ABSTRACT
For many tissues, single resident stem cells grown in vitro under appropriate three-dimensional conditions can produce outgrowths known as organoids. These tissues recapitulate much of the cell composition and architecture of the in vivo organ from which they derive, including the formation of a stem cell niche. This has facilitated the systematic experimental manipulation and single-cell, high-throughput imaging of stem cells within their respective niches. Furthermore, emerging technologies now make it possible to engineer organoids from purified cellular and extracellular components to directly model and test stem cell-niche interactions. In this Review, we discuss how organoids have been used to identify and characterize stem cell-niche interactions and uncover new niche components, focusing on three adult-derived organoid systems. We also describe new approaches to reconstitute organoids from purified cellular components, and discuss how this technology can help to address fundamental questions about the adult stem cell niche.
KEY WORDS: Cell culture, Engineer, Model, Niche, Organoid, Stem
Summary: This Review article discusses how organoids have been used to model and characterize stem cell-niche interactions and how new engineering approaches enable systematic study of the stem cell niche.
Introduction
Precise control over stem cell differentiation and tissue architecture is essential for development, organogenesis and tissue homeostasis. Recently, organoids have emerged as key in vitro models of these processes, as advances in three-dimensional (3D) culture techniques have enabled the expansion of single stem cells into self-organizing tissues that functionally recapitulate key aspects of their in vivo tissue of origin. These aspects include the presence of multiple differentiated cell types, self-organization into a stereotyped tissue architecture, and activation of developmental gene expression programs (Camp et al., 2015; Clevers, 2016; Lancaster and Knoblich, 2014). The term organoid can refer to outgrowths from primary tissue explants (as in the mammary field) or to clonal outgrowths from single cells (Simian and Bissell, 2017). In this Review, we focus in particular on stem cell-derived organoids (Fig. 1A) as a model system to interrogate the stem cell niche. These organoids can be derived from embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs), or tissue-resident adult stem cells. Organoids grown from pluripotent ESCs or iPSCs mimic embryonic developmental processes, whereas those derived from adult stem cells can be used to model tissue homeostasis and its disruption during disease progression. Together, such organoids, whether derived from pluripotent or adult stem cells, represent a diversity of organotypic cultured tissues that each recapitulate aspects of brain, retina, stomach, prostate, liver or kidney structure (Clevers, 2016; Lancaster and Knoblich, 2014).
Fig. 1.
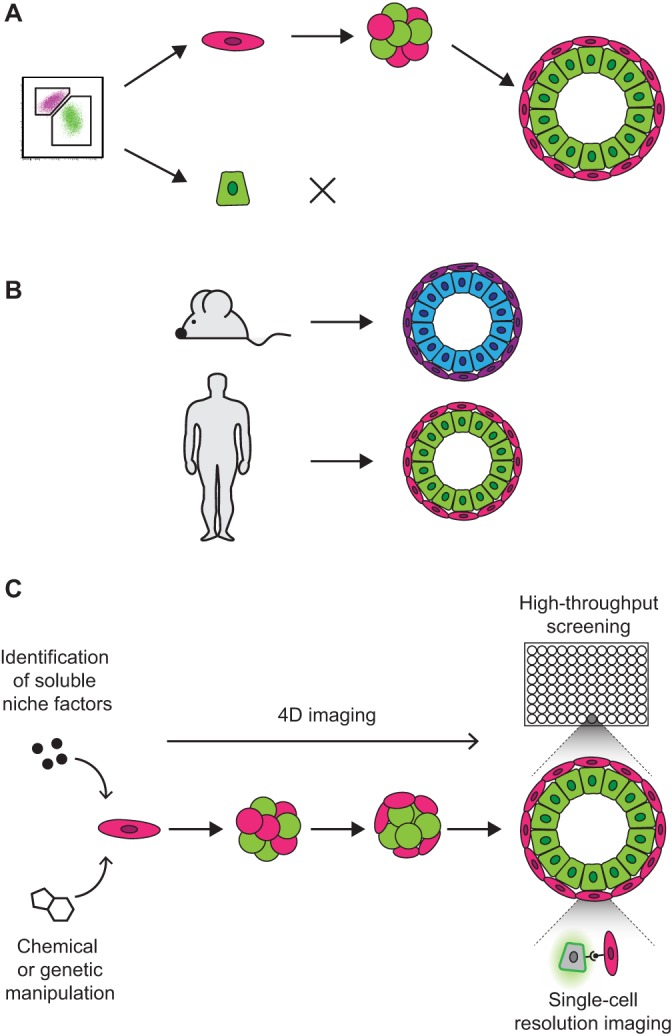
Advantages of organoid models for studying adult stem cells. (A) Organoids grown clonally from single cells can be used to prospectively identify adult stem cell populations based on the capacity of a cell to form organoids. (B) Organoids can be derived from human cells as well as non-human cells such as mouse or zebrafish, which allows modeling of human-specific stem cell biology and the identification of differences between human and non-human tissues. (C) In vitro culture allows in-depth experimental perturbation and imaging of stem cells in their surrounding niche. Different approaches include tightly controlled chemical or genetic manipulation, 3D imaging of live tissues over time (4D imaging), high-throughput combinatorial screening, and single-cell resolution imaging to analyze specific cell-cell interactions.
As well as providing an easily accessible in vitro platform for understanding development and disease, organoids, especially those derived from adult stem cells, provide a convenient means to investigate stem cell-niche interactions (Box 1). The stem cell niche can be defined as the local environment that surrounds a stem cell, which directly influences stem cell behavior and fate (Scadden, 2014). Indeed, some evidence suggests that in many cases the stem cell niche – rather than the stem cell itself – is the functional unit that controls cell fate. For example, transplantation into the mammary gland microenvironment reprograms single neural stem cells into mammary epithelial cells that can regenerate the mammary epithelial tree (Booth et al., 2008). The individual components that comprise the stem cell niche depend on the specific tissue, but include factors such as other differentiated cell types, signaling molecules, extracellular matrix (ECM) components, the 3D shape and arrangement of cells, and mechanical forces such as tension, rigidity and even fluid flow. Although many important niche components have been identified for different adult stem cell populations throughout the body, there are still many unknowns. In particular, it has been difficult to dissect the precise mechanism by which individual components regulate the niche owing to their interdependence. While in vivo animal studies have proven invaluable in defining the concept of the stem cell niche and identifying key stem cell-niche interactions, organoids serve as a complementary approach that could provide a better-controlled and higher-throughput platform to assess the contributions of individual niche components. Additionally, organoids can be used to study uniquely human stem cell-niche interactions (Fig. 1B), which will further our understanding of human tissue homeostasis, disease and regeneration.
Box 1. Key advantages of in vitro organotypic systems.
In vitro organoid systems have a number of key advantages when it comes to modeling stem cell biology. These include the fact that organoids can be grown as clonal outgrowths from single cells, and that they can be derived from human cells. In organoids grown from single cells, putative adult stem cell populations can be identified based on a cell's organoid-forming capacity (Fig. 1A) (Jung et al., 2011; Lim et al., 2009; Rock et al., 2009). Moreover, organoids grown from human adult stem cells directly model human-specific stem cell biology and can identify differences between human and non-human tissues (Fig. 1B) (Jung et al., 2011; Karthaus et al., 2014; Lim et al., 2009). Another key advantage of organoid culture is that it allows in-depth experimental perturbation and imaging of stem cells in their surrounding niche. This reductionist model can be used to identify molecules that are sufficient for stem cell maintenance and differentiation, complementing in vivo models that identify those that are necessary. Finally, in vitro systems allow tight temporal control over chemical and genetic manipulation of the stem cell niche and facilitate both single-cell resolution and high-throughput 3D imaging over time (Fig. 1C) (Sasai, 2013). Together, these features of organoids, in parallel with in vivo models, will help to reveal the underlying principles that guide tissue formation, maintenance and breakdown during development and regeneration.
In this Review, we focus on adult stem cell-derived organoids and use the examples of the mammary gland, airway epithelium and intestine to describe how organoids have enabled intensive and systematic study of signals from the adult stem cell niche that control cell self-renewal and differentiation (Fig. 1C). We further describe some emerging technologies that now enable reconstitution of the stem cell niche from purified cellular and ECM components. These engineering-based approaches can provide tight control over individual niche components and parameters such as initial organoid composition and size, which, under most commonly used organoid culture conditions, can be heterogeneous. This increased experimental control will be essential for understanding how tissue architecture shapes the complex molecular and mechanical signals that underlie cellular decision-making in the niche – decisions that allow stem cells to maintain tissue homeostasis and that go awry during disease.
Defining the stem cell niche
Organoids are a powerful tool for studying the stem cell niche, as they allow systematic chemical and genetic perturbation combined with in toto imaging of 3D tissues over time (Fig. 1C) (Sasai, 2013). In this section, we focus on three tissues for which adult stem cell-derived organoids have been used to dissect stem cell-niche interactions. Mammary gland organoids have helped to uncover how short-range Wnt signaling in the mammary epithelial stem cell niche is directly coupled with long-range hormonal signaling via the bloodstream. Organoid models of the airway epithelium have helped uncover how specific cues in the niche can direct stem cell maintenance or differentiation. Intestinal organoids have helped identify the necessary versus sufficient components of the stem cell niche and to directly image signaling gradients within the niche. We have chosen these examples to highlight key features of organoid systems that have helped to identify crucial components of the stem cell niche, and how these components regulate stem cell behavior and fate.
