Abstract
The actin cytoskeleton coordinates numerous cellular processes required for plant development. The functions of this network are intricately linked to its dynamic arrangement, and thus progress in understanding how actin orchestrates cellular processes relies on critical evaluation of actin organization and turnover. To investigate the dynamic nature of the actin cytoskeleton, we used a fusion protein between green fluorescent protein (GFP) and the second actin-binding domain (fABD2) of Arabidopsis (Arabidopsis thaliana) fimbrin, AtFIM1. The GFP-fABD2 fusion protein labeled highly dynamic and dense actin networks in diverse species and cell types, revealing structural detail not seen with alternative labeling methods, such as the commonly used mouse talin GFP fusion (GFP-mTalin). Further, we show that expression of the GFP-fABD2 fusion protein in Arabidopsis, unlike GFP-mTalin, has no detectable adverse effects on plant morphology or development. Time-lapse confocal microscopy and fluorescence recovery after photobleaching analyses of the actin cytoskeleton labeled with GFP-fABD2 revealed that lateral-filament migration and sliding of individual actin filaments or bundles are processes that contribute to the dynamic and continually reorganizing nature of the actin scaffold. These new observations of the dynamic actin cytoskeleton in plant cells using GFP-fABD2 reveal the value of this probe for future investigations of how actin filaments coordinate cellular processes required for plant development.
The actin cytoskeleton coordinates numerous cellular processes required for plant development, including targeted-vesicle transport, to establish hormone gradients (Muday, 2000), polarity determination to direct localized-wall biosynthesis (Baluska et al., 2003), and organelle positioning in response to environmental cues (Takagi, 2003) or to achieve unbiased organelle inheritance (Sheahan et al., 2004). Actin has no known enzymatic activity, and thus its participation in such diverse cellular processes resides in its ability to provide a dynamic filamentous scaffold upon which many regulatory and motor proteins interact (Staiger et al., 2000, and refs. therein). Actin cytoskeleton function, therefore, is intricately linked to its dynamic structure, and thus progress in understanding how actin orchestrates cellular processes will depend in part on critical evaluation of actin organization and turnover in plant cells.
The recent discovery of the Arp2/3 complex as a regulator of plant cell morphogenesis (Le et al., 2003; Li et al., 2003; Mathur et al., 2003a, 2003b; El-Assal et al., 2004) provides a pertinent example of structure function relationships relevant to the plant actin cytoskeleton. The Arp2/3 complex in nonplant systems nucleates the formation of dendritic actin networks by promoting nucleation and growth of new actin filaments (AFs) to the sides of existing AFs (Svitkina and Borisy, 1999; Blanchoin et al., 2000). Filament nucleation by this mechanism facilitates plasma membrane protrusion at the leading edge of migrating cells and other motility related phenomena (for review, see Welch and Mullins, 2002). The ability to image detailed AF organization in these systems has contributed substantially to understanding how the Arp2/3 complex functions (e.g. Svitkina and Borisy, 1999). In contrast, while genetic evidence clearly establishes a role for the plant Arp2/3 complex in cellular morphogenesis, some disagreement exists as to the precise effects different mutations in Arp2/3 components have on actin organization (Le et al., 2003; Li et al., 2003) and thus how Arp2/3 may regulate such processes.
Unlike microtubules, the plant actin cytoskeleton can be difficult to preserve by conventional fixation and embedding techniques (Kost et al., 1999; Vitha et al., 2000). Microinjection of fluorescent-phalloidin conjugates has provided novel insights into the dynamic nature of AFs (Schmit and Lambert, 1990) but is not applicable to most cell types and may stabilize actin dynamics. These difficulties have led to the use of various green fluorescent protein (GFP) fusions to image the actin cytoskeleton in live plant cells (Kost et al., 1999; Klahre et al., 2000; Timmers et al., 2002). The most commonly used is GFP tagged to the carboxy-terminal actin-binding domain (ABD) of mouse talin (GFP-mTalin; Kost et al., 1998), or recently human talin (GFP-hTalin; Takemoto et al., 2003; Kwok and Hanson, 2004). The GFP-mTalin fusion, however, does not label all identifiable actin arrays in plant cells (Le et al., 2003; El-Assal et al., 2004), and some images of actin networks labeled with this probe suggest that the GFP-mTalin fusion may cause artificial aggregation of actin networks (e.g. see Jedd and Chua, 2002; Oikawa et al., 2003). These observations suggest GFP talin fusions may not accurately depict actin organization and turnover in plant cells.
To explore alternatives for GFP tagging of the plant actin cytoskeleton, we have used a fusion between GFP and ABD2 of the Arabidopsis (Arabidopsis thaliana) fimbrin AtFIM1 (fABD2). AtFIM1 binds plant AFs both in vitro (Kovar et al., 2000) and in vivo (Kovar et al., 2001), and both protein sequence analysis (McCurdy and Kim, 1998) and in vitro binding data (Kovar et al., 2000) suggest that AtFIM1 contains two independent ABDs. Recently, we used this GFP-fABD2 fusion to demonstrate detailed features of the actin cytoskeleton in protoplasts derived from mesophyll tissue of stably transformed Nicotiana tabacum (Sheahan et al., 2004). This fusion protein labeled a highly dynamic and dense actin cytoskeleton in protoplasts, suggesting that it reveals more structural detail than alternative GFP-fusion constructs. GFP-fABD2 has also been used to analyze actin cytoskeletal organization in Arabidopsis lines with modified levels of actin-interacting protein 1 (Ketelaar et al., 2004), and recently a series of GFP fusion constructs exploiting the ABDs of AtFIM1 has been used to image the actin cytoskeleton in Arabidopsis roots (Wang et al., 2004).
We report here quantitative, dynamic analysis of the plant actin cytoskeleton by stable expression of GFP-fABD2 in different species and cell types. In comparison to GFP-mTalin (Kost et al., 1998), GFP-fABD2 consistently labels finer and denser networks of cortical and subcortical actin, and does so without adverse effects on plant morphology or development. Fluorescence recovery after photobleaching (FRAP) in cells from GFP-fABD2-expressing lines revealed that AF bundles are highly dynamic structures, composed of AFs capable of sliding in opposite directions. Our study provides new insights into the complexity of the actin cytoskeleton and its dynamic properties in plant cells, and we conclude that GFP-fABD2 currently provides the best available GFP-fusion protein for investigating dynamic actin networks in live cells.
RESULTS
Expression of GFP-fABD2 Highlights Elaborate Actin Networks without Adverse Effect on Plant Morphology
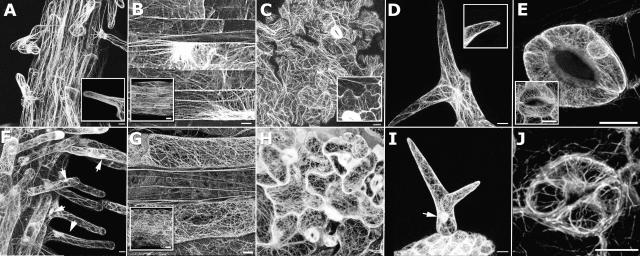
Expression of GFP-fABD2 in stably transformed Arabidopsis plants revealed dense networks of free and bundled AFs in the cell types examined (Fig. 1). Such arrays were present as dense filamentous networks in the cortex (Fig. 1, B–D) and perinuclear region (Fig. 1, B and D) of hypocotyl, pavement epidermal, and trichome cells, or as masses of bundled filaments in transvacuolar strands of pavement epidermal cells (Fig. 1C) and cortex of mature root epidermal cells (Fig. 1A). Guard cells had notably higher fluorescence intensities compared with surrounding epidermal cells, and AFs were more difficult to visualize. However, intricate cortical and perinuclear meshworks (Fig. 1E) or radially aligned AF bundles (Fig. 1E, inset) were commonly observed. AF bundles were frequently arrayed longitudinally along the axis of expansion in elongating cells while in diffuse-growing pavement epidermal cells, bundles were arrayed more randomly. In contrast to previous findings (Li et al., 2003), diffuse fluorescence, interpreted to represent fine AF arrays, in lobed regions of expanding pavement epidermal cells from both Arabidopsis (Fig. 1C, inset) and N. tabacum (data not shown) was rare and appeared to be present equally in regions of lobe expansion as it was in constrictions. Similar diffuse labeling was frequently observed at the ends of growing root hairs (Fig. 1A, inset) and trichome tips (Fig. 1D, inset). We interpret this diffuse fluorescence to represent fine AF arrays; however, our attempts to further resolve this fluorescence by conventional confocal microscopy were unsuccessful.
