ABSTRACT
We uncovered a crucial role for the Aurora kinase A (AURKA)–Twist1 axis in promoting epithelial-to-mesenchymal transition (EMT) and chemoresistance in pancreatic cancer. Twist1 is the first EMT-specific target of AURKA that was identified using an innovative screen. AURKA phosphorylates Twist1 at three sites, which results in its multifaceted regulation – AURKA inhibits its ubiquitylation, increases its transcriptional activity and favors its homodimerization. Twist1 reciprocates and prevents AURKA degradation, thereby triggering a feedback loop. Ablation of either AURKA or Twist1 completely inhibits EMT, highlighting both proteins as central players in EMT progression. Phosphorylation-dead Twist1 serves as a dominant-negative and fully reverses the EMT phenotype induced by Twist1, underscoring the crucial role of AURKA-mediated phosphorylation in mediating Twist1-induced malignancy. Likewise, Twist1-overexpressing BxPC3 cells formed large tumors in vivo, whereas expression of phosphorylation-dead Twist1 fully abrogated this effect. Furthermore, immunohistochemical analysis of pancreatic cancer specimens revealed a 3-fold higher level of Twist1 compared to that seen in healthy normal tissues. This is the first study that links Twist1 in a feedback loop with its activating kinase, which indicates that concurrent inhibition of AURKA and Twist1 will be synergistic in inhibiting pancreatic tumorigenesis and metastasis.
KEY WORDS: AURKA, Twist1, Pancreatic cancer, Drug discovery, Therapy, EMT
Summary: We show that the AURKA–Twist1 axis regulates EMT, xenograft growth and drug resistance in pancreatic cancer. This is the first mechanism linking AURKA activity with tumor metastasis through Twist1.
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is an exceptionally aggressive disease (Alian et al., 2014). Accumulating evidence suggests that the acquisition of epithelial-to-mesenchymal transition (EMT) phenotypes in pancreatic tumors is an important underlying cause for extensive tumor invasion, early metastasis, drug resistance and tumor recurrence (Castellanos et al., 2013). The EMT transcriptional program is primarily utilized by normal cells during embryogenesis and is silenced after birth; however, cancer cells exploit EMT developmental machinery to facilitate aggressive metastatic behavior. The mechanisms leading to the EMT phenotype are not fully understood, which has hindered the development of effective targeted therapies and the sensitive biomarkers needed to improve treatment outcomes for pancreatic cancer patients.
AURKA kinase is overexpressed in a majority of pancreatic tumors (Li et al., 2003; Rojanala et al., 2004). AURKA knockdown increases taxane chemosensitivity and hinders in vivo tumorigenesis (Hata et al., 2005). A recent study revealed a positive role of AURKA in promoting EMT in breast cancer and pancreatic cancer through experiments using the AURKA inhibitor Alisertib (also known as MLN8237); however, the molecular mechanism by which AURKA promotes EMT remains unclear (D'Assoro et al., 2014; Wang et al., 2015). In this study, we identified a crucial role of AURKA in promoting EMT in cells and in vivo. To uncover the molecular mechanism, we employed an innovative chemical genetic approach and identified Twist-related protein 1 (Twist1) as a direct substrate of AURKA. Twist1 is a basic helix-loop-helix (bHLH) transcription factor. Twist1 triggers EMT by transcriptional activation or inhibition of E-box-regulated target genes (Qin et al., 2012), which led us to hypothesize that AURKA may promote EMT via Twist1. Importantly, Twist1 levels have not been analyzed in human pancreatic cancer tissues.
RESULTS
Twist1 is a direct substrate of AURKA
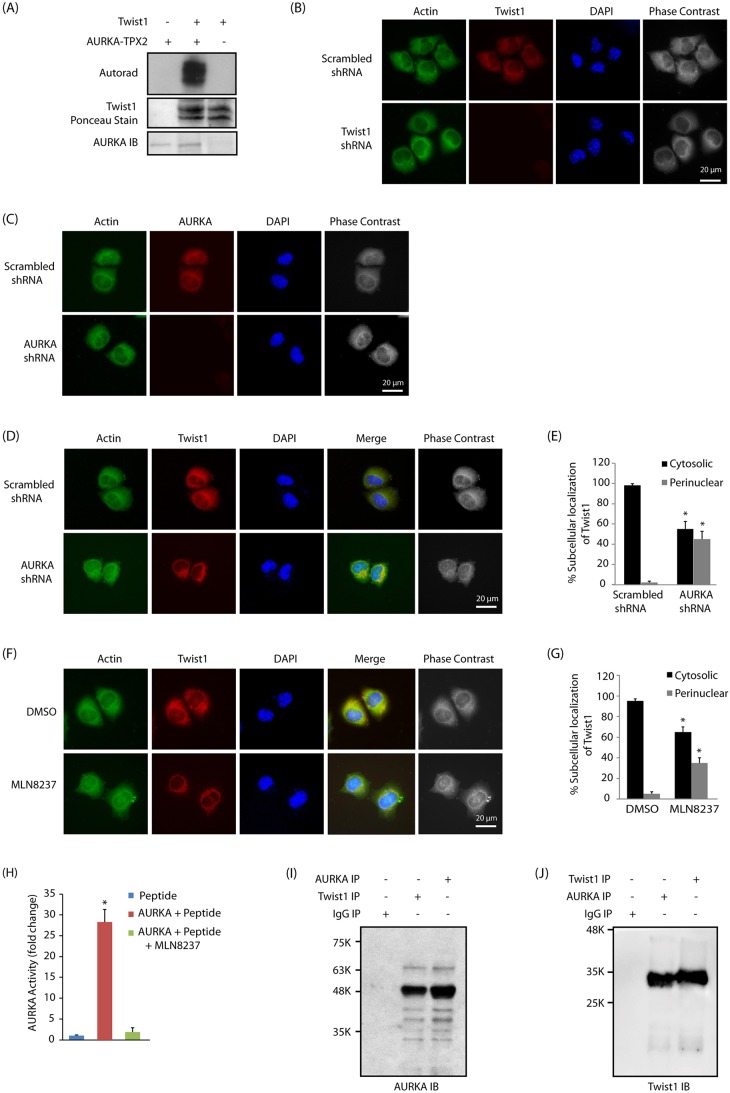
The chemical genetic approach involves an engineered kinase, which, in the presence of a radioactive orthogonal ATP analog, specifically transfers the radioactive tag (32P) to its substrates (Shah et al., 1997; Shah and Shokat, 2002, 2003; Shah and Vincent, 2005; Kim and Shah, 2007; Sun et al., 2008a,b; Chang et al., 2011, 2012; Shi et al., 2016). Using analog-sensitive AURKA (AA-as7) and [32P]phenethyl-ATP, we previously identified PHLDA1 and LIMK2 as its direct substrates (Johnson et al., 2011, 2012). In this study, we focused on Twist1, which was identified by us as a direct target of AURKA. As proteomics screening often leads to false positives, we subjected Twist1 to an in vitro kinase assay with AURKA, which confirmed the result obtained from the chemical genetic screen (Fig. 1A).
Fig. 1.
Twist1 is an AURKA substrate. (A) Twist1 is directly phosphorylated by AURKA. Lane 1 includes recombinant His6–AURKA–TPX2 with [32P]ATP, while lane 2 includes His6–Twist1, His6–AURKA–TPX2 and [32P]ATP. Lane 3 shows Twist1 incubated with [32P]ATP. IB, immunoblot. (B) Subcellular localization of Twist1 in BxPC3 cells treated with scrambled or Twist1 shRNA for 30 h. (C) AURKA localization in BxPC3 cells treated with scrambled or AURKA shRNA for 30 h. (D) AURKA regulates Twist1 subcellular localization in BxPC3 cells, as AURKA depletion results in a somewhat perinuclear localization of Twist1. More than 100 cells were analyzed from multiple random frames. Representative data are shown. Actin was used as a cytoplasmic marker. (E) Histogram showing the percentage of BxPC3 cells displaying cytosolic and perinuclear localization in response to AURKA shRNA. (F) AURKA regulates Twist1 subcellular localization through its kinase activity. Representative data are shown. (G) Histogram showing the percentage of BxPC3 cells displaying cytosolic and perinuclear localization in response to MLN8237. Values shown are mean±s.e.m. of three independent experiments. *P<0.05 compared with control (two-way ANOVA). (H) MLN8237-mediated inhibition of AURKA was analyzed using an in vitro kinase assay using AURKA optimal peptide substrate. MLN8237 was used at 1 µM concentration. *P<0.05 (two-way ANOVA). (I) AURKA and Twist1 associate in BxPC3 cells. Twist1 was immunoprecipitated (IP) from BxPC3 cells and AURKA binding analyzed (lane 2). AURKA (lane 3) and IgG (lane 1) were used as positive and negative controls, respectively. (J) AURKA and Twist1 associate in BxPC3 cells. AURKA was immunoprecipitated and Twist1 binding analyzed (lane 2). Each experiment was done at least three independent times. Representative data are shown.
AURKA associates with Twist1 and regulates its subcellular localization and levels
To determine whether Twist1 is a physiological substrate of AURKA in BxPC3 cells, we examined the subcellular location of Twist1 and AURKA, both of which displayed predominantly cytoplasmic localization (Fig. 1B,C). Notably, when AURKA levels were knocked down using AURKA shRNA or inhibited using MLN8237, Twist1 attained somewhat perinuclear localization (compare actin staining versus Twist1 staining), suggesting AURKA and Twist1 not only colocalize, but that AURKA also regulates the subcellular localization of Twist1 in BxPC3 cells via its kinase activity (Fig. 1D–G). As noted above, MLN8247 is a highly potent inhibitor of AURKA, which was confirmed using an in vitro kinase assay, which revealed >95% inhibition of AURKA kinase activity at 1 µM concentration (Fig. 1H). We also isolated a Twist1 immune complex and analyzed AURKA levels, which showed AURKA and Twist1 to be in the same complex (Fig. 1I). Conversely, pulldown of AURKA revealed that Twist1 was present in the immune complex (Fig. 1J). To examine whether AURKA-mediated regulation of Twist1 was a common mechanism in pancreatic cancer cells, we conducted similar experiments in Panc1 cells, which also revealed that AURKA associates and regulates Twist1 subcellular localization (Fig. S1A–D). Thus, these results demonstrate that Twist1 associates with AURKA and that its subcellular localization is regulated by AURKA.
