Significance
The molecular mechanism for sealing newly formed nuclear envelopes was unclear until the recent discovery that endosomal sorting complexes required for transport III (ESCRT-III) proteins mediate this process. Cmp7p (CHMP7), in particular, was identified as an early acting factor that recruits other ESCRT-III proteins to the nuclear envelope. A fundamental aspect of the varied roles of ESCRT factors is their recruitment by site-specific adaptors, yet the central question of how the ESCRT machinery is targeted to nuclear membranes has remained outstanding. Our study identifies the inner nuclear membrane protein LEM2 as a key, conserved factor that recruits CHMP7 and downstream ESCRT-III proteins to breaches in the nuclear envelope.
Keywords: LEM2, CHMP7, VPS4, ESCRT-III, nuclear envelope
Abstract
Endosomal sorting complexes required for transport III (ESCRT-III) proteins have been implicated in sealing the nuclear envelope in mammals, spindle pole body dynamics in fission yeast, and surveillance of defective nuclear pore complexes in budding yeast. Here, we report that Lem2p (LEM2), a member of the LEM (Lap2-Emerin-Man1) family of inner nuclear membrane proteins, and the ESCRT-II/ESCRT-III hybrid protein Cmp7p (CHMP7), work together to recruit additional ESCRT-III proteins to holes in the nuclear membrane. In Schizosaccharomyces pombe, deletion of the ATPase vps4 leads to severe defects in nuclear morphology and integrity. These phenotypes are suppressed by loss-of-function mutations that arise spontaneously in lem2 or cmp7, implying that these proteins may function upstream in the same pathway. Building on these genetic interactions, we explored the role of LEM2 during nuclear envelope reformation in human cells. We found that CHMP7 and LEM2 enrich at the same region of the chromatin disk periphery during this window of cell division and that CHMP7 can bind directly to the C-terminal domain of LEM2 in vitro. We further found that, during nuclear envelope formation, recruitment of the ESCRT factors CHMP7, CHMP2A, and IST1/CHMP8 all depend on LEM2 in human cells. We conclude that Lem2p/LEM2 is a conserved nuclear site-specific adaptor that recruits Cmp7p/CHMP7 and downstream ESCRT factors to the nuclear envelope.
Eukaryotic genomes are secluded within the nucleus, an organelle with a boundary that comprises the double-membraned nuclear envelope (NE) (1). The inner and outer bilayers of the NE are perforated by annular channels that contain nuclear pore complexes (NPCs), each a massive assembly that regulates the trafficking of macromolecules like mRNA and proteins between the cytoplasm and nucleoplasm. The evolution of the envelope, among other roles, helped safeguard genome duplication and mRNA transcription from parasitic nucleic acids (2). The isolation of nucleoplasm from cytoplasm, however, presents a challenge during cell division when duplicated chromosomes must be separated for daughter-cell inheritance.
Chromosome inheritance depends on assembly of a mitotic spindle, which pulls chromosomes toward opposite sides of the duplicating cell. Spindle assembly begins when two microtubule-organizing centers (MTOCs) nucleate polymerization of antiparallel arrays of microtubules to capture daughter chromosomes. Despite functional conservation throughout Eukarya, the mechanisms by which spindle microtubules breach the NE to gain access to metaphase chromosomes vary markedly (3–6). In vertebrates and other organisms that have an “open mitosis,” the NE disassembles completely, so that nucleoplasmic identity is lost. Certain protists and fungi, by contrast, maintain NE integrity throughout a “closed mitosis” (3, 5).
The fission yeast Schizosaccharomyces pombe and the budding yeast Saccharomyces cerevisiae integrate their MTOC, known as the spindle pole body (SPB), into the NE so that microtubule assembly can occur in the nucleoplasm (3, 4, 7, 8). In budding yeast, duplication of the SPB is coupled with NE remodeling so that mother and daughter SPBs reside within the envelope throughout the cell cycle (4, 9). Fission yeast, by contrast, restrict SPB access to the nucleoplasm during mitosis only (7, 8). Upon mitotic entry, a fenestration through the NE opens transiently, and the mother and daughter SPBs are incorporated (3, 5, 6, 8, 10). For every cell cycle, therefore, the fission yeast NE must open and reseal twice: once when the SPBs are inserted and again when the SPB is ejected from the envelope after a successful cell cycle.
Incorporating NPCs and SPBs into the NE requires certain factors and mechanisms in common, including membrane-remodeling activities (6, 11–15). We and others have previously reported strong genetic interactions between transmembrane nucleoporins, SPB components, and endosomal sorting complexes required for transport (ESCRT) genes—portending a role for certain ESCRT proteins in nuclear membrane remodeling (16, 17). In general, ESCRT components are recruited to different target membranes by site-specific adaptors that ultimately recruit the membrane-remodeling ESCRT-III subunits and their binding partners, including VPS4-family adenosine triphosphatases (ATPases) (18–20). We previously showed that certain ESCRT-III mutants and vps4Δ cells displayed an apparent overamplification of SPBs (or defective fragments) in fission yeast and that the severity of this SPB phenotype in fission yeast waned over time, suggesting possible genetic suppression (16). In budding yeast, Webster et al. reported that, without ESCRT-III/Vps4 activity, misassembled NPCs accumulate in a compartment they named the SINC (for storage of improperly assembled NPCs) (21). They also showed that LEM family (Lap2-Emerin-Man1) inner nuclear membrane (INM) proteins Heh1p and Heh2p in budding yeast associate with defective NPC assembly intermediates (but not with mature NPCs) and that Heh1/2 proteins may recruit ESCRT-III and Vps4 activities to malformed NPCs to clear them from the NE (21).
In mammals, VPS4 depletion induces nuclear morphology defects (22), and several recent reports have demonstrated that ESCRT pathway proteins are recruited transiently to seal gaps in reforming mammalian nuclear membranes during anaphase (23, 24) and to rupture sites in the nuclei of interphase mammalian cells (25, 26). Depletion of ESCRT factors delays sealing of the reforming NE and impairs mitotic spindle disassembly (23, 24). Moreover, depletion of SPASTIN, another meiotic clade VPS4-family member and ESCRT-III–binding enzyme (27), also delays spindle disassembly and envelope resealing (24, 28). Similar effects were seen upon depletion of several ESCRT-III proteins, including the poorly characterized ESCRT factor CHMP7, which has features of both ESCRT-II and -III proteins (29). These observations support a model in which ESCRT-III and VPS4 proteins and SPASTIN together coordinate microtubule severing with the closure of annular gaps in the NE. This model is conceptually similar to the mechanism of cytokinetic abscission, where SPASTIN disassembles the residual microtubules that pass between daughter cells, while ESCRT-III and VPS4 proteins constrict the midbody membrane to the point of fission (19, 28, 30).
Here, we address the key question of what upstream factor(s) serves as the membrane-specific adaptor that facilitates CHMP7 recruitment to function in sealing NE breaches. To identify factors in this pathway, we returned to the genetically tractable fission yeast system. We report that deletion of vps4 in S. pombe leads to severe defects in nuclear membrane morphology and nuclear integrity, with secondary defects in NPCs and SPB dynamics. Remarkably, these phenotypes are suppressed spontaneously when cells acquire loss-of-function mutations in cmp7, an ortholog of human CHMP7, or in lem2, a LEM domain INM protein and ortholog of human LEM2. We also show that, in human cells, recruitment of CHMP7 and downstream ESCRT-III proteins to the reforming NE during anaphase depends on LEM2, most likely through a direct interaction between CHMP7 and the C-terminal nucleoplasmic domain of LEM2. Together, these observations implicate LEM2 as a nucleus-specific adaptor that recruits ESCRT pathway activities to remodel the NE during both open and closed mitoses across Eukaryotes.
Results
vps4∆ Fission Yeast Cells Grow Very Slowly, and Loss of Either cmp7 or lem2 Rescues Growth.
The AAA ATPase VPS4 has a primary role in disassembling ESCRT-III polymeric structures in the different settings where the ESCRT pathway mediates membrane remodeling. To determine whether and how reported phenotypes that result from deletion of VPS4 were suppressed over time (16), we monitored the growth of individual colonies after sporulation and tetrad dissection of vps4Δ/+ diploid cells. Growth rates of vps4Δ spores were dramatically slower than wild-type (WT) spores (Fig. 1A). This growth defect spontaneously reverted over time, so that when mutant spores were streaked on rich medium, some vps4Δ colonies exhibited growth rates comparable to WT colonies (Fig. 1B). To identify potential suppressor mutations, we sequenced complete genomes for 12 strains that spontaneously reverted to WT growth rates and compared them with genomes of both WT and apparently unsuppressed vps4Δ strains. The analysis revealed that 7 of the 12 suppressors had different loss-of-function mutations in the ESCRT-II/-III hybrid gene, cmp7 (Table S1). The remaining five each had equivalent independent mutations in a LEM domain family member, lem2, within what appears to be a slippery poly-T track (Table S1). These 12 mutant alleles were further confirmed by Sanger sequencing, and none of the suppressors were found to harbor mutations in both cmp7 and lem2 simultaneously.
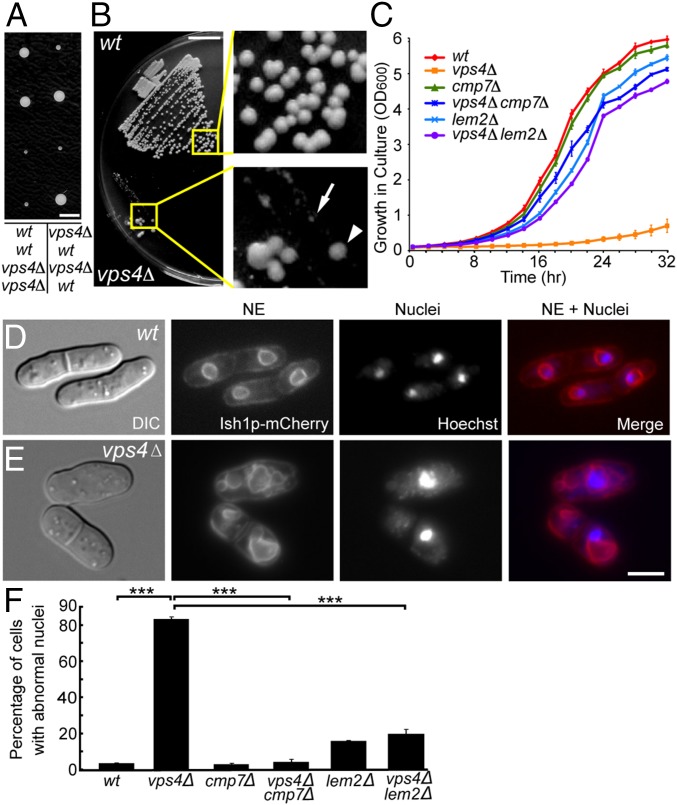
Fig. 1.
vps4Δ cells grow slowly and have severe NE defects, which are suppressed by loss of cmp7 or lem2. (A) Tetrad dissection of vps4Δ/+ diploids, with genotypes labeled below the image. (Scale bar: 0.25 cm.) (B) Spontaneous suppressors (arrowhead) of vps4Δ (arrow) appearing after 3 d on rich medium, with colony sizes comparable to those of WT cells. (Scale bar: 1 cm.) (C) Growth curves of each genotype showing optical densities of yeast cultures, starting at OD600nm 0.06, for 32 h in 2-h intervals. Three independent isolates for each genotype were measured. The plots show mean ± SEM. (D) WT cells, showing normal NE morphologies (Ish1p-mCherry) and DNA (Hoechst staining) within the nuclei. (E) vps4Δ, showing various NE morphology defects including excess NE, fragmented NE, and DNA that appears to be outside of the nucleus. (Scale bar: 5 μm.) (F) Deletion of either cmp7 or lem2 rescues these NE morphology defects. Here and throughout, three independent isolates/strains were imaged for each genotype, and n represents the total number of scored cells. Mean ± SEM, WT: 1 ± 1%, n = 115, 121, 139; vps4Δ: 85 ± 5%, n = 62, 69, 86; cmp7Δ: 2.0 ± 0.4%, n = 151, 166, 147; vps4Δcmp7Δ: 1.1 ± 0.7%, n =161, 118, 145; lem2Δ: 8 ± 3%, n= 131, 103, 128; vps4Δlem2Δ: 12.2 ± 0.7%, n = 99, 176, 111. Two-tailed Student t tests were used here and throughout. ***P < 0.001.
Table S1.
Spontaneous suppressor alleles of vps4Δ identified in this study
| Suppressor | Suppressor mutation alleles | Mutation | Total no. of valid reads | No. of supporting reads | Percentage of supporting reads, % | Residues affected | Length of peptide translated, amino acids |
| A. cmp7* | |||||||
| 1 | cmp7-1 | Frameshift | 26 | 25 | 96 | 24–449 | 24 |
| 2 | cmp7-2 | Nonsense | 35 | 35 | 100 | W49*(stop) | 48 |
| 3 | cmp7-3 | Nonsense | 20 | 19 | 95 | Q139*(stop) | 138 |
| 4 | cmp7-4 | Frameshift | 15 | 15 | 100 | 280–449 | 284 |
| 5 | cmp7-5 | Frameshift | 10 | 10 | 100 | 126–449 | 155 |
| 6 | cmp7-6 | Frameshift | 31 | 31 | 100 | 115–449 | 148 |
| 7 | cmp7-7 | Frameshift | 19 | 19 | 100 | 285–449 | 287 |
| B. lem2† | |||||||
| 8 | lem2-1‡ | Frameshift | 20 | 20 | 100 | 550–668 | 561 |
| 9 | lem2-1 | Frameshift | 17 | 17 | 100 | 550–668 | 561 |
| 10 | lem2-1 | Frameshift | 23 | 23 | 100 | 550–668 | 561 |
| 11 | lem2-1 | Frameshift | 22 | 22 | 100 | 550–668 | 561 |
| 12 | lem2-1 | Frameshift | 24 | 23 | 95 | 550–668 | 561 |
Whole-genome sequencing of WT (n = 2), vps4Δ mutant (n = 2), and spontaneous suppressors (n = 12) uncovered mutant alleles responsible for rescuing growth defects of vps4Δ.
WT cmp7 encodes a peptide of 449 amino acids.
WT lem2 encodes a peptide of 668 amino acids.
The lem2-1 mutant allele had a single T inserted within a series of 7 T residues corresponding to nucleotides 1,643–1,649 (relative to the lem2 ATG).
To determine whether these potentially suppressive alleles rescued the growth of vps4Δ cells, we engineered cmp7Δ/+ and lem2Δ/+ genotypes within our vps4Δ/+ diploid background and isolated lem2Δ, cmp7Δ, and vps4Δ single mutants and their corresponding double mutants via sporulation and tetrad dissection. Quantitative growth rates in liquid culture for biological and technical triplicates demonstrated that cmp7Δ single mutants had WT growth rates and that the vps4Δcmp7Δ double-mutant cells grew much faster than vps4Δ single-mutant cells (albeit slightly more slowly than WT cells; Fig. 1C). Similarly, lem2Δ single mutants displayed a modest growth deficit compared with WT, but vps4Δlem2Δ double-mutant cells again grew much faster than vps4Δ cells (Fig. 1C). Thus, our unbiased whole-genome sequencing and targeted double-mutant studies demonstrate that cmp7 and lem2 are bona fide vps4 suppressors and that the spontaneous mutations phenocopy null alleles.
vps4∆ Cells Have NE Defects, Which Are Suppressed by Loss of cmp7 or lem2.