Mammary gland: direct coordination of local Wnt and systemic hormone signals
A key feature of organoid models is that in vitro growth allows decoupling of local signals within the niche from long-range signals, such as those that would impinge on the tissue from the bloodstream or surrounding stroma in vivo. Joshi and colleagues showed that systemic signals in the form of the steroid hormones estrogen and progesterone regulate stem cell dynamics in vivo but, surprisingly, that mammary stem cells themselves do not express receptors for progesterone and estrogen (Joshi et al., 2010). In this study, treatment of ovariectomized mice with estrogen and progesterone restricted mammary luminal differentiation and increased the proportion of organoid-forming cells in the mammary gland. These studies suggest that additional and locally acting paracrine signals could provide a link between systemic hormone levels and mammary stem cell function. Among locally acting paracrine signaling molecules, Wnt has been found to control stem cell activity in multiple tissues (Clevers et al., 2014) and, indeed, evidence suggests that Wnt is also a key component of the mammary stem cell niche. Wnt ligand treatment was found to promote mammary stem cell self-renewal, allowing serial, long-term expansion of mammary organoids (Cai et al., 2014; Zeng and Nusse, 2010). Although these studies identified systemic (estrogen) and local (Wnt) signals that regulate mammary stem cell function, how these signals are coordinated remained unclear.
Recent work with mammary organoids has helped to uncover the mechanism by which hormone receptor-negative mammary stem cells respond to hormones through local relay of Wnt molecules (Cai et al., 2014). Combined treatment with progesterone and estrogen induced the expression of Wnt4 and the Wnt agonist R-spondin 1 in luminal cells, which the authors showed is important for mammary stem cell self-renewal. Furthermore, they demonstrated that, in the absence of exogenous Wnt, hormone treatment supports expansion of mammary organoids from mammary stem cells only in the presence of luminal cells. Together, these data are consistent with a model in which systemic steroid hormone signaling induces local Wnt signaling from daughter luminal cells to restrict luminal differentiation and promote self-renewal of parent stem cells. This study illustrates how in vitro organoid systems serve as an important complement to in vivo experiments, since it would be difficult to distinguish between direct or secondary effects of hormone treatment without using tissue-specific conditional knockout mouse lines, which is both costly and time consuming. Organoid culture therefore provides a simplified experimental system in which to quickly and directly test the effects of specific niche signals on discrete cell types, which can then be confirmed by in-depth animal studies.
Airway epithelium: from single-cell imaging to high-throughput interrogation of the stem cell niche
As a complement to animal studies, organoid culture allows both imaging of the stem cell niche at single-cell resolution and large-scale screens to identify novel niche components. Recent studies have used these features to identify juxtacrine and secreted factors that direct airway epithelial cell fate in mouse tracheosphere and human bronchosphere organoids (Danahay et al., 2015; Rock et al., 2011; Tadokoro et al., 2014). Single-cell resolution imaging of stem cells within their developing niche helped to identify Notch as a feedforward signal that controls daughter cell differentiation (Pardo-Saganta et al., 2015; Rock et al., 2011). Using cells expressing a Notch activity reporter, Rock and colleagues analyzed Notch activity in developing 2- to 16-cell mouse tracheospheres following initial stem cell division. They observed that single basal stem cells that express the Notch ligands Jagged and Delta divide either symmetrically to produce two basal daughter cells or asymmetrically to produce one basal daughter cell and one luminal daughter cell in which Notch signaling is active. Further in vivo experiments demonstrated that Notch activation is required for luminal differentiation of basal stem cells (Rock et al., 2011). Using a series of sophisticated cell ablation, lineage tracing and conditional knockout experiments, Pardo-Saganta and colleagues further dissected this parent-daughter signaling pathway in vivo and identified a contact-dependent feedforward signal from basal cells to their progeny that maintains a pool of differentiated secretory progenitor cells (Pardo-Saganta et al., 2015). Together, these studies demonstrate how relatively simple in vitro organoid models allow sophisticated single-cell analyses that inform the design of more complicated animal studies interrogating cell type-specific niche signals.
In contrast to these low-throughput, high-resolution studies, two recent papers exemplify the usefulness of high-throughput screens using organoids (Danahay et al., 2015; Tadokoro et al., 2014). Tadokoro and colleagues used airway basal stem cells from transgenic reporter mice in which EGFP is driven by the Foxj1 promoter to rapidly screen for potential niche signals that bias stem cell differentiation towards a ciliated cell fate. Stem cells were seeded at clonal density into 96-well plates and treated with individual factors such as cytokines or inhibitors of key signaling pathways. Tracheospheres were screened by fluorescence microscopy for EGFP at multiple time points, and quantified at final time points by fluorescence-activated cell sorting (FACS) of dissociated organoids or immunostaining for known markers. These screens, combined with in vivo experiments, identified IL6/STAT3 as a key signal that promotes differentiation into ciliated cells (Tadokoro et al., 2014). Using a similar strategy, Danahay and colleagues (Danahay et al., 2015) performed a high-throughput screen using human bronchosphere organoids, seeding stem cells at clonal density into 384-well plates and screening a collection of 4876 secreted recombinant human proteins. As promoter-reporter strategies are more difficult to implement in primary human culture systems than in mouse, the authors used a multiplex RNA quantification assay to identify changes in the expression of cell type-specific markers following two weeks of growth. Using this strategy, they identified 420 proteins that led to a 2-fold or greater change in expression of at least one cell type marker, including 69 that altered stem cell fate decisions. These screens identified the epidermal growth factor (EGF) family of ligands as a class of signals that promote stem cell self-renewal, and a class of inflammatory cytokines including IL13 and IL17A that promote differentiation into secretory goblet cells. Similar to the mammary gland, long-range signaling is coupled to short-range signaling to regulate stem cell differentiation in the lung, as additional in vitro and in vivo experiments demonstrated that local Notch signaling was required for goblet cell differentiation following IL13 or IL17A treatment (Danahay et al., 2015).
Together, these studies demonstrate how organoids facilitate the study of the stem cell niche at multiple levels, from single-cell interrogation of cell-cell juxtacrine cues to high-throughput screening for key paracrine signals. Moreover, they have provided insight into the question of how long-range signals are integrated with short-range cell-cell interactions to direct tissue homeostasis and disease. One logical extension to these types of studies would be to perform combinatorial screens to identify functional combinations of secreted proteins with ECM molecules or juxtacrine cues present on other cell types within the niche.
Intestinal epithelium: direct manipulation and visualization of local Wnt signaling
Intestinal organoids were among the first to be derived from adult stem cells, rather than tissue explants or pluripotent ESCs or iPSCs. Using key insights from mouse studies, early experiments confirmed that niche signals such as Wnt/R-spondin that control crypt proliferation in vivo (Kim et al., 2005; Korinek et al., 1998; Kuhnert et al., 2004; Pinto et al., 2003) are also required for in vitro maintenance of intestinal stem cells in organoid cultures (Sato et al., 2009). The intestinal organoid field has also led the way in the manipulation of organoids at the single-cell level, for example by reconstructing aggregates containing both stem cells and purified Paneth cells. These methods have enabled direct manipulation and imaging of Wnt signaling in the intestinal stem cell niche, identifying multiple sources of Wnt and providing insight into how its activity is restricted to the base of the crypt. In an elegant series of reaggregation experiments, Sato et al. (2011) demonstrated that Wnt3-producing Paneth cells, together with the soluble factors EGF, R-spondin, and noggin, can serve as a minimal in vitro niche for intestinal stem cells. Reconstituted heterotypic aggregates or cell doublets containing both Lgr5+ stem cells and Paneth cells displayed greatly increased efficiency of organoid formation compared with homotypic aggregates or doublets, and exogenous Wnt ligand treatment rescued organoid formation in Lgr5+ singlets or homotypic doublets (Sato et al., 2011). Interestingly, although Wnt secretion by Paneth cells is required for in vitro maintenance of Lgr5+ stem cells, it is dispensable in vivo. Further work has demonstrated that surrounding mesenchymal cells serve as an alternative source of Wnt both in vitro and in vivo (Farin et al., 2012; Valenta et al., 2016). This study highlights one aspect of organoid models that is both an advantage and potential drawback. Organoid models provide a reductionist system that can be used to easily identify niche signals that are sufficient for stem cell maintenance. However, these experiments cannot fully recapitulate the complexity of in vivo biology, and therefore might identify necessary components that are dispensable in vivo. Key results from in vitro studies should therefore be confirmed in animal models, when possible. In this case, a combination of in vitro and in vivo work demonstrated that although Wnt itself is necessary, Wnt secretion from either Paneth cells or the mesenchyme is sufficient in vivo for maintenance of the intestinal stem cell niche (Farin et al., 2012; Goss et al., 2009; Valenta et al., 2016).
