Significance
In the initiation of protein synthesis, a preinitiation complex (PIC) of the 40S ribosomal subunit, initiation factors, and initiator tRNAi scans the mRNA leader for an AUG codon in favorable context; and AUG recognition evokes a closed conformation of the PIC with more tightly bound tRNAi. uS3 (Rps3 in yeast) is a protein in the 40S mRNA entry channel, whose function during initiation was unknown. Substituting uS3 arginine residues in contact with mRNA reduces initiation at suboptimal start codons (UUG and AUG in poor context), weakens ribosome–mRNA interaction specifically at the entry channel, and destabilizes tRNAi binding selectively at UUG codons. Thus, uS3 promotes mRNA:40S interaction at the entry channel to enhance initiation accuracy.
Keywords: translation, initiation, uS3, ribosome, yeast
Abstract
The eukaryotic 43S preinitiation complex (PIC) bearing Met-tRNAiMet in a ternary complex (TC) with eukaryotic initiation factor (eIF)2-GTP scans the mRNA leader for an AUG codon in favorable “Kozak” context. AUG recognition provokes rearrangement from an open PIC conformation with TC bound in a state not fully engaged with the P site (“POUT”) to a closed, arrested conformation with TC tightly bound in the “PIN” state. Yeast ribosomal protein Rps3/uS3 resides in the mRNA entry channel of the 40S subunit and contacts mRNA via conserved residues whose functional importance was unknown. We show that substitutions of these residues reduce bulk translation initiation and diminish initiation at near-cognate UUG start codons in yeast mutants in which UUG selection is abnormally high. Two such substitutions—R116D and R117D—also increase discrimination against an AUG codon in suboptimal Kozak context. Consistently, the Arg116 and Arg117 substitutions destabilize TC binding to 48S PICs reconstituted in vitro with mRNA harboring a UUG start codon, indicating destabilization of the closed PIN state with a UUG–anticodon mismatch. Using model mRNAs lacking contacts with either the mRNA entry or exit channels of the 40S subunit, we demonstrate that Arg116/Arg117 are crucial for stabilizing PIC–mRNA contacts at the entry channel, augmenting the function of eIF3 at both entry and exit channels. The corresponding residues in bacterial uS3 promote the helicase activity of the elongating ribosome, suggesting that uS3 contacts with mRNA enhance multiple phases of translation across different domains of life.
Accurate identification of the translation initiation codon in mRNA by ribosomes is crucial for expression of the correct cellular proteins. This process generally occurs in eukaryotic cells by a scanning mechanism wherein the small (40S) ribosomal subunit first recruits charged initiator tRNA (Met-tRNAiMet) in a ternary complex (TC) with eukaryotic initiation factor (eIF)2-GTP in a reaction stimulated by eIFs 1, 1A, 3, and 5. The resulting 43S preinitiation complex (PIC) attaches to the 5′ end of the mRNA and scans the 5′ UTR with TC bound in a metastable state, “POUT,” suitable for inspecting successive triplets for complementarity with the anticodon of Met-tRNAiMet in the P site, to identify the AUG start codon. Nucleotides surrounding the AUG, particularly at the −3 and +4 positions (the Kozak context), further influence the efficiency of start-codon selection. In the scanning PIC, eIF2 can hydrolyze GTP, dependent on GTPase-activating protein eIF5, but Pi release is blocked by eIF1, whose presence also impedes stable binding of Met-tRNAiMet in the “PIN” state. Start-codon recognition triggers dissociation of eIF1 from the 40S subunit, allowing Pi release from eIF2-GDP•Pi and TC binding in the PIN state of the 48S PIC (Fig. 1A). Subsequent dissociation of eIF2-GDP and other eIFs from the 48S PIC enables eIF5B-catalyzed subunit joining and formation of an 80S initiation complex with Met-tRNAiMet base-paired to AUG in the P site (reviewed in ref. 1).
Fig. 1.
Rps3/uS3 plays a critical role in promoting mRNA binding at the 40S entry site and stabilizing the preinitiation complex at the start codon. (A) Model describing known conformational rearrangements of the PIC during scanning and start-codon recognition. (A, i) eIF1 and the scanning enhancers (SEs) in the C-terminal tail (CTT) of eIF1A stabilize an open conformation of the 40S subunit to which TC rapidly binds. Rps3 (uS3) is located on the solvent-exposed surface of the 40S subunit near the entry channel; the bulk of eIF3 binds on the solvent-exposed surface, with a prominent domain at the mRNA exit channel. (A, ii) The 43S PIC in the open conformation scans the mRNA for the start codon with Met-tRNAiMet bound in the POUT state. eIF2 can hydrolyze GTP to GDP•Pi, but release of Pi is blocked. (A, iii) On AUG recognition, Met-tRNAiMet moves from the POUT to the PIN state, clashing with eIF1 and the CTT of eIF1A, provoking displacement of the eIF1A CTT from the P site, dissociation of eIF1 from the 40S subunit, and Pi release from eIF2. The N-terminal tail (NTT) of eIF1A, harboring scanning inhibitor (SI) elements, adopts a defined conformation and interacts with the codon–anticodon helix. (Top) Arrows summarize that eIF1 and the eIF1A SE elements promote POUT and impede transition to the PIN state, whereas the SI element in the NTT of eIF1A stabilizes the PIN state. Results presented here indicate that uS3/Rps3 residues R116/R117, in contact with mRNA at the entry channel, stabilize the PIN state and also promote PIC interaction with mRNA at the entry channel, augmenting the role of eIF3 in PIC–mRNA interactions at the exit channel (adapted from ref. 1). (B) Alignment of a portion of uS3 sequences from diverse eukaryotes and Escherichia coli using Clustal Omega (www.ebi.ac.uk/Tools/msa/clustalo/). Boundaries of secondary structure on the top line refer to the Saccharomyces protein; symbolic summary of sequence conservation applies only to the upper seven eukaryotic sequences. Six conserved residues of Rps3 at the mRNA entry channel analyzed in this study are highlighted in black or pink. (C) Position of uS3/Rps3 in the yeast 48S PIC, and locations of conserved residues at the entry channel. The solvent-exposed surface of the partial yeast 48S PIC [Protein Data Bank (PDB) ID code 3J81] is depicted (Left) in cartoon format highlighting uS3 (green), mRNA (black), Met-tRNAiMet (pink), and rRNA residues of h18 or h34 that comprise the entry channel latch (blue). The boxed region is amplified (Right) where the six uS3/Rps3 residues analyzed here, which interact with mRNA or the rRNA latch, are highlighted in black or magenta (only for ease of visualization), shown in stick format, and labeled. Other ribosomal proteins and eIFs 1, 1A, and 2 are hidden for clarity.
eIF1 plays a dual role in the scanning mechanism, promoting rapid TC loading in the POUT conformation while blocking rearrangement to PIN at both near-cognate start codons (e.g., UUG) and cognate (AUG) codons in poor Kozak context; hence eIF1 must dissociate from the 40S subunit for start-codon recognition (Fig. 1A). Consistent with this, structural analyses of partial PICs reveal that eIF1 and eIF1A promote rotation of the 40S head relative to the body (2, 3), thought to be instrumental in TC binding in the POUT conformation, but that eIF1 physically clashes with Met-tRNAiMet in the PIN state (2, 4), and is both deformed and displaced from its 40S location during the POUT-to-PIN transition (3). Mutations that weaken eIF1 binding to the 40S subunit reduce the rate of TC loading and elevate initiation at near-cognate codons or AUGs in poor context as a result of destabilizing the open/POUT conformation and favoring rearrangement to the closed/PIN state during scanning (5, 6). Moreover, decreasing wild-type (WT) eIF1 abundance reduces initiation accuracy, whereas overexpressing eIF1 suppresses initiation at near cognates or AUGs in poor context (5, 7–10). In fact, cells exploit the mechanistic link between eIF1 abundance and initiation accuracy to autoregulate eIF1 expression: The AUG codon of the eIF1 gene (SUI1 in yeast) occurs in poor context, and the frequency of its recognition is inversely related to eIF1 abundance (5, 10).
