Abstract
We investigated the molecular bases for resistance to several classes of herbicides that bind tubulins in green foxtail (Setaria viridis L. Beauv.). We identified two α- and two β-tubulin genes in green foxtail. Sequence comparison between resistant and sensitive plants revealed two mutations, a leucine-to-phenylalanine change at position 136 and a threonine-to-isoleucine change at position 239, in the gene encoding α2-tubulin. Association of mutation at position 239 with herbicide resistance was demonstrated using near-isogenic lines derived from interspecific pairings between green foxtail and foxtail millet (Setaria italica L. Beauv.), and herbicide sensitivity bioassays combined with allele-specific PCR-mediated genotyping. Association of mutation at position 136 with herbicide resistance was demonstrated using herbicide sensitivity bioassays combined with allele-specific PCR-mediated genotyping. Both mutations were associated with recessive cross resistance to dinitroanilines and benzoic acids, no change in sensitivity to benzamides, and hypersensitivity to carbamates. Using three-dimensional modeling, we found that the two mutations are adjacent and located into a region involved in tubulin dimer-dimer contact. Comparison of three-dimensional α-tubulin models for organisms with contrasted sensitivity to tubulin-binding herbicides enabled us to propose that residue 253 and the vicinity of the side chain of residue 251 are critical determinants for the differences in herbicide sensitivity observed between organisms, and that positions 16, 24, 136, 239, 252, and 268 are involved in modulating sensitivity to these herbicides.
Microtubules are cytoskeletal polymers essential for the survival of all eukaryotes. They are crucial for mitosis, cell transport, and cell motility. Microtubules are primarily made of repeated α,β-tubulin heterodimers that bind head to tail to form linear protofilaments. The dynamic character of microtubules is essential for their functions (for review, see Nogales, 1999). In plants, microtubules construct four arrays that are crucial for cell cycle completion (for review, see Wasteneys, 2002). α- and β-tubulins are among the most conserved proteins throughout evolution. In all multicellular eukaryotes, α- and β-tubulins are encoded by multigene families. These give rise to various tubulin isotypes, which are differentially expressed and modified during growth and development (e.g. Carpenter et al., 1992).
Given their role in key cell processes, microtubules turned out to be the target site for a number of drugs such as antitumor agents, fungicides, and herbicides. These drugs most often display different affinities for tubulins from metazoan, animal, algal, and plant cell (for review, see Anthony and Hussey, 1999). This allowed the relatively safe use of tubulin-binding herbicides in the field. A number of molecules, representing various chemical classes, has thus been developed and used as herbicides. They consist of dinitroanilines (e.g. trifluralin), benzoic acids (e.g. chlorthal-dimethyl), phosphoroamidates (e.g. amiprophos-methyl), pyridines (e.g. dithiopyr), benzamides (e.g. pronamide), and carbamates (e.g. chlorpropham). These herbicides have a severe effect on shoot and root elongation and development, causing both shoots and roots to be stunted with a characteristic swollen tip (Anthony and Hussey, 1999). Among these herbicides, dinitroanilines and phosphoroamidates have been shown to act by binding to the same site(s) on the α,β-tubulin dimer (Murthy et al., 1994; Blume et al., 2003). The incorporation of dimer herbicide complexes into growing tubulin protofilaments was proposed to cause microtubule disruption (Hugdahl and Morejohn, 1993). The mode of action of carbamate and benzamide herbicides, although distinct from that of dinitroanilines (Vaughan and Vaughn, 1987; Eleftheriou and Bekiari, 2000), remains fairly less well understood.
The repeated use of one class of herbicides with a single target site generally results in the selection of resistant weed populations. Interestingly, in spite of the use of tubulin-inhibiting herbicides since the end of the 1970s, only a few weed species evolved resistance toward these molecules (Anthony and Hussey, 1999). Furthermore, resistance has been shown to be controlled by a single, nuclear, recessive gene in the only two species investigated so far: goosegrass (Eleusine indica L. Gaertn.; Zeng and Baird, 1997, 1999) and green foxtail (Setaria viridis L. Beauv.; Jasieniuk et al., 1994). Both species are highly selfing. Resistance to tubulin-inhibiting herbicides, and especially dinitroanilines, has been extensively studied in a single higher plant: goosegrass. Two distinct categories of resistant mutants, named R (resistant) and I (intermediate, moderately resistant), have been identified in this weed (for review, see Anthony and Hussey, 1999). The molecular basis for resistance to dinitroanilines was found to be a Thr-239-Ile change in the α-tubulin TUA1 gene in the R mutant (Anthony et al., 1998; Yamamoto et al., 1998) and a Met-268-Thr change in the same gene in the I mutant (Yamamoto et al., 1998). In spite of comparison with the known three-dimensional structure of the α,β-tubulin dimer from pig obtained by Nogales et al. (1998), and although a recent modeling study proposed a binding site for tubulin-inhibiting herbicides in plants (Blume et al., 2003), the details of the interaction of tubulin-inhibiting herbicides with plant tubulins remain incompletely understood.
In an effort to enhance our knowledge of herbicide-tubulin interactions, we investigated the molecular basis of resistance to dinitroanilines in green foxtail. We herein report the full sequences of four genes encoding α- and β-tubulin in green foxtail. We identified single amino acid changes at positions 136 and 239 in the α2-tubulin gene, which are involved in herbicide resistance. Using protein modeling, we discuss the structural consequences of these changes for tubulin herbicide interactions.
RESULTS
Herbicide Sensitivity Assay
To assess the effect of tubulin-inhibiting herbicides, we focused on the shoots of foxtail seedlings, which were reported to display a greater sensitivity to dinitroanilines than the roots (Waldin et al., 1992). Foxtail millet (Setaria italica L. Beauv.) lines D97-3 (resistant to trifluralin) and D97-4 (sensitive to trifluralin) showed uniform shoot emergence. Both lines exhibited a dose-dependent inhibition of shoot elongation for all eight herbicides tested (data not shown). As judged by the values of herbicide concentrations inhibiting 50% shoot elongation (IC50), D97-3 and D97-4 seedlings exhibited identical sensitivity to the benzamide herbicide pronamide (Table I). D97-3 seedlings displayed a 2.1- to 7.7-fold resistance to all four dinitroanilines tested and to the benzoic acid chlorthal-dimethyl. By contrast, D97-3 seedlings were about twice as susceptible to the two carbamate herbicides tested as D97-4 seedlings (Table I) and were thus considered to be hypersensitive. Full-shoot growth inhibition could never be obtained for seedlings from either lines with chlorthal-dimethyl, most likely because of the limited aqueous solubility limit of this compound, which prevented concentrations higher than 15 to 20 μm being achieved.
Table I.
Differential sensitivity of foxtail lines, cultivars, and populations to tubulin-inhibiting herbicides
| Herbicides
|
Lines (Foxtail Millet)
|
D97-3/D97-4a
|
Discriminating Dose
|
Populations (Green Foxtail)
|
Cultivars (Foxtail Millet)
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| D97-4 | D97-3 | Oak River | Deloraine | D96-174 | Ouges99 | Amende4 | Jigu11 | |||
| μm | ||||||||||
| Benfluralin | 50 Sb | 50 R | 6.9 | 0.2 | 50 R | 48 R, 2 S | 50 S | 50 S | 50 S | 50 S |
| Butralin | 50 S | 50 R | 2.1 | 0.5 | 50 R | 47 R, 3 S | 50 S | 50 S | 50 S | 50 S |
| Pendimethalin | 50 S | 50 R | 3.7 | 0.4 | 50 R | 46 R, 4 S | 50 S | 50 S | 50 S | 50 S |
| Trifluralin | 50 S | 50 R | 7.7 | 0.2 | 50 R | 48 R, 2 S | 50 S | 50 S | 50 S | 50 S |
| Chlorthal-dimethyl | 50 S | 50 R | 4.6 | 5.0 | 50 R | 47 R, 3 S | 50 S | 50 S | 50 S | 50 S |
| Pronamide | 50 S | 50 S | 1.0 | 6.0 | 50 S | 50 S | 50 S | 50 S | 50 S | 50 S |
| Carbetamide | 50 S | 50 HS | 0.4 | 10.0 | 50 HS | 46 HS, 4 S | 50 S | 50 S | 50 S | 50 S |
| Chlorpropham | 50 S | 50 HS | 0.5 | 3.5 | 50 HS | 48 HS, 2 S | 50 S | 50 S | 50 S | 50 S |
For each line, cultivar or population, 50 seedlings were assayed per herbicide.
Ratio of IC50 for line D97-3 to IC50 for line D97-4.
S, Sensitive; R, resistant; HS, hypersensitive.
