Abstract
Three different nuclear genes encode the essential iron-sulfur subunit of mitochondrial complex II (succinate dehydrogenase) in Arabidopsis (Arabidopsis thaliana), raising interesting questions about their origin and function. To find clues about their role, we have undertaken a detailed analysis of their expression. Two genes (SDH2-1 and SDH2-2) that likely arose via a relatively recent duplication event are expressed in all organs from adult plants, whereas transcripts from the third gene (SDH2-3) were not detected. The tissue- and cell-specific expression of SDH2-1 and SDH2-2 was investigated by in situ hybridization. In flowers, both genes are regulated in a similar way. Enhanced expression was observed in floral meristems and sex organ primordia at early stages of development. As flowers develop, SDH2-1 and SDH2-2 transcripts accumulate in anthers, particularly in the tapetum, pollen mother cells, and microspores, in agreement with an essential role of mitochondria during anther development. Interestingly, in contrast to the situation in flowers, only SDH2-2 appears to be expressed at a significant level in root tips. Strong labeling was observed in all cell layers of the root meristematic zone, and a cell-specific pattern of expression was found with increasing distance from the root tip, as cells attain their differentiated state. Analysis of transgenic Arabidopsis plants carrying SDH2-1 and SDH2-2 promoters fused to the β-glucuronidase reporter gene indicate that both promoters have similar activities in flowers, driving enhanced expression in anthers and/or pollen, and that only the SDH2-2 promoter is active in root tips. These β-glucuronidase staining patterns parallel those obtained by in situ hybridization, suggesting transcriptional regulation of these genes. Progressive deletions of the promoters identified regions important for SDH2-1 expression in anthers and/or pollen and for SDH2-2 expression in anthers and/or pollen and root tips. Interestingly, regions driving enhanced expression in anthers are differently located in the two promoters.
The mitochondrial electron transport chain of eukaryotes consists of four major multimeric enzyme complexes, one of which is succinate:ubiquinone oxidoreductase (succinate dehydrogenase; EC 1.3.5.1), commonly referred to as complex II. This important membrane-associated complex is a functional part of both the citric acid cycle and the aerobic respiratory chain, catalyzing the oxidation of succinate to fumarate and the reduction of ubiquinone to ubiquinol. Complex II has been well characterized in bacteria, fungi, and mammals and is known to be the simplest of all the complexes of the electron transport chain, with four subunits (Lemire and Oyedotun, 2002; Yankovskaya et al., 2003). It contains two peripheral membrane proteins, a flavoprotein (SDH1) and an iron-sulfur protein (SDH2), and two small integral membrane proteins (SDH3 and SDH4). The succinate-binding site is formed by the SDH1 polypeptide, which is linked covalently to a FAD molecule acting as acceptor of a hydride ion at an early step of succinate oxidation. This flavoprotein subunit interacts with the SDH2 subunit that contains three nonheme iron-sulfur centers acting as conductors of electrons from the flavoprotein to the membrane. The two small integral proteins anchor the SDH1-SDH2 subcomplex to the matrix side of the inner membrane and contain a b-type heme and the ubiquinone-binding site (Yankovskaya et al., 2003). Interestingly, plant complex II appears to contain additional subunits of unknown function (Eubel et al., 2003).
Complex II subunits are all encoded in the nuclear genome in animals and fungi (Scheffler, 1998). Although in some angiosperms and Marchantia polymorpha SDH3 and SDH4 are present in the mitochondrial DNA (Burger et al., 1996; Adams et al., 2001), we have found that in the model plant Arabidopsis (Arabidopsis thaliana), SDH1, SDH2, SDH3, and SDH4 are nuclear encoded (Figueroa et al., 2001, 2002). Surprisingly, we found that three nuclear genes, designated SDH2-1 (At3g27380), SDH2-2 (At5g40650), and SDH2-3 (At5g65165), encode the iron-sulfur subunit in Arabidopsis. The three proteins are likely functional as complex II subunits since they are highly conserved compared with their homologs in other organisms and they contain the Cys motifs involved in binding the three iron-sulfur clusters essential for electron transport. Furthermore, they bear target peptides able to sustain active import into isolated plant mitochondria (Figueroa et al., 2001). To our knowledge, there is only one additional report describing more than one SDH2 gene in a eukaryotic organism: the sheep nematode Haemonchus contortus (Roos and Tielens, 1994). In this organism, two SDH2 genes are differentially expressed in the larval and adult stages, a fact that may be related to a switch in energy metabolism during development.
The presence of multiple SDH2 genes in Arabidopsis raises interesting questions about their origin and function. Duplication events have been frequent during Arabidopsis genome evolution (e.g. Blanc et al., 2000). One may speculate that genes arising from a very recent duplication event are functionally redundant and have similar expression patterns, whereas genes diverging for a long time have acquired new functions and/or different expression patterns. In this context, it is interesting to point out that Arabidopsis SDH2-1 and SDH2-2 genes likely arose via a relatively recent duplication event, whereas separation with SDH2-3 would be more ancient (Figueroa et al., 2001). Whereas SDH2-3 transcripts were not detected by northern-blot hybridization, SDH2-1 and SDH2-2 expression was detected in all organs examined, with the highest mRNA levels in flowers (Figueroa et al., 2001). Analogous enhanced expression in flowers has been observed for other plant nuclear genes encoding mitochondrial proteins (e.g. Huang et al., 1994; Heiser et al., 1996; Felitti et al., 1997) and for mitochondrial transcripts (e.g. Monéger et al., 1992; Smart et al., 1994). This may be related to the fact that sporogenesis is one of the highest energy-requiring process in plants. Furthermore, it is known that the number of mitochondria per cell increases in anthers to face this high-energy demand (Lee and Warmke, 1979; Huang et al., 1994).
These observations suggest that mitochondrial biogenesis requires the coordinate expression of nuclear and mitochondrial genes, but mechanisms underlying this regulation remain largely unknown. Regarding nuclear genes that encode mitochondrial proteins, few studies have been performed to characterize gene promoters (Zabaleta et al., 1998; Satoh et al., 2002; Thirkettle-Watts et al., 2003) or to analyze cell-specific expression (Smart et al., 1994; Ribichich et al., 2001). To characterize the regulation of nuclear genes encoding mitochondrial proteins and clarify further the roles of multiple SDH2 genes in complex II biogenesis, our group has undertaken a detailed analysis of SDH2 expression. In this study we analyze the expression of SDH2 transcripts by in situ hybridization, with emphasis in the process of flower development. Furthermore, transgenic Arabidopsis plants carrying progressive deletions of SDH2 promoters fused to the β-glucuronidase (GUS) reporter gene allowed us to define promoter regions involved in tissue-specific expression.
RESULTS
Expression of SDH2 Genes in Organs from Arabidopsis Adult Plants
SDH2 mRNA steady-state levels in flowers, leaves, inflorescences, stems, and roots were examined by the sensitive, semiquantitative reverse transcription (RT)-PCR Multiplex procedure. The 18S rRNA was simultaneously analyzed as an internal standard.
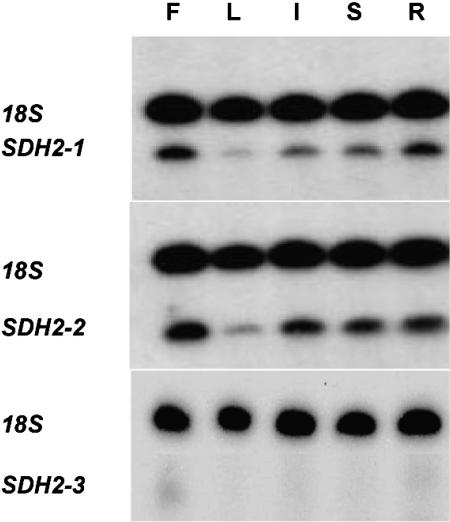
Similar expression patterns were obtained for SDH2-1 and SDH2-2 mRNAs, showing that both genes are transcribed in all organs from adult plants (Fig. 1). The highest and lowest mRNA levels were found in flowers and leaves, respectively. In contrast, SDH2-3 transcripts could not be detected by this sensitive method, confirming previous results obtained by northern-blot hybridization analysis (Figueroa et al., 2001). These results prompted us to further characterize SDH2 gene expression at the cellular level.