Figure 1.
Expression of GFP-fABD2 and GFP-mTalin in Arabidopsis. Confocal projections showing GFP-fABD2 (A–E) or GFP-mTalin (F–J) expression in roots (A and F), hypocotyls (B and G), leaf epidermis (C and H), trichomes (D and I), and stomata (E and J). A, Longitudinal cables in mature root epidermal cells and hairs; inset shows diffuse, irresolvable fluorescence in growing root hair tip. B, Longitudinal cables and transverse perinuclear arrays in hypocotyls that have ceased growth; inset shows similar arrangement of AFs in immature hypocotyls. C, Pavement epidermal cells show randomly arrayed transvacuolar bundles; inset shows expanding epidermal cells. D, Longitudinal bundles in young trichomes; inset shows diffuse tip fluorescence. E, Intricate cortical and simple perinuclear arrays in guard cells; inset shows transversely oriented AF bundles. F and I, Helical arrays of filaments in root hairs and trichomes; nucleoplasmic labeling is evident (arrows). G, H, and J, Randomized networks of short, curved filaments in hypocotyl, leaf epidermis, and stomata. G, Mature hypocotyl tissue; inset shows immature hypocotyl. All images are confocal projections composed of 85 (A and F), 63 (B and G), 54 (D and I), 35 (B inset and G inset), 15 (E, J, and E inset), or 10 (C, H, C inset, and D inset) optical sections. Scale bar = 10 μm (A–J) or 20 μm (B and G, inset).
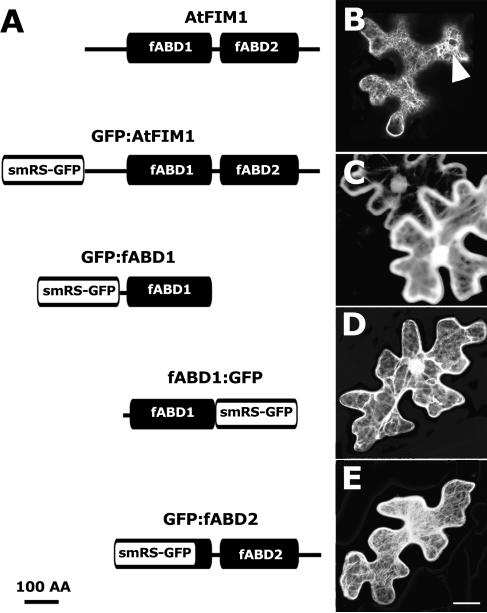
In addition to GFP-fABD2, we tested GFP fusions to the amino terminus of full-length AtFIM1 (GFP-AtFIM1) and to either the carboxy or amino terminus of the first ABD (fABD1) of AtFIM1 (fABD1-GFP, GFP-fABD1; Fig. 2A). Transient expression of GFP-AtFIM1 in N. tabacum leaf epidermal cells showed predominantly filamentous labeling, but labeling became cytoplasmic in regions where expression levels were locally higher (Fig. 2B). Expression of either fABD1-GFP or GFP-fABD1 consistently resulted in cytoplasmic labeling (Fig. 2, C and D), in contrast to GFP-fABD2, which labeled distinctly filamentous structures in all but the most highly expressing cells (Fig. 2E). Similar results were obtained for GFP-AtFIM1 and GFP-fABD2 with stably expressing N. tabacum mesophyll protoplasts, confirming these findings (data not shown). We therefore conclude that, of the constructs tested, GFP-fABD2 provides the best GFP fusion protein for consistent labeling of the actin cytoskeleton.
Figure 2.
GFP fusions to full-length AtFIM1, fABD1, or fABD2 and transient expression in N. tabacum leaf epidermal cells. A, Schematic diagram of AtFIM1 with two ABDs (fABD1 and fABD 2) and amino- or carboxy- terminal fusions o f GFP with full-length AtFIM1, fABD1, or fABD2. B, Low-level GFP-AtFIM1 expression showing filamentous labeling with overexpression and cytoplasmic fluorescence in localized regions (arrow). C and D, Cytoplasmic localization of GFP-fABD1 and fABD1-GFP, respectively. E, Visualization of filamentous structures with GFP-fABD2. Image in B is single-optical section with depth of 2.5 μm. Images in C to E are confocal projections composed of 45 (C and D) or 30 (E) optical sections. Scale bar = 20 μm.
Our analysis of Arabidopsis plants stably transformed with the GFP-mTalin fusion protein (Kost et al., 1998) revealed broadly similar structures to those in GFP-fABD2 plants (Fig. 1, F–J); however, close examination revealed that AFs in GFP-mTalin plants displayed increased bundling and were more randomly arranged (compare Fig. 1, B, G, and insets). AF networks in these plants generally displayed increased branching, being composed of shorter, more convoluted filaments, while in root hairs and trichomes, longitudinal AF bundles tended to form into helical arrays (Fig. 1, F and I). Plants expressing GFP-mTalin also showed fluorescence localized to the nucleoplasm (arrows in Fig. 1, F and I), whereas cells expressing GFP-fABD2 exhibited neither organellar nor nucleoplasmic staining. Notably, due to small cell size, tissue organization, and low levels of expression, we were unable to satisfactorily visualize AFs in the root meristem of plants stably transformed with either GFP-fABD2 or GFP-mTalin (data not shown). This observation is consistent with the generally poor visualization of actin networks seen in meristematic cells by Wang et al. (2004) using fimbrin-based fusion proteins.
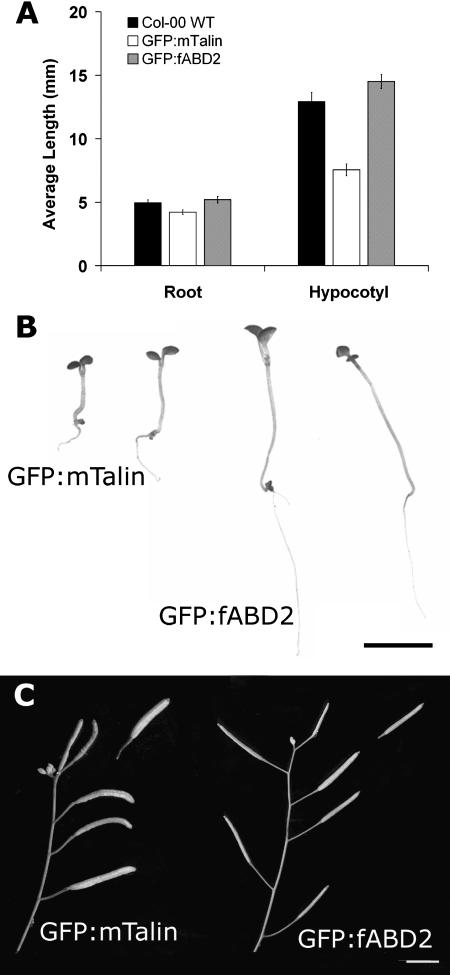
We compared plant development in transgenic Arabidopsis seedlings expressing either GFP-mTalin or GFP-fABD2. Although reported to have no effects on plant morphology or development (Kost et al., 1998), plants expressing GFP-mTalin in our experiments showed retarded growth compared to wild-type or GFP-fABD2 plants. In 4-d-old etiolated seedlings, plants expressing GFP-mTalin showed retarded hypocotyl elongation but only slightly reduced root growth (Fig. 3A). However, 3 d after transfer to light, differences in the growth of GFP-fABD2- and GFP-mTalin-expressing plants were clear, with reduced growth in all organs in GFP-mTalin plants (Fig. 3B). Mature GFP-mTalin-expressing plants were stunted and had short inflorescences and stout siliques (Fig. 3C) that carried only 45% ± 7% (mean ± se, n = 4) of the seed of GFP-fABD2-expressing siliques. Further analysis of epidermal cells from hypocotyls, inflorescences, siliques, and leaves indicated that cells from plants expressing GFP-mTalin had smaller plan areas compared with GFP-fABD2 plants (Table I). Cell perimeter was also reduced in GFP-mTalin plants at 69% ± 4% (mean ± se; n = 385), the length of cell peripheries in GFP-fABD2-expressing plants (Table I). Most distinct, however, was the difference in the length of cells, with GFP-mTalin-expressing plants having cells on average 66% ± 4% (mean ± se; n = 385) the length of GFP-fABD2 cells (Table I). Average cell width in the two lines, however, was similar. Stunted growth in GFP-mTalin plants is therefore primarily due to reduced cell elongation.
