Significance
Understanding of myelin diseases and development of new treatment options are at least partly hampered by the limited availability of human oligodendrocytes. Induced pluripotent stem cells (iPSC) may be an ideal tool to circumvent this problem; however, rapid and efficient protocols to generate oligodendrocytes from human iPSC are currently lacking. The induction of the transcription factors SOX10, OLIG2, and NKX6.2 in iPSC-derived neural progenitors accelerates oligodendroglial differentiation significantly resulting in up to 70% of O4+ oligodendrocytes within 28 d. These oligodendrocytes myelinate the CNS during development and after demyelination, and are suitable for pharmacological screens and disease modeling. The strategy presented herein will markedly facilitate the studying of human myelin diseases and the development of screening platforms for drug discovery.
Keywords: human iPSC, oligodendrocytes, forward patterning, myelination, disease modeling
Abstract
Rapid and efficient protocols to generate oligodendrocytes (OL) from human induced pluripotent stem cells (iPSC) are currently lacking, but may be a key technology to understand the biology of myelin diseases and to develop treatments for such disorders. Here, we demonstrate that the induction of three transcription factors (SOX10, OLIG2, NKX6.2) in iPSC-derived neural progenitor cells is sufficient to rapidly generate O4+ OL with an efficiency of up to 70% in 28 d and a global gene-expression profile comparable to primary human OL. We further demonstrate that iPSC-derived OL disperse and myelinate the CNS of Mbpshi/shi Rag−/− mice during development and after demyelination, are suitable for in vitro myelination assays, disease modeling, and screening of pharmacological compounds potentially promoting oligodendroglial differentiation. Thus, the strategy presented here to generate OL from iPSC may facilitate the studying of human myelin diseases and the development of high-throughput screening platforms for drug discovery.
Oligodendrocytes (OL) play a key role in myelin-related diseases, including multiple sclerosis (MS), leukodystrophies, as well as periventricular leukomalacia, and there is an increasing awareness of their potential role in neurodegenerative diseases (e.g., multiple system atrophy and amyotrophic lateral sclerosis) or traumatic spinal cord injury (1–6). OL form and maintain the myelin sheaths that insulate axons and organize the distribution of axonal voltage-gated ion channels, a prerequisite for conduction of action potentials and trophic support of axons. Demyelination in MS contributes to axonal damage and disease progression (7). Immunosuppressive or immunomodulatory therapies, including complete ablation of the immune system by radiation and chemotherapy, prevent new inflammatory lesions that underlie clinical relapses but do not arrest disease (8). Therapies promoting remyelination represent a promising new treatment strategy to protect and restore axonal integrity and neurologic function (4). The development of such therapeutics is hampered, at least in part, by the limited availability of human OL. Stem cell technologies are a promising tool to circumvent this problem. Availability of OL derived from human induced pluripotent stem cells (iPSC) would permit studies to delineate mechanisms regulating repair by endogenous myelin lineage cells and provide a source of autologous cells for replacement therapy. Such cells would also provide new opportunities to identify pathological mechanisms underlying de- or dysmyelinating diseases. However, to date only a few protocols have resulted in the successful generation of human stem cell-derived OL. Furthermore, these protocols require long culture periods (70–150 d) to obtain O4+ OL and show limited efficiency (9–12).
Here, we describe an efficient strategy that facilitates and optimizes the generation of human O4+ OL from human iPSC-derived neural progenitor cells (NPC) (13). This approach yields up to 70% O4+ OL within 28 d of differentiation, using a combination of three transcription factors (TF), namely SOX10, OLIG2, and NKX6.2. Furthermore, 30% of the O4+ iPSC-derived OL (iOL) differentiate into mature myelin basic protein-positive (MBP+) OL within 7 additional days. The global gene-expression pattern of O4+ OL is comparable to that of human primary OL (pOL). The induced human iOL produce myelin-like structures around nanofibers or iPSC-derived neurons. After transplantation into MBP-deficient shiverer mice (Shi/Shi Rag2−/−) iOL disperse widely and myelinate host axons in the developing central nervous system (CNS), as well as the adult demyelinated spinal cord. Furthermore, iOL can be exploited for disease modeling and to test the potential of pharmacological compounds in promoting oligodendroglial differentiation.
Results
Identification of OL Lineage Inducing TF in Human NPC.
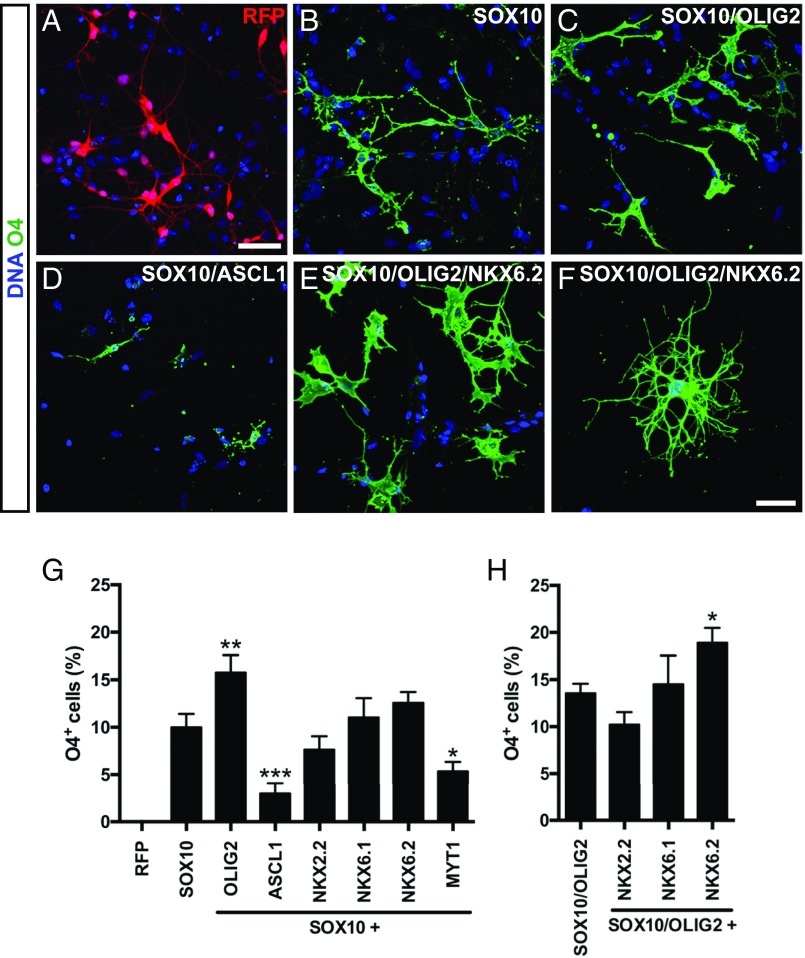
Human PSC present a valuable source for the generation of myelinogenic OL for research and autologous cell-replacement therapies (9–12). Currently, available protocols are technically challenging and time-consuming, limiting the application of these cells for research and regenerative medicine. In these protocols, NPC are rapidly and efficiently derived from human PSC, which is followed by the rate-limiting step of oligodendroglial specification and differentiation. Therefore, we first aimed to identify TF accelerating the oligodendroglial specification and differentiation from human iPSC-derived NPC. We performed literature data mining and selected a set of seven TF, which are enriched in OL compared with other neural lineages (2, 14), and are required for oligodendroglial specification (15, 16): ASCL1, MYT1, NKX2.2, NKX6.1, NKX6.2, OLIG2, and SOX10. Coding sequences for these proteins, as well as red fluorescent protein (RFP), were individually cloned into a tetracycline (tet)-inducible lentiviral vector. Human iPSC-derived NPC, which can be frozen and cost-efficiently expanded, as previously described (17), were transduced with a combination of lentiviruses expressing one of the TF candidates and the reverse tet-controlled transactivator (rtTA). Among all TF candidates, only SOX10 was capable of inducing O4 (9.99 ± 0.81%), a highly specific marker of late-stage oligodendroglial progenitor cells (OPC) and OL, after 14 d of exposure (Fig. 1B). Controls, including NPC either uninfected or infected with RFP only, did not yield any O4+ cells (Fig. 1A). We subsequently determined the oligodendroglial induction capacity of SOX10 in combination with any of the remaining six TF. We identified OLIG2 as a factor that substantially increased the SOX10-mediated oligodendroglial lineage commitment (Fig. 1 C and G), whereas ASCL1 and MYT1 significantly decreased the number of O4+ cells (Fig. 1 D and G). The combination of SOX10 and OLIG2 with NKX6.2, a TF associated with oligodendroglial maturation, significantly increased further the portion of O4+ cells (Fig. 1 E, F, and H). Thus, we concluded that the ectopic expression of SOX10, OLIG2, and NKX6.2 (subsequently referred to as SON) was the most efficient combination of TF tested here to induce OL from iPSC-derived NPC.
Fig. 1.
Screening for oligodendroglial lineage inducing TF in human NPC. Human iPSC-derived NPC were infected with individual OL-specific TFs or RFP control virus. (A–F) OL-lineage commitment of infected iPSC-derived NPC was analyzed 14 d after transgene induction by immunostaining using the OL-specific antibody O4 (green). Nuclei were counterstained with Hoechst (blue). (A) Control cultures did not express the O4 epitope. (B) SOX10 was the only tested TF inducing O4+ OL. (C) Addition of OLIG2 enhanced the OL-lineage commitment, whereas (D) ASCL1 led to a decreased number of O4+ iOL. (E) Coexpression of SOX10, OLIG2, and NKX6.2 increased the number of O4+ cells, (F) accompanied by the appearance of iOL with a more mature oligodendroglial morphology. (Scale bars, 50 µm in A–E and 25 µm in F.) (G and H) Quantification of O4+ iOL over all cells with indicated TF combinations 2 wk after transgene induction. Data are presented as mean of replicates from three independent experiments + SD. One-way ANOVA with Bonferroni’s multiple comparisons test was used as statistical test (*P < 0.05, **P < 0.01, ***P < 0.001).
Ectopic Expression of TF Accelerates Oligodendrogenesis from iPSC–NPC.
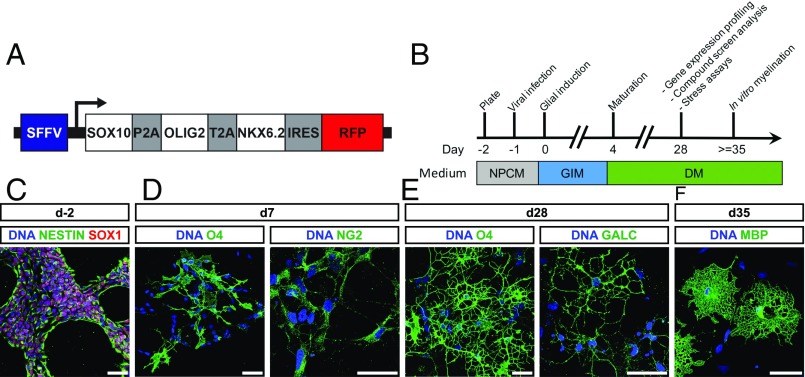
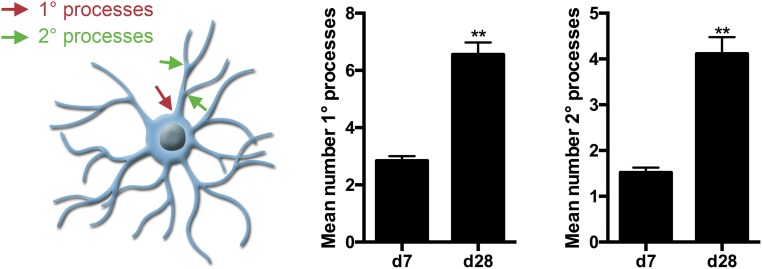
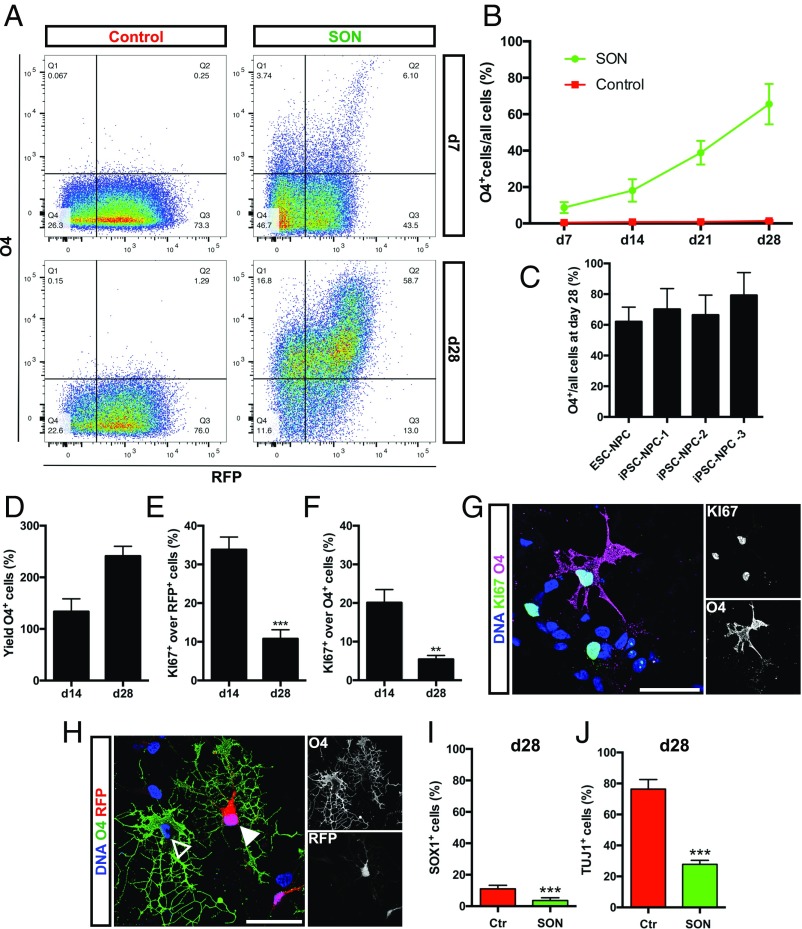
To further enhance the generation of human iOL, we generated a polycistronic lentiviral expression vector containing SON and RFP as a reporter gene under control of the retroviral spleen focus-forming virus (SFFV) promoter (Fig. 2A). iPSC-derived NPC (13) were infected with SON/RFP-expressing lentivirus (Fig. 2B). After induction of SON, a two-step differentiation protocol was sufficient to derive increasing numbers of iOL over 28 d (Fig. 2B). One day after viral transduction, NPC medium was replaced by glial induction medium (GIM) containing different supplements, including PDGF, SAG (smoothened agonist), and thyroid hormone (Triiodo-l-Thyronine, T3; 10 ng/µL). After 4 d, this medium was replaced by differentiation medium (DM) containing higher concentrations of T3 (60 ng/µL) and lacking PDGF and SAG. To ensure the reproducibility of our protocol, all experiments were performed with four independent NPC lines derived from three different iPSC lines and one embryonic stem cell (ESC) line. All NPC lines homogeneously expressed the neural stem cell marker SOX1 and NESTIN (Fig. 2C). Seven days after SON induction, OL cultures comprised mainly NG2+ glial progenitor cells (82.26 ± 2.36%) together with O4+ cells (8.7 ± 3.0%) (Fig. 2D). At day 28 after SON induction, the majority of cells expressed the OL marker O4 (65.5 ± 11.1%) and presented with a more complex morphology identified by an increased number of primary and secondary processes (Fig. S1). Additionally, cells started to express more mature oligodendroglial markers, like GALC (19.19 ± 1.46%) and MBP (30.37 ± 7.87% of O4+ cells) by days 28 and 35, respectively (Fig. 2 E and F; see also Fig. 5 D and E).
