Abstract
FHY3 (far-red elongated hypocotyls 3) and FAR1 (far-red-impaired response) are two homologous proteins essential for phytochrome A controlled far-red responses in Arabidopsis (Arabidopsis thaliana). There are 12 additional FHY3/FAR1-related genes in the Arabidopsis genome. The predicted sizes of this family of proteins range from 531 amino acids to 851 amino acids, and they share 12.0% to 82.4% amino acid identities over their entire lengths. In addition, most FRS proteins contain one to three coiled-coil domains and one or two putative nuclear localization signals. Semiquantitative reverse transcription-polymerase chain reaction analyses revealed that all FRS genes except FRS10 are expressed in all tissues examined, including rosette leaves, cauline leaves, inflorescence stems, flowers, and siliques. Analyses of gene specific promoter∷GUS fusion reporter gene expression revealed that all FRS genes except FRS1 are expressed in hypocotyls, and their expression in hypocotyl is induced by far-red light treatment. Transient expression of green fluorescent protein tagged FRS fusion proteins in onion (Allium cepa) epidermal cells revealed that all FRS proteins are targeted into the nucleus. T-DNA knockout frs6 and frs8 mutants flowered early under both long-day and short-day conditions (with much more drastic effects under short-day conditions), suggesting that FRS6 and FRS8 regulate flowering time. In addition, FRS9 RNAi transgenic plants showed a specific hypersensitivity to red light inhibition of hypocotyl elongation and light-regulated gene expression, indicating that FRS9 is a specific negative regulator of phyB signaling mediating seedling deetiolation. In summary, our results support the notion that FRS family members play distinct roles in light control of Arabidopsis development, most likely by regulating nuclear gene expression.
As sessile organisms, plants utilize sophisticated sensory systems to monitor their ambient environments and undergo adaptive growth. Light is one of the major environmental signals that influences many aspects of plant growth and development, including seed germination, seedling deetiolation, phototropism, stomata and chloroplast movement, stem elongation, circadian rhythms, and flowering (Quail, 2002; Wang and Deng, 2003, 2004).
Plants use three major classes of photoreceptors to monitor variations in direction, duration, quantity, and wavelength of light. The cryptochromes and phototropins sense the blue/UV-A region of the spectrum, whereas the phytochromes perceive primarily the red (R) and far-red (FR) wavelengths. Of these photoreceptors, phytochromes are the best characterized. In Arabidopsis (Arabidopsis thaliana), phytochromes are encoded by five distinct genes (PHYA–PHYE) belonging to a small gene family. The gene products of PHYB to PHYE have similar functions in regulating light responses under continuous R and white light, with phyB playing a predominant role. phyA is primarily responsible for the very low fluence responses and for the FR light-dependent high-irradiance responses (HIRs), including inhibition of hypocotyl elongation, opening of apical hook, expansion of cotyledons, accumulation of anthocyanin, and FR preconditioned blocking of greening (Whitelam et al., 1993; Deng and Quail, 1999; Neff et al., 2000; Briggs and Olney, 2001).
Molecular genetic studies have identified two loci, FHY3 and FAR1, as two positive regulators specifically for phyA-mediated HIR responses in response to FR light. Their loss-of-function mutants display elongated hypocotyls specifically under continuous FR light (FRc). Although fhy3 and far1 mutants display similar defects in hypocotyl elongation and anthocyanin accumulation, fhy3 has a more pleiotropic effect on phyA signaling. For instance, apical hook and cotyledon opening, and FRc preconditioned block of greening are affected by fhy3 but not by far1 (Hudson et al., 1999; Wang and Deng, 2002). Interestingly, FHY3 and FAR1 encode two homologous proteins, and overexpression of FAR1 or FHY3 can suppress each other's loss-of-function mutant phenotype (Wang and Deng, 2002). Furthermore, a genome expression study demonstrated that the transgenes FHY3 and FAR1 driven by the strong constitutive cauliflower mosaic virus (CaMV) 35S promoter could essentially restore the genome expression profiles abolished by the far1-2 and fhy3-1 mutations, respectively (Wang et al., 2002). These observations suggest that FHY3 and FAR1 play both overlapping and distinct roles in phyA signaling.
The Arabidopsis genome project revealed that FHY3 and FAR1, together with FRS1 to FRS12 (for FAR1-related sequences), comprise a 14-member gene family (Arabidopsis Genome Initiative, 2000). The sequence homologies and conserved secondary structure among the family members suggest that this group of proteins could be involved in distinct signaling processes in response to various biotic and abiotic stresses. In addition, FRS-like proteins have also been identified in other plant species, including monocotyledonous plants, indicating that this family of proteins is conserved throughout the evolution of the plant kingdom. Therefore, they may play essential roles unique to plant growth and development (Hudson et al., 1999; Wang and Deng, 2002). In this report, we conducted a detailed molecular analysis of this gene family, their expression patterns, and subcellular localization of the encoded proteins. We also present evidence for differential roles of members of this gene family in mediating distinct photo-responses.
RESULTS
Identification of the FHY3/FAR1 Gene Family
We used full-length peptide sequences of both FAR1 and FHY3 as the query sequences in our BLAST searches against the Arabidopsis genome sequences at the National Center for Biotechnology Information (NCBI) Web site (http://www.ncbi.nlm.nih.gov/) and identified 12 additional sequences that share significant homologies with both FAR1 and FHY3 throughout most of their lengths (Table I). These homologous genes are distributed on all five chromosomes of Arabidopsis with the exception of FRS5/FRS9, which are arranged in tandem on chromosome IV. To carry out functional studies of the additional 12 FRS genes, we sought to obtain their full-length cDNA clones. For FRS6, FRS7, and FRS11, full-length cDNAs were available when we initiated this work and were obtained from the Arabidopsis Biological Resources Center (ABRC). For other novel FRS genes (FRS1, FRS2, FRS3, FRS4, FRS5, FRS8, FRS9, FRS10, and FRS12), we generated their cDNA clones using reverse transcription (RT)-PCR. For each gene, a pair of primers with added restriction sites at their 5′ ends was designed to cover the longest open reading frame based on genome annotation. We obtained RT-PCR products for all novel FRS genes except FRS10. These PCR products were subcloned into the pCR-TOPO2.1 vector (Invitrogen, Carlsbad, CA) and validated by sequencing. All of these cDNA clones, with the exception of FRS1, are consistent with the annotation of the Arabidopsis genome project and available cDNA or expressed sequence tag sequences. Sequence analysis of our FRS1 cDNA and other reported cDNA sequences indicated that this gene was misannotated in the number and locations of introns (Fig. 1) and that our cDNA clone was missing approximately 80 amino acids of C-terminal sequences. We performed a separate RT-PCR to generate the missing C-terminal cDNA fragment and ligated it to the original FRS1 cDNA clone (representing the N-terminal portion) to create a real full-length cDNA clone for FRS1. In the case of FRS10, we have been unable to obtain RT-PCR products using various tissue sources (cotyledons, hypocotyls, roots, rosette leaves, cauline leaves, inflorescence stems, flowers, and siliques), suggesting that the FRS10 transcript might be unstable, or expressed at a very low level, or only expressed under certain specific conditions. Therefore, the deduced amino acid sequence of FRS10 (At5g28530) was used in our molecular analysis (Table I).
