Significance
In the 15 y since metabolite-binding riboswitches were first experimentally validated, only 4 examples of riboswitch classes with altered specificity have been confirmed by experiments out of ∼30 distinct structural architectures. In contrast, evolutionary changes in ligand specificity of proteins are routinely reported. To further investigate the propensity for natural adaptation of riboswitch specificity, we developed a structural bioinformatics method to systematically search for variant riboswitches with altered ligand recognition. This search method yielded evidence for altered specificity within five riboswitch classes, including validation of a second riboswitch class that senses 2′-deoxyguanosine.
Keywords: 2′-deoxyguanosine, aptamer, c-di-GMP, glycine, guanine
Abstract
Riboswitches are RNAs that form complex, folded structures that selectively bind small molecules or ions. As with certain groups of protein enzymes and receptors, some riboswitch classes have evolved to change their ligand specificity. We developed a procedure to systematically analyze known riboswitch classes to find additional variants that have altered their ligand specificity. This approach uses multiple-sequence alignments, atomic-resolution structural information, and riboswitch gene associations. Among the discoveries are unique variants of the guanine riboswitch class that most tightly bind the nucleoside 2′-deoxyguanosine. In addition, we identified variants of the glycine riboswitch class that no longer recognize this amino acid, additional members of a rare flavin mononucleotide (FMN) variant class, and also variants of c-di-GMP-I and -II riboswitches that might recognize different bacterial signaling molecules. These findings further reveal the diverse molecular sensing capabilities of RNA, which highlights the potential for discovering a large number of additional natural riboswitch classes.
Riboswitches are structured noncoding RNA domains that regulate gene expression in response to the selective binding of small-molecule or ion ligands. The discovery of numerous classes of riboswitches has helped reveal how RNAs can form exquisitely precise ligand-binding pockets using only the four common RNA nucleotides (1–4). Furthermore, each discovery links the riboswitch ligand to the protein products of the genes under regulation. Recent riboswitch findings have exposed unique facets of biology, such as the widespread molecular mechanisms that confer fluoride (5) or guanidine (6) resistance, that maintain metal ion homeostasis (7–9), and that control important bacterial processes such as sporulation, biofilm formation, and chemotaxis (10–14). Thus, the identification of additional riboswitch classes promises to offer insights into otherwise hidden biological processes and their regulation.
Riboswitch variants have been reported previously, wherein the ligand-binding “aptamer” domain has mutated to accommodate a different metabolite or signaling compound. The identification of such RNAs provides rare opportunities to study how small changes in RNA sequence can lead to major changes in small-molecule ligand affinity. There have been seven examples, either experimentally validated or suspected, of ligand specificity changes reported to date. These include guanine aptamer variations present in riboswitches for adenine (15) and 2′-deoxyguanosine (2′-dG) (16), c-di-GMP-I aptamer variations that result in riboswitches that bind the recently discovered bacterial signaling molecule c-AMP-GMP (13, 14), and coenzyme B12 aptamer changes (17, 18) that yield riboswitches selective for aquocobalamin (19). Three additional ligand specificity changes are suspected. Namely, some molybdenum cofactor riboswitches appear to exploit an altered aptamer structure to selectively recognize tungsten cofactor (20), certain flavin mononucleotide (FMN) riboswitches carry binding site mutations that alter ligand specificity (21, 22), and a large number of guanidine-I riboswitches carry mutations in the binding pocket and sense an as-yet-unknown ligand (6).
Several variant riboswitches share a number of characteristics that could have been exploited in a bioinformatic search for such RNAs. We chose to apply three important properties common to the guanine/adenine (15) and c-di-GMP-I/c-AMP-GMP (13, 14) riboswitch sets, among other variants. The first of these properties is that some variant riboswitches with altered ligand specificity will remain somewhat close in both sequence and structure to the predominant or “parent” class. For example, the initial collections of representatives for guanine (23) and c-di-GMP-I (originally called GEMM) (10, 24) riboswitches were uncovered by comparative sequence analyses, and unknowingly included less common examples of the variant riboswitches that were eventually proven to exhibit altered ligand specificity. Thus, the “purine” riboswitch entry in the Rfam Database (25) includes both guanine and adenine variants. Similarly, the Rfam “GEMM” sequence alignment includes c-di-GMP-I and c-AMP-GMP riboswitches. We therefore hypothesize that other existing Rfam sequence alignments could also include unrecognized examples of riboswitches that respond to a ligand that is different from the ligand sensed by the parent riboswitch class.
Although an obvious strategy to identify variant riboswitches would be to select any representatives whose sequences differ from most others in the class, we cannot rely exclusively on sequence variation because most nucleotide changes will not lead to altered ligand specificity. Therefore, we turned to the second common characteristic of these particular parent/variant riboswitch sets, which is that the change in specificity is strongly associated with a change at a single nucleotide position that interacts with the ligand. For example, “nucleotide 74” in riboswitches for both guanine and adenine forms a Watson/Crick base pair to the ligand. Given this important role for nucleotide 74, as numbered according to the Bacillus subtilis xpt aptamer construct reported previously (15), we call this position a “key” nucleotide.
When nucleotide 74 is a cytidine, guanine is specified as the ligand (23, 26). Alternatively, when this nucleotide is a uridine, adenine is specified as the ligand (15, 27). Similarly, in riboswitches for c-di-GMP-I and c-AMP-GMP, “nucleotide 20” is typically a guanosine for recognition of the c-di-GMP ligand, and adenosine for recognition of the c-AMP-GMP ligand (13, 14). Nucleotide 20 establishes ligand specificity at least in part by forming a Hoogsteen interaction with its target compound (28, 29). Therefore, evaluating sequence alignments of riboswitches for variation at nucleotide positions that serve critical roles in binding pockets will help reveal variant riboswitch candidates that have altered ligand specificity.
The third common characteristic of riboswitch sets with altered ligand specificity is their association with distinct groups of genes. For example, riboswitches that sense guanine commonly associate with purine biosynthesis and import (23), whereas adenine riboswitches frequently regulate genes for the degradation or export of adenine (15). Similarly, although c-di-GMP-I riboswitches regulate a great diversity of genes important for various physiological processes in bacteria, they only rarely associate with genes for cytochrome c (10). In stark contrast, c-AMP-GMP riboswitches commonly regulate cytochrome c genes (13, 14). Thus, gene associations that are unusual or that cannot be easily rationalized based on the putative ligand identity might be explained by a specificity change in the associated riboswitches.
To exploit these observations, we constructed a bioinformatics pipeline that searches for riboswitch RNAs associated with protein-coding regions that are unusual for the predominant members of a given riboswitch class, and that carry unusual nucleotides within the ligand-binding pocket. These efforts, when applied to 28 parental riboswitch classes, have revealed the existence of unique variant groups derived from the guanine, c-di-GMP-I, c-di-GMP-II, and glycine classes, as well as additional representatives of a variant FMN riboswitch class discovered previously. Biochemical analysis of representatives of the variant guanine, glycine, and FMN riboswitch RNAs reveal that they indeed reject the predominant ligands of their parent riboswitch classes. Moreover, the guanine riboswitch variants were found to function as distinct sensors for the ligand 2′-dG, whereas the natural ligands for the remaining variants remain unsolved.
