ABSTRACT
Lungs are composed of a system of highly branched tubes that bring air into the alveoli, where gas exchange takes place. The proximal and distal regions of the lung contain epithelial cells specialized for different functions: basal, secretory and ciliated cells in the conducting airways and type II and type I cells lining the alveoli. Basal, secretory and type II cells can be grown in three-dimensional culture, with or without supporting stromal cells, and under these conditions they give rise to self-organizing structures known as organoids. This Review summarizes the different methods for generating organoids from cells isolated from human and mouse lungs, and compares their final structure and cellular composition with that of the airways or alveoli of the adult lung. We also discuss the potential and limitations of organoids for addressing outstanding questions in lung biology and for developing new drugs for disorders such as cystic fibrosis and asthma.
KEY WORDS: Lung organoids, Stem cells, Lung progenitors, Plasticity
Summary: This Review article explores the latest advances in both adult and embryonic stem cell-derived lung organoid culture, and discusses how these systems can be used to understand homeostasis and regeneration.
Introduction
The main function of the lungs is to enable efficient gas exchange between the air and the blood. For this purpose, they are composed of a complex three-dimensional (3D) system of tubes that terminate in hundreds of millions of highly vascularized distal sacs (Fig. 1). During development, the lungs arise from the anterior foregut as two small rudimentary endodermal buds surrounded by mesoderm and a vascular plexus (Morrisey and Hogan, 2010). The epithelium undergoes extensive branching morphogenesis to give rise to the conducting airways known as bronchi (if they are supported by cartilage) and bronchioles (if they are not). The bronchioles open into the air sacs, known as alveoli, where gas exchange takes place. The epithelium lining the airways is composed mainly of multiciliated cells and secretory cells, including Club and goblet cells. Together, these specialized components produce a thin surface layer of liquid that contains mucins and glycoproteins and serves to moisten the air, provide antimicrobial activity, and move particles directionally out of the lungs. In the larger airways of the mouse lung and throughout most of the human lung the so-called mucociliary epithelium contains basal cells that function as progenitors of the multiciliated and secretory populations. By contrast, the air sacs are lined by two other distinct cell types: specialized alveolar type II cells (AEC2s) that secrete surfactants and other proteins; and very thin, delicate type I cells (AEC1s) that provide an extensive surface area for gas exchange with the surrounding capillaries. The mesoderm of the embryonic lung gives rise to numerous specialized cell populations that interact closely with the conducting airways, such as cartilage, smooth muscle, and fibroblasts, the alveolar epithelium, which includes myofibroblasts and lipofibroblasts, and the vasculature, which includes pericytes and vascular smooth muscle cells. Other important cell populations of the lung are the outer mesothelial layer and immune cells. The latter comprises T cells, mast cells, eosinophils, dendritic cells and distinct populations of macrophages that either reside permanently in the alveoli or interstitium or that traffic in and out of the lung in response to injury or infection (Tan and Krasnow, 2016). Immune cells are not the only source of pro- and anti-inflammatory cytokines in the lung; the epithelial cells themselves are known to produce numerous cytokines directly in response to injury or pathogens and they contribute to the impressive innate immunity functions of the lung (Whitsett and Alenghat, 2015).
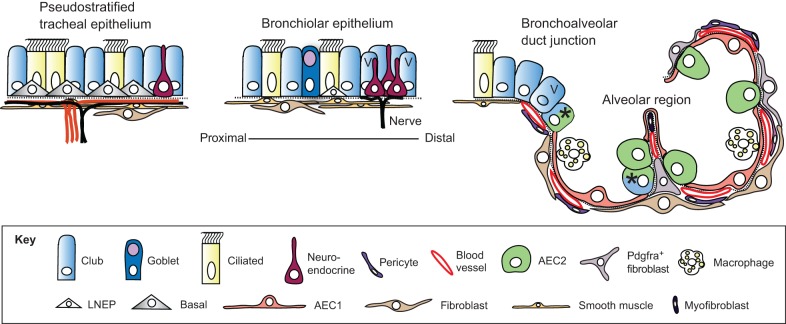
Fig. 1.
Epithelial cell types of the mouse lung. Schematic of the major cell types in different regions of the mouse lung (Hogan et al., 2014). Goblet cells are much more abundant in human versus mouse airways. Basal cells expressing detectable levels of Trp63 and Krt5 are only present in trachea and main stem bronchi. Lineage-negative epithelial progenitors (LNEPs) have been proposed for the distal airways (Vaughan et al., 2015), which in the mouse are known as bronchioles as they lack associated cartilage. There is evidence that the Club cell population is heterogeneous, with a few cells in the bronchioalveolar duct junction (BADJ) and alveoli (marked with an asterisk) co-expressing Scbg1a1 and Sftpc (Kim et al., 2005; Rawlins et al., 2009). In addition, some Club cells in the vicinity of neuroendocrine bodies and the BADJ are resistant to killing by naphthalene. These ‘variant' Club cells (marked with V) can restore the population after damage (Giangreco et al., 2002). In the alveolar region, the two major epithelial cell types are type II (AEC2) and type I (AEC1) cells. The latter are closely apposed to capillary endothelial cells. Also present are a variety of stromal cells, including Pdgfra+ fibroblasts and lipofibroblasts (the latter located close to AEC2 cells), myofibroblasts and pericytes. Image modified from Rock and Hogan (2011).
Under ideal environmental conditions, such as those encountered by laboratory mice in specific pathogen-free units, cell turnover in the lung is very low. However, in real life, the human lung is directly exposed to many airborne hazards. Among these are pollutants, such as tobacco and biofuel smoke, and pathogens such as bacteria, mycobacteria and viruses. These agents, as well as others such as the anticancer drug bleomycin and X rays, can inflict considerable damage on the lungs. Consequently, respiratory diseases, as well as lung cancer, are a major cause of morbidity in vulnerable human populations (www.who.int/respiratory/en/). Fortunately, there are innate mechanisms that can be called into play to repair epithelial damage, and in laboratory animals these are usually remarkably efficient. Over the past few years there have been exciting advances in our understanding of the regenerative processes activated in different regions of the lung in response to various injuries, and of the relative roles of either undifferentiated stem/progenitor cells or specialized cells that can proliferate and undergo phenotypic reprogramming (transdifferentiation) (Hogan et al., 2014; Tata and Rajagopal, 2017). Despite these advances there is still much to be learned, in particular about the identity of stem/progenitor cells in the human lung and how deficient repair may contribute to pathological conditions such as chronic obstructive pulmonary disease (COPD), emphysema, familial and idiopathic pulmonary fibrosis (IPF) and bronchiolitis obliterans syndrome (BOS). As we shall see, organoids hold great promise in this area and in the quest for new drugs and therapies that enhance endogenous repair. Lung organoids also have considerable potential in the search for new treatments for diseases such as asthma, in which there is an overabundance of mucus-secreting cells as a result of the chronic release of cytokines in response to allergens (see www.nature.com/ni/multimedia/lung), and cystic fibrosis (CF), a genetic condition that leads to an increase in the viscosity of the mucus layer over the surface of the epithelium and to greater risks of bacterial infection and cellular stress.
In the context of this Review, ʻlung organoids' refers to self-assembling structures generated from lung epithelial progenitor cells cultured in 3D, with or without mesenchymal support cells. These organoids do not yet recapitulate all of the complex structures and cellular interactions of the different regions of the lung, especially the highly vascularized and delicate alveolar region. Nevertheless, over the past decade they have become an indispensable tool for basic and translational research. This Review highlights discoveries and advances made using lung organoids derived from three of the epithelial stem/progenitor cell populations of the adult lung: basal cells, airway secretory Club cells (previously known as Clara cells), and AEC2 cells. We also briefly review the current status of lung organoids derived from embryonic and induced pluripotent stem cells. These contain both lung epithelium and mesoderm and, together with cell lines derived from human fetal lung, have the potential to provide important information about human lung development as well as disease. We consider how lung organoids can be used to address questions in lung biology, such as the mechanisms by which endogenous lung progenitors effect repair, and how these might be enhanced by small molecules or drugs. Finally, we discuss some of the major limitations in lung organoid culture and how they might be overcome in the future.
Basal progenitor cells
Basal cells make up ∼30% of the pseudostratified mucociliary epithelium, lining most of the conducting airways of the human lung and the trachea and main stem bronchi in the mouse. Basal cells adhere closely to the basal lamina and do not extend to the lumen, unlike the more columnar multiciliated and secretory cells, and the minor populations of neuroendocrine and tuft cells that make up the rest of the epithelium (Fig. 1) (Hogan et al., 2014). The luminal cells are connected apically by junctional complexes and play a crucial role in forming a selectively permeable barrier between the external and internal environments of the lung. Genes characteristically expressed by basal cells include those encoding the transcription factor Trp63, the cytokeratin Krt5, integrin alpha 6 (Itga6), podoplanin (Pdpn; also known as T1alpha) and the transmembrane nerve growth factor receptor (Ngfr; also known as p75) (Hackett et al., 2011; Rock et al., 2009; Watson et al., 2015).
