ABSTRACT
Tissue, organ and organoid cultures provide suitable models for developmental studies, but our understanding of how the organs are assembled at the single-cell level still remains unclear. We describe here a novel fixed z-direction (FiZD) culture setup that permits high-resolution confocal imaging of organoids and embryonic tissues. In a FiZD culture a permeable membrane compresses the tissues onto a glass coverslip and the spacers adjust the thickness, enabling the tissue to grow for up to 12 days. Thus, the kidney rudiment and the organoids can adjust to the limited z-directional space and yet advance the process of kidney morphogenesis, enabling long-term time-lapse and high-resolution confocal imaging. As the data quality achieved was sufficient for computer-assisted cell segmentation and analysis, the method can be used for studying morphogenesis ex vivo at the level of the single constituent cells of a complex mammalian organogenesis model system.
KEY WORDS: Kidney; Organ culture; Time-lapse; Organoid, Imaging
Summary: Time-lapse confocal imaging of organoids and embryonic tissues through fixed z-direction culture allows long-term single-cell resolution live imaging of tissue growth and morphogenesis.
INTRODUCTION
High-resolution confocal imaging is an important means of studying the cellular behavioural dynamics of organ morphogenesis, a process that involves division, migration, death and changes in shape of the cells. Yet, we have a poor understanding of how these cellular events establish the shapes of the eventual organs. The rapid development of microscopic imaging (Huisken et al., 2004) and automated image analysis technologies (Eliceiri et al., 2012) has provided the basis for achieving a better understanding of these processes, as has been shown in certain model organisms (Khan et al., 2014).
In the traditionally used Trowell setup, the explant is placed on a filter supported by a metal grid (Auerbach and Grobstein, 1958; Grobstein, 1955; Kispert et al., 1998; reviewed in Rak-Raszewska et al., 2015; Saxén, 1987), and more recently, commercial inserts have also been used for this purpose (Costantini et al., 2011). For high-resolution time-lapse imaging, however, more sophisticated ways of organizing the organotypic culture experiments using an on-stage microscope incubator are required. The branching morphogenesis and patterning of the ureteric bud (UB) has been monitored using Transwell inserts (Watanabe and Costantini 2004) and the low-volume method (Lindström et al., 2014), and there have been reports on UB cell behaviour (Packard et al., 2013) and the movement of progenitor cells during UB branching morphogenesis (Riccio et al., 2016) based on the use of Transwell inserts. The same system was used to investigate the effect of β-catenin levels during nephron patterning (Lindström et al., 2015) and for tracking the movements of cap mesenchymal cells (Combes et al., 2016). In all of these applications, however, the imaging of individual cells was limited, hindering the full use of these elegant models.
We report here a novel technique for culturing embryonic kidneys and organoids in a fixed z-direction culture (FiZD) set up in a restricted space between a Transwell filter and a glass coverslip. This technology provides high-quality single-cell-resolution time-lapse imaging and permits computer-assisted analysis of the assembly of complex cellular structures.
RESULTS AND DISCUSSION
The main disadvantage of traditional culture methods for conducting time-lapse imaging is poor light microscopy image quality. The reasons for this are the long focal length, the thickness of the tissue, and the air-liquid interface or Transwell membrane between the specimen and the objective (Fig. 1A).
Fig. 1.
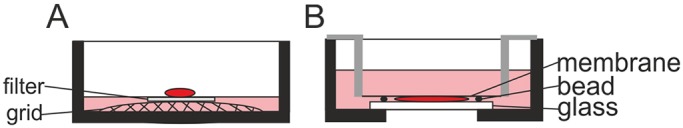
Fixed z-direction system. (A) In the Trowell system the explant is placed on a filter supported by a grid holding the explant at the air-liquid interface. (B) In FiZD culture the explant is between a glass surface and a Transwell insert. Spacer beads are used to adjust the thickness of the tissue.
This led us to consider whether restricting the ability of the sample to grow in the z-direction by use of a porous membrane (Fig. 1B) would enable better imaging quality. This proved to be optimal for supporting organogenesis and providing a high imaging quality (Fig. 2).
Fig. 2.
