Significance
Our study shows that deep subseafloor sediments are populated by descendants of rare members of surface sediment microbial communities that become predominant during burial over thousands of years. We provide estimates of mutation rates and strength of purifying selection in a set of taxonomically diverse microbial populations in marine sediments and show that their genetic diversification is minimal during burial. Our data suggest that the ability of subseafloor microbes to subsist in the energy-deprived deep biosphere is not acquired during burial but that these microbes were already capable of living in this unique environment. These findings represent a significant step toward understanding the bounds for life in the deep biosphere and its connection to life in the surface world.
Keywords: marine sediment, bacteria, metagenomics, evolution, single-cell genomics
Abstract
Bacterial and archaeal communities inhabiting the subsurface seabed live under strong energy limitation and have growth rates that are orders of magnitude slower than laboratory-grown cultures. It is not understood how subsurface microbial communities are assembled and whether populations undergo adaptive evolution or accumulate mutations as a result of impaired DNA repair under such energy-limited conditions. Here we use amplicon sequencing to explore changes of microbial communities during burial and isolation from the surface to the >5,000-y-old subsurface of marine sediment and identify a small core set of mostly uncultured bacteria and archaea that is present throughout the sediment column. These persisting populations constitute a small fraction of the entire community at the surface but become predominant in the subsurface. We followed patterns of genome diversity with depth in four dominant lineages of the persisting populations by mapping metagenomic sequence reads onto single-cell genomes. Nucleotide sequence diversity was uniformly low and did not change with age and depth of the sediment. Likewise, there was no detectable change in mutation rates and efficacy of selection. Our results indicate that subsurface microbial communities predominantly assemble by selective survival of taxa able to persist under extreme energy limitation.
Bacteria and archaea inhabiting the subsurface seabed account for more than half of all microbial cells in the oceans (1, 2). The subsurface communities are cut off from fresh detrital organic matter deposited on the sea floor. The energy available for cellular maintenance and growth decreases rapidly with depth and age of the sediment (3–5). As a consequence, the microbial abundance and cell-specific metabolic rates decrease by orders of magnitude already within the top few meters of sediment (3, 6–8). However, even hundreds of meters below the seafloor sediments are populated by microbes that actively turn over their biomass (3, 8). The growth characteristics of these microbial populations are unlike anything studied in pure culture as estimates suggest that the generation times of subsurface cells are tens to hundreds of years (3, 7, 8). As a model for how these deep biosphere communities are assembled, it was suggested that the microbes found in the subsurface seabed represent descendants of surface communities that were buried in the past (9–12). So far this model has not been tested systematically. Furthermore, it is not known if subsurface microorganisms during the burial evolve unique genetic traits conferring adaptation to this environment (13). Studies of adaptation and evolution in free-living microorganisms reported high genetic heterogeneity and rapid evolutionary change within natural populations. However, these studies have focused on dynamic or mixed environments with large population sizes and rapid growth, e.g., biofilms or planktonic communities (14–16) that are very different from the subsurface seabed environment.
According to “Drake’s rule” (17), DNA-based organisms only experience one fitness-affecting mutation per 300 genomes replicated. Therefore, in the highly structured subsurface sediment environment, for which generation times dramatically increase as burial proceeds (8), we predict that only a handful of adaptive mutations will arise during burial and that these will not be able to sweep through populations. Alternatively, depending on the strength of environmental selection, cells present in the subsurface seabed may accumulate mutations as a result of reduced potential for DNA repair due to extreme energy limitation. Here we test these hypotheses by exploring the microbial community assembly and evolutionary processes in the transition from shallow to deep marine sediment. Our findings have implications for understanding microbial life in one of the largest, yet most inaccessible, ecosystems on Earth: the marine deep biosphere.