The stability of the codon–anticodon duplex is an important determinant of initiation accuracy, as the rate of the POUT-to-PIN transition is accelerated and the PIN state is stabilized in the presence of AUG versus non-AUG start codons (11). Favorable Kozak context might also contribute to PIN stability (5, 12), but the stimulatory effect of optimum context on initiation rate is not well-understood. It seems to require the α-subunit of eIF2 (12), and structural analyses of partial mammalian 43S (13) and yeast 48S PICs (3) place eIF2α domain 1 near the key −3 context nucleotide in the exit channel of the 40S subunit. The conserved β-hairpin of 40S protein uS7 (Rps5 in yeast) also occurs in this vicinity in a yeast partial 48S (py48S) PIC (3). We have shown that the β-hairpin of yeast Rps5 is important for both efficient and accurate translation initiation in vivo and the stability of PIN complexes reconstituted in vitro (14). Approximately 7 additional mRNA nucleotides upstream of the −3 position occupy the 40S exit channel, and there is evidence that these 40S–mRNA interactions plus additional contacts between segments of eIF3 and mRNA nucleotides protruding from the exit channel also enhance PIC assembly at the start codon (15–18).
eIF3 is a multisubunit complex that binds directly to the 40S subunit and promotes both recruitment and stable association of TC and mRNA with the PIC, and enhances scanning and accurate start-codon recognition in vivo (19). Recent structural analyses (20–22), combined with earlier biochemical and genetic studies (19), reveal that different subunits/domains of yeast eIF3 interact with the PIC at multiple sites, effectively encircling the PIC and interacting with both the mRNA entry and exit pores on the solvent-exposed surface, as well as with the decoding center on the interface surface of the 40S subunit. The major point of contact involves binding of a heterodimer of the proteasome-COP9-initiation factor (PCI) domains of the eIF3a and c subunits to the 40S solvent side, below the platform and exit channel pore. A second contact occurs near the entry channel and involves segments of the eIF3a, b, g, and i subunits; at least some of these interactions appear to be dynamic, as alternative contacts with eIFs anchored in the decoding center have also been observed (20–22). We recently presented evidence that the PCI domain of eIF3a mediates a key stabilizing interaction between the PIC and mRNA at the exit channel (18). Using a panel of m7G-capped, unstructured model mRNAs to reconstitute 48S PICs, we determined that the eIF3a PCI domain interaction at the exit channel is functionally redundant with mRNA–PIC interactions at the entry channel, and is essential for stable 48S PIC assembly when the mRNA is truncated in a way that leaves the entry channel largely empty. Other eIF3 domains/subunits contribute to the functionally redundant contacts at the entry channel, but are not essential even when the opposite exit channel is unoccupied by mRNA. This suggests that other components of the PIC, including elements of the ribosome itself, participate in stabilizing mRNA binding at the entry channel.
In fact, the cryo-EM structure of a partial yeast 48S PIC revealed predicted contacts between mRNA residues located 6 to 12 nt downstream (3′) of the AUG codon with particular amino acids of ribosomal protein Rps3/uS3 at the mRNA entry channel of the 40S subunit. Several Rps3 residues also appear to interact with 18S rRNA residues that form the “latch” on the entry channel, a noncovalent interaction between rRNA nucleotides in helices 18 and 34 (3). Interestingly, whereas the latch is closed in the yeast 48S complexes with AUG in the P site and in crystal structures of other partial PICs (2, 4, 23), the latch is open in a recent cryo-EM structure of a PIC formed with mRNA containing an AUC codon, owing to upward movement of the head away from the body of the 40S subunit. The P site is also widened in this open PIC conformation such that the tRNAiMet is not fully engaged with rRNA residues in the body that contribute to the highly stable PIN conformation observed in the corresponding AUG complex (22). Hydroxyl radical probing of yeast PICs reconstituted with AUG or AUC mRNAs also revealed a more open conformation of the P site and less constricted mRNA entry channel in the AUC complex (24), consistent with a scanning-conducive conformation of the 40S subunit when a near-cognate triplet occupies the P site. Although Rps3 residues appear to interact with the mRNA and with rRNA residues of the entry channel latch, there is no functional evidence that these predicted contacts are important for the efficiency or fidelity of start-codon recognition. Here we provide strong genetic and biochemical evidence that these Rps3 residues enhance the stability of the PIN state and promote recognition of poor initiation sites in vivo, and also mediate stabilizing mRNA interactions with the PIC at the entry channel that are functionally redundant with eIF3-dependent PIC–mRNA interactions at the exit channel.
Results
RPS3 Mutations Restore Discrimination Against Near-Cognate UUG Codons in a Hypoaccurate eIF5 Sui− Mutant in Vivo.
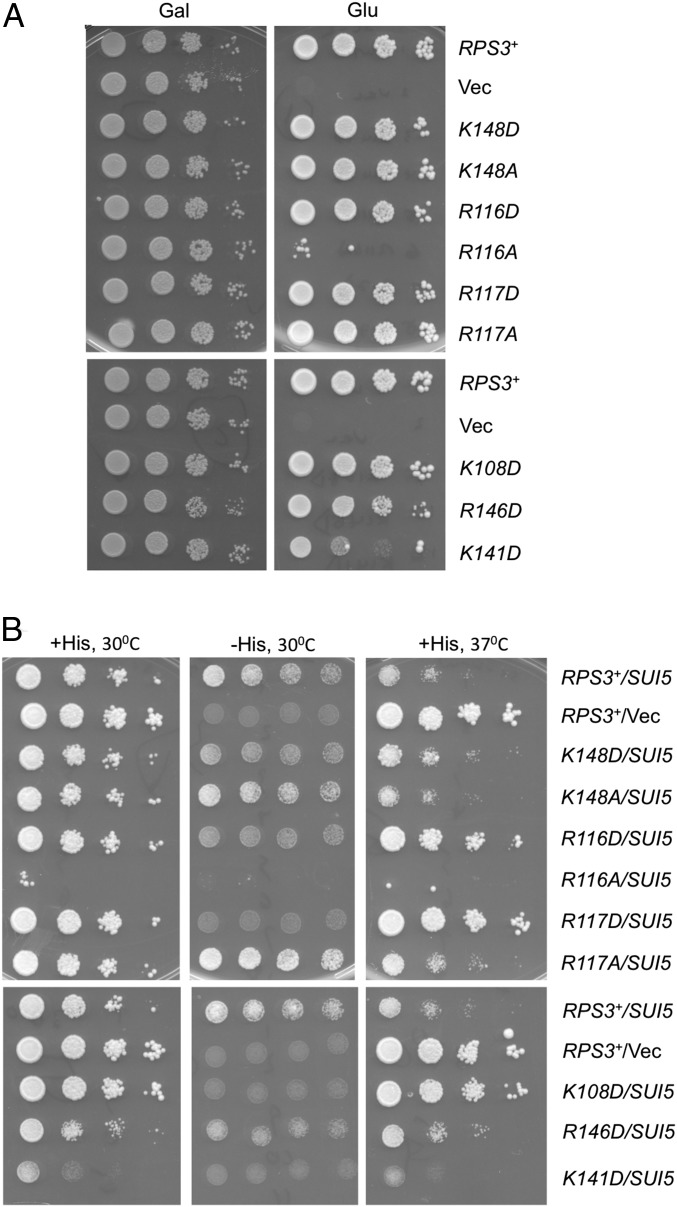
To examine the role of Rps3 in the mRNA entry channel, we introduced single substitutions into six highly conserved basic residues found in proximity to either mRNA nucleotides or rRNA residues in helix 18 (h18) or h34 of the entry channel latch in the cryo-EM structure of the partial yeast 48S PIC, with Met-tRNAiMet base-paired with AUG in the PIN state (3). These include Arg-116 and Arg-117 at the N terminus of helix α5, and Lys-141, Arg-146, and Lys-148 in the loop between β-strands 4 and 5 (Fig. 1 B and C). We also substituted Lys-108 (K108) at the C terminus of α4, based on its proximity to the entry channel and predicted involvement, along with R116/R117, in ribosome helicase activity (25, 26). Residues were substituted with Ala to shorten the side chain, or with acidic residues to alter side-chain charge (SI Appendix, Table S1). The mutations were generated in an RPS3 allele under its own promoter on a low-copy plasmid and examined in a yeast strain with WT chromosomal RPS3 under a galactose-inducible promoter (PGAL1). Mutant phenotypes were scored following a switch from galactose to glucose, where PGAL1-RPS3+ expression is repressed. Mutations K141A and R146A were lethal and prevented growth on glucose medium, whereas others conferred slow-growth (Slg−) phenotypes that were very strong for R116A, moderate for K141D, and slight for R146D (Fig. 2; summarized in SI Appendix, Table S1).
Fig. 2.
