Abstract
Due to their capability of eliciting a form of posttranscriptional gene silencing (termed virus-induced gene silencing or VIGS), plant viruses are increasingly used as reverse-genetics tools for functional characterization of plant genes. RNA viruses have been shown to trigger silencing in a variety of host plants, including members of Solanacae and Arabidopsis (Arabidopsis thaliana). Several factors affect the silencing response, including host range and viral tropism within the plant. The work presented here demonstrates that a modified tobacco rattle virus (TRV) vector retaining the helper protein 2b, required for transmission by a specific vector nematode, not only invades and replicates extensively in whole plants, including meristems, but also triggers a pervasive systemic VIGS response in the roots of Nicotiana benthamiana, Arabidopsis, and tomato (Lycopersicon esculentum). This sustained VIGS response was exemplified by the silencing of genes involved in root development (IRT1, TTG1 [transparent testa glabra], RHL1 [root hairless1], and β-tubulin), lateral root-meristem function (RML1 [root meristemless1]), and nematode resistance (Mi). Roots of silenced plants exhibit reduced levels of target mRNA and phenocopy previously described mutant alleles. The TRV-2b vector displays increased infectivity and meristem invasion, both key requirements for efficient VIGS-based functional characterization of genes in root tissues. Our data suggest that the TRV helper protein 2b may have an essential role in the host regulatory mechanisms that control TRV invasion.
Multicellular organisms have developed complex mechanisms involved in the survey of their genome integrity. Operating at the RNA level, posttranscriptional gene silencing (PTGS) is one of the most adaptable and specific of these mechanisms, culminating in a sequence-specific degradation of so-called aberrant RNA. This process is widely conserved between kingdoms and is referred to either as queling in fungi (Cogoni and Macino, 2000), RNA interference in nematodes, or cosuppression in plants (Napoli et al., 1990). In plants, PTGS can be triggered by transgenes (Napoli et al., 1990) and viruses (Covey et al., 1997; Ratcliff et al., 1997). PTGS functions as an endogenous defense mechanism against virus invasion by directly targeting viral genome integrity and consequently lowering the titer of the invading virus (Voinnet, 2001). This mechanism can be redirected to target endogenous host mRNAs by introducing corresponding plant cDNA fragments within the viral genome, providing a means to down-regulate host gene expression (Kumagai et al., 1995). This approach, termed virus-induced gene silencing (VIGS), allows rapid transient knockdown of host gene expression and is more flexible than PTGS-based approaches that necessitate stable transformation of antisense or inverted-repeat sequences (Wesley et al., 2001).
Due to their relative flexibility, plant viruses have emerged as powerful tools for functional studies and are increasingly exploited for this purpose. Several RNA or DNA viruses with different specificities and host range have been developed as potential VIGS vectors, primarily in the permissive host Nicotiana benthamiana (for review, see Robertson, 2004). These include RNA viruses, such as tobacco mosaic virus (Kumagai et al., 1995; Lacomme et al., 2003), potato virus X (PVX; English et al., 1996; Angell and Baulcombe, 1997; Lu et al., 2003), tobacco rattle virus (TRV; Ratcliff et al., 2001; Liu et al., 2002b), and DNA viruses such as tomato golden mosaic virus (Peele et al., 2001). Recent developments have extended the use of such vectors, originally developed in N. benthamiana, to model plants such as Arabidopsis (Arabidopsis thaliana; Turnage et al., 2002) and crops such as tomato (Lycopersicon esculentum; Liu et al., 2002b), potato (Solanum tuberosum; Faivre-Rampant et al., 2004), and barley (Holzberg et al., 2002; Lacomme et al., 2003). This range of virus-host systems has demonstrated the potential for the functional characterization of endogenous genes in a variety of organs and tissues. Indeed, there are numerous examples of endogenous genes that have been successfully silenced in foliar tissues (e.g. phytoene desaturase [pds; Ratcliff et al., 2001] or cellulose synthase [Burton et al., 2000]), flower primordia (FLO/LFY; Ratcliff et al., 2001), meristematic tissue (proliferating cell nuclear antigen; Peele et al., 2001), and potato tubers (pds; Faivre-Rampant et al., 2004). However, root functions have remained an elusive target for many virus-based vectors due to the inefficient invasion of root systems by many viruses.
Virus-host range, infectivity, and in planta movement are key factors in conditioning the VIGS response. The role of double-stranded (ds)RNA molecules appears to be general to other forms of PTGS, acting as initiators of a silencing signal that generates a systemic VIGS response or by acting as a systemic signal itself. dsRNAs are precursors of small 21- to 25-nucleotide dsRNA sequences that are incorporated into a silencing complex to mediate the sequence-specific degradation of RNA (Voinnet, 2001). Two distinct pathways (transitivity-dependent or -independent) distinguish between mechanisms involved in transgene or endogenous gene silencing. The latter is subject to short-distance transitivity effects (Himber et al., 2003). This could explain the fact that VIGS of an endogenous gene necessitates concomitant and close proximity of replicating recombinant virus and a threshold of dsRNA molecules eliciting VIGS (Lacomme et al., 2003).
It is believed that VIGS efficiency relies mainly on the capacity of the virus to invade the host and replicate to a sufficient level in targeted tissues (Faivre-Rampant et al., 2004). This suggests that viral tropism would reflect the VIGS response and that it should be stronger in the tissue where viral replication is more favored.
Viral invasion and silencing within the root system has seldom been studied (Palauqui et al., 1997; Dalmay et al., 2000; Sonoda and Nishiguchi, 2000; Valentine et al., 2002; Saedler and Baldwin, 2004). Previous investigations of tobacco mosaic virus movement and repression within N. benthamiana, together with the silencing of the 35S∷gfp transgene in lateral roots, have suggested a role for the root meristems in silencing in roots (Valentine et al., 2002), while silencing of a gfp transgene in Arabidopsis roots has been demonstrated using a PVX:GFP (green fluorescent protein) amplicon (Dalmay et al., 2000).
In this study, we investigated the utility of TRV as a vector for silencing in the roots of a range of species, with emphasis on those genes involved in cell division, differentiation, and host resistance during invasion by nematodes. TRV was chosen as it is known to invade roots successfully (MacFarlane and Popovich, 2000), enters meristematic tissues (Matthews, 1991), and has been used as a vector for the expression of foreign proteins within plant roots (MacFarlane and Popovich, 2000). Tobraviruses such as TRV can be transmitted between plants via vector nematodes and require a viral helper protein encoded by the RNA2-2b gene (Vassilakos et al., 2001; Vellios et al., 2002a). All previous VIGS vectors of TRV were derived from a construct based on the TRV RNA2 in which the 2b gene was deleted (Ratcliff et al., 2001; Liu et al., 2002b). Here we show that inclusion of the 2b gene greatly improves the capacity of TRV to invade roots and meristematic tissues of N. benthamiana and Arabidopsis, suggesting a novel role for the 2b helper protein, namely in conferring root tropism to viral infection. The resultant TRV-2b vector triggers VIGS efficiently in roots, as exemplified by the silencing of genes controlling root development in N. benthamiana and Arabidopsis, and the nematode gene Mi that confers resistance to root-knot nematodes in cultivated tomato. Our results show that the TRV-2b vector offers great utility in knockout studies of regulatory genes that influence root development and resistance.