Fig. 2.
SON induce a rapid and efficient oligodendroglial lineage commitment. (A) Schematic presentation of the lentiviral expression vector used for the polycistronic expression of SON. (B) Schematic summary of the differentiation protocol using NPCM, GIM, and DM. (C–F) Representative immunofluorescence images of different NPC and OL markers during differentiation. Nuclei were counterstained with Hoechst (blue). (C) iPSC-derived NPC homogenously expressed the neural progenitor marker NESTIN (green) and SOX1 (red). (D) Seven days after transgene induction, NG2+ and O4+ oligodendroglial lineage cells were detected. (E) By day 28, iOL expressed the O4-epitope, the more mature OL marker GALC, and presented with a branched morphology. (F) Further maturation led to the emergence of MBP+ mature iOL forming myelin sheaths. (Scale bars, 50 µm.)
Fig. S1.
iOL develop increased numbers of primary and secondary processes during differentiation. To analyze morphological changes of O4+ iOL during differentiation, mean numbers of primary and secondary processes per cell body were quantified at day 7 and day 28 of differentiation. Mean numbers of primary and secondary processes significantly increased during differentiation, demonstrating a more complex morphology at day 28. Data are presented as mean of replicates from three independent differentiation experiments, each using an independent iPSC-derived NPC line + SD. Process numbers of at least 100 O4+ cells were quantified per differentiation. Student’s t test was performed for statistical analysis (**P < 0.01).
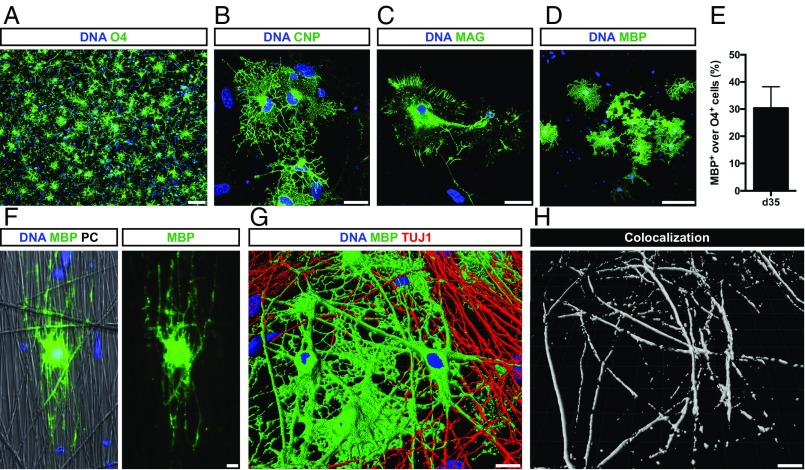
Fig. 5.
iOL differentiate into mature OL and ensheath iPSC-derived neurons in vitro. (A) Thirty-five days after transgene induction, O4+ iOL presented a branched morphology typical for mature OL and (B–D) expressed the mature oligodendroglial markers CNP, MAG, and MBP. (E) Quantification of mature MBP+ iOL over all O4+ iOL. Data are presented as mean of replicates from four independent differentiation experiments + SD. (F) Immunostaining of iOL 14 d after replating on 3D nanofiber scaffolds illustrating the formation of myelin sheaths around nanofibers. Nuclei are counterstained with Hoechst. (G and H) Human in vitro myelination assay: coculture of O4+ iOL purified at day 21 by MACS with iPSC-derived neurons for 3 wk. (G) Three-dimensional reconstruction of confocal images for MBP (green) and the neuronal marker TUJ1 (red), suggesting wrapping of axons. Nuclei were counterstained with Hoechst (blue). (H) Three-dimensional illustration of MBP and TUJ1 colocalization (white) from the same detail. [Scale bars, 100 µm (A), 20 µm (B and C), 50 µm (D), and 10 µm (F and H).]
To assess the kinetics, efficiency, and yield of SON-mediated oligodendroglial lineage specification, we conducted weekly flow cytometry analyses of the O4 epitope expression during differentiation (Fig. 3A). As a control, NPC were infected with RFP expressing lentivirus and cultured under the same differentiation conditions. All NPC lines tested were found to perform similarly with respect to iOL generation, starting from 8.7 ± 3.0% O4+ cells at day 7 to 65.5 ± 11.1% O4+ cells by day 28 (Fig. 3B). In contrast, only 1.4 ± 0.5% O4+ cells were identified in RFP-transduced cell cultures (Fig. 3B). The protocol was highly efficient and reproducible among all cell lines, illustrated by the quantification of O4+ cells at day 28, ranging from 62.1 ± 9.5% (ESC–NPC) to 79.0 ± 14.8% (iPSC–NPC-3) (Fig. 3C).
Fig. 3.
Quantification of oligodendroglial lineage cells after SON induction. (A) Representative flow cytometry analyses for the expression of O4 and RFP in control and SON cultures 7 and 28 d after transgene induction. (B) Quantification of O4+ cells in control and SON cultures 1 to 4r wk after transgene induction. Data are presented as mean of replicates from four independent experiments, each using NPC derived from an independent human PSC line + SD. (C) Quantification of O4+ iOL at day 28 derived from one human ESC and three independent iPSC lines. Data are presented as mean of replicates from three to five independent differentiation experiments per cell line + SD. (D) Quantification of O4+ iOL yields at days 14 and 28 after transduction. Data are presented as mean of replicates from three independent differentiation experiments, each using an independent iPSC-derived NPC line + SD. (E) Quantification of Ki-67+ transgene-expressing cells (RFP) and of (F) Ki-67+/O4+ iOL at days 14 and 28 after induction. Data are presented as mean of replicates from three independent differentiation experiments + SD (**P < 0.01, ***P < 0.001). (G) Immunostaining of iOL for O4 (purple) and the proliferation marker Ki-67 (green) at day 14 after transduction. (H) Representative immunofluorescence image of O4+ iOL (green) 28 d after transduction either expressing (filled arrowhead) or silencing (empty arrowhead) the transgenes. (Scale bars, 40 μm.) (I) Quantification of SOX1+ iPSC-derived NPC and (J) TUJ1+ neurons in control and SON cultures at day 28. Data are presented as mean of replicates from three independent differentiation experiments + SD. Student’s t test was performed for statistical analysis (***P < 0.001).
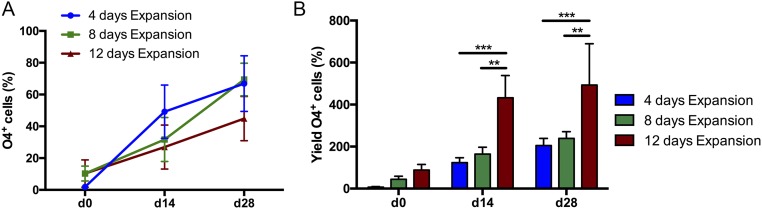
Yields of O4+ iOL (total O4+ cell number/starting NPC cell number) ranged from 133.70 ± 24.83% at day 14 to 241.20 ± 19.07% at day 28 (Fig. 3D). These data suggest an expansion of SON-expressing cells during our differentiation protocol. Identification of proliferative cells using Ki-67 revealed a proliferation rate of 35% among RFP+ cells at day 14, which declined to 10% by day 28, confirming that the transgene-expressing cell population further expanded during differentiation (Fig. 3E). Interestingly, the proliferation capability was retained in 20% of O4+ iOL at day 14 and diminished to 5% by day 28 (Fig. 3 F and G). Next, we tested whether iOL cultures can be expanded by a prolonged exposure to a glial expansion medium (GEM) postlentiviral transduction. Compared with GIM, GEM additionally contained FGF2 (5 ng/mL) and completely lacked IGF-1. SON-transduced NPC were cultured in GEM for either 4, 8, or 12 d and subsequently transferred to DM for an additional 28 d. FACS analyses of the iOL cultures after 0, 14, and 28 d in DM revealed that although exposure to GEM for 12 d reduced the differentiation efficiency (Fig. S2A), yields of O4+ iOL were significantly increased (Fig. S2B).
Fig. S2.
Impact of prolonged expansion of SON transduced NPC in GEM on oligodendroglial differentiation efficiencies and yields. SON-transduced NPC were expanded in GEM for either 4 (blue), 8 (green), or 12 d (red) and subsequently transferred to DM for an additional 28 d. FACS analyses at days 0, 14, and 28 of differentiation revealed a trend to decreased differentiation efficiency after 12 d of expansion (red curve) (A), whereas yields of O4+ cells (red columns) were significantly increased (B). Data are presented as mean of replicates from three independent experiments, each using an independent iPSC-derived NPC line + SD. Two-way ANOVA with Tukey’s multiple comparisons test was performed for statistical analysis (**P < 0.01, ***P < 0.001).
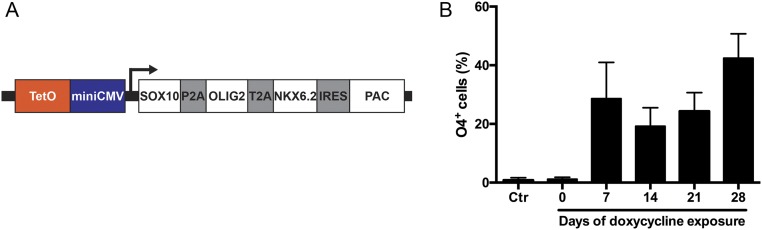
Furthermore, flow cytometry analyses exhibited the presence of an O4+/RFP− cell population in SON-transduced cultures (Fig. 3A), which comprised up to 50% of the O4+ cell population and which could be confirmed by immunocytochemical (ICC) analysis (Fig. 3H), suggesting transgene silencing in a subset of iOL. To analyze whether iOL become independent from transgene expression during differentiation, we generated a tet-inducible lentiviral expression vector containing SON (Fig. S3A). Quantification of O4+ cells at day 28 of differentiation revealed that induced expression of SON for 7 d is already sufficient to obtain a stable and transgene independent oligodendroglial lineage commitment (Fig. S3B). Interestingly, longer expression of SON (14 and 21 d) did not significantly enhance the differentiation efficiency. Constitutive tet-controlled expression of SON for 28 d demonstrated the highest differentiation efficiency (Fig. S3B), suggesting that a subset of O4+ cells was still dependent on ectopic expression of SON.
Fig. S3.
Analysis of iOL transgene dependency using a tet-inducible lentiviral expression construct. (A) Tet-inducible SON was generated by cloning a polycistronic SON cassette into pHAGE, a third-generation lentiviral expression vector. An IRES-PAC cassette was introduced following the SON expression cassette allowing optional puromycin selection. (B) NPC were cotransduced with lentiviruses expressing tet-inducible SON and lentiviruses expressing rtTA. Expression of SON was induced by doxycycline for different time periods including 0 d as a negative control and 28 d as a positive control. Oligodendroglial lineage commitment was assessed by FACS analyses at day 28. Transduced NPC populations without doxycycline exposure (0) demonstrated comparable differentiation efficiencies as nontransduced NPC populations (Ctr), confirming the tight control of SON expression. Constitutive expression of SON (28 d) resulted in an increased differentiation efficiency compared with transient SON expression (7, 14, 21 d), indicating that a subpopulation of O4+ iOL was dependent on the ectopic expression of SON.
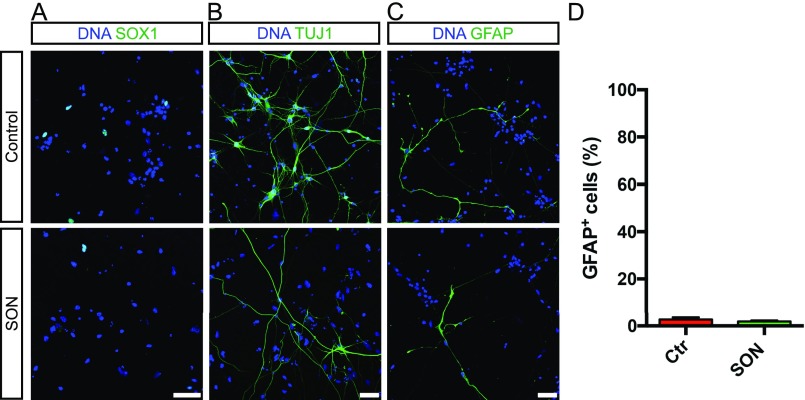
Next, we determined the influence of SON overexpression on the lineage commitment of NPC. ICC analysis of SON infected NPC cultures compared with RFP-infected control cultures at day 28 revealed a decreased number of SOX1+ NPC (Fig. 3I and Fig. S4A) and a significant switch from neuronal to oligodendroglial cell fate (Fig. 3J and Fig. S4B). In contrast, the astroglial lineage commitment was not affected (Fig. S4 C and D).
Fig. S4.
ICC analysis of the lineage commitment of SON-transduced and control NPC at day 28. The neural lineage commitment of control and SON-transduced NPC was assessed by SOX1 (A), TUJ1 (B), and GFAP (C) immunostaining at differentiation day 28. (Scale bars, 50 µm.) (D) Quantification of GFAP+ cells revealed no differences in the differentiation efficiencies in control and SON-transduced NPC populations. Data are presented as mean of replicates from three independent differentiation experiments + SD. Student’s t test was performed for statistical analysis.
Global Gene-Expression Profiling Demonstrates That iOL Resemble Primary Human Adult OL.
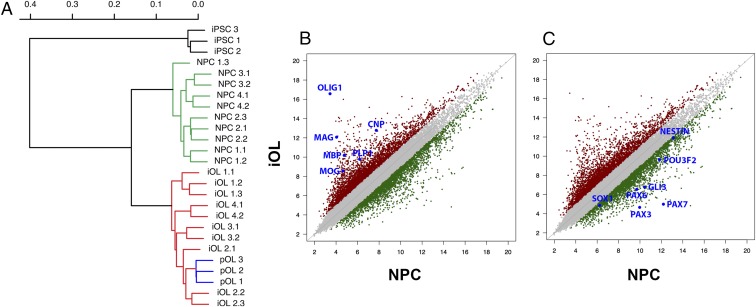
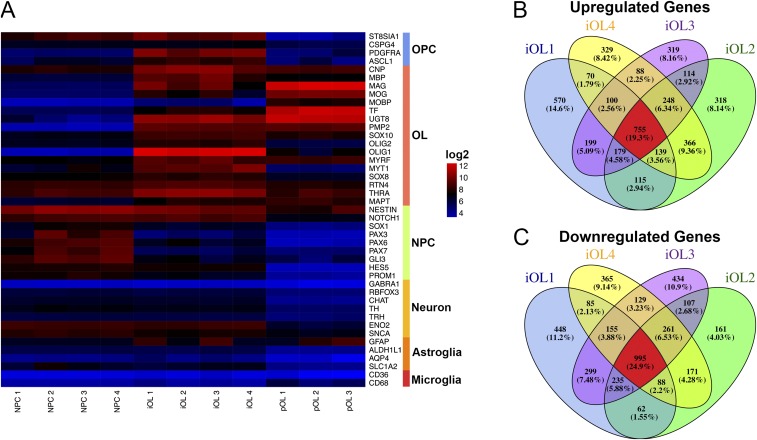
To further characterize the cellular identity of iOL, we compared the global gene-expression profiles of O4+ iOL purified at day 28 of differentiation with human MBP+ pOL-derived from surgically resected brain samples from adult patients (Fig. S5), as well as with iPSC-derived NPC before induction of SON. As a negative control, we used gene-expression values of undifferentiated iPSC. The unbiased hierarchical clustering clearly demonstrated that iOL and pOL exhibit highly comparable gene-expression signatures and form a distinct cluster significantly segregating from NPC and iPSC (Fig. 4A). When we compared neural lineage-specific gene sets, we identified a strong up-regulation of OL-specific genes, such as OLIG1, MOG, and MBP in iOL compared with NPC, whereas NPC-related genes, including SOX1, PAX6, and PAX7, were down-regulated in iOL (Fig. 4 B and C). Because we compared O4-sorted iOL with MBP+ pOL, it is not surprising that iOL expressed some OPC-specific genes, such as PDGFRA and ST8SIA1, to a higher extent than pOL, indicating a more immature cell identity of iOL (Fig. S6A). Coherently, pOL exhibit a stronger expression of some mature OL-specific genes, such as MAG and MOBP, compared with O4+ iOL. To further analyze the influence of ectopic SON expression on the oligodendroglial lineage commitment of NPC, we determined differentially expressed genes in iOL compared with the original NPC population. This analysis revealed 755 commonly up- and 955 commonly down-regulated genes among all iOL cell lines (Fig. S6 B and C). Gene Ontology (GO) terms associated with up-regulated genes in iOL include categories such as “cell adhesion,” “myelin sheath,” “axon ensheathment,” “myelin,” and “regulation of action potential.” Conversely, GO terms associated with down-regulated genes include categories such as “cell cycle,” “DNA replication,” “mitosis,” and “nucleoplasm” (Table S1).
