = 1.05–1.33, Fig. 5), allele model (T vs. C: OR = 1.10, 95% CI = 1.01–1.20, Fig. 6). Low to moderate between study heterogeneity was detected (I2 = 15.8%, P = 0.293 for TT vs. CC; I2 = 46.2%, P = 0.046 for CT vs. CC; I2 = 58.3%, P = 0.025 for TT + CT vs. CC; I2 = 7.7%, P = 0.369 for TT vs. CT + CC; I2 = 47.0%, P = 0.057 for T vs. C).
Table 2. Meta-analysis of all studies and subgroups.
| Overall and subgroups | Number of Studies | Heterogeneity | Model | Meta-analysis | ||
|---|---|---|---|---|---|---|
| P | I2 (%) | OR (95%CI) | P for OR | |||
| Homozygous model (TT vs. CC) | ||||||
| Overall | 10 | 0.293 | 15.8 | Fixed | 1.30 (1.16–1.44) | <0.001 |
| HWE (Yes) | 9 | 0.354 | 9.7 | Fixed | 1.32 (1.18–1.47) | <0.001 |
| Sensitivity analysisa | 9 | 0.251 | 20.9 | Fixed | 1.29 (1.16–1.44) | <0.001 |
| Asians | 3 | 0.454 | 0.0 | Fixed | 1.48 (1.24–1.78) | <0.001 |
| Caucasians | 5 | 0.581 | 0.0 | Fixed | 1.21 (1.06–1.38) | 0.004 |
| Mixed | 2 | 0.07 | 69.5 | Fixed | 1.05 (0.33–3.38) | 0.928 |
| Hospital-based | 4 | 0.159 | 39.3 | Fixed | 1.30 (1.07–1.57) | 0.008 |
| Population-based | 6 | 0.381 | 5.6 | Fixed | 1.30 (1.14–1.47) | <0.001 |
| Heterozygous model (CT vs. CC) | ||||||
| Overall | 10 | 0.046 | 46.2 | Random | 1.13 (1.03–1.25) | 0.013 |
| HWE (Yes) | 9 | 0.039 | 50.9 | Random | 1.18 (1.05–1.32) | 0.005 |
| Sensitivity analysisa | 10 | 0.164 | 30.7 | Fixed | 1.10 (1.03–1.16) | 0.003 |
| Asians | 3 | 0.841 | 0.0 | Random | 1.38 (1.17–1.61) | <0.001 |
| Caucasians | 5 | 0.715 | 0.0 | Random | 1.06 (0.99–1.13) | 0.096 |
| Mixed | 2 | 0.029 | 79.0 | Random | 1.42 (0.48–4.13) | 0.525 |
| Hospital-based | 4 | 0.299 | 18.2 | Random | 1.08 (0.95–1.22) | 0.219 |
| Population-based | 6 | 0.020 | 62.7 | Random | 1.20 (1.02–1.41) | 0.031 |
| Dominant model (TT + CT vs. CC) | ||||||
| Overall | 7 | 0.025 | 58.3 | Random | 1.21 (1.04–1.42) | 0.014 |
| HWE (Yes) | 7 | 0.025 | 58.3 | Random | 1.21 (1.04–1.42) | 0.014 |
| Sensitivity analysisa | 6 | 0.103 | 45.3 | Fixed | 1.13 (1.05–1.21) | 0.001 |
| Asians | 2 | 0.827 | 0.0 | Random | 1.44 (1.17–1.78) | 0.001 |
| Caucasians | 3 | 0.410 | 0.0 | Random | 1.10 (1.02–1.19) | 0.012 |
| Mixed | 2 | 0.012 | 84.3 | Random | 1.30 (0.39–4.35) | 0.669 |
| Hospital-based | 3 | 0.104 | 55.8 | Random | 1.21 (0.97–1.50) | 0.085 |
| Population-based | 4 | 0.025 | 68.0 | Random | 1.26 (0.92–1.72) | 0.157 |
| Recessive model (TT vs. CT + CC) | ||||||
| Overall | 8 | 0.437 | 0.0 | Fixed | 1.18 (1.05–1.33) | 0.006 |
| HWE (Yes) | 8 | 0.437 | 0.0 | Fixed | 1.18 (1.05–1.33) | 0.006 |
| Sensitivity analysisa | 7 | 0.333 | 12.7 | Fixed | 1.18 (1.05–1.33) | 0.006 |