For all herbicides but pronamide, doses representing one-half the IC50 value for the seedlings from the most resistant line were found to adequately enable discrimination of resistant and sensitive seedlings on the basis of shoot length and morphology. These doses were used to assess the sensitivity of the four green foxtail populations and of the two millet lines (Table I). Population Oak River displayed a homogenous resistance pattern identical to that of line D97-3. Population Deloraine appeared to be heterogeneous, with about 95% seedlings displaying a resistance pattern identical to that of line D97-3, and 5% seedlings displaying a resistance pattern identical to that of line D97-4. The remaining lines and populations displayed a homogenous resistance pattern identical to that of line D97-4 (Table I). No differences in growth inhibition between seedlings from the different populations, cultivars, and lines were observed using pronamide doses representing the IC50 value (6 μm) obtained for lines D97-3 and D97-4.
Foxtail Tubulin Genes
Two α- and two β-tubulin genes were isolated from a D96-174 green foxtail plant using degenerate and inverse PCR. A single sequence was obtained for each of the four genes, showing that this plant was homozygous at each of the four gene loci.
The two β-tubulin genes consisted of open reading frames of 1,861 (β1) and 1,522 (β2) bp that both encoded a polypeptide of 448 amino acids. Both coding sequences were interrupted by two introns located between the first and second nucleotides in codons 132 and 222, two positions highly conserved in higher plants (Snustad et al., 1992). The first intron was 404 bp long in gene β1 and 81 bp long in gene β2, thus explaining the amplification of two distinct DNA fragments using β-tubulin-specific, degenerate PCR primers. The two remaining introns were each about 100 bp long. Among all tubulin sequences in the GenBank/EMBL/DNA Data Bank of Japan (DDBJ) database, the β1 and β2 proteins were most similar to two rice tubulins, accessions D30716 (98.8% identity with β1) and NM187634 (97.9% identity with β2). β1 and β2 proteins were 95.7% identical.
The two α-tubulin genes consisted of open reading frames of 2,333 (α1) and 2,502 (α2) bp that encoded polypeptides of 450 and 451 amino acids, respectively. Although intron positions vary in α-tubulin genes, even within a given species (Kopczak et al., 1992) both coding sequences were interrupted by two introns located before the first nucleotide in codons 31 and 233 and by an intron located between the first and second nucleotides in codon 110. The intron before codon 31 was 762 and 946 bp long in genes α1 and α2, respectively, while all other introns were about 100 bp long. Among all tubulin sequences in the GenBank/EMBL/DDBJ database, the α1 protein was most similar to maize α3-tubulin (accession M60171; 94.8% identity), while the α2 protein was most similar to goosegrass TUA1 protein (accession AF008120; 99.7% identity). α1 and α2 proteins were 92.6% identical.
For all four genes, intron excision was verified using reverse transcription-PCR with gene-specific primer pairs (Supplemental Table I, available at www.plantphysiol.org) followed by sequencing. All sequences have been deposited in the GenBank/EMBL/DDBJ database under accession numbers AJ586804 (α1), AJ586805 (α2), AJ586806 (β1), and AJ586807 (β2).
Identification of Nonsynonymous Substitutions in Tubulin Sequences
The full coding sequences of the two α- and β-tubulin genes, including introns, were then determined for one trifluralin-resistant plant each from populations Oak River and Deloraine (green foxtail) and from line D97-3, and for one trifluralin-sensitive foxtail plant from population Ouges99 (green foxtail), cultivars Amende4 and Jigu11 (foxtail millet) and line D97-4. All plants contained two identical copies for each of the four genes. A total of eight sequences for each gene were thus available for comparison. In the following text, the reference sequence for each of the four genes is the sequence from the green foxtail plant from population D96-174. All nucleotide and amino acid positions referred to in this paper correspond to those in these sequences.
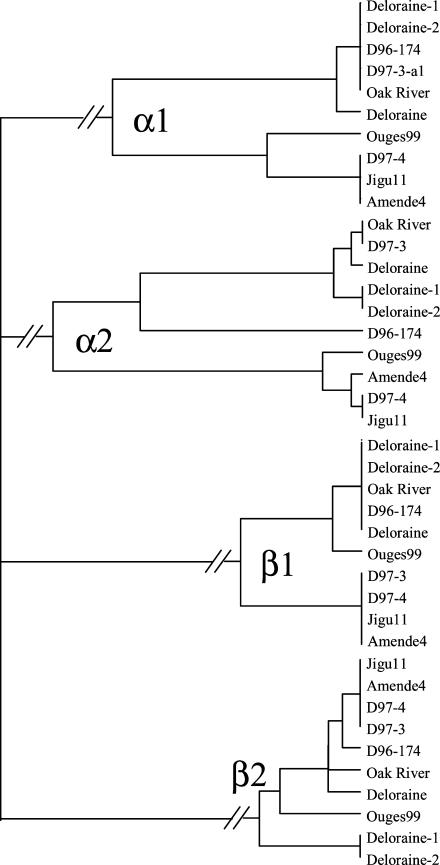
The alignments of β1- and β2-tubulin sequences were 1,862 and 1,522 bp long, respectively. The eight β1 and the eight β2 sequences comprised a total of three and five haplotypes, respectively (Fig. 1). No nonsynonymous single-nucleotide polymorphisms (SNPs) were identified among these sequences. Trifluralin-resistant and sensitive plants from lines D97-3 and D97-4 contained the same β1 and β2-tubulin haplotypes as the two plants from foxtail millet parental cultivars.
Figure 1.
Phylogenetic tree of foxtail α- and β-tubulin sequences, including introns. The tree was constructed using the maximum parsimony method as implemented in the Mega2.1 software (www.megasoftware.net).
The alignment of α1-tubulin sequences was 2,333 bp long. The eight α1 sequences comprised a total of four haplotypes (Fig. 1). A single nonsynonymous change was recorded. It was caused by a T-to-G transversion at the last position in codon 443, resulting in an Asp-to-Glu change in the resistant plant from population Deloraine and in all sensitive plants but that from population D96-174. Because of its presence in both resistant and sensitive plants, this Asp-to-Glu change was unlikely to be involved in resistance to tubulin-inhibiting herbicides in green foxtail.
The alignment of α2-tubulin sequences was 2,501 bp long. The eight α2 sequences comprised a total of six haplotypes (Fig. 1). A single nonsynonymous change was recorded. It was caused by a C-to-T transition at the second position in codon 239, resulting into a Thr-to-Ile change. Thr-239 was found in all sensitive plants, while Ile-239 was found in all resistant plants. The trifluralin-resistant plant from line D97-3 contained the same α1- and α2-tubulin haplotypes as the parental resistant Oak River green foxtail plant. In contrast, the trifluralin-sensitive plant from line D97-4 contained the same α1- and α2-tubulin haplotypes as the parental-sensitive Jigu11 foxtail millet plant.
A Second Nonsynonymous Mutation in Foxtail α2-Tubulin
To check whether the Thr-to-Ile change at position 239 in foxtail α2-tubulin was consistently associated with resistance to herbicides, a bidirectional allele-specific PCR assay simultaneously detecting the Ile-239 and Thr-239 α2-tubulin alleles was used to genotype seedlings after their sensitivity to trifluralin had been assessed using a seed bioassay. Two hundred seedlings from the heterogeneous population Deloraine and 50 seedlings from each of the 7 remaining lines and populations and from each of the 5 F2 hybrid foxtail populations segregating for trifluralin resistance (D00-096, D00-100, D00-109, D00-110, and D00-112) described in “Materials and Methods” were investigated. In total, 800 seedlings were thus genotyped (Table II). All 268 seedlings containing two Ile-239 α2-tubulin alleles were resistant to trifluralin. However, although Ile-239 α-tubulin has been demonstrated to be a recessive-resistance gene in goosegrass (Zeng and Baird, 1997; Anthony et al., 1998; Yamamoto et al., 1998), 5 seedlings containing a single Ile-239 α2-tubulin allele were resistant to trifluralin (Table II). Furthermore, 80 seedlings containing two Thr-239 α2-tubulin alleles were also resistant to trifluralin (Table II). Those 85 seedlings were all issued from population Deloraine. The four tubulin genes from two trifluralin-resistant seedlings in this population that contained two Thr-239 α2-tubulin alleles were thus sequenced. A single haplotype for each of the four genes was found in both seedlings. Their α1- and β1-tubulin haplotypes were identical to haplotypes previously found in other green foxtail seedlings (Fig. 1). Their β2-tubulin haplotype was new and did not contain nonsynonymous SNPs. Their α2-tubulin haplotype contained a single new nonsynonymous SNP. It was a C-to-T transition at the first position in codon 136, resulting into a Leu-to-Phe change. Residue 136 is Leu in most known α-tubulin sequences, except those from a few protozoans (Met-136), from pig (Ser-136), and from basidiomycetous fungi (Phe-136).
Table II.