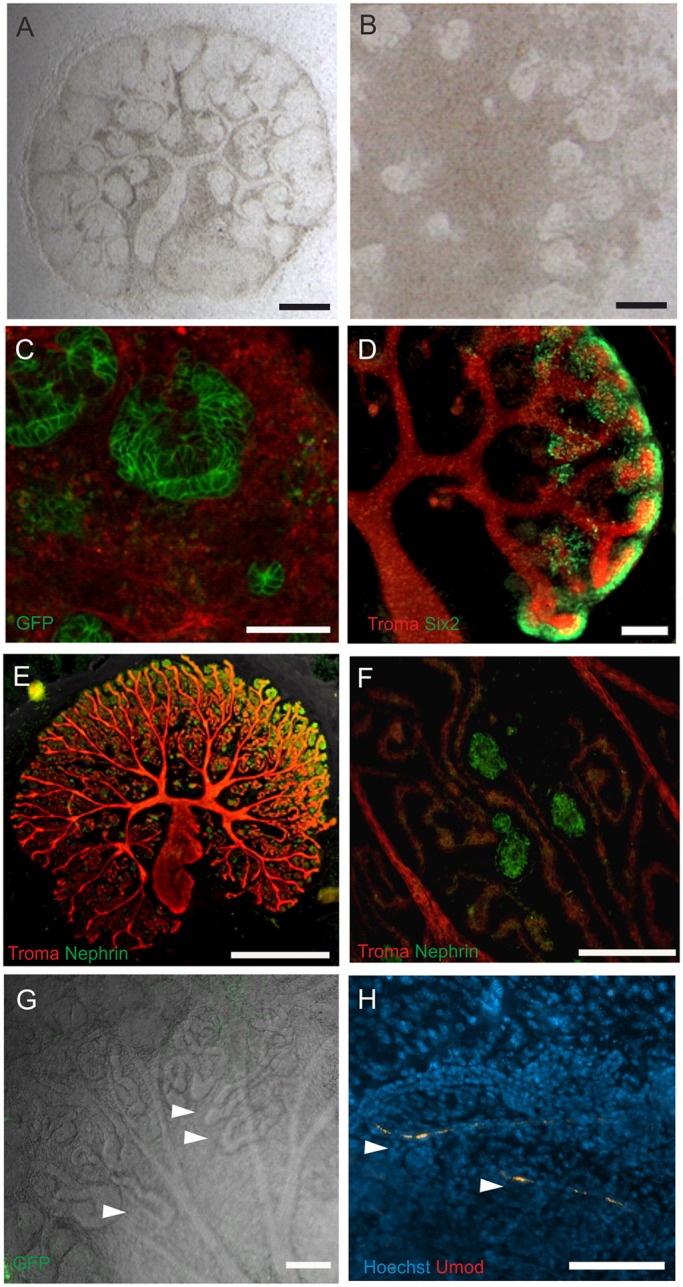
Kidney morphogenesis progresses well in FiZD culture. Embryonic kidneys (A,D-G) and organoids (B,C,H) were grown in FiZD culture. (A) Brightfield micrograph of an intact embryonic kidney, and (B) kidney organoid cultured for 7 days. (C) Snapshot of the time-lapse image stack depicting Wnt4Cre-activated GFP expression in the assembling nephrons on the fourth day of FiZD culture. (D) Six2 and Troma-I (Krt8) staining highlight nephron precursors and UB bifurcations, respectively, in a 7-day FiZD culture. (E) Kidney rudiment FiZD-cultured for 12 days. Troma-I and Nephrin staining depict the UB bifurcations and podocytes, respectively. (F) High-power magnification of the Nephrin+ podocytes and Troma-I+ UB in E. (G) Frame from the time-lapse image stack of Flk-GFP endothelial cells. Henle's loop-like structures (arrowheads). (H) Umod+ loops of Henle (arrowheads) in an organoid cultured for 7 days. Umod and Hoechst stainings. Scale bars: 100 µm in A-D,F-H; 1000 µm in E.
The z-direction was regulated by polyester beads that serve as spacers (Fig. 1B). We found empirically that a spacer of <20 µm diameter did not prevent mechanical destruction of the samples (Fig. S2A-F) and 40 µm beads gave greater variation in sample viability (Fig. S2G-L), whereas 70 µm beads provided the best conditions for both high-quality imaging and optimal culture conditions (Fig. S2M-R). These observations indicate that the diameter of the beads must be optimized for each experiment according to the size and properties of the sample.
The formation of the renal vesicle, comma-shaped and S-shape bodies, and eventually segmented nephrons with Bowman's capsule and loops of Henle were also observed in intact kidneys and organoids in FiZD culture (Fig. 2A-H; Movies 1-3,6-10).
