Abstract
Background
After fertilization, embryo development involves differentiation, as well as development of the fetal body and extra-embryonic tissues until the moment of implantation. During this period various cellular and molecular changes take place with a genetic origin, e.g. the elongation of embryonic tissues, cell-cell contact between the mother and the embryo and placentation. To identify genetic profiles and search for new candidate molecules involved during this period, embryonic gene expression was analyzed with a custom designed utero-placental complementary DNA (cDNA) microarray.
Methods
Bovine embryos on days 7, 14 and 21, extra-embryonic membranes on day 28 and fetuses on days 28 were collected to represent early embryo, elongating embryo, pre-implantation embryo, post-implantation extra-embryonic membrane and fetus, respectively. Gene expression at these different time points was analyzed using our cDNA microarray. Two clustering algorithms such as k-means and hierarchical clustering methods identified the expression patterns of differentially expressed genes across pre-implantation period. Novel candidate genes were confirmed by real-time RT-PCR.
Results
In total, 1,773 individual genes were analyzed by complete k-means clustering. Comparison of day 7 and day 14 revealed most genes increased during this period, and a small number of genes exhibiting altered expression decreased as gestation progressed. Clustering analysis demonstrated that trophoblast-cell-specific molecules such as placental lactogens (PLs), prolactin-related proteins (PRPs), interferon-tau, and adhesion molecules apparently all play pivotal roles in the preparation needed for implantation, since their expression was remarkably enhanced during the pre-implantation period. The hierarchical clustering analysis and RT-PCR data revealed new functional roles for certain known genes (dickkopf-1, NPM, etc) as well as novel candidate genes (AW464053, AW465434, AW462349, AW485575) related to already established trophoblast-specific genes such as PLs and PRPs.
Conclusions
A large number of genes in extra-embryonic membrane increased up to implantation and these profiles provide information fundamental to an understanding of extra-embryonic membrane differentiation and development. Genes in significant expression suggest novel molecules in trophoblast differentiation.
Background
Embryogenesis in the period from fertilization to implantation involves various morphological, cellular, and biochemical changes related to genomic activity [1]. These changes include the elongation of embryonic tissues, cell-cell contact between the mother and the embryo, and placentation. The embryo begins to form the placenta around day 20 of gestation in the bovine [2,3], while embryonic trophoblast and endometrial cells tightly unite to form placentomes on day 30 [4,5]. The bovine embryos at the blastocyst stage are 100 to 200 μm in diameter, but become 20 to 30 cm long by the time of implantation. Embryonic cells undergo both proliferation and differentiation to form the fetus and placenta throughout early embryogenesis. Reprogramming of the genome may be completed and reset during these steps, with embryonic development progressing on to temporal and spatial gene expression [6,7]. The literature data indicate that a number of specific genes are expressed during the period from blastogenesis through implantation, such as interferon-τ (IFN-τ) [8,9], matrix metalloproteinases (MMPs) [10], heparanase [11], and retinoid X receptors [12]. Vast numbers of genes change their expression levels during this period to support the complex mechanisms of embryogenesis and implantation. Although numerous molecules participate in trophoblast differentiation and placentation, the precise molecular and genetic pathways which lead to the formation of placenta remain difficult to clarify. Nevertheless, recent data from animal cloning technology where a somatic or stem cell nucleus is transplanted to an enucleated unfertilized egg, strongly suggested that inappropriate expression of genes is possibly the main cause of early embryo loss [13,14]. These studies have shown that aberrant gene expression patterns were associated with placental abnormalities in several species. Thus, it is clear that the differentiation of trophoblastic cells and the formation of placenta are tightly controlled by mechanisms which precisely govern switch on and off of the appropriate set of genes during embryogenesis. Therefore, a detailed gene expression profile in the pre-implantation embryo provides insights into the molecular mechanisms that are vital to farther our knowledge of the embryogenesis and implantation processes. Unlike traditional molecular methods that have been used to look at a single gene at one time point, a complementary DNA (cDNA) microarray is an efficient tool for analyzing tens of thousands of genes in a single experiment and to identify groups of genes that are critical during early development.
We have developed a bovine utero-placental specific cDNA microarray that contains various genes known to play key roles in extra-embryonic membrane development in the bovine [15]. To our knowledge, there has been no report to date that has evaluated the differential gene expression profile in the embryo as well as in extra-embryonic cells of the pre-implantation embryo in the bovine. Thus, we applied our cDNA microarray to detect gene expressions comprehensively during embryogenesis and implantation in the bovine. Furthermore we attempted to address the expression changes of the key genes that are involved in trophoblastic cell proliferation and differentiation. We first compared the gene expression profiles among blastocyst-stage embryos, elongated-stage embryos, and the trophoblastic tissue at the peri-implantation stage. We further characterized gene expression profiles between the trophoblastic tissue and embryo just after implantation. Comparison of expression differences of specific gene(s) provides important information on the potential role of the particular gene at specific time points of trophoblast development. Approaches such as this help identifying specific genes that are up- or down-regulated during trophoblast development.
Although there is no defined standard method that has been developed for the analysis of microarray data, application of k-means [16,17] and hierarchical clustering [18] methods has been shown to yield reliable data. These two methods have been successfully used to obtain information from the embryo, cytotrophoblast, and uterine tissue during the early stages of pregnancy in humans [19-22] and mice [6,23,24]. We therefore applied k-means and hierarchical clustering methods to identify genes involved in trophoblastic differentiation.
Materials and Methods
Animals and sample collection
Totally five Japanese black cows were slaughtered on Days 19, 21, 27 (two) and 28 of gestation (the day of artificial insemination was designated as Day 0), and collected embryonic membrane and fetus. Embryos were collected non-surgically on Days 7 and 14 of gestation from superovulated Japanese black cows. Namely, a follicle-stimulating hormone (FSH; Antrin-R10, Kawasaki Pharmaceutical, Kawasaki, Japan) was injected twice daily at doses of 5 mg, 3 mg, and 2 mg on the first, second, and third day of treatment, respectively. Luteolysis was induced by injecting prostaglandin F2α (cloprostenol 750 μg/cow; Fujita Pharmaceutical, Tokyo, Japan) on three days after the initial injection of FSH. Artificial insemination was performed both on the day of estrus (Day 0 of gestation) and the next morning, using frozen semen from Japanese black bulls. Twelve blastocysts collected on Day 7 were pooled to represent the Day 7 embryo (abbreviated as Day 7E), two expanded and elongated embryos collected on Day 14 were pooled to represent Day 14E, two embryos collected on Days 19 and 21 were used individually but designated as Day 21E (n = 2). Extra-embryonic membranes collected on Days 27 (two samples) and 28 (one sample) were used as Day 28 extra-embryonic membranes (28EEM) (n = 3), and two fetuses, excluding the extra-embryonic membranes, collected on Days 27 and 28 were used as the Day 28 fetus (28F) (n = 2). All samples were snap frozen in liquid nitrogen immediately after collection and stored at -80°C until RNA extraction. The days of collection were selected depending on the macro morphological changes of the embryos during the peri-implantation periods; bovine embryo passes into the uterus around a morula stage and develop to a blastocyst stage by Day 7. Embryo begins to elongate on Day 14 and form a long embryo of 20 to 30 cm in length on Day 20 just before implantation occurs. The embryo is easily identifiable around Day 30 around which placentome formation is visible [3,5]. All procedures for these animal experiments were carried out in accordance with the guidelines and ethics approved by the Animal Ethics Committee of the National Institute of Agrobiological Sciences for the use of animals.
Sample RNA preparation
RNA extraction
The total RNA was isolated from the embryos using ISOGEN (NipponGene, Toyama, Japan) according to the manufacturer's instructions. Briefly, Samples were mixed and homogenized with ISOGEN. The aqueous phase was collected after the centrifugation. This phase was washed by chloroform for the RNA purification. The aqueous phase was collected after the centrifugation again. The total RNA pellet in the aqueous phase was obtained using the isopropanol sedimentation. Total RNA was directly used for the T7-based linear amplification for the cDNA microarray analysis. Messenger RNA (mRNA) were reverse transcribed by using the T7-oligo dT primer in the T7-based linear amplification system.
T7-based linear amplification
Linear-amplified antisense RNA (aRNA) was used for the cDNA microarray experiment. Because the amount of total RNA extracted was small from Day 7 blastocysts for the microarray analysis, mRNA from Day 7 blastocysts was amplified along with all other mRNA samples that were collected from Days 14 to 28 of gestation to eliminate technical variations and to efficiently compare the array data.
