Abstract
The genus Wolbachia is an archetype of maternally inherited intracellular bacteria that infect the germline of numerous invertebrate species worldwide. They can selfishly alter arthropod sex ratios and reproductive strategies to increase the proportion of the infected matriline in the population. The most common reproductive manipulation is cytoplasmic incompatibility (CI), which results in embryonic lethality in crosses between infected males and uninfected females. Females infected with the same Wolbachia strain rescue this lethality. Despite more than 40 years of research1 and relevance to symbiont-induced speciation2,3, as well as control of arbovirus vectors4,5,6 and agricultural pests7, the bacterial genes underlying CI remain unknown. Here, we use comparative and transgenic approaches to demonstrate that two differentially transcribed, codiverging genes in the eukaryotic association module of prophage WO8 from Wolbachia strain wMel recapitulate and enhance CI. Dual expression in transgenic, uninfected males of Drosophila melanogaster crossed to uninfected females causes embryonic lethality. Each gene additively augments embryonic lethality in infected males crossed to uninfected females. Lethality associates with embryonic defects that parallel those of wild type CI and is notably rescued by wMel-infected embryos in all cases. The discovery of cytoplasmic incompatibility factor genes cifA and cifB pioneers genetic studies of prophage WO-induced reproductive manipulations and informs Wolbachia’s ongoing utility to control dengue and Zika transmission to humans.
We hypothesized that the genes responsible for CI (Extended Data Fig. 1a) are present in all CI-inducing Wolbachia strains but absent or divergent in non-CI strains; we also predicted that these genes would be expressed in the gonads of infected insects. To elucidate CI candidates, we determined the core genome shared by the CI-inducing Wolbachia strains wMel (from D. melanogaster), wRi (from D. simulans), wPip (Pel strain from Culex pipiens), and wRec (from D. recens), while excluding the pan-genome of the mutualistic strain wBm (from Brugia malayi). This yielded 113 gene families representing 161 unique wMel genes (Fig. 1a, Supplementary Table 1). We further narrowed this list by comparing it to (i) homologs of genes previously determined by comparative genomic hybridization to be absent or divergent in the strain wAu9, a non-CI strain, (ii) homologs to genes highly expressed at the RNA level in wVitA-infected Nasonia vitripennis ovaries, and (iii) homologs detected at the protein level in wPip (Buckeye)-infected C. pipiens ovaries. We included ovarian data with the reasoning that CI genes might be generally expressed in infected reproductive tissues, or that the CI inducer and rescue genes might be the same. Remarkably, only two genes, wMel locus tags WD0631 and WD0632, were shared among all four gene subsets (Fig. 1b, Supplementary Table 2–4). Notably, the homolog of WD0631 in the Wolbachia strain wPip was found at the protein level in the fertilized spermathecae of infected mosquitoes, lending support to its role in reproductive manipulation10.
Figure 1. Comparative analyses reveal WD0631 and WD0632 in the eukaryotic association module of prophage WO as candidate CI genes.
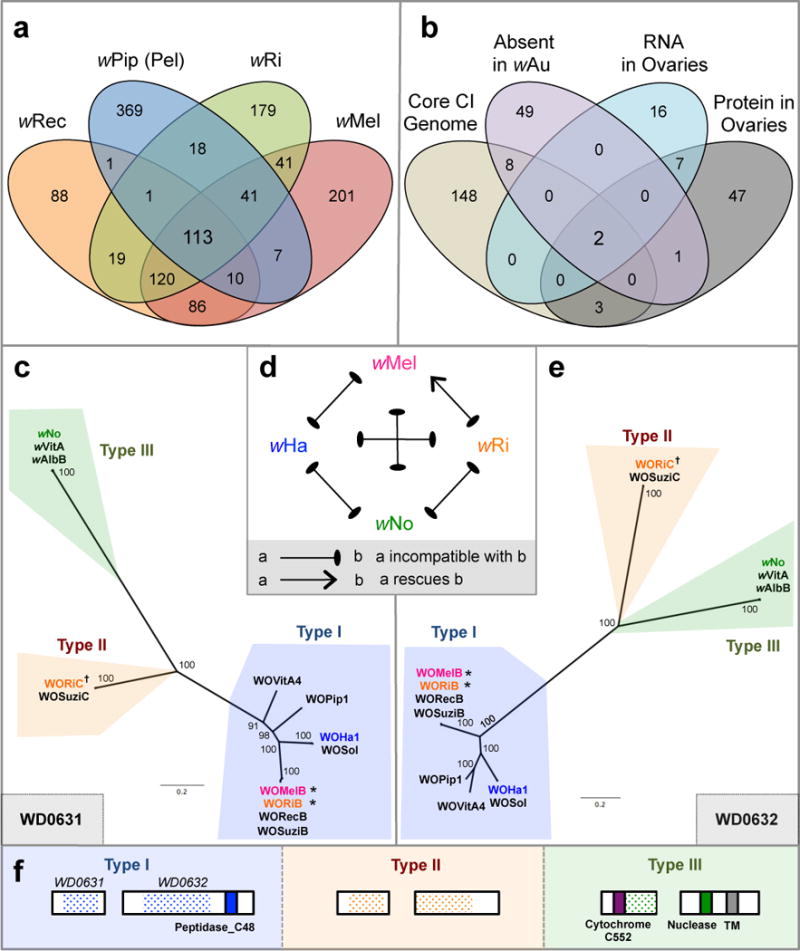
(a) Venn diagram illustrating the number of unique and shared gene families from four CI-inducing Wolbachia strains. (b) Venn diagram illustrating the number of unique and shared wMel genes matching each criteria combination. Bayesian phylogenies of (c) WD0631 (e) and WD0632 and their homologs, based on a core 256-aa alignment of WD0631 reciprocal BLASTp hits and a core 462-aa alignment of WD0632 reciprocal BLASTp hits. When multiple similar copies exist in the same strain, only one copy is shown. Consensus support values are shown at the nodes. Both trees are based on the JTT+G model of evolution and are unrooted. (d) CI patterns correlate with WD0631/WD0632 sequence homology. wRi rescues wMel and both share a similar set of homologs (*). The inability of wMel to rescue wRi correlates with a type (†) that is present in wRi but absent in wMel. Likewise, bidirectional incompatibility of all other crosses correlates to divergent homologs. This diagram was adapted from Bossan et. al55. (f) Protein architecture of the WD0631 and WD0632 types is conserved for each clade and is classified according to the WD0632-like domain. TM = transmembrane. Dotted shading represents the region of shared homology used to construct phylogenetic trees. For (c) and (e), the WO-prefix indicates a specific phage WO haplotype and the w-prefix refers to a “WO-like island,” a small subset of conserved phage genes, within that specific Wolbachia strain.
We analyzed the evolution and predicted protein domains of these two genes and found that their homologs are always paired within the eukaryotic association module of prophage WO8, and they codiverged into three distinct phylogenetic groups that we designate type I, II, and III (Fig. 1c, e, Supplementary Table 5). These relationships are not evident in the phylogeny of the Wolbachia cell division gene ftsZ, which exhibits the typical bifurcation of supergroup A and B Wolbachia (Extended Data Fig. 1b), or in the phylogeny of prophage WO baseplate assembly gene gpW (Extended Data Fig. 1c). This suggests that homologs of WD0631 and WD0632 evolve under different evolutionary pressures than genes in the core Wolbachia genome or in a structural module of phage WO.
Type I genes are the most prevalent amongst ten sequenced Wolbachia strains, and are always associated with large prophage WO regions that are often missing tail genes (Extended Data Fig. 2); it is unclear if these WO regions forge fully intact or defective interfering particles. The functions of type I WD0631 homologs are unknown, though type I WD0632 homologs contain weak homology to a putative Peptidase_C48 domain (wMel, NCBI conserved domain E-value 6.69e-04, Fig. 1f), a key feature of Ulp1 (ubiquitin-like-specific protease) proteases10. Type II genes are located within more complete phage haplotypes (Extended Data Fig. 2), but the WD0632 homologs are truncated and lack recognized protein domains (Fig. 1f). Notably, all Wolbachia strains that contain type II homologs contain at least one pair of intact type I homologs. Type III genes possess WD0631 homologs with a weakly predicted cytochrome C552 domain involved in nitrate reduction (wNo, NCBI conserved domain E-value 3.79e-03), while type III WD0632 homologs contain weak homology to the PD-(D/E)XK nuclease superfamily (wNo, NCBI conserved domain E-value 1.15e-03) and to a transmembrane domain predicted by the transmembrane hidden Markov model11 (Fig. 1f). Finally, a putative type IV group encoding a C-terminal PD-(D/E)XK nuclease superfamily (NCBI conserved domain E-value 3.69e-03) was identified in Wolbachia strains wPip and wAlbB, but not included in phylogenetic analyses because the WD0632 homologs are highly divergent (28% identity across 17% of the protein) and do not appear in reciprocal BLASTp analyses. The functions of type III and IV domains are not well understood, but a homolog of the putative nuclease domain was previously found in a selfish genetic element that mediates embryonic lethality in Tribolium beetles12. Uncertain annotations and substantial unknown sequence in these genes necessitate caution in extrapolating definitive gene functions. Importantly, the region of shared homology among the WD0632-like proteins (Fig. 1f) is outside the putative C-terminal Peptidase_C48 domain, suggesting that the unannotated regions could represent an ancestral CI sequence core that warrants closer inspection.