Figure 1.
RT-PCR Multiplex analysis of SDH2 transcripts in adult organs. Total RNA was obtained from flowers (F), leaves (L), inflorescences (I), stems (S), and roots (R). SDH2 transcripts were analyzed by RT-PCR Multiplex as described in “Materials and Methods” with 26 amplification cycles for SDH2-1 and SDH2-2 and 32 cycles for SDH2-3. Radioactive PCR products corresponding to SDH2 transcripts and 18S rRNA were fractionated on urea-polyacrilamide gels and visualized by autoradiography.
Cell-Specific Accumulation of SDH2 Transcripts
We have used RNA in situ hybridization to analyze developmental regulation of SDH2 expression and to ascertain if SDH2-1 and SDH2-2 expression patterns show differences suggesting functional divergence of the genes. We focus on transcripts in Arabidopsis flowers and root tips, since enhanced mitochondrial biogenesis has been reported to occur in meiotic and tapetal cells during anther development and in root tips (Lee and Warmke, 1979; Fujie et al., 1993). Before in situ RNA hybridizations, we verified that cross hybridization does not occur when using RNA probes from the 3′ untranslated regions (UTR) of SDH2-1, SDH2-2, and SDH2-3.
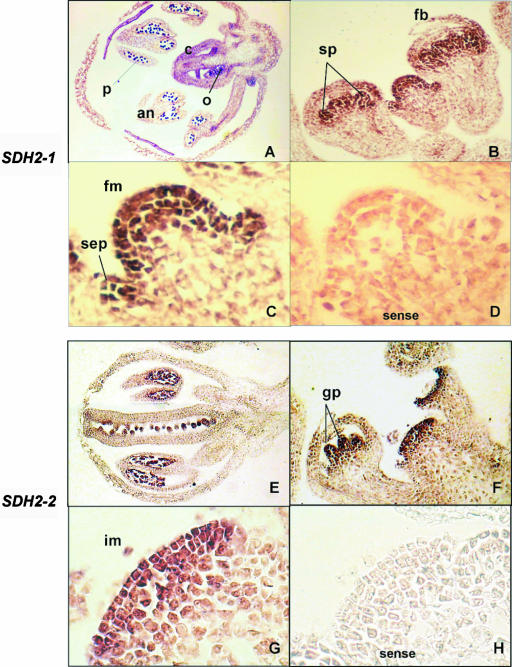
The SDH2-3 probe was unable to detect transcripts in either floral tissues or root tips (data not shown). In contrast, expression of SDH2-1 and SDH2-2 occurs in all tissues of Arabidopsis inflorescences (Fig. 2). In flower buds from stages 3 to 7 (Bowman, 1994), high expression was observed in floral and inflorescence meristems and in sepal, stamen, and gynoecium primordia (Fig. 2, B, C, F, and G). Later in development (floral stages 10–12), transcripts were detected in sepals, petals, anthers, and carpels, being more abundant in microspores and ovules (Fig. 2, A and E).
Figure 2.
Expression of SDH2 genes in developing flower buds of Arabidopsis. SDH2-1 (A–C) and SDH2-2 (E–G) DIG-labeled specific antisense RNA probes were hybridized to longitudinal sections of flower buds at early (B, C, F, and G) and later (A and E) developmental stages. C and G are enlargements of B and F, respectively. D and H, Control sense strand probes hybridized to sections equivalent to those shown in C and G. an, Anther; c, carpel; fb, flower bud; fm, flower meristem; gp, gynoecium primordia; im, inflorescence meristem; o, ovary; p, pollen; sep, sepal primordium; sp, stamen primordia.
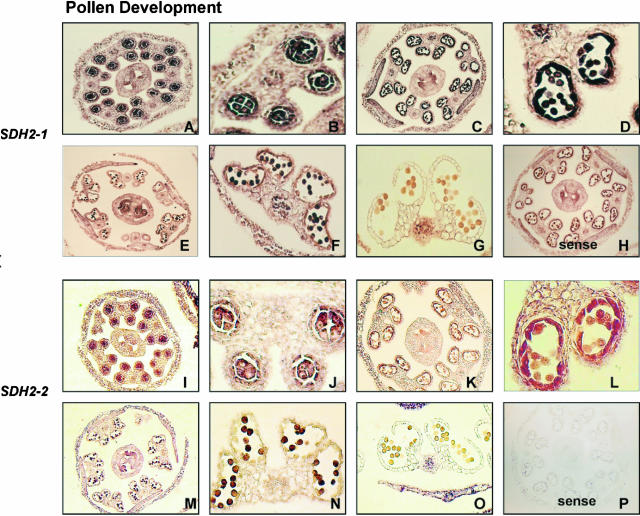
Since pollen development is known to be a high energy-requiring process in plants, we performed in situ hybridization to transverse sections through anthers at different floral stages (Fig. 3). Expression of SDH2-1 and SDH2-2 transcripts occurs in premeiotic anthers, with pollen mother cells and tapetum showing the highest abundance of both mRNAs (Fig. 3, A, B, I, and J). Transcript levels persist in the tapetum and microspores in stage 10 flower buds, where anthers are at a postmeiotic stage and microspores have been released from the tetrads (Fig. 3, C, D, K, and L). Thereafter, transcript levels remain high in microspores but markedly decline in the degenerating tapetum (Fig. 3, E, F, M, and N). Finally, no significant SDH2 expression was seen within the anthers at dehiscence (floral stage 13; Fig. 3, G and O). Altogether, these results show that SDH2-1 and SDH2-2 genes are regulated in a very similar way during flower development.
Figure 3.
Higher abundance of SDH2-1 and SDH2-2 transcripts in meiocyte and tapetal cells. DIG-labeled antisense SDH2-1 (A–G) and SDH2-2 (I–O) probes were hybridized to transverse anther sections at different stages of development. A, B, I, and J, Premeiotic anthers, floral stage 9 (Bowman, 1994); C, D, K, and L, postmeiotic anthers, microspores have been released from the tetrads (floral stage 10); E, F, M, and N, floral stage 12, mitotic division of microspores and tapetum degeneration have occurred; G and O, dehiscent anthers, floral stage 13. B, D, F, J, L, and N are enlargements of A, C, E, I, K, and M, respectively. H and P, Control hybridizations of sense probes equivalent to those shown in C and K, respectively.
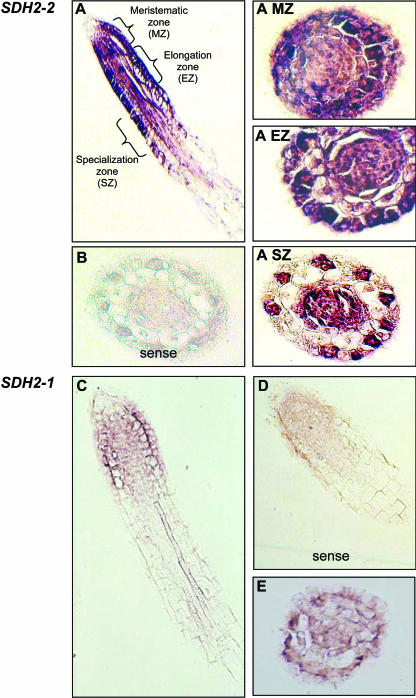
Interestingly, SDH2-1 and SDH2-2 expression patterns are clearly different in root tip sections (Fig. 4). Whereas SDH2-1 transcripts were barely detected (Fig. 4, C–E), SDH2-2 is strongly expressed (Fig. 4, A and B). Moreover, cell-specific patterns were found for this gene. Indeed, SDH2-2 transcript levels were high in all cell layers of the meristematic zone (e.g. epidermis, cortex, endodermis, and stele; Fig. 4A, subsection MZ). In the elongation zone, transcript abundance decreases selectively in the cortex (Fig. 4A, subsection EZ). With increasing distance from the root tip, transcripts became highly localized to the vascular cylinder, endodermis, and, interestingly, to the epidermal cells that will give rise to the root hairs (recognized by their location over the anticlinal walls in the cortex; Fig. 4A, subsection SZ).
