Abstract
Expression of green fluorescent protein (GFP) linked to an actin binding domain is a commonly used method for live cell imaging of the actin cytoskeleton. One of these chimeric proteins is GFP-mTalin (GFP fused to the actin binding domain of mouse talin). Although it has been demonstrated that GFP-mTalin colocalizes with the actin cytoskeleton, its effect on actin dynamics and cell expansion has not been studied in detail. We created Arabidopsis (Arabidopsis thaliana) plants harboring alcohol inducible GFP-mTalin constructs to assess the effect of GFP-mTalin expression in vivo. We focused on the growing root hair as this is a model cell for studying cell expansion and root hair tip growth that requires a highly dynamic and polar actin cytoskeleton. We show that alcohol inducible expression of GFP-mTalin in root hairs causes severe defects in actin organization, resulting in either the termination of growth, cell death, and/or changes in cell shape. Fluorescence recovery after photobleaching experiments demonstrate that the interaction of GFP-mTalin and actin filaments is highly dynamic. To assess how GFP-mTalin affects actin dynamics we performed cosedimentation assays of GFP-mTalin with actin on its own or in the presence of the actin modulating protein, actin depolymerizing factor. We show that that GFP-mTalin does not affect actin polymerization but that it does inhibit the actin depolymerizing activity of actin depolymerizing factor. These observations demonstrate that GFP-mTalin can affect cell expansion, actin organization, and the interaction of actin binding proteins with actin.
The actin cytoskeleton is a dynamic structure, present throughout all eukaryotic cells. Processes such as cell growth, signaling, transport, and cell division depend on an intact and functional actin network. Since the actin cytoskeleton is involved in many processes and is very dynamic, understanding how actin reorganization occurs is a goal in cell biology. Two methods have been used to study actin reorganizations in plant cells: the microinjection of fluorescently labeled phallotoxins that bind the filamentous actin (e.g. Staiger et al., 1994; Valster et al., 1997) and the use of green fluorescent protein (GFP) and actin binding domain chimeric proteins. Numerous studies have used expression of GFP, linked to the actin binding domain of actin binding proteins, talin, plastin, and fimbrin to visualize the actin cytoskeleton in living cells (e.g. Kost et al., 1998; Fu et al., 2001; Timmers et al., 2002; Ketelaar et al., 2004; Sheahan et al., 2004). A chimeric protein, consisting of GFP, fused to the actin binding domain of mouse talin (GFP-mTalin) has been shown to decorate all the F-actin that is visualized with fluorescently labeled phalloidin in BY2 tobacco tissue culture cells (Kost et al., 1998).
Although expression of GFP fused to actin binding domains is a commonly used and convenient method to study the actin cytoskeleton in live cells, the effect of the expression of such proteins on actin organization and dynamics has never been studied in detail. The expression of actin binding chimeric proteins that are larger than an actin monomer could block or compete with endogenous actin binding proteins or even stabilize actin. Therefore, it is reasonable to suggest that changes in the dynamics or the organization of the actin cytoskeleton take place and that these changes may cause problems with cell growth, cell division, transport, or signaling.
In this paper, we asked whether there are limitations in the use of GFP-mTalin for in vivo actin visualization. We have used alcohol-inducible GFP-mTalin lines to monitor the effect of the chimeric protein on actin organization and cell expansion in root hairs, a model cell-type for studying polar cell expansion. We complemented these analyses with in vitro actin binding studies. Recombinant GFP-mTalin was used to assess its effect on actin polymerization in the absence and presence of the known actin modulating protein, actin depolymerizing factor (ADF). Here we demonstrate that expression of GFP-mTalin causes severe defects in actin organization and cell expansion in Arabidopsis (Arabidopsis thaliana) root hairs and that GFP-mTalin inhibits the actin depolymerizing activity of the endogenous actin binding protein ADF in vitro.
RESULTS
Alcohol Inducible GFP-mTalin
An N-terminal fusion of soluble-modified GFP to the actin-binding domain of mouse talin (mTn) placed under the control of an ethanol inducible promoter in a binary vector was generated. The ethanol inducible ALC switch system was based on the ALCR transcription factor and alcA promoter of Aspergillus nidulans as reported by Salter et al. (1998) and Caddick et al. (1998). The sequence of the actin binding domain of mTn (McCann and Craig, 1997) and the linker sequence between soluble-modified GFP and mTn was identical to those in the construct reported by Kost et al. (1998). The predicted molecular mass of the chimeric GFP-mTalin protein is 48.8 kD. Three independent transgenic Arabidopsis lines were generated and analyzed. Before induction with ethanol, the transgenic lines were indistinguishable from wild-type plants.