Next, we sought to discover the cellular defects that correlated with the slow growth of vps4Δ cells and to test whether those defects were also rescued by cmp7Δ or lem2Δ. In light of our prior work on mitotic and SPB defects in vps4∆ cells, we first examined NE and SPB morphology and dynamics. More than 80% of vps4Δ cells from recently dissected vps4Δ haploid spores displayed severe NE morphology defects. These defects were rescued by loss of either cmp7 or lem2 (Fig. 1 D–F). Moreover, ∼30% of vps4Δ cells displayed clearly asymmetric SPB segregation errors and anucleate daughter cells (Fig. S1 A and B). In this case, loss of cmp7 rescued these phenotypes, but loss of lem2 did not. Indeed, even lem2Δ single-mutant cells displayed similar SPB segregation defects (Fig. S1C). Thus, defects in NE morphology and integrity were the features that correlated best with the slow-growth phenotype, suggesting that these defects were primarily responsible for the vps4Δ slow-growth phenotype.
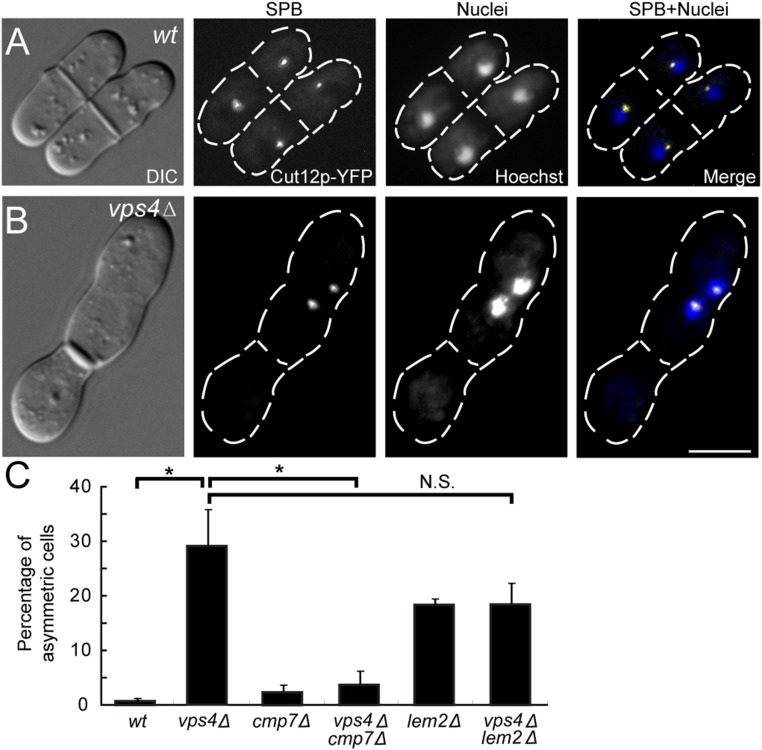
Fig. S1.
vps4Δ and lem2Δ exhibit asymmetric SPB segregation. (A) WT cells showing symmetric SPB segregation. (B) vps4Δ showing asymmetric SPB segregation. (C) Quantified percentages of asymmetric SPB segregation events in each genotype. Cells with septa were selected and scored for SPB separation patterns. Nuclei were stained with Hoechst. WT: 0.7 ± 0.3%, n = 103, 101, 101; vps4Δ: 29 ± 7%, n = 88, 103, 107; cmp7Δ: 2 ± 1%, n = 105, 103, 101; vps4Δcmp7Δ: 4 ± 2%, n = 117, 100, 104; lem2Δ: 18 ± 1%, n = 102, 103, 100; vps4Δlem2Δ: 18 ± 4%, n = 102, 103, 101. *P < 0.05. N.S., not significant. (Scale bar: 5 μm.)
vps4Δ Cells Display a Series of Mitotic Errors Associated with NE Defects.
Live cell imaging using NE (Ish1p-mCherry) and SPB (Cut12p-YFP) markers enabled us to monitor the development and consequences of NE defects in mutant vs. WT cells. Abnormal NE morphologies or asymmetric and even failed karyokinesis were observed in the majority of cells (Fig. 2), and only ∼30% of vps4Δ cells displayed normal, symmetric karyokinesis (Fig. 2A). An apparent proliferation or overgrowth of Ish1p-marked membranes was a particularly common defect in vps4Δ cells. Approximately 25% of mutant cells displayed these long-lived NE “outgrowths” that we later determined were karmellae (see below). In cells with karmellae, daughter SPBs often failed to separate normally or displayed extensive delays in separation (Fig. 2B). Indeed, separation of duplicated SPBs was significantly prolonged in vps4Δ cells, whether or not they exhibited abnormal NE malformations (Fig. S2). Together, these observations suggest that Vps4p plays a central role in regulating NE morphology in fission yeast, particularly during SPB extrusion or insertion through the NE, and perhaps during karyokinesis.
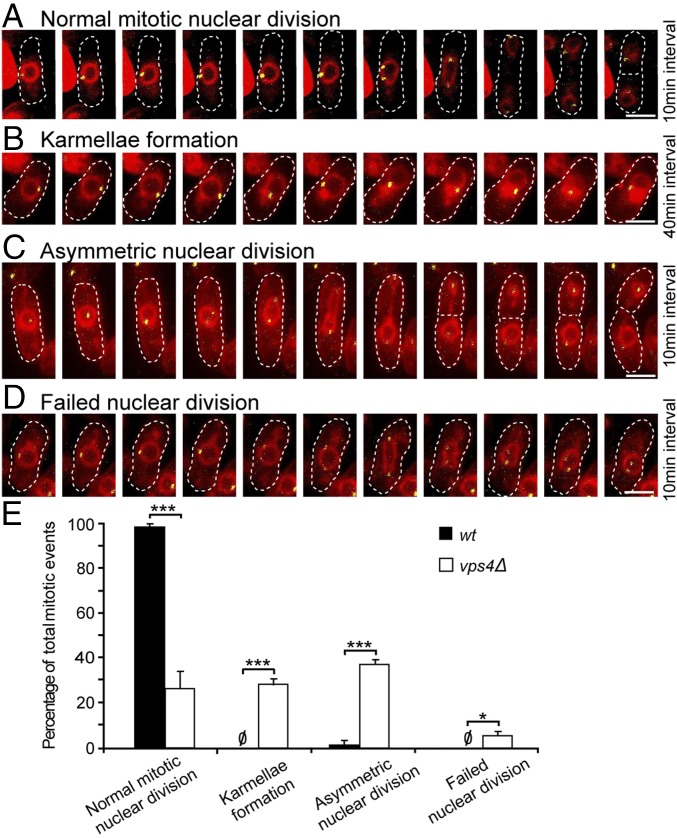
Fig. 2.
vps4Δ cells display a series of mitotic defects associated with failure of nuclear membrane maintenance. (A) Time lapse of normal mitotic karyokinesis in vps4Δ cells. In all cases, the nuclear membrane is marked by Ish1p-mCherry, and the SPB is marked by Cut12p-YFP. The total length of time is 100 min. (B) Karmellae formation correlates with defective separation of duplicated SPB. The total length of time is 400 min. (C) Asymmetric nuclear division. Total length of time is 100 min. (D) Failed nuclear division. Total length of time is 100 min. (E) Quantification of NE morphology during mitosis in WT and vps4Δ. Normal mitotic nuclear division, WT: 99 ± 1%, vps4Δ: 27 ± 8%; karmellae formation, WT: 0%, vps4Δ: 29 ± 3%; asymmetric nuclear division, mean ± SEM, WT: 1 ± 1%, vps4Δ: 38 ± 2%; failed nuclear division, WT: 0%, vps4Δ: 6 ± 2%. WT, n = 23, 21, 18; vps4Δ, n = 43, 51, 84. *P < 0.05; ***P < 0.001. (Scale bars: 5 μm.) (A–D) Dashed lines correspond with the cell wall.
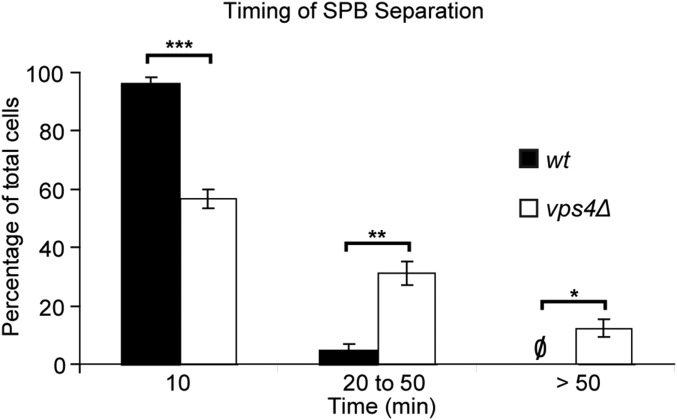
Fig. S2.
SPB separation is delayed in vps4Δ mutants. A 16-h time-lapse imaging (10-min intervals) was conducted on WT and vps4Δ cells at 32 °C, and the times required for SBP separation were scored. Data were binned and plotted by mean frequency ± SEM: WT: 10 min 96 ± 2%; 20–50 min 4 ± 2%; >50 min 0%; n = 23, 22, 22; vps4Δ: 10 min 57 ± 4%; 20–50 min 31 ± 4%; >50 min 12 ± 3%; n = 30, 30, 30. *P < 0.05; **P < 0.01; ***P < 0.001.
vps4Δ Nuclei Leak and Their Integrity Is Largely Restored by Loss of cmp7 or lem2.
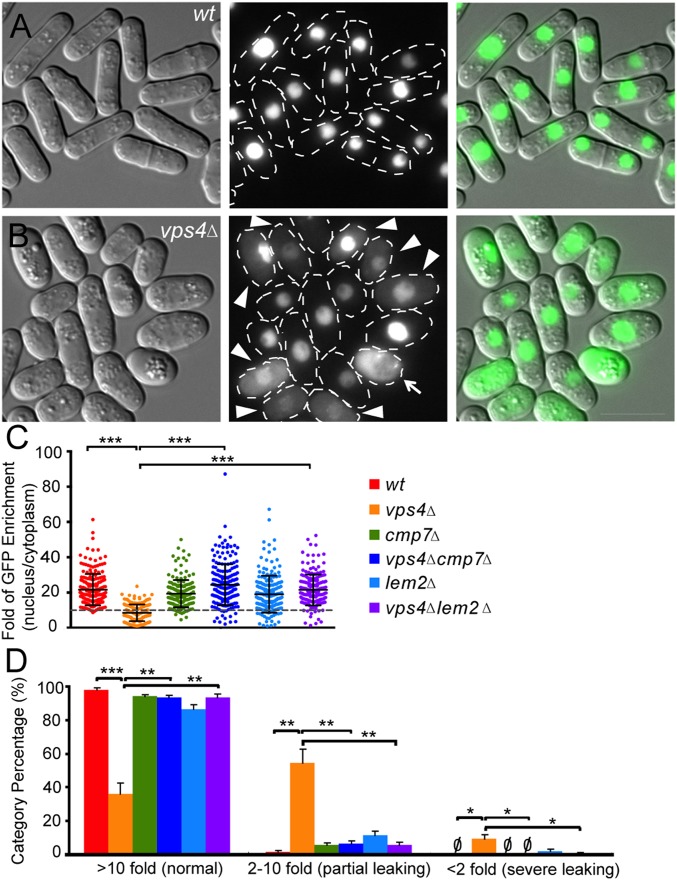
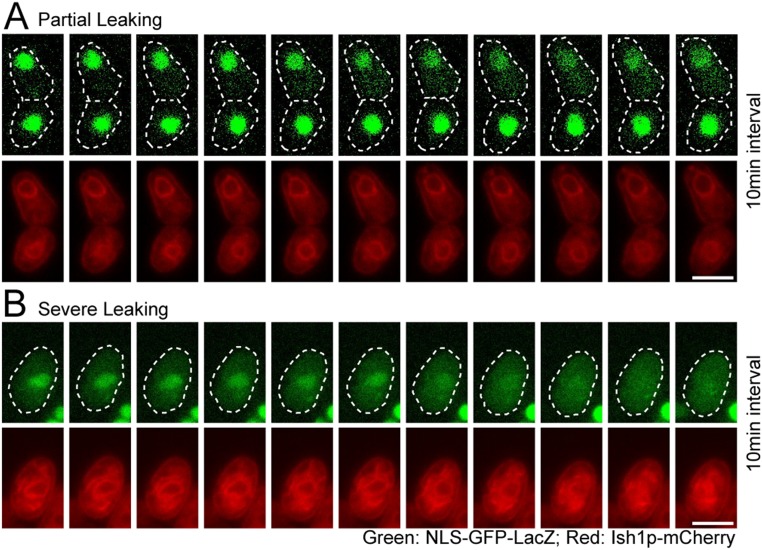
Our observations, together with recent reports that the ESCRT pathway closes holes in the mammalian NE (23, 24), prompted us to test the integrity of vps4Δ nuclei. Image analysis revealed that a large nuclear import cargo, NLS-GFP-LacZ, was enriched within nuclei by >10-fold in 98% of WT cells (Fig. 3A). By contrast, ∼55% of vps4Δ cells displayed <10-fold nuclear enrichment (partial leaking; Fig. 3B, arrowheads), and ∼10% of vps4Δ cells displayed <2-fold nuclear enrichment of NLS-GFP-LacZ (severe leaking; Fig. 3B, arrow; Fig. 3 C and D, quantification). Remarkably, loss of cmp7 or lem2 rescued this abnormal nuclear integrity phenotype to a large extent, although a small minority of single and double cmp7 or lem2 cells still displayed partial leaking (Fig. 3 C and D). Live cell imaging also revealed that the extent of nuclear integrity loss correlated with NE morphology defects (Fig. S3). Cells that initially displayed normal GFP reporter localization and normal NE morphology gradually accumulated cytoplasmic signal over the course of tens of minutes (Fig. S3A). Cells with abnormal NE morphology at the beginning of the experiment, by contrast, lost nuclear GFP completely over the time course (Fig. S3B). Thus, cytoplasmic GFP resulted from loss of nuclear integrity rather than from defects in nuclear import.
Fig. 3.
vps4Δ cells have leaky nuclei and nuclear integrity is restored by loss of either cmp7 or lem2. (A) GFP signals in the nuclear lumen of WT cells expressing NLS-GFP-LacZ. (B) vps4Δ cells expressing NLS-GFP-LacZ with moderate (arrowheads) or severe (arrow) nuclear leaking. (Scale bar: 10 μm.) (C) Nuclear enrichment of NLS-GFP-LacZ (nucleus/cytoplasm). Mean ± SD, WT: 21.6 ± 8.9, n = 205; vps4Δ: 8.6 ± 4.7, n = 180; cmp7Δ: 19.4 ± 7.7, n = 175; vps4Δcmp7Δ: 24.5 ± 11.7, n = 202; lem2Δ: 19.1 ± 10.4, n = 198; vps4Δlem2Δ: 21.5 ± 8.8, n = 189. (D) Percentage of cells in different nuclear leaky phenotype categories. Mean ± SEM, normal (>10-fold GFP nuclear enrichment) WT: 97.9 ± 1.3%, vps4Δ: 36.2 ± 6.5%, cmp7Δ: 94.1 ± 1.1%, vps4Δcmp7Δ: 93.2 ± 1.6%, lem2Δ: 86.6 ± 2.3%, vps4Δlem2Δ: 93.4 ± 2.0%; partial leaking (2- to 10-fold GFP nuclear enrichment) WT: 1.6 ± 0.9%, vps4Δ: 54.4 ± 8.1%, cmp7Δ: 5.9 ± 1.1%, vps4Δcmp7Δ: 6.8 ± 1.6%, lem2Δ: 11.5 ± 2.3%, vps4Δlem2Δ: 5.9 ± 1.4%; severe leaking (<2-fold GFP nuclear enrichment) WT: 0%, vps4Δ: 9.4 ± 2.4%, cmp7Δ: 0%, vps4Δcmp7Δ: 0%, lem2Δ: 2.0 ± 1.3%, vps4Δlem2Δ: 0.7 ± 0.7%; WT: n = 82,56,67, vps4Δ: n = 60,59,61, cmp7Δ: n = 62,63,50, vps4Δcmp7Δ: n = 82,59,61, lem2Δ: n = 68,61,69, vps4Δlem2Δ: n = 76,50,63. *P < 0.05; **P < 0.01; ***P < 0.001.
