Summary
Insulin-producing pancreatic β-cells in mice can slowly regenerate from glucagon-producing α-cells in settings like β-cell loss, but the basis of this conversion is unknown. Moreover it remains unclear if this intra-islet cell conversion is relevant to diseases like type 1 diabetes (T1D). We show that the α-cell regulators Aristaless-related homeobox (Arx) and DNA methyltransferase 1 (Dnmt1) maintain α-cell identity in mice. Within 3 months of Dnmt1 and Arx loss, lineage tracing and single cell RNA sequencing revealed extensive α-cell conversion into progeny resembling native β-cells. Physiological studies demonstrated that converted α-cells acquire hallmark β-cell electrophysiology, and show glucose-stimulated insulin secretion. In T1D patients, subsets of Glucagon-expressing cells show loss of DNMT1 and ARX, and produce Insulin and other β-cell factors, suggesting that DNMT1 and ARX maintain α-cell identity in humans. Our work reveals pathways regulated by Arx and Dnmt1 sufficient for achieving targeted generation of β-cells from adult pancreatic α-cells.
Introduction
Restoration of lost or diseased cells is a focus for intensive efforts in developmental and regenerative biology. Pancreatic islets are a paradigm for investigating organ restoration, reflecting growth in our understanding of development and maturation by the principal islet cell types (which include Insulin+ β-cells, Glucagon+ α-cells and Somatostatin+ δ-cells). Understanding mechanisms maintaining islet cell fate and function is important for addressing the urgent challenge of restoring islet β-cell and α-cell function compromised in diseases like type 1 diabetes (T1D). Prior studies have demonstrated that mouse α-cells or δ-cells can convert into insulin-producing cells following extreme experimental (>99%) β-cell ablation; in the case of α-cells, about 1% convert toward an insulin-producing fate without detectable proliferation over a period of 6–7 months (Thorel et al., 2010; Chera et al., 2014). However, the genetic or epigenetic basis of this conversion, including the extent or heterogeneity of reprogramming by individual adult α-cells has not been elucidated. Thus it remains unknown whether α-cell gene targeting in adult mice could enhance conversion into β-cells.
Maintenance of fate and function by adult cells likely reflects both genetic and epigenetic mechanisms (Morris and Daley, 2013). Prior studies demonstrate that the transcription factors MAFA, NKX6.1, and PDX1, the proinsulin-processing enzyme PCSK1/3, and - in mice - the glucose transporter encoded by Slc2a2 are essential regulators of β-cell fate and mature function (Arda et al., 2013). By contrast, mouse and human islet α-cells require Aristaless-related homeobox (Arx) to specify α-cell fate and to maintain production of hallmark factors like glucagon (Collombat et al., 2003; Collombat et al., 2007; Kordowich et al., 2011; Papizan et al., 2011; Itoh et al., 2010; Mastracci et al., 2011). Ectopic expression of Pdx1, Nkx6.1 or Pax4 in α-cells may be sufficient to induce β-cell features in fetal or neonatal α-cells (Yang et al., 2011; Collombat et al., 2009; Schaffer et al 2013).
Surprisingly, studies of Arx inactivation in adult mouse glucagon-producing pancreatic cells have not detected clear evidence of direct α-to-β cell conversion (Courtney et al., 2013; Wilcox et al., 2013). In a prior study of Dox-induced Arx inactivation in mice (Courtney et al., 2013), lineage-tracing reflected a schedule of constitutive Dox exposure, and did not distinguish ductal cell from α-cell progeny. This study concluded that Arx loss in adult mice induced a program of β-cell neogenesis resembling embryonic islet development, where ductal cells expressed the embryonic islet regulator Neurogenin3 then Glucagon and Insulin. In other work, continuous Arx inactivation from embryonic stages led to development of polyhormonal cells (Wilcox et al., 2013). Thus, it remains unclear whether targeted Arx inactivation specifically in adult mouse α-cells could induce loss of α-cell features and acquisition of β-cell properties. In humans with T1D, blunted glucagon output in the setting of severe hypoglycemia is a frequent complication, and suggests that islet α-cell fate and/or function may be attenuated by disease (Cryer et al., 2003; Pietropaolo et al., 2013). However, the molecular basis of this α-cell dysfunction remains unclear.
Regulation of islet epigenetics by DNA methylation appears to be an important regulatory mechanism during α- and β-cell differentiation and maturation (Papizan et al., 2011; Avrahami et al., 2015; Dhawan et al., 2011; Dhawan et al., 2015), and prior studies report an unexpected degree of similarity in gene expression and chromatin modifications of α-cells and β-cells in mice and humans (Arda et al., 2016; Bramswig et al., 2013; Benitez et al., 2014; Moran et al., 2012). Adult α-cells and other islet cells express enzymes like DNA methyltransferase 1 (DNMT1) suggesting a requirement for these factors in maintaining α-cell fate (Avrahami et al., 2015; Dhawan et al., 2011; Benitez et al., 2014). Although DNMT1 activity is best understood in the context of maintaining epigenetic ‘memory’ in proliferating cells, recent studies demonstrate DNMT1 function in non-dividing cells (Dhawan et al., 2011). However, direct testing of in vivo DNMT1 requirements in α-cells has not been described.
Here we report that simultaneous inactivation of Arx and Dnmt1 in mouse α-cells promotes efficient conversion of α-cells into progeny resembling β-cells in multiple ways, including Insulin production, global gene expression, hallmark electrophysiology and insulin secretion in response to glucose stimulation. Studies of Glucagon+ cells in islets from a subset of humans with T1D similarly reveal loss of ARX and DNMT1, with gain of β-cell features.
Results
Altered cell fates after Arx loss in adult mouse α-cells
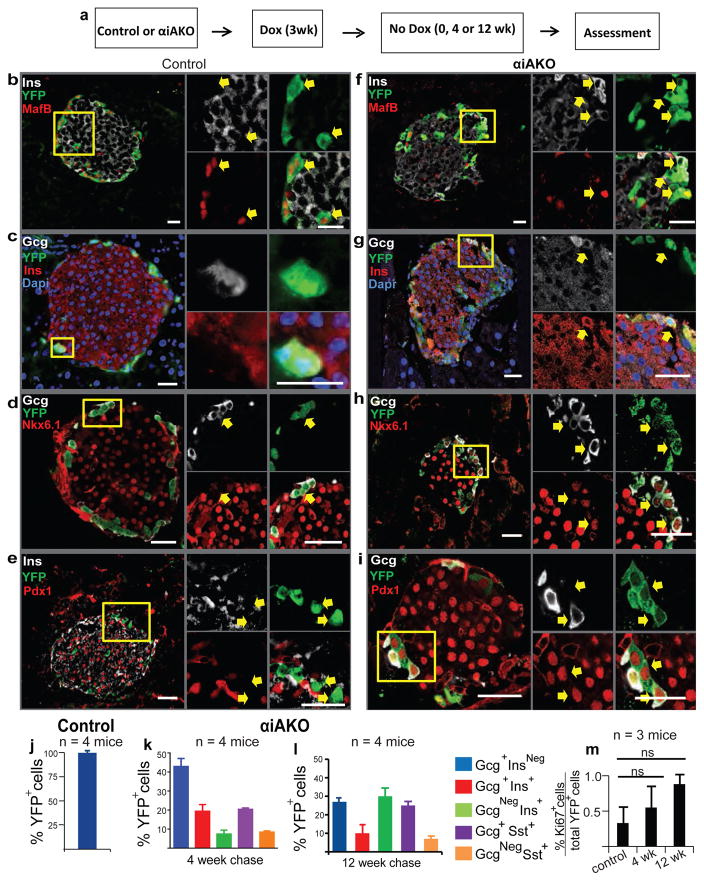
To determine if Arx loss in vivo directly alters adult α-cell fate, we developed systems for simultaneous in vivo Arx inactivation and lineage tracing in mouse α-cells (Experimental Procedures, Figure S1a). We used previously-described mice (Thorel et al., 2010) harboring a Doxycycline inducible Glucagon (Gcg) driven-reverse tet Transactivator (Gcg-rtTA) to direct Cre recombinase expression from a Tet-O-Cre transgene in Gcg+ α-cells: Cre then activates lineage-independent YFP transgene expression from the Rosa26 locus. Intercrosses generated α cell inducible Arx Knock Out (αiAKO) mice (Figure S1a) harboring a Cre recombinase-sensitive floxed Arx allele (Marsh et al., 2009), and the three alleles described above. Briefly, in αiAKO islets Dox exposure should stimulate Cre recombinase expression specifically in Gcg+ α-cells: Cre then inactivates the floxed Arx allele, and activates YFP transgene expression from the Rosa26 locus.
