ABSTRACT
The tumor suppressor retinoblastoma protein (RB) regulates S-phase cell cycle entry via E2F transcription factors. Knockout (KO) mice have shown that RB plays roles in cell migration, differentiation and apoptosis, in developing and adult brain. In addition, the RB family is required for self-renewal and survival of human embryonic stem cells (hESCs). Since little is known about the role of RB in human brain development, we investigated its function in cerebral organoids differentiated from gene-edited hESCs lacking RB. We show that RB is abundantly expressed in neural stem and progenitor cells in organoids at 15 and 28 days of culture. RB loss promoted S-phase entry in DCX+ cells and increased apoptosis in Sox2+ neural stem and progenitor cells, and in DCX+ and Tuj1+ neurons. Associated with these cell cycle and pro-apoptotic effects, we observed increased CCNA2 and BAX gene expression, respectively. Moreover, we observed aberrant Tuj1+ neuronal migration in RB-KO organoids and upregulation of the gene encoding VLDLR, a receptor important in reelin signaling. Corroborating the results in RB-KO organoids in vitro, we observed ectopically localized Tuj1+ cells in RB-KO teratomas grown in vivo. Taken together, these results identify crucial functions for RB in the cerebral organoid model of human brain development.
KEY WORDS: Retinoblastoma, Human embryonic stem cells, Organoid, Brain, CRISPR/Cas9, Cell cycle
Summary: In human cerebral organoids, depletion of the tumor suppressor retinoblastoma protein disrupts proliferation, promotes entry into S-phase, and causes increased apoptosis and aberrant neuronal migration.
INTRODUCTION
The retinoblastoma protein (RB) is a tumor suppressor, encoded by a gene (RB1) located in chromosome 13q14.2 in humans, which was discovered in 1986 in retinoblastoma tumors (Friend et al., 1986). Later, mutations in the RB1 gene were found in a wide variety of human tumors (Giacinti and Giordano, 2006). Initially, RB was described as regulating the G1/S transition of the cell cycle through E2F-mediated transcriptional regulation in many tissues, including the nervous system (Classon and Harlow, 2002; McClellan and Slack, 2006; Julian et al., 2016; MacPherson et al., 2003; Naser et al., 2016). Recently, studies of brain-specific Rb1-KO mice revealed other roles of the Rb1 gene that extend beyond cell cycle control, such as regulation of neuronal differentiation and migration (Andrusiak et al., 2011; Ghanem et al., 2012; McClellan et al., 2007; Christie et al., 2014; Ferguson et al., 2005). Moreover, the lack of RB promotes apoptosis in some cell types, but not in others, through p53-dependent or -independent pathways (Macleod et al., 1996; Vandenbosch et al., 2016; Yu et al., 2012).
In human, the lack of RB has been associated with structural brain abnormalities, such as macrocephaly and hypoplastic corpus callosum (Mitter et al., 2011; Rodjan et al., 2010), suggesting that RB may be required in human brain development. As human brains have a marked expansion of cerebral cortex with a unique outer subventricular zone compared with rodents (Hoerder-Suabedissen and Molnár, 2015; Kelava and Lancaster, 2016; Hansen et al., 2010), RB may affect human brain development in a different manner than it has been described in mice. Although the inactivation of RB family proteins causes G2/M arrest and cell death in human embryonic stem cells (ESCs) (Conklin et al., 2012), little is known about the role of RB during human brain development. In the past, ethical considerations and challenges in obtaining human samples have hampered the expansion of knowledge of human brain development. However, advancement of novel technologies, such as growth of three-dimensional human cerebral organoids (Lancaster et al., 2013; Pasca et al., 2015; Eiraku et al., 2011; Kadoshima et al., 2013) and CRISPR/Cas9-mediated gene editing (Ran et al., 2013), opens up the possibility of recapitulating the process of human brain development, including the rapid expansion of cerebral cortex (Lancaster and Knoblich, 2014b).
Here, to analyze the role of RB in an in vitro model of human brain development, we established a human RB1-KO ESC line using CRISPR/Cas9-mediated gene editing and we differentiated wild-type and RB1-KO ESCs into cerebral organoids. Our results show that the lack of RB enlarged the volume of organoids, possibly owing to the increased entry of neuroblasts in S phase. Moreover, RB1-KO organoids had increased apoptosis of neural stem cells (NSCs), neuroblasts and neurons, as well as aberrant neuronal migration. Altogether, our data suggest that RB not only regulates cell cycle progression, but also controls cell survival and neuronal migration during human brain development.
RESULTS
Generation of RB1-KO human ESCs
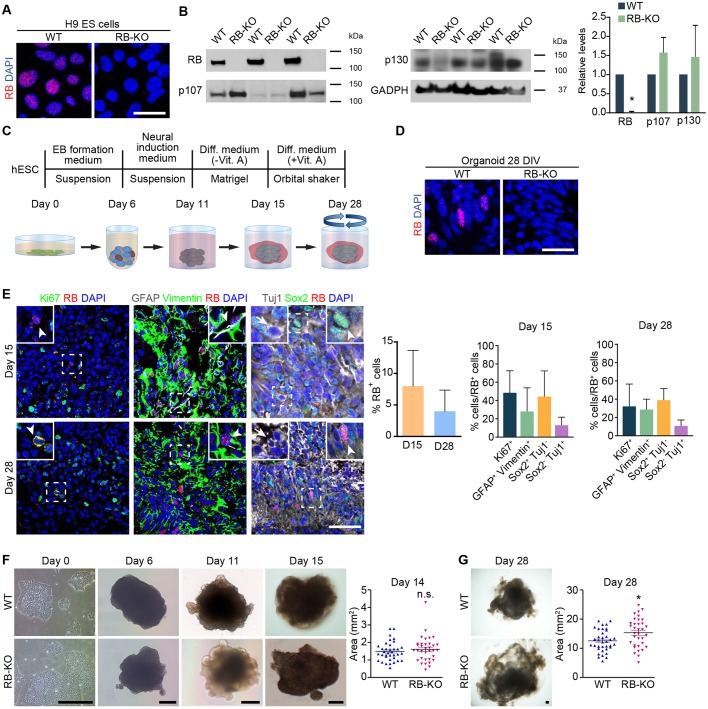
To analyze the role of RB during human brain development, we generated two clones of human ESC lines deficient for RB using CRISPR/Cas9 and differentiated them to organoids and NSCs. First, we analyzed the expression of RB in wild-type and RB1-KO cells by immunocytochemistry and by western blot (Fig. 1 and Fig. S1). Although the majority of wild-type ESCs expressed RB, we did not find any expression of RB in RB1-KO cells (Fig. 1A and Fig. S1A). Similar results were found by western blot. We detected a band of 106 kDa in wild-type ESCs, but did not observe any expression of RB in KO ESCs, although both samples showed the same levels of GAPDH (Fig. 1B). RB levels were also significantly reduced in another clone of RB1-KO ESCs (Fig. S1B). We also analyzed the expression of additional members of the RB family (p107 and p130) in RB1-KO cells and found no differences in the relative levels of both proteins by western blot (Fig. 1B).
Fig. 1.