Fig. S3.
Time-lapse analysis of NLS-GFP-LacZ localization in vps4Δ. (A) Time-lapse images showing normal nuclear localization of NLS-GFP-LacZ converting to partial cytoplasmic localization. A pair of newly divided cells is shown over a time course of 100 min with 10-min interval. (B) Time lapse images showing partial cytoplasmic localization of NLS-GFP-LacZ converting to a homogenous distribution throughout the cell. A single cell is shown over a time course of 100 min with 10-min intervals. (Scale bars: 5 μm.)
vps4Δ NEs Are Persistently Fenestrated and Have Karmellae and Disorganized Tubular Extensions.
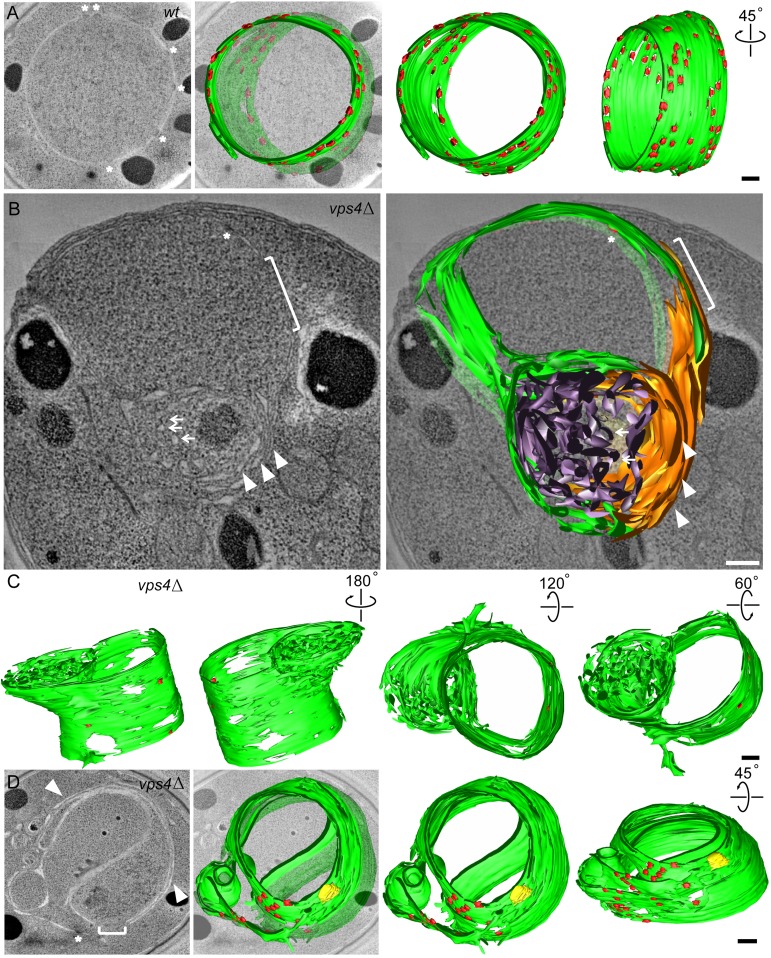
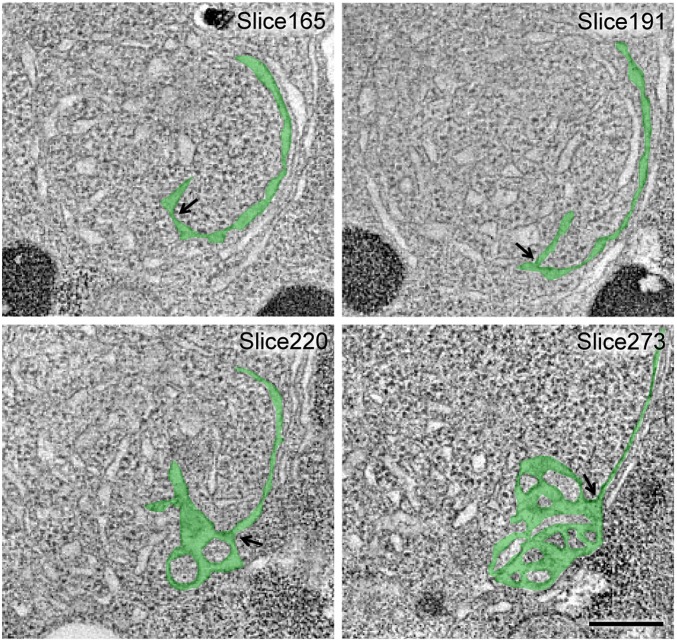
We used electron tomography of high-pressure frozen and freeze-substituted cells to examine whether we could detect morphological defects in the NE that could account for the loss of nuclear integrity. Serial 400-nm sections were imaged and reconstructed to generate 3D volumes of >1-μm thickness. WT nuclei had evenly spaced inner and outer lipid bilayers with embedded NPCs evenly distributed around the periphery (Fig. 4A and Movie S1). vps4Δ nuclei, by contrast, displayed at least four structural abnormalities. First, large fenestrations through both the inner and outer nuclear membranes were observed (Fig. 4B, bracket). Second, karmellae or concentric layers of membrane were present around certain regions of the mutant nuclei (Fig. 4B, arrowheads, and Fig. 5B). Third, extensive whorls of disorganized tubulo-vesicular membranes that were topologically continuous with the adjacent karmellae were apparent (Fig. 4B, arrows, and Fig. S4). Fourth, the total number of NPCs appeared to be decreased (Fig. 4B, asterisk), and the NPCs that were present were localized to regions that were largely free of karmellae and tubulo-vesicular structures (Fig. 4D). The 3D reconstruction of these features confirmed the presence of very large gaps (>400 nm) in the NE and topologically continuous karmellae and whorls of tubular extensions (Fig. 4C and Movies S2–S4). Persistent fenestrations explain the loss of nuclear integrity in vps4Δ cells, whereas karmellae and other disorganized membrane extensions may underlie the kinetic delays, asymmetries, and outright failures of SPB separation and karyokinesis.
Fig. 4.
vps4Δ NEs have persistent fenestrations, karmellae, and disorganized tubular extensions. (A, Left) Single tomographic slice showing the WT NE. NPCs are marked by asterisks. Segmented NE (green) with NPCs (red) reconstructed from 150 tomographic slices is shown in a merged view (A, Left Center), a front view (A, Right Center), and a side view (A, Right). (B, Left) Single tomographic slice showing the vps4Δ NE. The following features and defects are highlighted: a fenestration (bracket), an NPC (asterisk), karmellae layers (white arrowheads), and a disorganized whorl of tubules (arrows). (B, Right) Segmented model of NE reconstructed from 100 tomographic slices is shown in a merged view with karmellae in gold and a whorl of tubules in purple. (C) A segmented model of vps4Δ NE reconstructed from 200 tomographic slices is shown from the front (Left), back (Left Center), top (Right Center), and bottom (Right). (D, Left) Single vps4Δ tomographic slice showing NPCs are absent from karmellae region. The following features and defects are highlighted: a fenestration (bracket), an NPC (asterisk), and karmellae layers (white arrowheads). Segmented NE (green) with NPCs (red) and SPB (yellow) reconstructed from 150 tomographic slices shown in a merged view (D, Left Center), a front view (D, Right Center), and a side view (D, Right). (Scale bars: 200 nm.)
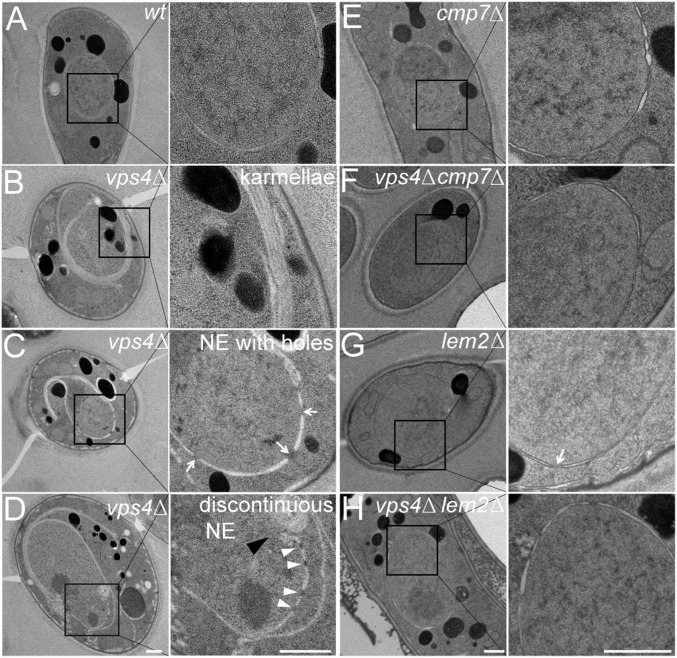
Fig. 5.
vps4Δ NEs have persistent fenestrations, karmellae, and disorganized extensions, and these defects are suppressed by loss of cmp7 or lem2. (A) WT cells with normal NE. (B–D) vps4Δ cells with a variety of NE phenotypes, including karmellae (B), multiple holes (arrows) (C), and discontinuities (black arrowhead) that can contain apparently intranuclear NPCs (D, arrowheads). (E–H) cmp7Δ and lem2Δ single mutants (E and G) vs. vps4Δcmp7Δ and vps4Δlem2Δ double mutants (F and H). (Scale bars: 500 nm.)
Fig. S4.
Karmellae and tubular extension are connected NE structures in vps4Δ. Selected tomographic slices of vps4Δ show connected karmellae and tubular extensions. The relevant membranes are shaded in light green, and the junction is marked by black arrows. (Scale bar: 200 nm.)
vps4Δ NE Defects Are Suppressed by Loss of cmp7 or lem2.
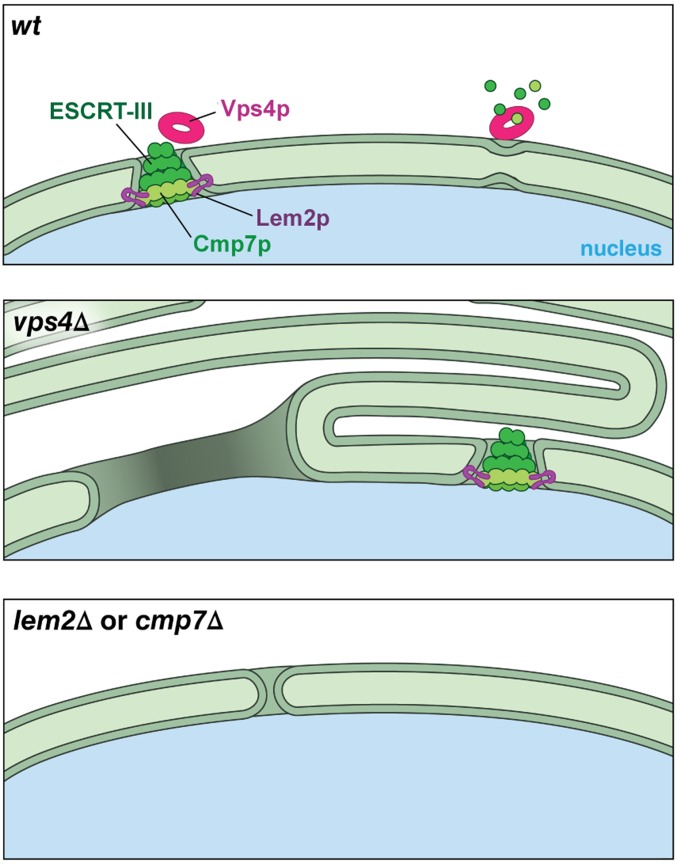
Thin-section electron microscopy of single- and double-mutant cells demonstrated that karmellae formation in vps4Δ cells was completely suppressed by loss of cmp7 or lem2 (Fig. 5). Remarkably, double-mutant cells displayed WT-like nuclei, although a few examples of probable nuclear fenestrations were observed in lem2Δ single-mutant cells (Fig. 5G). These observations indicate that, in the absence of Vps4p, karmellae formation depends on both Lem2p and Cmp7p. Similarly, overexpressing Lem2p in S. pombe, or compromising nuclear import of Heh2p, an ortholog of Lem2p in S. cerevisiae, induces the formation of similar abnormalities (31, 32). Together, these results suggest that, in the absence of vps4, unregulated Lem2p activity drives formation of toxic NE malformations via a pathway that also requires Cmp7p (Fig. S5).
Fig. S5.
A cartoon model for the NE phenotypes in vps4Δ, lem2Δ, and cmp7Δ in S. pombe. (Top) In WT, the exposed Lem2p recruits Cmp7p to initiate an ESCRT-III–mediated NE sealing event. That process is regulated and/or completed by the AAA ATPase, Vps4p. (Middle) in vps4Δ, unrestricted Lem2p/Cmp7p activities induce the formation of karmellae and large gaps in NE so that the proper progress of mitosis is disrupted. (Bottom) In lem2Δ or cmp7Δ, mutant cells tolerate a limited number of small breaches and therefore show little or no growth defect.
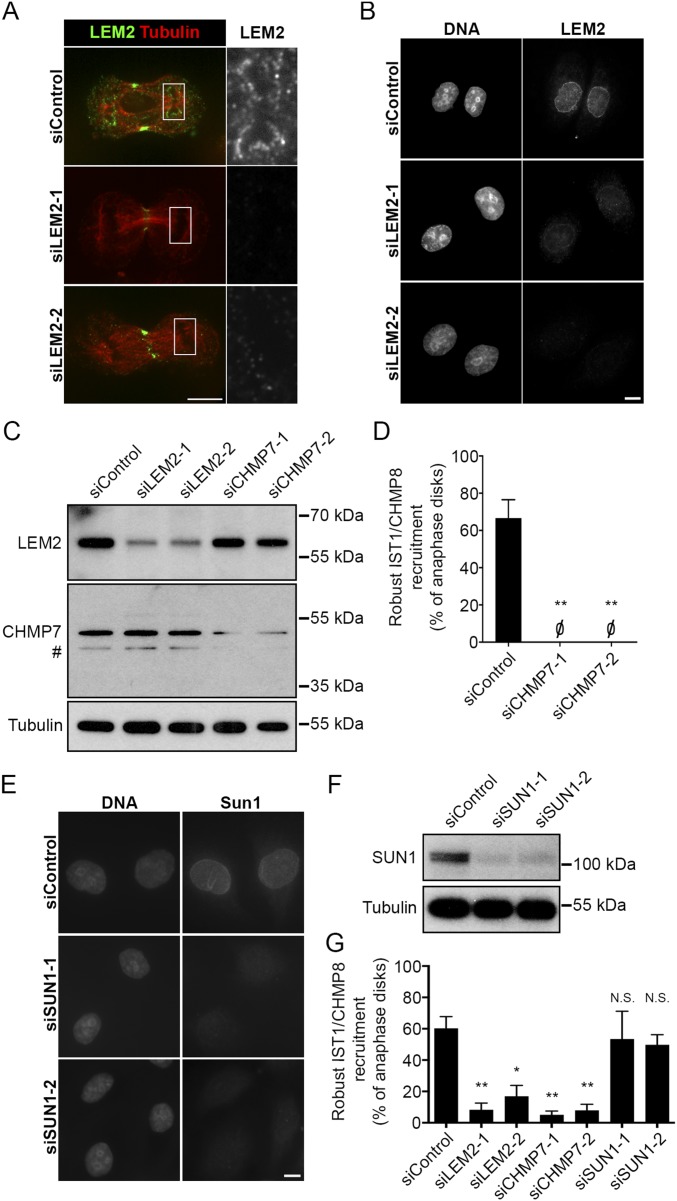
LEM2 Recruits CHMP7 to the Reforming NE During Anaphase in Mammalian Cells.