Expression Levels Differ Moderately between Lines
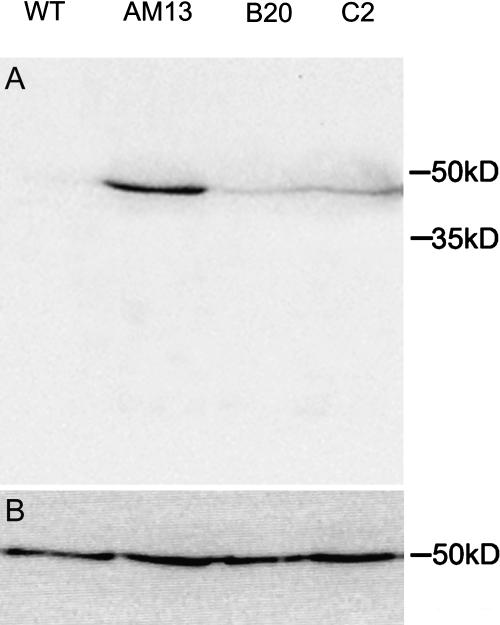
Four-week-old plants of three independent transgenic lines grown in soil were tested for GFP-mTalin expression. Total protein samples were prepared from whole plants 48 h after ethanol induction and fractionated on one-dimensional gels prior to western blotting. The blots were probed both with an antibody against GFP, to analyze the expression levelsm and an antibody against tubulin as a loading control. Figure 1 shows that there is some variation in the expression levels of GFP-mTalin between the lines with line AM13 showing the highest expression.
Figure 1.
Western blot with anti-GFP antibody (A) and anti-tubulin antibody (B) on GFP-mTalin expressing lines. Plants were induced for 48 h with 1% ethanol and show different expression levels of GFP-mTalin per transgenic line.
GFP-mTalin Decorates the Actin Cytoskeleton
To observe the GFP-mTalin expression in roots, Arabidopsis plants were grown with their roots in a thin layer of solidified growth medium (Wymer et al., 1997) covered by biofoil. Adding liquid medium containing 0.05% to 1% ethanol induced the expression of the GFP-mTalin. In control root hairs, the concentrations of ethanol that were used for induction did not cause any toxic effects, as has been demonstrated previously (Ketelaar et al., 2004).
In GFP-mTalin expressing cells in the roots, the first fluorescent cells appeared approximately 15 min after induction. The number of cells that were fluorescing increased over the next 15 min. After approximately 30 min, the fluorescence level in any induced cell did not appear to increase further. However, fluorescence occassionally appeared in newly formed cells over time. For analysis of the effects of GFP-mTalin expression, we focused on root hairs. The root hairs used for the analysis described below were induced for 30 to 120 min.
The organization of the actin cytoskeleton in root hairs that had terminated growth before induction was similar to that previously described in Ketelaar et al. (2002). Thick longitudinal bundles of F-actin run through the cortical cytoplasm and loop through the apex (Fig. 2A; Ketelaar et al., 2002, 2003). Root hairs that were growing when GFP-mTalin expression was induced with ethanol responded to GFP-mTalin induction in four different ways: (1) The tip of the root hairs swelled as the root hairs terminated growth. In these root hairs, the organization of the actin cytoskeleton was similar to that in root hairs that had terminated growth before induction with ethanol (Fig. 2B). (2) The root hairs exploded at the tip at a certain stage of their development after fluorescence appeared. In these root hairs, the actin configuration was not recordable (compare Fig. 3, A and B). (3) The root hairs continued to grow but partially lost their polarity, resulting in hairs with a widened diameter (up to 3 times the normal root hair width). The actin cytoskeleton in this class of root hairs was distorted (Fig. 2C). In the cortex, there was a dense network of helical oriented F-actin (Fig. 2D), whereas many actin bundles ramified the central vacuole (Fig. 2E). (4) The root hairs continued to grow normally. When examined in the microscope, the actin cytoskeleton in these root hairs was not fluorescently labeled, indicating that no GFP-mTalin was expressed. Table I gives the percentages of root hairs in each class in wild-type and GFP-mTalin plants.
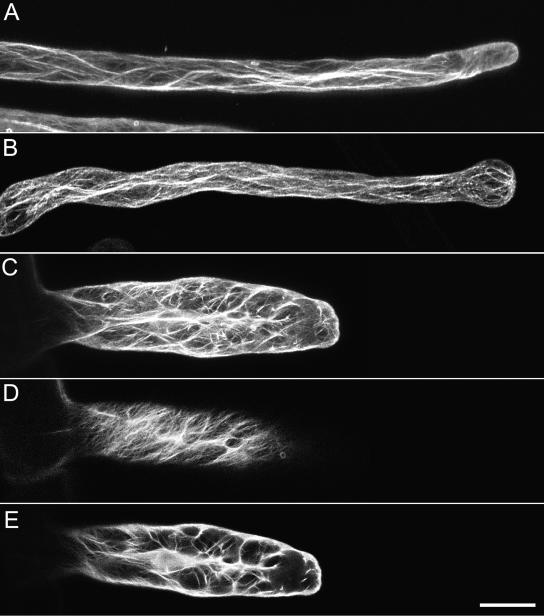
Figure 2.
