Abstract
In diffusely growing plant cells, cortical microtubules play an important role in regulating the direction of cell expansion. Arabidopsis (Arabidopsis thaliana) spiral2 (spr2) mutant is defective in directional cell elongation and exhibits right-handed helical growth in longitudinally expanding organs such as root, hypocotyl, stem, petiole, and petal. The growth of spr2 roots is more sensitive to microtubule-interacting drugs than is wild-type root growth. The SPR2 gene encodes a plant-specific 94-kD protein containing HEAT-repeat motifs that are implicated in protein-protein interaction. When expressed constitutively, SPR2-green fluorescent protein fusion protein complemented the spr2 mutant phenotype and was localized to cortical microtubules as well as other mitotic microtubule arrays in transgenic plants. Recombinant SPR2 protein directly bound to taxol-stabilized microtubules in vitro. Furthermore, SPR2-specific antibody and mass spectrometry identified a tobacco (Nicotiana tabacum) SPR2 homolog in highly purified microtubule-associated protein fractions from tobacco BY-2 cell cultures. These results suggest that SPR2 is a novel microtubule-associated protein and is required for proper microtubule function involved in anisotropic growth.
The microtubule is a dynamic filamentous polymer consisting of heterodimers of highly conserved α- and β-tubulin subunits and plays key roles in cell morphogenesis, polarity establishment, cell division, intracellular organelle transport, and other cellular processes in all eukaryotic cells (Desai and Mitchison, 1997). Despite the conserved polymer structure of microtubules, the organization and dynamics of microtubule arrays are remarkably diverse and change during the cell cycle, in different cell types, and in response to environmental and developmental stimuli. As the plant cell cycle progresses, microtubules are organized into the preprophase band, the mitotic spindle, the cytokinetic phragmoplast, the perinuclear array, and the cortical array (Wasteneys, 2002). Cortical microtubule arrays in interphase cells are arranged in a random meshwork or adopt a more ordered distribution (Cyr and Palevitz, 1995). In rapidly elongating cells with diffuse growth, cortical arrays are arranged perpendicular to the cell's growth axis and are thought to dictate the growth anisotropy (Sugimoto et al., 2000). Physical stimuli, such as gravity and light, and plant hormones are known to rearrange cortical microtubule arrays within an hour from one predominant orientation to another (Shibaoka, 1994; Wymer and Lloyd, 1996).
The dynamics and array organization of microtubules are likely controlled by stabilizing and destabilizing factors, as well as other microtubule regulators (Hashimoto, 2003). Microtubule-associated proteins (MAPs) were originally defined as proteins that are copurified with microtubules in vitro from cell extracts and are generally used to refer to proteins that modulate microtubule functions by directly binding to the polymer. Biochemical purification, genetic studies, and homology cloning based on animal and fungal sequences have been used to identify several plant MAPs and microtubule regulators. Proteins belonging to the MAP-65 family localize to areas of overlapping antiparallel microtubules, such as in the mitotic spindle and the phragmoplast (Smertenko et al., 2000). In Arabidopsis (Arabidopsis thaliana) ple mutants that are defective in one MAP-65 member, cytokinesis in the root cells is abnormal due to the distorted and probably dysfunctional phragmoplast (Müller et al., 2004). After this MAP family was first recognized in plants, related midzone-specific MAPs with conserved domain structures were identified in fungi and mammals (Mollinari et al., 2002; Schuyler et al., 2003). Arabidopsis MOR1 is a member of the evolutionally conserved XMAP215/TOG MAP family and localizes to cortical arrays and the phragmoplast. Cortical microtubules are rapidly depolymerized in temperature-sensitive mor1 mutants upon transfer to a restrictive temperature (Whittington et al., 2001), whereas null gem1 alleles of the MOR1 gene are defective in cytokinesis during pollen development (Twell et al., 2002). Mutations in a katanin microtubule-severing protein lead to various cellular defects, including reduced anisotropic growth, abnormal cell specification in root epidermis, and altered regulation of gibberellin biosynthesis and responses in Arabidopsis (Bouquin et al., 2003 and refs. therein). Arabidopsis microtubule-end-binding protein homologs are found to localize to plus and minus ends of cortical microtubules (Chan et al., 2003) and to endomembrane networks (Mathur et al., 2003). Besides these plant homologs of conserved microtubule regulators, some MAPs identified in plants do not appear to have overall sequence homology to animal and fungal MAPs. Such apparent plant-specific MAPs include tobacco (Nicotiana tabacum) MAP-190, which binds to both microtubules and actin microfilaments (Igarashi et al., 2000), and maize TAN1, which is required to guide phragmoplasts to the former preprophase band site during leaf development (Smith et al., 2001). Since the microtubule surface appears to be recognized by various sequence motifs in MAPs (Hashimoto, 2003), there may be other unique MAPs that are specific to the plant lineage.
Molecular genetic analyses of helical growth mutants of Arabidopsis have been contributing to our understanding of anisotropic growth regulation. Epidermal cells of longitudinally elongating tissues in these twisted mutants are skewed and, importantly, in several helical growth mutants the orientation of helices is fixed in either a right- or a left-handed direction (Hashimoto, 2002). Cortical microtubule arrays in rapidly elongating root epidermal cells are arranged in prominent right-handed helices in left-handed mutants, lefty1 and lefty2 (Thitamadee et al., 2002), and in left-handed helices in a right-handed spiral1 (spr1) mutant (Furutani et al., 2000), in contrast to the transverse arrays in wild-type cells. Molecular cloning of these loci revealed that α-tubulins in lefty mutants have dominant negative mutations at the tubulin intradimer interface (Thitamadee et al., 2002) and SPR1 is a 12-kD plant-specific microtubule-localized protein (Nakajima et al., 2004; Sedbrook et al., 2004). These studies indicate that defects in microtubule function can lead to helical growth phenotypes with a fixed orientation and that novel factors involved in the cortical microtubule organization may be identified by analyzing such helical growth mutants.
spiral2 (spr2) has been isolated as an Arabidopsis right-handed helical mutant with prominent counterclockwise twisting in leaf petioles and flower petals, when viewed from above (Furutani et al., 2000), and is allelic to two classic twisting mutants, tortifolia1 (Bürger, 1971) and convoluta (Relichova, 1976). Addition of low doses of the microtubule-depolymerizing drug propyzamide to the culture medium induces left-handed helical growth in wild-type Arabidopsis seedlings, but this twist-inducing effect of propyzamide is somewhat less effective in spr2 than the wild type, indicative of a mild defect in microtubule function in spr2 (Furutani et al., 2000). It is also noted that transgenic expression of a green fluorescent protein (GFP)-α-tubulin chimeric protein generates right-handed twisting resembling the spr2 phenotype (Ueda et al., 1999). Genetic studies suggest that SPR1 and SPR2 act on a similar process but via separate pathways (Furutani et al., 2000).
In this study, we cloned the SPR2 gene using a map-based strategy. SPR2 is a member of a thus far uncharacterized HEAT-repeat-containing protein family and binds to microtubules both in vivo and in vitro. We demonstrate that a defect in SPR2 MAP results in a right-handed helical growth phenotype.
RESULTS
Phenotypes and Sensitivity to Microtubule-Interacting Drugs
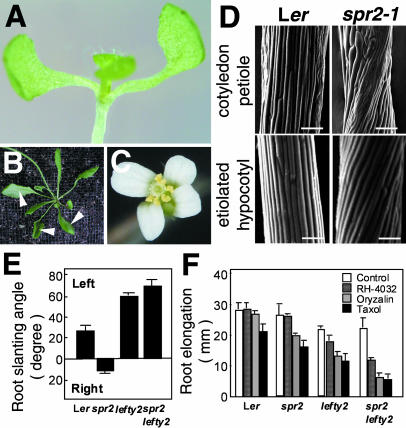
We have previously isolated spr2-1 in the Landsberg erecta (Ler) background and reported that spr2 is allelic to a classic twisting mutant convoluta (Furutani et al., 2000). Further genetic screening identified two additional alleles, spr2-2 and spr2-3, from ethyl methanesulfonate-mutagenized M2 seedlings of the Columbia ecotype (Col). Because all spr2 alleles showed very similar phenotypes (data not shown), only spr2-1 is shown in Figure 1. The twisting phenotype of spr2 was most apparent in lateral organs, such as cotyledon, rosette leaf, and petal (Fig. 1, A–C). When the plant is viewed from above, these organs are twisted in a counterclockwise direction. The counterclockwise twisting of rosette leaf and cotyledon was caused by right-handed helical growth of epidermal cell files in petioles (Fig. 1D). Epidermal cell files of hypocotyl and stem in dark-grown plants also formed constitutive right-handed helices (Fig. 1D; Furutani et al., 2000).