Consistent with a role in CI, the degree of relatedness and presence/absence of homologs of WD0631 and WD0632 between Wolbachia strains correlates with known patterns of bidirectional incompatibility (Fig. 1d). Among the strains wRi, wHa, and wNo, only wRi rescues wMel-induced CI in same-species crosses13,14. We postulate that this is due to wRi and wMel sharing highly related type I homologs (99% amino acid identity), and thus likely sharing a rescue factor, while wRi also has a type II homolog that may explain its ability to induce CI against wMel. Meanwhile, bidirectionally incompatible pairs are highly divergent, with only 29–68% amino acid identity (Extended Data Fig. 3a). Additionally, variation in CI strength between strains correlates with the number of copies of the WD0631/WD0632 pair (Extended Data Fig. 3b). Strains with only one copy, such as wMel, have a comparatively weak CI phenotype, while those with two or three copies, such as wRi and wHa, cause strong CI14.
Given the various lines of evidence that associate these two genes with CI, we next examined the functional role of WD0631 and WD0632 in CI. For comparison, the following control genes were also used: WD0034, which is predicted to encode a PAZ (Piwi, Argonaut, and Zwille) domain containing protein (NCBI conserved domain E-value 1.85e–18), WD0508, a prophage gene annotated as a putative transcriptional regulator with two helix-turn-helix domains (NCBI conserved domain E-value 9.29e-12) in the Octomom genomic region, and WD0625, a prophage gene annotated as a DUF2466 with a JAB1/MPN/Mov34 metalloenzyme (JAMM) domain (NCBI conserved domain E-value 1.60e-41). We first examined the expression of these genes in the testes of wMel-infected, one-day-old and seven-day-old D. melanogaster males. Since CI strength decreases significantly in aged males15, we predicted that a CI factor would be expressed at a lower level in seven-day-old males versus one-day-old males that both emerged on day one of the cross. Indeed, WD0631 and WD0632 show a significantly lower transcription level in aged males (Fig 2). Moreover, WD0631 exhibits 18.6- and 83.0-fold higher expression than WD0632 for young and aged males, respectively (Fig. 2). Coupled with RNA-seq expression data16 and operon predictor algorithms, evidence suggests that these genes are not generally acting as an operon in wMel. Both prophage-encoded control genes, WD0508 and WD0625, also exhibited this age-dependent expression pattern, but the non-prophage gene WD0034 did not (Fig. 2). WD0640, which encodes prophage WO structural protein gpW, was also reduced in older males, suggesting that prophage genes in general are relatively downregulated in seven-day-old testes (Fig 2). The phenomenon of decreased CI in older males is not due to decreases in Wolbachia titer over time, as the copy number of Wolbachia groEL relative to D. melanogaster rp49 increases as males age, and there is no significant difference in absolute Wolbachia gene copies between one-day-old and seven-day-old males (Extended Data Fig. 4a,b). Since CI expression also correlates with male development time, we examined gene expression in early emerging “older brothers” (emerged on day one) and later emerging “younger brothers” (emerged on day five). Expression was statistically equivalent for WD0631 (Extended Data Fig. 4c), and slightly reduced in younger brothers for WD0632 (Extended Data Fig. 4d). These results are consistent with a small younger brother effect17, though we did not observe a statistically significant effect on CI penetrance (Extended Data Fig. 4e).
Figure 2. Relative expression of CI candidate and prophage WO genes decreases as males age.
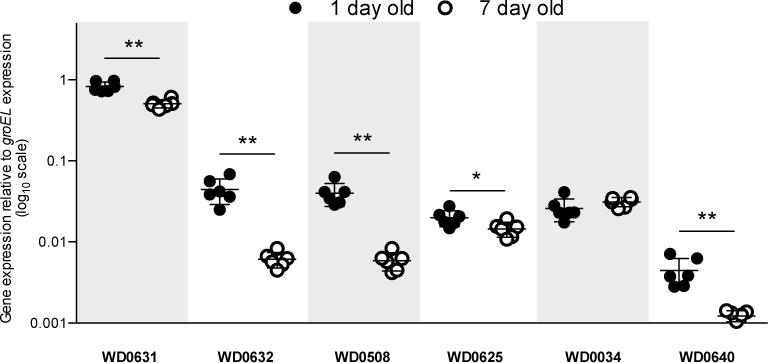
RNA expression in one-day-old (1d) versus seven-day-old (7d) testes, normalized to expression of groEL in wMel-infected D. melanogaster testes from the fastest developing males. Values denote 2−(Delta Ct). Bars indicate mean and standard deviation. * = P<0.05, ** = P<0.01 by Mann-Whitney U test. This experiment has been performed once. Exact p-values are provided in Supplementary Table 7.
To directly test involvement of these genes in CI, we generated transgenic D. melanogaster expressing candidate genes using an upstream activating sequence (UAS), since Wolbachia itself cannot be genetically transformed. We utilized a nanos-Gal4 driver line for tissue-specific expression in the germline18,19. We assessed CI by measuring the percentage of embryos that hatched into larvae. While wild type (WT) CI between infected males and uninfected females led to significantly reduced hatch rates, expressing each of four candidate transgenes in uninfected (fastest-developing, day one) males did not affect hatch rates when crossed to uninfected females (Fig. 3a, Extended Data Fig. 5a). These genes also had no effect on sex ratios (Extended Data Fig. 5b, 6a). There are no phenotypic effects despite confirmed expression of each transgene in the testes (Extended Data Fig. 7a–d).
Figure 3. Dual expression of WD0631 (cifA) and WD0632 (cifB) is necessary to induce CI-like defects.
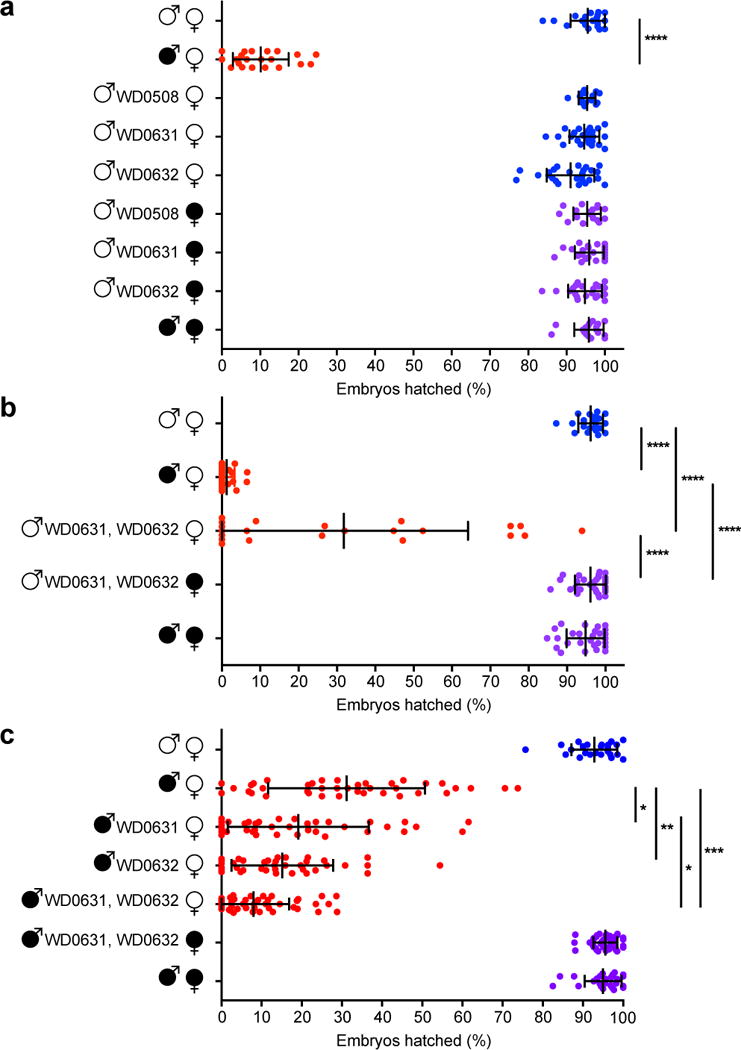
Hatch rate assays used the fastest developing males that were aged either (a, b) one-day or (c) two- to four-days in parental crosses. Parental infection status is designated with filled symbols for wMel-infected parents or open symbols for uninfected parents. Transgenic flies are labeled with their transgene to the right of their male/female symbol. Unlabeled symbols represent WT flies. Data points are colored according to the type of cross, with blue indicating no CI, red for CI crosses, and purple for rescue crosses with wMel-infected females. Bars indicate mean and standard deviation. * = P<0.05, ** = P<0.01, *** = P<0.001, **** = P<0.0001 by ANOVA with Kruskal-Wallis test and Dunn’s multiple test correction. Statistical comparisons are between all groups (panels a and b); or between CI crosses only (panel c). All experiments were performed at least twice, except for the increase of wild-type CI by WD0631, which has been performed once. Exact p-values are provided in Supplementary Table 7.