Over 90% of Gcg+ cells were labelled with YFP in 2 month-old control Gcg-rtTA, Tet-O-Cre, Rosa26-YFP animals exposed to Dox for 3 weeks, or in αiAKO animals exposed to Dox for 3 weeks, followed by a 4 or 12 week ‘chase’ period without Dox (Figure 1a). We have previously found extremely low (0.1–0.2%) non-specific labeling of YFP+ cells in control mice when Doxycycline is given after birth (Thorel et al., 2010). Loss of Arx protein in αiAKO mouse α-cells was confirmed by immunostaining (Figure S1b,c). We did not detect differences in glycemia during ad libitum feeding or after overnight fasting in control and αiAKO mice (Table S4). After 0, 4 or 12 weeks without Dox, we sacrificed mice and immunostained the pancreas to assess islet cell fates. By 4 weeks, 50% of YFP+ cells without Arx showed evidence of failure to maintain α-cell identity. This included loss of α-cell gene products like Glucagon or MafB (Figure 1f,b,i,k Figure S2f–t). Notably, the majority of cells (60%) co-expressing Glucagon or MafB also produced gene products not observed in normal α-cells, like Insulin, Pdx1 or Nkx6.1 (Figure 1f–i, Figure S2), Somatostatin, and rarely, Ghrelin or Pancreatic Polypeptide (Figure 1k; Figure S3a–c, e–g). Quantification of YFP+ cell phenotypes revealed that 20% co-expressed α-cell (Gcg, MafB) and β-cell gene products (Ins, Pdx1, Nkx6.1; Figure 1g–i, 1k, Figure S2i,j). Only 6% of YFP+ cells produced Insulin without Glucagon. After 12 weeks off Dox, this population of YFP+ Ins+ GcgNeg cells increased to 29% of YFP+ cells, but none expressed the mature β-cell marker MafA (Figure S3d,h). By contrast, YFP+ cells remained 99.8% Gcg+ InsNeg in control mice (Figure 1c,j). In control and αiAKO mice, we scored production of the proliferation marker Ki67 and did not detect changes of YFP+ cell proliferation after 4 and 12 weeks off Dox (Figure 1j, Supplementary Table 2). However, this does not exclude the possibility that α-cell proliferation occurred at earlier times after Arx deletion. We also did not detect induction of Neurogenin3 (Neurog3), which encodes a bHLH transcription factor expressed in fetal pancreatic endocrine progenitor cells (Gradwohl et al., 2000; Gu et al., 2002). Thus, loss of Arx led to failure of adult mouse α-cells to maintain their differentiated fates, leading to time-dependent adoption of alternate islet cell fates, mainly a population of poly-hormonal cells, and a smaller fraction resembling islet β, δ, ε and PP-cells. These findings and approaches differ from those reported by Courtney and colleagues (Courtney et al., 2013) who constitutively inactivated Arx in adult Gcg+ cells, and observed islet hyperplasia with a large increase in Ins+ GcgNeg cell number accompanied by reactivation of Neurog3 and without durable increases of polyhormonal Gcg+ cells expressing Sst, Ghrelin or PP (see Discussion).
Figure 1. Loss of α-cell identity after deletion of Arx.
(a) Schematic showing experimental design for Dox treatment of knock out and control animals.
(b–i) Immunostaining showing expression of α and β-cell markers MafB, Ins, Gcg, Nkx6.1, Pdx1 with YFP in control and αiAKO mice 4 weeks after Dox treatment. Yellow boxes show specific area of islet enlarged and represented by arrows on the right to demonstrate gene expression within specific cells or sets of cells. Scale bars represent 25 μm.
(j–l) Quantification of α-to-β-cell conversion in control mice at the end of 3 weeks of Dox treatment (Time 0) (j) and αiAKO mice at the end of a 4 week chase and 12 week chase (k,l). (m) Quantification of Ki67+ YFP+ cells at the end of a 4 week chase and 12 week chase compared to controls (N=3 mice). Bar graph data are represented as mean ± S.D. Dox, Doxycycline. N=4 mice per time point.
α-cell identity is unaltered by loss of endogenous Dnmt1
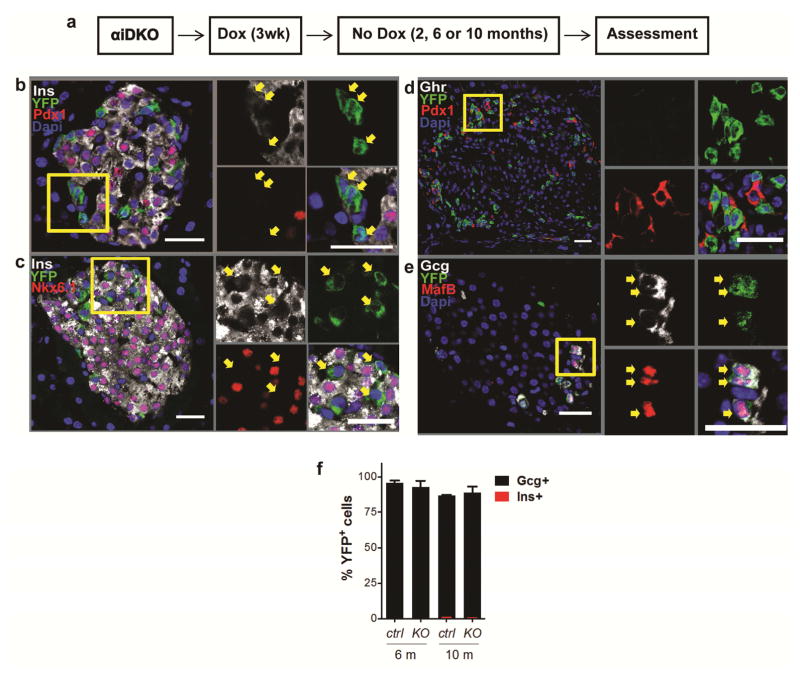
Our finding that over 60% of α-cells failed to express Insulin after Arx loss suggested that α-cell fate was likely maintained by additional regulatory factors. DNA methyltransferase 1 (Dnmt1) is an enzyme that functions to regulate genome-wide gene expression by methylating cytosine residues within regulatory regions of genes. Others have reported that Dnmt1 is important for islet cell fate maintenance across species (Dhawan et al., 2011; Dhawan et al., 2015; Bramswig et al., 2013, Anderson et al., 2009). Thus, we postulated that removal of Dnmt1 might compromise α-cell fate. To test this, we constructed mice harboring the alleles Gcg-rtTA, Tet-O-Cre, Dnmt1f/f Rosa26-YFP (“αiDKO mice”; Figure 2a). After Dox exposure we detected loss of Dnmt1 in YFP+ cells; however, YFP+ cells produced no detectable Insulin, Pdx1, or Nkx6.1, even 10 months after DOX removal (Figure 2b,c). Instead, YFP+ cells maintained expression of glucagon and MafB, and did not produce detectable Sst or Ghr (Figure 2d–f). Thus, unlike targeted Arx loss in α-cells, targeted Dnmt1 inactivation did not discernably alter α-cell fate.
Figure 2. α-cells maintain their fate in the absence of Dnmt1.
(a) Schematic showing experimental design for Dox treatment of knock out and control animals.
(b–e) Immunostaining showing expression of α, β, δ, and ε-cell markers Gcg, MafB, Ins, Pdx1, Nkx6.1, Sst, and Ghrelin with YFP in αiDKO mice 10 months after Dox treatment. Yellow boxes show specific area of islet enlarged and represented by arrows on the right to demonstrate gene expression within specific cells or sets of cells. Scale bars represent 25 μm.
(f) Percentage of YFP+ cells that express glucagon or insulin in knockout and control mice. Bar graph data are represented as mean ± S.D. Dox, Doxycycline. N=4 mice per time point.