The actin configuration of root hairs expressing GFP-mTalin. Root hairs were induced for 2 h with 1% ethanol. A shows a root hair that had terminated growth before induction (Z-stack of 20 optical slices, Z-step 1 μm). B is a root hair that was growing during induction and responded by terminating growth after swelling (Z-stack of 20 optical slices, Z-step 1 μm). C displays a root hair that continued to grow after induction of ethanol. Root hairs that develop when expressing GFP-mTalin have a much wider diameter than control root hairs. D and E are partial projections of the root hair in C. D is an optical section of the cortical localization of GFP-mTalin and E is an optical section through the center of the cell. The complete Z-stack consisted of 20 optical slices, Z-step 1.5 μm; D and E are projections of five subsequent slices. Bar = 20 μm.
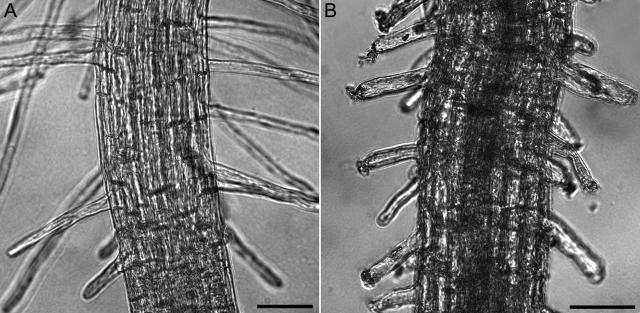
Figure 3.
The response of growing wild-type and GFP-mTalin root hairs to induction with 1% ethanol. A is an image of the zone of growing root hairs of a wild-type plant, 2 h after induction with ethanol. B is an image of the zone of growing root hairs of a GFP-mTalin 2 h after induction with ethanol. Bars = 50 μm.
Table I.
Percentages of root hairs in GFP-mTalin lines and wild-type Arabidopsis that swell and stop growing (1), explode at the tip (2), partially loose polarity (3), and continue to grow normally (4) upon induction with 1% ethanol in the medium
| Genotype
|
Response
|
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| Wild type | 9.8 | 2.9 | 0 | 87.3 |
| AM13 | 43.2 | 45.9 | 5.4 | 5.4 |
| B20 | 39.2 | 43.1 | 5.9 | 11.8 |
| C2 | 38.5 | 38.5 | 3.8 | 19.2 |
At least 8 plants and 15 root hairs per plant were analyzed per line.
The Interaction between GFP-mTalin and F-Actin Is Dynamic
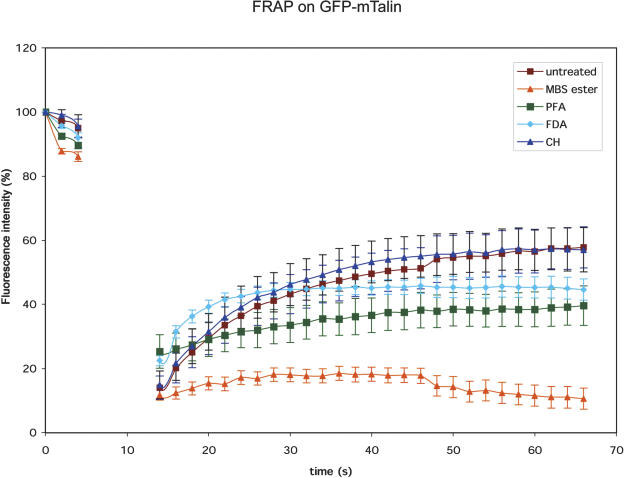
To analyze the binding dynamics of GFP-mTalin with actin, we performed fluorescence recovery after photobleaching (FRAP) experiments on induced, fully grown root hair cells. A small square was photobleached by five repetitive scans at full laser power and the reappearance of fluorescence was studied. Figure 4 shows an area of a GFP-mTalin expressing root hair cell during a FRAP experiment. After photobleaching, we observed a partial recovery of the fluorescence in the bleached area within seconds. The new fluorescence originated from the sides of the photobleached area. These data can be explained either by a dynamic flux of GFP-mTalin off and on the actin filaments or an increase in decoration density of actin with newly synthesized GFP-mTalin. To exclude the latter possibility, we treated roots with 10 μg/mL of the translation blocker cycloheximide for 15 min prior to the FRAP experiment. The cycloheximide treatment did not cause significant changes in recovery rate, indicating that the observed recovery is indeed a dynamic GFP-mTalin flux. As additional controls, we used cytoplasmic fluorescein in noninduced cells (loaded as 0.05% fluorescein di-acetate [FDA]) and the cross-linkers m-maleimidobenzoyl N-hydroxysuccinimide ester (MBS-ester, 1 mm; Sonobe and Shibaoka, 1989) and paraformaldehyde (2%) in induced cells (Fig. 5). When cells were treated with MBS-ester or paraformaldehyde, GFP dissociated from the filamentous structures and became cytoplasmic. Treating the cells with paraformaldehyde led to an increase in background fluorescence caused by aldehyde induced autofluorescence (thus increasing the total fluorescence level), and both cross-linkers inhibited cytoplasmic streaming, causing a decrease in recovery speed after photobleaching. The recovery speed of cytoplasmic fluorescein after photobleaching is only slightly faster than GFP-mTalin over the first seconds of imaging. The decrease in intensity of the fluorescein over time is caused by its higher sensitivity to photobleaching than GFP. The photobleaching caused by the high frequency of scans also explains the failure of the GFP-mTalin to recover fully.