Fig. S5.
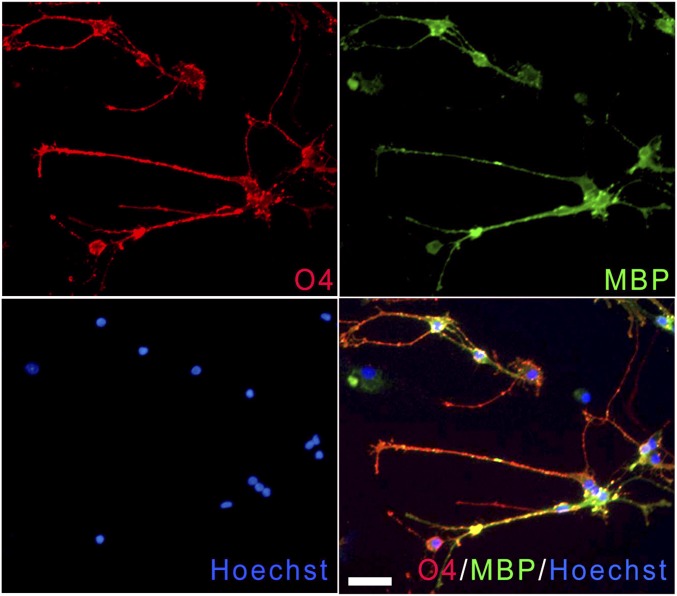
ICC analysis of pOL used for whole-genome expression analysis. Representative immunofluorescence image of pOL obtained from adults undergoing surgical resections as treatment for nontumor-related intractable epilepsy after 6 d in vitro. The vast majority of cells were O4+ (red) and MBP+ (green). Nuclei were counterstained with Hoechst (blue). (Scale bar, 50 µm.)
Fig. 4.
Global transcriptional profiling of iOL. (A) Hierarchical clustering of whole-genome expression profiles of iPSC (black), iPSC-derived NPC (green), iOL (28 d after transduction) (red), and human adult OL pOL (blue) revealed a strong correlation between iOL and pOL. (B and C) Pairwise scatterplot analysis of log2-adjusted global gene-expression values of iPSC-derived NPC and their corresponding iOL (n = 10). Genes presenting with a <twofold difference in gene expression are illustrated in gray. (B) Characteristic OL-enriched genes were up-regulated in iOL, whereas (C) characteristic NPC-enriched genes were down-regulated.
Fig. S6.
Global transcriptional profiling of iOL. (A) Heatmap illustrating gene expression for cell-type–enriched genes comparing iPSC-derived NPC, iOL and pOL. Each biological replicate of NPC and iOL presents the mean of two to three independent experiments. (B and C) Venn diagram showing the overlap of genes significantly up-regulated (B) or down-regulated (C) in four biological independent iOL cell lines compared with their corresponding iPSC-derived NPC population. Each iOL cell line presents the mean of replicates from two to three independent experiments.
Table S1.
Gene ontology analysis performed for up- and down-regulated genes in iOL compared with iPSC-derived NPC
| GO term | Genes | P value |
| Up-regulated | ||
| Cell adhesion | 66 | 4.1E-12 |
| Regulation of action potential | 13 | 1.1E-6 |
| Lipoprotein | 41 | 1.3E-5 |
| Myelin sheath | 6 | 7.9E-5 |
| Axon ensheathment | 8 | 3.0E-4 |
| Ensheathment of neurons | 8 | 3.1E-4 |
| Myelination | 6 | 6.3E-3 |
| Down-regulated | ||
| DNA replication | 62 | 2.9E-32 |
| Nucleoplasm | 124 | 2.2E-27 |
| Cell cycle | 113 | 1.2E-25 |
| DNA repair | 60 | 1.6E-20 |
| Mitosis | 51 | 2.7E-19 |
These results indicate that ectopic expression of SON induces an oligodendroglial gene-expression profile comparable to native human adult OL.
iOL Differentiate into Mature MBP-Expressing OL in Vitro and Produce Myelin-Like Sheaths.
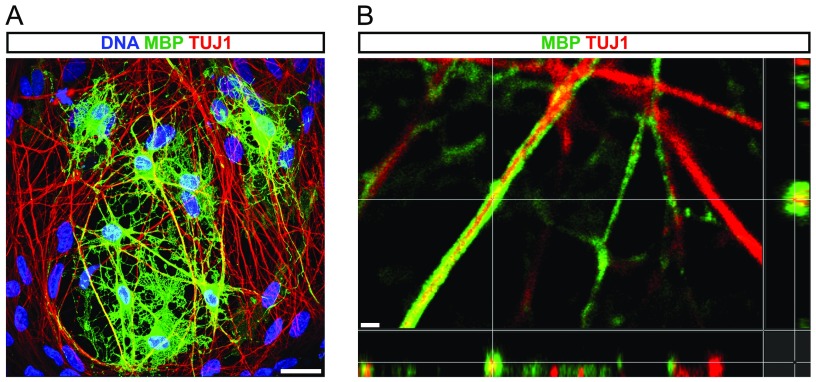
Next, we assessed the terminal differentiation potential of iOL in vitro. At day 35, iOL cultures contained many highly branched O4+ cells (Fig. 5A), as well as mature OL-expressing CNP (2′,3′-cyclic nucleotide 3′-phosphodiesterase) (32.82 ± 2.80%) (Fig. 5B). Additionally, 30.37 ± 7.87% of O4+ cells differentiated into mature MBP+ iOL with myelin-like sheaths (Fig. 5 D and E) of which 36.62 ± 8.93% coexpressed MAG (Fig. 5C). To evaluate the myelinogenic capability of iOL in vitro, we purified O4+ iOL using magnetic cell separation (MACS) at day 21 and cultured them for 14 d on 3D cell culture surfaces with aligned nanofibers. ICC analysis of mature MBP+ iOL in these cultures revealed the extension of multiple processes along the nanofibers, with some of these extensions wrapping around the nanofibers (Fig. 5F). Evidence for ensheathment of axons in vitro was evaluated in cocultures of O4+ iOL with iPSC-derived neurons. After 3 wk, the cultures exhibited myelin-like sheaths surrounding the axons, identified by confocal analysis of MBP and TUJ1 expression (Fig. S7A). Three-dimensional reconstruction of confocal optical sections in high magnification showed colabeling of neuronal processes (TUJ1) with MBP (Fig. 5 G and H), which was further evaluated by orthogonal projections clearly displaying the formation of MBP+ structures around neuronal processes (Fig. S7B). Control cultures completely lacked these MBP+ structures. These data clearly illustrate the capability of iOL to mature into MBP+ OL and to ensheath neuronal processes in vitro.
Fig. S7.
Confocal analysis of in vitro myelination assays. (A) Confocal immunofluorescence image of iOL cocultured with iPSC-derived neurons for 21 d. The image illustrates the colocalization of MBP (green) with neuronal processes visualized by TUJ1 (red). Nuclei were counterstained with Hoechst. No MBP expression was detectable in control cultures. (B) Orthogonal projection illustrates the formation of MBP+ (green) sheaths around TUJ1+ (red) neuronal processes. (Scale bars, 25 µm in A and 1 µm in B.)
iOL Myelinate the Developing Brain and Remyelinate the Demyelinated Spinal Cord of Dysmyelinating Mice.
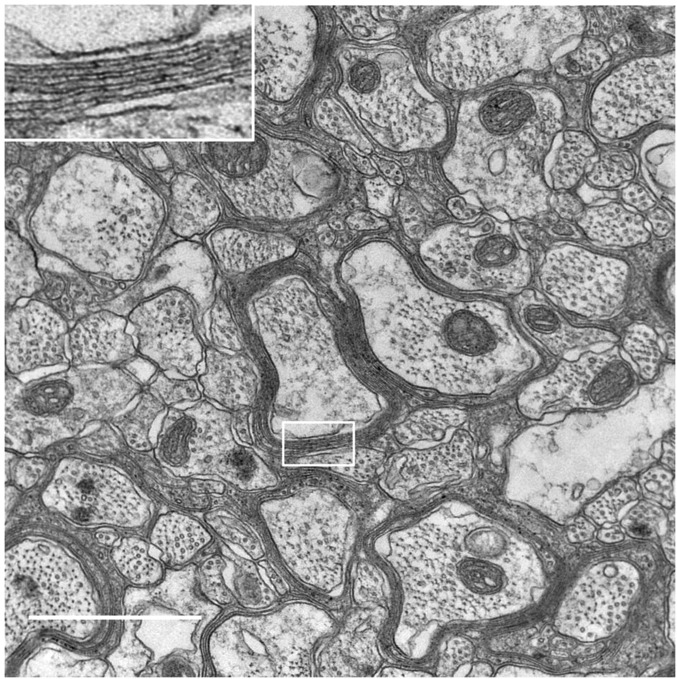
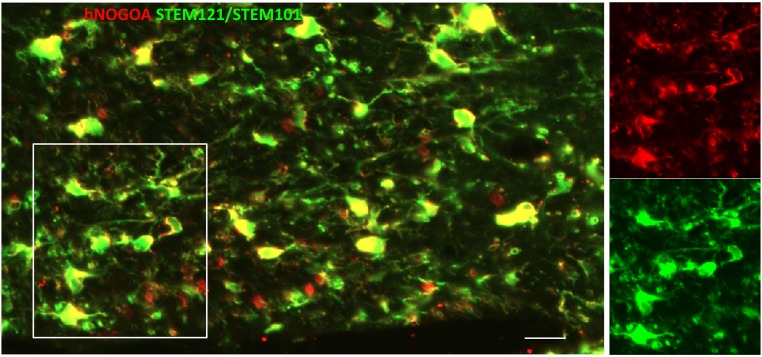
The differentiation of iOL into myelin-forming OL was further validated by grafting day 14 MACS-purified O4+ iOL in the immune- and MBP-deficient Shi/Shi Rag2−/− mouse CNS. To facilitate the identification of transplanted cells, iOL coexpressed RFP as a reporter. These mice carry a deletion of seven exons of the MBP gene and completely lack MBP protein expression. Because of the lack of endogenous MBP expression in Shi/Shi Rag2−/− mice, myelin generated by transplanted cells can be easily identified by MBP immunohistochemistry and by detection of myelin compaction, as well as the presence of the major dense line by electron microscopy (EM) (18, 19). To address developmental myelination, cells were grafted bilaterally and rostrally to the corpus callosum of newborn mice. Analysis of sagittal sections 16 wk postgrafting (wpg) indicated the presence of numerous MBP+ myelin profiles associated with RFP+ and human nuclei-positive (STEM101+) cells (Fig. 6 A and B). Higher magnification using confocal microscopy showed that iOL, identified by combined human nuclei (STEM101+) and cytoplasmic (STEM121+) antigens to highlight their cellular contours, extended processes frequently connected to MBP+ myelin, thus further validating the donor origin of the myelin (Fig. 6 C and G). MBP+ myelin generated by grafted iOL wrapped around SM312+ host axons (Fig. 6D). The presence of human-derived myelin surrounding SM312+ axons was further validated combining MBP detection with human NOGO-A, which identifies late stages of mature OL (Fig. 6 E and F). The human-derived myelin internodes were often associated with the paranodal marker CASPR flanking Ankyrin-G nodal structures, demonstrating functionality of the human cell-derived myelin (Fig. 6H). Some of the animals were also used for ultrastructural analysis of myelin compaction. In control Shi/Shi Rag2−/− mice, myelin sheaths were thin and noncompacted (Fig. S8). In mice that received iOL, numerous normal compacted myelin sheaths with alternating major dense lines and intermediate lines were observed (Fig. 6 I–K), validating unambiguously that iOL have the capacity to differentiate into functional myelin-forming cells in vivo. We quantified the percentage of myelinated axons over axons with a diameter >1 µm in grafted (n = 3) vs. nongrafted mice (n = 3) by EM analyses. We did not detect a difference in the percentage of myelinated axons (grafted mice: 69.7 ± 3.28% vs. nongrafted mice 73.9 ± 5.1%; P value: 0.2892). This result is most likely because of the competition between endogenous and grafted cells and the relatively early time point after transplantation of human iOL, which are known to differentiate more slowly than rodent cells. However, in the corpus callosum of transplanted mice, 12% of the axons were myelinated by grafted cells, as determined by the percentage of axons with compacted myelin over the total numbers of myelinated axons. Moreover, the g-ratio in grafted mice was significantly smaller compared with nongrafted mice (76.8 ± 1.8 vs. 89.5 ± 1.36; P value: 0.0003).