Table I.
Summary of FHY3/FAR1 gene family
| Gene Name | Locus | Protein ID | Amino Acid | MW | Coiled-Coil Domain | NLS | cDNA Clone |
|---|---|---|---|---|---|---|---|
| kD | |||||||
| FAR1 | At4g15090 | AAD51282 | 827 | 95.4 | 1 | + | RAFL09-68-G16, Hudson et al. (1999) |
| FHY3 | At3g22170 | NP_188856 | 839 | 96.0 | 1 | + | RAFL21-40-L21, BX825360, BX840685, CK111774, T20465, BE529279, AV528578, AV529810, AV523823, Wang and Deng (2002) |
| FRS1 | At4g19990 | NP_193732 | 687 | 80.3 | 2 | − | AV555016, this work |
| FRS2 | At2g32250 | NP_180784 | 807 | 92.0 | 2 | + | RAFL16-06-P03, RAFL07-55-M09, BX820631, BX837725, BX834611, AV565526, BE525295, BE527036, BE525980, this work |
| FRS3 | At2g27110 | NP_565636 | 851 | 96.3 | None | + | RAFL09-09-M18, RAFL15-44-E06, N37149, BX839696, T14215, AV522646, AV528393, AV528316, AV522858, this work |
| FRS4 | At1g76320 | NP_177759 | 670 | 78.5 | 1 | + | RAFL25-49-E06, BX838539, BX834442, BX837506, AV557793, this work |
| FRS5 | At4g38180 | NP_195531 | 788 | 90.5 | 1 | + | RAFL09-61-A11, BX834891, this work |
| FRS6 | At1g52520 | NP_175661 | 703 | 81.6 | 1 | + | RAFL05-20-M15, RAFL21-86-F16, RAFL24-26-K23, RAFL25-17-H18 |
| FRS7 | At3g06250 | NP_566278 | 764 | 87.8 | 1 | + | RAFL09-18-D04, BX835891, BX839909, AV528281 |
| FRS8 | At1g80010 | NP_178118 | 696 | 79.6 | None | − | This work |
| FRS9 | At4g38170 | NP_195530 | 531 | 60.8 | 3 | − | RAFL19-54-J19, BX838072, BX827635, this work |
| FRS10 | At5g28530 | AAF88018 | 685 | 78.5 | 1 | + | None |
| FRS11 | At1g10240 | NP_563865 | 680 | 77.7 | 2 | + | RAFL09-33-I16, RAFL16-61-E10 |
| FRS12 | At5g18960 | NP_197397 | 788 | 90.5 | 1 | + | RAFL16-82-P07, BX833504, AV535499, AV535209, AV440408, T14014, this work |
MW, Molecular weight; NLS, nuclear localization signal.
Figure 1.
FRS1 gene structure.
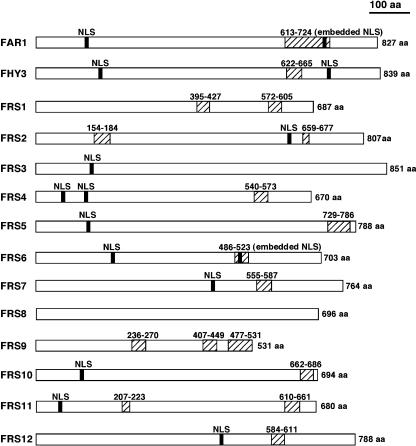
The predicted sizes of the FRS family of proteins range from 531 amino acids to 851 amino acids. Interestingly, FHY3 and FAR1 share the highest homology to each other (47.3% amino acid identity and 79.4% similarity), and they comprise a branch of this gene family (Fig. 2). Other FRS proteins share 17.6% to 37.5% amino acid identities with FHY3 over their entire lengths. It is likely that FRS7 and FRS12 are products of a recent gene duplication event (82.4% amino acid identity and 93.3% similarity; Table II). FRS1 and FRS4 are most similar to each other and they also form a branch of the gene family (31.8% amino acid identity and 59.1% similarity). FRS6 and FRS8 share the highest homology and form another branch (42.0% amino acid identity and 74.5% amino acid similarity). In addition, the tandem arrangement and high homology between FRS5 and FRS9 suggest that they are a pair of recently duplicated genes (33.4% identity and 56.7% similarity). All FRS proteins except FRS3 and FRS8 contain one to three putative coiled-coil domains (predicted with the COILs program at http://www.ch.embnet.org/software/COILS_form.html). Further, most of the FRS proteins also possess one or two putative nuclear localization signals (NLSs) of four-residue pattern (predicted with the PSORT program at http://psort.nibb.ac.jp/form.html). The putative NLSs are located either in the C-terminal portion in some FRS proteins (FRS2, FRS7, and FRS12) or in the N-terminal half of other molecules (FRS3, FRS4, FRS5, FRS10, and FRS11). FAR1, FHY3, and FRS6 possess two putative NLSs, with one located in the N-terminal half and the other in the C-terminal half (Fig. 3). Three of the proteins (FRS1, FRS8, and FRS9) lack putative NLSs. It is worth noting that FRS8 possesses neither coiled-coil domains nor NLSs, whereas FRS9 is significantly smaller than the other FRS proteins and contains three putative coiled-coil domains (Fig. 3).
Figure 2.
A phylogenetic tree of the FRS gene family based on amino acid sequences. The plot was obtained by the Cluster Method of the MegAlign program (DNAstar, Madison, WI).
Table II.