Intestinal organoids have also facilitated dissection of the spatial regulation of Wnt activity. In mice, Paneth cells and Wnt activity are both localized at the base of crypts, and live imaging of organoids has demonstrated that stem cell progeny self-organize into similar crypt-like structures (Sato et al., 2009, 2011). Organoids grown in the presence of exogenous Wnt display global rather than restricted Wnt activity and adopt a rounded morphology, suggesting that local Wnt activity maintains crypt architecture (Sato et al., 2009). Using Paneth cells from mice with a functional HA-tagged knock-in allele of Wnt3, Farin et al. (2016) directly visualized Wnt diffusion in reaggregated organoids containing both Wnt3HA/HA and wild-type cells. Transfer of HA-tagged Wnt was primarily restricted to a one-cell diameter distance away in control organoids and accumulated around Wnt-producing cells following cell cycle arrest. This observation is consistent with a mechanism of membrane-bound Wnt transfer via cell division, rather than the free diffusion of Wnt. This study therefore revealed a direct mechanism for spatial restriction of Wnt activity to the crypt base (Farin et al., 2016). The use of organoids allowed the simple mixing of wild-type and HA-tagged Wnt3 knock-in cells from two different mice to directly visualize Wnt transfer between producing and receiving cells. Although similar experiments would be possible in vivo, they would require the development of tissue-specific HA-tagged Wnt3 conditional knock-in mouse strains. Furthermore, in vitro culture allowed the authors to produce spherical, rather than crypt-like, organoids by treatment with exogenous Wnt, greatly simplifying image acquisition and quantification. Nonetheless, given that previous studies have identified multiple sources of Wnt in vivo (Farin et al., 2012; Valenta et al., 2016), additional animal studies to dissect Wnt transfer from Paneth cells versus mesenchymal sources will be necessary to fully understand how the Wnt gradient is maintained in vivo.
Engineering the stem cell niche: a bottom-up approach to controlling initial organoid size, shape and composition
Most current protocols for guiding stem cells to form organoids with functional niches rely on the ability of stem cells to differentiate into a number of more mature cell types, as well as the ability of these cells to self-organize into the correct tissue architecture. These protocols build upon now classic methods in developmental biology that used dissociation and reaggregation of cells from various tissues to understand how cells self-sort during morphogenesis (Bernfield and Fell, 1967; Townes and Holtfreter, 1955; Weiss and Taylor, 1960). Together with more recent data, these reaggregation studies suggest that cells isolated from adult tissues can retain a ʻmemory' of their developmental or homeostatic program that allows them to self-organize through local interactions, recapitulating aspects of the tissue architecture from which they are derived (Cerchiari et al., 2015a; Chanson et al., 2011; Runswick et al., 2001).
While the capacity to self-organize leads to the formation of reproducible and tightly regulated tissue architectures in vivo, the process can be considerably more variable in vitro, particularly at length scales larger than a few hundred microns (Gracz et al., 2015). Such variability suggests that key chemical, physical and/or spatial cues that guide the progress of self-organization in vivo may be lacking after cell reaggregation in vitro. In these cases, the tools and techniques of engineering could facilitate the more robust formation and analysis of organoids. New engineering technologies such as microwell arrays, droplet-based microfluidics, 3D bioprinting, chemically programmed tissue assembly (see Glossary, Box 2) and chemically defined ECMs mean that it is now feasible to engineer organoids in such a way as to precisely define their initial size, composition and spatial organization (Allazetta and Lutolf, 2015; Ellison et al., 2016; Magin et al., 2016; Murphy and Atala, 2014; Todhunter et al., 2015; Gjorevski et al., 2016). Control over these culture parameters has the potential to open up new experimental approaches for understanding development, regeneration and disease by providing greater spatiotemporal control over organoid culture in general and the stem cell niche in particular (Fig. 2).
Box 2. Glossary.
3D bioprinting: A technique using a droplet-, laser- or extrusion-based printing device in conjunction with control of a 2D stage to precisely place cell or ECM ‘inks’ additively in a desired pattern.
Chemically programmed tissue assembly: Directed cell adhesion using artificial and bio-orthogonal adhesion molecules on cell surfaces. In this Review, we are specifically referring to modification with single-stranded DNA molecules, which encode interactions by Watson-Crick base pairing.
Click chemistry: A fast, biocompatible and bio-orthogonal chemical reaction joining two modular chemical moieties. In this specific context, we refer to the reaction of modified lipids containing ketones or oxyamines on two different cell surfaces to form a covalent oxime bond.
Droplet-based microfluidics: Precise manipulation of discrete microscale droplets of aqueous fluid in an immiscible non-aqueous medium (or vice versa).
Intaglio-void/embed-relief topographic (InVERT) molding: A technique in which cells are deposited into a recessed pattern and embedded in a hydrogel. The cells and gel are then removed from the pattern and inverted, creating recesses into which the next type of cell can be deposited.
Microraft array: A variant of microwell arrays (see below) in which individual wells are detachable for isolation and subsequent study.
Microwell array: Geometrically spaced concave recesses, microscale in diameter, into which cells are flowed or allowed to sediment to form cell aggregates of defined shape and size.
Photolithography: A microfabrication technique in which photomasks and ultraviolet light are used to generate microscale patterns in a photoresist material. In this context, photoresist wafers are typically used as inverse patterns to make microarray molds from another material such as PDMS.
RGD peptide: A synthetic integrin-recognition sequence comprising arginine, glycine and aspartic acid. Commonly used as an adhesion ligand in engineered ECMs.
Fig. 2.
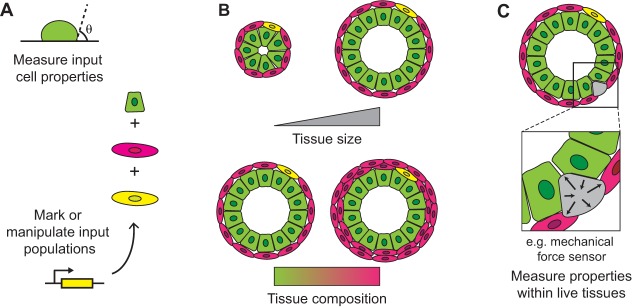
Advantages of engineered organoids for studying the stem cell niche. (A) Constructing organoids from purified cellular components allows the direct measurement of input cell properties and labeling of different input populations. (B) In addition to the advantages of classical organoid models (outlined in Fig. 1), controlled organoid engineering provides tight experimental control over the numbers and types of cells in the resulting tissue. (C) Engineered organoids can incorporate non-cellular material, such as sensors that dynamically measure properties such as mechanical forces and signaling pathway activation within live tissues.
Ideally, engineered organoids should be made from purified components, allowing direct measurement and manipulation of the physical (e.g. shape, mechanics, cellular composition) and chemical (e.g. molecular composition) properties of input cells, ECM and medium. For example, engineering strategies provide a simple and precise means for the marking and tracking of input cell populations as they move and differentiate within organoids (Fig. 2A). Furthermore, they provide a facile means of activating or inhibiting chemical and physical cues in specific cell types by targeting gain- or loss-of-function perturbations to only the intended cell type prior to cell reaggregation (Sato et al., 2011). Similar experiments using standard organoid cultures grown from individual stem cells or tissue explants would require sophisticated cell type-specific chemical or genetic targeting strategies. Combined with new and powerful technologies for genome editing and spatiotemporal control of gene expression (e.g. CRISPR/Cas9 and optogenetics), engineering approaches will allow the manipulation and de novo construction of synthetic signaling circuits to facilitate our understanding of cell-cell communication within and around the niche.
An engineering approach to organoids will also allow the generation of tissues with defined initial compositions, shapes and spatial organization (Discher et al., 2009). By enabling control over the starting number and types of cells in the organoid (Fig. 2B), these approaches provide a highly reproducible system that can be used to understand how tissue composition affects stem cell differentiation and cellular plasticity, and how stem cells self-organize within a tissue. Precise control over the cellular inputs for organoid culture can also identify the cell types and matrix components that comprise the ʻminimal niche' sufficient for tissue self-renewal (Gjorevski et al., 2016; Sato et al., 2011). Furthermore, combinations of technologies, such as microfluidics and chemically programmed assembly, make it possible to incorporate sensors that dynamically measure properties such as mechanical forces within live tissues (Fig. 2C) (Campàs et al., 2014). These advantages will enable systematic modeling, quantification and testing of stem cell-niche interactions to more fully define the systems-level cellular and molecular networks that control tissue homeostasis.
In this section of our Review, we summarize some emerging approaches for precisely controlling organoid cultures, including the use of microwells, microfluidics, bioprinting, chemically programmed assembly and engineered ECMs (Fig. 3). These new approaches for the generation and study of organoids will help define the necessary and sufficient components of the stem cell niche, provide more quantitative insight into the role of plasticity and dedifferentiation in maintaining tissue homeostasis, and contribute to our emerging understanding of how the chemical and physical properties of the niche direct stem cell self-organization and behavior.
Fig. 3.