| Asians | 3 | 0.165 | 44.5 | Fixed | 1.21 (1.01–1.44) | 0.036 |
| Caucasians | 3 | 0.674 | 0.0 | Fixed | 1.17 (0.99–1.37) | 0.058 |
| Mixed | 2 | 0.156 | 50.4 | Fixed | 0.81 (0.26–2.51) | 0.709 |
| Hospital-based | 3 | 0.453 | 0.0 | Fixed | 1.26 (1.02–1.57) | 0.003 |
| Population-based | 5 | 0.309 | 16.6 | Fixed | 1.15 (1.00–1.32) | 0.057 |
| Allele model (T vs. C) | ||||||
| Overall | 8 | 0.057 | 47.0 | Random | 1.10 (1.01–1.20) | 0.05 |
| HWE (Yes) | 7 | 0.054 | 51.5 | Random | 1.14 (1.02–1.26) | 0.018 |
| Sensitivity analysisa | 7 | 0.115 | 39.6 | Fixed | 1.09 (1.04–1.15) | 0.001 |
| Asians | 2 | 0.199 | 39.5 | Random | 1.22 (1.03–1.44) | 0.023 |
| Caucasians | 4 | 0.399 | 1.4 | Random | 1.08 (1.02–1.14) | 0.010 |
| Mixed | 2 | 0.012 | 84.2 | Random | 1.08 (0.41–2.83) | 0.871 |
| Hospital-based | 4 | 0.073 | 53.3 | Random | 1.10 (0.98–1.24) | 0.115 |
| Population-based | 4 | 0.089 | 53.9 | Random | 1.11 (0.93–1.32) | 0.251 |
aRemoving the study of Canbay 2009.
HWE = Hardy-Weinberg equilibrium; OR = odds ratio; CI = confidence interval.
Figure 2. Forest plot for the homozygous model (TT vs. CC).
a = BRCA1 carriers; b = BRCA2 carriers.
Figure 3. Forest plot for the heterozygous model (CT vs. CC).
a = BRCA1 carriers; b = BRCA2 carriers.
Figure 4. Forest plot for the dominant model (TT + CT vs. CC).
Figure 5. Forest plot for the recessive model (TT vs. CT + CC).
Figure 6. Forest plot for the allele model (T vs. C).
a = BRCA1 carriers; b = BRCA2 carriers
Subgroup analysis showed increased cancer susceptibility under all tested genetic models (TT vs. CC: OR = 1.48, 95% CI = 1.24–1.78, Fig. 2; CT vs. CC: OR = 1.38, 95% CI = 1.17–1.61, Fig. 3; TT + CT vs. CC: OR = 1.44, 95% CI = 1.17–1.78, Fig. 4; TT vs. CT + CC: OR = 1.21, 95% CI = 1.01–1.44, Fig. 5; T vs. C: OR = 1.22, 95% CI = 1.03–1.44, Fig. 6) for Asians and under three genetic models for Caucasians (TT vs. CC: OR = 1.21, 95% CI = 1.06–1.38, Fig. 2; TT + CT vs. CC: OR = 1.10, 95% CI = 1.02–1.19; T vs. C: OR = 1.08, 95% CI = 1.02–1.14) (Table 2). Pooled results of studies with controls in HWE were similar to the overall results. Sensitivity analysis was performed by removing the study of Canbay 2009 and the results were similar to the overall results under all the genetic models (Table 2).
Publication bias
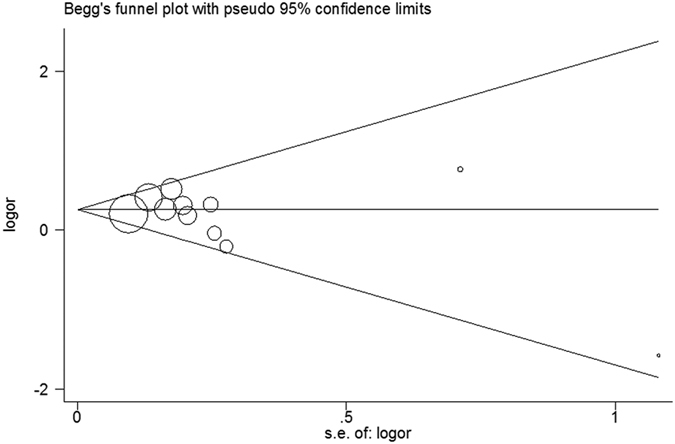
According to funnel plots and Egger’s test, no publication bias was observed in this meta-analysis (homozygous model: P = 0.339 for Egger’s test, Fig. 7).
Figure 7. Begg’s funnel plot based on homozygous model (TT vs. CC).