Certain RPS3 alleles suppress both the His+ and Slg− phenotypes conferred by eIF5 mutation SUI5 in his4-301 cells. (A) Certain RPS3 alleles confer Slg− phenotypes in otherwise WT cells. Serial dilutions of the following PGAL-RPS3 his4-301 strain (HD2738) harboring the indicated plasmid-borne RPS3 allele or empty vector (Vec) were spotted on SGAL-Leu or SC-Leu medium and incubated for 3 to 4 d at 30 °C: RPS3+ (HD2754), vector (HD2755), K148D (HD2765), K148A (HD2764), R116D (HD2767), R116A (HD2766), R117D (HD2769), R117A (HD2768), K108D (HD3120), R146D (HD2772), and K141D (HD2779). (B) Serial dilutions of the his4-301 strains in A also containing sc SUI5 plasmid p4281 or empty TRP1 vector YCplac22 (Vec) were spotted on SC medium lacking Leu and Trp (SC-L-W) (+His plates) or SC-L-W-H supplemented with 0.0015 mM histidine (0.5% of the standard supplement; −His plates) and incubated at 30 or 37 °C for 3 to 5 d.
To assess changes in initiation fidelity, the RPS3 mutations were tested for the ability to suppress the histidine auxotrophy of his4-301, a mutant allele lacking the AUG start codon, by increasing initiation at the third, in-frame UUG codon and thereby restoring expression of the histidine biosynthetic enzyme His4. Suppression of the histidine auxotrophy conferred by his4-301 in this manner is designated the Sui− (suppressor of initiation codon mutation) phenotype (27). None of the RPS3 mutations allowed detectable growth on glucose medium lacking histidine, suggesting that they do not increase utilization of UUG start codons. Accordingly, we tested them for the Ssu− (suppressor of Sui−) phenotype, signifying suppression of the increased UUG initiation on his4-301 mRNA and attendant His+ phenotype conferred by a known Sui− mutation, in this case the dominant mutation SUI5 encoding the G31R variant of eIF5 (28). The His+ phenotype of plasmid-borne SUI5 was diminished to varying extents by the K148D, R116D, R117D, K108D, and R146D alleles of RPS3 (Fig. 2B and SI Appendix, Table S1). Although K141D also suppresses growth on −His medium, it has a similar effect on +His medium, making it difficult to assess whether the His+ phenotype was suppressed. SUI5 also confers a Slg− phenotype in histidine-replete medium, particularly at elevated temperature (37 °C), and this phenotype was also diminished strongly by R116D, R117D, and K108D, and to a lesser extent by K148D and R146D (Fig. 2B and SI Appendix, Table S1). [Considering that R146D confers a Slg− phenotype in otherwise WT cells (Fig. 2A), its ability to improve growth at 37 °C relative to WT RPS3 in the SUI5 background (Fig. 2B) indicates a greater ability to suppress the growth defect of SUI5 than is evident from judging growth of SUI5 strains alone.] These results suggest that a subset of Rps3 substitutions mitigate the effect of SUI5 in reducing the accuracy of start-codon recognition.
The effect of SUI5 on the fidelity of start-codon selection can be quantified by an increase in the expression of a HIS4-lacZ reporter containing a UUG start codon, compared with the expression levels observed when the same cells are transformed with empty vector (Fig. 3A, columns 1 and 2). Supporting the interpretation that specific Rps3 substitutions dampen this effect, we found that all five RPS3 mutations that reduce the His+/Sui− phenotype of SUI5 (Fig. 2B) also suppress the elevated expression of a HIS4-lacZ reporter containing a UUG start codon that is conferred by SUI5 (Fig. 3A, column 1 vs. 2 to 7). SUI5 additionally produces a modest increase in expression of the matching reporter with an AUG initiation codon (Fig. 3B, columns 1 and 2), an effect that is similarly reversed by the RPS3 mutations (Fig. 3B, column 1 vs. 3 to 7). Despite this last effect, all five RPS3 mutations confer a marked reduction in the UUG:AUG initiation ratio for the two reporters (Fig. 3C, column 1 vs. 3 to 7). These results demonstrate that the Rps3 substitutions restore the strong preference, typical of WT cells, for AUG versus UUG start codons in SUI5 mutant cells, thus conferring strong Ssu− phenotypes.
Fig. 3.
RPS3 alleles suppress the elevated UUG:AUG initiation ratio of HIS4-lacZ reporters conferred by SUI5. (A and B) Transformants of his4-301 strains with the indicated RPS3 alleles and either SUI5 plasmid p4281 or vector and harboring the HIS4-lacZ reporters shown schematically with UUG (A) or AUG (B) start codons (plasmids p367 and p391, respectively) were cultured in SD+His medium to an A600 of 1.0 to 1.2, and β-galactosidase specific activities were measured in whole-cell extracts in units of nanomoles of o-nitrophenyl-β-d-galactopyranoside cleaved per min per mg of total protein. Mean activities with SEMs (shown as error bars) were determined from six independent transformants. (C) Mean ratios (with SEMs) of expression of the UUG versus AUG reporter were calculated from the results in A and B. Asterisks indicate statistically significant differences between each strain and the RPS3/SUI5 strain determined by a two-tailed, unpaired Student’s t test (*P < 0.05; **P < 0.01).
RPS3 Ssu− Mutations Increase Discrimination Against the eIF1 AUG Codon in Suboptimal Context.
In addition to reducing initiation at near-cognate UUG codons in Sui− mutants, previously identified Ssu− substitutions in eIF1 and eIF1A were shown to intensify discrimination against the AUG start codon of the SUI1 gene encoding eIF1, which occurs in poor Kozak context. This feature of SUI1 initiation underlies negative autoregulation of eIF1 synthesis, dampening the ability to overexpress eIF1 in WT cells, as the excess eIF1 suppresses initiation at the SUI1 start codon (5). As observed previously for Ssu− substitutions in eIF1 and eIF1A (5), we observed that four of the five RPS3 Ssu− substitutions reduce steady-state expression of eIF1, with the strongest reductions seen for R116D and R117D and lesser effects for R146D and K148D (Fig. 4 A and B). R116D and R117D also decreased the expression of the WT SUI1-lacZ fusion containing the native, poor context of the SUI1 AUG, but did not diminish expression of the SUI1-opt-lacZ reporter, in which the native SUI1 context is replaced with an AUG in optimum context with A nucleotides at the upstream −1 to −3 positions (Fig. 4C). Accordingly, R116D and R117D significantly increase the SUI1-opt-lacZ:SUI1-lacZ initiation ratio (Fig. 4C). Thus, in addition to suppressing UUG initiation, R116D and R117D clearly discriminate against the SUI1 AUG codon in poor context. In contrast, R146D and K148D reduce eIF1 expression but have little effect on SUI1-lacZ expression, perhaps indicating that their ability to discriminate against the native SUI1 initiation region, which is smaller in magnitude compared with R116D/R117D (Fig. 4 A and B), requires other sequence or structural features not maintained in the SUI1-lacZ reporter. The fact that the RPS3 Ssu− mutants exhibit reduced expression of native eIF1 implies that their increased discrimination against the suboptimal context of the SUI1 AUG codon is strong enough to overcome the increase in eIF1 synthesis expected from the autoregulatory mechanism governing SUI1 translation (5, 10).
Fig. 4.
Certain RPS3 Ssu− alleles exacerbate discrimination against the AUG start codon of the SUI1 gene encoding eIF1. (A) Strains described in Fig. 2A with the indicated RPS3 alleles were cultured in SD+His+Trp+Ura medium to an A600 of ∼1.0 and WCEs were subjected to Western analysis using antibodies against eIF1 or Hcr1 (as loading control). Two amounts of each extract differing by a factor of two were loaded in successive lanes. (B) eIF1 expression, normalized to that of Hcr1, was obtained for each strain by quantifying the Western signals in A, and mean values (±SEM) were calculated from three biological replicates. Asterisks indicate significant differences between mutant and WT as judged by a Student’s t test (P < 0.005). (C) Strains from A also harboring the SUI1-lacZ (pPMB24) or SUI1-opt-lacZ (pPMB25) reporter were cultured and assayed for β-galactosidase expression as described in Fig. 3, except using SD+His+Trp medium. Mean expression levels and SEMs calculated from six transformants of each strain are plotted, and relative (Rel) mean expression levels normalized to that of the WT strain are listed below the histogram, along with the expression ratios for the SUI1-lacZ versus SUI1-opt-lacZ reporters. Asterisks indicate significant differences between mutant and WT as judged by a two-tailed, unpaired Student’s t test (*P < 0.05; **P < 0.01).
RPS3 Ssu− Mutations Reduce Bulk Translation Initiation Without Affecting 40S Subunit Abundance.
