Oxidative DNA damage is an implacable consequence of aerobic metabolism and often exacerbated in inflammatory processes that use reactive oxygen species (ROS) both as signaling molecules and as chemical warfare against pathogens. An extensive body of work, recently reviewed in ref. 1, has highlighted the deleterious consequences of oxidative DNA damage, which involves oxidized nucleobases that, if left unrepaired, are either mutagenic or strong replication blockers. Most oxidative DNA damage is efficiently processed by DNA repair pathways, primarily base excision repair (BER), the molecular details of which are generally well understood (2). However, an emerging area of research posits that certain oxidative DNA lesions and their associated repair complexes are intermediates in a signaling transduction cascade that uses ROS as secondary messengers to ultimately effect transcriptional regulation (3–7). In PNAS, Fleming et al. (8) reinforce these notions by describing a compelling mechanism by which 8-oxoguanine (OG), a canonical oxidative DNA damage product, when occurring in guanine-rich, G-quadruplex–forming promoter sequences, directly up-regulates transcription of the downstream gene.
Evidence for ROS acting as signaling molecules has been around for more than a decade (9). In the hypoxia field, it has been increasingly appreciated that the expression of hypoxia-inducible genes [e.g., vascular endothelial growth factor (VEGF)] depends, in part, on a controlled oxidative DNA damage and repair cycle (3, 6). More intriguingly, even in normoxic conditions, the expression of estrogen- or androgen-responsive genes also requires DNA oxidation and repair (10, 11). Fleming et al. provide an overarching mechanistic framework for these studies, by highlighting a connection between DNA repair and the ability of certain DNA sequences to fold into G quadruplexes (G4s).
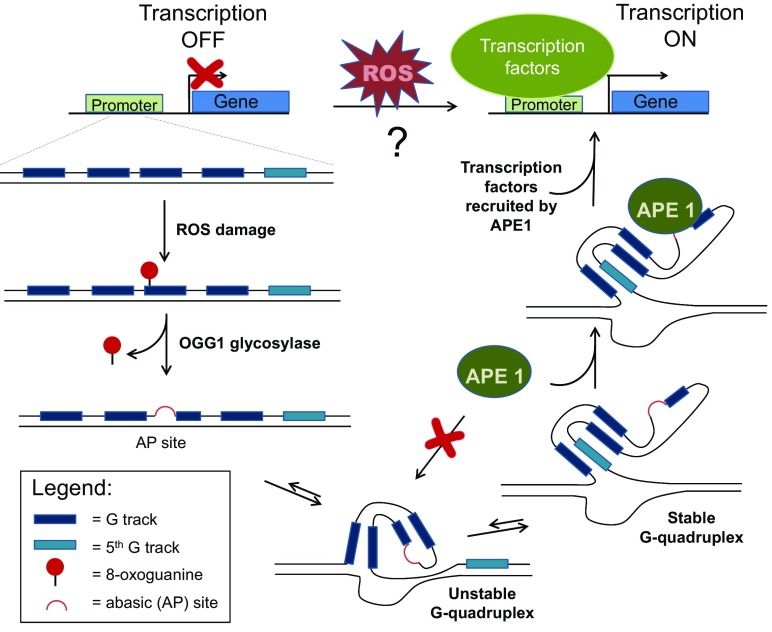
Non-Watson–Crick secondary structures of nucleic acids, and in particular, G4 structures in DNA is an area of extremely active research (12–16). Although G4 structures were initially described in the context of telomeres, recent genomic data revealed tens of thousands of sequences throughout the genome (14, 17) that, in principle, could form G4 structures, called potential G-quadruplex–forming sequences (PQS). Many of these sequences, when folded as G4s, can bind with high-affinity cellular proteins or transcription factors (16), but the relevance and physiological role of such interactions are still actively investigated. In PNAS, Fleming et al. advance the hypothesis that a subset of these PQS can function as sensors of oxidative stress (8). Using the VEGF PQS sequence as a model, the authors provide a mechanistic connection between two previously known, but seemingly disparate characteristics of the VEGF promoter: (i) oxidative lesions (i.e., OG) in the promoter region lead to an increased expression level of VEGF; (ii) the promoter features a PQS composed of five guanine tracks (G tracks). Specifically, Fleming et al. propose that OG formation in the G-rich regions of the VEGF PQS recruits 8-oxoguanine DNA glycosylase-1 (OGG1), which removes OG and generates an abasic (AP) site. When present in a G track, the AP site destabilizes the G4 fold and induces the formation of a new G4 that involves the fifth G track, and loops out the G track containing the AP site. This new conformation facilitates the binding of apurinic/apyrimidinic endonuclease 1 (APE1) to the AP site; APE1 is then well positioned to stimulate binding of SP1 transcription factor, which activates transcription (Fig. 1).