As WD0631 and WD0632 are adjacent, coevolving genes, we reasoned that dual expression of WD0631 and WD0632 might be required to induce CI. Indeed, expression of both transgenes in the same males significantly reduced hatch rates by 68% compared to uninfected WT crosses (Fig. 3b), with no effect on sex ratios (Extended Data Fig. 6b). Roughly half of the crosses with transgenic males yielded hatch rates within the range observed in WT CI (3.8 ± 5.6% hatch rate). Interestingly, there was a strong positive correlation between hatch rate and clutch size when both transgenes were expressed (rs=0.7; p=0.0003), but not in WT CI, suggesting that dilution of transgene products across larger clutches may explain variation in transgene-induced CI. It is also possible that full transgene induction of CI requires other factors, or that transgenes are not expressed at the ideal time or place for complete CI, though transgene expression in adult testes was confirmed (Extended Data Fig. 7c,d). Importantly, transgene-induced lethality is fully rescued by wMel-infected females (Fig. 3b), indicating that these genes produce probable CI factors rather than artifacts that reduce hatch rates through off target effects that would not be rescued. We therefore name these genes cytoplasmic incompatibility factors, cifA and cifB, for WD0631 and WD0632, respectively. Type II, III, and IV homologs are designated cif-like until experimental evidence demonstrates they recapitulate CI.
To test if WD0631 and WD0632 transgenes act additively with Wolbachia to enhance WT CI levels, wMel-infected male flies expressing either transgene were aged two to four days to lower WT CI penetrance before crossing with uninfected females. In support of transgene-induced enhancement of CI, hatch rates in these aged males decreased significantly compared to WT CI crosses of the same age (Fig. 3c), with no effect on sex ratios (Extended Data Fig. 6c). In this context, wherein aged flies cause a weaker level of WT CI, the transgenes appear to add to the quantity of CI effectors in wMel-infected tissues, causing stronger CI overall. This effect was not observed when control transgenes WD508 or WD0625 were expressed individually in wMel-infected males (Extended Data Fig. 8a,b). Dual expression of WD0631 and WD0632 in wMel-infected flies reduced hatch rates further than either gene alone, yet was still fully rescued by wMel-infected females (Fig. 3c). Adding WD0625 to WD0632 in aged wMel-infected males did not increase CI beyond WD0632 alone (Extended Data Fig. 8b), and had no effect on hatch rates in one-day-old uninfected males (Extended Data Fig. 8c). Finally, none of these gene combinations affected offspring sex ratios (Extended Data Fig. 9). Taken together, these findings support the conclusion that both WD0631 and WD0632 are necessary to induce the CI phenotype, and that they do not represent an artifact of the transgenic system.
To rule out the possibility that transgene-induced enhancement of CI in infected lines is due to increased Wolbachia titers, we quantitated amplicons of single copy genes from Wolbachia and D. melanogaster. Although there were some differences in Wolbachia titers between infected transgenic lines (Extended Data Fig. 10a–c), the variation did not correlate with induction or magnitude of CI, signifying that decreased offspring viability was due to the direct effect of transgenes rather than changes in Wolbachia density. Most notably, densities significantly increased in infected flies expressing the control Octomom transgene WD0508 (Extended Data Fig. 10a) that did not enhance CI (Extended Data Fig. 8a).
Next we tested if transgene-induced CI associates with canonical cytological defects observed in Wolbachia-induced CI. Although CI is typically thought to cause failure of the first mitotic division20,21, nearly half of the embryonic arrest in incompatible crosses occurs during advanced developmental stages in Drosophila simulans22,23, Aedes polinesiensis24 and Culex pipiens25. We examined embryos from control and experimental crosses after one to two hours of development and binned their cytology into one of six phenotypes. While a few embryos in each cross were unfertilized (Fig. 4a), most embryos in compatible crosses were either in normal late-stage preblastoderm (Fig. 4b) or syncytial blastoderm stages (Fig. 4c)26. In WT CI, embryos exhibited three defects: arrest of cellular division after two to three mitotic divisions (Fig. 4d), later stage arrest associated with chromatin bridging, as is classically associated with strong CI in D. simulans21 (Fig. 4e), or arrest associated with regional failure of division in one embryo region (Fig. 4f). After blindly scoring embryo cytology, we determined that arrest phenotypes (d, e, and f) were significantly more common in the offspring of dual WD0631/WD0632 transgenic males mated to uninfected females, but these abnormalities were rescued in embryos from wMel-infected females (Fig. 4g). These effects were not seen with control gene WD0508 or with singular expression of WD0631 or WD0632 (Fig. 4h). These data again validate that transgene-induced CI, as measured through cytological defects, recapitulates WT CI. Most of the embryos arrest after two to three mitotic divisions.
Figure 4. Dual expression of WD0631 (cifA) and WD0632 (cifB) recapitulates CI-associated embryonic defects.
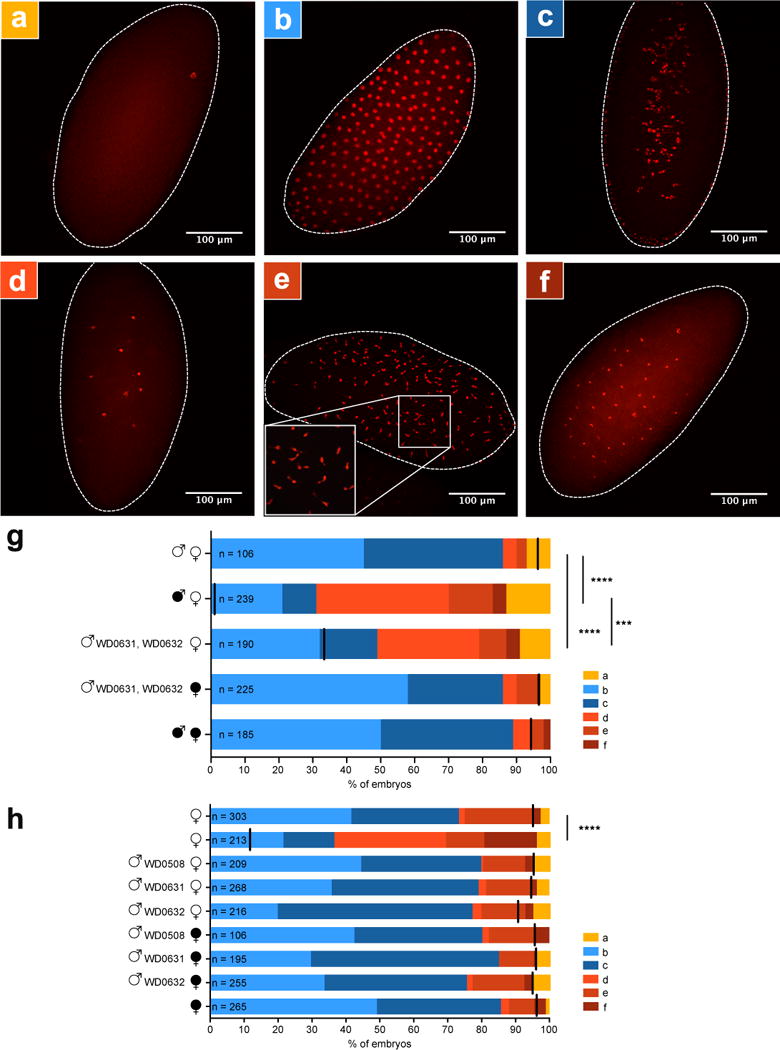
Representative embryo cytology is shown for (a) unfertilized embryos, (b) normal multi-nucleated embryos at one hour of development, (c) normal embryos near two hours of development in which nuclei begin to migrate to the periphery of the cytoplasm, and three different mitotic abnormalities: (d) failure of nuclear division after two to three mitoses, (e) chromatin bridging, and (f) regional mitotic failure. (g) The number of embryos with each cytological phenotype resulting from the indicated crosses. Infection status is designated with filled symbols for wMel-infected parents or open symbols for uninfected parents. Transgenic flies are labeled with their transgene to the right of their male/female symbol. Unlabeled symbols represent WT flies. Black lines on each graph indicates mean hatch rate for the cross. *** = P<0.001, **** = P<0.0001 by two-tailed Fisher’s exact test comparing normal (phenotypes b and c) to abnormal (phenotypes a, d, e, and f) for each cross. (h) Quantitation of cytological defects in control crosses. Cytology for (g) has been performed twice and cytology for (h) has been performed once. Exact p-values are provided in Supplementary Table 7.