Results and Discussion
Strategy to Detect Variant Riboswitches.
To reveal undiscovered riboswitch classes, we used a computational pipeline (Fig. 1A) that exploits three characteristics common to several known examples of closely related riboswitch sets that recognize different molecules. In the current study, this method has been applied to all riboswitch classes for which structural information was available by using Rfam sequence alignments and atomic-resolution structural models corresponding to each initial riboswitch class (SI Appendix, Table S1, and Dataset S1 at breaker.yale.edu/variants).
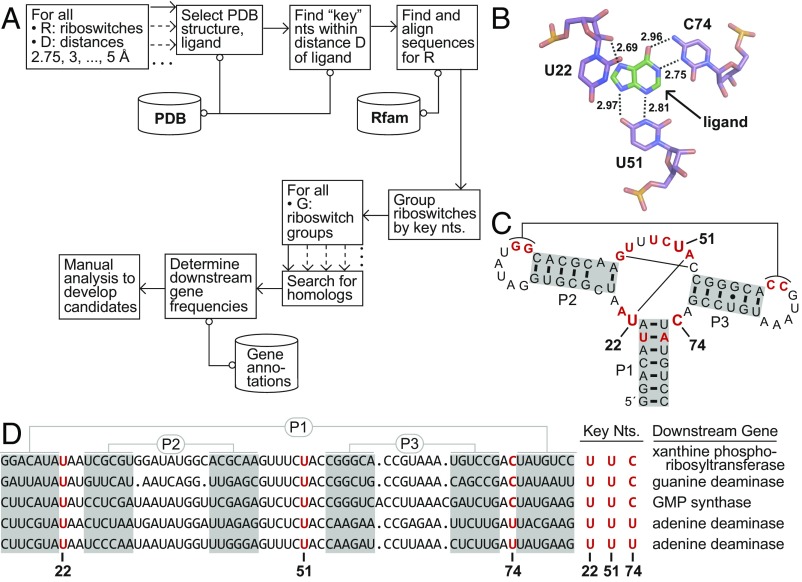
Fig. 1.
Bioinformatic search method illustrated using guanine and adenine riboswitch examples. (A) Schematic depiction of a process to detect riboswitches with altered ligand specificities (see text for details). (B) Atomic-resolution model of the ligand-binding pocket of a guanine riboswitch aptamer bound to the guanine analog hypoxanthine (26) (PDB ID code 4ef5). Three “key” nucleotides (U22, U51, and C74) of the aptamer carry atoms that are within 3 Å of a ligand atom (see dashed lines). The same three key nucleotides would have been identified if the natural ligand guanine were docked. (C) Key nucleotides at positions 22, 51, and 74 mapped onto the sequence and secondary-structure model for the guanine riboswitch aptamer whose X-ray structure was used to conduct this analysis (26). Nucleotides in red identify positions that are conserved in 97% or greater of the known guanine riboswitch aptamers. Thin lines identify long-range base pairs. (D) Alignment of the sequence of the guanine riboswitch aptamer in B and C with two additional guanine and adenine aptamers, arranged from Top to Bottom, respectively. Adenine riboswitches, known to carry a C-to-U mutation at position 74, commonly regulate adenine deaminase genes that are not regulated by these three guanine riboswitches. The complete analysis for this collection of riboswitches included 3,462 guanine and 187 adenine riboswitches.
First, we expect that some riboswitch representatives with altered ligand specificity will be sufficiently close in both sequence and structure to a previously established riboswitch class that they will appear in collections of riboswitch sequences. To exploit this characteristic, we need only to access published sequence alignments for each riboswitch class, or public databases such as Rfam (25).
Second, we exploit the fact that ligand specificity changes will commonly be caused by sequence alterations within the ligand-binding pocket of each parent riboswitch class. To exploit this characteristic, we need to identify any nucleotides that are near the ligand when it is docked to the aptamer domain of the riboswitch. To achieve this objective, one or more atomic-resolution structures for each parent riboswitch class of interest are selected (Fig. 1B and SI Appendix, Table S1), and the distances between each nucleotide of the aptamer and the ligand are determined. These distances are defined as the nearest distance of any atom within each nucleotide to any atom within the ligand.
This collection of distance values is used to define a list of key nucleotides whose identities might affect ligand specificity. We used a range of distance thresholds to establish the list of key nucleotides because the optimal distance threshold to use would likely vary from case to case based on the resolution of the structure model and with the type of molecular interaction formed between the riboswitch aptamer and its ligand. Thus, for a given distance threshold (e.g., 3 Å), a computer finds the nucleotides with at least one riboswitch atom within this distance of a ligand atom. These key nucleotides (Fig. 1C) are then mapped onto their positions within the sequence alignment to determine possible specificity-determining nucleotides of all riboswitch sequences in the alignment (Fig. 1D). Thus, riboswitches are classified into different “groups,” so that the riboswitches in each group have the same key nucleotides.
Unfortunately, the identification of riboswitch groups based solely on the identification of key nucleotides yields too many candidates for subsequent experimental validation. To focus our experiments on the most promising groups, we exploited the third characteristic of surprising gene associations compared with the parent riboswitch class. All groups corresponding to sequence variations at key sites are computationally analyzed to determine whether they associate with genes and biological processes that are unusual compared with the group corresponding to the predominant riboswitch class. A demonstration of this analysis is presented (Fig. 1D) for a sequence alignment originally expected (23) to include only guanine riboswitches (key nucleotides U22, U51, and C74, abbreviated U,U,C, respectively). However, RNA representatives in this collection also include adenine riboswitches (key nucleotides U,U,U). These variant RNAs carry the C74U mutation and associate with adenine metabolism genes, which hint at their true biological function as adenine riboswitches.
Moreover, it seemed likely that some members of a particular variant riboswitch group might contain additional sequence changes relative to the consensus of the parent class. Thus, although the alignment model used to generate the Rfam sequence list might find representatives of the variant group, it might not be a good model for discovering all such variants. As a consequence, we designed our system to perform automated homology searches for each variant riboswitch group to expand both the number of representatives and the information regarding distinctive gene associations. In one example described in more detail below, the initial group of variants was represented by only six members. After conducting automated homology searches incorporating the sequence features characteristic of the initial group, a total of 19 variant representatives were identified. Subsequent manual homology searches revealed a total of 31 members. Such additional sequences and their associated genes can then be used to help assess whether the variant group merits experimental validation efforts.
Purine Riboswitch Variants.
Guanine riboswitch aptamers have three key nucleotides within 3 Å of the ligand (Figs. 1 and 2A). Our analysis revealed seven additional groups of purine riboswitch variants that differ from the parent guanine riboswitch in at least one of these three positions (Fig. 2B), including a previously validated group that exhibits altered ligand binding. Specifically, guanine riboswitches (U,U,C) were readily differentiated from the published variant riboswitch class that selectively binds adenine (U,U,U) (15). However, this analysis did not uncover the previously validated class of guanine riboswitch variants that bind 2′-dG (16) because these RNAs are sufficiently different from guanine and adenine riboswitches so as to not fall within the Rfam definitions for this riboswitch class.