We addressed the question of how renal vasculature structures labelled with GFP reporter emerge in the FiZD setup of intact kidney (Movie 1), which was in line with previously published data (Halt et al., 2016).
We employed mT/mG;HoxB7Cre transgenic mice to study the degree of UB development, as highlighted by the HoxB7Cre-activated GFP expression in the epithelium of the UB (Movie 2).
FiZD was also used for studying nephron development in mT/mG;Wnt4Cre organoids. The metanephric mesenchyme was induced to undergo nephrogenesis by transient exposure to a GSK-3α/β inhibitor. In such a setting the behaviour of individual GFP+ cells could be monitored in the images captured, allowing the analysis of cell movements and nephron patterning (Movie 3).
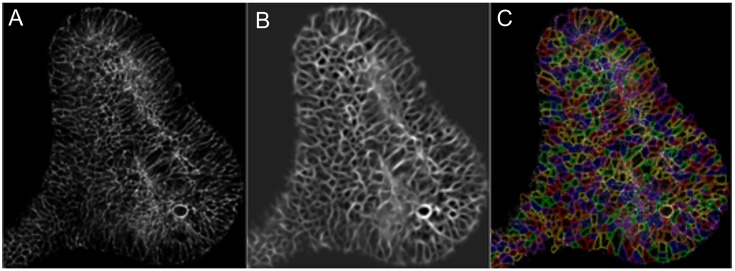
We next explored whether the data would allow computer-assisted cell segmentation. Confocal microscopic micrographs of GFP+ UB cells before (Fig. 3A) and after deconvolution (Fig. 3B) to improve the image resolution enabled cell segmentation (Fig. 3C). The GFP allowed tracking of the UB cell membranes as a result of the HoxB7Cre-mediated GFP activation (Movie 4).
Fig. 3.
FiZD culture-generated image data stacks can be subjected to computer-assisted cell segmentation. Time-lapse images were captured at 5 min intervals and processed with a program developed in Matlab. (A) Frame of the 3D image stack. (B) Ridge-enhanced image after deconvolution. (C) Cell boundaries highlighted by the cell segmentation program.
The computer-assisted image analysis enabled several key morphogenetic parameters to be examined simultaneously (Fig. 4A,B). The data derived from the FiZD served to identify the speed and direction of UB cell migration as presented in a wind rose plot (Movie 5), as used in certain systems (Stegmaier et al., 2016). By analysing segmentation data it is possible to study cellular behavioural dynamics in detail during morphogenesis, including the processes that take place during the development of a nephron.
Fig. 4.
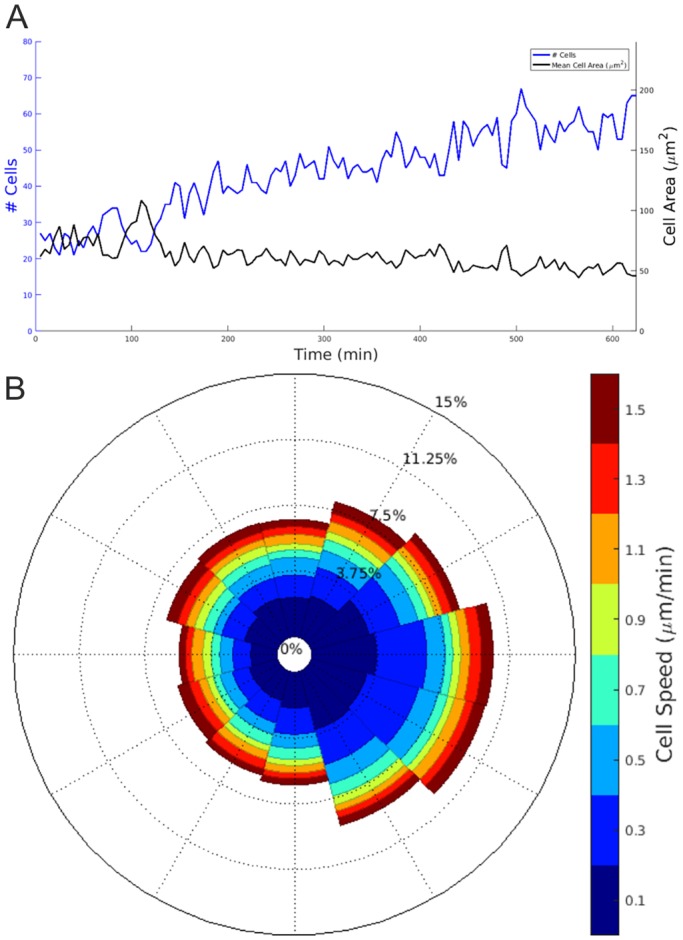
Segmented FiZD culture-based image data enable analysis of kidney morphogenetic parameters. (A) Increase in the number of UB tip cells in the right-hand side of the growing kidney (blue), and the mean cell area (black). (B) Wind rose plot illustrating the direction of UB tip cell migration. The lengths of the spokes of the wind rose plot indicate the proportions of cells moving in a given direction and the thicknesses of the colour bands within a spoke indicate their speed distribution. Both wind rose and cell count plots show that the kidney is growing towards the right, as the number of cells in the right half is increasing and more cells are moving towards the right. The data correspond to the single z-projection presented in Movie 3.