We used a MessageAmp aRNA kit (Ambion, Austin, TX, USA) for the T7-based linear amplification of mRNA. Synthesis and purification of cDNA and aRNA were carried out according to the manufacturer's instructions. The aRNA amplification procedure consisted of (i) first-strand cDNA synthesis (reverse transcription of mRNA), (ii) second-strand cDNA synthesis, (iii) cDNA purification, (iv) in vitro transcription, and (v) aRNA purification. The aRNA amplification for Day 7 blastocysts was done with two steps purification. The comparison between first and second round amplification was r = 0.96 (data not shown).
cDNA microarray
Bovine utero-placental cDNA microarray
For the series of present experiments we used a utero-placental custom cDNA microarray developed in our laboratory, as described previously [13,15]. Briefly, a cDNA library was constructed from mRNA isolated from endometrial (caruncular and intercaruncular endometrium) and placental tissues (cotyledonary and intercotyledonary fetal membrane) of Japanese Black cows on days 0 and 10 of the estrous cycle, and days 30, 60, 100 and 245 of gestation, respectively. PCR products of about 4,000 clones from the cDNA library which was normalized by hit-picking method [15] were spotted onto glass slides robotically. Simultaneously, the clones were sequenced by using the MegaBACE 1000 DNA Sequencing System (Amersham Pharmacia Biotech, Piscataway, NJ). The array contained 3,955 spots that were clustered into 1,738 unique genes on the basis of sequence analysis. The sequenced clones were compiled and annotated by basic local alignment search tool (BLASTn), and the clone that was positioned at the top of the hierarchical tree of genes by GenBank database matching was selected. The 1,738 annotated unique genes represented 816 known genes, 530 expressed sequencing tags (ESTs) and 392 unknown novel genes. The unknown sequences were submitted to DDBJ as ESTs. The DDBJ and GenBank accession numbers of genes on this custom microarray are BP106801 to BP113049 and AB098745 to AB099150, respectively. An additional 35 genes that were not included in the cDNA library but also spotted onto the cDNA microarray were used for analysis since these genes have been shown to be characteristically expressed during gestation in bovines and humans [9,25-29].
cDNA microarray hybridization
We have previously reported our microarray hybridization procedures [15]. Briefly, two μg of amplified aRNA was reverse transcribed in the presence of cyanine 3 (Cy3) or Cy5 fluorescence dye (Amersham Biosciences, Buckinghamshire, UK) using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA) to make the hybridization probes. The reaction mixture was incubated for 2 hr at 42°C. The labeled probes were concentrated in a Microcon filter device (Millipore, Bendford, MA, USA), diluted in 15 μl hybridization solution (3.4 × SSC, 0.3% SDS, 20 μg poly (A), and 20 μg yeast tRNA), and applied to the microarray. Identical samples were labeled separately with either Cy3- or Cy5-dye. Thus, two hybridization reactions could be carried out with the same sample. The arrays were sequentially washed with 2 × SSC/0.5% SDS, 0.2 × SSC/0.5% SDS, and 0.2 × SSC solutions after 16 hr incubation at 65°C. The arrays were dried by centrifugation at 1,000 × g. Hybridization images were immediately scanned by a GenePix 4000B laser scanner (Axon Instrument, Union City, CA, USA) and analyzed with GenePix Pro 4.0 computer software. The data were viewed as a scatter plot between Cy3 and Cy5 intensities. Each color intensity value was individually normalized, and the average intensity value of each spot (gene) obtained from different hybridization reactions of the same sample was used for data analysis.
Data normalization of the cDNA microarray
Data normalization was performed by following the procedures described previously [30,31]. The local background intensity of each array spot was smoothed by local weight regression (Lowess) and subtracted from the spot intensity data. The subtracted intensity data were subjected to non-parametric regression and local variance normalization since non-parametric regression can reduce intensity-dependent biases. The accuracy is improved over that of linear regression if the points in the scatter plot of Cy3 versus (vs.) Cy5 are not distributed around a straight line. Minimum Information About a Microarray Experiment (MIAME; http://www.mged.org/Workgroups/MIAME/miame.html) compliance was met by depositing all the data in the Gene Expression Omnibus (GEO) repository http://www.ncbi.nlm.nih.gov/geo. The GEO accession numbers are as follows. Platform: GPL1221; Samples: GSM23324, GSM26510, GSM26511, GSM23327, GSM23328, GSM23329, GSM23330, GSM23331, and GSM23332; Series: GSE1414.
Cluster analysis of the cDNA microarray data
Data for individual genes were obtained by averaging the intensity values of analogous spots on the microarray. Data were log2 transformed and used for cluster analysis. The TIGR (The Institute for Genome Research) MultiExperiment Viewer (MeV) program http://www.tigr.org/software/tm4/ was used to derive the k-means and for hierarchical tree clustering analysis [32]. The general expression patterns of the 1,773 individual genes (including 35 manually spotted genes) were investigated by k-means algorithm. Data for each gene were represented by an eight-dimensional vector. K-means clustering was performed by division into 12 centroid centers. The distance between gene vectors was calculated by the cosine coefficient (vector angle). Hierarchical tree clustering was performed on the annotated and EST genes that demonstrated up-regulation when the comparison was made between Day 28EEM vs. Day 28F. Genes that displayed two-fold (and greater) differences were selected and the normalized expression of each sample was utilized for the cluster analysis. The distance between gene vectors was also calculated by the cosine coefficient. We adopted average linkage clustering in the hierarchical tree clustering method. Data used for the cluster analyses were as follows: Day 21E (n = 2; r = 0.84; P < 0.05); Day 28EEM (n = 3; r ≥ 0.90; P < 0.05); and Day 28F (n = 2; r = 0.90; P < 0.05). Other samples were used independently for cluster analysis.
Real-time RT-PCR analysis
Gene expression profiles derived from microarray analyses were confirmed quantitatively by real-time RT-PCR analysis. The selected genes for RT-PCR included two key known genes (placental lactogen-Ala (PL-Ala) and prolactin-related protein-1 (PRP-I)) and four apparently up-regulated ESTs, and compared their expression from Day 7 through Day 28. The ESTs selected have the GenBank accession numbers AW464053, AW465434, AW462349, and AW485575.
The details of real-time RT-PCR procedures have been described in previous reports [11,33]. Briefly, fifty ng total RNA was reverse transcribed into cDNA for 30 min at 48°C by MultiScribe™ reverse transcriptase with an Oligo dT primer, dNTP mixture, MgCl2 and an RNase inhibitor. Primer pairs and oligonucleotide probes labeled with a reporter fluorescence dye at the 5' end and a quencher fluorescence dye at the 3' end were designed using Primer Express computer software (Applied Biosystems, Foster City, CA, USA). The primers and probes for the selected genes are listed in Table 1. The thermal cycling conditions included an initial incubation of samples at 50°C for two minutes and at 95°C for ten minutes, followed by forty cycles with each cycle at 95°C for fifteen seconds and at 60°C for one minute. The cycle threshold value (CT) is related to the initial quantity of the target gene in each sample determined in real time using an ABI Prism 7700 sequence detector (Applied Biosystems). The relative difference in the initial amount of each mRNA species (or cDNA) was determined by comparing the CT values between the samples at different stages. Bovine GAPDH was used as endogenous control. The standard curve for each gene was generated by serial dilution of plasmid containing PL-Ala, PRP-I, four ESTs, or GAPDH cDNA to quantify mRNA concentrations. A ratio of PL-Ala, PRP-I and four ESTs mRNA to GAPDH mRNA was calculated to adjust for any variation in the RT-PCR reaction. All values are presented as mean ± SD. Data were analyzed initially by ANOVA and followed by either Tukey-Kramer multiple comparison test. P-values of <0.05 were considered significant.
Table 1.
Oligonucleotide primers and TaqMan probes used for real-time RT-PCR analysis
| Gene | Primer or TaqMan probe | Sequence | Position |
| PL-Ala (J02840) | Forward | 5' GCAACATTGGTGGCTAGCAA 3' | 262-281 |
| Reverse | 5' GCCCTCGCCAAACTGTTTATTA 3' | 339-317 | |
| Probe | 5' CTATAGGCTCGCCAGGGAAATGTTCAATGA 3' | 285-314 | |
| PRP-I (J02944) | Forward | 5' CAGACAGGTTTATGAATGCCGC 3' | 458-479 |
| Reverse | 5' CGCAGGCAGTAGAACAGGTTAT 3' | 541-520 | |
| Probe | 5' TCCTCTGCATCATCTAGTCACGGAGCTG 3' | 483-511 | |
| AW464053 | Forward | 5' AATATGCCCAGGGCAAACTG 3' | 296-315 |
| Reverse | 5' TCGGGAGTTTGGAGGGAATT 3' | 368-349 | |
| Probe | 5' TCAATGCCATCAAGAGCTGCCACAC 3' | 323-347 | |
| AW465434 | Forward | 5' ACATCTCCCTGAAAGTGAACCC 3' | 287-308 |
| Reverse | 5' TCCATCCTTGCAGAAGTCTCCT 3' | 369-348 | |
| Probe | 5' CCCTGGAAGCTCATCTGCAATGTAAAGC 3' | 316-343 | |
| AW462349 | Forward | 5' GCGTGGATGGTGTCCTACTTCTA 3' | 216-238 |
| Reverse | 5' GCCACAACGAGAAACAGGAAA 3' | 301-281 | |
| Probe | 5' TGTCTGTTTGCCTTTACTGGTGAGCCCT 3' | 240-267 | |
| AW485575 | Forward | 5' CCTCTGATGAAAGATTGGGAACAG 3' | 195-218 |
| Reverse | 5' AAGTGCCAGAGATCTTGGCCT 3' | 285-265 | |
| Probe | 5' TTCTCCAAACCAACCACCACCAGCTG 3' | 230-255 | |
| GAPDH (U85042) | Forward | 5' AAGGCCATCACCATCTTCCA 3' | 178-197 |
| Reverse | 5' CCACTACATACTCAGCACCAGCAT 3' | 253-230 | |
| Probe | 5' AGCGAGATCCTGCCAACATCAAGTGG 3' | 200-225 |
Results
T7-based linear amplification
We confirmed the reliability of this amplification method by comparing unamplified mRNA vs. amplified mRNA (aRNA) derived from the same bovine placental tissue after hybridization into the utero-placental cDNA microarray. We observed a significant correlation (r ≥ 0.81, P < 0.05; data not shown) between the unamplified mRNA and amplified aRNA. Confirmation experiments were conducted twice with reverse labeling (for a total of four times). Previous reports have also described substantial cDNA microarray population correlation between unamplified mRNA and amplified aRNA [34,35]. The correlation of our amplification was equivalent to previous studies [34,35]. Thus, amplified aRNAs can be used as representative materials in cDNA microarray experiments.