Lineage tracing and other studies have shown that in the adult mouse lung there is only very slow turnover of the mucociliary epithelium and replacement of luminal cells from basal cells and their immediate progeny (Ghosh et al., 2013, 2011; Hong et al., 2004; Rock et al., 2011, 2009; Watson et al., 2015). However, following cell damage by agents typically used experimentally – for example naphthalene, which kills secretory Club cells, or SO2 gas, which kills all luminal cells – or viral infection there are rapid changes in the behavior and proliferation of the basal cells so that they quickly regenerate the epithelium and restore barrier function. Recently, genetic techniques have been used to kill very selectively most basal cells. In response, some Club secretory cells undergo reprogramming to become Krt5+ Trp63+ basal cells that can function as stem cells in vivo (Pardo-Saganta et al., 2015; Tata et al., 2013). Taken together, these injury/repair studies have revealed remarkable and rather unexpected flexibility in the way in which basal cells, luminal precursors and differentiated secretory cells can work together to maintain and repair the pseudostratified mucociliary epithelium of the mouse airway. Organoids provide an in vitro model for the regeneration of the mucociliary epithelium from basal cells. They can therefore be used to test regenerative mechanisms proposed from in vivo studies and to screen for drugs, small molecules and molecular pathways that can regulate cellular plasticity and lineage outcomes, as well as crucial epithelial cell functions.
In the human lung, TRP63+ KRT5+ basal cells are present throughout the airways, extending down to bronchioles of ∼1 mm in diameter. There can be considerable variation in their abundance and organization between and within lungs, even from normal donors, with regions of hyperplasia and metaplasia interspersed with normal histology (Ghosh et al., 2011; Rock et al., 2010). Genetic lineage tracing is not possible in the airways of the human lung. Nevertheless, a very elegant substitute has been developed, based on analysis of the size and cellular composition of clonal patches of cells carrying mutations in the gene for mitochondrial cytochrome oxidase (Teixeira et al., 2013). The results predict the existence of a multipotent progenitor population of basal cells that maintains the secretory and ciliated cell populations through the stochastic replacement of lost cells. Various methods have been developed for isolating and growing these basal cells from different regions of the normal human respiratory system, including nasal epithelium, ʻlarge airways', which include the trachea, primary bronchi and intralobar bronchi down to about the third or fourth generation, and from bronchial brushings (Hackett et al., 2011; Randell et al., 2011). The most efficient methods for expanding and cloning TRP63+ KRT5+ human basal cells involves culturing them either on irradiated mouse 3T3-J2 fibroblasts in the presence of the Rho kinase inhibitor Y-27632 (Butler et al., 2016; Kumar et al., 2011; Suprynowicz et al., 2012), or with a Rho kinase inhibitor together with inhibitors of Smad-dependent signaling through the BMP and TGFβ pathways and an activator of Wnt signaling (Mou et al., 2016). The progenitor properties and differentiation capacity of these basal cells can then be followed in organoid cultures.
Organoids from mouse basal cells
The first organoids derived from mouse tracheal basal cells were called tracheospheres. These were clonal, as shown by mixing basal cells constitutively expressing red or green fluorescent proteins (Rock et al., 2009). Typically, flow cytometry is used to isolate the cells from protease-dissociated tissue, based on the surface expression of Ngfr, Itga6 or a carbohydrate that binds the lectin GSIβ4 (Rock et al., 2011; Tata et al., 2013). The cells are seeded into medium containing growth factor-reduced Matrigel and cultured in either transwell inserts or multiwells under conditions in which they do not adhere to the substrate. This can be achieved using a relatively high concentration of Matrigel (50%) or by suspending the cells in a low concentration (2-5%) of gel on top of a cushion of higher concentration (25-40%) (Fig. 2). In the latter condition the cells sink into the lower layer, and some spheres may fuse and therefore not be clonal.
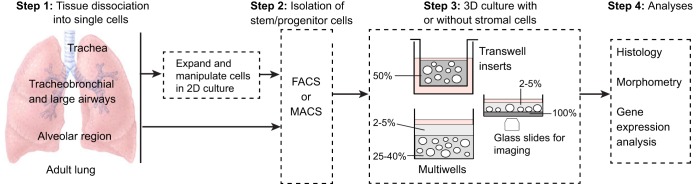
Fig. 2.
Overview of the derivation of lung organoids. Cells isolated from different regions of the adult mouse and human lung have been used for 3D culture. If intact pieces of lung are used, rather than bronchial brushings for example, the tissue is dissociated using proteases (step 1). Primary cells are isolated using FACs or MACs (magnetic bead sorting) (step 2) and can be seeded directly into Matrigel (gray, percentage indicated). In the case of basal cells, the number of undifferentiated cells can be increased by culturing them in 2D before transferring to 3D. This enables genetic manipulation and the selection and cloning of specific mutants. Methods for expanding AEC2s in 2D have not yet been reported. In Step 3, single-cell suspensions are seeded into 3D culture in inserts or multiwells, with or without mesenchymal cells (Table 1). Methods include suspending the cells in 50% Matrigel (Rock et al., 2009) or in a low concentration of Matrigel and layering this over a higher concentration into which the cells sink (Butler et al., 2016; Danahay et al., 2015; Tata et al., 2013). For live imaging, cultures can be established in glass-bottomed wells coated with a thin layer of dense Matrigel. Cells sink through the upper layer and accumulate at the interface so that they remain in the same plane for imaging (Rock et al., 2011). For histological analysis, cultures are fixed in the Matrigel. For quantification of different cell types or passaging stem cells, the Matrigel can be removed using dispase and spheres dissociated with trypsin.
Most of the various culture media used to date (Table 1) are not chemically defined but consist of a medium with ∼1 mM calcium and supplements such as bovine pituitary extract (BPE), insulin, transferrin and selenium (ITS), cholera toxin (CTX) and retinoic acid (RA). The most important additive is epidermal growth factor (EGF), which promotes growth. In some protocols the medium is switched after a few days to one with a lower concentration of EGF to slow proliferation and promote differentiation. Rho-associated protein kinase (ROCK) inhibitor Y-27632 is usually added for the first 48 h to promote cell survival. After ∼7 days, each sphere has developed a single lumen and there is evidence for differentiation of luminal cells (Fig. 3A). Colony forming efficiency (CFE), which is calculated as the number of spheres that grow compared with the total number of starting progenitor cells, is typically ∼3-10%. At 14 days, which is when the spheres are typically harvested, sphere diameters range from 150-500 µm. Immunohistochemistry shows that the majority of spheres of >300 µm diameter have an outer layer of Trp63+ Krt5+, Krt14+, Ngfr+ basal cells and an inner population of Krt8+ columnar ciliated and secretory cells (Fig. 3B,C). At this time, about half of the luminal cells have cilia and express the ciliated cell-specific transcription factor Foxj1. Although the cilia are motile, there is no evidence for coordination between cells, and manifestation of planar cell polarity (Vladar et al., 2012) has not been explored. The other half of the differentiated luminal cells are secretory cells. For unknown reasons these only express very low levels of the secretoglobin Scgb1a1 (also known as CCSP or CC10), which is normally expressed by secretory Club cells at high levels in vivo. They do, however, express other proximal Club cell markers, namely Scgb3a2 and an antimicrobial peptide known as Splunc1 (palate, lung and nasal epithelium clone; officially known as Bpifa1) (Musa et al., 2012; Tadokoro et al., 2014) (Fig. 3B,C). Importantly, expression of Splunc1 and Muc5AC can be dramatically upregulated at the expense of ciliated cell-specific genes by addition of the cytokine IL13 to the culture medium (Fig. 3D,D′). Thus, there is no doubt that single basal cells can give rise to both ciliated and secretory cells in this assay. Differentiation of neuroendocrine cells is rarely seen.
Table 1.