Trifluralin sensitivity of and amino acid present at positions 136 and 239 on each copy of the gene encoding α2-tubulin in 800 seedlings from 13 lines, cultivars, or populations
| Codon 136 | Leu/Leu | Leu/Leu | Leu/Leu | Phe/Leu | Phe/Leu | Phe/Phe |
|---|---|---|---|---|---|---|
| Codon 239 | Thr/Thr | Thr/Ile | Ile/Ile | Thr/Ile | Thr/Thr | Thr/Thr |
| Sensitive | 341 | 104 | 0 | 0 | 2 | 0 |
| Resistant | 0 | 0 | 268 | 5 | 0 | 80 |
A bidirectional allele-specific PCR assay simultaneously detecting the Leu-136 and Phe-136 α2-tubulin alleles was therefore developed and used to genotype the same seedlings as above (Table II). Phe-136 α2-tubulin alleles were exclusively found in seedlings from population Deloraine. Resistant seedlings in this population contained two Phe-136, Thr-239 α2-tubulin alleles (80 seedlings), two Leu-136, Ile-239 α2-tubulin alleles (99 seedlings), or one Leu-136, Ile-239 α2-tubulin allele and one Phe-136, Thr-239 α2-tubulin allele (5 seedlings; Table II). From the analysis of the 800 seedlings summarized in Table II, it appeared that all seedlings containing two mutant alleles (353 seedlings) were resistant to trifluralin. All 341 seedlings containing only Leu-136, Thr-239 α2-tubulin alleles were sensitive to trifluralin, as were all 106 seedlings containing a single mutant-α2-tubulin allele.
Herbicide sensitivity bioassays were performed at the discriminating dose with the seven herbicides used in this study using 100 seeds from population Deloraine per herbicide. All seedlings were subsequently analyzed using the two allele-specific PCR assays (Table III). Seedlings containing two Phe-136, Thr-239 α2-tubulin alleles exhibited the same cross-resistance pattern as seedlings containing two Leu-136, Ile-239 α2-tubulin alleles, i.e. resistance to the four dinitroaniline herbicides and to chlorthal-dimethyl, sensitivity to pronamide, and hypersensitivity to the two carbamate herbicides (Table III).
Table III.
Herbicide sensitivity of and amino acid present at positions 136 and 239 on each copy of the gene encoding α2-tubulin in the green foxtail population Deloraine
| Herbicides
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dinitroanilines
|
Benzoic Acid
|
Carbamates
|
|||||||||||||
| Genotype at Codon
|
Benfluralin
|
Butralin
|
Pendimethalin
|
Trifluralin
|
Chlorthal-Dimethyl
|
Carbetamide
|
Chlorpropham
|
||||||||
| 136 | 239 | Ra | S | R | S | R | S | R | S | R | S | HS | S | HS | S |
| Leu/Leu | Thr/Thr | 0 | 6 | 0 | 6 | 0 | 5 | 0 | 6 | 0 | 4 | 0 | 5 | 0 | 6 |
| Leu/Leu | Thr/Ile | 0 | 1 | 0 | 2 | 0 | 1 | 0 | 1 | 0 | 3 | 0 | 2 | 0 | 1 |
| Leu/Leu | Ile/Ile | 49 | 0 | 47 | 0 | 50 | 0 | 48 | 0 | 49 | 0 | 50 | 0 | 48 | 0 |
| Phe/Leu | Thr/Ile | 3 | 0 | 2 | 0 | 2 | 0 | 3 | 0 | 2 | 0 | 2 | 0 | 3 | 0 |
| Phe/Leu | Thr/Thr | 0 | 1 | 0 | 2 | 0 | 1 | 0 | 2 | 0 | 1 | 0 | 2 | 0 | 2 |
| Phe/Phe | Thr/Thr | 40 | 0 | 41 | 0 | 41 | 0 | 40 | 0 | 41 | 0 | 39 | 0 | 40 | 0 |
One hundred seedlings were assayed per herbicide.
R, Resistant; S, sensitive; HS, hypersensitive.
α2-Tubulin Modeling
For a detailed evaluation of the effects of amino acid substitutions at positions 136 and 239 in foxtail α2-tubulin, we reconstructed a three-dimensional model of this molecule using a model built into a 3.5-Å density map obtained by electron crystallography of pig zinc-induced tubulin sheets (Research Collaboratory for Structural Bioinformatics Protein DataBase [PDB; http://www.rcsb.org/pdb] accession no. 1JFF; Löwe et al., 2001) as a template. Because of poor density map resolution in the region located in loop H1-B2, near pig α-tubulin N terminus (Löwe et al., 2001), this template did not enable us to model the α-tubulin region ranging from Met-36 to His-61. The unrefined, 3.7-Å density map-based pig tubulin model (PDB accession no. 1TUB; Nogales et al., 1998) was thus used as a template to generate a first foxtail α2-tubulin three-dimensional model that enabled us to ascertain that loop H1-B2 was not involved in the three-dimensional conformation of α-tubulin region around residues 136 and 239 (data not shown). The refined 3.5-Å density map-based α-tubulin model was then used as a template for modeling because it included the correction of numerous imperfect orientations of amino acid side chains and of several significant steric clashes that were present in the original 3.7-Å model (Löwe et al., 2001).
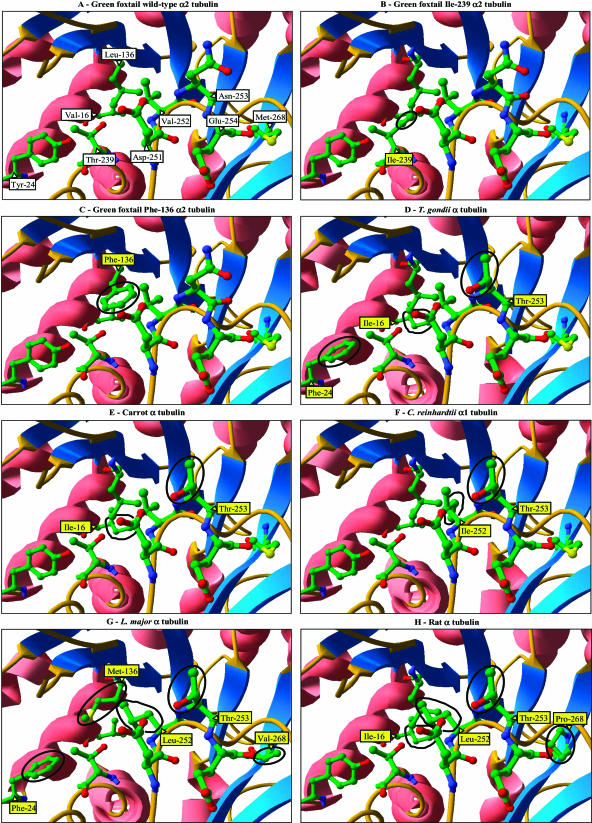
Alignment of primary amino acid sequences of PDB accession 1JFF and foxtail α2-tubulin was visually checked for inconsistency. The sequence similarity between foxtail tubulin and the template sequence was 82.0%, which was largely sufficient to achieve a satisfying model (Schwede et al., 2003). In the three-dimensional model obtained, residues Leu-136 and Thr-239 are adjacent (Fig. 2). They are located within the cavity formed by residues Arg-243, Asp-251, Val-252, and Asn-253, which were proposed to form the binding site for tubulin-inhibiting herbicides in goosegrass (Blume et al., 2003). Residues Leu-136 and Thr-239 are located close to residue Asp-251, which is absolutely conserved in all known α-tubulin sequences from eukaryotes. This residue is crucial for β-α-tubulin interaction during dimer-dimer assembly into tubulin protofilaments (Nogales et al., 1998; Nogales, 1999). Interestingly, residues 136 and 239 are located in the neighborhood of residue Tyr-24, which is involved in resistance to dinitroanilines and phosphoroamidates in the alga Chlamydomonas reinhardtii P.A. Dangeard (James et al., 1993). These two classes of herbicides share common binding site(s) on α,β-tubulin dimers with the dinitroaniline herbicides (Murthy et al., 1994; Blume et al., 2003). Residue Met-268, which is involved in resistance to tubulin-inhibiting herbicides in goosegrass (Yamamoto et al., 1998), is located close to residue Glu-254. This residue, like residue Asp-251, is crucial for dimer-dimer interaction and absolutely conserved in all known α-tubulin sequences from eukaryotes (Nogales et al., 1998; Nogales, 1999).
Figure 2.