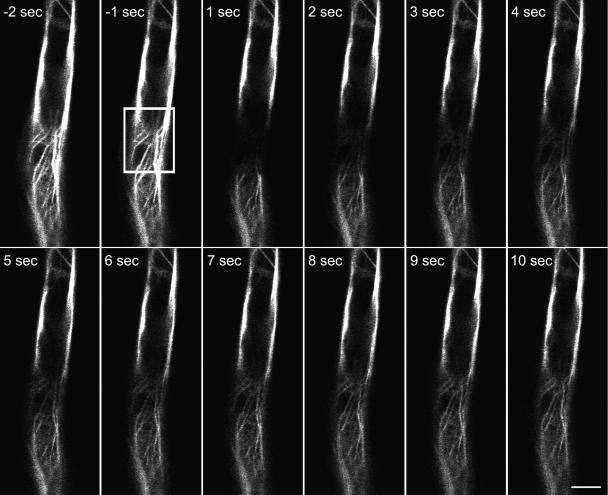
Figure 4.
Rapid recovery takes place after local photobleaching of GFP-mTalin. Confocal images (single optical sections) of a fully expanded root hair cell were captured with 1-s intervals. After two initial scans, the area within the white box was bleached as described in “Materials and Methods.” Recovery takes place in seconds, as the fluorescence reappears, emerging from the sides of the bleached box. Bar = 10 μm.
Figure 5.
FRAP reveals a dynamic flux of GFP-mTalin off and on F-actin. First three scans (single optical sections in one focal plane) were performed, whereafter a box of approximately 10 μm in length was bleached by five consecutive scans at full laser power (see Fig. 4). This interval corresponds with the data-less area in the graph. The recovery after FRAP takes place slightly slower than the cytoplasmic streaming speed (FDA control), but much faster than in cells that have been fixed with MBS-ester or paraformaldehyde (PFA). The reappearance of fluorescence continues after cycloheximide (CH) treatment, indicating that the appearing fluorescence is generated by release of existing GFP-mTalin from F-actin.
GFP-mTalin Does Not Stabilize Actin, But Inhibits Actin Depolymerization by ADF in Vitro
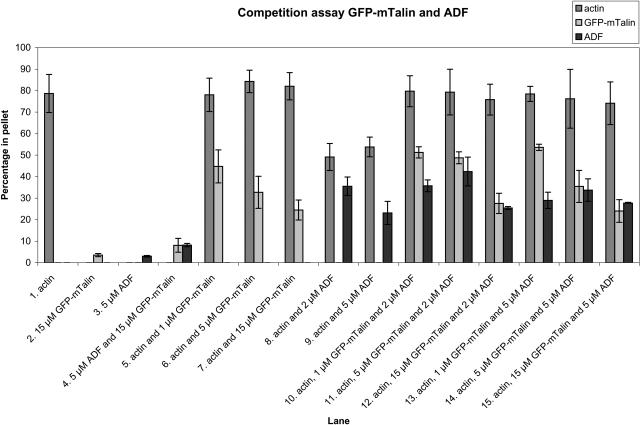
We expressed GFP-mTalin and AtADF2 (At3g46000) in Escherichia coli and analyzed the actin binding capacities of these proteins on their own or together in F-actin cosedimentation assays (Allwood et al., 2002). When 5 μm rabbit muscle actin was polymerized for 30 min at room temperature, the percentage of the total amount of actin that polymerized and appeared in the pellet was approximately 80% (Fig. 6, lane 1). When GFP-mTalin was added to final concentrations of 1, 5, and 15 μm during actin polymerization, part of it cosedimented with the polymerized actin (Fig. 6, lanes 5–7). Very little sedimentation of 15 μm GFP-mTalin on its own occurred (Fig. 6, lanes 2 and 4). At the higher concentrations of GFP-mTalin, a lower percentage appeared in the pellet, possibly because of the saturation of binding sites on the F-actin. The addition of GFP-mTalin did not change the percentage of actin that would polymerize, and this result is similar to previously documented results (McCann and Craig, 1997). We conclude that GFP-mTalin binds to actin, but does not affect the amount of the actin polymerization.