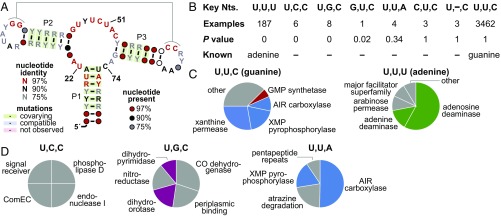
Fig. 2.
Guanine riboswitch variants. (A) Consensus sequence and secondary-structure model for riboswitches that recognize purines, which are dominated by guanine riboswitches. Three nucleotides located within 3 Å of ligand atoms and representing the key ligand-binding nucleotides are identified at positions 22, 51, and 74. “R” represents a purine, and “Y” represents a pyrimidine. (B) Distinct groups of purine riboswitches identified by the computational method used in this study. Groups corresponding to guanine and adenine riboswitches are indicated as “Known.” “Key Nts.” are presented from 5′ to 3′ (positions 22, 51, and 74), where the dash indicates a nucleotide deletion. “Examples” report the number of distinct sequence representatives within each group before the automated homology search was conducted. “P value” reports the arithmetic average of two P value estimates (Materials and Methods). This P value average estimates how dissimilar the group’s genes are to genes associated with guanine riboswitches. A P value average close to zero indicates that the two sets of genes differ significantly, and therefore the group is a promising candidate (e.g., the adenine riboswitch group). P value averages near 1 indicate similar genes, and thus little evidence for an altered ligand. Groups are sorted by average P value from zero (best candidates) to 1 (worst). The 2′-dG riboswitches in M. florum are not listed because they are not detected by the Rfam search. (C) Gene associations of groups corresponding to already-validated guanine (U,U,C) and adenine (U,U,U) riboswitches. The pie charts reflect the relative abundance of the five most common gene classes (excluding those encoding hypothetical proteins) associated with the group. Red, clear association with guanine; green, clear association with adenine; blue, general association with purine metabolism; purple, pyrimidine metabolism; gray, other genes. (D) Gene associations of other groups of purine riboswitches. Annotations are as described in C. The G,U,C, C,U,C, and U,–,C groups are not listed because only one or zero of their associated genes code for proteins that match known conserved domains, and therefore functions cannot easily be predicted.
If adenine riboswitches had not been discovered previously, they would have easily been detected using our method. A total of 187 examples with the key nucleotides U,U,U was identified, making this the most common group that varies from the guanine riboswitch parent class (Fig. 2B). Furthermore, the genes most commonly associated with the adenine riboswitch group are very rarely found downstream of guanine riboswitches (Fig. 2C). For example, the most common adenine riboswitch gene class encodes adenosine deaminase. This gene class is regulated by ∼58% of adenine riboswitches, vs. only 0.1% of guanine riboswitches. Thus, the U,U,U group of RNAs would have made an excellent candidate for a variant riboswitch class that has undergone a change in ligand specificity.
The U,C,C group has several features that indicate that it is also an excellent variant riboswitch candidate. Its associated genes (Fig. 2D) are never observed to be downstream of guanine riboswitches or any of the other variant groups. Moreover, these genes are not directly involved in basic metabolism, which is unlike the vast majority of genes associated with guanine and adenine riboswitches. There are also numerous differences in conserved sequence features in the U,C,C group compared with guanine and adenine riboswitches (Fig. 3A). Notably, the U51C substitution that distinguishes this group from guanine riboswitches was proven (30) to be structurally important for ligand binding by the previously discovered (16) rare purine riboswitch variant that binds 2′-dG. The U,C,C RNAs also have a longer junction between P3 and P1, which contains position 74. This junction comprises up to five nucleotides for the U,C,C group vs. only two for guanine and adenine riboswitches. Additionally, nucleotide position 47 is highly conserved as a U residue in guanine riboswitches and is predicted to closely approach the ligand in atomic-resolution structures (26, 27, 31). However, this position is not well conserved in U,C,C-group RNAs. Finally, the A–U base pair at the top of stem P1 is strongly conserved in guanine riboswitches and contributes to ligand specificity (32) but differs in the U,C,C group. Collectively, the sequence features and gene associations distinguishing U,C,C-group RNAs from guanine and adenine riboswitches strongly suggest that a ligand specificity change has occurred.
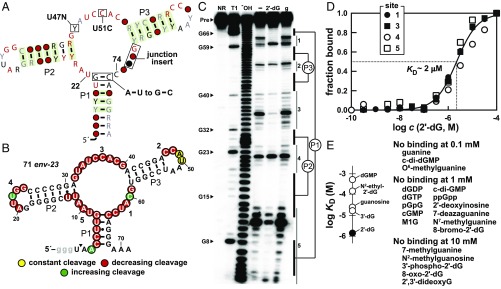
Fig. 3.
Selective recognition of 2′-dG by a U,C,C group of guanine riboswitch variants. (A) Consensus sequence and secondary structure of a putative purine riboswitch variant identified via the bioinformatics strategy described in this report. Boxed annotations indicate differences from guanine riboswitches. Other annotations are as described for Fig. 2A. (B) Sequence and secondary structure of the 71 env-23 RNA. Regions of constant, increasing, and decreasing internucleotide cleavage were determined from the in-line probing data presented in C. The arrowhead indicates the start of this data. Lowercase g letters identify guanosine nucleotides encoded by the template to facilitate efficient RNA production by in vitro transcription. Numbers 1 through 5 identify regions that undergo 2′-dG–dependent structure modulation. (C) Denaturing (8 M urea) PAGE analysis of in-line probing reactions of 5′-32P-labeled 71 env-23 RNA in the presence of 100 µM deoxyguanosine (2′-dG), 100 µM guanine (g), or in the absence of ligand (‒). NR, T1, and −OH indicate no reaction, partial digestion with RNase T1 (cleaves after G residues), and partial digestion with hydroxide (cleaves after every residue). Several RNase T1 product bands are labeled. Regions undergoing structural modulation (1 through 5) and predicted stems (P1 through P3) are indicated. (D) Plot of the fraction of RNA bound to ligand vs. the logarithm of the molar concentration (c) of 2′-dG. Data are derived from SI Appendix, Fig. S1. Included are a theoretical binding curve expected for a one-to-one interaction between ligand and RNA for the indicated KD value. (E) Plot of the dissociation constants measured for various analogs of 2′-dG for the 71 env-23 RNA (Left). List of compounds that resulted in no structural modulation of the 71 env-23 RNA upon addition at the indicated concentrations (Right). 3-(2-Deoxy-β-d-erythro-pentofuranosyl)pyrimido[1,2-a]purin-10(3H)-one is abbreviated M1G.
Variant Riboswitches That Sense 2′-dG.
We experimentally examined two typical members of the U,C,C group to assess the hypothesis that they have altered ligand specificity relative to their parent guanine riboswitches. The first RNA construct, called 71 env-23 (Fig. 3B), includes 71 nt encompassing a U,C,C group member from an environmental sequence sample. This RNA was subjected to in-line probing, which is a structure analysis method that enables the detection of riboswitch folding changes upon recognition of a cognate ligand (33, 34). By testing a variety of potential ligands that are structurally related to guanine, we found that the 71 env-23 RNA is capable of recognizing 2′-dG, but not guanine (Fig. 3C).