Results and Discussion
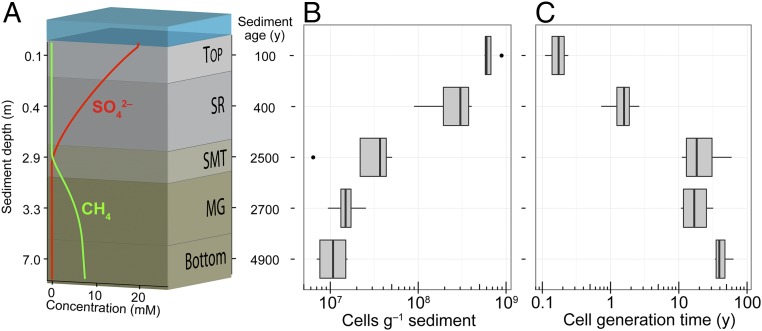
We combined PCR amplicon sequencing with single-cell and metagenomic sequencing to investigate how microbial communities assemble and evolve across sediment depths at four stations in Aarhus Bay, Denmark (Materials and Methods). The bay has a well-described sedimentary history with up to 10-m thick organic-rich deposits that represent the past 8,700 y of stable marine sedimentation (18). The sampled depths varied between stations, but were selected to sample the five same biogeochemical zones at each station, namely: Top, the bioturbated surface sediment; SR, the underlying zone of anaerobic sulfate reduction; SMT, the sulfate–methane transition where sulfate-dependent anaerobic methane oxidation takes place; MG, the methanogenic zone where biogenic methane is accumulating; and Bottom, the deepest sampled part of the sediments (methanogenic zone) (Fig. 1A and SI Appendix, Table S1). Although the depth distribution of sediment age and microbial carbon mineralization rates show little variation across stations in Aarhus Bay (19), the depth of the SMT zone varies between stations due to differences in the thickness of the Holocene mud layer forming the marine sediment (19). Across the sampled sediment depths (0–7 m) the microbial cell density decreased from 109 to 107 cells⋅g−1 wet weight (Fig. 1B) and the estimated mean generation time of cells increased from months to tens of years (Fig. 1C). The generation times were estimated as the turnover time of cellular carbon by applying a model combining cell-specific carbon oxidation rates (inferred from experimentally measured sulfate reduction rates and cell abundances), estimated growth yield, and the carbon content of cells (Materials and Methods).
Fig. 1.
Vertical biogeochemical zonation and distribution of microbial biomass and turnover in Aarhus Bay sediments. (A) Biogeochemical zonation. Top, surface sediment; SR, upper sulfate-rich sediment; SMT, sulfate–methane transition zone; MG, methanogenic sediment; Bottom, deep methanogenic zone. Pore water concentrations of sulfate and methane and sediment depth and age relate to Station M29A (SI Appendix, Table S1). Sediment depth and age axes are not drawn to scale. (B) Microbial cell abundances determined by qPCR quantification of 16S rRNA genes in extracted DNA assuming that bacterial and archaeal cells on average harbor 4.1 and 1.6 gene copies, respectively. (C) Average generation time for cells as estimated from cell-specific rates of carbon oxidation, cellular carbon content, and growth yield (Materials and Methods). In the box-and-whisker plots, the middle line represents the median value for four different sampling stations and horizontal lines represent minimum and maximum values. The box stretches from the lower to the upper quartile and dots show outliers (>1.5 times the corresponding quartile).
We used high-throughput 16S rRNA gene PCR amplicon sequencing to compare microbial community compositions in samples from the different geochemical zones. The community composition [expressed in operational taxonomic units (OTUs) defined by a 97% sequence similarity cutoff] changed strongly with depth, with the surface and sulfate-rich zones each supporting distinct communities whereas the deeper zones had more similar composition and lower richness (SI Appendix, Fig. S1). Within this dataset we successfully traced OTUs across the geochemical zones in the sediment and discovered that a small number of OTUs (79 OTUs) were present throughout all sampled depths of the sediment column (Fig. 2A). This core set of persisting OTUs represented less than 10% of the total OTU richness (SI Appendix, Table S1). The persisting OTUs showed low relative abundance in the Top layer (Fig. 2B) but collectively made up a large proportion of the total microbial community in the deeper layers (up to 40–50%, Fig. 2B). We confirmed the trend of persisting OTUs at two stations by PCR amplifying and sequencing the dsrB gene, which encodes the dissimilatory sulfite reductase in sulfate-reducing microorganisms (SI Appendix, Fig. S2). The dsrB gene evolves faster than the 16S rRNA gene (20). However, even when using a 98% similarity cutoff to cluster all dsrB sequence reads into OTUs, we observed OTUs persisting across depths and making up a large proportion of the reads in the subsurface dsrB sequence libraries (SI Appendix, Fig. S2). We consistently observed the same persisting OTUs at different sampling stations (SI Appendix, Fig. S3 A–C), indicating that the community change was due to environmental selection and not random sampling. These results offer strong empirical evidence for an assembly of subsurface communities by selection of those populations from the surface community that can effectively conserve energy for cellular maintenance and growth in deep sediments (9, 11, 12). Approximately half of the microbial communities in the deeper sediment layers represents OTUs that did not persist across sediment depth (Fig. 2B). To assess whether these OTUs represent a unique subsurface community, we identified 855 OTUs that were exclusively present in the MG and Bottom zones compared with the Top and SR zones of each of the four stations (SI Appendix, Fig. S3D). Many of these OTUs were of low relative abundance and specific to individual stations, yet 380 OTUs were shared between the MG and Bottom zones of at least three of the four stations (SI Appendix, Fig. S3 E and F). This indicates the presence of a core subsurface community, which accounted for 8–14% of the reads in the subsurface sequence libraries.