The low-volume method provided better quality images than Trowell culture (Sebinger et al., 2010) and was compared with the FiZD method. The FiZD and low-volume experiments were organized in one 6-well plate, with confocal imaging of intact kidney cultures and organoids (Fig. S3, Movies 6,7,10). To provide an objective comparison, the culture medium was changed daily in both types of culture, even though this is not required for the FiZD system. The results demonstrate the benefits of the FiZD system: better overall imaging quality, easy medium change without affecting the sample, reduced thickness of the sample and perfect stability of the image. Additionally, several samples could be assembled simultaneously in one FiZD setup with precise control of their position, whereas the low-volume method does not allow more than one sample per silicon chamber (note the fusion of what were initially two organoids in Movie 10, right panel). The FiZD method was also compared with the Transwell culture method (Movie 11). However, due to the low signal the laser power had to be doubled and this was toxic to the kidney rudiment (Fig. S4). To present the FiZD time-lapse data at the highest possible resolution, we made a short time-lapse of the developing nephrons (Movie 8), 3D structural analysis of the same developing nephron was provided by displaying all the z-layers at one point in time (Movie 9).
The capacity of the kidney to develop under FiZD culture conditions could be attributed to the conditions under which this occurs during normal nephrogenesis, when the nephron takes shape during its assembly. It has been shown that the viscoelasticity of the tissue, and thus also its rigidity, depends on the composition and crosslinking of the extracellular matrix components and their binding to cells (Forgacs et al., 1998; Phillips and Steinberg, 1978).
As judged by several organogenesis indicators presented here, kidney morphogenesis advances well under FiZD conditions, and at the same time the FiZD setup provides a superior capacity for image quality due to the reduction in tissue thickness. Here, multiple tissue samples can be cultured simultaneously and organs can be fixed in defined positions prior to culture. The culture medium can be changed or supplemented with given factors in a manner that does not disturb the development of the tissue or its capacity to retain its initial position. Moreover, the relatively large volume of the wells and lower laser power enables long-term imaging. Most importantly, the FiZD is well suited for microscope stage incubation system culture and provides an excellent platform for high-resolution confocal imaging, automatic cell segmentation, tracking and image quantification that would also be suitable for use with other organotypic cultures (Prunskaite-Hyyryläinen et al., 2016) and organoids.
MATERIALS AND METHODS
Mouse models and dissection of kidneys
The embryonic kidneys were dissected from wild-type CD-1 embryos or crosses between Tie1Cre (Gustafsson et al., 2001), Hoxb7Cre (Yu et al., 2002), Wnt4Cre (Shan et al., 2010), Flk1-GFP (Licht et al., 2004), GFP (Hadjantonakis et al., 1998) or tomato floxed Rosa26 Green fluorescent protein (GFP) (mT/mG) reporter mice (Muzumdar et al., 2007), as described by Junttila et al. (2015). Animal care and procedures were in accordance with Finnish national legislation for the use of laboratory animals, the European convention for the protection of vertebrate animals (ETS 123) and EU Directive 86/609/EEC.
