Abstract
A widely held view of influenza virus infection is that the viral receptor consists of cell surface carbohydrate sialic acid, which can be present as glycoprotein or glycolipid. Here, we examined influenza virus entry and infection in Lec1 cells, a mutant CHO cell line deficient in terminal N-linked glycosylation caused by a mutation in the N-acetylglucosaminyltransferase I (GnT1) gene. We show that influenza virus cannot infect Lec1 cells, despite having full capacity to undergo virus binding and fusion. Lec1 cells also show no virus replication defect, and infection was restored in Lec1 cells expressing wild-type GnT1. Viruses were apparently arrested at the level of internalization from the plasma membrane and were not endocytosed. Lec1 cells were refractory to infection by several strains of influenza virus, including H1 and H3 strains of influenza A, as well as influenza B virus. Finally, cleavage of N-glycans from wild-type CHO cells markedly reduced infection by influenza virus. We suggest that influenza virus specifically requires N-linked glycoprotein for entry into cells, and that sialic acid, although acting as an efficient attachment factor, is not sufficient as an influenza virus receptor in vivo.
Keywords: fusion, receptor, Lec1, internalization
Influenza virus is an enveloped negative-sense RNA virus that is a major public health problem worldwide. In the United States, the virus is responsible for 20,000 deaths annually, with the frequent emergence of new and potentially deadly strains of the virus (1). As with all viruses, influenza virus needs to penetrate target cells to cause infection. For enveloped viruses like influenza, the principal route of entry takes place by a combination of receptor binding and fusion. In the case of influenza virus, these events have generally been well characterized from a biochemical and biophysical perspective (2); however, many of the cell biological aspects of virus entry remain unclear.
Sialic acid was first identified as being responsible for binding of influenza ≈50 years ago (3), and it has since become established that the influenza A and B viruses can bind to sialic acid residues that are present on either glycoprotein or glycolipid (2). The specific conformation of the sialic acid linkage (α, 2–3 vs. α, 2–6) has also been established to control species tropism of the virus (4, 5). The crystal structure of the viral hemagglutinin (HA) shows that it binds sialic acid substrates via surface pockets at the membrane-distal region of HA (2). In vitro assays, examining hemagglutination of red blood cells, have shown that certain gangliosides can act as influenza virus receptors (5).
After initial interaction with its receptor, studies in Madin–Darby canine kidney (MDCK) cells and other tissue culture cells show that the virus enters by receptor-mediated endocytosis. Studies of influenza A virus entry using morphological analysis by electron microscopy (6–8) reveal virions in both coated and noncoated vesicles. The virus can use nonclathrin endocytosis for productive entry and infection (9), and fusion can be observed from smooth-surfaced vesicles as well as coated vesicles (7). Studies from several different groups have shown that the virus fuses out of a low-pH compartment, with fusion occurring at pH 5.0–5.5 in vitro (10, 11). Once fusion has occurred from the endosomal compartment, the uncoated virus is released into the cytoplasm and the genomic ribonucleoproteins enter the nucleus (12, 13).
To dissect the relative role of glycolipids vs. glycoproteins for influenza virus infection in vivo, a previous study used GM95 cells, which have been shown to be deficient in the production of glycolipids, with no effect on glycoproteins. GM95 cells were shown to be efficiently infected with influenza virus, indicating that glycolipids are not essential for influenza infection in vivo (14). Here, we examined entry of influenza viruses into Lec1 cells, which are deficient in terminal N-linked glycosylation. Lec1 cells were isolated by selection of CHO cells with the cytotoxic lectin Phaseolus vulgaris leukoagglutinin (L-PHA) (15), which binds to complex carbohydrate structures, such as tri- and tetraantennary glycopeptides containing outer galactose residues and an α-linked mannose residue substituted at positions C-2 and C-6 (16). Lec1 cells have been characterized as having a defect in the N-acetylglucosaminyltransferase I (GnT1) gene (17, 18). As such, they are unable to synthesize mature N-glycans containing terminal branches ending with galactose and sialic acid due to the absence of the glycosyltransferase GnT1, the outcome of which is that they are deficient in receptor sialo-N-glycans (19), although they are not deficient in glycosphingolipids and should not be deficient in O-linked glycans (P. Stanley, personal communication).
We show that influenza virus undergoes efficient binding, fusion, and replication in Lec1 cells, but cannot initiate infection. The deficiency in virus entry appears to be at the level of virus internalization.
Methods
Cells, Viruses, and Infections. CHO, Lec1, Pro-5, and HeLa cells (American Type Culture Collection) were maintained in α modification of MEM (αMEM) (Cellgro) containing 10% FBS, 100 units/ml penicillin, and 10 μg/ml streptomycin and passaged twice weekly.