Figure 3.
Expression of GFP-mTalin affects plant growth and morphology. A, Root and hypocotyl elongation of 4-d-old, dark-grown seedlings; elongation is retarded in GFP-mTalin, but not GFP-fABD2 plants. B, Retarded growth in 7-d-old GFP-mTalin but not GFP-fABD2 plants. C, Altered silique morphology in GFP-mTalin plants. Error bars shown in A, se (n = 20); scale bar = 5 mm (B and C).
Table I.
Epidermal cell characteristics in GFP-fABD2- and GFP-mTalin-expressing Arabidopsis plants
| Measurement | GFP-fABD2 | GFP-mTalin |
|---|---|---|
| Plan Area (μm2) | 2,639 ± 95 | 2,127 ± 98 |
| Cell Perimeter (μm) | 363 ± 9 | 249 ± 8 |
| Cell Length (μm) | 139 ± 3 | 92 ± 3 |
| Cell Width (μm) | 26 ± 1 | 28 ± 1 |
| Circularity | 0.285 ± 0.006 | 0.454 ± 0.008 |
Epidermal cells from hypocotyls, inflorescences, siliques, and leaves of cleared GFP-fABD2 and GFP-mTalin-expressing plants were imaged, and measurements of individual cells obtained. Values are means ± se, n = 385.
GFP-mTalin expression affected diffuse-growth processes in leaves, hypocotyls, inflorescences, and siliques. However, no significant differences were found between GFP-mTalin- and GFP-fABD2-expressing root hairs, suggesting that tip-growth processes remain unaffected, consistent with the finding that GFP-mTalin expression does not perturb pollen tube growth in N. tabacum (Kost et al., 1998). In contrast to our general finding that GFP-mTalin expression causes reduced cell elongation, expression of GFP-fABD2 in Arabidopsis had no measurable effect on plant growth, suggesting that the GFP-fABD2 fusion protein interferes minimally with native AF function.
Estimation of GFP levels in GFP-fABD2- and GFP-mTalin-expressing plants, based on quantification of total GFP fluorescence in individual cells (Table II), indicated that GFP-mTalin expression levels were roughly twice that of GFP-fABD2 in the plants used in this study (Table II). In agreement with this analysis, semiquantitative immunoblotting using an anti-GFP antibody and whole 7-d-old seedlings revealed that the abundance, relative to α-tubulin, of GFP-mTalin was 1.6- ± 0.9-fold (mean ± se; n = 3) greater than GFP-fABD2 in the respective transgenic lines (see Supplemental Fig. 1, available at www.plantphysiol.org). We were unable to generate stable lines of Arabidopsis expressing GFP-fABD2 to levels comparable to that seen in GFP-mTalin plants. However, when GFP-fABD2 was transiently expressed in N. tabacum pavement epidermal cells to levels comparable to GFP-mTalin plants, as determined by measurement of total cellular GFP fluorescence, GFP-fABD2 accumulated in the cytoplasm (data not shown). These observations suggest fundamental differences in the mechanisms of GFP-mTalin and GFP-fABD2 binding in plant cells, and the consequent effects this may have on plant development. Our conclusion that expression of GFP-fABD2 does not perturb actin functionality is supported by the observation that chloroplast repositioning during N. tabacum protoplast culture proceeds at similar rates in both wild-type and GFP-fABD2-expressing cells (Sheahan et al., 2004).
Table II.
GFP fluorescence levels in GFP-fABD2- and GFP-mTalin-expressing Arabidopsis plants
| Actin Label
|
GFP Level
|
||
|---|---|---|---|
| Root hairs | Hypocotyl Epidermis | Leaf Epidermis | |
| Fluorescence Intensityμm−3 | |||
| GFP-fABD2 | 9.7 ± 0.4 | 5.7 ± 0.1 | 10.1 ± 0.5 |
| GFP-mTalin | 22.7 ± 0.6 | 11.5 ± 0.7 | 16.6 ± 0.5 |
Quantification of GFP fluorescence in individual root hairs and epidermal cells from hypocotyls and leaves of GFP-fABD2- and GFP-mTalin-expressing plants. Values are means ± se, n ≥ 20.
GFP-fABD2 Labels Actin Networks in Diverse Cell Types from Different Species
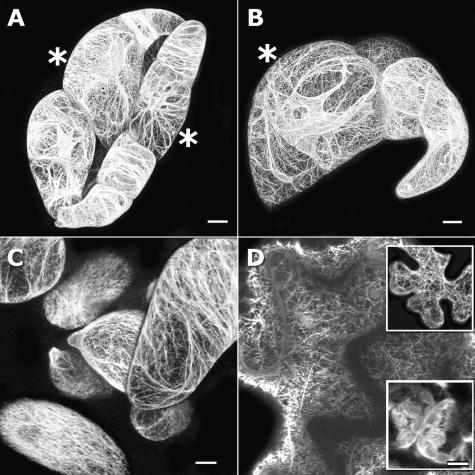
We investigated the usefulness of the GFP-fABD2 fusion protein to label actin networks in diverse cell types from different species. In both stably transformed N. tabacum BY-2 and Medicago truncatula suspension-cultured cells, we observed intricate networks of nucleus-associated AF arrays and dense, at times transversely oriented, cortical networks (Fig. 4, A and C). In BY-2 cells, transverse bundles of cortical actin were more prominent in smaller, expanding cells compared with older, expanded cells (Fig. 4A). In cells predicted to be entering division, actin was arranged prominently as complex transvacuolar strands linking the subcortical-perinuclear region with the cortex (Fig. 4, A and B). This observation indicates that reorganization of the actin cytoskeleton occurs during the transition from cell expansion to division. In mesophyll tissue from N. tabacum and Lycopersicon esculentum, dense cortical arrays and chloroplast actin baskets were evident (Fig. 4D and insets). These results demonstrate that expression of the GFP-fABD2 fusion protein provides detailed visualization of the actin network in live cells, across different species and cell types.
Figure 4.
GFP-fABD2 labels rich AF networks in different cell types from diverse species. A and B, Dense cortical and perinuclear AF arrays in cultured N. tabacum BY-2 cells. C, Cortical AF arrays in M. truncatula suspension cultures. D, Cortical AF arrays in N. tabacum (L. esculentum; top inset) leaf epidermis, and chloroplast baskets in underlying mesophyll (bottom inset). Cells predicted to be entering division are indicated (*). All images are confocal projections composed of 75 (A), 71 (B), 15 (C and D top inset), or 4 (D and D bottom inset) optical sections. Scale bar = 20 μm (A and B) or 10 μm (C and D).