Embryonic kidney organoids
An organoid can be defined as an in vitro 3D aggregate derived from primary tissue (Auerbach and Grobstein, 1958; Junttila et al., 2015; Unbekandt and Davies, 2010; Vainio et al., 1992), embryonic stem cells (Eiraku et al., 2008) or induced pluripotent stem cells (iPSCs) (Morizane et al., 2015; Takasato et al., 2015), which are capable of self-renewal and self-organization, and exhibit similar organ functionalities as the tissue of origin (Fatehullah et al., 2016). In our experiments we used organoids derived from primary cell cultures isolated from embryonic metanephric mesenchyme, as described by Junttila et al. (2015), except that 10 µM BIO (6-bromoindirubin-3′-oxime, Sigma) was used for induction.
Embryonic kidney organoids derived from frozen primary cells
The organoids in the experiments presented in Fig. 2H and Movie 10 were made using frozen primary cells. Dissociated embryonic day (E)11.5 mesenchyme or intact E13.5 mT/mG kidneys were suspended in 20% DMSO/80% FBS and frozen in cell-freezing containers at −80°C and transferred the next day to liquid nitrogen. Upon usage the cell vial was quickly warmed, the cells washed twice with medium and then pelleted (4 min at 1380 g) to make organoids, as in the usual protocol. The intact kidneys were dissociated during the freezing/thawing process. When whole kidneys were used the cell suspension included UB cells, which induced the nephrogenesis.
Assembly of the tissue culture
The FiZD conditions were set up in 6-well CellStar plates (Greiner Bio-One) (Fig. S1). A hole (20 mm diameter) was drilled at the bottom of wells and glass coverslips (24×24 mm) were glued to the upper side of the bottom with either dental wax or Histoacryl glue (Braun, 1050052). The coverslips were cleaned with ethanol and sonication to promote affixation of the organ rudiments to the coverslip glass (supplementary Materials and Methods). The culture plates were rinsed with ethanol, distilled water and dried in a UV hood. Kidney organoids incubated overnight in a microfuge tube or E11.5 to E13.5 embryonic kidneys were arranged on the lower side of the Transwell insert membrane (Corning, 3450) and the excess volume of PBS was aspirated. Polystyrene beads (Corpuscular, 100263-10) were mixed with Matrigel on ice and a few microliters were added to the samples with care not to disturb their position on the filter. Matrigel can aid fixation of the specimen in a defined position, but it is not required (see Fig. 2B,D with Matrigel, and Fig. 2H without) and it did not influence morphogenesis. Next, the insert was turned so that the samples were face down, positioned in a well, and pressed gently into place. The rim of the insert was melted with a heated glass capillary at three points to fix the insert to the plate (Fig. S1). The well was filled with 2 ml DMEM (Gibco, 41965-039), 10% foetal calf serum (Gibco, 10500-064) and 1% penicillin/streptomycin (Sigma, P4333) and the plate was kept at 37°C and 5% CO2. The culture setups were repeated a minimum of five times for each type of experiment. For the low-volume culture system we followed the method described in Sebinger et al. (2010), repeating the experiment 15 times. For imaging of the kidney cultures on the top of the filter (Costantini et al., 2011) the standard 6-well glass-bottomed plates were used (BD Falcon).
Time-lapse image capture
The culture plates were inserted into an on-stage incubator on a Zeiss LSM780 confocal microscope at 37°C and 5% CO2. The time-lapse images were captured at 5-20 min intervals and processed with ZEN 2012 (blue edition) (Zeiss), Huygens Professional (Scientific Volume Imaging) and Fiji (Schindelin et al., 2012).
Whole mount immunostaining
The explants were transferred to 4% PFA for 20 min, washed in PBS, and stored at 4°C. For immunostaining, the samples were blocked for 1 h in 0.1% Triton-X100/1% BSA/10% goat serum/0.02 M glycine PBS at room temperature. Primary antibodies against Krt8 (1:100; clone Troma-I, AB_531826, Developmental Studies Hybridoma Bank), Nephrin (also known as Nphs1) (1:1000; a gift from Prof. Karl Tryggvason, Duke-NUS Medical School, Singapore), Six2 (1:200; 11560, Proteintech), Umod (1:20; LS-C150268, LSBio) and Pax2 (1:100; PRB-276P, Covance) were used for immunostaining.
After an overnight incubation at 4°C the samples were washed six times for 30 min in PBS and goat anti-rat Alexa Fluor 546 (1:1000; A11081, Molecular Probes) and goat anti-rabbit Alexa Fluor 488 (1:1000; A11008, Molecular Probes) or donkey anti-sheep NorthernLights NL557 (1:200; NL010, R&D Systems) were incubated 1 h at room temperature and washed several times with PBS. A Zeiss LSM780 microscope and Zeiss Axiolab were used for analysis and image capture.