This study identifies, for the first time, two differentially transcribed and codiverging prophage WO genes that recapitulate and enhance CI. These rapidly evolving genes are not chromosomal Wolbachia genes per se, but rather occur widely in the eukaryotic association module of prophage WO8. This module notably contains genes with amino acid sequence homologous to eukaryotes or annotated to interact with animal cells, though WD0631 and WD0632 do not appear to have eukaryotic homology. CI can therefore be categorized as a prophage WO-induced phenotype rather than a Wolbachia-induced phenotype. We name the genes and close homologs “cytoplasmic incompatibility factors”, cifA and cifB, for WD0631 and WD0632, respectively. The cif name is conservatively grounded in phenotype and makes no assumptions regarding mechanism, which is notable because there are unannotated core regions throughout the cif genes that could have as much bearing on mechanism as the annotated domains.
The discovery of cifA and cifB genes that functionally recapitulate and enhance CI is the first inroad in solving the genetic basis of reproductive parasitism, a phenomenon induced worldwide in potentially millions of arthropod species27. These prophage WO genes have implications for microbe-assisted speciation, because they likely underlie CI-induced hybrid lethality observed between closely related species of Nasonia and Drosophila28,29. Finally, cifA and cifB are important for arthropod pest and vector control strategies, as they could be an alternative or adjunct to current Wolbachia-based paradigms aimed at controlling agricultural pests or curbing arthropod-borne transmission of infectious diseases4–7.
METHODS
Comparative genomics and transcriptomics
MicroScope30 was used to select the set of genes comprising the core genomes of CI-inducing Wolbachia strains wMel [NC_002978.6]31, wRi [NC_012416.1]32, wPip (Pel) [NC_010981.1]33, and the recently sequenced wRec [GCA_000742435.1]34, while excluding the pan-genome of the mutualistic strain wBm [NC_006833.1]35, using cutoffs of 50% amino acid identity and 80% alignment coverage. For the “absent in wAu” criterion, wAu microarray data were obtained from the original authors9 and genes that were present in CI-inducing strains wRi and wSim but absent or divergent in the non-CI strain wAu were selected.
For ovarian transcriptomics, one-day old females from wVitA infected-Nasonia vitripennis 12.1 were hosted as virgins on Sarcophaga bullata pupae36 for 48 hours to stimulate feeding and oogenesis. Females were then dissected in RNase-free 1× PBS buffer, and their ovaries were immediately transferred to RNase-free microcentrifuge tubes in liquid nitrogen. Fifty ovaries were pooled for each of three biological replicates. Ovaries were manually homogenized with RNase-free pestles, and their RNA was extracted using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol for purification of total RNA from animal tissues. After RNA purification, samples were treated with RQ1 RNase-free DNase (Promega), and ethanol precipitation was performed. PCR of RNA samples with Nasonia primers NvS6KQTF4 and NVS6KQTR437 confirmed that all samples were free of DNA contamination. RNA concentrations were measured with a Qubit 2.0 Fluorometer (Life Technologies) using the RNA HS Assay kit (Life Technologies), and approximately 5 μg of total RNA from each sample was used as input for the MICROBEnrich Kit (Ambion) in order to enrich for Wolbachia RNA in the samples. Bacterial-enriched RNA was then ethanol-precipitated, and rRNA was depleted from the samples using the Ribo-Zero Magnetic kit (Illumina) according to manufacturer’s protocol. Approximately 1.5 μg of enriched, rRNA-depleted RNA for each replicate was shipped to the University of Rochester Genomics Research Center for sequencing. Library preparation was performed using the Illumina ScriptSeq v2 RNA-Seq Library Preparation kit, and all samples were run multiplexed on a single lane of the Illumina HiSeq2500 (single-end, 100 bp reads). Raw sequence reads were trimmed and mapped to the wVitA genome (PRJNA213627) in CLC Genomics Workbench 8.5.1 using a minimum length fraction of 0.9, a minimum similarity fraction of 0.8, and allowing one gene hit per read. With all three replicates combined, a total of 364,765 reads out of 41,894,651 (0.87%) mapped to the wVitA genome with the remaining reads mapping to the N. vitripennis host genome (GCF_000002325.3). All Wolbachia genes with greater than or equal to five RNA-seq reads, with the exception of the 16S and 23S RNA genes, were selected. For non-wMel data sets, the closest homologs in wMel were found using BLASTp in Geneious Pro v5.5.638.
Protein extraction and mass spectrometry
Protein was extracted from Culex pipiens tissues as described previously10. Ovaries from 30 wPip (Buckeye)-infected mosquitoes were dissected in 100% ethanol and collected in a 1.5 ml tube filled with 100% ethanol. Pooled tissues were sonicated at 40 mA for 10 seconds in a Kontes GE 70.1 ultrasonic processor, and trichloroacetic acid (TCA) was added to a final concentration of 10% (v/v). After centrifugation at 13,000 rpm in a microcentrifuge, pellets were washed with acetone:water (9:1), dried, and stored at −20°C. Samples were directly submitted to the University of Minnesota’s Center for Mass Spectrometry and Proteomics for iTRAQ (isobaric tagging for relative and absolute quantification) analysis. Proteins were sorted according to their relative abundance as determined by the number of spectra from the single most abundant peptide. Because proteins can often produce varying amounts of detectable tryptic peptides depending upon protein size and lysine/arginine content, we counted only the single most abundant peptide for each protein. This quantification is justified by a previous report39 showing that the two most abundant proteins are the Wolbachia surface protein (WSP; WP_007302328.1) and another putative membrane protein (WP0576; WP_012481859.1). Only proteins with at least three unique peptides (95% confidence) detected were reported, and using this criterion the false discovery rate was zero.
Evolutionary analyses
WD0631 (NCBI accession number AAS14330.1) and WD0632 (AAS14331.1) from wMel were used as queries to perform a BLASTp search of NCBI’s nonredundant (nr) protein sequence database with algorithm parameters based on a word-size of six and BLOSUM62 scoring matrix40. Homologs were selected based on the satisfaction of three criteria: (i) E-value ≤ 10−20, (ii) query coverage greater than 60%, and (iii) presence in fully sequenced Wolbachia and/or phage WO genomes. FtsZ and gpW proteins were identified for all representative Wolbachia and phage WO genomes, respectively. Protein alignments were performed using the MUSCLE plugin41 in Geneious Pro v8.1.738; the best models of evolution, according to the corrected Akaike Information Criteria (AICc)42, were estimated using the ProtTest server43; and phylogenetic trees were built using the MrBayes plugin44 in Geneious. Putative functional domains were identified using NCBI’s BLASTp, Wellcome Trust Sanger Institute’s PFAM database45, a transmembrane hidden Markov model11 and EMBL’s Simple Modular Architecture Research Tool (SMART)46. WD0631/WD0632 protein homology (% aa identity) was based on a 1:1 BLASTp analysis for each pair. Prophage/WO-like island association for each pair of genes was based on prophage regions identified in a previous study8.
Gene expression assays and Wolbachia titers
For the male age effect, native expression of CI candidates was tested with RT-qPCR on replicate pools of 20 pairs of testes from the fastest developing, virgin males that were aged one day or seven days. RNA was extracted with the Qiagen RNeasy mini kit, DNase treated with TURBO DNase (Life Technologies) and cDNA was generated with Superscript III Reverse Transcriptase (Invitrogen). Primer sequences are listed in Supplementary Table 6. Quantitative PCR was performed on a Bio-Rad CFX-96 Real-Time System using iTaq Universal SYBR Green Supermix (Bio-Rad). 30 cycles of PCR were performed against positive controls (extracted DNA), negative controls (water), RNA, and cDNA with the following conditions: 95°C 2 min, 30× (95°C 15 sec, 56°C 30 sec, 72°C 30 sec), 72°C 5 min. Delta Ct values between the target gene and housekeeping gene groEL were used to determine relative gene expression. These experiments were performed once with multiple replicates for each condition.
For experiments on the younger brother effect, replicate pools of 20 pairs of testes were collected from the fastest developing virgin males that emerged on the first day (older brothers) or fifth day (younger brothers). Male siblings for the younger brother effect analysis were also collected concurrently for hatch rates described in the Hatch Rate Assays section by crossing the wMel-infected males to 3–5 day old wMel-infected or uninfected females. RNA was extracted using the Direct-zol RNA MiniPrep Kit (Zymo), DNase treated with DNA-free (Ambion, Life Technologies), cDNA was generated with SuperScript VILO (Invitrogen), and RT-qPCR was run using iTaq Universal SYBR Green (Bio-Rad). Primers, PCR conditions, and analysis are the same as for the male age effect above. These experiments have been performed once with multiple replicates for each condition.