α-cells with combined Arx and Dnmt1 loss resemble β-cells
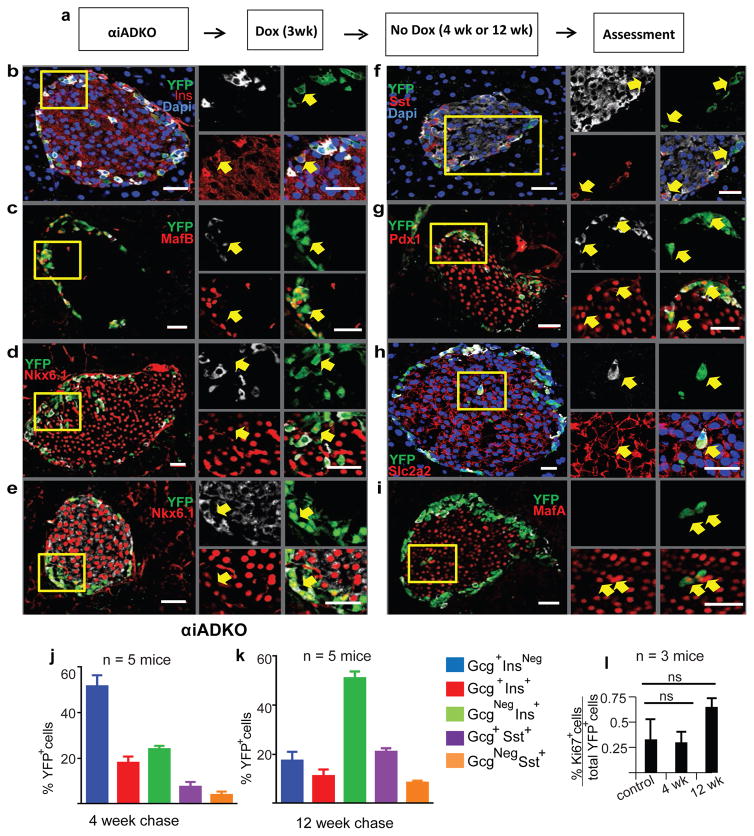
To test whether simultaneous loss of Arx and Dnmt1 might alter the pattern of α-cell conversion in adult mice, we intercrossed mice to produce progeny permitting Dox-dependent α-cell inactivation of Dnmt1 and Arx combined with Rosa26-YFP lineage tracing (Experimental Procedures; Figure S1a). Mice with the six alleles Gcg-rtTA, Tet-O-Cre, Arxf/Y Dnmt1f/f Rosa26-YFP (hereafter, “αiADKO mice”) and controls were exposed to Dox for 3 weeks followed by 4 or 12 weeks without Dox (Figure 3a). Loss of Arx or Dnmt1 in α-cells, labelled with over 90% efficiency by YFP, was confirmed by immunostaining (Figure S1b-e, Figure S4u–y).
Figure 3. Expression of β-cell genes in murine α-cells lacking Dnmt1 and Arx.
(a) Schematic showing experimental design for Dox treatment of knock out and control animals.
(b–i) Immunostaining showing expression of α, β, and δ-cell markers Ins, Gcg, MafB, Nkx6.1, Sst, Pdx1, Slc2a2 and MafA with YFP in αiADKO mice 12 weeks after Dox treatment. Yellow boxes show specific area of islet enlarged and represented by arrows on the right to demonstrate gene expression within specific cells or sets of cells. Scale bars represent 25 μm.
(j–k) Quantification of α-to-β-cell conversion in αiADKO mice at the end of a 4 week chase and 12 week chase.
(l) Quantification of Ki67+ YFP+ cells at the end of a 4 week chase and 12 week chase compared to controls (N=3 mice). Bar graph data are represented as mean ± S.D. Dox, Doxycycline. N=4 mice per time point.
After 4 weeks, immunostaining revealed that 50% of YFP+ cells failed to maintain α-cell identity, showing either loss of α-cell products like Glucagon and MafB, or coexpression of Glucagon and MafB with gene products characteristic of β-cells like Insulin, Nkx6.1 and Pdx1 (Figure 3b–d,g,I, Figure S4a–t), or δ-cells (Somatostatin: Figure 3f,j). At 4 weeks, 23% of YFP+ cells in αiADKO mice co-expressed α and β-cell gene products like Glucagon and Insulin, and 16% expressed β-cell gene products like Pdx1 and Insulin without detectable Glucagon or MafB (Figure 3j, Figure S4a–t). Only 14% of YFP+ cells produced Sst (Figure 3j): by contrast, we did not detect production of PP or Ghrelin in YFP+ cells. After 12 weeks, immunostaining revealed that 82% of αiADKO YFP+ cells had lost their α-cell fate. 50% of YFP+ cells had lost the expression of either Gcg or MafB or both and produced β-cell factors including Insulin, Pdx1 and Nkx6.1 (Figure 3b–g,k, Figure S4a–t). Like in αiAKO mice, we did not detect changes in Ki67 or Neurog3 in αiADKO mice (Figure 3l, Figure S4z‴).
By 12 weeks, we also observed some YFP+ Ins+ GcgNeg cells expressing the glucose transporter Slc2a2 and MafA, markers and regulators of native β-cells (Figure 3h,i,). 27% of YFP+ cells in αiADKO mice produced Sst (Figure 3f,k) and 10% co-expressed Gcg and Insulin (Figure 3k). Thus, using lineage tracing and conditional genetics to inactivate Arx and Dnmt1, we observed evidence of extensive direct α-cell conversion into progeny resembling β-cells.
Single Cell RNA-Seq reveals that converted α-cells closely resemble native β-cells
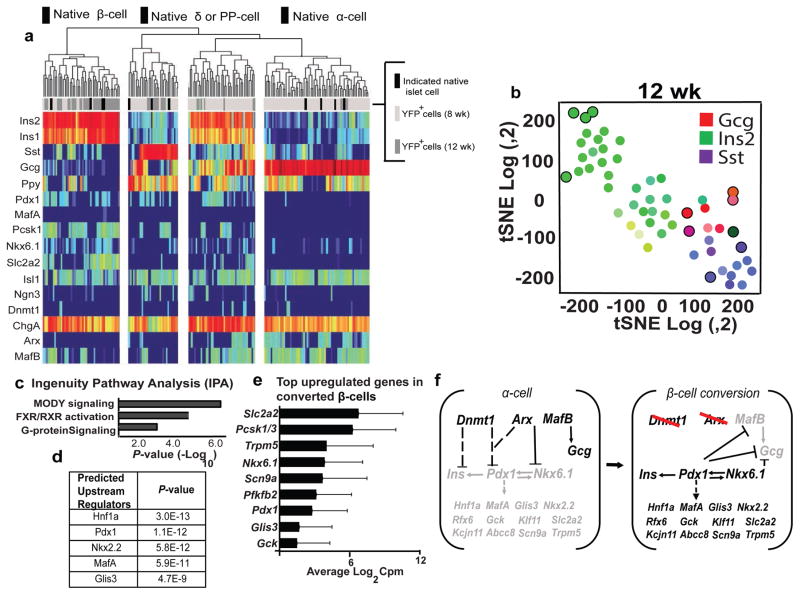
To investigate further the extent of α-cell conversion toward fates resembling β-cells, we performed single cell RNA-Seq (scRNA-Seq) after purifying YFP+ cells and control (YFPNeg) cells from αiADKO mice and control mice by Fluorescent Activated Cell Sorting (FACS). After Dox exposure, we obtained 127 YFP+ cells at 8 weeks (‘early’: light grey bars Figure 4a), and 44 YFP+ cells at 12 weeks (‘late’: dark grey bars, Figure 4a) in addition to YFPNeg control native β-cells, α-cells, or δ-cells (black bars, Figure 4a). All YFP+ cells expressed the YFP transgene and pan-endocrine genes like Chromogranin A and Chromogranin B, demonstrating the islet endocrine origin of these cells. These did not express endocrine precursor markers such as Ngn3 mRNA (Figure 4a), similar to prior findings with α-cell conversion after β-cell ablation (Thorel et al 2010; Chera et al 2014). Consistent with our immunohistological analysis, YFP+ cells from both early and late collections clustered into three major populations after t-Distributed Stochastic Neighbor Embedding (tSNE) dimensionality-reduction analysis: 1) cells that are similar to normal α-cells 2) cells that are similar to normal β-cells, and 3) cells that express other islet hormones such as Sst (Figure 4b, data not shown).
Figure 4. Single Cell RNA-Seq analysis of mouse α-cells undergoing loss of identity.
(a) Heat map generated from single cells obtained from two αiADKO mice 8 and 12 weeks after Dnmt1 and Arx deletion (N=2 mice). Black bars represent normal islet cells, light grey and dark grey bars represent individual YFP+ cells obtained from αiADKO mice at the 8 week and 12 week time-points respectively.
(b) tSNE plot generated from single cells obtained from αiADKO mice 12 weeks after Dnmt1 and Arx deletion. Black circles represent normal islet cells. Green circles represent individual YFP+ cells obtained from αiADKO mice. The color inside each circle indicates whether the cell expressed Gcg (red), Ins (green) or Sst (Purple). Intermediate colors represent intermediate cell states.
(c,d) Key predicted pathways and gene regulators generated by Ingenuity Pathway Analysis (IPA) comparing single cells obtained from αiADKO mice 8 weeks and 12 weeks after Dnmt1 and Arx deletion, and progressing from an α-cell state to a β-cell state.
(e) Top upregulated genes in converted α-cells represented as the average of log2 counts per million (Cpm) values from single cells expressing Insulin alone obtained from αiADKO mice at the 12 week time-point. The average Insulin expression in these cells is 16.1 (±0.37) cpm.
(f) A model for activation of a β-cell gene regulatory network in cells undergoing α-to-β-cell conversion. Dashed lines and arrows represent indirect transcriptional relationships between regulators and targets. Solid lines and arrows represent established direct transcriptional relationships between regulators and targets. Repressed genes are in grey font, while induced genes are in black font.