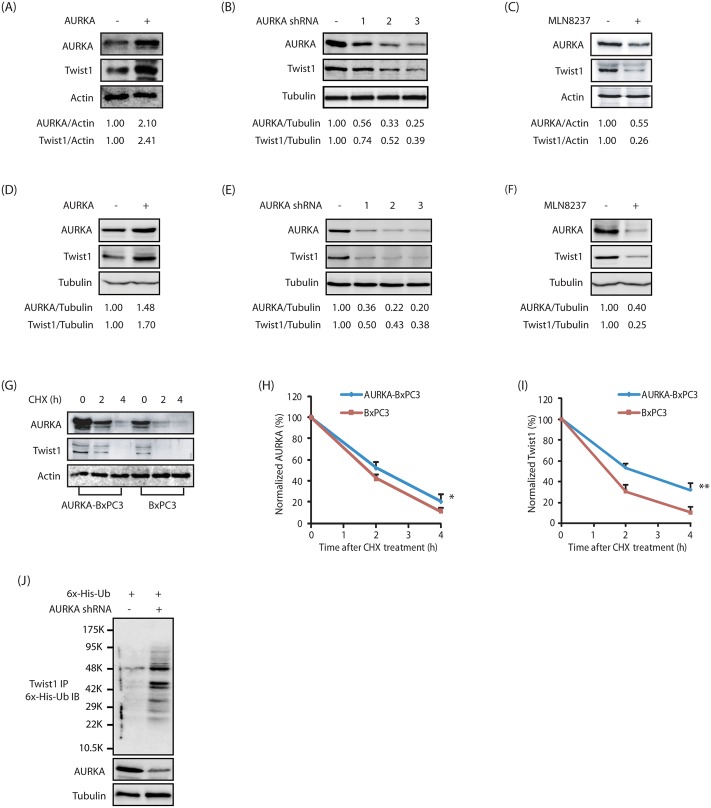
AURKA-mediated phosphorylation prompted us to investigate its effect on Twist1 levels. Overexpression of AURKA increased Twist1 level, and its knockdown or inhibition decreased it, suggesting that AURKA positively regulates Twist1 via phosphorylation both in BxPC3 and Panc1 cells (Fig. 2A–F). Importantly, MLN8237 treatment also reduced AURKA levels as reported previously (Fig. 2C,F; Johnson et al., 2012).
Fig. 2.
AURKA positively regulates Twist1 protein levels. (A) AURKA overexpression increases Twist1 levels in BxPC3 cells. AURKA and Twist1 levels were analyzed in wild-type HA–AURKA-expressing BxPC3 and vector-infected cells. (B) AURKA ablation depletes Twist1 in BxPC3 cells. Cells were infected with scrambled shRNA, or AURKA-shRNA-1, -2 or -3, and AURKA and Twist1 levels analyzed. (C) AURKA positively regulates Twist1 through its kinase activity. BxPC3 cells were either treated with DMSO or MLN8237. (D) AURKA overexpression increases Twist1 levels in Panc1 cells. (E) AURKA ablation depletes Twist1 in Panc1 cells. (F) AURKA inhibition depletes Twist1 levels in Panc1 cells. The indicated ratios from a representative experiment are given below the blots in A–F. (G) AURKA inhibits Twist1 degradation. AURKA-BxPC3 and BxPC3 cells were treated with cycloheximide (CHX, 10 μM) for 2 and 4 h, and AURKA and Twist1 levels analyzed. (H) Graphical representation of AURKA degradation rate in cells treated as in G. The results of densitometric scanning are shown graphically with the AURKA signal normalized to actin signal. AURKA half-life is ∼2 h. *P<0.05 (the significance of the difference between means determined by a Duncan's multiple-range test). (I) Graphical representation of Twist1 degradation rate. Twist1 half-life is ∼2 h. **P<0.01 determined as in H. (J) AURKA stabilizes Twist1 by inhibiting its ubiquitylation. Cells were co-transfected with AURKA shRNA along with His6–ubiquitin (6x-His-Ub). Twist1 was immunoprecipitated and ubiquitylated proteins were analyzed as described in the Materials and Methods. Each experiment was performed at least three independent times. Representative data are shown.
AURKA inhibits Twist1 degradation
Twist1 is an unstable protein with a half-life of ∼2 h (Lander et al., 2011). As AURKA directly phosphorylates Twist1, we hypothesized that AURKA might increase Twist1 levels by inhibiting its degradation via phosphorylation. Thus, BxPC3 and AURKA-overexpressing BxPC3 (AURKA-BxPC3) cells were treated with cycloheximide to prevent further protein synthesis, and the degradation profile of Twist1 was examined. AURKA overexpression reduced Twist1 degradation, suggesting that AURKA regulates the level of Twist1 by inhibiting its degradation (Fig. 2G–I). To investigate whether Twist1 degradation is mediated through a ubiquitin or non-ubiquitin mechanism, we transfected His6–ubiquitin into BxPC3 and AURKA-depleted BxPC3 cells, and analyzed the ubiquitylation of Twist1. Knockdown of AURKA led to increased ubiquitylation of Twist1 (Fig. 2J), thus confirming that AURKA stabilizes Twist1 levels by inhibiting its ubiquitin-mediated degradation.
Twist1 positively regulates AURKA levels in a reciprocal feedback loop
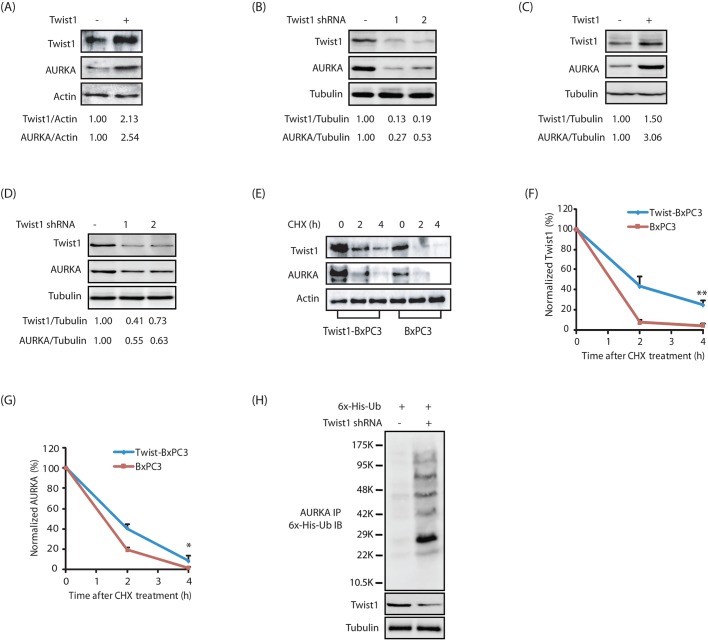
Several AURKA substrates regulate the activity and levels of AURKA in a feedback loop. We have previously demonstrated that PHLDA1 decreases AURKA level, whereas LIMK2 increases it (Johnson et al., 2011, 2012). Consequently, we investigated whether Twist1 displays a similar effect on AURKA levels. Twist1 overexpression led to a strong increase in AURKA levels, whereas its knockdown decreased AURKA levels both in BxPC3 and Panc1 cells (Fig. 3A–D). We next examined whether Twist1 increases AURKA levels by inhibiting its degradation. AURKA and Twist1 levels were examined in BxPC3 and Twist1-overexpressing BxPC3 (Twist1-BxPC3) cells treated with cycloheximide. Twist1 overexpression significantly reduced the degradation of AURKA, suggesting that Twist1 stabilizes AURKA protein levels (Fig. 3E–G). We further investigated the ubiquitylation of AURKA in BxPC3 and Twist1-depleted BxPC3 cells, and found that there was increased ubiquitylation of AURKA upon Twist1 knockdown (Fig. 3H). Taken together, these results suggest that Twist1 increases AURKA levels by preventing its degradation, thereby eliciting a positive-feedback loop.
Fig. 3.
Twist1 positively regulates AURKA protein levels. (A) Overexpression of wild-type HA-tagged Twist1 increases AURKA levels in BxPC3 cells. (B) Twist1 ablation depletes AURKA in BxPC3 cells. (C) Twist1 overexpression increases AURKA levels in Panc1 cells. (D) Twist1 knockdown depletes AURKA in Panc1 cells. The indicated ratios from a representative experiment are given below the blots in A–D. (E) Twist1 inhibits AURKA degradation. Twist1-BxPC3 and BxPC3 cells were treated with cycloheximide (CHX) for 2 h and 4 h, and AURKA and Twist1 levels analyzed. (F) Graphical representation of Twist1 degradation rate. Twist1 half-life is ∼2 h. **P<0.01 (the significance of the difference between means as determined by Duncan's multiple-range test). (G) Graphical representation of AURKA degradation rate. AURKA half-life is ∼2 h. *P<0.05 determined as in F. (H) Twist1 stabilizes AURKA by inhibiting its ubiquitylation. Each experiment was performed at least three independent times. Representative data are shown.
AURKA phosphorylates Twist1 at S123, T148 and S184
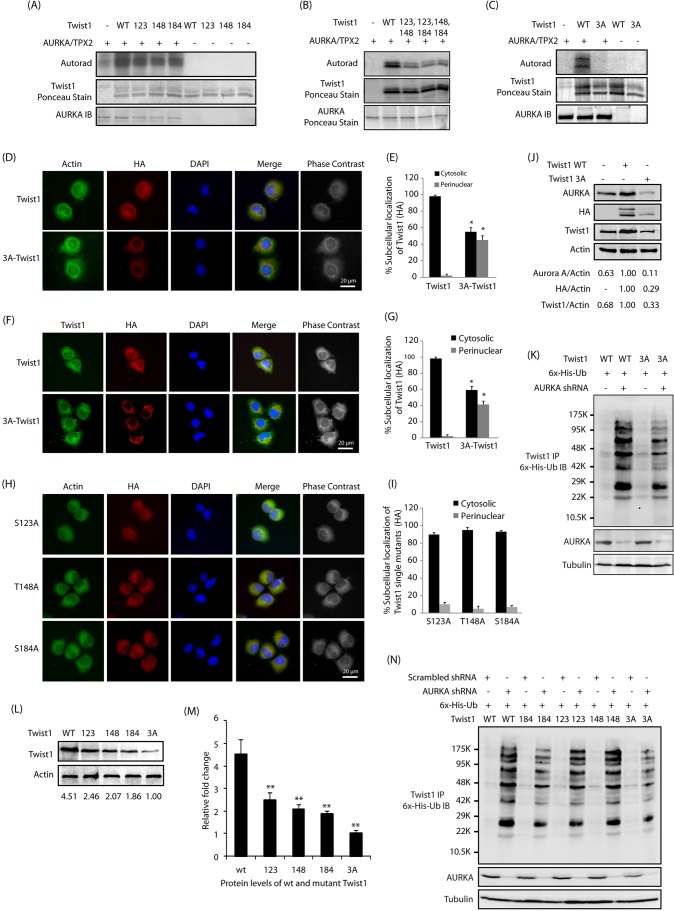
AURKA prefers to phosphorylate an R/K/N-R-X-S/T-Φ consensus sequence where Φ denotes a hydrophobic residue except proline (Ferrari et al., 2005). This preference suggested S123, T148 and S184 as potential AURKA sites on human Twist1 (the corresponding murine Twist1 sites are S127, T152 and S188). Human Twist1 shares 96% sequence identity with murine Twist1 (Wolf et al., 1991). The corresponding phosphorylation-resistant single mutants (S123A, T148A and S184A, human numbering) were generated and their phosphorylation status analyzed in vitro by using the recombinant AURKA–TPX2 complex. We found that AURKA phosphorylates all the three sites on Twist1 (Fig. 4A). We also generated corresponding phosphorylation-dead double mutants of Twist1 (S123A, T148A-Twist1; S123A, S184A-Twist1; and T148A, S184A-Twist1), and analyzed their phosphorylation using an in vitro kinase assay. As shown in Fig. 4B, all double mutants of Twist1 revealed reduced phosphorylation compared to wild-type Twist1, suggesting that AURKA phosphorylates all three sites. The corresponding phosphorylation-dead triple mutant (denoted as 3A) was not phosphorylated by AURKA, confirming that S123, T148 and S184 are the only AURKA sites on Twist1 (Fig. 4C).