For gene expression in Extended Data Figure 7, six pools of six pairs of testes were dissected from parents utilized in hatch rate assays from a repeat of Figure 3a and Extended Figure 5. In samples designated “High CI” and “No CI”, the males correspond to crosses that had lower or normal hatch rates, respectively. For all other samples, the flies utilized were chosen at random. RNA was extracted using the same method as the younger brother experiment above. 30 cycles of PCR were performed against positive controls (extracted DNA), negative controls (water), RNA, and cDNA with PCR conditions described above. Gel image size and brightness were adjusted in some cases for clarity. These experiments have been performed once.
For the Wolbachia titers, pools of testes were dissected from 15 males in ice-cold PBS. For Extended Data Fig. 10a–c, brothers of those used in the corresponding hatch rates were utilized. DNA was extracted using the Gentra Puregene Tissue kit (Qiagen). qPCR was done as described above. Absolute quantification was achieved by comparing all experimental samples to a standard curve generated on the same plate. Primers are listed in the Supplementary Table 6. qPCR conditions: 50°C 10 min, 95°C 5 min, 40× (95°C 10 sec, 55°C 30 sec), 95°C 30 sec. To obtain a more accurate Wolbachia:host cell ratio, it was assumed that each host cell has two copies of rp49 and each Wolbachia cell has one copy of groEL. These experiments have been performed once but with a sample size of eight for each condition.
Fly rearing
D. melanogaster were reared on a standard cornmeal and molasses based media. Stocks were maintained at 25°C while virgin flies were stored at room temperature. During virgin collections, stocks were kept at 18°C overnight and 25°C during the day. All flies were kept on a 12-hour light/dark cycle. Wolbachia uninfected lines were generated through tetracycline treatment for three generations. Briefly, tetracycline was dissolved in ethanol and then diluted in water to a final concentration of 1mg/mL. 1mL of this solution was added to 50mL of media (final concentration of 20ug/mL). Freshly treated media was used for each generation. Infection status was confirmed with PCR using Wolb_F and Wolb_R3 primers47, and flies were reared on untreated media for at least three additional generations to allow for mitochondrial recovery before being utilized48.
Transgenic flies
Each CI candidate gene was cloned into the pTIGER plasmid for transformation and expression in D. melanogaster49. pTIGER, a pUASp-based vector designed for germline expression, exhibits targeted integration into the D. melanogaster genome using PhiC31 integrase50 and tissue-specific, inducible expression through the Gal4-UAS system51. Cloning was performed using standard molecular biology techniques and plasmids were purified and Sanger-sequenced for confirmation before injection. At least 200 D. melanogaster embryos were injected per gene by Best Gene, Inc (Chino Hills, CA), and transformants were selected based on w+ eye color. All transgenic lines were made in the yw D. melanogaster background, and each was an isofemale line derived from the offspring of a single transformant. Homozygous lines were maintained when possible, or heterozygous flies were maintained when homozygous transgenics were inviable (WD0625/CyO). WD0508 and WD0631 insertion was carried out with the y1 M{vas-int.Dm}ZH-2A w*; P{CaryP}attP40 line. WD0625 was inserted into BSC9723 with the genotype: y1 M{vas-int.Dm}ZH-2A w*; PBac{y+-attP-3B}VK00002. WD0632 insertion was done using BSC8622 with the genotype: y1 w67c23; P{CaryP}attP2.
Hatch rate and sex ratio assays
Parental females were either infected or uninfected y1w* flies (wMel-infected or uninfected) and aged for 2–6 days before crossing. Uninfected y1w* flies were generated as described for transgenic lines. Parental males were created by crossing nanos-Gal4 virgin females (wMel-infected or uninfected) with either y1w* or UAS-candidate gene-transgenic, uninfected males. Only the first males emerging between 0–30 hours from these crosses were used in CI assays to control for the younger-brother effect associated with CI17. To test if CI can be increased by transgenes, virgin, day one males were aged for 2–4 days before crossing to reduce the level of WT CI. Within experiments, care was taken to match the age of males between experimental and control crosses. 32–64 individual crosses were used for each crossing condition. The flies used were chosen at random from the desired group based on age, sex, and genotype. These sample sizes are based on previous studies of CI in D. melanogaster that detected significant differences between treatment groups52.
To perform the hatch rate assays, a male and female pair was placed in an 8oz, round bottom, polypropylene Drosophila stock bottle. A grape juice-agar plate with a small amount of yeast mix smeared on top was placed in the bottle opening and affixed with tape. To create grape juice-agar plates, 12.5g of agar is mixed in 350mL of deionized water and autoclaved. In a separate flask, 10mL of ethanol is used to dissolve 0.25g tegosept (methyl 4-hyrdoxybenzoate). 150mL of Welch’s grape juice is added to the tegosept mix, combined with the agar, and poured into lids from 35×10mm culture dishes (CytoOne).
Hatch rate bottles were placed in a 25°C incubator overnight (~16 hours). After this initial incubation the grape plates were discarded and replaced with freshly yeasted plates. After an additional 24 hours, the adult flies were removed and frozen for expression analysis and the embryos on each plate were counted. The counting was not blinded. These plates were then incubated at 25°C for 36 hours before the number of unhatched embryos were counted. Larvae from each pair of flies were moved from these plates using a probe and placed in vials of standard fly media with one vial being used for each individual grape plate to be assayed for sex ratios at adulthood. A total of 10–20 vials were used for each cross type. Any crosses with fewer than 25 embryos laid were discarded from the hatching analysis while vials with fewer than 10 adults emerging were discarded from the sex ratio analysis. Statistical analysis and outlier removal, utilizing the ROUT method, were performed using Graphpad Prism v6 software.
Embryo cytology
Embryos were collected in a fashion similar to hatch rate assays except bottles contained 60–80 females and 15–20 males. All flies used were brothers and sisters of those used during corresponding hatch rates. Embryo collections and hatch rates were performed side-by-side. After initial mating overnight, fresh grape plates with yeast were provided and were removed after 60 minutes. The embryo-covered plates were then placed in the incubator at 25°C for a further 60 minutes to ensure each embryo was at least 1–2 hours old. Embryos were then moved to a small mesh basket and dechorionated in 50% bleach for 1–3 minutes. These were then washed in embryo wash solution (0.7% NaCl, 0.05% Triton X-100) and moved to a small vial containing ~2 mL heptane. An equal amount of methanol was added to the vial and then vigorously shaken for 15 seconds. After the embryos settled, the upper heptane layer and as much methanol as possible were removed, and the embryos were moved into ~500 uL fresh methanol in a 1.5 mL microcentrifuge tube. Embryos were stored overnight at 4°C. The old methanol was then removed and replaced with 250 uL of fresh methanol along with 750 uL of PBTA (1× PBS, 1% BSA, 0.05% Triton X-100, 0.02% sodium azide). After inverting the tube several times, the solution was removed and replaced with 500 uL PBTA. Embryos were then rehydrated for 15 minutes on a rotator at room temperature. After rehydrating, the PBTA was replaced with 100 uL of a 10 mg/mL RNase A (Clontech Labs, Inc) solution and incubated at 37°C for 2 hours. The RNase was then removed and embryos were washed several times with PBS followed by a final wash with PBS-Azide (1× PBS, 0.02% sodium azide). After removing the PBS-Azide, embryos were mounted on glass slides with ProLong Diamond Antifade (Life Technologies) spiked with propidium iodide (Sigma-Aldrich) to a final concentration of 1 μg/mL. Imaging was performed at the Vanderbilt Cell Imaging Shared Resource using a Zeiss LSM 510 META inverted confocal microscope. All scores were performed blind (researcher was not aware of which slide represented which cross) and image analysis was done using ImageJ software53. Matched scoring, where embryos are derived from a side-by-side hatch rate, has been performed once for conditions shown in Figure 4h and twice for 4g.
Statistical analyses
All statistical analyses were performed using GraphPad Prism software (either Prism 6 or online tools). When comparing gene expression levels or Wolbachia titers between two sets of data, we used a two-tailed, non-parametric Mann-Whitney U test since it does not require a normal distribution of the data. For comparisons between more than two data sets, we used the non-parametric Kruskal-Wallis one-way analysis of variance test that, if significant, was followed by a Dunn’s test of multiple comparisons. This allowed for robust testing between all data groups while avoiding multiple test bias. For the cytology studies, embryos were classified as either “normal” or “CI-like” in a 2×2 contingency table, and statistical differences between the groups were calculated using a Fisher’s Exact Test.
Data availability
wVitA transcriptome data is deposited in the Sequence Reads Archives with Bioproject PRJNA319204 and BioSample SAMN04881412. wPip-infected ovarian proteome data was deposited at the Proteome Xchange Consortium via the PRIDE54 partner repository with the dataset identifier PXD004047. All other source data is available as supplemental information with this publication.