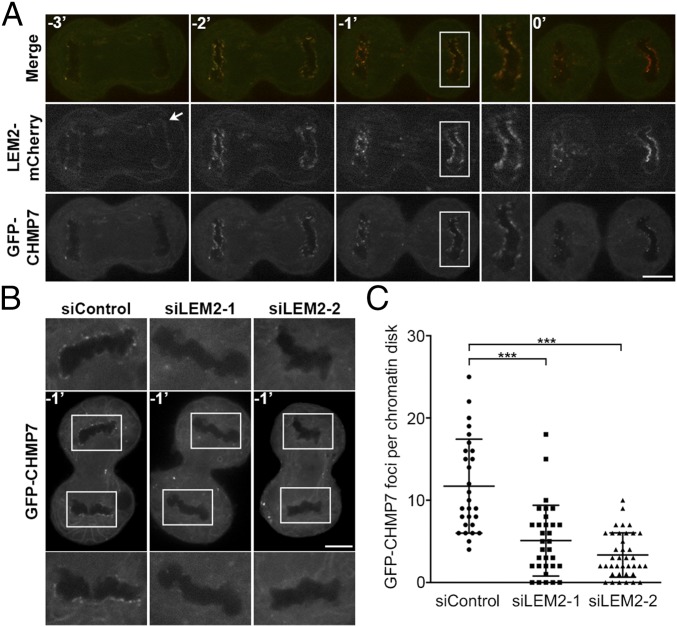
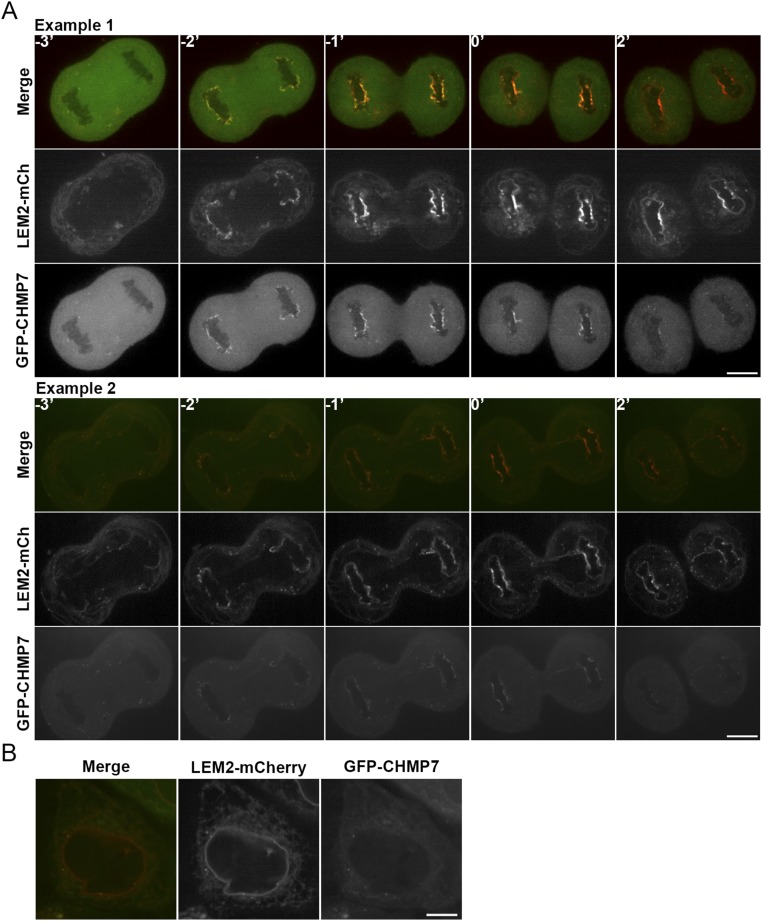
LEM2 and its homologs are two-pass membrane proteins that reside in the INM (32, 33). A LEM2 homolog in budding yeast, Heh2, has been previously implicated in ESCRT-dependent surveillance of defective NPCs (21). These studies and our observations suggested that LEM2-like proteins in both budding and fission yeast may be site-specific nuclear membrane adaptors that recruit and/or activate Cmp7p/CHMP7. Although a role for CHMP7 in NE closure has been reported in mammalian cells (24), its localization during the process of nuclear assembly had not been determined. To confirm that CHMP7 is indeed present at this site and to track the spatial relationship between LEM2 and CHMP7 in human cells, we assessed the dynamics of LEM2-mCherry and GFP-CHMP7 localization by live imaging of HeLa cells. Like other NE proteins that reside in the endoplasmic reticulum during mitosis (34), LEM2-mCherry was found in a reticular network as cells emerged from metaphase. Early in anaphase, LEM2-mCherry rapidly accumulated around condensed chromatin disks—first at distal ends, then concentrating at the central or “core” region of the anaphase disk, and then broadening to a distributed nuclear rim pattern. The latter pattern has been reported (35). CHMP7 recruitment to the chromatin surface coincided closely in time with the appearance of LEM2, resulting initially in their robust concentration at the same regions of the chromatin disks (Fig. 6A, Movie S5, and Fig. S6A). The spatiotemporal specificity of the CHMP7 and LEM2 recruitment during early anaphase contrasts with their lack of colocalization after cleavage furrow ingression (Fig. 6A, Movie S5, and Fig. S6A) and their even more distinct spatial distributions during interphase, when GFP-CHMP7 was no longer present at the chromatin surface, and LEM2-mCherry decorated the nuclear rim as expected (Fig. S6B).
Fig. 6.
Live imaging of LEM2-dependent recruitment of CHMP7 to reforming nuclei during mammalian anaphase. (A) Montage of a representative cell expressing LEM2-mCherry and GFP-CHMP7 progressing through anaphase before complete furrow ingression (designated as t = 0′). LEM2-mCherry makes initial contacts with chromatin (white arrow), and GFP-CHMP7 localizes to sites of LEM2-mCherry enrichment (t = −2′, −1′). (B) Illustrative cells, at t = −1′, treated with siRNA as indicated and expressing GFP-CHMP7. (C) Quantification of GFP-CHMP7 peaks at chromatin disks of cells treated with siRNA (mean ± SD; siControl: 12 ± 6%, n = 28; siLEM2-1: 5 ± 4%, n = 32; siLEM2-2: 3 ± 3%, n = 38). ***P < 0.001. (Scales bar: 10 μm.)
Fig. S6.
LEM2 and CHMP7 colocalization is restricted to a brief window specifically during anaphase. (A) Additional montages of cells expressing LEM2-mCherry and GFP-CHMP7 progressing through anaphase before complete furrow ingression (designated as t = 0′). (B) Representative image of an interphase cell expressing LEM2-mCherry and GFP-CHMP7. (Scale bars: 10 μm.)
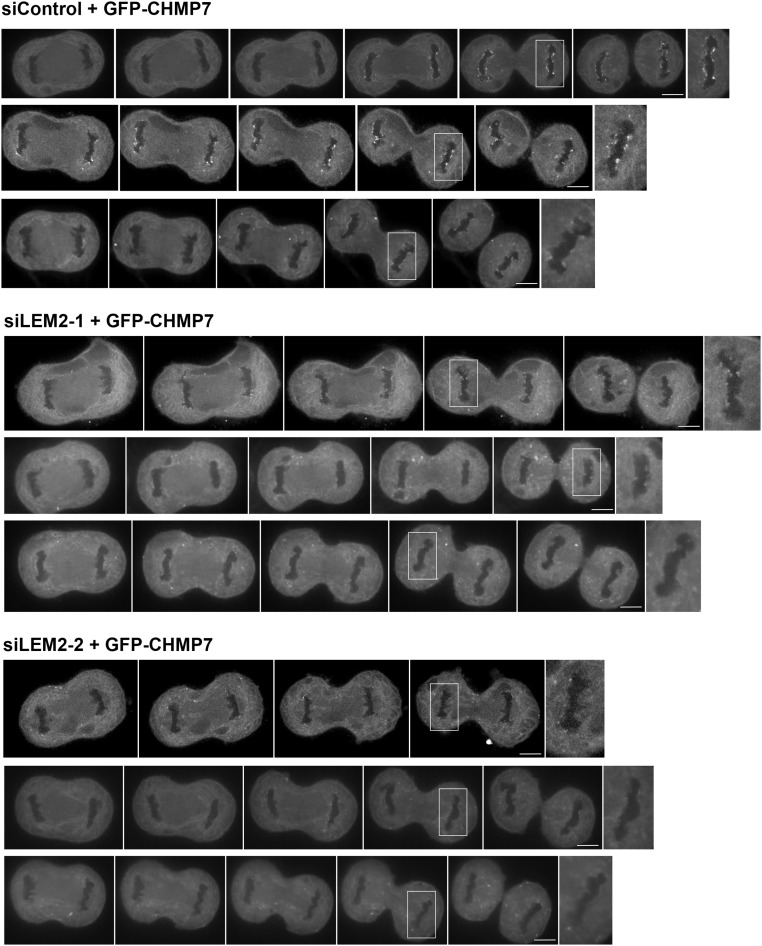
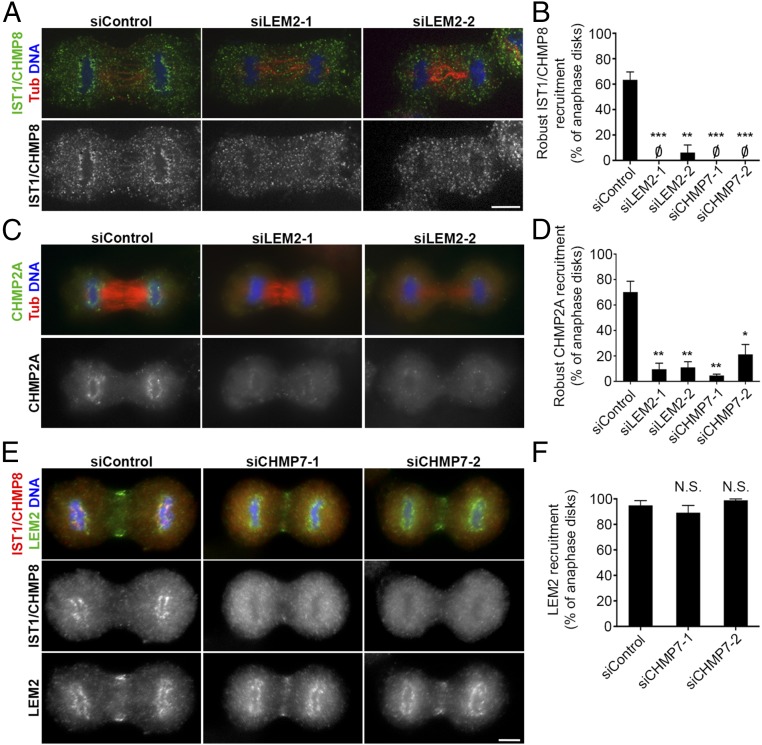
We next tested whether LEM2 is required for CHMP7 recruitment to reforming nuclei. After depletion of endogenous LEM2 with two independent siRNA oligos (Fig. S7 A–C), GFP-CHMP7 recruitment to anaphase chromatin was notably attenuated (Fig. 6 B and C and Fig. S8). These results were also recapitulated in a cell line expressing H2B-mCherry, confirming that CHMP7 foci are in close apposition to the chromatin surface and that this localization is impaired after LEM2 depletion (Movies S6 and S7). To further evaluate the role of LEM2 in recruiting the ESCRT pathway to the reforming NE, we also tracked the localization of IST1/CHMP8 and CHMP2A, two ESCRT-III proteins that were shown to be recruited to the surface of condensed chromatin disks to promote coordinated NE closure and disassembly of spindle microtubules (24) (also see Movie S8). siRNA depletion of CHMP7 abrogated robust IST1/CHMP8 and CHMP2A recruitment to chromatin, as assayed by detection of endogenous protein by fixed-cell immunofluorescence (Fig. 7 B and D, Fig. S7 D and G, and Tables S2 and S3), consistent with the reported role for CHMP7 in the recruitment of CHMP4B to assembling nuclei (24). Importantly, siRNA-mediated depletion of LEM2 also strongly attenuated robust IST1/CHMP8 and CHMP2A recruitment (Fig. 7 A–D and Tables S2 and S3). By contrast, depletion of the abundant INM protein SUN1 did not alter IST1/CHMP8 localization (Fig. S7 E–G), suggesting a specific requirement for LEM2 in this pathway. To clarify the epistatic relationship between CHMP7 and LEM2, we also depleted CHMP7 and measured LEM2 recruitment to chromatin disks during anaphase. Loss of CHMP7 had no effect on levels of LEM2 recruitment (Fig. 7 E and F and Table S4).
Fig. S7.
Control experiments for knockdown analysis. (A) Endogenous LEM2 detected at anaphase chromatin or disks. Spinning disk confocal microscopy of representative anaphase cells 72 h after treatment with siLEM2-1 or -2 confirmed that detection of signal at the chromatin surface is specific. Signal detected at the midzone with LEM2 antibody is nonspecific, persisting after knockdown of LEM2 with two independent oligos. (B) Widefield microscopy of representative interphase cells comparing detection of endogenous LEM2 at the nuclear rim 72 h after treatment with siControl or LEM2-specific oligos. (C) Specific depletion of LEM2 and CHMP7 with respective oligos used for RNAi is confirmed by immunoblot. # indicates a smaller protein product likely derived from CHMP7. (D) IST1/CHMP8 recruitment, assessed and graphed as described in Fig. 7B, performed in parallel with LEM2 analysis in Fig. 7F (siControl: 67 ± 10%, n = 104, 14, 40; siCHMP7-1: 0 ± 0%, n = 68, 26, 24; siCHMP7-2: 0 ± 0%, n = 24, 10, 48). (E) Widefield microscopy of representative interphase cells comparing detection of endogenous SUN1 at the nuclear rim 72 h after treatment with siControl or SUN1-directed oligos. (F) Specific depletion of SUN1 by RNAi confirmed by immunoblot. (G) IST1/CHMP8 recruitment, assessed and graphed as described in Fig. 7B, performed in parallel with CHMP2A analysis in Fig. 7D (siControl: 60 ± 8%, n = 80, 42, 48; siLEM2-1: 8.2 ± 4%, n = 80, 82, 30; siLEM2-2: 17 ± 7%, n = 120, 82, 78; siCHMP7-1: 5 ± 3%, n = 76, 50, 58; siCHMP7-2: 8 ± 4%, n = 58, 46, 48; siSUN1A: 53 ± 18%, n = 58, 18, 20; siSUN1B: 50 ± 7%, n = 78, 44, 36). All graphs plot mean ± SEM. *P < 0.05; **P < 0.01. N.S., not significant. Scale bars, 10 μm.
Fig. S8.
LEM2 plays a role in the recruitment of CHMP7 to anaphase chromatin disks. Additional montages of cells expressing GFP-CHMP7 and treated with siRNA, as indicated, progressing through anaphase are shown. Scale bars, 10 μm.
Fig. 7.