Fig. 4.
AURKA-mediated phosphorylation of Twist1 at S123, T148 and S184 regulates its protein stability and subcellular localization. (A) AURKA phosphorylates Twist1 at S123, T148 and S184. Phospho-dead single mutants of Twist1 (S123A, T148A and S184A, denoted 123, 148 and 184) were subjected to an in vitro kinase assay using AURKA–TPX2. IB, immunoblot; WT, wild-type. (B) Phosphorylation of Twist1 double mutants by AURKA. (C) S123, T148 and S184 are the only AURKA sites on Twist1. (D) AURKA regulates the subcellular localization of Twist1 via phosphorylation. BxPC3 cells stably expressing HA-tagged wild-type and 3A-Twist1 were fixed and immunostained with anti-actin and anti-HA antibody. More than 100 cells were analyzed from multiple random frames. Representative data are shown. (E) Histogram showing the percentage of BxPC3 cells as in D displaying cytosolic and perinuclear localization of ectopically expressed HA-tagged Twist1. Values shown are mean±s.e.m. of three independent experiments. *P<0.05 with respect to controls (two-way ANOVA). (F) Differential localization of wild-type and 3A Twist1 mutant in BxPC3 cells. (G) Histogram showing the percentage of BxPC3 cells expressing wild-type Twist1 and 3A-Twist1 showing cytosolic and perinuclear localization of Twist1. Values shown are mean±s.e.m. of three independent experiments. *P<0.05 with respect to controls (two-way ANOVA). (H) AURKA regulates the subcellular localization of Twist1 via phosphorylation of all three sites. Representative data are shown. (I) Histogram showing the percentage of single mutant Twist1-BxPC3 cells displaying cytosolic and perinuclear localization. Values shown are mean±s.e.m. of three independent experiments. *P<0.05 with respect to controls (two-way ANOVA). (J) AURKA promotes Twist1 levels through phosphorylation. (K) AURKA inhibits Twist1 ubiquitylation by phosphorylating S123, T148 and S184 sites. AURKA depletion in WT-Twist1-expressing cells causes higher levels of ubiquitylation than its depletion in 3A-Twist1-expressing cells. (L) AURKA increases Twist1 levels through phosphorylation at S123, T148 and S184. The ratio of Twist1 to actin from a representative experiment is given below the blots. (M) Graphical representation of Twist1 levels in BxPC3 cells expressing either wild-type Twist1, S123A Twist1, T148A Twist1, S184A Twist1 or 3A-Twist1. Mean±s.e.m. values of wild-type and mutant Twist1 levels from three independent experiments are depicted in the graph. **P<0.01 compared to control (one-way ANOVA). (N) AURKA inhibits Twist1 ubiquitylation by phosphorylating S123, T148 and S184 sites. Each experiment was done at least three independent times. Representative data are shown.
AURKA regulates the subcellular localization of Twist1 via phosphorylation
We next investigated the subcellular localization of HA-tagged wild-type and 3A-Twist1 in BxPC3 cells using HA antibody. While wild-type Twist1 showed predominantly cytoplasmic localization, the 3A-Twist1 mutant predominantly showed perinuclear localization (Fig. 4D,E, compare actin versus HA staining). We confirmed this finding by double staining these cells for Twist1 (total Twist1) and HA (for ectopically expressed Twist1) antibodies. Overexpressed wild-type Twist1 had a diffuse cytoplasmic localization, but ∼50% of 3A-Twist1 cells displayed a specific perinuclear localization of 3A-Twist1, as assessed by HA staining (Fig. 4F,G). We also ectopically expressed HA-tagged phosphorylation-resistant single mutants in BxPC3 cells and analyzed their subcellular localization using actin staining as a control for cytoplasm. Surprisingly, all single mutants of Twist1 showed predominantly cytoplasmic localization, suggesting that inhibition of phosphorylation at all sites contributes to its perinuclear localization (Fig. 4H,I).
AURKA prevents the degradation of Twist1 by phosphorylating S123, T148 and S184
To investigate whether AURKA-mediated phosphorylation of Twist1 prevents its degradation, we stably expressed wild-type and 3A-Twist1 in BxPC3 cells and analyzed their protein levels. Wild-type Twist1 expression levels were significantly higher than those of 3A-Twist1, suggesting that AURKA-mediated phosphorylation of Twist1 is required to stabilize its protein levels. Furthermore, as 3A-Twist1 was expressed at a lower level, it resulted in a concomitant decrease in AURKA level due a disruption of the proposed signaling feedback loop (Fig. 4J).
As AURKA increases Twist1 levels by inhibiting its ubiquitylation, we examined whether the triple mutant was resistant to AURKA-mediated protein stabilization. We transiently depleted AURKA from Twist1-BxPC3 and 3A-Twist-BxPC3 cells and analyzed the relative ubiquitylation of Twist1. While wild-type Twist1 was significantly degraded via ubiquitylation upon AURKA knockdown, 3A-Twist1 showed little ubiquitylation, confirming that AURKA-mediated phosphorylation is responsible for the observed increase in its half-life (Fig. 4K).
Deletion of C-terminal domain of Twist1 (amino acids 143–202) has been shown to prevent its ubiquitylation (Lander et al., 2011). As two of the AURKA-mediated phosphorylation sites on Twist1 were at the C-terminus (T148 and S184), we investigated their individual contribution to protein stabilization. It is known that simultaneous phosphorylation of Twist1 at T121 and S123 by the protein inhibitor of κB kinase β results in its cytoplasmic retention and subsequent degradation (Zhong et al., 2013). That study, however, did not analyze the individual contributions of the T121 and S123 phosphorylation sites. As AURKA phosphorylates S123 (and not T121), we also wanted to investigate its impact on Twist1 protein stability. Thus, we infected HA-tagged wild-type and phospho-resistant single mutants of Twist 1 individually in BxPC3 cells, and examined their protein levels. Mutation of either of the three sites resulted in reduced protein levels, with the triple mutant showing minimal levels, suggesting that phosphorylation of each of the three sites contribute to the increase of Twist1 levels (Fig. 4L,M).
As AURKA increases Twist1 levels by inhibiting its ubiquitylation, we examined whether each of these three sites affected protein levels via increased protein stability. The mutant Twist1-expressing cell lines exhibited a similar steady state decrease in Twist1 levels to that in wild-type-Twist1 expressing cells as shown in Fig. 4L,M. We next transiently depleted AURKA in these cells using AURKA shRNA, which revealed substantial ubiquitylation of wild-type Twist1, and a little decrease for each single phosphorylation-resistant mutant (Fig. 4N). The phosphorylation-dead triple mutant (3A-Twist1) exhibited the least ubiquitylation upon AURKA depletion, confirming its independence from AURKA-mediated phosphorylation. Collectively, these results demonstrate that AURKA-mediated phosphorylation of Twist1 at each of the three sites increase protein stability. Our results also provide a potential molecular mechanism explaining the instability of C-terminally deleted Twist1, which houses two of the AURKA-mediated phosphorylation sites (T148 and S184).
AURKA-mediated phosphorylation of Twist1 favors homodimerization versus heterodimerization with E12 and Hand2
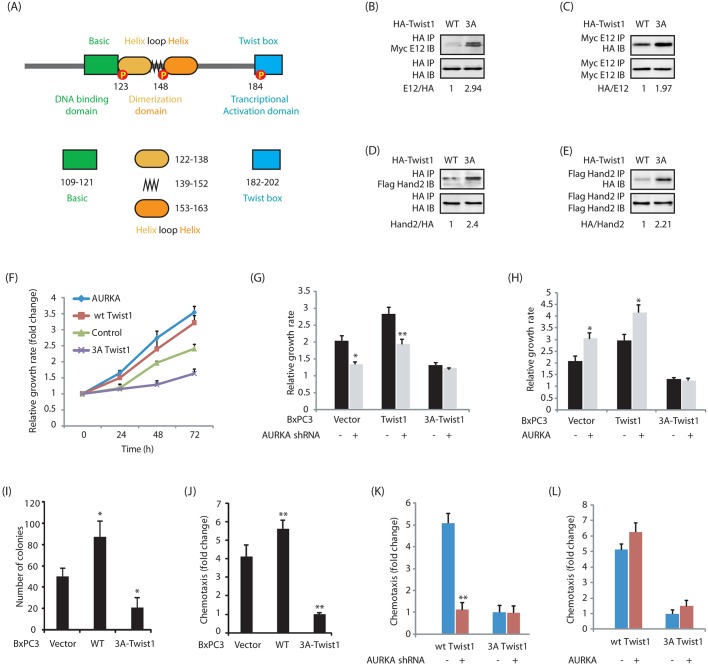
The function of Twist1 in cells is influenced by its spatiotemporal expression, phospho-regulation, protein–protein interactions and dimer choice. Twist1 either forms a homodimer or heterodimers with other helix-loop-helix (HLH)-containing transcription factors (Franco et al., 2011). The DNA-binding basic region of Twist1 is from 109–121, the HLH is from 122–163 and Twist1 box is 182–202 (Qin et al., 2012; Fig. 5A). Dimerization occurs via the HLH domains, which allows the basic region to interact with the DNA in a bipartite fashion. This DNA-binding domain specifically recognizes the E-box consensus sequence present in the regulatory regions of many mesenchymal-lineage-specific genes.
