ABSTRACT
Cells have developed different mechanisms to respond to stress, including the formation of cytoplasmic foci known as stress granules (SGs). SGs are dynamic and formed as a result of stress-induced inhibition of translation. Despite enormous interest in SGs due to their contribution to the pathogenesis of several human diseases, many aspects of SG formation are poorly understood. SGs induced by different stresses are generally assumed to be uniform, although some studies suggest that different SG subtypes and SG-like cytoplasmic foci exist. Here, we investigated the molecular mechanisms of SG assembly and characterized their composition when induced by various stresses. Our data revealed stress-specific differences in composition, assembly and dynamics of SGs and SG-like cytoplasmic foci. Using a set of genetically modified haploid human cells, we determined the molecular circuitry of stress-specific translation inhibition upstream of SG formation and its relation to cell survival. Finally, our studies characterize cytoplasmic stress-induced foci related to, but distinct from, canonical SGs, and also introduce haploid cells as a valuable resource to study RNA granules and translation control mechanisms.
KEY WORDS: Haploid cell, Translational control, Stress response, Stress granules, Translation initiation, eIF2α
Highlighted Article: Haploid cells were used to study stress-specific differences in the dynamics and composition of stress granule-like foci, the translation-related mechanisms of their assembly and their connection to cell survival.
INTRODUCTION
During stress, translational control allows for the rapid and efficient reprogramming of gene expression, allowing the cell to adapt and survive (Yamasaki and Anderson, 2008). Cells exposed to various stresses respond with the immediate shutoff of general protein synthesis, mediated by the inhibition of translation initiation via two key regulatory mechanisms. The first mechanism involves phosphorylation of the α-subunit of the eukaryotic initiation factor 2 (eIF2α, also known as EIF2S1) by one or more stress-activated eIF2α kinases (eIF2αKs) (reviewed in Holcik and Sonenberg, 2005; Ivanov et al., 2011; Sonenberg and Hinnebusch, 2009). eIF2 is a component of the ternary complex that delivers initiator tRNA to translationally-competent pre-initiation complexes assembled at the 5′-ends of mRNAs (Jackson et al., 2010). Phosphorylation of eIF2α at S51 prevents GDP/GTP exchange on eIF2 and, consequently, it fails to deliver initiator tRNA to ribosomes for start codon recognition, ultimately leading to a reduction in global translation initiation. In mammalian cells, four different eIF2αKs are known (Donnelly et al., 2013), each of which is activated by a distinct set of endogenous or exogenous stimuli. Heme-regulated initiation factor 2α kinase (HRI; also known as eIF2αK1) monitors the synthesis of globin chains in order to balance them with available heme levels during erythrocyte maturation (McEwen et al., 2005), and also senses oxidative stress. Protein kinase RNA-activated (PKR; also known as eIF2αK2) is a double-stranded RNA-dependent eIF2αK activated by viral infection, heat shock and ultraviolet irradiation (Srivastava et al., 1998). PKR-like endoplasmic reticulum (ER) kinase (PERK; also known as eIF2αK3) is activated by disruption of protein homeostasis in the ER lumen (Harding et al., 2000a,b). General control non-derepressible 2 (GCN2; also known as eIF2αK4) monitors amino acid levels and is activated by amino acid deprivation (Wek et al., 1995).
The second stress-sensitive mechanism of translation initiation involves the assembly of the eIF4F (i.e. eIF4E–eIF4G–eIF4A) complex that recognizes the cap (m7GTP) structure at the 5′-end of an mRNA (Jackson et al., 2010). Formation of this complex is under the stringent control of the phosphoinositide 3-kinase (PI3K) mammalian target of rapamycin (mTOR) kinase cascade (Laplante and Sabatini, 2013). Under growth conditions, the mTOR cascade results in phosphorylation of eIF4E-binding proteins (4E-BPs) to maintain its inactive phosphorylated form (p-4E-BPs). Stress-induced inactivation of mTOR leads to dephosphorylation of p-4E-BPs and their conversion into an active form that prevents the assembly of eIF4F and inhibits translation initiation (Jackson et al., 2010). It should also be noted that, beyond these two global translational control mechanisms, specific RNA-binding proteins contribute to the regulation of translation of the selected subset of mRNAs.
As a consequence of stress-induced translational arrest, mRNAs released from disassembled polysomes can be actively routed into discrete cytoplasmic foci known as stress granules (SGs) (Kedersha et al., 1999). SGs are microscopically visible foci composed of messenger ribonucleoproteins (mRNPs), including mRNA, small 40S ribosomal subunits, mRNA-associated translation initiation complexes and RNA-binding proteins (Anderson and Kedersha, 2006, 2008; Anderson et al., 2015; Kedersha et al., 2013). SGs are dynamic entities that are in equilibrium with actively translating polysomes; thus, translational control is tightly connected to SG assembly and disassembly. SG proteins not only determine the fate of specific transcripts that shuttle in and out of SGs, but also modulate various signaling cascades in stressed cells. Dysregulation of SG dynamics is implicated in the pathogenesis of a number of human diseases including cancer, inflammatory, neurodegenerative and neuromuscular diseases (Anderson and Kedersha, 2002, 2009; Anderson et al., 2015; Aulas and Vande Velde, 2015; Buchan, 2014; Ivanov and Anderson, 2013; Kedersha et al., 2013; Panas et al., 2016).
Phosphorylated eIF2α (p-eIF2α)- and mTOR-mediated pathways play complementary ‘checkpoint’ roles in general translational control, but also allow specialized control for specific subsets of mRNA. For example, phosphorylation of eIF2α selectively enhances the translation of some stress-responsive mRNAs bearing upstream open reading frames (uORFs) preceding the start AUG codon of the coding ORF (e.g. ATF4 in mammals or GCN4 transcripts in yeast; Holcik and Sonenberg, 2005; Yamasaki and Anderson, 2008). Similarly, specific mRNAs that bear an internal ribosome entry site (IRES) in their 5′-untranslated region (5′-UTR) escape 4E-BP-mediated inhibition, since their translation initiation is independent of eIF4F assembly on the mRNA cap structures. IRESs are commonly found in viruses and are used as a means to ensure that viral transcripts are still translated during periods of time when host translation is inhibited (Pestova et al., 2001). IRES-like structures can also be found in human transcripts, including transcripts that encode apoptosis-related and stress-responsive proteins, although whether these structures are bona fide IRESs is still a matter of debate (Shatsky et al., 2014, 2010). The molecular mechanisms of non-canonical translation have only started to be dissected.
To study the molecular mechanisms of translational control during stress, we sought a stress-responsive cellular model that can reliably be used for biochemical studies in vitro, and is also genetically tractable for various genetic (e.g. by RNAi or CRISPR/Cas9) and pharmacological manipulations. Commercially available in vitro translation (IVT) systems based on rabbit reticulocyte lysate (RRL) are widely used to study mammalian translation mechanisms. Although this system is robust and easy to use, it is artificial, non-human and cannot be genetically manipulated. More importantly, it does not replicate the stimulatory synergistic effects of the cap structure and poly(A) tail for mRNA translation (Michel et al., 2000). Other IVT systems utilize cytoplasmic extracts from mouse embryonic fibroblasts (MEFs) or mouse Krebs-2 ascites cells. In comparison to RRL, these systems faithfully recapitulate certain aspects of in vivo mRNA translation [e.g. 5′-cap and 3′-end poly(A) tail synergy] (Michel et al., 2000). However, these systems are murine and derived from specialized cells (thus not recapitulating many aspects of somatic cells, e.g. the stress response) and difficult to genetically manipulate. Finally, diverse human cell lines can be used to study translational control mechanisms under stress (Terenin et al., 2013). These cells can be genetically manipulated and used for preparation of translationally competent cell extracts. However, they are genetically heterogeneous (e.g. they often contain extra chromosomes or large sections of chromosomes), often refractory to efficient gene silencing (e.g. primary cells) and not convenient for microscopic studies [e.g. suspension cells for the detection of SG proteins by immunostaining, or transcripts by fluorescence in situ hybridization (FISH)] (Kedersha and Anderson, 2007).