Extended Data
Extended Data Figure 1. CI and the evolution of Wolbachia and prophage WO genes.
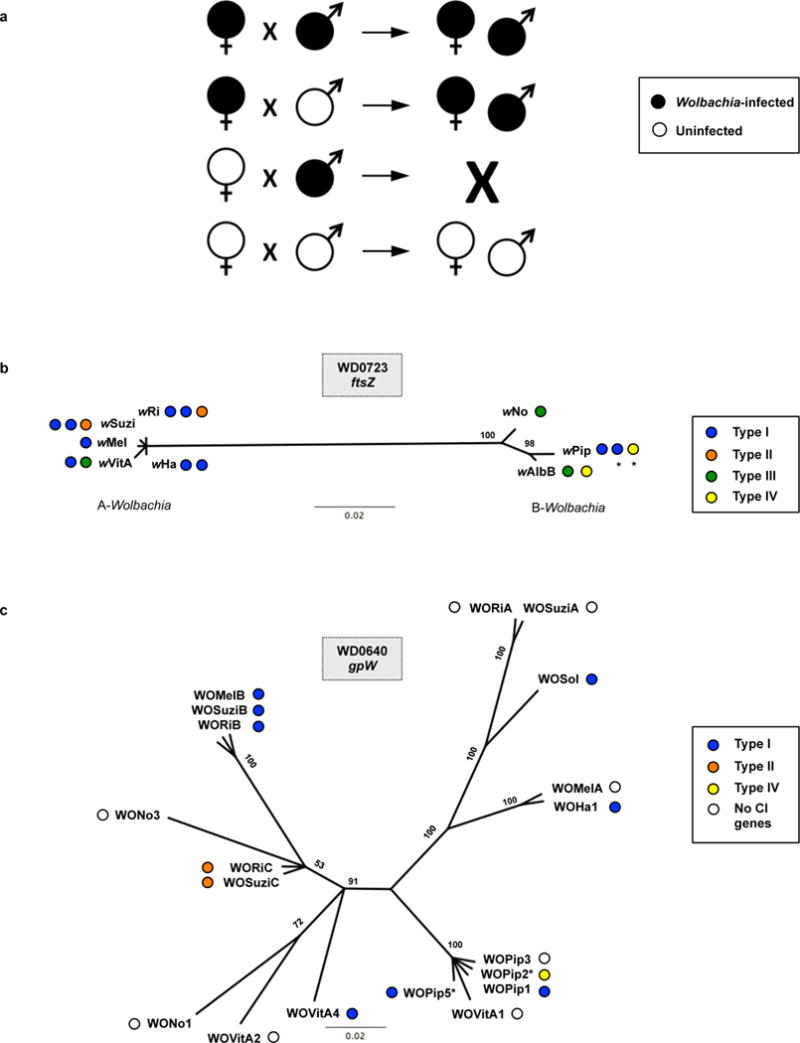
(a) Diagram shows the effect of parental Wolbachia infection on progeny viability and infection status. CI (embryonic inviability) occurs in crosses between Wolbachia-infected males and uninfected females. Wolbachia-infected females mated to infected males rescue the inviability. (b) Bayesian phylogenies based on a 393-aa alignment of WD0723, the wMel ftsZ gene, and its homologs and (c) a 70-aa alignment of WD0640, the phage WO gpW gene, and its homologs. Trees are based on JTT+G and CpRev+I models of evolution, respectively, and are unrooted. Consensus support values are shown at the nodes. (*) indicates that the CI genes are not included in Figure 1. The WOPip5 homolog is truncated while the WOPip2 and second wAlbB homologs are highly divergent from WD0632.
Extended Data Figure 2. WD0631/WD0632 homologs always associate with the eukaryotic association module in prophage WO regions.

CI gene homologs are labeled and colored pink. Structural modules are labeled as baseplate, head or tail. The WD0611-WD0621 label highlights a conserved gene cluster that is often associated with the CI genes. Only one phage haplotype is shown per Wolbachia strain when multiple copies of the same type are present.
Extended Data Figure 3. Wolbachia CI patterns correlate with WD0631/WD0632 homolog similarity and copy number.

(a) The % amino acid (aa) identity between each WD0631/WD0632 homolog correlates with Wolbachia compatibility patterns. The only compatible cross, wMel males × wRi females, features close homology between WOMelB and WORiB. All other crosses are greater than 30% divergent and are bidirectionally incompatible. Each “% aa identity” value is based on the region of query coverage in a 1:1 BLASTp analysis. (b) CI strength, protein architecture and clade type are listed for each of the Wolbachia strains shown in Figure 1d. (*) indicates the proteins are disrupted and not included in comparison analyses.
Extended Data Figure 4. Wolbachia titers, the male age effect, and the younger brother effect.

(a) Relative Wolbachia titers in wild type lines do not decrease with age. DNA copy number of the wMel groEL gene is shown normalized to D. melanogaster rp49 gene copy number in testes at the indicated ages. (b) Absolute Wolbachia titers do not decrease with male age. (c, d) In wMel-infected males, WD0631 gene expression is equal between older (first day of emergence) and younger brothers (fifth day of emergence) while WD0632 gene expression is slightly higher in early emerging brothers. (e) There is no statistical difference in CI penetrance between older and younger brothers. Bars show mean and standard deviation. * = P<0.05, *** = P<0.001, **** = P<0.0001 by ANOVA with Kruskal-Wallis test and Dunn’s multiple test correction for (a), (b), and (e), and two-tailed Mann-Whitney U test used for (c), and (d). Exact p-values are provided in Supplementary Table 7. These experiments have been performed once.
Extended Data Figure 5. WD0625 transgene expression does not induce CI-like defects.
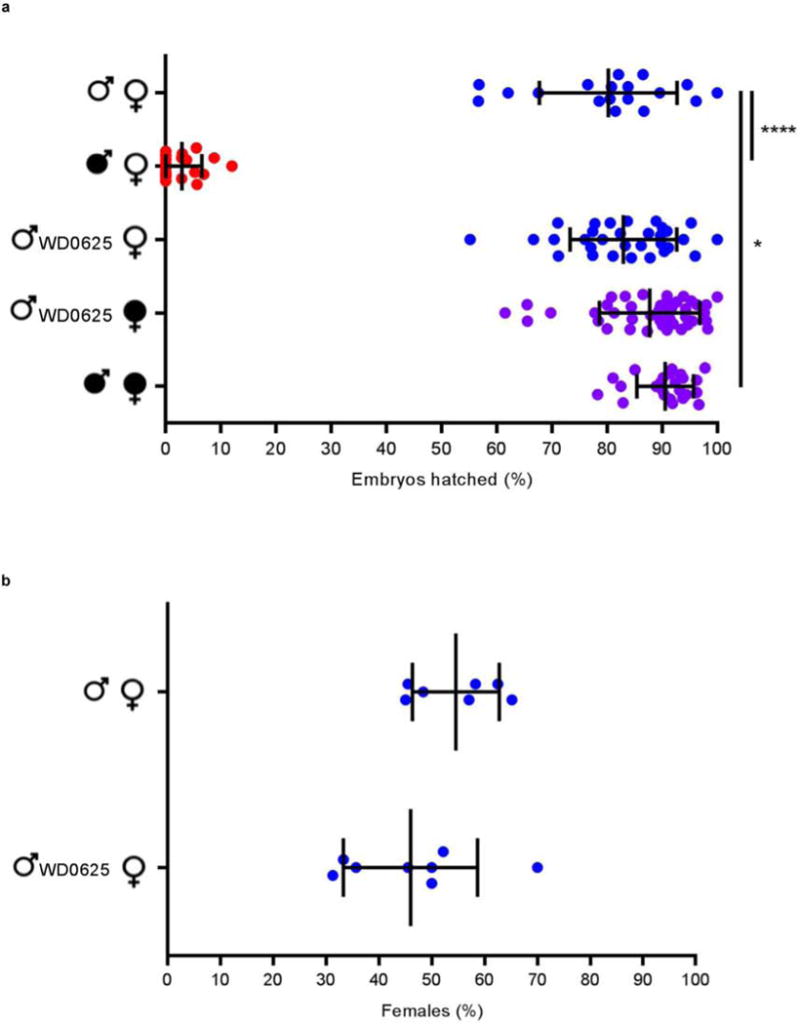
Expression of control gene WD0625 in one-day-old uninfected males does not affect (a) embryo hatch rates or (b) sex ratios. Infection status is designated with filled in symbols for a wMel-infected parent or open symbols for an uninfected parent. Transgenic flies are labeled with their transgene to the right of their male/female symbol. Unlabeled symbols represent wild type flies. Data points are colored according to the type of cross, with blue indicating no CI, red indicating a CI cross, and purple indicating a rescue cross with wMel-infected females. Bars indicate mean and standard deviation. * = P<0.05, *** = P<0.001 by ANOVA with Kruskal-Wallis test and Dunn’s multiple test correction. Exact p-values are provided in Supplementary Table 7. This experiment has been replicated three times.