RESULTS AND DISCUSSION
TRV-2b-Derived Vectors Efficiently Invade Meristems of N. benthamiana and Arabidopsis
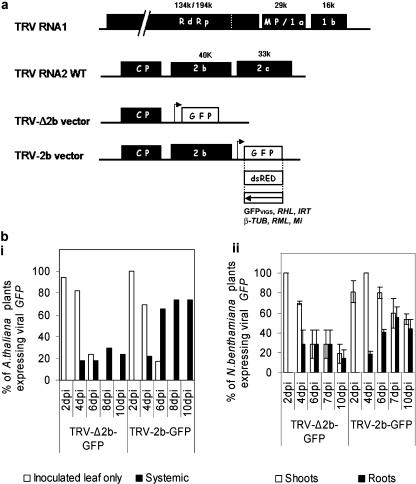
The relative ability of TRV-2b-GFP and TRV-Δ2b-GFP vectors (Fig. 1a; “Materials and Methods”) to invade and spread systemically in shoots and roots of N. benthamiana and Arabidopsis was investigated. Viral movement was monitored by observation of GFP fluorescence (Fig. 1b). TRV-2b-GFP was able to establish a stronger, more invasive infection in both species than the TRV-Δ2b vector (Fig. 1b). A higher percentage of infected plants became systemically infected (Fig. 1b, i and ii) when inoculated with TRV-2b-GFP (60% N. benthamiana, 74% Arabidopsis) compared with TRV-Δ2b-GFP (23% N. benthamiana, 30% Arabidopsis). GFP was also clearly visible in N. benthamiana roots as previously described (MacFarlane and Popovich, 2000).
Figure 1.
Invasion of roots by TRV. a, Genomic organization of wild-type TRV RNA1 and TRV RNA2, and TRV-derived vectors used for expression of foreign protein and VIGS with modified RNA2 deleted from 2b and 2c protein or only 2c protein for, respectively, TRV-Δ2b and TRV-2b vectors. The TRV ORFs are represented in black boxes (not to scale) and correspond to the RNA-dependent RNA polymerase (RdRp), 1a or movement protein (MP), 1b or 16-K protein, coat protein (CP), 2b and 2c proteins. The size of the encoded proteins is indicated. Arrow represents the position of the duplicated subgenomic CP promoter driving the expression of inserted coding cDNA (GFP or DsRED represented in white boxes) or noncoding cDNA for silencing represented in white box and the name of corresponding target genes. b, Movement of TRV-Δ2b-GFP and TRV-2b-GFP vectors over 10 d. Subsection i, Virus movement in Arabidopsis shoots. (subsection i, TRV-Δ2b-GFP, n = 17; TRV-2b-GFP, n = 23). Virus movement in whole N. benthamiana plants (subsection ii, TRV-Δ2b-GFP, n = 26 ± se; TRV-2b-GFP, n = 45 ± se). GFP fluorescence was viewed using a stereofluorescence microscope (MZFLIII; Leica, Deerfield, IL). Filter set GFP3, excitation 450/90 nm; emission 500/50 nm).
Nevertheless, a higher percentage of infected plants expressed GFP in roots at 7 d postinoculation (dpi; Fig. 1b, ii) in plants inoculated with the TRV-2b vector (55%) compared with the TRV-Δ2b-vector (29%). The pattern of TRV infection in roots was explored further using TRV-2b-GFP, TRV-Δ2b-GFP, and TRV-2b-DsRED constructs (the latter expresses a red fluorescent protein encoded by DsRED cDNA). Virus-encoded GFP expression was first seen within 3 to 4 dpi in N. benthamiana roots as small streaks just behind meristems (data not shown). Within a further 24 h, the whole of the root system was infected when using TRV-2b-GFP, with meristems exhibiting the highest level of fluorescence (Fig. 2a, i). TRV-Δ2b-GFP exhibited a similar root tropism but recovery occurred quicker than in TRV-2b-GFP-infected plants (Fig. 2a, ii). DsRED expression within Arabidopsis roots was first seen in root meristematic regions by approximately 7 dpi (Fig. 2a, ii). Vibraslice sections of the N. benthamiana root meristem (Fig. 2b, i and ii) and shoot meristem (Fig. 2b, iii) and optical sections of Arabidopsis root meristems (Fig. 2b, iv) confirmed invasion of the meristems by the TRV-2b constructs. In roots of N. benthamiana, all cell types within the meristems appeared to be infected except the root cap, which was only occasionally infected (Fig. 2b, i and ii). Occasional infection of the root cap may occur due to invasion of the quiescent center, followed by cell turnover of quiescent cells into root cap initials. In Arabidopsis, the outer root cap appeared to be more accessible to the virus (Fig. 2b, iv and v). These results indicate that TRV-2b vectors spread and sustain a more prolonged systemic expression of GFP both in roots and shoots of Arabidopsis and N. benthamiana than the corresponding TRV-Δ2b vectors.
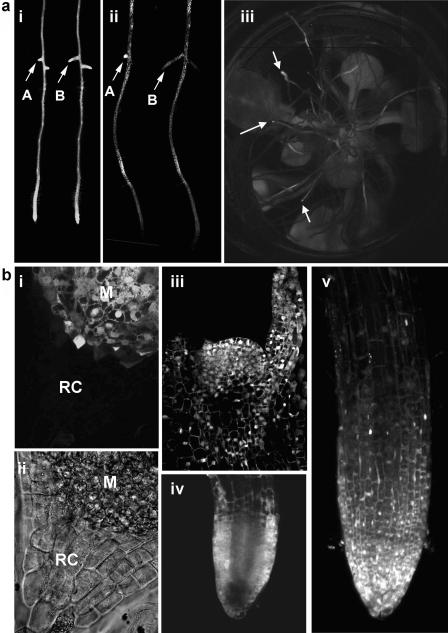
Figure 2.
Distribution of TRV-2b construct in roots. a, Invasion of the root systems of N. benthamiana by TRV-2b-GFP (subsection i, same root observed at 4 and 6 dpi; arrows indicate developing lateral roots A and B at these two time points, respectively) or TRV-Δ2b-GFP (ii, same root observed at 4 and 6 dpi; arrows indicate developing lateral roots A and B as before). Note the much higher fluorescence at 6 dpi in lateral root B infected with TRV-2b-GFP (subsection i, arrow B) compared with TRV-Δ2b-GFP (subsection ii, arrow B). Arabidopsis roots infected by TRV-2b-DsRED (subsection iii, roots at 7 dpi viewed through the bottom of a multiwell petri dish). In Arabidopsis, DsRED fluorescence was most visible in the meristematic regions of the root (subsection iii, arrows). b, Shoot and root meristem invasion by TRV-2b constructs. Vibraslice section of N. benthamiana root meristem infected with TRV-2b-GFP showing the high level of infection of the root meristem (M) with much reduced invasion of the root cap (RC; subsection i). Subsection ii, Transmission image of subsection i. Vibraslice section of the shoot apical meristem of N. benthamiana showing a high level of TRV-2b-GFP invasion (subsection iii). TRV-2b-DsRED infection of Arabidopsis root meristem, optical section (subsection iv), and stacked image (subsection v) showing extensive infection of meristem (subsection iv) with limited infection of the internal root cap.