Figure 4.
Localization of SDH2 transcripts in root tip sections by in situ hybridization. SDH2-2 (A–A, subsection SZ) and SDH2-1 (C and E) antisense probes were hybridized to longitudinal (A and C) and transverse (A, subsections MZ–SZ, and E) sections of root tips. B and D, Control hybridizations of SDH2-2 (B) and SDH2-1 (D) sense probes to sections equivalent to those shown in A, subsection SZ, and C, respectively.
Analysis of the SDH2 Promoters
To investigate regulatory-control regions of nuclear genes encoding complex II iron-sulfur subunit, upstream sequences were fused to the GUS gene (uid) and their expression tested in transgenic plants. The constructs containing the putative promoter regions, the 5′ UTR, and the first codon in frame with the reporter gene are depicted in Figure 5. Arabidopsis plants were transformed by the floral dip method, and the presence of transgenes was confirmed by PCR. For each construct, histochemical GUS staining patterns were analyzed in at least 12 independent transformed lines to define a major relevant expression pattern. As expected, variations in intensity were observed among lines carrying the same construct, likely due to positional effects resulting from random T-DNA insertion.
Figure 5.
Schematic representation of SDH2 reporter gene fusions. The gray boxes represent the different promoter regions used in P5 to P1 constructs. Promoter sizes (in bp) are shown on the right and are relative to the transcription initiation site. White and black boxes represent the 5′ UTR and GUS-coding region, respectively.
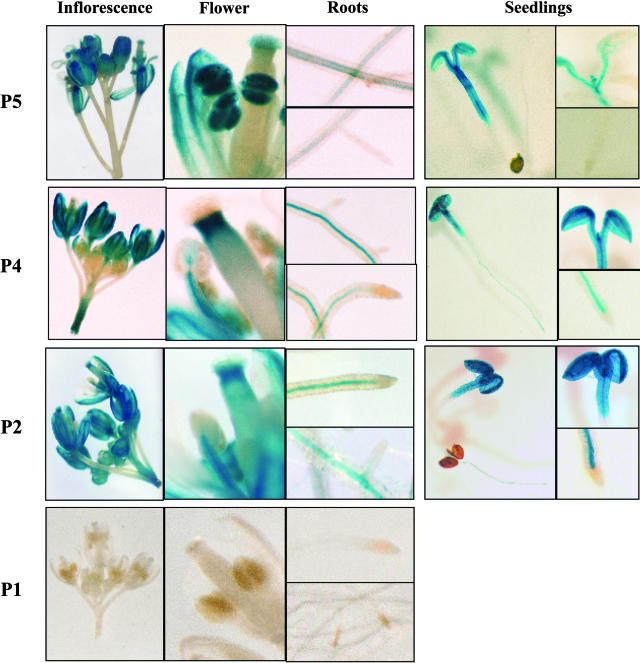
Results of GUS staining in transgenic seedlings and plants carrying SDH2-1 constructs are shown in Figure 6. A 1.6-kb upstream region (P5) appears to mimic the expression pattern found by in situ hybridization, driving strong GUS expression in flowers, especially in anthers and/or pollen. Weak GUS activity was detected in root vascular tissue but not in root tips or emergent lateral roots. In 7-d-old seedlings, GUS activity appeared strong in cotyledons, hypocotyl, and primordia of first new true leaves.
Figure 6.
GUS expression patterns in transgenic plants carrying SDH2-1 promoter fusions. Histochemical localization of GUS activity is shown for 7-d-old seedlings and adult plants carrying P5, P4, P2, and P1 constructs. P5 construct drives high expression in flowers, particularly in anthers, cotyledons, and hypocotyl, and weak expression in roots. In plants with the P4 construct, no GUS activity was detected in anthers, and root vascular tissue shows higher GUS staining. In plants with the P2 construct, GUS staining is similar to that of the P4 construct.
Interestingly, deletion of the region between −1,598 and −385 (relative to the transcription initiation site) abolished GUS expression exclusively in anthers (Fig. 6, P4 construct), suggesting the presence of specific cis-elements involved in transcriptional activation in anthers and pollen. Stronger expression was observed in the root vascular system in comparison to P5 construct, while no GUS activity was detected in root tips or emergent lateral roots. Additional deletion of the promoter to −89 (P2 construct) did not affect GUS staining patterns, but removal of the region between −89 and −11 (P1 construct) abolished GUS expression in all tissues. These results indicate that the region between −89 and −11 contains the elements required to drive expression of SDH2-1 in most tissues.
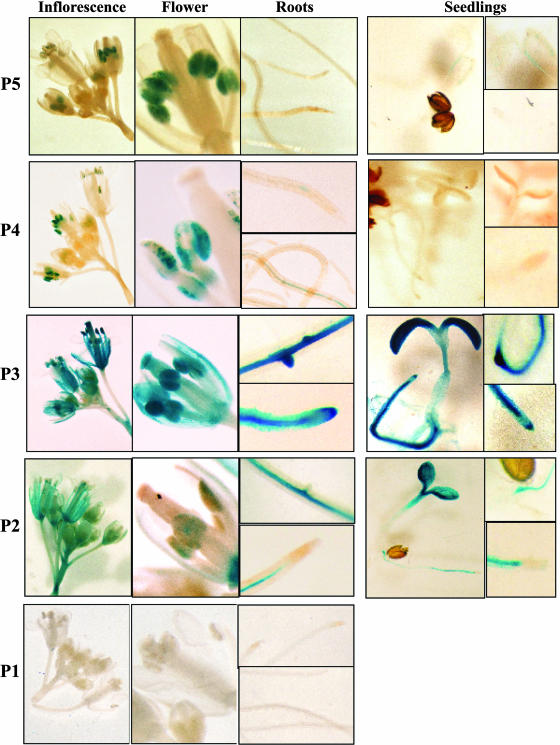
Histochemical analysis of transgenic plants and seedlings carrying SDH2-2 promoter constructs is depicted in Figure 7. The P3 construct, containing 254 bp of the promoter region, mimics the expression pattern observed by in situ hybridization; it drove GUS expression in flowers and roots and higher expression was found in anthers, root tips, and emergent lateral roots. In 7-d-old seedlings, GUS activity was strong in cotyledons, root tips, root vascular tissue, and emerging lateral roots. Larger fragments of this promoter (P5 and P4 constructs) gave unexpected results. Although some transgenic lines showed GUS staining patterns similar to those observed for P3 construct, in most lines GUS activity was present exclusively in anthers (Fig. 7). This result, which confirmed the strong activity of the SDH2-2 promoter in anthers, suggests the presence of negative regulatory elements upstream of −254 (see “Discussion”).
Figure 7.
GUS expression patterns in transgenic plants carrying SDH2-2 promoter fusions. Seven-day-old seedlings and adult plants containing P5, P4, P3, P2, and P1 constructs were stained as described in “Materials and Methods.” P3 construct drives high GUS expression in flowers (especially in anthers), in roots (including root tips and lateral root primordia), and cotyledons. Deletion of the upstream region to produce P2 drastically reduced GUS staining in anthers and root tips and lateral primordia. In plants with P5 and P4 constructs, strong GUS staining was observed only in anthers.
Deletion of the region between −254 and −92 of the SDH2-2 promoter drastically reduced GUS expression in anthers and root tips (Fig. 7, P2 construct). These results suggest that this region contain cis-elements required for expression in root tips and pollen grains. Further deletion of the SDH2-2 promoter (P1 construct) resulted in undetectable GUS activity, demonstrating that the region between −91 and −10 confers basal constitutive expression in most tissues.