Fig. 2.
Persisting OTUs and potential for population growth. (A) Abundance of 16S rRNA gene sequence OTUs persisting across different biogeochemical sediment zones. Top, total number of OTUs in the top zone. All other zones: number of OTUs from the Top zone that persist at each of these zones. Bottom: Number of OTUs persisting throughout all zones. (B) Relative abundance in each zone of the OTUs persisting throughout all zones. (C) Cumulative number of cell generations during burial, estimated from cell-generation times (Fig. 1C) and sediment age (Materials and Methods). Bar plots in A and B show mean ± SD for four sampling stations. Top, surface sediment; SR, upper sulfate-rich sediment; SMT, sulfate–methane transition zone; MG, methanogenic zone; Bottom, deep methanogenic zone.
The dominant persisting 16S rRNA gene OTUs belong to bacterial and archaeal lineages usually considered key components of the deep marine biosphere, such as Atribacteria, Dehalococcoidia, Bathyarchaeota, and Marine Benthic Group-B archaea (SI Appendix, Fig. S3). However, we find that the absolute abundance of these lineages peaks in the Top or SR zone (SI Appendix, Fig. S5). These lineages lack cultured representatives; however, our limited knowledge of their physiology points to a range of organotrophic metabolic strategies for generating ATP (SI Appendix, Table S2). Although the overall microbial abundance decreased strongly from the surface to the sulfate reduction zone and below (Fig. 1B), the persisting OTUs did not follow this pattern. They either showed little change with depth or increased in absolute abundance in the sulfate-reduction zone (SI Appendix, Figs. S3C and S5B), suggesting that this zone is where the assembly of the subsurface microbial community mostly takes place. In agreement, our estimates of generation times suggest that the upper zones of the sediment sustain much faster microbial growth rates than the deeper zones and thus offer a potential for more dynamic changes in microbial community composition (Figs. 1C and 2C).
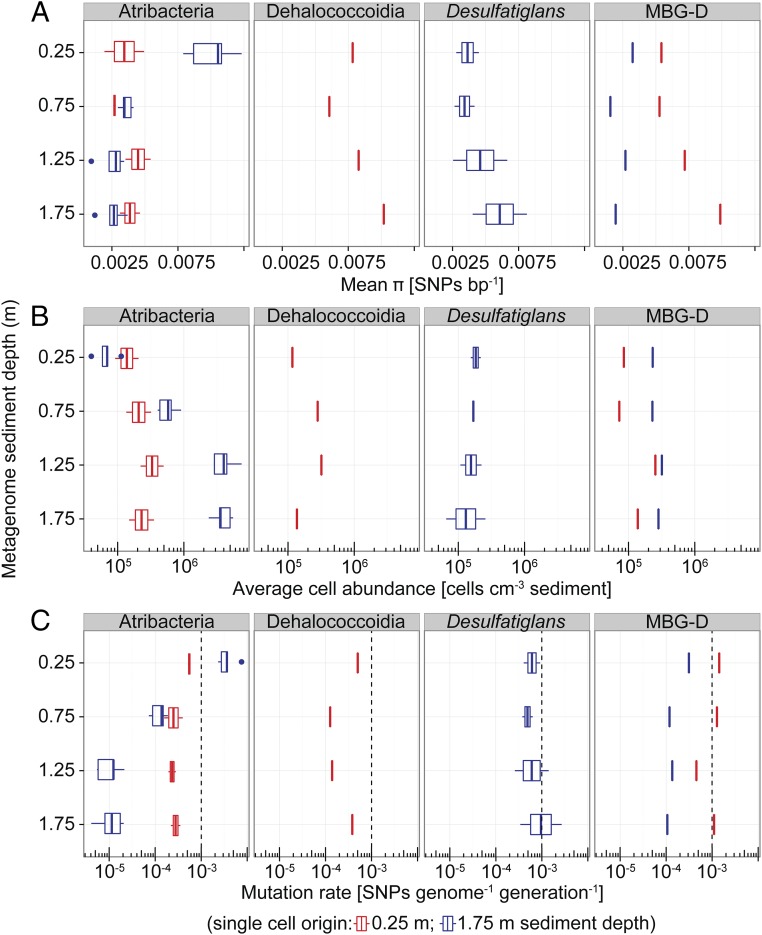
Do the persisting populations undergo adaptive evolution in response to the decreasing energy availability with sediment depth, or are they already adapted to live under extreme energy limitation as previously hypothesized (9)? To answer this central question, we used metagenomics and single-cell genomics to assess the extent and nature of genomic change in four dominant persisting lineages with sediment depth and age. We extracted cells from the upper part of the sulfate reduction zone (0.25 m depth) and from the methanogenic zone (1.75 m) at station M5 (SI Appendix, Table S1), sorted individual cells randomly by flow cytometry, and amplified and sequenced each of their genomes. We obtained 17 single-cell amplified genomes (SAGs) representing four different taxonomic lineages, which all harbor persisting populations (SI Appendix, Table S3 and Fig. S6). We also constructed metagenomic DNA sequence libraries from 0.25, 0.75, 1.25, and 1.75 m sediment depth of station M5 (SI Appendix, Table S1). We then mapped the reads of each library onto predicted genes in the assembled SAGs with a conservative nucleotide identity cutoff of 95%, which represents the recognized species boundary in prokaryotic systematics (21). From these data we could measure the genetic diversity in the persisting lineages represented by the SAGs and evaluate their mode of evolution. We first calculated genomic nucleotide diversity (π) as the average number of single-nucleotide polymorphism positions (SNPs) per base pair for each lineage and depth. The observed nucleotide diversity was low [≤0.0075 SNPs bp−1; notably, the sequencing error rate was >10-fold lower (Materials and Methods)] and mostly varied within less than twofold over depth depending on the population (Fig. 3A). These results indicate that absolute levels of diversity in persisting populations are similar across sediment depth. We also found that, within lineages, individuals from different sediment depths shared a high proportion of identical SNPs within their genomes (40–60%; SI Appendix, Fig. S7A). It is highly unlikely that populations in different subsurface zones have exchanged cells or genes because the clay sediments show no advective fluid or gas flow, and the energy limitation prevents dispersal of cells by motility (7). We thus conclude that populations separated in time (thousands of years) and space (meters) throughout the sediment column are genetically very similar because they hardly accumulated any mutations during burial.
Fig. 3.
Genetic diversity and rates of evolution within subsurface microbial populations. (A) Genome nucleotide diversity (π) as measured by mapping metagenome reads onto genomes of single cells derived from 0.25 m or 1.75 m sediment depth and quantifying the number of SNPs. Taxonomic names indicate the identities of the single-cell genomes (Atribacteria, n = 7; Dehalococcoidia, n = 1; Desulfatiglans, n = 2; MBG-D, n = 2; SI Appendix, Table S3). (B) Abundance of the populations represented by the single cells as inferred by multiplying the fraction of reads within a metagenomic library that mapped to the SAGs by microscopic total cell counts from the respective sediment depths. (C) Mutation rates (µg) calculated according to µg = π/2Ne, assuming that effective population size (Ne) equals the total population size shown in B and using π and the estimated genome sizes to infer SNPs genome−1 (Materials and Methods). The dashed lines show, for comparison, the µg of growing E. coli cultures. Box-and-whisker plots are as defined in Fig. 1.