Fig. 5.
Impact of Twist1 phosphorylation on its oncogenic functions. (A) Domain stricture of Twist1. (B–E) The 3A-Twist1 mutant favors heterodimerization with E12 and Hand2. E12 and Hand2 retrovirus were infected in BxPC3 cells expressing either HA-tagged wild-type (WT) Twist1 or 3A-Twist1. After 36 h, Twist1 was immunoprecipitated (IP) and E12 and Hand2 binding analyzed by immunoblotting (IB). Similarly, Myc–E12 and Flag–Hand2 were immunoprecipitated, and Twist1 binding analyzed by using anti-HA antibody. The indicated ratios for a representative experiment are given below the blots. (F) Twist1 promotes cell proliferation in BxPC3 cells. (G) AURKA depletion decreases cell proliferation in BxPC3 and Twist1-BxPC3 cells, but not in phospho-dead 3A-Twist1-BxPC3 cells. *P<0.05, **P<0.01 compared to scrambled shRNA controls (two-sample t-test). (H) AURKA overexpression increases cell proliferation in vector-expressing BxPC3 and Twist1-BxPC3 cells, but not in 3A-Twist1-BxPC3 cells. An MTT assay was performed after 48 h. *P<0.05 compared to vector infected controls (two-independent sample t-test). (I) Twist1 promotes colony formation in a soft agar assay. *P<0.05 compared to vector-expressing control (two-sample t-test). (J) Twist1 promotes cell motility. **P<0.01 compared to vector-expressing control (two-sample t-test). (K) AURKA depletion inhibits cell motility in Twist1-BxPC3 cells, but not in phospho-dead 3A-Twist1-BxPC3 cells. Twist1-BxPC3 and 3A-Twist1-BxPC3 cells were infected with either scrambled shRNA or AURKA shRNA lentivirus. The cell migration test was performed after 30 h. **P<0.01 compared to scrambled shRNA control (two-sample t-test). (L) AURKA overexpression increases cell motility in Twist1-BxPC3 cells, but not in 3A-Twist1-BxPC3 cells. Twist1-BxPC3 and 3A-Twist1-BxPC3 cells were infected with either vector or AURKA retrovirus. Cell migration was performed after 30 h.
During development, phosphorylation of Twist1 by protein kinase A (PKA) at T121 and S123 favors its dimerization with E12 (TCF3) and Hand2 transcription factors and disruption of this phospho-regulation leads to Saethre–Chotzen syndrome (Firulli and Conway, 2008). Thus, we next investigated the consequences of Twist1 phosphorylation on its ability to form homodimers and heterodimers with E12 and Hand2 transcription factors. Wild-type Twist1 and 3A-Twist1 were either co-expressed with Myc-tagged E12 or Flag-tagged Hand2 in BxPC3 cells, and their binding analyzed. Our data shows that the 3A mutant prefers to form heterodimers with both E12 and Hand2 as compared to wild-type Twist1 (Fig. 5B–E). Taken together, these results show that AURKA-mediated phosphorylation of Twist1 also changes the partner preference for Twist1, presumably due to its increased levels.
A Twist1 and AURKA feedback loop promotes highly aggressive pancreatic cancer phenotypes
We next investigated the impact of AURKA-mediated phosphorylation on Twist1 on cell proliferation. Ectopic expression of either AURKA or Twist1 increased cellular proliferation (Fig. 5F). In contrast, expression of 3A-Twist1 led to a significantly diminished proliferation rate, which was lower than that in BxPC3 cells. Furthermore, AURKA knockdown in BxPC3, Twist1-BxPC3 and 3A-Twist1-BxPC3 cells considerably reduced proliferation in BxPC3 and Twist1-BxPC3 cells, but not in 3A-Twist1-BxPC3 cells, indicating that the Twist1-mediated increase in cell proliferation is predominantly due to AURKA (Fig. 5G). We further examined this hypothesis by overexpressing AURKA, which increased cell proliferation in BxPC3 and wild-type Twist1-BxPC3 cells, but not in 3A-Twist1-BxPC3, thereby confirming that AURKA-mediated phosphorylation of Twist1 is crucial for the increased growth rate (Fig. 5H).
The effect of Twist1 was next evaluated under anchorage-independent conditions. Twist1 expression led to a substantial increase in colony formation in cells (Fig. 5I). By contrast, expression of 3A-Twist1 acted as a dominant-negative and only a minimal number of colonies formed in the soft agar assay. These findings show that AURKA-mediated phosphorylation of Twist1 is crucial for promoting cell proliferation both under attached and anchorage-independent conditions.
The Twist1 and AURKA feedback loop enhances cell motility
A substantial increase in cell motility was observed upon Twist1 expression (Fig. 5J). This finding is not surprising as Twist1 is a known driver of EMT phenotypes. Interestingly, 3A-Twist1 overexpression significantly impaired chemotaxis compared to vector-expressing BxPC3 cells, suggesting that 3A-Twist1 acts as dominant-negative and inhibits cell motility.
We also depleted AURKA in these cells, which considerably reduced the cell motility in Twist1-BxPC3 cells, but not in 3A-Twist1-BxPC3 cells, suggesting that the Twist1-triggered increase in cell motility is due to positive AURKA-mediated regulation (Fig. 5K). Similarly, AURKA overexpression increased migration in Twist1-BxPC3 cells, but not in 3A-Twist1-BxPC3 cells (Fig. 5L). Taken together, these results corroborate that AURKA-mediated phosphorylation of Twist1 is crucial for cell motility.
AURKA and Twist1 in EMT
A critical role of Twist1 in promoting EMT and drug resistance is well established. However, currently there are no known small molecules inhibitors for Twist1. Our goal was to determine the impact of AURKA in promoting Twist1-induced oncogenic signaling. In such case, AURKA inhibition provides a potent tool to manage Twist1-triggered cancer aggression.
BxPC3 cells exhibit an epithelial cell phenotype and express high levels of E-cadherin. Overexpression of Snail (also known as SNAI1) in these cells induces EMT, leading to metastasis in vivo (Nishioka et al., 2010). Therefore, BxPC3 cells provided an apt system to investigate the consequences of Twist1 phosphorylation in potentially inducing the EMT phenotype.
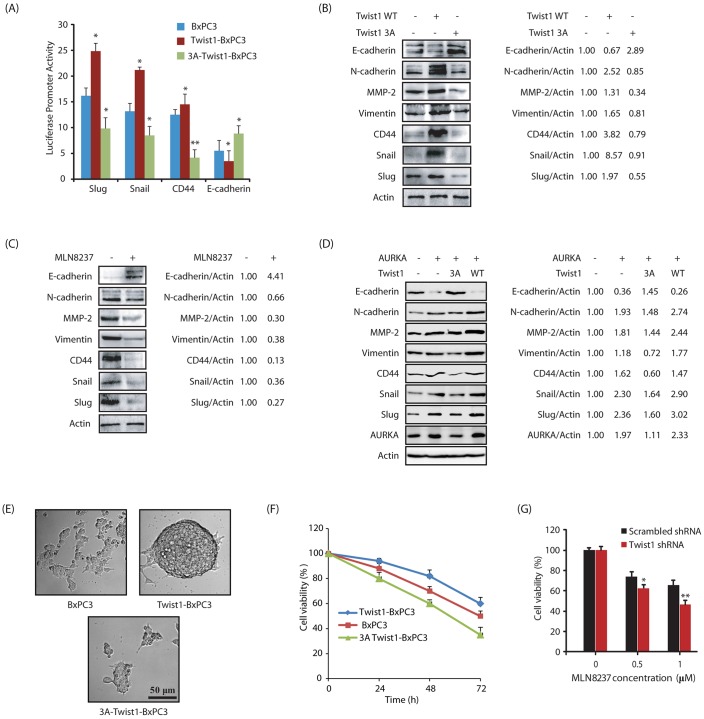
Twist1 expression downregulates E-cadherin, but upregulates the EMT markers CD44, Slug (also known as SNAI2) and Snail (Fu et al., 2011; Li and Zhou, 2011; Casas et al., 2011). Thus, we initially tested their promoter activities using the corresponding luciferase plasmids. As expected, ectopic expression of wild-type Twist1 in BxPC3 cells increased the promoter activities of CD44, Slug and Snail, but decreased the activity of E-cadherin compared to vector-expressing BxPC3 cells (Fig. 6A). More importantly, ectopic expression of 3A-Twist1 not only prevented the increase in promoter activities of CD44, Slug and Snail, it resulted in even less activity than in parental cells. Furthermore, expression of 3A-Twist1 significantly increased E-cadherin expression, thereby underscoring a crucial role of AURKA in Twist1-induced aggressive oncogenic phenotypes (Fig. 6A). We confirmed these findings by analyzing the protein levels of N-cadherin, CD44, Slug, Snail and E-cadherin in BxPC3, Twist1-BxPC3 and 3A-Twist1-BxPC3 cells. As expected, N-cadherin, CD44, Slug and Snail levels increased significantly upon Twist1 expression, but were prevented in the presence of 3A-Twist1 in BxPC3 cells (Fig. 6B). In addition, E-cadherin levels decreased in Twist1-overexpressing cells, but showed a substantial enhancement in 3A-Twist1-expressing cells. We also analyzed the levels of vimentin and matrix metalloproteinase-2 (MMP-2), both of which increase in EMT events and promote metastasis. Twist1 overexpression increased both vimentin and MMP-2 levels, whereas 3A-Twist1 expression abrogated their expression (Fig. 6B). Collectively, these results validate the idea that AURKA-mediated phosphorylation plays a central role in Twist1-mediated EMT phenotype.
Fig. 6.
