ABSTRACT
The intestine plays a central role in digestion, nutrient absorption and metabolism, with individual regions of the intestine having distinct functional roles. Many examples of region-specific gene expression in the adult intestine are known, but how intestinal regional identity is established during development is a largely unresolved issue. Here, we have identified several genes that are expressed in a region-specific manner in the developing human intestine. Using human embryonic stem cell-derived intestinal organoids, we demonstrate that the duration of exposure to active FGF and WNT signaling controls regional identity. Short-term exposure to FGF4 and CHIR99021 (a GSK3β inhibitor that stabilizes β-catenin) resulted in organoids with gene expression patterns similar to developing human duodenum, whereas longer exposure resulted in organoids similar to ileum. When region-specific organoids were transplanted into immunocompromised mice, duodenum-like organoids and ileum-like organoids retained their regional identity, demonstrating that regional identity of organoids is stable after initial patterning occurs. This work provides insights into the mechanisms that control regional specification of the developing human intestine and provides new tools for basic and translational research.
KEY WORDS: Human, Intestine, Organoid, Patterning, Pluripotent stem cells
Summary: Human embryonic stem cell-derived intestinal organoids can be patterned into duodenum-like or ileum-like tissue, recapitulating in vivo human development.
INTRODUCTION
The mature gastrointestinal tract is a highly compartmentalized organ with distinct regions that serve specific roles in digestion, absorption, hormone secretion and immunity. Many studies have detailed the complex reciprocal interactions between the endoderm-derived epithelium and the mesoderm-derived stroma for establishing regional identity along the anterior-posterior axis of the gut tube in the early embryo (Dessimoz et al., 2006; Duluc et al., 1994, 2001; Kedinger et al., 1998; Ratineau et al., 2003; Sherwood et al., 2011; Wells and Melton, 2000). However, very little is known about the mechanisms in place after intestinal specification that pattern the duodenum, jejunum and ileum, which make up the regions of the small intestine from proximal to distal.
One of the earliest requirements for intestinal development is the establishment of embryonic posterior identity (Wells and Melton, 1999; Zorn and Wells, 2007). Although the embryo receives anterior-posterior (A-P) positional information prior to and during gastrulation, posterior endoderm identity is not irreversibly specified until after gastrulation (Ho and Kimmel, 1993; Horb and Slack, 2001). At this stage, there are several signaling pathways that promote the posterior patterning of the vertebrate embryo. The WNT and FGF signaling pathways play a central role in establishing the posterior axis of the vertebrate embryo, and Caudal homeobox (Cdx) genes, which are transcription factors that regulate intestinal specification, development and maintenance of regional identity, are targets of these signaling pathways (Beck and Slack, 1999; Beland et al., 2004; Bradley et al., 2000; Cox and Hemmati-Brivanlou, 1995; Dale et al., 1992; Dessimoz et al., 2006; Domingos et al., 2001; Erter et al., 2001; Gao et al., 2009; Grainger et al., 2010; Greco et al., 1996; Gregorieff et al., 2004; Hollyday et al., 1995; Huelsken et al., 2000; Ikeya and Takada, 2001; Isaacs et al., 1998; Keenan et al., 2006; Kiecker and Niehrs, 2001; Lekven et al., 2001; Lickert et al., 2000; Liu et al., 1999; Marvin et al., 2001; McLin et al., 2007; Northrop and Kimelman, 1994; Parr and McMahon, 1995; Pownall et al., 1996; Satoh et al., 2006; Verzi et al., 2010, 2013).
FGF and/or WNT signaling controls posterior fate and intestinal lineage commitment in mouse or human pluripotent stem cells (PSCs) (Cao et al., 2011, 2015; Hannan et al., 2013; Sherwood et al., 2011; Tamminen et al., 2015), and a combination of WNT and FGF signaling can induce an intestinal fate in human definitive endoderm (DE), which gives rise to human intestinal organoids (McCracken et al., 2011; Spence et al., 2011). Although it is clear that hPSC-derived intestinal organoids are small intestinal in identity (Finkbeiner et al., 2015b; Spence et al., 2011; Watson et al., 2014), their exact regional identity (duodenum, jejunum, ileum) is unclear. Here, we sought to clarify the regional identity of organoids, and to determine whether organoids could be patterned into different regions of the intestinal tract. Given that the posterior endoderm of the developing embryo is exposed to higher concentrations and longer durations of growth factors (Arnold and Robertson, 2009), we hypothesized that the duration of exposure to active FGF and WNT signaling would control regional intestinal identity. In this study, we take advantage of hESC-derived intestinal organoids to test this hypothesis. Our results demonstrate that the duration of exposure to active WNT/β-catenin (using CHIR99021, a GSK3β inhibitor) and FGF (FGF4) signaling results in gene and protein expression profiles that are consistent with tissue that has been patterned into proximal (duodenum) or distal (ileum) small intestine, respectively. To validate our findings, we used both candidate gene expression analysis to compare organoids with the human fetal intestine and an unbiased transcriptome-level approach. To determine whether ‘patterned’ organoids retain their regional identity in vivo, we transplanted organoids into mice, which allows maturation of organoids into adult-like tissue (Finkbeiner et al., 2015b; Watson et al., 2014).
Taken together, our findings shed light on the mechanisms that control regional identity of the developing human intestine. Regionally specified organoids provide a platform for uncovering the genes and signaling pathways that are responsible for common congenital malformations or for functional adaptation of the adult gastrointestinal tract following injury. We suggest that the impact of regional identity is an important consideration in such studies.
RESULTS
Regional identity markers in human fetal intestine
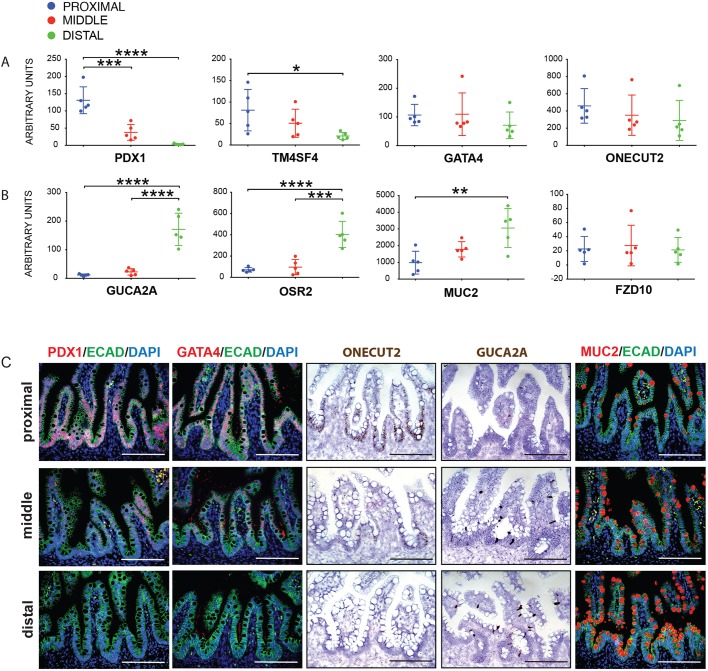
Many genes with differential regional identity in the small intestine have been identified in the adult human and mouse intestine, where gene expression reflects adult region-specific intestinal function, as well as in the developing murine intestine. However, in the developing/fetal human intestine, genes corresponding to adult function are expressed at very low levels (Finkbeiner et al., 2015b), and therefore adult-stage markers do not faithfully identify regional identity in the embryo. Fortunately, several regional identity markers have been described in the fetal mouse intestine (Battle et al., 2008; Dusing et al., 2001; Gao and Kaestner, 2010; Sherwood et al., 2009). To identify a cohort of markers that are regionally expressed in the human intestine, we assessed expression of genes and proteins orthologous to those enriched in embryonic mouse regions by qRT-PCR, in situ hybridization and immunofluorescent staining (Fig. 1; n=5, independent biological samples ranging from 14-19 weeks of gestation). Intestines from human fetuses were obtained and divided into thirds, corresponding to the proximal, middle and distal regions of the small intestine. We observed that PDX1 and TM4SF4 were enriched in the proximal intestine, similar to the embryonic mouse proximal intestine (Sherwood et al., 2011), whereas the expression of GATA4 and ONECUT2 showed non-statistically significant trends of higher expression in the proximal human intestine (Fig. 1A). Of note, some individual samples had more pronounced region-specific expression of GATA4 and ONECUT2 when technical replicates were examined (Fig. S1), and regional-specific expression was confirmed for ONECUT2 using in situ hybridization (Fig. 1C), suggesting that region-specific gene expression may be dynamic over time, or may vary significantly between biological specimens; however, additional studies at each time point will be required to more conclusively assess biological variation or time-dependent changes. Guca2a, Osr2, Muc2, Fzd10, Cib2 and several Hox genes have higher distal gene expression levels in mice (Gao and Kaestner, 2010; Sherwood et al., 2009). In the human fetal intestine, GUCA2A, OSR2 and MUC2 showed increased expression in the distal small intestine (Fig. 1B), along with HOXB6 (Fig. S1). We confirmed proximal enrichment of PDX1 and ONECUT2 and distal enrichment of MUC2 and GUCA2A using immunofluorescence and in situ hybridization (Fig. 1C). Together, these data identify a cohort of molecular markers that are regionally expressed in the human fetal small intestine and demonstrate that some of these regional identifiers are conserved between the mouse and human fetal small intestine.