RB is expressed in stem cells and progenitor cells during organoid development. We generated a RB1-deficient H9 ESC line using CRISPR/Cas9. (A) Representative wild-type and KO ESCs in culture immunostained against RB and counterstained with DAPI. (B) Western blot was performed to detect RB, p107, p130 and GADPH proteins in samples from wild-type and RB1-KO ESCs; the graph shows their relative levels. (C) Experimental scheme to generate organoids from H9 ESCs after 28 DIV. (D) Representative wild-type and KO organoids at 28 DIV sectioned and immunostained against RB and counterstained with DAPI. (E) Representative wild-type organoids at 15 and 28 DIV sectioned and immunostained against Ki67 and RB (arrowheads); GFAP, Vimentin and RB (arrowheads); Sox2 and RB (arrowheads); and Tuj1 and RB (arrows), and counterstained with DAPI. The graphs show the percentage of RB+ cells in the organoid at 15 and 28 DIV, and the RB+ cells that colocalized with the aforementioned markers. (F) Representative wild-type and KO organoids grown during cerebral organoid induction from ESCs to 15 DIV organoids. The graphs show the mean area of wild-type and KO organoids at 14 DIV. (G) Representative wild-type and KO organoids at 28 DIV. The graph shows the mean area of wild-type and KO organoids at 28 DIV. The results are mean±s.e.m. of 34-36 organoids from three or four independent experiments (*P<0.05, Student's t-test; n.s., not significant). Scale bars: 25 µm in A,D; 50 µm in E; 250 µm in F,G.
RB expression in wild-type organoids
Next, we generated cerebral organoids using a protocol very similar to the one described previously by Lancaster et al., which does not involve the use of growth factors to allow spontaneous organization of neural tissues (Lancaster et al., 2013) (Fig. 1C). In wild-type organoids, 5-10% of the cells expressed RB at 15 and 28 days in vitro (DIV) (Fig. 1E), whereas we did not detect any RB+ cells in organoids derived from RB1-KO ESCs (Fig. 1D and Fig. S1C). Next, we analyzed the expression of RB in radial glial cells (RGCs) (GFAP+ vimentin+), NSCs (Sox2+), proliferating cells (Ki67+) and neurons (Tuj1+) in wild-type organoids by immunohistochemistry. In wild-type organoids at 15 DIV, 28-48% of the RB+ cells were Sox2+, Ki67+ or GFAP+ vimentin+ cells, whereas a smaller proportion (13%) was Tuj1+ (Fig. 1E). Similar results were found in organoids at 28 DIV (Fig. 1E). These results support the conclusion that RB expression in cerebral organoids is highest during NS and progenitor cell stages and is downregulated in neuronal stages.
Loss of RB produces an increase in organoid size and S-phase entry
As RB is expressed in NS and progenitor cells, we hypothesized that RB may play a role in regulating organoid growth. The organoids derived from KO ESCs were slightly bigger than the organoids derived from wild-type ESCs at 28 DIV (P<0.01, Fig. 1G). Similar results were found in the second clone (Fig. S1D). However, we did not find any statistically significant difference in organoid size at 14 DIV (Fig. 1F).
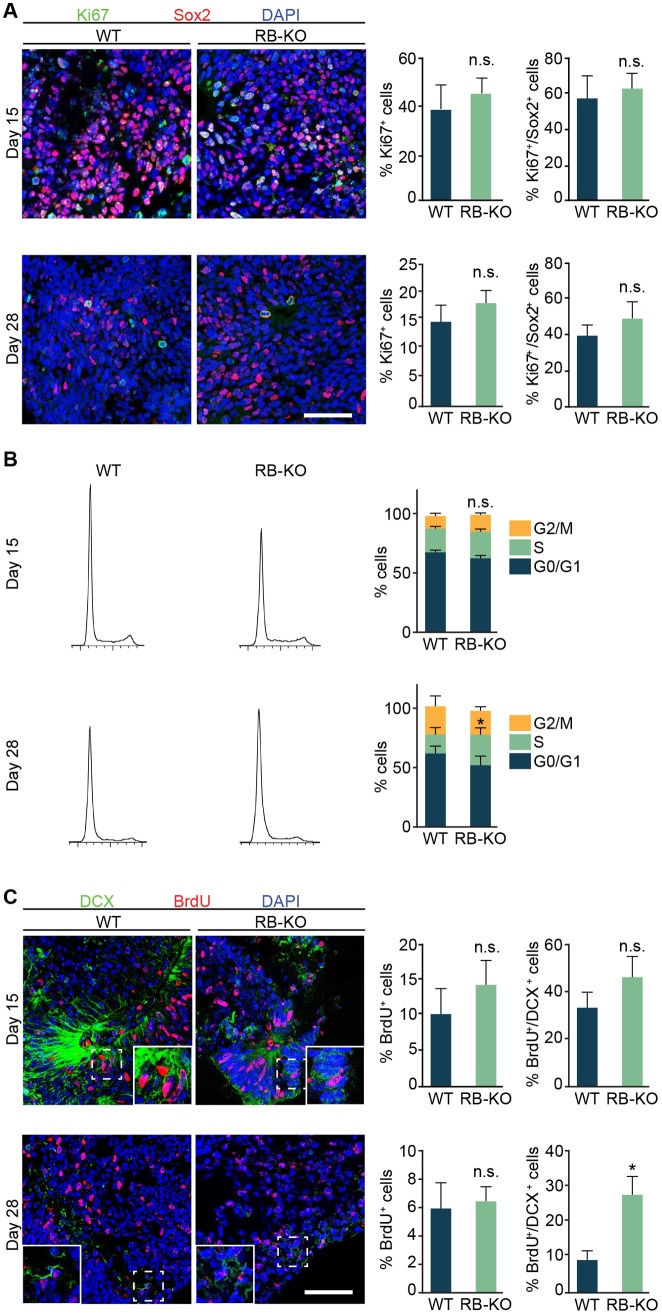
The increased size of RB1-KO organoids and the known role of RB in cell cycle regulation (Classon and Harlow, 2002; McClellan and Slack, 2006) prompted us to analyze the effect of the lack of RB in cell proliferation during organoid development. Our analysis of the percentage of Ki67+ and Ki67+ Sox2+ cells at 15 and 28 DIV showed no difference in RB1-KO organoids compared with wild type (Fig. 2A). To further analyze the role of RB in cell cycle progression, we dissociated the organoids into individual cells and stained with propidium iodide (PI) (Fig. 2B). At 15 DIV, the percentage of cells in each phase of the cell cycle was similar in organoids derived from wild-type and KO ESCs. However, at 28 DIV, although RB1-KO organoids did not have a significant change in the percentage of cells in G0/G1 (P>0.05, n=14) or G2/M (P>0.05, n=14), there was a significant increase (36%) in the percentage of cells in S phase compared with wild-type organoids (P<0.05, n=14; Fig. 2B). Nevertheless, organoids derived from the second RB1-KO human ESC clone did not show significant change in any of the cell cycle phases (Fig. S1E). To identify the cells in S phase in the organoids from the first RB1-KO clone, we added BrdU and immunostained the organoids with markers of NSCs (Sox2), progenitor cells (Pax6) and neuroblasts or immature neurons (DCX). The percentage of BrdU+ cells was similar in organoids from both genotypes at 15 and 28 DIV (Fig. 2C). Although we found no differences in the percentage of BrdU+ cells either in Sox2+ or Pax6+ cells (Fig. S2), we found a threefold increase in the percentage of BrdU+ DCX+ cells in RB1-KO organoids at 28 DIV (Fig. 2C). Our data suggest that RB deficiency produces an increase in organoid size by promoting S-phase entry of DCX+ cells.
Fig. 2.