Amino acid identities (above slash) and similarities (below slash) over the alignment lengths between predicted full-length sequences of FRS proteins
| Protein Name | FAR1 | FRS1 | FRS2 | FRS3 | FRS4 | FRS5 | FRS6 | FRS7 | FRS8 | FRS9 | FRS10 | FRS11 | FRS12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FHY3 | 47.3/79.4 | 33.2/63.3 | 37.5/69.6 | 27.1/61.0 | 35.6/61.5 | 28.5/62.7 | 21.4/54.5 | 20.1/48.0 | 23.6/52.7 | 19.4/45.0 | 18.3/46.9 | 17.6/48.1 | 20.0/47.9 |
| FAR1 | 34.5/61.7 | 37.3/70.0 | 27.6/61.8 | 37.7/65.7 | 29.2/64.3 | 23.9/54.8 | 20.7/47.6 | 23.7/54.8 | 20.3/44.5 | 19.3/49.0 | 20.7/54.2 | 21.3/45.5 | |
| FRS1 | 28.7/60.0 | 21.4/54.9 | 31.8/59.1 | 24.0/54.7 | 20.5/46.4 | 12.0/46.5 | 18.8/46.5 | 20.1/49.9 | 12.1/49.6 | 13.6/50.1 | 12.2/43.9 | ||
| FRS2 | 25.0/59.7 | 31.9/58.3 | 26.2/56.4 | 20.7/48.0 | 18.6/42.6 | 20.7/47.8 | 18.5/45.0 | 17.0/43.3 | 16.1/48.4 | 14.1/49.9 | |||
| FRS3 | 23.5/51.5 | 35.9/63.4 | 19.6/50.1 | 23.4/45.3 | 20.0/50.8 | 28.9/49.1 | 18.0/45.2 | 17.5/47.7 | 23.2/44.8 | ||||
| FRS4 | 28.4/62.0 | 24.4/55.1 | 22.2/45.8 | 25.3/55.2 | 21.6/50.9 | 19.1/47.6 | 21.7/54.6 | 21.8/45.5 | |||||
| FRS5 | 25.6/58.2 | 24.6/52.0 | 24.4/57.1 | 33.4/56.7 | 22.7/52.0 | 23.6/56.0 | 24.9/51.8 | ||||||
| FRS6 | 23.4/59.0 | 42.0/74.5 | 17.7/41.0 | 22.4/53.0 | 21.5/57.7 | 23.6/54.9 | |||||||
| FRS7 | 21.7/56.5 | 20.2/38.3 | 20.8/53.1 | 19.6/52.6 | 82.4/93.3 | ||||||||
| FRS8 | 16.4/40.0 | 21.2/54.9 | 23.7/54.6 | 21.8/53.3 | |||||||||
| FRS9 | 12.3/40.6 | 14.7/39.1 | 19.3/37.5 | ||||||||||
| FRS10 | 37.8/71.3 | 19.9/50.1 | |||||||||||
| FRS11 | 19.0/51.7 |
Pair-wise analyses were performed using ClustalW with a gap-opening penalty of 10 and a gap extension penalty of 0.2 (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_clustalw.html).
Figure 3.
Diagram of predicted molecular structure of FRS proteins. Vertical bars, Putative NLSs. Slashed boxes, Putative coiled-coil domains.
In addition to the coiled-coil domains and putative NLS, Hudson et al. (2003) reported that FAR1, FHY3, and FRS4 (AAF16668) are related to Mutator-like elements (MULEs; Lisch, 2002). Sequence analyses indicate that all predicted FRS polypeptides share significant homology with the transposase domain of type II MuDR family transposons such as Jittery (AAF66982) and MuDRA (S59141) of maize. In addition, all FRS proteins also share similarities with type II MuDR family transposons in the putative zinc-binding motif with conserved Cys and His residues (Fig. 4). The zinc-binding motif of transposon is involved in the nicking and transesterification steps of the integration process (Haren et al., 1999). The functional implications for these conserved domains in FRS proteins are currently unknown.
Figure 4.
Amino acid sequence alignment between all FRS members. Identical and similar amino acid residues are shaded in black and gray, respectively. The Mutator transposase-homologous domain is indicated by a line above the sequences. The asterisks indicate the conserved Cys and His residues of the potential zinc-binding motif.
Expression Patterns of the FRS Genes
To understand the respective roles of the FRS genes in plant development, we conducted a semiquantitative RT-PCR analysis to determine the tissue-specific expression patterns for all FRS genes except FRS10 for which no RT-PCR product was detected. The FRS genes with detected RT-PCR products are expressed in all organs examined, including rosette leaves, cauline leaves, inflorescence stems, flowers, and siliques (Fig. 5A).
Figure 5.
Expression patterns of the FRS genes. A, RT-PCR showing that all FRS genes (except FRS10) are expressed in all organs examined here (rosette leaf, cauline leaf, stem, flower, and silique). B, Histochemial staining of FRS∷GUS reporter gene transgenic lines. The left sections are histochemical staining of FR light-grown plants and the right sections are histochemical staining of dark-grown plants. 35S, transgenic plants harboring 35S promoter driven GUS reporter gene. WT, Wild-type nontransgenic plants (Col ecotype).
To extend the observations made with the RT-PCR analyses, we have generated FRS∷GUS reporter lines for each of the FRS gene, in which the β-glucuronidase (GUS) gene was fused to each of the FRS promoters (FRS∷GUS). Promoter regions (with 5′-untranslated regions included) up to 2 kb from these genes were cloned into the binary vector pCAMBIA3301 (CAMBIA, Australia, http://www.cambia.org/) to replace the 35S promoter and drive the GUS gene expression. These FRS∷GUS reporter gene constructs were introduced into Columbia (Col) wild-type plants via agrobacterium-mediated transformation (Clough and Bent, 1998). Several independent homozygous transformant lines were established for each of the reporter genes. Histochemical staining analyses of GUS activity revealed that all these FRS genes except FRS1 are clearly expressed in hypocotyls. Comparison of dark versus FR light-grown seedlings showed that FRS gene expression in hypocotyls is induced by FR light treatment. FRS5, FRS7, FRS8, FRS10, FRS11, and FRS12 are also expressed in cotyledons of light-grown seedlings. In addition, FRS2, FRS3, FRS4, FRS6, FRS8, and FRS9 are also expressed in roots, whereas FRS1, FRS5, FRS10, FRS11, and FRS12 have barely detectable or no detectable level of expression in this tissue (Fig. 5B). These results are consistent with a potential role of these FRS genes in regulating light control of Arabidopsis seedling development. Strikingly, the FRS10∷GUS reporter gene is strongly expressed in hypocotyls and cotyledons, despite the fact that we have not been able to detect FRS10 transcript from these tissues using RT-PCR (data not shown), suggesting that the native FRS10 transcript might be highly unstable.