Figure 1.
spr2 shows right-handed helical growth and increased sensitivity to microtubule-interacting drugs. A, Cotyledon twisted in a counterclockwise direction in a 10-d-old spr2-1 seedling. B, Rosette leaves twisted in a counterclockwise direction in a 1-month-old spr2-1 plant. Arrowheads indicate the abaxial side of leaves facing up. C, Petals twisted in a counterclockwise direction in a spr2-1 flower. D, Scanning electron micrographs of cotyledon petioles and etiolated hypocotyls of 7-d-old Ler and spr2-1 seedlings. Bars = 100 μm. E, Root-slanting angle of 7-d-old seedlings grown on vertically placed agar plates. Left and right indicate that roots grow toward the left or right side of the plates. F, Inhibition of root growth by microtubule-interacting drugs. After seedlings were grown for 7 d on agar plates containing 0.1 μm oryzalin, 10 nm RH-4032, or 1 μm taxol, lengths of primary roots were measured. More than 30 roots were measured for each treatment and genotype in E and F.
To test whether spr2 is compromised in terms of microtubule function, we examined the effects of microtubule-interacting drugs on the growth of primary roots. When grown on a vertically placed hard agar plate, wild-type roots (Ler ecotype) grew slightly toward the left side of the plate, whereas spr2 roots grew slightly toward the right (Fig. 1E), indicating that the spr2 mutation affected the root as well. When seedlings were grown on a medium containing a low dose of the microtubule-depolymerizing drug oryzalin (0.1 μm) or the microtubule-stabilizing drug taxol (1 μm), growth of wild-type primary roots was inhibited slightly by oryzalin and by 25% by taxol (Fig. 1F). At these drug concentrations, spr2 roots were affected somewhat more strongly than wild-type roots.
To substantiate the drug response, we studied the effect of the spr2 mutation in the lefty2 tubulin mutant background (Thitamadee et al., 2002). The spr2lefty2 double mutant showed left-handed helical growth in all twisting organs where twisting angles were slightly increased compared to lefty2 (Fig. 1F; data not shown), suggesting that lefty2 is epistatic to spr2 and that its helical growth phenotype is slightly enhanced by the spr2 mutation. When spr2lefty2 seedlings were treated with 0.1 μm oryzalin, 1 μm taxol, or 10 nm RH-4032, another potent microtubule-depolymerizing drug (Young and Lewandowski, 2000), the root growth was severely inhibited by more than 70% by oryzalin and taxol and by 50% by RH-4032 (Fig. 1E). At the same drug concentrations, growth of lefty2 primary roots was inhibited by less than 45% by oryzalin and taxol and only by 20% by RH-4032. These results indicate that the spr2 mutation markedly enhanced sensitivities to microtubule-interacting drugs in the lefty2 mutant background.
Map-Based Cloning of SPR2
By crossing spr2-1 with wild-type Col, the SPR2 locus had been mapped between molecular markers PG11 and mi123 on chromosome 4 (Furutani et al., 2000). For further genetic mapping, cleaved-amplified polymorphic sequence markers were developed based on sequence information on the bacterial artificial chromosome (BAC) clones covering this region. By analyzing 2,518 chromosomes, the SPR2 locus was delimited to a 168-kb region containing 44 putative open reading frames (ORFs; Fig. 2A). We sequenced some of these ORFs and found a 1-bp deletion in At4g27060 from the spr2-1 genomic DNA (Fig. 2B). When 5′-RACE was used to examine the 5′ end of the transcript, we found that the predicted ORF of At4g27060 in the database lacks 38 amino acids at the N terminus. The corrected DNA sequence was deposited in GenBank.
Figure 2.
Map-based cloning of SPR2. A, Recombination mapping of spr2-1. Mapping with the PCR-based markers localized the SPR2 locus to a region spanned by BACs F10M23, T24A18, and M4I22. B, A schematic diagram of SPR2. SPR2 contains four exons (thick boxes) separated by three introns (lines). The ORF in the exons is shown in black. Mutant lesions are indicated for five spr2 alleles. C, Molecular complementation of spr2. The pBI-SPR2 vector in which SPR2 cDNA was expressed with the 5′ and 3′ regions of the SPR2 gene was introduced into the spr2-2 mutant.
The 1-bp deletion in the third exon of At4g27060 in spr2-1 would cause a frameshift in the encoded protein after amino acid 550. Further sequence analysis revealed that the other three spr2 alleles all carried mutations in this gene. In spr2-2, the 5′ exon-intron boundary of the third exon was changed from AG:AT to AA:AT. This change destroys the splicing consensus at the acceptor site. In spr2-3, the C at 1,585 was changed to a T, resulting in the substitution of Glu-528 with a stop codon. We also found a 2-bp deletion in the first exon of At4g27060 in the convoluta allele, which would cause a frameshift after amino acid 76. A line with a T-DNA insertion in the third exon of At4g27060 (designated as spr2-4) was identified in the SALK T-DNA collection and showed a right-handed helical growth phenotype characteristic of spr2 mutants when the T-DNA was present in the homozygous state. These five spr2 alleles showed very similar phenotypes, indicating that these are all null alleles.
A full-length cDNA of At4g27060 was assembled, fused to 1.3-kb 5′-flanking and 1.4-kb 3′-flanking genomic regions, and introduced into spr2-2 by an Agrobacterium-mediated transformation method. Thirteen kanamycin-resistant transgenic lines were generated and clear complementation of the spr2 mutant phenotype was confirmed in 11 of them (Fig. 2C). We therefore conclude that At4g27060 is the SPR2 gene.
SPR2 Encodes a Novel Protein with HEAT Repeats
SPR2 is predicted to encode a protein of 864 amino acids with an estimated molecular mass of 94 kD and a pI of 5.47 (Fig. 2B). A BLAST search of databases identified similar proteins of unknown function in Arabidopsis, potato (Solanum tuberosum), and rice (Oryza sativa; Fig. 3A) and multiple expressed sequence tag sequences in various plants, but no homologous proteins in nonplant organisms. The Arabidopsis protein encoded by At1g50890 and potato HIP2 share the highest similarity to SPR2 over the entire length (48% and 50% identity to SPR2, respectively; Fig. 3A). Potato HIP2 had been isolated as a host protein interacting with a potyviral multifunctional protein helper-component proteinase by a yeast (Saccharomyces cerevisiae) two-hybrid screening (Guo et al., 2003). Sequence analysis using a repeat-finding method (Andrade et al., 2000) identified repeat motifs in SPR2 and its homologs. Nine repeats are predicted in SPR2, At1g50890, and HIP2 in the regions highly conserved among them (Fig. 3B), and seven of them are clustered in the N-terminal region. The repeat unit consists of 38 to 41 amino acid residues and is composed of two predicted α-helical structures separated by a hinge region (Fig. 3C). Alignment of the SPR2-repeat motifs showed that the SPR2 repeat is characteristic of the HEAT repeat. Canonical HEAT-repeat motifs have seven conserved hydrophobic residues in two α-helices, in which four are often Leu and one is frequently Val (Fig. 3C; Andrade et al., 2001). Although structurally related Armadillo motifs also share these residues, the signature residues for HEAT motifs are Asp right after the first α-helix and Arg/Lys in the N terminus of the second α-helix, which are both conserved in the SPR2-repeat motifs. The HEAT-repeat sequences and their predicted numbers in the four less-related SPR2 homologs (At2g07170, At1g27210, At1g59850, and At5g62580) are divergent from those in SPR2 and At1g50890.
Figure 3.