Automatic image analysis
Embryonic UBs labelled with HoxB7Cre-activated GFP, highlighting the cell boundaries, were segmented and tracked using a program developed in Matlab (MathWorks). Hessian ridge enhancement (Hodneland et al., 2009) was used to enhance the cell membrane intensity, fill the membrane gaps and remove cytoplasmic fluorescence. H-minima transformation (Soille, 1999) was used to filter out local minima and the Watershed transformation (Meyer, 1994) for cell segmentation. The centroids of the segmented cells in the first frame were used to initialize the cell tracks and the Hungarian algorithm (Munkres, 1957) was used to associate detections with tracks. For details on segmentation of image data and preparation of wind rose plots, see the supplementary Materials and Methods.
Acknowledgements
We thank Paula Haipus, Hannele Härkman and Johanna Kekolahti-Liias for technical assistance, Prof. Karl Tryggvason (Duke-NUS Medical School, Singapore) for the anti-nephrin antibody, Vera Eremina (The Lunenfeld-Tanenbaum Research Institute, Mount Sinai Hospital, Toronto, Ontario, Canada) and Susan E. Quaggin (The Lunenfeld-Tanenbaum Research Institute, Mount Sinai Hospital, Toronto, Ontario, Canada; Feinberg Cardiovascular Research Institute and Division of Nephrology and Hypertension, Northwestern University, Chicago, Illinois; and Human Health and Nutritional Sciences, University of Guelph, Guelph, Ontario, Canada) for the Flk1-GFP samples and Antti Viklund for the processing of the videos. The anti-Krt8 (Troma-I) antibody was obtained from the Developmental Studies Hybridoma Bank.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
I.S. conceived and designed the project and experimental strategies. U.S. and I.S. performed the cultures, the time-lapse analysis, their processing and the biomarker analyses and were responsible for writing the paper. S.U.A. and J.H. performed the image analysis and A.D. and I.S. compiled the HoxB7Cre data sets. S.C. generated Hoxb7cre mouse embryos A.R.-R. assisted in writing the manuscript. J.S. developed the Wnt4Cre mouse line, V.-P.R. assisted in the image analysis, and S.J.V. led the project, provided the infrastructure and finalized the writing of the paper.
Funding
This work was supported financially by the Suomen Akatemia (Academy of Finland) (206038, 121647, 250900, 260056; Centre of Excellence grant 2012-2017 251314), Munuaissäätiö – Finnish Kidney and Liver Association, the Sigrid Juséliuksen Säätiö, Novo Nordisk, Syöpäjärjestöt (Cancer Society of Finland), the European Community's Seventh Framework Programme (FP7/2007-2013; grant FP7-HEALTH-F5-2012-INNOVATION-1 EURenOmics 305608), and H2020 Marie Skłodowska-Curie Actions Innovative Training Network “RENALTRACT” Project ID 642937. Deposited in PMC for immediate release.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.142950.supplemental
References
- Auerbach R. and Grobstein C. (1958). Inductive interaction of embryonic tissues after dissociation and reaggregation. Exp. Cell Res. 15, 384-397. 10.1016/0014-4827(58)90039-9 [DOI] [PubMed] [Google Scholar]
- Combes A. N., Lefevre J. G., Wilson S., Hamilton N. A. and Little M. H. (2016). Cap mesenchyme cell swarming during kidney development is influenced by attraction, repulsion, and adhesion to the ureteric tip. Dev. Biol. 417, 98-107. 10.1016/j.ydbio.2016.06.028 [DOI] [PubMed] [Google Scholar]
- Costantini F., Watanabe B., Lu B., Chi X. and Srinivas S. (2011). Dissection of embryonic mouse kidney, culture in vitro, and imaging of the developing organ. Cold Spring Harb. Protoc. 2011:pdb.prot5613 10.1101/pdb.prot5613 [DOI] [PubMed] [Google Scholar]
- Eiraku M., Watanabe K., Matsuo-Takasaki M., Kawada M., Yonemura S., Matsumura M., Wataya T., Nishiyama A., Muguruma K. and Sasai Y (2008). Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3, 519-532. 10.1016/j.stem.2008.09.002 [DOI] [PubMed] [Google Scholar]