The above data show that both genes contain cis-elements for anther and/or pollen expression (upstream of −384 for SDH2-1 and downstream of −254 for SDH2-2) and that only SDH2-2 contains cis-elements for expression in root tips and emergent lateral primordia (between −254 and −92). These observations may be related to the fact that anther development is a higher energy-demanding process.
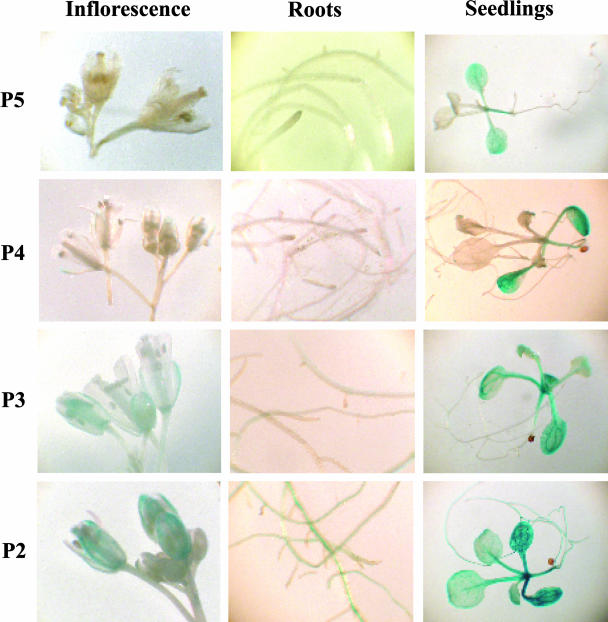
A histochemical analysis of transgenic plants carrying SDH2-3 promoter constructs has been performed (Fig. 8). Larger fragments (P5 and P4 constructs) do not drive GUS expression in flowers and roots from adult plants, in agreement with northern and RT-PCR Multiplex results (Figueroa et al., 2001; Fig. 1). Weak GUS activity was observed in young seedlings, although SDH2-3 transcripts were not detected in RT-PCR Multiplex experiments (data not shown). Thus, the SDH2-3 promoter may have some activity in young seedlings, yet transcripts may be unstable. Progressive deletions of this promoter unveil some GUS activity in flowers, roots, and seedlings, suggesting the presence of negative regulatory elements.
Figure 8.
GUS expression patterns in transgenic plants carrying SDH2-3 promoter fusions. Histochemical localization of GUS activity is shown for adult plants and 13-d-old seedlings carrying P5, P4, P3, and P2 constructs. No GUS activity was detected in flowers or roots from adult plants carrying P5 and P4 constructs, whereas weak GUS expression was observed in P3 and P2 constructs. Weak GUS activity was also observed in young P5 seedlings, and this activity increases progressively with promoter deletions (P4–P2 constructs).
Isolation of a T-DNA Insertional Mutation of the SDH2-1 Gene
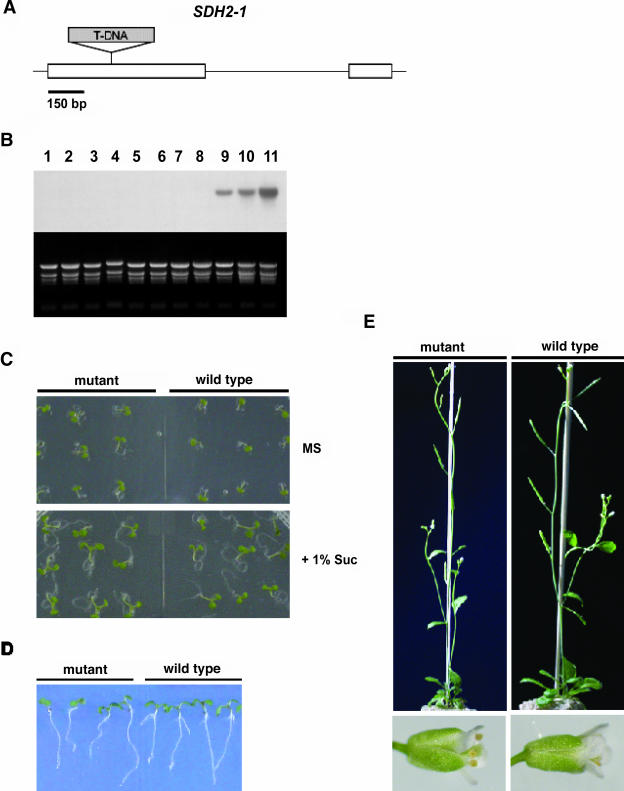
We searched the SALK and GARLIC sequence-indexed mutant collections for T-DNA insertions in SDH2 coding regions. Only three different mutant lines that carry T-DNA insertions in the SDH2-1 gene were identified. The SALK_031100 mutant, harboring a T-DNA insertion in exon 1, was obtained from the Arabidopsis Biological Resource Center (ABRC; Ohio State University, Columbus) and further characterized. The T-DNA was confirmed to be in the first exon of the SDH2-1 coding region, between codons 93 and 94 (Fig. 9A). Disruption at this position, which is upstream from regions encoding the three conserved Cys clusters involved in binding the iron-sulfur centers, is expected to result in a null mutation.
Figure 9.
Isolation of a T-DNA insertion mutant allele of SDH2-1 in Arabidopsis. A, Scheme of the SDH2-1 gene. The position of the T-DNA insertion within SDH2-1 in the SALK_031100 line is shown. White boxes denote exons while black lines indicate UTRs and the intron. The T-DNA insert is not drawn to scale and its left border is to the left of the figure. B, Northern-blot analysis of SDH2-1 gene expression in plants either homozygous (lanes 1–8) or hemizygous (lanes 9 and 10) for the T-DNA insertion and in wild-type Col-0 plants (lane 11). Each lane was loaded with 20 μg of total RNA isolated from leaves. The blot was hybridized with the SDH2-1-specific probe. Ethidium bromide-stained rRNAs served as a loading control. The SDH2-1 transcript was present in total RNA from wild-type and hemizygous mutant plants (at a lower level) but absent from homozygous mutant plants. C and D, Growth of homozygous mutant and wild-type plants. Seven-day-old seedlings were grown on Murashige and Skoog plates with or without Suc (C) and on Murashige and Skoog plates placed vertically to analyze root growth (D). E, Six-week-old homozygous mutant and wild-type plants.
Homozygous SALK_031100 mutant plants were obtained as described in “Materials and Methods.” Northern-blot analysis confirmed that it is a null allele since no SDH2-1 transcripts were detected in these homozygous plants (Fig. 9B, lanes 1–8). The sdh2-1 mutant showed no apparent defective phenotypes as to growth and development, at least under the growth conditions used (Fig. 9, C–E). Particularly, it did not exhibit any obvious phenotype during reproductive growth (i.e. onset of flowering, number, and morphology of flowers, fertility, seed set) when compared to wild-type plants. These results indicate that under the growth conditions used, the loss of SDH2-1 has little or no impact on Arabidopsis growth and development, suggesting functional redundancy between SDH2-1 and SDH2-2.
DISCUSSION
SDH2-1 and SDH2-2 Arose Via a Relatively Recent Duplication Event
Three nuclear genes encode the iron-sulfur subunit of complex II in Arabidopsis (Figueroa et al., 2001). This unexpected finding raises interesting questions about their origin and function. Several observations suggest that Arabidopsis SDH2-1 and SDH2-2 arose via a relatively recent duplication event. First, both genes present a similar exon-intron structure, with a single intron located at a conserved position. Second, they encode nearly identical proteins (96% identity). And third, this similarity extends into the sequence encoding the putative N-terminal mitochondrial-targeting amino acid sequence. In contrast, separation with SDH2-3 gene would be more ancient since it has a different exon-intron structure, with four introns. Moreover, SDH2-3 encodes a protein that is only 67% similar to SDH2-1 and SDH2-2 and has a different N-terminal presequence.