Extended Data Figure 6. Expression of transgenes does not alter sex ratios.

Graphs correspond to the same crosses as Figure 3. Infection status is designated with filled in symbols for a wMel-infected parent or open symbols for an uninfected parent. Transgenic flies are labeled with their transgene to the right of their gender symbol. Unlabeled gender symbols represent WT flies. Data points are colored according to the type of cross, with blue indicating no CI, red indicating a CI cross, and purple indicating a rescue cross with wMel-infected females. Bars indicate mean and standard deviation. Statistics include a Kruskal-Wallis tests and Dunn’s multiple test corrections. Extended Data Figures 6a and 6c have been performed once, while Extended Data Figure 6b has been performed twice.
Extended Data Figure 7. Transgenes are expressed in testes.

(a, b) WD0508 and WD0625 transgenes are expressed in testes as evident by PCR performed against cDNA generated from dissected males utilized in Figure 3a and Extended Data Fig. 5a, respectively. (c, d) WD0631 and WD0632 transgenes are expressed in the testes from transgenic males specifically inducing high CI, no CI, or rescued CI. Testes were removed from males used in a replicate of Figure 3b. This experiment has been performed once.
Extended Data Figure 8. Transgenic expression of WD0508, WD0625, and WD0625/WD0632 does not enhance or induce CI.

(a) The WD0508 transgene alone does not enhance CI in two- to four-day-old infected males. (b) The WD0625 transgene alone does not enhance CI either; conversely, WD0632 does enhance CI as previously shown in Figure 3C. The WD0625 transgene together with WD0632 does not enhance CI further than WD0632 alone. (c) WD0625/WD0632 dual expression cannot induce CI in uninfected one-day-old males. Infection status is designated with filled in symbols for a wMel-infected parent or open symbols for an uninfected parent. Transgenic flies are labeled with their transgene to the right of their male/female symbol. Unlabeled symbols represent WT flies. Data points are colored according to the type of cross, with blue indicating no CI, red indicating a CI cross, and purple indicating a rescue cross with wMel-infected females. Bars indicate mean and standard deviation. ** = P<0.01, *** = P<0.001, **** = P<0.0001 by ANOVA with Kruskal-Wallis test and Dunn’s multiple test correction. Exact p-values are provided in Supplementary Table 7. These experiments have been done twice (8a, 8c), three times (8b, WD0625, WD0632), or once (8b, WD0625/WD0632).
Extended Data Figure 9. Transgenic expression of control genes does not affect sex ratios.

All flies are from same crosses shown in Extended Data Figure 8, except for 9c which comes from a replicate experiment. Infection status is designated with filled in symbols for a wMel-infected parent or open symbols for an uninfected parent. Transgenic flies are labeled with their transgene to the right of their male/female symbol. Unlabeled symbols represent WT flies. Data points are colored according to the type of cross, with blue indicating no CI, red indicating a CI cross, and purple indicating a rescue cross with wMel-infected females. Bars indicate mean and standard deviation. Statistics performed by ANOVA with Kruskal-Wallis test and Dunn’s multiple test correction. These experiments have been done twice (9b) or once (9a,c).
Extended Data Figure 10. There is variation in Wolbachia titers in transgenic lines.
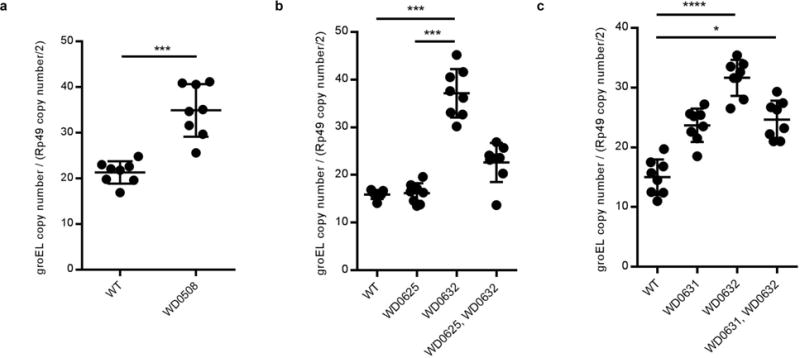
(a–c) Relative Wolbachia titers are higher in WD0508, WD0631, and WD0632 transgenic lines in comparison to wild type lines. This does not occur in the WD0625 transgenic line nor does there appear to be an additive effect. DNA copy number of wMel groEL gene is shown normalized to D. melanogaster rp49 gene copy number in testes of the indicated strains. Bars show mean and standard deviation. * = P<0.05, *** = P<0.001, **** = P<0.0001 for two-tailed Mann-Whitney U test (a) and Kruskal-Wallis test with Dunn’s multiple test correction (b–c). Exact p-values are provided in Supplementary Table 7. These experiments have been done once.
Supplementary Material
Acknowledgments
This work was supported by NIH R21 HD086833 and NSF IOS 1456778 to Seth R.B., NSF DEB-1501398 and NIH 5T32GM008554 training grant support to D.P.L., NIH T32GM07347 training grant support for J.A.M. to the Vanderbilt Medical Scientist Training Program, and NIH AI081322 to A.M.F. Imaging was performed in part through the use of the VUMC Cell Imaging Shared Resource (supported by NIH grants CA68485, DK20593, DK58404, DK59637, and EY08126). We thank Dr. Kristin Jernigan and Paul Snider for help with preliminary studies and Andrew Brooks for assistance with figure preparation.
Footnotes
SUPPLEMENTARY INFORMATION
This manuscript includes 10 extended data figures and 7 Supplementary Tables.
AUTHOR CONTRIBUTIONS
D.P.L. performed gene expression and hatch rate assays, embryo cytology, and assayed for transgene and infection status of flies. J.A.M. performed comparative genomics analyses, generated transgenic flies, and drafted the manuscript. Sarah R.B. performed evolutionary and bioinformatic analyses. J.O. performed hatch rates, assayed sex ratios, collected flies for all experiments, and assayed for transgene and infection status of flies. J.I.P conducted younger brother effect experiments and performed embryo cytology. J.D.S performed hatch rate assays, collected flies for parallel embryo cytology, and assayed for transgene and infection status of flies. E.M.L collected flies and performed hatch rate assays. L.J.F. obtained the wVitA transcriptome. J.F.B. obtained the wPip proteome. Seth R.B. supervised the work and contributed to all experimental designs, data analysis, and data interpretation. All authors participated in manuscript preparation, editing, and final approval.
The authors have no competing financial interests.