- Eliceiri K. W., Berthold M. R., Goldberg I. G., Ibáñez L., Manjunath B. S., Martone M. E., Murphy R. F., Peng H., Plant A. L., Roysam B. et al. (2012). Biological imaging software tools. Nat. Methods 9, 697-710. 10.1038/nmeth.2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatehullah A., Tan S. H. and Barker N. (2016). Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 18, 246-254. 10.1038/ncb3312 [DOI] [PubMed] [Google Scholar]
- Forgacs G., Foty R. A., Shafrir Y. and Steinberg M. S. (1998). Viscoelastic properties of living embryonic tissues: a quantitative study. Biophys. J. 74, 2227-2234. 10.1016/S0006-3495(98)77932-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobstein C. (1955). Tissue disaggregation in relation to determination and stability of cell type. Ann. N. Y. Acad. Sci. 60, 1095-1107. 10.1111/j.1749-6632.1955.tb40091.x [DOI] [PubMed] [Google Scholar]
- Gustafsson E., Brakebusch C., Hietanen K. and Fässler R. (2001). Tie-1-directed expression of Cre recombinase in endothelial cells of embryoid bodies and transgenic mice. J. Cell Sci. 114, 671-676. [DOI] [PubMed] [Google Scholar]
- Hadjantonakis A. K., Gertsenstein M., Ikawa M., Okabe M. and Nagy A. (1998). Generating green fluorescent mice by germline transmission of green fluorescent ES cells. Mech. Dev. 76, 79-90. 10.1016/S0925-4773(98)00093-8 [DOI] [PubMed] [Google Scholar]
- Halt K. J., Pärssinen H. E., Junttila S. M., Saarela U., Sims-Lucas S., Koivunen P., Myllyharju J., Quaggin S., Skovorodkin I. N. and Vainio S. J. (2016). CD146+ cells are essential for kidney vasculature development. Kidney Int. 90, 311-324. 10.1016/j.kint.2016.02.021 [DOI] [PubMed] [Google Scholar]
- Hodneland E., Bukoreshtliev N. V., Eichler T. W., Tai X. C., Gurke S., Lundervold A. and Gerdes H.-H. (2009). A unified framework for automated 3-d segmentation of surface-stained living cells and a comprehensive segmentation evaluation. IEEE Trans. Med. Imaging 28, 720-738. 10.1109/TMI.2008.2011522 [DOI] [PubMed] [Google Scholar]
- Huisken J., Swoger J., Del Bene F., Wittbrodt J. and Stelzer E. H. (2004). Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007-1009. 10.1126/science.1100035 [DOI] [PubMed] [Google Scholar]
- Junttila S., Saarela U., Halt K., Manninen A., Pärssinen H., Lecca M. R., Brändli A. W., Sims-Lucas S., Skovorodkin I. and Vainio S. J. (2015). Functional genetic targeting of embryonic kidney progenitor cells ex vivo. J. Am. Soc. Nephrol. 26, 1126-1137. 10.1681/ASN.2013060584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan Z., Wang Y. Z., Wieschaus E. F. and Kaschube M. (2014). Quantitative 4D analyses of epithelial folding during Drosophila gastrulation. Development 141, 2895-2900. 10.1242/dev.107730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispert A., Vainio S. and McMahon A. P. (1998). Wnt-4 is a mesenchymal signal for epithelial transformation of metanephric mesenchyme in the developing kidney. Development 125, 4225-4234. [DOI] [PubMed] [Google Scholar]
- Licht A. H., Raab S., Hofmann U. and Breier G. (2004). Endothelium-specific Cre recombinase activity in flk-1-Cre transgenic mice. Dev. Dyn. 229, 312-318. 10.1002/dvdy.10416 [DOI] [PubMed] [Google Scholar]
- Lindström N. O., Chang C.-H., Valerius T. M., Hohenstein P. and Davies J. A. (2014). Node retraction during patterning of the urinary collecting duct system. J. Anat. 226, 13-21. 10.1111/joa.12239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström N. O., Lawrence M. L., Burn S. F., Johansson J. A., Bakker E. R., Ridgway R. A., Chang C. H., Karolak M. J., Oxburgh L., Headon D. J. et al. (2015). Integrated β-catenin, BMP, PTEN, and Notch signalling patterns the nephron. Elife 3, e04000 10.7554/eLife.04000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F. (1994). Topographic distance and watershed lines. Signal Process. 38, 113-125. 10.1016/0165-1684(94)90060-4 [DOI] [Google Scholar]