SPR2 is a novel, HEAT-repeat-containing protein. A, Phylogenetic tree of SPR2 and its relatives. Full-length amino acid sequences were aligned with the ClustalW method (http://www.ebi.ac.uk/clustalw/#), and the unrooted phylogram was generated using TreeView (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). The GenBank protein ID number is Q8H513 for the rice SPR2 homolog and CAD45375 for potato HIP2 (Guo et al., 2003). B, Alignment of deduced amino acid sequences of SPR2, its closest Arabidopsis homolog (At1g50890), and potato HIP2. Residues identical in a least two sequences are shaded in gray, and dashes indicate gaps introduced to maximize the alignment. Nine HEAT-repeat motifs are underlined. Asterisks above the N-terminal sequences (amino acids 1–37 in SPR2) indicate Ser and Thr residues. Peptide fragments of HIP2 that were identified by GC-MS/MS analysis in Figure 8C are boxed. C, Alignment of nine predicted HEAT-repeat motifs in SPR2. HEAT-repeat motifs consist of two α-helices containing moderately conserved amino acid residues at the indicated positions (Andrade et al., 2001). Residues conforming to the consensus are shaded in black, and similar residues are shaded in gray. Gaps were introduced in the hinge region between the two α-helices to maximize the alignment.
The extreme N-terminal region (amino acids 1–37) of SPR2 is rich in Ser and Thr (11 and 6 out of 37 residues, respectively; Fig. 3B). A Ser/Thr-rich N terminus was also found in At1g50890 and HIP2 but not in other homologs. A motif search with the PSORT program (http://psort.nibb.ac.jp) predicted that this N-terminal region is a targeting signal to plastids, but cellular localization studies (see below) demonstrated that SPR2 is not targeted to plastids.
SPR2 Expression
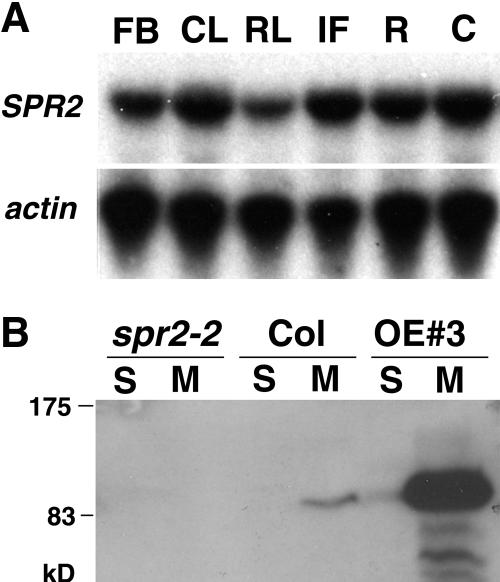
We did not detect significant levels of the SPR2 transcript in wild-type plants by RNA gel-blot analysis. The low abundance of SPR2 sequences in the Arabidopsis expressed sequence tags and massively parallel signature sequencing databases (http://www.arabidopsis.org/Blast/ and http://mpss.udel.edu/at) supports that the SPR2 expression level is low in wild-type Arabidopsis plants. Therefore, we carried out a reverse transcription (RT)-PCR analysis using primers specific to SPR2 but not to its homologs in Arabidopsis. SPR2 expression was detected in flower bud, cauline leaf, rosette leaf, inflorescence stem, root, and cotyledon at similar levels (Fig. 4A), indicating that SPR2 expression is not restricted to any particular organ.
Figure 4.
SPR2 expression and protein accumulation. A, RT-PCR analysis of SPR2 expression in various tissues of wild-type Col plants. FB, flower bud; CL, cauline leaf; RL, rosette leaf; IF, inflorescence stem; R, root; C, cotyledon. Actin gene expression was used as a control. B, Immunoblot analysis with SPR2-specific antibody. Soluble and microsomal proteins (10 μg each) were analyzed in 7-d-old seedlings of Col, spr2-2, and a SPR2-overexpressing line (OE 3).
Next, we raised an anti-SPR2 polyclonal antibody against an N-terminal region of recombinant SPR2 and used it to detect SPR2 protein in Arabidopsis seedlings (Fig. 4B). The antibody recognized a band of about 90 to 95 kD in wild-type seedlings but not in spr2-2 seedlings, indicating its specificity. In wild-type seedlings, more SPR2 protein was recovered in the microsomal fraction than in the soluble fraction.
SPR2 Overexpression
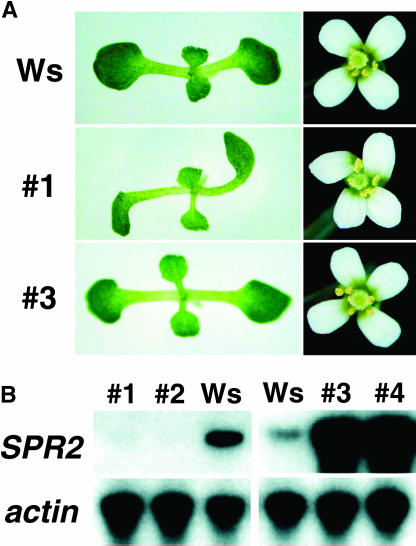
Since the expression level of SPR2 appears to be low in wild-type plants, we examined the consequences of overexpression for cell morphology and growth. SPR2 was fused to a tandem tag of hemaglutinin and hexa-His at its C terminus and expressed under the transcriptional control of the cauliflower mosaic virus (CaMV) 35S promoter in Arabidopsis plants. When the fusion protein was expressed in spr2-2, the helical mutant phenotype was effectively complemented with the transgene expression (data not shown), indicating that this SPR2 fusion protein is functional in plants. We transformed wild-type Wassilewskija plants with the construct and obtained 18 kanamycin-resistant transgenic lines. Ten lines showed right-handed helical growth characteristic of the spr2 mutant, while the remaining eight lines did not differ from wild-type plants in growth or cell morphology and did not show any twisting phenotypes (Fig. 5A). Two lines each from the twisting and wild-type growth groups were analyzed further. In both twisting lines, expression levels of SPR2 were severely reduced, probably by cosuppression (Fig. 5B). In contrast, SPR2 expression was considerably increased in the two lines that showed wild-type growth (Fig. 5B). The SPR2 protein level was also highly increased in these lines (see Fig. 4B). These results indicate that SPR2 overexpression does not induce helical growth or other morphological abnormalities.
Figure 5.
SPR2 overexpression and cosuppression. SPR2 cDNA was expressed under the control of the CaMV 35S promoter in Wassilewskija plants. Transgenic lines were numbered from 1 to 4. A, Seedling and flower phenotypes. B, RT-PCR analysis of SPR2 expression in 7-d-old seedlings. Lines 1 and 2 are cosuppression lines, whereas lines 3 and 4 are overexpression lines. Actin gene expression was used as a control.
SPR2-GFP Protein Associates with Microtubules in Vivo
To examine the intracellular localization, SPR2 was fused to GFP at its C terminus and was expressed under the transcriptional control of the CaMV 35S promoter. When expressed in spr2-2, the SPR2-GFP fusion gene rescued the spr2 mutant phenotype (data not shown), suggesting the functional integrity of this fusion protein. Confocal microscopic analysis revealed that SPR2-GFP fluorescence aligned along filamentous structures in the cortical region of leaf epidermal cells (Fig. 6A). When the epidermal cells were optically sectioned at deeper focal planes, no obvious fluorescent structures were observed in the cytoplasmic regions, but strong fluorescent dots were seen underneath the plasma membrane (Fig. 6B). At high magnification, the cortical filamentous labeling was found in either continuous or linear punctuate patterns (Fig. 6C). In a single cell, both patterns are often present in varying preference to either pattern. Similar filamentous labeling was also observed in leaf trichome (Fig. 6D), epidermal cells of etiolated hypocotyl (Fig. 6E), and epidermal cells of the differentiated region in root (Fig. 6F).
Figure 6.
Intracellular localization of SPR2-GFP protein in vivo. SPR2-GFP fusion protein was stably expressed under the control of the CaMV 35S promoter in spr2-2 plants and the spr2 mutant phenotype was rescued in the transgenic plants. Seven-day-old Arabidopsis seedlings were analyzed with confocal microscopy. A, Leaf epidermis. B, Leaf epidermis with a focal plane adjusted to the middle of the cells' thickness. C, Magnified picture of a portion of the leaf epidermal cells shown in A. D, Leaf trichome. E, Epidermis of etiolated hypocotyl. F, Root epidermis at the differentiation zone. G, Effect of propyzamide on the SPR2-GFP localization. Hypocotyls of transgenic seedlings were treated with propyzamide at 50 μm for 1 h. H, Root meristematic region. Mitotic spindles (sp) and phragmoplasts (ph) were labeled with SPR2-GFP. Bars = 25 μm in A and B, 5 μm in C, and 10 μm in D to H.
