Significance
In transcription-coupled repair (TCR), nucleotide excision repair occurs most rapidly in the template strand of actively transcribed genes. TCR has been observed in a limited set of genes directly assayed in Escherichia coli cells. In vitro, Mfd translocase performs reactions necessary to mediate TCR: It removes RNA polymerase blocked by a template strand lesion and rapidly delivers repair enzymes to the lesion. This study applied excision repair sequencing methodology to map the location of repair sites in different E. coli strains. Results showed that Mfd-dependent TCR is widespread in the E. coli genome. Results with UvrD helicase demonstrated its role in basal repair, but no overall role in TCR.
Keywords: Mfd, UvrD, transcription-coupled repair, XR-seq, mutagenesis
Abstract
We used high-throughput sequencing of short, cyclobutane pyrimidine dimer-containing ssDNA oligos generated during repair of UV-induced damage to study that process at both mechanistic and systemic levels in Escherichia coli. Numerous important insights on DNA repair were obtained, bringing clarity to the respective roles of UvrD helicase and Mfd translocase in repair of UV-induced damage. Mechanistically, experiments showed that the predominant role of UvrD in vivo is to unwind the excised 13-mer from dsDNA and that mutation of uvrD results in remarkable protection of that oligo from exonuclease activity as it remains hybridized to the dsDNA. Genome-wide analysis of the transcribed strand/nontranscribed strand (TS/NTS) repair ratio demonstrated that deletion of mfd globally shifts the distribution of TS/NTS ratios downward by a factor of about 2 on average for the most highly transcribed genes. Even for the least transcribed genes, Mfd played a role in preferential repair of the transcribed strand. On the other hand, mutation of uvrD, if anything, slightly pushed the distribution of TS/NTS ratios to higher ratios. These results indicate that Mfd is the transcription repair-coupling factor whereas UvrD plays a role in excision repair by aiding the catalytic turnover of excision repair proteins.
Transcription-coupled repair (TCR) is the transcription-dependent enhancement of the rate of repair of the transcribed strand (TS) (or “template strand”) of a transcription unit relative to the nontranscribed strand (NTS) (or “coding strand”) and nontranscribed regions of the genome. The phenomenon was discovered first in mammalian cells (1, 2) and subsequently in Escherichia coli (3) and other organisms (4). In mammalian cells, it was discovered that TCR is dependent on the CSB protein, the CSA WD40 protein (5), and the mammalian nucleotide excision repair factors. However, reconstitution of mammalian TCR is ongoing, and therefore the mechanism of TCR in eukaryotes remains poorly understood. In contrast to mammalian cells and eukaryotic cells in general, the mechanism of TCR in E. coli is reasonably well-understood. First, an E. coli cell-free extract system was used to accomplish TCR in vitro (6), and then, using this assay, a protein was purified that was necessary and sufficient to reconstitute TCR with purified excision repair proteins UvrA, UvrB, UvrC, RNA polymerase (RNAP), DNA polI, and DNA ligase, plus appropriate transcription/repair substrates. Genetic analysis revealed that the E. coli transcription repair-coupling factor (TRCF) is encoded by the mfd gene (7, 8), and therefore we use the terms TRCF and Mfd interchangeably. The mfd gene was originally identified as the gene required for reducing the rate of UV mutagenesis in E. coli cells held under conditions of amino acid starvation (9, 10). Further genetic work let to the suggestion that the mutation frequency decline effect was due to the repair of dipyrimidine UV photoproducts located in the anticodon of tRNA genes (11, 12). Thus, the biochemical reconstitution system of TCR in E. coli supported the model proposed for the mutation frequency decline phenomenon.
The mfd gene has been cloned, and the Mfd protein (TRCF) has been purified and characterized in some detail (13). It has homology to the excision repair protein UvrB and to the RecG protein (14). It contains helicase motifs I to VI but no helicase activity; rather, it functions as an ATP-dependent translocase (7, 15–17). Biochemical reconstitution with a completely defined system comprising damaged DNA (UV, psoralen, or cisplatin damage), UvrA, UvrB, and UvrC excision repair proteins, RNAP, and the necessary cosubstrates (ATP, rNTPs, dNTPs), as well as protein–protein and protein–DNA interaction studies, revealed that RNAP stops at a lesion site (18) [cyclobutane pyrimidine dimer (CPD), cisplatin- or psoralen-DNA adduct] and led to the following model: RNAP stops at a lesion site in the TS but not the NTS and, in the absence of Mfd, inhibits repair of the lesion. When present, Mfd binds to the stalled RNAP–RNA–DNA ternary complex, and the translocation activity of Mfd moves both proteins toward the damage, which causes collapse of the transcription bubble, leading to displacement of the stalled RNAP along with the truncated transcript (15–17). The DNA–Mfd–tethered RNAP complex recruits the UvrA2 UvrB1 complex to the damage site, which leads to dissolution of the DNA–Mfd–tethered RNAP complex (19). The net effect is to accelerate the damage recognition step of excision repair and therefore the rate of excision repair of the TS because the damage recognition step is the rate-limiting step in excision repair. Subsequent detailed biochemical work has confirmed this model, and structural studies (20) have provided detailed mechanistic insight into Mfd-mediated transcription repair coupling. Most recently, an elegant study using a series of single molecule assays determined the rate constants for the various steps of TCR (19), providing direct evidence for the Mfd-UvrA2-UvrB1 proposed in the original model (7) and the quantitative data for the enhancement of repair rate by this complex.
Although ensemble biochemical and structural studies and single molecule assays have confirmed and refined the original model for the role of Mfd in TCR, some recent studies have implicated the transcription elongation factor NusA (21) and the replication and repair helicase UvrD (22) in transcription-coupled repair, and it has been suggested that Mfd-catalyzed TCR is one of several pathways for TCR in E. coli. Because these latter studies were based largely on genetic data and indirect readouts for TCR, we wished to determine the extent to which Mfd contributes to TCR in E. coli by using the eXcision repair-sequencing (XR-seq) method to map excision repair of the entire E. coli genome in strains with defined genetic mutations.
Results
XR-seq Method.
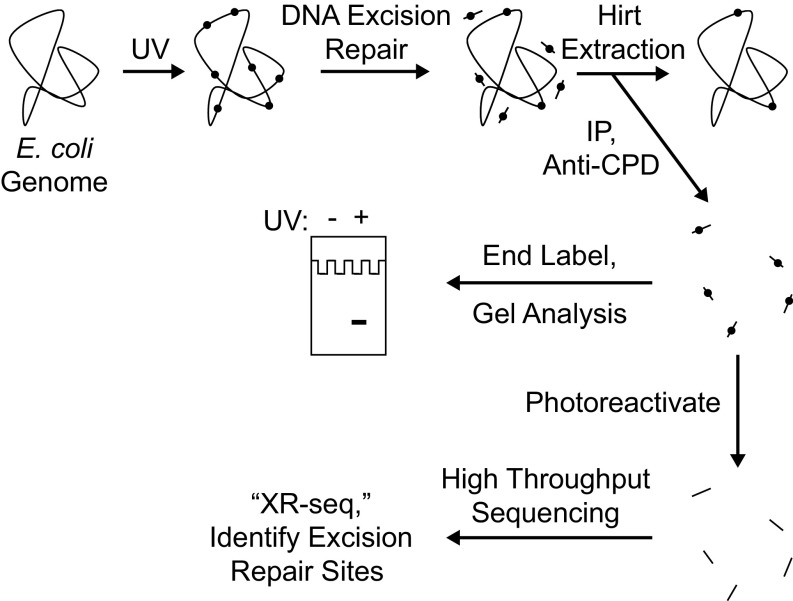
Recently, we developed a method for isolating the oligonucleotides removed by excision repair in mammalian cells (23), and, using next generation sequencing (NGS) methods, we generated nucleotide resolution repair maps for UV- and cisplatin-induced DNA damage in human cells (24, 25). In this study, we adapted this method to analyze genome-wide repair, and thus genome-wide transcription-coupled repair in E. coli, and used various mutants to analyze the contributions of candidate proteins to TCR. The XR-seq method as applied to CPD repair in E. coli is shown schematically in Fig. 1. Briefly, cells are irradiated with UV, and, after incubation for a period for repair, cells are collected by centrifugation and small oligonucleotides carrying the excised CPDs are separated by the Hirt procedure and immunoprecipitated with anti-CPD antibodies to separate them from small oligomers released from the genome by nonspecific nucleases during sample processing. Then, a fraction of the isolated oligonucleotides are 3′ terminally labeled with 32P for visual analysis of the products, and the bulk are ligated with adapters, reimmunoprecipitated with the anti-CPD antibody, and subjected to photoreactivation with Anacystis nidulans photolyase to remove the photodimers, which is followed by PCR amplification and gel purification, and then the sequences of the excised oligomers are determined by NGS and aligned with the E. coli chromosome.