Here, we utilize the near-haploid human HAP1 cell line derived from chronic myelogenous leukemia cells (Carette et al., 2011) as a tool to study translational control and stress responses. The genome of these cells has been fully sequenced and thus the cells are ideal for genetic manipulations such as gene deletion or site-specific mutagenesis (Carette et al., 2011), both of which are easier to facilitate by the absence of a second allele. We describe the properties of a HAP1-derived in vitro mRNA translation system, characterize HAP1-derived sublines with genetically ablated eIF2αKs HRI, PKR, PERK or GCN2, as well as HAP1 knock-in cells containing a S51A mutation in the eIF2α-encoding gene (S51A HAP1). We determined the utility of HAP1 cells for monitoring SG dynamics in response to various stresses, and reveal previously unappreciated aspects of SG assembly and inhibition of translation. Specifically, we found that some stresses are strictly dependent on eIF2α phosphorylation for SG formation while others are not. Interestingly, rocaglamide A (RocA), an agent that inhibits translation in an eIF2α-independent manner through interference with the RNA helicase eIF4A, induces formation of cytoplasmic foci that are positive for core SG markers but negative for poly(A) mRNAs. Similarly, treatment with NaCl also does not require eIF2α phosphorylation to inhibit translation and induces the assembly of poly(A)-positive cytoplasmic foci that compositionally resemble canonical SGs (Kedersha et al., 2016). However, NaCl-induced foci are refractory to drugs that distinguish bona fide SGs from other RNA granules. Of the stresses that are dependent on p-eIF2α, some [e.g. heat shock and proteasome inhibitor MG132 (Mazroui et al., 2007)] activate more than one eIF2αK whereas others [e.g. sodium arsenite (SA) or thapsigargin (Thaps) (Kedersha and Anderson, 2007)] activate a single eIF2αK. UV light triggers phosphorylation of eIF2α, but this is not required for SG formation, and UV only partially inhibits translation. Moreover, UV-induced SGs are not canonical as they lack translation initiation factors eIF4G and eIF3 and only weakly recruit poly(A) mRNAs [note that such foci may contain deadenylated mRNA, as is the case of processing bodies (PBs)]. These data indicate that mammalian cells assemble distinct types of SGs and SG-like granules in a stress-specific manner. It also cautions against declaring stress-induced foci to be SGs based only on one or two SG markers. Our data also demonstrate that HAP1-derived cells and cell-free systems are valuable tools for studying aspects of protein synthesis, translation-coupled stress responses and mechanisms of RNA granule assembly. They constitute an ideal system for genetic and/or chemical screens to investigate translational control.
RESULTS
Monitoring protein synthesis in HAP1 cells
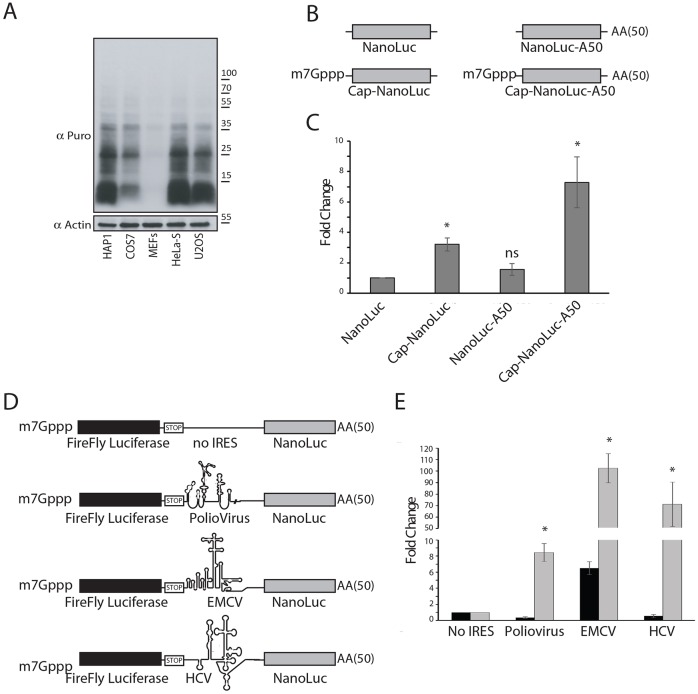
HAP1 cells have been used in genetic screens and ‘loss-of-function’ studies for various purposes (Elling and Penninger, 2014). To determine whether these cells can also be used for studies related to protein synthesis and RNA granule assembly, we first examined the efficiency of general translation in HAP1 cells using RiboPuromycylation, an assay that labels nascent peptide chains on elongating ribosomes (Panas et al., 2015). We compared HAP1 cells to other cell lines that are widely used to study mRNA translation, and found that HAP1 cells efficiently support de novo protein synthesis at efficiencies comparable to those observed in HeLa-S, COS7 and U2OS cells, and exceeding those of MEFs (Fig. 1A).
Fig. 1.
HAP1 cells are suitable for cellular and in vitro translation assay. (A) HAP1, COS7, MEFs, HeLa-S and U2OS cells were subjected to RiboPuromycylation to compare levels of basal translation. An anti-puromycin antibody (Puro) was used to visualize de novo synthesized proteins. Actin is a loading control. A representative image is shown (n=3). (B) Schematic of NanoLuc-based luciferase mRNA reporters with cap structures (Cap-NanoLuc), a poly(A) tail (NanoLuc-A50), a cap and poly(A) (Cap-NanoLuc-A50) or without a 5′-cap or 3′-poly(A) tail (NanoLuc). (C) IVT system based on HAP1 lysates was used to assess 5′-cap or 3′-poly(A) tail synergy using in vitro transcribed reporters from B. Relative translation efficiency of NanoLuc mRNA is set as 1. n≥3. *P<0.05; ns, not significant compared with NanoLuc (unpaired Student's t-test). (D) Schematic of bicistronic constructs used. The first ORF encoding firefly luciferase is translated in cap-dependent manner, the second ORF encoding NanoLuc luciferase is translated in IRES-dependent (for Polio virus, EMCV or HCV IRESs) manner. Control reporter (no IRES) contains no IRES element between the ORF encoding firefly and NanoLuc luciferases. (E) IVT system based on HAP1 lysates was used to assess translation of in vitro transcribed bicistronic reporters compared with the relative translation efficiency of firefly luciferase (black columns) or NanoLuc luciferase (gray) mRNAs is set as 1, respectively. The relative translation of IRESs over No IRES is shown. n≥3; *P<0.05; unpaired Student's t-test. Quantitative results are mean±s.e.m.
Cell-free IVT systems utilizing cellular extracts are commonly used to dissect mechanisms of protein synthesis. Different systems have been developed, yet many do not fully recapitulate all aspects of translational control and regulation that are known to exist. To determine whether HAP1 cells can be used for IVT, we assessed the translational competence of HAP1 extracts using a variety of reporters. In mammalian cells, mRNA translation is dramatically enhanced by the presence of a 5′-cap and 3′-poly(A) tail. We tested whether these elements exhibit these properties in HAP1 extracts by using an mRNA reporter encoding NanoLuc luciferase (NanoLuc) (Fig. 1B). Addition of a 5′-cap or poly(A) tail to uncapped mRNA potently enhances its translation (Fig. 1C, compare NanoLuc-A50 and Cap-NanoLuc to Nano-Luc). Importantly, the presence of both the 5′-cap and poly(A) tail stimulate translation synergistically (Fig. 1C, compare Cap-NanoLuc-A50 to NanoLuc-A50 and Cap-NanoLuc), in agreement with accepted models. Also of note, HAP1 extracts do not require pre-treatment with nucleases (e.g. micrococcal nuclease), which eliminates endogenous cellular transcripts and results in a non-competitive and artificial system.