AURKA-mediated phosphorylation of Twist1 is crucial for EMT, the CSC phenotype and drug resistance. (A) Expression of wild-type (WT) Twist1 increases CD44, Slug and Snail promoter activities and decreases E-cadherin promoter activity, whereas expression of 3A-Twist1 decreases CD44, Slug and Snail promoter activities and increases E-cadherin promoter activity. Results are for three independent experiments performed in triplicate. *P<0.05, **P<0.01 compared to control BxPC3 cells (one-way ANOVA). (B) Twist1 expression increases the levels of EMT and CSC markers while decreasing E-cadherin levels. 3A-Twist1 expression decreases the levels of EMT and CSC markers while increases E-cadherin. (C) AURKA inhibition decreases the levels of EMT and CSC markers. (D) AURKA overexpression increases the expression of EMT and CSC markers, while the expression of 3A Twist1 in the AURKA-expressing cells rescues the phenotype. The indicated ratios for a representative experiment are given next to the blots in B–D. (E) Twist1 overexpression increases the sphere-forming ability in BxPC3 cells. (F) Twist1 overexpression increases drug resistance in BxPC3 cells. BxPC3 cells and BxPC3 cells expressing wild-type Twist1 and 3A-Twist1, were plated in 96-well plates overnight. Then gemcitabine (1 µM) was added and cells were cultured for another 24, 48 or 72 h. (G) Twist1 shRNA-mediated depletion sensitizes BxPC3 cells to AURKA inhibition with MLN8237 (1 µM, treated for 48 h). *P<0.05, **P<0.01 compared to scrambled shRNA control (two-sample t-test).
In parallel, we also analyzed the levels of E-cadherin, N-cadherin, CD44, vimentin, MMP-2, Slug and Snail in BxPC3 cells treated either with DMSO control or MLN8237. AURKA inhibition reduced the levels of N-cadherin, CD44, vimentin, MMP-2, Slug and Snail, but increased E-cadherin levels in a pancreatic cancer cell line (Fig. 6C).
To further investigate whether 3A-Twist1 can rescue the EMT phenotype induced by AURKA overexpression, we investigated the levels of E-cadherin, N-cadherin, CD44, vimentin, MMP-2, Slug and Snail in BxPC3 cells, AURKA-BxPC3 cells and AURKA-BxPC3 cells co-expressing either 3A-Twist1 or wild-type Twist1. Ectopic expression of AURKA increased the expression of all EMT markers with a concurrent decrease in E-cadherin levels as expected (Fig. 6D). More importantly, co-expression of 3A-Twist1 significantly rescued the increase in EMT markers caused by AURKA overexpression, confirming that Twist1 phosphorylation is one of the key mechanisms by which AURKA promotes EMT.
We further conducted a sphere-forming assay to gauge the self-renewal capacity of BxPC3, Twist1-BxPC3 and 3A-Twist1-BxPC3 cells. Under ultra-low attachment conditions, cancer stem cells (CSCs) grow in suspension and form independent spheres or colonies. When subjected to these conditions, BxPC3 mainly formed aggregates of cells but showed no pancreatosphere formation (Fig. 6E). In contrast, overexpression of Twist1 induced the formation of large pancreatosphere, confirming that Twist1 overexpression causes a CSC phenotype. Importantly, 3A-Twist1-BxPC3 cells also showed no pancreatosphere formation, thereby confirming that AURKA-mediated phosphorylation of Twist1 is crucial for pancreatosphere formation.
Significance of Twist1 phosphorylation in drug resistance and cell viability
Because EMT contributes to drug resistance, we examined gemcitabine sensitivity in BxPC3 cells, and observed a ∼50% loss in cell viability. Twist1 expression offered resistance to gemcitabine (∼35% loss), whereas 3A-Twist1 expression rendered these cells highly sensitive to gemcitabine-induced toxicity (∼70% loss) (Fig. 6F). These results prompted us to investigate whether Twist1 ablation sensitizes BxPC3 cells to AURKA inhibition. Twist1-depleted cells exhibited higher sensitivity to AURKA inhibition than scrambled shRNA-expressing cells, suggesting that concurrent inhibition of AURKA and Twist1 is likely to act synergistically in targeting highly chemoresistant PDAC (Fig. 6G).
AURKA-mediated Twist1 phosphorylation in crucial for tumorigenesis in vivo
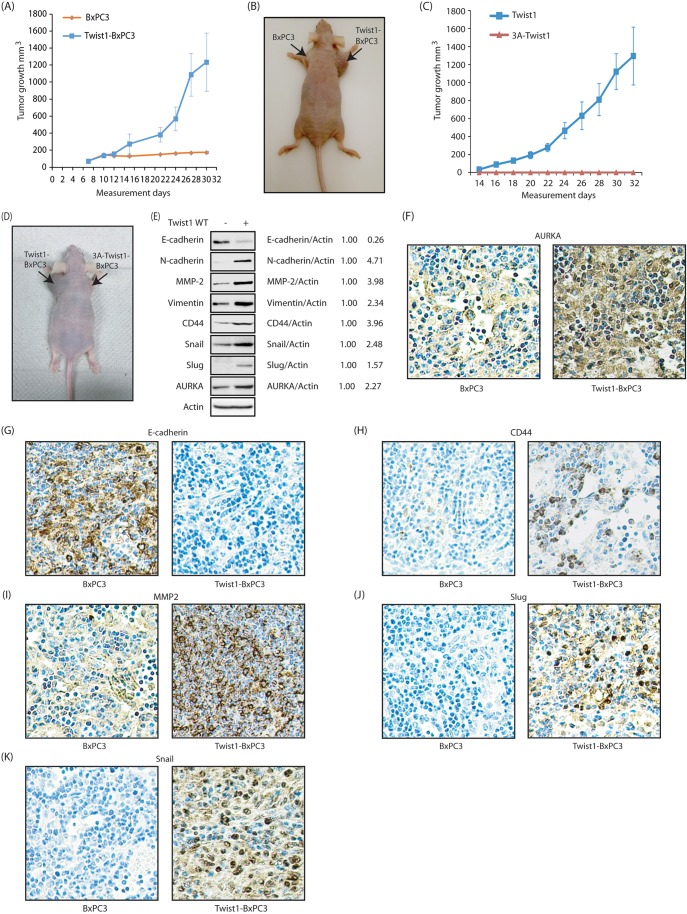
The potential of Twist 1 phosphorylation to induce tumorigenesis in a mouse xenograft model was next analyzed. Athymic nude mice were subcutaneously inoculated with either BxPC3 or Twist1-BxPC3 cells on the left and right shoulders, respectively. The tumors became measurable after 12–14 days, and were measured every 2 days. While Twist1-BxPC3 cells formed large tumors, BxPC3 cells showed much smaller tumor formation (∼200 mm3 size), confirming that Twist1 overexpression indeed increases tumorigenesis (Fig. 7A,B).
Fig. 7.
The Twist1–AURKA axis regulates EMT in vivo. (A) Twist1 overexpression increases tumorigenesis in vivo. Five athymic nude mice were inoculated with BxPC3 cells and Twist1-BxPC3 cells on left and right shoulder, respectively. (B) Athymic nude mouse were injected with control BxPC3 cells and Twist1-BxPC3 cells on the left and right shoulder, respectively. The pictures were taken 30 days following inoculation. A representative image is shown. (C) Effect of 3A-Twist1 expression on subcutaneous tumor growth in athymic nude mice. Three nude mice were inoculated with Twist1-BxPC3 cells and 3A-Twist1-BxPC3 cells on the left and right shoulder, respectively. (D) Athymic nude mouse injected with Twist1-BxPC3 cells and 3A-Twist1-BxPC3 cells on left and right shoulder. The pictures were taken 32 days following inoculation. A representative image is shown. (E) Immunoblot analysis to show the expression of levels of AURKA, EMT and CSC markers in tumors of athymic nude mouse injected with control BxPC3 and Twist1-BxPC3 cells. (F) AURKA immunohistochemistry of BxPC3 and Twist1-BxPC3 xenografts. (G) E-cadherin immunohistochemistry of BxPC3 and Twist1-BxPC3 xenografts. (H) CD44 immunohistochemistry of BxPC3 and Twist1-BxPC3 xenografts. (I) MMP2 immunohistochemistry of BxPC3 and Twist1-BxPC3 xenografts. (J) Slug immunohistochemistry of BxPC3 and Twist1-BxPC3 xenografts. (K) Snail immunohistochemistry of BxPC3 and Twist1-BxPC3 xenografts.
To investigate the consequences of 3A-Twist1 expression on tumorigenesis, we next subcutaneously inoculated Twist1-BxPC3 and 3A-Twist1-BxPC3 cells on the left and right shoulders, respectively. While Twist1-BxPC3 cells formed large tumors, 3A-Twist1-BxPC3 cells showed absolutely no tumor formation (Fig. 7C,D). These results further confirm that phosphorylation of Twist1 by AURKA is crucial for its tumorigenic potential in vivo.
Twist1-BxPC3 xenografts express high levels of EMT markers
We next examined the levels of E-cadherin and EMT markers in BxPC3 and Twist1-BxPC3 xenografts. Tumors were isolated following euthanization of the mice, and immediately frozen in liquid nitrogen. Twist1-BxPC3 xenografts showed increased expression of N-cadherin, CD44, vimentin, MMP-2, Slug and Snail, but reduced expression of E-cadherin (Fig. 7E).
We also observed higher levels of AURKA in Twist1-BxPC3 tumors compared to BxPC3 tumors, which confirms that Twist1 expression indeed has a positive impact on AURKA levels in vivo (Fig. 7E).
We then compared the levels of AURKA and EMT markers in BxPC3 and Twist1-BxPC3 xenografts using immunohistochemistry. Tumors were isolated following euthanization of the mice, and paraffin-embedded slides were generated. AURKA was highly expressed in Twist1-BxPC3 xenografts compared to BxPC3 xenografts, further confirming the Twist1–AURKA feedback loop in vivo (Fig. 7F). Twist1-BxPC3 xenografts did not express E-cadherin (Fig. 7G), but showed substantial expression of CD44, MMP2, Slug and Snail, compared to BxPC3 xenografts, further confirming that Twist1 promotes EMT in pancreatic cancer in vivo (Fig. 7H–K).