GFP-fABD2 Highlights Dynamic Networks of AFs
We analyzed the dynamic nature of the actin network with time-lapse imaging (see supplemental videos), Latrunculin B (LatB) washout experiments, and FRAP. Time-lapse imaging of epidermal cells in leaves, hypocotyls, and roots of 7-d-old plants revealed that the AF network labeled with GFP-fABD2 underwent continual reorganization. Closer analysis established that the highly dynamic nature of this network arose predominantly from the small movements of a large number of filaments or bundles. Transvacuolar bundles abutting the cell periphery underwent lateral migrations, while regions where bundles of AFs intersected, termed focal junctions, moved rapidly around the cell (Supplemental Video 1). Measurement of focal junction movement in pavement epidermal cells using centroid tracking revealed irregular, yet directional movement, with speeds ranging from 80 to 800 nm s−1 (mean, 248 ± 38; n = 20) and velocities from 25 to 600 nm s−1 (mean, 140 ± 32; n = 20). Both lateral AF movement and dynamic movements of focal junctions were difficult to observe in plants expressing GFP-mTalin (Supplemental Video 2), suggesting that this fusion protein stabilizes an otherwise highly dynamic network.
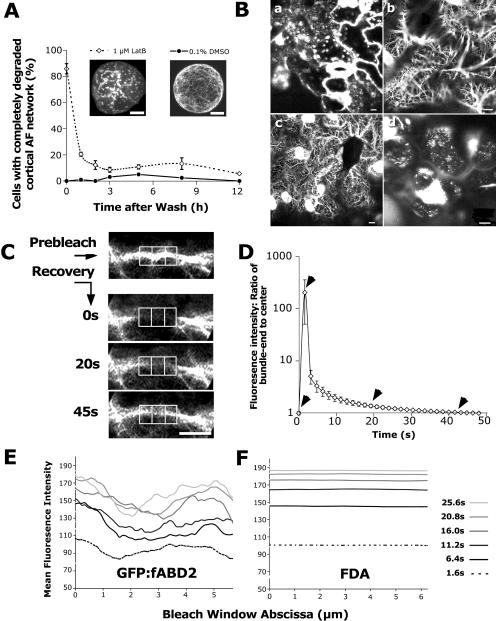
To estimate the contribution of filament turnover to dynamic processes, we used LatB washout experiments. Treatment of GFP-fABD2-expressing N. tabacum protoplasts for 24 h with 1 μm LatB caused complete depolymerization of AFs in most cells, with GFP-fABD2 visualized as localized accumulations of fluorescence in the cell cortex (Fig. 5A and insets). Monitoring recovery of the actin network following LatB washout indicated that most cells had at least a partially repolymerized network within 1 h of washing (Fig. 5A). Treatment of GFP-fABD2-expressing Arabidopsis seedlings with 2 μm LatB for 8 h also caused an almost complete depolymerization of the actin network in near-fully expanded pavement epidermal cells (Fig. 5B, section a). Recovery following thorough washing was swift, and within 1.5 h, most cells had actin networks (Fig. 5B, section c) similar to dimethyl sulfoxide (DMSO)-treated controls (Fig. 5B, section b). These networks were dynamic, as indicated by time-lapse imaging (Supplemental Video 3); however, bundles of AFs appeared somewhat thicker and shorter than those in DMSO controls. Parallel analysis of GFP-mTalin plants similarly indicated an almost complete depolymerization of the actin network in the presence of LatB. In contrast, however, reassembly of the actin network following LatB washout was substantially retarded, with few cells exhibiting polymerized filaments but many exhibiting short, rod-like structures (Fig. 5B, section d). Time-lapse analysis indicated that these rods moved slightly, but overall, dynamic movement was substantially reduced compared to GFP-fABD2 plants (Supplemental Video 3). These results suggest that filament assembly/disassembly processes contribute significantly to observed actin network reorganization and, further, that GFP-mTalin expression impedes the repolymerization of AFs.
Figure 5.
Expression of GFP-fABD2 reveals a dynamic framework of AFs. A, Recovery from LatB treatment in washed N. tabacum protoplast populations; rapid recovery of filamentous actin within 1.5 h of washing. Insets show half-cell (approximately 15 μm), confocal projections of protoplasts treated with 1 μm LatB or 0.1% DMSO for 24 h. B, Recovery from LatB treatment in washed Arabidopsis leaf epidermis; 8 h of LatB treatment (a) caused near complete depolymerization of the network, contrasting normal arrays in 0.1% DMSO-treated controls (b). Substantial recovery of the actin network 1.5 h after LatB washout in GFP-fABD2-expressing plants (c) but not in plants expressing GFP-mTalin (d). C, Fluorescence recovery along bleached AF bundles occurs rapidly and in a pattern resembling the prebleach image; images show typical fluorescence recovery in bleached bundle over 45 s, with left, center, and right boxes used in recovery analysis shown. Fluorescence recovers to 84% of prebleach intensity by 45 s in this example. D, Detailed analysis of fluorescence recovery following bleaching of bundled AFs; graph shows recovery ratio for one end compared with the center; arrows on graph correspond to time point for images displayed on right (n = 7). E, Plotted mean fluorescence intensity over time along bleach window in a GFP-fABD2-tagged transvacuolar actin bundle. F, Plotted mean fluorescence intensity over time along bleach window in an FDA-stained transvacuolar strand; fluorescence recovery is uniform over the bleach window with FDA, but is irregular and occurs fastest at the edges of the bleach window in GFP-fABD2-expressing cells. Insets in A are confocal projections (half-cell) and images in B are single-plane images with a z-depth of 2.5 μm. Scale bar = 10 μm (A–C).
We used FRAP in 7-d-old Arabidopsis seedlings to further investigate the contribution of filament movement to actin network dynamics. Assuming that photobleaching of GFP does not alter the affinity of GFP-fABD2 for AFs, fluorescence recovery of actin bundles in these experiments is predicted to come from two sources; firstly, the exchange of bleached GFP-fABD2 subunits bound to bundled AFs, and secondly, the lateral movement of AFs carrying unbleached GFP-fABD2 into the bleach window. In all cells examined, fluorescence (corrected for bleaching caused by laser scanning during image collection) recovered within 50 s to between 80% to 90% of the prebleach value in a pattern resembling the prebleach image (Fig. 5C), demonstrating that bleaching did not alter actin structure.
To understand the mechanisms of fluorescence recovery, we first analyzed FRAP in expanding epidermal cells of 7-d-old plants expressing cytoplasmic yellow fluorescent protein (YFP) or in cells stained with fluorescein diacetate (FDA). We compared the first-order rate constant (k) and recovery half-times (t1/2) for YFP and GFP-fABD2 fluorescence recovery by fitting exponential curves to the recovery data. Generally, fluorescence recovery rate in YFP-expressing cells was at least twice that of cells expressing GFP-fABD2 (Table III), indicating that recovery of GFP-fABD2 is limited by the exchange of bleached units on AFs with those in the cytoplasm. Fluorescence recoveries were similar in different cell types of GFP-fABD2-expressing plants (Table III), suggesting that the dynamic nature of actin networks are comparable in different cell types.
Table III.
Rate constants and half-times for FRAP
| Label | Cell Type | k | t1/2 |
|---|---|---|---|
| s−1 | s | ||
| YFP | Hypocotyl | 0.16 ± 0.03 | 6.3 ± 0.8 |
| FDA | Hypocotyl | 0.15 ± 0.021 | 5.9 ± 1.1 |
| GFP-mTalin | Hypocotyl | 0.05 ± 0.005 | 13.4 ± 1.2 |
| GFP-fABD2 | Hypocotyl | 0.05 ± 0.006 | 13.4 ± 1.0 |
| Root hair | 0.05 ± 0.007 | 16.1 ± 3.0 | |
| Guard cell | 0.05 ± 0.003 | 14.7 ± 0.9 | |
| Leaf epidermal | 0.06 ± 0.008 | 13.2 ± 1.3 |
Arabidopsis cells expressing GFP-fABD2, GFP-mTalin, or YFP, or nonexpressing cells labeled with FDA were photobleached, the recovery plotted, and an exponential function fitted. The k and t1/2 were derived from the exponential function. Values are means ± se, n ≥ 12.