The cortical filamentous labeling suggests an association of SPR2-GFP with cortical microtubules. To confirm this, we treated the leaf tissues with the microtubule-destabilizing drug propyzamide (Fig. 6G). This treatment caused the disappearance of filamentous fluorescent patterns, leaving fluorescent foci at the cell cortex. Similar results were obtained with oryzalin (data not shown).
In addition to cortical microtubules, SPR2-GFP was also localized to mitotic spindles and phragmoplast microtubules in dividing cells of the transgenic Arabidopsis roots (Fig. 6H). Cortical microtubules, preprophase bands, mitotic spindles, and phragmoplasts were also stained with SPR2-GFP in stably transformed tobacco BY-2 cells (data not shown).
SPR2 Binds to Microtubules in Vitro
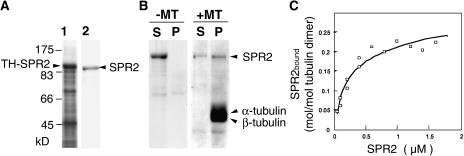
To test whether SPR2 directly binds to microtubules, we carried out an in vitro microtubule-cosedimentation assay. Full-length SPR2 was fused at the N terminus with thioredoxin and poly-His tags (TH-SPR2) and was expressed in Escherichia coli. Chromatography and the cleavage of N-terminal tags with a protease resulted in a homogeneous preparation of recombinant SPR2 in a native form (Fig. 7A). Taxol-stabilized microtubules were prepared from MAP-free tubulins of tobacco BY-2 cells. When the purified SPR2 was centrifuged in the absence of microtubules, the protein remained in the supernatant. However, a significant proportion of SPR2 was recovered in the pellet when it was incubated with an excess of microtubules (Fig. 7B).
Figure 7.
Recombinant SPR2 binds to taxol-stabilized microtubules in vitro. A, Bacterial expression and purification of recombinant SPR2 protein. Full-length SPR2 was fused with thioredoxin and poly-His tags (TH-SPR2), overexpressed in E. coli., and purified to near homogeneity after enzymatic removal of the tags. Crude soluble extract of E. coli and purified SPR2 (without tags) were separated by SDS-PAGE and stained with Coomassie Brilliant Blue. B, Cosedimentation of recombinant SPR2 with taxol-stabilized microtubules. Taxol-stabilized microtubules were prepared from purified tobacco BY-2 tubulins. After recombinant SPR2 was incubated with or without taxol-stabilized microtubules, proteins were centrifuged and analyzed by SDS-PAGE and Coomassie Brilliant Blue staining. The positions of SPR2, α-tubulin, and β-tubulin are indicated by arrowheads. S, Supernatant fraction; P, pellet fraction; and ±MT, presence or absence of microtubules in the assay mixture. C, Quantitative analysis of the binding between SPR2 and microtubules. Various concentrations of purified SPR2 were mixed with taxol-stabilized microtubules (6 μg) and then subjected to cosedimentation analysis, as in B. Assuming there is one SPR2-binding site on the tubulin dimer, the equation q = (qmax × c)/(Kd + c) was fitted. In this equation, q is the amount of protein bound to the tubulin dimer and c is the concentration of free SPR2 in solution. Kd and qmax, which represent the dissociation constant and the amount of bound SPR2 at the saturated level, were calculated from the fitting.
To determine the stoichiometry and affinity of SPR2 toward microtubules, various amounts of SPR2 were centrifuged with a constant quantity of microtubules, and a binding curve was obtained (Fig. 7C). Recombinant SPR2 bound to microtubules in a concentration-dependent and saturable manner. The binding to microtubules was saturated at a stoichiometry of 0.26 ± 0.02 mol of SPR2 per mole of tubulin dimer if we assume that SPR2 binds uniformly to microtubule side walls. The dissociation constant Kd of SPR2 was determined to be 0.23 ± 0.04 μm.
Tobacco SPR2 Homolog Is Enriched in Purified MAP Fractions
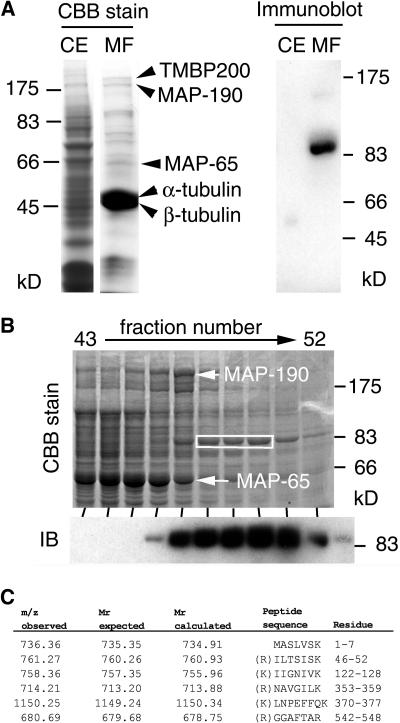
To examine whether endogenous SPR2-like protein is associated with microtubules in plants other than Arabidopsis as well, we searched for the presence of a tobacco SPR2 homolog in purified MAP fractions. Vacuole-free miniprotoplasts were prepared from cultured tobacco BY-2 cells, and microtubules were purified from the protein-rich protoplast extracts through two rounds of microtubule polymerization and depolymerization using taxol (Igarashi et al., 2000). The purified fraction was highly enriched with α- and β-tubulins, as well as several copurified proteins (Fig. 8A, left). Mass spectrometric analysis identified three major copurified bands as known tobacco MAPs: TMBP200 (Yasuhara et al., 2002), MAP-190 (Igarashi et al., 2000), and a mixture of MAP-65a and MAP-65c (Smertenko et al., 2000; data not shown). Immunoblot analysis showed that a protein of about 90 kD cross-reacted with anti-SPR2 antibody in the microtubule protein fraction but was undetectable in the crude cell extract (Fig. 8A, right), indicating that a tobacco protein immunologically related to SPR2 was copurified with microtubules.
Figure 8.
Immunological and mass spectrometric identification of tobacco SPR2 homolog in MAP fractions purified from tobacco BY-2 cells. A, Crude protein extract from tobacco miniprotoplasts (CE) and proteins in a purified MAP fraction (MF) were separated by SDS-PAGE and stained with Coomassie Brilliant Blue (left) or subjected to immunoblotting with an anti-SPR2 antibody (right). Arrowheads on the left indicate the positions of TMBP200, MAP-190, MAP-65, α-tubulin, and β-tubulin. B, After proteins in the tobacco MAP fraction were fractionated with anion-exchange chromatography, eluted fractions (43–52) were separated by SDS-PAGE and stained with Coomassie Brilliant Blue (upper image) or analyzed by immunoblotting with an anti-SPR2 antibody (IB; lower image). The positions of MAP-190 and MAP-65 are indicated by arrows. Tobacco SPR2-related protein was abundant in fractions 47 to 51. Coomassie Brilliant Blue-stained bands containing the SPR2-related protein in fractions 48 to 50 (white box) were excised and subjected to LC-MS/MS analysis. C, Peptide sequences identified by LC-MS/MS analysis. Mass values of precursor ions (m/z observed), expected peptide mass values (molecular mass expected), and mass values calculated from the peptide sequences in the database (molecular mass calculated) are shown. Amino acid residues of the identified peptides are numbered based on the HIP2 sequence and are also shown in boxes in Figure 3B. All peptides had an Arg or a Lys at the last residue position and at the position immediately prior to the initial peptide residues in the intact HIP2 sequence (except the N-terminal peptide), as expected for peptides digested with trypsin.
To unambiguously identify this SPR2-related protein, tobacco proteins in the purified MAP fraction were further fractionated by anion-exchange chromatography, and eluted fractions were analyzed by general protein staining and immunoblotting. Tobacco SPR2-related protein was eluted right after the MAP-65 and MAP-190 fractions (Fig. 8B) and before the tubulin fractions (data not shown). The amount of this protein appeared to be less than that of MAP-65 or MAP-190. The SPR2-related protein bands in fractions 48 to 50 were excised from the polyacrylamide gel and then subjected to liquid chromatography (LC)-tandem mass spectrometry (MS/MS) analysis. When the mass spectrum data were used to screen the National Center for Biotechnology Information (NCBI) nonredundant protein database, the best fit was found in the amino acid sequence of potato HIP2, a closely related homolog of SPR2. Six peptide sequences matched perfectly with the expected peptide sequences from tryptic digests of HIP2 (Figs. 8C and 3B). Taken together, we conclude that a tobacco SPR2 homolog was copurified with microtubules from tobacco BY-2 cells.