RT-PCR Multiplex analysis gave a similar transcript distribution for SDH2-1 and SDH2-2 in organs from adult plants (Fig. 1). In contrast, RT-PCR Multiplex and in situ hybridization failed to detect SDH2-3 transcripts. Moreover, SDH2-3 promoter appears to be inactive at least in adult plants (Fig. 8, P5 and P4 constructs). These results suggest that SDH2-3 does not contribute significantly to complex II biogenesis. It has to be pointed out, however, that SDH2-3 is an active gene since transcripts were detected in RT-PCR and 3′RACE experiments (Figueroa et al., 2001). Therefore, SDH2-3 appears to be expressed at a very low level, and further work will be necessary to unveil its role.
SDH2-1 and SDH2-2 Are Highly Expressed in Anthers, a High Energy-Demanding Organ
Increased SDH2-1 and SDH2-2 expression was observed in floral and inflorescence meristems, in sex organ primordia, and in anthers. In anthers, transcripts are highly localized in the tapetum, which has a high biosynthetic activity to support microspore development, as well as in microspore mother cells. This remarkable similarity in SDH2-1 and SDH2-2 expression during flower development suggests that these genes may be redundant.
Our results are in agreement with an essential role for mitochondria during anther development. This role was first suggested by the phenomenon of cytoplasmic male sterility, in which mitochondrial defects specifically disrupt pollen formation (Hanson, 1991). Active biogenesis of mitochondria has been shown to occur in meiotic and tapetal cells during early anther development in maize, where numbers of mitochondria increase 20- to 40-fold (Lee and Warmke, 1979). Consistently, several studies have reported increases in abundance of transcripts for mitochondrial proteins during flower development (e.g. Monéger et al., 1992; Huang et al., 1994; Smart et al., 1994; Heiser et al., 1996; Felitti et al., 1997). However, most studies of tissue-specific or developmental regulation have relied on the analysis of total RNA preparations from different plant organs, and differences within specialized cells could not be detected. Cell-specific expression of several mitochondrial genes in developing anthers has been documented (Smart et al., 1994). Few studies have used the in situ hybridization approach to localize transcripts of nuclear-encoded mitochondrial proteins in specialized cells. The accumulation of transcripts encoding the β-subunit of ATP synthase and cytochrome c has been reported in meiocytes and tapetal cells at premeiosis, and only in tapetal cells at later stages (Balk and Leaver, 1998; Ribichich et al., 2001). These results and those obtained by Smart et al. (1994) for mitochondrial genes overlap with our data on SDH2-1 and SDH2-2 expression in flowers, suggesting a coordinate regulation of gene expression in mitochondrial biogenesis.
SDH2-2 Is Specifically Expressed in Roots
Interestingly, and in contrast to the situation described in flowers, SDH2-1 and SDH2-2 are differentially expressed in root tips. Indeed, only SDH2-2 appears to be expressed at a significant level, suggesting that SDH2-1 and SDH2-2 are only partially redundant in Arabidopsis. It is tempting to speculate that different roles may be evolving for these genes after their divergence from a common ancestor. Active mitochondrial biogenesis has been shown to occur in the root apical meristem of Arabidopsis and other species (e.g. Kuroiwa et al., 1992; Fujie et al., 1993). The analysis of root cell-specific expression of either mitochondrial transcripts (Li et al., 1996) or nuclear transcripts encoding mitochondrial proteins (Ribichich et al., 2001) showed that transcript levels correlated with cell division activity, in agreement with the high levels of SDH2-2 transcripts found in the meristematic zone of Arabidopsis root tips. In addition, our results showed a cell-specific transcript accumulation in the epidermis of the root specialization zone, where only trichoblasts, which will give rise to root-hair cells, showed significant SDH2-2 expression. This observation may be related to the fact that trichoblasts display a greater cell-division rate, a reduced cell length, and a greater cytoplasmic density than immature nonhair cells (atrichoblasts; Scheres et al., 2002, and refs. therein).
Consistent with the essential role of succinate dehydrogenase, SDH2 transcripts were detected in every organ examined. Nevertheless, our results provide compelling evidence that the three nuclear genes encoding the iron-sulfur subunit of complex II are differentially expressed and that the expression of SDH2-1 and SDH2-2 is regulated in a cell- and developmental-specific manner.
SDH2 Promoters Have cis-Elements Important for Anther and/or Pollen and Root Tip Expression
To gain further insight into the mechanisms of SDH2-1 and SDH2-2 gene regulation, we have used regions upstream of these genes to drive the expression of the GUS reporter protein in transgenic Arabidopsis plants, and we have defined important regions for promoter activity. GUS staining patterns obtained for SDH2-1 (P5 construct) and SDH2-2 (P3 construct) in flowers and roots parallel those observed by in situ hybridization, suggesting that control of the expression of these nuclear complex II genes is achieved mainly through transcriptional regulation (Figs. 6 and 7). When larger fragments of the SDH2-2 upstream region (P5 and P4 constructs) were fused to GUS, staining was observed mainly in anthers. These results suggest the presence of negative regulatory elements upstream of −254 and that GUS assay was less sensitive than in situ hybridization, since down-regulation of GUS expression may prevent its detection except in tissues with higher expression (anthers). However, it has to be pointed out that our constructs were obtained before annotation of the genes located upstream of SDH2-1 and SDH2-2, and that P5 constructs encompass neighboring genes (Fig. 10A). Thus, P5 SDH2-2 construct contains the whole At5g40645 gene, which is transcribed divergently from SDH2-2. The presence of this gene in chromosome 5, and its absence in chromosome 3, may explain why GUS activity of longer promoter constructs does not parallel (SDH2-2) or parallel (SDH2-1) in situ hybridization patterns. Alternatively, since no GUS activity was observed with the 1.6-kb SDH2-2 upstream region in tissues where transcripts were detected by in situ hybridization, additional positive elements may exist downstream from the translation initiation site or further upstream of the 1.6-kb region.
Figure 10.
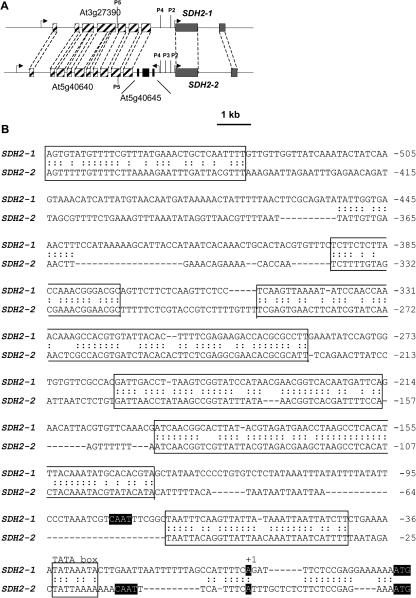
Comparison between SDH2-1 and SDH2-2 promoter sequences. A, Schematic representation of the Arabidopsis nuclear loci containing SDH2-1 (At3g27380) and SDH2-2 (At5g40650) genes. Gray boxes represent SDH2 exons and hatched boxes the exons of two similar genes located upstream from SDH2-1 and SDH2-2. In chromosome 5, a three-exon gene (At5g40645) is inserted between SDH2-2 and At5g40640. Curved arrows indicate transcriptional orientations. The 5′ ends of upstream regions fused to GUS in P5 to P2 constructs are indicated by vertical lines. B, The sequences upstream from SDH2-1 and SDH2-2 ATG initiation codons are compared. Putative TATA boxes and similarity stretches between both promoters are boxed, a colon (:) indicating nucleotide identity. ATG translation initiation codons, transcription initiation sites (+1) mapped by 5′RACE (Figueroa et al., 2001) and putative CAAT boxes are indicated by white letters on black background. Sequence numbers are in relation to transcription initiation site.