We next asked whether HAP1 extracts support translation driven by different viral IRESs (Fig. 1D). We utilized bicistronic mRNA reporters in which the first ORF encoding firefly luciferase is driven in a cap-dependent (canonical) manner, while the second ORF encoding NanoLuc luciferase is driven by an IRES. HAP1 lysates efficiently translate luciferase driven by hepatitis C virus (HCV), encephalomyocarditis virus (EMCV) and poliovirus (PV) IRESs, with translation efficiencies comparable to those of the capped and poly(A) mRNAs (Fig. 1E). Taken together, our data indicate that HAP1 cells and a HAP1-derived IVT system can be used to study many aspects of translation regulation.
HAP1 cells form canonical SGs in response to sodium arsenite
As translation inhibition is coupled with the formation of SGs (Kimball et al., 2003), we monitored SG dynamics in HAP1 cells exposed to oxidative stress. Cells were treated with SA, the most commonly used agent to induce oxidative stress and a robust SG-inducing agent, for 1 h and then returned to regular media. SA-treated HAP1 cells assemble SGs, which disassemble when stress is removed, in accordance with the transient nature of SGs (Fig. S1A). SG formation correlates with translation inhibition, as measured by RiboPuromycylation and phosphorylation of eIF2α during SA treatment. SGs disassembly parallels restoration of protein synthesis and dephosphorylation of eIF2α following stress release (Fig. S1B). As seen in other mammalian cell lines, HAP1 cells form canonical SA-induced SGs, containing the SG-nucleating proteins G3BP1 and Caprin1, translation initiation factors eIF4G and eIF3b, poly(A)-binding protein cytoplasmic 1 (PABPC1), and TIA-1 (Fig. S1C). However, we were not able to detect TDP-43 (also known as TARDBP) in SA-induced SGs (Fig. S1C). FISH using an oligo(dT) probe shows poly(A) mRNA colocalized with the SG markers G3BP1 (Fig. S1C, left image). Taken together, this data indicates that SA-treated HAP1 cells form canonical SGs that coincide with translation inhibition.
Assessing compositional diversity of SGs and SG-like foci in response to different stress stimuli
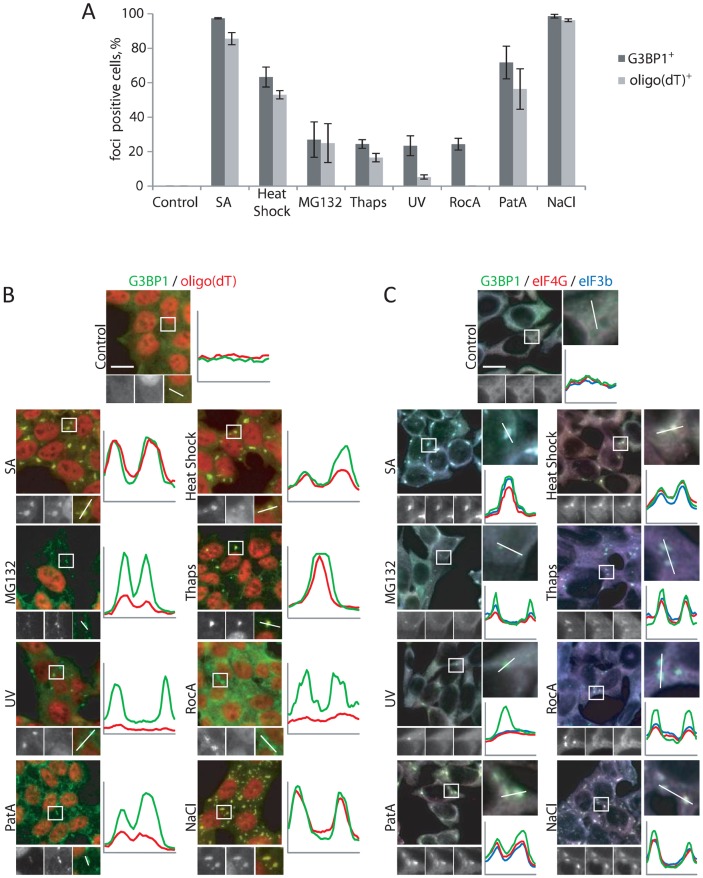
The list of SG-promoting stimuli and cell lines that are used for SG studies is extensive and continues to grow (Aulas and Vande Velde, 2015). Most studies presume that SGs are uniformly the same despite differences in stressors or cell type. However, some evidence shows that their composition varies according to the stress (Kedersha et al., 1999). These differences have not been systematically assessed in a unified system. We therefore employed a panel of HAP1 cells to investigate the stress-specific compositional diversity of SGs. Oxidative stress (SA) (Kedersha et al., 1999), heat shock (Kedersha et al., 1999), ER stress (Thaps) (Kimball et al., 2003), proteasome inhibition (MG132) (Mazroui et al., 2007), hyperosmotic stress caused by NaCl (Kedersha et al., 2016), UV radiation (Kwon et al., 2007), inhibition of eIF4A through Pateamine A (PatA) (Dang et al., 2006) and RocA (Kedersha et al., 2016) all induce G3BP1-positive foci in HAP1 cells. The frequency of G3BP1-positive foci induction is variable and stress-dependent, ranging from 20% of cells treated with Thaps, MG132 and UV to over 90% in the case of SA and hyperosmotic stress (Fig. 2A,B). As poly(A) mRNA is a defining component of bona fide SGs (Kedersha et al., 1999), we investigated the various stress-induced G3BP1-positive foci using FISH for poly(A) mRNA with an oligo(dT) probe. Most of the stresses promote recruitment of poly(A) mRNA to G3BP1-positive foci (Fig. 2A,B). However, RocA- and UV-induced G3BP1-positive foci contain little or no polyadenylated mRNA (Fig. 2A,B), suggesting that these G3BP1-positive foci are not canonical SGs. As histone mRNAs contain a stem-loop instead of a poly(A) tail, we assessed the SGs for stem-loop-binding protein (SLBP), a protein that binds to the histone mRNA stem-loop (Marzluff et al., 2008). Only MG132, Thaps and osmotic stress recruit SLBP to SGs (Fig. S2), indicating the presence of histone RNA in these foci. Whether RocA and UV foci contain other non-polyadenylated mRNA remains to be determined.
Fig. 2.
Composition of stress-induced foci. (A) HAP1 cells were subjected to treatment with SA (200 µM, 1 h), heat shock (44°C, 1 h), the proteasome inhibitor MG132 (100 µM, 1 h), the endoplasmic reticulum stressor Thaps (4 µM, 2 h), the eIF4A inhibitors RocA (2 µM, 2 h) and pateamine A (PatA, 0.5 µM, 1 h), or were subjected to osmotic stress by treatment with NaCl (0.2 M, 1 h). Unstressed cells (Control) were used as a control. Cells were examined for the presence of he core SG marker G3BP1 (immunofluorescence using G3BP1-specific antibody) and poly(A) mRNAs [FISH using oligo(dT) probe]. G3BP1- or oligo(dT)-positive cells were quantified. Results are mean±s.e.m. (n=3). (B) Representative images of HAP1 cells stained with G3BP1 (green) and oligo(dT) (red) after the cells had been subjecting to the specific stresses. The boxed region is enlarged and line scans used to assess colocalization (separate colors shown on graphics) of markers. (C) Representative images of HAP1 cells stained with G3BP1 (green), eIF4G (red) and eIF3B (blue) after the cells had been subjected to the specific stresses or left untreated (Control). The boxed region is enlarged and line scans used to assess colocalization (separate colors shown on graphics) of the markers.
We also assessed the presence of other protein SG markers including eIF3b, eIF4G, PABP and TIA-1 (Fig. 2C, Fig. S2). We find that all tested stresses induced recruitment of TIA-1 and G3BP1 to stress-induced foci (Fig. 2C; Fig. S2, and data not shown), whereas SGs induced in response to RocA, MG132 and UV contain less eIF3b and eIF4G (Fig. 2C; Fig. S2).
We also monitored formation of PBs, cytoplasmic RNA granules distinct from SGs (Anderson and Kedersha, 2006; Anderson et al., 2015; Stoecklin and Kedersha, 2013), under different stresses using two different PB markers (Hedls, also known as EDC4, and Dcp1a) (Fig. S3A,B). PBs are present in HAP1 cells both under control conditions and under stress (Fig. S3A). SA and, to a lesser extent, NaCl promote PB formation, while heat shock, MG132, UV and RocA treatments decrease the number of PB-positive cells. In contrast, Thaps and PatA do not influence PB formation (Fig. S3B). Interestingly, under UV treatment, PBs are only present in the cells lacking any SG-like foci (as assessed by TIA-1 staining).