The lack of RB produces enlarged organoids and promotes S entry. (A) Representative wild-type and KO organoids at 15 and 28 DIV sectioned and immunostained against Ki67 and Sox2 and counterstained with DAPI. The graphs show the percentage of Ki67+/DAPI cells and the percentage of Ki67+/Sox2+ cells at both ages. (B) Plots on the left show representative wild-type and KO organoid cell cycle profiles at 15 and 28 DIV and the graphs on the right show the percentage of cells in each cell cycle phases at both time points. (C) Representative wild-type and KO organoids at 15 and 28 DIV sectioned and immunostained against BrdU and DCX, and counterstained with DAPI. The graphs show the percentage of BrdU+/DAPI cells and the percentage of BrdU+/DCX+ cells at both ages. The results are mean±s.e.m. of three to seven organoids from three independent experiments (*P<0.05, Student's t-test; n.s., not significant). Scale bars: 50 µm.
RB deletion increases cell death in organoids
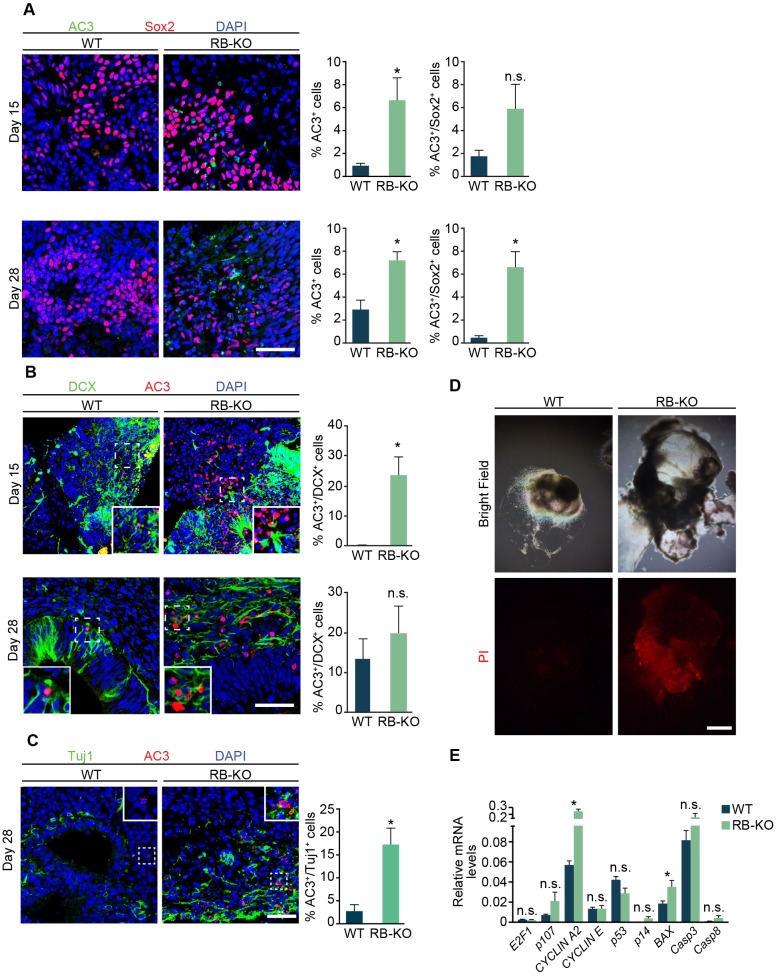
RB loss has been related to an increase of cell death in different areas of the mouse brain during development (Macleod et al., 1996; Vandenbosch et al., 2016; Yu et al., 2012). We therefore analyzed whether the absence of RB promotes cell death using a marker of apoptosis (cleaved caspase 3, AC3). At 15 and 28 DIV, RB1-KO organoids showed a sevenfold and 2.5-fold increase in the number of AC3+ cells, respectively (P<0.05, Fig. 3A) compared with wild-type organoids. Although there was not a significant increase in cell death in Sox2+ cells at 15 DIV, at 28 days, the KO organoids showed a significant increase in Sox2+ cell death (P<0.05, Fig. 3A). In addition, the RB1-KO organoids had a 90-fold increase in the percentage of AC3+ DCX+ cells at 15 DIV and sixfold increase in AC3+ Tuj1+ neurons at 28 DIV (P<0.05, Fig. 3B,C). These results were also confirmed in the second clone, where we found a significant increase of AC3+ Sox2+ and AC3+ Tuj1+ cells (Fig. S1F,G). Additionally, we stained wild-type and RB1-KO organoids at 28 DIV with PI to determine whether the accumulation of cell death found in RB1-KO organoids could contribute to their increased size. We adopted a method of PI staining in live organoids that labels dead or dying cells (Hsieh et al., 2004). We found a larger number of PI+ cells in RB1-KO organoids at 28 DIV compared with wild-type organoids; this increase was associated with the formation of many cysts in RB1-KO organoids (Fig. 3D). To corroborate the role of RB in cell survival and proliferation, and gain mechanistic insight into the nature of effector genes, we performed RT-qPCR to survey the expression of E2F targets involved in cell cycle regulation and apoptosis (Fig. 3E). We found an increase in the relative cyclin A2 (CCNA2) and BAX mRNA levels in RB1-KO organoids compared with wild type, whereas we did not find significant differences in the expression of RB family member genes or of other genes related to the cell cycle and apoptosis. Altogether, our data revealed that RB is essential for the cell survival in human cerebral organoids.
Fig. 3.
RB loss stimulates cell death in cerebral organoids. (A) Representative wild-type and KO organoids at 15 and 28 DIV sectioned and immunostained against AC3 and Sox2, and counterstained with DAPI. The graphs show the percentage of AC3+/DAPI cells and AC3+/Sox2+ cells at both time points. (B) Representative wild-type and KO organoids at 15 and 28 DIV sectioned and immunostained against AC3 and DCX, and counterstained with DAPI. The graphs show the percentage of AC3+/DCX+ cells at both time points. (C) Representative wild-type and KO organoids at 28 DIV sectioned and immunostained against AC3 and Tuj1, and counterstained with DAPI. Insets in wild-type and KO organoids show an enlarged image of AC3+ and Tuj1+ cells in the RB-KO organoid, which were reduced in the wild-type organoid. The graph shows the percentage of AC3+/Tuj1+ cells. (D) Bright-field and red fluorescence images showing representative PI-treated wild-type and KO organoids at 28 DIV. (E) Relative mRNA levels of genes of the RB family and genes involved in cell cycle regulation and apoptosis measured by RT-qPCR. The results are mean±s.e.m. of five or six organoids from three independent experiments (*P<0.05, Student's t-test; n.s., not significant). Scale bar: 50 µm in A-C; 200 µm in D.
Loss of RB augments proliferation of NSCs in vitro
The RB family of proteins have been described as critical factors for the self-renewal and survival of human ESCs (Conklin et al., 2012). Given that human cerebral organoids are very heterogeneous and complex, to examine the role of RB in a more homogenous population of NSCs, we induced the differentiation of wild-type and KO-ESCs into NSCs (‘neurospheres’) (Zhang et al., 2001) (Fig. S3A). At 6 DIV, the RB1-KO cultures had a higher number of neurospheres than did wild type, whereas the neurosphere size was similar in both cultures (Fig. S3B). To evaluate whether the increase in the number of neurospheres was due to an augmentation in cell proliferation, we fixed and stained dissociated neurospheres with PI. Surprisingly, cells from RB1-KO cultures did not show a significant change in G0/G1, S or G2/M phase compared with wild-type cells (P>0.05, n=8; Fig. S3C), although there was an increasing trend in the percentage of cells in S phase. Our results evaluating the growth of RB1-KO neurospheres is consistent with the phenotype of RB1-KO organoids, i.e. the lack of RB promotes the S entry.