FRS Protein Subcellular Localization
Previously, FAR1 and FHY3 were shown to be targeted to the nucleus of onion (Allium cepa) epidermal cells and Arabidopsis cells, respectively (Hudson et al., 1999; Wang and Deng, 2002). To determine the subcellular localization of the 12 additional FRS proteins, we have generated green fluorescent protein (GFP)-FRS fusion protein constructs for each FRS gene (with the exception of FRS10 for which no cDNA clone is available) in the pRTL2-S65TGFP vector that contains the strong CaMV35S promoter to drive transient gene expression (Restrepo et al., 1990; von Arnim et al., 1998). We conducted transient expression assays to determine the fusion protein localization and light regulation in onion epidermal cells. After gene delivery by particle bombardment, onion cells were incubated for 24 to 48 h under both dark and light conditions, and then the GFP localization patterns were examined by microscopy. Consistent with the presence of putative NLSs in FRS2, FRS3, FRS4, FRS5, FRS6, FRS7, FRS11, and FRS12, their GFP-fusion proteins are localized into the nucleus. Most strikingly, we found that FRS1, FRS8, and FRS9 are also targeted into the nucleus despite their lack of predicted NLSs. It should be noted that FRS1 seems to be nuclear enriched with residual amount of cytoplasmic distribution, whereas other FRS proteins seem to be exclusively nuclear localized (Fig. 6). Further, the nuclear localization of these FRS proteins is independent of the light treatment (data not shown).
Figure 6.
Subcellular localization of GFP-FRS fusion proteins in onion epidermal cells. The top sections show GFP localization of the fusion proteins. The lower sections show DAPI staining of the corresponding images. Arrowheads indicate the positions of nuclei. Bar represents approximately 50 μm.
Identification of T-DNA Insertion Mutants in FRS6 and FRS8
For functional analysis of the FRS gene family, we searched the T-DNA insertional pools generated by The Salk Institute Genome Analysis Laboratory (SIGnAL) and The Syngenta Arabidopsis Insertion Library (SAIL, formerly GARLIC) for T-DNA insertion loss/reduction-of-function frs mutants (Sessions et al., 2002; Alonso et al., 2003). We used both antibiotic selection and PCR/sequencing to confirm the presence and determine the number of T-DNAs. For PCR confirmation, we amplified the T-DNA flanking region for each T-DNA line using a left-border primer and a gene-specific primer. We have also backcrossed original lines with corresponding wild-type ecotype (Col) to remove other possible background mutations and to reaffirm the cosegregation of the T-DNA insertion with the observed mutant phenotypes (see below).
Homozygous mutant lines for two independent alleles of frs6, designated frs6-1 (Salk_019743) and frs6-2 (Salk_114017), and two independent alleles of frs8, designated frs8-1 (Salk_122261) and frs8-2 (Salk_077996), have been identified from the SIGnAL population. In all four lines, T-DNAs were inserted in the predicted coding regions and presumably would cause null mutations (Fig. 7A). RT-PCR analysis showed that accumulation of their corresponding message RNAs was abolished in these mutant lines (Fig. 7B). Photobiology experiments were conducted to determine the effects of altering FRS6 and FRS8 gene expression on light control of Arabidopsis development. Although all of these homozygous mutant seedlings deetiolated normally in response to different light conditions (white light, FR, R, blue, and darkness), they all display a slight but reproducible early flowering phenotype (about 1 d earlier) under long-day conditions (LD; 16 h light/8 h darkness; Fig. 8, A and B). Under short-day conditions (SD; 8 h light/16 h darkness), these mutants flowered much earlier (approximately 10 d earlier) than wild-type control plants (Fig. 8, C and D). Consistent with this, both frs6 and frs8 mutant plants also possess fewer rosette leaves than wild-type plants at bolting (Fig. 8, B and D), suggesting that FRS6 and FRS8 have a similar role in controlling flowering time.
Figure 7.
Analyses of frs6 and frs8 T-DNA insertion mutants. A, Diagram of the genomic structures of FRS6 and FRS8 genes and locations of T-DNA insertions. B, RT-PCR analysis showing that message RNAs of FRS6 and FRS8 were abolished in their respective frs homozygous mutants.
Figure 8.
Early flowering phenotype of frs6 and frs8 mutants. A and B, frs6 and frs8 mutants flower early under LD conditions. C and D, frs6 and frs8 mutants flower early under SD conditions. B and D show days to flower and the rosette leaf numbers of various plants at bolting under LD and SD conditions, respectively (based on the opening of first flower). Bar in A represents approximately 1 cm. Bar in C represents approximately 5 cm. Bars in B and D represent sds.
RNAi Silencing of FRS9
T-DNA insertion mutants for FRS9 were not available until recently. Therefore, we attempted to use the RNA interference (RNAi) gene silencing technology to eliminate or reduce mRNA accumulation (Wesley et al., 2001). To achieve gene-specific silencing, two inverted repeat sequences (approximately 350 bp each) of FRS9 were generated by PCR and inserted into the pHANNIBAL vector (Wesley et al., 2001), and a NotI fragment containing this repeat sequence was cloned into the pART27 binary vector (Gleave, 1992). This construct was transformed into Col wild-type Arabidopsis and several homozygous transgenic lines were established. Photobiology experiments showed that multiple homozygous FRS9 RNAi lines exhibit a hypersensitive response specifically to continuous R light (Fig. 9, A–C). This phenotypic alternation seems to be specifically caused by RNAi silencing of FRS9, as FRS9 mRNA transcript level is undetectable, whereas the expression of FRS5, the closest homolog of FRS9, is not affected in the RNAi plants (Fig. 9D). To provide molecular evidence for the observed phenotypic abnormality of FRS9 RNAi plants, northern-blot analyses were conducted to compare the expression of CAB and RBCS, two representative light-responsive genes. After 12 h of R light irradiation, the induction of both CAB and RBCS is clearly more enhanced in the FRS9 RNAi transgenic plants than the wild-type control plants (Fig. 9E). Together, these results suggest FRS9 acts as a negative regulator specific to phyB signaling.
Figure 9.
Phenotype of FRS9 RNAi transgenic plants. A, Four independent RNAi lines of FRS9 show hypersensitive response to continuous R light. Bar represents approximately 1 mm. B, FRS9 RNAi plants (represented by the transgenic line 4-2) are specifically hypersensitive to R light and display normal responses to FR and blue light. Bar represents approximately 1 mm. C, Quantification of hypocotyl lengths of seedlings grown in continuous darkness or R light for 4 d. D, RT-PCR showing that FRS9 mRNA accumulation is abolished in FRS9 RNAi plants (represented by the transgenic line 4-2), whereas accumulation of FRS5 mRNA is not affected. E, CAB and RBCS gene expression is enhanced in FRS9 RNAi plants compared with wild-type plants (grown in darkness for 4 d then transferred to continuous R light for 12 h). Col, Wild-type Col ecotype plants. 1, Col grown in darkness for 5 d; 2 and 3 are Col and FRS9 RNAi plants (line 4-2) grown in darkness for 4 d then transferred to continuous R light for 12 h.