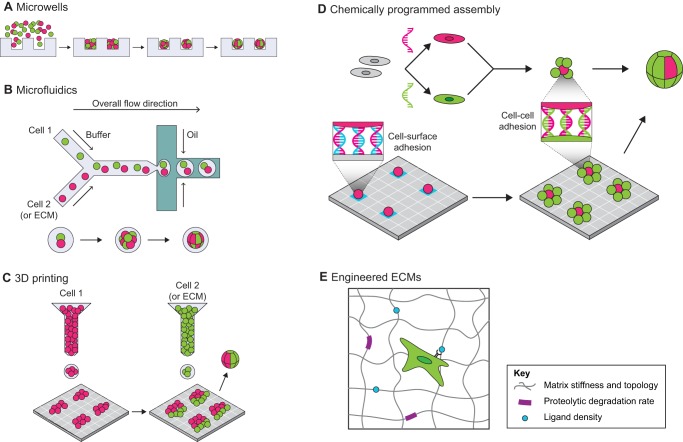
Technologies to reconstitute organoids from purified cell populations. (A) Microwells. Cells can be centrifuged, flowed or injected into arrays of microwells to produce organoids that conform to the size and shape of the microwell. (B) Microfluidics. Individual cells or ECM components can be captured in aqueous droplets and combined to produce precisely sized spheroids that are amenable to high-throughput recovery and analysis. (C) 3D bioprinting. Cells suspensions or ECM components can be used as a printable ‘ink’, with control in the x, y plane over individual components. Multiple ‘inks’ can be loaded into the printer to create complex, patterned tissues. (D) Chemically programmed assembly. Cell surfaces can be chemically modified with single-stranded DNA or other bio-orthogonal molecules to program adhesion to surfaces or other cells, independent of endogenous cellular machinery. This technique can achieve single-cell resolution to create organoids with precisely controlled cell-cell interactions. (E) Engineered ECMs. Polymer hydrogels such as PEG can be tuned over a range of stiffnesses and topologies by varying monomer concentration, molecular weight and degree of crosslinking. Cell adhesion ligands (e.g. RGD integrin-recognition sequences) or proteolytically degradable sequences can be engineered back into the system.
Microwells provide control over organoid shape and size
Photolithographically (see Glossary, Box 2) defined microwells are one relatively simple technique to control initial organoid shape and size by providing a well-defined environment in which to aggregate cells. Wells are commonly microfabricated from weakly adhesive materials such as polydimethylsiloxane (PDMS) or non-adhesive materials such as agarose, and can be used to produce uniform cell aggregates of discrete sizes by varying well depth, diameter and seeding density (Fig. 3A) (Choi et al., 2010; Karp et al., 2007; Napolitano et al., 2007). Following condensation, spheroids can be transferred into ECMs for 3D culture. This general strategy is even compatible with forming reproducible embryoid bodies from human ESCs (Ungrin et al., 2008). For cell aggregates that require immediate ECM contact for appropriate condensation and self-organization, microwells can be directly stamped into matrices such as collagen and subsequently overlaid with additional collagen to create fully embedded tissues (Nelson et al., 2006). This technique also facilitates the production of organoids with more complicated shapes, such as branching patterns. For softer ECMs that cannot be directly stamped, Cerchiari et al. (2015b) developed a technique using gelatin as a degradable scaffold to produce microwells that could then be removed by buffer exchange and replaced by matrices such as Matrigel or fibrin, maintaining the positional fidelity of the original wells. Both of these latter methods allow the production of organoids of non-spherical shapes that can be used to dissect how morphogen gradients are set up and maintained in the stem cell niche.
One notable variation on microwells, termed microraft arrays (MRAs; see Glossary, Box 2), utilizes a translucent polystyrene array to facilitate live in-well imaging and controlled release of individual wells for further examination such as gene expression analysis (Gracz et al., 2015; Wang et al., 2010). This method has enabled high-throughput live imaging of intestinal stem cells interacting with other cells in the niche to show that Wnt signaling from Paneth cells is contact dependent (Gracz et al., 2015). Another approach, known as intaglio-void/embed-relief topographic (InVERT) molding (see Glossary, Box 2), uses intaglio/relief-based cell deposition – originally a type of printing technique – to seed one cell population within a recessed surface (e.g. microwells), embed the patterned cells within a hydrogel, and deposit a second cell population in the ʻvoids' around the first pattern. This technique has been used to study the interactions between iPSC-derived hepatocytes and surrounding non-parenchymal cells (Stevens et al., 2013). Overall, microwell approaches facilitate the rapid production of well-defined organoids and are therefore particularly well suited for screens identifying soluble factors exchanged within the niche, and for live imaging studies of niche dynamics that require large numbers of highly reproducible tissues to obtain sufficient statistical power.
Microfluidics approaches guide organoid size and shape
Similar to microwell approaches, microfluidics techniques can be used to assemble spheroids of desired size in a cell adhesion-dependent manner (Fig. 3B). Recently, microfluidic platforms have been developed to capture cells in aqueous droplets within a carrier oil. These techniques produce spheroids of a tailored size from suspended cells by controlling the size of the droplets and the density of cells in the droplet-forming solution. An advantage of this technique over microwells is the speed of droplet generation and the ease of automation, which allows high-throughput assembly of cell aggregates (Allazetta and Lutolf, 2015; Chan et al., 2013; Tumarkin et al., 2011).
In contrast to microwells, which rely on probabilistic cell loading, recent advances in droplet-based microfluidics can achieve single-cell droplet loading and can load droplets with combinations of cell types with a precision that exceeds Poisson limitations (Collins et al., 2015; Edd et al., 2008; Schoeman et al., 2014). These devices can also be used to capture precise cell pairs, facilitating the dynamic dissection of cell-cell interactions from the initiation of contact (Dura et al., 2016). Droplet microfluidic platforms can also be used to investigate signals from the ECM. Cell-containing droplets can be fused with droplets containing diverse ECM components, or cells themselves can be directly encapsulated within microgels containing matrix components such as collagen, synthetic integrin-recognition sequences such as arginine-glycine-aspartic acid (RGD) peptides (see Glossary, Box 2) (Allazetta and Lutolf, 2015; Chan et al., 2013; Ma et al., 2013) or tunable alginate gels (Khavari et al., 2016; Mao et al., 2016).
In organoid cultures, microfluidic devices can also be used to deliver soluble signals that mimic in vivo signaling gradients (Attayek et al., 2016; Ellison et al., 2016). Based on these features, microfluidic platforms might be particularly well suited for large-scale combinatorial screens to identify cell and matrix components that comprise the stem cell niche. Future advances in microfluidics techniques will provide automated, high-throughput interrogation of stem-cell niche components, specifically how different ratios of cell types, specific cell-cell interactions, cell-ECM interactions and ECM mechanical properties direct stem cell maintenance and differentiation.
3D bioprinting enables increased spatial control and complexity
Although both microwell and microfluidics approaches can be used to produce organoids with controlled numbers and proportions of purified cell populations, these methods provide relatively little spatial control over the organization of biomaterials in the extracellular environment. The rapidly developing field of 3D bioprinting provides a potential solution to this limitation. In this suite of techniques (reviewed extensively by Gudapati et al., 2016; Murphy and Atala, 2014; Truby and Lewis, 2016) biomaterials including cell and ECM components are deposited in layers to rapidly pattern multicomponent objects with defined x, y and z coordinates (Fig. 3C). Newer approaches allow even more complex structures to be generated in fluidizing granular baths that can support intricate 3D structures during printing, which are then gently melted away prior to culture (Bhattacharjee et al., 2015; Hinton et al., 2015). These technologies provide control over the initial positions of different biomaterials and, to a more limited extent, cell subtypes within the resolution allowed by the print head and stage. Theoretically, small organoids themselves can also be used as a printable ink, allowing higher-order structures to be printed (Livoti and Morgan, 2010; Rago et al., 2009; Tejavibulya et al., 2011). These and related 3D printing platforms, although still limited in their number of applications in the study of the stem cell niche, will ultimately provide increased control over tissue architecture spanning multiple length scales – a major challenge for directing the growth of tissues and organoids.
Chemically programmed assembly: precise control of cell-cell interactions in 3D
In contrast to the systems described above, which utilize endogenous cell adhesion machinery to form condensed cellular aggregates, there are a number of techniques for aggregate formation that control adhesion synthetically (Chandra et al., 2006; Dutta et al., 2011; Hamon et al., 2011; Hsiao et al., 2009; Konagaya and Iwata, 2016; Zhao et al., 2011). For example, liposomes can be used as a delivery system to modify the cell surface by incorporating bio-orthogonal lipids that drive covalent cell-cell adhesion via click chemistry (see Glossary, Box 2). This technique has been combined with microfluidics approaches to assemble spheroids containing two cell types (O'Brien et al., 2015). An alternative approach uses DNA as a synthetic adhesion molecule to produce 3D tissues with programmable connectivity (Fig. 3D) (Chandra et al., 2006; Gartner and Bertozzi, 2009; Hsiao et al., 2009; Selden et al., 2012; Teramura et al., 2010; Weber et al., 2014). In a recent advance, DNA-labeled cells were assembled onto a complementarily labeled glass surface that functioned as a spatial template, and the resulting aggregates were subsequently embedded into a 3D matrix to create tissues at single-cell resolution with defined size, shape, composition and initial spatial organization (Todhunter et al., 2015). Using DNA-programmed assembly of cells (DPAC), it is also possible to incorporate components of the mesenchyme, such as fibroblasts, allowing the engineering of a stem cell niche that captures stromal contributions. These techniques allow precise control over individual cell-cell interactions and will enable direct examination and manipulation of juxtacrine cell-cell and cell-ECM cues within the stem cell niche.
Although engineering techniques provide the possibility for increased control over organoid culture, they also come with potential pitfalls. Prior to using any of these engineering techniques, it is necessary to process tissue samples into single cells and purify the desired cell populations away from undesired cellular components. Therefore, there is a time lag before cells are placed in culture to become organoids. During this lag time, microenvironmental cues that may be necessary for appropriate localization and function within a tissue are absent. For example, it has been observed that trypsinization can cleave integral surface proteins, potentially perturbing cell function, at least temporarily (Huang et al., 2010). Furthermore, in certain contexts cells display broader differentiation potential in vitro or in transplantation models that might not be relevant for undamaged tissues operating in a more typical homeostatic regime (Rios et al., 2014; Van Keymeulen et al., 2011; Wuidart et al., 2016). Given these considerations, researchers must be vigilant of potential artifacts introduced by processing steps in any of these techniques, and findings will likely need to be validated in intact tissue or in vivo animal models when possible.