RB is essential for proper neuronal migration in organoids
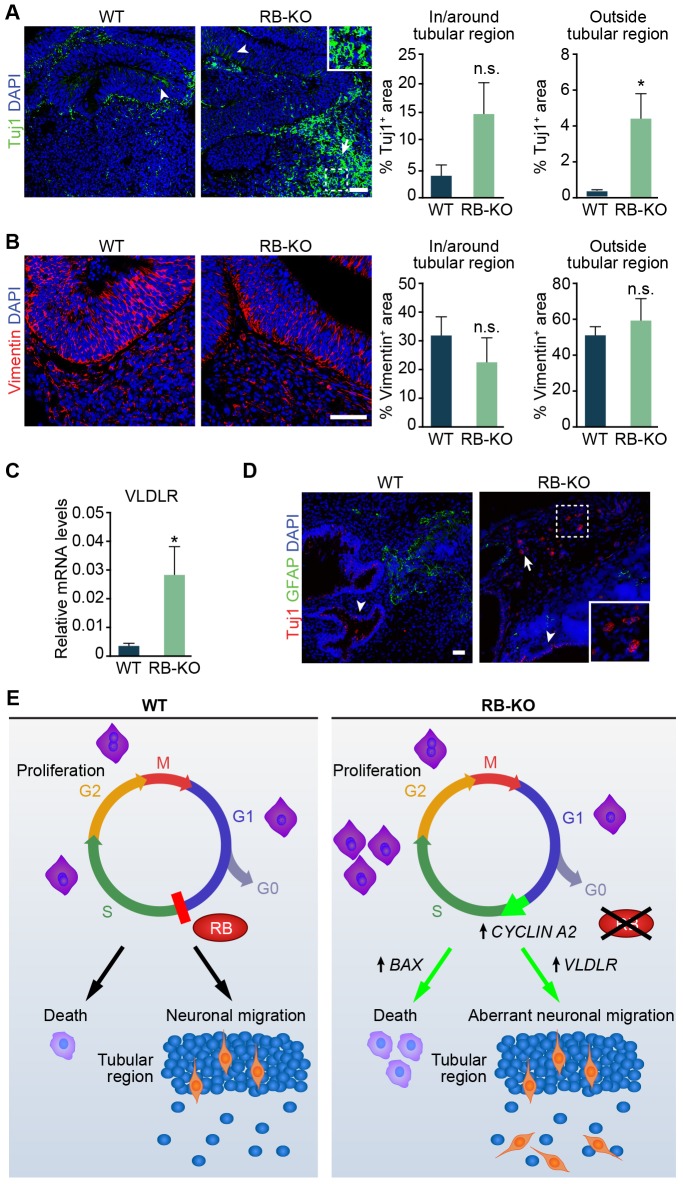
As RB is also expressed in a subset of Tuj1+ neuronal cells and RB plays important roles during mouse neurogenesis (Ferguson et al., 2005; Ghanem et al., 2012; Naser et al., 2016), we also analyzed sections from 28 DIV wild-type and RB1-KO organoids stained for Tuj1. In both sets of organoids, Tuj1+ cells were present in or around the tubular region (Fig. 4A), However, whereas in the wild-type organoid Tuj1+ cells were rarely observed outside the tubular structure, we estimated that 4% of the area outside the tubular region was covered by Tuj1+ cells in RB1-KO organoids (P<0.05, Fig. 4A). Consistent with aberrant neuronal migration in RB1-KO organoids, we found Tuj1+ cells with their corresponding nuclei outside the tubular region. We observed a similar increase in the Tuj1+ area outside the tubular region in the organoids derived from the second RB1-KO clone, although this result was not statistically significant (Fig. S1H). As neural progenitors use a RGC scaffold to migrate (Brunne et al., 2013) and to rule out the possibility that the aberrant positioning of Tuj1+ cells in RB1-KO organoids was due to a defect in RGC morphology, we stained sections from wild-type and RB1-KO organoids using an antibody against vimentin. However, the vimentin pattern was similar both inside and outside the tubular region in organoids from both genotypes (Fig. 4B). In addition, we analyzed the mRNA expression of very low density lipoprotein receptor (VLDLR) and apolipoprotein E receptor 2 (ApoER2, also known as LRP8), two receptors of the reelin pathway that are widely involved in neuronal migration in mice and humans (Cheng et al., 2011; Teixeira et al., 2012). The RB1-KO organoids showed an increase in relative VLDLR mRNA levels compared with wild type (P<0.05, Fig. 4C), whereas ApoER2 levels were similar in both genotypes (data not shown). Finally, to provide an in vivo corollary of the RB1-KO organoid phenotype, we injected wild-type and KO ESCs subcutaneously in immunodeficient mice and analyzed the formation of teratomas in vivo. Both teratomas formed from wild-type and RB1-KO ESCs presented neural tube-like structures and had Tuj1+ and GFAP+ cells. However, whereas in wild-type and RB1-KO ESC-derived teratomas, Tuj1+ cells were found in or around neural tube-like structures (Fig. 4D), in RB1-KO teratomas, we also detected Tuj1+ cells in ectopic locations distant from tubular regions (Fig. 4D). These results suggest that the lack of RB induces aberrant neuronal migration in cerebral organoids, possibly owing to an upregulation of the VLDLR gene. These results also suggest that RB1-KO neuronal migration defects can be modeled in both cerebral organoid cultures and teratomas formed in mice.
Fig. 4.
RB loss results in aberrant neuronal migration. (A) Representative wild-type and KO organoids at 28 DIV sectioned and immunostained against Tuj1 and counterstained with DAPI. Arrowheads in the wild-type and KO organoid show Tuj1+ cells in or around the tubular region. An arrow in the KO organoid shows ectopic Tuj1+ cells outside the tubular region. Inset in the KO organoid shows an enlarged image of Tuj1+ cells with corresponding nuclei outside the tubular region. The graphs show the percentage of Tuj1-stained area in or around and outside the tubular region. (B) Representative wild-type and KO organoids at 28 DIV sectioned and immunostained against Vimentin and counterstained with DAPI. The graphs show the percentage of Vimentin-stained area in or around and outside the tubular region. (C) Relative VLDLR mRNA levels measured by RT-qPCR. (D) Representative wild-type and KO teratomas sectioned and immunostained against Tuj1 and GFAP, and counterstained with DAPI. Arrowheads in the wild-type and KO teratoma show Tuj1+ cells in or around the tubular region. An arrow in the KO teratoma shows ectopic Tuj1+ cells outside the tubular region. Inset in the KO teratoma shows an enlarged image of Tuj1+ cells with corresponding nuclei outside the tubular region. (E) A model of the role of RB during human brain development. In wild-type cells, RB regulates entry into S-phase. However, loss of RB promotes an accumulation of cells in S-phase and an increase in cell death that is associated with upregulation of CCNA2 and BAX. RB deficiency also leads to aberrant neuronal migration associated with an increase in expression of VLDLR. The results in A-D are mean±s.e.m. of three to five organoids from two to four independent experiments (*P<0.05, paired Student's t-test, n.s., not significant). Scale bars: 50 µm; 25 µm in insets.