Ribbon diagram view of the surface of the region involved in dimer-dimer tubulin contact in three-dimensional models for α-tubulins. Sequences used for modeling are wild type, Leu-136, Thr-239 foxtail α2-tubulin (A); mutant, Leu-136, Ile-239 foxtail α2-tubulin (B); mutant, Phe-136, Thr-239 foxtail α2-tubulin (C); T. gondii α-tubulin (GenBank/EMBL/DDBJ accession no. M20024; D); carrot α-tubulin (GenBank/EMBL/DDBJ accession no. AY007250; E); C. reinhardtii α1-tubulin (GenBank/EMBL/DDBJ accession no. M11447; F); L. major α-tubulin (GenBank/EMBL/DDBJ accession no. AL359683; G); and rat α-tubulin (GenBank/EMBL/DDBJ accession no. V01227; H). On the figure, α-helixes are pink, β-sheets are blue, and the areas with no particular structure are orange. Atom colors are green, C; red, O; blue, N; yellow, S. Side chains of residues not discussed in the text and hydrogen atoms are not shown for clarity. Amino acid positions are labeled at the base of the corresponding side chain. Amino acid positions containing side chains different from those in wild type, Leu-136, Thr-239 foxtail α2-tubulin are in yellow boxes, and the resulting structural differences are circled.
The Leu-to-Phe substitution at position 136 does not alter hydrophobicity at this position (not shown). Yet, it causes an aliphatic chain in Leu-136 that is oriented toward the inside of the tubulin molecule to be replaced with an aromatic group in Phe-136 that protrudes close to the carboxyl group of Asp-251 (Fig. 2C). The Thr-to-Ile substitution at position 239 increases hydrophobicity at this position (not shown) because of the replacement of a hydroxyl group with an aliphatic chain that also protrudes close to the carboxyl group of Asp-251 (Fig. 2C).
Comparison of α-Tubulin Three-Dimensional Structures
In this work, we assessed foxtail sensitivity to herbicides using a whole-seedling-based bioassay. A broad variation in herbicide IC50 values may be observed for the same organism when different kinds of assays are used. The most suitable way of assessing sensitivity to herbicides is an in vitro tubulin herbicide-binding assay. However, few such studies have been published to date, and the results of in vitro assays may vary greatly depending on the binding buffer used (Hugdahl and Morejohn, 1993). Furthermore, we found no demonstration of tubulin-binding herbicide metabolism in the literature. For comparison with our data, we thus considered studies that used whole-organism growth or whole-cell proliferation, in the case of rat (Rattus norvegicus), to measure herbicide effect upon organisms for which α-tubulin sequences are available in databases (Table IV). Ten organisms, which could roughly be classified as highly sensitive (green foxtail and goosegrass), moderately sensitive (tobacco [Nicotiana tabacum], C. reinhardtii, and four protozoan species), highly insensitive (carrot [Daucus carota]), and fully insensitive (rat) to dinitroanilines were considered (Table IV). No α-tubulin sequences were available for the protozoan Tetrahymena thermophila. However, because protozoans seem to lack diversity in their tubulins (Silflow, 1991), a sequence from the closely related species Tetrahymena pyriformis was used for modeling. For each organism, three-dimensional models were reconstructed as described above for each distinct α-tubulin isotype sequence available in the databases. Four models (including the two mutant α2-tubulins) were built for green foxtail, four for tobacco, three for rat and goosegrass, and one for each of the other organisms. We then investigated structural differences in α-tubulin three-dimensional models from the 10 organisms in the region involved in dimer-dimer contact, which contained the two variable amino acid residues identified in green foxtail. All α-tubulin three-dimensional models from a given organism displayed near identical structures in this region (data not shown). This, and the very high similarity observed between α-tubulin models from green foxtail, goosegrass, and tobacco; between those from C. reinhardtii and T. thermophila; and between those from Leishmania major and Trypanosoma brucei are the reason why only eight models are illustrated on Figure 2.
Table IV.
Sensitivity to dinitroanilines of and amino acid present at specific positions in α-tubulins from 10 eukaryotic organisms
| Species
|
Class
|
Range of Dinitroaniline IC50
|
Amino Acid Present at Position
|
References for IC50 values
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 16 | 24 | 136 | 239 | 252 | 253 | 268 | ||||
| Green foxtail | Monocotyledon | 0.02–0.2 μm | Val | Tyr | Leu | Thr | Val | Asn | Met | Waldin et al. (1992); This work |
| Goosegrass | Monocotyledon | 0.02–0.2 μm | Val | Tyr | Leu | Thr | Val | Asn | Met | Waldin et al. (1992) |
| Tobacco | Dicotyledon | 0.9–1.0 μm | Val | Tyr | Leu | Thr | Val | Asn | Met | Anthony et al. (1999) |
| Carrot | Dicotyledon | >10 μm | Ile | Tyr | Leu | Thr | Val | Thr | Met | Vaughan and Vaughn (1988) |
| C. reinhardtii | Alga | 1.0–18.5 μm | Val | Tyr | Leu | Thr | Ile | Thr | Met | Schibler and Huang (1991),James et al. (1993) |
| T. gondii | Protozoan | >0.5–<2.5 | Ile | Phe | Leu | Thr | Val | Thr | Met | Morrissette et al. (2004) |
| T. thermophila | Protozoan | <7.5 μm | Val | Phe | Leu | Thr | Ile | Thr | Met | Stargell et al. (1992) |
| L. major | Protozoan | 4.0 μm | Val | Phe | Met | Thr | Leu | Thr | Val | Chan et al. (1993) |
| T. brucei | Protozoan | 6.0–7.0 μm | Val | Phe | Leu | Thr | Leu | Thr | Val | Chan et al. (1993) |
| Rat | Mammal | Not sensitive | Ile | Tyr | Leu | Thr | Leu | Thr | Pro | Morejohn et al. (1987) |
IC50 values are based upon whole-organisms bioassays using wild-type lines or populations (cell-based bioassay for rat), and correspond to dinitroaniline concentrations inhibiting 50% organism growth. Because no sequence from Tetrahymena thermophila is available, a sequence from the closely related species Tetrahymena pyriformis was used for comparison.
Models for α-tubulin from the grasses foxtail and goosegrass and from tobacco displayed very similar three-dimensional structures (data not shown). Two areas of structural differences were identified in all other α-tubulin models when compared to models for those plants (Fig. 2). The first one was a substantial difference in the distribution of polar chains at the surface of the α-tubulin molecule model that was due to the presence of a Thr-253 residue instead of an Asn-253 residue in grasses and tobacco (Fig. 2; Table IV). The distances between the most external oxygen atom in the carboxyl group in Asp-251 and the nitrogen and oxygen atoms in the amide group in Asn-253 were 10.5 Å and 7.12 Å, respectively, while this distance was 3.88 Å with the oxygen atom in the hydroxyl group in Thr-253.
The second area of structural differences was located close to residue Asp-251 side chain. In α-tubulin sequences for the alga C. reinhardtii and the protozoan T. thermophila, an Ile residue occurred at position 252 instead of a Val residue in grasses and tobacco. The resulting structural difference was tiny because the longer aliphatic chain in Ile-252 side chain was oriented toward the inside of the α-tubulin model. In contrast, in the rat and in the protozoan L. major sequences, position 252 is occupied by a Leu residue, the protruding side chain of which encumbered the vicinity of Asp-251 side chain in a manner approaching what was observed for Phe-136 side chain in foxtail Leu-136-Phe mutant α2-tubulin model (Fig. 2). Furthermore, the presence of an Ile residue at position 16 in the sequences from the protozoan Toxoplasma gondii, rat, and carrot caused the vicinity of Asp-251 side chain to be encumbered in a manner approaching what was observed for Ile-239 side chain in foxtail Thr-239-Ile mutant α2-tubulin model (Fig. 2). At the level of the dimer-dimer contact region, α-tubulin models for other organisms thus differed from those for grasses and tobacco by (1) occurrence of a Thr-253 residue instead of an Asn-253 residue, and (2) encumbering aliphatic chains around the carboxyl group in Asp-251 side chain, which was also observed in both foxtail mutant α2-tubulin models.
Other structural differences included the presence of a Phe residue at position 24 in the sequences from the four protozoan species (Tyr-24 in all other sequences), and the presence of either a Val (L. major and T. brucei) or a Pro (rat) residue at position 268 (Met in all other sequences). Both differences locally increased hydrophobicity. The difference at position 24 caused a hydrophobic side chain to be located close to residue Thr-239 in α-tubulin models for the four protozoan species instead of the hydroxyl group in Tyr-24 side chain that is found in other organisms. Both differences at position 268 resulted in the absence of a sulfur atom located in a 12- to 15-Å range from the polar side chains of residues Asp-251 and Asn-253 in grasses and tobacco.
Searching the GenBank/EMBL/DDBJ database for full α-tubulin sequences yielded 134 distinct α-tubulin isotypes. Aligning revealed that residue Asp-251 is absolutely conserved in all α-tubulin sequences available so far. The seven other amino acid positions discussed above and listed in Table IV are fairly conserved within and between classes of organisms. Consensus sequences for those positions are Val-16, Tyr-24, Leu-136, Thr-239, Val-252, Asn/Thr-253, and Met-268 for higher plants but carrot (Ile-16; Table IV); Val-16, Tyr-24, Leu-136, Thr-239, Val/Ile-252, Thr-253, and Met/Val-268 for algae; Val/Ile-16, Phe-24, Leu/Ile/Met-136, Thr-239, Ile/Leu/Val-252, Thr-253, and Met/Val-268 for protozoans; and Ile-16, Tyr-24, Leu-136, Thr-239, Leu-252, Thr-253, and Pro-268 for mammals except pig (Ser-136). The sequences we used for three-dimensional modeling are thus representative for the variability existing in α-tubulins at the eight amino acid positions discussed.