TRV-2b Triggers a Pervasive VIGS Response in N. benthamiana and Arabidopsis
We evaluated the efficacy of the TRV-2b vectors for silencing in roots by inoculating with the TRV-2b-GFPVIGS construct N. benthamiana expressing a GFP transgene from the 35S promoter and an Arabidopsis enhancer trap endoplasmic reticulum-targeted GFP line N9135 (Nottingham Arabidopsis Stock Center [NASC]; Haseloff, 1999), which displayed GFP fluorescence throughout the plant (Fig. 3a). Silencing of the GFP transgene was observed in the leaves of both species by approximately 5 to 10 dpi (data not shown). Silencing extended into the majority of the shoot apical meristem of Arabidopsis (Fig. 3a). Silencing of GFP in roots was also observed after 4 to 10 dpi in N. benthamiana (data not shown) and after 10 dpi in Arabidopsis (Fig. 3b, iv–vi). A comparison of the timing of the onset of silencing in roots with TRV-2b invasion was explored in transgenic 35S∷GFP N. benthamiana using a TRV-2b-DsRED construct (Fig. 3c, i). Within 24 h of root meristem invasion with TRV-2b-DsRED, the root meristem had grown away from the initial viral invasion point leaving a visible constriction in the root diameter that expressed a high level of DsRED fluorescence (Fig. 3c, ii; arrow A). All new root tissue expressed a much reduced level of DsRED (Fig. 3c, ii; arrow B). After 12 dpi, some DsRED expression was still visible in an area around the quiescent center within the root meristem (Fig. 3c, ii). Silencing of the 35S∷GFP using TRV-2b-GFPVIGS was also first visible soon after viral invasion in new root growth (data not shown).
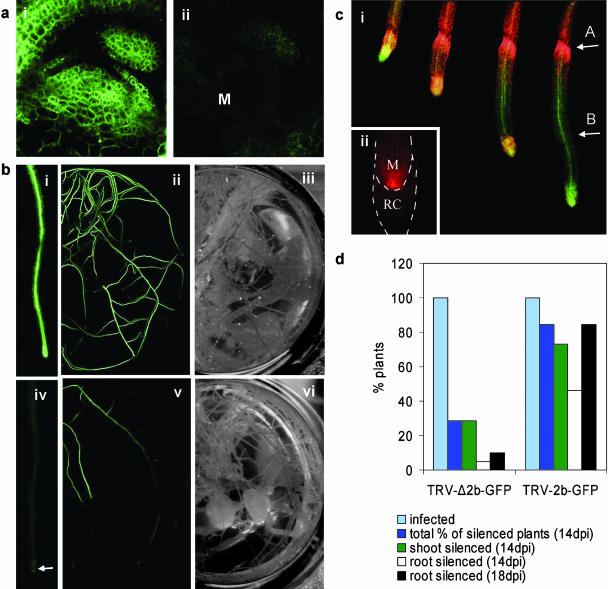
Figure 3.
Virus-induced gene silencing of GFP transgene and suppression of viral replication in Arabidopsis and N. benthamiana. a, VIGS in the shoot meristem of Arabidopsis N9135 line. Control GFP expression (subsection i), VIGS after TRV-2b-GFPVIGS vector infection (subsection ii; M, shoot meristem region). b, VIGS of GFP transgene in roots of Arabidopsis. Nonsilenced control plants GFP expression (subsections i–iii). Roots exhibiting silencing of the GFP (subsections iv–vi). Arrow indicates positions of root meristem (B, subsection iv). c, Virus replication suppression in N. benthamiana roots. Plants inoculated with TRV-2b-DsRED and in the background 35S∷GFP expression, 5–8 dpi (subsection i). Note the reduced level of viral replication in the new root tissue produced after the virus has invaded the meristem. Root meristem at 13 dpi with detectable viral DsRED fluorescence indicating active viral replication and recovery (subsection ii; M, meristem; RC, root cap). Arrow A indicates the zone of constriction due to TRV invasion; arrow B indicates new root growth exhibiting suppression of viral replication. d, Relative number of plants exhibiting silencing of GFP transgene observed on whole Arabidopsis plants infected with TRV-Δ2b-GFPVIGS (n = 17) or TRV-2b-GFPVIGS (n = 23) vectors. Images in subsection iii and subsection vi are similar to subsection ii and subsection v, respectively, from underneath petri dish to visualize all roots.
The TRV-2b virus construct effectively invades root meristems and appears to be able to maintain pockets of infection within the root meristem. This suggests that TRV-2b evades a host resistance mechanism and may therefore have the future opportunity for reinvasion, possibly causing cycles of silencing and viral replication.
Efficient induction of VIGS in plants is believed to require a threshold level of expression of the virus at, or near, the targeted cells (Faivre-Rampant et al., 2004). The improved level of systemic root invasion obtained with the TRV-2b-GFP may lead to an increase in this construct's effectiveness as a silencing vector over the TRV-Δ2b vector. This was indeed found, with TRV-2b-GFP inducing greater silencing in both shoots (85% TRV-2b versus 10% TRVΔ2b) and roots of Arabidopsis compared to TRV-Δ2b-GFP (Fig. 3d). At 18 dpi, 80% of Arabidopsis infected with TRV-2b-GFP exhibited root silencing compared with 4% of TRV-Δ2b-infected plants (Fig. 3d). This efficient invasion appears to trigger a strong silencing reaction that can spread extensively throughout the roots.
Silencing of Endogenous Genes Associated with Root Development
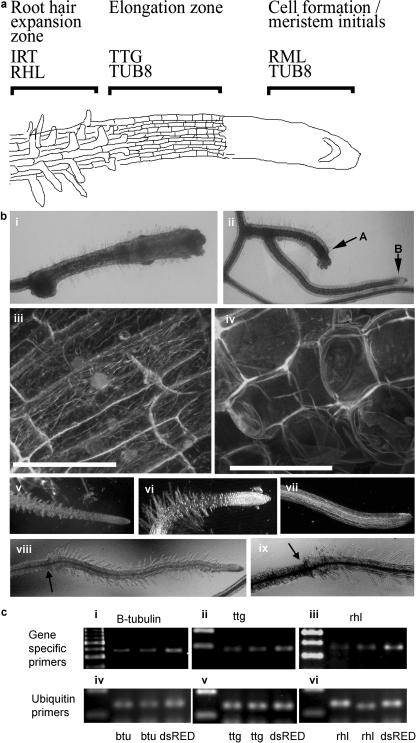
To investigate the efficacy of the TRV-2b VIGS vector in silencing endogenous plant genes involved in root development, a TRV-2b VIGS vector harboring fragments of the Arabidopsis genes, IRT1 (iron-regulated metal transporter; Korshunova et al., 1999), TTG1 (transparent testa glabra; Galway et al., 1994), RHL1 (root hairless1; Schneider et al., 1998), RML1 (root meristemless1; Cheng et al., 1995), and N. benthamiana β-tubulin were engineered. Expression levels in wild-type plants are either high (β-tubulin, RML1) or moderate (IRT1, TTG1, RHL1) for all the target genes. Further, despite some tissue-specific variations, expression is observed across the whole root (Birnbaum et al., 2003). The N. benthamiana β-tubulin cDNA fragment that was subcloned into TRV displays a high degree of identity to the Arabidopsis TUB8 gene, therefore allowing efficient silencing in both species. Figure 4a shows the main areas of the root meristem where silencing phenotypes were anticipated for each gene.