Approximately 20% of the YFP+ cells at 8 weeks exclusively express Insulin and other β-cell genes. The majority of the YFP+ cells from this time-point maintained mRNA expression of the α-cell gene MafB (Figure 4a). In addition to these distinct populations, YFP+ cells that exhibit features of two or more islet cell types (including polyhormonal mRNA expression) occurred more frequently at 8 weeks than at later times. At 12 weeks, RNA-Seq revealed that nearly 80% of the YFP+ cells expressed Ins1 and Ins2, and the majority of these did not express mRNAs encoding other islet hormones. Moreover, just 7% of YFP+ cells exclusively expressed Gcg at 12 weeks. Thus, α-cells in αiADKO mice appeared to preferentially convert toward β-cell fates. Ingenuity Pathway Analysis (IPA) of these datasets identified pathways in converted α-cells that are crucial for β-cell identity and function including Maturity Onset Diabetes of the Young (MODY) signaling factors (Hnf1a, Pdx1 and Gck) (Figure 4c,d). RNA-Seq confirmed that a subset of the converted α-cells expressed many of these regulators (like Pdx1, Nkx6.1, Glis3) and their known downstream targets like, Scn9a, Gck, Slc2a2 which are established effectors of β-cell function (Figure 4e). Together, single cell RNA-Seq and our analysis provide unprecedented genome-scale evidence for the range, trajectory and extent of gene expression changes in α-cells directly converting toward β-cells after conditional Arx and Dnmt1 inactivation.
Electrophysiological resemblance of converted α-cells and native β-cells
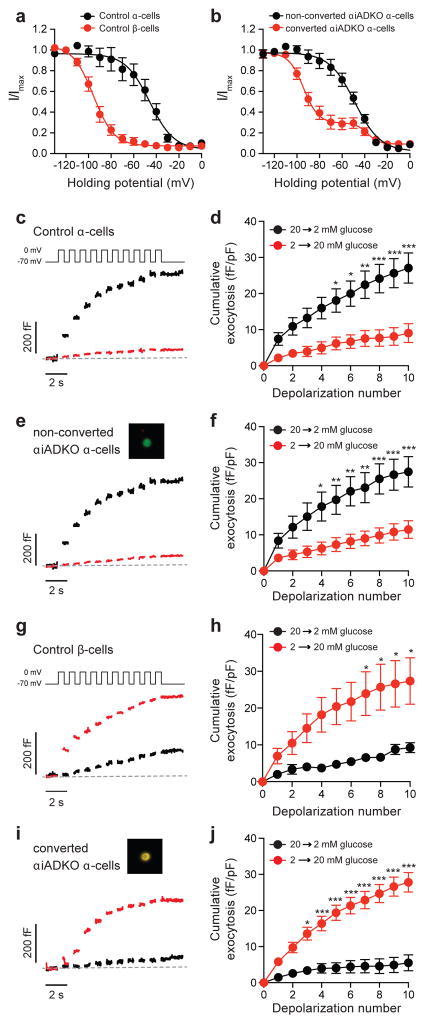
In electrophysiological studies, mouse α- and β-cells have long been distinguished by characteristic differences in the voltage-dependent inactivation of Na+ channels (Gopel et al., 2000) and more recently by opposing glucose-dependent exocytotic responses to serial membrane depolarization (Ferdaoussi et al., 2015; Dai et al., 2014). Our single cell RNA-Seq studies reveal upregulation of genes mediating the β-cell Na+ current (Scn9a) and contributing to β-cell glucose sensing (Slc2a2) in the converted α-cells, but not in the unconverted α-cells from αiADKO mice (Figure 4e). We postulated that converted α-cells lose the electrophysiological response features of α-cells and acquire β-cell responses. To assess this, we dispersed islets into single cells from αiADKO mice after 12 weeks of Dox treatment and measured Na+ inactivation and glucose-dependent capacitance responses in YFP+ cells. We find that Na+ current inactivation is half maximal at −46 and −96 mV in α- and β-cells, respectively, from control mice (n=13 and 9 cells; Figure 5a). Non-converted α-cells from the αiADKO mice (InsNeg,YFP+) maintained their right-shifted Na+ current inactivation, which was half-maximal at −49 mV (n=21 cells; Figure 5b). However, Na+ current inactivation in converted α-cells from the αiADKO mice (Ins+,YFP+), resembled that of native β-cells, where most channels (~70%) inactivated half-maximally at −94.2 mV (n=27 cells; Figure 5b).
Figure 5. Electrophysiological properties of converted α-cells.
(a) Voltage-dependent inactivation of Na+ channels is left-shifted in control β-cells (red; n=9) compared with control α-cells (black; n=13). Total mice = 3.
(b) Inactivation of Na+ currents in non-converted (InsNeg,YFP+) αiADKO α-cells (black; n=21, mice = 3) was identical to the control α-cells, while the majority (~70%) of Na+ current inactivation in the converted (Ins+,YFP+) αiADKO β-cells (red; n=27, mice = 3) was left-shifted similar to that observed in β-cells.
(c–j) Single-cell exocytosis was measured by monitoring capacitance increases in response to a series of membrane depolarizations following transition from 20 to 2 mM glucose (black) or from 2 to 20 mM glucose (red). Representative traces (c,e,g) and averaged data (d,f,h) are shown. In control α-cells (c,d), exocytosis was amplified by lowering glucose (n=21, mice = 2) and suppressed by raising glucose (n=16, mice = 2). Identical results were observed (e,f) in non-converted (InsNeg,YFP+; inset) αiADKO α-cells (n=13 and 24, mice = 3). In control β-cells (g,h), exocytotic response was amplified by raising glucose (n=8) and suppressed by lowering glucose (n=6, mice=2). In converted (i,j; Ins+,YFP+ ; inset) αiADKO α-cells, however, the response was reversed to recapitulate a β-cell phenotype such that raising glucose amplified the exocytotic response (n=23, mice = 3) while lowering glucose suppressed it (n=14). * P<0.05; ** P<0.01; *** P>0.001.
Glucose-stimulation amplifies the exocytotic response to membrane depolarization in β-cells (Ferdaoussi 2015) but suppresses the response in α-cells (Dai 2014). Accordingly, exocytosis in α-cells from the control mice is suppressed by a rise in glucose (n=16 cells) and amplified by a drop in glucose (n=21; Figure 5c–d), and this is also observed in the non-converted (InsNeg,YFP+) α-cells from the αiADKO mice (n=24 and 13 cells; Figure 5e–f). The converted α-cells (Ins+,YFP+) from the αiADKO mice, however, again resembled native β-cells in which the exocytotic response was suppressed by a drop in glucose (from 20 to 2 mM; n=14 cells), and amplified by a rise in glucose (from 2 to 20 mM; n=23 cells; Figure 5g–j). Together, these studies reveal a striking functional switch from α-cell to β-cell phenotypes in converted mouse β-cells after conditional deletion of Dnmt1 and Arx.
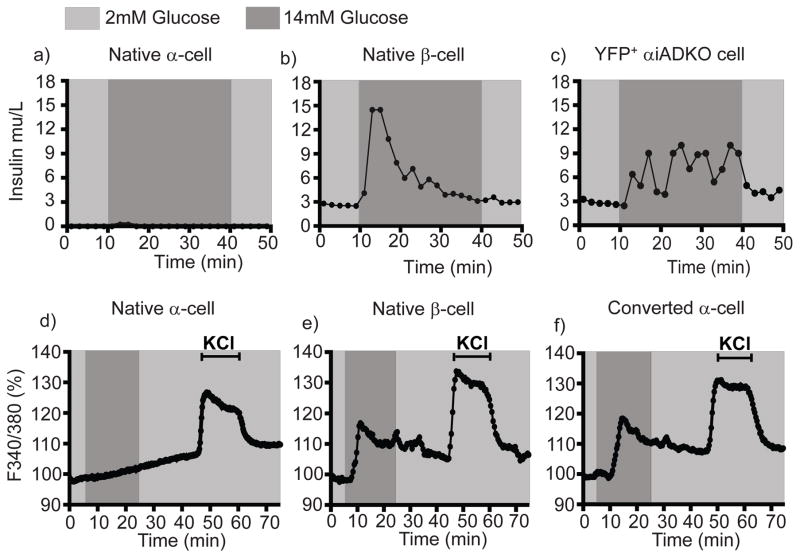
Glucose-dependent insulin secretion by converted α-cells and native β-cells
Our electrophysiological studies and findings suggested that converted α-cells might also functionally resemble native β-cells by (1) increasing their intracellular calcium ([Ca2+]i) upon glucose stimulation and (2) by secreting insulin in response to glucose, two tightly coupled functions. To assess this, we dispersed islets into single cells from control (see Methods) and αiADKO mice after Dox treatment, FACS-purified the YFP+ cells, then measured calcium influx and insulin secretion kinetics in response to glucose by relevant FACS-purified cells using a microfluidics perifusion system (Adewola et al 2010: Xing et al 2016). As expected, GFP+ β-cells from MIP-GFP mice secreted insulin, but not glucagon, in response to glucose stimulation, and Venus+ cells from Glucagon-Venus mice secreted glucagon when challenged by glucose reduction (Figure 6a,b; Figure S5a,b). By contrast, converted α-cells from αiADKO mice secreted insulin in response to high glucose (Figure 6c). Compared to native β-cells, the level of insulin secretion by converted αiADKO α-cells was lower but prolonged after glucose challenge (Figure 6a,c). Glucagon secretion by pooled YFP+ αiADKO α-cells and Venus+ control α-cells from Glucagon-Venus mice was similar (Figure S5c), consistent with our finding that a subset of DOX-exposed αiADKO α-cells maintain Glucagon expression and electrophysiological features of native α-cells.