Each the RPS3 Ssu− mutations except K148D conferred moderate reductions in the ratio of polysomes to 80S monosomes, measured in cycloheximide-stabilized extracts resolved by sedimentation through sucrose gradients (Fig. 5), suggesting a reduced rate of bulk translation initiation relative to elongation in the mutants. No perturbation to the ratio of free 40S to free 60S subunits was evident in these gradients except for a modest reduction in the R116D mutant (Fig. 5); even in this mutant, however, there was no significant decrease in the ratio of bulk 40S to 60S subunits in extracts prepared without cycloheximide and magnesium, wherein polysomes and 80S monosomes dissociate into subunits (SI Appendix, Fig. S1). These findings suggest that the alterations in accuracy of start-codon selection observed in these mutants arise from altered 40S function, and not from abnormalities in expression of Rps3, 40S biogenesis, or stability of mature 40S subunits. This is consistent with the fact that scanning occurs only after assembly and attachment of 43S PICs to the mRNA, and so the fidelity of start-codon recognition during scanning should not be influenced by the concentration of 43S PICs.
Fig. 5.
RPS3 Ssu− alleles reduce bulk translation initiation. (A–F) Strains from Fig. 2A were cultured in SC-Leu at 30 °C to an A600 of ∼1.0, and cycloheximide was added (50 μg/mL) before harvesting. WCEs were separated by sucrose density gradient centrifugation and scanned at 254 nm. Mean (±SEM) polysomes:monosomes (P/M) and free 40S:60S ratios from three biological replicates are indicated. Asterisks indicate significant differences between mutant and WT as judged by a two-tailed, unpaired Student’s t test (*P < 0.05; **P < 0.01).
Rps3 Ssu− Substitutions R116D and R117D Destabilize the PIN Conformation of the 48S PIC at UUG Codons in Vitro.
Among the Rps3 substitutions tested, R116D and R117D conferred the broadest and most pronounced genetic defects, reducing initiation at UUG start codons in a Sui− mutant (Ssu− phenotype) and also diminishing initiation at the eIF1 AUG codon in its native, poor context. These phenotypes suggest that they destabilize the PIN state of the 48S PIC and thereby exacerbate the effects of suboptimal initiation sequences. We tested this hypothesis by analyzing their effects on the rate of TC dissociation from PICs reconstituted in vitro. To this end, we purified 40S subunits from rps3Δ::kanMX deletion strains harboring either plasmid-borne R116D, R117D, or WT RPS3 as the only source of Rps3. The 48S PICs were formed by incubating WT TC (assembled with [35S]Met-tRNAiMet and nonhydrolyzable GTP analog GDPNP) with saturating amounts of eIF1, eIF1A, an uncapped unstructured model mRNA with either an AUG or UUG start codon in poor context (5′-UCUAUGC-3′ or 5′-UCUUUGC-3′, respectively), and either WT or mutant 40S subunits. 48S PICs containing [35S]Met-tRNAiMet were chased with excess unlabeled TC, incubated for increasing time periods, and then resolved via native gel electrophoresis to separate 40S-bound and unbound fractions of TC. In agreement with previous findings (11, 29, 30), little to no TC dissociation occurred from WT PICs formed with the AUG-containing mRNA [mRNA(AUG)] over the time course of the experiment (Fig. 6), whereas appreciable dissociation was observed from WT PICs assembled on UUG-containing mRNA (koff 0.19 ± 0.07 h−1; Fig. 6). Neither Rps3 substitution appreciably alters the kinetics of TC dissociation from PICs assembled on mRNA(AUG), although R117D generally conferred a moderate increase in the extent of dissociation (Fig. 6, AUG mRNAs). By contrast, both the extent and rate of TC dissociation were substantially increased for PICs assembled on mRNA(UUG) using either R116D or R117D mutant 40S subunits compared with WT subunits (Fig. 6). From previous work, it was determined that TC bound in the POUT state is too unstable to remain associated with the PIC during the native gel electrophoresis used to separate PIC-bound from unbound TC in this assay. It was also deduced that most or all of the WT complexes formed with mRNA(AUG) achieve a highly stable state from which no TC dissociation occurs on the timescale of the experiments. A smaller fraction of complexes formed with mRNA(UUG) accesses this highly stable state, and the remainder dissociates with a measurable off-rate. Thus, the extent of dissociation reflects the ratio of PICs in PIN versus the distinct, hyperstable conformation, and the rate of dissociation reflects the stability of the PIN conformation (11, 30). Accordingly, the results in Fig. 6 indicate that Rps3 substitutions R116D and R117D diminish rearrangement to the hyperstable state at near-cognate UUG codons, increasing the fraction of complexes from which TC dissociates, and also destabilize the PIN state at UUG codons, increasing koff for the mRNA(UUG) complexes.
Fig. 6.
Rps3 Ssu− substitutions R116D and R117D destabilize the PIN conformation of the 48S PIC at UUG codons in vitro. (A) Analysis of TC dissociation from 43S–mRNA complexes assembled with WT, Rps3-R116D, or Rps3-R117D 40S subunits and either mRNA(AUG) or mRNA(UUG). Representative curves are shown for each measurement. (B) The end points and koff values (±SEMs) determined from between four and nine replicate experiments (numbers in parentheses); ND, dissociation was too limited in most replicate determinations to permit koff calculations. Asterisks indicate significant differences between mutant and WT as judged by a two-tailed, unpaired Student’s t test (*P < 0.05; **P < 0.01).
Rps3 Ssu− Substitutions R116D and R117D Destabilize mRNA Binding at the 40S Entry Channel.
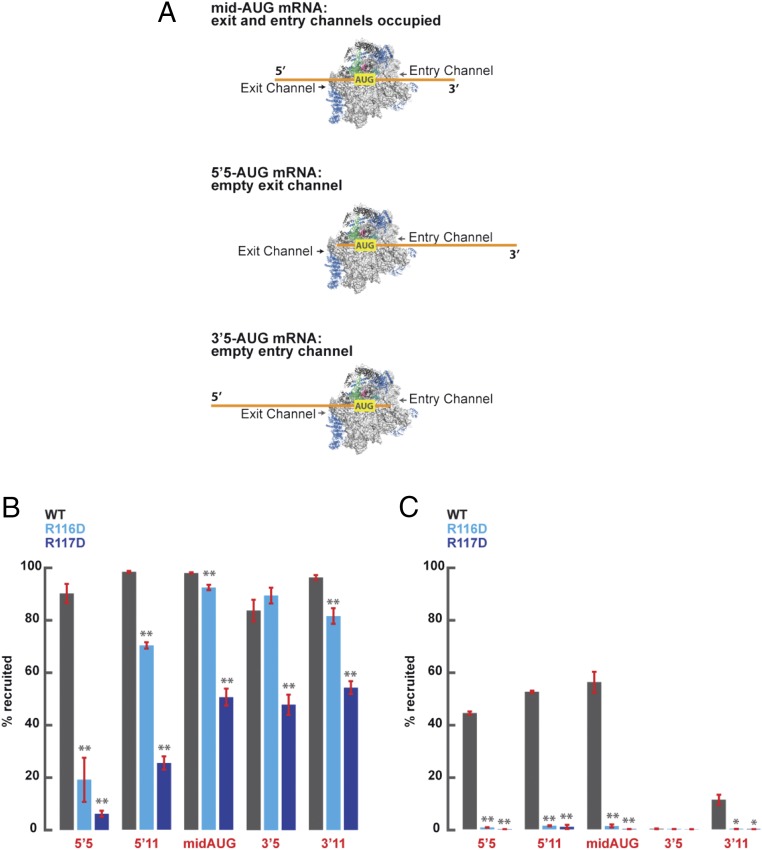
We recently analyzed the role of eIF3 subunits and domains in mediating stable interactions of the PIC with mRNA nucleotides located at the entry or exit channel of the 40S subunit by reconstituting PICs with different model mRNAs designed to incompletely occupy either the entry or exit channel of the 40S subunit (18). Capped model mRNAs designated “5′5-AUG” and “5′11-AUG” contain only 5 or 11 nt located 5′ of the AUG start codon, respectively, but >30 nt 3′ of the AUG. Hence, with the AUG positioned in the 40S P site, both mRNAs should fully occupy the entry channel and protrude from the entry channel opening on the solvent-exposed surface of the 40S subunit but contain only 2 or 8 nt in the exit channel beyond the 3 nt occupying the E site (positions −3, −2, and −1 relative to the AUG; Fig. 7A). Because the exit channel accommodates ∼10 nt, it is largely empty for PICs assembled on 5′5-AUG mRNA but nearly filled for those assembled on 5′11-AUG mRNA, although neither mRNA will protrude outside the exit channel pore. Conversely, model mRNAs “3′5-AUG” and “3′11-AUG” contain >30 nt upstream of the AUG codon, and thus should fully occupy the exit channel but will contain only 2 or 8 nt in the entry channel in addition to the 3 nt that occupy the A site (positions +4 to +6) when the AUG codon is in the 40S P site (Fig. 7A). Thus, the ∼9-nt-long entry channel should be largely empty in PICs assembled on 3′5-AUG but fully occupied in PICs assembled on 3′11-AUG mRNA; neither mRNA protrudes beyond the entry channel pore. As a positive control, we also examined “mid-AUG” mRNA, containing residues both 5′ and 3′ of the AUG sufficient to fully occupy both the entry and exit channels and extend outside the openings of both channels (Fig. 7A). Radiolabeled versions of these mRNAs were used to assay 48S PIC assembly in the reconstituted system, at saturating concentrations of 40S subunits, TC (assembled with GDPNP), eIF1, eIF1A, eIF5, eIF4A, eIF4B, the eIF4E–eIF4G complex, and eIF3, using native gel electrophoresis to separate 43S PIC-bound mRNA from unbound mRNA. Although the eIF4 group of factors is dispensable for 48S PIC assembly for the uncapped model mRNAs used for the TC dissociation assays in Fig. 6, they were included to ensure rapid and complete recruitment of the capped mRNAs used here (18).