- Morizane R., Lam A. Q., Freedman B. S., Kishi S., Valerius M. T. and Bonventre J. V. (2015). Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 33, 1193-1200. 10.1038/nbt.3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkres J. (1957). Algorithms for the assignment and transportation problems. J. Soc. Ind. Appl. Mathematics 5, 32-38. 10.1137/0105003 [DOI] [Google Scholar]
- Muzumdar M. D., Tasic B., Miyamichi K., Li L. and Luo L. (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593-605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Packard A., Georgas K., Michos O., Riccio P., Cebrian C., Combes A. N., Ju A., Ferrer-Vaquer A., Hadjantonakis A. K., Zong H. et al. (2013). Luminal mitosis drives epithelial cell dispersal within the branching ureteric bud. Dev. Cell 27, 319-330. 10.1016/j.devcel.2013.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips H. M. and Steinberg M. S (1978). Embryonic tissues as elasticoviscous liquids. J. Cell Sci. 30, 1-20. [DOI] [PubMed] [Google Scholar]
- Prunskaite-Hyyryläinen R., Skovorodkin I., Xu Q., Miinalainen I., Shan J. and Vainio S. J. (2016). Wnt4 coordinates directional cell migration and extension of the Müllerian duct essential for ontogenesis of the female reproductive tract. Hum. Mol. Genet. 25, 1059-1073. 10.1093/hmg/ddv621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak-Raszewska A., Hauser P. V. and Vainio S. (2015). Organ in vitro culture: what have we learned about early kidney development? Stem Cells Int. 2015, 959807 10.1155/2015/959807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio P., Cebrian C., Zong H., Hippenmeyer S. and Costantini F. (2016). Ret and Etv4 promote directed movements of progenitor cells during renal branching morphogenesis. PLoS Biol. 14, e1002382 10.1371/journal.pbio.1002382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxén L. (1987). Organogenesis of the Kidney. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebinger D. D., Unbekandt M., Ganeva V. V., Ofenbauer A., Werner C. and Davies J. A. (2010). A novel, low-volume method for organ culture of embryonic kidneys that allows development of cortico-medullary anatomical organization. PLoS ONE 5, e10550 10.1371/journal.pone.0010550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan J., Jokela T., Skovorodkin I. and Vainio S. (2010). Mapping of the fate of cell lineages generated from cells that express the Wnt4 gene by time-lapse during kidney development. Differentiation 79, 57-64. 10.1016/j.diff.2009.08.006 [DOI] [PubMed] [Google Scholar]
- Soille P. (1999). Morphological Image Analysis: Principles and Applications, pp. 170-171. Berlin: Springer-Verlag. [Google Scholar]
- Stegmaier J., Amat F., Lemon W. C., McDole K., Wan Y., Teodoro G., Mikut R. and Keller P. J. (2016). Real-time three-dimensional cell segmentation in large-scale microscopy data of developing embryos. Dev. Cell 36, 225-240. 10.1016/j.devcel.2015.12.028 [DOI] [PubMed] [Google Scholar]
- Takasato M., Er P. X., Chiu H. S., Maier B., Baillie G. J., Ferguson C., Parton R. G., Wolvetang E. J., Roost M. S., Lopes S. M. et al. (2015). Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 536, 238 10.1038/nature17982 [DOI] [PubMed] [Google Scholar]
- Unbekandt M. and Davies J. A. (2010). Dissociation of embryonic kidneys followed by reaggregation allows the formation of renal tissues. Kidney Int. 77, 407-416. 10.1038/ki.2009.482 [DOI] [PubMed] [Google Scholar]
- Vainio S., Jalkanen M., Bernfield M. and Saxén L. (1992). Transient expression of syndecan in mesenchymal cell aggregates of the embryonic kidney. Dev. Biol. 152, 221-232. 10.1016/0012-1606(92)90130-9 [DOI] [PubMed] [Google Scholar]
- Watanabe T. and Costantini F. (2004). Real-time analysis of ureteric bud branching morphogenesis in vitro. Dev. Biol. 271, 98-108. 10.1016/j.ydbio.2004.03.025 [DOI] [PubMed] [Google Scholar]
- Yu J., Carroll T. J. and McMahon A. P. (2002). Sonic hedgehog regulates proliferation and differentiation of mesenchymal cells in the mouse metanephric kidney. Development 129, 5301-5312. [DOI] [PubMed] [Google Scholar]