Fig. 1.
The ability of ROS generated during aerobic metabolism, hypoxia, or oxidative stress to turn on the expression of certain genes has been previously documented. However, the mechanistic details of this process are unclear. In PNAS, Fleming et al. describe a mechanism by which oxidative DNA damage can activate transcription of genes (e.g., VEGF, NTHL1) that contain potential G-quadruplex (G4)–forming sequences with five G tracks in their promoters. ROS damage in the G-rich regions of the promoter generates 8-oxoguanine, which is removed by the BER glycosylase OGG1 to form an AP site. The rearrangement of the DNA strand into a G4 structure prevents APE1 lyase, the second BER enzyme, to access the AP site. However, a more stable G4 can form by involving the fifth G track and looping out the AP site, which can now be bound by APE1. Through its regulatory domains, APE1 then recruits transcription factors to initiate transcription.
The workhorse for the study is a luciferase reporter plasmid that encodes two luciferase genes: (i) a Renilla luciferase (Rluc) with a synthetically accessible promoter; (ii) a constitutive firefly luciferase as an internal standard. After incorporating synthetic oligonucleotides containing OG (or other lesions or controls) at defined sites in the Rluc promoter, the reporter plasmid is transfected into relevant cells and the level of expression of the two luciferase genes is evaluated at 48 h from luminescence measurements. The assay is very robust, as highlighted by the use throughout the work of 95% confidence intervals as error bars, instead of the common SD error bars.
Fleming et al. found that a unique OG lesion, placed at five different positions in the VEGF PQS promoter element, led to a two to three times increase in transcription over 48 h. The positions chosen reflected the most oxidation-prone guanines in the PQS region (18). The transcription induction depended on the OGG1 glycosylase; OGG1 KO cells showed no transcriptional changes with any of the constructs. OGG1 initiates BER by removing OG, generating an AP site. Indeed, transcriptional activation was restored when tetrahydrofuran, a stable AP site analog, was used instead of OG, even in OGG1 KO cells. During BER, the AP site is processed by APE1 [also known as redox effector factor (Ref-1)], a DNA lyase that nicks the DNA 5′ to the AP site. The authors demonstrate that the transcription induction required APE1 binding to the AP site; siRNA down-regulation of APE1 levels or cotreatment with an APE1 inhibitor abrogated the transcription activation. The inhibitor used blocks the lyase activity of APE1 but not its binding to the AP site, suggesting that the binding and residence time of APE1 at the AP site within the promoter region was responsible for transcription activation. Further support for this hypothesis was provided by using constructs containing poorly cleavable phosphorothioate backbones 5′ to the AP sites; in this case, the transcriptional activation was substantially higher than that observed with canonical AP substrates.
The second part of the study investigated the contribution of G4 formation in the VEGF promoter element to transcriptional activation, and its mechanistic connection to OG repair by BER. The authors acknowledge the significant challenges of this undertaking because it is known that SP1, the primary transcription factor that binds this promoter element, can bind with similar affinity to both duplex and G4 DNA (19). Two modified sequences were tested, both with or without OG modifications: one that still bound SP1 as a duplex but did not form a G4, and a double-negative control (no SP1 binding, no G4 formation). The latter promoter sequence yielded, as expected, no transcriptional activation. The former sequence, however, also produced a negative result, highlighting the importance of G4-forming ability for transcription activation, particularly when OG residues were present.