Fig. 7.
Rps3 Ssu− substitutions R116D and R117D destabilize PIC–mRNA interaction at the 40S entry channel. (A) Schematic representation of 48S PICs formed in the presence of the mid-AUG, 5′5-AUG, and 3′5-AUG mRNAs. Whereas the mid-AUG mRNA programs a complex in which both the mRNA entry and exit channels are occupied, the 5′5-AUG and 3′5-AUG mRNAs program complexes that leave either the exit or entry channels largely unoccupied, respectively. (B) The maximal extent of recruitment observed for capped model mRNAs in the presence of all factors, including eIF3, to PICs assembled with 40S subunits containing WT (gray), R116D (light blue), or R117D (dark blue) Rps3. (C) The maximal extent of recruitment observed for model mRNAs in the presence of all factors, with the exception of eIF3, to PICs assembled with 40S subunits containing WT, R116D, or R117D Rps3. Asterisks indicate significant differences between mutant and WT as judged by a two-tailed, unpaired Student’s t test (*P < 0.05; **P < 0.01).
Consistent with our previous study, 80 to 100% of each of these mRNAs can be driven into 48S PICs when eIF3 is present together with all other components and WT 40S subunits (Fig. 7B, gray bars), whereas omitting eIF3 reduces the extent of recruitment of 5′5-AUG, 5′11-AUG, and mid-AUG mRNAs by ∼40 to 60%, drastically impairs recruitment of 3′11 mRNA, and abolishes recruitment of 3′5 mRNA (Fig. 7C, gray bars). Similar results previously led us to conclude that eIF3 mediates interactions of the PIC with mRNA residues at the exit channel, which become crucial for mRNA recruitment when the entry channel is largely empty (as for 3′5-AUG mRNA) or when no mRNA residues protrude from the entry channel pore (for 3′11-AUG mRNA). eIF3 also enhances PIC–mRNA interactions at the entry channel, increasing recruitment of mRNAs that fully occupy the entry channel even when eIF3–mRNA interactions at the exit channel are impaired, as with 5′5 mRNA (cf. Fig. 7 B and C, 5′5 mRNA, gray bars) (18). The fact that mRNA nucleotides at the entry channel can compensate for the absence of eIF3-mediated PIC–mRNA interactions at the exit channel implies the existence of compensatory PIC–mRNA interactions at the entry channel. We hypothesized here that these deduced PIC–mRNA interactions include the contacts between Rps3 and mRNA. Indeed, the Rps3 residues studied here are located in proximity to mRNA nucleotides both within and just outside the entry channel opening in the cryo-EM structure of py48S (3) (Fig. 1C).
Remarkably, when WT 40S subunits were replaced with R116D mutant subunits in reactions containing eIF3, we observed a dramatic reduction in recruitment of 5′5 mRNA, which leaves the exit channel largely empty (Fig. 7B, cyan bars). This indicates that Arg-116 in Rps3 is crucial for PIC–mRNA interactions at the entry channel, and that these interactions are essential for mRNA recruitment when PIC–mRNA interactions at the opposite, exit, channel are missing. Supporting this interpretation, the magnitude of the defect using R116D subunits is progressively diminished when the exit channel is filled by using 5′11-AUG mRNA and then when mRNA protrudes from the exit channel pore by using mid-AUG mRNA. The defect is also eliminated entirely using 3′5-AUG or 3′11-AUG mRNAs (Fig. 7B, cyan bars), which lack mRNA bases at the entry channel but fully occupy and protrude from the exit channel. These last findings are consistent with our previous observation that interactions at the entry channel are redundant with those at the exit channel (18), and also suggest that R116D does not impair other aspects of 40S structure or function that would compromise stable 48S PIC assembly with these mRNAs beyond the loss of mRNA–40S interactions at the entry channel.
Similar results were obtained when WT 40S subunits were replaced with R117D mutant subunits in reactions containing eIF3, including the nearly complete loss of 5′5 mRNA recruitment, which is partially rescued with 5′11 mRNA and more completely rescued with mid-AUG, 3′5, and 3′11 mRNAs (Fig. 7B, blue bars). In contrast to the results for R116D, however, the latter three mRNAs do not fully rescue recruitment to the level observed with WT or Rps3-R116D 40S subunits (Fig. 7B, blue vs. cyan/gray bars). This last distinction might indicate that the R117D substitution impairs another aspect of stable 48S PIC assembly in addition to mRNA interactions at the entry channel.
Finally, in reactions lacking eIF3, we found that both Rps3 substitutions essentially abolished the appreciable mRNA recruitment retained in the absence of eIF3 for the mRNAs that completely occupy the entry channel (5′5, 5′11, and mid-AUG; Fig. 7C, blue bars). These findings fully support our conclusion that Rps3–mRNA interactions involving R116/R117 are essential for the eIF3-independent interaction of the PIC with mRNA at the entry channel. These results further provide strong evidence that direct contacts of Rps3 residues R116/R117 with mRNA nucleotides at the entry channel cooperate with eIF3 interactions at both the exit and entry channels to stabilize mRNA binding to 43S PICs.
Discussion
In this study, we obtained genetic and biochemical evidence implicating Rps3 residues at the 40S mRNA entry channel in promoting bulk translation initiation and maintaining wild-type accuracy of start-codon recognition in vivo, stabilizing the PIN conformation of TC binding to the PIC, and stabilizing interactions of the PIC with mRNA nucleotides at the entry channel that augment eIF3-mediated PIC–mRNA interactions at the entry and exit channels. In the recent py48S cryo-EM structure (3), mRNA in the entry channel was visualized up to residue +12, where it emerges from the entry channel pore on the solvent-exposed surface of the 40S subunit, with nucleotides +8 to +12 in proximity to basic residues of Rps3 (Fig. 1C). We found that Asp substitutions of three such residues, R116, R117, and R146, and the nearby basic residue K108, moderately reduced the rate of bulk translation initiation, as indicated by a decreased ratio of polysomes to monosomes, without decreasing the ratio of bulk 40S to 60S subunit abundance. These substitutions dramatically suppressed the increased utilization of UUG start codons in HIS4 mRNA engendered by the Sui− variant of eIF5 encoded by SUI5, thus conferring the Ssu− phenotype. The R116, R117, R146, and K148 substitutions also reduced expression of eIF1, an effect observed previously for Ssu− mutations of eIF1 and eIF1A and attributed to increased discrimination against the poor Kozak context of the eIF1 start codon (5). We established that this mechanism also applies to the Rps3-R116D and -R117D substitutions by showing that they reduce expression of a SUI1-lacZ fusion containing the native, poor context for the (eIF1) AUG start codon but not that of a modified SUI1-lacZ reporter containing optimum Kozak context. It is unclear, however, whether this mechanism underlies the relatively weaker reductions in eIF1 levels conferred by the K148D and R146D substitutions. Thus, R116D and R117D increase discrimination against the eIF1 AUG start codon in its native, poor context as well as reduce recognition of near-cognate UUG codons, whereas K148D and R146D only strongly suppress UUG initiation. Our previous genetic analyses of residues of the β-hairpin of uS7/Rps5, located in the 40S mRNA exit channel, provide a precedent for altered 40S contacts with mRNA that increase discrimination against a mismatch in the start codon–anticodon duplex at UUG codons without altering the influence of Kozak context (14). As discussed further below, this might indicate that these two aspects of start-codon recognition have distinct molecular mechanisms.