Discussion
As single studies may have inadequate statistical power to precisely estimate the association between p27 gene rs34330 polymorphism and cancer susceptibility, we performed this meta-analysis, which is a quantitative approach, to precisely estimate the true effects of gene polymorphism on cancer susceptibility25,26. The present meta-analysis included ten case-control studies involving 11 214 cancers and more than 8776 controls and suggested that p27 gene rs34330 polymorphism may increase the susceptibility to cancer, especially in Asian populations.
Cyclins and CDKs play crucial role in the cell cycle during cellular proliferation. These proteins regulate transitions between G1, S, G2 and M phases of the cell cycle, especially the transition from G1 to S27,28. Results from experimental studies suggested that CDKIs could inhibit cellular proliferation through suppressing the kinase activity of the cyclin-CDK complexes and block the transition from G1 to S29. Therefore, CDKIs have been identified as tumor suppressor proteins. The p27, which is a member of the CDKIs, has been postulated as a tumor suppressor gene27. In 2012, a meta-analysis has been performed to investigate the association between rs2066827 polymorphism of p27 gene and cancer susceptibility, and they suggested that the p27 gene rs2066827 polymorphism did not associate with the overall cancer susceptibility in the general population14. No meta-analysis was conducted investigating the association between other polymorphisms of p27 gene and cancer susceptibility.
It has been postulated that the rs34330 polymorphism, located in the 5′-untranslating region of p27 gene, might be correlated with a reduced production of p27 protein16; and certain evidence indicated that rs34330 polymorphism could alter the transcription of p2721. In recent decades, this mutation has been widely investigated concerning human cancer susceptibility and the current evidence is still inconclusive. Therefore, we performed the present meta-analysis, for the first time to the best of our knowledge, to summarize the true association between p27 gene rs34330 polymorphism and cancer susceptibility, and we found an elevated risk of cancer development associated with this polymorphism for overall populations. In this meta-analysis, certain evidence of low to moderate degree of between-study heterogeneity was detected in three genetic models (heterozygous, dominant and allele model) and no evidence of heterogeneity was found in homozygous model and recessive model. The low to moderate between-study heterogeneity indicated acceptable credibility of the results of this meta-analysis. Moreover, the results of sensitivity analysis were robust when we excluded the studies with controls not in HWE. The results of subgroup analysis according to ethnicity showed that the elevated risk associated with p27 gene rs34330 polymorphism were more predominant in Asians than Caucasians. The results indicated that ethnicity might play an important role in cancer susceptibility; however the underlying mechanism is unclear.
Previously, two meta-analyses investigated the association of CDKN1B gene polymorphisms (including rs34330 and rs2066827) and CDKN1B rs2066827 polymorphism with susceptibility to breast cancer, respectively30,31. The previous meta-analyses found that p27 gene rs2066827 polymorphism was not associated with breast cancer susceptibility. One meta-analysis30 comprehensively evaluated the association between polymorphisms of p27 gene and breast cancer susceptibility, and they found that p27 gene rs34330 polymorphism might be associated with breast cancer susceptibility. In this previous meta-analysis30, the included studies with rs34330 polymorphism were four case-control studies, which were also included in our meta-analysis. Moreover, due to the quantity of included studies on different type of cancer, we did not performed subgroup analysis according to type of cancer.
In this study, certain limitations should be taken into consideration. First, confounding factors, such as selection bias and measurement bias, might distort the credibility of the result32. Second, this meta-analysis only included Asian and Caucasian populations. Therefore, the external validity is relatively limited. Third, we could not eliminate the possibility of publication bias even though we detected no evidence of publication bias through Begg’s funnel plot and Egger’s linear regression method. In addition, the sample size and number of included studies is relatively small for gene-susceptibility investigation.
In conclusion, our meta-analysis suggests that the p27 gene rs34330 polymorphism might increase the cancer susceptibility, especially in Asians. Further well-designed and large sample size studies are warranted to verify the conclusion.
Methods
Literature search
A thorough literature search was performed using PubMed database up to June 2015. We used the following search terms to identify all potential relevant studies: (p27 OR p27Kip1 OR CDKN1B OR “cyclin-dependent kinase inhibitor 1B”) AND (cancer OR carcinoma OR adenocarcinoma OR neoplasm OR tumor) AND (gene OR allele OR polymorphism OR variation OR variant OR mutation). In addition, reference lists of retrieved articles were manually searched. No restriction was applied.