Twist1 expression in pancreatic cancer tissues
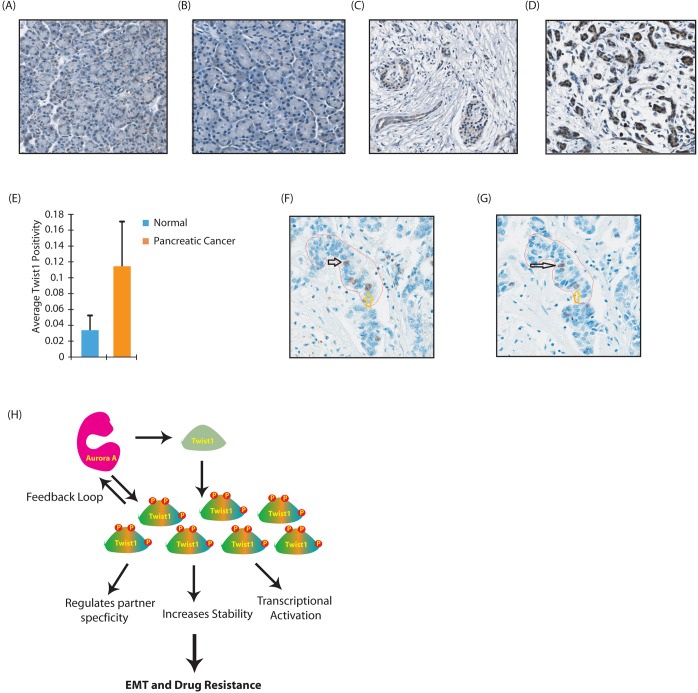
Twist1 levels have not been analyzed in human pancreatic cancer tissues. We analyzed Twist1 levels using immunohistochemistry in normal pancreas, chronic pancreatic and stage 4 ductal pancreatic carcinoma patient tissues. The immunostaining of Twist1 was observed mainly in the ductal epithelial carcinoma cells in the carcinoma cells in the pancreas (Fig. 8A–D). There was no staining in the stroma in any of the groups. There was also no staining of the tumor stroma in the pancreas in this study. The positive pixel algorithm in the Twist1 showed that the immunostaining was specific and localized, with minimal background staining. The immunostaining was seen as positive brown staining localized to ductal epithelial carcinoma cells and lightly seen in some of the normal acinar pancreas. Notably, pancreatic cancer had a 3-fold increase in Twist1 levels compared to the normal acinar pancreas, underscoring an important role of Twist1 in pancreatic cancer pathogenesis (Fig. 8E).
Fig. 8.
Twist1 is overexpressed in human pancreatic cancer tissues and colocalizes with AURKA. (A) Twist 1 immunohistochemistry in human normal pancreas tissues viewed at 15.0×. (B) Twist 1 immunohistochemistry in normal human pancreas tissues viewed at 15.1×. (C) Twist 1 immunohistochemistry in human pancreatic cancer tissues viewed at 15.0×. (D) Twist 1 immunohistochemistry in human pancreatic cancer tissues viewed at 15.1×. (E) Graph showing average positivity for the two groups of normal pancreas and pancreatic cancer. Pancreatic cancer had a 3-fold increase in Twist1 compared to the normal acinar pancreas. The number of tissues for the normal group and pancreatic cancer group was 5 and 12 respectively. P=0.000533 (Student's t-test for a two-tailed distribution with unequal variances). (F,G) Representative examples showing Twist1 immunohistochemistry (F) and AURKA immunohistochemistry (G) from the exact same tissue cores. These images show Twist1 and AURKA colocalization in pancreatic adenocarcinoma tissues (arrows). Imaged at 20×. (H) Proposed model showing AURKA-Twist1 axis in promoting EMT and chemoresistance in pancreatic cancer.
To further examine the clinical significance of AURKA-mediated Twist1 regulation, we evaluated 27 additional cases of pancreatic carcinoma and stained adjacent sections (5 µm apart in depth) with both anti-Twist1 and -AURKA antibodies. AURKA was localized predominantly in the nuclei of the tumor cells. Twist1 was present both in the cytoplasm and nuclei of tumor cells. Most importantly, colocalization of Twist1 and AURKA was observed in approximately one third of the cases, which underscores the clinical significance of AURKA–Twist1 axis in promoting pancreatic carcinoma (Fig. 8F,G).
DISCUSSION
Twist1 expression is essential for embryonic development; however, its expression is downregulated after birth and is restricted to quiescent adult stem cells located in mesoderm-derived mesenchymal tissues. In several cancers, the Twist1 gene is reactivated and its expression correlates with poor prognosis and high-grade metastatic tumors (Puisieux et al., 2006). Twist1 levels and activity are also upregulated in patients treated with repeated neoadjuvant chemotherapy, which is correlated with extreme chemoresistance (Mizukami et al., 2014). Thus, there is a critical need to develop therapies that target Twist1 alone or in combination to eliminate these ‘stem cell-like’ tumor cells in multiple cancers, including those of the pancreas.
Several mechanisms have been identified by which Twist1 is transcriptionally upregulated in cancer. However, only a few studies have recognized its regulation at the protein level. Active Ras or TGFβ stabilizes Twist1 levels by triggering its phosphorylation at S68 by MAPKs [p38, c-Jun N-terminal kinases (JNK), and extracellular signal-regulated kinases 1/2] (Hong et al., 2011). Similarly, IL-6 inhibits Twist1 degradation through promoting its phosphorylation at S18 and S20 by casein kinase 2 (Su et al., 2011). c-Src also phosphorylates Twist1, increasing its transcriptional activity (Bourguignon et al., 2010). Akt family proteins phosphorylate Twist1 at S42 causing increased EMT and resistance to DNA damage (Vichalkovski et al., 2010; Xue et al., 2012). PKA phosphorylates Twist1 at T121 and S123, which increases its oncogenic potential and promotes heterodimerization with E12 (Gajula et al., 2015). Taken together, these findings show that phosphorylation of Twist1 can affect its protein levels, subcellular localization, transcriptional activity and dimer choice, thereby leading to diverse consequences.
In this study, we identified two mechanisms by which Twist1 is regulated by AURKA: (1) through an increase in levels and (2) through mediating its phosphorylation. Although Twist1 is stabilized via phosphorylation at each of the three sites identified, it appears that phosphorylation also affects its partner selection and transcriptional activity independently of its levels. The phosphorylation-dead Twist1 mutant prevents the promoter activation of various EMT-inducing proteins while concurrently failing to repress the E-cadherin promoter, suggesting that inhibition of Twist1 phosphorylation impairs its transcriptional activity. Consequently, 3A-Twist1 serves as a dominant-negative that quelled the inherent aggressiveness of the parental BxPC3 cells. This is not surprising as S123 and T148 sites reside on the HLH domain of Twist1, while S184 is within the Twist box. Twist1 homodimerizes or heterodimerizes via its HLH domain and its transcriptional activity resides in the Twist box (Laursen et al., 2007) (Fig. 5A). Thus, Twist1 phosphorylation at S123 and T148 may regulate its partner binding, while S184 may directly impact its transcriptional activity. Importantly, S123 and T148 sites are conserved in E12, but not Hand2, suggesting that AURKA may also phosphorylate E12, which may further impact on the ability of Twist1 to select its partner upon phosphorylation. In addition, modulation in Twist1 levels due to AURKA may also favor homodimerization irrespective of phosphorylation. Thus, multiple layers of regulation likely exist for Twist1-mediated signaling. Future studies are needed to unravel these mechanisms both under physiological and pathological conditions.
Slug and Snail are highly expressed in PDAC tissues, but not in normal pancreas (Hotz et al., 2007). Snail and Slug repress E-cadherin, leading to tumor invasion (Peinado et al., 2004). N-cadherin and MMP-2 upregulation also cause enhanced metastasis. CD44 is a CSC marker that is highly expressed in PDAC, which is further enriched upon chemotherapy leading to cancer relapse. Targeting CD44 using the corresponding antibody in nude mice harboring gencitabane-resistant pancreatic patient-derived xenografts resulted in complete tumor eradication (Molejon et al., 2015). This finding strongly suggests that targeting CD44 is an effective strategy to target highly chemoresistant PDAC. Thus, our results showing AURKA as an upstream regulator of Snail, Slug, N-cadherin, MMP-2 and CD44 highlight the clinical potential of AURKA-targeted drugs in eradicating EMT and CSCs in PDAC. We also show 3-fold higher levels of Twist1 in human pancreatic cancer specimens compared to normal pancreas.
Although several kinases that regulate Twist1 have been identified to date, this is the first study that shows Twist1 in a reciprocal feedback loop with its activating kinase (Fig. 8H). This finding is extremely important for two reasons. First, AURKA inhibition provides a potent tool to reduce Twist1 levels and inhibit its downstream signaling, which is highly desirable for PDAC therapy. Twist1 inhibitors are currently not available, presumably due to its non-druggable nature. Second, the reciprocal loop between Twist1 and AURKA ensures that their concurrent inhibition will be highly synergistic in inhibiting PDAC tumorigenesis, chemoresistance and metastasis when Twist1 inhibitors become available.
In conclusion, we unraveled a novel mechanism of Twist1 regulation that is triggered by AURKA in a phosphorylation-dependent manner (Fig. 8H). AURKA overexpression is associated with enhanced chemoresistance in cancers of multiple origins. Our discovery of Twist1 as an AURKA substrate provides the molecular mechanism by which AURKA promotes chemoresistance and stem cell formation in pancreatic cancer via Twist1 and vice versa.
MATERIALS AND METHODS
Validated antibodies against AURKA (H-130), actin (C-2), and Twist1 (H-81) were purchased from Santa Cruz Biotech (Santa Cruz, CA). Snail (40084), Slug (40088), N-cadherin (39429) and CD44 (39435) antibodies were purchased from One World Lab. Antibodies against Vimentin (bs-0756R), E-cadherin (bs-10009R) and MMP-2 (bs-4599R) were purchased from Bioss. All antibodies are validated and were used at a 1:1000 dilution. BxPC3, Panc1, Phoenix and HEK293T cells were purchased from ATCC and used within 2 months. MLN8237 was purchased from Adooq.
Twist1 shRNA
Twist1 and AURKA shRNAs were cloned into pLKO.1 TRC vector, which was a gift from David Root (Broad Institute of MIT and Harvard, USA). Stable cells were generated using puromycin selection. The sequences are as follows: Twist1 shRNA1 (forward), 5′-CCGGAAGATGGCAAGCTGCAGCTATCTCGAGATAGCTGCAGCTTGCCATCTTTTTTTG-3′; Twist1 shRNA1 (reverse), 5′-AATTCAAAAAAAGATGGCAAGCTGCAGCTATCTCGAGATAGCTGCAGCTTGCCATCTT-3′; Twist1 shRNA2 (forward), 5′-CCGGGGTACATCGACTTCCTCTACCCTCGAGGGTAGAGGAAGTCGATGTACCTTTTTG-3′; Twist1 shRNA2 (reverse), 5′-AATTCAAAAAGGTACATCGACTTCCTCTACCCTCGAGGGTAGAGGAAGTCGATGTACC-3′; AURKA shRNA1 (forward), 5′-CCGGGGCTTTGGAAGACTTTGAAATCTCGAGATTTCAAAGTCTTCCAAAGCCTTTTTG-3′; AURKA shRNA1 (reverse), 5′-AATTCAAAAAGGCTTTGGAAGACTTTGAAATCTCGAGATTTCAAAGTCTTCCAAAGCC-3′. AURKA shRNA2 (forward), 5′-CCGGGCACCACTTGGAACAGTTTATCTCGAGATAAACTGTTCCAAGTGGTGCTTTTTG-3′; AURKA shRNA2 (reverse): 5′-AATTCAAAAAGCACCACTTGGAACAGTTTATCTCGAGATAAACTGTTCCAAGTGGTGC-3′. AURKA shRNA3 (forward), 5′-CCGGGCCAATGCTCAGAGAAGTACTCTCGAGAGTACTTCTCTGAGCATTGGCTTTTTG-3′. AURKA shRNA3 (reverse), 5′-AATTCAAAAAGCCAATGCTCAGAGAAGTACTCTCGAGAGTACTTCTCTGAGCATTGGC-3′.