Compendium of protocols for growing organoids from basal cells
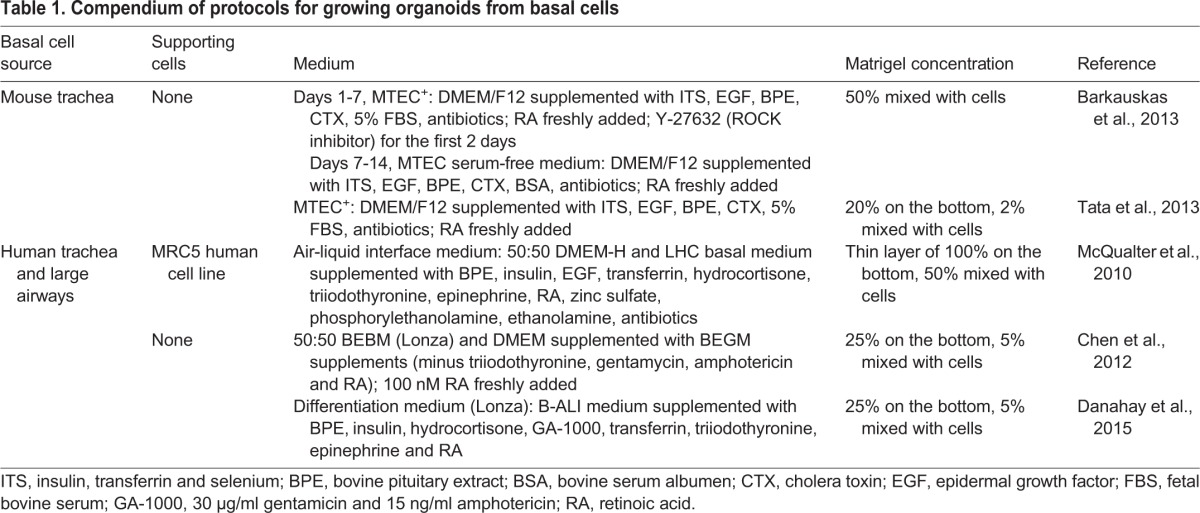
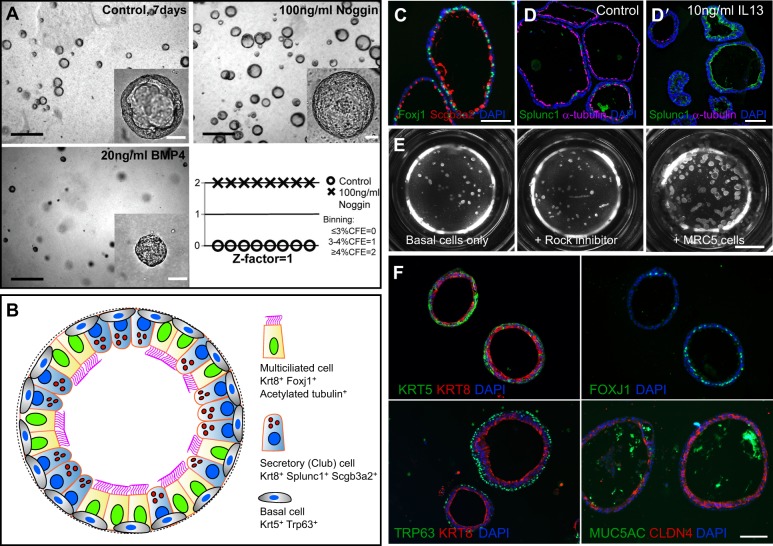
Fig. 3.
Basal cell-derived organoids. (A) Basal cell-derived organoids have been used for high- and medium-throughput screens (Danahay et al., 2015; Tadokoro et al., 2014). An example shows mouse tracheospheres after 7 days of culture, with and without 100 ng/ml noggin (a BMP inhibitor) and 20 ng/ml BMP4. The bottom right panel shows the results of scoring colony forming efficiency (CFE) in eight control wells versus eight wells with added noggin as one of the three values shown. The results were highly reproducible, giving a z factor of 1 (Zhang et al., 1999). It is important to establish such reproducibility before embarking on a large screen because conditions such as the position of a well in the tray, and changes in temperature and pH while changing the medium, can affect differentiation. (B) Schematic of a typical mouse tracheosphere after ∼14 days of culture, showing the relative position of basal versus luminal cells and markers of ciliated versus secretory cell types. (C) Section through clonal mouse tracheospheres cultured for 14 days and stained with antibody to Scgb3a2 (Club cells) and Foxj1 (ciliated cells). (D,D′) Sections of tracheospheres cultured without (D) or with (D′) 10 ng/ml IL13 and stained with DAPI and antibody to acetylated tubulin (cilia, red) and Splunc1 (Club cells, green). Note the dramatic increase in the number of secretory cells at the expense of ciliated cells in the presence of the cytokine. (E) Organoids (bronchospheres) derived from human basal cells cultured for 21 days without added factors (left), with the Rho kinase inhibitor Y-27632 (center), and with human lung fibroblasts (MRC5 line) (right). (F) Sections of human bronchospheres cultured for 21 days with MRC5 fibroblasts, stained with DAPI and markers for basal cells (KRT5, TRP63), luminal cells (KRT8, CLDN4), ciliated cells (FOXJ1) and secretory cells (MUC5AC). Scale bars: 100 µm in A insets, C,D′,F; 1 mm in A; 2 mm in E. Panel A was generated by Jason Rock; D,D′ by Tomomi Tadokoro.
At present it is unclear whether all basal cells and their proposed lineage-biased progenitors (Watson et al., 2015) can potentially give rise to tracheospheres with the same probability, or whether only a subset has this capacity. In the future, live imaging of developing tracheospheres, already shown to be feasible (Rock et al., 2011) (Fig. 2), could be used to follow in real time the asymmetric versus symmetric divisions of individual basal cells and their progeny, coincident with dynamic changes in the activity of Notch and other key signaling pathways.
Mouse tracheospheres have been used to screen for small molecules and drugs that regulate basal cell proliferation and differentiation. Such screens are highly relevant to finding new therapies for lung diseases in which the proportion of ciliated versus secretory cells is disturbed. For example, in patients suffering from asthma, COPD and CF, all of which are associated with inflammation, immune cytokine production and cellular stress, the proportion of mucus-producing cells is greatly increased at the expense of multiciliated cells (Rock et al., 2010). Mucus-secreting cells can be generated in two ways: either directly from Scgb1a1+ Club cells without cell proliferation (Evans et al., 2004; Pardo-Saganta et al., 2015) or from basal cells. Using organoids opens up the possibility of high-throughput screening for compounds that regulate the fate of basal cells, and whether they differentiate into ciliated versus secretory lineages. Although this screening is possible using air-liquid interface (ALI) cultures, in which both multiciliated and secretory cells are generated from basal cells, many more samples can be assayed quickly and quantitatively in a multiwell format. One of the first such screens involved using basal cells from Foxj1-GFP transgenic mice, which enabled the detection of differentiated ciliated cells using immunofluorescence microscopy (Tadokoro et al., 2014). Among the compounds that increased the proportion of GFP+ cells at the expense of Club cells was the cytokine IL6, which is expressed by tracheal stromal cells and immune cells following various injuries. A similar assay was used to screen for compounds that increase CFE and the total cell number in spheres without regard for cell types. This led to the discovery that inhibitors of the BMP and TGFβ signaling pathways increased both CFE and cell proliferation (as assessed by EdU incorporation), resulting in larger diameter spheres with more cells (Fig. 3A). With the BMP inhibitors dorsomorphin and DMH1, no change in the proportion of ciliated and secretory cells was seen, although other studies have suggested that inhibition of BMP signaling inhibits basal cell differentiation (Mou et al., 2016; Tadokoro et al., 2016). Addition of BMP4 ligand results in small spheres with mostly basal cells and few Krt8+ cells (Fig. 3A).
Organoids from human basal cells
Organoids have been obtained from basal cells isolated and expanded from human lungs (see Box 1). Different names have been given to the organoids depending on whether the basal cells are derived from the trachea (tracheospheres) or large airways (bronchospheres). As with mouse basal cells, the culture media for human basal cells are not yet fully defined (Table 1), but contain EGF as the major mitogen. CFE is ∼10% and can be increased to ∼20% by adding Rho kinase inhibitor and/or a human fibroblast cell line such as MRC5 cells (Fig. 3E). Under standard conditions the organoids contain TRP63+ KRT5+ basal cells, functional multiciliated cells and secretory goblet (MUC5AC+, MUC5B+) cells (Butler et al., 2016; Danahay et al., 2015; Hild and Jaffe, 2016; Rock et al., 2009) (Fig. 3F). Since basal cells also exist in the nasal epithelium, it should be possible to derive organoids, or ʻnasospheres', from these cells, which would be a particularly convenient approach for generating organoids from patients for eventual drug screening. It is likely that these organoids would give variable results, depending on where in the nose the basal cells are isolated from, since in one published case the luminal cells were reported to differentiate into squamous epithelial cells (Kumar et al., 2011).
Box 1. Obtaining human lung samples.
Human lung epithelial cells (mostly TRP63+ basal cells) isolated from large and small airways are commercially available from companies such as Lonza or Epithelix. The cells will have been expanded over a few passages from primary cultures. Samples from diseased lungs (COPD, asthmatic, CF) are also available. Investigators should obtain as much information as possible about their origin, including donor sex, age, smoking history, time since diagnosis, medications, and disease classification (in the case of COPD). Investigators should also be aware that there is considerable variability in cell growth rates and efficiency of differentiation even among cells from normal donors, and at least three to five different lots should be tested. An alternative to obtaining human lung epithelial cells commercially is to obtain donated normal lung tissue with institutional review board approval directly from hospital clinics, in particular academic centers with large lung transplant programs. Investigators should be aware of variability in handling, for example the time the sample is kept in ice-cold saline before processing, and should also know whether the samples come from donor lungs deemed unsuitable for lung transplant (in which case some areas may be contused, infected or otherwise damaged) or from trimmings of transplanted lungs. In either case, there can be variability between donors. If desired, diseased samples can be obtained from a number of sources: (1) lung explants; (2) bronchial brushings or endobronchial biopsies performed during bronchoscopy; or (3) lung resection samples. In the case of lung explant, investigators should be aware that this tissue comes from patients with end-stage disease and could be very different to that from an individual with earlier stage disease. Regardless of the source there can be tremendous regional variability within the lung, and stem cells isolated from a less affected region may have different properties than cells isolated from a more severely affected area. This is also true for samples obtained from the nasal passages by either brushing or curettage. Depending on the position from which the samples are taken and the disease status, samples may be more likely to undergo squamous versus mucociliary differentiation. In all cases with donated human lung samples, a consistent and proscribed isolation protocol should be followed.