Figure 6. Insulin secretion and Calcium signaling properties of converted α-cells.
(a–c) Temporal insulin secretion profiles of 5000 YFP+ cells from Glucagon-Venus mice (N=3 mice) (a), MIP-GFP mice (N=3 mice) (b) and αiADKO mice (N=4 mice) (c) perfused with basal (2mM Glucose) and stimulatory (14mM Glucose) at the indicated time-points.
(d–f) Intracellular calcium profiles indicated by Fura-2 ratio of the fluorescence intensities (340/380nm) of Venus+ cells and GFP+ from Glucagon-Venus mice (N=3 mice) (d), MIP-GFP mice (N=3 mice) (e), and αiADKO mice (N=5 mice) (f) perfused with basal (2mM Glucose) and stimulatory (14mM Glucose or 30mM KCl) solutions at the indicated time-points. In a–f, cells are FACS-purified from the indicated mice.
Glucose-stimulated insulin secretion in native β-cells is tightly coupled to glucose metabolism, membrane depolarization and transient intracellular increases of [Ca2+]i. We assessed [Ca2+]i changes in isolated control α- and β-cells and in YFP+ αiADKO cells during exposure to basal (2.8 mM) and high (14 mM) glucose concentrations, or to potassium chloride (KCl) a general membrane depolarizer. Single cell calcium imaging revealed that KCl provoked increased [Ca2+]i in β-cells from MIP-GFP mice, α-cells from Glucagon-Venus mice, and YFP+ cells from αiADKO mice (Figure 6d–f). After exposure to 14 mM glucose, we observe an average increase of [Ca2+]i in native β-cells and YFP+ αiADKO cells, but not in native α-cells (Figure 6d–f, dark grey bars). Together, our physiological studies revealed that converted mouse α-cells acquired multiple cardinal functional features of normal β-cells, supporting our molecular findings.
Evidence of altered Glucagon+ cell fates in pancreas from T1D subjects
Impaired glucagon responses to hypoglycemia in T1D (Cryer et al 2003; Pietropaolo et al 2013) have suggested that islet α-cell fates may be altered in T1D. To determine whether changes, including loss of islet DNMT1 and ARX, might occur in human T1D, we used immunohistochemistry to analyze cell-enriched transcription factor and hormone expression in pancreata from control (Figure S6a–f) and T1D donors (Figure S6g–m). As expected, previously healthy control subjects aged 4, 7 and 26 years (Table 1) produced Insulin (INS), PDX1, and NKX6.1 exclusively in β-cells, Glucagon (GCG) and ARX in α-cells and Somatostatin (SST) in δ-cells (Figure S6a–e). DNMT1 (Figure S6f) was expressed in a subset of α- and β-cells (Figure S6e). There was no detectable co-expression in controls of Insulin with Glucagon, Somatostatin or ARX, or Glucagon with PDX1 or NKX6.1 (Figure S6a–e, quantification in Figure S6n–r). In samples from donors with T1D for 4, 5, 7, 23 or 33 years (Figure S7i,j, Figure S7b–f, Figure S7a–f), we observed pronounced loss of INS+ cells. However, the expression of a variety of pan-endocrine markers including PAX6, NKX2.2 and Chromogranin A (CHGA) was maintained in hormone+ cells (H.C. and S.K., unpubl. results).
Table 1.
List of human pancreas samples
| Age | Sex | Disease | Disease Duration | # Insulin+ cells scored |
|---|---|---|---|---|
| 27 | Male | Normal | Not applicable | Not applicable |
| 7 | Male | Normal | Not applicable | Not applicable |
| 4 | Male | Normal | Not applicable | Not applicable |
| 9 | Female | Type 1 Diabetes | 5 | 19 |
| 8 | Female | Type 1 Diabetes | 4 | 11 |
| 22 | Male | Type 1 Diabetes | 7 | 122 |
| 32 | Male | Type 1 Diabetes | 23 | 96 |
| 38.5 | Male | Type 1 Diabetes | 32.5 | 73 |
In T1D islets from donors with 4–5 years’ disease duration, we detected additional abnormal GCG+ cells: 10% of remaining GCG+ cells lacked ARX or produced characteristic β-cell factors like PDX1 or NKX6.1 (Figure S6g,i,j,n,p,q, Figure S7b,c,f). Moreover, bi-hormonal GCG+ INS+ cells were also observed in 2% of islets from donors with T1D for 4 or 5 years (Figure S6h,o, Figure S7d), which correlated with loss of DNMT1 in these cells (Figure S6m, yellow and white arrows, Figure S6s). In samples from subjects with longer T1D duration, approximately 5% of remaining GCG+ cells lacked ARX or co-expressed NKX6.1. However, GCG+ PDX1+ or bihormonal GCG+ INS+ cells were not detected in these samples (Figure S7a–c, f). Thus, our studies of T1D islets from five donors revealed: (1) loss of the hallmark α-cell features and gain of the β-cell features in a fraction of GCG+ cells, and (2) GCG+ INS+ expression in cells lacking ARX and DNMT1.
Discussion
Dissecting and controlling the mechanisms governing cell fate is a central challenge for developmental and regenerative biology (Kim et al., 2016). We investigated α-cells in mice affording conditional genetics, lineage-tracing, single cell RNA-Seq and functional analyses, and in humans with T1D and β-cell destruction. To determine the genetic mechanism by which insulin-producing cells might be spontaneously regenerated from α-cells, we inactivated two genes, Arx and Dnmt1 in adult pancreatic α-cells and found this was sufficient for direct, efficient conversion of islet α-cells into progeny resembling β-cells. We investigated islet cell identity in the human T1D pancreas and discovered changes of multiple regulators in Glucagon+ islet cells, including loss of ARX and DNMT1. We speculate that such changes could underlie α-cell dysfunction in T1D.
Directing effective conversion of non β-cells into insulin-producing cells could be crucial for achieving regenerative goals. Studies here revealed efficient formation of insulin-expressing cells within 3 months by 50–80% of α-cells after targeted inactivation of Arx and Dnmt1. Converted α-cells resembled native β-cells in their electrophysiology and ability to secrete insulin in response to glucose stimulation. Thus, our histology, lineage-tracing, single cell RNA-Seq analysis, electrophysiological and hormone studies provided an unprecedented assessment of the trajectory of cells undergoing α-to-β cell conversion. Formation of Insulin-producing cells after targeted inactivation of Arx alone produced fewer GlucagonNeg Insulin+ cells and more poly-hormonal cell types, while Dnmt1 inactivation alone was insufficient to induce Insulin+ α-cells.
Prior studies postulated that effective somatic cell conversion may require at least two steps of re-programming: a ‘priming’ step to poise genes for alternative expression, and loss (or gain) of a cell type-specific ‘master’ regulator (Efe et al., 2011; Shu et al., 2013). The modest or ineffective α-cell conversion following Dnmt1 loss alone, or Arx loss alone is consistent with this hypothesis. We speculate that Dnmt1 loss could constitute a priming step while loss of Arx – a master regulator of α-cell fate – is a required concurrent step to achieve α-to-β cell conversion. The mechanisms by which deletion of Dnmt1 contribute to loss of α-cell identity are not known. However, it seems likely that the reduction in promoter/enhancer methylation within transcriptional control sequences of key β-cell genes such as Insulin, Pdx1 and Nkx6.1 (Avrahami et al 2015., Akinci et al., 2012; Park et al., 2008) combined with loss of Arx permits activation of these genes (Papizan et al 2011).
In αiADKO mice, production of insulin-producing cells from α-cells occurred without induction of embryonic islet regulators like Neurog3. In a prior study of Dox-induced Arx inactivation in mice (Courtney et al., 2013), lineage-tracing reflected a schedule of constitutive Dox exposure, and did not distinguish ductal cell from α-cell progeny. In other work, continuous Arx inactivation from embryonic stages led to the development of polyhormonal cells (Wilcox et al., 2013). Future studies with complementary lineage-tracing methods would reinforce the findings and conclusions of our study.