Differential gene expression profiles determined by cluster analysis
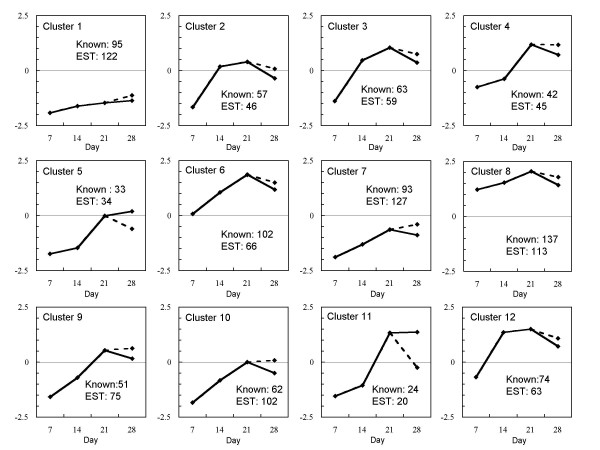
Sixty-six genes out of a total of 1,773 were exempted from cluster analysis due to low expression intensity values. Consequently, 1,707 genes, including 833 previously annotated, were classified into 12 categories by k-means clustering, as illustrated in Fig. 1. Twelve k-means cluster profiles were summarized into three types, in which the gene-expression intensity (i) increased only from Day 7 to Day 14 and thereafter either remained constant or decreased slightly (clusters 2, 3, 12); (ii) Continued to increase until Day 21 (clusters 1, 6, 7, 8, 9, and 10); and (iii) progressively increased until Day 21 (clusters 4, 5, and 11). Clusters 1 and 7 contained more than 200 genes that included annotated and ESTs, whereas clusters 4, 5, and 11 contained fewer than 100 genes. Clusters 5 and 11 contained mostly members of the cytokine family, and clusters 4 and 5 contained ECM-related genes. In particular, trophoblast-specific genes, such as placental lactogens (PLs), prolactin-related proteins (PRPs), and pregnancy-associated glycoproteins (PAGs), were found in cluster 11.
Figure 1.
K-means clusters of the expression patterns of 1707 genes in a bovine embryo. Lines refer to the k-means of gene expression on Days 7E to 21E, Day 28EEM and Day 28F. The solid line refers to the k-means center of gene expression on Days 7E to 21E and Day 28EEM. The dotted line refers to the k-means center of Day 21E to Day 28F.
Temporal specific gene expression in extra-embryonic membranes
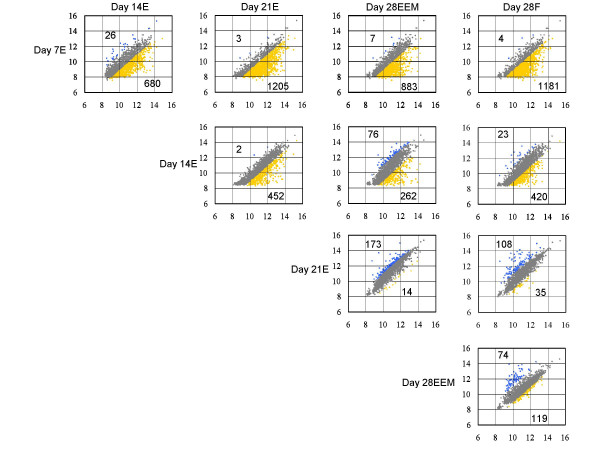
Pair-wise comparisons were made to identify genes displaying differential expression; the results are given in Fig. 2. All data considered significant exhibited an increase (up-regulation; yellow) or decrease (down-regulation; blue) of at least two-fold. Top 20 known genes of the most or less expression over two-fold differences were listed in Tables 2 to 5. All data of individual gene changes are available in an additional file on same web site of this paper or http://genome.nias-k.affrc.go.jp/lrbt/Suppl_Table1-4.pdf as supplemental tables (Suppl. Tables 1–4 in Additional files – see http://www.rbej.com/imedia/9437445795174588/sup1.pdf).
Figure 2.
Gene expression correlations of bovine peri-implantation embryo between Day 7E vs. Day 14E, Day 7E vs. Day 21E, Day 7E vs. Day 28EEM, Day 7E vs. Day 28F, Day 14E vs. Day 21E, Day 14E vs. Day 28EEM, Day 14E vs. Day 28F, Day 21E vs. Day 28EEM, Day 21E vs. Day 28F, and Day 28EEM vs. Day 28F. The yellow areas highlight a greater than two-fold gene expression difference (up-regulated) between the X-axis and Y-axis samples. The blue areas highlight a greater than two-fold gene expression difference (down-regulated) between the X-axis and Y-axis samples. The gray areas highlight a 0.5- to 2-fold gene expression difference between the X-axis and Y-axis samples.
Table 2.
Top 20 known genes of the most or less expression over two-fold differences between Day 7E vs. Day 14E – see also additional file 1
| Accession No. | Gene name | D14E/D7E | K-means | Classification |
| Day 14E/Day 7E down-regulated genes (<0.5) | ||||
| L07872 | Homo sapiens Jk-recombination signal binding protein | 0.19 | 8 | Apoptosis & Cell cycle |
| NM_001294 | Homo sapiens CLPTM1 | 0.23 | 4 | Cytokine family |
| NM_001021 | Homo sapiens ribosomal protein S17 (RPS17) | 0.25 | 8 | Ribosomal |
| M20866 | Sus scrofa cofilin | 0.25 | 1 | Cytoskeleton |
| M21683 | Sus scrofa nonhistone protein HMG1 | 0.30 | 8 | DNA binding protein |
| AF020508 | Bos taurus PAG-6 | 0.36 | 11 | Cytokine family |
| AF020507 | Bos taurus PAG-5 | 0.39 | 11 | Cytokine family |
| X91755 | Bos taurus cathepsin L | 0.42 | 8 | Oncogene & Tumor inhibitor |
| NM_005722 | Homo sapiens ARP2 actin-related protein 2 homolog (yeast) | 0.43 | 8 | Cytoskeleton |
| AF210381 | Bos taurus DDVit1 | 0.44 | 4 | ECM & related |
| AF020506 | Bos taurus PAG-4 | 0.46 | 1 | Cytokine family |
| U21660 | Bos taurus phosphatidylcholine transfer protein | 0.47 | 4 | |
| L02897 | Dog nonerythroid beta-spectrin | 0.47 | 9 | |
| AF166124 | Homo sapiens selenoprotein X | 0.49 | 8 | |
| Day 14E/Day 7E up-regulated genes (2<) | ||||
| X14926 | Mus musculus calreticulin | 24.00 | 12 | |
| AB009282 | Homo sapiens cytochrome b5 | 16.48 | 12 | |
| NM_004494 | Homo sapiens hepatoma-derived growth factor (HDGF) | 13.52 | 12 | Oncogene & Tumor inhibitor |
| AF000137 | Bos taurus connective tissue growth factor precursor (CTGF) | 12.18 | 12 | Cytokine family |
| NM_000365 | Homo sapiens triosephosphate isomerase 1 (TPI1) | 10.52 | 12 | |
| X89984 | Homo sapiens BCL7A protein | 10.31 | 12 | Cytoskeleton |
| AF217197 | Homo sapiens FBP interacting repressor (FIR) | 9.80 | 12 | Transcriptional regulator |
| NM_005720 | Homo sapiens ARPC1B | 9.45 | 3 | Cytoskeleton |
| NM_001404 | Homo sapiens EEF1G | 9.43 | 12 | Transcriptional regulator |
| AB003094 | Bos taurus ferritin L subunit | 9.38 | 12 | |
| X56503 | Sus scrofa casein kinase II beta subunit (CKII beta) | 8.55 | 12 | Enzyme |
| X13684 | Bos taurus glutathione peroxidase (gpx1) | 8.39 | 3 | Enzyme |
| NM_002436 | Homo sapiens membrane protein, palmitoylated 1 (55 kD) | 8.33 | 12 | Membrane protein |
| NM_005022 | Homo sapiens profilin 1 (PFN1) | 8.09 | 12 | Cytoskelton |
| X01809 | Bos taurus cathepsin 3' terminus | 8.08 | 12 | Oncogene & Tumor inhibitor |
| AF207664 | Homo sapiens matrix metalloprotease (ADAMTS1) | 8.03 | 3 | ECM & related |
| X56597 | Homo sapiens humFib fibrillarin | 7.97 | 12 | |
| AF182001 | Bos taurus D4-GDP-dissociation inhibitor (D4-GDI) | 7.82 | 3 | |
| NM_007317 | Homo sapiens kinesin-like 4 (KNSL4) | 7.66 | 2 | |
| M21044 | Bos taurus MHC class I BoLA | 7.65 | 2 | Cytokine family |
All data of individual gene changes are available in Suppl. Table 1 of the Additional file.
Table 5.