Expression plasmids and constructs
pBABE-puro-mTwist is Addgene plasmid #1783, deposited by Robert Weinberg (Whitehead Institute for Biomedical Research, Boston, USA; Yang et al., 2004). HA-tagged Twist1 was cloned into the TAT-HA vector at the BamHI and EcoRI sites. HA-tagged Twist1 mutants were generated using site-directed mutagenesis.
Expression and purification of AURKA, TPX2 and Twist1
AURKA and His6–TPX2 was generated as reported previously (Johnson et al., 2011). His6-tagged wild-type and mutant Twist1 were expressed in E. coli and purified as described previously (Sun et al., 2008a).
In vitro kinase assays
The AURKA–TPX2 complex (on beads) was pre-incubated with 100 μM of ATP for 15 min in a kinase buffer to activate AURKA and reduce background phosphorylation. The beads were washed twice with kinase buffer, and subjected to a kinase assay with His6-tagged wild-type and mutant Twist1 proteins using [32P]ATP. MLN8237-mediated inhibition of AURKA was investigated using AURKA peptide substrate as reported previously (Johnson et al., 2011).
Immunofluorescence
BxPC3, HA-Twist1-BxPC3, HA-3A-Twist1-BxPC3 or Panc1 cells were grown on poly-L-lysine-coated coverslips for 24 h, fixed with 4% formaldehyde in PBS for 15 min at room temperature, and then washed three times with PBS. For AURKA inhibition, cells were treated with MLN8237 (1 µM) for 16 h, followed by fixation. For AURKA knockdown, unsynchronized cells were either treated with scrambled or AURKA shRNA for 30 h. The cells were fixed and then blocked in 1% fetal bovine serum (FBS), 2% bovine serum albumin (BSA) and 0.1% Triton X-100 in PBS for 1 h at 25°C. Cells were labeled with antibodies (for AURKA or Twist1) for 3 h in PBS, followed by incubation with fluorescein-isothiocyanate- or Texas-Red-conjugated secondary antibody. Cells were visualized using Nikon Eclipse E600 microscope (Nikon Instruments, Melville, NY).
Immunohistochemistry
Immunostaining
Unstained slides of tumor xenograft, normal pancreas, chronic pancreatic and stage 4 ductal pancreatic carcinoma patient tissues were deparaffinized in xylene and rehydrated using graded alcohols and water. Antigen retrieval was performed as described previously (Johnson et al., 2011). Control sections were treated with an isotype control using the same concentration as primary antibody to verify the staining specificity.
For analyzing the potential colocalization of AURKA and Twist1 in pancreatic carcinoma, 27 additional cases of human pancreatic adenocarcinoma were evaluated with both anti-Twist 1 and -AURKA antibodies through tissue microarrays. Stains were very clean and no background staining was observed. Informed consent was obtained for all tissue donors and all clinical investigation have been conducted according to the principles expressed in the Declaration of Helsinki.
Whole-slide digital imaging
The Aperio whole-slide digital imaging system was used for imaging at either 15×, 15.1× or 20×. The photo microscopy images were captured from the whole slide images.
Automated image quantitation
Computer-assisted morphometric analysis of digital images was carried out using the Aperio Image scope software of the Aperio whole-slide imaging system called the positive pixel algorithm. This was used to quantify the Twist1 positivity in normal and cancer tissues.
Dual-luciferase assay
Luciferase assays were conducted as described previously (Shi et al., 2016). Briefly, BxPC3 cells were plated in a 96-well plate at a density of 3000 cells/well. After 12 h, the cells were transfected with 50 ng/well of Snail, Slug, CD44 or E-cadherin promoter-driven luciferase plasmids using lipofectamine (Invitrogen). Cells were simultaneously co-transfected with 50 ng/well of the pRL-SV40 Renilla luciferase plasmid (Promega) as an internal control. After 48 h, firefly and Renilla luciferase activities were measured with the dual-luciferase kit (Thermo-Scientific). The firefly luciferase signal was normalized to the Renilla luciferase signal to account for variations in transfection efficiency. All experiments were performed in triplicate, three independent times.
Pancreatosphere assay
Cells were plated in ultralow attachment plates (Corning Costar Corp., Cambridge, MA) and grown in a serum-free RPMI1640 and Nutrient Mixture F-12 medium supplemented with B27 (1:50, Gibco), 20 ng/ml epithelial growth factor (EGF), 20 ng/ml basic fibroblast growth factor (R&D System, Minneapolis, MN), 5 μg/ml bovine insulin, and penicillin-streptomycin. In the sphere culture conditions, BxPC3 cells were grown for 5 days and pancreatospheres were visualized under the microscope.
Soft agar colony formation
BxPC3 , stable Twist1-BxPC3 and 3A-Twist1-BxPC3 cell lines were plated in RPMI containing 0.3% agar and 10% fetal bovine serum in six-well plates as reported before (Shah and Vincent, 2005). Transformed colonies were counted after 3 weeks.
Ubiquitylation assay
The ubiquitylation assay was conducted as described previously (Johnson et al., 2011).
Chemotaxis assay
Cell migration was determined as reported previously (Shah and Vincent, 2005). The assays were performed in triplicate, four independent times. To allow for comparison between multiple assays, the data were normalized and expressed as fold change with respect to the number of cells present on the membrane.
MTT assay
The MTT assay was conducted as reported previously (Sun et al., 2009). Experiments were repeated three times in triplicate wells to ensure the reproducibility. Gemcitabine was used at 1 µM concentration for drug resistance assay.
In vivo xenograft in nude mice
All the animal experiments were done in accordance with institutional guidelines of the Purdue Animal Care and Use Committee. Female athymic nude mice at 4–5 weeks of age were obtained from Taconic Laboratories and injected as reported previously (Johnson et al., 2012). Mice bearing tumors did not display any weight loss compared with control mice at the time of killing. The animals were killed 32 days following tumor induction, and the tumor tissues were isolated and frozen in liquid nitrogen immediately or placed in paraformaldehyde for histology.
Statistical analysis
Bar graphs results are plotted as the mean±s.e.m. Significance was evaluated using a two-tailed Student's t-test or one- or two-way ANOVA and is displayed as: *P<0.05, **P<0.01, ***P<0.001.
Acknowledgements
We thank Dr Anthony Firulli (Department of Pediatrics, Indiana University, USA) for Hand2 and E12 plasmids.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
K.S. conceived experiments, wrote the manuscript, and secured funding. J.W., K.N., K.V., L.C., M.J. performed experiments. G.S. conceived experiments and provided expertise and feedback.
Funding
This work was supported by the National Institutes of Health (R03 CA 166912 to K.S.). K.S. also gratefully acknowledges the Biological Evaluation facility and support from the Purdue University Center for Cancer Research (NIH grant P30 CA023168) for in vivo experiments. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.196790.supplemental
References
- Alian O. M., Philip P. A., Sarkar F. H. and Azmi A. S. (2014). Systems biology approaches to pancreatic cancer detection, prevention and treatment. Curr. Pharm. Des. 20, 73-80. 10.2174/138161282001140113124643 [DOI] [PubMed] [Google Scholar]
- Bourguignon L. Y. W., Wong G., Earle C., Krueger K. and Spevak C. C. (2010). Hyaluronan-CD44 interaction promotes c-Src-mediated twist signaling, microRNA-10b expression, and RhoA/RhoC up-regulation, leading to Rho-kinase-associated cytoskeleton activation and breast tumor cell invasion. J. Biol. Chem. 285, 36721-36735. 10.1074/jbc.M110.162305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos J. A., Merchant N. B. and Nagathihalli N. S. (2013). Emerging targets in pancreatic cancer: epithelial-mesenchymal transition and cancer stem cells. Onco. Targets Ther. 6, 1261-1267. 10.2147/OTT.S34670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas E., Kim J., Bendesky A., Ohno-Machado L., Wolfe C. J. and Yang J. (2011) Snail2 is an essential mediator of Twist1-induced epithelial mesenchymal transition and metastasis. Cancer Res. 71, 245-254. 10.1158/0008-5472.CAN-10-2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K.-H., Multani P. S., Sun K.-H., Vincent F., de Pablo Y., Ghosh S., Gupta R., Lee H.-P., Lee H.-g., Smith M. A. et al. (2011). Nuclear envelope dispersion triggered by deregulated Cdk5 precedes neuronal death. Mol. Biol. Cell 22, 1452-1462. 10.1091/mbc.E10-07-0654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K.-H., Vincent F. and Shah K. (2012). Deregulated Cdk5 triggers aberrant activation of cell cycle kinases and phosphatases inducing neuronal death. J. Cell Sci. 125, 5124-5137. 10.1242/jcs.108183 [DOI] [PubMed] [Google Scholar]