DISCUSSION
Since the discovery of the tumor suppressor gene RB1, many studies have demonstrated its role in regulating the G1-to-S transition in various cell types (Classon and Harlow, 2002; McClellan and Slack, 2006; Julian et al., 2016; MacPherson et al., 2003; Naser et al., 2016). Moreover, KO mouse models have revealed the role of RB during neuronal differentiation and migration, as well as in the regulation of cell death in the embryo and adult brain (Andrusiak et al., 2011; Ghanem et al., 2012; McClellan et al., 2007; Christie et al., 2014; Ferguson et al., 2005; Macleod et al., 1996; Vandenbosch et al., 2016; Yu et al., 2012). The finding that lack of the RB1 gene is associated with structural brain abnormalities in human (Mitter et al., 2011; Rodjan et al., 2010), and inactivation of RB family proteins affects cell cycle and cell death in human ESCs (Conklin et al., 2012), suggest that RB might have a role during human brain development. However, the function of RB function in human development is still a mystery. Here, using human cerebral organoids and CRISPR/Cas9 gene editing, we showed that lack of RB promotes S-phase entry of DCX+ cells and increases cell death. Moreover, RB deficiency promotes aberrant migration of neuronal cells in cerebral organoids as well as in teratomas in vivo. Based on these results, we present a model suggesting a crucial role for RB in human brain development (Fig. 4E).
Role of RB in cell proliferation and cell death
Our data demonstrate that RB deletion produces enlarged organoids, possibly owing to an increase of cell proliferation and an accumulation of dead cells. This increase in cell proliferation is in agreement with results described in primary tumors, such as human glioblastoma, and in mouse cells, such as retinal progenitor cells, and in different regions of the nervous system, including the central and peripheral nervous system and lens (Classon and Harlow, 2002; McClellan and Slack, 2006; Julian et al., 2016; MacPherson et al., 2003; Naser et al., 2016; Cen et al., 2012; Zhang et al., 2004). Although we did not find any differences in the percentage of Ki67+ cells in wild-type compared with RB1-KO organoids, the cell cycle analysis and BrdU-incorporation assay revealed an increase in S-phase entry of DCX+ cells in RB1-KO organoids at 28 DIV. Surprisingly, although RB was highly expressed in Sox2+ NS and progenitor cells in organoids and we found a tendency towards a higher percentage of NSCs in S-phase in vitro, we detected no difference in the percentage of BrdU+ Sox2+ neural stem cells between wild-type and RB1-KO organoids. Nevertheless, this increase in organoid size was only significant in the organoids at 28 DIV and not at earlier stages (15 DIV). It could be possible that in both wild-type and RB1-KO organoids at 15 DIV, the ratio of cell death was similar to that of cell proliferation. Consistent with the lack of difference in organoid size between wild-type and RB1-KO organoids at 15 DIV, the percentage of AC3+/Sox2+ NS and progenitor cells at 15 DIV was not significantly different in RB1-KO organoids compared with wild type, although there was a trend towards more AC3+ cells in the RB1-KO at this time point. In addition to the increase in the NSC death, we also found more apoptotic neuroblasts and neurons, represented by AC3+ DCX+ cells and AC3+ Tuj1+ cells at 15 and 28 DIV, respectively. Similar results were previously observed in mice (Yu et al., 2012; Vandenbosch et al., 2016). In addition, there was no significant change in S-phase entry in RB1-KO organoids compared with wild type at 15 DIV, which is in line with no difference in organoid size between wild type and RB1-KO at this stage. Consistently, we did not find any differences in the cell cycle in ESCs or embryoid bodies (EBs) (data not shown), which are earlier stages that precede organoid induction.
Interestingly, at 28 DIV, although RB1-KO organoids were larger compared with wild type, there was a significant increase in S-phase entry. Supporting these results, RB1-KO organoids showed an increase in the expression of CCNA2, which regulates the G1- to S-phase transition (Macleod et al., 1996). What is surprising is that there was also increased cell death in RB1-KO organoids compared with wild type at 28 DIV, possibly due to an increase of the pro-apoptotic gene BAX (Drew et al., 2016). In line with our results, it has been described that loss of RB in mice promotes apoptosis in the developing and adult brain (Macleod et al., 1996; Vandenbosch et al., 2016; Yu et al., 2012). However, other studies have shown that an increase in cell proliferation due to the lack of RB is not followed by enhanced cell death (MacPherson et al., 2003). Our results suggest that the cell death is the primary effect of the RB loss in human cerebral organoids. Subsequently, RB loss produces an S-entry in the surviving cells. In fact, in the organoids derived from second RB1-KO clone we found more AC3+ Sox2+ and AC3+ Tuj1+ cells compared with wild-type organoids, without any effect on the cell cycle. We speculate that the low level of RB detected in the second clone by western blot (Fig. S1B) might be sufficient to maintain cell proliferation in RB1-KO organoids, although it did not prevent cell death. Moreover, the increase in S-phase of DCX+ cells found at 28 DIV is insufficient to explain the bigger organoid size at that age. However, this increase in the organoid size could be the consequence of increased cell death at an earlier stage. In the brain, dead cells are removed by phagocytosis; however, this process does not appear to occur in organoids in vitro, so cell debris and other internal components might accumulate in cysts and contribute to the larger size in RB1-KO organoids. We speculate that in organoids maintained longer than 28 DIV, the ratio of cell proliferation to cell death may be even larger, thus further increasing the difference in size between wild-type and KO organoids. In addition, the DCX+ cells that escape cell death and enter into S-phase could correlate with the formation of retinoblastoma or other tumors described in humans. Although we found no tumors in our RB1-KO organoids, additional studies using other 3D cultures, such as optical cups (Eiraku et al., 2011), are needed to understand the role of RB during tumor formation.
Effect of RB in neuronal migration
Moreover, the neurons that survived in the RB1-KO organoids occupied aberrant positions. Although in the wild-type organoids Tuj1+ neurons were found mainly in the tubular region, in RB1-KO organoids we observed Tuj1+ neurons outside this region. Aberrant neuronal migration was also found in Rb1-KO mice (Andrusiak et al., 2011; McClellan et al., 2007; Naser et al., 2016). This aberrant migration could be due to a disruption in the radial pattern of RGCs (Brunne et al., 2013). However, we did not find any morphological changes in the RGCs in KO organoids compared with wild-type organoids. Another possibility is that the increase in cell proliferation in KO organoids forces NSCs and neuroblasts to occupy other positions outside the normal niche, which give rise to aberrant neurons. The ectopic neurons may also contribute to the increased size of RB1-KO organoids. The aberrant neuronal migration in KO organoids could be partially due to the increased level of VLDLR. The VLDLR gene encodes a canonical receptor of reelin, which is widely involved in neuronal migration in mice and humans (Cheng et al., 2011; Teixeira et al., 2012); therefore, additional studies are needed to determine whether expression of VLDLR is a cause or consequence of aberrant migration. Previous microarray analyses carried out with Rb1-deficient mouse osteoblasts (Sosa-García et al., 2010) and Sertoli cells (Nalam et al., 2009) have also revealed that Vldlr expression is induced upon the loss of Rb1 expression in these cell types. These data suggest the possibility of conserved regulatory mechanisms across different tissues by which RB regulates VLDLR expression to control cell migration or cholesterol homeostasis. Additionally, after subcutaneous injection in immunodeficient mice, RB1-KO ESCs formed teratomas with Tuj1+ cells distant from the neural tube-like structures, which were rarely observed in wild-type ESC-derived teratomas. Despite the lack of neuronal morphology in Tuj1+ cells within KO teratomas, this result supports the role of RB in proper cell migration during human brain development. This ectopic migration, in addition to the cell proliferation increase mentioned above, could explain the macrocephaly described in individuals who lack RB (Mitter et al., 2011; Rodjan et al., 2010). Clearly, more questions remain, such as whether VLDLR is strictly required for the ectopic migration observed in RB1-KO organoids. Although we treated wild-type and RB1-KO organoids with an anti-reelin antibody or with the receptor-associated protein (RAP), which blocks VLDLR, among others, we did not detect a clear effect in neuronal migration (data not shown). However, with these experiments, we could not rule out completely that the reelin pathway might be involved in the phenotype observed in RB1-KO organoids. We also do not know if the source of ectopic neurons could come from different tubule structures in the same organoids. Studies using organoid protocols to minimize organoid heterogeneity and variability (Pasca et al., 2015; Qian et al., 2016) to examine the effect of RB loss may address this issue.