As with the mouse, organoids from human basal cells have been used to screen for cytokines and other proteins that affect the ratio of ciliated and secretory cells and might therefore be potential therapeutic agents for disorders in which the balance is disrupted, for example chronic asthma. One such study involved plating cells in 384-well trays and analyzing almost 5000 different compounds (Danahay et al., 2015). The results identified a number of proteins that promote mucus cell production, including IL13, and showed that antibodies to NOTCH2 were very effective in inhibiting the proportion of secretory relative to ciliated (FOXJ1+) cells. Currently, screens using human rather than mouse organoids are limited by the paucity of easily scored fluorescent reporters for assaying gene expression. This should change as it becomes more feasible to manipulate basal cells genetically using CRISPR/Cas9.
In assays with human basal cells, it is important to recognize that there is considerable variability in the kinetics of proliferation and differentiation of basal cells from different lung donors. Samples from at least three to five different donors are therefore typically used in quantitative assays. Taking this variability into account is important when organoids are used to address outstanding questions in human lung biology. Among these questions are whether the chronic inflammatory conditions prevalent in disorders such as asthma, smoking-associated COPD, and CF result in epigenetic changes in basal cells. Such changes might make them inherently more likely to differentiate into secretory rather than multiciliated cells, or into squamous versus mucociliary epithelium, even when pathological conditions revert to normal (Shaykhiev et al., 2013). Another question under investigation is whether basal cells isolated from different positions along the proximal-distal axis of the human airways have inherently different potentials to give rise to ciliated versus secretory lineages, or even alveolar lineages under certain conditions (Kumar et al., 2011).
Human 3D cultures are well suited to exploit CRISPR/Cas9 gene editing technology to identify genes that regulate important airway functions such as barrier formation, selective permeability, fluid transport, innate immunity and ciliogenesis (Chu et al., 2015; Gao et al., 2015). Recent studies, for example, identified a central role for the transcription factor grainyhead-like 2 (GRHL2) in coordinating barrier function and differentiation, and identified the transcription factor ZNF750 as a new component of the ciliogenesis pathway in the human lung (Gao et al., 2015). In these studies, however, basal cells were not cloned after transfection, and cell populations carrying a mixture of different mutant GRHL2 alleles were tested. Although conditions have been developed in which single basal cells can be cloned in 2D culture (Mou et al., 2016), it remains to be rigorously tested whether each clone retains full differentiation capacity in organoid culture after expansion. Finally, there is great potential in using nasospheres to screen for small molecules and drugs that may regulate or compensate for the activity of mutant forms of the cystic fibrosis transmembrane conductance regulator (CFTR), as measured by fluid transport and sphere diameter. Since nasal basal cells can be isolated with minimal invasion, such an approach might be used in the future to individualize the treatment of patients suffering from CF.
Airway secretory cells
‘Secretory cells' refers here to the columnar, non-ciliated, non-neuroendocrine cells present in the airway epithelium of the lung. The two main classes are Club cells and goblet cells (see Fig. 1). Mature Club cells synthesize proteins such as secretoglobins (Scgb1a1, Scgb3a2) and Splunc1, which are stored in apical dense granules. Goblet cells, which are much more numerous in the human lung than in that of the laboratory mouse, synthesize mucins such as Muc5AC and Muc5B, and these are stored in large electron-lucent vesicles. The proportion of Club, goblet and ciliated cells varies somewhat along the proximal-distal axis of the mouse intralobar airways, with more ciliated and goblet cells proximally than distally.
Lineage-tracing studies in the mouse have shown that, at steady state, cells in the bronchioles that express Scgb1a1 can self-renew over the long term and give rise to ciliated cells, establishing their credentials as a stem cell population (Rawlins et al., 2009). Club cells can also directly differentiate into mucus-secreting goblet cells in response to cytokines such as IL13, especially in more proximal regions of the lung. Importantly, as summarized briefly in the legend to Fig. 1, there is extensive evidence that airway Club cells are a heterogeneous population that displays considerable phenotypic plasticity in response to viral and bacterial infections and agents that damage either the airway or alveolar epithelium. For example, lineage-tracing studies after damage to the alveolar region by the chemotherapeutic drug bleomycin have shown that Scgb1a1-expressing cells in the distal bronchioles proliferate and give rise to progeny in the alveoli with characteristics of AEC2s and AEC1s (Rock et al., 2011; Barkauskas et al., 2013; Tropea et al., 2012).
In such pathological conditions, which involve the production of numerous inflammatory cytokines as well as hypoxia, the contributions of different signaling pathways to changes in cell behavior are hard to disentangle. Theoretically, organoid culture provides a model system for testing the effect of individual cytokines and growth factors on the proliferation and differentiation of secretory cells, and for identifying subpopulations of Club cells with enhanced regenerative potential – that is, a higher CFE and with greater plasticity. Such populations could be exploited for therapeutic purposes. This ideal, however, is confounded by the current paucity of surface markers that can be used to both rigorously purify subsets of Club cells and to localize them unambiguously to specific regions of the mouse and human lung.
Organoids from mouse airway secretory cells
Two different methods have been used to isolate secretory Club cells by FACS for organoid culture studies: isolation based on the expression of surface markers; and lineage tracing using an Scgb1a1-CreER knock-in allele (Rawlins et al., 2009) with a fluorescent reporter allele. Using the first approach, McQualter and colleagues sorted lung epithelial cells on the basis of being CD45 (Ptprc)neg, CD31 (Pecam1)neg, EpCAMhigh, CD49f (Itga6)pos, CD104 (Itgb4)pos and CD24low (McQualter et al., 2010). This population includes some, but not all, Scgb1a1-expressing cells. When placed in 50% Matrigel in a relatively simple ʻbasal' medium, these cells gave rise to spheres, but only when co-cultured with primary EpCAMneg Sca1 (Ly6a)+ lung stromal cells. The spheres were divided into three general categories based on morphology after ∼14 days culture: large and rounded with a single lumen (type A, 46%); small, dense and lobular (type C, 35%); and ʻmixed', with multiple bud-like protrusions (type B, 19%). Immunohistochemistry and RT-PCR studies showed that, in addition to Scgb1a1+ cells, type A and B colonies contained Trp63+ cells, Foxj1+ ciliated cells, and Muc5AC+ secretory cells that were absent from type C colonies. By contrast, the type C colonies contained predominantly Sftpc+ AEC2-like cells. Mixed colonies also contained Sftpc+ AEC2 cells, predominantly at the tips of the buds. Broadly similar results were obtained by combining Scgb1a1-CreER lineage traced cells with a mouse lung stromal cell line (MLg) and SB431542, a TGFβ inhibitor, during the initial culture period (Chen et al., 2012). One drawback to both approaches is that it is not known whether the cells that gave rise to the large cystic spheres are normally located in a different region of the lung from those that gave rise to spheres containing AEC2s. This question was addressed in part by isolating EpCAMpos CD24low cells from mice carrying an Sftpc-GFP transgene (Chen et al., 2012). GFPhigh cells, which gave rise predominantly to type C colonies, were assumed on the basis of the in vivo localization of GFP to be derived from the very terminal bronchioles, whereas the GFPneg cells that gave rise to type A colonies were all proximal. GFPlow cells, which gave rise to mixed colonies, were assumed to come from distal bronchioles (Fig. 1). These results lend some support to the idea that there are intrinsic differences between subpopulations of Club cells in their ability to transdifferentiate into basal cells versus AEC2s.
Finally, organoid culture has been used to test the response of a small subpopulation of Scgb1a1+ Club cells in the distal bronchioles that also express Sftpc (known as dual-positive, bronchioalveolar stem cells or BASCs) to factors made by lung endothelial cells (Lee et al., 2014; Tropea et al., 2012). These experiments suggest that BASCs have the potential to differentiate into both AEC2s and airway cells, and that the alveolar differentiation can be specifically enhanced by thrombospondin secreted by endothelial cells. Currently, the standard way to isolate BASCs involves FACS, and is based on their expression of EpCAM and Sca1. Going forward, new technologies should enable these cells to be more rigorously purified based on co-expression of the genes encoding Scgb1a1 and Sftpc, so that additional markers can be found to distinguish them from other Club cells. This will allow a more detailed comparison of their potentially unique responses to cytokines and other factors produced by both endothelial cells and fibroblasts following lung injury.
Alveolar type II cells
The alveolar epithelium is composed of two distinct epithelial cell types. Type II cells (AEC2s) are cuboidal and characterized by the production of pulmonary surfactant proteins (e.g. Sftpc, Sftpb) and the lamellar bodies and machinery associated with their production and secretion (e.g. Lamp3 and Lyz2) (Fig. 1). By contrast, type I cells (AEC1s) are large squamous cells that cover most of the surface area of the alveoli and are closely apposed to a fine network of capillaries. AEC1s typically express advanced glycosylation end product-specific receptor (Ager), Pdpn and the transcription factor Hopx.