Recruitment of CHMP2A and IST1/CHMP8 during mammalian nuclear reformation depends on LEM2, whose targeting is independent of CHMP7. (A) Confocal images illustrating IST1/CHMP8 localization in anaphase B cells after siControl, siLEM2-1, or siLEM2-2 treatment. (B) Quantification of IST1/CHMP8 recruitment to chromatin disks during anaphase B. IST1/CHMP8 recruitment was scored as robust, weak, or no chromatin-associated foci. The robust category was graphed and statistical analysis was performed, comparing the siControl dataset to each depletion condition dataset (siControl: 63 ± 6%, n = 18, 58, 24; siLEM2-1: 0 ± 0%, n = 40, 44, 12; siLEM2-2: 6 ± 6%, n = 34, 22, 6; siCHMP7-1: 0 ± 0%, n = 42, 20, 12; siCHMP7-2: 0 ± 0%, n = 22, 20, 2). (C) Widefield images illustrating CHMP2A localization in anaphase B cells after siControl, siLEM2-1, or siLEM2-2 treatment. (D) CHMP2A recruitment to chromatin disks at anaphase B, assessed as in B (siControl: 70 ± 9%, n = 48, 48, 52; siLEM2-1: 9 ± 5%, n = 108, 86, 38; siLEM2-2: 11 ± 5%, n = 98, 52, 42; siCHMP7-1: 4 ± 1%, n = 78, 102, 47; siCHMP7-2: 21 ± 8%, n = 112, 58, 56). Analysis of parallel samples confirmed that LEM2 and CHMP7 depletion profoundly disrupted IST1/CHMP8 recruitment as before and is shown in Fig. S7G. (E) Widefield images of cells costained for IST1/CHMP8 and LEM2 illustrates the differential sensitivity of their localization at anaphase chromatin disks after siCHMP7-1 or siCHMP7-2 treatment. Signal detected at the midzone with LEM2 antibody is likely nonspecific (Fig. S7A). (F) Quantification of LEM2 recruitment to chromatin disks at anaphase B. Images were used for blind scoring the presence of chromatin-associated LEM2 (siControl: 95 ± 4%, n = 28, 35, 39; siCHMP7-1: 90 ± 6%, n = 18, 72, 45; siCHMP7-2: 99 ± 1%, n = 35, 56, 7). Analysis of parallel samples confirmed that CHMP7 depletion profoundly disrupted IST1/CHMP8 recruitment as before and is shown in Fig. S7D. *P < 0.05; **P < 0.01; ***P < 0.001. N.S., not significant. (Scale bars: 10 μm.) All graphs plot mean ± SEM.
Table S2.
Quantification of IST1 recruitment in mammalian cells
| IST1 | Fig. 7B | Fig. S7G | Fig. S7D | |||||||||
| Exp. 1 | Exp. 2 | Exp. 3 | Mean | Exp. 4 | Exp. 5 | Exp. 6 | Mean | Exp. 7 | Exp. 8 | Exp. 9 | Mean | |
| siControl | ||||||||||||
| Robust | 56 | 76 | 58 | 63 | 45 | 67 | 69 | 60 | 81 | 71 | 48 | 67 |
| Weak | 39 | 19 | 38 | 32 | 40 | 26 | 25 | 30 | 19 | 29 | 45 | 31 |
| Absent | 5 | 5 | 4 | 5 | 15 | 7 | 6 | 9 | 0 | 0 | 7 | 2 |
| siLEM2-1 | ||||||||||||
| Robust | 0 | 0 | 0 | 0 | 0 | 15 | 10 | 8 | ||||
| Weak | 53 | 52 | 17 | 41 | 60 | 39 | 47 | 49 | ||||
| Absent | 47 | 48 | 83 | 59 | 40 | 46 | 43 | 43 | ||||
| siLEM2-2 | ||||||||||||
| Robust | 0 | 18 | 0 | 6 | 5 | 16 | 29 | 17 | ||||
| Weak | 32 | 50 | 100 | 61 | 34 | 46 | 35 | 38 | ||||
| Absent | 68 | 32 | 0 | 33 | 61 | 38 | 36 | 45 | ||||
| siCHMP7-1 | ||||||||||||
| Robust | 0 | 0 | 0 | 0 | 0 | 8 | 7 | 5 | 0 | 0 | 0 | 0 |
| Weak | 2 | 0 | 0 | 1 | 8 | 22 | 29 | 20 | 3 | 4 | 0 | 2 |
| Absent | 98 | 100 | 100 | 99 | 92 | 70 | 64 | 75 | 97 | 96 | 100 | 98 |
| siCHMP7-2 | ||||||||||||
| Robust | 0 | 0 | 0 | 0 | 0 | 13 | 10 | 8 | 0 | 0 | 0 | 0 |
| Weak | 9 | 0 | 0 | 3 | 10 | 50 | 50 | 37 | 0 | 0 | 0 | 0 |
| Absent | 91 | 100 | 100 | 97 | 90 | 37 | 40 | 56 | 100 | 100 | 100 | 100 |
| siSun1A | ||||||||||||
| Robust | 36 | 89 | 35 | 53 | ||||||||
| Weak | 55 | 11 | 60 | 42 | ||||||||
| Absent | 9 | 0 | 5 | 5 | ||||||||
| siSun1B | ||||||||||||
| Robust | 49 | 61 | 39 | 50 | ||||||||
| Weak | 45 | 32 | 56 | 44 | ||||||||
| Absent | 6 | 7 | 6 | 6 | ||||||||
Numbers reported are the percentage of chromatin disks scored as robust, weak, or absent for IST1 for each experiment (Exp.).
Table S3.
Quantification of CHMP2A recruitment in mammalian cells
| CHMP2A | Fig. 7D | |||
| Exp. 4 | Exp. 5 | Exp. 6 | Mean | |
| siControl | ||||
| Robust | 73 | 83 | 54 | 70 |
| Weak | 23 | 17 | 38 | 26 |
| Absent | 4 | 0 | 8 | 4 |
| siLEM2-1 | ||||
| Robust | 12 | 16 | 0 | 9 |
| Weak | 70 | 63 | 61 | 65 |
| Absent | 18 | 21 | 39 | 26 |
| siLEM2-2 | ||||
| Robust | 19 | 4 | 10 | 11 |
| Weak | 64 | 50 | 52 | 55 |
| Absent | 16 | 46 | 38 | 33 |
| siCHMP7-1 | ||||
| Robust | 6 | 5 | 2 | 4 |
| Weak | 26 | 38 | 53 | 39 |
| Absent | 68 | 57 | 45 | 57 |
| siCHMP7-2 | ||||
| Robust | 5 | 28 | 30 | 21 |
| Weak | 38 | 60 | 57 | 52 |
| Absent | 56 | 12 | 13 | 27 |
Numbers reported are the percentage of chromatin disks scored as robust, weak, or absent for CHMP2A for each experiment (Exp.).
Table S4.
Quantification of LEM2 recruitment in mammalian cells
| LEM2 | Fig. 7F | |||
| Exp. 7 | Exp. 8 | Exp. 9 | Mean | |
| siControl | ||||
| Present | 100 | 97 | 87 | 95 |
| Absent | 0 | 3 | 13 | 5 |
| siCHMP7-1 | ||||
| Present | 100 | 81 | 87 | 89 |
| Absent | 0 | 19 | 13 | 11 |
| siCHMP7-2 | ||||
| Present | 100 | 96 | 100 | 99 |
| Absent | 0 | 4 | 0 | 1 |
Numbers reported are the percentage of chromatin disks scored as present or absent for LEM2 for each experiment (Exp).
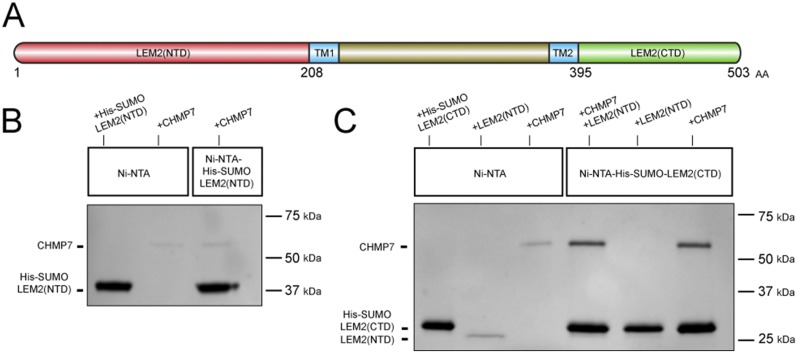
Finally, to determine whether LEM2 can bind directly to CHMP7, we purified full-length human CHMP7 as well as the soluble N-terminal domain (NTD) vs. C-terminal domain (CTD) of human LEM2 to homogeneity. After immobilization on Ni-nitrilotriacetic acid (Ni-NTA) beads via a fused His-SUMO tag, the C-terminal domain—but not the N-terminal domain—bound full-length CHMP7 (Fig. S9). Thus, our imaging, siRNA depletion, and biochemical data all support the idea that LEM2 binds CHMP7 directly and serves as an adaptor that recruits CHMP7 and other downstream ESCRT-III proteins, including IST1/CHMP8 and CHMP2A, to the reforming NE during anaphase.
Fig. S9.
The CTD of LEM2 binds full-length CHMP7 in vitro. (A) Protein domain structure of human LEM2. TM, transmembrane domain. (B and C) In vitro binding assay of full-length human CHMP7 with immobilized human His-SUMO-LEM2(NTD) (amino acids 1–208; B) and immobilized His-SUMO-LEM2(CTD) (amino acids 398–503; C). Samples are SDS-buffer eluates analyzed by SDS/PAGE and Coomassie staining. Note that CHMP7 showed weak unspecific binding to the resin (B, lane 2) that was not increased by immobilized His-SUMO-LEM2(NTD) (B, lane 3). In contrast, immobilized His-SUMO-LEM2(CTD), increased CHMP7 binding significantly (C, compare lanes 3 and 4). Finally, LEM2 (NTD) did not bind immobilized LEM2 (CTD) (C, lane 5), and the presence of LEM2 (NTD) did not enhance CHMP7 binding to immobilized His-SUMO-LEM2 (CTD) (C, compare lanes 4 and 6).
Discussion
Pioneering work on the ESCRT pathway in budding yeast led to our understanding of its roles in multivesicular body biogenesis (18), while work in human cells is leading to a broader view of ESCRT roles at a variety of different target membranes (20). Our results indicate that the role of the ESCRT pathway in closing holes in the NE is evolutionarily ancient (ref. 16 and this work). The different nuclear ESCRT functions (NPC surveillance, NE reformation, and SPB insertion/removal) can now be unified by the hypothesis that all of these processes generate fenestrations in the NE that are closed by the action of the ESCRT machinery (refs. 21, 23–26, and 36 and this study). Conserved activities include roles for nucleus-specific adaptors of the LEM domain family and the ESCRT-II/-III hybrid protein CHMP7/Cmp7p (refs. 21 and 29 and this study). This conserved pathway may likewise underlie the requirement for Src1, a LEM-domain protein in Aspergillus nidulans, in the formation of stable nuclei (37). This body of work suggests that LEM2 plays a specific, initiating role in coordinating membrane remodeling events, particularly during nuclear assembly, in addition to the other roles it plays as a NE resident during interphase (31, 35, 38–43).
Two LEM family members, Heh1p and Heh2p, are involved in ESCRT recruitment in S. cerevisiae, so multiple LEM family proteins may also be involved in recruiting ESCRT-III activities in mammalian cells. In this regard, we found that knockdown of LEM2 in HeLa cells led to a less severe IST1/CHMP8 recruitment phenotype than knockdown of CHMP7 (Tables S2 and S3). It will therefore be of great interest to determine whether additional LEM-domain family members, which are present at the nascent NE (44, 45), also serve as ESCRT recruitment factors in human cells. Like other ESCRT-III proteins, CHMP7 can also interact with membranes, and this activity contributes to its targeting (46), so further biochemical studies will be required to elucidate how the dynamic interplay between LEM2, CHMP7, and lipids regulates the recruitment and activity of the ESCRT pathway at the nascent NE. It will additionally be informative to determine the mechanistic basis of vps4Δ phenotypes in fission yeast and the underlying toxicity of unrestricted Lem2p-Cmp7p activity. We speculate that in the absence of Vps4p, Lem2p–Cmp7p complexes stabilize nuclear membrane fenestrations aberrantly and/or promote NE remodeling events that—when unregulated by Vps4p—result in karmellae, large gaps, and other malformations that ultimately compromise cell division (Fig. S5). Similarly, overexpression of Lem2p in fission yeast induces NE malformations (31), and impairment of Heh1/2p nuclear import in budding yeast induces karmellae formation (32). We predict that both of these phenotypes depend on CHMP7/Cmp7p proteins and other downstream ESCRT pathway activities. Studies of these and related activities in the future will benefit from the facile S. pombe genetic system for investigating how the ESCRT pathway senses and seals breaches of the envelope—a nuclear membrane integrity pathway that is conserved from yeast to human.
Materials and Methods
Yeast Strains and Growth Medium.
S. pombe, diploid strain SP286 (h+/h+, leu1-32/leu1-32, ura4-D18/ura4-D18, ade6-M210/ade6-M216) was used for all haploid constructions. All other strains used in this study are listed in Tables S1 and S5. Cells were routinely grown in YE5 rich medium [yeast extract 0.5% (wt/vol), glucose 3.0% (wt/vol) supplemented with 225 mg/L histidine, leucine, uracil, and lysine hydrochloride, and 250 mg/L adenine] or Edinburgh Minimal Media (EMM; Sunrise Sciences Products) with supplements described above (EMM5). Sporulation of diploids was induced by culturing cells in EMM5 without glutamate (EMMG; Sunrise Sciences Products) and supplemented as above but without uracil. Dominant drug selection was performed with YE5 supplemented with G418 disulfate (KSE Scientific) at a concentration of 200 mg/L, hygromycin B, Streptomyces sp. (Calbiochem) at a concentration of 100 mg/L and ClonNAT at a concentration of 100 mg/L. To prevent caramelization, YE5 was routinely prepared by leaving out glucose during autoclaving and adding it before inoculation.
Table S5.