Top 20 known genes of the most or less expression over two-fold differences between Day 28EEM vs. Day 28F.
| Accession No. | Gene name | D28F/D28EEM | K-means | Classification |
| Day 28F/Day 28EEM down-regulated genes (<0.5) | ||||
| M73961 | Ovis aries PAG-1 | 0.14 | 6 | Cytokine family |
| X89984 | Homo sapiens BCL7A protein | 0.14 | 12 | Cytoskeleton |
| X15975 | Bos taurus PRP-V | 0.15 | 11 | Cytokine family |
| AF079545 | Ovis aries placental lactogen precursor (PL) | 0.16 | 8 | Cytokine family |
| AF020509 | Bos taurus PAG-7 | 0.19 | 11 | Cytokine family |
| J02840 | Bos taurus placental lactogen (bPL-Ala) | 0.20 | 5 | Cytokine family |
| X59504 | Bos taurus PRP-VI | 0.20 | 11 | Cytokine family |
| S72871 | Homo sapiens GATA-2 transcription factor | 0.20 | 8 | Transcriptional regulator |
| AF192336 | Bos taurus PAG-19 | 0.22 | 11 | Cytokine family |
| AF020508 | Bos taurus PAG-6 | 0.23 | 11 | Cytokine family |
| J02944 | Bos taurus PRP-I | 0.24 | 11 | Cytokine family |
| L06151 | Bos taurus PAG-2 | 0.25 | 11 | Cytokine family |
| AF020512 | Bos taurus PAG-10 | 0.27 | 5 | Cytokine family |
| AF020514 | Bos taurus PAG-12 | 0.28 | 11 | Cytokine family |
| AF192334 | Bos taurus PAG-17 | 0.30 | 11 | Cytokine family |
| AL133034 | Homo sapiens clone DKFZp727K171 | 0.31 | 11 | |
| Z11742 | Bos taurus annexin XI | 0.31 | 5 | Apoptosis & Cell cycle |
| U89321 | Homo sapiens nucleophosmin phosphoprotein (NPM) | 0.32 | 5 | |
| X17614 | Bos taurus 3 beta hydroxy-5-ene steroid dehydrogenase/delta 5-delta4 isomerase | 0.32 | 5 | Enzyme |
| AF192333 | Bos taurus PAG-16 | 0.32 | 11 | Cytokine family |
| Day 28F/Day 28EEM up-regulated genes (2<) | ||||
| L34261 | Bos taurus palmitoyl-protein thioesterase | 4.32 | 6 | Enzyme |
| AF000137 | Bos taurus connective tissue growth factor precursor (CTGF) | 2.85 | 12 | Cytokine family |
| NM_004458 | Homo sapiens FACL4 transcript variant 1 | 2.84 | 7 | Enzyme |
| NM_006491 | Homo sapiens NOVA1 transcript variant 3 | 2.82 | 1 | Oncogene & Tumor inhibitor |
| NM_006838 | Homo sapiens methionine methionyl aminopeptidase 2 | 2.72 | 4 | Enzyme |
| AB028449 | Homo sapiens helicase-MOI | 2.65 | 9 | DNA binding protein |
| AL080102 | Homo sapiens clone DKFZp564N1916 | 2.64 | 9 | |
| AF057300 | Homo sapiens truncated RAD50 protein | 2.63 | 3 | |
| AB043994 | Bos taurus MMP-2 | 2.56 | 7 | ECM & related |
| NM_006719 | Homo sapiens transcript variant ABLIM-m | 2.53 | 1 | Cytoskeleton |
| AF195417 | Homo sapiens DEAD-box protein abstrakt (ABS) | 2.51 | 2 | |
| Z25531 | Bos taurus repeat region DNA | 2.41 | 3 | |
| AF113682 | Homo sapiens clone FLB3436 PRO0868 | 2.40 | 4 | |
| S76474 | Homo sapiens trkB | 2.39 | 1 | Cytokine family |
| Y16533 | Ovis aries IGF-II | 2.39 | 7 | Cytokine family |
| M86739 | Bos taurus neuropeptide Y receptor | 2.38 | 7 | Cytokine family |
| AF043937 | Homo sapiens DHAPAT | 2.37 | 9 | Enzyme |
| AF144763 | Bos taurus TIMP-1 protein | 2.36 | 3 | ECM & related |
| NM_005563 | Homo sapiens stathmin 1/oncoprotein 18 (STMN1) | 2.36 | 4 | Oncogene & Tumor inhibitor |
| AF198487 | Homo sapiens transcription factor LBP-1b | 2.34 | 4 | Transcriptional regulator |
All data of individual gene changes are available in Suppl. Table 4 of the Additional file.
Day 7 to Day 14
A comparison between Day 7E and Day 14E clearly indicates that only 26 genes were down-regulated (14 annotated and 12 ESTs), with many genes (680) being up-regulated from Day 7 through Day 14E. The most significant genes during this period are listed in Table 2 and Suppl. Table 1 (see Additional files). They included 22 cytokine-related molecules, 44 enzymes, 21 transcriptional regulators, 16 oncogenes or tumor suppressor genes, 12 apoptosis or cell-cycle molecules, 10 heat-shock proteins (HSP), 9 cell-adhesion molecules, and 8 membrane proteins. A striking expression was found for several genes during this period, i.e. calrecticulin (X14926), which is a 55 kd calcium binding protein of the ER lumen, and caldesmon (X89984), an actin-binding protein, were two of the most significantly expressed genes. PAGs and IFN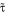 were also strongly expressed. Other cytokine molecules, such as HDGF (NM_00494), had their expression accented. Cell-function-related genes such as integrin, mucin, ADAMTS, keratin, and cytokeratin were found to be significantly increased in their expression.
were also strongly expressed. Other cytokine molecules, such as HDGF (NM_00494), had their expression accented. Cell-function-related genes such as integrin, mucin, ADAMTS, keratin, and cytokeratin were found to be significantly increased in their expression.
Day 14 to Day 21
A total of 452 genes were significantly up-regulated from Day 14 through Day 21, and half of these (226 genes) were annotated by BLASTn (Fig. 2). Only two genes were down-regulated, and they contained one annotated gene, myotrophin. The expression of various PL-related genes, including PRPs, was up-regulated during this period. The expression of extra-matrix-related genes, such as collagens, proteoglycans, MMPs, extracellular MMP inducer (EMMPRIN), and heparanase, and cell adhesion molecules, such as mucin, integrins, ezrin, and certain cytokines such as insulin-like growth factors (IGFs) and epidermal growth factor receptor (EGFR), were up-regulated. IFN-τ expression was emphasized. The individual gene expression data is provided in Table 3 and Suppl. Table 2 (see Additional files).
Table 3.
Top 20 known genes of the most or less expression over two-fold differences between Day 14E vs. Day 21E.
| Accession No. | Gene name | D21E/D14E | K-means | Classification |
| Day 21E/Day 14E down-regulated genes (<0.5) | ||||
| U21661 | Rattus norvegicus myotrophin | 0.28 | 2 | Cytokine family |
| Day 21E/Day 14E up-regulated genes (2<) | ||||
| AF004877 | Homo sapiens pro-alpha 2(I) collagen (COL1A2) | 16.08 | 4 | ECM & related |
| U21660 | Bos taurus phosphatidylcholine transfer protein | 10.13 | 4 | |
| X15112 | Bos taurus cytochrome c oxidase subunit VIb (AED) | 10.01 | 4 | |
| X59504 | Bos taurus prolactin-like protein (PRP-VI) | 9.62 | 11 | Cytokine family |
| AB008683 | Bos taurus alpha2(I) collagen (COL1A2) | 9.27 | 4 | ECM & related |
| AF105429 | Ovis aries H19 | 7.81 | 4 | |
| X65210 | Bos taurus microsatellite DNA | 7.62 | 11 | |
| X15975 | Bos taurus PRP-V | 7.59 | 11 | Cytokine family |
| S74761 | Bos taurus water channel protein CHIP29 | 7.44 | 9 | Membrane protein |
| AF196320 | Bos taurus Interferon-tau1C | 7.39 | 2 | Cytokine family |
| NM_001613 | Homo sapiens actin, alpha 2 (ACTA2) | 6.98 | 4 | Cytoskeleton |
| L06151 | Bos taurus PAG-2 | 6.86 | 11 | Cytokine family |
| M32303 | Bos taurus metalloproteinase inhibitor | 6.82 | 11 | ECM & related |
| NM_001553 | Homo sapiens IGFBP7 | 6.62 | 9 | Cytokine family |
| M21683 | Sus scrofa nonhistone protein HMG1 | 6.58 | 8 | DNA binding protein |
| AF020508 | Bos taurus PAG-6 | 6.54 | 11 | Cytokine family |
| AF125041 | Ovis aries decorin | 6.42 | 4 | |
| J02944 | Bos taurus PRP-1 | 6.40 | 11 | Cytokine family |
| AB004800 | Sus scrofa S100C protein | 5.68 | 9 | Cytokine family |
| AF192336 | Bos taurus PAG-19 | 5.47 | 11 | Cytokine family |
All data of individual gene changes are available in Suppl. Table 2 of the Additional file.
Day 21 to Day 28
A comparison between Day 21E and Day 28EEM revealed that the expression of 14 genes increased slightly in Day 28EEM and 11 of these were annotated, which included PAGs, PLs, and IL-1. A total of 173 genes were significantly down-regulated in Day 28EEM, including 109 annotated genes; these genes were found mainly in k-means clusters 2, 3, 6, 8, and 12. The most significantly down-regulated gene during this critical period for implantation was IFN-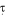 . Down-regulation of certain MMP or matrix-related genes was also found (see Table 4 and Suppl. Table 3 in Additional files) in the top 50 genes of lowest expression.
. Down-regulation of certain MMP or matrix-related genes was also found (see Table 4 and Suppl. Table 3 in Additional files) in the top 50 genes of lowest expression.
Table 4.