Figure 4.
Silencing of endogenous genes associated with root development. a, Diagrammatic view of the Arabidopsis root meristem, indicating meristematic zones where mutant or silencing phenotypes are observed. b, VIGS of endogenous genes. VIGS of β-tubulin in N. benthamiana (subsections i and ii) and transgenic 35S∷α-tubulin-GFP plants (subsection iv). Transgenic 35S∷α-tubulin-GFP β-tubulin-silenced plants exhibit changes in cell shape and in the microtubule structure with no effect on the overall GFP brightness (subsection iv) in comparison to the unsilenced control (subsection iii). Recovery from VIGS of β-tubulin in a root meristem 28 dpi (subsection ii; arrow A, silenced root; arrow B, recovered lateral root). Scale bar represents 100 μm. VIGS phenotypes visible on Arabidopsis plants inoculated with TRV-2b-TTG (subsection vi), TRV-2b-RHL (subsection vii), TRV-2b-IRT (subsection viii and ix; arrows indicate zone of constriction as in Fig. 2c, subsection i), TRV-2b-DsRED is used as a control (subsection v). c, Semiquantitative RT-PCR on Arabidopsis-silenced and control roots using β-tubulin (TUB8; subsection i), TTG1 (subsection ii), and RHL1 (subsection iii) specific primers, showing reduced expression in plants inoculated with VIGS vectors harboring the β-tubulin (subsection i), TTG1 (subsection ii), or RHL1 (subsection iii) sequences compared with control plants infected with TRV-2b-DsRED virus (right lane on each image). RNA was extracted from the whole roots. Semiquantitative RT-PCR using ubiquitin primers as internal control for samples used in gene-specific RT-PCRs (subsection iv, β-tubulin; subsection v, TTG1; subsection vi, RHL1).
As a consequence of silencing the β-tubulin gene in N. benthamiana with the TRV-2b-βTu construct, the root meristems became disorganized and stunting of root growth was observed (Fig. 4b, i and ii; arrow A). This was further explored by inoculating N. benthamiana plants expressing an α-tubulin∷GFP fusion protein (Gillespie et al., 2002) with the TRV-2b-βTu construct (Fig. 4b, iv). Confocal images showed there was no overall decrease in the level of α-tubulin∷gfp fusion in stunted roots (Fig. 4b, iii) when compared with controls (Fig. 4b, iv) as would be expected. α-Tubulin∷GFP fusion protein was seen clearly to be incorporated in microtubules in control plants (Fig. 4b, iii).
However, plants in which the β-tubulin gene was silenced displayed severe defects in cell shape and microtubules were absent (Fig. 4b, iv). Observations of the silencing phenotypes of β-tubulin on plants were continued until 28 dpi. Roots at this stage appeared to circumvent the severe silencing phenotype by producing new lateral meristems further back along the root (Fig. 4b, ii; arrow B). Occasionally, the silenced meristems recovered from the silencing phenotype (data not shown).
VIGS was also explored in Arabidopsis, with phenotypes observed in roots after 7 to 14 dpi (Fig. 4b, vi–ix). Roots of silenced plants phenocopy previously described mutant alleles, with TRV-2b-TTG-infected plants producing ectopic root hairs (Fig. 4b, vi), and those inoculated with TRV-2b-RHL producing a reduced number of root hairs (Fig. 4b, vii). Plants inoculated with TRV-2b-IRT also showed a change in root hair phenotype, with root hairs becoming more extended than the control (Fig. 4b, viii and ix). This phenotype has been associated with iron deficiency (Schmidt and Schikora, 2001). Changes in root phenotype were often marked by a root constriction, usually associated with viral invasion into the root meristem (Fig. 4b, viii and ix; arrow), after which the new growth exhibited the silencing phenotype. This is particularly apparent in Figure 4b, viii.
The effect of silencing was analyzed at the RNA level. RNA was isolated from the whole root system of Arabidopsis plants inoculated with TRV-2b-DsRED, TRV-2b-βTu, TRV-2b-TTG, and TRV-2b-RHL. Semiquantitative PCR was performed, with the results shown in Figure 3c. Lower RNA levels for β-tubulin, TTG1, and RHL1 (Fig. 4c, i–iii, respectively) were found in the silenced plants that exhibited altered root phenotypes (lanes 1 and 2, left to right) compared with the RNA levels found in the control TRV-2b-DsRED-inoculated plants (lane 3, right). The overall level of RNA included was ascertained using reverse transcription (RT)-PCR using primers against the ubiquitin gene (Fig. 4c, iv–vi).
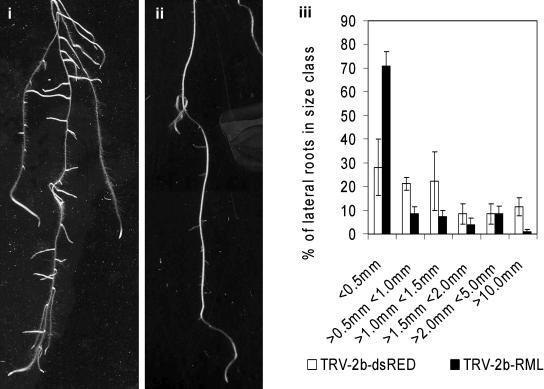
The development of lateral roots may be divided into a series of stages, including dedifferentiation of the pericycle, ordered cell division and cell differentiation to generate lateral root primordia, emergence (via cell expansion and activation of the lateral root meristem), and continued growth of the organized lateral root (Malamy and Benfey, 1997). Plants with mutations in the RML1 gene initially produce normal root development in the embryo (Cheng et al., 1995). However, these roots undergo limited cell division after germination producing roots <1 mm in length. Lateral roots can be produced, but these do not expand to lengths >1 mm. To evaluate silencing of RML1 in Arabidopsis, plants were inoculated with TRV-2b-RML or TRV-2b-DsRED. Plants exhibited a short lateral root phenotype (Fig. 5, ii) compared with TRV-2b-DsRED-inoculated plants (Fig. 5, i). Measurement of the lateral root lengths showed a significant shift of root-to-length ratios in these plants toward extremely short roots, with more than 70% being <0.5 mm in length (Fig. 5, iii) compared with <30% of laterals of plants inoculated with TRV-2b-DsRED.
Figure 5.