DISCUSSION
SPR2 Binds to Microtubules in Vivo and in Vitro
In this study, we showed that SPR2-GFP fusion protein decorated microtubules in Arabidopsis cells and that recombinant SPR2 bound to taxol-stabilized microtubules in vitro. These results establish that SPR2 is a bona fide MAP from plants. In addition, tobacco SPR2 homolog was concentrated in MAP fractions highly purified from tobacco BY-2 cells.
When SPR2-GFP was overexpressed in Arabidopsis and tobacco cells, it decorated not only cortical microtubules but also preprophase bands, spindle microtubules, and phragmoplast microtubules. In such microtubule structures, we did not find any prominent labeling of either plus or minus ends of microtubules. Thus, SPR2 may not distinguish between the distinct microtubule structures found in interphase and mitotic cells and may not be recruited to microtubule ends. However, the data on the distribution of SPR2-GFP should be interpreted with caution. SPR2 transcripts are not abundant in Arabidopsis plants, and tobacco SPR2 homolog is present in relatively small amounts in purified MAP fractions from tobacco BY-2 cells, compared to major MAPs such as TMBP200, MAP-190, and MAP-65. Although the complementation of spr2 mutant phenotypes by overexpressed SPR2-GFP indicates that a fraction of the expressed fusion protein should function at normal cellular target sites, excess SPR2 molecules might associate with microtubule structures and microtubule regions for which endogenous SPR2 normally shows low affinity and may thus mask the true in vivo localization patterns. When mammalian plus-end labeling MAP CLIP170 was expressed transiently in cowpea protoplasts or stably in tobacco BY-2 cells as a construct fused to GFP, microtubules were labeled along their entire length as expression levels increased (Dhonukshe and Gadella, 2003). The immunolocalization of endogenous SPR2 in wild-type plants should give a more in-depth characterization of the association of SPR2 with microtubules.
We observed that, in some cells, SPR2-GFP accumulated in discrete punctate patterns along the microtubules. Our time-lapse observation indicates that these SPR2-GFP dots on microtubules do not represent plus ends of microtubules (data not shown). Interestingly, a similar linear punctate labeling of microtubules was sometimes observed for the GFP fusion of an Arabidopsis end-binding protein, AtEB1a (Chan et al., 2003). We need to confirm such punctate-labeling patterns of SPR2 for the wild-type endogenous protein.
SPR2 Defines a New MAP Family with HEAT Repeats
SPR2 and its homologs contain tandemly repeated sequences that show characteristics of the HEAT motif. HEAT repeats are found in a wide variety of eukaryotic proteins, including those from which it derives its name (Huntingtin, elongation factor 3, a subunit of protein phosphatase 2A, and lipid kinase TOR1; Andrade and Bork, 1995). Although amino acid sequences of the HEAT motif are highly divergent, there is a consensus pattern of conserved hydrophobic and other characteristic residues (Andrade et al., 2001), which are generally found in nine repeat motifs in SPR2. Crystal structures of three HEAT-repeat proteins, a subunit of protein phosphatase 2A (Groves et al., 1999), importin β1 (Cingolani et al., 1999), and β2 (Chook and Blobel, 1999), revealed that each motif consists of a pair of antiparallel α-helices that form a helical hairpin. In HEAT-repeat proteins, neighboring repeats stack together into a single domain with a continuous hydrophobic core, forming an elongated superhelix. HEAT repeats appear to function as flexible joints that can wrap around target substrates and as scaffolding on which other molecular components may assemble (Neuwald and Hirano, 2000).
It is noteworthy that HEAT repeats also occur in two classes of proteins that associate with tubulins or microtubules. First, XMAP215/TOG (Ohkura et al., 2001) and distantly related MAST/ORBIT families (Inoue et al., 2000; Lemos et al., 2000) are evolutionary conserved MAPs that contain a large number of HEAT motifs. Electron microscopic analysis suggests that XMAP215 is a long flexible molecule that may be composed entirely of HEAT repeats and could span up to seven to eight tubulin dimers along a microtubule protofilament (Cassimeris et al., 2001). The C-terminal part of XMAP215 and its Arabidopsis homolog MOR1 contains a microtubule-binding domain that is mainly responsible for targeting to centrosomes or microtubules (Popov et al., 2001; Twell et al., 2002), while the evolutionary conserved N-terminal XMAP215 domain consisting of HEAT repeats stabilizes microtubules (Popov et al., 2001). It remains unknown whether the N-terminal HEAT-repeat domain directly associates with microtubules or exerts its microtubule stabilization effect by interacting with destabilizing factors. Two temperature-sensitive mor1 alleles have single amino acid substitutions in the most N-terminal HEAT motif, suggesting the functional significance of this HEAT motif in stabilizing cortical microtubules (Whittington et al., 2001). Second, more than six repeated HEAT motifs are found in tubulin-folding cofactor D (Neuwald and Hirano, 2000). The fission yeast homolog of cofactor D colocalizes with microtubules and binds to taxol-stabilized microtubules (Hirata et al., 1998). Thus, in addition to a role in the folding of β-tubulin, cofactor D has been suggested to function as a MAP.
Because SPR2 does not contain any known microtubule-binding domains found in established MAPs, it may be recruited to microtubules mainly through the HEAT-repeat motifs. In vitro binding stoichiometry data indicate that one SPR2 molecule spans four tubulin dimers when bound to microtubules if it binds uniformly to microtubule side walls. Mutational analysis of the SPR2 sequence should reveal the significance of the HEAT repeat in microtubule binding in vitro and in vivo.
Alternatively, but not mutually exclusively, the SPR2 HEAT repeat may be used as a scaffold to interact with molecules other than microtubules. A potato SPR2 homolog HIP2 has been isolated as an interaction partner of potyvirus helper-component proteinase (HCpro; Guo et al., 2003). HCpro is a mutifunctional protein involved in diverse viral activities, such as autoproteolytic cleavage of its C terminus from the polyprotein, RNA binding, genome replication and symptom expression, aphid transmission, viral movement in plants, and suppression of RNA silencing (Maia et al., 1996; Kasschau and Carrington, 1998). Plant viruses move toward the plasmodesmata via the cytoskeletal network and spread to neighboring cells. Association of the viral genome with microtubules is well established in tobacco mosaic virus. Current models propose that the movement protein of tobacco mosaic virus tracks microtubules toward the plasmodesmata (Boyko et al., 2000; Más and Beachy, 2000) or that the microtubule network is used to provide a route toward a cellular degradation site of the movement protein (Gillespie et al., 2002). The proposed HCpro-HIP2 interaction might link the potyvirus activities to the microtubule cytoskeleton.
SPR2 and Helical Growth
Mutations in the SPR2 gene cause right-handed helical growth in epidermal cells of elongating tissues, most notably in hypocotyls, petioles, and petals. Now that SPR2 is found to be a MAP, the mutant phenotype is understood in the context of microtubule functions. It should be noted, however, that we are yet to directly demonstrate microtubule abnormalities in spr2 mutants. This is partly because spr2 mutants show relatively mild defects in growth anisotropy, especially in the root where a strict relationship between the orientation of cortical microtubule arrays and direction of cell expansion is observed in the rapid elongation zone (e.g. Sugimoto et al., 2000). In the petiole epidermal cells, microtubule patterns significantly deviate from a predominantly transverse orientation and are often different in neighboring cells. In these cells, we could not observe a clear difference in cortical microtubule patterns between the wild type and spr2 (T. Shoji and T. Hashimoto, unpublished data). Microtubule defects in spr2 mutants may be sought for the in vivo dynamics of cortical microtubules rather than gross orientation of microtubule arrays.