Yeast strains constructed for this study
| Strain ID | Genotypes | Used for experiments in |
| UBSP424 | h+, cut12-YFP::kanMX6, leu1-32, ura4-D18, ade6-M210 | Figs. 1, 4, and 5 and Figs. S1 and S2 |
| UBSP425 | h+, cut12-YFP::kanMX6, leu1-32, ura4-D18, ade6-M210 | Fig. 1 and Fig. S1 |
| UBSP426 | h+, cut12-YFP::kanMX6, leu1-32, ura4-D18, ade6-M210 | Fig. 1 and Fig. S1 |
| UBSP461 | h+, vps4Δ::natMX6, cut12-YFP::kanMX6, leu1-32, ura4-D18, | Fig. 1 and Figs. S1 and S2 |
| ade6-M216 | ||
| UBSP462 | h+, vps4Δ::natMX6, cut12-YFP::kanMX6, leu1-32, ura4-D18, | Fig. 1 and Fig. S1 |
| ade6-M210 | ||
| UBSP463 | h+, vps4Δ::natMX6, cut12-YFP::kanMX6, leu1-32, ura4-D18, | Fig. 1 and Fig. S1 |
| ade6-M210 | ||
| UBSP464 | h+, vps4Δ::natMX6, cut12-YFP::kanMX6, leu1-32, ura4-D18, | Figs. 4 and 5 and Fig. S4 |
| ade6-M210 | ||
| UBSP468 | h+, vps4Δ::natMX6, cmp7Δ::hphMX4, cut12-YFP::kanMX6, | Figs. 1 and 5 and Fig. S1 |
| leu1-32, ura4-D18, ade6-216 | ||
| UBSP469 | h+, vps4Δ::natMX6, cmp7Δ::hphMX4, cut12-YFP::kanMX6, | Fig. 1 and Fig. S1 |
| leu1-32, ura4-D18, ade6-216 | ||
| UBSP470 | h+, vps4Δ::natMX6, cmp7Δ::hphMX4, cut12-YFP::kanMX6, | Fig. 1 and Fig. S1 |
| leu1-32, ura4-D18, ade6-216 | ||
| UBSP480 | h+, cmp7Δ::hphMX4, cut12-YFP::kanMX6, leu1-32, ura4-D18, ade6-M210 | Figs. 1 and 5 and Fig. S1 |
| UBSP481 | h+, cmp7Δ::hphMX4, cut12-YFP::kanMX6, leu1-32, ura4-D18, | Fig. 1 and Fig. S1 |
| ade6-M216 | ||
| UBSP482 | h+, cmp7Δ::hphMX4, cut12-YFP::kanMX6, leu1-32, ura4-D18, ade6-M210 | Fig. 1 and Fig. S1 |
| UBSP570 | h+, lem2Δ::hphMX4, cut12-YFP::kanMX6, leu1-32, ura4-D18, ade6-M210 | Figs. 1 and 5 and Fig. S1 |
| UBSP571 | h+, lem2Δ::hphMX4, cut12-YFP::kanMX6, leu1-32, ura4-D18, ade6-M216 | Fig. 1 and Fig. S1 |
| UBSP572 | h+, lem2Δ::hphMX4, cut12-YFP::kanMX6, leu1-32, ura4-D18, ade6-M210 | Fig. 1 and Fig. S1 |
| UBSP590 | h+, vps4Δ::natMX6, lem2Δ::hphMX4, cut12-YFP::kanMX6, | Figs. 1 and 5 and Fig. S1 |
| leu1-32, ura4-D18, ade6-M216 | ||
| UBSP591 | h+, vps4Δ::natMX6, lem2Δ::hphMX4, cut12-YFP::kanMX6, | Fig. 1 and Fig. S1 |
| leu1-32, ura4-D18, ade6-M216 | ||
| UBSP592 | h+, vps4Δ::natMX6, lem2Δ::hphMX4, cut12-YFP::kanMX6, | Fig. 1 and Fig. S1 |
| leu1-32, ura4-D18, ade6-M216 | ||
| UBSP652 | h+, ish1-mCherry::hphMX4, cut12-YFP::kanMX6, leu1-32, | Figs. 1 and 2 |
| ura4-D18, ade6-M210 | ||
| UBSP653 | h+, ish1-mCherry::hphMX4, cut12-YFP::kanMX6, leu1-32, | Figs. 1 and 2 |
| ura4-D18, ade6-M210 | ||
| UBSP654 | h+, ish1-mCherry::hphMX4, cut12-YFP::kanMX6, leu1-32, | Figs. 1 and 2 |
| ura4-D18, ade6-M210 | ||
| UBSP670 | h+, vps4Δ::natMX6, ish1-mCherry::hphMX4, | Figs. 1 and 2 |
| cut12-YFP::kanMX6, leu1-32, ura4-D18, ade6-M216 | ||
| UBSP694 | h+, vps4Δ::natMX6, ish1-mCherry::hphMX4, | Figs. 1 and 2 |
| cut12-YFP::kanMX6, leu1-32, ura4-D18, ade6-M216 | ||
| UBSP697 | h+, vps4Δ::natMX6, ish1-mCherry::hphMX4, | Figs. 1 and 2 |
| cut12-YFP::kanMX6, leu1-32, ura4-D18, ade6-M216 | ||
| UBSP829 | h+, vps4Δ::natMX6, cmp7Δ::hphMX4, ish1-mCherry::ura4(+), | Fig. 1 |
| cut12-YFP::kanMX6, leu1-32, ade6-M216 | ||
| UBSP830 | h+, vps4Δ::natMX6, cmp7Δ::hphMX4, ish1-mCherry::ura4(+), | Fig. 1 |
| cut12-YFP::kanMX6, leu1-32, ade6-M216 | ||
| UBSP809 | h+, cmp7Δ::hphMX4, ish1-mCherry::natMX6, | Fig. 1 |
| cut12-YFP::kanMX6, leu1-32, ura4-D18, ade6-M210 | ||
| UBSP810 | h+, cmp7Δ::hphMX4, ish1-mCherry::natMX6, | Fig. 1 |
| cut12-YFP::kanMX6, leu1-32, ura4-D18, ade6-M210 | ||
| UBSP811 | h+, cmp7Δ::hphMX4, ish1-mCherry::natMX6, | Fig. 1 |
| cut12-YFP::kanMX6, leu1-32, ura4-D18, ade6-M216 | ||
| UBSP801 | h+, lem2Δ::hphMX4, ish1-mCherry::natMX6, | Fig. 1 |
| cut12-YFP::kanMX6, leu1-32, ura4-D18, ade6-M210 | ||
| UBSP803 | h+, lem2Δ::hphMX4, ish1-mCherry::natMX6, | Fig. 1 |
| cut12-YFP::kanMX6, leu1-32, ura4-D18, ade6-M216 | ||
| UBSP804 | h+, lem2Δ::hphMX4, ish1-mCherry::natMX6, | Fig. 1 |
| cut12-YFP::kanMX6, leu1-32, ura4-D18, ade6-M216 | ||
| UBSP807 | h+, vps4Δ::natMX6, lem2Δ::hphMX4, ish1-mCherry::kanMX6, | Fig. 1 |
| leu1-32, ura4-D18, ade6-M216 | ||
| UBSP808 | h+, vps4Δ::natMX6, lem2Δ::hphMX4, ish1-mCherry::kanMX6, | Fig. 1 |
| leu1-32, ura4-D18, ade6-M216 | ||
| UBSP831 | h+, int::Pnmt1-SV40NLS-GFP-LacZ::leu1(+), ura4-D18, | Fig. 3 |
| ade6-M216 | ||
| UBSP832 | h+, int::Pnmt1-SV40NLS-GFP-LacZ::leu1(+), ura4-D18, ade6-M216 | Fig. 3 |
| UBSP833 | h+, int::Pnmt1-SV40NLS-GFP-LacZ::leu1(+), ura4-D18, ade6-M216 | Fig. 3 |
| UBSP896 | h+, vps4Δ::natMX6, ish1-mCherry::phpMX4, | Fig. 3 and Fig. S3 |
| int::Pnmt1-SV40NLS-GFP-LacZ::leu1(+), ura4-D18, | ||
| ade6-M210 | ||
| UBSP900 | h+, vps4Δ::natMX6, ish1-mCherry::phpMX4, | Fig. 3 |
| int::Pnmt1-SV40NLS-GFP-LacZ::leu1(+), ura4-D18, ade6-M210 | ||
| UBSP903 | h+, vps4Δ::natMX6, ish1-mCherry::phpMX4, | Fig. 3 and Fig. S3 |
| int::Pnmt1-SV40NLS-GFP-LacZ::leu1(+), ura4-D18, ade6-M210 | ||
| UBSP843 | h+, vps4Δ::natMX6, cmp7Δ::hphMX4, | Fig. 3 |
| int::Pnmt1-SV40NLS-GFP-LacZ::leu1(+), ura4-D18, ade6-M216 | ||
| UBSP844 | h+, vps4Δ::natMX6, cmp7Δ::hphMX6, | Fig. 3 |
| int::Pnmt1-SV40NLS-GFP-LacZ::leu1(+), ura4-D18, ade6-M216 | ||
| UBSP845 | h+, vps4Δ::natMX6, cmp7Δ::hphMX4, | Fig. 3 |
| int::Pnmt1-SV40NLS-GFP-LacZ::leu1(+), ura4-D18, ade6-M216 | ||
| UBSP839 | h+, cmp7Δ::hphMX4, int::Pnmt1-SV40NLS-GFP-LacZ::leu1(+), | Fig. 3 |
| ura4-D18, ade6-M216 | ||
| UBSP840 | h+, cmp7Δ::hphMX4, int::Pnmt1-SV40NLS-GFP-LacZ::leu1(+), | Fig. 3 |
| ura4-D18, ade6-M216 | ||
| UBSP841 | h+, cmp7Δ::hphMX4, int::Pnmt1-SV40NLS-GFP-LacZ::leu1(+), | Fig. 3 |
| ura4-D18, ade6-M216 | ||
| UBSP835 | h+, lem2Δ::hphMX4, int::Pnmt1-SV40NLS-GFP-LacZ::leu1(+), | Fig. 3 |
| ura4-D18, ade6-M210 | ||
| UBSP836 | h+, lem2Δ::hphMX4, int::Pnmt1-SV40NLS-GFP-LacZ::leu1(+), | Fig. 3 |
| ura4-D18, ade6-M210 | ||
| UBSP837 | h+, lem2Δ::hphMX4, int::Pnmt1-SV40NLS-GFP-LacZ::leu1(+), | Fig. 3 |
| ura4-D18, ade6-M216 | ||
| UBSP842 | h+, vps4Δ::natMX6, lem2Δ::hphMX4, | Fig. 3 |
| int::Pnmt1-SV40NLS-GFP-LacZ::leu1(+), ura4-D18, ade6-M216 | ||
| UBSP847 | h+, vps4Δ::natMX6, lem2Δ::hphMX4, | Fig. 3 |
| int::Pnmt1-SV40NLS-GFP-LacZ::leu1(+), ura4-D18, ade6-M216 | ||
| UBSP848 | h+, vps4Δ::natMX6, lem2Δ::hphMX4, | Fig. 3 |
| int::Pnmt1-SV40NLS-GFP-LacZ::leu1(+), ura4-D18, ade6-M216 |
Yeast Transformation.
Log-phase yeast cells were incubated in 0.1 M lithium acetate (pH 4.9) for 2 h, at a concentration of 5 × 108 cells per mL. A total of 100 μL of this cell suspension was then mixed with 0.5–1 μg of DNA and 290 μL of 50% (wt/vol) PEG 8000 and incubated overnight. Cells were recovered in 0.5× YE5 medium overnight before plating. All steps were conducted at 32 °C.
Sporulation, Random Spore Analysis, and Tetrad Dissection.
Sporulation of S. pombe diploids for tetrad dissection or random spore analysis was induced by first transforming pON177 (h− mating plasmid with ura4 selectable marker, a gift from Megan King, Yale School of Medicine, New Haven) into the parental strain. Transformants were selected on EMM5 without uracil (EMM5-uracil) for 3 d at 32 °C followed by induction in 0.5 mL of EMMG without uracil (EMMG-uracil) for 36–48 h at 25 °C. Sporulation was confirmed by microscopy. Ascus walls of tetrads were digested with 2% (vol/vol) β-glucuronidase (Sigma-Aldrich) overnight at 25 °C. This overnight treatment also eliminated nonsporulated diploids. Enrichment of spores was verified by microscopy. Fivefold to 10-fold serial dilutions were made, and spores were plated on YE5 supplemented with specific antibiotics.
For tetrad dissection, 10-fold serial dilutions of β-glucuronidase–treated tetrads were placed onto YE5 plates as a narrow strip, and digestion was monitored at room temperature. Tetrads were picked and microscopically dissected along predesignated lines of the same YE5 plate. Spores were allowed to germinate and grow at 32 °C.
Knockout Cassettes and Plasmids.
vps4 deletion cassette.
The vps4Δ::natMX6 template (16) was amplified to create an amplicon covering 756 bp upstream of the ATG and 499 bp downstream of the stop codon. This 2,439-bp amplicon was transformed into SP286 and plated on YE5 + ClonNAT to select for heterozygous vps4Δ::natMX6/+ diploids.
cmp7 deletion cassette.
A fragment including 420 bp upstream of the ATG and 377 bp downstream of the stop codon was amplified and cloned into BamHI/BglII and EcoRV/SpeI of pAG32 (a pFA6-derived plasmid containing hphMX4, which confers resistance to hygromycin B, was a gift from David Stillman, Department of Pathology, University of Utah). The cmp7Δ::hphMX4 fragment was amplified and transformed into heterozygous vps4Δ::natMX6/+ diploid intermediate strain and selected on YE5 supplemented with both ClonNAT and hygromycin B.
lem2 deletion cassette.
A 3.1-kb lem2 genomic fragment spanning 600 bp upstream of the ATG to 500 bp downstream of the stop codon was subcloned into pGEM vector. The ORF region was replaced with a hphMX4 hygromycin resistance cassette to make the final knockout construct (pMGF130). The lem2Δ::hphMX4 cassette was amplified and transformed into heterozygous vps4Δ::natMX6/+ diploids and selected on YE5 supplemented with both ClonNAT and hygromycin B.
cut12-YFP cassette.
A 3.5-kb synthetic DNA fragment was created that spanned 550-bp C-terminal of cut12 fused with YFP followed by kanMX6 and a 500-bp fragment downstream of the cut12 stop codon. The fragment was transformed into heterozygous diploid strains, and the resulting diploids were selected on YE5 supplemented with G418 disulfate.
ish1-mCherry cassettes.
A 1-kb ish1 genomic DNA fragment corresponding to 500 bp upstream and 500 bp downstream of the ish1 stop codon was subcloned into the pGEM vector. A flexible linker (GGTGGSGGT) and mCherry fusion cassettes were assembled, followed by the yADH1 terminator and MX4/6 drug resistance or auxotrophic markers. These cassettes were integrated at the native ish1 locus to make the final fusion constructs: ish1-mCherry::natMX6 (pMGF170), ish1-mCherry::kanMX6 (pMGF169), ish1-mCherry::hphMX4 (pMGF157), and ish1-mCherry::ura4(+) (pMGF172).
pDual-SV40NLS-GFP-LacZ construct.
A SV40NLS-GFP-LacZ fragment was amplified from pREP3X (provided by Shelley Sazer, Baylor College of Medicine, Houston) and subcloned downstream of the Pnmt1 promoter in the pDual vector. The final construct (pMGF173) was integrated at the leu1 locus.
Pcmv(Δ5)-GFP-CHMP7 and pCMV(Δ5)-LEMD2-mCherry.
CHMP7 and LEMD2 cDNA were amplified and subcloned downstream of Pcmv(Δ5) (47). GFP+linker (see as above) was inserted before CHMP7 (pMGF182), and linker+mCherry was inserted after LEMD2 (pMGF196) to make the fusions.
Isolation of vps4Δ and Suppressors.
Isolation and handling of vps4Δ haploids.
Individual petite colonies from random spore analysis plates of YE5+ClonNAT were selected, resuspended in 200 μL of YE5 medium, plated onto two YE5+ClonNAT plates, and incubated at 32 °C for 3 d. Isolates that grew across one plate without apparent suppression were frozen with glycerol without further culturing by scraping and resuspension in YE5 with 15% (wt/vol) glycerol. Cells from the other plate were scraped, and genomic DNA was immediately extracted for Illumina sequencing (see below).
Isolation of vps4Δ suppressors.
vps4Δ isolates were restreaked from glycerol stocks and cultured on YE5 at 32 °C. Large colonies (apparent suppressors) were picked, resuspended in 200 μL of YE5 medium, and plated onto two YE5 plates. After 2 d, cells were harvested as described above for glycerol stocks and genomic DNA extraction.
Yeast Genomic DNA Extraction, Illumina Sequencing, and Analysis.
Genomic DNA extraction.
Frozen pellets of WT, vps4Δ, or suppressor cells (200 μL) were thawed on ice. A total of 250 μL of resuspension buffer (20 mM Tris⋅HCl, pH 8.0, 100 mM EDTA, and 0.5 M β-mercaptoethanol) and 50 μL of lyticase (50 units) were then added to remove the cell wall. Genomic DNA was extracted by using phenol/chloroform/isoamyl alcohol, precipitated with ethanol, and treated with RNase, followed by DNeasy Blood Tissue purification according to the manufacturer’s protocol (Qiagen catalog no. 69504).
Illumina sequencing.
Libraries were constructed by using the Illumina TruSeq DNA Sample Preparation Kit (catalog nos. FC-121-2001 and FC-121-2002). Briefly, genomic DNA was sheared in a volume of 52.5 μL by using a Covaris S2 Focused-ultra-sonicator with the following settings: intensity, 5.0; duty cycle, 10%; cycles per burst, 200; treatment time, 120 s. Sheared DNA was converted to blunt-ended fragments and size-selected to an average length of 275 bp by using AMPure XP (Beckman Coulter catalog no. A63882). After adenylation of the DNA, adapters containing a T-base overhang were ligated to the A-tailed fragments. Adapter-ligated fragments were enriched by PCR (eight cycles) and purified with AMPure XP. The amplified libraries were qualified on an Agilent Technologies 2200 TapeStation by using a D1K ScreenTape assay (catalog no. 5067-5363), and quantitative PCR was performed by using the Kapa Biosystems Kapa Library Quant Kit (catalog no. KK4824) to define the molarity of adapter-modified molecules. Molarities of all libraries were subsequently adjusted to 10 nM, and equal volumes were pooled in preparation for Illumina sequencing.