Fig. 1.
Identification of regionally expressed molecular markers in the human fetal intestine. (A) Genes known to be enriched in the proximal developing mouse intestine, including PDX1, GATA4, TM4SF4 and ONECUT2 were examined in different regions of the human fetal intestine (n=5 individual biological specimens; proximal, blue; middle, red; distal, green). (B) Genes know to be enriched in the distal developing mouse intestine, including GUCA2A, OSR2, MUC2 and FZD10 were examined in different regions of the human fetal intestine (n=5 individual biological specimens; proximal, blue; middle, red; distal, green). (C) Enrichment of PDX1 and GATA4 protein as assessed by immunofluorescence and of ONECUT2 mRNA as assessed by in situ hybridization was confirmed in the proximal region of the fetal intestine, whereas GUCA2A mRNA and MUC2 protein were enriched in the distal fetal intestine when assessed by in situ hybridization and immunofluorescence, respectively. Scale bars: 200 μm.
Prolonged WNT/FGF signaling distalizes hESC-derived intestinal organoids
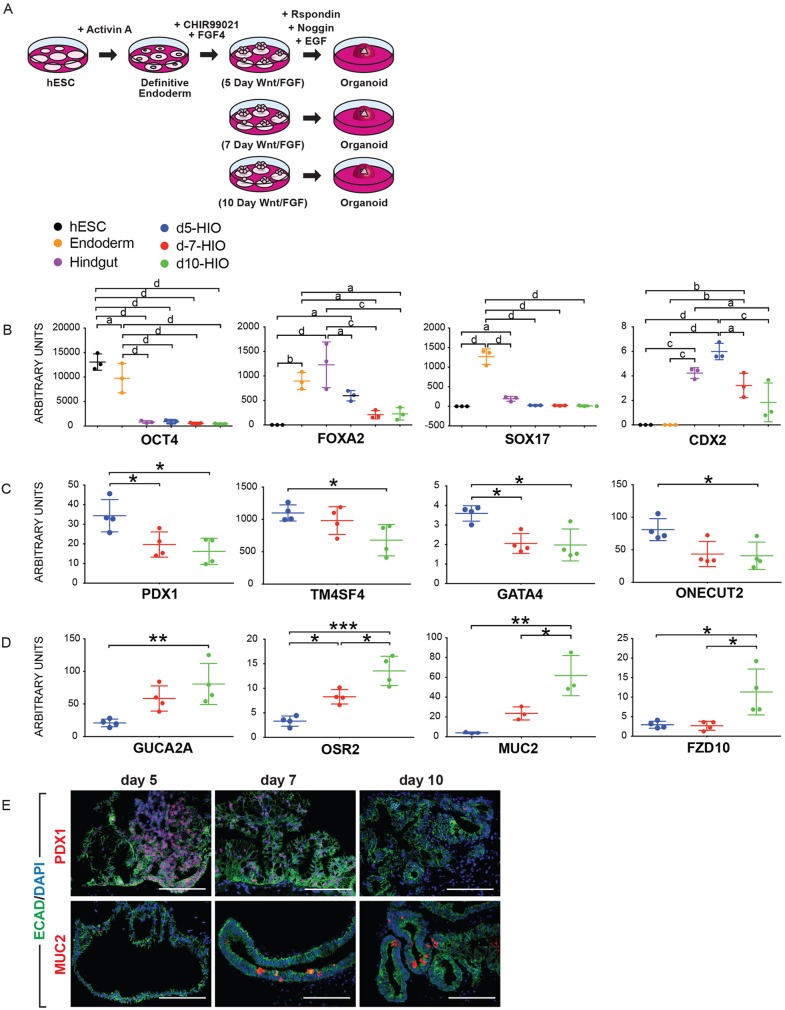
To examine the effects of WNT and FGF signaling on developing human intestinal tissue, we took advantage of the hESC-derived intestinal organoid culture system (Spence et al., 2011). hESCs were exposed to activin A for 3 days to induce endoderm, which was then exposed to CHIR99021/FGF4-enriched media to induce mid/hindgut spheroid formation, as previously described (Finkbeiner et al., 2015a,b; Xue et al., 2013). Spheroids began to bud from monolayer cultures after 4 days and continuously generated new spheroids for over 10 days; however, incubation beyond 10 days resulted in far fewer spheroids (data not shown). Spheroids were collected from the cultures after 5 days (d5), 7 days (d7) or 10 days (d10), and embedded into matrigel (Fig. 2A). Spheroids were expanded into larger human intestinal organoids for 30-35 days in intestinal growth medium containing EGF, noggin and R-spondin 2 (Fig. 2A). qRT-PCR performed on tissues collected at progressive stages of organoid differentiation, including undifferentiated hESCs, definitive endoderm, hindgut tissue after 4 days of CHIR99021/FGF4 and organoids generated after d5, d7 and d10, showed the expected stage-specific mRNA expression of pluripotency genes (OCT4), endoderm genes (FOXA2, SOX17) and the mid-hindgut and intestinal specification gene CDX2 (Fig. 2B).
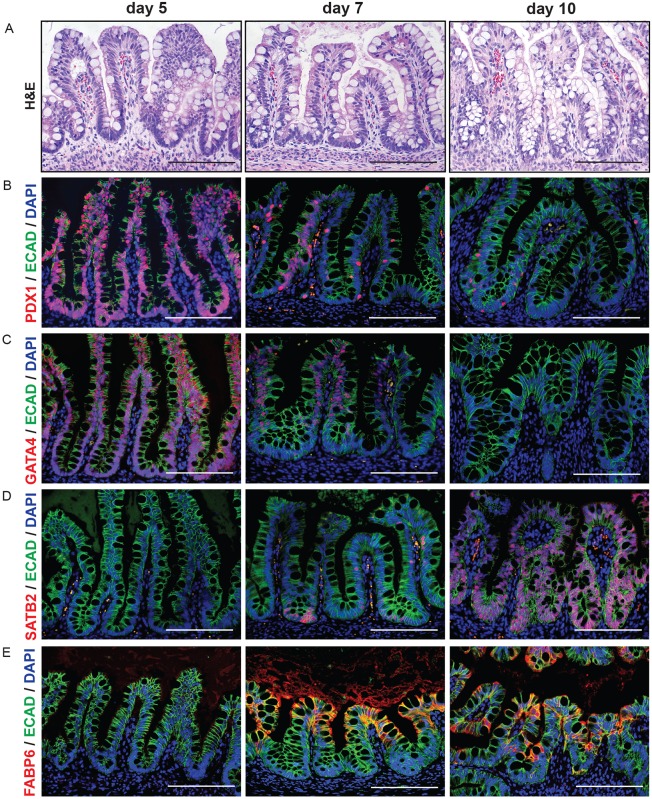
Fig. 2.