In FRAP experiments using GFP-fABD2, we did not observe translocation of the bleach window, however the size of the bleached region progressively decreased (Fig. 5C). Similar observations of YFP-expressing and FDA-stained cells indicated that the bleach window sometimes translocated in the direction of net cytoplasmic streaming, but that fluorescence recovery was reasonably uniform across the bleach window (data not shown). To understand the contribution of diffusion to the observed recovery patterns, we analyzed how fluorescence recovered along bleached AF bundles in hypocotyl epidermal cells. By comparing the ratio of fluorescence recovery at the ends of the bleached bundle (left and right boxes in Fig. 5C) with the central region of the bundle (center box in Fig. 5C), we found that fluorescence recovered along the whole bundle, but recovery initially occurred faster and to a greater extent at either end of the bundle (Fig. 5D). We also compared the mean fluorescence intensity plotted along the horizontal axis of the bleach window over time for both GFP-fABD2-expressing cells and FDA-stained cells (Fig. 5, E and F). This analysis confirmed that fluorescence recovery in bleached AF bundles was greater and faster at each edge of the bleached AF bundle (Fig. 5E). In addition, recovery appeared to be directional in several cases, in that recovery was slightly faster at one end of the bleached bundle compared to the other (Fig. 5E). In contrast, recovery of mean fluorescence intensity was comparatively uniform across the bleach window in FDA-stained cells (Fig. 5F). By comparing recovery in GFP-fABD2 bleached AF bundles and FDA-stained cytoplasmic strands, we found that the ratio of change in fluorescence intensity between the edge and center of the bleach window over time was on average 5.9-fold greater in GFP-fABD2-expressing cells. Thus, diffusion alone cannot account for the more rapid recovery of fluorescence at the edges of bleached AF bundles, suggesting that rapid-edge recovery may be the result of lateral sliding in opposite directions of GFP-fABD2-labeled filaments or bundles into the bleach window. Consistent with potential filament sliding, bleached GFP-fABD2-labeled AFs moving out of the bleach window caused fluorescence intensities immediately outside the bleach window to decrease an additional 0.7 ± 0.1-fold (mean ± se, n = 7) more than could be accounted for by bleaching caused by laser scanning during image collection.
Comparison of fluorescence recovery in bleached GFP-fABD2- and GFP-mTalin-labeled AF bundles indicated that the ratio of change in fluorescence intensity between the edge and the center of the bleach window over time was, on average, 1.3-fold greater in GFP-fABD2-expressing cells, suggesting that filament sliding occurs to a greater extent in these cells. Similar fluorescence-t1/2 in GFP-fABD2- and GFP-mTalin-expressing cells (Table III) therefore indicate that GFP-mTalin is likely to have a faster rate of exchange from AFs than does GFP-fABD2.
DISCUSSION
Progress in understanding functions of the actin cytoskeleton will be aided by improved techniques for dynamic visualization of actin organization in live plant cells. Advances in this area have emerged from the increasing use of GFP fusions to various actin-binding proteins, including GFP fusions to plastin (mammalian fimbrin; Timmers et al., 2002) and ABDs from mouse (Kost et al., 1998) and human talin (Takemoto et al., 2003). We evaluated the potential of different AtFIM1-based GFP fusions to label dynamic actin networks in different higher plant species.
Of the three fusion proteins tested, only GFP-fABD2 consistently labeled dynamic AFs over a range of expression conditions. Amino- or carboxy-terminal GFP fusions to ABD1 of AtFIM1 accumulated within the cytoplasm, without evidence of filamentous labeling, while GFP fused to full-length AtFIM1 (GFP-AtFIM1) only labeled AFs clearly when expressed at low levels. Several undetermined factors presumably might contribute to the different outcomes observed for these fusion constructs. In vitro binding assays predict two different actin-binding activities for AtFIM1 (Kovar et al., 2001), and recent biochemical analysis of L-plastin, a human fimbrin homolog, shows that ABD2 binds with lower affinity to actin than does ABD1 (Lebart et al., 2004; but see Nakano et al., 2001). Thus, the potential lower affinity ABD2 of AtFIM1 may provide superior characteristics for noninvasive labeling of AFs. Furthermore, recent analysis of the crystal structure of AtFIM1 suggests that the carboxy-terminal extension of AtFIM1, which is retained in the GFP-fABD2 and GFP-AtFIM1 fusion proteins (see Fig. 2), may be important for stabilizing AF interactions (Klein et al., 2004). Our observations agree with recent work by Wang et al. (2004), who show that different GFP fusions, including ABD2 of AtFIM1, produce filamentous labeling when expressed in N. tabacum or Arabidopsis, whereas fusion to ABD1 of AtFIM1 yields cytoplasmic labeling.
Expression of GFP-fABD2 in Arabidopsis revealed elaborate and intricate actin networks in the cell types examined. Cortical AFs were organized into linear arrays of predominantly longitudinally aligned AFs in elongating tissues such as hypocotyl and root epidermis. In pavement epidermal cells, cortical actin tended to have a more random alignment. Subcortical actin bundles radiated from the perinuclear AF basket through the vacuole were common to all cell types examined. Similar structures were observed in N. tabacum and L. esculentum cells, and in BY-2 and M. truncatula suspension cultures expressing GFP-fABD2.
Expression of GFP-fABD2 in Arabidopsis did not disrupt plant growth or morphology when compared with nontransgenic controls. In contrast, however, plants expressing GFP-mTalin showed reduced cell elongation, resulting in plants with stunted growth and reduced seed set. Actin networks visualized by the GFP-mTalin construct differed to those visualized by GFP-fABD2, being composed of comparatively shorter, more branched and convoluted filament networks with increased bundling and greatly reduced dynamism.
Our observations indicate that the altered morphological and growth phenotypes of GFP-mTalin-expressing plants are likely to be caused by changes in actin network structure, suggesting that overexpression of certain ABDs can alter function of the actin network. Similarly, Wang et al. (2004) found that overexpression of the BD1/2-GFP construct altered actin structure and plant growth. The suppressed recovery of AFs following LatB washout and reduced dynamic behavior of actin networks in GFP-mTalin-expressing plants indicate that GFP-mTalin may affect plant growth by interfering with AF polymerization. Evidence from profilin mutants suggests that hypocotyl elongation requires a dynamic actin network, since underexpression of profilin decreases hypocotyl cell elongation (Ramachandran et al., 2000). In plants, anisotropic growth but not polarity determination requires dynamic AFs (Smith, 2003) and, typically, fine AF arrays are associated with rapid, polar cell growth (Waller et al., 2002). Disruption of the actin network with LatB (Baluska et al., 2001) or by overexpression of a reproductive actin (ACT1; Kandasamy et al., 2002) creates plants with a dwarf phenotype but no significant changes in morphology. AFs probably direct vesicular transport, but may also influence cell shape by determining microtubule alignment (Kost et al., 1999).