SDH2-1 and SDH2-2 expression patterns were similar in flowers, root vascular system, seedling cotyledons, and hypocotyl, suggesting a coordinated control of their expression. This can be taken as an indication that both genes carry similar cis-regulatory elements in their promoters and led us to compare the sequences of the upstream regions (Fig. 10B). In agreement with a relatively recent duplication event at the origin of these genes, stretches of sequence similarity were found, which may be responsible for their common expression in most tissues. However, the loss-of-function experiments delimited different regions in the SDH2-1 promoter (upstream of −385) and the SDH2-2 promoter (−254 through −92) important for expression in anthers and pollen. Moreover, these experiments defined a promoter region (−254 through −92), which is important for expression in root tips and lateral root primordia only in SDH2-2. Considered together, these results show that, in spite of sequence similarities, SDH2-1 and SDH2-2 promoters have functionally diverged during evolution since they contain different elements for anther and/or pollen expression and only the SDH2-2 promoter contains elements for root tip expression.
To our knowledge, gene promoters from citric acid cycle enzymes have not been analyzed in plants, and only one study has characterized promoters from nuclear genes encoding mitochondrial respiratory complex subunits (Zabaleta et al., 1998). These authors report that expression of Arabidopsis complex I genes is specifically enhanced at the transcriptional level in anthers and pollen, and defined in detail the promoter regions involved in this transcriptional regulation. These regions have specific motifs important for pollen expression (Twell et al., 1991; Eyal et al., 1995; Rogers et al., 2001). Here we report the first analysis, to our knowledge, of promoters from genes encoding subunits of a complex involved in both the citric acid cycle and the aerobic-respiratory chain.
Our results indicate that the first approximately 90 bp from SDH2-1 and SDH2-2 promoters, which include putative TATA and CAAT boxes (Fig. 10B), contain sequences essential for transcription in all plant tissues but not sufficient for expression in anthers (SDH2-1 and SDH2-2) and root tips and primordia (SDH2-2). Thus, elements required for anther- or root-specific expression appear to be physically separated from basal elements. In the SDH2-1 promoter, the region upstream of −385 was found to be required for pollen expression and to exert a negative effect on root vascular expression. The presence of analogous negative-regulatory elements has been recently reported for the promoters of the nuclear genes encoding mitochondrial alternative oxidases (Thirkettle-Watts et al., 2003).
The region between −254 and −92 of the SDH2-2 promoter was found to enhance expression in pollen and root tips (Fig. 7). No pollen elements highly similar to those characterized in tomato LAT genes or Arabidopsis complex I genes are present in this region, the best match being found between the LAT56/59 box (GAAA/TTTGTGA) and the sequence from −169 to −160 of the SDH2-2 promoter (complementary strand, AAAATCGTGA). Further work is needed to ascertain the functional role of this or other sequences.
SDH2-1 Disruption Has No Impact on Arabidopsis Growth
We have shown that two out of three genes encoding the complex II iron-sulfur subunit are expressed at a significant level in Arabidopsis adult plants. While these two genes are highly expressed during flower development, particularly in the tapetum and microspores, only SDH2-2 appears to be active in root tips. These results support the view that SDH2-1 and SDH2-2 have acquired different regulatory mechanisms since their divergence.
To gain further insight into the role of these genes, a search for SDH2 insertional mutants was performed. Three different mutant lines harboring T-DNA insertions in SDH2-1 were identified in the SALK and GARLIC mutant collections. Homozygous mutant plants were obtained for the SALK_031100 line, which carries the T-DNA insert between codons 93 and 94. This insertion should result in a null mutation, and northern analysis confirmed that no functional SDH2-1 mRNA is produced. Nevertheless, no visible phenotypic differences were seen between sdh2-1 mutant and wild-type plants under normal growth conditions. Therefore, these results suggest that there is functional redundancy between SDH2-1 and SDH2-2. The lack of altered phenotype in the sdh2-1 mutant carrying the intact SDH2-2 gene, as well the expression profiles (SDH2-2 is expressed wherever SDH2-1 is expressed), strongly suggest that SDH2-1 biological role overlaps with that of SDH2-2. Since we have not identified SDH2-2 insertion mutants in the sequence-indexed T-DNA insertion collections, we do not know if mutant sdh2-2 plants carrying the intact SDH2-1 gene will show any defective phenotype.
MATERIALS AND METHODS
Plant Growth and Transformation
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (Col-0) plants were grown hydroponically at 22°C under a 16-h-light/8-h-dark cycle (Gibeaut et al., 1997). Agrobacterium tumefaciens-mediated transformation of Arabidopsis plants was accomplished using the floral dip protocol (Clough and Bent, 1998). Seeds of the T1 generation were selected for resistance to hygromycin. Seeds were surface sterilized with a solution composed of 50% (v/v) commercial bleach and 0.05% (v/v) Tween 20 for 10 min, then rinsed three times with sterile distilled water. Seeds were placed on one-half-concentrated Murashige and Skoog medium solidified with 0.8% (w/v) agar and supplemented with 25 μg mL−1 hygromycin. Seeds were cold treated at 4°C for 48 h in darkness, then germinated. Plants were grown in a controlled growth chamber at 22°C under a 16-h-light/8-h-dark cycle. At least 12 independent transgenic lines were obtained for each construct and transgene presence verified by PCR.
Isolation of RNA and RT-PCR Multiplex
Total RNA was isolated from Arabidopsis samples using the RNeasy Plant mini kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. For RT-PCR Multiplex, total RNA (2 μg) was treated with 2 units of RQ1 RNase-free DNase (Promega, Madison, WI) at 37°C for 35 min in 40 mm Tris-HCl, pH 8, 10 mm MgSO4, 1 mm CaCl2. cDNA synthesis was performed on 0.2 μg of total RNA, with random hexamers as primers and according to the instructions of the SuperScript First-Strand Synthesis system (Invitrogen, Carlsbad, CA).
PCR Multiplex experiments were performed amplifying simultaneously a SDH2 cDNA and the cDNA corresponding to 18S rRNA using the QuantumRNA 18S Internal Standards kit (Ambion, Austin, TX) according to supplier's instructions. Appropriate number of cycles (exponential phase of amplification) and ratio of competimers to normal 18S rRNA primers were determined for each SDH2 cDNA. Hot-start PCR amplifications were performed in 50 μL, with TITANIUM Taq DNA polymerase (BD Biosciences CLONTECH, Palo Alto, CA) and 0.5 μL of [α-32P] dCTP (3,000 Ci/mmol, 10 mCi/mL, Amersham Biosciences, Buckinghamshire, UK). The following SDH2 primer pairs, located in the 3′ UTRs, were used: for SDH2-1, 5′-AACAGATCACACACATCAAGCA-3′ and 5′-TAGGTTTAAGTTTACTCGAGTTT-3′; for SDH2-2, 5′-AGGAAAGACTTGAAGCCATTGA-3′ and 5′-CTAATTCTGATAGTTGTGACAGC-3′; for SDH2-3, 5′-GAGAGGCTACAGGCAATAACTGAG-3′ and 5′-AATGAGTATCCAGAGAGAGAGAGAGAG-3′. PCR products were fractionated by electrophoresis on denaturing 6%-polyacrilamide gels and analyzed by autoradiography.
In Situ Hybridization
Paraffin-embedded Arabidopsis inflorescence and root sections were used for in situ hybridization with digoxigenin (DIG)-labeled RNA probes. Briefly, tissues were fixed in formaldehyde-acetic acid (4% [w/v] paraformaldehyde, 0.1 m NaOH; 5% [v/v] acetic acid, and 50% [v/v] ethanol) at room temperature for 6 h, dehydrated through an ethanol series, and then embedded in paraffin (Paraplast Plus, fusion temperature of 56°C, Oxford Labware, St. Louis). Tissue sections 7 μm thick were spread on a drop of water on slides coated with 3-aminopropyltriethoxy-silane (Sigma-Aldrich, St.Louis) and baked overnight at 45°C. After removing paraffin with xylene, sections were rehydrated by an ethanol series and treated for 30 min at 37°C with 1 μg mL−1 of proteinase K in 100 mm Tris-HCl pH 7.5, 50 mm EDTA. After two washes in water, sections were incubated three times for 5 min in 1× phosphate-buffered saline (10 mm sodium phosphate, pH 7.2, 0.13 m NaCl) and once for 10 min in 2× sodium chloride/sodium phosphate/EDTA (SSPE; 10 mm sodium phosphate, 0.15 m NaCl, 1 mm EDTA, pH 7.4). Then, tissue sections were prehybridized 1h at room temperature in 200 μL of hybridization solution/slide (50% [v/v] formamide, 4× SSPE, 10% [w/v] dextran sulfate, 1x Denhardt's solution, 250 μg mL−1 yeast tRNA, 250 μg mL−1 salmon sperm ssDNA) and hybridized overnight at 45°C in a moist chamber with 200 μL/slide of the hybridization solution containing 250 ng mL−1 of labeled riboprobe.