Human intestinal organoids are patterned by FGF and WNT signaling. (A) Schematic of experimental design showing spheroids generated in culture over increasing periods of time. (B) Expression of OCT4, FOXA2, SOX17 and CDX2 during differentiation in undifferentiated hESCs, in endoderm and hindgut (4 days after FGF4/CHIR99021), and in organoids derived from d5, d7 and d10 cultures (d5, blue; d7, red; d10, green). (C) Markers shown to be enriched in the human fetal duodenum (Fig. 1), including PDX1, GATA4, TM4SF4 and ONECUT2 were examined in d5, d7 and d10 organoids. (D) Markers shown to be enriched in the human fetal ileum (Fig. 1), including MUC2, OSR2, MUC2 and FZD10 were examined in d5, d7 and d10 organoids. (E) Immunofluorescence demonstrated that PDX1 protein expression was enriched in d5 organoids, whereas MUC2 protein expression was enriched in d7 and d10 organoids. Scale bars: 200 μm.
We next evaluated the effects of exposing human DE to CHIR99021 and FGF4 for different lengths of time, and examined the expression of region-specific markers of the developing human intestine identified in Fig. 1. Similar to the human fetal intestine, we found that d5 human organoids had significantly higher expression of the proximal identity marker genes PDX1, TM4SF4, GATA4 and ONECUT2 compared with d7 and d10 organoids (Fig. 2C). Conversely, distally enriched intestinal genes GUCA2A, OSR2 and MUC2 were expressed at significantly higher levels in d10 organoids compared with d5 and d7 organoids (Fig. 2D). Interestingly, whereas FZD10 was not regionally expressed in the distal human fetal intestine, it was significantly higher in d10 organoids (Fig. 2D). Although further exploration of this observation is warranted, one possible explanation for this discrepancy is that organoids may represent an earlier stage of development than the human fetal intestine used in this study. Immunostaining confirmed that PDX1 was more abundant in d5 organoids, and that MUC2 was more abundant in d7 and d10 organoids (Fig. 2E).
To further validate our findings, we conducted a series of additional experiments (Fig. S2). We treated endoderm for 5 days with FGF4 plus CHIR99021 (500 ng/ml FGF4, 2 μM CHIR99021), and then varied the concentration of CHIR99021 for the next 5 days, or alternatively, removed CHIR99021 and added IWP2, a WNT inhibitor, or the FGF and ERK inhibitors SU5402 or U0126, respectively (Fig. S2). Spheroids from all conditions were embedded in matrigel and expanded into organoids for 30 days, and were then compared for regional gene expression. As expected, d5 and d10 organoids demonstrated region-specific gene expression when grown in standard conditions (as in Fig. 2, 500 ng/ml FGF4+2μM CHIR99021). However, when CHIR99021 concentrations were reduced, d10 organoids expressed much higher levels of proximal markers but this did not lead to reduced posterior marker gene expression. Interestingly, when WNT signaling was blocked between d5 and d10 (FGF4+IWP2), expression of proximal genes was enhanced and distal gene expression was reduced when compared with d5 and d10 organoids, respectively. These data suggest that, during distalization, one of the roles of WNT/β-catenin may be to repress proximal genes while inducing posterior genes (Fig. S2). In total, these experiments show that prolonged exposure to high levels of CHIR99021 is required for expression of distal intestinal markers.
Regional identity is maintained over time in vitro
It is unclear whether organoids change over time in culture, and it is also unknown whether regional identity might also be determined by the length of time spent in culture. To test this, we generated d5, d7 and d10 organoids, and examined them after 1 month and after 90 days in culture (compare Fig. 2 with Fig. S3). Although some individual regional identity markers changed over time in culture, the trends were similar at the two different time points: d5 organoids had more abundant proximal marker expression with low distal marker expression whereas d10 organoids had low proximal marker expression with enriched distal marker expression.
Whole-transcriptome profiling by RNA sequencing demonstrates that organoids are patterned into proximal and distal intestine
Because the expression patterns of several markers of human fetal proximal-distal intestinal identity suggested that d5 organoids are similar to human duodenum and d10 organoids are similar to human fetal ileum (Figs 1 and 2), we next took an unbiased approach to confirm these results. We conducted RNA-sequencing (RNAseq) at different stages of differentiation, including undifferentiated hESCs (H9 hESC line), DE and organoids grown for 30 days from each stage of spheroid formation [5 day organoids (OD5), 7 day organoids (OD7) and 10 day organoids (OD10)].
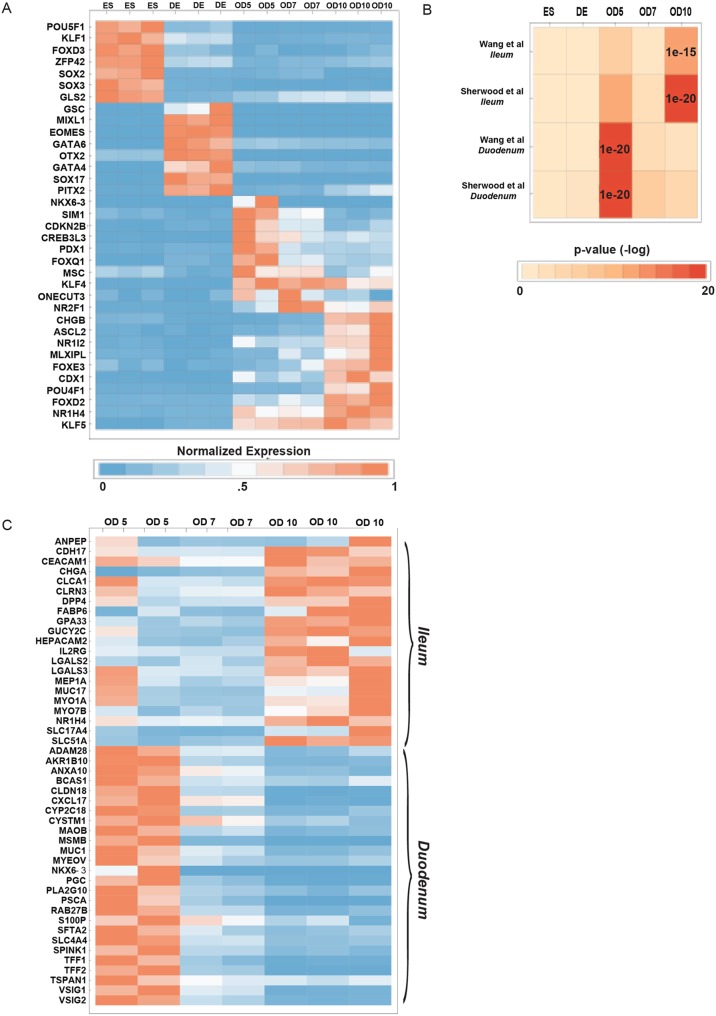
As a first step towards an unbiased assessment of the hypothesis that d5, d7 and d10 organoids are patterned into proximal or distal intestine, we determined unique stage-specific gene expression patterns in each of our RNAseq datasets. Using non-negative matrix factorization (NNMF) (Brunet et al., 2004), we identified gene expression programs that were highly enriched at one stage among the various conditions (Fig. 3A, Table S1). Each program corresponds to a cohort of genes that are statistically enriched at only one stage. In order to determine whether d5 gene expression programs corresponded to duodenal genes and d7/10 gene expression programs corresponded to ileal genes, we conducted a hypergeometric test to compare organoid gene expression programs (enriched gene sets) against regional identity gene sets of the fetal mouse and human intestine previously identified by microarray (Sherwood et al., 2009; Wang et al., 2015) (Fig. 3B). In this analysis, it was important to compare gene expression programs with data obtained from fetal, as opposed to adult, intestine because, as we have recently demonstrated, organoids resemble fetal intestine and do not express many of the genes found in the adult organ (Finkbeiner et al., 2015b). This analysis revealed that d5 gene sets had statistically significant overlap with genes expressed in the duodenum of the mouse (Sherwood et al., 2009; ‘Sherwood duo’) and human (Wang et al., 2015; ‘Wang duo’) (P<1.0×10−18 for both), whereas the d10 gene set had statistically significant overlap with genes expressed in the ileum of the mouse (Sherwood et al., 2009; ‘Sherwood ileum’; P<1.0×10−18) and human (Wang et al., 2015; ‘Wang ileum’; P<2.22×10−16) (Fig. 3B, Table S2). Importantly, we could not resolve whether d7 organoids are similar to jejunum based on Sherwood et al. and Wang et al., because the gene sets exclusively expressed in this region were very small and did not allow us to perform the statistical comparisons with confidence. Thus, we have limited our conclusions to d5 and d10 organoids. Overlapping genes identified in the duodenum/d5 organoid and ileum/d10 organoids comparisons (Fig. 3B) were further plotted as a heatmap (Fig. 3C). As a control, we also compared d5 and d10 gene expression programs against genes that are enriched in the human fetal colon (Wang et al., 2015). Here, we found no statistically significant overlap, adding confidence to our conclusion that d5 and d10 organoids are most similar to human duodenum and ileum, respectively (Fig. S4).