Engineered ECMs provide control over mechanical and biochemical properties of the stem cell niche
The ECM is a key component of organoid culture that, across multiple tissues, supports phenotypes not seen in 2D culture on plastic. The most commonly used ECMs are derived from animal sources. For example, Matrigel is produced from Engelbreth-Holm-Swarm tumors grown in mice. It is primarily composed of laminin, collagen IV and entactin, but also contains a poorly defined cocktail of growth factors and trace amounts of hundreds of other proteins (Hughes et al., 2010; Kleinman and Martin, 2005). Similarly, other matrices such as collagen I are typically purified from animal sources or cultured cells. Because these matrices can be heterogeneous and poorly defined, lot-to-lot variability can affect experimental reproducibility (Hughes et al., 2010). Therefore, there is significant interest in synthesizing well-defined ECM mimics that support similar 3D growth and developmental phenotypes to those observed for Matrigel or other purified ECMs (Fig. 3E). These rationally designed matrices will significantly enhance reproducibility and enable new experimental approaches. Most notably, engineered matrices will allow the systematic and independent perturbation of ECM properties such as stiffness, viscosity, porosity, protease cleavage sites, ligand type and ligand density (Magin et al., 2016). Manipulation of these ECM properties will allow researchers to identify how mechanical and biochemical signals interact at the stem cell niche to control self-renewal and differentiation.
To address this need, many labs have used biochemically inert crosslinked hydrogels such as polyethylene glycol (PEG) or alginate to encapsulate cells in 3D (Magin et al., 2016; Seliktar, 2012). These hydrogels can be tuned over a wide range of stiffnesses and topologies by varying monomer concentration, molecular weight and degree of crosslinking. Moreover, these hydrogels provide an inert starting material, allowing specific concentrations and combinations of bioactive molecules, such as integrin-recognition sequences, to be rationally engineered back into the system (Zhu, 2010). For example, Enemchukwu and colleagues designed a PEG-based ECM mimetic with independent, tunable control over matrix stiffness, RGD ligand density and proteolytic degradation rate (Enemchukwu et al., 2015).
Synthetic matrices are very tractable systems for interrogating the spatiotemporal dynamics of cell-matrix interactions. For example, hydrogels can be designed with different rates of stress relaxation over time (Darnell et al., 2016). Photodegradable (Kloxin et al., 2009) or photoactivatable (Mosiewicz et al., 2014; Sunyer et al., 2012) crosslinkers can be used to pattern matrix stiffness within a gel or change stiffness over time. By coupling bioactive ligands to the hydrogel with a photodegradable linker, similar strategies can be used to manipulate the biochemical properties of the matrix in space and time (Kloxin et al., 2009). This is important, as work from several labs has suggested that the mechanical properties of the stem cell niche can control self-renewal and differentiation. Engineered matrices have been essential for isolating the effects of mechanical cues on stem cell activity, independent of biochemical signals. For example, mesenchymal stem cells can be driven towards osteogenic lineages via contact with a stiff substrate (Engler et al., 2006), and experiments using collagen-coupled hydrogels demonstrated that this is independent of collagen density (Wen et al., 2014).
Perhaps the greatest challenge for efforts to engineer ECM has been identifying a material capable of supporting the spectrum of cell behaviors necessary for single stem cells to survive, divide, differentiate, and ultimately self-organize into organoid-like structures. While many efforts have optimized the properties of synthetic ECM towards supporting single-cell behaviors (Chaudhuri et al., 2016; Enemchukwu et al., 2015; Green and Elisseeff, 2016), fewer reports have incorporated the full spectrum of chemical and physical properties necessary to support organoid growth and differentiation from stem cells. In a recent landmark study, Gjorevski and colleagues used a minimal, molecularly defined synthetic matrix to define the ECM properties that drive different stages of organoid formation from intestinal stem cells (Gjorevski et al., 2016). The authors systematically modulated matrix stiffness and degradation kinetics, while simultaneously grafting bioactive molecules onto PEG hydrogels to measure the individual effects of these proteins on intestinal stem cell survival, proliferation and self-organization. Interestingly, they found that the requisite matrix stiffness, degradation kinetics and ligand composition changed with development stage. Stem cell self-renewal and expansion required a stiff matrix modified with RGD peptides, but stem cell differentiation and organoid formation required a softer matrix containing laminin-111. To satisfy these requirements and design a matrix that supports all stages of organoid formation, the authors synthesized a laminin- and RGD-containing matrix that dynamically softens over time via the inclusion of hydrolytically degradable PEG monomers.
For future studies, it is exciting to consider how the various approaches described above could be combined. For example, tunable multicomponent matrices combined with high-throughput microfluidic platforms would allow rapid screening of key mechanical and biochemical properties within the stem cell niche. Similarly, engineered ECMs combined with bioprinting or chemically programmed assembly techniques could be used to produce sophisticated custom matrices with defined 3D architecture that more closely mimic stem cell niches and the dynamic changes that occur during different developmental stages, aging and disease.
Conclusions
The use of organoid culture has helped to identify important niche components and has led to a deeper understanding of how the niche controls stem cell activity. The ability to manipulate organoid formation in vitro has enabled careful dissection of niche requirements, as demonstrated by reaggregation experiments that identified Paneth cells as a key member of the intestinal stem cell niche (Sato et al., 2011). Such experiments in other tissues will be useful for identifying the cell types that comprise the ʻminimal niche', that is, the components that are necessary and sufficient for stem cell maintenance and tissue self-renewal. In addition to identifying feedback pathways from daughter to parent cells, organoids can also help to identify feedforward pathways between parent and daughter cells that help direct and maintain differentiation, as discussed in the case of Notch signaling in airway basal stem cells (Pardo-Saganta et al., 2015; Rock et al., 2011). Controlled manipulation of organoid composition, including the numbers of stem cells and specific numbers and types of differentiated daughter cells, will allow interrogation of the feedforward and feedback loops that control cell fate in other tissues.
Emerging methods in organoid engineering will accelerate progress in our understanding of the stem cell niche by providing powerful means of interrogating organoids at the single cell level, as well as a means to guide the morphogenesis of organoids with significantly greater precision. This is crucial if we are to be able to use organoids in any meaningful way to address fundamental questions in developmental and regenerative biology (Huch et al., 2017). These emerging technologies will facilitate studies that aim to address key concepts in stem cell biology, such as the influence of the niche on cell plasticity, as well as how the physical properties of the stem cell niche direct stem cell self-organization and, in turn, stem cell fate. For example, progenitors that are restricted to the enterocyte lineage during normal intestinal homeostasis can dedifferentiate to become stem cells and repopulate the crypt following damage (Tetteh et al., 2016). Similarly, in the prostate both luminal and basal cells can produce in vitro organoids containing both lineages, but only basal cells give rise to all lineages when transplanted in vivo (Goldstein et al., 2008; Karthaus et al., 2014). Together, these data clearly demonstrate that cell plasticity is context dependent. In vitro lineage tracing of single cells in organoids of precisely defined composition combined with in vivo models will help define the principles that govern dedifferentiation or interconversion between different cell identities and the long-term maintenance of tissue architecture and composition. As such, we predict that new engineering approaches will allow researchers to address fundamental questions that have been difficult to answer with less well-controlled organoid systems, or with in vivo lineage tracing experiments.
One intriguing possibility is that the mechanical forces that drive self-organization play a key role in directing cell fate, as stem cell localization is often highly stereotyped within a tissue. In the intestine, Lgr5+ stem cells and supporting Paneth cells are located at the crypt base, and progenitors become committed to secretory or enterocyte lineages as they migrate away from the base. This same architecture is observed in intestinal organoids, where stem cell progeny self-organize into a crypt-like morphology containing Lgr5+ stem cells and Paneth cells, with a transitional zone containing transit amplifying cells and regions of mature secretory and absorptive cells (Sato et al., 2009, 2011). In the mammary gland, stem cells are enriched in the myoepithelial subpopulation and are thought to reside in a basal or suprabasal location (Lim et al., 2009; Smith, 2003). Interestingly, basal cells normally divide parallel to the basement membrane, but deletion of β1-integrin from the basal compartment leads to random orientation of the mitotic spindle, an increased number of basal cell progeny located in the luminal compartment, and a loss of stem cells (Taddei et al., 2008). Together, these results suggest that specific localization within the niche is important for stem cell maintenance. In vitro experiments using engineered organoids will allow direct measurement of the physical properties of individual cell types that comprise the niche, along with transcriptional and proteomic identification of key molecules that drive these properties, to enable systematic perturbation of niche organization. Furthermore, chemically programmed assembly methods can be used to directly manipulate cell positioning within the niche, and engineered ECMs can be used to manipulate mechanical, chemical and adhesive properties of the niche. These techniques will play a crucial role in determining how mechanical cues are coordinated with spatially confined chemical cues to control stem cell activity and tissue homeostasis.
In this Review, we have described how organoid systems allow systematic and in-depth interrogation of the stem cell niche, and how state-of-the-art approaches for building synthetic niches will enable more direct quantification, modeling and testing of the processes that control stem cell activity. As a powerful complement to in vivo animal models, these current and emerging technologies will provide crucial insight into human tissue homeostasis and disease that could not be studied by other means.