Top 20 known genes of the most or less expression over two-fold differences between Day 21E vs. Day 28EEM.
| Accession No. | Gene name | D28EEM/D21E | K-means | Classification |
| Day 28EEM/Day 21E down-regulated genes (<0.5) | ||||
| AF196320 | Bos taurus Interferon-tau1C | 0.07 | 2 | Cytokine family |
| AF033096 | Avena sativa nonphototropic hypocotyl 1 (NPH1-1) | 0.22 | 12 | |
| L10240 | Homo sapiens EMMPRIN | 0.24 | 7 | ECM & related |
| U21660 | Bos taurus phosphatidylcholine transfer protein | 0.27 | 4 | |
| X15112 | Bos taurus cytochrome c oxidase subunit VIb (AED) | 0.28 | 4 | |
| L34261 | Bos taurus palmitoyl-protein thioesterase | 0.29 | 6 | Enzyme |
| M26576 | Homo sapiens alpha-1 collagen type IV | 0.29 | 6 | ECM & related |
| NM_001747 | Homo sapiens capping protein, gelsolin-like (CAPG) | 0.32 | 2 | Cytoskeleton |
| U46064 | Sus scrofa aldehyde reductase (ALR1) | 0.33 | 3 | Enzyme |
| AF020513 | Bos taurus PAG-11 | 0.33 | 12 | Cytokine family |
| M11120 | Rat 28S rRNA | 0.33 | 2 | |
| NM_004718 | Homo sapiens COX7A2L | 0.34 | 8 | Enzyme |
| AF144763 | Bos taurus TIMP-1 | 0.35 | 3 | ECM & related |
| AF034607 | Homo sapiens chloride channel ABP | 0.36 | 8 | Membrane protein |
| NM_005556 | Homo sapiens keratin 7 (KRT7) | 0.36 | 2 | Cytoskeleton |
| AJ243656 | Methanobacterium thermoautotrophicum ehbA-Q | 0.37 | 2 | |
| M83104 | Bos taurus cytochrome b5 reductase | 0.37 | 2 | Enzyme |
| X59693 | Bos taurus ubiquinol-cytrochrome-c reductase (subunit II) | 0.38 | 6 | Enzyme |
| D84557 | Homo sapiens HsMcm6 | 0.38 | 12 | |
| M77234 | Homo sapiens ribosomal protein S3a | 0.38 | 6 | Ribosomal |
| Day 28EEM/Day 21E up-regulated genes (2 <) | ||||
| AF020508 | Bos taurus PAG-6 | 3.45 | 11 | Cytokine family |
| X01912 | Goat epsilon I beta-globin | 3.00 | 9 | |
| AF004133 | Sus scrofa adipocyte membrane protein | 2.66 | 1 | Membrane protein |
| M73961 | Ovis aries PAG-1 | 2.35 | 6 | Cytokine family |
| J02840 | Bos taurus placental lactogen (PL-Ala) | 2.21 | 5 | Cytokine family |
| AF192336 | Bos taurus PAG-19 | 2.17 | 11 | Cytokine family |
| AB005148 | Bos taurus interleukin 1 (IL-1) receptor antagonist | 2.13 | 1 | Cytokine family |
| D10989 | Bos taurus endothelin ETB receptor | 2.12 | 1 | |
| X59504 | Bos taurus PRP-VI | 2.08 | 11 | Cytokine family |
| M80328 | Bos taurus PL-Val | 2.01 | 5 | Cytokine family |
| Z11742 | Bos taurus annexin XI | 2.00 | 5 | Apoptosis & Cell cycle |
All data of individual gene changes are available in Suppl. Table 3 of the Additional file.
Comparison between extra-embryonic membranes on Day 28 and the fetus on Day 28
A total of 119 genes, including ESTs, decreased significantly in expression, with only 52 annotated genes among them in Day 28EEM (Fig. 2). However, in Day 28EEM, 74 up-regulated genes contained members of the cytokine family and molecules that play a role in cell-to-cell interactions, such as PLs, PRPs, PAGs, MHCs, and mucin, as indicated in Fig. 3. The up-regulated genes included 35 ESTs, which exhibited expression profiles similar to the cytokines during the pre-implantation period in extra-embryonic membranes. In addition to these trophoblast-specific genes, which are known to be specific to extra-embryonic membranes, other interesting genes, such as dickkopf-1, which is a Wnt signal regulation molecule, grancalcin, which is a calcium binding protein, two actin-related proteins (NM_005720, X89984), and cell proliferation related genes (NM_006429, U89321), were specifically found in extra-embryonic membranes when compared to the fetus. The actual roles of these genes in extra-embryonic cell development are as yet unknown. Individual data are provided in Table 5 and Suppl. Table 4 in Additional file 1.
Figure 3.
Hierarchical tree cluster of differentially expressed genes in the bovine embryo during the implantation period (Days 7 to 28 of gestation). Two-fold and greater differences in gene expression between Day 28EEM vs. Day 28F are selected in the expression profiles from all samples of Day 7E, 14E, 21E, 28EEM and 28F. Normalized and log2 transformed expression data were used for the clustering analysis. The yellow cells indicate up-regulated genes in the median of the total value; the blue cells indicate down-regulated genes in the median of the total value. The black cells indicate no changes in expression.
Candidate extra-embryonic membrane specific genes
Genes related to trophoblast cell differentiation were anticipated to change their expression when the comparison was made between Day 28EEM and Day 28F. Most of the annotated genes of the 74 genes that increased over two-fold in Day 28EEM were PL-, PRP-, and PAG-related genes. The expression of 35 ESTs showed profiles similar to those of the annotated genes during the pre-implantation period, as determined by three hierarchical analyses (Fig. 3). An attempt was made to find new genes related to extra-embryonic cell lineages. Four ESTs (AW464053, AW465434, AW462349, and AW485575) were selected and analyzed by real-time RT-PCR. The quantitative data coincided with the microarray data as shown in Figs. 3 and 4. The relative intensities of these ESTs, PL-Ala and PRP-I were similar to those of real-time RT-PCR.
Figure 4.
Real-time RT-PCR analysis of PL-Ala, PRP-I, and ESTs (GenBank accession No. AW464053, AW465434, AW462349, and AW485575) mRNA in bovine embryos. Gene expression on Days 7E to 21E, and 28EEM or 28F is provided. The expression of each mRNA was normalized to the expression of GAPDH mesured in same RNA preparation. The expression refers "means ± SD". Values with different letters are significantly different (P < 0.05).
Discussion
The major reproductive wastage in farm animals is early embryo loss, i.e. the anomalous development of embryos and/or an aberration of placentation [36]. Various technologies, such as artificial insemination, embryo transfer, and cloning, have been applied to bovine reproduction [13,37]. Precise knowledge of the gene expression profile during peri-implantation is necessary to reduce early losses and to improve the reproductive efficiency of these new technologies [6,38-40]. However, little is known about the complex molecular regulation of embryos and extra-embryonic membrane development in cattle. Thus, the genes to be profiled include new, functional gene candidates. We suggest an assessment method or key gene to help clarify the complex mechanisms in early embryo and trophoblast cell proliferation and differentiation.
Various genes changes during embryonic development, particularly in extra-embryonic membranes, with specific morphological changes occur in bovine: a remarkable elongation of the embryonic membrane, fusion between fetal membrane and caruncular epithelium, endometrial reorganization [3,41,42]. Specific genes, like IFN-τ, PLs, PRPs, PAGs, IGFs, and IGFBPs have been studied as they take important roles around implantation in bovine [3,43-48]. However these molecules are essential for development of the embryo and the formation of extra-embryonic membranes, determination of their specific roles are still difficult to prove.
Microarray analyses provided the time-dependant genes profiles in accord with the progress of gestation during the peri-implantation period. Most genes that expressed from the blastocyst stage to early placentation were up-regulated, while only 14 of the identified genes exhibited down-regulation (Fig 1). For example, selenoprotein X (AF166124) exerts a stimulative function on cell proliferation, particularly in the blastocyst, since its expression decreased on Day 14 compared to Day 7. This result coincided with that in silico work [49].
We selected genes related to heat-shock proteins (HSP) to interpret comprehensive genes expression, because they were frequently found in the list of significant genes and previous data had revealed their importance in early embryo development [50]. Ten HSP-related genes out of 333 annotated genes were up-regulated over two-fold on Day 14E in comparison to Day 7E. Similar genes were detected, i.e. seven on Day 21E and three on Day 28EEM. These results suggest a specific importance for HSPs in the early development of embryos. HSPs may disturb the coordination between the conceptus and endometrium in bovines and be related to the induction of early embryo loss [51]. Aberration of HSP90 expression in mice causes a placental abnormality that is closely related to trophoblast cell differentiation and proliferation [52].
Intensive IFN-τ expression was found during the implantation period (Days 14E to 21E). The trophoblast elongates and grows into a thread-like structure during this period and produces IFN-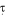 which inhibits luteolysis and plays a role in embryonic survival by mediating embryo-maternal crosstalk [8,43,53-56]. IFN-τ not only has an anti-luteolytic function enabling the establishment of gestation in cows, but apparently also participates in various other functions necessary for embryo survival [56,57]. It has been reported that IFN-τ has an immunoregulative effect [58] and also interferon modulates the expression of MHC class I antigen in mouse trophoblast cell cultures [59]. It is interesting that MHC I and its related genes, MHC II and β-microglobulin, exhibited increased expression in the present study only during the implantation period.
which inhibits luteolysis and plays a role in embryonic survival by mediating embryo-maternal crosstalk [8,43,53-56]. IFN-τ not only has an anti-luteolytic function enabling the establishment of gestation in cows, but apparently also participates in various other functions necessary for embryo survival [56,57]. It has been reported that IFN-τ has an immunoregulative effect [58] and also interferon modulates the expression of MHC class I antigen in mouse trophoblast cell cultures [59]. It is interesting that MHC I and its related genes, MHC II and β-microglobulin, exhibited increased expression in the present study only during the implantation period.