Nevertheless, moderate defects of microtubule function in spr2 can be inferred from the enhanced inhibition of root growth by microtubule-interacting drugs in the spr2 mutant background (Fig. 1F). The expression of a GFP-α-tubulin fusion protein in transgenic Arabidopsis plants results in spr2-like phenotypes (Ueda et al., 1999), and a similar right-handed helical growth mutant spr1 is caused by mutations in a microtubule-localized protein (Nakajima et al., 2004). These results suggest that relatively moderate microtubule defects in expanding interphase cells can lead to right-handed helical growth. One possible explanation for the relatively mild microtubule defects in spr2 mutants is that other MAPs compensate for the function of SPR2. The Arabidopsis genome contains one protein highly homologous to SPR2 and four other HEAT-repeat-containing proteins in the SPR2 family (Fig. 3A). Simultaneous knockouts in these SPR2-related genes may reveal the full functional spectrum of SPR2-type MAPs in microtubule organization and cell morphology.
lefty tubulin mutants (Thitamadee et al., 2002) and wild-type seedlings treated with low doses of microtubule-interacting drugs (Furutani et al., 2000) show consistent left-handed helical growth, whereas mutations in SPR2 MAP (this study) and SPR1 microtubule-localized protein (Nakajima et al., 2004) result in right-handed twisting in elongating cells. What determines the direction of growth is an interesting question. In left-handed helical growth, cortical microtubule arrays may be somewhat destabilized and further destabilization may lead to random orientation and microtubule fragmentation (Abe et al., 2004). We believe that microtubule defects are qualitatively different in cells undergoing either left-handed or right-handed helical growth. To understand the microtubule status in more depth, it will be necessary to evaluate microtubule dynamics in various helical growth mutants. Techniques are available for in vivo imaging of dynamic microtubule behavior using GFP-fused tubulins or MAPs (Chan et al., 2003; Dhonukshe and Gadella, 2003; Shaw et al., 2003; Nakamura et al., 2004; Vos et al., 2004). The biochemical characterization of SPR2 protein based on in vitro microtubule dynamics should also advance our understanding of the control of growth anisotropy.
MATERIALS AND METHODS
Plant Growth and Genetic Crosses
Seedlings of Arabidopsis (Arabidopsis thaliana) were grown on agar plates as described (Furutani et al., 2000) unless otherwise noted. Oryzalin, propyzamide, and taxol (pactitaxel from Taxus brevifilia) were purchased from Wako (Osaka), while RH-4032 [3,5-dichloro-N-(3-chloro-1-ethyl-1-methyl-2-oxopropyl-9-benzamide] was a gift from D.H. Young (Rohm and Haas, Philadelphia). spr2-1 (Ler background) and convoluta (S95 background) alleles had been described (Furutani et al., 2000). spr2-2 and spr2-3 alleles were newly isolated from ethyl methanesulfonate-mutagenized M2 seeds of the Columbia (Col) background (Lehle Seeds, Round Rock, TX). spr2-4 is a T-DNA-tagged allele generated at the Salk Institute Genomic Analysis Laboratory.
For allelism tests, the newly isolated spr2-2 and spr2-3 were crossed with spr2-1, and phenotypic complementation was confirmed in the F1 seedlings. The double mutant between spr2-1 and lefty2 was selected in F2 populations according to their molecular lesions, and the F3 plants were used for analysis. For mapping, spr2-1 (Ler) was crossed with a wild-type plant (Col) and selfed to obtain an F2-mapping population.
Microscopy
Gross morphology was observed using an Olympus stereoscope SZX12 equipped with a digital camera DP70 or DP10 (Olympus, Tokyo). We used a replica method to observe epidermal cell files by scanning electron microscopy. Molds were made by pressing the tissues into polyvinylsiloxane impression material (Extrude, Kerr, Romulus, MI) and were filled with epoxy glue (Araldite, Ciba-Geigy, Research Triangle Park, NC). After being coated with platinum, the replica images were examined with an S4700 scanning electron microscope (Hitachi, Tokyo).
Confocal imaging was performed with a Zeiss Axioplan microscope equipped with a confocal head LSM510 (Zeiss, Jena, Germany) and an argon ion laser. GFP was excited at a 488-nm laser line, and emitted fluorescence was collected through a 525/30 bandpass filter.
Molecular Analysis
spr2-1 homozygous seedlings were selected from the F2-mapping population and their DNA was used to map the SPR2 locus using cleaved-amplified polymorphic sequence and simple sequence length polymorphic markers (http://www.arabidopsis.org/aboutcaps.html). Two PCR-based markers were newly designed to detect polymorphisms between Col and Ler near the SPR2 locus. Marker names, primer sequences, restriction enzymes used, and the ecotype cut by the enzyme are as follows: F10M23.1, 5′-CAAGTCTTCAACAGATGATG-3′, 5′-CCACATCTAACCCACTAGAC-3′, no restriction (simple sequence length polymorphic marker); and M4I22.1, 5′-CCGTTCTCTAATTGCATGTC-3′, 5′-AGCATTTTGTTGACCGACTC-3′, MboI, Ler. Candidate genes in the fine-mapped region were amplified from genomic DNA of Ler and spr2-1 by PCR and were directly sequenced to detect mutations.
Both ends of SPR2 cDNA were determined using 5′- and 3′-RACE systems (Gibco BRL, Cleveland). Full-length SPR2 cDNA was cloned from Col wild-type seedlings by RT-PCR using the SuperScript First-Strand Synthesis System (Gibco BRL). From BAC clone T24A18, the 5′-flanking region (1,340 bp), and the 3′-flanking region (1,436 bp) of SPR2 were cloned by PCR. The cloned fragments were sequenced to verify that there were no unexpected mutations.
For pBI-SPR2, SPR2 cDNA was placed between the 5′- and 3′-flanking regions of SPR2 in pBI 101 (CLONTECH Laboratories, Palo Alto, CA). For pBI-35S-SPR2HH, SPR2 cDNA with a 3′-terminal tag (encoding N-GGLVGGYPYDVPDYAGVEHHHHHH-C) was subcloned immediately downstream of the CaMV 35S promoter in pBI121 (CLONTECH Laboratories). For pBI-SPR2-GFP, SPR2 cDNA was subcloned with a linker (N-GGLVRPPAGRGGGGAT-C) in-frame just upstream of GFP in pTH-2 (Niwa, 2003), and the SPR2-GFP portion was excised and placed downstream of the CaMV 35S promoter in pBI121. For bacterial expression, full-length SPR2 and an N-terminal region (amino acids 1–438) of SPR2 were subcloned in pET32b and pET40b (Novagen, Madison, WI) to give pET-SPR2 and pET-SPR2N, respectively. Detailed information on these constructs is available on request.
Total RNA was isolated using the RNeasy kit (Qiagen, Valencia, CA) and converted to cDNA with the SuperScript First-Strand Synthesis System (Gibco BRL). For semiquantitative RT-PCR, we calibrated the amounts of cDNA templates by comparing the amounts of PCR products from a dilution series of the templates. Aliquots of first-strand cDNA were amplified by PCR for 3 min at 94°C, followed by 30 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C, and finally a 10-min incubation at 72°C. The PCR primers were as follows: 5′-TGGTGCAGTCCCTCGTCCAACC-3′ and 5′-GTTCCATTGGAACTCCAAACGT-3′ for SPR2 amplification, and 5′-ATGAAGATTAAGGTCGTGGCA-3′ and 5′-TCCGAGTTTGAAGAGGCTAC-3′ for ACT8 amplification. After separation on agarose gel, the PCR products were blotted on membranes and detected by Southern hybridization using an Alkphos Direct Labeling and Detection System (Amersham Biosciences, Piscataway, NJ).
Plant Transformation
Binary vectors pBI-SPR2, pBI-35S-SPR2HH, and pBI-SPR2-GFP were used to transform Agrobacterium tumefaciens strain MP90 and then introduced into Arabidopsis plants by floral dipping. T1 plants were selected for kanamycin resistance, and the integration of the transgenes into the plant genome was confirmed by PCR. T2 and T3 plants homozygous for the transgene were used for analysis.
Suspension cultured tobacco (Nicotiana tabacum) BY-2 cells were maintained at 27°C in a Murashige and Skoog medium containing 3% Suc, 0.2 mg/mL of KH2PO4, 0.1 mg/mL of myoinositol, 1.0 mg/L of thiamine, and 0.9 μm 2,4-dichlorophenoxyacetic acid. The binary vector pBI-SPR2-GFP was introduced into tobacco BY-2 cells using A. tumefaciens strain LBA4404, essentially as described (An, 1987). Kanamycin-resistant calli were transferred to a liquid medium to establish suspension cultures.