Single cell RNA-Seq evaluation of converted α-cells revealed rapid, extensive and significant induction of gene expression networks known to regulate β-cell fate and function, confirming our immunohistological findings (Arda et al., 2013; Maestro et al., 2007; Boj et al., 2010; Hunter et al., 2011) (Figure 5g). Our scRNA-Seq analysis also revealed differences between these converted α-cells and native β-cells reflecting the observed differences in hormone secretion. For example, only a subset of the converted α-cells expressed β-cell regulators like MafA, Pdx1, Nkx6.1 and Slc2a2. This heterogeneity of β-cell gene expression within the population of converted α-cells suggests that other β-cell gene regulators may require activation to promote a more complete conversion towards a β-cell fate.
Electrophysiological and hormone secretion assessments of cells undergoing α-to-β-cell conversion confirmed our single cell RNA-Seq predictions that a majority of the converted α-cells are physiologically similar to normal β-cells. We showed that Na+ channel inactivation, a classical electrophysiological marker for distinguishing mouse α-cells versus β-cells, shifts from an α-cell phenotype to a β-cell phenotype in converted α-cells. Moreover, we showed that converted α-cells, like normal β-cells, secrete insulin and show a clear switch to high glucose-amplification of exocytosis, in contrast to unconverted α-cells which secrete glucagon and have exocytotic responses blunted by high glucose (Gylfe et al., 2016). We postulate that loss of Arx and Dnmt1 in α-cells triggers the upregulation of gene regulatory networks controlling metabolic sensing pathways that are intrinsic to β-cells. Our findings suggest that additional extrinsic signaling modulation could additively enhance the pace and quality of α-to-β cell conversion achieved after targeted Arx and Dnmt1 inactivation. Thus, identifying signaling pathways that regulate Arx and Dnmt1 could be useful for directing α-cells toward alternate fates.
Although mouse studies by us and by others provide evidence that murine α-cells can undergo conversion towards a β-cell fate, the relevance of these findings to humans was unclear. Bi-hormonal human pancreatic cells including Glucagon+ Insulin+ cells in subjects with diabetes (Piran et al., 2014; Yoneda et al., 2013) or in cultured islets (Bramswig et al., 2013) have been documented. However the molecular or regulatory features underlying development of these abnormal Glucagon+ cells in T1D has not been described, reflecting inherent difficulties of pancreas procurement in humans with specific diseases. The finding of ARX and DNMT1 loss or reduction in Glucagon+ Insulin+ cells from two subjects with T1D corroborates prior studies in mice and human cell lines suggesting that Arx or Dnmt1 establish and/or maintain α-cell fate and function (Collombat et al., 2003; Collombat et al., 2007; Collombat et al., 2009; Avrahami et al., 2015; Gage et al., 2015). Moreover, the abnormal expression of non α-cell factors such as NKX6.1 and PDX1 in a subset of Glucagon+ cells is consistent with prior reports of impaired α-cell function in T1D (Cryer et al., 2003; Pietropaolo et al., 2013). Here, we found Glucagon+ Insulin+ cells in the pancreata of two T1D donors less than 10 years of age and with 4–5 years of disease, but not in three samples from older donors with longer disease duration. The possibility that formation or maintenance of bi-hormonal Glucagon+ Insulin+ DNMT1Neg cells in humans with T1D might depend on subject age, duration of disease, or other variables requires additional studies with more sampling. A previous study has reported the presence of residual Insulin C-peptide in subjects with T1D of several decades’ duration (Keenan et al, 2010). Since insulin or glucagon levels in our younger subjects are not known, the physiological relevance of these bi-hormonal cells remains unclear. Nevertheless, our findings suggest a potential molecular basis for compromised human α-cell function in at least a subset of T1D patients. In summary, studies of human and mouse α-cells here advance the understanding of α-cell defects in T1D, and promote the plausibility of targeted intra-islet cell conversion for regenerative goals.
Experimental Procedures
Human Tissues
De-identified normal human pancreas specimens and pancreas specimens from type 1 diabetic donors were obtained from the International Institute for the advancement of Medicine (IIAM) and the network of pancreatic organ donors (nPOD) (Table 1).
Mouse studies
Glucagon-rtTA, Tet-o-Cre, R26-YFP, mice have been described previously (Thorel et al., 2010). We generated 4 types of mice, all harboring the α-cell lineage tracing system (Glucagon-rtTA, Tet-o-cre, R26-YFP): 1) Control (with only the α-cell lineage tracing system), 2) αiAKO (with the addition of floxed alleles to inactivate Arx - either Arxf/Y or Arxf/f) 3) αiADKO (with the addition of floxed alleles to inactivate both Arx –Arxf/Y or Arxf/f, and Dnmt1 - Dnmt1f/f ), 4) αiDKO (with addition of floxed alleles to inactivate Dnmt1 – Dnmt1f/f). To achieve lineage labeling and inactivation of Arx, Dnmt1 or both, Doxycycline (DOX; Sigma) was administered via the drinking water prepared freshly every 2 days at 2 mg/mL for a total exposure of 3 weeks. After DOX removal, mice were maintained for an additional 4 weeks or 12 weeks without DOX treatment before sacrifice. Glucagon-Venus mice in which cells that express proglucagon are labeled by the yellow fluorescent protein Venus have been previously described (Reimann et al., 2008).
Immunohistology and Confocal Microscopy
Human pancreas sections were stained with antibodies against a panel of endocrine as described previously (Chen et al., 2011). In brief, slides were washed with PBS, blocked with normal donkey serum (5%) and primary antibodies were applied (Table S5). Antigen retrieval was performed for specified antibodies (Table S5) using Antigen retrieval solution (S1699; DAKO; Carpinteria, CA) according to the manufacturer’s instructions. Signal amplification using a biotin-streptavidin system (SP-2002; Vector Laboratories; Burlingame, CA), and tyramide signal amplification (T30955; ThermoFisher; Grand Island NY) was performed for specified antibodies (Table S5). For all mouse sections, in addition to the antibodies above, anti-YFP (yellow fluorescent protein) was used to detect YFP in lineage-marked α-cells, anti-Neurog3, Ki67, PPY, Ghrelin, and Glut2 antibodies were used. Fluorescent secondary antibodies used were from Jackson ImmunoResearch (West Grove, PA), or Molecular Probes (Eugene, OR) (Table S6). Stained sections were mounted with VECTASHIELD Mounting Medium with Dapi (H-1200, Vector Laboratories) and visualized using a Leica SP2 inverted confocal laser scanning microscope (Supplemental Methods).
Immuno-morphometry
For human sections, 60–100 islets were analyzed per staining and sample. Cells coexpressing GCG and a marker representative of another endocrine cell type were quantified as a percentage of the total number of GCG+ cells counted. For mouse sections, islets were counted from 4 mice per genotype (Tables S1–3). These analyses were non-randomized. The number of α-cells undergoing conversion into other endocrine cell types (where ‘conversion’ is defined as the expression of β, δ, or other endocrine cell genes in YFP+ cells) was quantified as a percentage of total YFP+ cells counted. Two-tailed student’s t-test was used to determine whether the difference in the percentage of Ki67+ YFP+ cells between knock out and control mice was statistically significant (P-value > 0.05).
Statistical Analysis
Where indicated in the legends, graphed data in the figures are represented as mean ± Standard Deviation (S.D.) for immunomorphometry analyses.
Flow Cytometry
Isolated mouse islets were dissociated into single cells and processed as described (Supplemental Methods). We collected cells from YFP+, lineage-traced population and YFPNeg, non-labeled population on a special order 5-laser FACS Aria II directly into 96-well containing 4 μL of lysis buffer with dNTPs37 for downstream single-cell RNA-Seq assays.
Single-Cell RNA-Seq and Data Analysis
Single-cell RNA-Seq libraries were generated as described (Picelli et al., 2014). Briefly, single cells were lysed, followed by reverse transcription, pre-amplification, DNA purification and analysis for successful amplification products. Barcoded sequencing libraries were prepared, libraries were pooled and sequenced on the Illumina NextSeq instrument (Supplemental Methods). Transcript counts were obtained using HT-Seq (Anders et al., 2015) and mm10 UCSC exon/transcript annotations. Pairwise distances between cells were estimated using Pearson correlation of overdispersed genes as described (Fan et al., 2016). Subsequent hierarchical clustering was done using hclustfunction in R, and dimension reduction was performed using the t-SNE method on pairwise distances (Van der Maaten and Hinton, 2008). Data were also analyzed with QIAGEN Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity). The GEO accession number is GSE79457.