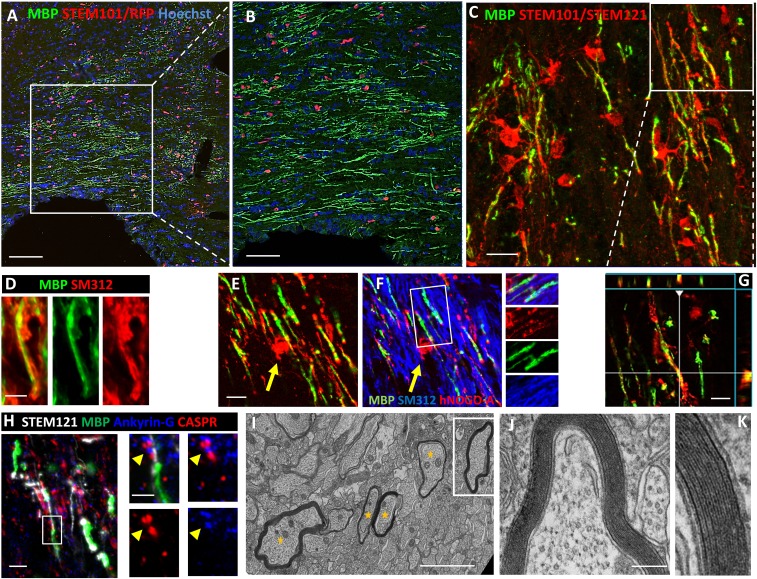
Fig. 6.
iOL give rise to functional myelin following engraftment in brains of newborn mice. (A) Transplantation of iOL into the corpus callosum of newborn Shi/Shi Rag2−/− mice resulted in extensive generation of MBP+ myelin (green) by human cells expressing RFP and staining positive for the human nuclei marker STEM101 (red) 16 wpg. (B) Higher magnification of the boxed area in A. (C) Human OL revealed by combined human nuclei STEM101 and cytoplasmic STEM121 (red) markers send multiple processes connected with MBP+ myelin. Orthogonal view of the boxed area illustrating the colocaliziation of cytoplasmic STEM121 with MBP is depicted in G. (D) Colabeling of MBP (green) and axonal neurofilament (SM312, red), highlights wrapping of host axons by donor-derived myelin. (E and F) Mature human NOGO-A+ oligodendrocyte (red, yellow arrow) connected to MBP+ myelin (green) wrapped around host axons (blue). The small panels in F illustrate the merged and single fluorochromes images represented in the boxed area. Note that unlike MBP, NOGO-A is expressed in the cell cytoplasm (cell body, processes, and paranodal loops) but not in compact myelin. (H) Human-derived (STEM121+, white)/MBP+ myelin (green) integrate into axo-glial elements expressing Ankyrin-G nodal protein (blue, yellow arrow) and CASPR paranodal proteins (red). The boxed area is enlarged to illustrate a typical node defined by a STEM121+ grafted cell and its MBP+ myelin internode, with Ankyrin-G+ aggregate flanked by two CASPR+ domains. (I–K) EM images demonstrate that human-derived myelin undergoes final maturation via compaction and formation of the major dense line. Axons surrounded by compact myelin are indicated by yellow stars. J and K are higher magnifications of boxed axon in I. n = 4 for immunostaining, n = 3 for EM. [Scale bars, 100 µm (A), 50 µm (B), 20 µm (C), 5 µm (D), 10 µm (E and F), 5 µm (H), 2 µm (I), and 200 nm (J).]
Fig. S8.
EM analysis of the corpus callosum of nongrafted shiverer mice. In the corpus callosum of adult shiverer mice, 73% of myelin-competent axons (>1 µm diameter) are ensheathed by thin and noncompacted myelin. Inset shows a 3.5-fold higher magnification of noncompacted myelin. (Scale bar, 1 µm.)
To address the ability of iOL to remyelinate demyelinated axons, RFP+ iOL were grafted into the dorsal funiculus of adult Shi/Shi Rag2−/− mice spinal cord. Although Shi/Shi Rag2−/− mice are hypomyelinated, numerous axons are surrounded by MBP− noncompacted myelin (20) (Fig. S8). To optimize the number of demyelinated axons accessible for remyelination, mice were injected with lysophosphatidyl-choline (LPC) 48 h prior grafting to induce demyelination (21, 22). Twelve weeks postgrafting, immunolabeling of serial cross-sections for nucleic STEM101 and cytoplasmic RFP, together with MBP, revealed MBP+ donut-like myelin structures, indicating that grafted iOL not only colonized and remyelinated the lesion site, but also myelinated the entire neuraxis, including ventral and dorsal white and gray matter (Fig. S9 A, C, and D).
Fig. S9.
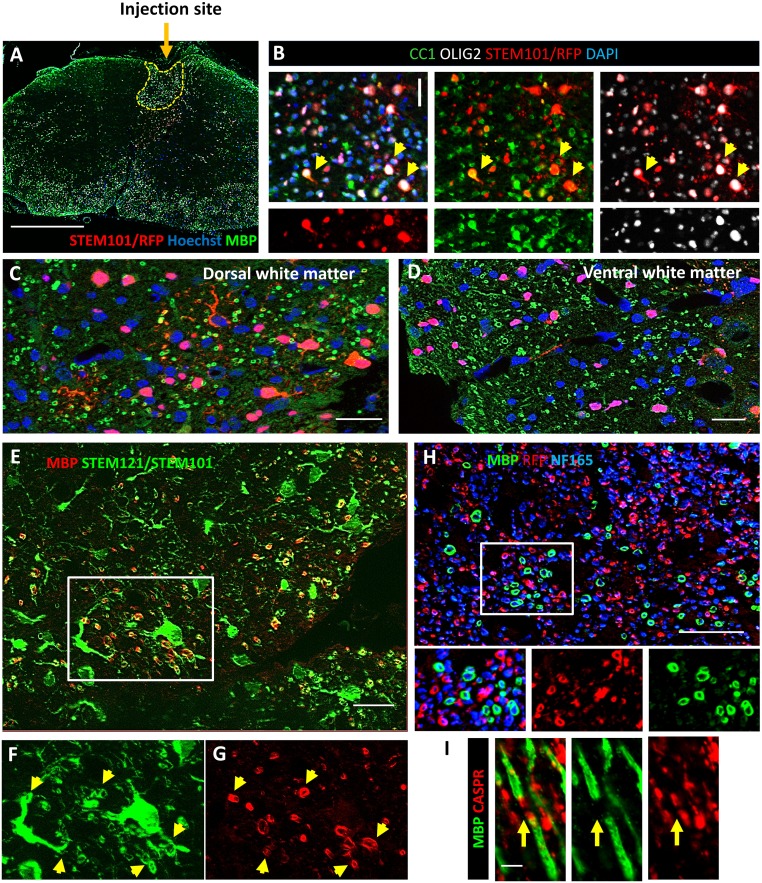
Functional differentiation of iOL into bona fide mature re/myelinating OL following transplantation in adult demyelinated mice. (A) Coronal section illustrating widespread distribution of iOL (red) derived MBP+ myelin (green) after engraftment into the dorsal funiculus (highlighted by dotted line) of the adult demyelinated Shi/Shi Rag2−/− spinal cord 12 wpg. (C and D) Higher magnification illustrating that grafted RFP+/ STEM101+ (red) human cells not only remyelinated the lesion site (C) by producing high amounts of MPB+ myelin (green), but also myelinated axons throughout the spinal cord white and gray matters, including the ventral column (D). (B) The majority of RFP+/ STEM101+ (red) human cells found in the dorsal funiculus of adult mice were CC1+ (green)/OLIG2+ (white) mature OL (yellow arrowheads highlight some of them). (E–G) Colabeling for human cytoplasmic/human nuclei (STEM121/STEM101 in green) revealed that many human cells (E and G) were connected to multiple MBP+ donut-shaped myelin-like structures (red, E and F) indicated by yellow arrowheads in F and G. (H) MBP+ myelin sheaths (green) surrounding NF165+ host axons (blue) in the dorsal funiculus. It also shows that in demyelinated spinal cord many axons are wrapped by human cells (RFP+) not yet expressing MBP, indicating a prospective larger remyelination potential of the grafted iOL. The boxed area is illustrated at higher magnification in the small panels below. (I) Longitudinal view of functional human-derived MBP+ myelin (green) integrated into nodes of Ranvier revealed by paranodal marker CASPR (red) in adult spinal cord. n = 4 mice for all staining. (Scale bars, 200 µm in A, 20 µm in B–H, and 5 µm in I.)
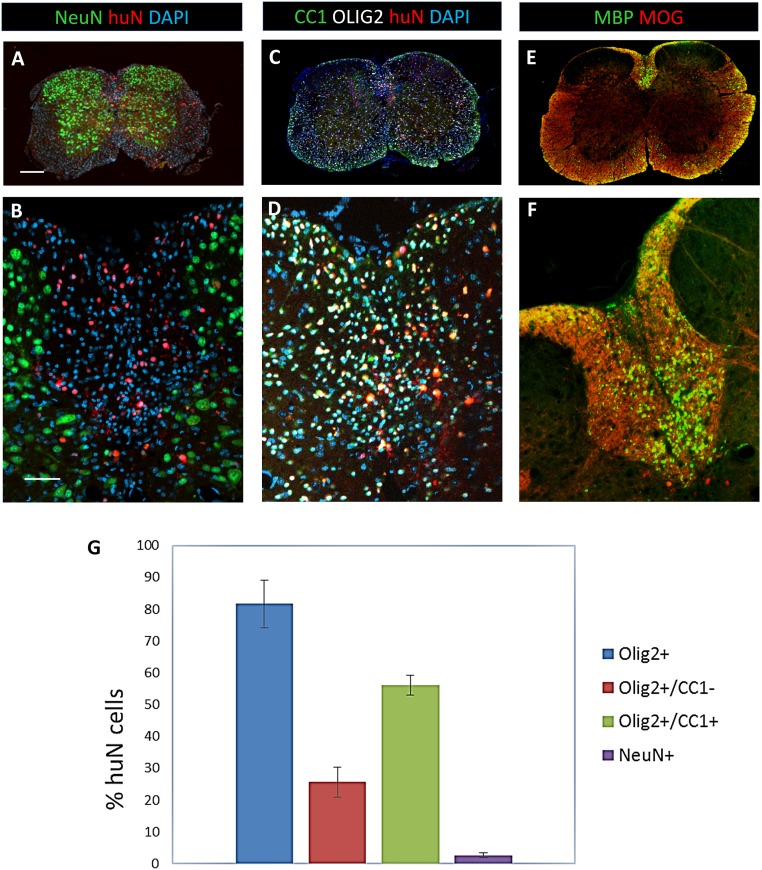
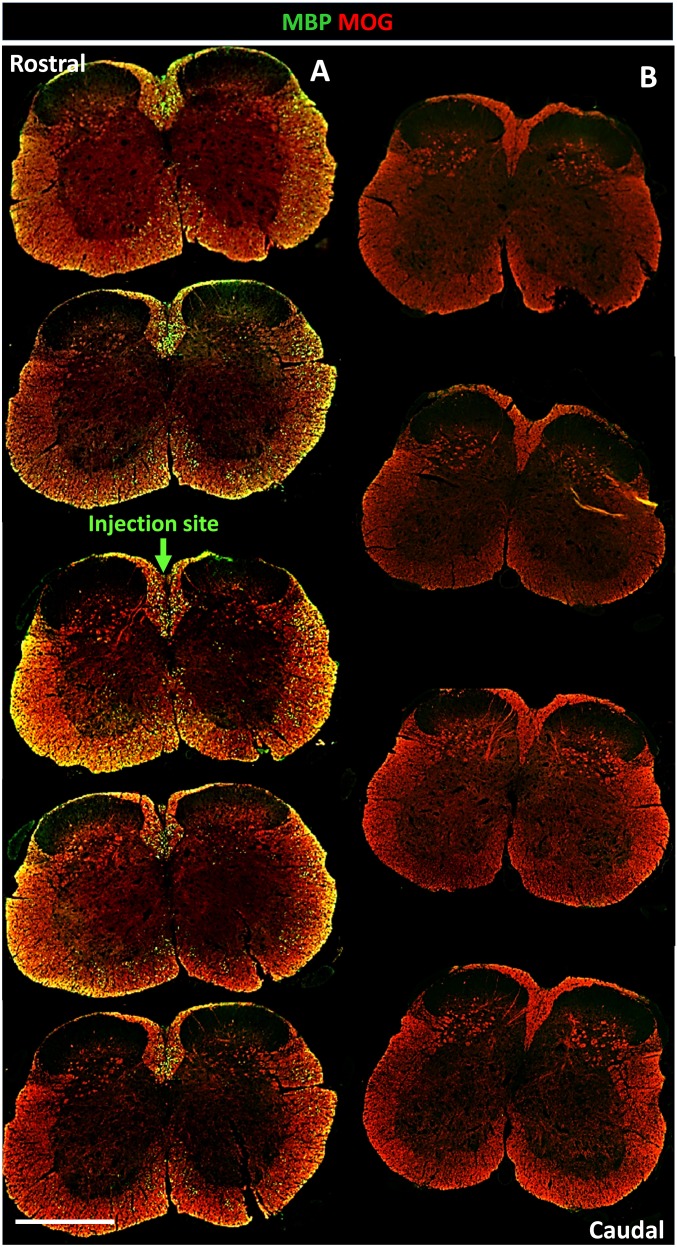
The differentiation of human grafted cells into various neural cell types was assessed by colabeling of human nucleic STEM101 and RFP with cell-specific markers of oligodendroglial cells (OLIG2, CC1), neurons (NeuN), and astrocytes (GFAP) (Figs. S9B and S10). Quantification of the various populations at the lesion site indicated that about 81% of the grafted human cells generated OLIG2+ oligodendroglial cells with 25.64% ± 4.65% OPCs (OLIG2+/CC1−) and 56.06% ± 3.11% mature OL (OLIG2+/CC1+) (Fig. S10G). The remaining population (19%) differentiated into NeuN+ neurons (2.66% ± 0.66%) and GFAP+ astrocytes which could not be quantified because of highly interdigitated processes, but which by deduction represent most likely 17% of the grafted cells. That the majority of the grafted cells differentiated into mature OL was further confirmed by widespread coexpression of human cytoplasmic STEM121 with human NOGO-A (Fig. S11). The extent of human-derived myelination in the spinal cord of Shi/Shi Rag2−/− mice was evaluated by immunolabeling of MBP and MOG, and further confirmed the widespread, integrated, and high amount of human cell-derived myelin (Fig. S12). Analysis of the MBP+ area expressed by the human cells (exogenous myelin) per MOG+ area, identifying endogenous as well as exogenous myelin at the lesion site (dorsal funiculus), showed that at 12 wpg, 12.95% ± 2.02% of the total myelin was derived from the grafted cells. MBP was detected over 18.14 ± 6.39 mm in the grafted mice (Fig. S12). MBP expression was specific for grafted cells, as no MBP expression was found in sections remote from the lesion (Fig. S12). Higher magnification showed that processes extended by iOL were frequently connected to MBP+ donut-shaped myelin internodes, thus validating the exogenous source of the myelin (Fig. S9 E–G). Although most NF165+ axons were surrounded by RFP+ processes, fewer of them coexpressed MBP, indicating that myelination was still ongoing (Fig. S9H). MBP+ myelin structures were often colabeled for the paranodal protein CASPR, as viewed on longitudinal sections (Fig. S9I), indicating the formation of nodes of Ranvier and suggesting that the iOL-derived newly formed myelin was also functional in the adult demyelinated spinal cord.
Fig. S10.
The majority of human cells grafted in dorsal funiculus were mature OL. (A and B) Illustration that most of huNuclei+ grafted cells (red) were negative for the neuronal marker NeuN. (C and D) The majority of the huNuclei+ grafted cells differentiated into CC1+/Olig2+ mature OL. Nuclei are stained by Dapi in A–D. (E and F) Donor-derived MOG+/MBP+ myelin (green) integrated among MOG+/MBP− endogenous myelin (red) in the demyelinated dorsal funiculus. (G) Percentage of the different cell types generated by the huNuclei+ grafted cells. n = 3 mice. (Scale bars, 200 µm in A, C, and E and 50 µm in B, D, and F.) B, D, and F are higher magnifications of dorsal funiculus of A, C, and E.
Fig. S11.