Relationship between SG-like foci formation and translation
Formation of SGs is coupled to general translation repression. Both eIF2α-dependent and -independent stimuli can inhibit translation initiation and promote SG formation (Dang et al., 2006; Kedersha et al., 1999). Ribosomes must run off transcripts before mRNPs are assembled into SGs, as drugs that preserve polysomes [e.g. cycloheximide (CHX) treatment] or promote their disassembly [e.g. puromycin (Puro) treatment] inhibit or promote SG formation, respectively (Kedersha et al., 2000). Accordingly, we assessed whether stress-specific formation of SGs and SG-like foci is dependent on eIF2α phosphorylation and is in dynamic equilibrium with translating polysomes.
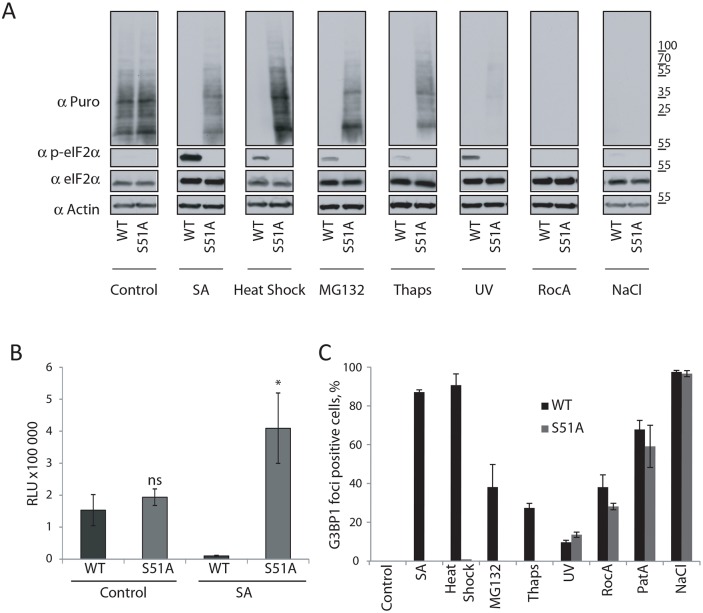
CRISPR/Cas9 was used to create HAP1 cells that bear only a non-phosphorylatable variant of eIF2α (S51A) (Fig. S4A, upper panel). These cells do not demonstrate any detectable differences in morphology, rate of growth or viability under unstressed conditions compared with wild-type (WT) HAP1 cells (data not shown). WT and S51A HAP1 cells express similar levels of eIF2α protein (Fig. 3A, control), demonstrate comparable levels of basal translation in vivo (Fig. 3A, control) and in IVT lysates (Fig. 3B, control). Treatment of the cells with SA, which triggers eIF2α phosphorylation in WT but not S51A cells, potently inhibits translation in lysates prepared from WT but not S51A HAP1 cells (Fig. 3B, right panel).
Fig. 3.
Relationship between stresses, eIF2α phosphorylation, translation inhibition and stress foci formation. (A) WT or eIF2αS51A (S51A) HAP1 cells were subjected to treatment with SA (200 µM, 1 h), heat shock (44°C, 1 h), MG132 (100 µM, 1 h), Thaps (4 µM, 2 h), RocA (2 µM, 2 h), PatA (0.5 µM, 1 h) or NaCl (0.2 M, 1 h). Cells were pulsed with puromycin for 5 min and lysed. Cell lysates were subjected to western blotting using antibodies for puromycin (Puro), p-eIF2α, total eIF2α and actin. Representative images are shown (n≥3). (B) IVT assay of NanoLuc mRNA reporter based on cell lysates prepared from WT and S51A HAP1 cells that were treated with SA or were left untreated (Control). Relative luciferase units are shown. n=3. *P≤0.05 compared with SA-treated WT (unpaired Student's t-test). (C) WT and S51A HAP1 cells were assessed for G3BP1-positive foci by immunofluorescence using G3BP1. Percentage of cells with G3BP1-positive foci is shown. n≥3. Quantitative results are mean±s.e.m.
We then compared WT and S51A HAP1 cells after subjecting them to the panel of stress-inducing conditions. All stresses dramatically repress translation in WT cells as indicated by RiboPuromycylation (Fig. 3A). The level of translation repression correlates with the ability of SA, heat shock, MG132, Thaps and UV to trigger eIF2α phosphorylation in WT cells. As expected from other studies, RocA and osmotic stress do not affect p-eIF2α levels (Fig. 3A), but still inhibit translation. While SA, heat shock, MG132 and Thaps do not effectively repress translation in S51A cells and are thus p-eIF2α-dependent, UV inhibits translation in S51A cells despite triggering strong eIF2α phosphorylation in WT cells. Thus, UV appears to inhibit translation using both p-eIF2α-dependent and -independent mechanisms. RocA and osmotic stress also repress translation in S51A cells (Fig. 3A) confirming that these stresses inhibit translation in a p-eIF2α-independent manner.
We also monitored formation of SGs and SG-like foci in WT and S51A HAP1 cells (Fig. 3C) after subjecting them to the panel of stress conditions. WT, but not S51A cells, readily promote SG formation in response to SA (Figs 3C and 4A–C), in agreement with published data. Similar analysis demonstrated that SA, heat shock, MG132 and Thaps induce formation of stress-induced foci in a p-eIF2α-dependent manner, whereas UV, RocA, PatA and osmotic stress induce the formation of stress-induced foci in a p-eIF2α-independent manner (Fig. 3C). In summary, our data indicate that formation of stress-induced foci generally correlates with stress-induced translational repression.
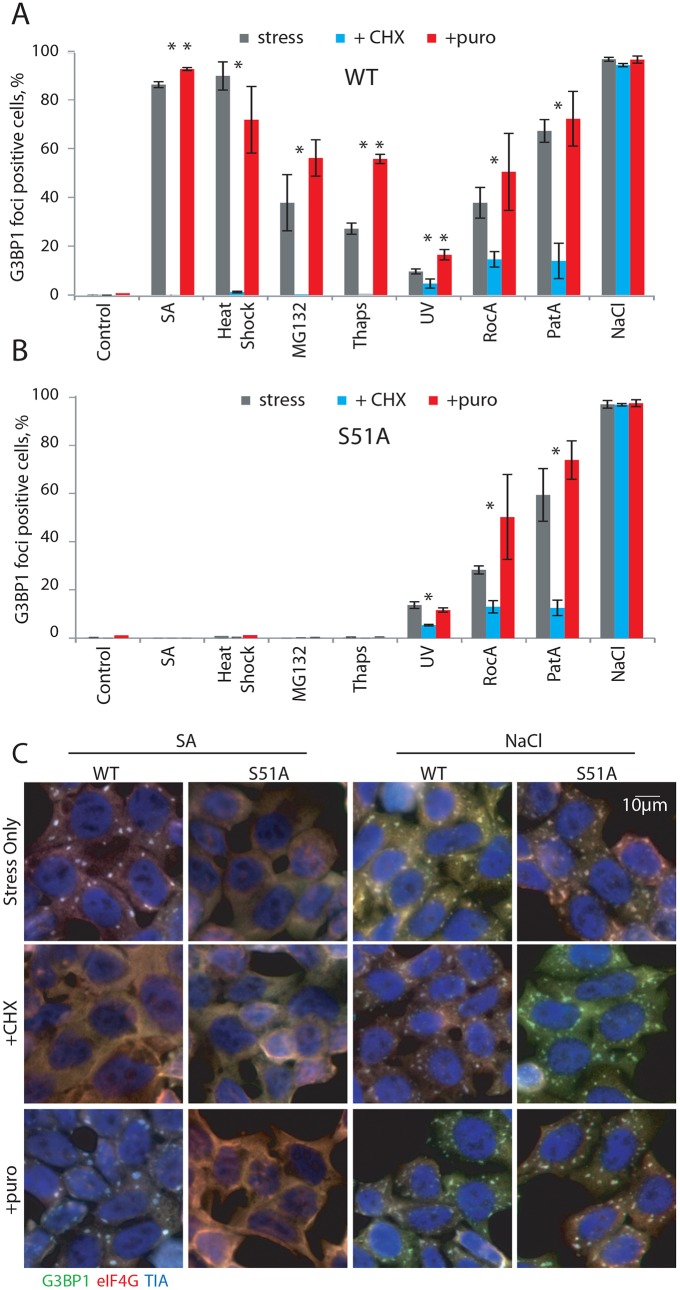
Fig. 4.