Electrophysiological studies
Islets from control or αiADKO mice were dispersed to single cells and plated overnight on 35-mm dishes as previously (Dai et al., 2011). Cells were patch-clamped in the whole-cell voltage-clamp configuration and Na+ channels were activated by a depolarization to 0 mV following holding potentials ranging from −140 to 0 mV. Single cell exocytosis was measured as described previously (Ferdaoussi et al., 2015). Briefly, cells were pre-incubated at either 2 or 20 mM glucose for 1 hour and transferred to bath solution (Supplemental methods) with either 20 or 2 mM glucose ~10–30 minutes prior to patch-clamping. Exocytosis was elicited by a series of ten 500-ms membrane depolarizations from −70 to 0 mV and monitored as increases in cell capacitance. Following the experiments cells were immunostained for insulin and YFP to identify β-cells (Ins+ only), α-cells (YFP+ only) or converted α-cells (Ins+,YFP+). Statistical analysis of exocytosis data was by 2-way ANOVA followed by Bonferroni post-test (P<0.05 considered significant).
Hormone secretion and Calcium Imaging
Hormone secretion and calcium imaging studies were performed as previously described (Adewola et al., 2010; Xing et al., 2016, Supplemental Methods). Briefly, islets from MIP-GFP, Glucagon-Venus, and αiADKO mice were dispersed into single cells and GFP+, Venus+ or YFP+ cells were collected by FACS as described above. For calcium imaging, the sorted cells were incubated in Kreb’s Ringer Buffer (KRB) with 2mM glucose and 5μM Fura-2/AM (Molecular Probes, CA) for 30 minutes, then loaded into a temperature equilibrated microfluidic device mounted on an inverted epifluorescence microscope. KRB with 14 mM glucose or 2mM glucose with 30mM KCl was administered to the cells for 20 minutes and 15 minutes respectively. Dual-wavelength Fura-2/AM was excited at 340 and 380 nm (shift in excitation wavelength occurs upon binding Ca2+), and fluorescent emission was detected at 510 nm. Intracellular Ca2+ concentration was expressed as a ratio of fluorescent emission intensity (% F340/F380). The fluorescence signal was expressed as a change in percentage after being normalized to basal intensity levels established before stimulation. For hormone secretion studies, 5000 GFP+ cells from MIP-GFP mice, Venus+ cells from Glucagon-Venus mice or YFP+ cells from αiADKO mice were collected by FACS and loaded onto the microfluidic device. To measure insulin secretion, YFP+ cells were incubated in basal KRB with 2mM glucose for 30 mins and then stimulated with KRB containing 14 mM glucose for 30 mins followed by 2 mM glucose for 10 mins. To measure glucagon secretion, YFP+ cells were incubated in KRB with 11.2 mM glucose for 30 min and then stimulated with KRB with 2 mM glucose for 30 minutes. Ultrasensitive Rodent Insulin or Glucagon ELISAs (Mercodia, Uppsala, Sweden) were used to measure perfusate insulin or glucagon levels.
Supplementary Material
Acknowledgments
We thank Dr. H.E. Arda for discussions for FACS experiments, Dr. Y. Hang for help with islet isolations, members of the Kim lab, especially Dr. S. Park, for advice, and Drs. J. A. Golden, and L. Jackson-Grusby for mouse strains. We also thank N. Neff and G. Mantalas for assistance with sequencing. H.C. was supported by a T32 training award to the Endocrinology Division, Dept. of Medicine, Stanford University School of Medicine, and fellowships from the Stanford Child Health Research Institute, and the JDRF. C.D. was supported by Stanford University Vice-Provost Undergraduate Education grants. N.D. was supported by a grant from the Institute of Genetics and Genomics of Geneva. Effort in the Quake group was supported by National Institutes of Health grants U01-HL099999 and U01-HL099995, California Institute of Regenerative Medicine grant GC1R-06673, Center of Excellence for Stem Cell Genomics, The Wallenberg Foundation Postdoctoral Scholarship Program at Stanford and the Howard Hughes Medical Institute (HHMI), in the Herrera group by grants from JDRF, the Swiss National Science Foundation, the U.S. NIH Beta-cell Biology Consortium (BCBC), and the European Union (IMIDIA), and in the Kim group by HHMI, the H.L. Snyder Foundation, the Elser Trust, by grants from JDRF, the NIH BCBC (UO1DK089532 and UO1DK089572) and NIH Human Islet Resource Network (UC4DK104211).
Footnotes
Author Contributions. H.C. and N.D. performed mouse and human pancreas studies with X.G. and C.D., K.L. J.W., X.G., and K.L. assisted H.C with tissue collection and analysis from mice or humans. H.C, X.G., C.D. and N.D. performed immunohistochemistry and confocal microscopy. K.L. performed FACS analysis. M.E. and S.Q. performed and analyzed RNA-Seq from sorted cells. S.C. performed Ingenuity Pathway Analysis on the single cell RNA-Seq data. X.Q and P.E.M performed electrophysiological studies. Y.W., Y.X. and J.O. performed calcium imaging and hormone measures from dispersed cells. F.T., N.D. and P.L.H. generated mouse strains. H.C and S.K.K. conceived the experiments, and H.C, P.L.H and S.K.K wrote the manuscript. None of the authors of this manuscript have a financial interest related to this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adewola AF, Lee D, Harvat T, Mohammed J, Eddington DT, Oberholzer J, Wang Y. Microfluidic perifusion and imaging device for multi-parametric islet function assessment. Biomed Microdevices. 2010;12(3):409–17. doi: 10.1007/s10544-010-9398-1. [DOI] [PubMed] [Google Scholar]
- Arda HE, Li L, Tsai J, Torre EA, Rosli Y, Peiris H, Spitale RC, Dai C, Gu X, Qu K, Wang P, Wang J, Grompe M, Scharfmann R, Snyder MS, Bottino R, Powers AC, Chang HY, Kim SK. Age-dependent pancreatic gene regulation reveals mechanisms governing human β-cell function. Cell Metab. 2016;23(5):909–20. doi: 10.1016/j.cmet.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinci E, Banga A, Greder LV, Dutton JR, Slack JM. Reprogramming of pancreatic exocrine cells towards a beta (β) cell character using Pdx1, Ngn3 and MafA. Biochem J. 2012;442(3):539–550. doi: 10.1042/BJ20111678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HT-Seq – a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RM, et al. Loss of Dnmt1 catalytic activity reveals multiple roles for DNA methylation during pancreas development and regeneration. Dev Biol. 2009;334(1):213–23. doi: 10.1016/j.ydbio.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arda HE, Benitez CM, Kim SK. Gene regulatory networks governing pancreas development. Dev Cell. 2013;25(1):5–13. doi: 10.1016/j.devcel.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avrahami D, et al. Aging-Dependent Demethylation of Regulatory Elements Correlates with Chromatin State and Improved β-cell Function. Cell Metab. 2015;22(4):619–32. doi: 10.1016/j.cmet.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez CM, et al. An integrated cell purification and genomics strategy reveals multiple regulators of pancreas development. Plos Genet. 2014;10(10):e1004645. doi: 10.1371/journal.pgen.1004645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boj SF, Petrov D, Ferrer J. Epistasis of transcriptomes reveals synergism between transcriptional activators Hnf1a and Hnf4a. Plos Genet. 2010;6(5):e1000970. doi: 10.1371/journal.pgen.1000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramswig NC, Everett LJ, Schug J, Dorrell C, Liu C, Luo Y, Streeter PR, Naji A, Grompe M, Kaestner KH. Epigenomic plasticity enables human pancreatic α to β cell reprogramming. J Clin Invest. 2013;123(3):1275–84. doi: 10.1172/JCI66514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Gu X, Liu Y, Wang J, Wirt SE, Bottino R, Schorle H, Sage J, Kim SK. PDGF signaling controls age-dependent proliferation in pancreatic beta-cells. Nature. 2011;478:349–355. doi: 10.1038/nature10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chera S, et al. Diabetes Recovery By Age-Dependent Conversion of Pancreatic δ-Cells Into Insulin Producers. Nature. 2014;514:503–507. doi: 10.1038/nature13633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat P, Mansouri A, Hecksher-Sorenson J, Serup P, Krull J, Gradwohl G, Gruss P. Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev. 2003;17(20):2591–603. doi: 10.1101/gad.269003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat P, Hecksher-Sorenson J, Krull J, Berger J, Riedel D, Herrera PL, Serup P, Mansouri A. Embryonic endocrine pancreas and mature beta cells acquire alpha and PP cell phenotypes upon Arx misexpression. J Clin Invest. 2007;117(4):961–70. doi: 10.1172/JCI29115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat P, Xu X, Ravassard P, Sosa-Pineda B, Dussaud S, Billestrup N, Madsen OD, Serup P, Heimberg H, Mansouri A. The ectopic expression of Pax4 in the mouse pancreas converts progenitor cells into alpha and subsequently beta cells. Cell. 2009;138(3):449–62. doi: 10.1016/j.cell.2009.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney M, et al. The inactivation of Arx in pancreatic α-cells triggers their neogenesis and conversion into functional β-like cells. Plos Genet. 2013;9(10):e1003934. doi: 10.1371/journal.pgen.1003934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryer PE, Davis SN, Shamoon H. Hypoglycemia in Diabetes. Diabetes Care. 2003;26(6):1902–12. doi: 10.2337/diacare.26.6.1902. [DOI] [PubMed] [Google Scholar]