In summary, our data reveal that RB plays vital roles in cerebral organoids. Lack of RB produces an increase in neuroblast proliferation, and NSC and neuronal death. In addition, RB deficiency causes aberrant neuronal migration. Our data demonstrate that human cerebral organoids are a powerful tool with which to study human brain development in a dish and opens up future avenues to model RB dysfunction in human neurological disease.
MATERIALS AND METHODS
Culture of wild-type and RB1-KO human ESCs
Human H9 ESC line (WA09, WiCell Research Institute, Madison, WI, USA) was obtained from WiCell at passage 15 with verified normal karyotype and contamination free. H9 ESCs were cultivated in mTeSRTM1 medium (catalog # 05851, Stemcell Technologies) based on feeder-free culture protocols on six-well plates (catalog # 3506, Corning) coated with growth factor-reduced Matrigel (catalog # 356230, BD Biosciences) and adding ROCK inhibitor (final concentration 10 µM, catalog # S-1049, Selleck Chemicals). The cells were maintained with daily medium change without ROCK inhibitor until they reached about 70% confluency. Then, they were detached using versene solution (catalog # 15040-066, Thermo Fisher Scientific) and seeded. RB1-KO human H9 ESCs were generated using CRISPR/Cas9 (Ran et al., 2013) and two different clones were obtained. Details regarding the generation of RB1-KO human ESCs will be published elsewhere (J.W.S., unpublished).
Neurosphere induction
We induced neurospheres from human ESCs after EB formation as previously described with slight modifications (Zhang and Zhang, 2010). Human ESCs were detached using versene and accutase (catalog # A6964, Sigma), dissociated into single cells and suspended at 60 cells/µl in 150 µl of the human EB medium (DMEM/F12, 3% FBS, 20% knockout serum replacement, 1× non-essential amino acids, 1× Glutamax, and 2-mercaptoethanol) in 96-well clear round-bottom ultra low attachment microplates (catalog #7007, Corning). The medium was supplemented with ROCK inhibitor (final concentration 50 μM) and basic FGF (final concentration 4 ng/ml), and half of medium was replaced after 48 h. After 4 days, individual EBs were transferred to individual wells in 24-well ultra-low attachment multiple well plates (catalog # 3473, Corning), filled with 500 µl of neural induction medium (NIM), composed of DMEM-F12 with 1× N2 supplement (catalog # 17502048, Thermo Fisher Scientific), 1× Glutamax, 1× non-essential amino acids and heparin (1 μg/ml). After 3 days of culture in NIM, cell clusters were transferred to 6-well plates and attached to the bottom of wells coated with laminin (catalog # 23017, Invitrogen Life Technologies) and filled with 2 ml of NIM. After 8 days, the formed neural rosettes were detached and the cells were cultured in suspension condition in NIM containing B27 supplement lacking vitamin A (catalog # 12587010, Thermo Fisher Scientific) and half of the medium was replaced every other day. After 6 days, neurospheres were counted and the diameter was measured before passaging or subsequent analyses.
Human cerebral organoid induction
Human ESCs were detached and subjected to EB induction using the same protocol as described for neurosphere induction. After 4 days, half of media were replaced by human EB medium without ROCK inhibitor and basic FGF. After 2 days, EBs were transferred into NIM, similar to that used for neurosphere culture, and embedded in matrigel after 5 days. Organoids were induced following previously published methods using an orbital shaker (Lancaster et al., 2013; Lancaster and Knoblich, 2014a). The organoid size was analyzed by measuring the area using ImageJ software. Some cerebral organoids were treated by BrdU (Sigma # B9285) at a final concentration of 30 µM every 2 hours and fixed 10 h after first BrdU treatment.
PI incorporation analysis
To visualize dead or dying cells in wild-type and RB-KO organoids at 28 DIV, live organoids were incubated with PI (catalog # P-4864-10 Ml, Sigma) at a final concentration of 2.5 µg/ml for 10 min and analyzed immediately by fluorescence microscopy under a 2× objective lens.
Cryosectioning and immunostaining of organoids
Each human cerebral organoid was fixed in 4% paraformaldehyde overnight at 4°C, dehydrated by 30% sucrose in PBS and embedded in OCT Compound (catalog # 23-730-571, Thermo Fisher Scientific). Cryostat sections (14 µm) were cut and mounted onto slides (Thermo Fisher Scientific). Mounted sections were incubated for 1 h at room temperature with blocking solution (3% normal donkey serum+0.3% Triton X-100 in TBS) and incubated with primary antibodies (Table S1) diluted in blocking solution overnight at 4°C. After three washes with TBS, corresponding fluorophore-conjugated secondary antibodies (Table S2) diluted in the blocking solution were added and incubated for 2 h at room temperature and followed by DAPI staining. Finally, stained slides were rinsed by TBS three times, mounted and analyzed using a Nikon A1R confocal microscope equipped with four laser lines (405, 488, 561 and 633 nm) under 20× and 40× objective lenses. Serial Z-stack images were obtained and collapsed to obtain a maximum intensity projection of lines. BrdU-treated organoids were incubated with 2 N HCl for 30 min at RT and then with 0.1 M sodium borate (pH 8.5) for 10 min at room temperature. After rinsing with TBS three times, standard immunostaining were performed as described above.
Quantitative analysis of immunostained sections
To determine the type of cells that expressed RB at 15 and 28 DIV, we counted all the RB+ cells in two randomly selected fields (318.2 µm×318.2 µm each) in two immunostained slices per organoid and calculated the percentage of marker-positive cells out of the total RB+ cells. To analyze the number of Ki67+, Ki67+/Sox2+, BrdU+, BrdU+/Sox2+, BrdU+/Pax6+, BrdU+/DCX+, AC3+, AC3+/Sox2+ or AC3+/DCX+ cells at 15 and 28 DIV, we randomly selected 10 fields (46.6 µm×46.6 µm each) in two immunostained slices per organoid and calculated the percentage of marker-positive cells out of the total number of DAPI+ or Sox2+ cells. To analyze the number of AC3+/Tuj1+ in organoids at 28 DIV, we randomly selected 10 fields (318.2 µm×318.2 µm each) in two immunostained slices per organoid and calculated the percentage of AC3+ cells out of the total Tuj1+ cells. To estimate the area of Tuj1+ or Vimentin+ cells in or around or outside the tubular region, we randomly selected four fields (1254.4 µm×940.8 µm for Tuj1 and 633.17 µm×633.17 µm for Vimentin) in four immunostained slices per organoid. We used ImageJ software to trace Tuj1+ or Vimentin+ areas within each field and calculated percentage of marker-positive area. The area within 60 µm of the four borders of each picture was excluded from quantification as it could be in or around other tubes outside the picture. Within each field, if the traced marker-positive area was found less than 20 µm from the outer DAPI+ border of an identified tubular region or within the tubular region, we defined this marker-positive area as in or around the tubular region. If the marker-positive area was found more than 20 µm from the outer DAPI+ border of an identified tubular region, we defined this marker-positive area as outside the tubular region. If marker-positive areas were found in a field that lacked visible tubular regions, we defined this area as outside the tubular region. The number of organoids used for marker quantification is included in the figure legends.