Translation-dependent dynamics of SGs and stress-induced foci. (A,B) WT (A) and S51A (B) HAP1 cells were treated with SA, heat shock, MG132, Thaps, UV, RocA, PatA or NaCl as previously described (Fig. 3C). 30 min before collection cells were treated with CHX (50 µg/ml) or puromycin (20 µg/ml). Cells were assessed by G3BP1 staining and plotted as percentage on the graphs according to color code: gray, stress only; blue, stress with CHX; red, stress with puromycin. Results are mean±s.e.m., n≥3. *P≤0.05 compared with stress alone (unpaired Student's t-test). (C) Representatives images of cells from B stained with G3BP1, eIF4G and TIA-1.
We next used CHX or puromycin treatments to determine whether stress-induced foci are in equilibrium with polysomes. As expected, CHX effectively promotes enforced disassembly of SA-induced SGs, even in the continued presence of SA in WT cells (Fig. 4A,C, +CHX). Similarly, CHX effectively induces the disassembly of other p-eIF2α dependent SGs (MG132-, Thaps-, heat shock-induced), and partially disassembles PatA-, RocA- and UV-induced foci (Fig. 4A,B, +CHX), but does not affect osmotic-induced foci (Fig. 4A–C). While formation of SA-induced (at lower concentration, data not shown) and Thaps-induced SGs display statistically significant increases upon puromycin treatment, puromycin does not significantly increase MG132-, UV-, RocA- and PatA-induced SG assembly (Fig. 4A,B). Again, osmotic-induced foci are not affected by puromycin treatment (Fig. 4A–C).
HAP1 cells with knockout of specific eIF2αKs
MEFs obtained from mouse models with knockouts (KO) of individual eIF2αKs have been used to study cellular stress responses, mechanisms of translation inhibition and SG formation. However, such KO MEFs are non-human and heterogeneous (coming from different genetic backgrounds). Here, we used commercially available HAP1 variants lacking each of the known eIF2αKs – denoted ΔHRI, ΔPKR, ΔPERK and ΔGCN2 (eIF2αK1–eIF2αK4) to study the specificity and redundancy of each eIF2αK in response to specific stresses.
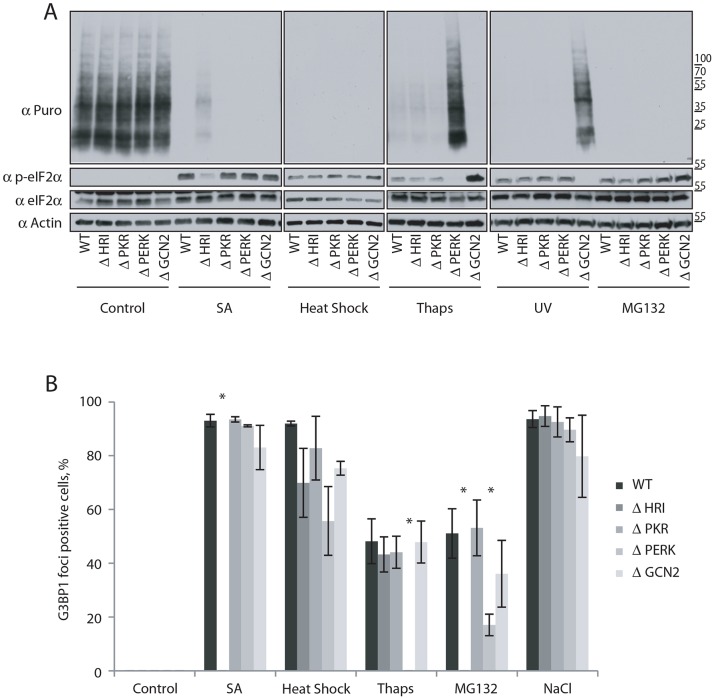
We verified the genomic alterations by sequencing the locus of interest (Fig. S4A), and also confirmed the lack of PKR, PERK and GCN2 protein expression by western blotting (Fig. S4B). Unfortunately, we could not find an HRI-specific antibody that consistently worked (data not shown). All KO cell lines exhibit comparable levels of general translation, and demonstrate the absence of eIF2α phosphorylation under basal conditions (Fig. 5A, control panel). We then treated KO cells with the stresses that cause eIF2α phosphorylation (SA, heat shock, Thaps, MG132 and UV) and monitored levels of translation in these cells. In response to SA, all the cell lines display inhibited translation, with the exception of ΔHRI cells. Similarly, translation inhibition by Thaps and UV requires a single eIF2αK, PERK and GCN2, respectively (Fig. 5A). In contrast, MG132 and heat shock efficiently inhibit translation in every KO cell line, suggesting they activate more than one eIF2αK or have other eIF2α-independent effects on translation. Similarly, no general translation and p-eIF2α level differences were observed between the different eIF2αK-KO and WT cell lines in the case of heat shock or MG132 treatment (Fig. 5A).
Fig. 5.
Determination of the eIF2αK activated in response to stresses. (A) WT, ΔHRI, ΔPKR, ΔPERK and ΔGCN2 mutant cells were exposed to SA, heat shock, Thaps, MG132, UV or NaCl as described in previous figures. Cells were pulsed with puromycin for 5 min before lysis. Whole-cell extract was analyzed by western blotting for puromycin, p-eIF2α, total eIF2α and actin, n≥3. (B) Cells positive for G3BP1 staining were analyzed as in Fig. 2B. Results are mean±s.e.m., n≥3. *P≤0.05 compared with WT (unpaired Student's t-test).
We next assessed the ability of KO cells to induce SGs or SG-like foci in a stress-specific manner (Fig. 5B). For SA and Thaps, the kinase involved in translation repression is also involved in SG formation (HRI and PERK, respectively). NaCl, used here as a control, induces hyperosmotic stress and inhibits translation in a p-eIF2α-independent manner (Bevilacqua et al., 2010) and does not require any eIF2αK to promote SG formation. MG132 elicits reduced SG formation in both ΔHRI and ΔPERK cell lines, consistent with its activation of more than one eIF2αK (Fig. 5B). After heat shock, none of the KO cells demonstrate a significant difference in their ability to induce SGs (Fig. 5B), consistent with the data in Fig. 5A. This suggests that heat shock activates multiple eIF2αKs, or perhaps inactivates the phosphatases that dephosphorylate p-eIF2α. Moreover, heat shock may also work through alternative p-eIF2α-independent pathways that rely on the activation of 4E-BPs (Sukarieh et al., 2009).