The locations and extents of 71 env-23 RNA structural modulation observed upon addition of 2′-dG was similar to that previously observed for guanine (23) and adenine (15) riboswitches. This suggests that the U,C,C-group RNAs adopt the same general architecture as the parent riboswitch class, but have exploited mutations in the key nucleotides to selectively respond to 2′-dG as their natural target. By conducting in-line probing reactions with 71 env-23 RNA and a range of ligand concentrations (SI Appendix, Fig. S1), we determined that the dissociation constant (KD) for 2′-dG is ∼2 μM (Fig. 3D). Although guanine binds its cognate riboswitch aptamer with an affinity that is about 3 orders of magnitude better, this variant riboswitch binds 2′-dG with an affinity that is similar to that observed for several 2′-dG riboswitches reported previously (hereafter called 2′-dG-I riboswitches) (16). Likewise, the 71 env-23 representative of U,C,C-group RNAs discriminates against guanine, guanosine, and many other close analogs of 2′-dG by at least an order of magnitude (Fig. 3E).
To further investigate the function of U,C,C RNAs, a second representative of this group called 71 env-16 (SI Appendix, Fig. S2A) was prepared that included 71 nt encompassing the variant motif from Gracillimonas tropica DSM 19535. Again, RNA structure changes occur in response to increasing concentrations of 2′-dG as revealed by in-line probing (SI Appendix, Fig. S2B), to yield a KD of ∼1 μM (SI Appendix, Fig. S2C). These findings likewise support the conclusion that U,C,C-group RNAs function as members of a class of riboswitches for 2′-dG that are distinct from the parent guanine riboswitch class.
When classifying members of the U,C,C group, we also took into consideration gene associations. Genes located immediately downstream from U,C,C-group riboswitches that have an assigned function (Fig. 2D and Dataset S2 at breaker.yale.edu/variants) are predicted to encode a signal receiver domain, endonuclease I, phospholipase D, and ComEC. Protein products of the signal receiver domain and phospholipase D genes typically participate in signal transduction (35), and the precise role of this signaling is unclear. Interestingly, endonuclease I and ComEC function on DNA substrates. Analysis of the protein sequence of the endonuclease suggests that it is secreted from cells (36). Thus, it is possible that a lack of 2′-dG could be mitigated by salvaging deoxyribonucleotides using secreted endonucleases, and that the expression of such genes would be desirable as the cellular concentration of 2′-dG declines. Moreover, ComEC is a competence protein involved in importing foreign DNA (37). Perhaps cells deficient in 2′-dG activate production of ComEC to import DNA polymers as a source of premade DNA monomers.
In contrast, previously discovered 2′-dG-I riboswitches clearly associate with genes whose protein products participate in 2′-dG production or transport. For example, a previously discovered 2′-dG-I riboswitch from Mesoplasma florum controls ribonucleotide reductase (16), which synthesizes deoxyribonucleotides and therefore has a clear metabolic connection to 2′-dG.
Despite the uncertainties noted above, we speculate that, like 2′-dG-I riboswitches, RNAs in the U,C,C group also might sense 2′-dG as the natural ligand. Therefore, we call members of the U,C,C group 2′-dG-II riboswitches. Notably, the bacterium M. florum, which carries multiple examples of 2′-dG-I riboswitches, is classified in the phylum Tenericutes, whereas 2′-dG-II riboswitches are found in the phylum Bacteroidetes. In addition, members of the U,C,C group have a distinct identity for the key nucleotides compared with 2′-dG-I riboswitches (C,C,C), although the overall architecture and certain sequences for the two RNAs are similar (SI Appendix, Fig. S3). The U,C,C group also differs from 2′-dG-I riboswitches in the junction between stems P3 and P1, which is longer and carries additional conserved nucleotides in the U,C,C variant group (Fig. 3A). Additionally, the base pair at the top of P1 is A–U in 2′-dG-I riboswitches, like guanine and adenine riboswitches and unlike 2′-dG-II aptamers, which carry a G–C at this position. However, a G–C base pair in this position is well tolerated by a mutant guanine riboswitch construct tested previously (38), suggesting that this variation in 2′-dG-II riboswitches might contribute only modestly to the ligand-binding differences observed.
That these 2′-dG riboswitch types are in highly diverged organisms and have differences in sequence features might indicate that they have evolved from guanine riboswitches via two distinct evolutionary events. Atomic-resolution structural studies could help to further determine how 2′-dG-II riboswitches exploit the sequence variations to accommodate a new ligand and whether these distinct riboswitch classes for 2′-dG might have emerged by taking two independent evolutionary paths.
Evidence for Additional Ligand Changes from Parent Guanine Riboswitches.
Another candidate riboswitch variant is the U,G,C group. Representatives of this group associate with pyrimidine-related genes that are rarely if ever regulated by guanine riboswitches. However, the fact that these and other pyrimidine-related genes are sometimes regulated by guanine riboswitches, and the fact that pyrimidine biosynthesis is known to be regulated by purines (39), provide reasons to expect that these RNAs might still recognize guanine.
In-line probing was used to assess the ligand-binding specificity of a representative U,G,C-group RNA. However, we did not observe recognition of any of a number of compounds, including guanine (SI Appendix, Fig. S4). Our in-line probing results demonstrate that this RNA is adopting the same general secondary structure as observed for guanine, adenine, and 2′-dG riboswitches described above. Therefore, the negative binding results are not due to comprehensive misfolding of the RNA construct chosen for analysis. Thus, a broader search, perhaps involving both biochemical and genetic approaches, will be needed to identify a potential natural ligand for this riboswitch variant.
A member of the U,U,A group of guanine riboswitch variants (Fig. 2) was the final guanine riboswitch-derived candidate we examined. Because there were only four U,U,A-group RNAs identified, and because they control genes that are similar to those controlled by guanine riboswitches, they represent a borderline candidate. Unfortunately, in-line probing experiments revealed that the representative U,U,A-group RNA chosen for analysis is misfolded under our reaction conditions. Therefore, we could not determine from these data whether the construct rejects guanine, and if so, what compound might serve as its natural ligand. Other variant groups are even rarer and therefore were not experimentally examined in this study. Given the rarity of these other groups, as well as U,U,A RNAs, it is possible that these sequences represent false positives and do not function as riboswitches.
Glycine Riboswitch Variants.