Fig. 1.
Scheme for analyzing repair of the E. coli genome. Cells are irradiated with UVC (254 nm) and then incubated for a period to allow nucleotide excision repair. Cells are then harvested and cooled on ice, and then the Hirt procedure (23) is used to separate relatively small DNA molecules from genomic DNA and cellular debris. From the preparation of small DNA molecules, DNA molecules containing cyclobutane pyrimidine dimers (CPDs) are then isolated by immunoprecipitation with an anti-CPD–specific antibody. The isolated products may be analyzed in two ways. To detect the overall excision of damage from the genome, samples may be directly end-labeled with 32P and subjected to denaturing polyacrylamide gel electrophoresis. To map the sites of DNA excision repair throughout the genome at nucleotide resolution, products may be analyzed by high-throughput sequencing after they are ligated to adaptors, immunoprecipitated again with anti-CPD antibody, photoreactivated, amplified by PCR, and gel-purified (25).
Excision Assay.
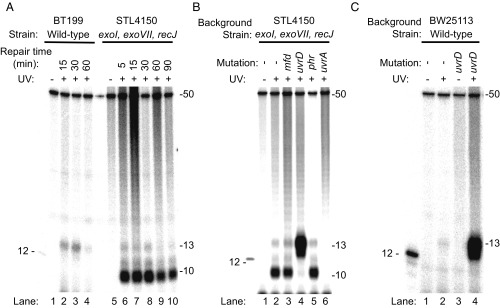
Fig. 2 shows sequencing gel analysis of the end-labeled, excised oligomers. In preliminary experiments with WT cells, we discovered that the excised oligonucleotides were degraded rather rapidly. Therefore, we carried out time course experiments using both a WT strain and an E. coli strain, STL4150 (26), defective in all major ssDNA exonucleases (exoI, exoVII, recJ). Fig. 2A (lanes 5 to 10) shows that, in the triple exonuclease mutant strain, the major species is a 10-mer whereas, in WT cells, it is a 13-mer (and 12-mer) (Fig. 2A, lanes 1–4), which shows that these three exonucleases play an important role in processing the excised oligomer and that, in the absence of these exonucleases, the excised oligomer is degraded to a 10-mer by 3′ to 5′ exonucleases that stop at a nucleotide 3′ to the photoproduct. We thus demonstrate that the 12- and 13-mer excision product generated by the purified UvrA, UvrB, and UvrC proteins (27) is of the same size as the product generated in WT cells in vivo and also provide an explanation for why, in the seminal papers describing nucleotide excision repair in E. coli cells, the thymine CPDs were found primarily in 4- to 6-nt-long oligomers, because the excised oligomer is rapidly degraded by the numerous ssDNA-specific exonucleases present in E. coli, in addition to the nucleases deleted in STL4150 (28).
Fig. 2.
Analysis of nucleotide excision repair products from E. coli by excision assay. Cells were irradiated with 100 J/m2 (A and B) or 120 J/m2 (C) and then incubated for the indicated times (in A) or for 5 min (B and C). Repair products containing a CPD were isolated from cells, end-labeled with 32P, and resolved by denaturing polyacrylamide gel electrophoresis. The gel images show a predominantly 13-mer size excision product obtained from WT cells, which is consistent with results from in vitro analysis (27). This 13-mer size excision product is simultaneously generated and degraded in vivo, and a reduction in the amount of the 13-mer product seen after 30 min is consistent with an ∼30-min time course to complete nucleotide excision repair in E. coli cells (3). Strain STL4150 lacks the major ssDNA exonucleases, and the images in A and B show that, in STL4150 cells, there is limited degradation of the 13-mer past the 10-mer stage. In uvrD mutant cells (B and C), there is an elevated level of 13-mer product. The UvrD protein is the major helicase in E. coli. In nucleotide excision repair, UvrD catalyzes the displacement of the damage-containing 13-mer excision product and initiates displacement of the UvrB and UvrC proteins from the genome. Consequently, in these cells, the 13-mer remains annealed to the genome where it is resistant to nucleases. This accumulation of the 13-mer is observed even though uvrD mutant cells excise the 13-mer slowly because turnover of UvrB and UvrC is slow. The 12-mer marker DNA shown in the images contains a CPD and was end-labeled with polynucleotide kinase. The 50-mer (1 fmol) was included in each sample before end-labeling, as an internal control.
In Fig. 2 B and C, we analyzed the excision products in exonuclease-deficient (STL4150) and WT (BW25113) E. coli strains mutated in genes implicated in various aspects of nucleotide excision repair. Mfd and UvrD have been proposed to play direct roles in TCR. Phr (photolyase) stimulates excision repair in the absence of transcription (29), Phr is inhibited by RNAP stalled at a CPD (30), and, finally, UvrA is known to be essential for all types of excision repair. The most striking feature of the results in Fig. 2 is the fact that, even though the uvrD mutation confers significantly reduced repair in vivo (31), in both Fig. 2B (STL4150 background) and Fig. 2C (BW25113 background), the excision product is, first, much more abundant in uvrD mutants than in all other mutants and is, second, almost exclusively 12 to 13 nt in length. This seemingly paradoxical result is in fact readily explained by the role of UvrD in nucleotide excision repair as established by in vitro experiments: After the dual incisions by UvrC in the UvrB–UvrC–DNA complex, the UvrB-UvrC heterodimer, along with the excised oligomer, remains bound to the duplex (no catalytic turnover), and addition of UvrD helicase initiates displacement of UvrB and UvrC, along with the excised oligomer, to enable the Uvr(A)BC excision nuclease to act catalytically (32–34) and at the same time makes the excised fragment available for degradation by ssDNA nucleases.
The results in Fig. 2 also show that phr– cells excised at a level comparable with parental cells. A stimulatory effect of photolyase on excision has been demonstrated in vitro (29), but, in vivo, it has been seen only in repair-deficient and photolyase-overexpressing E. coli cells (35). Similarly, the results in Fig. 2B show no substantial loss in excision overall associated with mutation of mfd. Mfd enhances repair in vitro, but, in vivo, the mfd mutation confers only modest UV sensitivity in WT cells whereas mfd confers substantial sensitivity in recA– cells (7).
XR-seq of CPDs in the E. coli Genome.
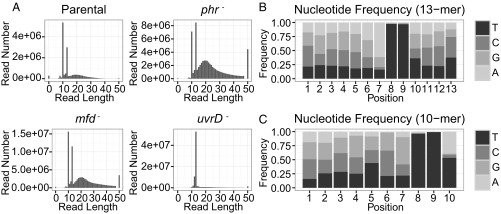
It has been shown that the maximal TCR (ratio of TS/NTS repair) in the lac operon occurs at relatively early time points and that repair of both strands is nearly complete after ∼30 min under conditions comparable with our repair assays (3). With this result in mind, we incubated STL4150 cells and mutant derivatives in the dark for 5 min after UV treatment and then harvested them for analysis of repair of CPDs by XR-seq. Two independent experiments gave results that were consistent with one another, with exceptions noted below. Fig. 3A shows that the reads from the uvrD− mutant were principally 12 to 13 nt in length, which is consistent with the sequencing gel analysis (Fig. 2B). The remaining reads are considered background. The greatest number of reads (about 5 × 107) was obtained from uvrD mutant cells, consistent with the strong signal in Fig. 2B. The background seems relatively small with uvrD– cells because there was a stronger signal and less relative degradation with the uvrD– cells. Reads obtained from the other strains were principally 10 or 13 nt (plus background), also consistent with results in Fig. 2B, and consistent with partial, processive 3′ to 5′ exonucleolytic degradation of the 13mer that stops at +1 from the photodimer and produces the 10mer. Sequence analysis revealed that TT was the predominant di-nucleotide at positions 8 to 9 from the 5′ end of the excised fragment for both the 12- to13-nt class and the 10-nt class of captured oligomers (Fig. 3 B and C). Thus, these experiments revealed that CPDs are removed by incising 7 nt 5′ and 3 to 4 nt 3′ to the photodimer, in agreement with the in vitro experiments with purified Uvr(A)BC excision nuclease (27). We selected approximately 3 million 13-nt reads from each genotype for further analysis.
Fig. 3.
Length distribution and nucleotide composition of the reads. (A) Length distribution of the isolated excised fragments for four strains. The lengths of the principal excised products are 10, 12, or 13 nt. The remaining reads are considered as background. (B and C) Position-specific nucleotide frequencies of the 13-mer (B) and 10-mer (C) reads from parental strain STL4150. Results are from the first of two experiments.