DISCUSSION
In this work, we investigated the molecular bases for resistance to tubulin-inhibiting herbicides in green foxtail, using the resistant populations Oak River and Deloraine from Canada that have been selected by trifluralin in oilseed crops (Morrison et al., 1989). In both populations, resistant plants were resistant to dinitroanilines and chlorthal-dimethyl, sensitive to pronamide, and hypersensitive to carbamates (Table I). The moderate levels of resistance observed, ranging from 2.7 to 7.7, are consistent with those obtained in a previous study using seed bioassay to assess herbicide sensitivity in Oak River population (Waldin et al., 1992). Grasses have been shown to be more sensitive to dinitroanilines than dicotyledonous species (Cleary and Hardham, 1988). Indeed, the selectivity of these herbicides in dicotyledonous crops is based upon dicotyledonous crops being able to withstand herbicide doses a few fold higher than those sufficient to kill grass weeds (Cleary and Hardham, 1988; see also tobacco and grasses IC50 values in Table IV). Thus, the moderate levels of resistance to dinitroanilines and chlorthal-dimethyl observed in resistant plants from populations Oak River and Deloraine are clearly sufficient to enable their surviving under field rate applications of those herbicides.
In an effort to identify mutant tubulin gene(s) that confer resistance to tubulin-inhibiting herbicides in green foxtail, we cloned genes encoding α- and β-tubulins in this plant by PCR using degenerate primers. Two α-tubulin and two β-tubulin genes were identified. The number of tubulin genes found in green foxtail is low with respect to what has been found in Arabidopsis (Arabidopsis thaliana; at least six α- and nine β-tubulin genes; Kopczak et al., 1992; Snustad et al., 1992) or even in the more phylogenetically closely related goosegrass (at least three α- and four β-tubulin genes; Yamamoto et al., 1998; Yamamoto and Baird, 1999). It is however consistent with a previous study that identified two α-tubulin and two β-tubulin isotypes in green foxtail (Waldin et al., 1992). This study, and the high level of conservation observed between tubulin gene and protein sequences, enabled us to assume we identified most α- and β-tubulin genes in green foxtail, even if the possibility that genes encoding divergent α- or β-tubulins may also exist in this plant cannot be ruled out.
Comparative sequencing of all four tubulin genes in foxtail plants resistant or sensitive to trifluralin identified no nonsynonymous SNP in β-tubulin genes. In particular, position 350, the mutation site for colchicine and dinitroaniline cross resistance in the green alga C. reinhardtii, was Lys in all foxtail β-tubulin sequences, as it is in sensitive lines of C. reinhardtii (Lee and Huang, 1990). Similarly, position 241, which might be involved in dinitroaniline resistance in bluegrass (Poa annua), was Arg in all foxtail β-tubulin sequences, as it is in sensitive bluegrass accessions (Lowe et al., 2001). Those data put together indicated that the β-tubulins we analyzed are not involved in resistance to tubulin-inhibiting herbicides in the green foxtail populations studied herein.
When considering α-tubulins, two amino acid changes at positions 136 and 239 in α2-tubulin discriminated resistant and sensitive plants. The first change consisted of a Thr-to-Ile replacement at position 239. Residue Thr-239 is absolutely conserved in all known α-tubulin sequences. Sequencing and allele-specific PCR showed that all plants in green foxtail population Oak River contained two Ile-239 α2-tubulin alleles. All plants in line D97-3 inherited both two copies of an Ile-239 α2-tubulin allele identical to that from the Oak River plant used for sequencing experiments (Fig. 1) and the resistance pattern displayed by Oak River green foxtail plants (Table I). Besides, in goosegrass R biotype, a Thr-to-Ile change at position 239 in the TUA1 protein, the known α-tubulin protein most similar to foxtail α2-tubulin (99.7% identity), was demonstrated by Anthony et al. (1998) and Yamamoto et al. (1998) to be the cause for the resistance to dinitroanilines, the unchanged sensitivity to pronamide, and the hypersensitivity to carbamates previously found by Vaughn et al. (1987). A Thr-to-Ile change at position 239 was also shown to cause resistance to a dinitroaniline herbicide in artificial mutants of T. gondii (Morrissette et al., 2004). These data indicate that an Ile-239 α2-tubulin allele very likely confers resistance to tubulin-inhibiting herbicides in green foxtail as well.
The second nonsynonymous substitution identified in α2-tubulin caused a Leu-to-Phe change at position 136. Residue Leu-136 is absolutely conserved in all known α-tubulin sequences. Allele-specific PCR showed that the presence of a single Phe-136 α2-tubulin allele was associated with sensitivity to all herbicides assayed, while the presence of two Phe-136 α2-tubulin alleles was associated with resistance to dinitroanilines and chlorthal-dimethyl, sensitivity to pronamide, and hypersensitivity to carbamates (Table III). This resistance pattern was also observed for the few seedlings containing one Phe-136, Thr-239 α2-tubulin alleles and one Leu-136, Ile-239 α2-tubulin allele (Table III). A Leu-to-Phe change at position 136 was shown to cause resistance to a dinitroaniline herbicide in artificial mutants of T. gondii (Morrissette et al., 2004). Besides, amino acid substitutions at positions 136 and 239 resulted in adjacent structural changes in α2-tubulin three-dimensional models constructed using the most refined α-tubulin three-dimensional structure available as a template (Fig. 2). Both changes occurred within the binding site for tubulin-inhibiting herbicides predicted for goosegrass (Blume et al., 2003). On the basis of these data, we conclude that the Leu-to-Phe change at position 136 and the Thr-to-Ile change at position 239 in the gene encoding α2-tubulin are both associated with the same cross-resistance patterns, and that the corresponding mutant α2-tubulin alleles are the most likely candidates for the status of recessive resistance genes in green foxtail.
Our work enabled the identification of two alleles of the gene encoding α2-tubulin in green foxtail that are associated with cross resistance to these molecules. The allele-specific PCR assays that enable detection of resistant α2-tubulin alleles in green foxtail plants will be of great help to manage resistance to dinitroaniline-like herbicides, a class of compounds broadly used in row crop, vegetable, fruit, and ornamental production, as well as for turf. DNA-based assays are of particular interest in such cases where resistance genes are recessive and can consequently spread within and between weed populations without causing immediate changes in sensitivity.
Because dinitroanilines and phosphoroamidates share the same site(s) of action (Murthy et al., 1994; Blume et al., 2003) and because we showed that mutations associated with resistance to dinitroanilines were associated with cross-resistance to the benzoic acid chlorthal-dimethyl, all these compounds will be referred to hereafter as dinitroaniline-like herbicides. Three sets of data emerge from our work and the literature. First, comparison of three-dimensional structure models for α-tubulins from various organisms and from herbicide-resistant green foxtail mutants identified amino acid positions that most likely play a key role in sensitivity to dinitroaniline-like herbicides (Table IV; Fig. 2). The first key amino acid is the Asn/Thr residue at position 253. Asn-253 only exists in organisms most sensitive to dinitroaniline-like herbicides (grasses and tobacco), while Thr-253 is found in less sensitive (C. reinhardtii, protozoans, and carrot) or fully insensitive (rat) organisms. Interestingly, residue 253 in α-tubulins is involved in dimer-dimer contact in protofilaments. It plays an essential role for the specificity and/or strength of dimer-dimer contact via a salt bridge with the absolutely conserved residue Gly-98 in β-tubulins (Nogales, 1999). The second key amino acid is residue Asp-251, which is absolutely conserved in all known tubulin sequences. This residue is involved in interactions with GTP and is therefore crucial for dimer-dimer polymerization (Nogales, 1999). Amino acid differences at positions 16, 136, 239, and 252 observed between grasses and tobacco on one side, and resistant grass mutants and the other organisms in Table IV on the other side, all resulted in a higher obstruction of the vicinity of Asp-251 side chain in the less sensitive organisms (Fig. 2). In addition, the highest level of obstruction is observed in the three-dimensional model for α-tubulin from the least sensitive organism, i.e. rat (Fig. 2). Second, two other positions are involved in sensitivity to dinitroaniline-like herbicides. The first one is position 24, that is, Tyr in all organisms studied here but protozoans, where it is Phe (Table IV). A Tyr-to-His change at position 24 in the already moderately dinitroaniline-like herbicide sensitive alga C. reinhardtii conferred a 2-fold decrease in sensitivity to dinitroanilines (James et al., 1993). In contrast with mutations at positions 136 and 239, this change also caused a 10-fold decrease in sensitivity to pronamide (James et al., 1993), a herbicide with separate site(s) of action (Vaughan and Vaughn, 1987). Similarly, the Met-to-Thr change at position 268 found in goosegrass I biotype by Yamamoto et al. (1998) conferred a decrease in sensitivity to dinitroaniline-like herbicides lower than what was observed for the R goosegrass biotype (Vaughn et al., 1990) that contained a Thr-to-Ile change at position 239. Unlike the changes at position 136 or 239, the change at position 268 did not alter sensitivity to carbamates (Vaughn et al., 1990). The changes at positions 239 and 268 were shown to have additive effects (Anthony et al., 1999). These data suggest that residues 24 and 268 play a role in herbicide sensitivity that is minor with respect to the role played by residues discussed above. Third, molecular modeling indicated that two electronegative domains spaced about 10 Å apart are crucial for tubulin-binding activity of dinitroaniline-like herbicides (Ellis et al., 1994).