Fig. 3.
Bioinformatic identification of stage-enriched genes and comparison with published datasets. (A) Non-negative matrix factorization was used to identify stage-enriched genes. A normalized and curated heatmap shows representative genes (the full list is in Table S1). ES, human embryonic stem cells; DE, definitive endoderm; OD5, day 5 organoids; OD7, day 7 organoids; OD10, day 10 organoids. (B) Enriched genes in d5, d7 and d10 organoids were compared with published lists of genes whose expression is regionally restricted to the duodenum or the ileum. A hypergeometric test was used to determine the level of significance of overlapping gene sets. (C) Heatmap of representative genes found to overlap in patterned organoids and in published datasets shows enrichment for ileal genes in d10 organoids and for duodenal genes in d5 organoids.
Confirming regional identity markers in human fetal tissue
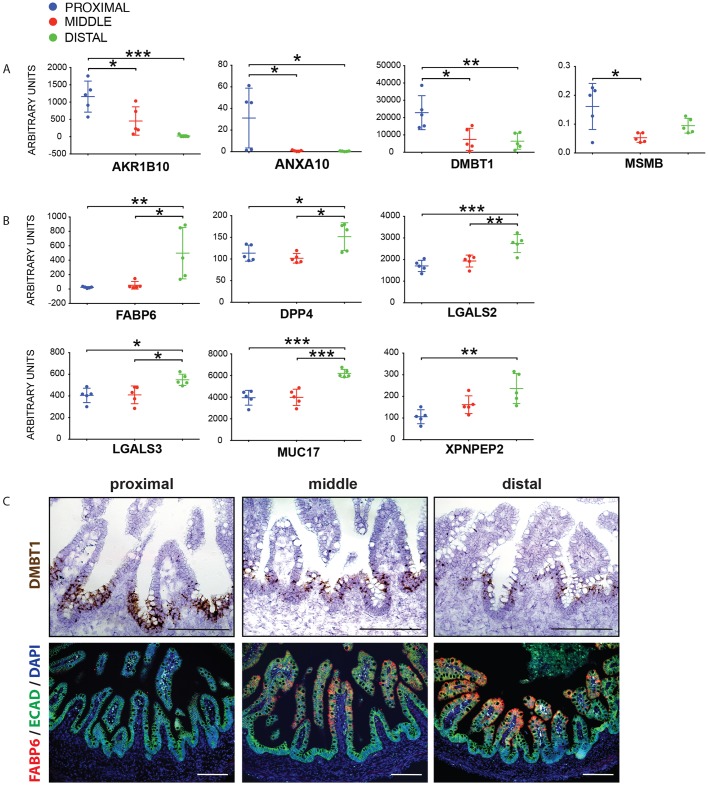
The analyses presented thus far demonstrate that increased culture time is well correlated with progressive distalization of intestinal organoids. However, we also needed to grapple with several caveats at this point, including: (1) organoids grown in vitro lack many cell types found in the endogenous intestine (e.g. vasculature, enteric neurons, immune cells); (2) genes regionally expressed in the human intestine were identified in tissue grown in culture that supported growth of the epithelium only and that was enriched for stem/progenitor cells but had few differentiated cell types (Wang et al., 2015); and (3) several of the regionally expressed genes were identified in mouse as opposed to human (Sherwood et al., 2009). Therefore, we set out to further validate our findings by investigating expression of genes identified in Fig. 3 (Table S2) in full thickness human fetal intestine. In total, our results had identified 33 ‘duodenal’ genes and 26 ‘ileal’ genes that were consistent across datasets. Of note, several of the genes identified in our unbiased analysis were identified in our candidate approach (Fig. 1), including PDX1, TM4SF4, ONECUT2 and GUCA2A. We further confirmed that four additional duodenal genes were enriched in the proximal human fetal intestine compared with the distal, and six ileal genes were enriched in the distal compared with the proximal human fetal intestine (Fig. 4, Fig. S5). Although other genes did not show enrichment across the different regions of the human fetal intestine (Figs S6 and S7), it is important to note that examination of technical replicates between intestine regions of individual samples often showed region-specific gene expression (compare Fig. S6 with Fig. S7). Thus, it is possible that our approach has identified additional region-specific markers but that our analysis was not sufficiently powered at individual gestational stages to draw strong conclusions. In addition, it is interesting that many of the genes enriched in the human fetal intestine in a regional manner were regulated in a graded, as opposed to a binary ‘on/off’, manner, and only a few genes appeared to be expressed exclusively in one region or another.
Fig. 4.
Validation of region-enriched genes in human fetal tissue. (A) qRT-PCR showing genes enriched in the human fetal proximal small intestine (n=5 individual biological specimens; proximal, blue; middle, red; distal, green). (B) qRT-PCR showing genes enriched in the human fetal distal small intestine. (C) In situ hybridization of DMBT1 showing stronger expression in the proximal small intestine, and immunohistochemistry of FABP6 showing more abundant protein staining in the middle/distal regions of the human small intestine. Scale bars: 200 μm.
Regional identity is maintained in patterned organoids transplanted in vivo
Organoids grown in vitro are similar to the fetal intestine, and upon transplantation into immunocompromised mice, they become more mature at the cellular, molecular and functional levels (Finkbeiner et al., 2015b; Watson et al., 2014). Transplantation also causes morphological remodeling such that the simple epithelium of the organoid in vitro gains crypt-villus structure similar to that of the native adult human intestine (Finkbeiner et al., 2015b; Watson et al., 2014). Thus, in order to confirm that the patterned organoids maintained their regional identity when transplanted in vivo, we generated d5, d7 and d10 organoids, and transplanted them under the kidney capsule of immunocompromised NSG mice (n=5 per group; Fig. S8). Organoids were allowed to engraft and mature for 10 weeks, and were then harvested for histological and immunohistochemical analysis (Fig. 5, Figs S9 and S10). Low-magnification Hematoxylin and Eosin staining revealed crypt-villus architecture in d5, d7 and d10 transplanted organoids (Fig. 5A, Fig. S9A). As expected, proliferation, marked by Ki67, was restricted to the crypt domains in all transplanted tissues (Fig. S9). In addition, CDX2 and the brush border enzymes sucrose isomaltase (SI) and dipeptidyl peptidase IV (DPPIV), which are expressed throughout the small intestine, were similarly expressed in all regionalized organoids (Fig. S10). By contrast, many proteins known to be expressed in the proximal small intestine (duodenum) were restricted to the d5 organoids, with very little or no expression in d10 organoids (Middendorp et al., 2014; Uhlén et al., 2010), including PDX1, GATA4, FABP2 and LCT (Fig. 5B,C, Fig. S9C). Similarly, proteins previously shown to be enriched in the ileum were more abundant in the d10 organoids, including the transcription factor SATB2, which is expressed at high levels in the adult colon and at lower levels in the distal ileum and FABP6 (Fig. 5D,E) (Uhlén et al., 2010; Wang et al., 2015). In addition, goblet cells, marked by MUC2, were much more abundant in d7 and d10 organoids (Fig. S9E,F).