In the mouse lung, alveolar cell turnover is slow at steady state. However, early studies suggested that AEC2s can proliferate and behave as alveolar stem cells during repair after injury, repopulating both AEC2s and AEC1s (Evans and Bils, 1969; Evans et al., 1973). Recent genetic lineage-tracing studies in the mouse have further established that AEC2s proliferate and give rise to AEC1s in vivo, especially in response to tissue remodeling after injury (Barkauskas et al., 2013; Desai et al., 2014; Jain et al., 2015). In addition to epithelial cells, alveoli contain multiple stromal cell types, capillary endothelium and associated pericytes, as well as interstitial and alveolar macrophages (Fig. 1).
Given the clinical importance of respiratory disorders such as emphysema and idiopathic pulmonary fibrosis, which affect the structure and function of the gas exchange region, we need to know much more about the basic biology of alveolar stem cells and their niche. For example, little is known about the potential heterogeneity of the AEC2 population and whether specific subsets, such as those located near the periphery of the lung and the pleural surface, have higher proliferative and regenerative capacity than those in the interior. We also understand relatively little about the molecular crosstalk between alveolar epithelium and the different types of mesenchymal, endothelial and immune cells that reside close to them in the alveolar region. For example, how are trophic signals disrupted in the setting of aging and disease states? Can small molecules and drugs alleviate or reverse pathological changes once they have occurred? These and other questions are being addressed using organoid cultures under conditions in which AEC2s, in combination with support cells, both proliferate and differentiate into AEC1s.
Organoids from AEC2 cells
The two methods most commonly used to isolate murine AEC2s via FACS for organoid culture are genetic lineage tracing with a fluorescent reporter and the use of antibodies to bind surface markers (Table 2). The lineage-tracing approach, combining the Sftpc-CreER knock-in and Rosa26-lox-stop-lox–tdTomato (Rosa-tdTm) alleles, was used in the first studies that generated ʻalveolospheres' (Barkauskas et al., 2013). There are several advantages to using this general method. For example, the original location of the AEC2s in the lung can be determined by immunohistochemistry (Fig. 4A). Second, the proliferation and fate of the lineage-labeled cells can be followed in culture in the presence of unlabeled stromal support cells or contaminating epithelial cells (Fig. 4C). Disadvantages include the fact that the generation of the mice requires expensive breeding, and tamoxifen treatment is needed to activate the reporter. By contrast, isolation of AEC2s with cell surface markers can be applied to any mouse strain and these mice do not require exposure to tamoxifen. The main disadvantage of this approach is the fact that the precise initial location of the isolated cells in the lung cannot be determined by immunohistochemistry.
Table 2.
Mouse AEC2 isolation strategies
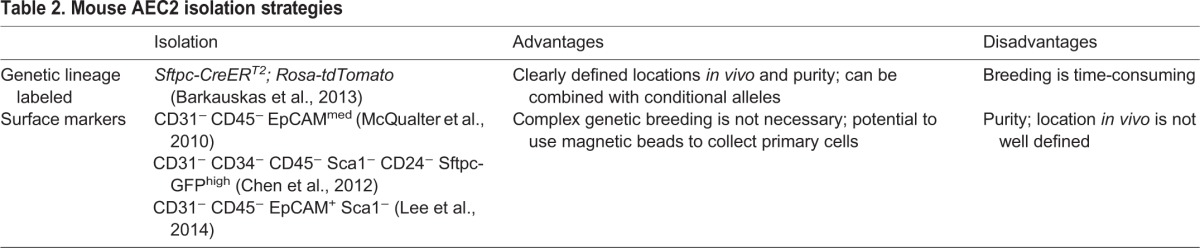
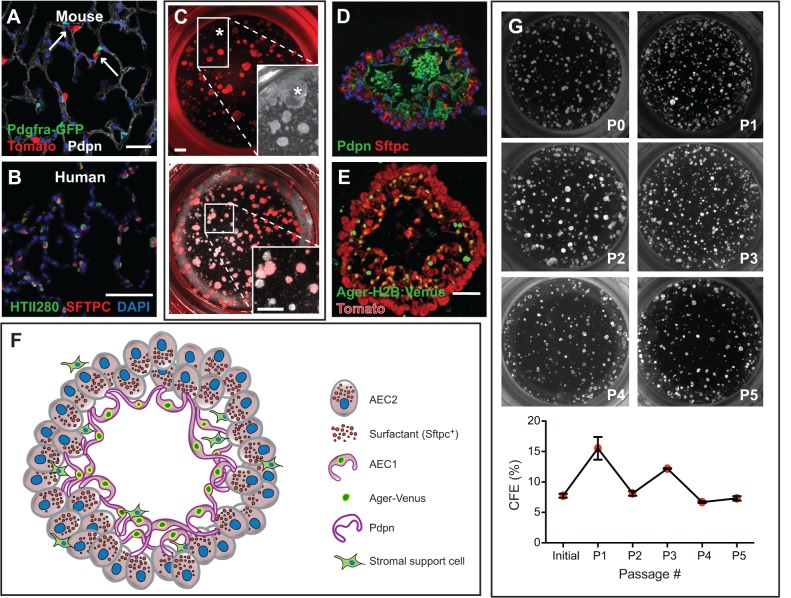
Fig. 4.
AEC2-derived alveolosphere culture. (A) Sftpc-CreERT2/+; R26R-tdTomato/+; Pdgfra-GFP/+ mice were injected with tamoxifen, leading to lineage labeling of ∼80% of AEC2s (Tomato+). Arrows point to Pdgfra-GFP+ lipofibroblasts in close proximity to lineage-labeled AEC2s. (B) HTII280 is a surface marker coexpressed with SFTPC to mark AEC2s in human lung. (C) Lineage-labeled AEC2s and GFP+ fibroblasts from the mouse lung in A were isolated by FACS and placed into the alveolosphere culture system in a ratio of 1:10, respectively. The asterisk in the top panel and in the brightfield inset marks a large, lobular, non-lineage-labeled sphere that is likely to have derived from a non-AEC2 epithelial cell. Without the lineage label it would have been incorrectly assumed that this sphere derived from an AEC2. The bottom panel and higher magnification inset shows an example of several non-lineage-labeled alveolospheres (lacking a fluorescent signal) that are likely to be derived from AECs that had not undergone recombination of the reporter allele. (D) Section of an alveolosphere showing Sftpc+ AEC2s on the outside and AEC1s (Pdpn+) on the inside. (E) Section of an alveolosphere showing lineage-labeled (Tomato+) Ager-H2B:Venus+ AEC1s on the inside. Green cells that are not lineage labeled are Pdgfra-GFP+ stromal cells. (F) Schematic illustrating the main cellular components of an alveolosphere. Currently, the precise way in which the AEC2s and AEC1s are connected to each other is not known. (G) Lineage-labeled AEC2s can be isolated and passaged (at day 14) at least five times without significant loss of CFE. Scale bars: 25 μm in A; 50 μm in B,E; 500 μm in C.
Human AEC2s are typically isolated with the use of a monoclonal antibody, HTII280, that is specific for human AEC2s (Gonzalez et al., 2010) (Fig. 4B). From dissociated human lung, AEC2s are defined as being propidium iodide staining (PI)neg, CD31neg, CD45neg, EPCAMpos, HTII280pos. These cells can be isolated by either FACS or by magnetic bead sorting (MACS). Increasingly, investigators are relying on MACS as this is more gentle to the cells than FACS, resulting in enhanced cell survival and organoid growth. Because HTII280 can reliably be used to stain human lung sections, this antibody can also be used to verify the location of HTII280+ cells in normal and diseased lung (Fig. 4B).
To date, some kind of support cell is required for the generation of alveolospheres. This necessitates using a culture medium in which both cell populations will survive, as it appears that close proximity of the epithelial and mesenchymal cells is required (McQualter et al., 2010). Several different support cell populations have been used with mouse organoids (Table 3). These include Pdgfra+ fibroblasts, which in the mouse lung include lipofibroblasts that lie in close proximity to AEC2s (Barkauskas et al., 2013) (Fig. 4A), EpCAMneg Sca1pos primary lung mesenchymal cells (McQualter et al., 2010), the MLg cell line (Chen et al., 2012) and lung endothelial cells (Lee et al., 2014). The effects of combining different cell types and immune/macrophage subpopulations are under investigation. So, too, is the consequence of using cells isolated before or after injury, or from old versus young mice, or those carrying specific mutations associated with human alveolar disease (Alder et al., 2015).
Table 3.
Compendium of alveolosphere protocols
When genetically lineage-labeled mouse AEC2s are used to initiate alveolosphere cultures, the 3D structures that arise contain Ager+, Pdpn+, Hopx+ AEC1s in the interior and Sftpc+ cells on the outside (Barkauskas et al., 2013; Jain et al., 2015) (Fig. 4D-F). This configuration does not strictly reproduce the structure of alveoli in the adult lung (Fig. 1) and it is still not clear how the AEC1s and AEC2s are polarized and interconnected by junctional complexes. To address these questions, the dynamics of AEC1 formation are being studied using live imaging to follow the morphogenesis of the spheres. In addition, an Ager-H2B:Venus knock-in allele is being used to quantify AEC1 differentiation under various conditions (Fig. 4E) and, in the long term, to develop high-throughput screens for small molecules and drugs that promote AEC2 differentiation. The long-term self-renewal of AEC2s can be quantified by dissociating spheres after 14 days of culture, resorting AEC2s and reseeding them with fresh stromal cells. As shown in Fig. 4G, this assay demonstrates that mouse AEC2s retain stem cell function for at least five passages.