DISCUSSION
Molecular Features of FRS Proteins
The founding members of the FRS gene family, FAR1 and FHY3, were independently identified as two essential signal transducers for phyA-mediated FR-HIR responses (Hudson et al., 1999; Wang and Deng 2002). Other FRS proteins share 17.6% to 37.5% amino acid identities with FHY3 over their entire lengths. Similar to FHY3 and FAR1, all other FRS proteins except FRS3 and FRS8 contain one to three putative coiled-coil domains. Further, most of the FRS proteins possess one or two putative NLSs. The overall sequence similarities and the conserved secondary structure suggest that some of the FRS proteins may have overlapping function in mediating light signaling like FHY3 and FAR1. The presence of coiled-coil domain(s) in most FRS proteins suggests that they may form homo- and/or hetero-dimers or interact with diverse partners to control a broad range of signaling processes. Indeed, evidence for homodimerization of FAR1 and FHY3 and their interaction with other proteins has been reported previously (Wang and Deng, 2002; Hudson et al., 2003). Further, overexpressing the C-terminal portion of FHY3 (C473-839, which contains a coiled-coil domain) in a wild-type background causes an apparent loss of responses to FRc light, suggesting a dominant-negative effect most likely caused by hetero-interaction of the partial FHY3 fragment with other FRS proteins through their coiled-coil domains (Wang and Deng, 2002).
The sequence homologies between FRS proteins and the MULEs suggest that the FRS gene family might be derived from a common ancestor of a MULE-type mobile element. However, it is unlikely that they possess transposition capacity, as these genes lack the characteristic terminal inverted repeats and other transposon-associated features (Hudson et al., 2003). The nuclear localization of FAR1 and its demonstrated ability to activate a reporter gene expression in yeast and Arabidopsis has lead to the proposition that FAR1 might act as a transcriptional regulator (Hudson et al., 1999; Hudson et al., 2003). Genome profiling analyses have shown that both FAR1 and FHY3 are required for phyA-mediated gene expression by FR light (Wang et al., 2002). Since all FRS proteins are localized into the nucleus and all share similarities with transposases of the MULE family, these FRS proteins may define a novel class of transcriptional regulators with transposase homology. Transposases of the Mutator family are capable of regulating the expression of their own genes and the associated genes via direct DNA binding (Barkan and Martienssen, 1991; Benito and Walbot, 1997; Raizada et al., 2001). It will be interesting to determine whether FRS proteins can bind DNA directly and to identify their direct target genes in future studies.
Previously, both FAR1 and FHY3 have been demonstrated to be targeted into the nucleus (Hudson et al., 1999; Wang and Deng, 2002), consistent with the presence of putative NLSs in both proteins. Here we showed that 11 additional FRS proteins are also targeted into the nucleus of onion epidermal cells and this pattern is independent of light control, although it should be noted that FRS1 is only nuclear enriched with clear residual cytoplasmic distribution. Strikingly, FRS1, FRS8, and FRS9 are predicted to lack putative NLSs, thus their nuclear localization suggests that they may either use nontypical NLSs for nuclear import or they may interact with their partners (such as other NLS-containing FRS proteins) and be imported into nucleus as protein complexes.
Distinct Roles of FRS Proteins in Mediating Light Control of Arabidopsis Development
Our RT-PCR analyses showed that with the exception of FRS10, all FRS genes are expressed in all major organs examined, including rosette leaves, cauline leaves, stems, flowers, and siliques. We have not been able to recover RT-PCR products for FRS10 using various tissue sources, consistent with the fact that no expressed sequence tag clones or any type of cDNA clones have been documented for this gene. However, FRS10∷GUS reporter gene shows strong expression in hypocotyls and cotyledons (Fig. 5B). These observations suggest that the endogenous FRS10 transcript might be highly unstable. Further studies, such as nuclear run-on assay, are required to clarify this issue. Histochemical staining analyses of FRS∷GUS fusion reporter genes revealed that all these FRS genes except FRS1 are clearly expressed in hypocotyls. Comparison of dark versus FR light-grown seedlings showed that FRS gene expression in hypocotyls is induced by FR light treatment. FRS5, FRS7, FRS8, FRS10, FRS11, and FRS12 are also expressed in cotyledons of light-grown seedlings (Fig. 5B). These results are consistent with a potential role of these FRS genes in regulating light control of Arabidopsis development. Functional analyses of FRS6, FRS8, and FRS9 genes supported this notion. frs6 and frs8 mutants display an early flowering phenotype under both LD and SD conditions, with more drastic effect under SD conditions. Both phytochromes and cryptochromes play important roles in flowering time regulation (Guo et al., 1998; Somers et al., 1998). Among these photoreceptors, phyA promotes flowering as Arabidopsis phyA mutant flowers later than wild-type plants in LD (Johnson et al., 1994; Neff and Chory, 1998). phyB plays an inhibitory role in floral initiation. The Arabidopsis phyB mutant flowers earlier than wild-type plants in both LD and SD conditions, with a more pronounced phenotype in SD than in LD conditions (Goto et al., 1991; Mockler et al., 1999; Lin, 2000). The early flowering phenotype of frs6 and frs8 mutants under both LD and SD conditions and their more pronounced phenotype in SD conditions suggest that FRS6 and FRS8 likely act as positive regulators in phyB signaling pathway controlling flowering time. Interestingly, FRS6 and FRS8 polypeptides have the highest homology to one another and form a separate branch of the FRS gene family (42.0% amino acid identity and 74.5% amino acid similarity), supporting the functional similarity of these two genes. On the other hand, the R light hypersensitivity exhibited by the FRS9 RNAi transgenic plants indicates that FRS9 is a negative regulator of phyB signaling mediating seedling deetiolation. Together, our results support the notion that FRS gene family members play differential roles regulating distinct photo-responses.