Sequencing data analysis.
Raw reads were aligned to the S. pombe genome, obtained from Ensembl Fungi, by using NovoCraft Novoalign, allowing for no repeats (-r None) and base calibration (-k). Alignments were converted to Bam-formatted files by using samtools (samtools.sourceforge.net). Sequence pileups were generated with samtools pileup, and variants were called by using the bcftools utility (options -c -g -v). Variants were filtered by using the included varFilter Perl script included with samtools and written out as a vcf file. To distinguish unique variants in each strain from common variants, sample vcf files were intersected with one another by using the Perl script intersect_SNPs (https://github.com/tjparnell). Variants were annotated with the Perl script locate_SNPs (https://github.com/tjparnell) by using a GFF3 gene annotation file obtained from Ensembl. From the resulting table, variants were further filtered by the fraction of reads supporting the alternate allele, the presence of codon changes, and visual inspection in a genome browser. The summary statistics are reported in Table S1.
Fluorescence Microscopy.
All yeast strains were cultured in either YE5 or EMM5, if the desired protein was induced by the nmt1 promoter. Cells were imaged after reaching log phase. Hoechst staining was conducted at a concentration of 1 μg/mL in water for 15 min. Images were collected on a Zeiss Axio Observer Z1 microscope by using a 100× oil-immersion objective.
HeLa cells were fixed in −20 °C methanol for 10 min. The primary antibodies used for immunodetection were rabbit α-IST1/CHMP8 (48), α-LEM2 (HPA017340; Sigma), rat α-tubulin (YL1/2; Accurate Chemical & Scientific), rabbit α-CHMP2A (UT 634; Covance), and mouse α-IST1/CHMP8 (UT 697; Covance). Full-length CHMP2A and IST1/CHMP8 protein (49) were used as antigens to produce custom antibodies by Covance Immunology Services. The anti-IST1/CHMP8 antibody (UT 697) was affinity-purified (50) before use. After incubation with fluorescently labeled secondary antibodies (Thermo Fisher), coverslips were mounted by using DAPI ProLong Gold (Thermo Fisher) and imaged. For the purpose of illustration, images of anaphase B cells were acquired by spinning-disk confocal microscope and adjusted so that cytoplasmic IST1/CHMP8 intensity was comparable between samples. Images acquired by widefield microscopy at 100× were used to score the IST1/CHMP8 and CHMP2A phenotypes after minimal adjustment that was applied uniformly. For each graph, IST1/CHMP8 and CHMP2A localization to anaphase B chromatin masses were assessed in three independent experiments. Each chromatin disk (two per cell) was scored as having robust, weak, or no recruitment for IST1/CHMP8 or CHMP2A. Robust recruitment was characterized by distinctive foci organized at chromatin masses, whereas weak recruitment was characterized by less intense, often fewer, and less organized foci at the chromatin surface. Images of anaphase B cells from all treatments and experiments were randomized and quantified blindly by three scorers. The majority score was used in cases where the three scores differed. In an analogous set of experiments, anaphase B cells from all treatments were scored blindly for the absence or presence of chromatin-associated LEM2. As a positive control, anaphase B cells were scored, in parallel, as having robust, weak, or no IST1/CHMP8 recruitment.
Time-Lapse Light Microscopy Analysis.
WT and vps4Δ cells were grown in YE5 at 32 °C for 8 h and loaded into the CellASIC ONIX Microfluidic system (catalog no. EVE262, EMD Millipore), which immobilizes the cells in a single focal plane, maintains a constant temperature (32 °C), and pumps fresh medium over the cells. Images from multiple positions per chamber were captured every 10 min for 16 h. A lens heater was used to maintain constant temperature inside the chamber. Images were collected with an Andor Clara CCD camera attached to a Nikon Ti microscope using a 60× oil Nikon Apo Lambda S NA 1.4 lens. The samples were illuminated with a Lumencor Sola LED at 20% intensity, which was further reduced by the insertion of an ND8 filter. Exposure times ranged between 1 and 3 s for both YFP and mCherry channels. Five Z plane images separated by 1 μm were collected. Maximum intensity projection images were created to follow the Cut12p-YFP signals within a given cell. For Ish1p-mCherry signals, only the Z plane that bisected the nucleus was chosen for further image analysis.
For time-lapse colocalization experiments in HeLa cells, cells were plated on fibronectin-coated Mat-Tek dishes and incubated overnight. Cells were then transiently cotransfected with pCMV(Δ5)-GFP-CHMP7 and pCMV(Δ5)-LEM2-mCherry using Lipofectamine 3000 (Thermo Fisher) to coexpress GFP-CHMP7 and LEM2-mCherry under attenuated CMV promoters (47). For siRNA depletion and GFP-CHMP7 expression experiments, HeLa cells, either parental or stably expressing H2B-mCherry, were plated on fibronectin-coated Mat-Tek dishes in the presence of siRNA (siControl, siLEM2-1, or siLEM2-2, as described below). After 24 h, pCMV(Δ5)-GFP-CHMP7 was delivered by transient transfection with Lipofectamine 3000. In all live imaging experiments, cells were transiently transfected for 24 h before being arrested at G1/S and released, as described below. Twelve hours after release, cells were live-imaged by spinning disk confocal microscopy.
Quantification of Nuclear Enrichment of NLS-GFP-LacZ.
Fluorescence microscopy of fission yeast was performed as described above by using 0.2-s exposures for five Z-sections separated by 0.29-μm steps. Integrated pixel intensities for GFP were measured within a 100- × 100-pixel square box at both the center of nucleus and cytoplasm near the pole of the cell. The average background pixel intensity was also measured from cell-free regions of the image, and this value was subtracted from both the nuclear sum and the cytoplasmic sum. The fold nuclear GFP enrichment was calculated as the ratio of nuclear/cytoplasmic integrated intensity. A minimum of 150 cells were quantified for each genotype.
Quantification of CHMP7 Recruitment During Anaphase.
Live-cell fluorescence microscopy was performed as described above. Images of cells from 1 min before cleavage furrow ingression were selected for scoring. The “Find Maxima” function with variable noise tolerance values, as implemented in ImageJ, was used to identify CHMP7 foci around the contour of chromatin disks in each cell. The absolute number of those foci were recorded and scored blindly between siControl and siLEM2.
Electron Microscopy.
Yeast strains were grown to log phase and harvested by using gentle vacuum filtration onto filter paper. The cell pellet was scraped from the filter, mixed with cryoprotectant [20% (wt/vol) BSA in PBS], transferred to the well of a 100-µm specimen carrier (type A), and covered with the flat side of a type B specimen carrier (51). The loaded specimens were immediately frozen with a high-pressure freezer (EM-HPM100; Leica Microsystems). Frozen cell were transferred for freeze substitution (FS) to a precooled Leica AFS unit (Leica Microsystems) and processed in the following substitution solution: 97% anhydrous acetone (EMS, RT10016) and 3% of water, 2% osmium tetroxide, and 0.1% uranyl acetate. Substitution started at –90 °C for 72 h, followed by a gradual increase in temperature (5 °C/h) to −25 °C over 13 h, held at −25 °C for 12 h, then warmed (10 °C/h) to 0 °C over 2.5 h). The samples were removed from the AFS unit, washed six times with pure acetone, and gradually infiltrated and embedded in Epon12-araldite resin as follows: 50% (vol/vol) epon12-araldite/acetone overnight, 75% (vol/vol) epon12-araldite/acetone for 24 h, 100% epon12-araldite for 8 h, and polymerized at 60 °C for 48 h. Ultrathin sections (70 nm) were cut by using a diamond knife (Diatome), using a UC6 microtome (Leica Microsystems), transferred to Formvar- and carbon-coated mesh copper grids (Electron Microscopy Sciences; FCF-200-Cu) and poststained with 3% (wt/vol) uranyl acetate and Reynold’s lead citrate. The sections were viewed with a JEM-1400 Plus transmission electron microscope (JEOL, Ltd.) at 120 kV and images collected on a Gatan Ultrascan CCD (Gatan, Inc.).
Electron Tomography.
Blocks of embedded WT and vps4Δ S. pombe cells were trimmed to ∼100- × 200-μm faces. Serial semithick (400 nm) sections were cut with a UC6 Ultramicrotome (Leica Microsystems) by using a diamond knife (Diatome Ltd.). Ribbons of 10–20 sections were placed on Formvar-coated, copper-rhodium, 1-mm slot grids (Electron Microscopy Sciences) and stained with 3% (wt/vol) uranyl acetate and Reynold’s lead citrate. Colloidal gold particles (10 nm) were placed on both surfaces of the sections to serve as fiducial markers for subsequent tomographic image alignment, and the grids were carbon-coated to enhance stability in the electron beam.
Grids were placed in a dual-axis tomography holder (model 2040; EA Fischione Instruments, Inc.) and imaged with a Tecnai TF30-ST transmission electron microscope (FEI Company) equipped with a field emission gun and operating at 300 kV. Well-preserved cells displaying structures of interest were identified and tracked over 6–10 serial sections. The volume of the cell present in each section was then imaged as a dual-axis tilt series; for each set, the grid was tilted ± 64°, and images were recorded at 1° intervals. The grid was then rotated 90°, and a similar series were recorded about the orthogonal axis. Tilt-series datasets were acquired automatically by using the SerialEM software package (52), and images were recorded with a XP1000 CCD camera (Gatan, Inc.). Tomographic data were calculated and serial tomograms were joined together by using the IMOD software package (53–55). Tomograms were analyzed, segmented, and modeled by using IMOD. The NE was traced with closed contours in each tomogram. Modeled contours were smoothed and 3D surfaces were generated with tools in the IMOD software package. The “Z inc” value was set to three for all NE objects to further smooth their surface. The number of serial semithick (400-nm) sections used for model segmentation and display were three for WT (Movie S1), two for vps4Δ (Movie S2), four for vps4Δ (Movie S3), and three for the second vps4Δ (Movie S4).
siRNA-Mediated Depletion and Cell-Cycle Synchronization.
HeLa cells were plated on fibronectin-coated coverslips in the presence of 10 nM siRNA oligo, delivered by Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher). Specific sequences used were: siControl [siScr-1 (56)], siLEM2-1 [antisense sequence targeting nucleotides 78–98: UUGCGGUAGACAUCCCGGGdTdT (43)], siLEM2-2 [antisense sequence targeting nucleotides 1,297–1,317: UACAUAUGGAUAGCGCUCCdTdT (43)], siCHMP7-1 [CHMP7 650; sense sequence: GGGAGAAGAUUGUGAAGUUdTdT (22)], and siCHMP7-2 [CHMP7 613; sense sequence: CAGAAGGAGAAGAGGGUCAdTdT (22)], siSUN1A [SUN1; sense sequence targeting nucleotides 2,321–2,345 of SUN1, transcript variant 1 (NM_001130965.2): CCAUCCUGAGUAUACCUGUCUGUAUdTdT (57)], and siSUN1B [SUN1; sense sequence targeting nucleotides 865–883 of NM_001130965.2: UUACCAGGUGCCUUCGAAAdTdT (58)]. Culture medium containing 2 mM thymidine was then added for 24 h to arrest cells at G1/S. Cells were then rinsed thoroughly with PBS, followed by the addition of culture media. Twelve hours after release, cells were imaged live or fixed for microscopy. To verify efficacy and specificity of siRNA treatments, HeLa cells were plated in six-well dishes and subjected to similar conditions as above. Cell lysates were then harvested and analyzed with immunoblots. After incubation with primary antibodies [α-LEM2 (HPA017340; Sigma-Aldrich), α-CHMP7 (HPA036119; Sigma-Aldrich), α-tubulin (YL1/2), and α-SUN1 (EPR6554; Abcam)], reactivity was detected by using HRP-coupled secondary antibodies (Thermo Fisher) and chemiluminescence.
Protein Expression and Purification.
We individually expressed full-length human CHMP7 (Uniprot ID Q8WUX9) and the terminal domains of LEM2, encoded by LEMD2 (NTD 1-208, CTD 395–503; Uniprot ID Q8NC56) with a N-terminal His6-Sumo affinity tag by using a pCA528 vector (WISP08-174; DNASU Plasmid Repository) in BL21-(DE3)-RIPL Escherichia coli. The cells were grown in autoinduction medium ZYP-50529 (1.5 L cultures each). Cells were grown at 37 °C to OD 0.8 with vigorous shaking in baffled flasks, moved to 19 °C and grown for an additional 19 h. Cells were then harvested by centrifugation, and bacterial pellets were snap-frozen in liquid nitrogen. Subsequent purification steps were performed at 4 °C. Cells were thawed and lysed for 30 min with lysozyme in 2.5 times the pellet volume of lysis buffer followed by sonication. The supernatant was clarified by centrifugation (40,000 × g, 45 min).
CHMP7 purification.
A total of 20–30 g of bacterial pellet were lysed (50 mM Tris, pH 8.0, 1 M NaCl, 20 mM imidazole, 10% (wt/vol) glycerol, 5 mM beta-mercaptoethanol (BME), 0.1% Triton X-100, 2 µg/mL DNase1, and protease inhibitors (84 µM leupeptin, 0.3 µM aprotinin, 1 µM pepstatin, 100 µM phenylmethylsulfonyl fluoride (PMSF)), and incubated with Ni-NTA agarose beads (Qiagen) for 2 h. The bound protein was washed with 20 column volumes (CVs) of lysis buffer without lysozyme and protease inhibitors, 20 CVs of wash buffer [50 mM Tris (pH 8), 1 M NaCl, 20 mM Imidazole, 5% (wt/vol) glycerol, 5 mM BME, and 0.009% Triton X-100] and eluted in four steps (1 CV per step) with wash buffer supplemented with increasing imidazole concentrations (50, 150, 250, and 400 mM). The eluted protein was dialyzed for 2 h against gel filtration buffer [50 mM Tris, (pH 8.0), 150 mM KCl, 1 mM DTT, 5% (wt/vol) glycerol]. Ulp1 protease (0.75 mg per 30 mL) was added to the dialysis bag to remove the affinity tag, and the dialysis reaction was performed overnight. The cleaved protein was incubated with 2 mL of Ni-NTA agarose beads (Qiagen) to remove His6-Sumo and Ni-NTA agarose binding contaminants. The eluate was concentrated to 5 mL with a Viviaspin 20 [30,000 nominal molecular weight cutoff (MWCO), polyethersulfone (PES) membrane]. Monomeric Chmp7 was isolated by gel filtration chromatography. Monomeric Chmp7 was concentrated to 24–42 µM by using a Viviaspin 20 concentrator (30,000 nominal MWCO, PES membrane), aliquoted, and snap-frozen in liquid N2. For binding experiments, the protein was thawed on ice and cleared from aggregates by centrifugation for 20 min at 98,600 × g, using a TLA-55 rotor (Beckman Coulter). Yields were 300–500 µg of protein per 1 L of bacterial culture.
LEM2(NTD) purification.