Silencing of the RML1 gene in Arabidopsis root phenotype of plants exhibiting RML1 silencing (TRV-2b-DsRED [subsection i], TRV-2b-RML [subsection ii]). Lengths of the lateral roots were measured for TRV-2b-DsRED (n = 3) and TRV-2b-RML (n = 8) inoculated plants (subsection iii).
These data show that a wide variety of endogenous genes expressed in roots can be targeted and silenced. Furthermore, we have demonstrated that VIGS can facilitate the study of genes such as RML1 that cause extremely severe phenotypes early in plant development.
Silencing of the Mi Gene in Tomato
The potential of the TRV-2b vector to target root-associated functions was further tested with the Mi-1 (Mi) gene that confers resistance to the root-knot nematodes Meloidogyne javanica, Meloidogyne incognita, and Meloidogyne arenaria, as well as the aphid Macrosiphum euphorbiae in tomato (Milligan et al., 1998; Vos et al., 1998). No development of feeding sites occurs in resistant plants (such as tomato cv Rossol), and nematodes either leave the roots or fail to develop (Milligan et al., 1998). The Mi gene is expressed endogenously in uninfected roots and expression levels are maintained after inoculation with root-knot nematodes (Martinez de Ilarduya and Kaloshian, 2001). A cDNA fragment of the Mi gene was subcloned in antisense orientation into the TRV-2b vector to generate construct TRV-2b-Mi (Fig. 1a). VIGS efficiency was monitored by quantifying Mi mRNA accumulation levels using RT-PCR and real-time RT-PCR in both leaves and roots of tomato cv Rossol.
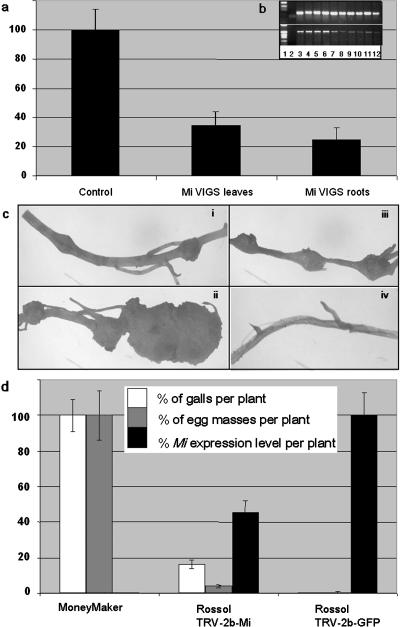
Samples were taken at 3 to 4 weeks postchallenge with either TRV-2b-GFP or TRV-2b-Mi VIGS constructs, the former used as a control of TRV infection. RT-PCR experiments detected a lower amount of Mi PCR product in both leaves and roots from Rossol tomato plants challenged by TRV-2b-Mi than in TRV-2b-GFP control plants (Fig. 6b). The levels of PDS mRNA, used as an internal RT-PCR control, were comparable in tested samples. To gain more insight about the relative expression levels in both tissues, real-time RT-PCR was performed. A decrease in normalized Mi mRNA of 34% ± 9% in leaves and of 25% ± 8% in root tissues was observed relative to controls (Fig. 6a). This result correlates with the previous observation that the TRV-2b vector is not only efficient in invading root tissues, but also in triggering a strong VIGS response in root tissue. The observed silencing of the Mi gene led us to assess this effect on nematode reproduction. TRV-2b-Mi and control TRV-2b-GFP Rossol tomato plants were challenged with M. javanica nematodes 3 weeks after TRV inoculation, as this timing corresponds to the observed silencing on plants challenged with the TRV-2b-Mi VIGS construct. Roots were scored 7 to 8 weeks after nematode inoculation, representing a total of 10 to 11 weeks postchallenge with the TRV-2b constructs. From all the plants tested, the average number of galls ranged from 17 to 69, and egg masses from 4 to 16 per silenced Rossol plant (Table I). This represents an average percentage of reproduction reaching 15% to 25% with respect to the susceptible tomato cv Money Maker (Fig. 6e). Galls and egg masses on the Rossol-silenced roots (Fig. 6c, iii) were typically smaller than on the Money Maker controls (Fig. 6c, i and ii), suggesting that nematode development was restricted and/or delayed relative to that on Money Maker. Neither galls nor egg masses were observed in control Rossol plants infected by TRV-2b-GFP (Fig. 6c, iv).
Figure 6.
VIGS of the Mi gene in tomato. a, Molecular characterization of Mi silencing in tomato leaves and roots. Real-time RT-PCR determination of normalized relative amounts of Mi mRNA levels in silenced and control roots or leaves of tomato plants challenged with TRV-2b-Mi or TRV-2b-GFP constructs (21 dpi). Values are expressed in percentage of normalized Mi mRNA-related TRV-2b-GFP control in a given tissue. Values represent the means of at least three to six different plants per construct ± se. b, Semiquantitative RT-PCR of leaves and roots of Mi-silenced and control tomato plants challenged with, respectively, TRV-2b-Mi or TRV-2b-GFP constructs (21 dpi; same set of samples used for real-time RT-PCR). Both RT-PCR products corresponding to Mi and PDS mRNA have been assessed. Presented here are PCR conditions corresponding to the log-linear phase of amplified PCR product in nonsilenced tissues. Lane 1 is 1-kb molecular mass marker; lane 2 is nontemplate control; lanes 3 to 4 and 5 to 6 are leaf root tissues from two independent control plants challenged with TRV-2b-GFP; lanes 7 to 8, 9 to 10, and 11 to 12 are leaf root tissues from three independent silenced plants challenged with TRV-2b-Mi. Top, PDS PCR product; bottom, Mi PCR product. c, Mi resistance-breaking phenotype in Rossol silenced roots. Upon nematode infection, roots of susceptible Money Maker plants display both small (subsection i) and large galls with abundant egg masses (subsection ii). Small galls were observed on Mi-silenced Rossol plants (subsection iii). No galls were observed in all control unsilenced Rossol plants infected with TRV-2b-GFP (subsection iv). d, Molecular characterization of Mi silencing in tomato roots and effect on nematode infection. Real-time RT-PCR determination of normalized relative amounts of Mi mRNA levels in silenced (n = 6) and control (n = 3) roots of tomato plants challenged with TRV-2b-Mi or TRV-2b-GFP constructs (49 to 56 dpi). Values are expressed in percentage of normalized Mi mRNA related to TRV-2b-GFP control plants (black). The averaged percentage of galls (white) and egg masses (gray) per plant from two independent experiments (Table I) is presented. Values represent the means of different plants per construct ± se (n ranging from 7–18). RNA was extracted from root segments of 2- to 3-cm lengths spanning the galls on TRV-2b-Mi roots.
Table I.