Because R116D and R117D conferred the broadest and most pronounced genetic defects, we purified mutant 40S subunits harboring these Rps3 substitutions and analyzed their effects on the stability of either TC or mRNA binding to the PIC in the yeast reconstituted system. We found that both R116D and R117D substantially increased the dissociation rate of TC from PICs reconstituted with a model unstructured mRNA containing a UUG start codon, without significantly affecting the corresponding AUG complex. These findings support the possibility that the suppression of UUG initiation conferred by these mutations in vivo (their Ssu− phenotype) arises from destabilization of the PIN state of TC binding in the presence of the inherently less stable codon–anticodon duplex containing a U–U mismatch that is formed by tRNAi at UUG codons. The discrimination against the SUI1 AUG in native, poor Kozak context engendered by the R116D/R117D substitutions cannot be explained by the same mechanism, as the model mRNAs used for koff measurements contain poor Kozak context for the AUG/UUG start codons; however, R116D/R117D had no significant effects on the stability of PICs formed with mRNA(AUG). Instead, R116D/R117D might reduce recognition of the SUI1 AUG in poor context by decreasing its dwell time in the P site during scanning—an important factor in determining the impact of poor context on initiation in the mammalian system (31)—by weakening PIC–mRNA interactions at the entry channel. If so, this effect might also contribute to the reduced recognition of UUG codons conferred by these Rps3 substitutions.
Using a complementary assay that employs model mRNAs that leave empty or only partially occupied the entry or exit channel of the 40S subunit within 48S PICs, we observed that both R116D and R117D dramatically destabilize mRNA interactions at the entry channel. We showed previously that eIF3 is crucial for stabilizing mRNA–PIC interactions at the exit channel, whereas substantial mRNA interactions at the entry channel can occur independent of eIF3 (18). Here we found that the Rps3-R116D and -R117D substitutions strongly destabilize 48S PICs assembled in the presence of eIF3 on 5′-truncated mRNAs lacking some or all of the eIF3-sensitive mRNA contacts in the exit channel (5′5-AUG and 5′11-AUG mRNAs). In the absence of eIF3, these Rps3 substitutions destabilize binding of PICs to all mRNA substrates except the one nearly devoid of mRNA in the entry channel (3′5 mRNA). However, this last mRNA does not bind stably to the PIC in the absence of eIF3 even when using WT 40S subunits, because it lacks contacts with the PIC at the entry channel. Our results indicate that these entry channel contacts include the direct interactions of Rps3 Arg116 and Arg117 with mRNA.
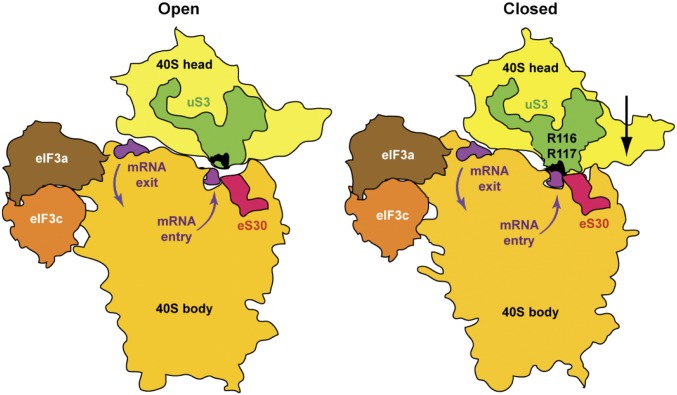
Previous studies have shown that elongating bacterial 70S ribosomes exhibit a helicase activity, driven by mRNA translocation through the ribosome, that enables efficient elongation through highly structured mRNA sequences, and that basic residues in 30S ribosomal proteins uS3 and uS4 at the opening of the mRNA entry channel are required for the helicase activity in vitro, including R131, R132, and K135 in uS3 (25). Remarkably, alignment of uS3 sequences from eukaryotic, archaebacterial, and bacterial sources indicates that residues R131 and R132 in bacterial uS3 align with R116 and R117 in yeast uS3/Rps3 (26) (Fig. 1B). It has been proposed that the conserved basic residues in uS3 and uS4 contribute to ribosome helicase function by interacting with the phosphate backbone of the complementary strands of an mRNA duplex positioned at the entry channel pore; these interactions would either stabilize the unwound strands generated by spontaneous melting of the duplex or function more directly to couple 30S head movement during translocation to mRNA duplex unwinding (25). Because the model mRNAs used in our experiments were designed to be devoid of secondary structure, it seems unlikely that Rps3 residues R116/R117 promote PIC–mRNA interactions at the entry channel by helping to melt secondary structures in the mRNA. Instead, they might simply provide electrostatic attraction for the mRNA phosphate backbone, helping to fix mRNA to the head of the 40S subunit, where Rps3 resides. In this view, basic residues in Rps30/eS30, also located at the entry channel opening, could similarly fix the mRNA to the 40S body. All of these interactions would occur simultaneously when the head moves closer to the body as a result of the transition from the open to the closed conformation of the PIC that is provoked by AUG recognition (22) (see model in Fig. 8). These pincers would thus help to clamp the mRNA into the now constricted entry channel, arresting scanning and helping to stabilize the closed conformation, in which the P site is fully formed and encloses the Met-tRNAiMet in the PIN state. Thus, by eliminating mRNA interactions at the entry channel, the Rps3-R116D/R117D substitutions would destabilize the closed conformation of the PIC and indirectly destabilize Met-tRNAiMet binding in the P site. Combining this destabilizing effect with the less stable codon–anticodon duplex formed at UUG codons could account for the increased rate of TC dissociation from PICs reconstituted with R116D/R117D mutant 40S subunits and mRNA(UUG) (Fig. 6), as well as the increased discrimination against UUG codons (Ssu− phenotype) conferred by these substitutions in vivo (Fig. 3).
Fig. 8.
Model depicting a conformational switch in the PIC between the open, scanning, and closed conformations on start-codon recognition that clamps mRNA at the entry channel between uS3/Rps3 and eS30/Rps30. Silhouettes of 40S subunits were traced from py48S-open (PDB ID 3JAQ) and py48S (PDB ID code 3J81) to depict the open and closed conformations of the PIC, respectively. PCI domains of eIF3a/c subunits were traced from py48S-closed (PDB ID code 3JAP) and mRNA (purple) was traced from py48S (PDB ID code 3J81), and docked at similar positions on both 40S silhouettes. Locations of R116/R117 are shown in black on the green silhouettes of uS3. Downward movement of the 40S head toward the body in the closed complex relative to its position in the open complex (22) (depicted by an arrow; Right) narrows the mRNA entry channel, engaging R116/R117 with mRNA at the entry channel pore. These contacts, and interactions of eS30 (red) with mRNA at the same location, help fix the mRNA in the entry channel and stabilize the closed/PIN state at the start codon. The eIF3a PCI domain stabilizes interaction of the PIC with mRNA at the exit channel (18), augmenting the function of uS3/Rps3 R116/R117 at the entry channel.
Finally, it is interesting that Rps3 amino acids Arg-146 and Lys-141 also interact with rRNA residues in helices 18 and 34 that are involved in the noncovalent interactions between rRNA residues in the 40S body and head. These interactions compose the latch over the mRNA entry channel, with Arg-146 interacting with h34 in both the open and closed conformations and Lys-141 interacting with both helices but with h18 only in the closed state (22). These Rps3–rRNA interactions should promote the closed conformation of the latch, which in turn should help to clamp mRNA in the exit channel and stabilize the closed arrangement of the 40S head relative to the body that locks Met-tRNAiMet into the P site. Thus, the diminished UUG initiation we observed for an acidic substitution of R146 might involve a weakened entry channel latch in addition to diminished Rps3–mRNA contacts in the entry channel. The strong growth defect observed for the K141D substitution is consistent with an even greater destabilization of the closed conformation of the PIC resulting from a destabilized latch. Given the collaboration of Rps3 with eIF3 in stabilizing mRNA at the entry channel, it will be interesting to learn whether the conserved basic residues in Rps3 functionally interact with segments of eIF3 subunits, in particular the eIF3a C-terminal domain, which has been implicated in PIC–mRNA interactions at the entry channel (18, 32) and in start-codon selection (19). It would also be worthwhile to examine whether the corresponding residues in bacterial uS3 promote the fidelity of initiation in bacterial cells.
Materials and Methods
Plasmid Constructions.