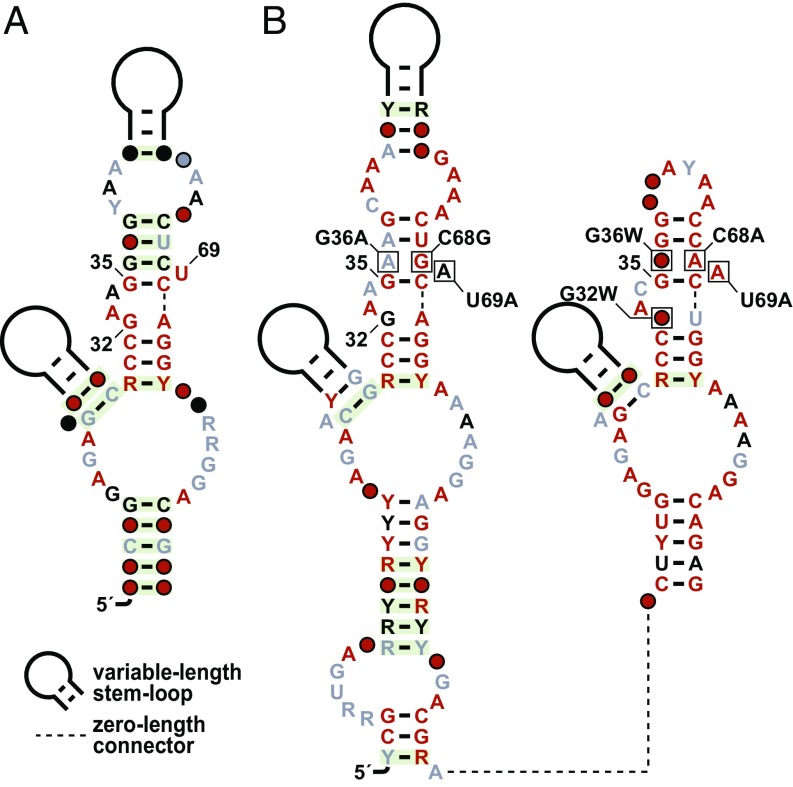
Another promising candidate identified in our bioinformatics search is derived from glycine riboswitches (40). Glycine riboswitch aptamers are commonly found in tandem arrangements. In some in vitro and in vivo assays, these tandem aptamers function cooperatively, such that glycine binding by one aptamer can improve the affinity for ligand binding at the other site (40, 41). Because both aptamers in such tandem arrangements bind glycine, the ligand-binding pockets have nearly identical conserved sequence features. Nucleotide positions near glycine in the parent riboswitch class were identified by computational analysis of atomic-resolution structures previously published (41, 42). These nucleotides, which are at positions 32, 35, and 69, as numbered for a previous glycine riboswitch construct (42), were chosen for subsequent bioinformatics analyses. The vast majority of representatives in the sequence alignment for this class carry the key nucleotides G,G,U (Fig. 4A), as do previously validated glycine riboswitches (40–42).
Fig. 4.
Key binding-site nucleotides and variants for glycine riboswitches. (A) Consensus sequence and secondary structure of glycine riboswitch aptamers, with key nucleotides G,G,U located at positions 32, 35, and 69. The secondary-structure model from Rfam has been adjusted based on crystallographic (41) and other (40, 58, 59) data. (B) Consensus sequence and secondary structure for tandem glycine riboswitch variants wherein key nucleotides have mutated. W refers to A or U nucleotides.
Upon conducting our bioinformatics analysis, we identified three variant groups with the key nucleotides G,G,A; A,G,A; or U,G,A that share a common U69A change and associate with the same set of genes as each other. Moreover, these genes are distinct from those typically regulated by glycine riboswitches. Therefore, we combined these variant groups to create a single group called D,G,A, where D represents any nucleotide except C. Sequence alignments of the D,G,A group revealed that several nucleotides adjacent to the key nucleotides also undergo mutation (Fig. 4B). In addition to the mutations at key sites, these RNAs carry mutations at an otherwise well-conserved G–C base pair between nucleotides G36 and C68 of the glycine aptamer. In a glycine riboswitch, this base pair largely forms one side of the binding pocket (42). However, in D,G,A-group RNAs, these two nucleotides form a U–A base pair, or form A·G, G·G, or A·A mismatches. Experimental mutation of the natural G–C base pair in a glycine riboswitch to either a C–G or A–U base pair results in a total loss of glycine binding (42). Therefore, the natural mutations at these sites presumably collaborate with mutations at key nucleotides to permit a ligand specificity change for the D,G,A riboswitches.
Most D,G,A RNAs are found upstream of predicted intrinsic transcription terminator hairpins (43, 44), and many of these terminator stems overlap the riboswitch aptamer and conflict with its structure. This arrangement suggests that, when the ligand binds and stabilizes the aptamer structure, it destabilizes the terminator hairpins, leading to increased gene expression. Importantly, D,G,A-group RNAs typically occur in tandem arrangements (Fig. 4B) similar to that observed for glycine riboswitches, wherein both ligand-binding sites conform to the combined variant group. In these arrangements, the 5′ aptamer is from the G,G,A group, and the 3′ aptamer is from the U,G,A or A,G,A groups. Such arrangements of dual D,G,A aptamers occur in 91 examples.
Interestingly, there are also 10 chimeric arrangements, in which a D,G,A aptamer occurs immediately adjacent to a typical G,G,U glycine aptamer. In these 10 instances, the downstream genes are characteristic of those controlled by glycine riboswitches. It appears that some bacteria use a D,G,A aptamer and a conventional glycine aptamer in a single mRNA leader to create a two-input logic gate (45) that responds to both glycine and the unidentified ligand of D,G,A aptamers.
When two D,G,A aptamers occur in tandem, they associate with genes that encode either saccharopine dehydrogenase or amino acid transporters classified in the COG0531 or COG1748 families, and other genes whose functions are not predicted. These D,G,A-associated genes are never observed to be regulated by glycine riboswitches from the canonical G,G,U group, or in any other group. Thus, RNAs from the D,G,A group are found upstream of unique gene classes and are observed in a wide range of organisms within the order Clostridiales. These findings strongly suggest that the observed mutations are not the result of random changes to glycine riboswitches that have simply become nonfunctional. That is, random changes would be unlikely to associate with two independent structural classes of genes that are never observed downstream of G,G,U glycine riboswitches, nor be present in distantly related Clostridiales bacteria.
Tandem Glycine Riboswitch Variants Reject Glycine.
In-line probing assays demonstrate that D,G,A RNAs do not bind glycine (SI Appendix, Fig. S5). This result is consistent with the findings of a previous study that analyzed the U69A mutation and found the resulting construct to be inactive for glycine binding (46). The lack of observed binding by D,G,A variants, in combination with our bioinformatic analysis, indicates that these RNAs have likely undergone a ligand specificity switch. However, the identity of the ligand for D,G,A riboswitches still remains unknown. Clues to the identity of this unknown ligand can be found in the genes regulated by D,G,A riboswitches. As noted above, a total of 10 D,G,A aptamers reside immediately adjacent to typical G,G,U glycine aptamers. In these 10 instances, the downstream genes are characteristic of those controlled by glycine riboswitches. Therefore, we speculate that the ligand for D,G,A riboswitches is somehow related to glycine metabolism.
Saccharopine dehydrogenase, whose genes are commonly associated with D,G,A variants, catalyzes one of the steps of lysine catabolism. Because glycine riboswitches are thought to direct excess glycine into the citric acid cycle (40), we originally hypothesized that the D,G,A variants might function analogously for lysine. In a preliminary effort to identify the natural ligand for D,G,A variants, we conducted additional in-line probing assays with lysine, a diversity of lysine derivatives, and other compounds related to glycine metabolism. However, we did not detect any evidence that the RNA is capable of recognizing the compounds tested (SI Appendix, Fig. S6). Despite this result, the D,G,A variant RNAs have all of the characteristics of an excellent variant riboswitch candidate, and therefore this class merits further investigation.
Numerous Variants of c-di-GMP Riboswitches Exist.