It should be noted that there is potential functional redundancy among this family of proteins, or they may have tissue and developmental stage-specific functions, thus masking the detection of an aberrant phenotype for some frs single mutants under the limited experimental conditions we tested. Future studies employing multiple alleles, double/higher order mutants, gain-of-function studies, and genome profiling changes caused by altering FRS gene expression will most likely reveal more information about the function of these FRS genes in light regulation of Arabidopsis development. Further, different members of this gene family could also be involved in signal transduction processes of other biotic or abiotic stresses such as phytohormones. The molecular analysis of this plant-unique gene family has laid a foundation for elucidating their biological functions as well as their biochemical mechanisms of action.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) materials are of Col ecotype background. T-DNA insertion mutants were obtained from ABRC (Columbus, OH). The seeds were surface-sterilized and sown on 1× Murashige and Skoog media plates with 1% Suc and cold-treated for 3 d at 4°C. Then the plates were exposed to white light for 24 h to stimulate seed germination before being transferred to FRc, R, blue light, or dark conditions for 4 d at 22°C. FR, R, and blue lights were supplied by LED light sources, with irradiance fluence rates of approximately 100, 1,200, and 250 uW/cm2, respectively (measured with International Light [Newburyport, MA] model IL1400A with sensor model SEL-033/F/W). White light was supplied by cool-white fluorescent lamps. Adult plants were grown in environmental chambers at 22°C and 60% humidity with 16 h of continuous fluorescence light unless otherwise specified.
PCR Genotyping
To identify the T-DNA insertion mutants, we amplified the T-DNA flanking region for each T-DNA line using a vector left-border primer (5′-CGGAACCACCATCAAACAGG-3′) and a gene-specific primer. The insertion site was confirmed by DNA sequencing.
RT-PCR
To generate full-length cDNA clones for FRS genes, RNAs were extracted from light-grown Arabidopsis seedlings with the RNeasy Plant Mini kit (Qiagen USA, Valencia, CA). First-stand cDNA was synthesized by StrataScript RT at 42°C for 30 min, and PCR was carried out by PfuTurbo DNA polymerase (Stratagene, La Jolla, CA) with the following program: an initial 95°C 1 min followed by 30 cycles of 95°C 30 s, 60°C 30 s, 68°C 7 min; and a final extension at 68°C for 10 min. Primers were designed to cover the whole open reading frame of each FRS gene with suitable restriction enzyme sites added at their 5′-ends to facilitate downstream cloning efforts. The primer pairs used for RT-PCR were: FRS1, 5′-GGTACCGGATCCATGTCGTCAGGAGAGTGTAGC-3′ and 5′-GTCGACACTAGTTTACTTTCCAGACTTCTTGCA-3′; FRS2, 5′-CTCGAGGGATCCATGGATGATGAAGATGTAGA-3′ and 5′-GACTCTAGATTAATTGGATAAGCGGTGATC-3′; FRS3, 5′-GTCGACCTGGATCCATGGATGTTCATTTGGTGGAAG-3′ and 5′-TCTAGATCAAAAGCGTTGCTTCTTTGC-3′; FRS4, 5′-CTCGAGTCAGATCTATGGAGTTCGAGACTCACGAA-3′ and 5′-TCTAGATCACCCAGGGGGATTGTTCTG-3′; FRS5, 5′-GTCGACAGATCTATGATGGATAATGAAGTGCTC-3′ and 5′-TCTAGATCACAGATTATCCTTCAAGCT-3′; FRS8, 5′-GTCGACGGATCCATGGAAGAGCAGCTGGTTGTT-3′ and 5′-TCTAGACTATGAAGGCTTCTCTGTCAC-3′; FRS9, 5′-GTCGACTCATGAGCAGGGTCGAGCATGT-3′ and 5′-TCTAGATCACTCTTTCAAGCTTAGTC-3′; FRS10, 5′-CTCGAGTCTATGGCGTTGAAGCCATTGAAC-3′ and 5′-TCTAGATCATGGCTGATACAAGCAATT-3′; and FRS12, 5′-GGTACCAGATCTATGGAGAGTGTAGATACTGAG-3′ and 5′-GCTAGCTCATCTCTGCCAACAAAGTTTC-3′. The RT-PCR products were cloned into the pCR-TOPO2.1 vector (Invitrogen) to generate pTOPO-FRS clones. A separate RT-PCR was performed to obtain the C-terminal fragment of FRS1 missed in our original pTOPO-FRS1 cDNA clone. The primers used were: 5′-GGAATGTGTACAGCTCAGAGA-3′ and 5′-AGTGGATCCGTCTAGTTCAAGTCTTTTACC-3′. A full-length cDNA clone of FRS1 was created by ligating the original FRS cDNA with the second RT-PCR product at the common BsrGI site.
For semiquantitative RT-PCR analysis of FRS gene expression, total RNAs were extracted from different organs of adult plants. RT-PCR was performed with the one-step RT-PCR system (Promega, Madison, WI) using the following program: 45 min at 48°C for RT, then 2 min at 94°C followed by 32 cycles of 30 s at 94°C; 1 min at 60°C; 2 min at 68°C, and a final extension at 68°C for 7 min. The actin-1 gene of Arabidopsis was used as a control. The primer pairs used were: FHY3, 5′-GCTGTGAGTGAACAGACCAG-3′ and 5′-CATCAGTCATGTAGGTTGGTG-3′; FAR1, 5′-GGATTCAGAGGAATGTCAAG-3′ and 5′-GTCTCCATAGACTCATCAGC-3′; FRS1, 5′-GATCAGACAGTGTGAACTCTG-3′ and 5′-GCTGTAACCGATTCTGACTC-3′; FRS2, 5′-TGGAGATGCAGGATAAGCAG-3′ and 5′-ACTGAACCCACTCATTCTCG-3′; FRS3, 5′-GAAGCCATCAAGTATGCTGAG-3′ and 5′-GACATCCATGTCTGCTGAGG-3′; FRS4, 5′-GTTGTCCTCCAAATGTCGGG-3′ and 5′-ACTCCTGGCATTGTGTTGTG-3′; FRS5, 5′-CGGAGATGTTGATGACGACG-3′ and 5′-GGTTGTCAGCATTCATCTGC-3′; FRS6, 5′-GCTGGAGAGACAATGGAGTC-3′ and 5′-CACGAAAGGTCCATCGACATG-3′; FRS7, 5′-GTCATATAGAGGAGGCTCAG-3′ and 5′-GGTACTACTGATGTGATTGC-3′; FRS8, 5′-CTATGTCTGGCTCTTCAGAG-3′ and 5′-GACATTGCTGAGACCTCGTC-3′; FRS9, 5′-TCTTGGGACTCTATCGTCAG-3′ and 5′-AGATACCGAGATGGAAGAGC-3′; FRS10, 5′-GAGTGATACGTTGGAGCTTC-3′ and 5′-GAGTGACAAGAAGATCCCAC-3′; FRS11, 5′-GGAGTTGGCAAAGGACTTAC-3′ and 5′-CCAACCCATATTCCAAGTGG-3′; FRS12, 5′-GAGGTGAAGGTAGTGTTGAG-3′ and 5′-TTGACATCCAATTCGACAGC-3′; and actin, 5′-CATCAGGAAGGACTTGTACGG-3′ and 5′-GATGGACCTGACTCGTCATAC-3′.