Genome-Wide Effects of Mfd, UvrD, and Phr Proteins on TCR.
Previous in vitro work has shown that photolyase, when bound to a CPD, stimulates excision repair of the CPD (29) and that RNAP stalled at a CPD interferes with this excision repair stimulatory activity of photolyase (30). RNAP also inhibits excision repair (in the absence or presence of photolyase) when stalled at a CPD in the template (18). It was found that Mfd (TRCF) overcomes the inhibitory effect of stalled RNAP and, in fact, in combination with RNAP, enhances the rate of repair of the TS by a factor of 4 to 5 (6, 7). Similarly, another more recent in vitro study has concluded that UvrD helicase stimulates the repair of the TS independently of Mfd and that the UvrD protein may in fact be the primary TRCF in E. coli (22). Thus, taking these reports at face value, we expected that (i) mutation of mfd would somewhat reduce the magnitude of TCR, (ii) mutation of uvrD would drastically reduce TCR, and (iii) mutation of phr would amplify the TCR effect because of a loss of a factor that can stimulate excision repair in the NTS more than the TS because stalled RNAP interferes with the repair stimulatory effect of photolyase while promoting TCR mediated by Mfd.
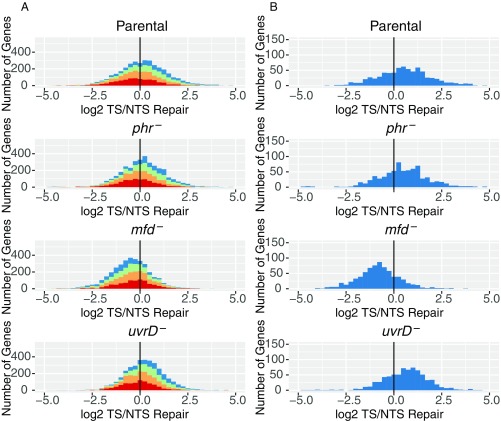
With these expectations, then, we analyzed our XR-seq data in the form of histograms and scatter plots. An analysis of frequency distribution histograms for the four E. coli strains is shown in Fig. 4. For these analyses, 3 million reads for each genotype were aligned to the genome. Aligned reads were then ascribed to the TS or NTS of each annotated gene in the genome. Genes are plotted on the x axis based upon the log2 transformation of their TS/NTS repair ratios. In Fig. 4A, annotated genes were divided into quartiles (red, orange, green, blue) to denote the relative transcription of each quartile, going successively from lowest (red) to highest (blue) transcription. Fig. 4B shows only the 25% most highly transcribed genes. Transcription levels were obtained from the literature (36), and transcript reads were obtained from cells growing exponentially in LB (OD600 = 0.4). In the XR-seq experiment, we also used exponentially growing cells (OD600 = 0.8). Of note, the published map for transcription (36) shows considerable antisense transcription, which, in addition to overlapping genes with opposite orientations, leads to more antisense transcripts than sense transcripts in many annotated genes. Restricting the analysis to genes with high ratios of TS/NTS transcription or high levels of transcription (Fig. 4B) focuses the analysis on the “simpler” genes with predominantly sense transcription.
Fig. 4.
Frequency distribution of Log2-transformed TS/NTS repair in all annotated genes. (A) E. coli genes in four different strains colored by sense strand transcription levels going from lowest quartile RNA-seq count colored in red, to orange, to green, to the highest transcription quartile, in blue. Theoretically, the more weakly transcribed genes are more likely to exhibit repair profiles strongly influenced by antisense transcription and by repair hotspots. The means for each sample are 1.16 (Parental), 1.14 (phr−), 0.68 (mfd−), and 1.17 (uvrD−), and all of them are different from 0 based on one sample t test with P values <0.01. In Exp. 2, means for each sample were 1.09, 1.14, 0.68, and 1.04. (B) Top 25% most transcribed genes are plotted. Means for parental, phr, mfd, and uvrD cells are 1.41, 1.46, 0.53, and 1.61 for Exp. 1 (plotted) and 1.41, 1.46, 0.54, and 1.21 in Exp. 2, respectively. The vertical black line represents the border where TS repair level is equal to NTS repair.
Fig. 4 reveals several interesting points. First, as seen in Fig. 4B, in the parental strain, even though in the majority of cases the TS is repaired at a faster rate than the NTS, it is also apparent that, for a subgroup of genes, the NTS is repaired more efficiently in all genetic backgrounds (Dataset S1). Second, the phr mutation seems to have no effect on the ratio of TS/NTS repair compared with parental, presumably because of the small number (10 to 20) of photolyase molecules per cell (37), compared with 100 to 1,000 RNAP molecules per cell. Third, in the mfd mutant, the TS repair/NTS repair ratio is drastically reduced at all transcription levels (Fig. 4A), and the NTS becomes the preferentially repaired strand (P < 2.2e−16), consistent with inhibition of TS repair by RNAP, which remains stalled at transcription-blocking lesions in the absence of Mfd. In contrast to expectations, in the uvrD mutant, TCR was either amplified compared with parental cells (P = 0.002, Exp. 1) or was no different from parental cells (P = 0.8, Exp. 2) instead of being diminished.
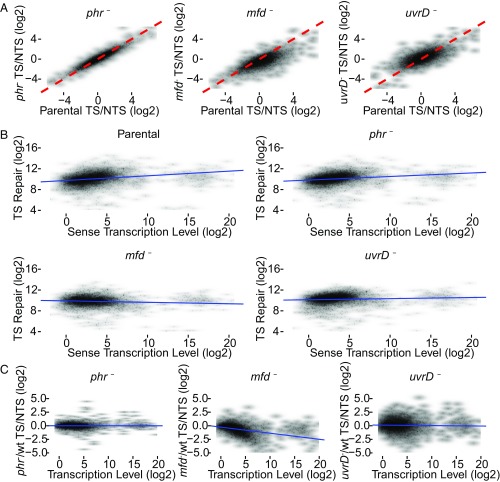
Fig. 5A shows the scatter plots, in which the TCR for each annotated gene (having >30 TT sites on TS) is represented. The three plots in Fig. 5A compare the level of TCR (TS/NTS repair ratio) in each gene of the mutant strain (y axis) with the TCR level of the same gene in the parental strain (x axis). The dashed red line indicates where TCR is equivalent in the mutant and parental strains. Substantial effects are seen especially with the mfd mutation: In the mfd plot, the shift of data points to the lower right (P < 2.2e−16, both experiments) is consistent with relatively less template strand repair in mfd− cells, due to loss of Mfd-stimulated repair and inhibition of TS repair by stalled RNAP. Note that the overall shift of data points downward in the left half of the plot (parental TS/NTS < 0) indicates that genes with high relative NTS repair in the parental strain have even higher relative NTS repair in the mfd− strain. Thus, Mfd promotes TCR even in genes with more repair on the NTS.
Fig. 5.
Scatter plots of TS/NTS repair signal in all annotated genes. (A) Log2-transformed TS/NTS repair ratio, which represents the TCR signal, is plotted for phr−, mfd−, and uvrD− strains on the y axis vs. the parental strain (x axis). The dashed red line is the line of equality (x = y) representing equal TCR signal in the two strains. In the phr− strain, although most genes exhibit mildly elevated TS/NTS compared with parental, the difference is not significant. In the mfd− strain, most genes have a lower value of TS/NTS repair compared with WT because blocked RNAP inhibits TS repair. The parental TS/NTS mean value is 1.71-fold higher than mfd− (P < 0.01). Note that the loss of Mfd is associated with more NTS repair (relative to parental cells) even in genes that, in the parental cell line, exhibit more NTS repair than TS repair (left half of plot). In uvrD− strains, the mean of TS/NTS is mildly but significantly higher than in WT (P = 8e−08) in this experiment, but note that there was no difference in Exp. 2. (B) The correlations between TS repair (fragments per 100 TT sites per million reads, on the y axis) and sense strand transcript levels (fragments per 1 kb per million reads, on the x axis). The correlations in WT, phr, and uvrD cells are significant, with correlation coefficients 0.36, 0.36, and 0.22, respectively (P values < 2e−16). In mfd, a mild negative correlation was observed (rho = −0.07, P = 2e−5). (C) The ratios of mutant TS/NTS repair over WT TS/NTS repair as a function of transcription level. The values on both axes are log2-transformed. Transcription levels are RNA-seq read counts that are compiled from the raw data produced by Thomason et al. (36) (SRR1173967). As opposed to other strains, in the mfd− strain, there is a strong correlation (P < 2e−16) with the Spearman’s rho of −0.55. The blue trend lines (B and C) were drawn based on simple linear regression models.