Fig. 5.
Organoids retain regionalization after maturation in vivo. (A) d5, d7, d10 organoids were harvested after maturation in vivo. Hematoxylin and Eosin staining reveals that transplanted tissue possesses villus- and crypt-like domains. (B) PDX1 is most highly enriched in d5 organoids. (C) GATA4 is most highly enriched in d5 organoids. (D) SATB2 is most highly enriched in d10 organoids. (E) FABP6 is enriched in both d7 and d10 organoids. Scale bars: 200 μm.
DISCUSSION
In this study, we provide evidence that the duration of exposure to FGF4 and CHIR99021 is able to specify the regional identity of organoids from human PSCs in vitro. This work sheds new light onto the regional identify of intestinal organoids grown using published methods (Finkbeiner et al., 2015b; McCracken et al., 2011; Spence et al., 2011; Watson et al., 2014), as it has been unclear whether these most closely represented a specific region of the intestine. Here, we show that organoids used in published work most closely represent duodenum, and we demonstrate that prolonged activation of FGF and WNT signaling patterns the intestine into increasingly distal tissue. The notion that duration of exposure to signaling pathway activation can influence identity came from our understanding that, as the embryo develops, the most caudal region of the embryo is exposed to higher levels of growth factor signaling relative to more proximal regions, and that this signaling is prolonged as high concentrations of ligands are maintained distally as the embryonic axis lengthens (Arnold and Robertson, 2009). However, whether or not intestinal patterning was dependent on growth factor signaling activity and time of exposure was unclear. Our findings provide evidence that variables, including concentration and length of exposure, are important for establishing intestinal regional identity.
Furthermore, the current studies also address several practical issues that arise from working with in vitro-derived tissues. One important issue that our work raises relates to understanding the strengths and limitations of different organoid model systems in order to ensure that they are employed in the correct context (Chen et al., 2014; Finkbeiner et al., 2015b, 2012; Leslie et al., 2014; Rodansky et al., 2015; Watson et al., 2014; Xue et al., 2013). For example, a number of reports have demonstrated that hPSC-derived organoids are small intestinal in nature and contain Paneth cells (Finkbeiner et al., 2015a,b; Spence et al., 2011; Watson et al., 2014), which are not present in the colon (Rothenberg et al., 2012). However, the regional identity of organoids generated using the same or similar methods to ours has been confused in other research, including recent reports stating that organoids are tissues ‘resembling human proximal colon’ (Engevik et al., 2015a,b). The current work underscores the observation that organoids, as originally described (Spence et al., 2011), are most similar to duodenum.
In addition to clarifying the nature of these in vitro-derived models, another important implication of the current work is the ability to begin generating different regions of the intestine for specific purposes. For example, a dangerous condition in human neonates called necrotizing enterocolitis (NEC) most often affects the ileum. When using organoids to model diseases such as NEC in vitro, or for generating tissue engineered small intestine for therapeutic purposes, it will be important to generate organoids that best match the appropriate region of the intestine (Finkbeiner et al., 2015a; Howell and Wells, 2011; Levin et al., 2013; Ullrich et al., 2012).
Given the inability to functionally explore developmental processes in human fetal tissue, using organoids as a surrogate model for human intestine development provides a path for better understanding of the developmental cues responsible for human fetal intestinal regionalization. Previously, little to nothing was known about this process in the human; this study provides the first evidence that the combined signaling activity of FGF and WNT plays a role in establishing regional intestinal identity. In the future, in vitro-derived regionalized small intestinal organoids can be used to study specific gene expressions programs and to better understand genes that play important roles in the development of congenital intestinal disease. From a regenerative medicine perspective, regionalized organoids will provide more reliable building blocks for replacement purposes.
MATERIALS AND METHODS
hESC lines, human tissue and mice
hESCs
All work with hESCs was reviewed and approved by the University of Michigan human pluripotent stem cell research oversight committee (HPSCRO). The hESC cell line H9 (WA09, NIH stem registry #0062) was obtained from the WiCell Research Institute. Karyotypically normal cell lines were used for all experiments.
Human tissue
Normal, de-identified human intestinal tissue was obtained from the University of Washington Laboratory of Developmental Biology, and was approved by the University of Michigan institutional review board.
Animal use
All mouse work was reviewed and approved by the University of Michigan Committee on Use and Care of Animals (UCUCA).
Differentiation of hESCs
Differentiation of hESCs and organoids was carried out as previously published, with minor modifications (Finkbeiner et al., 2015b; Leslie et al., 2014; McCracken et al., 2011). In brief, endoderm was generated by adding activin A (100 ng/ml) for 3 consecutive days in Roswell Park Memorial Institute 1640 (RPMI-1640) media supplemented with 0%, 0.2% and 2.0% HyClone FBS. FGF4 (500 ng/ml) plus CHIR99021 (2 μM) were then added to endoderm cultures and medium was replaced daily for 10 days. In our original protocol, spheroids were collected on d4 (McCracken et al., 2011; Spence et al., 2011), whereas in the current work, spheroids were collected on d5, d7 and d10, embedded in matrigel and overlaid with growth medium, as previously described (Finkbeiner et al., 2015b), containing RSPO2-conditioned medium (Bell et al., 2008), noggin-conditioned medium (Heijmans et al., 2013) and EGF. All experiments were conducted on organoids expanded for 30-35 days in vitro.
qRT-PCR
Briefly, RNA isolation was performed using MagMAXTM-96 Total RNA Isolation Kit (Ambion, AM1830). A SuperScript VILO cDNA synthesis kit (ThermoFisher, 11754250) was used to make cDNA from 200 ng RNA. cDNA levels were detected using QuantiTect SYBR Green (Qiagen, 608056). Relative gene expression was plotted as Arbitrary Units, using the following formula: [2^(housekeeping gene Ct-gene Ct)]×10,000. All primers sequences are listed in Table S3.
Histology, immunofluorescence and in situ hybridization
Immunofluorescence was carried out as previously described (Dye et al., 2015; Rockich et al., 2013) using antibodies outlined in Table S4. In situ hybridization was performed using the RNAscope 2.0 HD detection kit, and with commercially available mRNA probes outlined in Table S4, according to the standard protocol provided. All incubations were performed at 40°C in a HybEZ hybridization system oven (Advanced Cell Diagnostics, 310010). All immunostaining or in situ hybridization was conducted on at least three independent biological specimens (three independent human fetal samples or three independent organoids), and immunostaining images shown in the figures are representative images unless otherwise noted in the text.
Statistical analysis
For statistical analysis, data are expressed as the median of each sample set. Each data point in the plots represents an independent biological sample. For organoid experiments, each independent biological sample is comprised of three to five organoids pooled together. All organoid experiments were conducted on at least three independent biological replicates, and each experiment was repeated on at least two separate occasions (independent experiments). For human fetal tissue, all analysis was conducted on five independent biological replicates. One-way ANOVA was used for statistical analysis, except for Fig. S2, which used an unpaired t-test. Analyses were carried out with GraphPad Prism 5.0 software. Each data point is presented, with the middle line representing the mean, the error bars representing±s.e.m. In all figures, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, except for Fig. 2B where a=P<0.05, b=P<0.01, c=P<0.001, d=P<0.0001. For experiments in the supplemental data where individual human fetal samples are plotted (Figs S1, S5, S7), the intestine (n=1 biological sample per time point) was divided into proximal, middle and distal regions, and each region was measured and cut into 1 cm lengths. RNA was isolated from three to five different segments from each of the proximal, middle and distal regions (n=3-5 technical replicates).
RNA sequencing
RNA was isolated directly from tissue culture plates using the MagMAXTM-96 Total RNA Isolation Kit according to the manufacturer's protocol. RNA concentration and purity were assessed using Nanodrop spectrophotometer and bioanalyzed using Agilent RNA 6000 Nano Kit (260/280>1.7 and 260/230>1.7, RIN>8). Samples were stored at −80°C. cDNA libraries were generated using TrueSeq Kit (Illumina). Sequencing was then performed on HiSeq 2000 (Illumina, 100 bp, single-end reads). In all, 42 samples were run using two flow cells and four samples per lane. RNA sequencing data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-4168.