These data enabled us to propose an alpha model where dinitroaniline-like herbicides bind to α-tubulins from sensitive plants via polar interactions. The distances between the polar, carboxyl group in Asp-251 and the polar, amide group in Asn-253 being of around 10 Å in α-tubulin three-dimensional models for grasses and tobacco, dinitroaniline-like herbicides may bind to, or close to, Asp-251 and Asn-253 side chains. In the alpha model, occurrence of a Thr residue at position 253 would significantly reduce herbicide binding because the distance between the amide group in Asn-253 and the hydroxyl group in Thr-253 is of about 3.9 Å. An A-to-C transversion at the second codon position is sufficient to cause an Asn-to-Thr replacement. It has, however, never been observed in α-tubulins from plant mutants resistant to dinitroaniline-like herbicides studied to date. Given the crucial role of residue 253, such a mutation may have consequences deleterious for correct dimer-dimer assembly. For the same reason, amino acid replacements addressing the absolutely conserved amino acid residue Asp-251 are even more unlikely to occur, which may explain why indirect mutations occurred at positions 136 and 239 and may potentially occur at positions 16 and 252 in resistant grass mutants. The subsidiary role of positions 24 and 268 may be explained in the alpha model by the side chains of Tyr-24 and Met-268 being located at the border of the dinitroaniline-like herbicide-binding site. In sensitive organisms, polar interactions with the hydroxyl group in Tyr-24, which does not exist in His-24 C. reinhardtii resistant mutant and in protozoan (Table IV), and with the sulfur atom in Met-268, which does not exist in rat, in two protozoan species (Table IV), and in goosegrass I biotype (Yamamoto et al., 1998), would contribute to increase the stability of herbicide-tubulin dimer complexes.
The alpha model is consistent with another study where a dinitroaniline-like herbicide-binding site was predicted by docking a model of the herbicide molecule onto a model of goosegrass TUA1 α-tubulin (Blume et al., 2003). These authors identified amino acid residues Asp-251, Val-252, Asn-253, and Arg-243 as essential for the shape of dinitroaniline-like herbicide-binding site and for interaction with herbicide polar groups. They confirmed the role of the Thr-to-Ile substitution at position 239 in preventing efficient herbicide binding to α-tubulin, and they pointed out that residues Arg-2, Cys-4, His-8, Phe-138, and, interestingly, Leu-136, are involved in herbicide binding to α-tubulin, which strengthens the alpha model. Both the alpha model and the model for goosegrass (Blume et al., 2003) predict dinitroaniline-like herbicide binding at the very dimer-dimer contact site, thus differing from the model proposed for the protozoan T. gondii that predicts herbicide binding at a position mostly affecting lateral contacts between protofilaments (Morrissette et al., 2004). It is well established that dinitroaniline-like herbicides selectively bind to plant and protozoan microtubules (see Morrissette et al., 2004 for refs.). Predicted binding sites for these herbicides in goosegrass and T. gondii are distinct but partially overlapping, sharing residues Val/Lys-4, Thr-239, and Arg-243 (Blume et al., 2003; Morrissette et al., 2004). This, together with the differences in sensitivity and the structural differences in three-dimensional models observed between grasses and protozoans (Table IV; Fig. 2), suggest that dinitroaniline-like herbicides may bind in a slightly different manner to α-tubulins from grasses and protozoans.
It has been proposed that, in tubulin protofilaments, α,β dimers bind head to tail, with the α-tubulin subunit in a free dimer establishing a GTP-mediated link with the β-tubulin subunit in the last incorporated dimer (Nogales, 1999). Dinitroaniline-like herbicides have been shown to inhibit polymerization of tubulin dimers into protofilaments at a substoichiometric rate (Hugdahl and Morejohn, 1993). Tubulin filaments are in constant equilibrium between polymerization and depolymerization (for review, see Nogales, 1999). The model proposed by Hugdahl and Morejohn (1993) implies that dinitroaniline-like herbicides bind rapidly and reversibly to tubulin α,β dimers to form tubulin-herbicide complexes. Herbicide-dimer complexes would inhibit tubulin polymerization and/or depolymerize via a mechanism that remains to be elucidated (Hugdahl and Morejohn, 1993). In Arabidopsis, incorporation of tubulin dimers that were anomalous because of a mutation within the α-tubulin subunit into protofilaments was shown to decrease the stability of microtubule filaments and to increase their sensitivity to dinitroaniline-like herbicides (Thitamadee et al., 2002). It is possible that herbicide-dimer complexes may be integrated in protofilaments in spite of the presence of a molecule of herbicide bound at the very dimer-dimer contact site in the α-tubulin subunit. Herbicide-dimer complexes would then destabilize the protofilaments where they are integrated, thus increasing the depolymerization rate, and/or alter lateral contacts between protofilaments, as proposed in T. gondii (Morrissette et al., 2004).
One point is a bit tricky to explain using the alpha model. Only minute structural differences in side-chain orientation were observed when comparing the region around residues 251 and 253 in three-dimensional models for grasses with that in models for tobacco, whereas there is about 10-fold difference in herbicide sensitivity between these species (Table IV). A tobacco artificial β-tubulin mutant was shown to display decreased sensitivity to dinitroaniline-like herbicides (Blume et al., 1998). It is thus possible that differences at the β-tubulin level might also be involved in the differential sensitivity to dinitroaniline-like herbicides observed between organisms. Another explanation would be differential herbicide uptake and/or metabolization between monocotyledonous and dicotyledonous plants. A comparative in vitro tubulin-binding study is needed to make this point clear.
Comparative analyses of three-dimensional tubulin models in light of previous studies resulted in the identification of amino acid positions that are involved in sensitivity to dinitroaniline-like herbicides. Position 253 and the vicinity of the side chain of residue Asp-251, which is more or less obstructed depending on the nature of the amino acids present at positions 16, 136, 239, and 252, are clearly critical for herbicide sensitivity, while positions 24 and 268 are more likely playing a less essential role in herbicide sensitivity. These analyses enabled us to propose a model for dinitroaniline-like herbicide binding to plant α,β-tubulin dimers that is consistent with a previous study (Blume et al., 2003). Heterologous expression of artificial α-tubulin-mutant alleles in the system recently developed for goosegrass genetic engineering (Yemets et al., 2003), combined with tubulin-herbicide binding assays, could be a way of providing useful data to check the consistency of these models, therefore improving our knowledge of the detailed mode of action of dinitroaniline-like herbicides in order to perform rational herbicide design.
MATERIALS AND METHODS
Plant Material
We used interspecific hybridization to transfer resistance to trifluralin from a selected, homozygous, trifluralin-resistant plant from the green foxtail (Setaria viridis L. Beauv.) population Oak River (Manitoba, Canada; Morrison et al., 1989) into cultivated foxtail millet (Setaria italica L. Beauv.). Interspecific crossings were performed as described by Wang and Darmency (1997), using the trifluralin-sensitive foxtail millet cultivar Amende4 as the male parent. All F1 hybrid plants were trifluralin sensitive. One resistant plant in the F2 generation of self-pollination was selected on the basis of typical foxtail millet morphology. It was used to produce an F4 generation by two rounds of self-pollination. All F4 plants were trifluralin resistant. One resistant F4 plant with foxtail millet morphology was used as the female parent in an interspecific cross with the foxtail millet cultivar Jigu11. All hybrid F5 plants were susceptible. After two rounds of self-pollination, one trifluralin-resistant line (D97-3) and one trifluralin-sensitive line (D97-4) issued from the same F5 plant were selected for analysis. Analysis of self-pollination progenies from these two lines showed they were homozygous at the resistance/sensitivity locus. Lines D97-3 and D97-4 are thus an F7 generation from the original pairing. For our study, we also used the trifluralin-sensitive green foxtail population D96-174 (Manitoba, Canada), the trifluralin-sensitive green foxtail population Ouges99 (Burgundy, France), and a second trifluralin-resistant green foxtail population from Deloraine (Manitoba, Canada). Seeds from the three Canadian green foxtail populations were provided by Dr. I.N. Morrison (University of Manitoba, Winnipeg, Canada).