In the uvrD mutant, the trend is different from the mfd mutant (Fig. 5A): The mean shift of data points to the upper left in the uvrD plot is significant (P = 8e−08) in Exp. 1, but not in Exp. 2 (P = 0.75). The modest trend in higher overall TCR in the uvrD mutant again supports the view that UvrD helicase does not couple transcription to repair overall but that, in fact, in its absence, the trend toward enhanced TCR becomes more prominent. In both experiments, data points from uvrD− cells are widely distributed around the red line, indicating a heterogeneous effect of UvrD as discussed below. For phr– cells, the data points are narrowly located around the red line (Fig. 5A), suggesting no effect of Phr on TCR.
Effect of Transcription Rate on Repair.
To investigate the effect of transcription rate on repair, we plotted TS repair as a function of transcription levels, where levels of transcription are defined as the number of reads per kilobase (36). We observed a moderate but significant (P < 2e−16) correlation between transcription and TS repair in parental (rho = 0.36), phr (rho = 0.36), and uvrD (rho = 0.22) strains (Fig. 5B) (rho = 0.32, 0.36 and 0.12, respectively, in Exp. 2). Fig. 5 B and C shows that transcription clusters into two levels, low-medium and high. Interestingly, the plots for parental, phr−, and uvrD− mutant cells in Fig. 5B show that data points for the high transcription level cluster of genes are mostly below the trend line, suggesting that, above a certain level, increasing transcription rate is not associated with increasing repair. The possibility of inhibition of template strand repair by a high transcription level has been discussed (13). In mfd− cells, there was no positive correlation between TS repair and transcript level. In fact, a mild (rho = −0.07) but significant (P = 2e−5) negative correlation was observed. Thus, inhibition of repair by stalled RNAP could be enhanced by additional stalled RNAPs upstream, and/or the extent of inhibition could be limited by the time it takes for RNAP to arrive at a lesion.
We further analyzed the data by plotting in Fig. 5C the ratios of mutant TCR/parental TCR as a function of transcription level. In the phr mutant, compared with WT, we did not see an effect of transcription rate on TCR. In contrast, in the mfd versus WT comparison, there was a strong correlation between the level of transcription and the ratio of mfd−/parental TCR for low and medium abundance transcripts. Mfd is the only protein known to efficiently remove RNAP stalled at a lesion, and the stalled polymerase is known to inhibit repair. The data indicate that, in the absence of Mfd, up to a point, more transcription causes more repair inhibition. Finally, in comparing the uvrD mutant to WT, we found that the variations in TS/NTS repair ratios do not depend on the level of transcription.
Analysis of TCR at Individual Gene Resolution.
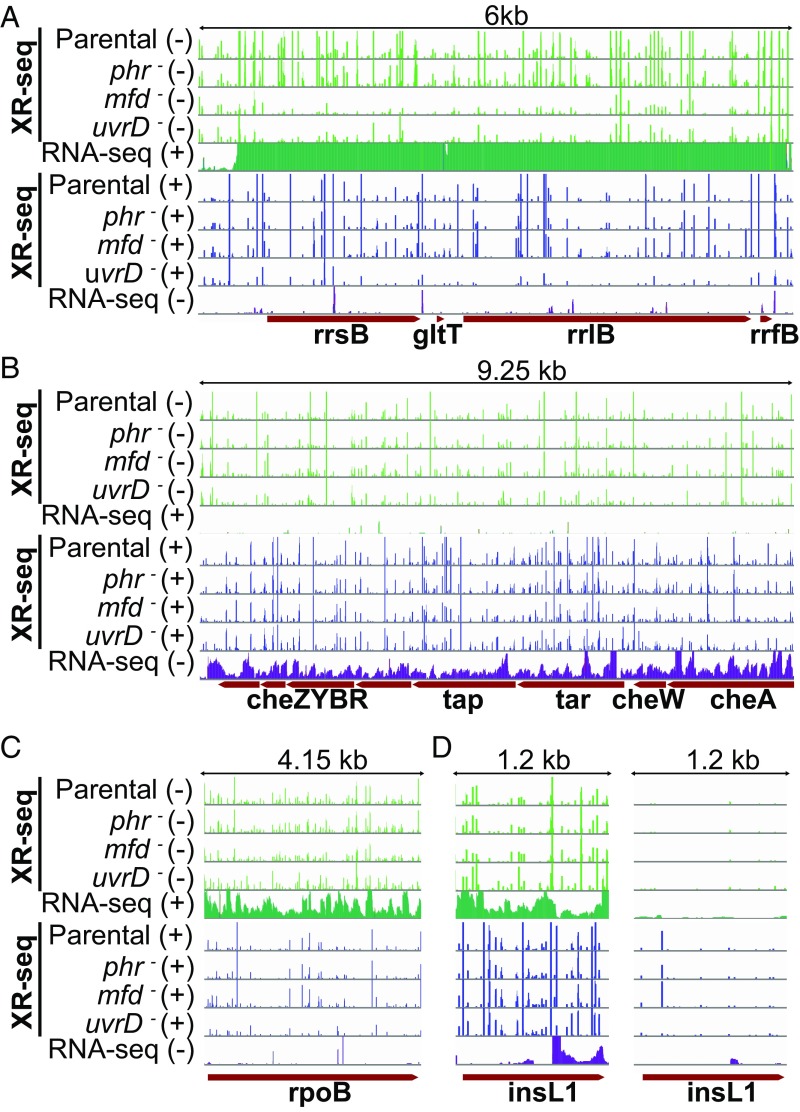
To gain further insight into TCR and the effects of various proteins on this repair mode, we illustrated the repair profiles of several genes in the form of screenshots, several of which are shown in Fig. 6. Fig. S1 shows repair profiles for each strain across the entire E. coli gene map. The screenshot in Fig. 6A shows a ribosomal RNA operon, which displays a very high level of transcription. Repair in this operon overall seems similar in parental and phr– cells although there is an increase at some repair sites in phr– cells (TS/NTS ratios in rrsB gene are 1.18 parental and 1.44 phr−). This operon illustrates, in mfd− cells, a dramatic reduction in TS repair (with some compensatory increase in rrsB NTS repair: TS/NTS is 0.06) and shows, in uvrD– cells, overall inhibition of repair of both strands while conserving TCR. This overall inhibition of repair suggests that, in parental cells, both strands are repaired rapidly as a “repair domain” (38). Notably, the elevated TCR ratio conserved in uvrD– cells (rrsB: 1.81) contributes to the heterogeneity in the effect of uvrD mutation on TCR compared with parental seen in Fig. 5A, uvrD panel. Fig. 6B illustrates a chemotaxis operon with a moderate level of transcription. Here, we saw a relatively small effect of the mfd mutation. As opposed to the rRNA operon, the rpoB gene in Fig. 6C displays an amplified TS repair in uvrD mutant cells.
Fig. 6.
XR-seq and RNA-seq patterns of exemplified operons and genes. In all panels, the first four green-colored rows represent the XR-seq reads of the four strains studied aligned with the minus strand of the reference genome (E. coli K-12 MG1655 genome with NCBI accession NC_000913.2). The fifth green bar shows the RNA-seq reads aligned with the plus strand. Blue bars represent the opposite strand. For the genes that are on the plus strand (A, C, and D), the green XR-seq reads correspond to the transcribed strand repair where green RNA-seq reads represent the RNA products that are due to the sense transcription. Blue XR-seq data illustrate repair of the coding (plus) strand, and blue RNA-seq reads represent the antisense transcription. The opposite is true for the genes on the minus strand (B). The y axis is scaled to show 105 counts for each bar except for the right panel of D, where the y axis is scaled up to 2,000. (A) The illustrated 6-kb genomic window is between 4,164 and 4,170 kb, which is an rRNA operon rrnB with 16S (rrsB), 23S (rrlB), and 5S (rrfB) rRNA genes, as well as the glutamate tRNA (gltT) gene. This operon was selected to represent a high level of transcription so that the Mfd effect is drastically visible. (B) Chemotaxis operon (1964.25 to 1973.5 kb) with moderate level of transcription. (C) The rpoB gene (4179.2 to 4183.35 kb) displays an amplified repair signal in the uvrD strain. (D) The insL1 gene (2512.3 to 2513.5 kb) with antisense transcription in the 3′ end (Left).