Cell cycle analysis
Organoids or neurospheres were dissociated to single cells by treatment with accutase for 10 min and fixed in 70% ethanol overnight. After washing with PBS, the samples were incubated for 30 min with RNase A (200 µg/ml), PI (20 µg/ml) and X-100 Triton (0.01%) in PBS and their DNA content was analyzed by flow cytometer (BD FACSAria) with 10,000 events per determination. Cell cycle profile was generated using Flowjo software (Tree Star).
RNA isolation, RT-PCR and quantitative PCR
Organoid RNA was extracted according to the protocol supplied with TRIzol reagent (catalog # 15596018, Thermo Fisher Scientific). The concentration and purity of the RNA samples were measured using Nano-drop (Thermo Fisher Scientific). The extracted RNA (400 ng) was reverse transcribed according to the protocol supplied with SuperScriptIII First-Strand Synthesis System for RT-PCR (catalog # 18080-051, Invitrogen Life Technologies). Quantitative real-time PCR (qRT-PCR) was carried out using ViiA (Applied Biosystems). Reactions were run in triplicate and expression of each gene was normalized to the geometric mean of GAPDH as a housekeeping gene and analyzed by using the ΔΔCT method. The primer sequences of each gene are listed in Table S3.
Western blot
Proteins were extracted using RIPA buffer with protease inhibitor (Roche). After quantification, proteins were resolved on 4-12% SDS-PAGE gels and transferred to a PVDF membrane. The membrane was incubated with primary antibodies (Table S1) after blocking for 1 h with 5% BSA/TBS-Tween. Immunoreactive bands were visualized by densitometry using HRP-conjugated secondary antibodies.
Teratoma formation
For teratoma formation, H9 wild-type and RB1-KO human ESCs were dissociated with versene, centrifuged at 1000 g for 4 min, resuspended in 200 μl of cold growth-factor-reduced Matrigel. Cells (5×105) were injected subcutaneously into NOD-SCID mice (NOD.CB17–Prkdcscid/NcrCrl strain code 394, Charles River). After 5-6 weeks, teratomas were removed, washed in PBS, cut into slices with a thickness of ≈5 mm and fixed in 10% neutral buffered formalin. After fixation, tissue slices were embedded in paraffin wax and sectioned by the Molecular Pathology Core of UT Southwestern Medical Center. The sections were then immunostained and analyzed by confocal microscopy in a manner similar to the organoid sections.
Statistical analysis
An unpaired two-tailed Student's t-test was used to compare the mean±s.e.m. values from the wild-type and RB1-KO cells with Welch's correction when the F-test indicated significant differences between the variances of both groups. Owing to the high variability of the results obtained from different cultures of neurospheres, we used paired two-tailed Student's t-test to compare the mean±s.e.m. values from the wild-type and RB1-KO cells. All analyses were carried out with GraphPad Prism software (version 5.0) and the differences were considered statistically significant when P<0.05.
Acknowledgements
We thank Becky Brulet for helpful advice on immunostaining, Joachim Herz for the gift of the RAP inhibitor and the anti-reelin antibody, and Jose Cabrera for graphical support. We also thank Sean S. Goetsch for help with RB-KO human ESC design and development, and teratoma studies.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
T.M., V.N.-E. and J.H. designed experiments and wrote the manuscript. T.M., V.N.-E. and S.K. performed and analyzed experiments. S.K. and J.W.S. generated the RB1-KO human ESCs using CRISPR/Cas9. All of the authors edited and provided comments on the manuscript.
Funding
This work was supported by grants from the National Institutes of Health (R01NS093992, R01NS089770, R01NS081203, R21NS090926 and K02AG041815 to J.H.; U01HL100401 and U54HD087351 to J.W.S.), the American Heart Association (15GRNT25750034 to J.H.), the U.S. Department of Defense (W81XWH-15-1-0399 to J.H.), the Texas Institute for Brain Injury and Repair, University of Texas Southwestern Medical Center (to J.H.) and the Cancer Prevention Research Institute of Texas (RP110486 to J.W.S.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.143636.supplemental
References
- Andrusiak M. G., Mcclellan K. A., Dugal-Tessier D., Julian L. M., Rodrigues S. P., Park D. S., Kennedy T. E. and Slack R. S. (2011). Rb/E2F regulates expression of neogenin during neuronal migration. Mol. Cell. Biol. 31, 238-247. 10.1128/MCB.00378-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunne B., Franco S., Bouché E., Herz J., Howell B. W., Pahle J., Müller U., May P., Frotscher M. and Bock H. H. (2013). Role of the postnatal radial glial scaffold for the development of the dentate gyrus as revealed by Reelin signaling mutant mice. Glia 61, 1347-1363. 10.1002/glia.22519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen L., Carlson B. L., Schroeder M. A., Ostrem J. L., Kitange G. J., Mladek A. C., Fink S. R., Decker P. A., Wu W., Kim J.-S. et al. (2012). p16-Cdk4-Rb axis controls sensitivity to a cyclin-dependent kinase inhibitor PD0332991 in glioblastoma xenograft cells. Neuro Oncol. 14, 870-881. 10.1093/neuonc/nos114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Tian Z., Sun R., Wang Z., Shen J., Shan Z., Jin L. and Lei L. (2011). ApoER2 and VLDLR in the developing human telencephalon. Eur. J. Paediatr. Neurol. 15, 361-367. 10.1016/j.ejpn.2011.03.011 [DOI] [PubMed] [Google Scholar]
- Christie K. J., Krishnan A., Martinez J. A., Purdy K., Singh B., Eaton S. and Zochodne D. (2014). Enhancing adult nerve regeneration through the knockdown of retinoblastoma protein. Nat. Commun. 5, 3670 10.1038/ncomms4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classon M. and Harlow E. (2002). The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer 2, 910-917. 10.1038/nrc950 [DOI] [PubMed] [Google Scholar]
- Conklin J. F., Baker J. and Sage J. (2012). The RB family is required for the self-renewal and survival of human embryonic stem cells. Nat. Commun. 3, 1244 10.1038/ncomms2254 [DOI] [PubMed] [Google Scholar]
- Drew L. J., Kheirbek M. A., Luna V. M., Denny C. A., Cloidt M. A., Wu M. V., Jain S., Scharfman H. E. and Hen R. (2016). Activation of local inhibitory circuits in the dentate gyrus by adult-born neurons. Hippocampus 26, 763-778. 10.1002/hipo.22557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M., Takata N., Ishibashi H., Kawada M., Sakakura E., Okuda S., Sekiguchi K., Adachi T. and Sasai Y. (2011). Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472, 51-56. 10.1038/nature09941 [DOI] [PubMed] [Google Scholar]
- Ferguson K. L., Mcclellan K. A., Vanderluit J. L., Mcintosh W. C., Schuurmans C., Polleux F. and Slack R. S. (2005). A cell-autonomous requirement for the cell cycle regulatory protein, Rb, in neuronal migration. EMBO J. 24, 4381-4391. 10.1038/sj.emboj.7600887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend S. H., Bernards R., Rogelj S., Weinberg R. A., Rapaport J. M., Albert D. M. and Dryja T. P. (1986). A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 323, 643-646. 10.1038/323643a0 [DOI] [PubMed] [Google Scholar]