We performed a cross between one trifluralin-sensitive plant from population Ouges99 and one plant in the trifluralin-resistant hybrid line D97-3. All F1 plants were trifluralin sensitive. Five F2 hybrid populations segregating for trifluralin resistance (D00-096, D00-100, D00-109, D00-110, and D00-112) were obtained from five distinct F1 plants by self-pollination and used for allele-specific PCR experiments.
Herbicide Sensitivity Bioassay
A rapid-seed bioassay was used to assess herbicide sensitivity of foxtail seedlings. Seeds were deposited on filter paper placed on 0.6-cm diameter glass beads in 13.5-cm diameter glass petri dishes containing 40 mL of herbicide solution. The dose of trifluralin-discriminating resistant and sensitive seedlings was 0.2 μm (Wang et al., 1997). A seedling was considered sensitive if its coleoptile was <0.6 cm in length and exhibited a swollen tip. To assess herbicide cross resistance and resistance-factor values, a dose response assay was performed using lines D97-3 and D97-4 with the following herbicides: butralin, benfluralin, pendimethalin, trifluralin (dinitroanilines), chlorthal-dimethyl (benzoic acid), pronamide (benzamide), carbetamide, and chlorpropham (carbamates). Herbicide solutions were prepared from the following commercial herbicides using deionized water: Amex (Nufarm, Gennevilliers, France; 480 g L−1 butralin), Bonalan (Dow AgroScience, Sofia Antipolis, France; 180 g L−1 benfluralin), Flurasan (Phytunion, Poitiers, France; 480 g L−1 trifluralin), Prowl (BASF Agro, Tassin-la-Demi-Lune, France; 400 g L−1 pendimethalin), Dacthal (Sipcam-Phyteurop, Levallois-Perret, France; 75% [w/w] chlorthal-dimethyl), Kerb (Dow Agrosciences; 400 g L−1 pronamide), Chlorpham TX (Sipcam-Phyteurop; 400 g L−1 chlorpropham), and Légurame liquide (FCS France, Guyancourt, France; 300 g L−1 carbetamide). Plates were incubated for 72 h at 27°C in darkness, and the length of the coleoptile was measured.
To obtain plant material for tubulin sequencing experiments, resistant or sensitive seedlings were collected from trifluralin plates, rinsed with deionized water, and cultivated in greenhouse as described by Wang and Darmency (1997) until they had three to four fully expanded leaves.
PCR Sequencing of Green Foxtail α- and β-Tubulin Genes
Total genomic DNA was extracted as described by Doyle and Doyle (1987) from a trifluralin-sensitive, four-leaf plant from population D96-174. All primers used in this work are shown in Supplemental Table I. PCR mixes were as described (Délye et al., 2002). Degenerate primers TaI1 and TaI1R targeted sequences encoding the highly conserved amino acid sequences VGNACWE and DWCPTGF, respectively, in plant α-tubulins. Degenerate primers TbI2 and TbI2R targeted sequences encoding the highly conserved amino acid sequences LEPGTMD and WYTGEGMD, respectively, in plant β-tubulins. Primers in each primer pair were used at a final concentration of 1.25 μm each. Cycling program consisted of 37 cycles with 7 s at 95°C, 15 s at 65°C, and 1 min at 72°C, followed by a final step of 10 min at 72°C.
Two amplicons of around 1,200 and 1,500 bp were obtained using primers TbI2 and TbI2R. They were purified from a 0.6% (w/v) agarose gel run at 50 V in 0.5 × tris-borate EDTA buffer using the Nucleospin Extract system (Macherey-Nagel, Hoerdt, France). Each amplicon contained a single DNA sequence, which enabled direct sequencing on both strands using specific primers. A single amplicon of about 2,000 bp was obtained using primers TaI1 and TaI1R and purified as above. This amplicon contained mixed DNA sequences. It was cloned into the vector pDrive (Qiagen USA, Valencia, CA), and 20 different inserts were sequenced using primer TaI1, revealing two distinct DNA sequences. For each sequence, three different clones were sequenced on both strands using specific primers.
Inverse PCR was used to clone the 5′ and 3′ ends of all four genes. Genes α2, β1, and β2 contained a unique VspI restriction site. Gene α1 contained a unique BcuI restriction site. A total of 500 μg DNA were digested with either BcuI or VspI (MBI Fermentas, Vilnius, Lithuania). Restriction mixes were heated at 75°C for 10 min and deposited upon a 0.025-μm dialysis membrane (Millipore SA, Molsheim, France) floating on sterile, distilled water. After 1 h dialysis, restricted DNA was circularized using T4 DNA ligase (MBI Fermentas) and used for PCR using gene-specific primers (Supplemental Table I) at a final concentration of 0.2 μm each. Cycling program consisted of 37 cycles with 7 s at 95°C, 15 s at 65°C, and 2 min at 72°C, followed by a final step of 10 min at 72°C. A single amplicon was obtained for each of the four genes, which was purified as above and sequenced on both strands using specific primers.
Each of the four tubulin genes was subsequently amplified from one trifluralin-resistant plant from each of line D97-3 and populations Oak River and Deloraine, and from one trifluralin-sensitive plant from each of population Ouges99, cultivars Amende4 and Jigu11, and line D97-4 using gene-specific primers (Supplemental Table I). All amplicons were sequenced on both strands using specific primers.
Genotyping by Allele-Specific PCR
We developed two bidirectional allele-specific PCR assays (Délye et al., 2002) to confirm that amino acid changes at positions 136 and 239 in foxtail α2 gene were associated with resistance to tubulin-inhibiting herbicides. Herbicide sensitivity of foxtail seedlings was assessed as before. DNA was extracted from all seedlings as described (Délye et al., 2002). Allele-specific primers were designed by using the fact that a 3′ mismatch does not prime in a PCR at a specific annealing temperature (Sommer et al., 1992). Primers Ta2SF and Ta2SLR specifically primed foxtail α2-tubulin sequences containing a T (Phe) or a C (Leu) at the first position in codon 136, respectively. They were used at a final concentration of 0.2 μm (Ta2SF) and 0.05 μm (Ta2SLR) together with primers Ta2S10 and Ta2S10R, which were used at a final concentration of 0.1 μm each. Primers Ta2ST and Ta2SIR specifically primed foxtail μ2-tubulin sequences containing a C (Thr) or a T (Ile) at the second position in codon 239, respectively. They were used together with primers Ta2S1 and Ta2S8R at a final concentration of 0.1 μm for each of the four primers. Allele-specific PCR mixes were as described (Délye et al., 2002). Cycling program consisted of one denaturation step of 30 s at 95°C, followed by 37 cycles of 5 s at 95°C, 10 s at 66°C, and 30 s at 72°C.
Each set of primers was designed to generate up to three distinct sizes of amplicons, depending on the genotype of the foxtail plant investigated. Primers targeting codon 136 will generate a 606-bp fragment from all foxtail plants (internal positive control), a 407-bp fragment from plants containing a Leu-136 codon, and a 242-bp fragment from plants containing a Phe-136 codon. Primers targeting codon 239 will generate a 632-bp fragment from all foxtail plants (internal positive control), a 406-bp fragment from plants containing a Thr-239 codon, and a 270-bp fragment from plants containing an Ile-239 codon.
Sequence Analysis and Protein Modeling
Sequence analysis and alignments were performed using BioEdit (Hall, 1999) and Multalin software (Corpet, 1988), respectively. α-Tubulin three-dimensional structures were modeled using the Swiss-Model server (Schwede et al., 2003). The template used for modeling was the refined three-dimensional structure of α-tubulin in the α,α-tubulin model from pig (PDB accession 1JFF; Löwe et al., 2001).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AJ586804 (α1), AJ586805 (α2), AJ586806 (β1), and AJ586807 (β2).
Supplementary Material
Acknowledgments
The authors are grateful to two anonymous reviewers for helpful comments to the discussion.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.037432.