Acknowledgements
There are many excellent recent studies in organoid and ECM engineering that we were unable to cover due to space constraints. We apologize to those authors whose work we have omitted. We also thank the reviewers for their helpful comments.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
Research in our laboratories is funded by the Damon Runyon Cancer Research Foundation (DRG-2239-15); Congressionally Directed Medical Research Programs (W81XWH-10-1-1-1023, W81XWH-13-1-0221); National Institutes of Health (DP2 HD080351-01); and National Science Foundation (DBI-1548297, MCB-1330864). Deposited in PMC for release after 12 months.
References
- Allazetta S. and Lutolf M. P. (2015). Stem cell niche engineering through droplet microfluidics. Curr. Opin. Biotechnol. 35, 86-93. 10.1016/j.copbio.2015.05.003 [DOI] [PubMed] [Google Scholar]
- Attayek P. J., Ahmad A. A., Wang Y., Williamson I., Sims C. E., Magness S. T. and Allbritton N. L. (2016). In vitro polarization of colonoids to create an intestinal stem cell compartment. PLoS ONE 11, e0153795-e23 10.1371/journal.pone.0153795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernfield M. R. and Fell P. E. (1967). Separation of the proliferating and differentiating cell populations of cultured embryonic pancreatic epithelium. Proc. Natl. Acad. Sci. USA 58, 2227-2234. 10.1073/pnas.58.6.2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee T., Zehnder S. M., Rowe K. G., Jain S., Nixon R. M., Sawyer W. G. and Angelini T. E. (2015). Writing in the granular gel medium. Sci. Adv. 1, e1500655-e1500655 10.1126/sciadv.1500655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth B. W., Mack D. L., Androutsellis-Theotokis A., McKay R. D. G., Boulanger C. A. and Smith G. H. (2008). The mammary microenvironment alters the differentiation repertoire of neural stem cells. Proc. Natl. Acad. Sci. USA 105, 14891-14896. 10.1073/pnas.0803214105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C., Yu Q. C., Jiang W., Liu W., Song W., Yu H., Zhang L., Yang Y. and Zeng Y. A. (2014). R-spondin1 is a novel hormone mediator for mammary stem cell self-renewal. Genes Dev. 28, 2205-2218. 10.1101/gad.245142.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp J. G., Badsha F., Florio M., Kanton S., Gerber T., Wilsch-Bräuninger M., Lewitus E., Sykes A., Hevers W., Lancaster M. et al. (2015). Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. USA 112, 15672-15677. 10.1073/pnas.1520760112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campàs O., Mammoto T., Hasso S., Sperling R. A., O'Connell D., Bischof A. G., Maas R., Weitz D. A., Mahadevan L. and Ingber D. E. (2014). Quantifying cell-generated mechanical forces within living embryonic tissues. Nat. Methods 11, 183-189. 10.1038/nmeth.2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerchiari A. E., Garbe J. C., Jee N. Y., Todhunter M. E., Broaders K. E., Peehl D. M., Desai T. A., LaBarge M. A., Thomson M. and Gartner Z. J. (2015a). A strategy for tissue self-organization that is robust to cellular heterogeneity and plasticity. Proc. Natl. Acad. Sci. USA 112, 2287-2292. 10.1073/pnas.1410776112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerchiari A., Garbe J. C., Todhunter M. E., Jee N. Y., Pinney J. R., LaBarge M. A., Desai T. A. and Gartner Z. J. (2015b). Formation of spatially and geometrically controlled three-dimensional tissues in soft gels by sacrificial micromolding. Tissue Eng. Part C Methods 21, 541-547. 10.1089/ten.tec.2014.0450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan H. F., Zhang Y., Ho Y.-P., Chiu Y.-L., Jung Y. and Leong K. W. (2013). Rapid formation of multicellular spheroids in double-emulsion droplets with controllable microenvironment. Sci. Rep. 3, 3462 10.1038/srep03462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra R. A., Douglas E. S., Mathies R. A., Bertozzi C. R. and Francis M. B. (2006). Programmable cell adhesion encoded by DNA hybridization. Angew. Chem. Int. Ed. Engl. 45, 896-901. 10.1002/anie.200502421 [DOI] [PubMed] [Google Scholar]
- Chanson L., Brownfield D., Garbe J. C., Kuhn I., Stampfer M. R., Bissell M. J. and LaBarge M. A. (2011). Self-organization is a dynamic and lineage-intrinsic property of mammary epithelial cells. Proc. Natl. Acad. Sci. USA 108, 3264-3269. 10.1073/pnas.1019556108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri O., Gu L., Klumpers D., Darnell M., Bencherif S. A., Weaver J. C., Huebsch N., Lee H.-P., Lippens E., Duda G. N. et al. (2016). Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat. Mater. 15, 326-334. 10.1038/nmat4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. Y., Chung B. G., Lee D. H., Khademhosseini A., Kim J.-H. and Lee S.-H. (2010). Controlled-size embryoid body formation in concave microwell arrays. Biomaterials 31, 4296-4303. 10.1016/j.biomaterials.2010.01.115 [DOI] [PubMed] [Google Scholar]
- Clevers H. (2016). Modeling development and disease with organoids. Cell 165, 1586-1597. 10.1016/j.cell.2016.05.082 [DOI] [PubMed] [Google Scholar]
- Clevers H., Loh K. M. and Nusse R. (2014). An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 346, 1248012-1248012 10.1126/science.1248012 [DOI] [PubMed] [Google Scholar]
- Collins D. J., Neild A., deMello A., Liu A.-Q. and Ai Y. (2015). The Poisson distribution and beyond: methods for microfluidic droplet production and single cell encapsulation. Lab. Chip 15, 3439-3459. 10.1039/C5LC00614G [DOI] [PubMed] [Google Scholar]
- Danahay H., Pessotti A. D., Coote J., Montgomery B. E., Xia D., Wilson A., Yang H., Wang Z., Bevan L., Thomas C. et al. (2015). Notch2 is required for inflammatory cytokine- driven goblet cell metaplasia in the lung. Cell Rep. 10, 239-252. 10.1016/j.celrep.2014.12.017 [DOI] [PubMed] [Google Scholar]
- Darnell M., Young S., Gu L., Shah N., Lippens E., Weaver J., Duda G. and Mooney D. (2016). Substrate stress-relaxation regulates scaffold remodeling and bone formation in vivo. Adv. Healthcare Mater., doi: 10.1002/adhm.201601185 10.1002/adhm.201601185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Discher D., Mooney B. and Zandstra P. (2009). Growth factors, matrices, and forces combine and control stem cells. Science 324, 1673-1677. 10.1126/science.1171643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dura B., Servos M. M., Barry R. M., Ploegh H. L., Dougan S. K. and Voldman J. (2016). Longitudinal multiparameter assay of lymphocyte interactions from onset by microfluidic cell pairing and culture. Proc. Natl. Acad. Sci. USA 113, E3599-E3608. 10.1073/pnas.1515364113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D., Pulsipher A., Luo W. and Yousaf M. N. (2011). Synthetic chemoselective rewiring of cell surfaces: generation of three-dimensional tissue structures. J. Am. Chem. Soc. 133, 8704-8713. 10.1021/ja2022569 [DOI] [PubMed] [Google Scholar]
- Edd J. F., Di Carlo D., Humphry K. J., Köster S., Irimia D., Weitz D. A. and Toner M. (2008). Controlled encapsulation of single-cells into monodisperse picolitre drops. Lab. Chip 8, 1262-1264. 10.1039/b805456h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison D., Mugler A., Brennan M. D., Lee S. H., Huebner R. J., Shamir E. R., Woo L. A., Kim J., Amar P., Nemenman I. et al. (2016). Cell-cell communication enhances the capacity of cell ensembles to sense shallow gradients during morphogenesis. Proc. Natl. Acad. Sci. USA 113, E679-E688. 10.1073/pnas.1516503113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enemchukwu N. O., Cruz-Acuña R., Bongiorno T., Johnson C. T., García J. R., Sulchek T. and García A. J. (2016). Synthetic matrices reveal contributions of ECM biophysical and biochemical properties to epithelial morphogenesis. J. Cell Biol. 212, 113-124. 10.1083/jcb.201506055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler A. J., Sen S., Sweeney H. L. and Discher D. E. (2006). Matrix elasticity directs stem cell lineage specification. Cell 126, 677-689. 10.1016/j.cell.2006.06.044 [DOI] [PubMed] [Google Scholar]
- Farin H. F., Van Es J. H. and Clevers H. (2012). Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 143, 1518-1529.e7. 10.1053/j.gastro.2012.08.031 [DOI] [PubMed] [Google Scholar]
- Farin H. F., Jordens I., Mosa M. H., Basak O., Korving J., Tauriello D. V. F., de Punder K., Angers S., Peters P. J., Maurice M. M. et al. (2016). Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature 530, 340-343. 10.1038/nature16937 [DOI] [PubMed] [Google Scholar]
- Gartner Z. J. and Bertozzi C. R. (2009). Programmed assembly of 3-dimensional microtissues with defined cellular connectivity. Proc. Natl. Acad. Sci. USA 106, 4606-4610. 10.1073/pnas.0900717106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjorevski N., Sachs N., Manfrin A., Giger S., Bragina M. E., Ordóñez-Morán P., Clevers H. and Lutolf M. P. (2016). Designer matrices for intestinal stem cell and organoid culture. Nature 539, 560-564. 10.1038/nature20168 [DOI] [PubMed] [Google Scholar]