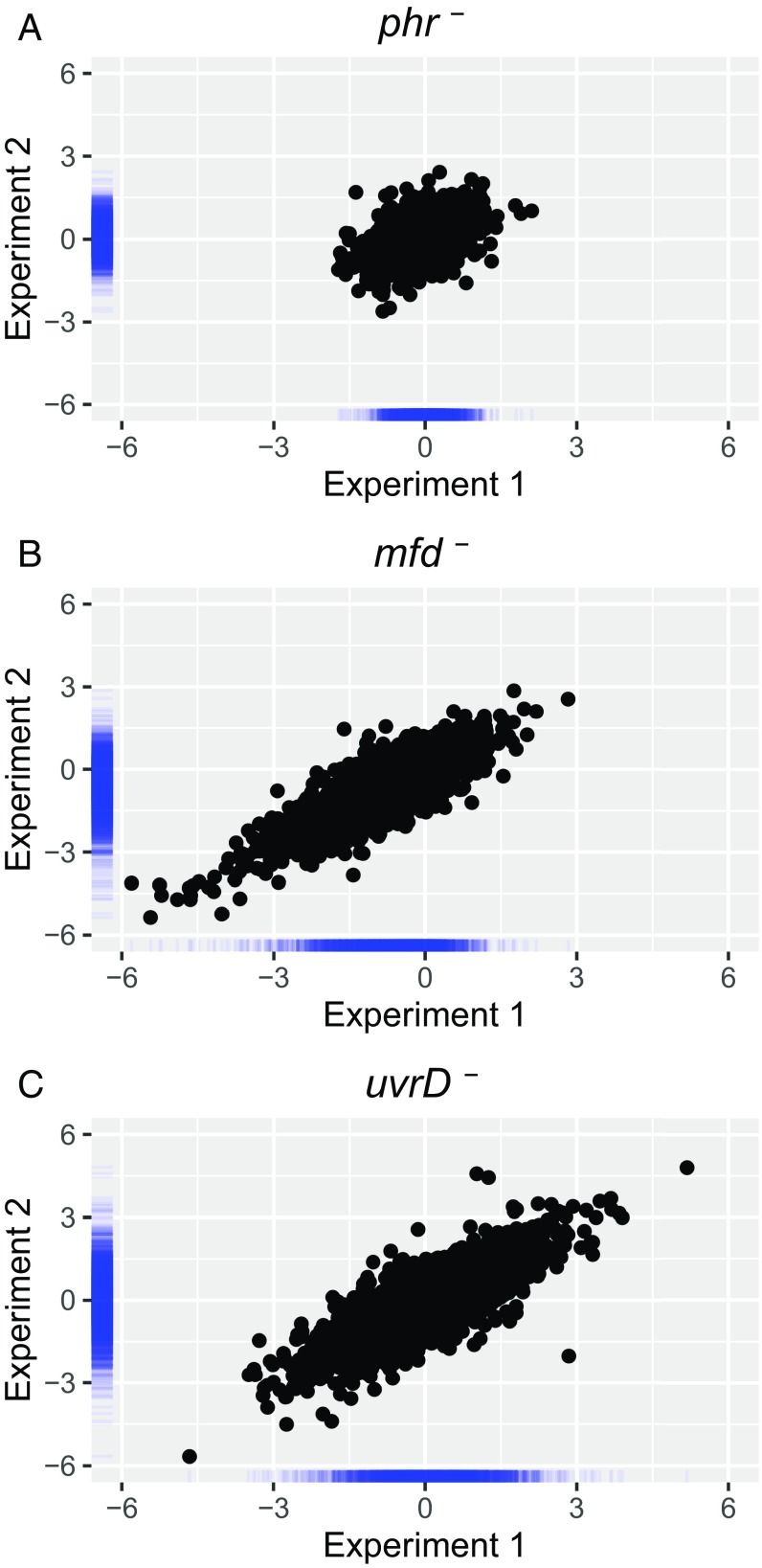
Fig. S1.
Reproducibility of the mutation effect on TCR (A) phr–, (B) mfd–, and (C) uvrD–. The x and y axes represent Exp. 1 and Exp. 2, respectively. Axis values are the log2-transformed ratio of mutant/Parental TS/NTS values. Pearson correlation coefficients are 0.43 (phr–), 0.85 (mfd–), and 0.81 (uvrD–). Blue bars along the axes represent density distribution of data points.
In addition to the genes in Fig. 6 B and C, which exhibit TCR patterns generally in line with the genome-wide trend, we were interested in the significant number of genes in which the coding (NTS) strand seems to be repaired more efficiently than the TS. Inspection of screenshots of a few representatives of this group and consideration of their transcription properties reveal that these genes may fall into one of two groups. In one group, there are a few very efficiently repaired hot spots in the NTS that dominate the repair landscape. In fact, when the TS repair versus NTS repair comparison is made, the rest of the gene repair is more efficient in the TS. In the second group are annotated genes with considerable levels of antisense transcription. In Fig. 6D, we illustrate antisense transcription in the insL1 gene. In the 3′ region, where there is antisense transcription, the mfd mutation seems to have no effect, as opposed to the 5′ end, where there is only sense strand transcription. In the same gene, we also saw a hotspot on the NTS, which is illustrated in Fig. 6D, Right, by replotting the data in the Left panel on a lower sensitivity scale. The hotspot dominated the total count, resulting in a low TS/NTS repair ratio for this gene. As evident, in genes with simple transcription patterns, the TS is repaired more efficiently than the NTS, and this preferential repair is abolished or reversed in mfd mutant cells and may be amplified in uvrD mutant cells, in agreement with overall genome-wide analysis of TCR. Thus, taken in its totality, our data show that the Mfd protein is responsible for TCR throughout the genome.
Discussion
Genetic Determinants of TCR.
Extensive functional and structural work has been carried out on RNAP, the Uvr proteins, and Mfd, which has provided considerable insight into the mechanistic aspects of TCR, culminating in analyses with single molecule assays that captured intermediates not detectable by ensemble experiments and that determined the rate constants for various steps and thus provided a quantitative explanation for rate enhancement seen in TCR (19).
However, some recent studies have led to suggestions that there are multiple pathways for TCR in E. coli. In one study, it was reported that a temperature-sensitive mutant of transcription elongation factor NusA conferred upon E. coli sensitivity to nitrofurazone and 4-nitroquinoline oxide, but not to UV radiation. In addition, evidence was presented for an interaction between NusA and UvrA, and, based on these and other findings, it was proposed that NusA participates in a TCR pathway independent of Mfd (21). It was also shown that UvrD causes RNAP to “back away” from a transcription-blocking lesion, exposing it to repair enzymes. Based largely upon an interaction identified between UvrD and UvrB (39, 40), and the UvrA–NusA interaction, it was suggested that UvrD and NusA function together in an Mfd-independent pathway. Furthermore, because uvrD mutants are more sensitive to UV than mfd mutants, it was proposed that the UvrD-mediated TCR was the major TCR pathway in E. coli (22). The data presented in this study do not support widespread UvrD-dependent TCR in E. coli but do support a widespread role of Mfd in TCR.
Role of UvrD.
Our data confirm the conclusion based on in vitro experiments (32–34) and CPD removal kinetics in vivo (31) that the role of UvrD in excision repair is to function as a helicase to displace the “excised” 12- to 13-nt-long oligomer carrying the damage along with the UvrB and UvrC proteins from the repair site. In the in vivo studies, the initial repair rates of WT and uvrD mutant cells were identical. However, repair then slowed in uvrD− but not WT cells. This finding was provided mechanistic explanation by in vitro experiments that revealed that, after dual incisions, the UvrB and UvrC proteins along with the excised oligomer carrying the dimer remain bound to the duplex and that UvrD helicase displaces the excision repair proteins and releases the excised oligomer to enable UvrB and UvrC to enter new catalytic cycles. The results of the in vivo excision assay in this study provide the strongest in vivo evidence for the proposed role of UvrD in nucleotide excision repair: We found that, although in WT cells and cells with phr or mfd mutations the excised oligomer is rapidly degraded to a 10-mer (and to smaller species not detectable by our 3′ labeling method), in uvrD mutant cells, the excised oligonucleotide retains its full size, consistent with being retained in the “excision gap” annealed to the genome and in complex with UvrB and UvrC proteins. The helicase function of UvrD explains the property of the uvrD mutant with regard to its overall effect on TCR: uvrD mutation does not diminish the genome-wide preferential repair of the TS because, after TCR, the UvrB–UvrC complex remains associated with the excised oligomer in the repair patch in the transcribed strand. The lack of turnover of these proteins results in proportionally less repair in the NTS compared with the WT strain.