Influenza A/WSN/33 (H1N1) was grown in 10-day-old embryonated eggs, or working stocks were produced in MDBK cells (20). Influenza A/Udorn/307/72 (H3N2) was provided by Brian Murphy (National Institutes of Health, Bethesda) and propagated in MDCK cells. Influenza B/Yamagata/78 was provided by Peter Palese (Mount Sinai School of Medicine, New York) and propagated in MDCK cells at 34°C. Vesicular stomatitis virus (VSV), strain Orsay (American Type Culture Collection), was propagated in BHK cells.
Infections were performed essentially as described (21). Briefly, viral stocks were diluted in binding medium containing 0.2% BSA. Unless described otherwise, virus was adsorbed for 60 min at 37°C. Cells were then maintained in growth medium containing 2% serum at 37°C before analysis. Binding experiments were carried out on ice. For experiments using N-glycanase, cell monolayers were treated with 100 units/ml N-glycanase (Glycopeptidase F; PNGase F) (Calbiochem) for 60 min at 37°C before analysis. Bafilomycin A (Calbiochem) was used at a concentration of 25 nM. For production of radioactively labeled virus, 1 mCi/ml 35S-methionine was added to cells from 6 to 24 h after infection, and influenza virus purified on sucrose gradients (1 Ci = 37 GBq). Influenza virus was biotinylated by using 100 μM sulfo-NHS-SS-biotin (Pierce) according to manufacturer's instructions, and biotin was cleaved with 100 mM Tris(2-carboxyethyl)phoshine hydrochloride (TCEP) (Calbiochem). Biotinylated virus was detected by using streptavidin conjugates labeled with Alexa Fluor 488 (Molecular Probes).
Transient transfection of the hamster GnT1 gene (provided by Paul Gleeson, University of Melbourne, Melbourne, VIC, Australia) was carried out by using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions.
Indirect Immunofluorescence Microscopy. Indirect immunofluorescence microscopy was performed as described (22). Influenza A virus nucleoprotein (NP) was detected by using monoclonal antibody H16 L10 4R5 (IgG2a) (American Type Culture Collection). Influenza B virus NP was detected by using monoclonal antibody 23A7 (provided by Wendy Barclay, University of Reading, Reading, U.K.). Secondary antibodies used were Alexa Fluor 488- or 568-labeled goat anti-mouse IgG (Molecular Probes). Human-specific anti-NuMA antibody (IgG2b) was purchased from Oncogene. Cell nuclei were stained with Hoechst dye 33258 (Molecular Probes). Cells were mounted in Mowiol and viewed on a Nikon Eclipse E600 fluorescence microscope using ×20 or ×60 objectives, and images were captured with a SPOT RT camera and spot 3.5 software before transfer into photoshop 7 (Adobe Systems, Mountain View, CA).
Flow Cytometry. For flow cytometry preparation, cells were scraped gently from the dish, washed in PBS, fixed in 3% paraformaldehyde/PBS, permeabilized in 0.1% Triton X-100 and blocked in 10% goat serum/PBS. To detect virus infection or binding, cells were incubated with the monoclonal antibody to influenza NP for 30 min, followed by Alexa Fluor 488-labeled goat anti-mouse IgG for 30 min. Lectin-binding assays used L-PHA, Sambucus nigra lectin (SNA), or Maackia amurensis lectin (Mal II) lectins (Vector Laboratories), either conjugated to FITC or biotin. Biotinylated lectins were localized with streptavidin conjugates labeled with Alexa Fluor 488 (Molecular Probes). VSV was detected by using monoclonal antibody P5D4 (provided by Ari Helenius, Eidgenössische Technische Hochschule, Zürich) and Alexa Fluor 488-labeled goat anti-mouse IgG (Molecular Probes). Cells were analyzed on a FACSCalibur cytometer by using cellquest 3.1F software (Becton Dickinson Immunocytometry Systems). Data analysis was performed with flow jo 4.6 software (Treestar, Ashland, OR). At least 104 cells were analyzed for each sample.
Virus–Cell Fusion Assay. Fusion assays were based on fluorescence dequenching of octadecyl rhodamine (R18)-labeled virus (23). Fifteen microliters of labeled virus [5 plaque-forming units (pfu) per cell] was bound to 2 × 106 cells at 4°C for 1 h in binding buffer (RPMI medium 1640 containing with 0.2% BSA, pH 6.8). Unbound virus was removed by washing with binding buffer, and cells were resuspended in 5 mM Hepes/5 mM Mes/5 mM succinate/150 mM NaCl (HMSS) buffer, pH 7.0/15 μM monensin at 37°C. Fusion of virus on the cell membrane was triggered by adding a predetermined amount of 250 mM HCl to obtain a final pH of 5.0. Fluorescence dequenching was measured by using a QM-6SE spectrofluorimeter (PTI, South Brunswick, NJ), with excitation and emission wavelengths set to 560 and 590 nm, respectively. Fusion efficiency was determined after addition of Triton X-100 (final concentration, 1%) to obtain 100% dequenching.