References
- Anthony RG, Hussey PJ (1999) Dinitroaniline resistance and the microtubule cytoskeleton. Trends Plant Sci 4: 112–116 [DOI] [PubMed] [Google Scholar]
- Anthony RG, Reichelt S, Hussey PJ (1999) Dinitroaniline herbicide-resistant transgenic tobacco plants generated by co-overexpression of a mutant α-tubulin and β-tubulin. Nat Biotechnol 17: 712–716 [DOI] [PubMed] [Google Scholar]
- Anthony RG, Waldin TR, Ray JA, Bright SWJ, Hussey PJ (1998) Herbicide resistance caused by spontaneous mutation of the cytoskeletal protein tubulin. Nature 393: 260–263 [DOI] [PubMed] [Google Scholar]
- Blume YB, Nyporko AY, Yemets AI, Baird WV (2003) Structural modeling of the interaction of plant α-tubulin with dinitroaniline and phosphoroamidate herbicides. Cell Biol Int 27: 171–174 [DOI] [PubMed] [Google Scholar]
- Blume YB, Strashnyuk NM, Smertenko AP, Solodushko VG, Sidorov VA, Gleba YY (1998) Alteration of β-tubulin in Nicotiana plumbaginifolia confers resistance to amiprophos-methyl. Theor Appl Genet 97: 464–472 [Google Scholar]
- Carpenter JL, Ploense SE, Snustad DP, Silflow CD (1992) Preferential expression of an α-tubulin gene of arabidopsis in pollen. Plant Cell 4: 557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MM-Y, Grogl M, Chen C-C, Bienen EJ, Fong D (1993) Herbicides to curb human parasitic infections: in vitro and in vivo effects of trifluralin on the trypanosomatid protozoans. Proc Natl Acad Sci USA 90: 5657–5661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary AL, Hardham AR (1988) Depolymerization of microtubule arrays in root tip cells by oryzalin and their recovery with modified nucleation patterns. Can J Bot 66: 2353–2366 [Google Scholar]
- Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16: 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Délye C, Wang T, Darmency H (2002) An isoleucine-leucine substitution in chloroplastic acetyl-Co A carboxylase from green foxtail (Setaria viridis L. Beauv.) is responsible for resistance to the cyclohexanedione herbicide sethoxydim. Planta 214: 421–427 [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL (1987) Isolation of DNA from fresh plant tissue. Focus 12: 13–15 [Google Scholar]
- Eleftheriou EP, Bekiari E (2000) Ultrastructural effects of the herbicide chlorpropham (CIPC) in root tip cells of wheat. Plant Soil 226: 11–19 [Google Scholar]
- Ellis JR, Taylor R, Hussey PJ (1994) Molecular modeling indicates that two chemically distinct classes of anti-mitotic herbicide bind to the same receptor site(s). Plant Physiol 105: 15–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98 [Google Scholar]
- Hugdahl JD, Morejohn LC (1993) Rapid and reversible high-affinity binding of the dinitroaniline herbicide oryzalin to tubulin from Zea mays L. Plant Physiol 102: 725–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James SW, Silflow CD, Stroom P, Lefebvre PA (1993) A mutation in the α1-tubulin gene of Chlamydomonas reinhardtii confers resistance to anti-microtubule herbicides. J Cell Sci 106: 209–218 [DOI] [PubMed] [Google Scholar]
- Jasieniuk M, Brûlé-Babel AL, Morrison IN (1994) Inheritance of trifluralin resistance in green foxtail (Setaria viridis). Weed Sci 42: 123–127 [Google Scholar]
- Kopczak SD, Haas NA, Hussey PJ, Silflow CD, Snustad DP (1992) The small genome of Arabidopsis contains at least six expressed α-tubulin genes. Plant Cell 4: 539–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee VD, Huang B (1990) Missense mutations at lysine 350 in β2-tubulin confer altered sensitivity to microtubule inhibitors in Chlamydomonas. Plant Cell 2: 1051–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe DB, Swire-Clark GA, McCarty LB, Whitwell T, Baird WV (2001) Biology and molecular analysis of dinitroaniline-resistant Poa annua L. Int Turfgrass Soc Res J 9: 1019–1025 [Google Scholar]
- Löwe J, Li H, Downing KH, Nogales E (2001) Refined structure of αβ-tubulin at 3.5 Å resolution. J Mol Biol 313: 1045–1057 [DOI] [PubMed] [Google Scholar]
- Morejohn LC, Bureau TE, Molè-Bajer J, Bajer AS, Fosket DE (1987) Oryzalin, a dinitroaniline herbicide, binds to plant tubulin and inhibits microtubule polymerization in vitro. Planta 172: 252–264 [DOI] [PubMed] [Google Scholar]
- Morrison IN, Todd BG, Nawolsky KM (1989) Confirmation of trifluralin-resistant green foxtail (Setaria viridis) in Manitoba. Weed Technol 3: 544–551 [Google Scholar]
- Morrissette NS, Mitra A, Sept D, Sibley LD (2004) Dinitroanilines bind α-tubulin to disrupt microtubules. Mol Biol Cell 15: 1960–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy JV, Kim H-K, Hanesworth VR, Hugdahl JD, Morejohn LC (1994) Competitive inhibition of high-affinity oryzalin binding to plant tubulin by the phosphoric amide herbicide amiprophos-methyl. Plant Physiol 105: 309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E (1999) A structural view of microtubule dynamics. Cell Mol Life Sci 56: 133–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH (1998) Structure of the αβ tubulin dimer by electron crystallography. Nature 391: 199–203 [DOI] [PubMed] [Google Scholar]
- Schibler MJ, Huang B (1991) The colR4 and colR15 β-tubulin mutations in Chlamydomonas reinhardtii confer altered sensitivities to microtubules inhibitors and herbicides by enhancing microtubule stability. J Cell Biol 113: 605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31: 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow CD (1991) Why do tubulin gene families lack diversity in flagellate/ciliate protists? Protoplasma 164: 9–11 [Google Scholar]
- Snustad DP, Haas NA, Kopczak SD, Silflow CD (1992) The small genome of Arabidopsis contains at least nine expressed β-tubulin genes. Plant Cell 4: 549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer SS, Groszbar AR, Bottema CDK (1992) PCR amplification of specific alleles (PASA) is a general method for rapidly detecting known single base-pair changes. Biotechniques 12: 82–87 [PubMed] [Google Scholar]
- Stargell LA, Heruth DP, Gaertig J, Gorovsky MA (1992) Drugs affecting microtubule dynamics increase α-tubulin mRNA accumulation via transcription in Tetrahymena termophila. Mol Cell Biol 12: 1443–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thitamadee S, Tuchihara K, Hashimoto T (2002) Microtubule basis for left-handed helical growth in Arabidopsis. Nature 417: 193–196 [DOI] [PubMed] [Google Scholar]
- Vaughn KC, Marks MD, Weeks DP (1987) A dinitroaniline-resistant mutant of Eleusine indica exhibits cross-resistance and hypersensitivity to antimicrotubule herbicides and drugs. Plant Physiol 83: 956–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn KC, Vaughan MA, Gossett BJ (1990) A biotype of goosegrass (Eleusine indica) with an intermediate level of dinitroaniline herbicide resistance. Weed Technol 4: 157–162 [Google Scholar]
- Vaughan MA, Vaughn KC (1987) Pronamide disrupts mitosis in a unique manner. Pestic Biochem Physiol 28: 182–193 [Google Scholar]
- Vaughan MA, Vaughn KC (1988) Carrot microtubules are dinitroaniline resistant. I. Cytological and cross-resistance studies. Weed Res 28: 73–83 [Google Scholar]
- Waldin TR, Ellis JR, Hussey PJ (1992) Tubulin-isotype analysis of two grass species-resistant to dinitroaniline herbicides. Planta 188: 258–264 [DOI] [PubMed] [Google Scholar]
- Wang T, Darmency H (1997) Dinitroaniline herbicide cross-resistance in resistant Setaria italica lines selected from interspecific crosses with S. viridis. Pestic Sci 49: 277–283 [Google Scholar]
- Wang T, Fleury A, Ma J, Darmency H (1997) Genetic control of dinitroaniline resistance in foxtail millet (Setaria italica). J Hered 87: 423–426 [Google Scholar]
- Wasteneys GO (2002) Microtubule organization in the green kingdom: chaos or self-order? J Cell Sci 115: 1345–1354 [DOI] [PubMed] [Google Scholar]
- Yamamoto E, Baird WV (1999) Molecular characterization of four β-tubulin genes from dinitroaniline susceptible and resistant biotypes of Eleusine indica. Plant Mol Biol 39: 45–61 [DOI] [PubMed] [Google Scholar]
- Yamamoto E, Zeng L, Baird WV (1998) α-tubulin missense mutations correlate with antimicrotubule drug resistance in Eleusine indica. Plant Cell 10: 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yemets AI, Klimkina LA, Tarassenko LV, Blume YB (2003) Efficient callus formation and plant regeneration of goosegrass. Eleusine indica (L.) Gaertn. Plant Cell Rep 21: 503–510 [DOI] [PubMed] [Google Scholar]
- Zeng L, Baird WV (1997) Genetic basis of dinitroaniline herbicide resistance in a highly resistant biotype of goosegrass (Eleusine indica). J Hered 88: 427–432 [Google Scholar]
- Zeng L, Baird WV (1999) Inheritance of resistance to anti-microtubule dinitroaniline herbicides in an “intermediate” resistant biotype of Eleusine indica (Poaceae). Am J Bot 86: 940–947 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.