Human iOL grafted in dorsal funiculus expressed human-specific mature oligodendrocyte marker NOGO-A. Triple-labeling of the human cells with human antinuclei STEM101, anticytoplasmic STEM 121, and anti-human NOGO-A revealed that most of the grafted cells expressed mature oligodendroglial marker NOGO-A (red). Note that unlike MBP, NOGO-A is expressed in the cell cytoplasm (cell body, processes, and paranodal loops) but not in compact myelin. Merged image in boxed area is detailed in small panels as NOGO-A (Upper) and STEM121/STEM101 (Lower). n = 3 mice. (Scale bar, 20 µm.)
Fig. S12.
Human myelin disperses widely and integrates very well among endogenous myelin in adult mice. (A) Serial coronal sections of the grafted adult Shi/Shi Rag2−/− mouse spinal cord stained for MOG (red) and MBP (green) reveal MOG+/MBP− endogenous and MOG+/MBP+ exogenous myelin, respectively, and illustrates the widespread distribution of human-derived MBP+ myelin along the rostro-caudal axis (between 10 and 22 mm in length) 12 wpg. (B) Illustration of more caudal sections expressing MOG only (internal negative controls). n = 3 mice. (Scale bar, 1,000 µm.)
Human iOL Respond to Compounds Promoting Oligodendroglial Differentiation.
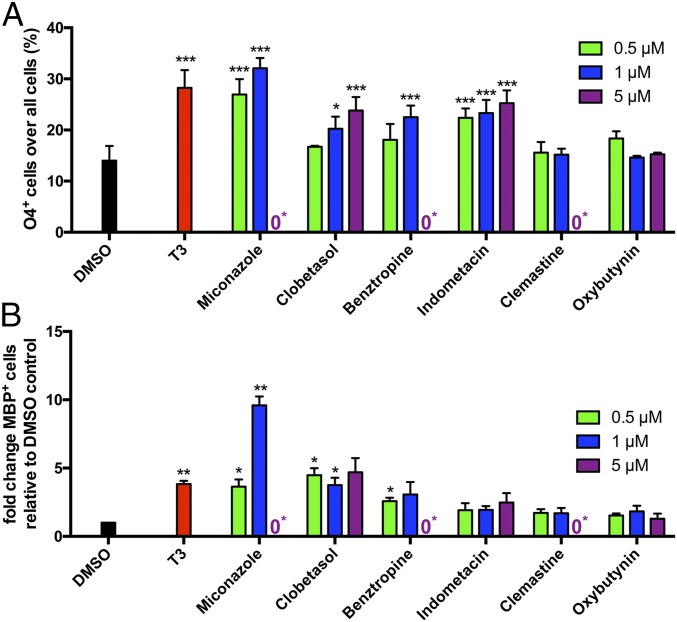
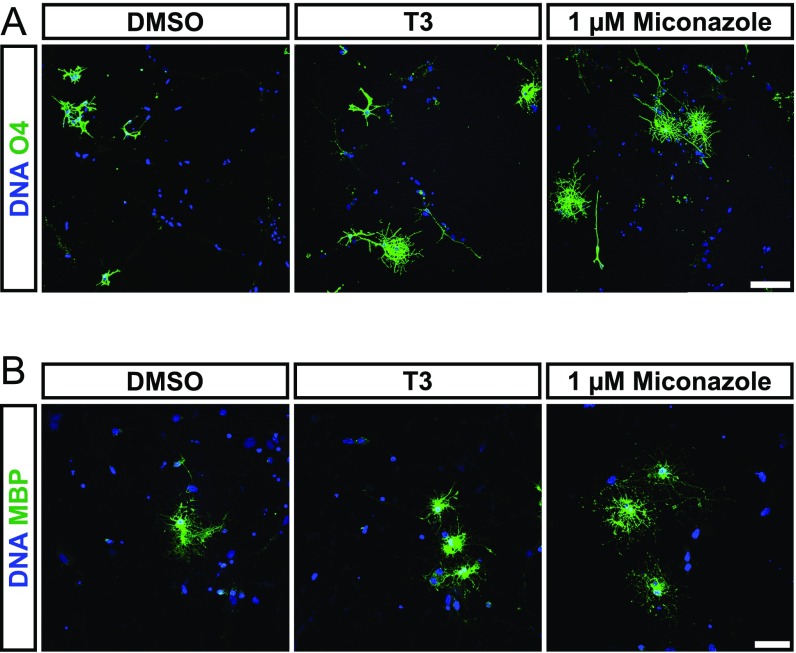
Identification of drugs inducing remyelination via promotion of oligodendroglial differentiation presents a promising approach for the treatment of demyelinating disorders like MS. Thus, we assessed whether iOL can be used to identify compounds promoting oligodendroglial differentiation. We selected six drug candidates (miconazole, clobetasol, benztropine, indometacin, clemastine, and oxybutynin), which have been previously described to promote differentiation or myelination of rodent OL (1, 23–25). iOL were cultured in a minimum growth medium lacking differentiation-promoting factors and were treated with either vehicle [0.01% (vol/vol) DMSO] as a negative control, T3 as a positive control, or the drug candidate dissolved in DMSO at three different concentrations (0.5, 1, 5 µM) (Fig. 7 and Fig. S13). In DMSO-treated control cultures, 14.01 ± 2.89% O4+ iOL were observed in minimum DM after 21 d of culture, whereas addition of T3 resulted in the doubling of O4+ cells (28.25 ± 3.47%). Several drug candidates performed as well as T3 and demonstrated a dose-dependent increase of O4+ cells (Fig. 7A). Interestingly, clemastine and oxybutynin failed to promote the formation of O4+ iOL. Furthermore, we observed a toxic influence of miconazole, benztropine, and clemastine on iOL at higher concentrations. Quantification of mature MBP+ iOL revealed a fourfold increase in the presence of T3 compared with DMSO control (Fig. 7B). The effect was most striking with 1 µM miconazole, inducing an almost 10-fold increase of MBP+ cells compared with DMSO control cultures. Clobetasol and benztropine enhanced the formation of MBP+ mature iOL comparable to T3-treated cultures. These data demonstrate that iOL can be used to identify compounds that promote differentiation into O4+ as well as maturation into MBP+ OL.
Fig. 7.
iOL are suitable to test the differentiation promoting effects of selected compounds. Quantification of (A) O4+ and (B) MBP+ iOL after treatment with either vehicle, T3, or the drug candidate dissolved in DMSO at three different concentrations (0.5, 1, 5 µM) for 21 d in minimum DM. Data are presented as mean of replicates from three independent experiments + SD. One-way ANOVA with Dunnett’s multiple-comparisons test was performed for statistical analysis comparing the mean of each sample with DMSO control (*P < 0.05, **P < 0.01, ***P < 0.001). 0* = Toxic culture condition.
Fig. S13.
ICC analysis of iOL in DM containing selected compounds. (A and B) Representative immunofluorescence images of iOL cultures treated with either vehicle [0.01% (vol/vol) DMSO], T3 as a positive control, or 1 µM of the drug candidate miconazole for 21 d in minimum DM. Oligodendroglial lineage commitment was assessed by (A) O4 (green) and (B) MBP (green) immunostaining; nuclei were counterstained with Hoechst (blue). (Scale bars, 50 µm.)
iOL from Patients Carrying the N279K Microtubule-Associated Protein Tau Mutation Have a Higher Susceptibility to Oxidative Stress-Induced Cell Death.
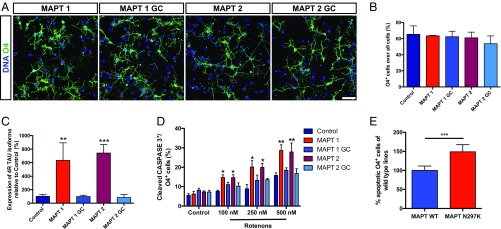
Next, we wanted to determine whether iOL can be used for in vitro disease modeling. The microtubule-associated protein tau (MAPT) is developmentally expressed in OL (26, 27) and mutations in MAPT have been associated with frontotemporal dementia with Parkinsonism linked to chromosome 17 (FTDP-17), a disease also characterized by pathological changes in white matter (28–30). Therefore, we generated iOL from two iPSC clones from one patient carrying the N279K MAPT mutation associated with FTDP-17 (17) and compared these to their isogenic controls (31). Additionally, we included another independent control iPSC line.
After 28 d of differentiation, O4+ iOL harboring the N279K mutation (MAPT-OL) were morphologically indistinguishable from their gene-corrected control cell lines (MAPT-GC-OL) (Fig. 8A) and featured similar differentiation efficiencies among all cell lines included (Fig. 8B). We next set out to investigate whether the N279K MAPT mutation induces an altered expression of tau isoforms in iOL. We purified O4+ iOL using FACS before RNA sample preparation. Analysis of MAPT expression revealed mutation-specific significantly higher levels of 4R tau compared with MAPT-GC-OL (Fig. 8C), which is in line with observations in iPSC-derived neurons and brains of FTDP-17 patients harboring this mutation (17, 32). FTDP-17 patients display widespread neurodegeneration because of increased cellular vulnerability. Therefore, we investigated whether MAPT-OL are more susceptible to oxidative stress induced by rotenone, an inhibitor of the mitochondrial complex I. Exposure of MAPT and MAPT-GC-OL for 48 h to rotenone revealed an enhanced vulnerability of MAPT-OL to oxidative stress identified by an increased number of cleaved CASPASE-3+ iOL in MAPT-cultures (Fig. 8D). This effect was obvious in all tested concentrations of rotenone (100, 250, and 500 nM), leading to an average increase of cell death of 48.9 ± 18.7% in MAPT-OL (Fig. 8E).
Fig. 8.
MAPT-OL exhibit mutation related phenotypes. (A) Immunostaining for O4 (green) demonstrating differentiation of iPSC carrying the N279K MAPT mutation (MAPT1, MAPT2) and genetic corrected controls (MAPT1 GC, MAPT2 GC) into iOL. Nuclei were counterstained with Hoechst (blue). (Scale bar, 25 µm.) (B) Flow cytometry-based quantification of O4+ iOL after 28 d of differentiation in MAPT mutation cultures, genetic corrected cultures, and an independent healthy control culture. Data are presented as mean of replicates from three independent experiments + SD. (C) Quantitative RT-PCR analysis on control, MAPT gene-corrected, and MAPT mutated iOL cultures for 4R tau isoforms containing exon 10. Expression levels were normalized to total tau expression and control lines. Data are presented as mean of replicates from three independent experiments + SD. One-way ANOVA with post hoc Tukey test was performed for statistical analyses (**P < 0.01, ***P < 0.001). (D) Quantification of cleaved CASPASE 3+ iOL in control and MAPT cultures after 48 h of either vehicle [0.01% (vol/vol) DMSO] or rotenone treatment. Data are presented as mean of replicates from three independent experiments + SD. One-way ANOVA with post hoc Tukey test was performed for statistical analysis (*P < 0.05, **P < 0.01). (E) All results combined after normalization by setting all control cultures to 100%, show that MAPT N279K causes a higher sensitivity to oxidative stress. Error bars present SD. Student’s t test was performed for statistical analysis (***P < 0.001).
Discussion
Previously established protocols using in vitro patterning to derive OL from murine iPSC require fewer than 30 d (33). Additionally, it has been shown that mouse fibroblasts can be directly converted into OPC by the forced expression of Sox10, Olig2, and Zfp536 or Nkx6.2, resulting in ∼15% O4+cells after 20 d (2, 14). However, the generation of human OL from iPSC or ESC is more challenging and characterized by much longer culture periods (70–150 d), limited efficiencies, and variable reproducibility. Although the protocols to generate human OL have been further optimized to reduce culture times and increase efficiency, they still require at least between 60 and 130 d of culture to generate OPC from iPSC/ESC-derived NPC and only a small percentage of cells become MBP+ mature OL (9–11). One of the first studies demonstrating the successful derivation of human OL from iPSC was published in 2013 (11) using a modified in vitro-patterning strategy from ESC-based protocols (34, 35). However, the transition from pre-OPC to OPC was hampered; by day 150, 20–40% of the cells expressed the OPC marker CD140a, whereas the late OPC marker O4 was only present in 5–10%. Interestingly, the application of low levels of oxygen (3%) during the differentiation of PSC to OL could greatly enhance oligodendroglial lineage commitment, resulting in around 40% O4+ cells at day 135 (10). The use of retinoic acid from the beginning of differentiation, followed by the application of sonic hedgehog agonist SAG could further increase the oligodendroglial lineage commitment, resulting in up to 70% of O4+ OPCs at day 75 (9). To accelerate oligodendroglial differentiation that is orchestrated by TF, we tested individual and combinations of TF previously shown to be involved in oligodendroglial differentiation in rodents (15, 36–40). Our results have defined a combination of three different TF that efficiently induces iOL and indeed overcomes the rate-limiting steps of the transition from a neurogenic to a glial progenitor and accelerates oligodendroglial differentiation. In line with an earlier study, in which overexpression of SOX10 alone was sufficient to induce oligodendroglial commitment (∼22% O4+ cells 14 d after transduction) in neural progenitors derived from the human fetal brain (16), in our experiments SOX10 was the only TF that induced expression of O4 in iPSC-derived NPC, demonstrating that SOX10 is one key TF to induce oligodendroglial lineage commitment. However, combination of SOX10 with OLIG2 and NKX6.2 further enhanced the commitment into the oligodendroglial lineage, resulting in a significantly higher percentage of O4+ cells 14 d after induction. Our protocol was highly efficient and reproducible, resulting in 50–70% O4+ cells and yields of 220–260% after 28 d using three different iPSC-derived NPC lines and a single ESC-derived NPC line.
The myelinating capacity of iOL was tested in vitro and in vivo. In vitro iOL ensheathed the neuronal processes of iPSC-derived neurons, as well as nanofibers confirming that physical properties of axons are sufficient to induce wrapping of axons, as it has been described for rodent OL (41, 42). The coculture of iOL with nanofibers facilitates the identification of compounds that exclusively promotes axon ensheathment without potentially modulating molecular axonal signaling. We did not detect CASPR accumulations, indicative for paranode formation in iPSC-derived neuron/iOL cocultures, suggesting that distinct axonal signaling cascades required for the formation of paranodes and nodes were not activated in iPSC-derived neurons. However, CASPR+ paranodes were easily detectable in vivo after transplantation into Shi/Shi Rag2−/− mice. The transplanted cells not only efficiently myelinated the forebrain in newborn Shi/Shi Rag2−/− mice, but also remyelinated the adult demyelinated spinal cord. Proof of the donor origin of the myelin was provided by immunohistochemistry using human-specific nuclei and cytoplasmic markers, allowing us to highlight the connection of donor cell processes to myelin internodes surrounding host axons, and the integration of these myelin internodes into nodal and paranodal elements. We provided additional proof of the donor origin of myelin by the detection at the ultrastructural level of thick, compact myelin with the major dense line, a hallmark of wild-type myelin compared with thin, noncompacted, and dense line-null shiverer myelin (18, 19). It is known that despite the lack of MBP, the number of axons ensheathed by noncompacted myelin increases progressively in shiverer mice over time. In the gracile fasciculus in the dorsal spinal cord, the number of myelinated axons increases from 56% 2 wk after birth to up to 77% at 20 wk (20). Injection of lysolecithin in the dorsal funiculus results in complete demyelination of these axons (22). (Re)myelination, although more efficient in the lesion, was not limited to the dorsal funiculus because MBP+ exogenous myelin was found throughout the spinal cord (over 22 mm) with numerous myelinated axons in gray matter and ventral white matter, suggesting that grafted cells dispersed widely in their adult host environment. Similar observations were reported for transplanted human fetal neural progenitors (21), indicating that O4+ iOL are as migratory as human fetal NPC in adult Shi/Shi Rag2−/− mice. Compared with the developing CNS, the adult CNS represents an environment with impaired tissue plasticity and trophic support. Whereas the colonization and remyelination potential of murine iPSC-derived NPC has already been described in this environment (22), this study demonstrates that human iPSC-derived oligodendroglial lineage cells have similar capabilities. However, one has to keep in mind that the endogenous cells are compromised because of the lack of MBP expression in contrast to the grafted cells. Therefore, the grafted cells may have a competitive advantage and not only remyelinate demyelinated axons but also replace the endogenous pool of myelinating cells. In the adult spinal cord, the majority of iOL differentiated into OL (81%), rarely into neurons (2%), and few into astrocytes (estimated to be 17%), confirming previous data obtained with iPS-derived O4+ cells (9) or O4+ human fetal cells (11) grafted in the newborn shiverer mouse brain. Interestingly iOL-derived neurons were observed only in gray matter surrounding the lesion, and iOL-derived astrocytes were detected only in the demyelinated area but not in the ventral or lateral white matter tracks, suggesting that their differentiation is highly regulated by cell-specific environmental cues.