Results
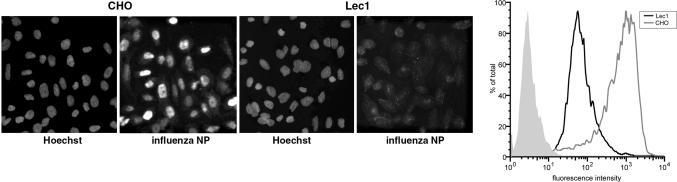
Influenza Virus Infection Is Severely Inhibited in Lec1 Cells. To determine whether Lec1 cells were infectable by influenza virus, we exposed Lec1 and CHO cells to influenza virus and carried out a single-hit infection assay (multiplicity of infection, 1; 5 pfu per cell) at an early time point of infection, based either on immunofluorescence microscopy or flow cytometry using an anti-NP antibody. When both techniques were used, CHO cells showed high levels of infection (≈95% infected), whereas Lec1 cells showed a dramatic decrease in virus infection (<1% infection); see Fig. 1. When we examined cells at 5 h after infection, only background cytoplasmic signal was present in Lec1 cells instead of the strong nuclear signal seen in CHO cells. A similar lack of infection was also observed in Lec1 cells at longer times of infection (e.g., 12 h).
Fig. 1.
Lec1 cells are not infected by influenza virus. (Left) CHO and Lec1 cells were infected with influenza virus (WSN) for 5 h and analyzed by immunofluorescence microscopy using anti-NP antibodies, with nuclei counterstained with Hoechst dye 33258. (Right) CHO (gray line) and Lec1 (black line) cells were also analyzed by flow cytometry. Mock-infected Lec1 cells are shown in the filled light gray trace.
To confirm that we had an authentic Lec1 phenotype, we examined L-PHA binding in Lec1 and CHO cells. Whereas wild-type CHO cells showed high levels of L-PHA binding as assayed by flow cytometry, Lec1 cells did not bind the lectin (see Fig. 6, which is published as supporting information on the PNAS web site). We also examined differences between Lec1 and Pro-5 (24), the parent cell of Lec1 that show no discernable differences with wild-type CHO. Essentially the same virus infectivity and lectin binding data were obtained when we compared the standard CHO cell line with the Pro-5 derivative (data not shown), and so we used CHO and Lec1 cells in all subsequent experiments. Based on the highly restricted infection we observed in Lec1 cells, these data suggest that N-linked glycoprotein(s) are specifically required, in addition to terminal sialic acid, for influenza virus infection.
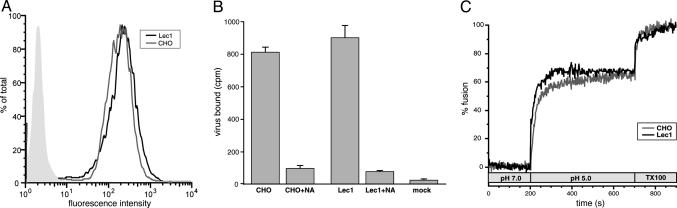
Influenza Virus Binds Efficiently to Lec1 Cells. The inability of influenza virus to infect Lec1 cells led us to consider the possibility that the sialic acid moiety required for influenza attachment to CHO cells depended on N-linked glycosylation. To investigate this, we performed virus-binding assays using Lec1 and CHO cells. Cells were exposed to a multiplicity of infection (MOI) (50 pfu per cell) of influenza virus on ice to allow virus binding, but not internalization. Virus binding was analyzed by flow cytometry after 90-min incubation on ice. Both Lec1 and CHO cells (Fig. 2A) showed comparable high levels of virus binding, indicating that the initial attachment and binding of influenza virus is not affected in Lec1 cells. We also carried out binding experiments with a lower MOI virus infection (5–10 pfu per cell) by using radioactively labeled virus (Fig. 2B). In this case, binding was essentially unchanged between Lec1 and CHO cells. Binding was also sensitive to neuraminidase treatment, indicating that the established sialic acid component of virus binding is still present on Lec1 cells
Fig. 2.