A major limitation in using organoids to study gene function in alveolar epithelium is the fact that neither mouse nor human AEC2s can be expanded efficiently in culture before seeding in Matrigel. Because current protocols for CRISPR/Cas9 genome editing require 2D growth and expansion of cells, ideally combined with single-cell cloning of specific mutants, this technique has yet to be applied in the alveolosphere culture system.
Lung organoids derived from embryonic and induced pluripotent stem cells
Lung tissues derived from human pluripotent stem cells (hPSCs), including embryonic stem cells and induced pluripotent stem cells (iPSCs), have the potential to make a powerful impact on our understanding and treatment of lung disease. Efforts are currently underway to generate populations of immature lung epithelial and mesenchymal progenitors that can be massively expanded – with the option to store in a cryobank – and then directed to differentiate efficiently into mature airway and/or alveolar tissue. There are many potential questions that could be addressed using such a resource. For example, airway epithelial cells could be produced from iPSCs derived from patients with chronic asthma or CF to test the idea that epigenetic changes in the progenitors affect their self-renewal and differentiation capacity (Mou et al., 2012; Vladar et al., 2016). CF iPSC-derived lung organoids could also provide a reliable and reproducible source of CF mutant cells for screening drugs that compensate for, or correct, patient-specific mutations (Wong et al., 2012). This would overcome the problem of variability in the behavior of primary lung progenitor cells derived from even healthy individuals.
In the case of alveolar tissue, the differentiation of hPSCs into distal lung progenitors would allow studies of mutations affecting surfactant genes (SFTPA, SFTPB, SFTPC) or telomerase (TERT) that in AEC2s can cause respiratory failure and interstitial lung disease (Whitsett et al., 2010). ʻOmics' – genome, transcriptome, proteome, metabolome, and so on – profiling of healthy versus patient-specific hPSC-derived distal epithelium would enable a better understanding of how mutant cells become dysregulated over time (Grün et al., 2015). Finally, a source of progenitor cells that can be expanded after manipulation by CRISPR/Cas9 gene editing and still reliably differentiate would enable this powerful technique to be used to test the function of specific human genes in airway and alveolar cell specification during development. Although the molecular mechanisms that drive lung development and repair are likely to be conserved in general between mouse and human, work with early human embryos has already revealed interspecies differences in the transcription factors or ligands that are crucial at certain stages (Madissoon et al., 2014). Therefore, it will be important to examine the expression and function of human genes in the lung in relevant models.
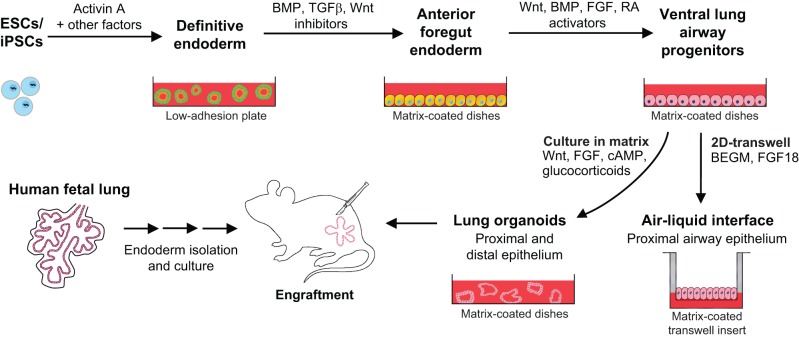
The primary challenge in realizing the above goals has been to direct the differentiation of hPSCs towards functional respiratory tissue that accurately resembles adult lung. The greatest progress in generating proximal and distal lung epithelial populations has been made by basing the differentiation protocols on the signaling pathways that direct embryonic lung development (Clevers, 2016; Dye et al., 2015; Huang et al., 2014; Longmire et al., 2012; Mou et al., 2012; Wong et al., 2012). A pioneering study first demonstrated the generation of anterior foregut endoderm (AFE) from hPSC-derived definitive endoderm (Green et al., 2011). To date, the most successful differentiation protocols first generate definitive endoderm, then AFE, followed by ventralization of the AFE via 3D culture using fibronectin or Matrigel substrates to yield immature, fetal-like lung and airway progenitors (Huang et al., 2015) (Fig. 5).
Fig. 5.
Derivation of lung organoids from hPSCs. Directed differentiation protocols vary as to the components of the growth medium, the extracellular coating, and the stages at which the cells are placed in a 3D environment. The schematic is based on results from three groups (Huang et al., 2014, 2015; Dye et al., 2015, 2016; Wong et al., 2012) (see main text). Human pseudoglandular and canalicular stage (weeks 6-19 of gestation) fetal lungs can also provide an epithelial cell source. A combination of in vitro growth and subsequent in vivo engraftment currently provides the best conditions for maturation of lung epithelium. Culture of ventral lung progenitors in 2D air-liquid interface transwells generates only proximal conducting airway epithelium. RA, retinoic acid; BEGM, bronchial epithelial growth medium (Fulcher et al., 2005).
Moving forward, one of the greatest obstacles is the development of protocols in which mature airway and alveolar cells are efficiently generated from their corresponding immature progenitors. Recent studies have shown that hPSC-derived lung organoids grown in a Matrigel-coated scaffold (serving as a bioartificial niche) and subsequently transplanted into mice generate more mature airway epithelium than previous methods (Dye et al., 2016). However, this particular way of transplanting lung organoids to induce cell differentiation and maturation does not generate alveolar cells, indicating a need for both proximal and distal cell-specific engraftment protocols. Air-liquid interface culture can be used to generate mature, polarized, pseudostratified proximal airway epithelium (Wong et al., 2012). This method of 2D-transwell culture exposes the apical side of the epithelium to the atmosphere and is a useful model of the airway microenvironment, but lacks the facility for in vivo transplantation for study of disease.
Finally, an alternative to using hPSCs to derive multipotential lung epithelium is to start with the fetal lung itself. Human embryonic lung from the pseudoglandular or canalicular stage (6-19 weeks gestation) may serve as the best source of immature cells with the potential to differentiate into both airway and alveolar cell types (Mondrinos et al., 2014; Rosen et al., 2015). However, use of fetal-lung derived organoids for cell therapy faces similar roadblocks as for hPSCs, and benefits derived from this cellular source must be weighed against the current challenges of obtaining suitable tissue, at least in some countries.
Future directions
This Review has surveyed some of the methods used to derive 3D organoids from different epithelial cell populations of the adult lung, including basal cells, secretory Club cells and AEC2 cells, as well as hPSCs, and the impact that this culture system has made on our understanding of lung biology. We have also outlined some of the potential future uses of organoids, especially those made from human cells, for both basic and translational research, including models of human disease and drug screening. However, for this potential to be fully realized there are a number of improvements that must be made to overcome significant limitations. These have been mentioned in the preceding text but, to reiterate, we highlight the three major issues again here.
First, to date none of the culture media used to derive organoids is chemically defined and they often contain complex supplements such as BPE or fetal bovine serum (FBS). The effect of parameters such as glucose levels and oxygen tension has also not been rigorously tested. Thus, we do not yet have a precise definition of the growth factors and small molecules and metabolites required for the long-term self-renewal and directed differentiation of lung epithelial stem and progenitor cells. In addition to defining these factors, we need to identify their sources in vivo and show, for example, which molecules are made by neighboring epithelial cells and which by mesenchymal cells in the stem cell niche. Progress is being made in establishing organoid cultures in which multiple stromal cell types are combined, and this will help to tease apart how the cell types interact in vivo and how they are affected by injury, inflammation and aging. It is also likely that extracellular matrix components and physical forces play key roles in regulating stem cell behavior and these parameters are also beginning to be explored using organoid culture.
Hand-in-hand with defining the culture requirements of adult lung epithelial stem cells is the need to establish efficient methods for cloning and expanding the clonal populations in 2D culture, while still maintaining their complete capacity for differentiation. As we have discussed, it is necessary to fully exploit the powerful technology of genome editing using CRISPR/Cas9, particularly for studying the role of specific genes in the self-renewal and differentiation of human lung stem cells and in generating models of human respiratory disease. hPSCs are currently more amenable to genome editing and thus the organoids generated from them have great potential for translational research, including drug discovery. The most difficult obstacle to overcome at present is to obtain full differentiation of hPSC cultures into specialized lung cell types, in particular AEC1 cells, and alveolar-like cellular arrangements.
Finally, progress in the utility of lung organoids requires the identification of more surface markers and/or reporters for isolating and purifying subpopulations of stem and progenitor cells and stromal support cells, in particular from the human lung. This will be important in understanding the functional heterogeneity of these cells and in developing protocols for directed cell differentiation and maturation. There is also the possibility that new classes of stem cells and support cells will be discovered, and organoid culture will provide one quantifiable method by which they can be compared with known populations. In summary, the array of different organoids that can be used to model various aspects of lung development, homeostasis, regeneration and disease represents an exciting new avenue for pursuing outstanding questions in lung – especially human lung – biology. With vigorous persistence in overcoming the challenges and careful analyses to interpret the results, we expect that organoid culture will become an indispensable tool for both basic and applied lung research.