The iPSC technology is an emerging tool for drug development. Promotion of remyelination represents until now an unmet treatment strategy for patients with MS. Without SON, less than 2% of NPC differentiated into O4+ OL. Importantly, SON transduced NPC cultured in minimal medium lacking T3 and growth factors (except dbcAMP), resulted in ∼14% of O4+ OL. Addition of T3 increased the efficiency to 28% and culturing in DM to 40% after 21 d (as shown in Figs. 3B and 7, respectively). These data demonstrate that SON is sufficient to induce an oligodendroglial cell fate; however, growth factors are necessary to further increase maturation of the cells. The increase in differentiation by exogenous factors is prerequisite to use iOL for pharmacological screens aimed at the identification of compounds promoting remyelination. In large compound screens using primary or iPSC-derived OL from rodents, a number of Food and Drug Administration-approved drugs has been identified that were able to promote oligodendroglial differentiation in vitro and remyelination in vivo (1, 24, 25). To determine whether iOL may be suitable for pharmacological screens, we cultured iOL in the presence of compounds identified in earlier rodent studies (1, 23–25). In contrast to these earlier studies, some but not all of these compounds increased the number of O4+ iOL in a dose-dependent manner and were at least as effective as T3, a known promoter of oligodendroglial differentiation. Furthermore, only a subset of these drugs enhanced the maturation of O4+ iOL into MBP+ mature OL, suggesting that the compounds affect different stages of oligodendroglial differentiation. Miconazole demonstrated the strongest effect on iOL; this is in line with an earlier publication by Najm et al., in which they reported a strong differentiation promoting effect of miconazole on OL (1). However, we observed a toxic effect with a fivefold-higher concentration, suggesting that miconazole might have a narrow range of efficacy. Our observations thus suggest that there are species-specific differences between rodent and human OL that could be relevant for drug screens aiming at identifying compounds that promote oligodendroglial differentiation.
To determine whether iOL are suitable for disease modeling, we characterized in a proof-of-concept study the phenotypes of iOL derived from a patient diagnosed with an inherited form of FTD. FTD is characterized by cortical degeneration of the frontal and temporal lobe that in 15–20% of patients with an inherited form of FTD is a result of mutations in the MAPT gene that encodes the MAPT located on chromosome 17q21. The neuropathology of FTDP-17 patients with mutations in the MAPT gene is characterized by tau+ inclusions in neurons and glia, including OL (for review, see ref. 43). Furthermore, extensive myelin pathology can be observed in patients with FTD (28–30). In OL tau regulates and stabilizes the microtubule network that is also involved in the transport of RNA granules, for example, those containing MBP mRNA. Knockdown of tau or mutated tau in rodent OL impairs process outgrowth and the differentiation into MBP+ myelinating mature OL (44, 45). Therefore, we assessed whether changes in OL may directly contribute to the white matter pathology observed in FTDP-17 patients. In iOL from patients with a N279K mutation in the MAPT gene, we observed as expected, significantly increased expression levels of the 4R tau isoform. Furthermore, we observed an increased susceptibility to cell death induced by respiratory stress compared with gene-corrected control cell lines, similar to that reported in iPSC-derived neurons from the same patient (17). These data suggest that MAPT mutations in OL may directly contribute to myelin pathology and thus to disease progression in patients with FTDP-17.
In summary, our studies demonstrate that a combination of three TFs, namely SOX10, OLIG2, and NKX6.2, greatly accelerates the generation of OL from iPSC-derived NPC and that these cells are suitable for disease modeling and pharmacological screens. Thus, our protocol should significantly facilitate the development of high-throughput screening platforms and the study of human myelin diseases and repair strategies using patient-derived iPSC.
Methods
Culturing of Human PSC.
The iPSC included in this study have previously been generated and characterized (13, 17). iPSC colonies were maintained on a layer of mitotically inactivated mouse embryonic fibroblasts in human ESC medium consisting of knockout DMEM (Invitrogen) with 20% (vol/vol) Knockout Serum Replacement (Invitrogen), 1 mM β-mercaptoethanol (Invitrogen), 1% nonessential amino acids (NEAA; Invitrogen), 1% penicillin/streptomycin/glutamine (PAA), freshly supplemented with 5 ng/mL FGF2 (Peprotech). PSC were split at ratios of 1:6–1:8 every 7 d by mechanic disaggregation with 1 mg/mL collagenase IV (Invitrogen).
Generation and Culturing of Human NPC.
NPC were derived from human PSC by treatment with small molecules as previously described (13, 17). Methodological details are provided in SI Methods.
Lentiviral Vector Construction and Production of Lentiviral Particles.
Details including the cloning strategies are provided in SI Methods and Fig. S14.
Fig. S14.
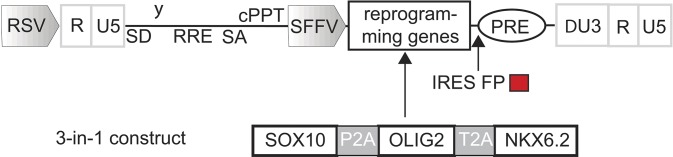
Schematic presentation of the polycistronic all-in-one SON lentiviral vector. The human cDNAs encoding SOX10, OLIG2, and NKX6.2 were linked by 2A self-cleavage sites and were inserted into a third-generation lentiviral expression vector equipped with the retroviral SFFV U3 promoter. For the visualization of transgene expression, an IRES-dTomato cassette was introduced following the SON expression cassette.
Transduction of NPC for TF Screening.
Human NPC were seeded with a density of 1 × 105 cells per well in 12-well plates, allowed to attach overnight, and transduced with equal volumes of concentrated Lenti-rtTA and 1-TF virus particle supplemented with 5 µg/mL protamine sulfate (Sigma) in fresh NPC expansion medium (NPCM). The 2-TF infections were done by mixing equivalent volumes of Lenti-rtTA, pLV-TetO-SOX10 and 1-TF virus particle for infection. For 3-TF infections, the volume of each virus was reduced by one quarter and equivalent volumes of Lenti-rtTA, pLV-TetO-SOX10, pLV-TetO-OLIG2, and 1-TF were mixed for NPC transduction. Further details are provided in SI Methods.
Oligodendroglial Differentiation.
For oligodendroglial differentiation, human NPC were seeded with a density of 1 × 105 cells per well in 12-well plates, allowed to attach overnight, and transduced with concentrated SON lentiviral particle and 5 µg/mL protamine sulfate in fresh NPCM. Viral medium was removed after 24 h and replaced with GIM. After 4 d, GIM was replaced by DM. Details are provided in SI Methods.
ICC.
Details of ICC are provided in SI Methods and Table S2.
Table S2.
Primary antibodies for ICC
| Antibody | Dilution | Source |
| Mouse anti-NESTIN | 1:300 | R&D (MAB1259) |
| Goat anti-SOX1 | 1:150 | R&D (AF3369) |
| Rabbit anti-NG2 | 1:200 | Millipore (AB5320) |
| Mouse anti-O4 | 1:100 | R&D (MAB1326) |
| Mouse anti-GALC | 1:100 | Millipore (MAB342) |
| Rat anti-MBP | 1:50 | Abcam (AB7349) |
| Rabbit anti–Ki-67 (SP6) | 1:250 | Abcam (AB16667) |
| Mouse anti-CNP | 1:250 | BioLegend (836401) |
| Mouse anti-MAG | 1:400 | Abcam (AB89780) |
| Mouse anti-TUBBIII (TUJ1) | 1:750 | Covance (MMS-435P) |
| Mouse anti-AT8 | 1:150 | Innogenetics (90206) |
Flow Cytometry Analysis.
Cells were enzymatically detached and singularized by accutase treatment for 10 min at 37 °C. Following washing with PBS, singularized cells were resuspended in PBS/0.5% BSA buffer and filtered through a 70-µm cell strainer (BD Falcon). After determination of cell number, cells were incubated with mouse IgM anti–O4-APC antibody (Miltenyi Biotec) following the manufacturer’s protocol. Stained cells were washed, resuspended in PBS/0.5% BSA buffer (5 × 106 cells/mL), and immediately sorted using a FACSAria cell sorter (BD Biosciences). Debris, doublets, and aggregates were excluded by appropriate gating using forward and side scatter. Additionally, DAPI was used for dead cell exclusion. Unstained cells were used to set background fluorescence and undifferentiated human NPC were used as negative controls.
Neuronal Differentiation, in Vitro Myelination Assay, and 3D Culture.
Human iPSC-derived NPC were differentiated into neurons as previously described (17). To assess the in vitro myelination capacity of iOLs, O4+ cells were purified at differentiation day 21 by magnetic cell sorting using anti-O4 MicroBeads (Miltenyi Biotec) following the manufacturer’s protocol. Purified iOLs were added to 21-d-old neuronal cultures derived from iPSC-derived NPC populations or nanofibers. Details are provided in SI Methods.
Isolation of Adult Human pOL.
Brain tissue was obtained from adults undergoing surgical resections as treatment for nontumor-related intractable epilepsy in accordance with the guidelines set by the Biomedical Ethics Unit of McGill University. As described previously (46), tissue specimens were enzymatically digested and placed on a linear 30% Percoll density gradient (Pharmacia Biotech). Further details are provided in SI Methods.
Whole-Genome Expression Analysis.
Details including the microarray data processing are provided in SI Methods.
Cell Transplantation.
Details of cell transplantation into Shi/Shi Rag2−/− dysmyelinating immunodeficient mice are provided in SI Methods. Animal experiments were performed according to European Community regulations and INSERM ethical committee (authorization 75-348; 20/04/2005) and were approved by the local Darwin ethical committee.
Immunohistochemistry and EM.
The methodology for immunohistochemistry and EM is described in detail in SI Methods.
Compound Screen.
To assess the sensitivity of iOL toward differentiation promoting drugs, iPSC-derived NPC were transduced with concentrated SON virus particle, as described above. Viral medium was removed after 24 h and replaced with GIM lacking T3. The end of the virus infection period was termed day 0. Further details including the differentiation with drug candidates are provided in SI Methods.
Generation and Characterization of N297K MAPT NPC and Isogenic Controls.
The N279K MAPT iPSC-derived NPC included in this study have previously been generated and characterized (17). Frozen NPC, termed FTDP-17-1-I and FDTP-17-1-II in the aforementioned publication were thawed at passages 10–14 and designated as MAPT-1 and MAPT-2 in this study.
Quantitative RT-PCR.
Details of RT-PCR including a list of primers are provided in SI Methods and Tables S3.
Table S3.
Primer for quantitative RT-PCR
| Primer | Sequence |
| hGAPDH_for | CTG GTA AAG TGG ATA TTG TTG CCA T |
| hGAPDH_rev | TGG AAT CAT ATT GGA ACA TGT AAA CC |
| hMAPT_total_for | CTC GCA TGG TCA GTA AAA GCA A |
| hMAPT_total_rev | GGG TTT TTG CTG GAA TCC TGG T |
| hMAPT_Exon10_for | CCA AGT GTG GCT CAA AGG AT |
| hMAPT_Exon12_rev | CCC AAT CTT CGA CTG GAC TC |
Stress-Induced Cell Death.
Details are described in SI Methods.
Quantifications.
Quantifications were performed as described in detail in SI Methods.
Statistics.
Data of at least three independent differentiation experiments are presented as mean + SD. Statistical significance was determined by Student’s t test and with one-way ANOVA, respectively.
SI Methods
Generation and Culturing of Human NPC.
Human PSC colonies from passages 10–15 were mechanically sectioned and enzymatically detached from mouse embryonic fibroblasts. Pieces of PSC colonies were collected by sedimentation, resuspended in ESC medium (without FGF) supplemented with 10 µM SB-431542 (Ascent Scientific), 1 µM dorsomorphin (Tocris), 3 µM CHIR99021 (CHIR; Axon Medchem), and 0.5 µM purmorphamine (Alexis), and subsequently cultured as embryoid bodies in Petri dishes. The medium was changed after 2 d to N2B27 medium consisting of equal parts DMEM-F12 (Invitrogen) and Neurobasal (Invitrogen) with 1:200 N2 supplement (Invitrogen), 1:100 B27 supplement lacking vitamin A (Invitrogen), 1% penicillin/streptomycin/glutamine, and with the same small-molecule supplements as mentioned before. On day 4, SB-431542 and dorsomorphin were withdrawn and 150 µM ascorbic acid (AA; Sigma) was added to the medium. On day 6, embryoid bodies were disintegrated into smaller pieces and plated on Matrigel-coated (Matrigel, growth factor reduced, high concentration; BD Biosciences) 12-well plates (Nunc) in NPCM consisting of N2B27 medium supplemented with 3 µM CHIR, 0.5 µM SAG (Cayman Chemical), and 150 µM AA. Cells were split once a week at ratios of 1:15–1:20 by treatment with accutase (Sigma). Regular tests for mycoplasma contamination using the MycoAlert mycoplasma detection kit (Lonza) were negative.
Lentiviral Vector Construction and Production of Lentiviral Particles.
The coding regions of SOX10, OLIG2, ASCL1, NKX2.2, NKX6.1, NKX6.2, MYT1, and RFP were amplified by PCR, validated by sequencing, cloned into pCR8/GW/TOPO (Invitrogen) according to the manufacturer’s instructions, and recombined into pLV-tetO-attR1/R2 (47) by LR clonase II (Invitrogen). pLV-tetO-attR1/R2 and Lenti-rtTA were kindly provided by Chad Cowan, Harvard University, Cambridge, MA. To construct the polycistronic lentiviral SON vector, we used a third-generation lentiviral vector (kindly provided by L. Naldini, Ospedale San Raffaele, Milan, Italy), which we further equipped with the retroviral SFFV U3 promoter and the reprogramming cassette (48). To be optionally able to excise the reprogramming cassette later, we incorporated a Flp recognition target site in the 3′ U3 region. The human cDNAs encoding SON were inserted to create a three-in-one vector, in which the transcription factor genes are coexpressed and linked by 2A self-cleavage sites (P2A, T2A). Furthermore, we introduced an internal ribosome entry site (IRES)-RFP cassette (encoding the dimeric RFP variant dTomato) for visualization of vector expression, as previously described (48) (Fig. S14). Tet-inducible SON was generated by cloning SON cassette into pHAGE, a third-generation lentiviral vector previously described (49). The construct was also equipped by IRES-pac cassette allowing puromycin selection (Fig. S3A). The constructs were validated by sequencing. Cloning details are available upon request. Lentiviral particles were produced by cotransfection of 293T cells with individual expression vectors in combination with the packaging plasmids psPAX2 (Addgene #12260) and pMD2.G (Addgene #12259). Additional reverse tetracycline transactivator lentivirus (FUW-M2rtTA, Addgene #20342) was generated to coinfect with tet-inducible lentiviruses. Virus-containing supernatants were harvested at 48 and 72 h posttransfection and filtered with 0.45-µm PVDF membrane (Millipore). Viral particles were subsequently concentrated by ultracentrifugation, resuspended in N2B27 medium, and stored at −80 °C.