A total of 18 g of bacterial pellet was lysed (25 mM Tris, pH 7.0, 500 mM KCl, 10 mM imidazole, 5 mM BME, 2 µg/mL DNase1, and protease inhibitors) and incubated with Ni-NTA agarose beads (Qiagen) for 2 h. The bound protein was washed with 40 CVs of lysis buffer without lysozyme and protease inhibitors and eluted in four steps (1 CV per step) with lysis buffer supplemented with increasing imidazole concentrations (50, 150, 250, and 400 mM). The eluted protein was dialyzed against SP column loading buffer (25 mM Tris, pH 7.0, 150 mM KCl, and 1 mM DTT), applied to a 5-mL HiTrap SP HP column (GE Healthcare), washed with loading buffer, and eluted with a gradient from 150 to 500 mM KCl. The eluate was concentrated to 5 mL with a Viviaspin 20 (10.000 KDa MWCO, PES), and monomeric His-Sumo-LEM2 (NTD) was isolated by gel filtration chromatography (25 mM Tris, pH 7.0, 150 mM KCl, and 1 mM DTT). Monomeric His-Sumo-LEM2 (NTD) was concentrated to 20 µM, and aliquots were snap-frozen in liquid N2. Yields were ∼2.5 mg of protein per 1 L of bacterial culture. For binding experiments, His-Sumo-LEM2 (NTD) was thawed on ice and spun for 10 min at 4 °C at 16,627 × g. The protein was used directly for binding or processed into untagged LEM2 (NTD). To obtain untagged protein, 250 µg of His-Sumo- LEM2 (NTD) were incubated with 30 µg of Ulp1 for 1 h at 4 °C and subsequently incubated with 100 µL of Ni-NTA agarose beads (Qiagen) to remove cleaved His6-Sumo. The beads were removed by centrifugation, and the supernatant was used for binding experiments.
LEM2(CTD) purification.
Cells were suspended in buffer with 50 mM Tris (pH 8.0), 150 mM KCl, and 1 mM DTT plus protease Inhibitor mixture and were lysed by freeze–thaw cycles, and the supernatant was harvested after 16,350 × g for 45 min. The lysate was incubated with Ni-beads (Qiagen) for 45 min, washed with 20 mM imidazole, and eluted with 750 mM imidazole. The eluted protein was dialyzed against gel-filtration buffer (50 mM Tris, pH 8.0, 150 mM KCl, and 1 mM DTT) and applied to 120 mL of Hiload Superdex 75PG (GE Healthcare). Fractions containing pure LEM2-CTD were collected and snap-frozen in liquid nitrogen. Yields were ∼2.5 mg of protein per 1 L of bacterial culture.
Protein Binding Experiments.
Binding experiments were performed at room temperature by using 40 µL of Ni-NTA agarose beads (Qiagen) in 0.8-mL centrifuge columns (Pierce). Proteins were mixed and incubated for 1 h. For control reactions, proteins were incubated with corresponding buffers to mimic binding conditions. The protein reactions were added to the beads, which were equilibrated with binding buffer (25 mM, Tris pH 7.0, 150 mM KCl, and 20 mM imidazole) and incubated for an additional 45 min. The resin was washed with 20 CVs of binding buffer. Excess His-Sumo-tagged-LEM2 (CTD or NTD) was eluted with 3 CVs of binding buffer supplemented with 150 mM imidazole. Note that the majority of LEM2 and bound CHMP7 did not elute at this step. Protein remaining on beads was eluted with two CVs of 2× SDS sample buffer and the eluate was analyzed with SDS/PAGE.
Note.
While our study was under review, corroborative evidence for these roles for CHMP7 in human cells (46) and LEM domain family proteins in budding yeast were also published by others (59).
Supplementary Material
Acknowledgments
We thank Drs. Brian Dalley and Tim Parnell for advice and expertise in whole-genome sequencing and analysis; Dr. Janet Iwasa for graphic illustration; Dr. Mark Smith for assistance with confocal microscopy; Sarah M. Pick for technical assistance with protein purification; Dr. Doug Mackay for advice; Dr. Shelley Sazer for a NLS-GFP-LacZ nuclear integrity reporter; Dr. Yasushi Hiraoka for an Ish1-GFP strain; and Drs. John McCullough, Jeremy Carlton, and Patrick Lusk for stimulating conversations about unpublished results. Light and 2D transmission electron microscopy were performed in the Health Sciences Cores at the University of Utah. Microscopy equipment was obtained by using NCRR Shared Equipment Grant 1S10-RR024761-01. Our research was also supported by the Searle Scholars Program (A.F.); NIH Grants 2P50-GM082545-06 (to W.I.S., A.F., M.S.L., and P.J.B.), 1DP2-GM110772-01 (to A.F.), and 1R01-GM112080 (to W.I.S.); and Huntsman Cancer Foundation and the Huntsman Cancer Institute Cancer Center Support Grant NIH P30CA042014 (to K.S.U., W.I.S., A.F., and the Genomics and Bioinformatics Shared Resource).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613916114/-/DCSupplemental.
References
- 1.Devos DP, Gräf R, Field MC. Evolution of the nucleus. Curr Opin Cell Biol. 2014;28:8–15. doi: 10.1016/j.ceb.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Madhani HD. The frustrated gene: Origins of eukaryotic gene expression. Cell. 2013;155(4):744–749. doi: 10.1016/j.cell.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heath IB. Variant mitoses in lower eukaryotes: Indicators of the evolution of mitosis. Int Rev Cytol. 1980;64:1–80. doi: 10.1016/s0074-7696(08)60235-1. [DOI] [PubMed] [Google Scholar]
- 4.Byers B, Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975;124(1):511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kubai DF. The evolution of the mitotic spindle. Int Rev Cytol. 1975;43:167–227. doi: 10.1016/s0074-7696(08)60069-8. [DOI] [PubMed] [Google Scholar]
- 6.Tamm T, et al. Brr6 drives the Schizosaccharomyces pombe spindle pole body nuclear envelope insertion/extrusion cycle. J Cell Biol. 2011;195(3):467–484. doi: 10.1083/jcb.201106076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCully EK, Robinow CF. Mitosis in the fission yeast Schizosaccharomyces pombe: A comparative study with light and electron microscopy. J Cell Sci. 1971;9(2):475–507. doi: 10.1242/jcs.9.2.475. [DOI] [PubMed] [Google Scholar]
- 8.Ding R, West RR, Morphew DM, Oakley BR, McIntosh JR. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol Biol Cell. 1997;8(8):1461–1479. doi: 10.1091/mbc.8.8.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams IR, Kilmartin JV. Spindle pole body duplication: A model for centrosome duplication? Trends Cell Biol. 2000;10(8):329–335. doi: 10.1016/s0962-8924(00)01798-0. [DOI] [PubMed] [Google Scholar]
- 10.Tallada VA, Tanaka K, Yanagida M, Hagan IM. The S. pombe mitotic regulator Cut12 promotes spindle pole body activation and integration into the nuclear envelope. J Cell Biol. 2009;185(5):875–888. doi: 10.1083/jcb.200812108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Witkin KL, Friederichs JM, Cohen-Fix O, Jaspersen SL. Changes in the nuclear envelope environment affect spindle pole body duplication in Saccharomyces cerevisiae. Genetics. 2010;186(3):867–883. doi: 10.1534/genetics.110.119149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muñoz-Centeno MC, et al. Saccharomyces cerevisiae MPS2 encodes a membrane protein localized at the spindle pole body and the nuclear envelope. Mol Biol Cell. 1999;10(7):2393–2406. doi: 10.1091/mbc.10.7.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaspersen SL, Winey M. The budding yeast spindle pole body: Structure, duplication, and function. Annu Rev Cell Dev Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- 14.Fischer T, et al. Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat Cell Biol. 2004;6(9):840–848. doi: 10.1038/ncb1163. [DOI] [PubMed] [Google Scholar]
- 15.Niepel M, Strambio-de-Castillia C, Fasolo J, Chait BT, Rout MP. The nuclear pore complex-associated protein, Mlp2p, binds to the yeast spindle pole body and promotes its efficient assembly. J Cell Biol. 2005;170(2):225–235. doi: 10.1083/jcb.200504140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frost A, et al. Functional repurposing revealed by comparing S. pombe and S. cerevisiae genetic interactions. Cell. 2012;149(6):1339–1352. doi: 10.1016/j.cell.2012.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costanzo M, et al. The genetic landscape of a cell. Science. 2010;327(5964):425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21(1):77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 19.McCullough J, Colf LA, Sundquist WI. Membrane fission reactions of the mammalian ESCRT pathway. Annu Rev Biochem. 2013;82:663–692. doi: 10.1146/annurev-biochem-072909-101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurley JH. ESCRTs are everywhere. EMBO J. 2015;34(19):2398–2407. doi: 10.15252/embj.201592484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Webster BM, Colombi P, Jäger J, Lusk CP. Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell. 2014;159(2):388–401. doi: 10.1016/j.cell.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morita E, et al. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc Natl Acad Sci USA. 2010;107(29):12889–12894. doi: 10.1073/pnas.1005938107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olmos Y, Hodgson L, Mantell J, Verkade P, Carlton JG. ESCRT-III controls nuclear envelope reformation. Nature. 2015;522(7555):236–239. doi: 10.1038/nature14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vietri M, et al. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature. 2015;522(7555):231–235. doi: 10.1038/nature14408. [DOI] [PubMed] [Google Scholar]
- 25.Raab M, et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 2016;352(6283):359–362. doi: 10.1126/science.aad7611. [DOI] [PubMed] [Google Scholar]
- 26.Denais CM, et al. Nuclear envelope rupture and repair during cancer cell migration. Science. 2016;352(6283):353–358. doi: 10.1126/science.aad7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monroe N, Hill CP. Meiotic clade AAA ATPases: Protein polymer disassembly machines. J Mol Biol. 2016;428(9 Pt B):1897–1911. doi: 10.1016/j.jmb.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang D, et al. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat Struct Mol Biol. 2008;15(12):1278–1286. doi: 10.1038/nsmb.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bauer I, Brune T, Preiss R, Kölling R. Evidence for a nonendosomal function of the Saccharomyces cerevisiae ESCRT-III-like protein Chm7. Genetics. 2015;201(4):1439–1452. doi: 10.1534/genetics.115.178939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Connell JW, Lindon C, Luzio JP, Reid E. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 2009;10(1):42–56. doi: 10.1111/j.1600-0854.2008.00847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez Y, Saito A, Sazer S. Fission yeast Lem2 and Man1 perform fundamental functions of the animal cell nuclear lamina. Nucleus. 2012;3(1):60–76. doi: 10.4161/nucl.18824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King MC, Lusk CP, Blobel G. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 2006;442(7106):1003–1007. doi: 10.1038/nature05075. [DOI] [PubMed] [Google Scholar]
- 33.Barton LJ, Soshnev AA, Geyer PK. Networking in the nucleus: A spotlight on LEM-domain proteins. Curr Opin Cell Biol. 2015;34:1–8. doi: 10.1016/j.ceb.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kutay U, Hetzer MW. Reorganization of the nuclear envelope during open mitosis. Curr Opin Cell Biol. 2008;20(6):669–677. doi: 10.1016/j.ceb.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brachner A, Reipert S, Foisner R, Gotzmann J. LEM2 is a novel MAN1-related inner nuclear membrane protein associated with A-type lamins. J Cell Sci. 2005;118(Pt 24):5797–5810. doi: 10.1242/jcs.02701. [DOI] [PubMed] [Google Scholar]
- 36.Campsteijn C, Vietri M, Stenmark H. Novel ESCRT functions in cell biology: Spiraling out of control? Curr Opin Cell Biol. 2016;41:1–8. doi: 10.1016/j.ceb.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 37.Liu HL, Osmani AH, Osmani SA. The inner nuclear membrane protein Src1 is required for stable post-mitotic progression into G1 in Aspergillus nidulans. PLoS One. 2015;10(7):e0132489. doi: 10.1371/journal.pone.0132489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tapia O, Fong LG, Huber MD, Young SG, Gerace L. Nuclear envelope protein Lem2 is required for mouse development and regulates MAP and AKT kinases. PLoS One. 2015;10(3):e0116196. doi: 10.1371/journal.pone.0116196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banday S, Farooq Z, Rashid R, Abdullah E, Altaf M. Role of inner nuclear membrane protein complex Lem2-Nur1 in heterochromatic gene silencing in S. pombe. J Biol Chem. 2016;291(38):20021–20029. doi: 10.1074/jbc.M116.743211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tange Y, et al. Inner nuclear membrane protein Lem2 augments heterochromatin formation in response to nutritional conditions. Genes Cells. 2016;21(8):812–832. doi: 10.1111/gtc.12385. [DOI] [PubMed] [Google Scholar]
- 41.Barrales RR, Forn M, Georgescu PR, Sarkadi Z, Braun S. Control of heterochromatin localization and silencing by the nuclear membrane protein Lem2. Genes Dev. 2016;30(2):133–148. doi: 10.1101/gad.271288.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huber MD, Guan T, Gerace L. Overlapping functions of nuclear envelope proteins NET25 (Lem2) and emerin in regulation of extracellular signal-regulated kinase signaling in myoblast differentiation. Mol Cell Biol. 2009;29(21):5718–5728. doi: 10.1128/MCB.00270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ulbert S, Antonin W, Platani M, Mattaj IW. The inner nuclear membrane protein Lem2 is critical for normal nuclear envelope morphology. FEBS Lett. 2006;580(27):6435–6441. doi: 10.1016/j.febslet.2006.10.060. [DOI] [PubMed] [Google Scholar]
- 44.Anderson DJ, Vargas JD, Hsiao JP, Hetzer MW. Recruitment of functionally distinct membrane proteins to chromatin mediates nuclear envelope formation in vivo. J Cell Biol. 2009;186(2):183–191. doi: 10.1083/jcb.200901106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asencio C, et al. Coordination of kinase and phosphatase activities by Lem4 enables nuclear envelope reassembly during mitosis. Cell. 2012;150(1):122–135. doi: 10.1016/j.cell.2012.04.043. [DOI] [PubMed] [Google Scholar]
- 46.Olmos Y, Perdrix-Rosell A, Carlton JG. Membrane binding by CHMP7 coordinates ESCRT-III-dependent nuclear envelope reformation. Curr Biol. 2016;26(19):2635–2641. doi: 10.1016/j.cub.2016.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morita E, Arii J, Christensen D, Votteler J, Sundquist WI. Attenuated protein expression vectors for use in siRNA rescue experiments. Biotechniques. 2012 doi: 10.2144/000113909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bajorek M, et al. Biochemical analyses of human IST1 and its function in cytokinesis. Mol Biol Cell. 2009;20(5):1360–1373. doi: 10.1091/mbc.E08-05-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCullough J, et al. Structure and membrane remodeling activity of ESCRT-III helical polymers. Science. 2015;350(6267):1548–1551. doi: 10.1126/science.aad8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.von Schwedler UK, et al. The protein network of HIV budding. Cell. 2003;114(6):701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 51.McDonald KL, Auer M. High-pressure freezing, cellular tomography, and structural cell biology. Biotechniques. 2006;41(2):137–143. doi: 10.2144/000112226. [DOI] [PubMed] [Google Scholar]
- 52.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152(1):36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 53.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116(1):71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 54.Mastronarde DN. Correction for non-perpendicularity of beam and tilt axis in tomographic reconstructions with the IMOD package. J Microsc. 2008;230(Pt 2):212–217. doi: 10.1111/j.1365-2818.2008.01977.x. [DOI] [PubMed] [Google Scholar]
- 55.Díez DC, Seybert A, Frangakis AS. Tilt-series and electron microscope alignment for the correction of the non-perpendicularity of beam and tilt-axis. J Struct Biol. 2006;154(2):195–205. doi: 10.1016/j.jsb.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 56.Mackay DR, Elgort SW, Ullman KS. The nucleoporin Nup153 has separable roles in both early mitotic progression and the resolution of mitosis. Mol Biol Cell. 2009;20(6):1652–1660. doi: 10.1091/mbc.E08-08-0883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Talamas JA, Hetzer MW. POM121 and Sun1 play a role in early steps of interphase NPC assembly. J Cell Biol. 2011;194(1):27–37. doi: 10.1083/jcb.201012154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li P, Noegel AA. Inner nuclear envelope protein SUN1 plays a prominent role in mammalian mRNA export. Nucleic Acids Res. 2015;43(20):9874–9888. doi: 10.1093/nar/gkv1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Webster BM, et al. Chm7 and Heh1 collaborate to link nuclear pore complex quality control with nuclear envelope sealing. EMBO J. 2016;35(22):2447–2467. doi: 10.15252/embj.201694574. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.