Effect of Mi silencing on nematode invasion on tomato
| Experiment 1
|
Experiment 2
|
|||||
|---|---|---|---|---|---|---|
| Galls | Egg Masses | n | Galls | Egg Masses | n | |
| Money Maker | 150 ± 9 | 134 ± 14 | 2 | 301 ± 40 | 284 ± 38 | 5 |
| Rossol TRV-2b-Mi | 17 ± 4 | 4 ± 1 | 10 | 69 ± 10 | 16 ± 4 | 8 |
| Rossol TRV-2b-GFP | 0 | 0 | 9 | 1 ± 1 | 2 ± 1 | 5 |
| Rossol Mock | 0 | 0 | 2 | 0 | 0 | 5 |
Mean number of galls and egg masses per plant observed in two independent nematode infection experiments on control or silenced tomato plants ±se (n = number of plants analyzed).
Colonized root tissue from at least six different TRV-2b-Mi-infected Rossol plants, and corresponding tissue from TRV-2b-GFP control plants, was harvested and submitted to semiquantitative and quantitative real-time RT-PCR. Despite the fact that these tissues were sampled at a time point corresponding to nearly 3 months postchallenge with TRV-2b constructs, significantly lower amounts of Mi PCR product was observed in VIGS plants compared to the control (Fig. 6d). The PDS mRNA level remained comparable in these samples (data not shown). Real-time RT-PCR was performed, and a decrease in normalized Mi mRNA to 45% ± 6.5% was observed in root tissues of Mi-silenced plants (Fig. 6e). Therefore, Mi silencing correlates with the Mi resistance-breaking phenotype and successful nematode development. These results indicate that, despite the relatively long experimental period necessary for optimal establishment of VIGS and nematode colonization, such an approach can be used for functional characterization of genes involved in nematode resistance.
TRV displays an increased tropism to the root vasculature by moving rapidly through the root system of infected plants, where it is ingested by specific vector nematodes such as Paratrichodorus pachydermus (Vassilakos et al., 2001). The 2b protein from TRV strain PpK20 (used in this study) is a 40-kD protein harboring a predicted conserved coiled-coil central domain, probably involved in virion-vector interaction. TRV-2b is known to act in trans to facilitate and confer specificity in TRV nematode transmission, and therefore is a true helper protein. Its role and function in vector transmission is not fully elucidated (Vassilakos et al., 2001). As proposed for the potyviral helper component proteinase (HC-Pro; Blanc et al., 1997, 1998), it is suggested that TRV-2b probably acts as a bridge linking the virion and vector mouthpart (Visser and Bol, 1999; Vellios et al., 2002b).
The results presented here suggest that the 2b ORF from TRV may have an additional role in host regulatory mechanisms that control TRV invasion. The presence of 2b does not modify TRV symptomatology but is required for extensive shoot and root meristem invasion. An attractive hypothesis is that 2b controls a host-silencing mechanism. However, a distinct ORF on TRV RNA1 encoding a 16-kD Cys-rich protein shows functional similarity to other well-characterized viral-borne silencing suppressors of PTGS, on the basis that its overexpression increases TRV symptomatology (Liu et al., 2002a). This argues for a novel role for 2b that differs from most other silencing suppressors identified to date (Roth et al., 2004) and indicates that this gene may confer tropism to the viral invasion process. The resultant TRV-2b vector displays an increased affinity for meristems that provides a key characteristic for VIGS-based functional characterization of genes in root tissues.
MATERIALS AND METHODS
Plant Material
All work involving virus-infected material was carried out in containment glasshouses under Scottish Executive Environment and Rural Affairs Department license GM/203/2004 and GM/210/2004. Arabidopsis (Arabidopsis thaliana) C24, Arabidopsis GAL4-VP16.GFP (NASC; Haseloff, 1999), Nicotiana benthamiana wild type, and α-tubulin∷gfp transgenics (Ueda et al., 1999; Gillespie et al., 2002) were surface sterilized and sown onto Murashige and Skoog media, 1% Suc, 0.8% agar (DIFCO Laboratories, Detroit), in 24- or 12-multiwell petri dishes, respectively. For virus movement experiments, N. benthamiana plants were grown in 10-cm-square petri dishes as previously described (Valentine et al., 2002). Arabidopsis seeds were surface sterilized by washing in 70% ethanol, 95% ethanol, and then allowing them to air dry. Nicotiana seeds were surface sterilized as previously described (Valentine et al., 2002). Petri dishes were sealed with Nescofilm (Bando Chemical, Kobe, Japan) and were vented slightly with micropore tape. Arabidopsis seeds were vernalized at 4°C for 3 to 5 d prior to transferring them to equal day (12-h light/12-h day) and 22°C growth conditions. N. benthamiana plants for use in infectious sap production and tomato (Lycopersicon esculentum) plant cultivars Rossol (Mi locus introgressed from Lycopersicon peruvianum conferring resistance to Meloidogyne javanica ) and MoneyMaker (susceptible to M. javanica), were grown in compost in controlled environment chambers with a 16-h photoperiod (22°C, light intensity ranging from 400–1,000 μE m−2 s−1).
Virus Constructs
The TRV-Δ2b-GFP vector was previously described (MacFarlane and Popovich, 2000). GFP expression is driven from a duplicated subgenomic coat protein (CP) promoter of a different tobravirus. The TRV-2b-GFP construct derived from TRV-Δ2b-GFP, where the 2b gene encompassing its native 2b promoter was cloned between the CP and the duplicated CP subgenomic promoter driving GFP expression (Vellios et al., 2002a). The modified TRV RNA2-2b vector (strain PpK20) corresponds to the previously described pK202b-GFP construct (Vellios et al., 2002a) and was used to engineer all the TRV-2b constructs. The GFP insert (Crameri et al., 1996) was excised by NcoI-EagI digestion and replaced by a DsRED ORF (CLONTECH, Palo Alto, CA) to generate the TRV-2b-DsRED construct. VIGS constructs were engineered by cloning GFP, RHL1, RML1, β-tubulin, TTG1, and Mi PCR fragments using gene-specific primers harboring NcoI and EagI restriction sites at their extremity. Details of the accession numbers and primer sequences used to clone the cDNA fragments are summarized in Supplemental Table I (available at www.plantphysiol.org). cDNA fragments were then inserted into TRV-2b VIGS vectors to generate, respectively, constructs TRV-2b-GFPVIGS, TRV-2b-RHL, TRV-2b-RML, TRV-2b-βTu, TRV-2b-TTG, and TRV-2b-Mi (Fig. 1a).
Virus Inoculation Procedures
N. benthamiana and tomato plants were inoculated with infectious TRV sap as previously described (Valentine et al., 2002). In vitro transcripts of RNA2 were obtained using a T7 transcription kit (mMessage mMachine T7 Transcription kit; Ambion, Austin, TX). These were mixed with total RNA isolated from plants infected with RNA1, plus bentonite solution, prior to inoculation onto source leaves of 6- to 8-week-old greenhouse-grown N. benthamiana. Infectious sap was produced from plants by grinding infected leaves in 10 mm sodium phosphate buffer, pH 7.2 (w/v), and stored at −20°C. For Arabidopsis inoculation, TRV infectious sap was centrifuged at 8,000g. The supernatant was collected and mixed with two volumes (v/v) of ice-cold absolute ethanol. Samples were incubated at −20°C for 1 h prior to centrifugation at 10,000g for 10 min. The supernatant was discarded and the pellet was washed briefly with 200 μL absolute ethanol. The excess ethanol was discarded and the pellet was allowed to dry slightly before mixing with a sterile solution of 0.05 m Gly, 0.03 m K2HPO4, 0.02% bentonite, and 1% celite at a ratio of 2:5 (v/v) to the original volume of supernatant. Arabidopsis plants aged 17 to 21 d were inoculated with virus suspension by rubbing onto 2 source leaves per plant using sterile cotton wool swabs. After inoculation, petri dishes were sealed with micropore tape and returned to the growth chamber at 22°C, with a 12-h photoperiod (light intensity 110 μE m−2 s−1).