The lack of UvrB-UvrC turnover produced another, more subtle, unexpected effect of UvrD. Anecdotally, we found that enhanced strand-specific repair carries over to enhanced repair of the NTS. This “domain”-level repair has been discussed as it relates to the phenomenon in mammalian cells (38). An example of this type of repair is illustrated by the screenshot of the highly transcribed rRNA operon in Fig. 6A. In this case, the overall repair of both strands of the operon is reduced in uvrD− cells although an elevated TCR signal (as TS repair/NTS repair) is still seen. Two factors may contribute to this effect: the accumulation of a high concentration of repair enzymes to actively transcribed genes by the high affinity of the Mfd intermediate to UvrA2UvrB1, and the turnover of the repair subunits, which are the limiting factors in global repair. Consistent with this interpretation, in the absence of UvrD and Uvr protein turnover, there are fewer highly and fewer weakly repaired genes: That is, repair across the genome is at a more even level, which is reflected in the relatively narrow distribution of TCR levels in the distribution plot of uvrD− cells (Fig. 4) and the gene repair maps (Dataset S1). By the same reasoning, the level of coding strand repair in mfd– cells should be reduced. However, Fig. 6A shows substantial coding strand repair in mfd– cells, which is likely compensatory, due to the low template strand repair and due to the use of the same total number of reads for each strain in this genome-wide analysis.
Another interesting aspect of the role of UvrD is shown by the distribution of data points about the red line in Fig. 5A, uvrD panel. As described earlier, a predominance of points above and to the left of the line is evidence for an anticoupling effect of UvrD. Furthermore, unlike phr− cells, and similar to mfd− cells, uvrD− cells demonstrate a wide distribution of data points around the red line, indicating that UvrD influences TCR differently in different genes. One reason for this observation may be the “evening out” effect of the uvrD mutation on repair mentioned above. In addition, as the major helicase in E. coli, UvrD is involved in numerous DNA transactions, including replication and repair pathways, and the absence of UvrD may be expected to influence localization of TCR as a secondary effect of disrupting other ongoing DNA metabolic processes.
Factors Influencing TCR.
To date, TCR, as TS/NTS repair ratios, has been measured in a number of systems. TCR ratios from in vitro studies, each using a different transcriptional unit, ranged from around 2 to 4 (6, 7, 39). Another in vitro study found an approximately threefold more rapid rate of repair of a template strand lesion in the presence versus the absence of RNAP and Mfd (19). In vivo, the TCR value for a region including most of the lac operon, at early time points, appeared to be over 4 (3). Also in vivo, a mutagenesis study of a region of the lacI gene suggested a high ratio of TS/NTS repair in the region studied (41). In contrast, our measurements of overall repair yielded mean values of 1.09 and 1.16 in parental cells measured in two experiments. The distribution of TCR covered a wide range (Fig. 4) and included numerous genes with TCR ratios in the 2 to 4 range and above; however, these represent a minority of the genes. The high levels of TCR that have been reported may reflect a number of factors. For one, the ratio of TT sites in the template/coding strand of annotated genes in E. coli is 1.3. Our data have been corrected to reflect repair per strand per TT per strand, which effectively lowers the TS/NTS repair ratio; however, this correction was not made in the other studies cited. Another possible source of bias in some of the studies cited is the use of simple transcription units and defined in vitro conditions that enable TCR experiments. In reality, in vivo, simple transcription units may be the exception. The widespread occurrence of antisense transcription (36) and ongoing DNA metabolism are complicating factors that may limit the overall occurrence of TCR in vivo. Other uncharacterized factors, such as DNA binding proteins (13), may also limit TCR. The overall level of global repair is expected to have a dampening effect on the TCR signal; this dampening effect is illustrated in mammalian cells by the greatly enhanced TCR signal in XPC cells, which lack global repair, compared with WT cells (25). By chance, there may be greater global repair in the NTS in E. coli cells. Technical factors may also impede our full assessment of TCR in vivo. These factors include the use of strains with different genetic backgrounds for measurements of transcription and repair, and alterations to gene expression induced by UV (42), which were not taken into account in the transcription measurements. Further experimentation and in-depth analyses are needed to clarify how the complex circumstances in vivo influence repair. It is worth noting that the TS/NTS repair ratio decreased overall by a factor of 1.71 in mfd− cells compared with parental cells (Figs. 4 and 5 A and B), and as noted above, the Mfd contribution to TCR was widespread among genes.
Our global repair data also reveal heterogeneity in TCR at individual sites within a given transcriptional unit. This heterogeneity could explain why a small region of the induced lac operon exhibited weak TCR (43) whereas a larger region containing the same sequences exhibited strong TCR after induction (3).
Model for TCR in E. coli.
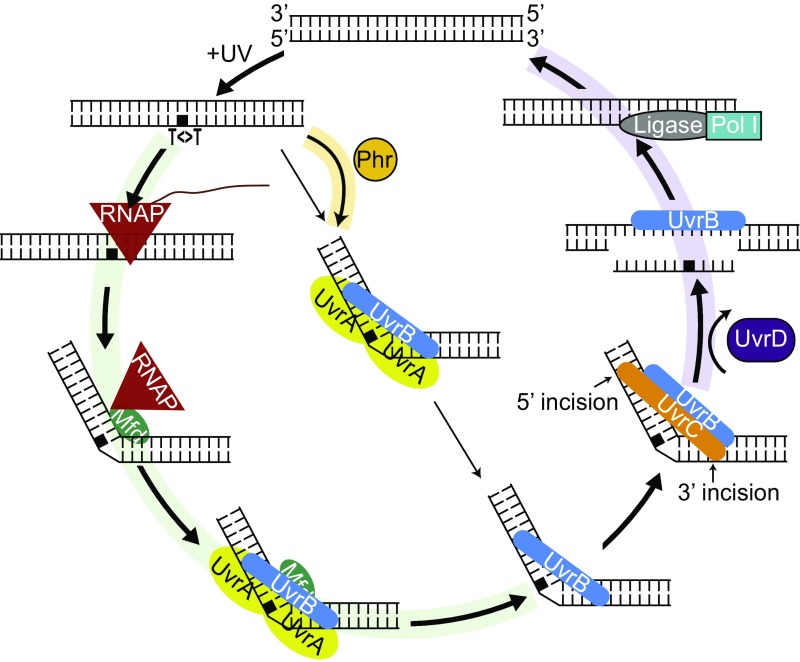
Fig. 7 shows an updated model for TCR in light of research on Mfd, including a recent kinetic study (19) and findings presented in this paper: RNAP stalls at damage sites in the template strand, and the stalled complex recruits Mfd at a relatively fast rate. Mfd, by virtue of its translocase action, releases the nascent transcript and dissociates RNAP from the template. RNAP remains tethered to the Mfd–DNA complex, in which Mfd assumes a conformation that recruits UvrA2B1 by binding to UvrA at about a 20- to 200-fold faster rate than the direct recruitment of UvrA2B1 to sites of damage (global repair). This recruitment is coupled with the loading of UvrB onto the transcription-blocking damage and release of RNAP, Mfd, and UvrA, which subsequently dissociate to component proteins. Then, UvrC binds to the UvrB–DNA complex and makes the dual incisions, which is followed by displacement of the excised oligomer and UvrB and UvrC from the repair site by the UvrD helicase. Fig. 7 also shows the consequences of various mutations on the coupling reaction: Phr (photolyase) repairs the photodimer in the presence of the blue light photon cosubstrate. In the absence of light, Phr binds to the photodimer and accelerates the rate of recognition by UvrA2UvrB1. In phr mutants, the overall repair rate is reduced, and this reduction is more prominent on the NTS because of the preferential recruitment of the repair proteins to the TS. Thus, the absence of photolyase leads to a lower repair rate of the NTS. However, a clear effect of the phr mutation was not observed because of the low amount of photolyase (10 to 20 molecules per cell). In the uvrD mutant, there is a tendency toward enhancement of TCR because the UvrBC proteins that are recruited to the TS by Mfd-mediated reaction are not released from the repair site after the dual incision and thus are unavailable to participate in NTS repair.
Fig. 7.
Schematic representation of the model for the nucleotide excision repair in E. coli controlled by Mfd and Uvr proteins in the dark. When there is an alternative path, the arrow widths indicate the pathway preference: The thicker arrows are preferred over the thinner ones. The green-shaded pathway indicates TCR, and, in the absence of Mfd, this pathway is blocked. The yellow-shaded arrow indicates the enhanced recruitment of UvrA2B1 with the aid of Phr, which is more prevalent with lesions in the NTS because TS lesions undergo TCR. The purple-shaded pathway is blocked in the absence of UvrD, and UvrBC complex doesn’t turn over.
Materials and Methods
Strains used in this study are listed in Table S1. PCR analyses were performed to confirm the gene deletions generated in MfdK33, UvrDK33, and PhrK33. Handling of cells post-UV was done in the absence of photoreactivating light.
Table S1.