After hybridization, slides were washed twice in 2× SSPE for 15 min each at room temperature. These washes were followed by four high-stringency washes (30 min each) at 60°C in 0.1× SSPE. For signal detection, sections were washed twice in 100 mm Tris-HCl, pH 7.5, 150 mm NaCl, incubated for 45 min in 1% (w/v) Blocking Reagent (Roche Diagnostics, Mannheim, Germany) and overnight at 8°C with an anti-DIG alkaline phosphatase-conjugated antibody (1:500 dilution in 1% [w/v] bovine serum albumin, 0.3% [v/v] Triton X-100, 100 mm Tris-HCl, pH 7.5, 150 mm NaCl; Roche Diagnostics). Three washes were performed for 30 min each in the same buffer and then signals were developed at room temperature using the 4-nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indolyl phosphate colorimetric system (Roche Diagnostics). After stopping the reaction with distilled water, sections were dried at room temperature and mounted in Entellan (Sigma-Aldrich). The slides were viewed by bright-field microscopy and photographed using a Nikon FX-35DX camera coupled to a Nikon labophot-2 microscope (Nikon, Tokyo).
To obtain the SDH2 gene-specific probes, DNA fragments from the 3′UTR were PCR amplified using the primer pairs described above. The PCR products were cloned into a pGEM-T plasmid (Promega), and the recombinant plasmids were sequenced. Sense and antisense riboprobes were labeled with digoxigenin-11-UTP using the DIG RNA Labeling kit (SP6/T7; Roche Diagnostics) and either NcoI- or SpeI-linearized plasmids.
Contructs for Transformation of Arabidopsis Plants
To construct GUS promoter fusions, 5′ upstream sequences of different sizes were obtained by PCR amplification of an Arabidopsis genomic DNA template. Different 5′ forward primers containing a BamHI site and a common 3′ reverse primer containing a NcoI site encompassing the SDH2 ATG initiation codon were used. The PCR products were cloned into pGEM-T plasmid (Promega). Recombinant plasmids were digested with BamHI and NcoI, and DNA fragments were purified by electrophoresis through agarose gels and ligated into pCAMBIA 1381 (CAMBIA, Canberra, Australia). For promoterless constructs two oligonucleotides containing the 5′UTR were annealed and cloned into pCAMBIA 1381. These promoterless (P1) constructs contain 10 and 9 upstream of the SDH2-1 and SDH2-2 transcription initiation sites, respectively. The structure of all constructs was verified by DNA sequencing. All constructs contain a promoter fragment (decreasing size from P5 to P1) and the 5′ UTR up to the first SDH2 codon in frame with GUS (Fig. 5), and were introduced into A. tumefaciens GV3101 by electroporation.
For SDH2-1, the reverse primer was 5′-CGTTCCATGGTTTTCCTCGGAGAA-3′ and the forward primers were 5′-TATAGGATCCTTGACTTTGATGTCTGTAT-3′ (P5), 5′-TATAGGATCCAAACGGGACGCAGTTC-3′ (P4), 5′-ATAAGGATCCAGTGGTGTGGTTCGCC-3′ (P3), and 5′-ATAAGGATCCAATCGTCAATTTCGGCTA-3′ (P2). The two oligonucleotides annealed to generate the promoter-less construct (P1) were 5′-GATCCAGCCATTTTCAGATTTCTCCGAGGAAAAC-3′ and 5′-CATGGTTTTCCTCGGAGAAATCTGAAAATGGCTG-3′. For SDH2-2, the reverse primer was 5′-TTCCATGGTCTCGGAGAAGAGAGC-3′, and the forward primers were 5′-TAAAGGATCCATTGTTGAATCTGAATC-3′ (P5), 5′-GAACGGATCCGCGTTTTCTGAAAG-3′ (P4), 5′-CCACGGGATCCACACACTTCTC-3′ (P3), and 5′-TACAAATACGGATCCATACATTTTTAC-3′ (P2). The oligonucleotides annealed to generate the promoter-less construct (P1) were 5′-GATCCTTTCATTTCATTTGCTCTCTTCTCCGAGAC-3′ and 5′-CATGGTCTCGGAGAAGAGAGCAAATGAAATGAAAG-3′. For SDH2-3, the reverse primer was 5′-ATTCCATGGTCTGTTCGCTTGATC-3′, and the forward primers were 5′-GCGTTGAGGATCCTCTAAACATATC-3′ (P5), 5′-TTGGATCCTTGACTTTTCTGTTGC-3′ (P4), 5′-CACGTTTGGATCCGACCCGATAC-3′ (P3), and 5′-ATTGGATCCACGTGTCATTTCCATG-3′ (P2). BamHI and NcoI restriction sites have been underlined.
Colorimetric GUS Assay
Samples for GUS staining were fixed in 90% (v/v) acetone for 10 min on ice, rinsed once in GUS buffer (100 mm sodium phosphate buffer, pH 7.0, 0.5 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, 0.1% [v/v] Triton X-100, 0.1% [w/v] sodium lauroyl sarcosine) and then incubated into GUS staining solution (GUS buffer plus 1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, CLONTECH) at 37°C overnight (Jefferson et al., 1987). After staining, samples were cleared by several incubation steps in 70% (v/v) ethanol to remove chlorophyll. Samples were visualized and photographed using a Nikon FX-35DX camera coupled to a Nikon SMZ 800 binocular stereoscopic microscope.
Identification of Insertional SDH2 Mutants
We searched the Arabidopsis Insertion Database (ATIDB, http://atidb.org/cgi-perl/index), the SALK T-DNA database (http://signal.salk.edu/cgi-bin/tdnaexpress), and the SAIL/GARLIC T-DNA database (http://www.tmri.org, now integrated into ATIDB; Sessions et al., 2002; Alonso et al., 2003). Either no T-DNA insertions were found in SDH2-2 or SDH2-3 coding regions or seeds were not available. In contrast, three SDH2-1T-DNA insertion lines, SALK_031100, GARLIC_379_C10, and GARLIC_1274_E11, were identified (Col-0 background). According to data provided by Salk Institute Genomic Analysis Laboratory (SIGnAL) and Torrey Mesa Research Institute, Syngenta (TMRI), the insertion positions were in the first exon (SALK_031100 and GARLIC_379_C10) and second exon (GARLIC_1274_E11). SALK_031100 seeds were obtained from ABRC. We verified the position of the T-DNA insert using PCR with the primers sdhb3 (5′-ACACGTAGCTATAATCCCCTGTGTCTCTA-3′) and LBb1 (5′-GCGTGGACCGCTTGCTGCAACT-3′; http://signal.salk.edu). This PCR product was sequenced, revealing that the T-DNA had inserted between codons 93 and 94.
Plants homozygous for this mutation were isolated. For genotyping, DNA was extracted from leaves using the Wizard Genomic DNA Purification kit according to the supplier's protocol (Promega). The genotype of plants was determined by PCR using primers flanking the insertion point for the wild-type allele (sdhb3 and 5′-AGGACAAGACGTGCTACAACA-3′) and a gene-specific (sdhb3) and left border-specific (LBb1) primer pair for the SALK_031100 insertion allele.