Plasmids used in this work are listed in SI Appendix, Table S3. Plasmid pDH412 was constructed as follows. The RPS3 gene, including 458 bp upstream of the ATG start codon and 200 bp downstream of the stop codon, was amplified by PCR using the two primers 5′-GAA TGC GGC CGC GAA GCA GTT ACA TCT CA-3′ and 5′-AAG TCG ACT TGT GTT GTA CAA AAC TT-3′ and genomic DNA from yeast WT strain BY4741 as template. The ∼1.4-kb amplicon was digested with NotI and SalI, and the resulting fragment was inserted between the NotI and SalI sites of plasmid pRS315 to produce pDH412. The DNA sequence of the entire open reading frame (ORF) was verified. pDH459 was constructed by inserting the NotI-SalI fragment containing RPS3 isolated from pDH412 between the NotI and SalI sites of pRS316. It was shown that both pDH412 and pDH459 fully rescue growth of PGAL-RPS3 strain HD2738 on glucose medium. Plasmids pDH424, pDH431, pDH425, pDH432, pDH482, pDH433, pDH13-37, pDH429, and pDH430 were derived from pDH412 by site-directed mutagenesis using the QuikChange XL Kit (Stratagene) and the corresponding primers in SI Appendix, Table S4, and the mutations were verified by sequencing the entire ORF and 5′ noncoding region.
Yeast Strain Constructions.
Yeast strains used in this work are listed in SI Appendix, Table S2.
Strain HD2738 was derived from H2995 by replacing the promoter of chromosomal RPS3 with the GAL1 promoter by one-step gene replacement, as follows. Primers 5′-CCT TTC CTG TAT AAT ATT CTT GCT GTA AAG TTT GTT TTT TTT ATG AAA AAA ACA TTT TCT TTT CTT GAG GAA TTC GAG CTC GTT TAA AC-3′ and 5′-GAA TTC GTT CAA TTC AGC GTA GAA GAC ACC GTC AGC GAC TAG CTT TCT TTT CTT AGA GAT TAA AGC GAC CAT TTT GAG ATC CGG GTT TT-3′ were used to amplify by PCR the appropriate DNA fragment from plasmid pFA6a-kanMX6-PGAL1 (p3218), which was used to transform strain H2995 to kanamycin resistance. The presence of PGAL1-RPS3 in the resulting strain (HD2738) was established by demonstrating that the lethality on glucose medium can be complemented by low-copy (lc) RPS3 LEU2 plasmid pDH412, and confirmed by PCR analysis of chromosomal DNA with the appropriate primers.
Strains HD2754, HD2755, HD2765, HD2764, HD2767, HD2766, HD2769, HD2768, HD3120, HD2772, and HD2779 were derived from HD2738 by transformation with a LEU2 plasmid containing the indicated RPS3 allele, or empty vector, as indicated in SI Appendix, Table S2. Strains HD2836, HD2911, HD2846, HD2868, HD2848, HD2861, HD2850, HD2849, HD3145, HD2841, and HD2860 were derived from the foregoing strains by transformation with single-copy (sc) TRP1 SUI5 plasmid p4281, or empty vector YCplac22, as indicated in SI Appendix, Table S2.
To produce strain HD2973, pDH459 (lc URA3 RPS3+) was introduced into the RPS3/rps3Δ diploid strain YSC1021-671817 purchased from Research Genetics. The Ura+ transformants were sporulated and subjected to tetrad analysis. HD2973 was identified as a KanR Ura+ ascospore clone incapable of growth on medium containing 5-fluoroorotic acid (5-FOA) unless transformed with RPS3+ LEU2 plasmid pDH412 but not with empty LEU2 vector pRS315. The presence of rps3Δ::kanMX in HD2973 was verified by PCR analysis of chromosomal DNA. The strains HD3240, HD3241, HD3242, HD3243, and HD3244 were derived from HD2973 by plasmid shuffling to replace pDH459 with the appropriate lc LEU2 plasmid containing the RPS3 allele indicated in SI Appendix, Table S2.
Biochemical Analysis of Yeast Cells.
Assays of β-galactosidase activity in whole-cell extracts (WCEs) were performed as described previously (33). For Western analysis, WCEs were prepared by trichloroacetic acid extraction as described (34), and immunoblot analysis was conducted as described previously (5) with antibodies against eIF1 (7) (35). The signal intensities were quantified using a LI-COR Odyssey infrared scanner.
For polysome analysis, strains HD2754 (RPS3+), HD2767 (R116D), HD2769 (R117D), HD3120 (K108D), HD2765 (K148D), or HD2767 (R146D) were grown on SC-Leu medium at 30 °C to A600 ∼1. Cycloheximide was added to 50 μg/mL 5 min before harvesting, and a WCE was prepared in breaking buffer [20 mM Tris⋅HCl, pH 7.5, 50 mM KCl, 10 mM MgCl2, 1 mM DTT, 5 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 1 cOmplete EDTA-free Protease Inhibitor Tablet (Roche) per 50 mL buffer]. Fifteen A260 units of WCE were separated by velocity sedimentation on a 4.5 to 45% (wt/vol) sucrose gradient by centrifugation at 39,000 rpm for 3 h in an SW41Ti rotor (Beckman). Gradient fractions were scanned at 254 nm to visualize ribosomal species. To analyze total 40S/60S profiles, the rps3Δ::kanMX deletion strains harboring plasmid-borne WT RPS3+ (HD3240) or rps3 alleles R116D (HD3241), R117D (HD3242), K108D (HD3243), K148D (HD3244) or R146D (HD3277) were grown in yeast extract peptone dextrose at 30 °C to A600 ∼1, but cycloheximide was omitted. WCEs were prepared in the absence of Mg+2, and 15 A260 units of WCE were resolved by velocity sedimentation through 5 to 30% (wt/vol) sucrose gradients at 40,000 rpm for 4 h in an SW41Ti rotor (Beckman).
Biochemical Analysis in the Reconstituted Yeast Translation System.
TC dissociation rates.
Initiation factors eIF1A, eIF1, and His6-tagged eIF2 were purified as described (36). WT and mutant 40S subunits were purified from the rps3Δ::kanMX deletion strains harboring plasmid-borne WT RPS3+ (HD3240), rps3-R116D (HD3241), or rps3-R117D (HD3242) as the only source of Rps3, as described previously (36). Model mRNAs with the sequences 5′-GGAA[UC]7UAUG[CU]10C-3′ and 5′-GGAA[UC]7UUUG[CU]10C-3′ were purchased from Thermo Scientific. Yeast tRNAiMet was synthesized from a hammerhead fusion template using T7 RNA polymerase, charged with [35S]methionine, and used to prepare radiolabeled eIF2–GDPNP–[35S]Met–tRNAiMet ternary complexes ([35S]TC), all as previously described (36). Yeast Met-tRNAiMet was purchased from tRNA Probes and used to prepare unlabeled TC in the same way. Dissociation rates of TC (koff) were measured by monitoring the amount of [35S]TC that remained bound to 40S–eIF1–eIF1A–mRNA complexes over time, in the presence of excess unlabeled TC (chase), using native gel electrophoresis to separate 40S-bound from unbound [35S]TC. 43S–mRNA complexes were preassembled for 2 h at 26 °C in reactions containing 40S subunits (20 nM), eIF1 (1 µM), eIF1A (1 µM), mRNA (10 µM), and [35S]TC (0.25 µM eIF2/0.1 mM GDPNP/1 nM [35S]Met-tRNAiMet) in 60 µL of 1× Recon buffer [30 mM Hepes⋅KOH, pH 7.4, 100 mM KOAc, 3 mM Mg(OAc)2, 2 mM DTT]. To initiate each dissociation reaction, a 6-µL aliquot of the preassembled 43S–mRNA complexes was mixed with 3 µL of threefold concentrated unlabeled TC chase (composed of 2 µM eIF2/0.3 mM GDPNP/0.9 µM Met-tRNAiMet), to yield a 300-fold excess of unlabeled versus labeled TC in the final dissociation reaction, and incubated for the prescribed period. A converging time course was used so that all dissociation reactions were terminated simultaneously by the addition of native-gel dye and loaded directly on a running native gel. The fraction of [35S]Met-tRNAi remaining in 43S complexes at each time point was determined by quantifying the 40S-bound and unbound signals by phosphorimaging, normalized to the ratio observed at the earliest time point, and the data were fit with a single exponential equation (11).
mRNA recruitment to 43S PICs.