Cell survival and SGs
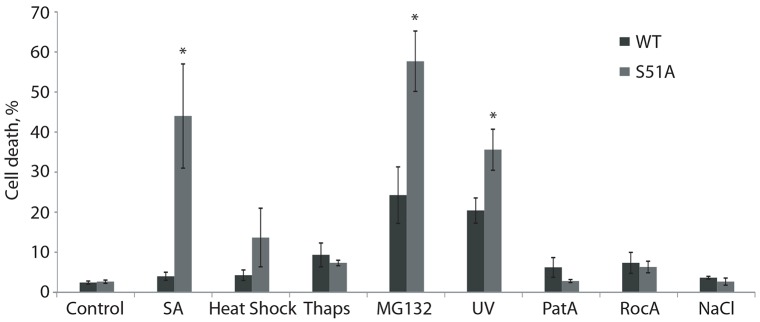
SGs are biomarkers of an adaptive response. If successful, adaptation to stress is pro-survival, leading to the prediction that defects in SG formation (either no formation or lack of secondary aggregation) may result in decreased cell survival (Aulas and Vande Velde, 2015). Other data suggest that some stresses (such as osmotic stress) can induce ‘defective’ SGs in cells unable to phosphorylate eIF2α, resulting in apoptosis instead of survival (Bevilacqua et al., 2010). We analyzed the stress-specific differences on the survival of WT and S51A HAP1 cells. Cells were subjected to acute stresses as described, and the cell medium was replaced and cells were allowed to recover for 24 h before assessing cell death through Trypan Blue exclusion. Stresses that induce stress granules independently of p-eIF2α (RocA, PatA and NaCl; Fig. 6) did not induce cell death in either WT or S51A cells. Of the p-eIF2α dependent stresses, SA, heat shock and Thaps did not induce cell death in WT HAP1 cells while MG132 and UV did. With the exception of Thaps, all p-eIF2α-dependent stresses promote significantly more cell death in S51A cells than in WT cells.
Fig. 6.
Stress-specific influences on cell death in WT and S51A HAP1 cells. WT or eIF2αS51A mutant cells were exposed to SA, heat shock, Thaps, MG132, UV, PatA, RocA or NaCl. After stress, the medium was changed and cell death was assessed 24 h later by Trypan Blue exclusion. Results are mean±s.e.m., n≥3. *P≤0.05 compared with WT (unpaired Student's t-test).
DISCUSSION
Historically, many SG studies have been performed in various cell lines of both human and murine origin. Although such studies are informative, the genetic diversity of murine cell lines contributes to the observed heterogeneity in the results. In addition, while some large flat cells (like osteosarcoma U2OS cells widely used for RNA granule studies) are ideal to study SGs using immunofluorescence, their aneuploidy complicates the use of genetic manipulations to dissect molecular pathways. We have used human haploid HAP1 cells, the genomes of which have been fully sequenced and are genetically tractable due to the presence of only one allele of each gene, to study SGs, mechanisms of translation and stress responses. In response to SA treatment, a classical trigger of SG formation, canonical SGs are robustly induced in nearly 100% of WT HAP1 cells (Fig. S1A,C). HAP1 SA-induced SGs are canonical in their composition, dependence on p-eIF2α and dynamic behavior. First, SA-induced SG composition is indistinguishable from that of SGs reported in other cell types (e.g. U2OS or HeLa cells). HAP1 SA-induced SGs contain poly(A) mRNA, translation initiation factors eIF4G, eIF3b, PABP and SG markers G3BP1, Caprin1 and TIA-1 (Fig. S1C). Upon stress removal, SA-induced SGs are efficiently dissolved, demonstrating their dynamic and stress-responsive nature. Second, formation of SA-induced SGs in HAP1 cells correlates with increased p-eIF2α and subsequent translational repression (Fig. S1B). Third, HAP1-derived IVT lysates support both canonical [5′-cap and 3′-poly(A) tail] and non-canonical (IRES-driven) translation initiation (Fig. 1). Crucially, a HAP1-based IVT system faithfully recapitulates 5′-cap and poly(A) tail synergy, an important aspect of in vivo mRNA translation (Fig. 1B). Moreover, mRNA reporters were efficiently translated in the presence of other competitive mRNAs, since our protocol for HAP1 extract preparation does not involve micrococcal nuclease treatment.
Although SGs are often assumed to be uniform entities formed under different stresses, their protein and mRNA composition varies. For example, HSP27 is only found in SGs induced by heat shock, but is absent in SA-induced SGs (Kedersha et al., 1999). Whereas most mRNA is recruited to heat shock-induced SGs, transcripts encoding HSP70 proteins are selectively excluded (Kedersha and Anderson, 2002). HSP90 mRNA transcripts are actively excluded from SA-induced SGs (Stöhr et al., 2006). Tristetraprolin (TTP, also known as ZFP36) is recruited to SGs induced by energy starvation (induced by FCCP) or nucleated by TTP overexpression (Stoecklin et al., 2004). However, TTP and its target mRNAs are actively exported from SA-induced SGs, a consequence of SA-induced activation of MAPKAPK2, which phosphorylates two sites on TTP resulting in recruitment of 14-3-3 proteins, and expulsion from SGs (Stoecklin et al., 2004). Taken together, those studies conclude that foci formed under different stress are different and the foci composition depends on the stress used to induce them. We thus employed a panel of HAP1 cells to investigate specific differences in SG composition in response to a panel of diverse stresses, including oxidative and hyperosmotic stress, heat shock, ER stress, UV radiation, proteasome inhibition (MG132) and eIF4A inhibition (PatA and RocA). Surprisingly, we found that some stresses, such as hyperosmotic stress, UV and RocA, induce foci that only partially resemble bona fide SGs in their composition and behavior.
First, we showed that UV- and RocA-induced stress foci do not fit the general definition of RNA granules as they do not contain poly(A) mRNAs, although they still could contain deadenylated mRNAs (similarly to what is observed in PBs) or transcripts with short poly(A) tails [intermediates of poly(A) shortening]. These foci also inefficiently recruit eIF3b and eIF4G, core SG components. In contrast, other canonical SG markers such as G3BP1 and TIA-1 are present in UV- and RocA-induced stress foci. Moreover, as UV- and RocA-induced foci are partially disassembled upon treatment with CHX and enhanced upon puromycin treatment (Fig. 4), they are in equilibrium with polysomes (see below). Thus they may contain mRNA not detectable via oligo(dT) FISH, possibly mRNA with short or no poly(A) tails. Our data reemphasize the importance of using more than one SG protein marker and FISH to detect the presence of poly(A) RNA to categorize, and thus understand, the nature of each subtype of stress-induced foci.
Second, SGs are dynamic entities in equilibrium with polysomes. Puromycin enhances SG formation by promoting premature disassembly of polysomes, whereas CHX inhibits elongation and prevents polysome disassembly, thus preventing SG formation and forcibly disassembling pre-existing SGs. All HAP1 stress-induced foci, except those induced by osmotic stress, were at least partially dissolved by CHX. To the best of our knowledge, osmotic SGs constitute the first SG-like foci (by composition) that cannot be reversibly induced to disassemble to any extent upon treatment with CHX. Despite their inability to be affected by CHX or puromycin after they are formed, NaCl-induced foci are prevented by pretreatment with CHX (Kedersha et al., 2016), suggesting that they are more stable than canonical SGs. As the proposed mechanism of their formation is driven by molecular crowding due to osmotic stress (Bounedjah et al., 2012) and as their formation is independent of p-eIF2α (Bevilacqua et al., 2010; Kedersha et al., 2016), their stability may indicate an energetic barrier to SG disassembly.
This study is the first to extensively compare many different stresses in the same cellular background. We provide evidence that stress-induced foci could be different in composition, in terms of equilibrium with polysomes and in relation to cell survival. We anticipate that the HAP1 system will provide valuable tools to elucidate RNA granule biology, morphology, and biochemistry.
MATERIAL AND METHODS
Cell culture, drug treatment
Wild type, ΔHRI (cat # HZGHC000141c011), ΔPKR (cat # HZGHC000338c010), ΔPERK (cat # HZGHC002033c003), ΔGCN2 (cat # HZGHC001245c030) and eIF2αS51A HAP1 cells (Horizon, custom service preparation) were maintained at 37°C in a CO2 incubator in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum (Sigma), HEPES (20 mM pH 7.0, Gibco) and 1% penicillin-streptomycin. U2OS, an osteosarcoma line, was obtained from the ATCC. HAP1 and U2OS cells were expanded and frozen down in individual aliquots for future use. Only one cell type is used in a tissue culture hood at any given time. This helps to avoid contamination with other cell lines. At monthly intervals, cells are screened for mycoplasma contamination. For SG induction, cells were grown to ∼70% confluency then treated with sodium arsenite (SA, 200 µM, 1 h), NaCl (0.2 M, 1 h), thapsigargin (Thaps, 4 µM, 2 h), rocaglamide A (RocA, 2 µM, 2 h), MG132 (100 µM, 1 h or 4 h), pateamine A (PatA, 0.5 µM, 1 h), UV (200 J/m2 using a Stratolinker then released 1 h), heat shock (44°C, 1 h). Puromycin (20 µg/ml) and CHX (50 µg/ml) treatment was performed for 30 min before collecting the coverslips or as described in the text. For RiboPuromycylation (Panas et al., 2015), cells were pulsed with puromycin (5 µg/ml) for 5 min before lysis.