List of strains
| Strain | Genotype | Source/derivation |
| BW25113 | ∆(araD-araB)567 ∆lacZ4787 (::rrnB-3) λ− rph-1 ∆(rhaD-rhaB) 568 hsdR514 | CGSC |
| JW1100-1 | BW25113 ∆mfd-739::kan | CGSC |
| JW3786-5 | BW25113 ∆uvrD769::kan | CGSC |
| JW0698-1 | BW25113 ∆phr-758::kan | CGSC |
| JW4019-2 | BW25113 ∆uvrA753::kan | CGSC |
| BT199 | F− thi-1 ∆(gpt-proA)62 thr-1 leuB6 kdgK51 rfbD1 ara-14 lacY1 galK2 lyl-5 mtl-1 tsx-33 supE44 rpsL31 rac− | V. Burdett, Duke University, Durham, NC |
| STL4150 | BT199 ∆xonA300:cat xseA18::amp recJ284::Tn10 | V. Burdett, Duke University, Durham, NC |
| MfdK33 | STL4150 ∆mfd-739::kan | Kanr transductant, P1 JW1100-1 X STL4150 |
| UvrDK33 | STL4150 ∆uvrD769::kan | Kanr transductant, P1 JW3786-5 X STL4150 |
| PhrK33 | STL4150 ∆phr-758::kan | Kanr transductant, P1 JW0698-1 X STL4150 |
| UvrAK33 | STL4150 ∆uvrA753::kan | Kanr transductant, P1 JW4019-2 X STL4150 |
CGSC, Coli Genetic Stock Center; Kanr, kanamycin resistant.
Excision Assays.
Overnight cultures were diluted 1/15 to 1/20 into nonselective LB medium and grown with shaking at 37 °C. Cells were transferred to R150 tissue culture dishes in volumes of 10 mL (Fig. 2 A and B) or 15 mL (Fig. 2C) and were irradiated at an OD600 of ∼0.8 at room temperature with 100 J/m2 (Fig. 2 A and B) or 120 J/m2 (Fig. 2C). Dishes were then transferred to 37 °C incubators for the times indicated in Fig. 2A, or for 5 min (Fig. 2 B and C). Cells were then harvested, chilled, and maintained on ice. Cells were pelleted at 4 °C, resuspended in ice-cold Tris (10 mM)-EDTA (1 mM) (pH 8.0), transferred to ice cold Eppendorf tubes, and pelleted at 4 °C, and the supernatants were removed. Pellets were resuspended with 320 to 340 μL of ice-cold Tris-EDTA. Then, 40 to 42 μL of room temperature 10% (wt/vol) SDS was added, and tubes were gently mixed and then incubated at room temperature for 20 to 25 min. A 100- to 105-μL volume of room temperature NaCl (5 M) was added, tubes were gently mixed, and suspensions were incubated at 4 °C overnight. The larger volumes of SDS and NaCl were used when 15 mL of cells was irradiated, and smaller volumes were used when 10 mL was irradiated. After centrifuging at high speed for 1 h in a microfuge at 4 °C, supernatants (about 380 μL each) were taken, and each was incubated with 12 μL of RNaseA (R4642; Sigma) for 1 h at 37 °C, and then with 12 μL of proteinase K (P8107S; NEB) for 1.5 h at 60 °C. Samples were then extracted twice with phenol/chloroform/isoamyl alcohol and precipitated with ethanol. Samples were immunoprecipitated with an anti-CPD antibody and washed as described (23), except wash buffer III contained 0.25 M LiCl. Extraction, precipitation, and labeling of the 3′ ends with cordycepin was as described (23), except labeling was for 2 to 3 h. Samples were then extracted with phenol/chloroform/isoamyl alcohol and precipitated with ethanol and resolved with a 16% (wt/vol) polyacrylamide sequencing gel. We note that the Hirt procedure removes UvrB and UvrC that remain associated with DNA after dual incision and thus causes release of the excised oligo from genomic DNA.
XR-seq Library Preparation.
We used STL4150 cells as the parental “wild-type” background strain, and STL4150 derivatives MfdK33 (mfd−), UvrDK33 (uvrD−), and PhrK33 (phr−). Cultures were inoculated as above, and cells were grown in nonselective LB medium at 37 °C with shaking and irradiated at an OD600 of ∼0.8 in 15-mL volumes in R150 tissue culture dishes with 120 J/m2 UVC (254 nm) at room temperature. At the dose rate used, irradiations lasted about 2 min. Dishes were then transferred to 37 °C incubators for a 5-min incubation and then were harvested and extracted by the Hirt procedure as above (23). For each cell line, 240 to 320 mL of cells were processed in this manner. Thus, amounts were scaled up around 20-fold compared with excision assays. Compared with excision assays, immunoprecipitations were scaled up 2.5-fold; thus, 12.5 μL of each resin (protein G and anti-rabbit dynabeads) and 2.5 μL of each antibody (anti-CPD and rabbit anti-mouse) were used for each cell line. Immunoprecipitation and harvesting of DNA was as above. The DNAs were then ligated to adaptors as described (25), and the immunoprecipitation was repeated. CPDs were then repaired with ∼400 nM of A. nidulans photolyase–MBP fusion protein and subjected to analytical scale PCR as described (25), so as to determine the minimum number of cycles needed for preparative scale amplification. The number of cycles used for preparative scale PCR was 15 (STL4150, MfdK33, PhrK33) and 13 (UvrDK33). Products were gel-purified as described (25), and DNA was eluted from gel slices by shaking at room temperature overnight in 300 μL of buffer (10 mM Tris, pH 8, 1 mM EDTA, 300 mM NaCl), followed by a second elution with 150 μL of the same buffer for 3 to 4 h. Pooled DNA was collected by precipitation, resuspended, and quantitated as described (25).
Sequencing and Analyses.
The WT sample was sequenced on a MiSeq platform, and the other three were sequenced in one HiSEq 2500 lane at the University of North Carolina High-Throughput Sequencing Facility. The STL4150 sample was resequenced in one HiSEq 2500 lane together with a set of STL4150, MfdK33, UvrDK33, and PhrK33 samples generated by repeating the entire experiment. Preparative PCR of this second set used 12 (UvrDK33) and 14 (STL4150, MfdK33, PhrK33) cycles. Reads were trimmed to remove flanking adapter sequences by cutadapt version 1.10 (44). Only the reads of 13-mer length were analyzed. We randomly sampled 2.9 million reads from each sample using python random module with the seed number 123. The reads were aligned to the E. coli str. K-12 substr. MG1655 genome [National Center for Biotechnology Information (NCBI) assembly accession no. NC_000913.2] by using bowtie (45) with the arguments –nomaqround, –phred33-quals-S, and –seed 123. For the less stringent alignment method, which was used to produce the screenshots, we added the arguments –all, –strata, and –best. Nucleotide distribution of the reads was obtained using bedtools (46), coupled by custom scripts. The reads having TT dinucleotide at the positions of 8 and 9 were used for further analysis. The alignment was separated into strand-specific files by using custom scripts. The gene intervals were retrieved from an NCBI genome annotation file and converted to bed format with custom scripts. Also, genes with <30 TT dinucleotides in the TS were omitted, and the number of reads per each strand per annotated gene was normalized to the number of TT dinucleotides per strand per annotated gene. The strand-specific repair signal for genes was computed with bedtools (46). The sequence data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE92734).
RNA-seq Data Analysis.
We used a publicly available RNA-seq dataset (36). We trimmed nonqualified sequences by using fastq-quality-trimmer from FASTX-Toolkit with the arguments –t 20 and –Q 33. We removed the poly-A tails and retrieved reads having at least 12 nt by using cutadapt version 1.10 (44) with the arguments –a A{100} and –minimum-length 12. We aligned the reads to the same reference genome (NC_000913.2) by using tophat (47) with the argument –library-type fr-firststrand. The aligned reads were separated by strand.
Visualization and Statistics.
Genomic distributions of the XR-seq and RNA-seq reads were visualized and explored using Integrative Genomics Viewer (48). Quantitative plots and data transformations were processed using R. The paired t test was used to test whether the means of two samples were different. Spearman’s rank correlation was used to measure the correlation between the repair ratios and transcription levels of the genes.
Supplementary Material
Acknowledgments
Strains were provided by Dr. V. Burdett (Dr. P. Modrich laboratory, Duke University) and by the E. coli Genetic Resources at the Yale Coli Genetic Stock Center (CGSC). We thank Dr. Roel Schaaper for useful comments.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE92734).
See Commentary on page 2791.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700230114/-/DCSupplemental.