Figure 6.
GFP-mTalin inhibits the actin depolymerizing activity of ADF. Schematic overview of cosedimentation assays of 5 μm actin with different concentrations of GFP-mTalin and ADF. Approximately 80% of polymerized actin sediments during centrifugation and appears in the pellet fraction (lane 1). ADF and GFP-mTalin do not sediment significantly on their own (lanes 2–4), whereas they do when mixed with actin (lanes 5–9). ADF has an actin depolymerizing effect as a lower percentage of actin appears in the pellet (lanes 8–9). When ADF and GFP-mTalin are incubated with actin simultaneously, ADF fails to depolymerize actin (lanes 10–15). Error bars display sds.
The percentage of ADF (both at 2 and 5 μm concentrations) that cosedimented with actin was much higher than the percentage of 5 μm ADF that appears in the pellet in the absence of actin (compare Fig. 6, lanes 3, and 8 and 9). However, the percentage of actin that is polymerized and thus appears in the pellet fraction was reduced to approximately 55%. When we assayed cosedimentation of the same concentrations of ADF with actin in the presence of GFP-mTalin (1, 5, and 15 μm), the percentage of actin in the pellet failed to drop as would be expected after ADF application for either concentration (Fig. 6, lanes 10–15). From these results it is concluded that the presence of GFP-mTalin inhibits the actin depolymerizing ability of ADF, although ADF binding to actin is not inhibited, as it still sediments with actin.
DISCUSSION
Here we show that GFP-mTalin expression in vivo causes defects in root hair expansion and that GFP-mTalin inhibits the actin severing activity of ADF. Root hairs were chosen for the analysis of the effects of GFP-mTalin expression on actin configuration and cellular development as growing root hairs have a highly dynamic actin cytoskeleton in the subapical area. Small changes in actin organization are therefore more likely to have a severe effect on root hair development than on development of intercalary growing cell types where the actin cytoskeleton is visually less dynamic. We generated ethanol inducible GFP-mTalin expressing Arabidopsis lines so that we could monitor the effect of GFP-mTalin expression as the GFP became visible. When GFP-mTalin expression was induced by the addition of ethanol to the medium, fluorescent filamentous structures were observed in the fully grown root hairs. These structures have been shown to be actin filaments by Kost et al. (1998). The overall organization of the actin network as visualized by GFP-mTalin was no different from the organization of actin filaments in fully grown root hairs visualized by immunofluorescence (Ketelaar et al., 2002, 2003). However, root hairs that were growing during induction responded differently when GFP-mTalin expression started. Most of the root hairs exploded or swelled at the tip, whereafter they terminated growth. In contrast, ethanol treatment did not affect most growing wild-type root hairs and these continued to grow. These data indicate that the dynamic, growing root hairs are more sensitive to GFP-mTalin expression than the fully grown root hairs. We suggest that the growing root hairs are unable to maintain the actin configuration that is required for the localized, polar, cell expansion when GFP-mTalin is present. The low percentage of growing root hairs that continue to grow when GFP-mTalin expression is induced widen up to 3 times the normal root hair diameter, indicating that growing root hairs are not able to maintain the actin configuration that is required for tip growth. The root hair phenotype that we observe in alcohol inducible GFP-mTalin lines upon alcohol induction is more severe than that of constitutively expressing plants. Possibly this is an expression level dependent difference, but it may also be caused by adaptation of the intracellular environment to GFP-mTalin expression in constitutively expressing lines. In growing root hairs, constitutively expressing GFP-mTalin, little filamentous actin can be observed (Baluška et al., 2000; Wang et al., 2004).
GFP-mTalin has been used to visualize actin in pollen tubes in a number of reports (e.g. Kost et al., 1998; Fu et al., 2001; Cheung and Wu, 2004). Only Cheung and Wu (2004) report GFP-mTalin induced effects in at least one-half of the pollen tubes but do not explain what these effects are. In other papers where GFP-mTalin is used in pollen tubes, no problems with expression of GFP-mTalin in pollen are reported. The species that were used in these reports (lily and tobacco) may be less sensitive than Arabidopsis to GFP-mTalin expression. In root hairs, two reports are available where the GFP-mTalin construct has been used. Jones et al. (2002) report the use of the GFP-mTalin construct to visualize the actin cytoskeleton in Arabidopsis root hairs by biolistic delivery. The root hairs they observed are likely to be fully grown (the actin configuration in their control root hair resembles the actin cytoskeleton in a fully grown root hair during induction of GFP-mTalin; Fig. 2A). Baluška et al. (2000) show root hairs of an Arabidopsis plant expressing GFP-mTalin at low magnification. Although a few thick actin bundles are visible throughout the root hairs, the GFP-mTalin localization does not resemble the pattern shown in this paper. The expression level in these root hairs may be so low that most of the actin fails to be decorated. The observations in pollen tubes point to at least some degree of sensitivity of pollen tube growth to GFP-mTalin. To our knowledge, there are no clear published data describing the effects and expression of GFP-mTalin in growing root hairs.