- Ghanem N., Andrusiak M. G., Svoboda D., Al Lafi S. M., Julian L. M., Mcclellan K. A., De Repentigny Y., Kothary R., Ekker M., Blais A. et al. (2012). The Rb/E2F pathway modulates neurogenesis through direct regulation of the Dlx1/Dlx2 bigene cluster. J. Neurosci. 32, 8219-8230. 10.1523/JNEUROSCI.1344-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacinti C. and Giordano A. (2006). RB and cell cycle progression. Oncogene 25, 5220-5227. 10.1038/sj.onc.1209615 [DOI] [PubMed] [Google Scholar]
- Hansen D. V., Lui J. H., Parker P. R. L. and Kriegstein A. R. (2010). Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554-561. 10.1038/nature08845 [DOI] [PubMed] [Google Scholar]
- Hoerder-Suabedissen A. and Molnár Z. (2015). Development, evolution and pathology of neocortical subplate neurons. Nat. Rev. Neurosci. 16, 133-146. 10.1038/nrn3915 [DOI] [PubMed] [Google Scholar]
- Hsieh J., Aimone J. B., Kaspar B. K., Kuwabara T., Nakashima K. and Gage F. H. (2004). IGF-I instructs multipotent adult neural progenitor cells to become oligodendrocytes. J. Cell Biol. 164, 111-122. 10.1083/jcb.200308101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julian L. M., Liu Y., Pakenham C. A., Dugal-Tessier D., Ruzhynsky V., Bae S., Tsai S.-Y., Leone G., Slack R. S. and Blais A. (2016). Tissue-specific targeting of cell fate regulatory genes by E2f factors. Cell Death Differ. 23, 565-575. 10.1038/cdd.2015.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoshima T., Sakaguchi H., Nakano T., Soen M., Ando S., Eiraku M. and Sasai Y. (2013). Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. USA 110, 20284-20289. 10.1073/pnas.1315710110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelava I. and Lancaster M. A. (2016). Stem cell models of human brain development. Cell Stem Cell 18, 736-748. 10.1016/j.stem.2016.05.022 [DOI] [PubMed] [Google Scholar]
- Lancaster M. A. and Knoblich J. A. (2014a). Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 9, 2329-2340. 10.1038/nprot.2014.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M. A. and Knoblich J. A. (2014b). Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345, 1247125 10.1126/science.1247125 [DOI] [PubMed] [Google Scholar]
- Lancaster M. A., Renner M., Martin C.-A., Wenzel D., Bicknell L. S., Hurles M. E., Homfray T., Penninger J. M., Jackson A. P. and Knoblich J. A. (2013). Cerebral organoids model human brain development and microcephaly. Nature 501, 373-379. 10.1038/nature12517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod K. F., Hu Y. and Jacks T. (1996). Loss of Rb activates both p53-dependent and independent cell death pathways in the developing mouse nervous system. EMBO J. 15, 6178-6188. [PMC free article] [PubMed] [Google Scholar]
- Macpherson D., Sage J., Crowley D., Trumpp A., Bronson R. T. and Jacks T. (2003). Conditional mutation of Rb causes cell cycle defects without apoptosis in the central nervous system. Mol. Cell. Biol. 23, 1044-1053. 10.1128/MCB.23.3.1044-1053.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcclellan K. A. and Slack R. S. (2006). Novel functions for cell cycle genes in nervous system development. Cell Cycle 5, 1506-1513. 10.4161/cc.5.14.2980 [DOI] [PubMed] [Google Scholar]
- Mcclellan K. A., Ruzhynsky V. A., Douda D. N., Vanderluit J. L., Ferguson K. L., Chen D., Bremner R., Park D. S., Leone G. and Slack R. S. (2007). Unique requirement for Rb/E2F3 in neuronal migration: evidence for cell cycle-independent functions. Mol. Cell. Biol. 27, 4825-4843. 10.1128/MCB.02100-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitter D., Ullmann R., Muradyan A., Klein-Hitpass L., Kanber D., Õunap K., Kaulisch M. and Lohmann D. (2011). Genotype-phenotype correlations in patients with retinoblastoma and interstitial 13q deletions. Eur. J. Hum. Genet. 19, 947-958. 10.1038/ejhg.2011.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalam R. L., Andreu-Vieyra C., Braun R. E., Akiyama H. and Matzuk M. M. (2009). Retinoblastoma protein plays multiple essential roles in the terminal differentiation of Sertoli cells. Mol. Endocrinol. 23, 1900-1913. 10.1210/me.2009-0184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naser R., Vandenbosch R., Omais S., Hayek D., Jaafar C., Al Lafi S., Saliba A., Baghdadi M., Skaf L. and Ghanem N (2016). Role of the Retinoblastoma protein, Rb, during adult neurogenesis in the olfactory bulb. Sci. Rep. 6, 20230 10.1038/srep20230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca A. M., Sloan S. A., Clarke L. E., Tian Y., Makinson C. D., Huber N., Kim C. H., Park J.-Y., O'rourke N. A., Nguyen K. D. et al. (2015). Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 12, 671-678. 10.1038/nmeth.3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Nguyen H. N., Song M. M., Hadiono C., Ogden S. C., Hammack C., Yao B., Hamersky G. R., Jacob F., Zhong C. et al. (2016). Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165, 1238-1254. 10.1016/j.cell.2016.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A. and Zhang F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281-2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodjan F., De Graaf P., Moll A. C., Imhof S. M., Verbeke J. I. M. L., Sanchez E. and Castelijns J. A. (2010). Brain abnormalities on MR imaging in patients with retinoblastoma. AJNR Am. J. Neuroradiol. 31, 1385-1389. 10.3174/ajnr.A2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa-García B., Gunduz V., Vázquez-Rivera V., Cress W. D., Wright G., Bian H., Hinds P. W. and Santiago-Cardona P. G. (2010). A role for the retinoblastoma protein as a regulator of mouse osteoblast cell adhesion: implications for osteogenesis and osteosarcoma formation. PLoS ONE 5, e13954 10.1371/journal.pone.0013954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira C. M., Kron M. M., Masachs N., Zhang H., Lagace D. C., Martinez A., Reillo I., Duan X., Bosch C., Pujadas L. et al. (2012). Cell-autonomous inactivation of the reelin pathway impairs adult neurogenesis in the hippocampus. J. Neurosci. 32, 12051-12065. 10.1523/JNEUROSCI.1857-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbosch R., Clark A., Fong B. C., Omais S., Jaafar C., Dugal-Tessier D., Dhaliwal J., Lagace D. C., Park D. S., Ghanem N. et al. (2016). RB regulates the production and the survival of newborn neurons in the embryonic and adult dentate gyrus. Hippocampus 26, 1379-1392. 10.1002/hipo.22613 [DOI] [PubMed] [Google Scholar]
- Yu Y., Ren Q.-G., Zhang Z.-H., Zhou K., Yu Z.-Y., Luo X. and Wang W. (2012). Phospho-Rb mediating cell cycle reentry induces early apoptosis following oxygen-glucose deprivation in rat cortical neurons. Neurochem. Res. 37, 503-511. 10.1007/s11064-011-0636-6 [DOI] [PubMed] [Google Scholar]
- Zhang X.-Q. and Zhang S.-C. (2010). Differentiation of neural precursors and dopaminergic neurons from human embryonic stem cells. Methods Mol. Biol. 584, 355-366. 10.1007/978-1-60761-369-5_19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.-C., Wernig M., Duncan I. D., Brüstle O. and Thomson J. A. (2001). In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 19, 1129-1133. 10.1038/nbt1201-1129 [DOI] [PubMed] [Google Scholar]
- Zhang J., Gray J., Wu L., Leone G., Rowan S., Cepko C. L., Zhu X., Craft C. M. and Dyer M. A. (2004). Rb regulates proliferation and rod photoreceptor development in the mouse retina. Nat. Genet. 36, 351-360. 10.1038/ng1318 [DOI] [PubMed] [Google Scholar]