We also found a number of variant riboswitch candidates among c-di-GMP-I (10) and c-di-GMP-II (11) riboswitch classes. Because c-di-GMP is a bacterial signaling compound, it is not surprising that a great diversity of genes is regulated by members of this riboswitch class. Our computational strategy to detect ligand changes is based partly on judging whether there is a difference in the types of genes controlled by a potential riboswitch variant compared with genes associated with the parent riboswitch group. Consequently, parent riboswitch classes that normally control highly diverse sets of genes could confound our analyses.
For example, it was difficult to identify the previously reported c-AMP-GMP riboswitch class (13) from among the parent c-di-GMP riboswitch alignment via our bioinformatics approach, although we were eventually able to do so. Importantly, although the variant riboswitches that sense c-AMP-GMP do control certain genes only very rarely or never controlled by c-di-GMP-I riboswitches, a number of other genes controlled by c-AMP-GMP riboswitches are also commonly controlled by c-di-GMP-I riboswitches. Thus, we would expect that any other variant cyclic dinucleotide riboswitches might also control a mix of typical and atypical c-di-GMP-I riboswitch-controlled genes.
Despite these expected difficulties, we identified three additional groups of c-di-GMP-I and five additional groups of c-di-GMP-II variants of interest (SI Appendix, Fig. S5). As with the analysis noted immediately above, these variant riboswitch candidates also occur upstream of a mix of unique genes and genes that are associated with canonical c-di-GMP-I riboswitches, making these candidate groups comparable to the c-AMP-GMP group. The genes most commonly associated with these eight variant groups are only rarely or never observed to be associated by known c-di-GMP-I or c-AMP-GMP riboswitches (SI Appendix, Tables S2 and S3). Because these eight candidate groups contain very few member sequences, it is possible that they simply are unusual variants of c-di-GMP–sensing riboswitches, or perhaps some are riboswitch mutants that no longer function. An intriguing alternate explanation is that these variant groups might be triggered by cyclic dinucleotides that are different from those sensed by known riboswitch classes, including potential signaling compounds not yet known to science.
Conclusions
The bioinformatics approach described herein constitutes a partially automated process to define changes to ligand-binding aptamer residues that have a high probability of modified riboswitch ligand specificity. This approach works best with an extensive list of representatives for a given riboswitch class and requires a quality high-resolution structural model of a riboswitch bound to its natural ligand. With these criteria met, our computational approach can systematically detect ligand changes across multiple riboswitch classes. Indeed, implementing this search strategy has resulted in multiple candidate riboswitch classes that have emerged by undergoing ligand specificity changes (Datasets S1 and S2 at breaker.yale.edu/variants).
We applied this strategy to 28 riboswitch classes and identified many distinct variant groups to reveal ligand-binding changes to five of these classes. These include the discovery of an RNA class called 2′-dG-II riboswitches, ligand specificity changes to variants of glycine, and potential ligand specificity changes to c-di-GMP-I and -II riboswitches. Variants of a fifth riboswitch class constitute additional examples of a rare variant of FMN riboswitches discovered previously (21, 22), increasing our confidence that these sequences in fact represent a separate riboswitch class. This unusual FMN riboswitch variant (SI Appendix, Fig. S7), which was first identified in Clostridium difficile, carries several mutations in key nucleotides at the binding site that cause it to reject binding the coenzyme (21). Although this variant has been shown to bind derivatives of FMN (22), its biologically relevant ligand remains a mystery.
We also identified variants of Ni/Co riboswitches (9), which would have represented a sixth additional parental class with evidence for ligand specificity changes, although analysis of a candidate suggests that these variants retain the ability to cooperatively bind Ni2+ and Co2+ (SI Appendix, Fig. S8). Identification of unusual riboswitches like these Ni/Co variants, which recognize the same ligand despite changes to key binding-site nucleotides, could represent interesting subjects for structural and functional analysis.
Experimentally validating “orphan riboswitches” whose ligands are unknown can be challenging (47, 48). Therefore, establishing the ligands for the variant riboswitch candidates generated by this or other bioinformatics-based search approaches might require considerable experimental effort. These efforts are further compounded by observation that the variant riboswitches uncovered in the current study are quite rare. This inherently reduces the number of known gene associations that otherwise could provide clues leading to the identification of the natural ligand. Regardless, our results suggest that numerous different classes of variant riboswitches with altered ligand-binding functions are present in nature. The ever-increasing collection of DNA sequence data could help to expose even rarer variants in the future and provide additional clues to aid in establishing ligand identities for existing candidates.
It is interesting to note that many of the variant groups we uncovered in this study were identified among members of parent riboswitch classes that had previously yielded variants with altered ligand binding. Specifically, these include guanine (with variants for adenine and 2′-dG) and c-di-GMP-I (c-AMP-GMP) riboswitch classes. This observation suggests that certain RNA structures might be more conducive to accruing mutations in the binding pocket to adapt to different ligands. However, there might alternatively be greater evolutionary utility to diversifying riboswitch aptamers that sense compounds that are structurally similar to guanine or to c-di-GMP, rather than for compounds similar to many other riboswitch ligands.
As noted, the previously known 2′-dG-I riboswitches from M. florum (16) are not detected by our method, because they are not predicted using Rfam’s existing search parameters. Improved homology search algorithms could thus help to discover other distal variants that currently elude searches. Also, the existing algorithms could be adjusted to include riboswitch-like sequences with weaker homology scores. However, such an approach will include more false riboswitch predictions and might thus lead to predictions of additional groups without ligand changes and pollute groups with members that are not riboswitches.
Variant riboswitches with altered ligand specificity are also known to exist that do not extensively alter binding-site nucleotides to modify their ligand-binding specificity. For example, the specificity of aquocobalamin vs. adenosylcobalamin riboswitches is to a large extent determined by nucleotides that are not close to the ligand in adenosylcobalamin riboswitches (19). This situation is also similar to the proposed distinction between molybdenum and tungsten cofactor riboswitches (20). Thus, more ligand variation could perhaps be detected by monitoring nucleotide changes outside of the ligand-binding core. However, applying our current method to all such nucleotides would likely lead to a large increase in false-positive predictions. Additional criteria would likely be needed to reduce these predictions to a manageable number. Regardless, there might be far more variant riboswitches that remain to be discovered that could be identified by developing even more powerful search approaches.
Materials and Methods
Databases.
Bacterial or archaeal genome sequences from RefSeq (49), version 63, were used, along with various metagenomes generally collected from IMG/M (50), the Human Microbiome Project (51), MG-RAST (52), or GenBank (53). Gene annotations were made in a previously described process (54) that classified conserved protein domains using the Conserved Domain Database (35). To find riboswitch structures, we used the Protein Data Bank (PDB) (55).
Riboswitch Analysis.
Alignments from Rfam, version 12.0 (25), were used to detect riboswitches, using the Infernal 1.1 software package (56) with the search parameters recommended by Rfam. For a given riboswitch class, Rfam-based searches were conducted on the genome and metagenome sequences as well as on nucleic acids in PDB. In some cases, crystallized RNAs had been modified in ways that resulted in their not being detected with Rfam’s parameters when we searched PDB sequences. In these cases, we lowered Rfam’s score threshold when searching PDB entries (SI Appendix, Table S1). PDB entries with a matching sequence are reported to the user, and the user manually selects a PDB entry to use, along with the appropriate chain. The sequence in this PDB entry is aligned along with the non-PDB riboswitches using Rfam’s parameters for the given riboswitch structure.