With our data it was possible to infer mutation rates (µg) in natural sediment populations assuming µg = π/2Ne (22, 23), where π is the observed nucleotide diversity and Ne the effective population size. For the calculations, we assumed that Ne equals the total population size, which we determined for each SAG from the relative abundance of mapped reads in the metagenomes and microscopic cell counts (Fig. 3B); see refs. 24 and 25 for a discussion of using the relative abundance of mapped reads in metagenomic libraries for estimating population sizes. We furthermore normalized for genome size (SI Appendix, Table S3) and the mean number of generations that separate the populations across sediment depth (Materials and Methods). For one atribacterial population, the inferred mutation rate decreased 100-fold with depth from 3·10−3 to 10−5 per genome per generation (Fig. 3C). For the other populations mutation rates were rather constant at 10−4–10−3 per genome per generation (Fig. 3C). Overall our data suggest that mutation rates and repair in the slow-growing and energy-limited subsurface microbes are not inherently different from those of fast-growing species because they are remarkably similar to rates reported by Drake for DNA-based genomes (17), for Escherichia coli cultures (26), and for natural biofilm communities (15). The low mutation rates of <10−3 per genome per generation inferred for some of the persisting subsurface populations indicate that their effective population size (Ne) is only a fraction of the measured total population size (1–90%, depending on the population).
In addition to the effective population size, the estimated mutation rates are sensitive to uncertainties in determination of SNP diversity and cellular generation times. The metagenome sequencing error rate was low and is unlikely to influence our results (SI Appendix). We used linear models and F tests and the type II sum-of-square test to exclude that differences in genetic diversity among lineages were simply explained by technical variables such as number of SAGs analyzed, the number of predicted genes, and the number of reads mapped to predicted genes (SI Appendix, Table S3). Differences in the number of reads mapped on SAGs do explain some of the diversity differences observed, but genuine differences among lineages remain (P = 0.016). Our generation time estimate is based on empirically determined parameters (Material and Methods); of these, cellular carbon content varies twofold (27) and sediment age by 2% (4), whereas rates of carbon mineralization were modeled from experimental data with a SD of 5% (19, 28). Finally, cellular growth yields likely vary by a factor of 2–3, according to pure culture studies and subsurface sediment modeling (3, 6, 8). These uncertainties overall result in an approximately fivefold variation of generation times, which are linearly linked to a corresponding variation in mutation rates.
It is an open question whether, in the subsurface, populations undergo adaptive evolution or if selection is even stringent enough to remove deleterious mutations and thus maintain gene functions. The stringency of selection within a population can be measured by the ratio of nonsynonymous (Pn) to synonymous (Ps) SNPs within protein-coding genes (29). Purifying selection maintains gene function by removing preferentially nonsynonymous mutations. Lower Pn/Ps ratios throughout a population indicate stronger purifying selection, whereas adaptive selection yields Pn/Ps ratios >1. We used the populations represented by the SAGs to compare the strength of purifying selection acting on these populations at 0.25 and 1.75 m sediment depth and asked the question whether the selection regime shifts with sediment depth. Pn/Ps ratios were generally well below 1 and thus indicative of stringent selection. In three of four taxonomic lineages, we could not detect a significant shift in genome-wide Pn/Ps ratios with depth (SI Appendix, Fig. S7B). In the Atribacteria, we observed a small, yet statistically significant, shift toward higher Pn/Ps ratios at 1.75 m depth, compatible with modest relaxation of purifying selection in deeper sediments. We expected that genes encoding key functions in subsurface sediment still undergo purifying or adaptive selection, whereas genes obsolete in this environment may experience relaxed purifying selection. We compared Pn/Ps across broad functional gene categories, but found no trend with depth (SI Appendix, Fig. S7B). Our data thus indicate that the selection regime remains mostly unchanged across depth in the subsurface sediments.
Although we explored evolutionary changes occurring within the upper 2 m of sediment, covering a time span of 2,000 y (or 500 generations), we expect that our findings can be generalized to deep, million-year-old subsurface sediments: because generation times increase successively upon burial, even in 1 million-year-old sediment layers [spanning over 6,000 generations (8)], most generations, and thus the greatest scope for adaptive evolution, is still within the top few meters, i.e., in the zone that we analyzed.
In conclusion, our study shows that subsurface microbial communities in nonadvective marine sediments predominantly assemble by selective survival of surface populations without detectable further diversification or adaptation in deeper sediments. Our results also indicate that mutation repairs, and therefore gene functions, are maintained in the subsurface sediment despite the extreme energy limitation, which is consistent with existing biogeochemical evidence for microbial metabolic activity throughout the marine deep biosphere.