Recombinant SPR2 Protein
The bacterial expression vector pET-SPR2 was introduced into Escherichia coli strain BL21(DE3) Codon-Plus (Novagen). Bacteria were grown in Luria-Bertani medium supplemented with 100 mg/L of ampicillin with vigorous shaking at 37°C. When the optical density of the culture at 600 nm reached 0.4, isopropylthio-β-d-galactoside was added at a concentration of 1 mm to induce protein expression and the culture continued for an additional 4 h. Bacteria were harvested and lysed in a phosphate buffer (50 mm NaH2PO4, pH 8.0, and 300 mm NaCl) containing 10 mm imidazole by repeating a freeze/thaw cycle three times followed by sonication. After centrifugation of the lysate, the supernatant was mixed with Ni-NTA agarose matrix (Qiagen) and gently rotated at 4°C for 1 h. After the mixture was loaded on a column, the column was washed with the phosphate buffer containing 20 mm imidazole and bound proteins were subsequently eluted with the phosphate buffer containing 100 mm imidazole. Partially purified protein was treated with enterokinase (Novagen) to remove the N-terminal thioredoxin and poly-His tags and then loaded onto a Mono Q Fast-Flow column (Amersham Biosciences). Proteins were eluted from the column using a linear gradient of NaCl from 0 to 1 m. Fractions containing the recombinant SPR2 protein were desalted and concentrated.
In Vitro Microtubule-Binding Assay
Tubulins were prepared from cultured tobacco BY-2 cells as described (Igarashi et al., 2000). Purified tubulins (10 μm in terms of the tubulin dimers) were polymerized at 37°C for 20 min in PME buffer (100 mm PIPES, pH 6.9, 0.5 mm MgSO4, 1 mm EGTA, 1 mm GTP, and 20 μm taxol). A constant quantity of taxol-stabilized microtubules (6 μg) was incubated with the recombinant SPR2 protein in PME buffer at 37°C for 15 min. Microtubules were pelleted by centrifugation at 100,000g for 10 min. Proteins in the pellet and the supernatant were separated by SDS-PAGE and stained with Coomassie Brilliant Blue. The gels were scanned with a CanoScan D2400U (Canon, Lake Success, NY) and the intensities of the stained bands were quantified using a BAStation (Fujifilm, Tokyo).
Immunoblot Analysis
The 6×-His-tagged truncated SPR2 (amino acids 1–438) was overexpressed in E. coli strain BL21 (DE3) containing pET-SPR2N, purified through an Ni-NTA agarose column (Qiagen) and a Q-Sepharose Fast Flow column (Amersham Biosciences), and used to produce polyclonal mouse and rabbit antisera.
Microsomal and soluble proteins were prepared from Arabidopsis seedlings grown in a liquid medium as described (Nakajima et al., 2004). MAPs were purified from tobacco BY-2 miniprotoplasts through two cycles of taxol-dependent polymerization and depolymerization as described (Igarashi et al., 2000). The purified tobacco MAPs were further fractionated through a Resource Q column (Amersham Biosciences) that had been equilibrated with 25 mm HEPES, pH 7.5, 1 mm MgCl2, and 1 mm EDTA, and the bound proteins were eluted using a linear gradient of NaCl from 0 to 1 m. Proteins in each elution fraction were separated by SDS-PAGE and transferred onto a Hybond-P membrane (Amersham Biosciences). An anti-SPR2 mouse antiserum was used at a 1:1,000 dilution, and signals were detected using the ECL-plus Western Blotting kit (Amersham Biosciences).
Immunoblot analysis of proteins prepared from tobacco BY-2 cells was done using an anti-SPR2 rabbit antiserum at a 1:1,000 dilution.
Mass Spectrometry
MAP fractions were prepared from tobacco BY-2 cells as described (Igarashi et al., 2000). Proteins in the purified tobacco MAP fractions were separated by SDS-PAGE, and the gel portions corresponding to tobacco SPR2 and other prominent bands were excised. After the reduction and alkylation treatments, proteins in the gel slices were digested with 12.5 μg/mL of modified trypsin (Roche, Indianapolis) at 37°C for 16 h as described (Shevchenko et al., 1996). Digested peptides were extracted with formic acid and acetonitrile. Peptide mixtures were separated and analyzed using a LC MAGIC 2002 (Michrom BioResources, Auburn, CA), which was connected directly to an electrospray ion-trapping tandem mass spectrometer LCQ-Advantage (Thermo Electron, San Jose, CA; Ohta et al., 2002). Mass spectrum data generated by the LC-MS/MS were used to search the NCBI nonredundant protein database with Mascot MS/MS Ion Search software (Matrix Science, Boston).
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requester.
Recent Developments
After this work was submitted, molecular cloning of TOR1, which is allelic to SPR2, was published (Buschmann H, Fabri CO, Hauptmann M, Hutzler P, Laux T, Lloyd CW, Schäffner AR [2004] Helical growth of the Arabidopsis mutant tortifolia1 reveals a plant-specific microtubule-associated protein. Curr Biol 14: 1515–1521).
Acknowledgments
We thank D.H. Young and Y. Niwa for providing RH-4032 and pTH-2, respectively, and M. Yoshimura, T. Yasuhara, and M. Kuwano for technical assistance. We would like to acknowledge the Arabidopsis Biological Resource Center for providing a T-DNA knockout allele, convoluta mutant, and DNA materials.
This work was supported in part by the Ministry of Education, Culture, Sports, Science and Technology (grant nos. 15031219 to T.H. and 15770029 to T.S.), and by the Sasakawa Scientific Research Grant from the Japan Science Society (to T.S.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.051748.
References
- Abe T, Thitamadee S, Hashimoto T (2004) Microtubule defects and cell morphogenesis in the lefty1lefty2 tubulin mutant of Arabidopsis thaliana. Plant Cell Physiol 45: 211–220 [DOI] [PubMed] [Google Scholar]
- An G (1987) Binary Ti vectors for plant transformation and promoter analysis. Methods Enzymol 153: 292–305 [Google Scholar]
- Andrade MA, Bork P (1995) HEAT repeats in the Huntington's disease protein. Nat Genet 11: 115–116 [DOI] [PubMed] [Google Scholar]
- Andrade MA, Petosa C, O'Donoghue SI, Müller CW, Bork P (2001) Comparison of ARM and HEAT protein repeats. J Mol Biol 309: 1–18 [DOI] [PubMed] [Google Scholar]
- Andrade MA, Ponting CP, Gibson TJ, Bork P (2000) Homology-based method for identification of protein repeats using statistical significance estimates. J Mol Biol 298: 521–537 [DOI] [PubMed] [Google Scholar]
- Bouquin T, Mattsson O, Naested H, Foster R, Mundy J (2003) The Arabidopsis lue1 mutant defines a katanin p60 ortholog involved in hormonal control of microtubule orientation during cell growth. J Cell Sci 116: 791–801 [DOI] [PubMed] [Google Scholar]
- Boyko V, Ferralli J, Ashby J, Schellenbaum P, Heinlein M (2000) Function of microtubules in intercellular transport of plant virus RNA. Nat Cell Biol 2: 826–832 [DOI] [PubMed] [Google Scholar]
- Bürger D (1971) Die morphologischen Mutanten des Göttinger Arabidopsis-Sortiments, einschließlich der Mutanten mit abweichender Samenfarbe. Arabidopsis Inf Serv 8: 36–42 [Google Scholar]
- Cassimeris L, Gard D, Tran PT, Erickson HP (2001) XMAP215 is a long thin molecule that does not increase microtubule stiffness. J Cell Sci 114: 3025–3033 [DOI] [PubMed] [Google Scholar]
- Chan J, Calder GM, Doonan JH, Lloyd CW (2003) EB1 reveals mobile microtubule nucleation sites in Arabidopsis. Nat Cell Biol 5: 967–971 [DOI] [PubMed] [Google Scholar]
- Chook X, Blobel Y (1999) Structure of the nuclear transport complex kanyopherin-beta2-Ran×GppNHp. Nature 399: 230–237 [DOI] [PubMed] [Google Scholar]
- Cingolani G, Petosa C, Weis K, Mueller CW (1999) Structure of importin-β bound to the IBB domain of importin-α. Nature 399: 221–229 [DOI] [PubMed] [Google Scholar]
- Cyr RJ, Palevitz BA (1995) Organization of cortical microtubules in plant cells. Curr Opin Cell Biol 7: 65–71 [DOI] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ (1997) Microtubule polymerization dynamics. Annu Rev Cell Dev Biol 13: 83–117 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Gadella TWJ Jr (2003) Alteration of microtubule dynamic instability during preprophase band formation revealed by yellow fluorescent protein-CLIP170 microtubule plus-end labeling. Plant Cell 15: 597–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani I, Watanabe Y, Prieto R, Masukawa M, Suzuki K, Naoi K, Thitamadee S, Shikanai T, Hashimoto T (2000) The SPIRAL genes are required for directional control of cell elongation in Arabidopsis thaliana. Development 127: 4443–4453 [DOI] [PubMed] [Google Scholar]