A nucleotide was classified as being close to the ligand, that is, being a key nucleotide, if any of its atoms is within a given distance of any ligand atom. The distances used were as follows: 2.5, 2.75, 3, 3.25, 3.5, 3.75, 4, 4.5, and 5 Å. Sometimes the same key nucleotides are determined for different distances, in which case the analysis is not repeated for redundant distances. If no nucleotides are within the distance (e.g., for 2.5 Å), the distance is skipped. Riboswitch sequences are divided into groups based on their key nucleotides.
All automated analysis is conducted on the nearest downstream gene from the riboswitch (i.e., the gene presumed to be regulated by the riboswitch). If the immediately downstream gene is farther than 700 bp, encoded in the opposite strand, or there is no downstream gene, the riboswitch is considered to have no regulated gene. To mitigate the effects of correlations between closely related riboswitch sequences, we applied the GSC algorithm in the Infernal package (56) to each riboswitch alignment. The resulting weights for each riboswitch were used to calculate gene frequencies.
Next, alignments of riboswitch groups are used as queries to automatically search for additional examples using Infernal, version 1.1 (56). If the riboswitch group represents a variant of the normal riboswitch, the search may uncover additional riboswitch sequences that were too diverged to find before. These searches can also uncover riboswitches corresponding to other groups. So, the newly predicted sequences are aligned to determine their key nucleotides, and only sequences with the appropriate key nucleotides for the group are retained. The search is performed on all intergenic regions (IGRs) contained in contigs that are at least 2 kb, to avoid low-quality IGRs in short contigs. IGRs that contain any degenerate nucleotides (letters other than A, C, G, or U) are skipped. Each IGR is extended by 50 bp on either side to account for inaccurate annotations of start codon positions.
It would be too computationally intensive to search for additional homologs of all of the riboswitch groups assembled for each of the distances chosen. We therefore first eliminated riboswitch groups with more than 500 sequences, reasoning that these already-large groups would not benefit much from finding additional members. For each riboswitch model, we selected up to 300 riboswitch groups for automated searches. Riboswitch groups were first sorted from smallest to largest core distances, and then from best to worst scores (see scoring below). The top 300 were selected. We tried performing additional rounds of automated searches but found that they did not noticeably improve results beyond the first automated search.
The third common property of previously known riboswitch groups with altered ligand specificity is that the groups are associated with different sets of genes. We therefore designed two strategies to automatically quantitate whether two sets of genes are significantly different, to focus our attention on the riboswitch groups that are most likely to reflect a change of ligand. In both strategies, we make the simplifying assumption that distinct conserved domains in the Conserved Domain Database (35) correspond to distinct biochemical functions. Both strategies thus attempt to quantitate the difference between the frequencies of conserved domains encoded by genes that are regulated by riboswitches in the group to be evaluated (group E) and the domain frequencies for the group containing the crystallized riboswitch (group C). We presume that riboswitches in the crystallized group bind the already-known ligand, because a crystallized RNA is likely to be well characterized. Therefore, if the two sets of genes are significantly different, group E is likely to contain riboswitches with altered ligand specificity. In the first method, based on relative entropy, the score is , where is a conserved domain, is the frequency of in the genes regulated by the crystallized group, and is the frequency in the evaluated group. If but , then is set to , a value that we chose by intuition. The second score is the negative of the logarithm of the likelihood of observing the frequencies of genes associated with group E, if the true distribution comes from group C, and is . We also calculated empirical P values by randomly sampling downstream conserved domains from the distribution , and computing the scores for each random sample. We found that these scores and P values were sometimes useful, although they did not reliably discriminate good from poor candidate groups. When riboswitch groups were sorted by scores, we used the minimum of P values of the two above statistics. Final decisions on promising riboswitch groups were made manually, and we found that the lower distances (e.g., ≤3 Å) tended to result in the best candidates.
Covariation in riboswitches was depicted based on the predictions of R-scape (57), version 0.2.1, with default parameters.
In-Line Probing.
DNA templates containing the RNA of interest whose expression was controlled by a T7 RNA polymerase (T7 RNAP) promoter were assembled by enzymatic (reverse transcriptase) extension of synthetic, overlapping single-stranded oligonucleotides. A list of oligonucleotides used in this study is in SI Appendix, Table S4. One or more G residues were added to the template in a position corresponding to the 5′ end of the RNA product to enable efficient transcription by T7 RNAP. Transcription was allowed to proceed for 4–16 h [80 mM Hepes-KOH (pH 7.5 at 23 °C), 24 mM MgCl2, 2 mM spermidine, 40 mM DTT; 100-µL reaction volume] after which the RNAs were purified via PAGE. The RNA was then excised from the gel and extracted via crush-soaking [200 mM NaCl, 10 mM Tris⋅HCl (pH 7.5 at 23 °C), 1 mM EDTA; 400-µL total volume] for 30 min. Following precipitation with ethanol and subsequent separation via centrifugation and removal of residual ethanol via rotary evaporation, the RNA was dephosphorylated (rapid alkaline phosphatase; Roche) and 5′-radiolabeled using T4 polynucleotide kinase [25 mM N-cyclohexyl-2-aminoethanesulfonic acid (pH 9.0 at 23 °C), 5 mM MgCl2, 3 mM DTT, 20 µCi of [γ-32P]ATP; 20-µL reaction volume] over 45 min. The RNA was then purified as described above. Approximately 5,000 cpm of RNA was incubated for 40 h at room temperature with the appropriate concentration of ligand in an in-line probing reaction mixture [20 mM MgCl2, 100 mM KCl, 50 mM Tris⋅HCl (pH 8.3 at 23 °C)]. The reaction products were then analyzed by PAGE and visualized using a phosphorimager. Dissociation constants were determined by varying the concentration of added ligand and quantifying the changes in band intensity at modulating sites. These data were then normalized between 0 and 1, plotted as fraction of RNA bound to ligand, and fit to a sigmoidal dose–response equation to determine the dissociation constant.
Supplementary Material
Acknowledgments
We thank members of the R.R.B. Laboratory for helpful discussions and Rob Bjornson for assistance with the Yale Life Sciences High Performance Computing Center (NIH Grant RR19895-02). M.E.S. was supported by NIH Cellular and Molecular Biology Training Grant T32GM007223. R.R.B. is a Howard Hughes Medical Institute Investigator and is also supported by NIH Grants GM022778 and DE022340.
Footnotes
The authors declare no conflict of interest.
Data deposition: The datasets reported in this paper have been deposited in breaker.yale.edu/variants (Datasets S1 and S2).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619581114/-/DCSupplemental.