In contrast to GFP-mTalin, which is expressed at high relative levels but continues to label highly bundled actin structures, cells showing similar levels of GFP fluorescence in GFP-fABD2-expressing cells caused cytoplasmic accumulation. This finding may reflect differences in the affinity of the carboxy-terminal ABD of talin and ABD2 of AtFIM1 for plant AFs, or reflect a reduced number of available binding sites for GFP-fABD2 given the presence of endogenous fimbrins. The tendency of GFP-mTalin to cause increased bundling may be due to a high density of binding along an AF causing a localized high concentration of GFP. GFP has a weak tendency to dimerize (Kd = 100 μm; Phillips, 1997) and thus under such conditions may cause cross-linking of AFs in vivo similar to the way McCann and Craig (1997) observed in vitro cross-linking of AFs when the single carboxy-terminal ABD of mTalin was fused to glutathione S-transferase. Interestingly, Wang et al. (2004) observed that plants expressing their independently created GFP-mTalin construct also displayed increased bundling and reduced dynamic behavior of AFs compared to AtFIM1-based GFP fusions.
Potential filament stabilization by GFP-mTalin expression in Arabidopsis plants may also occur, and indeed a carboxy-terminal 63-kD fragment of talin A has been used as a stabilizer of actin networks in Dictyostelium cells (Weber et al., 2002). The apparent inability of plants to regulate the binding of the talin ABD to AFs may also result in the failure of endogenous actin-binding proteins to interact correctly with AFs. Our findings highlight the delicate nature of the actin network in plant cells and emphasize the potential for GFP-fusion approaches to compromise the dynamic nature of actin networks. Based on the data presented, we conclude that the GFP-fABD2 fusion represents the best available probe for visualization of dynamic AFs in live plant cells, and emphasize that low levels of expression are vital for accurate imaging of the actin network in plants. Ultimately, GFP-actin fusions may provide the best approach to investigating dynamic turnover of the plant actin cytoskeleton. However, given the sensitive nature of plant actin networks demonstrated in this study and the large pool of unpolymerized actin in plant cells implicated in studies of pollen tubes (Gibbon et al., 1999) and soybean suspension cultures (Lee et al., 2003), it is likely that GFP-actin fusions may also encounter difficulties.
Several Processes Contribute to Actin Dynamics Observed in Plant Cells
Time-lapse imaging of GFP-fABD2-expressing cells revealed that several, simultaneous dynamic processes contribute to continual reorganization of the actin cytoskeleton. Large-scale, lateral movements of AF bundles occurred where transvacuolar AF bundles abutted cortical bundles and at the intersections of transvacuolar AF bundles (focal junctions) in the population of subcortical actin. Dynamic turnover of the actin network is also likely to contribute appreciably to overall cytoskeletal reorganization as implicated by LatB washout experiments. Detailed FRAP-based analysis suggested that AF bundles represent an actively moving collection of AFs, where either individual AFs or small bundles of AFs slide past one another, probably in opposite directions. Taken together, these results portray the plant actin network as a highly fluidic system of interacting filament-based structures. The ability of the actin cytoskeleton to change its form at several different levels of organization would presumably allow the cell to simultaneously perform the multitude of AF-dependent functions known in plants. Clearly, further analysis into the role of individual actin isovariants and the influence of specific actin-binding proteins on this dynamic behavior is required to understand these processes and their regulation at the molecular level.
MATERIALS AND METHODS
GFP-fABD2 Fusion Construct and Agrobacterium Transformation
Fusions between modified GFP and full-length AtFIM1 (McCurdy and Kim, 1998; GFP-AtFIM1), fABD1 (amino acids 101–370 of AtFIM1; fABD1-GFP and GFP-fABD1), and fABD2 of AtFIM1 (amino acids 325–687 of AtFIM1; GFP-fABD2) were tested (see Fig. 2A). The GFP-AtFIM1 and GFP-fABD2 constructs were supplied by Boris Voigt and Diedrik Menzel (Universität Bonn) in pCAT (see Ketelaar et al., 2004) and the fABD1-GFP and GFP-fABD1 constructs by Steven Pirlo (The University of Newcastle) in pART7 expression vector (Gleave, 1992). In the GFP-AtFIM1 and GFP-fABD2 constructs, the appropriate AtFIM1-derived DNA sequence was fused directly with GFP-encoding sequences without an intervening-linker sequence, while the fABD1-GFP and GFP-fABD1 constructs were created with a 5× Gly linker. The 35S:GFP-fABD2:35SpA or 35S:GFP-AtFIM1:35SpA region of pCAT was moved into pGEM-T by restriction digestion with SdaI and then into pART27 (Gleave, 1992) using NotI digestion. The 35S:GFP-fABD1:ocs3′ or 35S:fABD1-GFP:ocs3′ of pART7 was moved into pART27 using NotI digestion. Each recombinant pART27 vector was transferred into Agrobacterium tumefaciens strain LBA4404 by electroporation and bacteria selected on Luria-Bertani agar with 50 μg mL−1 kanamycin (Sigma, St. Louis).
Plant Transformation
The GFP-fABD2 fusion construct in pART27 was transformed into Arabidopsis (Arabidopsis thaliana; Col) by floral dipping (Clough and Bent, 1998) and transformants selected on 50 μg mL−1 kanamycin. Nicotiana tabacum was stably transformed using the leaf disc procedure of Horsch et al. (1985) as described by Sheahan et al. (2004). N. tabacum BY-2 cells were stably transformed by the method of Sullivan and Green (1996) with selection on 100 μg mL−1 kanamycin and 500 μg mL−1 carbenicillin (Sigma). The highly regenerable Medicago truncatula line 2HA (Rose et al., 1999) was stably transformed as described by Thomas et al. (1992). N. tabacum and Lycopersicon esculentum leaves were transiently transformed using the agroinfiltration method (Yang et al., 2000). Arabidopsis seed derived from the GFP-mTalin-expressing line reported by Kost et al. (1998) was used in this study. Madeline Rashbrooke (Australian National University) supplied transgenic Arabidopsis seed expressing cytoplasmic YFP.
Plant Growth
Arabidopsis plants were grown on vertical or horizontal plates containing 0.5× Murashige and Skoog salts, 0.8% (w/v) agar, and 1% (w/v) Suc. Surface-sterilized seeds were positioned on plates and vernalized for 2 d at 4°C before being moved to a growth room (25°C, 16/8 h photoperiod at 50 μmol photons m−2 s−1). Mesophyll protoplasts were isolated and cultured from axenic shoot cultures of transgenic GFP-fABD2 N. tabacum as described by Sheahan et al. (2004). N. tabacum and L. esculentum plants used for agroinfiltration were grown under glasshouse conditions as described for N. tabacum in Sheahan et al. (2004). Stably transformed BY-2 cultures were subcultured weekly and maintained in liquid Murashige and Skoog medium (plus 3% [w/v] Suc, 0.2 mg mL−1 2,4-dichlorophenoxyacetic acid, 255 mg L−1 KH2PO4, 100 μg mL−1 kanamycin, and 250 μg mL−1 Timentin [Glaxo SmithKline, Melbourne, Australia], pH 5.7) on a rotary shaker (130 rpm) at 27°C in darkness. M. truncatula suspension cultures were freshly established for each experiment using callus grown on plates containing P4 medium (Thomas et al., 1992). Callus was used to inoculate liquid Murashige and Skoog medium (plus 3% [w/v] Suc, 0.2 mg mL−1 2,4-dichlorophenoxyacetic acid, 100 μg mL−1 kanamycin, and 250 μg mL−1 Timentin, pH 5.7) and cultures grown as described for BY-2 cells.
Drug Treatments
Protoplasts were treated in culture for 24 h with 1 μm LatB (Merck, Rahway, NJ) or 0.1% (v/v) DMSO before washing three times in modified Nagata and Takebe medium (Thomas and Rose, 1983). Whole 14-d-old Arabidopsis plants were incubated in petri dishes with wetted filter paper containing 2 μm LatB. After 8 h, plants were washed thoroughly with distilled water and transferred to petri dishes containing filter paper wetted with distilled water. Arabidopsis seedlings were stained for 30 min with 0.5% (w/v) FDA (Calbiochem-Novabiochem, San Diego) in petri dishes containing filter paper wetted by distilled water.