Influenza virus shows no defect in binding or fusion with Lec1 cells. (A) Influenza virus (WSN) was bound to CHO (gray line) and Lec1 (black line) cells at 4°C for 90 min and analyzed by flow cytometry using anti-NP antibodies. Mock treated Lec1 cells are shown in the filled light gray trace. (B) Radioactive influenza virus (WSN) was bound to CHO and Lec1 cells at 4°C for 90 min, washed extensively, and analyzed by scintillation counting. Cells were also pretreated with 5 units/ml neuraminidase (NA) as a control. (C) R18-labeled influenza virus (WSN) was bound to CHO (gray line) and Lec1 (black line) cells at 4°C for 90 min. The temperature was raised to 37°C, and fusion was induced by dropping the pH to 5.0. Fusion was calibrated by induction of complete dequenching with Triton X-100.
To confirm that the restriction of influenza virus infection of Lec1 cells was not caused by a decrease in total cell-surface sialic acid, we performed a lectin-binding assay. We used two lectins specific for sialic acid residues: Sambucus nigra lectin (SNA; specific for α2–6 linkages) and Maackia amurensis lectin (Mal II or MAA; specific for α2–3 linkages) (25). Lectin binding was assayed by flow cytometry using saturating amounts of lectin, which showed that Lec1 cells have overall sialic acid levels equal to or greater than those of CHO cells (see Fig. 7, which is published as supporting information on the PNAS web site). These data are consistent with the presence of high levels of overall sialic acid present on Lec1 cells, and indicate that Lec1 cells are not defective in virus binding. Overall, these data indicate a specific requirement for N-linked sialoglycoprotein for influenza virus infection of host cells at a postbinding step.
Influenza Virus Fuses Efficiently with Lec1 Cells. Another possibility for the inability of influenza virus to infect Lec1 cells is that the virus envelope is unable to fuse with the target cells. To examine this, we performed fluorescence dequenching assays of R18-labeled influenza A/WSN/33 virus with both CHO and Lec1 cells. Fig. 2C shows the time course of a typical fusion assay using CHO and Lec1 cells. At neutral pH, we saw no significant dequenching, and addition of acid resulted in rapid dequenching up to a level that equated to ≈60% fusion in both cases. These data indicate that terminal N-linked glycosylation on host cells is not necessary for influenza virus–cell fusion, and that infection of Lec1 cells is restricted at another point in virus entry.
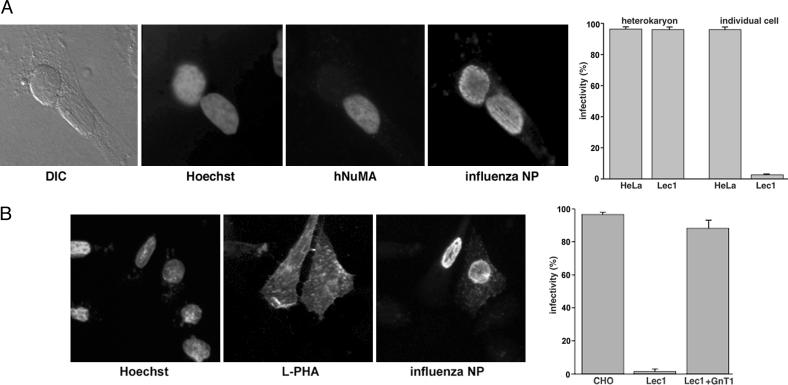
Influenza Virus Infection of Lec1 Cells Is Rescued in Heterokaryons of Heterologous Cells. We used a heterokaryon approach, as an additional way to investigate the molecular basis of the infection restriction in Lec1 cells. When Lec1 cells are fused to heterologous cells that show no influenza virus restriction (and can act as a source of “wild-type” membranes), all of the nuclei in the heterokaryon should show normal levels of infection. We used HeLa cells as heterologous cells for these experiments, due to the availability of human-specific antibody markers. Heterokaryons were produced between Lec1 cells and HeLa cells by polyethylene glycol-mediated fusion (26), and were subsequently infected with influenza virus. As expected, the HeLa cell nuclei show normal levels of influenza virus infection. In heterokaryons, Lec1 cell nuclei also showed high levels of virus replication (Fig. 3A). These data show that the restriction to influenza virus infection of Lec1 cells can be overcome by provision of a heterologous source of membranes. Our data also show that the nuclei of Lec1 cells show no inherent inability to support virus replication, adding further support to our hypothesis that Lec1 cells are defective in virus entry.
Fig. 3.