Inclusion criteria
All the studies included in the meta-analysis should meet the following inclusion criteria: (1) study design was case-control or cohort; (2) studies examined the association between p27 rs34330 polymorphism and cancer susceptibility; (3) studies presented detailed genotype counts or odds ratios (ORs) with corresponding 95% confidence intervals (CIs).
Data extraction
Two reviewers independently extracted the following information from all identified studies according to a standardized data collection form: author name, publication year, ethnicity, location, number of cases and controls, type of cancer, source of control, Hardy-Weinberg equilibrium (HWE) in controls, genotyping method, genetic data, and ORs with corresponding 95% CIs. Any disagreements were resolved by discussion.
Statistical analysis
The OR and 95% CI were used to measure the association between p27 rs34330 polymorphism and cancer susceptibility. The significance of pooled ORs was determined by Z test at the P < 0.05 level of significance. We estimated the effect of p27 rs34300 polymorphism on cancer susceptibility using homozygous, heterogeneous, dominant, recessive, and allele models. Heterogeneity test was performed with the use of Q statistic at the P < 0.10 level of significance since the chi-squire test has low power in the situation of a meta-analysis when numbers or sample sizes of included studies were small33. We also calculated the I2 metric, a quantitative measurement of heterogeneity among studies33. The summary ORs were pooled using a fixed-effects model when the studies included were homogeneous (P ≥ 0.1) and a random-effects model when statistical heterogeneity was detected (P < 0.1). Prespecified subgroup analyses according to HWE in controls, ethnicity, and source of control were performed to examine the impacts of these factors. Sensitivity analysis was performed by removing study which was an outlier. Potential publication bias was explored by Begg’s funnel plot and Egger linear regression test34. HWE in controls was examined using goodness-of-fit χ2 test. All analyses were performed using Stata 12.0 (Stata, College Station, TX). The two-sided P values less than 0.05 were considered statistically significant, except where extra specified.
Additional Information
How to cite this article: Cheng, X.-K. et al. Genetic association between the cyclin-dependent kinase inhibitor gene p27/Kip1 polymorphism (rs34330) and cancer susceptibility: a meta-analysis. Sci. Rep. 7, 44871; doi: 10.1038/srep44871 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
The authors declare no competing financial interests.
Author Contributions X.K.C. and X.Q.R. designed this study; X.J.W. and X.Q.R. performed the experiments; X.D.L. and X.Q.R. analyzed the data; X.K.C. and G.Z.Y. wrote the manuscript, all the authors reviewed the manuscript.
References
- Torre L. A. et al. Global cancer statistics, 2012. CA Cancer J Clin 65, 87–108 (2015). [DOI] [PubMed] [Google Scholar]
- Zeng X. T. et al. Meta-analysis on the association between toothbrushing and head and neck cancer. Oral Oncol 51, 446–451 (2015). [DOI] [PubMed] [Google Scholar]
- Gawade P. L. et al. Lifestyle, distress, and pregnancy outcomes in the Childhood Cancer Survivor Study cohort. Am J Obstet Gynecol 212, 47 e41–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen K. E. et al. The combined impact of adherence to five lifestyle factors on all-cause, cancer and cardiovascular mortality: a prospective cohort study among Danish men and women. Br J Nutr 113, 849–858 (2015). [DOI] [PubMed] [Google Scholar]
- Gao X., Wang J., Wang W., Wang M. & Zhang J. eNOS Genetic Polymorphisms and Cancer Risk: A Meta-Analysis and a Case-Control Study of Breast Cancer. Medicine (Baltimore) 94, e972 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng P. et al. Genetic Association Between NFKBIA -881A > G Polymorphism and Cancer Susceptibility. Medicine (Baltimore) 94, e1024 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng P. et al. Genetic association between PER3 genetic polymorphisms and cancer susceptibility: a meta-analysis. Medicine (Baltimore) 94, e568 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y. F. et al. Association between ABO gene polymorphism (rs505922) and cancer risk: a meta-analysis. Tumour Biol 36, 5081–5087 (2015). [DOI] [PubMed] [Google Scholar]