In a previous study, the authors demonstrated that the fifth G track of the VEGF PQS helped reconstitute a G4 structure when one of the four upstream G tracks contained a damaged base or AP site (18). The G track containing damage was looped and replaced by the fifth G track to form a new G4 structure. The current study demonstrates that the presence of the fifth G track contributes substantially to the transcriptional activation; additionally, as evidenced by DNA melting temperatures and circular dichroism spectra, an AP site destabilized G4 formation in a sequence with four G tracks; a fifth G track, however, restored the stability of the G4 fold. Nevertheless, some residual level of transcription activation was seen with the constructs containing only four G runs, suggesting that APE1 could still bind, but less tightly or only transiently. Furthermore, the formation of the G4 structure was important independently of APE1, perhaps as a scaffold for other transcription factors; in a non-G4 promoter sequence, with a noncleavable AP site, transcriptional activation was only 1.5 times, despite the long residence time of APE1.
Fleming et al. also include an abbreviated analysis of the endonuclease III-like protein 1 (NTHL1) PQS promoter sequence. NTHL1 is a broad-specificity glycosylase involved in the BER of many oxidized bases, primarily pyrimidines (20). Similar to the VEGF analysis, the introduction of OG or AP sites at two distinct positions in the NTHL1 PQS led to a substantial (four to seven times) increase in transcription. This result is significant because it suggests a direct way by which oxidative DNA damage may regulate the expression of DNA repair genes. Given the abundance of PQS in the promoter regions throughout the genome (12, 13), the generality of the proposed mechanism may be very likely. Supporting this view is the fact that Fleming et al. also provide a more comprehensive mechanistic explanation for the DNA oxidation and BER-dependent transcriptional activation reported in several cellular studies (4, 10, 11, 21). In each case, the target gene investigated turned out to contain a PQS in its promoter (8).
Although the mechanism of Fleming et al. is independent of the process that generates OG, the source of an essentially site-specific OG in a promoter PQS inside the cell remains an open question. Nonspecific oxidation of G by ROS generated from mitochondria or from inflammatory processes may be too erratic to constitute the primary mechanism. Chromatin-remodeling enzymes such as flavin-dependent lysine demethylases (e.g., LSD1) also generate the ROS hydrogen peroxide as part of their catalytic cycle (10, 11). As histone demethylation happens in the vicinity of DNA, such reactions may produce more localized OG residues that can participate in transcriptional activation. Additionally, the long-range charge transport properties of DNA may play a role. As shown by the Barton group (22), redox electrons can “tunnel” through the pi-stacked bases of intact DNA duplex over distances of more than 200 Å. Intriguingly, the exits from such “tunnels” are often tracks of guanines, with the 5′-most guanine in a G track being most susceptible to oxidation (22). This mechanism may allow, at least initially, the focusing of a diffuse oxidative insult to select guanines in PQS in the genome, which, in light of the Fleming et al. study (8), could initiate an oxidative stress-induced transcriptional response.
In sum, Fleming et al. describe a cogent mechanism by which oxidative stress, through oxidative DNA damage and BER, can directly stimulate transcription of genes that contain five G-track PQS in their promoters. However, more importantly, the proposed mechanism suggests the paradigm-shifting notion that OG, when occurring in certain PQS, may constitute an epigenetic marker for active transcription.
Footnotes
The author declares no conflict of interest.
See companion article on page 2604 in issue 10 of volume 114.