Influenza virus infection of Lec1 cells is rescued in heterokaryons or by expression of GnT1. (A) Heterokaryons were formed by PEG-mediated fusion between Lec1 and HeLa cells and infected with influenza virus (WSN) for 5 h before immunofluorescence microscopy using anti-NP antibodies. Nuclei were counterstained with Hoechst dye 33258, and HeLa cell nuclei were identified by using a human-specific NuMA antibody. Differential interference contrast (DIC) images of the heterokaryons are also shown. For quantitation, we scored the number of Lec1 and HeLa nuclei in heterokaryons that were infected, in comparison to those that we present as individual cells. The numbers of infected Lec1 and HeLa cell nuclei present in heterokaryons or present as individual cells are shown. (B) Lec1 cells were transfected with plasmids encoding the GnT1 gene. Expression was confirmed by the ability of cells to bind FITC-labeled L-PHA lectin. Infection was monitored by immunofluorescence microscopy using anti-NP antibodies and nuclei counterstained with Hoechst dye 33258.
Influenza Virus Infection of Lec1 Cells Is Rescued by Expression of GnT1. To confirm that the inability of influenza virus to infect Lec1 cells was solely due to the presence of terminal N-linked carbohydrate, we transiently expressed the wild-type GnT1 gene (17) in Lec1 cells and examined virus infection. GnT1 expression was confirmed by the ability of transfected cells to bind L-PHA. Influenza virus infection was fully restored in transfected Lec1 cells, in comparison to nontransfected cells (Fig. 3B). These data confirm that terminal N-linked carbohydrate is necessary for successful influenza virus infection.
Influenza Infection of Lec1 Cells Is Blocked During Virus Internalization. Because our data showed that Lec1 cell nuclei could replicate influenza virus efficiently, and that virus binding and fusion were not compromised, we focused on the very early events in virus entry. To examine the precise point in infection where infection might be blocked, we first examined infection in an assay where virus entry was arrested with bafilomycin A and then cells pulsed with low-pH buffer to induce infection (27). Under these conditions, we were unable to recover Lec1 infection (Fig. 8, which is published as supporting information on the PNAS web site), suggesting that influenza internalization and/or delivery into nascent endocytic vesicles is prevented in Lec1 cells.
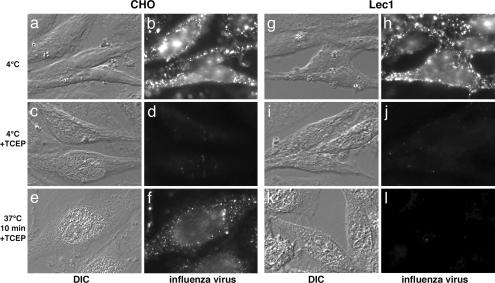
To examine internalization more directly, we infected cells with virus that had been biotinylated with a thiol-linked agent (28). Virus bound to the surface of CHO cells on ice could be detected with streptavidin (Fig. 4b), and this virus was essentially undetectable once the biotin groups had been cleaved with the cell-impermeable reducing agent TCEP (Fig. 4d). When internalization was induced at 37°C, the virus was protected from the action of TCEP and could be visualized in endosomes in CHO cells (Fig. 4f). However, in Lec1 cells, no internalized virus could be detected with this assay (Fig. 4l). These data show that influenza virus has an internalization block in Lec1 cells.
Fig. 4.
Lec1 cells are blocked for infection during influenza virus internalization. Biotinylated influenza virus was bound to the surface of CHO and Lec1 cells at 4°C (b and h) for 90 min, and surface biotin was cleaved with TCEP (d and j). Cells were then allowed to internalize virus for 10 min before TCEP treatment (f and l). Cells were analyzed by differential interference contrast (DIC) microscopy or by immunofluorescence microscopy using Alexa Fluor 488–streptavidin.
The Inability to Infect Lec1 Cells Is a Common Feature of Influenza Viruses. The WSN strain of virus used for our studies is a highly laboratory-adapted strain. To determine whether the requirement for N-linked sialoglycoprotein was a general feature of influenza viruses, we performed quantitation of immunofluorescence microscopy with several different strains of human influenza virus. We examined infection with A/WSN/33 (H1N1), A/Udorn/307/72 (H3N2), and B/Yamagata/78 viruses. In particular, we chose A/Udorn/307/72, because it is generally considered to be much less laboratory adapted than WSN. In all cases, the influenza viruses used failed to efficiently infect Lec1 cells, but gave high levels of infection in CHO cells (Fig. 9, which is published as supporting information on the PNAS web site). Therefore, we conclude that the restriction to entry and infection of Lec1 cells is not specific to the WSN strain of virus and appears to be a common feature of human influenza viruses.
Lec1 Cells Are Fully Infectable with VSV. To exclude the possibility that Lec1 cells have generalized defects in endocytosis and may therefore be refractory to infection with any enveloped virus, we tested the ability of Lec1 cells to become infected with VSV, a virus that also enters cells by endocytosis, followed by a pH-dependent fusion step (29). Flow cytometry showed that CHO and Lec1 cells had essentially the same of infectivity with VSV (Fig. 10, which is published as supporting information on the PNAS web site). These data confirm that the restriction for influenza infection of Lec1 cells is selective for this virus and does not occur for another virus that enters by pH-dependent endocytosis.