Materials and Methods
Please see SI Appendix for a detailed description of the materials and methods.
Sediment Sampling.
Sediment cores were collected from five stations in Aarhus Bay (SI Appendix, Table S1). The stations differed by the sediment depth of the sulfate–methane transition zone (the depth at which sulfate becomes depleted and methane begins to accumulate). The cores were subsampled for DNA extraction by a protocol, which removes extracellular DNA (30), and for whole-cell extraction for single-cell sorting. The biogeochemistry of the studied cores is reported elsewhere (19).
PCR Amplicon Sequence Libraries.
The 16S rRNA and dsrB gene fragments were PCR-amplified using universal primer pairs. The dsrB gene encodes the β-subunit of the dissimilatory sulfite reductase and represents a phylogenetic and functional marker gene for dissimilatory sulfate-reducing microorganisms (20). Barcoded PCR products were sequenced on an Ion Torrent PGM using 300-bp chemistry (Life Technologies). A mock 16S rRNA gene community was sequenced to test for contamination during sequencing procedures and to test the fidelity of our OTU clustering procedure.
Quantitative PCR.
SYBR-Green–based quantitative PCR (qPCR) was used to quantify total bacterial and archaeal 16S rRNA gene copies in extracted DNA using published general primer pairs. Furthermore, 16S rRNA gene copies of selected bacterial and archaeal lineages were quantified using custom-designed primer pairs.
Metagenome Library Construction.
Metagenomic sequence libraries were constructed from DNA extracted from 25, 75, 125, and 175 cm sediment depths of a single station by Illumina sequencing. For each library, sequencing error rates were quantified by analyzing the sequencing reads from the phiX174 phage genomic DNA added to all sequencing runs as an internal control as part of the standard Illumina sequencing protocol.
Generation of Single-Cell Amplified Genomes.
Single-cell sorting, whole-genome amplification, and 16S rRNA gene PCR screening of single cells were performed at the Bigelow Laboratory Single Cell Genomics Center (scgc.bigelow.org) using its established protocols as previously described (31). SAGs were sequenced on a MiSeq instrument with paired-end 2× 300-bp chemistry.
Estimation of Genomic Mutation Rates.
Per generation mutation rates (µg) were calculated according to π = 2Ne × µg (22, 23) assuming that effective population size (Ne) equals the total observed population size. The π values (SNPs base pair−1) were calculated by mapping metagenomic reads onto predicted genes in SAG assemblies following a stringent procedure for selecting genes and SNP positions for the analyses to minimize bias from differences in mapping coverage between sediment depths and between lineages (SI Appendix). Genomic mutation rates were then calculated from estimated genome sizes of SAGs and the estimated average number of microbial cell generations separating a given sediment depth from the surface sediment derived as described in Estimation of Rates of Microbial Biomass Turnover.
Pn/Ps Ratios.
The ratio of nonsynonymous to synonymous number of SNPs (Pn/Ps) was calculated by mapping metagenomic sequence reads from 25-cm and 175-cm sediment depths onto genes predicted in the SAG assemblies.
Estimation of Rates of Microbial Biomass Turnover.
The average rate by which cells turn over their biomass in subsurface sediments was estimated from measured rates of sulfate-dependent carbon oxidation, total cell abundance, and sediment age (8). Sulfate reduction is the main terminal electron-accepting process in coastal marine sediments and accounts for the entire net oxidation of organic matter to CO2 in the SR zone. Sulfate reduction rates therefore represent the metabolic activity of the entire anaerobic microbial food chain in this zone (6, 8). In marine sediments, including Aarhus Bay, sulfate reduction rates decrease with sediment depth according to a power law function that can be extrapolated below the SMT zone to reflect carbon mineralization rates in the methanogenic zone (i.e., by methanogenesis) (19, 32). We therefore used such a power law function determined for Aarhus Bay sediments (19) to calculate organic carbon oxidation rates. Cell-specific carbon oxidation rates were calculated from total cell abundances and were used to estimate biomass turnover of cells assuming a growth yield of 8% (8) and a cellular carbon content of 21.5 fg (27). The biomass turnover serves as a proxy for the generation times of cells assuming that the cells indeed are growing by cell division in the energy-depleted subseafloor as opposed to being in a nongrowth state where cells merely repair and sustain their biomolecules (9).