Cell-adhesion molecules and cytokines, such as the integrin family, have been emphasized their importance for initiation of implantation [60-62]. Recently, Ezrin, a cytoskeletal-membrane linker molecule belonging to the ezrin-radixin-moesin (ERM) family, proved its importance for implantation in mice [63], exhibiting greater expression than in the pre-implantation stage in bovine blastocysts in the present study. The importance of adhesion molecule involvement via the adhesion-regulating molecules (ARM-1), ICAM-1, LECAM-1, Lu-ECAM-1, calcium, and integrin-binding protein (CIB) was suggested by their expression increasing around the expected implantation starting day (Day 21E).
Numerous PAG, PL, and PRP genes, involved in the differentiation of trophoblastic cell lineage, were found among the 39 annotated genes of the 74 that exhibited over a two-fold difference between extra-embryonic membranes and the fetus on Day 28. The expression of most PAG family members increased towards Day 21E. Other binucleate cell-specific genes, such as PL and PRP family members, exhibited a coordinated expression with the PAG family of genes. These results confirm previous reports from this laboratory [3,13,33,64]. PAG-5, -15, -16, -17, and -19 were distributed to k-means cluster 11 in the present study, whereas PAG-8 was distributed to k-means cluster 3 (Fig. 1). The former group of PAG family members is apparently produced by binucleate cells, while those in the latter group may be produced by other cells of the trophectoderm [46].
Microarray data suggested various new candidate genes for extra-embryonic development even they have known function. Dickkopf-1 may inhibit Wnt proteins, which influence many aspects of embryonic development [65]. Annexins are a family of structurally related calcium-dependent phospholipid binding proteins [66]. Grancalcin is a Ca (2+)-binding protein [67]. Actin-related protein 2/3 (Arp2/3) complex may be related to the actin cytoskeleton [68]. Chaperonin containing t-complex polypeptide 1 (CCT complex) is essential for the maturation of cyclin E [69]. Nucleophosmin phosphoprotein (NPM) (U89321), is a major nucleolar protein that is 20 times more abundant in tumors or proliferating cells [70]. Finally, BCL7A exhibits homology with the actin-binding protein caldesmon [71].
The most straightforward explanation for present study is that EST genes that display similar expression patterns are functionally related [72]. AW464053, AW465434, AW462349, and AW485575 gene expression patterns were similar to the expression patterns of PL and PRP-I (Figs. 3 and 4). These ESTs were submitted from the cDNA library of Soares normalized bovine placenta (Lewin et al., unpublished). The EST of AW485575 was submitted from a library made from pooled tissue of Day 20 and Day 40 bovine embryos [73]. However, the function of all four ESTs remains obscure. The AW465434 gene has a 495 bp sequence and was found to be common for 21/21 bp (e-value = 1.1) in Homo sapiens in BAC clone RP11-1246C19 (AC102953). The AW462349 gene has 545 bp sequences and was found to be common for a slight 43/47 bp (e-value = 3 × 10-7) in Sus scrofa clone RP44-519O7 (AC096884). The AW485575 gene has 436 bp sequences and was found to be common for a slight 157/181 bp (e-value = 6 × 10-39) in Homo sapiens hypothetical protein MGC39389 (BC003531). The AW464053 gene expression patterns were similar to the expression pattern of PRP-I, and the gene sequence has a portion in common with an e-value = 1 × 10-102 and 4 × 10-31 (common sequence = 326/371, 87% and 105/116, 90%) for PRP-VI (X59504), or an e-value = 1 × 10-95 (common sequence = 351/408, 86%) for PRP-III (M27240 or NM_174160). We suggest this EST was new member of bPRP.
Conclusions
This study provides developmental expression changes of a large number of genes in the bovine embryo during the peri-implantation period, with particular focus on genes that express in extra-embryonic membranes. The new gene candidates that were discussed here may address a new set of annotated genes and ESTs for embryonic differentiation and development. Participation of several known genes like Ezrin, CIB, Dickkopf-1, Grancalcin and EST (AW464053, AW465434 etc) in extra-embryonic membrane differentiation and development is elucidated by this microarray analysis. Fundamental investigations of this sort contribute significantly to a better understanding of the effects of various cultural conditions and cellular and genetic manipulation of embryos, including in vitro fertilization, in vitro maturation, and embryo cloning technology.
Supplementary Material
Supplement tables list the whole genes which produced the expression difference in Fig. 2 and complement Tables 2–5 beneath (see text for details). The legends of supplemental tables are as follows: Supplement Table 1; Two-fold differentially expressed genes between Day 7E vs. Day 14E. Supplement Table 2; Two-fold differentially expressed genes between Day 14E vs. Day 21E. Supplement Table 3; Two-fold differentially expressed genes between Day 21E vs. Day 28EEM. Supplement Table 4; Two-fold differentially expressed genes between Day 28EEM vs. Day 28F.
Acknowledgments
Acknowledgements
This research was supported by grants from the Bio-oriented Technology Research Advancement Institution (BRAIN); and a grant-in-aid (HC-04-2261-1) from the Ministry of Agriculture, Forestry, and Fisheries of Japan.
Contributor Information
Koichi Ushizawa, Email: ushizawa@affrc.go.jp.
Chandana B Herath, Email: cherath@unimelb.edu.au.
Kanako Kaneyama, Email: kkane@affrc.go.jp.
Satoshi Shiojima, Email: shiojima@pharm.kyoto-u.ac.jp.
Akira Hirasawa, Email: akira_h@pharm.kyoto-u.ac.jp.
Toru Takahashi, Email: tatoru@affrc.go.jp.
Kei Imai, Email: k0imai@nlbc.go.jp.
Kazuhiko Ochiai, Email: g0101201@iwate-u.ac.jp.
Tomoyuki Tokunaga, Email: tom@affrc.go.jp.
Yukio Tsunoda, Email: tsunoda@nara.kindai.ac.jp.
Gozoh Tsujimoto, Email: gtsuji@pharm.kyoto-u.ac.jp.
Kazuyoshi Hashizume, Email: kazuha@iwate-u.ac.jp.
References
- Stanton JA, Macgregor AB, Green DP. Gene expression in the mouse preimplantation embryo. Reproduction. 2003;125:457–468. doi: 10.1530/rep.0.1250457. [DOI] [PubMed] [Google Scholar]
- King GJ, Atkinson BA, Robertson HA. Development of the bovine placentome from days 20 to 29 of gestation. J Reprod Fertil. 1980;59:95–100. doi: 10.1530/jrf.0.0590095. [DOI] [PubMed] [Google Scholar]
- Yamada O, Todoroki J, Kizaki K, Takahashi T, Imai K, Patel OV, Schuler LA, Hashizume K. Expression of prolactin-related protein I at the fetomaternal interface during the implantation period in cows. Reproduction. 2002;124:427–437. doi: 10.1530/rep.0.1240427. [DOI] [PubMed] [Google Scholar]
- Wooding FBP. Current topic: The synepitheliochorial placenta of ruminants: Binucleate cell fusions and hormone production. Placenta. 1992;13:101–113. doi: 10.1016/0143-4004(92)90025-o. [DOI] [PubMed] [Google Scholar]
- Wooding FBP, Flint AP. Placentation. In: Lamming GE, editor. In Marshall's Physiology of Reproduction. 4. Vol. 3. London: Chapman & Hall; 1994. pp. 233–460. [Google Scholar]
- Tanaka TS, Kunath T, Kimber WL, Jaradat SA, Stagg CA, Usuda M, Yokota T, Niwa H, Rossant J, Ko MSH. Gene expression profiling of embryo-derived stem cells reveals candidate genes associated with pluripotency and lineage specificity. Genome Res. 2002;12:1921–1928. doi: 10.1101/gr.670002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Inoue K, Ono R, Ogonuki N, Kohda T, Kaneko-Ishino T, Ogura A, Ishino F. Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development. 2002;129:1807–1817. doi: 10.1242/dev.00159. [DOI] [PubMed] [Google Scholar]
- Bazer FW, Spencer TE, Ott TL. Interferon tau: a novel pregnancy recognition signal. Am J Reprod Immunol. 1997;37:412–420. doi: 10.1111/j.1600-0897.1997.tb00253.x. [DOI] [PubMed] [Google Scholar]
- Flint AP, Guesdon FM, Stewart HJ. Regulation of trophoblast interferon gene expression. Mol Cell Endocrinol. 1994;100:93–95. doi: 10.1016/0303-7207(94)90285-2. [DOI] [PubMed] [Google Scholar]
- Whiteside EJ, Kan M, Jackson MM, Thompson JG, McNaughton C, Herington AC, Harvey MB. Urokinase-type plasminogen activator (uPA) and matrix metalloproteinase-9 (MMP-9) expression and activity during early embryo development in the cow. Anat Embryol. 2001;204:477–483. doi: 10.1007/s429-001-8004-7. [DOI] [PubMed] [Google Scholar]