RNA sequencing and data analysis
Raw mRNA-seq reads were aligned to hg19 exome constructed from UCSC gff files. Reads were aligned using Bowtie2 v 2.1.0 (Langmead and Salzberg, 2012) with the following options -D 25-R 3-N 1-L 20-i S 1 0.50 local. Transcript counts were normalized to fragments per kilobase of transcript per million mapped reads (FPKM) by applying UCSC transcript lengths and the number of collected reads per sample. Transcripts with a maximum FPKM of less than one across all samples were discarded.
Gene expression programs were assembled using non-negative matrix factorization (Lee and Seung, 1999). Prior to NNMF, the gene expression values were normalized by the mean gene expression value of each gene across all samples. This step removes expression bias and focuses analysis on gene expression variance during the process of differentiation. Following normalization, NNMF was performed both on the entire genome and on DNA-binding proteins as derived through PFAM annotations. DNA-binding proteins provide a reduced set of interpretable genes for comparison with existing literature. NNMF was performed with 10 replicates, and the highest scoring factorization was selected as the global dictionary of programs.
To assess enrichment of literature gene sets in NNMF programs, gene sets were assembled from Wang et al. (2015) and Sherwood et al. (2009), and gene set enrichment was performed as described previously (Mootha et al., 2003; Subramanian et al., 2005) to determine enrichment in NNMF programs. A gene was determined to be a member of an NNMF-derived program if the gene loading was two standard deviations above the mean loading in the NNMF program. Statistical significance for enrichments was calculated using the hyper-geometric test. Principal component analysis was performed using standard procedures on the FPKM and mean normalized gene expression data.
Kidney capsule transplantation
Kidney capsule transplantation was carried out as previously described (Finkbeiner et al., 2015b; Watson et al., 2014). All organoids were transplanted after being grown in vitro for 30-35 days. For each group (d5, d7, d10), organoids were transplanted into five different mice; each transplanted mouse was considered an independent biological specimen (n=5 per group) (Fig. S7). Transplanted organoids were harvested after 10 weeks.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Y.-H.T. and J.R.S. conceived the study. Y.-H.T., R.N., P.H.D., M.S.N., A.M.C. and M.T. conducted experiments. All authors analyzed data. R.N., M.T., O.D.K. and J.R.S. wrote the manuscript. All authors edited and approved final manuscript.
Funding
This research was performed as a project of the Intestinal Stem Cell Consortium – a collaborative research project funded by the National Institute of Diabetes and The National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and the National Institute of Allergy and Infectious Diseases (NIAID) (U01DK103141 to J.R.S and U01DK103147 to O.D.K.). This work was also supported, in part, by the NIDDK (K01DK091415), the NIAID (U19AI116482), the University of Michigan Center for Gastrointestinal Research (NIDDK P30DK034933) and the University of Michigan internal funds to J.R.S.; and by RN3 06525 from the California Institute of Regenerative Medicine to O.D.K. The University of Washington Laboratory of Developmental Biology was supported by a National Institutes of Health Award from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (5R24HD000836). Deposited in PMC for immediate release.
Data availability
RNA sequencing data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-4168.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.138453.supplemental
References
- Arnold S. J. and Robertson E. J. (2009). Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 10, 91-103. 10.1038/nrm2618 [DOI] [PubMed] [Google Scholar]
- Battle M. A., Bondow B. J., Iverson M. A., Adams S. J., Jandacek R. J., Tso P. and Duncan S. A. (2008). GATA4 is essential for jejunal function in mice. Gastroenterology 135, 1676-1686.e1. 10.1053/j.gastro.2008.07.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck C. W. and Slack J. M. (1999). A developmental pathway controlling outgrowth of the Xenopus tail bud. Development 126, 1611-1620. [DOI] [PubMed] [Google Scholar]
- Beland M., Pilon N., Houle M., Oh K., Sylvestre J.-R., Prinos P. and Lohnes D. (2004). Cdx1 autoregulation is governed by a novel Cdx1-LEF1 transcription complex. Mol. Cell. Biol. 24, 5028-5038. 10.1128/MCB.24.11.5028-5038.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S. M., Schreiner C. M., Wert S. E., Mucenski M. L., Scott W. J. and Whitsett J. A. (2008). R-spondin 2 is required for normal laryngeal-tracheal, lung and limb morphogenesis. Development 135, 1049-1058. 10.1242/dev.013359 [DOI] [PubMed] [Google Scholar]
- Bradley L., Sun B., Collins-Racie L., LaVallie E., McCoy J. and Sive H. (2000). Different activities of the frizzled-related proteins frzb2 and sizzled2 during Xenopus anteroposterior patterning. Dev. Biol. 227, 118-132. 10.1006/dbio.2000.9873 [DOI] [PubMed] [Google Scholar]
- Brunet J.-P., Tamayo P., Golub T. R. and Mesirov J. P. (2004). Metagenes and molecular pattern discovery using matrix factorization. Proc. Natl. Acad. Sci. USA 101, 4164-4169. 10.1073/pnas.0308531101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Gibson J. D., Miyamoto S., Sail V., Verma R., Rosenberg D. W., Nelson C. E. and Giardina C. (2011). Intestinal lineage commitment of embryonic stem cells. Differentiation 81, 1-10. 10.1016/j.diff.2010.09.182 [DOI] [PubMed] [Google Scholar]
- Cao L., Kuratnik A., Xu W., Gibson J. D., Kolling F., Falcone E. R., Ammar M., Van Heyst M. D., Wright D. L., Nelson C. E. et al. (2015). Development of intestinal organoids as tissue surrogates: cell composition and the epigenetic control of differentiation. Mol. Carcinog. 54, 189-202. 10.1002/mc.22089 [DOI] [PubMed] [Google Scholar]
- Chen Y.-J., Finkbeiner S. R., Weinblatt D., Emmett M. J., Tameire F., Yousefi M., Yang C., Maehr R., Zhou Q., Shemer R. et al. (2014). De novo formationof insulin-producing “‘neo-b cell islets’” from intestinal crypts. Cell Rep. 6, 1046-1058. 10.1016/j.celrep.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox W. G. and Hemmati-Brivanlou A. (1995). Caudalization of neural fate by tissue recombination and bFGF. Development 121, 4349-4358. [DOI] [PubMed] [Google Scholar]
- Dale L., Howes G., Price B. M. and Smith J. C. (1992). Bone morphogenetic protein 4: a ventralizing factor in early Xenopus development. Development 115, 573-585. [DOI] [PubMed] [Google Scholar]
- Dessimoz J., Opoka R., Kordich J. J., Grapin-Botton A. and Wells J. M. (2006). FGF signaling is necessary for establishing gut tube domains along the anterior-posterior axis in vivo. Mech. Dev. 123, 42-55. 10.1016/j.mod.2005.10.001 [DOI] [PubMed] [Google Scholar]
- Domingos P. M., Itasaki N., Jones C. M., Mercurio S., Sargent M. G., Smith J. C. and Krumlauf R. (2001). The Wnt/beta-catenin pathway posteriorizes neural tissue in Xenopus by an indirect mechanism requiring FGF signalling. Dev. Biol. 239, 148-160. 10.1006/dbio.2001.0431 [DOI] [PubMed] [Google Scholar]
- Duluc I., Freund J. N., Leberquier C. and Kedinger M. (1994). Fetal endoderm primarily holds the temporal and positional information required for mammalian intestinal development. J. Cell Biol. 126, 211-221. 10.1083/jcb.126.1.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duluc I., Hoff C., Kedinger M. and Freund J.-N. (2001). Differentially expressed endoderm and mesenchyme genes along the fetal rat intestine. Genesis 29, 55-59. [DOI] [PubMed] [Google Scholar]