Supplementary Material
Acknowledgments
We thank Britta Poulsen, Susanne Nielsen, Trine Bech Søgaard, and Lina Juzokaite for laboratory technical assistance; the crew of the R/V Aurora for help during sediment sampling; Sabine Flury and Hans Røy for sharing unpublished data on sulfate reduction rates; and Eva Fernández Cáceres for carrying out initial quality-filtering of metagenome sequence data. All metagenome sequencing was performed at the National Genomics Infrastructure sequencing platforms at the Science for Life Laboratory at Uppsala University, a national infrastructure supported by the Swedish Research Council (VR-RFI) and the Knut and Alice Wallenberg Foundation. This work was funded by Danish National Research Foundation Grant DNRF104; FP7 European Research Council (ERC) Advanced Grant 294200 (to B.B.J.); ERC Starting Grant PUZZLE_CELL (to T.J.G.E.); Swedish Foundation for Strategic Research Grant SSF-FFL5 (to T.J.G.E.); and grants from the Carlsberg Foundation, Julie-von-Moellen Foundation, and the Danish Agency for Science, Technology and Innovation (to A.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: PCR amplicon and metagenome sequence data have been deposited at the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under the BioProject accessions PRJNA308429 and PRJNA305566 and BioSample accessions: SAMN04395423, and SAMN04328853–SAMN04328860, respectively. Single-cell amplified genome assemblies were uploaded to the Integrated Microbial Genomes portal (https://img.jgi.doe.gov) with the following genome IDs: 2606217225, 2626541591, 2606217224, 2609459604, 2606217227, 2606217228, 2609459605, 2626541542, 2626541544, 2626541633, 2626541550, 2626541552, 2626541551, 2609459610, 2609459611, 2609459606, 2609459607, 2626541553, 2615840654 and 2615840655.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614190114/-/DCSupplemental.
References
- 1.Kallmeyer J, Pockalny R, Adhikari RR, Smith DC, D’Hondt S. Global distribution of microbial abundance and biomass in subseafloor sediment. Proc Natl Acad Sci USA. 2012;109(40):16213–16216. doi: 10.1073/pnas.1203849109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkes RJ, et al. A review of prokaryotic populations and processes in sub-seafloor sediments, including biosphere:geosphere interactions. Mar Geol. 2014;352:409–425. [Google Scholar]
- 3.Lomstein BA, Langerhuus AT, D’Hondt S, Jørgensen BB, Spivack AJ. Endospore abundance, microbial growth and necromass turnover in deep sub-seafloor sediment. Nature. 2012;484(7392):101–104. doi: 10.1038/nature10905. [DOI] [PubMed] [Google Scholar]
- 4.Langerhuus AT, et al. Endospore abundance and D:L-amino acid modelling of bacterial turnover in Holocene marine sediment (Aarhus Bay) Geochim Cosmochim Acta. 2012;99:87–99. [Google Scholar]
- 5.Middelburg JJ. A simple rate model for organic matter decomposition in marine sediments. Geochim Cosmochim Acta. 1989;53:1577–1581. [Google Scholar]
- 6.D’Hondt S, Rutherford S, Spivack AJ. Metabolic activity of subsurface life in deep-sea sediments. Science. 2002;295(5562):2067–2070. doi: 10.1126/science.1064878. [DOI] [PubMed] [Google Scholar]
- 7.Hoehler TM, Jørgensen BB. Microbial life under extreme energy limitation. Nat Rev Microbiol. 2013;11(2):83–94. doi: 10.1038/nrmicro2939. [DOI] [PubMed] [Google Scholar]
- 8.Jørgensen BB, Marshall IP. Slow microbial life in the seabed. Annu Rev Mar Sci. 2016;8:311–332. doi: 10.1146/annurev-marine-010814-015535. [DOI] [PubMed] [Google Scholar]
- 9.Lever MA, et al. Life under extreme energy limitation: A synthesis of laboratory- and field-based investigations. FEMS Microbiol Rev. 2015;39(5):688–728. doi: 10.1093/femsre/fuv020. [DOI] [PubMed] [Google Scholar]
- 10.Inagaki F, Okada H, Tsapin AI, Nealson KH. Microbial survival: The paleome: A sedimentary genetic record of past microbial communities. Astrobiology. 2005;5(2):141–153. doi: 10.1089/ast.2005.5.141. [DOI] [PubMed] [Google Scholar]