Nematode Inoculation and Scoring
Tomato plant cultivars Money Maker and Rossol were grown in compost in root trainers (Ronaash, Kelso, Roxburghshire, UK). Juveniles of M. javanica (L6/3; Tzortzakakis et al., 1999) were inoculated directly onto the roots of 4- to 5-week-old plants at a rate of 1,000 (experiment 1) or 1,500 second-stage juveniles (experiment 2) per plant. Forty-nine to 56 d after inoculation, the root systems were washed free of compost and stained in a solution of Phloxine B (Hartman and Sasser, 1985). The number of galls and visible egg masses were counted under a stereomicroscope.
Microscopy
For observations of shoot apical meristems, all leaves were removed at petiole bases before embedding the meristem in 3% agar (BD Biosciences, Oxford). Sections of 100 to 200 μm were cut using a vibraslice. Tissue slices were mounted in water for GFP observations. Longitudinal root meristem sections of N. benthamiana were cut using a razor blade. Transmission, GFP, and DsRED observations were recorded using either a Bio-Rad MRC 1000 confocal laser scanning microscope (Bio-Rad, Hemel, Hempstead, UK) using long-distance working lenses or a Coolview digital camera (Photonic Science, Robertsbridge, East Sussex, UK) attached to a Leica fluorescence stereomicroscope. Whole-root N. benthamiana roots were mapped in sections and collated using Photoshop software (Adobe, Mountain View, CA).
RNA Extraction and cDNA Synthesis
Arabidopsis endogenous plant gene fragments were obtained from Arabidopsis root cDNA. cDNA was obtained using an Ambion Poly(A) Pure/Micro Poly(A) Pure mRNA isolation kit (Ambion) as per the manufacturer's instructions, followed by first-strand cDNA synthesis by oligo(dT) priming using SuperScriptII RNaseH-Reverse Transcriptase (Invitrogen Life Technologies, Paisley, UK). Primers used for PCR and cloning are listed in Supplemental Table I.
RT-PCR and SYBR Real-Time RT-PCR Experiments
For semiquantitative and quantitative RT-PCR, total RNA was extracted from frozen tissue using the Qiagen RNeasy plant mini kit (Qiagen, Crawley, UK). Prior to cDNA synthesis, RNA samples were treated with RNase-free DNaseI (DNA-free kit; Ambion). One microgram (tomato) or 200 ng (Arabidopsis) of total RNA was used for first-strand cDNA synthesis by oligo(dT) priming using SuperScriptII RNase H-Reverse Transcriptase (Invitrogen Life Technologies). For RT-PCR analysis, primers that exclude the region of the cDNA cloned into the virus vectors to trigger silencing were used to ensure that only the endogenous mRNA is amplified (Supplemental Table I). PDS (tomato) or ubiquitin (Arabidopsis) cDNA (Supplemental Table I) was used as an internal constitutively expressed control. First-strand cDNA was used as a template for PCR amplification through 25, 30, 40, and 50 cycles. As 30 cycles for Mi and PDS and 35 cycles for all Arabidopsis PCR products was within the log-linear phase for PCR product amplification in the nonsilenced control samples (data not shown), these conditions were selected for comparison of relative accumulation of both internal control and target mRNAs in all samples. The primers used are described in Supplemental Table I. Similarly, for SYBR real-time RT-PCR (Taqman) experiments, primer pairs were designed outside the Mi cDNA fragment cloned into the TRV-2b vector and for the internal control pds cDNA using the Primer Express software supplied with the ABI PRISM 7700 Sequence Detection System (PE-Applied Biosystems, Foster City, CA) following the manufacturer's guidelines for primer design. The primers used for Taqman are described in Supplemental Table I. Primer concentrations giving the lowest threshold cycle (Ct) value were selected for further analysis. Detection of real-time RT-PCR products, calculations, and statistical analysis was performed as previously described (Lacomme et al., 2003).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permission will be the responsibility of the requester.
Supplementary Material
Acknowledgments
We thank Katarina Hrubikova for technical assistance.
This work was supported by the Large Scale Biology Corporation, by European Union funding from Nonema (QLK5–CT–1999–01501), and by the Scottish Executive Environment and Rural Affairs Department.
The online version of this article contains Web-only data.
References
- Angell S, Baulcombe DC (1997) Consistent gene silencing in plants expressing a replicating potato virus X RNA. EMBO J 16: 3675–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Blanc S, Ammar ED, Garcia-Lampasona S, Dolja VV, Llave C, Baker J, Pirone TP (1998) Mutations in the potyvirus helper component protein: effects on interaction with virions and aphid stylets. J Gen Virol 79: 3119–3122 [DOI] [PubMed] [Google Scholar]
- Blanc S, Lopez-Moya JJ, Wang RY, Garcia-Lampasona S, Thornbury DW, Pirone TP (1997) A specific interaction between coat protein and helper component correlates with aphid transmission of a potyvirus. Virology 231: 141–147 [DOI] [PubMed] [Google Scholar]
- Burton RA, Gibeaut DM, Bacic A, Findlay K, Roberts K, Hamilton A, Baulcombe DC, Fincher GB (2000) Virus-induced gene silencing of a plant cellulose synthase gene. Plant Cell 12: 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JC, Seeley KA, Sung ZR (1995) RML1 and RML2, Arabidopsis genes required for cell-proliferation at the root-tip. Plant Physiol 107: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C, Macino G (2000) Post-transcriptional gene silencing across kingdoms. Curr Opin Genet Dev 10: 638–643 [DOI] [PubMed] [Google Scholar]
- Covey SN, Al-Kaff NS, Langara A, Turner DS (1997) Plants combat infection by gene silencing. Nature 385: 781–782 [Google Scholar]
- Crameri A, Whitehorn EA, Tate E, Stemmer WP (1996) Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol 14: 315–319 [DOI] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Mueller E, Baulcombe DC (2000) Potato virus X amplicons in Arabidopsis mediate genetic and epigenetic gene silencing. Plant Cell 12: 369–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English JJ, Mueller E, Baulcombe DC (1996) Suppression of virus accumulation in transgenic plants exhibiting silencing of nuclear genes. Plant Cell 8: 179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faivre-Rampant O, Gilroy EM, Hrubikova K, Hein I, Millam S, Loake GJ, Birch P, Taylor M, Lacomme C (2004) Potato virus X-induced gene silencing in leaves and tubers of potato. Plant Physiol 134: 1308–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW (1994) The ttg gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev Biol 166: 740–754 [DOI] [PubMed] [Google Scholar]