The FRAP experiments on GFP-mTalin expressing root hairs show a recovery of fluorescence that originates from the periphery of the bleached area at a speed that is comparable to the speed of cytoplasmic streaming (fluorescein control). This recovery could either take place by sliding or by polymerizing of GFP-mTalin tagged actin into the bleached area. Alternatively, the interaction between F-actin and GFP-mTalin could be dynamic, leading to rapid dissociation and association of GFP-mTalin. It is unlikely that the thick bundles of actin filaments in fully grown root hairs are very dynamic (Miller et al., 1999; Ketelaar et al., 2003). Therefore, it is likely that the fluorescence recovery is caused by rapid association and dissociation of GFP-mTalin from F-actin. In vitro actin binding competition assays between the actin binding domain of talin and the whole talin protein, where both proteins compete for binding places (McCann and Craig, 1997), point to a similar conclusion. Moreover, the release of GFP-mTalin from the actin filaments by aldehyde or MBS-ester treatment further supports the idea that the interaction between GFP-mTalin and actin is highly dynamic.
We performed cosedimentation assays of GFP-mTalin with rabbit muscle actin to assess any change in actin polymerization caused by GFP-mTalin. McCann and Craig (1997) demonstrated that the actin binding domain of talin that we also used (the I/LWEQ motif) does not cause an increase in actin polymerization. Our results with the same motif linked to GFP are similar. However, competition studies with the I/LWEQ domain show that it competes with full-length talin for binding sites on actin filaments (McCann and Craig, 1997). Although the actin binding domain used in GFP-mTalin is derived from a mammalian actin-binding protein, it does bind to plant F-actin, and therefore it is likely that it competes with other actin binding proteins for a binding site. When ADF and GFP-mTalin were mixed with F-actin in a cosedimentation assay, the amount of actin in the pellet did not decrease. ADF on its own with F-actin caused a significant decrease in the amount of actin in the pellet. These observations show that GFP-mTalin inhibits the actin depolymerizing activity of ADF. It is likely that mTalin hinders the interaction of ADF with F-actin.
Generally, GFP-mTalin and other chimeric reporter proteins are in many cases the best, or even the only way to perform live cell imaging of the actin cytoskeleton. Our results demonstrate that although GFP-mTalin localizes to F-actin, the F-actin localization and/or dynamics may change due to the expression of GFP-mTalin. Fusion proteins such as GFP-mTalin offer a way to gain knowledge about in vivo localization and dynamic behavior of proteins, which cannot be obtained in another way. GFP-mTalin can be used as a useful marker for the actin cytoskeleton; however, if the actin cytoskeleton is highly dynamic and under control of actin binding proteins (ADF in particular), the data in this paper show that the expression of GFP-mTalin will affect actin dynamics to some degree. We suggest the use of alcohol inducible GFP-Talin expression when dynamics are being investigated and that this be done within a calculated timescale after induction: 15 to 30 min for root hairs is suggested from the data in this paper. This will minimize the adaptation of the cell to GFP-mTalin expression, so that one observes a more native situation. In addition, controls (such as immunostaining) should be used to demonstrate that observations are not artifacts, caused by overexpression of the GFP-mTalin construct and also a caveat should be added that GFP-mTalin can inhibit the activity of known actin modulating proteins.
MATERIALS AND METHODS
Construction of Alcohol Inducible GFP-mTalin Chimeric Gene
The C terminus of mouse talin (accession number X56123) was amplified to create an N-terminal XhoI site and a C-terminal SalI site (forward, 5′-TTCTCGAGATGATCCTAGAAGCTGCCAAGTCCATCGCTGCAGC-3′; reverse, 5′-TTGTCGACTTAGTGCTCGTCTCGAAGCTCTGAAGGCA-3′). Plant adapted GFP (accession no. U70496) was amplified incorporating an N-terminal SalI site and a C-terminal XhoI site (forward, 5′-TTGCATGCGTCGACATGAGTAAAGGAGAAGAACTT-3′; reverse, 5′-TTCTCGAGGCCTGCGCCTGCGCCTGCGCCTGCGCCTGCGCCGGATTTGTATAGTTCATCCATGCCATG-3′). The GFP C-terminal primer also contained a linker replacing the stop codon, as described in Kost et al. (1998). The two inserts were cloned together, in frame, into pGEM-T easy (Promega, Southampton, UK). The GFP-mTalin cassette was excised from pGEMT-easy and subcloned into the SalI site of an adapted version of the pACN vector (supplied by Syngenta; Roslan et al., 2001), in which the AlcA promoter-nos cassette can be excised with SpeI. The AlcA-GFP-mTalin-nos cassette was then subcloned into a specially adapted version of binSRNACatN (Syngenta, UK), in which constitutive expression of the AlcR gene was directed by the cauliflower mosaic virus 35S promoter. The final GFP-mTalin alcohol inducible binary vector was transferred to Agrobacterium tumefaciens strain GV3101 (pMP90) and used to transform Arabidopsis (Arabidopsis thaliana) ecotype Columbia by floral dipping (Clough and Bent, 1998).