- 11.Teske A. Marine deep sediment microbial communities. In: Rosenberg E, et al., editors. The Prokaryotes: Prokaryotic Communities and Ecophysiology. Springer; Berlin: 2013. pp. 123–138. [Google Scholar]
- 12.Walsh EA, et al. Bacterial diversity and community composition from seasurface to subseafloor. ISME J. 2016;10(4):979–989. doi: 10.1038/ismej.2015.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biddle JF, et al. Prospects for the study of evolution in the deep biosphere. Front Microbiol. 2012;2:285. doi: 10.3389/fmicb.2011.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kashtan N, et al. Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science. 2014;344(6182):416–420. doi: 10.1126/science.1248575. [DOI] [PubMed] [Google Scholar]
- 15.Denef VJ, Banfield JF. In situ evolutionary rate measurements show ecological success of recently emerged bacterial hybrids. Science. 2012;336(6080):462–466. doi: 10.1126/science.1218389. [DOI] [PubMed] [Google Scholar]
- 16.Bendall ML, et al. Genome-wide selective sweeps and gene-specific sweeps in natural bacterial populations. ISME J. 2016;10(7):1589–1601. doi: 10.1038/ismej.2015.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148(4):1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen JB, Bennike O. Geological setting as background for methane distribution in Holocene mud deposits, Aarhus Bay, Denmark. Cont Shelf Res. 2009;29(5-6):775–784. [Google Scholar]
- 19.Flury S, et al. Controls on subsurface methane fluxes and shallow gas formation in Baltic Sea sediment (Aarhus Bay, Denmark) Geochim Cosmochim Acta. 2016;188:297–309. [Google Scholar]
- 20.Müller AL, Kjeldsen KU, Rattei T, Pester M, Loy A. Phylogenetic and environmental diversity of DsrAB-type dissimilatory (bi)sulfite reductases. ISME J. 2015;9(5):1152–1165. doi: 10.1038/ismej.2014.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106(45):19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charlesworth B. Fundamental concepts in genetics: Effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 2009;10(3):195–205. doi: 10.1038/nrg2526. [DOI] [PubMed] [Google Scholar]
- 23.Wright S. Statistical genetics and evolution. Bull Am Math Soc. 1942;48(4):223–247. [Google Scholar]
- 24.Hingamp P, et al. Exploring nucleo-cytoplasmic large DNA viruses in Tara Oceans microbial metagenomes. ISME J. 2013;7(9):1678–1695. doi: 10.1038/ismej.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones MB, et al. Library preparation methodology can influence genomic and functional predictions in human microbiome research. Proc Natl Acad Sci USA. 2015;112(45):14024–14029. doi: 10.1073/pnas.1519288112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee H, Popodi E, Tang H, Foster PL. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc Natl Acad Sci USA. 2012;109(41):E2774–E2783. doi: 10.1073/pnas.1210309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braun S, et al. Cellular content of biomolecules in sub-seafloor microbial communities. Geochim Cosmochim Acta. 2016;188:330–351. [Google Scholar]
- 28.Røy H. Determination of dissimilatory sulfate reduction rates in marine sediment via radioactive 35S tracer. Limnol Oceanogr Methods. 2014;12(4):196–211. [Google Scholar]
- 29.Welch JJ, Eyre-Walker A, Waxman D. Divergence and polymorphism under the nearly neutral theory of molecular evolution. J Mol Evol. 2008;67(4):418–426. doi: 10.1007/s00239-008-9146-9. [DOI] [PubMed] [Google Scholar]
- 30.Lever MA, et al. A modular method for the extraction of DNA and RNA, and the separation of DNA pools from diverse environmental sample types. Front Microbiol. 2015;6:476. doi: 10.3389/fmicb.2015.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lloyd KG, et al. Predominant archaea in marine sediments degrade detrital proteins. Nature. 2013;496(7444):215–218. doi: 10.1038/nature12033. [DOI] [PubMed] [Google Scholar]
- 32.Jørgensen BB, Parkes JR. Role of sulfate reduction and methane production by organic carbon degradation in eutrophic fjord sediments (Limfjorden, Denmark) Limnol Oceanogr. 2010;55(3):1338–1352. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.