- D'Assoro A. B., Liu T., Quatraro C., Amato A., Opyrchal M., Leontovich A., Ikeda Y., Ohmine S., Lingle W., Suman V., et al. (2014). The mitotic kinase Aurora--a promotes distant metastases by inducing epithelial-to-mesenchymal transition in ERα(+) breast cancer cells. Oncogene 33, 599-610. 10.1038/onc.2012.628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S., Marin O., Pagano M. A., Meggio F., Hess D. El-Shemerly M., Krystyniak A. and Pinna L. A. (2005). Aurora-A site specificity: a study with synthetic peptide substrates. Biochem. J. 390, 293-302. 10.1042/BJ20050343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli A. B. and Conway S. J. (2008). Phosphoregulation of Twist1 provides a mechanism of cell fate control. Curr. Med. Chem. 15, 2641-2647. 10.2174/092986708785908987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Qin L., He T., Qin J., Hong J., Wong J., Liao L. and Xu J. (2011). The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res. 21, 275-289. 10.1038/cr.2010.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco H. L., Casasnovas J., Rodríguez-Medina J. R. and Cadilla C. L. (2011). Redundant or separate entities?--roles of Twist1 and Twist2 as molecular switches during gene transcription. Nucleic Acids Res. 39, 1177-1186. 10.1093/nar/gkq890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajula R. P., Chettiar S. T., Williams R. D., Nugent K., Kato Y., Wang H., Malek R., Taparra K., Cades J., Annadanam A. et al. (2015). Structure-function studies of the bHLH phosphorylation domain of TWIST1 in prostate cancer cells. Neoplasia 17, 16-31. 10.1016/j.neo.2014.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata T., Furukawa T., Sunamura M., Egawa S., Motoi F., Ohmura N., Marumoto T., Saya H. and Horii A. (2005). RNA interference targeting aurora kinase a suppresses tumor growth and enhances the taxane chemosensitivity in human pancreatic cancer cells. Cancer Res. 65, 2899-2905. 10.1158/0008-5472.CAN-04-3981 [DOI] [PubMed] [Google Scholar]
- Hong J., Zhou J., Fu J., He T., Qin J., Wang L., Liao L. and Xu J. (2011). Phosphorylation of serine 68 of Twist1 by MAPKs stabilizes Twist1 protein and promotes breast cancer cell invasiveness. Cancer Res. 71, 3980-3990. 10.1158/0008-5472.CAN-10-2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotz B., Arndt M., Dullat S., Bhargava S., Buhr H.-J. and Hotz H. G. (2007). Epithelial to mesenchymal transition: expression of the regulators snail, slug, and twist in pancreatic cancer. Clin. Cancer Res. 13, 4769-4776. 10.1158/1078-0432.CCR-06-2926 [DOI] [PubMed] [Google Scholar]
- Johnson E. O., Chang K. H., de Pablo Y., Ghosh S., Mehta R., Badve S. and Shah K. (2011). PHLDA1 is a critical negative regulator and effector of Aurora A kinase in breast cancer. J. Cell Sci. 124, 2711-2722. 10.1242/jcs.084970 [DOI] [PubMed] [Google Scholar]
- Johnson E. O., Chang K.-H., Ghosh S., Venkatesh C., Giger K., Low P. S. and Shah K. (2012). LIMK2 is a crucial regulator and effector of Aurora-A-kinase-mediated malignancy. J. Cell Sci. 125, 1204-1216. 10.1242/jcs.092304 [DOI] [PubMed] [Google Scholar]
- Kim S. and Shah K. (2007). Dissecting yeast Hog1 MAP kinase pathway using a chemical genetic approach. FEBS Lett. 581, 1209-1216. 10.1016/j.febslet.2007.02.032 [DOI] [PubMed] [Google Scholar]
- Lander R., Nordin K. and LaBonne C. (2011). The F-box protein Ppa is a common regulator of core EMT factors Twist, Snail, Slug, and Sip1. J. Cell Biol. 194, 17-25. 10.1083/jcb.201012085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen K. B., Mielke E., Iannaccone P. and Füchtbauer E. M. (2007). Mechanism of transcriptional activation by the proto-oncogene Twist1. J. Biol. Chem. 282, 34623-34633. 10.1074/jbc.M707085200 [DOI] [PubMed] [Google Scholar]
- Li D., Zhu J., Firozi P. F., Abbruzzese J. L., Evans D. B., Cleary K., Friess H. and Sen S. (2003). Overexpression of oncogenic STK15/BTAK/AURKA kinase in human pancreatic cancer. Clin. Cancer Res. 9, 991-997. [PubMed] [Google Scholar]
- Li J. and Zhou B. P. (2011) Activation of β-catenin and Akt pathways by Twist are critical for the maintenance of EMT associated cancer stem cell-like characters. BMC Cancer. 11, 49 10.1186/1471-2407-11-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami T., Kamachi H., Mitsuhashi T., Tsuruga Y., Hatanaka Y., Kamiyama T., Matsuno Y. and Taketomi A. (2014). Immunohistochemical analysis of cancer stem cell markers in pancreatic adenocarcinoma patients after neoadjuvant chemoradiotherapy. BMC Cancer 14, 687 10.1186/1471-2407-14-687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molejon M. I., Tellechea J. I., Loncle C., Gayet O., Gilabert M., Duconseil P., Lopez-Millan M. B., Moutardier V., Gasmi M., Garcia S. et al. (2015). Deciphering the cellular source of tumor relapse identifies CD44 as a major therapeutic target in pancreatic adenocarcinoma. Oncotarget 6, 7408-7423. 10.18632/oncotarget.3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka R., Itoh S., Gui T., Gai Z., Oikawa K., Kawai M., Tani M., Yamaue H. and Muragaki Y. (2010). SNAIL induces epithelial-to-mesenchymal transition in a human pancreatic cancer cell line (BxPC3) and promotes distant metastasis and invasiveness in vivo. Exp. Mol. Pathol. 89, 149-157. 10.1016/j.yexmp.2010.05.008 [DOI] [PubMed] [Google Scholar]
- Peinado H., Portillo F. and Cano A. (2004). Transcriptional regulation of cadherins during development and carcinogenesis. Int. J. Dev. Biol. 48, 365-375. 10.1387/ijdb.041794hp [DOI] [PubMed] [Google Scholar]
- Puisieux A., Valsesia-Wittmann S. and Ansieau S. (2006). A twist for survival and cancer progression. Br. J. Cancer. 94, 13-17. 10.1038/sj.bjc.6602876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q., Xu Y., He T., Qin C. and Xu J. (2012). Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms. Cell Res. 22, 90-106. 10.1038/cr.2011.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojanala S., Han H., Muñoz R. M., Browne W., Nagle R., Von Hoff D. D. and Bearss D. J. (2004). The mitotic serine threonine kinase, Aurora-2, is a potential target for drug development in human pancreatic cancer. Mol. Cancer Ther. 3, 451-457. [PubMed] [Google Scholar]
- Shah K. and Shokat K. M. (2002). A chemical genetic screen for direct v-Src substrates reveals ordered assembly of a retrograde signaling pathway. Chem. Biol. 9, 35-47. 10.1016/S1074-5521(02)00086-8 [DOI] [PubMed] [Google Scholar]
- Shah K. and Shokat K. M. (2003). A chemical genetic approach for the identification of direct substrates of protein kinases. Methods Mol. Biol. 233, 253-271. 10.1385/1-59259-397-6:253 [DOI] [PubMed] [Google Scholar]
- Shah K. and Vincent F. (2005). Divergent roles of c-Src in controlling platelet-derived growth factor-dependent signaling in fibroblasts. Mol. Biol. Cell 16, 5418-5432. 10.1091/mbc.E05-03-0263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah K., Liu Y., Deirmengian C. and Shokat K. M. (1997). Engineering unnatural nucleotide specificity for Rous sarcoma virus tyrosine kinase to uniquely label its direct substrates. Proc. Natl. Acad. Sci. USA 94, 3565-3570. 10.1073/pnas.94.8.3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C., Viccaro K., Lee H. G. and Shah K. (2016). Cdk5-FOXO3a axis: initially neuroprotective, eventually neurodegenerative in Alzheimer's disease models. J. Cell Sci. 129, 1815-1830. 10.1242/jcs.185009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y.-W., Xie T.-X., Sano D. and Myers J. N. (2011). IL-6 stabilizes Twist and enhances tumor cell motility in head and neck cancer cells through activation of casein kinase 2. PLoS ONE 6, e19412 10.1371/journal.pone.0019412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K.-H., de Pablo Y., Vincent F., Johnson E. O., Chavers A. K. and Shah K. (2008a). Novel genetic tools reveal Cdk5's major role in golgi fragmentation in Alzheimer's disease. Mol. Biol. Cell 19, 3052-3069. 10.1091/mbc.E07-11-1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K.-H., de Pablo Y., Vincent F. and Shah K. (2008b). Deregulated Cdk5 promotes oxidative stress and mitochondrial dysfunction. J. Neurochem. 107, 265-278. 10.1111/j.1471-4159.2008.05616.x [DOI] [PubMed] [Google Scholar]
- Sun K. H., Lee H. G., Smith M. A. and Shah K. (2009). Direct and indirect roles of cyclin-dependent kinase 5 as an upstream regulator in the c-Jun NH2-terminal kinase cascade: relevance to neurotoxic insults in Alzheimer's disease. Mol. Biol. Cell. 20, 4611-4619. 10.1091/mbc.E09-05-0433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vichalkovski A., Gresko E., Hess D., Restuccia D. F. and Hemmings B. A. (2010). PKB/AKT phosphorylation of the transcription factor Twist-1 at Ser42 inhibits p53 activity in response to DNA damage. Oncogene 29, 3554-3565. 10.1038/onc.2010.115 [DOI] [PubMed] [Google Scholar]
- Wang F., Li H., Yan X. G., Zhou Z. W., Yi Z. G., He Z. X., Pan S. T., Yang Y. X., Wang Z. Z., Zhang X. et al. (2015). Alisertib induces cell cycle arrest and autophagy and suppresses epithelial-to-mesenchymal transition involving PI3K/Akt/mTOR and sirtuin 1-mediated signaling pathways in human pancreatic cancer cells. Drug Des. Devel Ther. 9, 575-601. 10.2147/DDDT.S75221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf C., Thisse C., Stoetzel C., Thisse B., Gerlinger P. and Perrin-Schmitt F. (1991). The M-twist gene of Mus is expressed in subsets of mesodermal cells and is closely related to the Xenopus X-twi and the Drosophila twist genes. Dev. Biol. 143, 363-373. 10.1016/0012-1606(91)90086-I [DOI] [PubMed] [Google Scholar]
- Xue G., Restuccia D. F., Lan Q., Hynx D., Dirnhofer S., Hess D., Rüegg C. and Hemmings B. A. (2012). Akt/PKB-mediated phosphorylation of Twist1 promotes tumor metastasis via mediating cross-talk between PI3K/Akt and TGF-β signaling axes. Cancer Discov. 2, 248-259. 10.1158/2159-8290.CD-11-0270 [DOI] [PubMed] [Google Scholar]
- Yang J., Mani S. A., Donaher J. L., Ramaswamy S., Itzykson R. A., Come C., Savagner P., Gitelman I., Richardson A. and Weinberg R. A. (2004). Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117, 927-939. 10.1016/j.cell.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Zhong J., Ogura K., Wang Z. and Inuzuka H. (2013). Degradation of the transcription factor Twist, an oncoprotein that promotes cancer metastasis. Discov. Med. 15, 7-15. [PMC free article] [PubMed] [Google Scholar]