- Goldstein A. S., Lawson D. A., Cheng D., Sun W., Garraway I. P. and Witte O. N. (2008). Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. Proc. Natl. Acad. Sci. USA 105, 20882-20887. 10.1073/pnas.0811411106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss A. M., Tian Y., Tsukiyama T., Cohen E. D., Zhou D., Lu M. M., Yamaguchi T. P. and Morrisey E. E. (2009). Wnt2/2b and β-Catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Dev. Cell 17, 290-298. 10.1016/j.devcel.2009.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracz A. D., Williamson I. A., Roche K. C., Johnston M. J., Wang F., Wang Y., Attayek P. J., Balowski J., Liu X. F., Laurenza R. J. et al. (2015). A high-throughput platform for stem cell niche co-cultures and downstream gene expression analysis. Nat. Cell Biol. 17, 340-349. 10.1038/ncb3104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J. J. and Elisseeff J. H. (2016). Mimicking biological functionality with polymers for biomedical applications. Nature 540, 386-394. 10.1038/nature21005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudapati H., Dey M. and Ozbolat I. (2016). A comprehensive review on droplet-based bioprinting: past, present and future. Biomaterials 102, 20-42. 10.1016/j.biomaterials.2016.06.012 [DOI] [PubMed] [Google Scholar]
- Hamon M., Ozawa T., Montagne K., Kojima N., Ishii R., Yamaguchi S., Nagamune T., Ushida T. and Sakai Y. (2011). Avidin-biotin-based approach to forming heterotypic cell clusters and cell sheets on a gas-permeable membrane. Biofabrication 3, 034111 10.1088/1758-5082/3/3/034111 [DOI] [PubMed] [Google Scholar]
- Hinton T. J., Jallerat Q., Palchesko R. N., Park J. H., Grodzicki M. S., Shue H.-J., Ramadan M. H., Hudson A. R. and Feinberg A. W. (2015). Three-dimensional printing of complex biological structures by freeform reversible embedding of suspended hydrogels. Sci. Adv. 1, e1500758-e1500758 10.1126/sciadv.1500758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao S. C., Shum B. J., Onoe H., Douglas E. S., Gartner Z. J., Mathies R. A., Bertozzi C. R. and Francis M. B. (2009). Direct cell surface modification with DNA for the capture of primary cells and the investigation of myotube formation on defined patterns. Langmuir 25, 6985-6991. 10.1021/la900150n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.-L., Hsing H.-W., Lai T.-C., Chen Y.-W., Lee T.-R., Chan H.-T., Lyu P.-C., Wu C.-L., Lu Y.-C., Lin S.-T. et al. (2010). Trypsin-induced proteome alteration during cell subculture in mammalian cells. J. Biomed. Sci. 17, 36 10.1186/1423-0127-17-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M., Knoblich J. A., Lutolf M. P. and Martinez-Arias A. (2017). The hope and the hype of organoid research. Development 144, 938-941. [DOI] [PubMed] [Google Scholar]
- Hughes C. S., Postovit L. M. and Lajoie G. A. (2010). Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics 10, 1886-1890. 10.1002/pmic.200900758 [DOI] [PubMed] [Google Scholar]
- Joshi P. A., Jackson H. W., Beristain A. G., Di Grappa M. A., Mote P. A., Clarke C. L., Stingl J., Waterhouse P. D. and Khokha R. (2010). Progesterone induces adult mammary stem cell expansion. Nature 465, 803-807. 10.1038/nature09091 [DOI] [PubMed] [Google Scholar]
- Jung P., Sato T., Merlos-Suárez A., Barriga F. M., Iglesias M., Rossell D., Auer H., Gallardo M., Blasco M. A., Sancho E. et al. (2011). Isolation and in vitro expansion of human colonic stem cells. Nat. Med. 17, 1225-1227. 10.1038/nm.2470 [DOI] [PubMed] [Google Scholar]
- Karp J. M., Yeh J., Eng G., Fukuda J., Blumling J., Suh K.-Y., Cheng J., Mahdavi A., Borenstein J., Langer R. et al. (2007). Controlling size, shape and homogeneity of embryoid bodies using poly(ethylene glycol) microwells. Lab. Chip 7, 786-794. 10.1039/b705085m [DOI] [PubMed] [Google Scholar]
- Karthaus W. R., Iaquinta P. J., Drost J., Gracanin A., van Boxtel R., Wongvipat J., Dowling C. M., Gao D., Begthel H., Sachs N. et al. (2014). Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 159, 163-175. 10.1016/j.cell.2014.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavari A., Nydén M., Weitz D. A. and Ehrlicher A. J. (2016). Composite alginate gels for tunable cellular microenvironment mechanics. Sci. Rep. 6, 30854 10.1038/srep30854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.-A., Kakitani M., Zhao J., Oshima T., Tang T., Binnerts M., Liu Y., Boyle B., Park E., Emtage P. et al. (2005). Mitogenic influence of human R-Spondin1 on the intestinal epithelium. Science 309, 1256-1259. 10.1126/science.1112521 [DOI] [PubMed] [Google Scholar]
- Kleinman H. K. and Martin G. R. (2005). Matrigel: basement membrane matrix with biological activity. Semin. Cancer Biol. 15, 378-386. 10.1016/j.semcancer.2005.05.004 [DOI] [PubMed] [Google Scholar]
- Kloxin A. M., Kasko A. M., Salinas C. N. and Anseth K. S. (2009). Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science 324, 59-63. 10.1126/science.1169494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konagaya S. and Iwata H. (2016). Reproducible preparation of spheroids of pancreatic hormone positive cells from human iPS cells: an in vitro study. Biochim. Biophys. Acta 1860, 2008-2016. 10.1016/j.bbagen.2016.05.012 [DOI] [PubMed] [Google Scholar]
- Korinek V., Barker N., Moerer P., van Donselaar E., Huls G., Peters P. J. and Clevers H. (1998). Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 19, 379-383. 10.1038/1270 [DOI] [PubMed] [Google Scholar]
- Kuhnert F., Davis C. R., Wang H.-T., Chu P., Lee M., Yuan J., Nusse R. and Kuo C. J. (2004). Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc. Natl. Acad. Sci. USA 101, 266-271. 10.1073/pnas.2536800100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M. A. and Knoblich J. A. (2014). Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345, 1247125-1247125 10.1126/science.1247125 [DOI] [PubMed] [Google Scholar]
- Lim E., Vaillant F., Wu D., Forrest N. C., Pal B., Hart A. H., Asselin-Labat M.-L., Gyorki D. E., Ward T., Partanen A. et al. (2009). Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 15, 907-913. 10.1038/nm.2000 [DOI] [PubMed] [Google Scholar]
- Livoti C. M. and Morgan J. R. (2010). Self-assembly and tissue fusion of toroid-shaped minimal building units. Tissue Eng. Part A 16, 2051-2061. 10.1089/ten.tea.2009.0607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Natoli M., Liu X., Neubauer M. P., Watt F. M., Fery A. and Huck W. T. S. (2013). Monodisperse collagen–gelatin beads as potential platforms for 3D cell culturing. J. Mater. Chem. B 1, 5128-5136. 10.1039/c3tb20851f [DOI] [PubMed] [Google Scholar]
- Magin C. M., Alge D. L. and Anseth K. S. (2016). Bio-inspired 3D microenvironments: a new dimension in tissue engineering. Biomed. Mater. 11, 022001 10.1088/1748-6041/11/2/022001 [DOI] [PubMed] [Google Scholar]
- Mao A. S., Shin J.-W., Utech S., Wang H., Uzun O., Li W., Cooper M., Hu Y., Zhang L., Weitz D. A. et al. (2016). Deterministic encapsulation of single cells in thin tunable microgels for niche modelling and therapeutic delivery. Nat. Mater. 16, 236-243. 10.1038/nmat4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosiewicz K. A., Kolb L., van der Vlies A. J. and Lutolf M. P. (2014). Microscale patterning of hydrogel stiffness through light-triggered uncaging of thiols. Biomater. Sci. 2, 1640-1651. 10.1039/C4BM00262H [DOI] [PubMed] [Google Scholar]
- Murphy S. V. and Atala A. (2014). 3D bioprinting of tissues and organs. Nat. Biotechnol. 32, 773-785. 10.1038/nbt.2958 [DOI] [PubMed] [Google Scholar]
- Napolitano A. P., Dean D. M., Man A. J., Youssef J., Ho D. N., Rago A. P., Lech M. P. and Morgan J. R. (2007). Scaffold-free three-dimensional cell culture utilizing micromolded nonadhesive hydrogels. Biotechniques 43, 494-500. 10.2144/000112591 [DOI] [PubMed] [Google Scholar]
- Nelson C. M., Vanduijn M. M., Inman J. L., Fletcher D. A. and Bissell M. J. (2006). Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures. Science 314, 298-300. 10.1126/science.1131000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien P. J., Luo W., Rogozhnikov D., Chen J. and Yousaf M. N. (2015). Spheroid and tissue assembly via click chemistry in microfluidic flow. Bioconjug. Chem. 26, 1939-1949. 10.1021/acs.bioconjchem.5b00376 [DOI] [PubMed] [Google Scholar]
- Pardo-Saganta A., Tata P. R., Law B. M., Saez B., Chow R. D.-W., Prabhu M., Gridley T. and Rajagopal J. (2015). Parent stem cells can serve as niches for their daughter cells. Nature 523, 597-601. 10.1038/nature14553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Gregorieff A., Begthel H. and Clevers H. (2003). Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 17, 1709-1713. 10.1101/gad.267103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rago A. P., Chai P. R. and Morgan J. R. (2009). Encapsulated arrays of self-assembled microtissues: an alternative to spherical microcapsules. Tissue Eng. Part A 15, 387-395. 10.1089/ten.tea.2008.0107 [DOI] [PubMed] [Google Scholar]