Microscopy and Cell Measurements
The morphology of epidermal cells from wild type, GFP-fABD2- and GFP-mTalin-expressing inflorescences, siliques, and rosette leaves was analyzed using cleared intact tissues as described previously (Fu et al., 2002). Cleared tissues and hypocotyls of 7-d-old plants were stained with toluidine blue, washed, and observed with a light microscope (Axiophot, Zeiss, Sydney) equipped with a CCD camera (Axiocam MR, Zeiss). Images were captured using Axiovision 4.0 (Zeiss) and cell plan-area, length, width, perimeter, and circularity measurements performed in the public domain NIH ImageJ 1.31v program (developed at the United States National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/ij).
Confocal Imaging and Analysis
Arabidopsis seedlings, N. tabacum, and L. esculentum leaf sections were mounted in water under a coverslip while N. tabacum protoplasts, BY-2, and M. truncatula cells were embedded in agar by mixing 50 μL of cells with 100 μL of 0.5% (w/v) low-melt agarose (type VII; Sigma-Aldrich) in welled slides before mounting under a coverslip. Images of GFP-fABD2- and GFP-mTalin-expressing cells or FDA-stained cells were acquired as z-series with a 1-μm interval or time series with an interval of 1.6 to 6 s using a confocal laser-scanning microscope (LSM 510; Zeiss) equipped with a 40× C-Apochromat water-immersion objective (NA 1.2; Zeiss) and 488-nm argon laser (25 mW, 5%–10% power) and BP500-530IR filter. YFP was visualized using a 514-nm argon laser with BP560-615 filter.
To estimate GFP levels, a single-cell representative of each cell type measured from a GFP-mTalin plant was used to derive detector gain and offset settings such that there were no over- or under-exposed pixels when a whole-cell z-stack was collected. These settings were used to collect whole-cell z-stacks (2.5-μm optical slice) of individual cells in GFP-fABD2 and GFP-mTalin plants. GFP fluorescence intensity from individual cells was measured in ImageJ and data analyzed in Microsoft Excel.
Analysis of focal junction movement used centroid tracking in ImageJ. For FRAP experiments, the confocal pinhole was set to produce optical sections approximately 2.5-μm thick. Prebleach and recovery images were acquired at a rate of one image every 1.6 s for GFP-fABD2 and every 0.8 s for FDA and YFP. For photobleaching, all argon laser lines (476, 488, and 497 nm) were used simultaneously at 100% transmittance for 100 iterations to bleach an area approximately 100 μm2 (65 W m−2 for 5 s). Confocal images were stored using LSM software (version 3.2; Zeiss) and processed in Photoshop 7.0 (Adobe Systems, Mountain View, CA). Image analysis was performed in ImageJ and results tabulated, calculated, and plotted in Microsoft Excel. Determination of t1/2 and k followed the methods of Hush et al. (1994). Loss in fluorescence intensity caused by laser scanning during image collection was corrected for by adding the difference in fluorescence intensity calculated over the entire scan window between the first and selected postbleach image to the fluorescence intensity value obtained within the bleach window for the selected postbleach image. Fluorescence recovery across the bleach window was analyzed by dividing the bleach window into equal thirds (left, center, right) and comparing the ratio of fluorescence recovery for left/center or right/center, whichever was greater. The plot profile function of ImageJ was used to assess the change in fluorescence intensity pattern across the bleach window during recovery. Fluorescence intensities were plotted in Excel and a moving-average trend line (period = 8) fitted to the data to reduce plot noise and improve clarity of interpretation. The ratio of change in fluorescence intensity between the edge and center of the bleach window was calculated by dividing the gradient between the edge and center of the bleach window in the first postbleach trace by the gradient between the same positions in the final postbleach trace and averaging for at least six cells.
Immunoblotting
Protein was extracted from whole seedlings into 2× SDS sample buffer and equal total protein loaded and run on a 10% SDS-polyacrylamide gel before transfer to nitrocellulose overnight. Membranes were blocked for 1 h in 5% (w/v) skim milk powder in phosphate-buffered saline before incubation with a 1:500 dilution of anti-GFP (raised against native GFP; Peter Lewis, personal communication) or 1:500 anti-α-tubulin (Sigma; clone B-5-1-2). Washed membranes were then incubated with secondary antibody conjugated to alkaline phosphatase. Bands developed with Western Blue substrate (Promega, Madison, WI; no. S384B) were scanned and the integrated intensities from all bands quantified in ImageJ.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permission will be the responsibility of the requestor.
Supplementary Material
Acknowledgments
We thank Boris Voigt and Diedrik Menzel (Universität Bonn) for supplying pCAT:GFP-fABD2 and pCAT:GFP-AtFIM1 and Steven Pirlo (The University of Newcastle) for pART7:fABD1-GFP and pART7:GFP-fABD1. We are grateful to Madeleine Rashbrooke (Australian National University) and Geoffrey Wasteneys (University of British Columbia) for providing Arabidopsis seed expressing cytoplasmic YFP and to Stephen Dibley (The University of Newcastle) for subcloning. M.B.S. gratefully acknowledges Peter Lewis, Karen Davies, and Geoff Doherty (The University of Newcastle) for providing anti-GFP and for assistance with immunoblotting.
This work was supported by an Australian Research Council Centre of Excellence Grant to The University of Newcastle Node of the Centre of Excellence for Integrative Legume Research (to R.J.R.), and by the U.S. National Science Foundation (grant no. 0130576–MCB to C.J.S. and D.W.M.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.049411.
References
- Baluska F, Jasik J, Edelmann HG, Salajova T, Volkmann D (2001) Latrunculin B-induced plant dwarfism: plant cell elongation is F-actin-dependent. Dev Biol 231: 113–124 [DOI] [PubMed] [Google Scholar]
- Baluska F, Wojtaszek P, Volkmann D, Barlow P (2003) The architecture of polarized cell growth: the unique status of elongating plant cells. Bioessays 25: 569–576 [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD, Mullins RD (2000) Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr Biol 10: 1273–1282 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- El-Assal SE, Le J, Basu D, Mallery EL, Szymanski DB (2004) DISTORTED encodes an ARPC2 subunit of the putative Arabidopsis ARP2/3 complex. Plant J 38: 526–538 [DOI] [PubMed] [Google Scholar]
- Fu Y, Li H, Yang Z (2002) The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14: 777–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbon BC, Kovar DR, Staiger CJ (1999) Latrunculin B has different effects on pollen germination and tube growth. Plant Cell 11: 2349–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Hush JM, Wadsworth P, Callaham DA, Hepler PK (1994) Quantification of microtubule dynamics in living plant cells using fluorescence redistribution after photobleaching. J Cell Sci 107: 775–784 [DOI] [PubMed] [Google Scholar]
- Jedd G, Chua NH (2002) Visualization of peroxisomes in living plant cells reveals acto-myosin-dependent cytoplasmic streaming and peroxisome budding. Plant Cell Physiol 43: 384–392 [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, McKinney EC, Meagher RB (2002) Functional nonequivalency of actin isovariants in Arabidopsis. Mol Biol Cell 13: 251–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T, Allwood EG, Anthony R, Voigt B, Menzel D, Hussey PJ (2004) The actin-interacting protein AIP1 is essential for actin organization and plant development. Curr Biol 20: 145–149 [DOI] [PubMed] [Google Scholar]