- Dusing M. R., Florence E. A. and Wiginton D. A. (2001). Pdx-1 is required for activation in vivo from a duodenum-specific enhancer. J. Biol. Chem. 276, 14434-14442. [DOI] [PubMed] [Google Scholar]
- Dye B. R., Hill D. R., Ferguson M. A., Tsai Y.-H., Nagy M. S., Dyal R., Wells J. M., Mayhew C. N., Nattiv R., Klein O. D. et al. (2015). In vitro generation of human pluripotent stem cell derived lung organoids. eLife 4, 05098 10.7554/elife.05098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engevik M. A., Engevik K. A., Yacyshyn M. B., Wang J., Hassett D. J., Darien B., Yacyshyn B. R. and Worrell R. T. (2015a). Human Clostridium difficile infection: inhibition of NHE3 and microbiota profile. Am. J. Physiol. Gastrointest. Liver Physiol. 308, G497-G509. 10.1152/ajpgi.00090.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engevik M. A., Yacyshyn M. B., Engevik K. A., Wang J., Darien B., Hassett D. J., Yacyshyn B. R. and Worrell R. T. (2015b). Human Clostridium difficile infection: altered mucus production and composition. Am. J. Physiol. Gastrointest. Liver Physiol. 308, G510-G524. 10.1152/ajpgi.00091.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erter C. E., Wilm T. P., Basler N., Wright C. V. and Solnica-Krezel L. (2001). Wnt8 is required in lateral mesendodermal precursors for neural posteriorization in vivo. Development 128, 3571-3583. [DOI] [PubMed] [Google Scholar]
- Finkbeiner S. R., Zeng X.-L., Utama B., Atmar R. L., Shroyer N. F. and Estes M. K. (2012). Stem cell-derived human intestinal organoids as an infection model for rotaviruses. mBio 3, e00159-e00112 10.1128/mBio.00159-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner S. R., Freeman J. J., Wieck M. M., El-Nachef W., Altheim C. H., Tsai Y.-H., Huang S., Dyal R., White E. S., Grikscheit T. C. et al. (2015a). Generation of tissue-engineered small intestine using embryonic stem cell-derived human intestinal organoids. Biol. Open 4, 1462-1472. 10.1242/bio.013235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner S. R., Hill D. R., Altheim C. H., Dedhia P. H., Taylor M. J., Tsai Y.-H., Chin A. M., Mahe M. M., Watson C. L., Freeman J. J. et al. (2015b). Transcriptome-wide analysis reveals hallmarks of human intestine development and maturation in vitro and in vivo. Stem Cell Rep. 4, 1140-1155. 10.1016/j.stemcr.2015.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N. and Kaestner K. H. (2010). Cdx2 regulates endo-lysosomal function and epithelial cell polarity. Genes Dev. 24, 1295-1305. 10.1101/gad.1921510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N., White P. and Kaestner K. H. (2009). Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev. Cell 16, 588-599. 10.1016/j.devcel.2009.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger S., Savory J. G. A. and Lohnes D. (2010). Cdx2 regulates patterning of the intestinal epithelium. Dev. Biol. 339, 155-165. 10.1016/j.ydbio.2009.12.025 [DOI] [PubMed] [Google Scholar]
- Greco T. L., Takada S., Newhouse M. M., McMahon J. A., McMahon A. P. and Camper S. A. (1996). Analysis of the vestigial tail mutation demonstrates that Wnt-3a gene dosage regulates mouse axial development. Genes Dev. 10, 313-324. 10.1101/gad.10.3.313 [DOI] [PubMed] [Google Scholar]
- Gregorieff A., Grosschedl R. and Clevers H. (2004). Hindgut defects and transformation of the gastro-intestinal tract in Tcf4(−/−)/Tcf1(−/−) embryos. EMBO J. 23, 1825-1833. 10.1038/sj.emboj.7600191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan N. R. F., Fordham R. P., Syed Y. A., Moignard V., Berry A., Bautista R., Hanley N. A., Jensen K. B. and Vallier L. (2013). Generation of multipotent foregut stem cells from human pluripotent stem cells. Stem Cell Rep. 1, 293-306. 10.1016/j.stemcr.2013.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans J., de Jeude J., Koo B.-K. and Rosekrans S. L. (2013). ER stress causes rapid loss of intestinal epithelial stemness through activation of the unfolded protein response. Cell Rep. 3, 1128-1139. 10.1016/j.celrep.2013.02.031 [DOI] [PubMed] [Google Scholar]
- Ho R. K. and Kimmel C. B. (1993). Commitment of cell fate in the early zebrafish embryo. Science 261, 109-111. 10.1126/science.8316841 [DOI] [PubMed] [Google Scholar]
- Hollyday M., McMahon J. A. and McMahon A. P. (1995). Wnt expression patterns in chick embryo nervous system. Mech. Dev. 52, 9-25. 10.1016/0925-4773(95)00385-E [DOI] [PubMed] [Google Scholar]
- Horb M. E. and Slack J. M. W. (2001). Endoderm specification and differentiation in Xenopus embryos. Dev. Biol. 236, 330-343. 10.1006/dbio.2001.0347 [DOI] [PubMed] [Google Scholar]
- Howell J. C. and Wells J. M. (2011). Generating intestinal tissue from stem cells: potential for research and therapy. Regen. Med. 6, 743-755. 10.2217/rme.11.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsken J., Vogel R., Brinkmann V., Erdmann B., Birchmeier C. and Birchmeier W. (2000). Requirement for beta-catenin in anterior-posterior axis formation in mice. J. Cell Biol. 148, 567-578. 10.1083/jcb.148.3.567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeya M. and Takada S. (2001). Wnt-3a is required for somite specification along the anteroposterior axis of the mouse embryo and for regulation of cdx-1 expression. Mech. Dev. 103, 27-33. 10.1016/S0925-4773(01)00338-0 [DOI] [PubMed] [Google Scholar]
- Isaacs H. V., Pownall M. E. and Slack J. M. W. (1998). Regulation of Hox gene expression and posterior development by the Xenopus caudal homologue Xcad3. EMBO J. 17, 3413-3427. 10.1093/emboj/17.12.3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedinger M., Duluc I., Fritsch C., Lorentz O., Plateroti M. and Freund J. N. (1998). Intestinal epithelial-mesenchymal cell interactions. Ann. N. Y. Acad. Sci. 859, 1-17. 10.1111/j.1749-6632.1998.tb11107.x [DOI] [PubMed] [Google Scholar]
- Keenan I. D., Sharrard R. M. and Isaacs H. V. (2006). FGF signal transduction and the regulation of Cdx gene expression. Dev. Biol. 299, 478-488. 10.1016/j.ydbio.2006.08.040 [DOI] [PubMed] [Google Scholar]
- Kiecker C. and Niehrs C. (2001). A morphogen gradient of Wnt/beta-catenin signalling regulates anteroposterior neural patterning in Xenopus. Development 128, 4189-4201. [DOI] [PubMed] [Google Scholar]
- Langmead B. and Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357-359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. D. and Seung H. S. (1999). Learning the parts of objects by non-negative matrix factorization. Nature 401, 788-791. 10.1038/44565 [DOI] [PubMed] [Google Scholar]
- Lekven A. C., Thorpe C. J., Waxman J. S. and Moon R. T. (2001). Zebrafish wnt8 encodes two wnt8 proteins on a bicistronic transcript and is required for mesoderm and neurectoderm patterning. Dev. Cell 1, 103-114. 10.1016/S1534-5807(01)00007-7 [DOI] [PubMed] [Google Scholar]
- Leslie J. L., Huang S., Opp J. S., Nagy M. S., Kobayashi M., Young V. B. and Spence J. R. (2014). Persistence and toxin production by Clostridium difficile within human intestinal organoids results in disruption of epithelial paracellular barrier function. Infect. Immun. 83, 138-145. 10.1128/IAI.02561-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. E., Barthel E. R., Speer A. L., Sala F. G., Hou X., Torashima Y. and Grikscheit T. C. (2013). Human tissue-engineered small intestine forms from postnatal progenitor cells. J. Pediatr. Surg. 48, 129-137. 10.1016/j.jpedsurg.2012.10.029 [DOI] [PubMed] [Google Scholar]