- Lu L., Sun Y., Li Y. & Wan P. The polymorphism MMP1 -1607 (1G > 2G) is associated with a significantly increased risk of cancers from a meta-analysis. Tumour Biol 36, 1685–1693 (2015). [DOI] [PubMed] [Google Scholar]
- Polyak K. et al. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes Dev 8, 9–22 (1994). [DOI] [PubMed] [Google Scholar]
- Polyak K. et al. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell 78, 59–66 (1994). [DOI] [PubMed] [Google Scholar]
- Sherr C. J. Cancer cell cycles. Science 274, 1672–1677 (1996). [DOI] [PubMed] [Google Scholar]
- Sherr C. J. The Pezcoller lecture: cancer cell cycles revisited. Cancer Res 60, 3689–3695 (2000). [PubMed] [Google Scholar]
- Wei F. et al. p27(Kip1) V109G polymorphism and cancer risk: a systematic review and meta-analysis. Cancer Biother Radiopharm 27, 665–671 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. et al. Variant genotypes of CDKN1A and CDKN1B are associated with an increased risk of breast cancer in Chinese women. Int J Cancer 119, 2173–2178 (2006). [DOI] [PubMed] [Google Scholar]
- Wang W. et al. Genetic variants in cell cycle control pathway confer susceptibility to lung cancer. Clin Cancer Res 13, 5974–5981 (2007). [DOI] [PubMed] [Google Scholar]
- Driver K. E. et al. Association of single-nucleotide polymorphisms in the cell cycle genes with breast cancer in the British population. Carcinogenesis 29, 333–341 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y. et al. Genetic variants in cell cycle control pathway confer susceptibility to bladder cancer. Cancer 112, 2467–2474 (2008). [DOI] [PubMed] [Google Scholar]
- Spurdle A. B. et al. No evidence that CDKN1B (p27) polymorphisms modify breast cancer risk in BRCA1 and BRCA2 mutation carriers. Breast Cancer Res Treat 115, 307–313 (2009). [DOI] [PubMed] [Google Scholar]
- Canbay E. et al. CCND1 and CDKN1B polymorphisms and risk of breast cancer. Anticancer Res 30, 3093–3098 (2010). [PubMed] [Google Scholar]
- Landa I. et al. Allelic variant at -79 (C > T) in CDKN1B (p27Kip1) confers an increased risk of thyroid cancer and alters mRNA levels. Endocr Relat Cancer 17, 317–328 (2010). [DOI] [PubMed] [Google Scholar]
- Cai H. et al. Association of genetic polymorphisms in cell-cycle control genes and susceptibility to endometrial cancer among Chinese women. Am J Epidemiol 173, 1263–1271 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F. et al. Genetic variants of p21 and p27 and hepatocellular cancer risk in a Chinese Han population: a case-control study. Int J Cancer 132, 2056–2064 (2013). [DOI] [PubMed] [Google Scholar]
- Barbieri R. B. et al. Polymorphisms of cell cycle control genes influence the development of sporadic medullary thyroid carcinoma. Eur J Endocrinol 171, 761–767 (2014). [DOI] [PubMed] [Google Scholar]
- DerSimonian R. & Laird N. Meta-analysis in clinical trials. Control Clin Trials 7, 177–188 (1986). [DOI] [PubMed] [Google Scholar]
- Zeng X. et al. The methodological quality assessment tools for preclinical and clinical studies, systematic review and meta-analysis, and clinical practice guideline: a systematic review. J Evid Based Med 8, 2–10 (2015). [DOI] [PubMed] [Google Scholar]
- Kawamata N. et al. Molecular analysis of the cyclin-dependent kinase inhibitor gene p27/Kip1 in human malignancies. Cancer Res 55, 2266–2269 (1995). [PubMed] [Google Scholar]
- Sherr C. J. Mammalian G1 cyclins. Cell 73, 1059–1065 (1993). [DOI] [PubMed] [Google Scholar]
- Hunter T. Braking the cycle. Cell 75, 839–841 (1993). [DOI] [PubMed] [Google Scholar]
- Xiang H. et al. Association of CDKN1B gene polymorphisms with susceptibility to breast cancer: a meta-analysis. Mol Biol Rep 40, 6371–6377 (2013). [DOI] [PubMed] [Google Scholar]
- Jia Z. M., Liu Y. & Cui S. Y. Lack of association between cyclin-dependent kinase inhibitor 1B rs2066827 polymorphism and breast cancer susceptibility. Tumour Biol 35, 5527–5531 (2014). [DOI] [PubMed] [Google Scholar]
- Niu Y. M. et al. Systematic Review by Multivariate Meta-analyses on the Possible Role of Tumor Necrosis Factor-alpha Gene Polymorphisms in Association with Ischemic Stroke. Neuromolecular Med (2015). [DOI] [PubMed] [Google Scholar]
- Higgins J. P. & Thompson S. G. Quantifying heterogeneity in a meta-analysis. Stat Med 21, 1539–1558 (2002). [DOI] [PubMed] [Google Scholar]
- Egger M., Davey Smith G., Schneider M. & Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]