References
- 1.Yu Y, Cui Y, Niedernhofer LJ, Wang Y. Occurrence, biological consequences, and human health relevance of oxidative stress-induced DNA damage. Chem Res Toxicol. 2016;29(12):2008–2039. doi: 10.1021/acs.chemrestox.6b00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shafirovich V, Geacintov NE. Removal of oxidatively generated DNA damage by overlapping repair pathways. Free Radic Biol Med. 2016 doi: 10.1016/j.freeradbiomed.2016.10.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pastukh V, et al. An oxidative DNA “damage” and repair mechanism localized in the VEGF promoter is important for hypoxia-induced VEGF mRNA expression. Am J Physiol Lung Cell Mol Physiol. 2015;309(11):L1367–L1375. doi: 10.1152/ajplung.00236.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan L, et al. Oxidized guanine base lesions function in 8-oxoguanine DNA glycosylase-1-mediated epigenetic regulation of nuclear factor κB-driven gene expression. J Biol Chem. 2016;291(49):25553–25566. doi: 10.1074/jbc.M116.751453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aguilera-Aguirre L, et al. Whole transcriptome analysis reveals an 8-oxoguanine DNA glycosylase-1-driven DNA repair-dependent gene expression linked to essential biological processes. Free Radic Biol Med. 2015;81:107–118. doi: 10.1016/j.freeradbiomed.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillespie MN, Pastukh VM, Ruchko MV. Controlled DNA “damage” and repair in hypoxic signaling. Respir Physiol Neurobiol. 2010;174(3):244–251. doi: 10.1016/j.resp.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhakat KK, Mantha AK, Mitra S. Transcriptional regulatory functions of mammalian AP-endonuclease (APE1/Ref-1), an essential multifunctional protein. Antioxid Redox Signal. 2009;11(3):621–638. doi: 10.1089/ars.2008.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleming AM, Ding Y, Burrows CJ. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc Natl Acad Sci USA. 2017;114(10):2604–2609. doi: 10.1073/pnas.1619809114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunelle JK, et al. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab. 2005;1(6):409–414. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Perillo B, et al. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319(5860):202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 11.Yang S, et al. KDM1A triggers androgen-induced miRNA transcription via H3K4me2 demethylation and DNA oxidation. Prostate. 2015;75(9):936–946. doi: 10.1002/pros.22977. [DOI] [PubMed] [Google Scholar]
- 12.Varizhuk A, et al. The expanding repertoire of G4 DNA structures. Biochimie. 2017;135:54–62. doi: 10.1016/j.biochi.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Armas P, David A, Calcaterra NB. Transcriptional control by G-quadruplexes: In vivo roles and perspectives for specific intervention. Transcription. 2017;8(1):21–25. doi: 10.1080/21541264.2016.1243505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hänsel-Hertsch R, et al. G-quadruplex structures mark human regulatory chromatin. Nat Genet. 2016;48(10):1267–1272. doi: 10.1038/ng.3662. [DOI] [PubMed] [Google Scholar]
- 15.Valton A-L, Prioleau M-N. G-quadruplexes in DNA replication: A problem or a necessity? Trends Genet. 2016;32(11):697–706. doi: 10.1016/j.tig.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Mishra SK, Tawani A, Mishra A, Kumar A. G4IPDB: A database for G-quadruplex structure forming nucleic acid interacting proteins. Sci Rep. 2016;6:38144. doi: 10.1038/srep38144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gray LT, Vallur AC, Eddy J, Maizels N. G quadruplexes are genomewide targets of transcriptional helicases XPB and XPD. Nat Chem Biol. 2014;10(4):313–318. doi: 10.1038/nchembio.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleming AM, Zhou J, Wallace SS, Burrows CJ. A role for the fifth G-track in G-quadruplex forming oncogene promoter sequences during oxidative stress: Do these “spare tires” have an evolved function? ACS Cent Sci. 2015;1(5):226–233. doi: 10.1021/acscentsci.5b00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raiber E-A, Kranaster R, Lam E, Nikan M, Balasubramanian S. A non-canonical DNA structure is a binding motif for the transcription factor SP1 in vitro. Nucleic Acids Res. 2012;40(4):1499–1508. doi: 10.1093/nar/gkr882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prasad A, Wallace SS, Pederson DS. Initiation of base excision repair of oxidative lesions in nucleosomes by the human, bifunctional DNA glycosylase NTH1. Mol Cell Biol. 2007;27(24):8442–8453. doi: 10.1128/MCB.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antoniali G, et al. SIRT1 gene expression upon genotoxic damage is regulated by APE1 through nCaRE-promoter elements. Mol Biol Cell. 2014;25(4):532–547. doi: 10.1091/mbc.E13-05-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnold AR, Grodick MA, Barton JK. DNA charge transport: From chemical principles to the cell. Cell Chem Biol. 2016;23(1):183–197. doi: 10.1016/j.chembiol.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]