Transduction of NPC for TF Screening.
Viral medium was removed from human NPC after 24 h and replaced by N2B27 medium supplemented with 1 µM SAG, 10 ng/mL PDGF (Peprotech), 10 ng/mL NT3 (Peprotech), 10 ng/mL IGF-I (Peprotech), 200 µM AA, 1:1,000 Trace Elements B (Corning), and 60 ng/mL T3 (Sigma). The end of the virus infection period was termed day 0 and transgene expression was induced with 2 µg/mL doxycycline (Clontech) for 14 d. Medium was replaced every other day and cells were fixed in 4% paraformaldehyde (PFA; Sigma) in PBS (Invitrogen) for ICC analysis at day 14 of differentiation. Investigators were blinded for ICC analyses.
Oligodendroglial Differentiation.
GIM consisted of DMEM-F12 with 1:200 N2 supplement, 1:100 B27 supplement lacking vitamin A, 1% penicillin/streptomycin/glutamine, 1 µM SAG, 10 ng/mL PDGF, 10 ng/mL NT3, 10 ng/mL IGF-I, 200 µM AA, 1:1,000 Trace Elements B, 10 ng/mL T3. GIM was changed every other day and replaced at day 4 posttransduction by DM consisting of DMEM-F12 with 1:200 N2 supplement, 1:100 B27 supplement lacking vitamin A, 1% penicillin/streptomycin/glutamine, 60 ng/mL T3, 10 ng/mL NT3, 10 ng/mL IGF-I, 200 µM AA, 1:1,000 Trace Elements B, and 100 µM dbcAMP (Sigma). After 7–10 d of differentiation, cells were detached and singularized by treatment with accutase and reseeded at densities of 2.5 × 105 cells in 12-well plates and 1.5 × 105 cells in 24-well plates containing glass coverslips. DM was changed every other day.
ICC.
For ICC analysis, an equal volume of 4% PFA in PBS was added to the culture medium and cells were prefixed for 10 min at room temperature. After removing the supernatant, cells were fixed for an additional 15 min with 4% PFA in PBS and washed three times with PBS. Fixed cells were permeabilized by adding 0.2% Triton X-100 (Sigma) in PBS for 15 min at room temperature (this step was omitted for NG2, O4, and GalC staining). Subsequently, blocking was performed by incubating the cells with 5% normal goat serum (NGS; Gibco) and 5% FCS (Gibco) for 30 min at room temperature. Primary antibodies were applied overnight at 4 °C in blocking solution. Following three washing steps with PBS, cells were incubated with Alexa Fluor-conjugated secondary antibodies diluted in PBS for 1 h at room temperature. Cells were subsequently washed three times with PBS, including a Hoechst counterstaining for nuclei in the second washing step. Cells on glass coverslips were mounted in Dako Fluorescent Mounting Medium (Dako) and visualized on a Zeiss LSM700 confocal microscope. Cells on plastic cell culture plates were visualized on a Leica DMI6000 B inverted microscope. Primary antibodies used in this study are listed in Table S2.
Neuronal Differentiation.
For the derivation of human neurons, iPSC-derived NPC were cultured with N2B27 medium supplemented with 1 µM SAG (Cayman Chemical), 2 ng/mL BDNF (Peprotech), 2 ng/mL GDNF (Peprotech), and 100 µM AA (Sigma) for 6 d and afterward with N2B27 medium supplemented with 2 ng/mL BDNF (Peprotech), 2 ng/mL GDNF (Peprotech), 0.5 ng/mL TGF-β3 (Peprotech), 100 µM dbcAMP, and 100 µM AA. Additionally, 5 ng/mL Activin A (Sigma) was added to the medium from days 7–9. After 9 d of neuronal differentiation, cells were detached, singularized by treatment with accutase, and reseeded at densities of 2 × 105 per well in 24-well plates containing glass coverslips. After 21 d of differentiation, cells were used for coculture experiments. Transgene expression for the oligodendroglial differentiation experiments using the tet-inducible SON construct was induced with 1 µg/mL doxycycline (Clontech).
In Vitro Myelination Assay and 3D Culture.
To assess the in vitro myelination capacity of iOL, O4+ cells were purified at differentiation day 21 by magnetic cell sorting using anti-O4 MicroBeads (Miltenyi Biotec) following the manufacturer’s protocol. Purified iOL were added to 21-d-old neuronal cultures derived from iPSC-derived NPC populations with the aforementioned protocol at densities of 1 × 105 cells per well in Matrigel-coated 24-well plates containing glass coverslips. Cocultures were maintained in DM supplemented with 2 ng/mL BDNF and 2 ng/mL GDNF. After 14–28 d of coculture, cells were fixed in 4% PFA for ICC analysis.
For the 3D culture experiments, nanofiber chamber slides (Nanofiber Solutions) containing aligned nanofiber polymers were precoated with 10 µg/mL laminin (Sigma) and incubated with DM supplemented with 2 ng/mL BDNF and 2 ng/mL GDNF at 37 °C overnight. O4+ iOLs were purified after 21 d of differentiation using MACS and were reseeded at a density of 5 × 104 cells per chamber in DM supplemented with 2 ng/mL BDNF and 2 ng/mL GDNF. Half of the medium was changed every other day and cells were fixed in 4% PFA after 14 d for ICC analysis. All experiments have been repeated with at least three biological independent iOL populations.
Isolation of Adult Human pOL.
Microglia were separated and removed from the specimens by an initial adhesion step in which the total cell fraction was cultured for 24 h in noncoated flasks. The floating cell fraction was subjected to immunomagnetic bead selection with the A2B5 antibody (IgM) to select out progenitor cells (48). The nonselected fraction (referred to as pOL) was plated on poly-l-lysine–coated glass-chamber slides in defined medium (DFM) consisting of DMEM-F12 supplemented with N1 (Sigma), 0.01% BSA, 1% penicillin-streptomycin and B27 supplement lacking Vitamin A, 10 ng/mL PDGF, 10 ng/mL bFGF (Sigma), and 2 nM T3. After 6 d in DFM, cells were either lysed with Isol-RNA Lysis Reagent (Thermo Fisher) or fixed in 4% PFA for ICC analysis, demonstrating that ∼90% of cells were O4+ and MBP+ (Fig. S5). Expression of O1 (galactocerbroside) has been documented on these cells by flow cytometry.
Whole-Genome Expression Analysis.
Total RNA was quantified using a NanoDrop Spectrophotometer ND-1000 (NanoDrop Technologies) and its integrity was assessed using a 2100 Bioanalyzer (Agilent Technologies). Five nanograms of total RNA was used as input for cRNA synthesis with the Genechip WT Pico Reagent Kit (Affymetrix) according to the manufacturer’s protocol. The processed human iPSC-derived samples were hybridized onto GeneChip Human Transcriptome 2.0 Arrays (Affymetrix) and the primary human samples were hybridized onto Human Gene 2.0 ST Arrays (Affymetrix) for 16 h following the manufacturer’s protocol. Next, GeneChips were washed and stained using the GeneChip Hybridization, Wash and Stain Kit (Affymetrix) and the GeneChip Fluidics Station 450 (Affymetrix). The Arrays were scanned by the GeneChip Scanner 3000 7G (Affymetrix) and first data processing was performed by the GeneChip Command Console Viewer 3.2 (Affymetrix). iPSC-derived NPC and iOL RNA samples were processed at University Hospital Muenster, and pOL samples were processed by McGill University and the Genome Quebec Innovation Centre.
Microarray Data Processing.
Gene-expression data for iPSC samples were obtained from Gene Expression Omnibus (GSE61358) and used as a negative control. All microarray data from iPSC, pOL, iPSC-derived NPC, and iOL samples were processed using Bioconductor package “oligo.” Background subtraction, quantile gene-expression normalization, and summarization were performed using a robust multiarray average method implemented in the “oligo” package (50). Variance stabilization was performed using the log2 scaling. Differentially expressed genes among iOL and iPSC-derived NPC samples were identified through an unpaired one-way between-subject ANOVA. P values were corrected for multiplicity according to the Benjamini–Hochberg procedure with a threshold of 0.05 (false-discovery rate). Results were further filtered by fold-change magnitude (|fold change| ≥ 2). Graphics were obtained using ComplexHeatmap and VennDiagram R-packages. Hierarchical cluster of samples was performed with pvclust R-package using the one minus the sample correlation metric and complete-linkage clustering method. Probe mapping to the corresponding gene information was performed using the Bioconductor package Annotationdbi.
Cell Transplantation.
Shiverer mice were crossed to Rag2-null immunodeficient mice to generate a line of Shi/Shi Rag2−/− dysmyelinating immunodeficient mice. Mice were housed under standard conditions of 12-h light/ 12-h dark cycles with ad libitum access to dry food and water cycle at the Institut du Cerveau et de la Moelle épinière animal facility (PHENOPARC). Experiments were performed according to European Community regulations and INSERM ethical committee (authorization 75-348; 20/04/2005) and were approved by the local Darwin ethical committee. To assay iOL contributions to developmental myelination, newborn pups (n = 7) were cryoanesthetized before bilateral transplantation of RFP+ iOL, rostral to the corpus callosum. Injections (105 cellsµL each) were performed 1-mm caudally, 1-mm laterally from the bregma, and to a depth of 1 mm (51). Animals were killed at 16 wpg for immunohistological studies (n = 4) and EM (n = 3). To assay iOL involvement in remyelination, mice (n = 4) of 8–9 wk of age, were anesthetized by intraperitoneal injection of a mixture of 100 mg/kg ketamine (Alcyon) and 10 mg/kg xylazine (Alcyon). Focal demyelination was performed as previously described (51) by stereotaxic injection of 1 μL of 1% LPC (Sigma-Aldrich) in 1% PBS into the dorsal funiculus of the spinal cord at the level of the 13th thoracic vertebrae. Forty-eight hours after demyelination, mice received a single injection (1 µL, 105 cells/μL) of RFP+ iOL at the site of demyelination. All injections (LPC or cells) were performed at low speed (1 µL/2 min) using a stereotaxic frame equipped with a micromanipulator and a Hamilton syringe. Animals were killed at 12 wpg for immunohistological studies.
Immunohistochemistry.
For immunohistochemistry, mice were killed by transcardiac perfusion-fixation with 4% PFA in PBS and processed for freezing. After dissection, tissues were postfixed in the same fixative for 1 h, and then processed in 20% sucrose in 1× PBS overnight. Sagittal brain and serial cross spinal cord sections of 12-µm thickness were performed with a cryostat (CM3050S; Leica). Transplanted cells were identified by RFP expression and with anti-human cytoplasm (STEM121, 1:500; SC Proven) and anti-human nuclei (STEM101, 1:100; SC Proven) antigens. In vivo characterization of grafted cells was performed by immunostaining using the following antibodies: anti-MBP [(Abcam, ab7349) Rat, 1:100; (Millipore, MAB387) IgG2b, 1:100], anti-MOG (mouse IgG1 hybridoma, clone C18C5; 1:20; provided by C. Linnington, University of Glasgow, Glasgow, United Kingdom), anti-hGFAP (Covance; SMI21, IgG1, 1:1,000), anti-OLIG2 (Millipore AB 9610; rabbit, 1:400), anti-CC1(Millipore, OP80; IgG2b, 1:100), anti-NeuN (Abcam, ab177487; rabbit, 1:100). Identification of neurofilaments was performed by anti-NF200 (SMI 312, BioLegend, 837904; mixture IgG1-IgM, 1:200) or anti-NF165 (mouse IgG1 hybridoma 2H3, 1:20). Nodes of Ranvier were detected by anti-CASPR (1:1,000, rabbit polyclonal antibody, a generous gift from E. Peles, Weizmann Institute, Rehovot, Israel; from D. R. Colman, McGill University, Montreal, Canada, 1:200) and anti-AnkyrinG (Santa Cruz Biotechnology, sc-166602; IgG2b, 1:75). For MOG and MBP staining, sections were pretreated with ethanol (10 min, room temperature). For CASPR staining, slices were incubated in methanol (10 min, 20 °C) and saturated in the presence of 0.1% glycine (Research Organics). Secondary antibodies conjugated with FITC, TRITC (SouthernBiotech), or Alexa Fluor 647 (Life Technologies) were used, respectively, at 1:100 and 1:1,000. Nuclei were stained with Dapi (1 µg/mL; Sigma-Aldrich) (1:1,000). Tissue scanning, cell visualization and imaging were performed with a Carl Zeiss microscope equipped with ApoTome. 2.
Electron Microscopy.
For EM, grafted (n = 3) and nongrafted Shi/Shi Rag2−/− mice (n = 3) were perfused with 1% PBS followed by a mixture of 4% paraformaldehyde/5% glutaraldehyde (Electron Microscopy Science) in 1% PBS. After 2 h postfixation in the same solution, brains were cut sagitally in 100-µm-thick sections and fixed in 2% osmium tetroxide (Sigma-Aldrich) overnight. After dehydration, samples were embedded in Epon. Ultrathin sections (80 nm) of the medial part of the corpus callosum were examined and imaged with a HITACHI 120kV HT-7700 electron microscope.
Quantitative RT-PCR.
Total RNA was isolated from cell lysates using the RNeasy mini kit (Qiagen) according to the manufacturer’s protocol and including an on-column DNase digest (RNase free DNase Set; Qiagen). Quantification of total RNA was performed with Nanodrop ND1000 (Peqlab). cDNA was generated from isolated RNAs by reverse transcription using the High Capacity cDNA reverse Transcription Kit (Applied Biosystems). Quantitative RT-PCR was performed on an Applied Biosystems StepOne Plus real time cycler (Applied Biosystems) with the Power SYBR Green PCR master mix (Applied Biosystems). Specificity of the primers used for RT-PCR reactions was determined beforehand by agarose gelelectrophoresis. The melting curve of each sample was determined to ensure the specificity of the products. The quantitative RT-PCR conditions were 2 min at 50 °C, 10 min at 95 °C, 40 cycles of 15 s, at 95 °C, and 1 min at 60 °C. Relative expression levels were calculated using the 2−∆∆ct method and normalized to biological reference samples and using GAPDH as the housekeeping gene, unless otherwise noted. The primer sequences used in this study are listed in Table S3.
Compound Screen.
At day 5 of differentiation, cells were detached, singularized, and reseeded at densities of 5 × 104 cells in Matrigel-coated 48-well plates or 1.5 × 105 cells in 24-well plates containing Matrigel-coated glass coverslips. Cells were allowed to recover in minimum DM consisting of DMEM-F12 with 1:200 N2 supplement, 1:100 B27 supplement lacking vitamin A, 1% penicillin/streptomycin/glutamine, 200 µM AA, and 100 µM dbcAMP. After 24 h cells were treated with vehicle alone [0.01% (vol/vol) DMSO] as a negative control, 60 ng/mL T3 as a positive control, or with a drug candidate dissolved in DMSO at three different concentrations (0.5 µM, 1 µM, 5 µM) in minimum DM. The drug candidates comprised miconazole (Sigma), clobetasol (TCI), benztropine (Sigma), indometacin (Sigma), clemastine (Sigma), and oxybutynin (Sigma). Medium was changed every other day and cells were fixed in 4% PFA for ICC analysis after 21 d of treatment.