- Kizaki K, Nakano H, Nakano H, Takahashi T, Imai K, Hashizume K. Expression of heparanase mRNA in bovine placenta during gestation. Reproduction. 2001;121:573–580. doi: 10.1530/rep.0.1210573. [DOI] [PubMed] [Google Scholar]
- Mohan M, Malayer JR, Geisert RD, Morgan GL. Expression patterns of retinoid X receptors, retinaldehyde dehydrogenase, and peroxisome proliferator activated receptor gamma in bovine preattachment embryos. Biol Reprod. 2002;66:692–700. doi: 10.1095/biolreprod66.3.692. [DOI] [PubMed] [Google Scholar]
- Hashizume K, Ishiwata H, Kizaki K, Yamada O, Takahashi T, Imai K, Patel OV, Akagi S, Shimizu M, Takahashi S, Katsuma S, Shiojima S, Hirasawa A, Tsujimoto G, Todoroki J, Izaike Y. Implantation and placental development in somatic cell clone recipient cows. Cloning Stem Cells. 2002;4:197–209. doi: 10.1089/15362300260339485. [DOI] [PubMed] [Google Scholar]
- Zhang S, Kubota C, Yang L, Zhang Y, Page R, O'Neill M, Yang X, Tian XC. Genomic Imprinting of H19 in Naturally Reproduced and Cloned Cattle. Biol Reprod. 2004;71:1540–1544. doi: 10.1095/biolreprod.104.031807. [DOI] [PubMed] [Google Scholar]
- Ishiwata H, Katsuma S, Kizaki K, Patel OV, Nakano H, Takahashi T, Imai K, Hirasawa A, Shiojima S, Ikawa H, Suzuki Y, Tsujimoto G, Izaike Y, Todoroki J, Hashizume K. Characterization of gene expression profiles in early bovine pregnancy using a custom cDNA microarray. Mol Reprod Dev. 2003;65:9–18. doi: 10.1002/mrd.10292. [DOI] [PubMed] [Google Scholar]
- Hartigan JA. Clustering algorithms. New York: Wiley & Sons; 1975. [Google Scholar]
- Soukas A, Cohen P, Socci ND, Friedman JM. Leptin-specific patterns of gene expression in white adipose tissue. Genes Dev. 2000;14:963–980. [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudice LC, Telles TL, Lobo S, Kao L. The molecular basis for implantation failure in endometriosis: on the road to discovery. Ann NY Acad Sci. 2002;955:252–264. doi: 10.1111/j.1749-6632.2002.tb02786.x. [DOI] [PubMed] [Google Scholar]
- Martin J, Dominguez F, Avila S, Castrillo JL, Remohi J, Pellicer A, Simon C. Human endometrial receptivity: gene regulation. J Reprod Immunol. 2002;55:131–139. doi: 10.1016/S0165-0378(01)00140-1. [DOI] [PubMed] [Google Scholar]
- Chen HW, Chen JJ, Tzeng CR, Li HN, Chang SJ, Cheng YF, Chang CW, Wang RS, Yang PC, Lee YT. Global analysis of differentially expressed genes in early gestational decidua and chorionic villi using a 9600 human cDNA microarray. Mol Hum Reprod. 2002;8:475–484. doi: 10.1093/molehr/8.5.475. [DOI] [PubMed] [Google Scholar]
- Aronow BJ, Richardson BD, Handwerger S. Microarray analysis of trophoblast differentiation: gene expression reprogramming in key gene function categories. Physiol Genomics. 2001;6:105–116. doi: 10.1152/physiolgenomics.2001.6.2.105. [DOI] [PubMed] [Google Scholar]
- Reese J, Das SK, Paria BC, Lim H, Song H, Matsumoto H, Knudtson KL, DuBois RN, Dey SK. Global gene expression analysis to identify molecular markers of uterine receptivity and embryo implantation. J Biol Chem. 2001;276:44137–44145. doi: 10.1074/jbc.M107563200. [DOI] [PubMed] [Google Scholar]
- Tanaka TS, Jaradat SA, Lim MK, Kargul GJ, Wang X, Grahovac MJ, Pantano S, Sano Y, Piao Y, Nagaraja R, Doi H, Wood WH, III, Becker KG, Ko MSH. Genome-wide expression profiling of mid-gestation placenta and embryo using a 15,000 mouse developmental cDNA microarray. Proc Natl Acad Sci USA. 2000;97:9127–9132. doi: 10.1073/pnas.97.16.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamonsen LA. Role of proteases in implantation. Rev Reprod. 1999;4:11–22. doi: 10.1530/ror.0.0040011. [DOI] [PubMed] [Google Scholar]
- Kizaki K, Yamada O, Nakano H, Takahashi T, Yamauchi N, Imai K, Hashizume K. Cloning and localization of heparanase in bovine placenta. Placenta. 2003;24:424–430. doi: 10.1053/plac.2002.0909. [DOI] [PubMed] [Google Scholar]
- Luo H, Kimura K, Aoki M, Hirako M. Vascular endothelial growth factor (VEGF) promotes the early development of bovine embryo in the presence of cumulus cells. J Vet Med Sci. 2002;64:967–971. doi: 10.1292/jvms.64.967. [DOI] [PubMed] [Google Scholar]
- Huppertz B, Kertschanska S, Demir AY, Frank HG, Kaufmann P. Immunohistochemistry of matrix metalloproteinases (MMP), their substrates, and their inhibitors (TIMP) during trophoblast invasion in the human placenta. Cell Tissue Res. 1998;291:133–148. doi: 10.1007/s004410050987. [DOI] [PubMed] [Google Scholar]
- Krussel JS, Bielfeld P, Polan ML, Simon C. Regulation of embryonic implantation. Eur J Obstet Gynecol Reprod Biol. 2003;110:S2–9. doi: 10.1016/S0301-2115(03)00167-2. [DOI] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Callow MJ, Speed TP. Statistical method for identifying differentially expressed genes in replicated cDNA microarray experiment. Statistica Sinica. 2002;12:111–139. [Google Scholar]
- Herath CB, Shiojima S, Ishiwata H, Katsuma S, Kadowaki T, Ushizawa K, Imai K, Takahashi T, Hirasawa A, Tsujimoto G, Hashizume K. Pregnancy-associated changes in genome-wide gene expression profiles in the liver of cow throughout pregnancy. Biochem Biophys Res Commun. 2004;313:666–680. doi: 10.1016/j.bbrc.2003.11.151. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Patel OV, Yamada O, Kizaki K, Takahashi T, Imai K, Hashizume K. Quantitative analysis throughout pregnancy of placentomal and interplacentomal expression of pregnancy-associated glycoproteins-1 and -9 in the cow. Mol Reprod Dev. 2004;67:257–263. doi: 10.1002/mrd.20017. [DOI] [PubMed] [Google Scholar]
- Zhao H, Hastie T, Whitfield ML, Brresen-Dale A-L, Jeffrey SS. Optimization and evaluation of T7 based RNA linear amplification protocols for cDNA microarray analysis. BMC genomics. 2002;3:31–45. doi: 10.1186/1471-2164-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puskas LG, Zvara A, Hackler L, Jr, Van Hummelen P. RNA amplification results in reproducible microarray data with slight ratio bias. Biotechniques. 2002;32:1330–1334. doi: 10.2144/02326mt04. [DOI] [PubMed] [Google Scholar]
- Cross JC, Werb Z, Fisher SJ. Implantation and the placenta: key pieces of the development puzzle. Science. 1994;266:1508–1518. doi: 10.1126/science.7985020. [DOI] [PubMed] [Google Scholar]
- Holm P, Booth PJ, Schmidt MH, Greve T, Callesen H. High bovine blastocyst development in a static in vitro production system using SOFaa medium supplemented with sodium citrate and myo-inositol with or without serum-proteins. Theriogenology. 1999;52:683–700. doi: 10.1016/S0093-691X(99)00162-4. [DOI] [PubMed] [Google Scholar]
- Schultz RM, Davis W, Jr, Stein P, Svoboda P. Reprogramming of gene expression during preimplantation development. J Exp Zool. 1999;285:276–282. doi: 10.1002/(SICI)1097-010X(19991015)285:3<276::AID-JEZ11>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Lonergan P, Rizos D, Gutierrez-Adan A, Moreira PM, Pintado B, de la Fuente J, Boland MP. Temporal divergence in the pattern of messenger RNA expression in bovine embryos cultured from the zygote to blastocyst stage in vitro or in vivo. Biol Reprod. 2003;69:1424–1431. doi: 10.1095/biolreprod.103.018168. [DOI] [PubMed] [Google Scholar]
- Vigneault C, McGraw S, Massicotte L, Sirard MA. Transcription factor expression patterns in bovine in vitro-derived embryos prior to maternal-zygotic transition. Biol Reprod. 2004;70:1701–1709. doi: 10.1095/biolreprod.103.022970. [DOI] [PubMed] [Google Scholar]
- King GJ, Atkinson BA, Robertson HA. Development of the intercaruncular areas during early gestation and establishment of the bovine placenta. J Reprod Fertil. 1981;61:469–474. doi: 10.1530/jrf.0.0610469. [DOI] [PubMed] [Google Scholar]
- Grealy M, Diskin MG, Sreenan JM. Protein content of cattle oocytes and embryos from the two-cell to the elongated blastocyst stage at day 16. J Reprod Fertil. 1996;107:229–233. doi: 10.1530/jrf.0.1070229. [DOI] [PubMed] [Google Scholar]
- Farin CE, Imakawa K, Hansen TR, McDonnell JJ, Murphy CN, Farin PW, Roberts RM. Expression of trophoblastic interferon genes in sheep and cattle. Biol Reprod. 1990;43:210–218. doi: 10.1095/biolreprod43.2.210. [DOI] [PubMed] [Google Scholar]