- Lickert H., Domon C., Huls G., Wehrle C., Duluc I., CLEVERS H., Meyer B. I., Freund J. N. and Kemler R. (2000). Wnt/(beta)-catenin signaling regulates the expression of the homeobox gene Cdx1 in embryonic intestine. Development 127, 3805-3813. [DOI] [PubMed] [Google Scholar]
- Liu P., Wakamiya M., Shea M. J., Albrecht U., Behringer R. R. and Bradley A. (1999). Requirement for Wnt3 in vertebrate axis formation. Nat. Genet. 22, 361-365. 10.1038/11932 [DOI] [PubMed] [Google Scholar]
- Marvin M. J., Di Rocco G., Gardiner A., Bush S. M. and Lassar A. B. (2001). Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev. 15, 316-327. 10.1101/gad.855501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken K. W., Howell J. C., Spence J. R. and Wells J. M. (2011). Generating human intestinal tissue from pluripotent stem cells in vitro. Nat. Protoc. 6, 1920-1928. 10.1038/nprot.2011.410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLin V. A., Rankin S. A. and Zorn A. M. (2007). Repression of Wnt/(beta)-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development 134, 2207-2217. 10.1242/dev.001230 [DOI] [PubMed] [Google Scholar]
- Middendorp S., Schneeberger K., Wiegerinck C. L., Mokry M., Akkerman R. D. L., van Wijngaarden S., Clevers H. and Nieuwenhuis E. E. S. (2014). Adult stem cells in the small intestine are intrinsically programmed with their location-specific function. Stem Cells 32, 1083-1091. 10.1002/stem.1655 [DOI] [PubMed] [Google Scholar]
- Mootha V. K., Lindgren C. M., Eriksson K.-F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E. et al. (2003). PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267-273. 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- Northrop J. L. and Kimelman D. (1994). Dorsal-ventral differences in Xcad-3 expression in response to FGF-mediated induction in Xenopus. Dev. Biol. 161, 490-503. 10.1006/dbio.1994.1047 [DOI] [PubMed] [Google Scholar]
- Parr B. A. and McMahon A. P. (1995). Dorsalizing signal Wnt-7a required for normal polarity of D-V and A-P axes of mouse limb. Nature 374, 350-353. 10.1038/374350a0 [DOI] [PubMed] [Google Scholar]
- Pownall M. E., Tucker A. S., Slack J. M. and Isaacs H. V. (1996). eFGF, Xcad3 and Hox genes form a molecular pathway that establishes the anteroposterior axis in Xenopus. Development 122, 3881-3892. [DOI] [PubMed] [Google Scholar]
- Ratineau C., Duluc I., Pourreyron C., Kedinger M., Freund J.-N. and Roche C. (2003). Endoderm- and mesenchyme-dependent commitment of the differentiated epithelial cell types in the developing intestine of rat. Differentiation 71, 163-169. 10.1046/j.1432-0436.2003.t01-1-710203.x [DOI] [PubMed] [Google Scholar]
- Rockich B. E., Hrycaj S. M., Shih H.-P., Nagy M. S., Ferguson M. A. H., Kopp J. L., Sander M., Wellik D. M. and Spence J. R. (2013). Sox9 plays multiple roles in the lung epithelium during branching morphogenesis. Proc. Natl. Acad. Sci. USA 110, E4456-E4465. 10.1073/pnas.1311847110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodansky E. S., Johnson L. A., Huang S., Spence J. R. and Higgins P. D. R. (2015). Intestinal organoids: a model of intestinal fibrosis for evaluating anti-fibrotic drugs. Exp. Mol. Pathol. 98, 345-351. 10.1016/j.yexmp.2015.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg M. E., Nusse Y., Kalisky T., Lee J. J., Dalerba P., Scheeren F., Lobo N., Kulkarni S., Sim S., Qian D. et al. (2012). Identification of a cKit+ colonic crypt base secretory cell that supports lgr5+ stem cells in mice. Gastroenterology 142, 1195-1205.e6. 10.1053/j.gastro.2012.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh W., Gotoh T., Tsunematsu Y., Aizawa S. and Shimono A. (2006). Sfrp1 and Sfrp2 regulate anteroposterior axis elongation and somite segmentation during mouse embryogenesis. Development 133, 989-999. 10.1242/dev.02274 [DOI] [PubMed] [Google Scholar]
- Sherwood R. I., Chen T.-Y. A. and Melton D. A. (2009). Transcriptional dynamics of endodermal organ formation. Dev. Dyn. 238, 29-42. 10.1002/dvdy.21810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood R. I., Maehr R., Mazzoni E. O. and Melton D. A. (2011). Wnt signaling specifies and patterns intestinal endoderm. Mech. Dev. 128, 14 10.1016/j.mod.2011.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J. R., Mayhew C. N., Rankin S. A., Kuhar M. F., Vallance J. E., Tolle K., Hoskins E. E., Kalinichenko V. V., Wells S. I., Zorn A. M. et al. (2011). Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105-109. 10.1038/nature09691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S. et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545-15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamminen K., Balboa D., Toivonen S., Pakarinen M. P., Wiener Z., Alitalo K. and Otonkoski T. (2015). Intestinal commitment and maturation of human pluripotent stem cells is independent of exogenous FGF4 and R-spondin1. PLoS ONE 10, e0134551 10.1371/journal.pone.0134551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S. et al. (2010). Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 28, 1248-1250. 10.1038/nbt1210-1248 [DOI] [PubMed] [Google Scholar]
- Ullrich T., Tang Y.-W., Correa H., Garzon S. A., Maheshwari A., Hill M., Matta P., Krishnan M. K. and Weitkamp J.-H. (2012). Absence of gastrointestinal pathogens in ileum tissue resected for necrotizing enterocolitis. Pediatr. Infect. Dis. J. 31, 413-414. 10.1097/INF.0b013e318242534a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi M. P., Shin H., He H. H., Sulahian R., Meyer C. A., Montgomery R. K., Fleet J. C., Brown M., Liu X. S. and Shivdasani R. A. (2010). Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev. Cell 19, 713-726. 10.1016/j.devcel.2010.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi M. P., Shin H., San Roman A. K., Liu X. S. and Shivdasani R. A. (2013). Intestinal master transcription factor CDX2 controls chromatin access for partner transcription factor binding. Mol. Cell. Biol. 33, 281-292. 10.1128/MCB.01185-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Yamamoto Y., Wilson L. H., Zhang T., Howitt B. E., Farrow M. A., Kern F., Ning G., Hong Y., Khor C. C. et al. (2015). Cloning and variation of ground state intestinal stem cells. Nature 522, 173-178. 10.1038/nature14484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson C. L., Mahe M. M., Múnera J., Howell J. C., Sundaram N., Poling H. M., Schweitzer J. I., Vallance J. E., Mayhew C. N., Sun Y. et al. (2014). An in vivo model of human small intestine using pluripotent stem cells. Nat. Med. 20, 1310-1314. 10.1038/nm.3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J. M. and Melton D. A. (1999). Vertebrate endoderm development. Annu. Rev. Cell Dev. Biol. 15, 393-410. 10.1146/annurev.cellbio.15.1.393 [DOI] [PubMed] [Google Scholar]
- Wells J. M. and Melton D. A. (2000). Early mouse endoderm is patterned by soluble factors from adjacent germ layers. Development 127, 1563-1572. [DOI] [PubMed] [Google Scholar]
- Xue X., Ramakrishnan S., Anderson E., Taylor M., Zimmermann E. M., Spence J. R., Huang S., Greenson J. K. and Shah Y. M. (2013). Endothelial PAS domain protein 1 activates the inflammatory response in the intestinal epithelium to promote colitis in mice. Gastroenterology 145, 831-841. 10.1053/j.gastro.2013.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn A. M. and Wells J. M. (2007). Molecular basis of vertebrate endoderm development. Int. Rev. Cytol. 259, 49-111. 10.1016/S0074-7696(06)59002-3 [DOI] [PubMed] [Google Scholar]