Acknowledgements
We thank Drs Tomomi Tadokoro and Jason Rock for kindly providing unpublished data in Fig. 3 and Drs Purushothama Tata and Scott Randell for helpful discussion.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
B.L.M.H. acknowledges support from the National Institutes of Health (R37-HL071303, UO1-HL110967) and the Ellison Medical Foundation; C.E.B. acknowledges support from the Burroughs Wellcome Fund Career Award for Medical Scientists and the National Institutes of Health (KO8-HL122521). Deposited in PMC for release after 12 months.
References
- Alder J. K., Barkauskas C. E., Limjunyawong N., Stanley S. E., Kembou F., Tuder R. M., Hogan B. L. M., Mitzner W. and Armanios M. (2015). Telomere dysfunction causes alveolar stem cell failure. Proc. Natl. Acad. Sci. USA 112, 5099-5104. 10.1073/pnas.1504780112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas C. E., Cronce M. J., Rackley C. R., Bowie E. J., Keene D. R., Stripp B. R., Randell S. H., Noble P. W. and Hogan B. L. M. (2013). Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 123, 3025-3036. 10.1172/JCI68782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler C. R., Hynds R. E., Gowers K. H. C., Lee D. D. H., Brown J. M., Crowley C., Teixeira V. H., Smith C. M., Urbani L., Hamilton N. J. et al. (2016). Rapid expansion of human epithelial stem cells suitable for airway tissue engineering. Am. J. Respir. Crit. Care Med. 194, 156-168. 10.1164/rccm.201507-1414OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Matsumoto K., Brockway B. L., Rackley C. R., Liang J., Lee J.-H., Jiang D., Noble P. W., Randell S. H., Kim C. F. et al. (2012). Airway epithelial progenitors are region specific and show differential responses to bleomycin-induced lung injury. Stem Cells 30, 1948-1960. 10.1002/stem.1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H. W., Rios C., Huang C., Wesolowska-Andersen A., Burchard E. G., O'Connor B. P., Fingerlin T. E., Nichols D., Reynolds S. D. and Seibold M. A. (2015). CRISPR-Cas9-mediated gene knockout in primary human airway epithelial cells reveals a proinflammatory role for MUC18. Gene Ther. 22, 822-829. 10.1038/gt.2015.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. (2016). Modeling development and disease with organoids. Cell 165, 1586-1597. 10.1016/j.cell.2016.05.082 [DOI] [PubMed] [Google Scholar]
- Danahay H., Pessotti A. D., Coote J., Montgomery B. E., Xia D., Wilson A., Yang H., Wang Z., Bevan L., Thomas C. et al. (2015). Notch2 is required for inflammatory cytokine-driven goblet cell metaplasia in the lung. Cell Rep. 10, 239-252. 10.1016/j.celrep.2014.12.017 [DOI] [PubMed] [Google Scholar]
- Desai T. J., Brownfield D. G. and Krasnow M. A. (2014). Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature 507, 190-194. 10.1038/nature12930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye B. R., Hill D. R., Ferguson M. A. H., Tsai Y.-H., Nagy M. S., Dyal R., Wells J. M., Mayhew C. N., Nattiv R., Klein O. D. et al. (2015). In vitro generation of human pluripotent stem cell derived lung organoids. Elife 4, e05098 10.7554/elife.05098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye B. R., Dedhia P. H., Miller A. J., Nagy M. S., White E. S., Shea L. D. and Spence J. R. (2016). A bioengineered niche promotes in vivo engraftment and maturation of pluripotent stem cell derived human lung organoids. Elife 5, e19732 10.7554/elife.19732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M. J. and Bils R. F. (1969). Identification of cells labeled with tritiated thymidine in the pulmonary alveolar walls of the mouse. Am. Rev. Respir. Dis. 100, 372-378. 10.1164/arrd.1969.100.3.372 [DOI] [PubMed] [Google Scholar]
- Evans M. J., Cabral L. J., Stephens R. J. and Freeman G. (1973). Renewal of alveolar epithelium in the rat following exposure to NO2. Am. J. Pathol. 70, 175-198. [PMC free article] [PubMed] [Google Scholar]
- Evans C. M., Williams O. W., Tuvim M. J., Nigam R., Mixides G. P., Blackburn M. R., DeMayo F. J., Burns A. R., Smith C., Reynolds S. D. et al. (2004). Mucin is produced by clara cells in the proximal airways of antigen-challenged mice. Am. J. Respir. Cell Mol. Biol. 31, 382-394. 10.1165/rcmb.2004-0060OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher M. L., Gabriel S., Burns K. A., Yankaskas J. R. and Randell S. H. (2005). Well-differentiated human airway epithelial cell cultures. Methods Mol. Med. 107, 183-206. [DOI] [PubMed] [Google Scholar]
- Gao X., Bali A. S., Randell S. H. and Hogan B. L. M. (2015). GRHL2 coordinates regeneration of a polarized mucociliary epithelium from basal stem cells. J. Cell Biol. 211, 669-682. 10.1083/jcb.201506014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M., Brechbuhl H. M., Smith R. W., Li B., Hicks D. A., Titchner T., Runkle C. M. and Reynolds S. D. (2011). Context-dependent differentiation of multipotential keratin 14-expressing tracheal basal cells. Am. J. Respir. Cell Mol. Biol. 45, 403-410. 10.1165/rcmb.2010-0283OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M., Ahmad S., Jian A., Li B., Smith R. W., Helm K. M., Seibold M. A., Groshong S. D., White C. W. and Reynolds S. D. (2013). Human tracheobronchial basal cells. Normal versus remodeling/repairing phenotypes in vivo and in vitro. Am. J. Respir. Cell Mol. Biol. 49, 1127-1134. 10.1165/rcmb.2013-0049OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangreco A., Reynolds S. D. and Stripp B. R. (2002). Terminal bronchioles harbor a unique airway stem cell population that localizes to the bronchoalveolar duct junction. Am. J. Pathol. 161, 173-182. 10.1016/S0002-9440(10)64169-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R. F., Allen L., Gonzales L., Ballard P. L. and Dobbs L. G. (2010). HTII-280, a biomarker specific to the apical plasma membrane of human lung alveolar type II cells. J. Histochem. Cytochem. 58, 891-901. 10.1369/jhc.2010.956433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. D., Chen A., Nostro M.-C., d'Souza S. L., Schaniel C., Lemischka I. R., Gouon-Evans V., Keller G. and Snoeck H.-W. (2011). Generation of anterior foregut endoderm from human embryonic and induced pluripotent stem cells. Nat. Biotechnol. 29, 267-272. 10.1038/nbt.1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün D., Lyubimova A., Kester L., Wiebrands K., Basak O., Sasaki N., Clevers H. and van Oudenaarden A. (2015). Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 525, 251-255. 10.1038/nature14966 [DOI] [PubMed] [Google Scholar]
- Hackett N. R., Shaykhiev R., Walters M. S., Wang R., Zwick R. K., Ferris B., Witover B., Salit J. and Crystal R. G. (2011). The human airway epithelial basal cell transcriptome. PLoS ONE 6, e18378 10.1371/journal.pone.0018378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hild M. and Jaffe A. B. (2016). Production of 3-D airway organoids from primary human airway basal cells and their use in high-throughput screening. Curr. Protoc. Stem Cell Biol. 37, IE 9 1-IE 9 15. 10.1002/cpsc.1 [DOI] [PubMed] [Google Scholar]
- Hogan B. L. M., Barkauskas C. E., Chapman H. A., Epstein J. A., Jain R., Hsia C. C. W., Niklason L., Calle E., Le A., Randell S. H. et al. (2014). Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 15, 123-138. 10.1016/j.stem.2014.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K. U., Reynolds S. D., Watkins S., Fuchs E. and Stripp B. R. (2004). In vivo differentiation potential of tracheal basal cells: evidence for multipotent and unipotent subpopulations. Am. J. Physiol. Lung Cell. Mol. Physiol. 286, L643-L649. 10.1152/ajplung.00155.2003 [DOI] [PubMed] [Google Scholar]
- Huang S. X. L., Islam M. N., O'Neill J., Hu Z., Yang Y.-G., Chen Y.-W., Mumau M., Green M. D., Vunjak-Novakovic G., Bhattacharya J. et al. (2014). Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat. Biotechnol. 32, 84-91. 10.1038/nbt.2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. X. L., Green M. D., de Carvalho A. T., Mumau M., Chen Y.-W., D'Souza S. L. and Snoeck H.-W. (2015). The in vitro generation of lung and airway progenitor cells from human pluripotent stem cells. Nat. Protoc. 10, 413-425. 10.1038/nprot.2015.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R., Barkauskas C. E., Takeda N., Bowie E. J., Aghajanian H., Wang Q., Padmanabhan A., Manderfield L. J., Gupta M., Li D. et al. (2015). Plasticity of Hopx(+) type I alveolar cells to regenerate type II cells in the lung. Nat. Commun. 6, 6727 10.1038/ncomms7727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. F. B., Jackson E. L., Woolfenden A. E., Lawrence S., Babar I., Vogel S., Crowley D., Bronson R. T. and Jacks T. (2005). Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 121, 823-835. 10.1016/j.cell.2005.03.032 [DOI] [PubMed] [Google Scholar]