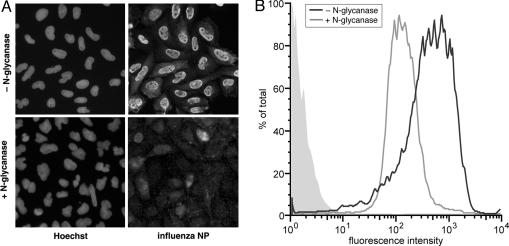
Treatment of Cells with N-Glycanase Prevents Influenza Virus Infection. To further examine a functional requirement for N-linked glycoprotein for influenza infection, we treated CHO cells with N-glycanase, an endoglycosidase that efficiently removes all forms of N-glycans from glycoproteins (30). CHO cells were treated with N-glycanase or were untreated, and then infected with influenza virus. Immunofluorescence microscopy and flow cytometry showed that infection was abrogated in cells treated with N-glycanase, with residual signal remaining presumably because of input viruses (Fig. 5). As a control, we tested the effects of N-glycanase pretreatment on infection on infection with VSV (Fig. 11, which is published as supporting information on the PNAS web site). In this case, there was no effect of enzyme treatment, showing a specific requirement for N-linked glycoproteins for influenza infection.
Fig. 5.
Treatment of cells with N-glycanase abrogates influenza virus infection. (A) CHO cells were treated with N-glycanase or untreated and were infected with influenza virus (WSN) for 5 h, followed by analysis by immunofluorescence microscopy using anti-NP antibodies, with nuclei counterstained with Hoechst dye 33258. (B) N-glycanase-treated (black line) and untreated (gray line) cells were also analyzed by flow cytometry. Mock-infected Lec1 cells are shown in the filled light gray trace.
Discussion
Here, we show that influenza virus infection is restricted at the level of virus entry in Lec1 cells, which lack complex N-linked glycosylation. Over the past few years, our laboratory has focused on the involvement of endocytic trafficking pathways for entry of influenza and other enveloped viruses. We have shown that influenza virus, unlike other enveloped viruses with less stringent pH requirements for fusion, requires entry into late endosomes for infection (31). More recently, we have shown a requirement for functional ubiquitylation (presumably of a receptor) for influenza virus sorting into late endosomes/multivesicular bodies (32). Our data on influenza virus endocytic trafficking clearly point to the use of a specific routing into the late endosome, which is presumably receptor-mediated. However, in the case of influenza virus, the only identified receptor is highly nonspecific in that the virus seems to bind sialic acid attached to either glycolipid or glycoprotein.
For most viruses, receptor interactions can be conveniently broken down into defined areas. First, many viruses may interact with cells initially by long-range, possibly nonspecific, electrostatic interactions, based on attraction of the negatively charged cell surface with virus particles. This is then followed by more specific, and higher affinity, interactions with coreceptors (33). Influenza virus is probably the best-known example of a virus that utilizes sialic acid as an initial receptor. Several other types of virus have also been shown to use sialic acid for interaction with the host cells (33); however, in many of these cases, sialic acid is sufficient for virus binding but not for infection. The data presented here confirm the large body of previous work showing that sialic acid mediates initial binding of influenza virus; however, our studies indicate that sialic acid is not sufficient for infection. A more specific secondary receptor appears to be necessary in vivo. Our results therefore corroborate those of Stray et al. (34), who showed influenza infection of desialated cells, albeit at relatively low efficiency.
The vast majority of assays to analyze sialic acid binding for influenza virus have relied on systems that are nonpermissive for virus infection (e.g., erythrocytes, liposomes, isolated molecules, etc.). Therefore, it remains possible that the receptors used by the virus to infect host cells are different to those mediating agglutination and binding to red blood cells. In particular, the Y98F mutation in the sialic acid binding pocket of HA (although clearly abrogating binding to red cells) does not affect virus infectivity (35). In addition, the recent data showing influenza virus infection of desialylated MDCK cells (34) confirm earlier reports suggesting that neuraminidase-treated CEM cells can still bind influenza virus, possibly via nonsialic acid receptors (36). In this case, binding was considered to occur at sites that were unfavorable for fusion. Kinetic modeling of virus binding and endocytosis with MDCK cells has also revealed two kinds of binding sites for influenza virus (37), with the possible presence of low-affinity binding sites that correspond to nonsialic acid-mediated virus–cell interactions.