Plasmid Construction
For FRS∷GUS reporter gene constructs, up to 2-kb upstream sequences of the predicted ATG start codons (including 5′-untranslated regions) were PCR amplified with gene-specific oligonucleotides containing proper restriction sites. PCRs were carried out on total genomic DNA extracted with the DNeasy Plant Mini kit (Qiagen). These generated promoter fragments were cloned into the binary vector pCAMBIA3301 (CAMBIA, http://www.cambia.org) to replace the 35S promoter and drive the GUS gene expression. The primer pairs used were: FHY3, 5′-GACAAGCTTCGATTTTACCTGAAGAGTGTGAG-3′ and 5′-GAGCCATGGCCATGACAAACCTATAGTCTCAGGC-3′; FAR1, 5′-GACAAGCTTGTGACTCAGAGCACAACTCTCGTAC-3′ and 5′-GATCCATGGGTATCAAAGTCTATACCATTTCGTG-3′; FRS1, 5′-GACCAATTGATCCTCGAAGCAGAGAGCTGCTATG-3′ and 5′-GATCCATGGACGACATTTCGAAATCAATACCTG-3′; FRS2, 5′-GACAAGCTTGAGAAGTGAGAAGATCCAATGTTTG-3′ and 5′-GATCCATGGCACTAGTTCCAACTCTGTTAACCG-3′; FRS3, 5′-GACAAGCTTCCAGACAAGATGGATTCGTAAGC-3′ and 5′-GATCCATGGCCTTCACTTAGTAGAGAAGTTC-3′; FRS4, 5′-GACCAATTGGAATGGTCTCTTGACAGCACAAGG-3′ and 5′-GATCCATGGGAGAAGCTTGATGAATCCACACC-3′; FRS5, 5′-GACAAGCTTCTTGAGAAGAAGGAATC-3′ and 5′-GATCCATGGCCTCCTTGTCTTCAGGGATGAATT G-3′; FRS6, 5′-GACGAATTCGCAAAAGAGTTGAACCTCAGCTGC-3′ and 5′-GATCCATGGTCTCCATTGGAGGTTTGGTTCCTC-3′; FRS7, 5′-GACCAATTGCCTTGTATCGTTGATCATCTATG-3′ and 5′-GATCCATGGTGTTCCCACAATTTAAACTGGTAGG-3′; FRS8, 5′-GACAAGCTTGTCCACCTTATACTGTCCTTGATG-3′ and 5′-GTCAGATCTACCATCTGAAGACCATCAAATTC-3′; FRS9, 5′-GATCAATTGCCAATACTCTCTCATAGCGTGGAG-3′ and 5′-GTCAGATCTACCATGCTTACTTCTTCCACTAATTCTG-3′; FRS10, 5′-GATTCTAGAAGGTCTTTACAATACGGTCCAAC-3′ and 5′-GATCCATGGATGGCTTCAACGCCATAAGACAAGG-3′; FRS11, 5′-GATTCTAGAAACACGTCTAGGTTCATGGATGCAC-3′ and 5′-GATCCATGGAGCTCCTAAGGTATATTCAGCAC-3′; and FRS12, 5′-GATGAATTCGCGTTATACGTGATTGCATTCGGTG-3′ and 5′-GATCCATGGTAGTAAGCTCAGTATCTACACTGC-3′.
For the GFP-FRS fusion protein constructs, the full-length cDNA fragment for each FRS gene was cloned into the pRTL2-S65TGFP vector that contains the strong CaMV 35S promoter to drive transient gene expression (Restrepo et al., 1990; von Arnim et al., 1998). Briefly, for FRS1, FRS2, FRS3, FRS4, FRS5, FRS8, and FRS12, their pTOPO-FRSs clones were digested with BglII/BamHI and XbaI/SpeI/NheI to release their cDNA fragments, and then the cDNA fragments were gel purified and cloned into the pRTL2-S65TGFP vector digested with BglII and XbaI. For FRS9, pTOPO-FRS9 was digested by SalI (blunted by Klenow fragment) and XbaI, and then the released cDNA fragment was cloned into the pRTL2-S65TGFP vector digested with BglII (blunted with Klenow fragment) and XbaI. For FRS6, FRS7, and FRS11, first their full-length cDNAs with modified restriction sites at both ends were PCR amplified from cDNA clones obtained from ABRC and cloned into the pGEM-T Easy vector (Promega) to generate corresponding pGEM-FRS clones. The primers used for PCRs were: FRS6, 5′-GATATCTCGGATCCTATGGAGAGAAGTGAGTCCGTTG-3′ and 5′-TCTAGAGTCGACCCCGGGACACTAGTCTTGTTTTTCTTCTAC-3′; FRS7, 5′-GGATCCATGGTTGTCAAAACTTATCC-3′ and 5′-GTCTCTAGATCATCTCTGCCAACACAG-3′; and FRS11, 5′-CTCGAGTCGGTACCAGATCTATGTCGGATGATCCTGGAC-3′ and 5′-GTCGACTCTAGATCAAAAATTCCTCTGCACAG-3′. The full-length cDNA fragments of FRS7 and FRS11 were released by digestion with BamHI/XbaI (for pGEM-FRS7) or BglII/SpeI (for pGEM-FRS11) and then cloned into BglII/XbaI digested pRTL2-S65TGFP vector. For FRS6, pGEM-FRS6 was digested with BamHI and XbaI, and the released cDNA fragment was cloned into the BamHI/XbaI sites of pRTL2-GFP-hCOP1 (Wang et al., 1999). The junction sites of all subcloning steps were confirmed by sequencing.
For RNAi silencing of FRS9, the first PCR fragment (348 bp) was amplified from FRS9 cDNA clone with primers S9-B (5′-CGCGGATCCGTGGATGCATCAACTACAATG-3′) containing a BamHI site, and S9-C (5′-TCCATCGATCTCAAACATTTGACAGCTGC-3) with a ClaI site. The PCR product was digested with BamHI and ClaI and then cloned into the BamHI/ClaI sites of the pHANNIBAL vector (Commonwealth Scientific and Industrial Research Organization [CSIRO]; http://www.csiro.au), generating pHANNIBAL-9BC. The second PCR fragment was amplified by primers S9-X (5′-GACCTCGAGGTGGATGCATCAACTACAATG-3′) containing an XhoI site, and S9-K (5′-CGGGGTACCCTCAAACATTTGACAGCTGC-3) containing a KhnI site. The PCR fragment was digested with XhoI and KpnI, and then inserted into XhoI/KpnI digested pHANNIBAL-9BC to generate pHANNIBAL-FRS9. A NotI fragment containing the inverted repeat sequences of FRS9 was released from pHANNIBAL-FRS9 and cloned into NotI-digested pART27 binary vector (Gleave, 1992), giving rise to pART-FRS9.