- Rios A. C., Fu N. Y., Lindeman G. J. and Visvader J. E. (2014). In situ identification of bipotent stem cells in the mammary gland. Nature 506, 322-327. 10.1038/nature12948 [DOI] [PubMed] [Google Scholar]
- Rock J. R., Gao X., Xue Y., Randell S. H., Kong Y.-Y. and Hogan B. L. M. (2011). Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell 8, 639-648. 10.1016/j.stem.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock J. R., Onaitis M. W., Rawlins E. L., Lu Y., Clark C. P., Xue Y., Randell S. H. and Hogan B. L. M. (2009). Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. USA 106, 12771-12775. 10.1073/pnas.0906850106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runswick S. K., O'Hare M. J., Jones L., Streuli C. H. and Garrod D. R. (2001). Desmosomal adhesion regulates epithelial morphogenesis and cell positioning. Nat. Cell Biol. 3, 823-830. 10.1038/ncb0901-823 [DOI] [PubMed] [Google Scholar]
- Sasai Y. (2013). Cytosystems dynamics in self-organization of tissue architecture. Nature 493, 318-326. 10.1038/nature11859 [DOI] [PubMed] [Google Scholar]
- Sato T., Vries R. G., Snippert H. J., van de Wetering M., Barker N., Stange D. E., van Es J. H., Abo A., Kujala P., Peters P. J. et al. (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262-265. 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- Sato T., van Es J. H., Snippert H. J., Stange D. E., Vries R. G., van den Born M., Barker N., Shroyer N. F., van de Wetering M. and Clevers H. (2011). Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415-418. 10.1038/nature09637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden D. T. (2014). Nice neighborhood: emerging concepts of the stem cell niche. Cell 157, 41-50. 10.1016/j.cell.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman R. M., Kemna E. W. M., Wolbers F. and van den Berg A. (2014). High-throughput deterministic single-cell encapsulation and droplet pairing, fusion, and shrinkage in a single microfluidic device. Electrophoresis 35, 385-392. 10.1002/elps.201300179 [DOI] [PubMed] [Google Scholar]
- Selden N. S., Todhunter M. E., Jee N. Y., Liu J. S., Broaders K. E. and Gartner Z. J. (2012). Chemically programmed cell adhesion with membrane-anchored oligonucleotides. J. Am. Chem. Soc. 134, 765-768. 10.1021/ja2080949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seliktar D. (2012). Designing cell-compatible hydrogels for biomedical applications. Science 336, 1124-1128. 10.1126/science.1214804 [DOI] [PubMed] [Google Scholar]
- Simian M. and Bissell M. J. (2017). Organoids: a historical perspective of thinking in three dimensions. J. Cell Biol. 216, 31-40. 10.1083/jcb.201610056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. H. (2003). Mammary epithelial stem cells. In Stem Cells Handbook (ed. Sell S.), pp. 437-444. New Jersey: Humana Press. [Google Scholar]
- Stevens K. R., Ungrin M. D., Schwartz R. E., Ng S., Carvalho B., Christine K. S., Chaturvedi R. R., Li C. Y., Zandstra P. W., Chen C. S. et al. (2013). InVERT molding for scalable control of tissue microarchitecture. Nat. Commun. 4, 1847 10.1038/ncomms2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyer R., Jin A. J., Nossal R. and Sackett D. L. (2012). Fabrication of hydrogels with steep stiffness gradients for studying cell mechanical response. PLoS ONE 7, e46107 10.1371/journal.pone.0046107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei I., Deugnier M.-A., Faraldo M. M., Petit V., Bouvard D., Medina D., Fässler R., Thiery J. P. and Glukhova M. A. (2008). Beta1 integrin deletion from the basal compartment of the mammary epithelium affects stem cells. Nat. Cell Biol. 10, 716-722. 10.1038/ncb1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro T., Wang Y., Barak L. S., Bai Y., Randell S. H. and Hogan B. L. M. (2014). IL-6/STAT3 promotes regeneration of airway ciliated cells from basal stem cells. Proc. Natl. Acad. Sci. USA 111, E3641-E3649. 10.1073/pnas.1409781111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejavibulya N., Youssef J., Bao B., Ferruccio T.-M. and Morgan J. R. (2011). Directed self-assembly of large scaffold-free multi-cellular honeycomb structures. Biofabrication 3, 034110 10.1088/1758-5082/3/3/034110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teramura Y., Chen H., Kawamoto T. and Iwata H. (2010). Control of cell attachment through polyDNA hybridization. Biomaterials 31, 2229-2235. 10.1016/j.biomaterials.2009.11.098 [DOI] [PubMed] [Google Scholar]
- Tetteh P. W., Basak O., Farin H. F., Wiebrands K., Kretzschmar K., Begthel H., van den Born M., Korving J., de Sauvage F., van Es J. H. et al. (2016). Replacement of lost Lgr5-positive stem cells through plasticity of their enterocyte-lineage daughters. Cell Stem Cell 18, 203-213. 10.1016/j.stem.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Todhunter M. E., Jee N. Y., Hughes A. J., Coyle M. C., Cerchiari A., Farlow J., Garbe J. C., LaBarge M. A., Desai T. A. and Gartner Z. J. (2015). Programmed synthesis of three-dimensional tissues. Nat. Methods 12, 975-981. 10.1038/nmeth.3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townes P. L. and Holtfreter J. (1955). Directed movements and selective adhesion of embryonic amphibian cells. J. Exp. Zool. 128, 53-120. 10.1002/jez.1401280105 [DOI] [PubMed] [Google Scholar]
- Truby R. L. and Lewis J. A. (2016). Printing soft matter in three dimensions. Nature 540, 371-378. 10.1038/nature21003 [DOI] [PubMed] [Google Scholar]
- Tumarkin E., Tzadu L., Csaszar E., Seo M., Zhang H., Lee A., Peerani R., Purpura K., Zandstra P. W. and Kumacheva E. (2011). High-throughput combinatorial cell co-culture using microfluidics. Integr. Biol. 3, 653-662. 10.1039/c1ib00002k [DOI] [PubMed] [Google Scholar]
- Ungrin M. D., Joshi C., Nica A., Bauwens C. and Zandstra P. W. (2008). Reproducible, ultra high-throughput formation of multicellular organization from single cell suspension-derived human embryonic stem cell aggregates. PLoS ONE 3, e1565 10.1371/journal.pone.0001565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenta T., Degirmenci B., Moor A. E., Herr P., Zimmerli D., Moor M. B., Hausmann G., Cantù C., Aguet M. and Basler K. (2016). Wnt ligands secreted by subepithelial mesenchymal cells are essential for the survival of intestinal stem cells and gut homeostasis. Cell Rep. 15, 911-918. 10.1016/j.celrep.2016.03.088 [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A., Rocha A. S., Ousset M., Beck B., Bouvencourt G., Rock J., Sharma N., Dekoninck S. and Blanpain C. (2011). Distinct stem cells contribute to mammary gland development and maintenance. Nature 479, 189-193. 10.1038/nature10573 [DOI] [PubMed] [Google Scholar]
- Wang Y., Phillips C., Xu W., Pai J.-H., Dhopeshwarkar R., Sims C. E. and Allbritton N. (2010). Micromolded arrays for separation of adherent cells. Lab. Chip 10, 2917-2924. 10.1039/c0lc00186d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber R. J., Liang S. I., Selden N. S., Desai T. A. and Gartner Z. J. (2014). Efficient targeting of fatty-acid modified oligonucleotides to live cell membranes through stepwise assembly. Biomacromolecules 15, 4621-4626. 10.1021/bm501467h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss P. and Taylor A. C. (1960). Reconstitution of complete organs from single-cell suspensions of chick embryos in advanced stages of differentiation. Proc. Natl. Acad. Sci. USA 46, 1177-1185. 10.1073/pnas.46.9.1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J. H., Vincent L. G., Fuhrmann A., Choi Y. S., Hribar K. C., Taylor-Weiner H., Chen S. and Engler A. J. (2014). Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat. Mater. 13, 979-987. 10.1038/nmat4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuidart A., Ousset M., Rulands S., Simons B. D., Van Keymeulen A. and Blanpain C. (2016). Quantitative lineage tracing strategies to resolve multipotency in tissue-specific stem cells. Genes Dev. 30, 1261-1277. 10.1101/gad.280057.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y. A. and Nusse R. (2010). Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell 6, 568-577. 10.1016/j.stem.2010.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Loh W., Droujinine I. A., Teo W., Kumar N., Schafer S., Cui C. H., Zhang L., Sarkar D., Karnik R. et al. (2011). Mimicking the inflammatory cell adhesion cascade by nucleic acid aptamer programmed cell-cell interactions. FASEB J. 25, 3045-3056. 10.1096/fj.10-178384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. (2010). Bioactive modification of poly(ethylene glycol) hydrogels for tissue engineering. Biomaterials 31, 4639-4656. 10.1016/j.biomaterials.2010.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]