- Gillespie T, Boevink P, Haupt S, Roberts AG, Toth R, Valentine T, Chapman S, Oparka KJ (2002) Functional analysis of a DNA-shuffled movement protein reveals that microtubules are dispensable for the cell-to-cell movement of tobacco mosaic virus. Plant Cell 14: 1207–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman KM, Sasser JN (1985) Identification of Meloidogyne species on the basis of differential host test and perineal pattern morphology. In KR Barker, CC Carter, JN Sasser, eds, An Advanced Treatise on Meloidogyne, Vol 2, Methodology. North Carolina State University Graphics, Raleigh, NC, pp 67–77
- Haseloff J (1999) GFP variants for multispectral imaging of living cells. Methods Cell Biol 58: 139–151 [DOI] [PubMed] [Google Scholar]
- Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O (2003) Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J 22: 4523–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzberg S, Brosio P, Gross C, Pogue GP (2002) Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J 30: 315–327 [DOI] [PubMed] [Google Scholar]
- Korshunova YO, Eide D, Clarke WG, Guerinot ML, Pakrasi HB (1999) The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol Biol 40: 37–44 [DOI] [PubMed] [Google Scholar]
- Kumagai MH, Donson J, della-Cioppa G, Harvey D, Hanley K, Grill LK (1995) Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc Natl Acad Sci USA 92: 1679–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacomme C, Hrubikova K, Hein I (2003) Enhancement of virus-induced gene silencing through viral-based production of inverted-repeats. Plant J 34: 543–553 [DOI] [PubMed] [Google Scholar]
- Liu H, Reavy B, Swanson M, MacFarlane S (2002. a) Functional replacement of the tobacco rattle virus cysteine-rich protein by pathogenicity proteins from unrelated viruses. Virology 298: 232–239 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2002. b) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Lu R, Malcuit I, Moffett P, Ruiz MT, Peart J, Wu AJ, Rathjen JP, Bendahmane A, Day L, Baulcombe DC (2003) High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J 22: 5690–5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane SA, Popovich AH (2000) Efficient expression of foreign proteins in roots from tobravirus vectors. Virology 267: 29–35 [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Martinez de Ilarduya O, Kaloshian I (2001) Mi-1.2 transcripts accumulate ubiquitously in resistant Lycopersicon esculentum. J Nematol 33: 116–120 [PMC free article] [PubMed] [Google Scholar]
- Matthews REF (1991) Plant Virology, Ed 3. Academic Press, San Diego
- Milligan SB, Bodeau J, Yaghoobi J, Kaloshian I, Zabel P, Williamson VM (1998) The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 10: 1307–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli C, Lemieux C, Jorgensen R (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2: 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palauqui JC, Elmayan T, Pollien JM, Vaucheret H (1997) Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J 16: 4738–4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peele C, Jordan CV, Muangsan N, Turnage M, Egelkrout E, Eagle P, Hanley-Bowdoin L, Robertson D (2001) Silencing of a meristematic gene using geminivirus-derived vectors. Plant J 27: 357–366 [DOI] [PubMed] [Google Scholar]
- Ratcliff F, Harrison BD, Baulcombe DC (1997) A similarity between viral defense and gene silencing in plants. Science 276: 1558–1560 [DOI] [PubMed] [Google Scholar]
- Ratcliff F, Martin-Hernandez AM, Baulcombe DC (2001) Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J 25: 237–245 [DOI] [PubMed] [Google Scholar]
- Robertson D (2004) VIGS vector for gene silencing: many targets, many tools. Annu Rev Plant Biol 55: 495–519 [DOI] [PubMed] [Google Scholar]
- Roth BM, Pruss GJ, Vance VB (2004) Plant viral suppressors of RNA silencing. Virus Res 102: 97–108 [DOI] [PubMed] [Google Scholar]
- Saedler R, Baldwin IT (2004) Virus-induced gene silencing of jasmonate-induced direct defences, nicotine and trypsin proteinase-inhibitors in Nicotiana attenuata. J Exp Bot 55: 151–157 [DOI] [PubMed] [Google Scholar]
- Schmidt W, Schikora A (2001) Different pathways are involved in phosphate and iron stress-induced alterations of root epidermal cell development. Plant Physiol 125: 2078–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K, Mathur J, Boudonck K, Wells B, Dolan L, Roberts K (1998) The ROOT HAIRLESS 1 gene encodes a nuclear protein required for root hair initiation in Arabidopsis. Genes Dev 12: 2013–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda S, Nishiguchi M (2000) Graft transmission of post-transcriptional gene silencing: target specificity for RNA degradation is transmissible between silenced and non-silenced plants, but not between silenced plants. Plant J 22: 1–8 [DOI] [PubMed] [Google Scholar]
- Turnage MA, Muangsan N, Peele CG, Robertson D (2002) Geminivirus-based vectors for silencing in Arabidopsis. Plant J 30: 107–114 [DOI] [PubMed] [Google Scholar]
- Tzortzakakis EA, Blok VC, Philips MS, Trudgill DL (1999) Variation in root-knot nematode (Meloidogyne spp.) in Crete in relation to control with resistant tomato and pepper. Nematology 1: 499–506 [Google Scholar]
- Ueda K, Matsuyama T, Hashimoto T (1999) Visualization of microtubules in living cells of transgenic Arabidopsis thaliana. Protoplasma 206: 201–206 [Google Scholar]
- Valentine TA, Roberts IM, Oparka KJ (2002) Inhibition of tobacco mosaic virus replication in lateral roots is dependent on an activated meristem-derived signal. Protoplasma 219: 184–196 [DOI] [PubMed] [Google Scholar]
- Vassilakos N, Vellios EK, Brown DJF, MacFarlane SA (2001) Tobravirus 2b protein acts in trans to facilitate transmission by nematodes. Virology 279: 478–487 [DOI] [PubMed] [Google Scholar]
- Vellios E, Brown DJF, MacFarlane SA (2002. b) Substitution of a single amino acid in the 2b protein of Pea early-browning virus affects nematode transmission. J Gen Virol 83: 1771–1775 [DOI] [PubMed] [Google Scholar]
- Vellios E, Duncan G, Brown D, MacFarlane S (2002. a) Immunogold localization of tobravirus 2b nematode transmission helper protein associated with virus particles. Virology 300: 118–124 [DOI] [PubMed] [Google Scholar]
- Visser PB, Bol JF (1999) Nonstructural proteins of Tobacco rattle virus which have a role in nematode-transmission: expression pattern and interaction with viral coat protein. J Gen Virol 80: 3272–3280 [DOI] [PubMed] [Google Scholar]
- Voinnet O (2001) RNA silencing as a plant immune system against viruses. Trends Genet 17: 449–458 [DOI] [PubMed] [Google Scholar]
- Vos P, Simons G, Jesse T, Wijbrandi J, Heinen L, Hogers R, Frijters A, Groenendijk J, Diergaarde P, Reijans M, et al (1998) The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat Biotechnol 16: 1365–1369 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abott D, Stoutjesdijk PA, et al (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.