- Gillespie T, Boevink P, Haupt S, Roberts AG, Toth R, Valentine T, Chapman S, Oparka KJ (2002) Functional analysis of a DNA-shuffled movement protein reveals that microtubules are dispensable for the cell-to-cell movement of Tobacco mosaic virus. Plant Cell 14: 1207–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves MR, Hanlon N, Turowski P, Hemmings BA, Barford D (1999) The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell 96: 99–110 [DOI] [PubMed] [Google Scholar]
- Guo D, Spetz C, Saarma M, Valkonnen JP (2003) Two potato proteins, including a novel RING finger protein (HIP1), interact with the potyviral multifunctional protein HCpro. Mol Plant Microbe Interact 16: 405–410 [DOI] [PubMed] [Google Scholar]
- Hashimoto T (2002) Molecular genetic analysis of left-right handedness in plants. Philos Trans R Soc Lond B Biol Sci 357: 799–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T (2003) Dynamics and regulation of plant interphase microtubules: a comparative view. Curr Opin Plant Biol 6: 568–576 [DOI] [PubMed] [Google Scholar]
- Hirata D, Masuda H, Eddison M, Toda T (1998) Essential role of tubulin-folding cofactor D in microtubule assembly and its association with microtubules in fission yeast. EMBO J 17: 658–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi H, Orii H, Mori H, Shimmen T, Sonobe S (2000) Isolation of a novel 190 kDa protein from tobacco BY-2 cells: possible involvement in the interaction between actin filaments and microtubules. Plant Cell Physiol 41: 920–931 [DOI] [PubMed] [Google Scholar]
- Inoue YH, Avides MDC, Shiraki M, Deak P, Yamaguchi M, Nishimoto Y, Matsukage A, Glover DM (2000) Orbit, a novel microtubule-associated protein essential for mitosis in Drosophila melanogaster. J Cell Biol 149: 153–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Carrington JC (1998) A counter-defensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95: 461–470 [DOI] [PubMed] [Google Scholar]
- Lemos CL, Sampaio P, Maiato H, Costa M, Omel'yanchuk LV, Liberal V, Sunkel CE (2000) Mast, a conserved microtubule-associated protein required for bipolar mitotic spindle organization. EMBO J 19: 3668–3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia IG, Haenni AL, Bernardi F (1996) Potyviral HC-pro: a multifunctional protein. J Gen Virol 77: 1335–1341 [DOI] [PubMed] [Google Scholar]
- Más P, Beachy RN (2000) Role of microtubules in the intracellular distribution of tobacco mosaic virus movement protein. Proc Natl Acad Sci USA 97: 12345–12349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kernebeck B, Srinivas BP, Hülskamp M (2003) A novel localization pattern for an EB1-like protein links microtubule dynamics to endomembrane organization. Curr Biol 13: 1191–1197 [DOI] [PubMed] [Google Scholar]
- Mollinari C, Kleman JP, Jian W, Schoehn G, Hunter T, Margolis RL (2002) PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J Cell Biol 157: 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Smertenko A, Wagner V, Heinrich M, Hussey PJ, Hauser MT (2004) The plant microtubule-associated protein AtMAP65-3/PLE is essential for cytokinetic phragmoplast function. Curr Biol 14: 412–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Furutani I, Tachimoto H, Matsubara H, Hashimoto T (2004) SPIRAL1 encodes a plant-specific microtubule-localized protein required for directional control of rapidly expanding Arabidopsis cells. Plant Cell 16: 1178–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Naoi K, Shoji T, Hashimoto T (2004) Low concentrations of propyzamide and oryzalin alter microtubule dynamics in Arabidopsis epidermal cells. Plant Cell Physiol 21: 1330–1334 [DOI] [PubMed] [Google Scholar]
- Neuwald A, Hirano T (2000) HEAT repeats associated with condensins, cohesins, and other complexes involved in chromosome-related functions. Genome Res 10: 1445–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y (2003) A synthetic green fluorescent protein for plant biotechnology. Plant Biotechnol 20: 1–11 [Google Scholar]
- Ohkura H, Garcia MA, Toda T (2001) Dis1/TOG universal microtubule adaptors—one MAP for all? J Cell Sci 114: 3805–3812 [DOI] [PubMed] [Google Scholar]
- Ohta S, Shiomi Y, Sugimoto K, Obuse C, Tsurimoto T (2002) A proteomics approach to identify proliferating cell nuclear antigen (PCNA)-binding proteins in human cell lysates. J Biol Chem 277: 40362–40367 [DOI] [PubMed] [Google Scholar]
- Popov AV, Pozniakovsky A, Arnal I, Antony C, Ashford AJ, Kinoshita K, Turnebize R, Hyman AA, Karsenti E (2001) XMAP215 regulates microtubule dynamics through two distinct domains. EMBO J 20: 397–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relichova J (1976) Some new mutants. Arabidopsis Inf Serv 13: 25–28 [Google Scholar]
- Schuyler SC, Liu JY, Pellman D (2003) The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. J Cell Biol 160: 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook JC, Ehrhardt DW, Fisher SE, Scheible W, Somerville CR (2004) The Arabidopsis SKU6/SPIRAL1 gene encodes a plus end-localized microtubule-interacting protein involved in directional cell expansion. Plant Cell 16: 1506–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SL, Kamyar R, Ehrhardt DW (2003) Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science 300: 1715–1718 [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Shibaoka H (1994) Plant hormone-induced changes in the orientation of cortical microtubules: alterations in the cross-linking between microtubules and the plasma membrane. Annu Rev Plant Physiol Plant Mol Biol 45: 527–544 [Google Scholar]
- Smertenko A, Saleh N, Igarashi H, Mori H, Hauser-Hahn I, Jiang CJ, Sonobe S, Lloyd CW, Hussey PJ (2000) A new class of microtubule-associated proteins in plants. Nat Cell Biol 2: 750–753 [DOI] [PubMed] [Google Scholar]
- Smith LG, Gerttula SM, Han S, Levy J (2001) TANGLED1: a microtubule binding protein required for the spatial control of cytokinesis in maize. J Cell Biol 152: 231–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Williamson RE, Wasteneys GO (2000) New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol 124: 1493–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thitamadee S, Tuchihara K, Hashimoto T (2002) Microtubule basis for left-handed helical growth in Arabidopsis. Nature 417: 193–196 [DOI] [PubMed] [Google Scholar]
- Twell D, Park SK, Hawkins TJ, Schuber D, Schmidt R, Smertenko A, Hussey PJ (2002) MOR1/GEM1 has an essential role in the plant-specific cytokinetic phragmoplast. Nat Cell Biol 4: 711–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K, Matsuyama T, Hashimoto T (1999) Visualization of microtubules in living cells of transgenic Arabidopsis thaliana. Protoplasma 206: 201–206 [Google Scholar]
- Vos JW, Dogterom M, Emons AMC (2004) Microtubules become more dynamic but not shorter during preprophase band formation. Cell Motil Cytoskeleton 57: 246–258 [DOI] [PubMed] [Google Scholar]
- Wasteneys GO (2002) Microtubule organization in the green kingdom: chaos or self-order? J Cell Sci 115: 1345–1354 [DOI] [PubMed] [Google Scholar]
- Whittington A, Vugrek O, Wei KJ, Hasenbein NG, Sugimoto K, Rashbrooke A, Wasteneys GO (2001) MOR1 is essential for organizing cortical microtubules in plants. Nature 411: 610–613 [DOI] [PubMed] [Google Scholar]
- Wymer C, Lloyd C (1996) Dynamic microtubules: implications for cell wall patterns. Trends Plant Sci 1: 222–228 [Google Scholar]
- Yasuhara H, Muraoka M, Shogaki H, Mori H, Sonobe S (2002) TMBP200, a microtubule bundling polypeptide isolated from telophase tobacco BY-2 cells is a MOR1 homologue. Plant Cell Physiol 43: 595–603 [DOI] [PubMed] [Google Scholar]
- Young DH, Lewandowski VT (2000) Covalent binding of the benzamide RH-4032 to tubulin in suspension-cultured tobacco cells and its application in a cell-based competitive-binding assay. Plant Physiol 124: 115–124 [DOI] [PMC free article] [PubMed] [Google Scholar]