Genotyping
Cells were washed and lysed in genotyping lysis buffer (10 mM Tris-HCl pH 7.5, 10 mM EDTA, 10 mM NaCl, 0.5% SDS, 10 µg/ml proteinase K) for 3 h at 60°C. Then precipitation buffer (150 mM NaCl, 70% ethanol) was added to the previous mix and incubated at room temperature for 30 min. Samples were centrifuged at 10,000 g for 15 min at 4°C. DNA was washed with 70% ethanol then centrifuged at 10,000 g for 5 min at 4°C. The final pellet was resuspended in elution buffer and genotypes were assessed using the following primers: HRI, 5′-GGTGTTAAAAGAACCCCTACAACAG-3′ and 5′-GTAAAGAGGGGGTTTCGTCATGTTA-3′; PKR, ACTGTTTGAGGTGACTGCTTAAATG-3′ and 5′-TTGAATGTAAGGGAACGTGTGAATG-3′; PERK, 5′-CTCTTGTGGCATAAATCAGT-3′ and 5′-AATGCCATAACTTTCCAGTC-3′; GCN2, 5′-GAACGAATGGAAGCTGAGT-3′ and 5′-AACATCTATTGCTGATGTAG-3′; and eIF2αS51A, 5′-ATGTTTGCTCACTTCGGCAA-3′ and 5′-CCATTTGCCCCATTTTCATGC-3′.
Western blotting
Following drug treatment, cells were washed with phosphate-buffered saline (PBS) and sonicated in lysis buffer (50 mM Hepes pH 7.6, 150 mM NaCl, 0.5% NP40 and 5% glycerol) with Halt phosphatase and protease inhibitors (Thermo Scientific). Laemmli's sample buffer supplemented with 100 mM dithiothreitol (DTT) was added to samples to 1× final concentration. Samples were boiled before being loaded on a 4–20% Tris-Glycine gel (BioRad) and transferred to nitrocellulose membrane. Membranes were blocked with Tris-buffered saline with 0.1% Tween-20 (TBS-T) with 5% milk for at least 30 min at room temperature. Antibodies were diluted in 5% normal horse serum in PBS. Primary antibodies were incubated overnight at 4°C and secondary antibodies for 1 h at room temperature. Antibody information is listed in Table S1. Antibody detection was performed using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific). Scanning and Photoshop was used to quantify western blots that were used in the RiboPuromycylation assay.
Immunofluorescence
3×105 HAP1 cells were grown on coverslips, washed with PBS and then fixed with −20°C methanol for 15 min. Some antibodies required fixation with 4% formaldehyde, and, for those, cells were permeabilized with 0.1% Triton X100 for 15 min (Table S1). Coverslips were blocked with 5% normal horse serum for at least 30 min. Primary antibodies were diluted in blocking buffer and incubated overnight at 4°C. Subsequently, secondary antibodies were added at a 1:250 dilution along with Hoechst 33342 for 1 h at room temperature. Cells were washed extensively and mounted with Vinol mounting medium.
In situ hybridization
For in situ hybridization, cells were fixed with 4% paraformaldehyde for 15 min then permeabilized with −20°C methanol for 15 min. Cells were incubated at least overnight in 70% ethanol at 4°C. The following day, cells were washed twice with 2× saline-sodium citrate (SSC), blocked in hybridization buffer (Sigma) for 30 min, then hybridization was performed using a biotinylated oligo(dT40) probe (2 ng/µl) diluted in hybridization buffer at 37°C. After extensive washes with 2× SSC at 37°C the probe was revealed using Cy-conjugated streptavidin (Jackson Immunoresearch Laboratories), followed by immunostaining as described above.
Microscopy
Wide-field fluorescence microscopy was performed using an Eclipse E800 microscope (Nikon) equipped with epifluorescence optics and a digital camera (Spot Pursuit USB). Image acquisition was performed with a 40× objective (PlanApo; Nikon).
Generation of line scans
Colocalization (line scans) was assessed using ImageJ. A line was drawn across the SG and intensity was measured over the line using the Plot Profile option and results were exported to Excel to generate graphs. The arbitrary intensity was plotted according to arbitrary distance for each channel.
Quantification
Quantification of the percentage of SG-positive cells was performed with ImageJ by counting the number of cells with at least two discrete G3BP1-positive foci from >200 cells per condition per experiment, from at least three independent experiments.
In vitro translation assay
To generate translation extracts, cells are trypsinized and washed in HBSS buffer and then resuspended in lysolecitin lysis buffer [20 mM HEPES-KOH pH 7.4, 100 mM KOAc, 2.2 mM Mg(OAc)2, 2 mM DTT and 0.1 mg/ml lysolecitin] for 1 min and centrifuge 10 s, 10,000 g at 4°C. The pellet was resuspended in hypotonic extraction buffer [20 mM HEPES-KOH pH 7.4, 10 mM KOAc, 1 mM Mg(OAc)2, 4 mM DTT with protease inhibitor] and incubated on ice for 10 min, then passed through a G-27 needle and centrifuged for 10 min at 10,000 g at 4°C.
Translation assays were performed by incubating translation extract with translation reaction buffer [20 mM HEPES-KOH pH 7.6, 1 mM DTT, 0.5 mM spermidine-HCl, 0.6 mM Mg(OAc)2, 8 mM creatine phosphate, 1 mM ATP, 0.2 mM GTP, 120 mM KOAc, 25 µM amino acid mix], 100 ng of reporter RNA and 2 U RNase inhibitor at 30°C for 1 h. Assays were read with GloMax EXPLORER (Promega).
Acknowledgements
We thank members of the Ivanov and Anderson laboratories for helpful discussions and feedback on the manuscript. We thank the Liu laboratory (Johns Hopkins School of Medicine, Baltimore) for proving Pateamine A and Prof. William Marzluff (University of North Carolina) for the anti-SLBP antibody.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
A.A., M.M.F., S.M.L. and N.K. helped design, perform and analyze the experiments. C.A.A. helped to conduct experiments. A.A., M.M.F. and S.M.L. helped with manuscript and image editing. P.I. conceived of the project, helped design the experiments, analyzed the data, and wrote the manuscript. P.A. co-wrote the manuscript.