- Anthony RV, Liang R, Kayl EP, Pratt SL. The growth hormone/prolactin gene family in ruminant placentae. J Reprod Fertil Suppl. 1995;49:83–95. [PubMed] [Google Scholar]
- Kessler MA, Duello TM, Schuler LA. Expression of prolactin-related hormones in the early bovine conceptus, and potential for paracrine effect on the endometrium. Endocrinology. 1991;129:1885–1895. doi: 10.1210/endo-129-4-1885. [DOI] [PubMed] [Google Scholar]
- Green JA, Xie S, Quan X, Bao B, Gan X, Mathialagan N, Beckers JF, Roberts RM. Pregnancy-associated bovine and ovine glycoproteins exhibit spatially and temporally distinct expression patterns during pregnancy. Biol Reprod. 2000;62:1624–1631. doi: 10.1095/biolreprod62.6.1624. [DOI] [PubMed] [Google Scholar]
- Pushpakumara PG, Robinson RS, Demmers KJ, Mann GE, Sinclair KD, Webb R, Wathes DC. Expression of the insulin-like growth factor (IGF) system in the bovine oviduct at oestrus and during early pregnancy. Reproduction. 2002;123:859–868. doi: 10.1530/rep.0.1230859. [DOI] [PubMed] [Google Scholar]
- Block J, Drost M, Monson RL, Rutledge JJ, Rivera RM, Paula-Lopes FF, Ocon OM, Krininger CE, 3rd, Liu J, Hansen PJ. Use of insulin-like growth factor-I during embryo culture and treatment of recipients with gonadotropin-releasing hormone to increase pregnancy rates following the transfer of in vitro-produced embryos to heat-stressed, lactating cows. J Anim Sci. 2003;81:1590–1602. doi: 10.2527/2003.8161590x. [DOI] [PubMed] [Google Scholar]
- Lescure A, Gautheret D, Carbon P, Krol A. Novel selenoproteins identified in silico and in vivo by using a conserved RNA structural motif. J Biol Chem. 1999;274:38147–38154. doi: 10.1074/jbc.274.53.38147. [DOI] [PubMed] [Google Scholar]
- Niemann H, Wrenzycki C. Alterations of expression of developmentally important genes in preimplantation bovine embryos by in vitro culture conditions: implications for subsequent development. Theriogenology. 2000;53:21–34. doi: 10.1016/S0093-691X(99)00237-X. [DOI] [PubMed] [Google Scholar]
- Putney DJ, Malayer JR, Gross TS, Thatcher WW, Hansen PJ, Drost M. Heat stress-induced alterations in the synthesis and secretion of proteins and prostaglandins by cultured bovine conceptuses and uterine endometrium. Biol Reprod. 1988;39:717–728. doi: 10.1095/biolreprod39.3.717. [DOI] [PubMed] [Google Scholar]
- Voss AK, Thomas T, Gruss P. Mice lacking HSP90beta fail to develop a placental labyrinth. Development. 2000;127:1–11. doi: 10.1242/dev.127.1.1. [DOI] [PubMed] [Google Scholar]
- Imakawa K, Anthony RV, Kazemi M, Marotti KR, Polites HG, Roberts RM. Interferon-like sequence of ovine trophoblast protein secreted by embryonic trophectoderm. Nature. 1987;330:377–379. doi: 10.1038/330377a0. [DOI] [PubMed] [Google Scholar]
- Thatcher WW, Binelli M, Burke J, Staples CR, Ambrose JD, Coelho S. Antiluteolytic signals between the conceptus and endometrium. Theriogenology. 1997;47:131–140. doi: 10.1016/S0093-691X(96)00347-0. [DOI] [Google Scholar]
- Ealy AD, Larson SF, Liu L, Alexenko AP, Winkelman GL, Kubisch HM, Bixby JA, Roberts RM. Polymorphic forms of expressed bovine interferon-tau genes: relative transcript abundance during early placental development, promoter sequences of genes and biological activity of protein products. Endocrinology. 2001;142:2906–2915. doi: 10.1210/en.142.7.2906. [DOI] [PubMed] [Google Scholar]
- Roberts RM, Ezashi T, Rosenfeld CS, Ealy AD, Kubisch HM. Evolution of the interferon-tau genes and their promoters, and maternal-trophoblast interactions in control of their expression. Reprod Suppl. 2003;61:239–251. [PubMed] [Google Scholar]
- Li Q, Zhang M, Kumar S, Zhu LJ, Chen D, Bagchi MK, Bagchi IC. Identification and implantation stage-specific expression of an interferon-alpha-regulated gene in human and rat endometrium. Endocrinology. 2001;142:2390–2400. doi: 10.1210/en.142.6.2390. [DOI] [PubMed] [Google Scholar]
- Roberts RM, Imakawa K, Niwano Y, Kazemi M, Malathy PV, Hansen TR, Glass AA, Kronenberg LH. Interferon production by the preimplantation sheep embryo. J Interferon Res. 1989;9:175–187. doi: 10.1089/jir.1989.9.175. [DOI] [PubMed] [Google Scholar]
- Zuckermann FA, Head JR. Expression of MHC antigens on murine trophoblast and their modulation by interferon. J Immunol. 1986;137:846–853. [PubMed] [Google Scholar]
- Bowen JA, Bazer FW, Burghardt RC. Spatial and temporal analyses of integrin and Muc-1 expression in porcine uterine epithelium and trophectoderm in vitro. Biol Reprod. 1997;56:409–415. doi: 10.1095/biolreprod56.2.409. [DOI] [PubMed] [Google Scholar]
- Xiang W, MacLaren LA. Expression of fertilin and CD9 in bovine trophoblast and endometrium during implantation. Biol Reprod. 2002;66:1790–1796. doi: 10.1095/biolreprod66.6.1790. [DOI] [PubMed] [Google Scholar]
- Kliem A, Tetens F, Klonisch T, Grealy M, Fischer B. Epidermal growth factor receptor and ligands in elongating bovine blastocysts. Mol Reprod Dev. 1998;51:402–412. doi: 10.1002/(SICI)1098-2795(199812)51:4<402::AID-MRD7>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Daikoku T, Wang H, Sato E, Dey SK. Differential expression of ezrin/radixin/moesin (ERM) and ERM-associated adhesion molecules in the blastocyst and uterus suggests their functions during implantation. Biol Reprod. 2004;70:729–736. doi: 10.1095/biolreprod.103.022764. [DOI] [PubMed] [Google Scholar]
- Nakano H, Takahashi T, Imai K, Hashizume K. Expression of placental lactogen and cytokeratin in bovine placental binucleate cells in culture. Cell Tissue Res. 2001;303:263–270. doi: 10.1007/s004410000316. [DOI] [PubMed] [Google Scholar]
- Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391:357–62. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- Benz J, Hofmann A. Annexins: from structure to function. Biol Chem. 1997;378:177–183. [PubMed] [Google Scholar]
- Teahan CG, Totty NF, Segal AW. Isolation and characterization of grancalcin, a novel 28 kDa EF-hand calcium-binding protein from human neutrophils. Biochem J. 1992;286:549–554. doi: 10.1042/bj2860549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RD, Pollard TD. Structure and function of the Arp2/3 complex. Curr Opin Struct Biol. 1999;9:244–249. doi: 10.1016/S0959-440X(99)80034-7. [DOI] [PubMed] [Google Scholar]
- Won KA, Schumacher RJ, Farr GW, Horwich AL, Reed SI. Maturation of human cyclin E requires the function of eukaryotic chaperonin CCT. Mol Cell Biol. 1998;18:7584–7589. doi: 10.1128/mcb.18.12.7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PK, Chan FY, Morris SW, Xie Z. Isolation and characterization of the human nucleophosmin/B23 (NPM) gene: identification of the YY1 binding site at the 5' enhancer region. Nucleic Acids Res. 1997;25:1225–1232. doi: 10.1093/nar/25.6.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zani VJ, Asou N, Jadayel D, Heward JM, Shipley J, Nacheva E, Takasuki K, Catovsky D, Dyer MJ. Molecular cloning of complex chromosomal translocation t(8;14;12)(q24.1;q32.3;q24.1) in a Burkitt lymphoma cell line defines a new gene (BCL7A) with homology to caldesmon. Blood. 1996;87:3124–3134. [PubMed] [Google Scholar]
- Kurella M, Hsiao L-L, Yoshida T, Randall JD, Chow G, Sarang SS, Jensen R, Gullans SR. DNA microarray analysis of complex biological Processes. J Am Soc Nephrol. 2001;12:1072–1078. doi: 10.1681/ASN.V1251072. [DOI] [PubMed] [Google Scholar]
- Smith TPL, Grosse WM, Freking BA, Roberts AJ, Stone RT, Casas E, Wray JE, White J, Cho J, Fahrenkrug SC, Bennett GL, Heaton MP, Laegreid WW, Rohrer GA, Chitko-McKown CG, Pertea G, Holt I, Karamycheva S, Liang F, Quackenbush J, Keele JW. Sequence evaluation of four pooled-tissue normalized bovine cDNA libraries and construction of a gene index for cattle. Genome Res. 2001;11:626–630. doi: 10.1101/gr.170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement tables list the whole genes which produced the expression difference in Fig. 2 and complement Tables 2–5 beneath (see text for details). The legends of supplemental tables are as follows: Supplement Table 1; Two-fold differentially expressed genes between Day 7E vs. Day 14E. Supplement Table 2; Two-fold differentially expressed genes between Day 14E vs. Day 21E. Supplement Table 3; Two-fold differentially expressed genes between Day 21E vs. Day 28EEM. Supplement Table 4; Two-fold differentially expressed genes between Day 28EEM vs. Day 28F.