It is possible that influenza virus binding and internalization proceeds by a series of distinct steps. In this scenario, initial low-affinity interactions between sialic acid and HA would allow tight interaction of the virus with the cell surface, but for productive infection, a secondary receptor(s) is then required. Our studies with Lec1 cells, which express high levels of sialic acid, show that they can bind normal levels of influenza virus. However, a postattachment block in infection is present in the absence of terminal carbohydrate on host cell N-linked glycoprotein. Virus entry in Lec1 cells is apparently blocked at the level of internalization and would be before actin-dependent stage I movement described by Zhuang and colleagues (38, 39) with influenza virus infection of live cells.
There are two basic possibilities for our observed block in virus entry in Lec1 cells: either there is a specific carbohydrate conformation that is universally (or at least commonly) present on N-linked glycoprotein and necessary for virus entry, or Lec1 cells show distinct differences in the expression profiles of their N-linked glycoproteins, with a distinct moiety being absent from their plasma (and/or endosomal) membranes. Overall, our data show that mutant Lec1 cells that have deficient terminal N-glycan processing are restricted for influenza entry at the level of virus internalization. Based on this finding, we suggest that influenza virus requires N-linked glycoprotein for entry into cells, and that sialic acid, although acting as an efficient attachment factor on cells, is not sufficient as an influenza virus receptor. In contrast to other viruses, such as HIV-1 and herpesviruses, where coreceptor requirements are required to trigger fusion (40, 41), influenza appears to require additional postattachment factors for successful endocytosis into host cells. Influenza may therefore show common features with reoviruses, where coreceptors are well established to mediate virus entry after initial sialic acid binding (42), or with adenoviruses, where integrin coreceptors are required for postbinding virus internalization (43). However, in the case of influenza virus, any such factor or coreceptor remains to be identified.
Supplementary Material
Acknowledgments
We thank Ruth Collins for helpful discussions throughout the course of this work, Xiangjie Sun for development of fusion and internalization assays, and Beverley Bauman and Damon Ferguson for excellent technical assistance. We also thank Peter Palese, Brian Murphy, Ari Helenius, Paul Gleeson, and Wendy Barclay for their kind contributions of reagents. This work was supported by a Career Investigator Grant from the American Lung Association and by National Institutes of Health Grant R01AI48678 (to G.R.W.).
Author contributions: G.R.W. designed research; V.C.C. performed research; G.R.W. analyzed data; and G.R.W. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HA, hemagglutinin; MDCK, Madin–Darby canine kidney; L-PHA, Phaseolus vulgaris leukoagglutinin; GnT1, N-acetylglucosaminyltransferase I; VSV, vesicular stomatitis virus; TCEP, Tris(2-carboxyethyl)phoshine hydrochloride; NP, nucleoprotein.
References
- 1.Cox, N. J. & Subbarao, K. (2000) Annu. Rev. Med. 51, 407–421. [DOI] [PubMed] [Google Scholar]
- 2.Skehel, J. J. & Wiley, D. C. (2000) Annu. Rev. Biochem. 69, 531–569. [DOI] [PubMed] [Google Scholar]
- 3.Gottschalk, A. (1959) in The Viruses: Biochemical Biological and Biophysical Properties, eds. Burnet, F. M. & Stanley, W. M. (Academic, New York), Vol. 3, pp. 51–61. [Google Scholar]
- 4.Steinhauer, D. A. & Wharton, S. A. (1998) in Textbook of Influenza, eds. Nicholson, K. G., Webster, R. G. & Hay, A. J. (Blackwell Science, Oxford), pp. 54–64.
- 5.Suzuki, Y., Ito, T., Suzuki, T., Holland, R. E., Jr., Chambers, T. M., Kiso, M., Ishida, H. & Kawaoka, Y. (2000) J. Virol. 74, 11825–11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patterson, S., Oxford, J. S. & Dourmashkin, R. R. (1979) J. Gen. Virol. 43, 223–229. [DOI] [PubMed] [Google Scholar]
- 7.Matlin, K. S., Reggio, H., Helenius, A. & Simons, K. (1981) J. Cell Biol. 91, 601–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshimura, A., Kuroda, K., Kawasaki, K., Yamashina, S., Maeda, T. & Ohnishi, S.-I. (1982) J. Virol. 43, 284–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sieczkarski, S. B. & Whittaker, G. R. (2002) J. Virol. 76, 10455–10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stegmann, T., Morselt, H. W. M., Scholma, J. & Wilschut, J. (1987) Biochem. Biophys. Acta 904, 165–170. [DOI] [PubMed] [Google Scholar]
- 11.Yoshimura, A. & Ohnishi, S. (1984) J. Virol. 51, 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin, K. & Helenius, A. (1991) J. Virol. 65, 232–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bui, M., Whittaker, G. & Helenius, A. (1996) J. Virol. 70, 8391–8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ichikawa, S., Nakajo, N., Sakiyama, H. & Hirabayashi, Y. (1994) Proc. Natl. Acad. Sci. USA 91, 2703–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stanley, P., Narasimhan, S., Siminovitch, L. & Schachter, H. (1975) Proc. Natl. Acad. Sci. USA 72, 3323–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cummings, R. D. & Kornfeld, S. (1982) J. Biol. Chem. 257, 11230–11234. [PubMed] [Google Scholar]
- 17.Puthalakath, H., Burke, J. & Gleeson, P. A. (1996) J. Biol. Chem. 271, 27818–27822. [DOI] [PubMed] [Google Scholar]
- 18.Sarkar, M., Hull, E., Nishikawa, Y., Simpson, R. J., Moritz, R. L., Dunn, R. & Schachter, H. (1991) Proc. Natl. Acad. Sci. USA 88, 234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robertson, M. A., Etchison, J. R., Robertson, J. S., Summers, D. F. & Stanley, P. (1978) Cell 13, 515–526. [DOI] [PubMed] [Google Scholar]
- 20.Mahy, B. W. J. (1985) Virology: A Practical Approach (IRL Press, Oxford).