Plant Transformation and Selection of Transgenic Plants
Each of the FRS∷GUS reporter gene constructs and the pART-FRS9 RNAi gene silencing constructs were electroporated into the Agrobacterium strain GV3101 and then introduced into Arabidopsis wild-type plants (Col ecotype) via a floral dip method (Clough and Bent, 1998). Transgenic plants were selected on germination plates containing 20 μg/mL glufosinate-ammonium (for FRS∷GUS reporter gene lines) or 50 μg/mL Kanamycin (for FRS9 RNAi lines). We selected about 20 T1 transgenic lines with a single T-DNA insertion and allowed them to self to produce T2 seeds. Phenotypic analyses were conducted with T2 plants and then confirmed in T3 generation. Histochemical GUS staining of FRS∷GUS reporter gene was conducted according to a described procedure (Jefferson et al., 1987).
Protein Localization Studies
Onion (Allium cepa) epidermal cells were transfected with pRTL2-GFP-FRSs constructs using helium biolistic gun transformation system (Bio-Rad, Hercules, CA) as described (Ang et al., 1998) and incubated in light or darkness for 24 to 48 h at 22°C. The subcellular localization of GFP fusion proteins was visualized with a fluorescence microscope.
RNA Gel-Blotting
Arabidopsis seedlings were grown either in darkness for 5 d, or in darkness for 4 d and then were transferred to red light for 12 h. Total RNA was extracted using RNeasy Plant Mini kit (Qiagen). Five micrograms total RNA per lane was size-fractionated on a formaldehyde agarose gel and subsequently transferred to a nylon membrane. After hybridization in 0.25 m sodium phosphate, 1 mm EDTA, 1% (w/v) casein, 7% (w/v) SDS at 65°C with random prime-labeled DNA probes (Roche, Indianapolis), membranes were washed two times each of 2× SSC, 0.1% SDS; 0.2× SSC, 0.1% SDS and 0.1× SSC, 0.1% SDS. CAB and RBCS probes were described previously (Wang and Deng, 2002).
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes.
Sequence data from this article (the new full-length cDNA sequence of FRS1) have been deposited with the EMBL/GenBank data libraries under accession number AY763412.
Acknowledgments
We thank Georg Jander and Elizabeth Estabrook for their reading and comments on the manuscript. We also thank Jacob Mace for his assistance with the subcellular localization studies of FRS proteins. We are grateful to the CSIRO Plant Industry for providing us with pHANNIBAL and pART27 vectors. Thanks are also due to ABRC for distributing seeds and cDNA clones.
This work was supported by set-up funds provided by Boyce Thompson Institute (to H.W.).
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW (1998) Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell 1: 213–222 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Barkan A, Martienssen RA (1991) Inactivation of maize transposon Mu suppresses a mutant phenotype by activating an outward-reading promoter near the end of Mu1. Proc Natl Acad Sci USA 88: 3502–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito MI, Walbot V (1997) Characterization of the maize Mutator transposable element MURA transposase as a DNA-binding protein. Mol Cell Biol 17: 5165–5175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs WR, Olney MA (2001) Photoreceptors in plant photomorphogenesis to date: five phytochromes, two cryptochromes, one phototropin, and one superchrome. Plant Physiol 125: 85–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Deng XW, Quail PH (1999) Signalling in light-controlled development. Semin Cell Dev Biol 10: 121–129 [DOI] [PubMed] [Google Scholar]
- Gleave AP (1992) A versatile binary vector system with a T-DNA organizational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Goto N, Kumagai T, Koornneef M (1991) Flowering responses to light-breaks in photomorphogenic mutants of Arabidopsis thaliana, a long-day plant. Physiol Plant 83: 209–215 [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Haren L, Ton-Hoang B, Chandler M (1999) Integrating DNA: transposases and retroviral integrases. Annu Rev Microbiol 53: 245–281 [DOI] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH (1999) The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev 13: 2017–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson ME, Lisch DR, Quail PH (2003) The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J 34: 453–471 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E, Bradley M, Harberd NP, Whitelam GC (1994) Photoresponses of light-grown phyA mutants of Arabidopsis. Plant Physiol 105: 141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C (2000) Photoreceptors and regulation of flowering time. Plant Physiol 123: 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D (2002) Mutator transposons. Trends Plant Sci 7: 498–504 [DOI] [PubMed] [Google Scholar]
- Mockler TC, Guo H, Yang H, Duong H, Lin C (1999) Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126: 2073–2082 [DOI] [PubMed] [Google Scholar]
- Neff MM, Chory J (1998) Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol 118: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Fankhauser C, Chory J (2000) Light: an indicator of time and place. Genes Dev 14: 257–271 [PubMed] [Google Scholar]
- Quail PH (2002) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3: 85–93 [DOI] [PubMed] [Google Scholar]
- Raizada MN, Brewer KV, Walbot V (2001) A maize MuDR transposon promoter shows limited autoregulation. Mol Genet Genomics 265: 82–94 [DOI] [PubMed] [Google Scholar]
- Restrepo MA, Freed DD, Carrington JC (1990) Nuclear transport of plant potyviral proteins. Plant Cell 2: 987–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Devlin PF, Kay SA (1998) Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282: 1488–1490 [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW, Stacey MG (1998) Cloning vectors for the expression of green fluorescence protein fusion proteins in transgenic plants. Gene 221: 35–43 [DOI] [PubMed] [Google Scholar]
- Wang H, Deng XW (2002) Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J 21: 1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Deng XW (2003) Dissecting phytochrome A dependent signaling network in higher plants. Trends Plant Sci 8: 172–178 [DOI] [PubMed] [Google Scholar]
- Wang H, Deng XW (2004) Phytochrome signaling mechanism. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. http://www.bioone.org/pdfserv/i1543-8120-018-01-0001.pdf [DOI] [PMC free article] [PubMed]
- Wang H, Kang D, Deng XW, Wei N (1999) Evidence for functional conservation of a mammalian homologue of the light-responsive plant protein COP1. Curr Biol 9: 711–714 [DOI] [PubMed] [Google Scholar]
- Wang H, Ma LG, Li JM, Zhao HY, Deng XW (2002) Analysis of far-red light regulated genome expression profiles of phytochrome A pathway mutants in Arabidopsis. Plant J 32: 723–733 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP (1993) Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5: 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]