Funding
This work is supported by the National Institutes of Health [GM111700 and CA168872 to P.A., NS094918 to P.I., GM119283 to S.M.L.]. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.199240.supplemental
References
- Anderson P. and Kedersha N. (2002). Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones 7, 213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P. and Kedersha N. (2006). RNA granules. J. Cell Biol. 172, 803-808. 10.1083/jcb.200512082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P. and Kedersha N. (2008). Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33, 141-150. 10.1016/j.tibs.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Anderson P. and Kedersha N. (2009). RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10, 430-436. 10.1038/nrm2694 [DOI] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. and Ivanov P. (2015). Stress granules, P-bodies and cancer. Biochim. Biophys. Acta 1849, 861-870. 10.1016/j.bbagrm.2014.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulas A. and Vande Velde C. (2015). Alterations in stress granule dynamics driven by TDP-43 and FUS: a link to pathological inclusions in ALS? Front. Cell Neurosci. 9, 423 10.3389/fncel.2015.00423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua E., Wang X., Majumder M., Gaccioli F., Yuan C. L., Wang C., Zhu X., Jordan L. E., Scheuner D., Kaufman R. J. et al. (2010). eIF2alpha phosphorylation tips the balance to apoptosis during osmotic stress. J. Biol. Chem. 285, 17098-17111. 10.1074/jbc.M110.109439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounedjah O., Hamon L., Savarin P., Desforges B., Curmi P. A. and Pastre D. (2012). Macromolecular crowding regulates assembly of mRNA stress granules after osmotic stress: new role for compatible osmolytes. J. Biol. Chem. 287, 2446-2458. 10.1074/jbc.M111.292748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J. R. (2014). mRNP granules: assembly, function, and connections with disease. RNA Biol. 11, 1019-1030. 10.4161/15476286.2014.972208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette J. E., Guimaraes C. P., Wuethrich I., Blomen V. A., Varadarajan M., Sun C., Bell G., Yuan B., Muellner M. K., Nijman S. M. et al. (2011). Global gene disruption in human cells to assign genes to phenotypes by deep sequencing. Nat. Biotechnol. 29, 542-546. 10.1038/nbt.1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y., Kedersha N., Low W.-K., Romo D., Gorospe M., Kaufman R., Anderson P. and Liu J. O. (2006). Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 281, 32870-32878. 10.1074/jbc.M606149200 [DOI] [PubMed] [Google Scholar]
- Donnelly N., Gorman A. M., Gupta S. and Samali A. (2013). The eIF2alpha kinases: their structures and functions. Cell. Mol. Life Sci. 70, 3493-3511. 10.1007/s00018-012-1252-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elling U. and Penninger J. M. (2014). Genome wide functional genetics in haploid cells. FEBS Lett. 588, 2415-2421. 10.1016/j.febslet.2014.06.032 [DOI] [PubMed] [Google Scholar]
- Harding H. P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M. and Ron D. (2000a). Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099-1108. 10.1016/S1097-2765(00)00108-8 [DOI] [PubMed] [Google Scholar]
- Harding H. P., Zhang Y., Bertolotti A., Zeng H. and Ron D. (2000b). Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5, 897-904. 10.1016/S1097-2765(00)80330-5 [DOI] [PubMed] [Google Scholar]
- Holcik M. and Sonenberg N. (2005). Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6, 318-327. 10.1038/nrm1618 [DOI] [PubMed] [Google Scholar]
- Ivanov P. and Anderson P. (2013). Post-transcriptional regulatory networks in immunity. Immunol. Rev. 253, 253-272. 10.1111/imr.12051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P., Kedersha N. and Anderson P. (2011). Stress puts TIA on TOP. Genes Dev. 25, 2119-2124. 10.1101/gad.17838411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R. J., Hellen C. U. T. and Pestova T. V. (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113-127. 10.1038/nrm2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N. and Anderson P. (2002). Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30, 963-969. 10.1042/bst0300963 [DOI] [PubMed] [Google Scholar]
- Kedersha N. and Anderson P. (2007). Mammalian stress granules and processing bodies. Methods Enzymol. 431, 61-81. 10.1016/S0076-6879(07)31005-7 [DOI] [PubMed] [Google Scholar]
- Kedersha N. L., Gupta M., Li W., Miller I. and Anderson P. (1999). RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147, 1431-1442. 10.1083/jcb.147.7.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Cho M. R., Li W., Yacono P. W., Chen S., Gilks N., Golan D. E. and Anderson P. (2000). Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151, 1257-1268. 10.1083/jcb.151.6.1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Ivanov P. and Anderson P. (2013). Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci. 38, 494-506. 10.1016/j.tibs.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Panas M. D., Achorn C. A., Lyons S., Tisdale S., Hickman T., Thomas M., Lieberman J., McInerney G. M., Ivanov P. et al. (2016). G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 212, 845-860. 10.1083/jcb.201508028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball S. R., Horetsky R. L., Ron D., Jefferson L. S. and Harding H. P. (2003). Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. Cell Physiol. 284, C273-C284. 10.1152/ajpcell.00314.2002 [DOI] [PubMed] [Google Scholar]
- Kwon S., Zhang Y. and Matthias P. (2007). The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 21, 3381-3394. 10.1101/gad.461107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M. and Sabatini D. M. (2013). Regulation of mTORC1 and its impact on gene expression at a glance. J. Cell Sci. 126, 1713-1719. 10.1242/jcs.125773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W. F., Wagner E. J. and Duronio R. J. (2008). Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 9, 843-854. 10.1038/nrg2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R., DI Marco S., Kaufman R. J. and Gallouzi I.-E. (2007). Inhibition of the ubiquitin-proteasome system induces stress granule formation. Mol. Biol. Cell 18, 2603-2618. 10.1091/mbc.E06-12-1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen E., Kedersha N., Song B., Scheuner D., Gilks N., Han A., Chen J.-J., Anderson P. and Kaufman R. J. (2005). Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 280, 16925-16933. 10.1074/jbc.M412882200 [DOI] [PubMed] [Google Scholar]
- Michel Y. M., Poncet D., Piron M., Kean K. M. and Borman A. M. (2000). Cap-Poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J. Biol. Chem. 275, 32268-32276. 10.1074/jbc.M004304200 [DOI] [PubMed] [Google Scholar]
- Panas M. D., Kedersha N. and McInerney G. M. (2015). Methods for the characterization of stress granules in virus infected cells. Methods 90, 57-64. 10.1016/j.ymeth.2015.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas M. D., Ivanov P. and Anderson P. (2016). Mechanistic insights into mammalian stress granule dynamics. J. Cell Biol. 215, 313-323. 10.1083/jcb.201609081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T. V., Kolupaeva V. G., Lomakin I. B., Pilipenko E. V., Shatsky I. N., Agol V. I. and Hellen C. U. T. (2001). Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. USA 98, 7029-7036. 10.1073/pnas.111145798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatsky I. N., Dmitriev S. E., Terenin I. M. and Andreev D. E. (2010). Cap- and IRES-independent scanning mechanism of translation initiation as an alternative to the concept of cellular IRESs. Mol. Cells 30, 285-293. 10.1007/s10059-010-0149-1 [DOI] [PubMed] [Google Scholar]
- Shatsky I. N., Dmitriev S. E., Andreev D. E. and Terenin I. M. (2014). Transcriptome-wide studies uncover the diversity of modes of mRNA recruitment to eukaryotic ribosomes. Crit. Rev. Biochem. Mol. Biol. 49, 164-177. 10.3109/10409238.2014.887051 [DOI] [PubMed] [Google Scholar]
- Sonenberg N. and Hinnebusch A. G. (2009). Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731-745. 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S. P., Kumar K. U. and Kaufman R. J. (1998). Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J. Biol. Chem. 273, 2416-2423. 10.1074/jbc.273.4.2416 [DOI] [PubMed] [Google Scholar]
- Stoecklin G. and Kedersha N. (2013). Relationship of GW/P-bodies with stress granules. Adv. Exp. Med. Biol. 768, 197-211. 10.1007/978-1-4614-5107-5_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G., Stubbs T., Kedersha N., Wax S., Rigby W. F. C., Blackwell T. K. and Anderson P. (2004). MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 23, 1313-1324. 10.1038/sj.emboj.7600163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöhr N., Lederer M., Reinke C., Meyer S., Hatzfeld M., Singer R. H. and Hüttelmaier S. (2006). ZBP1 regulates mRNA stability during cellular stress. J. Cell Biol. 175, 527-534. 10.1083/jcb.200608071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukarieh R., Sonenberg N. and Pelletier J. (2009). The eIF4E-binding proteins are modifiers of cytoplasmic eIF4E relocalization during the heat shock response. Am. J. Physiol. Cell Physiol. 296, C1207-C1217. 10.1152/ajpcell.00511.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenin I. M., Andreev D. E., Dmitriev S. E. and Shatsky I. N. (2013). A novel mechanism of eukaryotic translation initiation that is neither m7G-cap-, nor IRES-dependent. Nucleic Acids Res. 41, 1807-1816. 10.1093/nar/gks1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek S. A., Zhu S. and Wek R. C. (1995). The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell. Biol. 15, 4497-4506. 10.1128/MCB.15.8.4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S. and Anderson P. (2008). Reprogramming mRNA translation during stress. Curr. Opin. Cell Biol. 20, 222-226. 10.1016/j.ceb.2008.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]