- Kumar P. A., Hu Y., Yamamoto Y., Hoe N. B., Wei T. S., Mu D., Sun Y., Joo L. S., Dagher R., Zielonka E. M. et al. (2011). Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell 147, 525-538. 10.1016/j.cell.2011.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H., Bhang D. H., Beede A., Huang T. L., Stripp B. R., Bloch K. D., Wagers A. J., Tseng Y.-H., Ryeom S. and Kim C. F. (2014). Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell 156, 440-455. 10.1016/j.cell.2013.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longmire T. A., Ikonomou L., Hawkins F., Christodoulou C., Cao Y., Jean J. C., Kwok L. W., Mou H., Rajagopal J., Shen S. S. et al. (2012). Efficient derivation of purified lung and thyroid progenitors from embryonic stem cells. Cell Stem Cell 10, 398-411. 10.1016/j.stem.2012.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madissoon E., Töhönen V., Vesterlund L., Katayama S., Unneberg P., Inzunza J., Hovatta O. and Kere J. (2014). Differences in gene expression between mouse and human for dynamically regulated genes in early embryo. PLoS ONE 9, e102949 10.1371/journal.pone.0102949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQualter J. L., Yuen K., Williams B. and Bertoncello I. (2010). Evidence of an epithelial stem/progenitor cell hierarchy in the adult mouse lung. Proc. Natl. Acad. Sci. USA 107, 1414-1419. 10.1073/pnas.0909207107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondrinos M. J., Jones P. L., Finck C. M. and Lelkes P. I. (2014). Engineering de novo assembly of fetal pulmonary organoids. Tissue Eng. Part A 20, 2892-2907. 10.1089/ten.tea.2014.0085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisey E. E. and Hogan B. L. M. (2010). Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev. Cell 18, 8-23. 10.1016/j.devcel.2009.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou H., Zhao R., Sherwood R., Ahfeldt T., Lapey A., Wain J., Sicilian L., Izvolsky K., Lau F. H., Musunuru K. et al. (2012). Generation of multipotent lung and airway progenitors from mouse ESCs and patient-specific cystic fibrosis iPSCs. Cell Stem Cell 10, 385-397. 10.1016/j.stem.2012.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou H., Vinarsky V., Tata P. R., Brazauskas K., Choi S. H., Crooke A. K., Zhang B., Solomon G. M., Turner B., Bihler H. et al. (2016). Dual SMAD signaling inhibition enables long-term expansion of diverse epithelial basal cells. Cell Stem Cell 19, 217-231. 10.1016/j.stem.2016.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musa M., Wilson K., Sun L., Mulay A., Bingle L., Marriott H. M., LeClair E. E. and Bingle C. D. (2012). Differential localisation of BPIFA1 (SPLUNC1) and BPIFB1 (LPLUNC1) in the nasal and oral cavities of mice. Cell Tissue Res. 350, 455-464. 10.1007/s00441-012-1490-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Saganta A., Law B. M., Tata P. R., Villoria J., Saez B., Mou H., Zhao R. and Rajagopal J. (2015). Injury induces direct lineage segregation of functionally distinct airway basal stem/progenitor cell subpopulations. Cell Stem Cell 16, 184-197. 10.1016/j.stem.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randell S. H., Fulcher M. L., O'Neal W. and Olsen J. C. (2011). Primary epithelial cell models for cystic fibrosis research. Methods Mol. Biol. 742, 285-310. 10.1007/978-1-61779-120-8_18 [DOI] [PubMed] [Google Scholar]
- Rawlins E. L., Okubo T., Xue Y., Brass D. M., Auten R. L., Hasegawa H., Wang F. and Hogan B. L. M. (2009). The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell 4, 525-534. 10.1016/j.stem.2009.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock J. R. and Hogan B. L. M. (2011). Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu. Rev. Cell Dev. Biol. 27, 493-512. 10.1146/annurev-cellbio-100109-104040 [DOI] [PubMed] [Google Scholar]
- Rock J. R., Onaitis M. W., Rawlins E. L., Lu Y., Clark C. P., Xue Y., Randell S. H. and Hogan B. L. M. (2009). Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. USA 106, 12771-12775. 10.1073/pnas.0906850106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock J. R., Randell S. H. and Hogan B. L. M. (2010). Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis. Model. Mech. 3, 545-556. 10.1242/dmm.006031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock J. R., Gao X., Xue Y., Randell S. H., Kong Y.-Y. and Hogan B. L. M. (2011). Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell 8, 639-648. 10.1016/j.stem.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen C., Shezen E., Aronovich A., Klionsky Y. Z., Yaakov Y., Assayag M., Biton I. E., Tal O., Shakhar G., Ben-Hur H. et al. (2015). Preconditioning allows engraftment of mouse and human embryonic lung cells, enabling lung repair in mice. Nat. Med. 21, 869-879. 10.1038/nm.3889 [DOI] [PubMed] [Google Scholar]
- Shaykhiev R., Zuo W.-L., Chao I., Fukui T., Witover B., Brekman A. and Crystal R. G. (2013). EGF shifts human airway basal cell fate toward a smoking-associated airway epithelial phenotype. Proc. Natl. Acad. Sci. USA 110, 12102-12107. 10.1073/pnas.1303058110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suprynowicz F. A., Upadhyay G., Krawczyk E., Kramer S. C., Hebert J. D., Liu X., Yuan H., Cheluvaraju C., Clapp P. W., Boucher R. C. Jr. et al. (2012). Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells. Proc. Natl. Acad. Sci. USA 109, 20035-20040. 10.1073/pnas.1213241109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro T., Wang Y., Barak L. S., Bai Y., Randell S. H. and Hogan B. L. M. (2014). IL-6/STAT3 promotes regeneration of airway ciliated cells from basal stem cells. Proc. Natl. Acad. Sci. USA 111, E3641-E3649. 10.1073/pnas.1409781111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro T., Gao X., Hong C. C., Hotten D. and Hogan B. L. M. (2016). BMP signaling and cellular dynamics during regeneration of airway epithelium from basal progenitors. Development 143, 764-773. 10.1242/dev.126656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. Y. S. and Krasnow M. A. (2016). Developmental origin of lung macrophage diversity. Development 143, 1318-1327. 10.1242/dev.129122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata P. R. and Rajagopal J. (2017). Plasticity in the lung: making and breaking cell identity. Development 144, 755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata P. R., Mou H., Pardo-Saganta A., Zhao R., Prabhu M., Law B. M., Vinarsky V., Cho J. L., Breton S., Sahay A. et al. (2013). Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature 503, 218-223. 10.1038/nature12777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira V. H., Nadarajan P., Graham T. A., Pipinikas C. P., Brown J. M., Falzon M., Nye E., Poulsom R., Lawrence D., Wright N. A. et al. (2013). Stochastic homeostasis in human airway epithelium is achieved by neutral competition of basal cell progenitors. Elife 2, e00966 10.7554/eLife.00966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropea K. A., Leder E., Aslam M., Lau A. N., Raiser D. M., Lee J.-H., Balasubramaniam V., Fredenburgh L. E., Alex Mitsialis S., Kourembanas S. et al. (2012). Bronchioalveolar stem cells increase after mesenchymal stromal cell treatment in a mouse model of bronchopulmonary dysplasia. Am. J. Physiol. Lung Cell. Mol. Physiol. 302, L829-L837. 10.1152/ajplung.00347.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan A. E., Brumwell A. N., Xi Y., Gotts J. E., Brownfield D. G., Treutlein B., Tan K., Tan V., Liu F. C., Looney M. R. et al. (2015). Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature 517, 621-625. 10.1038/nature14112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar E. K., Bayly R. D., Sangoram A. M., Scott M. P. and Axelrod J. D. (2012). Microtubules enable the planar cell polarity of airway cilia. Curr. Biol. 22, 2203-2212. 10.1016/j.cub.2012.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar E. K., Nayak J. V., Milla C. E. and Axelrod J. D. (2016). Airway epithelial homeostasis and planar cell polarity signaling depend on multiciliated cell differentiation. JCI Insight 1, pii: e88027 10.1172/jci.insight.88027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J. K., Rulands S., Wilkinson A. C., Wuidart A., Ousset M., Van Keymeulen A., Göttgens B., Blanpain C., Simons B. D. and Rawlins E. L. (2015). Clonal dynamics reveal two distinct populations of basal cells in slow-turnover airway epithelium. Cell Rep. 12, 90-101. 10.1016/j.celrep.2015.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitsett J. A. and Alenghat T. (2015). Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat. Immunol. 16, 27-35. 10.1038/ni.3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitsett J. A., Wert S. E. and Weaver T. E. (2010). Alveolar surfactant homeostasis and the pathogenesis of pulmonary disease. Annu. Rev. Med. 61, 105-119. 10.1146/annurev.med.60.041807.123500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A. P., Bear C. E., Chin S., Pasceri P., Thompson T. O., Huan L.-J., Ratjen F., Ellis J. and Rossant J. (2012). Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat. Biotechnol. 30, 876-882. 10.1038/nbt.2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.-H., Chung T. D. and Oldenburg K. R. (1999). A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67-73. 10.1177/108705719900400206 [DOI] [PubMed] [Google Scholar]