Stress-Induced Cell Death.
To examine the effect of rotenone (Sigma) on the viability of iOL derived from either N279K MAPT or control cells, O4+ iOL were purified using the MACS technology after 21 d of differentiation and replated at a density of 8 × 103 cells per well into Matrigel-coated 96-well plates. After another 6 d in DM, cells were treated with either vehicle [0.01% (vol/vol) DMSO] or rotenone dissolved in DMSO at three different concentrations (100, 250, 500 nM). After 24 h, cells were fixed and cell toxicity was determined by ICC double-staining using antibodies against cleaved CASPASE 3 and O4.
Quantifications.
Cell differentiation in vitro.
Quantification of cell populations using ICC or FACS were obtained from at least three independent differentiation experiments. For ICC quantification, at least four randomly chosen fields were analyzed and investigators were blinded for quantification.
Cell differentiation in vivo.
Quantification of STEM101/RFP+ grafted cells expressing cell-specific markers in adult demyelinated spinal cord, was performed in the lesion core of the dorsal funiculus according to our previously published method (40). The lesion was defined by MOG and cell density by Hoechst staining of all cell nuclei on adjacent sections. For each animal (n = 3), eight transverse serial sections at a 120-µm interval per animal were analyzed. Cell counts were expressed as the percentage of total STEM101/RFP+ cells in the lesion.
MBP extension within the dorsal funiculus was performed on serial cross-sections of the grafted spinal cords (n = 3) scanned with an automated slide scanner equipped with fluorescence (Zeiss, Axioscan). MBP extension corresponds to the distance spanning between the most rostral and most caudal sections containing MBP in each animal × distance (120 µm) between two sections.
EM analyses.
The g-ratio in grafted versus nongrafted mice was determined as previously described (22). Briefly, the maximum and minimum diameters of a given axon and the maximum and minimum axon plus myelin sheath diameter were measured with the ImageJ software at a magnification of 62,000× for at least 70 myelinated axons per animal. Data were expressed as the mean of the maximal and minimal values for each axon for grafted (n = 3 mice) or nongrafted Shi/Shi Rag2−/− mice (n = 3). The percentage of myelinated axons in the corpus callosum of the adult grafted or nongrafted mice was evaluated for the axons larger than 1-µm diameter on 40 visual fields (magnification 62,000×) per animal.
Acknowledgments
We thank Jacqueline Trotter (Molecular Cell Biology, Department of Biology, Johannes Gutenberg University Mainz) for critical reading and helpful discussion of the manuscript, and Martina Sinn and Ingrid Gelker for excellent technical support. This study was supported by German Research Foundation Grant SFB-TR128-B7; Ku1477/6-1 (to T.K.); Interdisciplinary Clinical Research Center, Münster Grant IZKF; KuT3/012/15 (to T.K.); an operating grant from the Multiple Sclerosis Society of Canada (to J.A.); Cluster of Excellence REBIRTH (A.S.); Multiple Sclerosis Society of the United States Grant RGPA-1501-02553; and INSERM, the program “Investissements d’Avenir” ANR-10-IAIHU-06 and ANR-11-INBS-0011–NeurATRIS. S.M. is supported by a Du Pré grant from the Multiple Sclerosis International Foundation, Fondation des Treilles, Ecole de Neurosciences de Paris, and Electricité de France Foundation. A.M. is supported by the European Leukodystrophy Foundation.
Footnotes
Conflict of interest statement: M.E. and T.K. have a pending patent application for the oligodendroglial differentiation protocol.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE79914).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614412114/-/DCSupplemental.
References
- 1.Najm FJ, et al. Drug-based modulation of endogenous stem cells promotes functional remyelination in vivo. Nature. 2015;522(7555):216–220. doi: 10.1038/nature14335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang N, et al. Generation of oligodendroglial cells by direct lineage conversion. Nat Biotechnol. 2013;31(5):434–439. doi: 10.1038/nbt.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldman SA, Nedergaard M, Windrem MS. Glial progenitor cell-based treatment and modeling of neurological disease. Science. 2012;338(6106):491–495. doi: 10.1126/science.1218071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franklin RJ, ffrench-Constant C, Edgar JM, Smith KJ. Neuroprotection and repair in multiple sclerosis. Nat Rev Neurol. 2012;8(11):624–634. doi: 10.1038/nrneurol.2012.200. [DOI] [PubMed] [Google Scholar]
- 5.Sreedharan J, Brown RH., Jr Amyotrophic lateral sclerosis: Problems and prospects. Ann Neurol. 2013;74(3):309–316. doi: 10.1002/ana.24012. [DOI] [PubMed] [Google Scholar]
- 6.Kim JB, et al. Oct4-induced oligodendrocyte progenitor cells enhance functional recovery in spinal cord injury model. EMBO J. 2015;34(23):2971–2983. doi: 10.15252/embj.201592652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tallantyre EC, et al. Clinico-pathological evidence that axonal loss underlies disability in progressive multiple sclerosis. Mult Scler. 2010;16(4):406–411. doi: 10.1177/1352458510364992. [DOI] [PubMed] [Google Scholar]
- 8.Atkins H, Freedman M. Immune ablation followed by autologous hematopoietic stem cell transplantation for the treatment of poor prognosis multiple sclerosis. Methods Mol Biol. 2009;549:231–246. doi: 10.1007/978-1-60327-931-4_16. [DOI] [PubMed] [Google Scholar]
- 9.Douvaras P, et al. Efficient generation of myelinating oligodendrocytes from primary progressive multiple sclerosis patients by induced pluripotent stem cells. Stem Cell Rep. 2014;3(2):250–259. doi: 10.1016/j.stemcr.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stacpoole SR, et al. High yields of oligodendrocyte lineage cells from human embryonic stem cells at physiological oxygen tensions for evaluation of translational biology. Stem Cell Rep. 2013;1(5):437–450. doi: 10.1016/j.stemcr.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S, et al. Human iPSC-derived oligodendrocyte progenitor cells can myelinate and rescue a mouse model of congenital hypomyelination. Cell Stem Cell. 2013;12(2):252–264. doi: 10.1016/j.stem.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piao J, et al. Human embryonic stem cell-derived oligodendrocyte progenitors remyelinate the brain and rescue behavioral deficits following radiation. Cell Stem Cell. 2015;16(2):198–210. doi: 10.1016/j.stem.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reinhardt P, et al. Derivation and expansion using only small molecules of human neural progenitors for neurodegenerative disease modeling. PLoS One. 2013;8(3):e59252. doi: 10.1371/journal.pone.0059252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Najm FJ, et al. Transcription factor-mediated reprogramming of fibroblasts to expandable, myelinogenic oligodendrocyte progenitor cells. Nat Biotechnol. 2013;31(5):426–433. doi: 10.1038/nbt.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Z, et al. Induction of oligodendrocyte differentiation by Olig2 and Sox10: Evidence for reciprocal interactions and dosage-dependent mechanisms. Dev Biol. 2007;302(2):683–693. doi: 10.1016/j.ydbio.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, et al. Transcription factor induction of human oligodendrocyte progenitor fate and differentiation. Proc Natl Acad Sci USA. 2014;111(28):E2885–E2894. doi: 10.1073/pnas.1408295111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrlich M, et al. Distinct neurodegenerative changes in an induced pluripotent stem cell model of frontotemporal dementia linked to mutant TAU protein. Stem Cell Rep. 2015;5(1):83–96. doi: 10.1016/j.stemcr.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lachapelle F, et al. Transplantation of CNS fragments into the brain of shiverer mutant mice: Extensive myelination by implanted oligodendrocytes. I. Immunohistochemical studies. Dev Neurosci. 1983-1984;6(6):325–334. doi: 10.1159/000112359. [DOI] [PubMed] [Google Scholar]
- 19.Gansmuller A, et al. Transplantations of newborn CNS fragments into the brain of shiverer mutant mice: Extensive myelination by transplanted oligodendrocytes. II. Electron microscopic study. Dev Neurosci. 1986;8(4):197–207. doi: 10.1159/000112253. [DOI] [PubMed] [Google Scholar]
- 20.Inoue Y, Inoue K, Terashima T, Mikoshiba K, Tsukada Y. Developmental changes of oligodendroglia in the posterior funiculus of “Shiverer” mutant mouse spinal cord, with special reference to myelin formation. Anat Embryol (Berl) 1983;168(2):159–171. doi: 10.1007/BF00315814. [DOI] [PubMed] [Google Scholar]
- 21.Buchet D, Garcia C, Deboux C, Nait-Oumesmar B, Baron-Van Evercooren A. Human neural progenitors from different foetal forebrain regions remyelinate the adult mouse spinal cord. Brain. 2011;134(Pt 4):1168–1183. doi: 10.1093/brain/awr030. [DOI] [PubMed] [Google Scholar]
- 22.Mozafari S, et al. Skin-derived neural precursors competitively generate functional myelin in adult demyelinated mice. J Clin Invest. 2015;125(9):3642–3656. doi: 10.1172/JCI80437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Preisner A, et al. Non-steroidal anti-inflammatory drug indometacin enhances endogenous remyelination. Acta Neuropathol. 2015;130(2):247–261. doi: 10.1007/s00401-015-1426-z. [DOI] [PubMed] [Google Scholar]
- 24.Mei F, et al. Micropillar arrays as a high-throughput screening platform for therapeutics in multiple sclerosis. Nat Med. 2014;20(8):954–960. doi: 10.1038/nm.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deshmukh VA, et al. A regenerative approach to the treatment of multiple sclerosis. Nature. 2013;502(7471):327–332. doi: 10.1038/nature12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LoPresti P. Regulation and differential expression of tau mRNA isoforms as oligodendrocytes mature in vivo: Implications for myelination. Glia. 2002;37(3):250–257. doi: 10.1002/glia.10035. [DOI] [PubMed] [Google Scholar]
- 27.Gorath M, Stahnke T, Mronga T, Goldbaum O, Richter-Landsberg C. Developmental changes of tau protein and mRNA in cultured rat brain oligodendrocytes. Glia. 2001;36(1):89–101. doi: 10.1002/glia.1098. [DOI] [PubMed] [Google Scholar]
- 28.Lam BY, Halliday GM, Irish M, Hodges JR, Piguet O. Longitudinal white matter changes in frontotemporal dementia subtypes. Hum Brain Mapp. 2014;35(7):3547–3557. doi: 10.1002/hbm.22420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kovacs GG, et al. White matter tauopathy with globular glial inclusions: A distinct sporadic frontotemporal lobar degeneration. J Neuropathol Exp Neurol. 2008;67(10):963–975. doi: 10.1097/NEN.0b013e318187a80f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu PH, et al. Regional differences in white matter breakdown between frontotemporal dementia and early-onset Alzheimer’s disease. J Alzheimers Dis. 2014;39(2):261–269. doi: 10.3233/JAD-131481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hallmann AL, et al. Astrocyte pathology in a human neural stem cell model of frontotemporal dementia caused by mutant TAU protein. Sci Rep. 2017;7:42991. doi: 10.1038/srep42991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goedert M, Spillantini MG. Tau mutations in frontotemporal dementia FTDP-17 and their relevance for Alzheimer’s disease. Biochim Biophys Acta. 2000;1502(1):110–121. doi: 10.1016/s0925-4439(00)00037-5. [DOI] [PubMed] [Google Scholar]
- 33.Czepiel M, et al. Differentiation of induced pluripotent stem cells into functional oligodendrocytes. Glia. 2011;59(6):882–892. doi: 10.1002/glia.21159. [DOI] [PubMed] [Google Scholar]
- 34.Hu BY, Du ZW, Zhang SC. Differentiation of human oligodendrocytes from pluripotent stem cells. Nat Protoc. 2009;4(11):1614–1622. doi: 10.1038/nprot.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Izrael M, et al. Human oligodendrocytes derived from embryonic stem cells: Effect of noggin on phenotypic differentiation in vitro and on myelination in vivo. Mol Cell Neurosci. 2007;34(3):310–323. doi: 10.1016/j.mcn.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 36.Sugimori M, et al. Ascl1 is required for oligodendrocyte development in the spinal cord. Development. 2008;135(7):1271–1281. doi: 10.1242/dev.015370. [DOI] [PubMed] [Google Scholar]
- 37.Pozniak CD, et al. Sox10 directs neural stem cells toward the oligodendrocyte lineage by decreasing Suppressor of Fused expression. Proc Natl Acad Sci USA. 2010;107(50):21795–21800. doi: 10.1073/pnas.1016485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu QR, et al. Sonic hedgehog-regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25(2):317–329. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- 39.Jessberger S, Toni N, Clemenon GD, Jr, Ray J, Gage FH. Directed differentiation of hippocampal stem/progenitor cells in the adult brain. Nat Neurosci. 2008;11(8):888–893. doi: 10.1038/nn.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maire CL, et al. Directing human neural stem/precursor cells into oligodendrocytes by overexpression of Olig2 transcription factor. J Neurosci Res. 2009;87(15):3438–3446. doi: 10.1002/jnr.22194. [DOI] [PubMed] [Google Scholar]
- 41.Lee S, et al. A culture system to study oligodendrocyte myelination processes using engineered nanofibers. Nat Methods. 2012;9(9):917–922. doi: 10.1038/nmeth.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bechler ME, Byrne L, Ffrench-Constant C. CNS myelin sheath lengths are an intrinsic property of oligodendrocytes. Curr Biol. 2015;25(18):2411–2416. doi: 10.1016/j.cub.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghetti B, et al. Invited review: Frontotemporal dementia caused by microtubule-associated protein tau gene (MAPT) mutations: A chameleon for neuropathology and neuroimaging. Neuropathol Appl Neurobiol. 2015;41(1):24–46. doi: 10.1111/nan.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seiberlich V, et al. Downregulation of the microtubule associated protein tau impairs process outgrowth and myelin basic protein mRNA transport in oligodendrocytes. Glia. 2015;63(9):1621–1635. doi: 10.1002/glia.22832. [DOI] [PubMed] [Google Scholar]
- 45.Klein C, et al. Process outgrowth of oligodendrocytes is promoted by interaction of fyn kinase with the cytoskeletal protein tau. J Neurosci. 2002;22(3):698–707. doi: 10.1523/JNEUROSCI.22-03-00698.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui QL, et al. Response of human oligodendrocyte progenitors to growth factors and axon signals. J Neuropathol Exp Neurol. 2010;69(9):930–944. doi: 10.1097/NEN.0b013e3181ef3be4. [DOI] [PubMed] [Google Scholar]
- 47.Ahfeldt T, et al. Programming human pluripotent stem cells into white and brown adipocytes. Nat Cell Biol. 2012;14(2):209–219. doi: 10.1038/ncb2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warlich E, et al. Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Mol Ther. 2011;19(4):782–789. doi: 10.1038/mt.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sommer CA, et al. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27(3):543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 51.Marteyn A, et al. Modulation of the innate immune response by human neural precursors prevails over oligodendrocyte progenitor remyelination to rescue a severe model of Pelizaeus-Merzbacher disease. Stem Cells. 2016;34(4):984–996. doi: 10.1002/stem.2263. [DOI] [PubMed] [Google Scholar]