Homozygous mutant and wild-type Col-0 seeds were surface sterilized and placed on Murashige and Skoog medium solidified with 0.8% (w/v) agar. Seeds were cold treated at 4°C for 48 h in the dark and then germinated at 22°C under a 16-h-light/8-h-dark cycle. After 25 d on plates, plants were transferred and grown hydroponically at 22°C under the same light/dark conditions.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Acknowledgments
The authors are greatly indebted to Simon Litvak and Laura Tarragó-Litvak for their continued support and encouragement. We thank anonymous reviewers for their excellent suggestions and acknowledge the ABRC for providing the SALK_031100 mutant seeds.
This work was supported by Fondecyt-Chili (Cooperación Internacional grant nos. 1020930 and 7020930, and Ph.D. grant no. 2000073) and by Centre National de la Recherche Scientifique-France (Programme International de Coopération Scientifique grant no. 2179).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.049528.
References
- Adams KL, Rosenblueth M, Qiu Y-L, Palmer JD (2001) Multiple losses and transfers to the nucleus of two mitochondrial succinate dehydrogenase genes during angiosperm evolution. Genetics 158: 1289–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Balk J, Leaver CJ (1998) Cell-specific differences in the expression of a nuclear and mitochondrial transcript of ATP synthase during anther development. In IM Moller, P Gardeström, K Glimelius, E Glaser, eds, Plant Mitochondria: From Gene to Function. Backhuys Publishers, Leiden, The Netherlands, pp 57–61
- Blanc G, Barakat A, Guyot R, Cooke R, Delseny M (2000) Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12: 1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL (1994) Arabidopsis, An Atlas of Morphology and Development. Springer-Verlag, New York
- Burger G, Lang BF, Reith M, Gray MW (1996) Genes encoding the same three subunits of respiratory complex II are present in the mitochondrial DNA of two phylogenetically distant eukaryotes. Proc Natl Acad Sci USA 93: 2328–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Eubel H, Jänsch L, Braun HP (2003) New insights into the respiratory chain of plant mitochondria: supercomplexes and a unique composition of complex II. Plant Physiol 133: 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyal Y, Curie C, McCormick S (1995) Pollen specificity elements reside in 30bp of the proximal promoters of two pollen-expressed genes. Plant Cell 7: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felitti SA, Chan RL, Gago G, Valle EM, Gonzalez DH (1997) Expression of sunflower cytochrome c mRNA is tissue-specific and controlled by nitrate and light. Physiol Plant 99: 342–347 [Google Scholar]
- Figueroa P, León G, Elorza A, Holuigue L, Araya A, Jordana X (2002) The four subunits of mitochondrial respiratory complex II are encoded by multiple nuclear genes and targeted to mitochondria in Arabidopsis thaliana. Plant Mol Biol 50: 725–734 [DOI] [PubMed] [Google Scholar]
- Figueroa P, León G, Elorza A, Holuigue L, Jordana X (2001) Three different genes encode the iron-sulfur subunit of succinate dehydrogenase in Arabidopsis thaliana. Plant Mol Biol 46: 241–250 [DOI] [PubMed] [Google Scholar]
- Fujie M, Kuroiwa H, Kawano S, Kuroiwa T (1993) Studies on the behaviour of organelles and their nucleoids in the root apical meristem of Arabidopsis thaliana (L.) Col. Planta 189: 443–452 [DOI] [PubMed] [Google Scholar]
- Gibeaut DM, Hulett J, Cramer GR, Seemann JR (1997) Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiol 115: 317–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MR (1991) Plant mitochondrial mutations and male sterility. Annu Rev Genet 25: 461–486 [DOI] [PubMed] [Google Scholar]
- Heiser V, Brennike A, Grohmann L (1996) The plant mitochondrial 22kDa (PSST) subunit of respiratory chain complex I is encoded by a nuclear gene with enhanced transcript levels in flowers. Plant Mol Biol 31: 1195–1204 [DOI] [PubMed] [Google Scholar]
- Huang J, Struck F, Matzinger DF, Levings CS III (1994) Flower-enhanced expression of a nuclear-encoded mitochondrial respiratory protein is associated with changes in mitochondrion number. Plant Cell 6: 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa T, Fujie M, Kuroiwa H (1992) Studies on the behavior of mitochondrial DNA: synthesis of mitochondrial DNA occurs actively in a specific region just above the quiescent centre in the root meristem of Pelargonium zonale. J Cell Sci 101: 483–493 [Google Scholar]
- Lee SLJ, Warmke HE (1979) Organelle size and number in fertile and T-cytoplasmic male-sterile corn. Am J Bot 60: 141–148 [Google Scholar]
- Lemire BD, Oyedotun KS (2002) The Saccharomyces cerevisiae mitochondrial succinate:ubiquinone oxidoreductase. Biochim Biophys Acta 1553: 102–116 [DOI] [PubMed] [Google Scholar]
- Li X-Q, Zhang M, Brown GG (1996) Cell-specific expression of mitochondrial transcripts in maize seedlings. Plant Cell 8: 1961–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monéger F, Mandaron P, Niogret M-F, Freyssinet G, Mache R (1992) Expression of chloroplast and mitochondrial genes during microsporogenesis in maize. Plant Physiol 99: 396–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribichich KF, Tioni MF, Chan RL, Gonzalez DH (2001) Cell-type-specific expression of plant cytochrome c mRNA in developing flowers and roots. Plant Physiol 125: 1603–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers HJ, Bate N, Combe J, Sullivan J, Sweetman J, Swan C, Lonsdale DM, Twell D (2001) Functional analysis of cis-regulatory elements within the promoter of the tobacco late pollen gene g10. Plant Mol Biol 45: 577–585 [DOI] [PubMed] [Google Scholar]
- Roos MH, Tielens AGM (1994) Differential expression of two succinate dehydrogenase subunit-B genes and a transition in energy metabolism during the development of the parasitic nematode Haemonchus contortus. Mol Biochem Parasitol 66: 273–281 [DOI] [PubMed] [Google Scholar]
- Satoh R, Nakashima K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2002) ACTCAT, a novel cis-acting element for proline- and hypoosmolarity-reponsive expression of the ProDH gene encoding proline dehydrogenase in Arabidopsis. Plant Physiol 130: 709–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffler IE (1998) Molecular genetics of succinate:quinone oxidoreductase in eukaryotes. Prog Nucleic Acid Res Mol Biol 60: 267–315 [DOI] [PubMed] [Google Scholar]
- Scheres B, Benfey P, Dolan L (2002) Root development. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/101199/tab0009 http://www.aspb.org/publications/arabidopsis [DOI] [PMC free article] [PubMed]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart CJ, Monéger F, Leaver CJ (1994) Cell-specific regulation of gene expression in mitochondria during anther development in sunflower. Plant Cell 6: 811–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirkettle-Watts D, McCabe TC, Clifton R, Moore C, Finnegan PM, Day DA, Whelan J (2003) Analysis of the alternative oxidase promoters from soybean. Plant Physiol 133: 1158–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell D, Yamaguchi J, Wing RA, Ushiba J, McCormick S (1991) Promoter analysis of genes that are coordinately expressed during pollen development reveals pollen-specific enhancer sequences and shared regulatory elements. Genes Dev 5: 496–507 [DOI] [PubMed] [Google Scholar]
- Yankovskaya V, Horsefield R, Törnroth S, Luna-Chavez C, Miyoshi H, Léger C, Byrne B, Cecchini G, Iwata S (2003) Architecture of succinate dehydrogenase and reactive oxygen species generation. Science 299: 700–704 [DOI] [PubMed] [Google Scholar]
- Zabaleta E, Heiser V, Grohmann L, Brennicke A (1998) Promoters of nuclear-encoded respiratory chain complex I genes from Arabidopsis thaliana contain a region essential for anther/pollen-specific expression. Plant J 15: 49–59 [DOI] [PubMed] [Google Scholar]