GFP-mTalin Chimeric Gene Protein Expression in Escherichia coli
GFP-mTalin was amplified by PCR from cDNA of the Arabidopsis lines we generated with primers containing GATEWAY sequences (Invitrogen, Paisley, UK) and recombined into pDONR201 (Invitrogen, Paisley, UK) according to the manufacturer's guidelines. GFP-mTalin was than recombined into pGAT4, an ampicillin resistant pET based plasmid that adds an N-terminal 6×HIS to the protein. For expression in E. coli, the strain Rosetta (DE3; CLONTECH, via VWR, Lutterworth, UK) was used. Bacteria were cultured at 37°C to an OD600 of 0.6, whereafter they were transferred to 4°C and induced with isopropylthio-β-galactoside to a final concentration of 1 mm overnight.
HIS-tagged proteins were purified with the HisTrap purification system (Amersham Pharmacia, Uppsala) according to the manufacturer's protocol.
The purified proteins were dialyzed into phosphate-buffered saline, concentrated in a Vivaspin 6, 10,000 MWCO column (Vivascience, Lincoln, UK). AtADF2 was prepared as described previously (Allwood et al., 2002).
Cosedimentation Assays
Cosedimentation assays were performed as described previously (Allwood et al., 2002). Rabbit muscle actin was obtained from Cytoskeleton (Denver, CO) and all assays were performed at pH 6.5. Percentages of proteins that sedimented were calculated by comparing the intensity of the pellet and supernatant bands on SDS-page gel after Coomassie Blue staining by scanning the gels and analyzing the gray values in Photoshop 7.0 (Adobe Systems, Mountain View, CA).
Plant Material and Drug Treatments
Seeds were surface sterilized by rinsing them in 70% (v/v) ethanol for 1 min, followed by a 15 min treatment in 10% (v/v) bleach + 0.05% (v/v) Triton X-100 and three rinses in sterile distilled water. Subsequently, the seeds were germinated on coverslips with a thin layer of solid medium, covered with Biofoil (Vivascience, via Merck, Poole, UK). The composition of the medium was identical to the medium used by Wymer et al. (1997). A total of 0.7% (w/v) plant agar (Duchefa, Haarlem, The Netherlands) was used to solidify the medium. The slides with seedlings were contained in 70-mm petri dishes wrapped with parafilm.
Plants were cultured at 22°C at long daylight regime (16 h light, 8 h dark) for 5 d. Induction was carried out by flooding the coverslips with liquid medium, containing 1% (v/v) ethanol. Application of drugs was performed identically. Cycloheximide (Sigma, Poole, UK) was dissolved in methanol, paraformaldehyde (Merck, West Drayton, UK) was dissolved in water according to manufacturer's instructions, and FDA (Sigma, Poole, UK) and maleimido benzoyl N-hydroxysuccinimide ester (Sigma, Poole, UK) were dissoved in dimethyl sulfoxide. The amount of solvent never exceeded 1% (v/v) of the total volume of solutions.
Western Blotting
Four-week-old plants, grown individually in 3-inch pots, were induced for 48 h by watering them with 25 mL water with 1% (v/v) ethanol per pot. All the above ground tissues of three of these plants per line were harvested and immediately frozen in liquid nitrogen. Protein samples were prepared and western blotting was performed as described by Ketelaar et al. (2002). To probe for tubulin, we used 1:1,000 diluted anti α-tubulin clone DM1A (Sigma, Poole, UK) and to probe GFP-mTalin, we used 1:500 diluted anti-GFP ab6556 (Abcam, Cambridge, UK).
Microscopy
Confocal imaging and FRAP was performed on a Zeiss (Jena, Germany) LSM510 META system. The Argon-ion laser was used at 4% of the maximum power for imaging, combined with main dichroic beamsplitter HFT 488 nm and a BP 505- to 530-nm emission filter. For photobleaching, five scans at full laser power with all the lines of the Argon-ion laser (453, 470, 488, and 514 nm) were made.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Acknowledgments
We thank Ellen Allwood (University of Durham) for the generous gift of AtADF2 protein and Matt Spence (University of Durham) for technical assistance.
This work was supported by the Biotechnology and Biological Sciences Research Council, UK (to T.K., R.G.A., and P.J.H.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.050799.