References
- 1.Yen JH, Barr AR. New hypothesis of the cause of cytoplasmic incompatibility in Culex pipiens L. Nature. 1971;232:657–658. doi: 10.1038/232657a0. [DOI] [PubMed] [Google Scholar]
- 2.Brucker RM, Bordenstein SR. Speciation by symbiosis. Trends in Ecology and Evolution. 2012;27:443–451. doi: 10.1016/j.tree.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Shropshire JD, Bordenstein SR. Speciation by Symbiosis: the Microbiome and Behavior. MBio. 2016;7 doi: 10.1128/mBio.01785-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Connor L, et al. Open release of male mosquitoes infected with a wolbachia biopesticide: field performance and infection containment. PLoS Negl Trop Dis. 2012;6:e1797. doi: 10.1371/journal.pntd.0001797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker T, et al. The w Mel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature. 2011;476:450–453. doi: 10.1038/nature10355. [DOI] [PubMed] [Google Scholar]
- 6.Dutra Heverton Leandro C, et al. Wolbachia Blocks Currently Circulating Zika Virus Isolates in Brazilian Aedes aegypti Mosquitoes. Cell Host & Microbe. doi: 10.1016/j.chom.2016.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zabalou S, et al. Wolbachia-induced cytoplasmic incompatibility as a means for insect pest population control. Proc Natl Acad Sci U S A. 2004;101:15042–15045. doi: 10.1073/pnas.0403853101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bordenstein SR, Bordenstein SR. Eukaryotic association module in phage WO genomes from Wolbachia. Nat Commun. 2016;7:13155. doi: 10.1038/ncomms13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishmael N, et al. Extensive genomic diversity of closely related Wolbachia strains. Microbiology. 2009;155:2211–2222. doi: 10.1099/mic.0.027581-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beckmann JF, Fallon AM. Detection of the Wolbachia protein WPIP0282 in mosquito spermathecae: implications for cytoplasmic incompatibility. Insect Biochem Mol Biol. 2013;43:867–878. doi: 10.1016/j.ibmb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of Molecular Biology. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 12.Lorenzen MD, et al. The maternal-effect, selfish genetic element Medea is associated with a composite Tc1 transposon. Proc Natl Acad Sci U S A. 2008;105:10085–10089. doi: 10.1073/pnas.0800444105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zabalou S, et al. Multiple rescue factors within a Wolbachia strain. Genetics. 2008;178:2145–2160. doi: 10.1534/genetics.107.086488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poinsot D, Bourtzis K, Markakis G, Savakis C, Mercot H. Wolbachia transfer from Drosophila melanogaster into D. simulans: Host effect and cytoplasmic incompatibility relationships. Genetics. 1998;150:227–237. doi: 10.1093/genetics/150.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reynolds KT, Hoffmann AA. Male age, host effects and the weak expression or non-expression of cytoplasmic incompatibility in Drosophila strains infected by maternally transmitted Wolbachia. Genet Res. 2002;80:79–87. doi: 10.1017/s0016672302005827. [DOI] [PubMed] [Google Scholar]
- 16.Gutzwiller F, et al. Dynamics of Wolbachia pipientis Gene Expression Across the Drosophila melanogaster Life Cycle. G3 (Bethesda) 2015;5:2843–2856. doi: 10.1534/g3.115.021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamada R, Floate KD, Riegler M, O’Neill SL. Male development time influences the strength of Wolbachia-induced cytoplasmic incompatibility expression in Drosophila melanogaster. Genetics. 2007;177:801–808. doi: 10.1534/genetics.106.068486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rorth P. Gal4 in the Drosophila female germline. Mech Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- 19.White-Cooper H. Tissue, cell type and stage-specific ectopic gene expression and RNAi induction in the Drosophila testis. Spermatogenesis. 2012;2:11–22. doi: 10.4161/spmg.19088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serbus LR, Casper-Lindley C, Landmann F, Sullivan W. The genetics and cell biology of Wolbachia-host interactions. Annu Rev Genet. 2008;42:683–707. doi: 10.1146/annurev.genet.41.110306.130354. [DOI] [PubMed] [Google Scholar]
- 21.Landmann F, Orsi GA, Loppin B, Sullivan W. Wolbachia-mediated cytoplasmic incompatibility is associated with impaired histone deposition in the male pronucleus. PLoS Pathog. 2009;5:e1000343. doi: 10.1371/journal.ppat.1000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lassy CW, Karr TL. Cytological analysis of fertilization and early embryonic development in incompatible crosses of Drosophila simulans. Mech Dev. 1996;57:47–58. doi: 10.1016/0925-4773(96)00527-8. [DOI] [PubMed] [Google Scholar]
- 23.Callaini G, Riparbelli MG, Giordano R, Dallai R. Mitotic Defects Associated with Cytoplasmic Incompatibility in Drosophila simulans. Journal of Invertebrate Pathology. 1996;67:55–64. doi: http://dx.doi.org/10.1006/jipa.1996.0009. [Google Scholar]
- 24.Wright JD, Barr AR. Wolbachia and the normal and incompatible eggs of Aedes polynesiensis (Diptera: Culicidae) Journal of Invertebrate Pathology. 1981;38:409–418. doi: http://dx.doi.org/10.1016/0022-2011(8190109-9) [Google Scholar]
- 25.Duron O, Weill M. Wolbachia infection influences the development of Culex pipiens embryo in incompatible crosses. Heredity (Edinb) 2006;96:493–500. doi: 10.1038/sj.hdy.6800831. [DOI] [PubMed] [Google Scholar]
- 26.Bate M, Arias AM. The Development of Drosophila Melanogaster. Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- 27.Zug R, Hammerstein P. Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One. 2012;7:e38544. doi: 10.1371/journal.pone.0038544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaenike J, Dyer KA, Cornish C, Minhas MS. Asymmetrical reinforcement and Wolbachia infection in Drosophila. PLoS Biol. 2006;4:e325. doi: 10.1371/journal.pbio.0040325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bordenstein SR, O’Hara FP, Werren JH. Wolbachia-induced incompatibility precedes other hybrid incompatibilities in Nasonia. Nature. 2001;409:707–710. doi: 10.1038/35055543. [DOI] [PubMed] [Google Scholar]
METHODS REFERENCES
- 8.Bordenstein SR, Bordenstein SR. Eukaryotic association module in phage WO genomes from Wolbachia. Nat Commun. 2016;7:13155. doi: 10.1038/ncomms13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishmael N, et al. Extensive genomic diversity of closely related Wolbachia strains. Microbiology. 2009;155:2211–2222. doi: 10.1099/mic.0.027581-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beckmann JF, Fallon AM. Detection of the Wolbachia protein WPIP0282 in mosquito spermathecae: implications for cytoplasmic incompatibility. Insect Biochem Mol Biol. 2013;43:867–878. doi: 10.1016/j.ibmb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of Molecular Biology. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 17.Yamada R, Floate KD, Riegler M, O’Neill SL. Male development time influences the strength of Wolbachia-induced cytoplasmic incompatibility expression in Drosophila melanogaster. Genetics. 2007;177:801–808. doi: 10.1534/genetics.106.068486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vallenet D, et al. MicroScope: a platform for microbial genome annotation and comparative genomics. Database (Oxford) 2009;2009:bap021. doi: 10.1093/database/bap021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu M, et al. Phylogenomics of the reproductive parasite Wolbachia pipientis w Mel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2004;2:E69. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klasson L, et al. The mosaic genome structure of the Wolbachia w Ri strain infecting Drosophila simulans. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5725–5730. doi: 10.1073/pnas.0810753106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klasson L, et al. Genome evolution of Wolbachia strain w Pip from the Culex pipiens group. Molecular biology and evolution. 2008;25:1877–1887. doi: 10.1093/molbev/msn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metcalf JA, Jo M, Bordenstein SR, Jaenike J, Bordenstein SR. Recent genome reduction of Wolbachia in Drosophila recens targets phage WO and narrows candidates for reproductive parasitism. PeerJ. 2014;2:e529. doi: 10.7717/peerj.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foster J, et al. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 2005;3:e121. doi: 10.1371/journal.pbio.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Werren JH, Loehlin DW. Rearing Sarcophaga bullata fly hosts for Nasonia (parasitoid wasp) Cold Spring Harb Protoc. 2009;2009:pdb prot5308. doi: 10.1101/pdb.prot5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bordenstein SR, Bordenstein SR. Temperature affects the tripartite interactions between bacteriophage WO, Wolbachia, and cytoplasmic incompatibility. PLoS One. 2011;6:e29106. doi: 10.1371/journal.pone.0029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kearse M, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beckmann JF, Markowski TW, Witthuhn BA, Fallon AM. Detection of the Wolbachia-encoded DNA binding protein, HU beta, in mosquito gonads. Insect Biochem Mol Biol. 2013;43:272–279. doi: 10.1016/j.ibmb.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson M, et al. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hurvich CM, Tsai C-LA, Corrected Akaike. Information Criterion for Vector Autoregressive Model Selection. Journal of Time Series Analysis. 1993;14:271–279. doi: 10.1111/j.1467-9892.1993.tb00144.x. [DOI] [Google Scholar]
- 43.Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 44.Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finn RD, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Casiraghi M, Anderson TJ, Bandi C, Bazzocchi C, Genchi C. A phylogenetic analysis of filarial nematodes: comparison with the phylogeny of Wolbachia endosymbionts. Parasitology. 2001;122(Pt 1):93–103. doi: 10.1017/s0031182000007149. [DOI] [PubMed] [Google Scholar]
- 48.Chatzispyrou IA, Held NM, Mouchiroud L, Auwerx J, Houtkooper RH. Tetracycline antibiotics impair mitochondrial function and its experimental use confounds research. Cancer Res. 2015;75:4446–4449. doi: 10.1158/0008-5472.CAN-15-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferguson SB, Blundon MA, Klovstad MS, Schupbach T. Modulation of gurken translation by insulin and TOR signaling in Drosophila. J Cell Sci. 2012;125:1407–1419. doi: 10.1242/jcs.090381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Southall TD, Elliott DA, Brand AH. The GAL4 System: A Versatile Toolkit for Gene Expression in Drosophila. CSH Protoc. 2008;2008:pdb top49. doi: 10.1101/pdb.top49. [DOI] [PubMed] [Google Scholar]
- 52.LePage DP, Jernigan KK, Bordenstein SR. The relative importance of DNA methylation and Dnmt2-mediated epigenetic regulation on Wolbachia densities and cytoplasmic incompatibility. PeerJ. 2014;2:e678. doi: 10.7717/peerj.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vizcaino JA, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bossan B, Koehncke A, Hammerstein P. A new model and method for understanding Wolbachia-induced cytoplasmic incompatibility. PLoS One. 2011;6:e19757. doi: 10.1371/journal.pone.0019757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
wVitA transcriptome data is deposited in the Sequence Reads Archives with Bioproject PRJNA319204 and BioSample SAMN04881412. wPip-infected ovarian proteome data was deposited at the Proteome Xchange Consortium via the PRIDE54 partner repository with the dataset identifier PXD004047. All other source data is available as supplemental information with this publication.