References
- 1.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51(2):241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 2.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: Removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40(2):359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 3.Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342(6245):95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 4.Li W, Li S. Facilitators and repressors of transcription-coupled DNA repair in Saccharomyces cerevisiae. Photochem Photobiol. November 30, 2016 doi: 10.1111/php.12655. [DOI] [PubMed] [Google Scholar]
- 5.Venema J, Mullenders LH, Natarajan AT, van Zeeland AA, Mayne LV. The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc Natl Acad Sci USA. 1990;87(12):4707–4711. doi: 10.1073/pnas.87.12.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Selby CP, Sancar A. Gene- and strand-specific repair in vitro: Partial purification of a transcription-repair coupling factor. Proc Natl Acad Sci USA. 1991;88(18):8232–8236. doi: 10.1073/pnas.88.18.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260(5104):53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 8.Selby CP, Witkin EM, Sancar A. Escherichia coli mfd mutant deficient in “mutation frequency decline” lacks strand-specific repair: In vitro complementation with purified coupling factor. Proc Natl Acad Sci USA. 1991;88(24):11574–11578. doi: 10.1073/pnas.88.24.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Witkin EM. Radiation-induced mutations and their repair. Science. 1966;152(3727):1345–1353. doi: 10.1126/science.152.3727.1345. [DOI] [PubMed] [Google Scholar]
- 10.Witkin EM. Time, temperature, and protein synthesis: A study of ultraviolet-induced mutation in bacteria. Cold Spring Harb Symp Quant Biol. 1956;21:123–140. doi: 10.1101/sqb.1956.021.01.011. [DOI] [PubMed] [Google Scholar]
- 11.Bockrath RC, Palmer JE. Differential repair of premutational UV-lesions at tRNA genes in E. coli. Mol Gen Genet. 1977;156(2):133–140. doi: 10.1007/BF00283485. [DOI] [PubMed] [Google Scholar]
- 12.Bockrath R, Barlow A, Engstrom J. Mutation frequency decline in Escherichia coli B/r after mutagenesis with ethyl methanesulfonate. Mutat Res. 1987;183(3):241–247. doi: 10.1016/0167-8817(87)90006-x. [DOI] [PubMed] [Google Scholar]
- 13.Selby CP. Mfd protein and transcription-repair coupling in E. coli. Photochem Photobiol. January 18, 2017 doi: 10.1111/php.12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lloyd RG, Sharples GJ. Dissociation of synthetic Holliday junctions by E. coli RecG protein. EMBO J. 1993;12(1):17–22. doi: 10.1002/j.1460-2075.1993.tb05627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park JS, Marr MT, Roberts JW. E. coli Transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109(6):757–767. doi: 10.1016/s0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 16.Park JS, Roberts JW. Role of DNA bubble rewinding in enzymatic transcription termination. Proc Natl Acad Sci USA. 2006;103(13):4870–4875. doi: 10.1073/pnas.0600145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts J, Park JS. Mfd, the bacterial transcription repair coupling factor: Translocation, repair and termination. Curr Opin Microbiol. 2004;7(2):120–125. doi: 10.1016/j.mib.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 18.Selby CP, Sancar A. Transcription preferentially inhibits nucleotide excision repair of the template DNA strand in vitro. J Biol Chem. 1990;265(34):21330–21336. [PubMed] [Google Scholar]
- 19.Fan J, Leroux-Coyau M, Savery NJ, Strick TR. Reconstruction of bacterial transcription-coupled repair at single-molecule resolution. Nature. 2016;536(7615):234–237. doi: 10.1038/nature19080. [DOI] [PubMed] [Google Scholar]
- 20.Deaconescu AM, et al. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006;124(3):507–520. doi: 10.1016/j.cell.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 21.Cohen SE, et al. Roles for the transcription elongation factor NusA in both DNA repair and damage tolerance pathways in Escherichia coli. Proc Natl Acad Sci USA. 2010;107(35):15517–15522. doi: 10.1073/pnas.1005203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Epshtein V, et al. UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature. 2014;505(7483):372–377. doi: 10.1038/nature12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu J, et al. Nucleotide excision repair in human cells: Fate of the excised oligonucleotide carrying DNA damage in vivo. J Biol Chem. 2013;288(29):20918–20926. doi: 10.1074/jbc.M113.482257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu J, Lieb JD, Sancar A, Adar S. Cisplatin DNA damage and repair maps of the human genome at single-nucleotide resolution. Proc Natl Acad Sci USA. 2016;113(41):11507–11512. doi: 10.1073/pnas.1614430113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu J, Adar S, Selby CP, Lieb JD, Sancar A. Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 2015;29(9):948–960. doi: 10.1101/gad.261271.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Viswanathan M, Burdett V, Baitinger C, Modrich P, Lovett ST. Redundant exonuclease involvement in Escherichia coli methyl-directed mismatch repair. J Biol Chem. 2001;276(33):31053–31058. doi: 10.1074/jbc.M105481200. [DOI] [PubMed] [Google Scholar]
- 27.Sancar A, Rupp WD. A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell. 1983;33(1):249–260. doi: 10.1016/0092-8674(83)90354-9. [DOI] [PubMed] [Google Scholar]
- 28.Sancar A. Mechanisms of DNA repair by photolyase and excision nuclease (Nobel Lecture) Angew Chem Int Ed Engl. 2016;55(30):8502–8527. doi: 10.1002/anie.201601524. [DOI] [PubMed] [Google Scholar]
- 29.Sancar A, Franklin KA, Sancar GB. Escherichia coli DNA photolyase stimulates uvrABC excision nuclease in vitro. Proc Natl Acad Sci USA. 1984;81(23):7397–7401. doi: 10.1073/pnas.81.23.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selby CP, Drapkin R, Reinberg D, Sancar A. RNA polymerase II stalled at a thymine dimer: Footprint and effect on excision repair. Nucleic Acids Res. 1997;25(4):787–793. doi: 10.1093/nar/25.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuemmerle NB, Masker WE. Effect of the uvrD mutation on excision repair. J Bacteriol. 1980;142(2):535–546. doi: 10.1128/jb.142.2.535-546.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orren DK, Selby CP, Hearst JE, Sancar A. Post-incision steps of nucleotide excision repair in Escherichia coli: Disassembly of the UvrBC-DNA complex by helicase II and DNA polymerase I. J Biol Chem. 1992;267(2):780–788. [PubMed] [Google Scholar]
- 33.Orren DK, Sancar A. The (A)BC excinuclease of Escherichia coli has only the UvrB and UvrC subunits in the incision complex. Proc Natl Acad Sci USA. 1989;86(14):5237–5241. doi: 10.1073/pnas.86.14.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Husain I, Van Houten B, Thomas DC, Abdel-Monem M, Sancar A. Effect of DNA polymerase I and DNA helicase II on the turnover rate of UvrABC excision nuclease. Proc Natl Acad Sci USA. 1985;82(20):6774–6778. doi: 10.1073/pnas.82.20.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto K, Fujiwara Y, Shinagawa H. Evidence that the phr+ gene enhances the ultraviolet resistance of Escherichia coli recA strains in the dark. Mol Gen Genet. 1983;192(1-2):282–284. doi: 10.1007/BF00327679. [DOI] [PubMed] [Google Scholar]
- 36.Thomason MK, et al. Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J Bacteriol. 2015;197(1):18–28. doi: 10.1128/JB.02096-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kavakli IH, Sancar A. Analysis of the role of intraprotein electron transfer in photoreactivation by DNA photolyase in vivo. Biochemistry. 2004;43(48):15103–15110. doi: 10.1021/bi0478796. [DOI] [PubMed] [Google Scholar]
- 38.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9(12):958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 39.Manelyte L, et al. The unstructured C-terminal extension of UvrD interacts with UvrB, but is dispensable for nucleotide excision repair. DNA Repair (Amst) 2009;8(11):1300–1310. doi: 10.1016/j.dnarep.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahn B. A physical interaction of UvrD with nucleotide excision repair protein UvrB. Mol Cells. 2000;10(5):592–597. doi: 10.1007/s10059-000-0592-5. [DOI] [PubMed] [Google Scholar]
- 41.Oller AR, Fijalkowska IJ, Dunn RL, Schaaper RM. Transcription-repair coupling determines the strandedness of ultraviolet mutagenesis in Escherichia coli. Proc Natl Acad Sci USA. 1992;89(22):11036–11040. doi: 10.1073/pnas.89.22.11036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158(1):41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kunala S, Brash DE. Intragenic domains of strand-specific repair in Escherichia coli. J Mol Biol. 1995;246(2):264–272. doi: 10.1006/jmbi.1994.0082. [DOI] [PubMed] [Google Scholar]
- 44.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal. 2011;17(1):10–12. [Google Scholar]
- 45.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinlan AR. BEDTools: The Swiss-Army tool for genome feature analysis. Curr Protoc Bioinformatics. 2014;47:11.12.11–34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.