- Klahre U, Friederich E, Kost B, Louvard D, Chua N-H (2000) Villin-like actin-binding proteins are expressed ubiquitously in Arabidopsis. Plant Physiol 122: 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein MG, Shi W, Ramagopal U, Tseng Y, Wirtz D, Kovar DR, Staiger CJ, Almo SC (2004) Structure of the actin crosslinking core of fimbrin. Structure 12: 999–1013 [DOI] [PubMed] [Google Scholar]
- Kost B, Mathur J, Chua NH (1999) Cytoskeleton in plant development. Curr Opin Plant Biol 2: 462–470 [DOI] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua NH (1998) A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J 16: 393–401 [DOI] [PubMed] [Google Scholar]
- Kovar DR, Gibbon BC, McCurdy DW, Staiger CJ (2001) Fluorescently-labeled fimbrin decorates a dynamic actin filament network in live plant cells. Planta 213: 390–395 [DOI] [PubMed] [Google Scholar]
- Kovar DR, Staiger CJ, Weaver EA, McCurdy DW (2000) AtFIM1 is an actin filament crosslinking protein from Arabidopsis thaliana. Plant J 24: 625–636 [DOI] [PubMed] [Google Scholar]
- Kwok EY, Hanson MR (2004) In vivo analysis of interactions between GFP-labeled microfilaments and plastid stromules. BMC Plant Biol 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le J, El-Assal SE, Basu D, Saad ME, Szymanski DB (2003) Requirements for Arabidopsis ATARP2 and ATARP3 during epidermal development. Curr Biol 13: 1341–1347 [DOI] [PubMed] [Google Scholar]
- Lebart MC, Hubert F, Boiteau C, Venteo S, Roustan C, Benyamin Y (2004) Biochemical characterization of the L-plastin-actin interaction shows a resemblance with that of alpha-actinin and allows a distinction to be made between the two actin-binding domains of the molecule. Biochemistry 43: 2428–2437 [DOI] [PubMed] [Google Scholar]
- Lee S, Park J, Lee Y (2003) Phosphatidic acid induces actin polymerization by activating protein kinases in soybean cells. Mol Cells 15: 313–319 [PubMed] [Google Scholar]
- Li S, Blanchoin L, Yang Z, Lord EM (2003) The putative Arabidopsis arp2/3 complex controls leaf cell morphogenesis. Plant Physiol 132: 2034–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kernebeck B, Hulskamp M (2003. a) Mutation in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. Plant Cell 15: 1632–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kirik V, Kernebeck B, Srinivas BP, Hulskamp M (2003. b) Arabidopsis CROOKED encodes for the smallest subunit of the ARP2/3 complex and controls cell shape by region specific fine F-actin formation. Development 130: 3137–3146 [DOI] [PubMed] [Google Scholar]
- McCann RO, Craig SW (1997) The I/LWEQ module: a conserved sequence that signifies F-actin binding in functionally diverse proteins from yeast to mammals. Proc Natl Acad Sci USA 94: 5679–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurdy DW, Kim M (1998) Molecular cloning of a novel fimbrin-like cDNA from Arabidopsis thaliana. Plant Mol Biol 36: 23–31 [DOI] [PubMed] [Google Scholar]
- Muday G (2000) Maintenance of asymmetric cellular localization of an auxin transport protein through interaction with the actin cytoskeleton. J Plant Growth Regul 19: 385–396 [DOI] [PubMed] [Google Scholar]
- Nakano K, Satoh K, Morimatsu A, Ohnuma M, Mabuchi I (2001) Interactions among a fimbrin, a capping protein, and an actin-depolymerizing factor in organization of the fission yeast actin cytoskeleton. Mol Biol Cell 12: 3515–3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa K, Kasahara M, Kiyosue T, Kagawa T, Suetsugu N, Takahashi F, Kanegae T, Niwa Y, Kadota A, Wada M (2003) Chloroplast unusual positioning1 is essential for proper chloroplast positioning. Plant Cell 15: 2805–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips GN (1997) Structure and dynamics of green fluorescent protein. Curr Opin Struct Biol 7: 821–827 [DOI] [PubMed] [Google Scholar]
- Ramachandran S, Christensen HE, Ishimaru Y, Dong CH, Chao-Ming W, Cleary AL, Chua NH (2000) Profilin plays a role in cell elongation, cell shape maintenance, and flowering in Arabidopsis. Plant Physiol 124: 1637–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose RJ, Nolan KE, Bicego L (1999) The development of the highly regenerable seed line Jemalong 2HA for transformation of Medicago truncatula: implications for regenerabilty via somatic embryogenesis. J Plant Physiol 155: 788–791 [Google Scholar]
- Schmit AC, Lambert AM (1990) Microinjected fluorescent phalloidin in vivo reveals the F-actin dynamics and assembly in higher plant mitotic cells. Plant Cell 2: 129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan MB, Rose RJ, McCurdy DW (2004) Organelle inheritance in plant cell division: the actin cytoskeleton is required for unbiased inheritance of chloroplasts, mitochondria and endoplasmic reticulum in dividing protoplasts. Plant J 37: 379–390 [DOI] [PubMed] [Google Scholar]
- Smith LG (2003) Cytoskeletal control of plant cell shape: getting the fine points. Curr Opin Plant Biol 6: 63–73 [DOI] [PubMed] [Google Scholar]
- Staiger CJ, Baluška F, Volkmann D, Barlow PW (2000) Actin: A Dynamic Framework for Multiple Plant Cell Functions. Kluwer Academic Publishers, Dordrecht, The Netherlands
- Sullivan ML, Green PJ (1996) Mutational analysis of the DST element in tobacco cells and transgenic plants: identification of residues critical for mRNA instability. RNA 2: 308–315 [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Borisy GG (1999) Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol 145: 1009–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi S (2003) Actin-based photo-orientation movement of chloroplasts in plant cells. J Exp Biol 206: 1963–1969 [DOI] [PubMed] [Google Scholar]
- Takemoto D, Jones DA, Hardham AR (2003) GFP-tagging of cell components reveals the dynamics of subcellular re-organization in response to infection of Arabidopsis by oomycete pathogens. Plant J 33: 775–792 [DOI] [PubMed] [Google Scholar]
- Thomas MR, Rose RJ (1983) Plastid number and plastid structural changes associated with tobacco mesophyll protoplast culture and plant regeneration. Planta 158: 329–338 [DOI] [PubMed] [Google Scholar]
- Thomas MR, Rose RJ, Nolan KE (1992) Genetic transformation of Medicago truncatula using Agrobacterium with genetically modified Ri and disarmed Ti plasmids. Plant Cell Rep 11: 113–117 [DOI] [PubMed] [Google Scholar]
- Timmers AC, Niebel A, Balague C, Dagkesamanskaya A (2002) Differential localisation of GFP fusions to cytoskeleton-binding proteins in animal, plant, and yeast cells: green-fluorescent protein. Protoplasma 220: 69–78 [DOI] [PubMed] [Google Scholar]
- Vitha S, Baluska F, Braun M, Samaj J, Volkmann D, Barlow PW (2000) Comparison of cryofixation and aldehyde fixation for plant actin immunocytochemistry: aldehydes do not destroy F-actin. Histochem J 32: 457–466 [DOI] [PubMed] [Google Scholar]
- Waller F, Riemann M, Nick P (2002) A role for actin-driven secretion in auxin-induced growth. Protoplasma 219: 72–81 [DOI] [PubMed] [Google Scholar]
- Wang Y-S, Motes CM, Mohamalawari DR, Blancaflor EB (2004) Green fluorescent protein fusions to Arabidopsis fimbrin 1 for spatio-temporal imaging of F-actin dynamics in roots. Cell Motil Cytoskeleton 59: 79–93 [DOI] [PubMed] [Google Scholar]
- Weber I, Niewohner J, Du A, Rohrig U, Gerisch G (2002) A talin fragment as an actin trap visualizing actin flow in chemotaxis, endocytosis, and cytokinesis. Cell Motil Cytoskeleton 53: 136–149 [DOI] [PubMed] [Google Scholar]
- Welch MD, Mullins RD (2002) Cellular control of actin nucleation. Annu Rev Cell Dev Biol 18: 247–288 [DOI] [PubMed] [Google Scholar]
- Yang Y, Li R, Qi M (2000) In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J 22: 543–551 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.