- 21.Root, C. R., Wills, E. G., McNair, L. L. & Whittaker, G. R. (2000) J. Gen. Virol. 81, 2697–2705. [DOI] [PubMed] [Google Scholar]
- 22.Whittaker, G., Kemler, I. & Helenius, A. (1995) J. Virol. 69, 439–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert, J. M., Mason, D. & White, J. M. (1990) J. Virol. 64, 5106–5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanley, P., Caillibot, V. & Siminovitch, L. (1975) Cell 6, 121–128. [DOI] [PubMed] [Google Scholar]
- 25.Baum, L. G. & Paulson, J. C. (1990) Acta Histochem. Suppl. 40, 35–38. [PubMed] [Google Scholar]
- 26.Whittaker, G., Bui, M. & Helenius, A. (1996) J. Virol. 70, 2743–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mothes, W., Boerger, A. L., Narayan, S., Cunningham, J. M. & Young, J. A. T. (2000) Cell 103, 679–689. [DOI] [PubMed] [Google Scholar]
- 28.Pelkmans, L., Puntener, D. & Helenius, A. (2002) Science 296, 535–539. [DOI] [PubMed] [Google Scholar]
- 29.Matlin, K. S., Reggio, H., Helenius, A. & Simons, K. (1982) J. Mol. Biol. 156, 609–631. [DOI] [PubMed] [Google Scholar]
- 30.Fukuda, M. & Kobata, A. (1993) Glycobiology: A Practical Approach (IRL Press, Oxford).
- 31.Sieczkarski, S. B. & Whittaker, G. R. (2003) Traffic 4, 333–343. [DOI] [PubMed] [Google Scholar]
- 32.Khor, R., McElroy, L. & Whittaker, G. R. (2003) Traffic 4, 857–868. [DOI] [PubMed] [Google Scholar]
- 33.Dimitrov, D. S. (2004) Nat. Rev. Microbiol. 2, 109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stray, S., Cummings, R. D. & Air, G. M. (2000) Glycobiology 10, 649–658. [DOI] [PubMed] [Google Scholar]
- 35.Martin, J., Wharton, S. A., Lin, Y. P., Takemoto, D. K., Skehel, J. J., Wiley, D. C. & Steinhauer, D. A. (1998) Virology 241, 101–111. [DOI] [PubMed] [Google Scholar]
- 36.de Lima, M. C., Ramalho-Santos, J., Flasher, D., Slepushkin, V. A., Nir, S. & Duzgunes, N. (1995) Biochim. Biophys. Acta 1236, 323–330. [DOI] [PubMed] [Google Scholar]
- 37.Nunes-Correia, I., Ramalho-Santos, J., Nir, S. & Pedroso de Lima, M. C. (1999) Biochemistry 38, 1095–1101. [DOI] [PubMed] [Google Scholar]
- 38.Lakadamyali, M., Rust, M. J., Babcock, H. P. & Zhuang, X. (2003) Proc. Natl. Acad. Sci. USA 100, 9280–9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rust, M. J., Lakadamyali, M., Zhang, F. & Zhuang, X. (2004) Nat. Struct. Mol. Biol. 11, 567–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spear, P. G. (2004) Cell Microbiol. 6, 401–410. [DOI] [PubMed] [Google Scholar]
- 41.Berger, E. A., Murphy, P. M. & Farber, J. M. (1999) Ann. Rev. Immunol. 17, 657–700. [DOI] [PubMed] [Google Scholar]
- 42.Forrest, J. C. & Dermody, T. S. (2003) J. Virol. 77, 9109–9115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nemerow, G. R. (2000) Virology 274, 1–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.