References
- Allwood EG, Anthony RG, Smertenko AP, Reichelt S, Drobak BK, Doonan JH, Weeds AG, Hussey PJ (2002) Regulation of the pollen-specific actin-depolymerizing factor LlADF1. Plant Cell 14: 2915–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluška F, Salaj J, Mathur J, Braun M, Jasper F, Šamaj J, Chua NH, Barlow PW, Volkmann D (2000) Root hair formation: F-actin-dependent tip growth is initiated by local assembly of profilin-supported F-actin meshworks accumulated within expansin-enriched bulges. Dev Biol 227: 618–632 [DOI] [PubMed] [Google Scholar]
- Caddick MX, Greenland AJ, Jepson I, Krause K-P, Qu N, Riddell KV, Salter MG, Schuch W, Sonnewald U, Tomsett AB (1998) An ethanol inducible gene switch for plants used to manipulate carbon metabolism. Nat Biotechnol 16: 177–180 [DOI] [PubMed] [Google Scholar]
- Cheung AY, Wu HM (2004) Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell 16: 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Fu Y, Wu G, Yang ZB (2001) Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J Cell Biol 152: 1019–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MA, Shen JJ, Fu Y, Li H, Yang Z, Grierson CS (2002) The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14: 763–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T, Allwood EG, Anthony R, Voigt B, Menzel D, Hussey PJ (2004) The actin-interacting protein AIP1 is essential for actin organization and plant development. Curr Biol 14: 145–149 [DOI] [PubMed] [Google Scholar]
- Ketelaar T, de Ruijter NC, Emons AMC (2003) Unstable F-actin specifies the area and microtubule direction of cell expansion in Arabidopsis root hairs. Plant Cell 15: 285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T, Faivre-Moskalenko C, Esseling JJ, De Ruijter NCA, Grierson C, Dogterom M, Emons AMC (2002) Positioning of nuclei in Arabidopsis root hairs: an actin regulated process of tip growth. Plant Cell 14: 2941–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua N-H (1998) A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J 16: 393–401 [DOI] [PubMed] [Google Scholar]
- McCann RO, Craig SW (1997) The I/LWEQ module: a conserved sequence that signifies F-actin binding in functionally diverse proteins from yeast to mammals. Proc Natl Acad Sci USA 94: 5679–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DD, de Ruijter NCA, Bisseling T, Emons AMC (1999) The role of actin in root hair morphogenesis: studies with lipochito-oligosaccharide as a growth stimulator and cytochalasin as an actin perturbing drug. Plant J 17: 141–154 [Google Scholar]
- Roslan HA, Salter MG, Wood CD, White MRH, Croft KP, Robson F, Coupland G, Doonan J, Laufs P, Tomsett AB, et al (2001) Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. Plant J 28: 225–235 [DOI] [PubMed] [Google Scholar]
- Salter MG, Paine JA, Riddell KV, Jepson I, Greenland AJ, Caddick MX, Tomsett AB (1998) Characterisation of the ethanol-inducible alc gene expression system for transgenic plants. Plant J 16: 127–132 [Google Scholar]
- Sheahan MB, Rose RJ, McCurdy DW (2004) Organelle inheritance in plant cell division: the actin cytoskeleton is required for unbiased inheritance of chloroplasts, mitochondria and endoplasmic reticulum in dividing protoplasts. Plant J 37: 379–390 [DOI] [PubMed] [Google Scholar]
- Sonobe S, Shibaoka H (1989) Cortical fine actin filaments in higher plant cells visualized by rhodamine-phalloidin after pretreatment with m-maleimidobenzoyl N-hydroxysuccinimide ester. Protoplasma 148: 80–86 [Google Scholar]
- Staiger CJ, Yuan M, Valenta R, Shaw PJ, Warn RM, Lloyd CW (1994) Microinjected profilin affects cytoplasmic streaming in plant cells by rapidly depolymerizing actin microfilaments. Curr Biol 4: 215–219 [DOI] [PubMed] [Google Scholar]
- Timmers ACJ, Niebel A, Balague C, Dagkesaruanskaya A (2002) Differential localisation of GFP fusions to cytoskeleton-binding proteins in animal, plant, and yeast cells. Protoplasma 220: 69–78 [DOI] [PubMed] [Google Scholar]
- Valster AH, Pierson ES, Valenta R, Hepler PK, Emons AMC (1997) Probing the plant actin cytoskeleton during cytokinesis and interphase by profilin microinjection. Plant Cell 9: 1815–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YS, Motes CM, Mohamalawari DR, Blancaflor EB (2004) Green fluorescent protein fusions to Arabidopsis fimbrin 1 for spatio-temporal imaging of F-actin dynamics in roots. Cell Motil Cytoskeleton 59: 79–93 [DOI] [PubMed] [Google Scholar]
- Wymer CL, Bibikova TN, Gilroy S (1997) Cytoplasmic free calcium distributions during the development of root hairs of Arabidopsis thaliana. Plant J 12: 427–439 [DOI] [PubMed] [Google Scholar]