Preparation of initiation factors, charged Met-tRNAiMet, and 40S ribosomal subunits were purified as previously described (18, 36, 37). Model mRNAs described in ref. 18 were transcribed in vitro, capped with [α-32P]GTP (PerkinElmer) using vaccinia virus D1/D12 capping enzyme, and purified as described (37) with the modifications described in ref. 18. The extent of mRNA recruitment to reconstituted 43S PICs was determined using a previously described gel-shift assay (37). PICs were assembled in the presence of 1 µM eIF1, 1 µM eIF1A, 300 nM eIF2, 200 nM Met-tRNAiMet, 400 nM eIF3, 2 µM eIF4A, 300 nM eIF4B, 50 nM eIF4E•eIFG, 300 nM eIF5, and 30 nM 40S subunits in 1× Recon buffer [30 mM Hepes⋅KOH, pH 7.4, 100 mM KOAc, 3 mM Mg(OAc)2, 2 mM DTT] and incubated for 10 min at 26 °C. Reactions were initiated by the simultaneous addition of ATP•Mg2+ and the appropriate 32P-capped mRNA to final concentrations of 2 mM and 15 nM, respectively, and incubated for 2 h at 26 °C, at which point all reactions had proceeded to completion as judged by prior kinetic experiments (18). The free and PIC-bound mRNA fractions were resolved on a 4% native gel, run for 45 min at 200 V, and quantified by phosphorimaging analysis using a Typhoon FLA 9500 imager (GE Healthcare Life Sciences). The extent of recruitment was calculated as the fraction of total signal in the lane represented by PIC-bound mRNA.
Supplementary Material
Acknowledgments
We thank Jagpreet Nanda, Jyothsna Visweswaraiah, and Fan Zhang for advice on koff determinations. We are grateful to members of our laboratories and Tom Dever’s group for helpful suggestions. This work was supported in part by the Intramural Research Program of the NIH (A.G.H. and J.R.L.) and NIH Grant GM62128 (previously to J.R.L.). C.E.A. was further supported by an NIH minority supplement to NIH Grant GM62128 and by a Leukemia & Lymphoma Society Career Development Program Fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620569114/-/DCSupplemental.
References
- 1.Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 2.Lomakin IB, Steitz TA. The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature. 2013;500(7462):307–311. doi: 10.1038/nature12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hussain T, et al. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell. 2014;159(3):597–607. doi: 10.1016/j.cell.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331(6018):730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 5.Martin-Marcos P, Cheung YN, Hinnebusch AG. Functional elements in initiation factors 1, 1A, and 2β discriminate against poor AUG context and non-AUG start codons. Mol Cell Biol. 2011;31(23):4814–4831. doi: 10.1128/MCB.05819-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin-Marcos P, et al. Beta-hairpin loop of eukaryotic initiation factor 1 (eIF1) mediates 40 S ribosome binding to regulate initiator tRNAMet recruitment and accuracy of AUG selection in vivo. J Biol Chem. 2013;288(38):27546–27562. doi: 10.1074/jbc.M113.498642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valásek L, Nielsen KH, Zhang F, Fekete CA, Hinnebusch AG. Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol Cell Biol. 2004;24(21):9437–9455. doi: 10.1128/MCB.24.21.9437-9455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alone PV, Cao C, Dever TE. Translation initiation factor 2gamma mutant alters start codon selection independent of Met-tRNA binding. Mol Cell Biol. 2008;28(22):6877–6888. doi: 10.1128/MCB.01147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saini AK, Nanda JS, Lorsch JR, Hinnebusch AG. Regulatory elements in eIF1A control the fidelity of start codon selection by modulating tRNA(i)(Met) binding to the ribosome. Genes Dev. 2010;24(1):97–110. doi: 10.1101/gad.1871910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivanov IP, Loughran G, Sachs MS, Atkins JF. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1) Proc Natl Acad Sci USA. 2010;107(42):18056–18060. doi: 10.1073/pnas.1009269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolitz SE, Takacs JE, Lorsch JR. Kinetic and thermodynamic analysis of the role of start codon/anticodon base pairing during eukaryotic translation initiation. RNA. 2009;15(1):138–152. doi: 10.1261/rna.1318509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pisarev AV, et al. Specific functional interactions of nucleotides at key −3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev. 2006;20(5):624–636. doi: 10.1101/gad.1397906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashem Y, et al. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell. 2013;153(5):1108–1119. doi: 10.1016/j.cell.2013.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Visweswaraiah J, Pittman Y, Dever TE, Hinnebusch AG. The β-hairpin of 40S exit channel protein Rps5/uS7 promotes efficient and accurate translation initiation in vivo. eLife. 2015;4:e07939. doi: 10.7554/eLife.07939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozak M. A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr. 1991;1(2):111–115. [PMC free article] [PubMed] [Google Scholar]
- 16.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16(22):2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CU, Pestova TV. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008;27(11):1609–1621. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aitken CE, et al. Eukaryotic translation initiation factor 3 plays distinct roles at the mRNA entry and exit channels of the ribosomal preinitiation complex. eLife. 2016;5:e20934. doi: 10.7554/eLife.20934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valásek LS. ‘Ribozoomin’—Translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs) Curr Protein Pept Sci. 2012;13(4):305–330. doi: 10.2174/138920312801619385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erzberger JP, et al. Molecular architecture of the 40S⋅eIF1⋅eIF3 translation initiation complex. Cell. 2014;158(5):1123–1135. doi: 10.1016/j.cell.2014.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aylett CH, Boehringer D, Erzberger JP, Schaefer T, Ban N. Structure of a yeast 40S-eIF1-eIF1A-eIF3-eIF3j initiation complex. Nat Struct Mol Biol. 2015;22(3):269–271. doi: 10.1038/nsmb.2963. [DOI] [PubMed] [Google Scholar]
- 22.Llácer JL, et al. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol Cell. 2015;59(3):399–412. doi: 10.1016/j.molcel.2015.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weisser M, Voigts-Hoffmann F, Rabl J, Leibundgut M, Ban N. The crystal structure of the eukaryotic 40S ribosomal subunit in complex with eIF1 and eIF1A. Nat Struct Mol Biol. 2013;20(8):1015–1017. doi: 10.1038/nsmb.2622. [DOI] [PubMed] [Google Scholar]
- 24.Zhang F, Saini AK, Shin BS, Nanda J, Hinnebusch AG. Conformational changes in the P site and mRNA entry channel evoked by AUG recognition in yeast translation preinitiation complexes. Nucleic Acids Res. 2015;43(4):2293–2312. doi: 10.1093/nar/gkv028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120(1):49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 26.Graifer D, Malygin A, Zharkov DO, Karpova G. Eukaryotic ribosomal protein S3: A constituent of translational machinery and an extraribosomal player in various cellular processes. Biochimie. 2014;99:8–18. doi: 10.1016/j.biochi.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 27.Donahue TF, Cigan AM. Genetic selection for mutations that reduce or abolish ribosomal recognition of the HIS4 translational initiator region. Mol Cell Biol. 1988;8(7):2955–2963. doi: 10.1128/mcb.8.7.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang HK, Yoon H, Hannig EM, Donahue TF. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev. 1997;11(18):2396–2413. doi: 10.1101/gad.11.18.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin-Marcos P, et al. Enhanced eIF1 binding to the 40S ribosome impedes conformational rearrangements of the preinitiation complex and elevates initiation accuracy. RNA. 2014;20(2):150–167. doi: 10.1261/rna.042069.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong J, et al. Conserved residues in yeast initiator tRNA calibrate initiation accuracy by regulating preinitiation complex stability at the start codon. Genes Dev. 2014;28(5):502–520. doi: 10.1101/gad.236547.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci USA. 1990;87(21):8301–8305. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiu WL, et al. The C-terminal region of eukaryotic translation initiation factor 3a (eIF3a) promotes mRNA recruitment, scanning, and, together with eIF3j and the eIF3b RNA recognition motif, selection of AUG start codons. Mol Cell Biol. 2010;30(18):4415–4434. doi: 10.1128/MCB.00280-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moehle CM, Hinnebusch AG. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11(5):2723–2735. doi: 10.1128/mcb.11.5.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reid GA, Schatz G. Import of proteins into mitochondria. Yeast cells grown in the presence of carbonyl cyanide m-chlorophenylhydrazone accumulate massive amounts of some mitochondrial precursor polypeptides. J Biol Chem. 1982;257(21):13056–13061. [PubMed] [Google Scholar]
- 35.Valásek L, Hašek J, Nielsen KH, Hinnebusch AG. Dual function of eIF3j/Hcr1p in processing 20 S pre-rRNA and translation initiation. J Biol Chem. 2001;276(46):43351–43360. doi: 10.1074/jbc.M106887200. [DOI] [PubMed] [Google Scholar]
- 36.Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR. Reconstitution of yeast translation initiation. Methods Enzymol. 2007;430:111–145. doi: 10.1016/S0076-6879(07)30006-2. [DOI] [PubMed] [Google Scholar]
- 37.Mitchell SF, et al. The 5′-7-methylguanosine cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and to block an alternative pathway. Mol Cell. 2010;39(6):950–962. doi: 10.1016/j.molcel.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.