References
- 1.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garst AD, Edwards AL, Batey RT. Riboswitches: Structures and mechanisms. Cold Spring Harb Perspect Biol. 2011;3(6):a003533. doi: 10.1101/cshperspect.a003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serganov A, Patel DJ. Molecular recognition and function of riboswitches. Curr Opin Struct Biol. 2012;22(3):279–286. doi: 10.1016/j.sbi.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peselis A, Serganov A. Themes and variations in riboswitch structure and function. Biochim Biophys Acta. 2014;1839(10):908–918. doi: 10.1016/j.bbagrm.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker JL, et al. Widespread genetic switches and toxicity resistance proteins for fluoride. Science. 2012;335(6065):233–235. doi: 10.1126/science.1215063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson JW, Atilho RM, Sherlock ME, Stockbridge RB, Breaker RR. Metabolism of free guanidine in bacteria is regulated by a widespread riboswitch class. Mol Cell. 2017;65(2):220–230. doi: 10.1016/j.molcel.2016.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dambach M, et al. The ubiquitous yybP-ykoY riboswitch is a manganese-responsive regulatory element. Mol Cell. 2015;57(6):1099–1109. doi: 10.1016/j.molcel.2015.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price IR, Gaballa A, Ding F, Helmann JD, Ke A. Mn2+-sensing mechanisms of yybP-ykoY orphan riboswitches. Mol Cell. 2015;57(6):1110–1123. doi: 10.1016/j.molcel.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furukawa K, et al. Bacterial riboswitches cooperatively bind Ni2+ or Co2+ ions and control expression of heavy metal transporters. Mol Cell. 2015;57(6):1088–1098. doi: 10.1016/j.molcel.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sudarsan N, et al. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321(5887):411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329(5993):845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson JW, et al. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat Chem Biol. 2013;9(12):834–839. doi: 10.1038/nchembio.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson JW, et al. Control of bacterial exoelectrogenesis by c-AMP-GMP. Proc Natl Acad Sci USA. 2015;112(17):5389–5394. doi: 10.1073/pnas.1419264112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kellenberger CA, et al. GEMM-I riboswitches from Geobacter sense the bacterial second messenger cyclic AMP-GMP. Proc Natl Acad Sci USA. 2015;112(17):5383–5388. doi: 10.1073/pnas.1419328112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol. 2004;11(1):29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 16.Kim JN, Roth A, Breaker RR. Guanine riboswitch variants from Mesoplasma florum selectively recognize 2′-deoxyguanosine. Proc Natl Acad Sci USA. 2007;104(41):16092–16097. doi: 10.1073/pnas.0705884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nahvi A, Barrick JE, Breaker RR. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 2004;32(1):143–150. doi: 10.1093/nar/gkh167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weinberg Z, et al. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 2010;11(3):R31. doi: 10.1186/gb-2010-11-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson JE, Jr, Reyes FE, Polaski JT, Batey RT. B12 cofactors directly stabilize an mRNA regulatory switch. Nature. 2012;492(7427):133–137. doi: 10.1038/nature11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Regulski EE, et al. A widespread riboswitch candidate that controls bacterial genes involved in molybdenum cofactor and tungsten cofactor metabolism. Mol Microbiol. 2008;68(4):918–932. doi: 10.1111/j.1365-2958.2008.06208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blount KF. 2013. Methods for treating or inhibiting infection by Clostridium difficile. US Patent 13/576,989.
- 22.Blount KF, et al. 2012. Flavin derivatives. US Patent 13/381,809.
- 23.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113(5):577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 24.Weinberg Z, et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35(14):4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nawrocki EP, et al. Rfam 12.0: Updates to the RNA families database. Nucleic Acids Res. 2015;43(Database issue) D1:D130–D137. doi: 10.1093/nar/gku1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432(7015):411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 27.Serganov A, et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11(12):1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith KD, et al. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol. 2009;16(12):1218–1223. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulshina N, Baird NJ, Ferré-D’Amaré AR. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat Struct Mol Biol. 2009;16(12):1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edwards AL, Batey RT. A structural basis for the recognition of 2′-deoxyguanosine by the purine riboswitch. J Mol Biol. 2009;385(3):938–948. doi: 10.1016/j.jmb.2008.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porter EB, Marcano-Velázquez JG, Batey RT. The purine riboswitch as a model system for exploring RNA biology and chemistry. Biochim Biophys Acta. 2014;1839(10):919–930. doi: 10.1016/j.bbagrm.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilbert SD, Reyes FE, Edwards AL, Batey RT. Adaptive ligand binding by the purine riboswitch in the recognition of guanine and adenine analogs. Structure. 2009;17(6):857–868. doi: 10.1016/j.str.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5(10):1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods Mol Biol. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- 35.Marchler-Bauer A, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43(Database issue) D1:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bendtsen JD, Kiemer L, Fausbøll A, Brunak S. Non-classical protein secretion in bacteria. BMC Microbiol. 2005;5:58. doi: 10.1186/1471-2180-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergé M, Moscoso M, Prudhomme M, Martin B, Claverys JP. Uptake of transforming DNA in Gram-positive bacteria: A view from Streptococcus pneumoniae. Mol Microbiol. 2002;45(2):411–421. doi: 10.1046/j.1365-2958.2002.03013.x. [DOI] [PubMed] [Google Scholar]
- 38.Gilbert SD, Love CE, Edwards AL, Batey RT. Mutational analysis of the purine riboswitch aptamer domain. Biochemistry. 2007;46(46):13297–13309. doi: 10.1021/bi700410g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson HR, Turnbough CL., Jr Role of the purine repressor in the regulation of pyrimidine gene expression in Escherichia coli K-12. J Bacteriol. 1990;172(6):3208–3213. doi: 10.1128/jb.172.6.3208-3213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mandal M, et al. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306(5694):275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 41.Butler EB, Xiong Y, Wang J, Strobel SA. Structural basis of cooperative ligand binding by the glycine riboswitch. Chem Biol. 2011;18(3):293–298. doi: 10.1016/j.chembiol.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang L, Serganov A, Patel DJ. Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Mol Cell. 2010;40(5):774–786. doi: 10.1016/j.molcel.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol Cell. 1999;3(4):495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- 44.Yarnell WS, Roberts JW. Mechanism of intrinsic transcription termination and antitermination. Science. 1999;284(5414):611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- 45.Sudarsan N, et al. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314(5797):300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- 46.Ruff KM, Strobel SA. Ligand binding by the tandem glycine riboswitch depends on aptamer dimerization but not double ligand occupancy. RNA. 2014;20(11):1775–1788. doi: 10.1261/rna.047266.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyer MM, et al. Challenges of ligand identification for riboswitch candidates. RNA Biol. 2011;8(1):5–10. doi: 10.4161/rna.8.1.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell. 2011;43(6):867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Leary NA, et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Markowitz VM, et al. IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res. 2014;42(Database issue):D568–D573. doi: 10.1093/nar/gkt919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Methé BA, et al. Human Microbiome Project Consortium A framework for human microbiome research. Nature. 2012;486(7402):215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meyer F, et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benson DA, et al. GenBank. Nucleic Acids Res. 2013;41(Database issue) D1:D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weinberg Z, et al. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat Chem Biol. 2015;11(8):606–610. doi: 10.1038/nchembio.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29(22):2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rivas E, Clements J, Eddy SR. A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nat Methods. 2017;14(1):45–48. doi: 10.1038/nmeth.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sherman EM, Esquiaqui J, Elsayed G, Ye JD. An energetically beneficial leader-linker interaction abolishes ligand-binding cooperativity in glycine riboswitches. RNA. 2012;18(3):496–507. doi: 10.1261/rna.031286.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kladwang W, Chou FC, Das R. Automated RNA structure prediction uncovers a kink-turn linker in double glycine riboswitches. J Am Chem Soc. 2012;134(3):1404–1407. doi: 10.1021/ja2093508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.