- Dai XQ, Plummer G, Casimir M, Kang Y, Hajmrle C, Gaisano HY, Manning Fox JE, MacDonald PE. SUMOylation regulates insulin exocytosis downstream of secretory granule docking in rodents and humans. Diabetes. 2011;60(3):838–47. doi: 10.2337/db10-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai XQ, Spigelman AF, Khan S, Braun M, Manning Fox JE, MacDonald PE. SUMO1 enhances cAMP-dependent exocytosis and glucagon secretion from pancreatic α-cells. J Physiol. 2014;592(17):3715–26. doi: 10.1113/jphysiol.2014.274084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan S, Georgia S, Tschen SI, Fan G, Bhushan A. Pancreatic β cell identity is maintained by DNA methylation-mediated repression of Arx. Dev Cell. 2011;20(4):419–29. doi: 10.1016/j.devcel.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan S, Tschen SI, Zeng C, Guo T, Hebrok M, Matyevenko A, Bhushan A. DNA methylation directs functional maturation of pancreatic β cells. J Clin Invest. 2015;125(7):2851–60. doi: 10.1172/JCI79956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Gingeras TR. Mapping RNA-seq Reads with STAR. Curr Protoc Bioinformatics. 2015;3(51):11. doi: 10.1002/0471250953.bi1114s51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, Chen J, Ding S. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol. 2011;13(3):215–22. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- Fan A, et al. Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nat Methods. 2016;13(3):241–4. doi: 10.1038/nmeth.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdaoussi M, et al. Isocitrate-to-SENP1 signaling amplifies insulin secretion and rescues dysfunctional β cells. J Clin Invest. 2015;25(10):3847–60. doi: 10.1172/JCI82498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulp CT, Cho G, Marsh ED, Nasrallah IM, Labosky PA, Golden JA. Identification of Arx transcriptional targets in the developing basal forebrain. Hum Mol Genet. 2008;17(23):3740–60. doi: 10.1093/hmg/ddn271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage BK, Asadi A, Baker RK, Webber TD, Wang R, Itoh M, Hayashi M, Miyata R, Akashi T, Kieffer TJ. The role of ARX in Human Pancreatic Endocrine Specification. Plos One. 2015;10(12):e0144100. doi: 10.1371/journal.pone.0144100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopel SO, Kanno T, Barg S, Weng XG, Gromada J, Rorsman P. Regulation of glucagon release in mouse-cells by KATP channels and inactivation of TTX-sensitive Na+ channels. J Physiol. 2000;528:509–20. doi: 10.1111/j.1469-7793.2000.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradwohl G, Dierich A, LeMeur M, Guillemot F. Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci USA. 2000;97(4):1607–11. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G, Dubaushaite J, Melton DA. Direct evidence for the pancreatic lineage:NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129(10):2447–57. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- Gylfe E. Glucose control of glucagon secretion-‘There’s a brand-new gimmick every year’. Ups J Med Sci. 2016;121(2):120–32. doi: 10.3109/03009734.2016.1154905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter CS. Hnf1a (Mody3) regulates b-cell-enriched MafA transcription factor expression. Mol Endocrinol. 2011;25(2):339–47. doi: 10.1210/me.2010-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Takizawa Y, Hanai S, Okazaki S, Miyata R, Inoue T, Akashi T, Hayashi M, Goto Y. Partial loss of pancreas endocrine and exocrine cells of human ARX-null mutation: consideration of pancreas differentiation. Differentiation. 2010;80(2–3):118–22. doi: 10.1016/j.diff.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Jackson-Grusby L, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 27(1):31–9. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- Keenan HA, Sun JK, Levine J, Doria A, Aiello LP, Eisenbarth G, Bonner-Weir S, King GL. Residual insulin production and pancreatic β-cell turnover after 50 years of diabetes: Joslin Medalist Study. Diabetes. 2010;59(11):2846–53. doi: 10.2337/db10-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Baldwin K, Gao S, Bücker C, Parmar M, Benvenisty N. iPSCs: 10 years and counting. Cell. 2016;165(5):1041–1042. doi: 10.1016/j.cell.2016.05.027. [DOI] [PubMed] [Google Scholar]
- Kordowich S, Collombat P, Mansouri A, Serup P. Arx and Nkx2.2 compound deficiency redirects pancreatic alpha- and beta-cell differentiation to a somatostatin/ghrelin co-expressing cell lineage. BMC Dev Biol. 2011;11(52):1471–213. doi: 10.1186/1471-213X-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestro MA, Cardalda C, Boj SF, Luco RF, Servitja JM, Ferrer J. Distinct roles of Hnf1beta, Hnf1alpha, and Hnf4alpha in regulating pancreas development, beta-cell function and growth. Endocr Dev. 2007;12:33–45. doi: 10.1159/000109603. [DOI] [PubMed] [Google Scholar]
- Mastracci TL, Wilcox CL, Arnes L, Panea C, Golden JA, May CL, Sussel L. Nkx2.2 and Arx genetically interact to regulate pancreatic endocrine cell development and endocrine hormone expression. Dev Biol. 2011;359(1):1–11. doi: 10.1016/j.ydbio.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran I, et al. Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16(4):435–48. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan TH. Regeneration and Liability to Injury. Science. 1901;14:235–248. doi: 10.1126/science.14.346.235. [DOI] [PubMed] [Google Scholar]
- Morris SA, Daley GQ. A blueprint for engineering cell fate, current technologies to reprogram cell identity. Cell Res. 2013;1:33–48. doi: 10.1038/cr.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papizan JB, Singer RA, Tschen SI, Dhawan S, Friel JM, Hipkens SB, Magnuson MA, Bhushan A, Sussel L. Nkx2.2 repressor complex regulates islet β-cell specification and prevents β-to-α-cell reprogramming. Genes Dev. 2011;25(21):2291–305. doi: 10.1101/gad.173039.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Stoffers DA, Nicholls RD, Simmons RA. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J Clin Invest. 2008;118(6):2316–24. doi: 10.1172/JCI33655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli H, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9(1):171–81. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Pietropaolo M. Persistent C-peptide: what does it mean? Curr Opin Endocrinol Diabetes Obes. 2013;20(4):279–84. doi: 10.1097/MED.0b013e3283628610. [DOI] [PubMed] [Google Scholar]
- Piran R, Lee SH, Li CR, Charbono A, Bradley LM, Levine F. Pharmacological induction of pancreatic islet cell transdifferentiation: relevance to type 1 diabetes. Cell Death Dis. 2014;5:e1357. doi: 10.1038/cddis.2014.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman F, Habib AM, Tolhurst G, Parker HE, Rogers GJ, Gribble FM. Cell Metab. 2008;8(6):532–9. doi: 10.1016/j.cmet.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer AE, et al. Nkx6.1 controls a gene regulatory network required for establishing and maintaining pancreatic Beta cell identity. Plos Genet. 2013;1(9):e1003274. doi: 10.1371/journal.pgen.1003274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu J, et al. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell. 2013;153(5):963–75. doi: 10.1016/j.cell.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorel F, Nepote V, Avril I, Kohno K, Desgraz R, Chera S, Herrera PL. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature. 2010;464:1149–1154. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Maaten L, Hinton G. Visualizing data using t-SNE. Journal of Machine Learning Research. 2008;9:2579–2605. [Google Scholar]
- Wilcox CL, Terry NA, Walp ER, Lee RA, May CL. Pancreas α-cell specific deletion of mouse Arx leads to α-cell identity loss. Plos One. 2013;8(6):e66214. doi: 10.1371/journal.pone.0066214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YP, Thorel F, Boyer DF, Herrera PL, Wright CV. Context-specific α-to- β-cell reprogramming by forced Pdx1 expression. Genes Dev. 2011;25(16):1680–5. doi: 10.1101/gad.16875711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda S, et al. Predominance of β-cell neogenesis rather than replication in humans with an impaired glucose tolerance and newly diagnosed diabetes. J Clin Endocrinol Metab. 2013;95(5):2053–61. doi: 10.1210/jc.2012-3832. [DOI] [PubMed] [Google Scholar]
- Xing Y, Nourmohamadzadeh M, Elias JE, Chan M, Chen Z, McGarrigle JJ, Oberholzer J, Wang Y. A pumpless microfluidic device driven by surface tension for pancreatic islet analysis. Biomed Microdevices. 2016;18(5):80. doi: 10.1007/s10544-016-0109-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.