Significance
IFIT1 is an antiviral effector of host innate-immunity that selectively recognizes the 5′-end of viral mRNAs, which are often capped to mimic host mRNA, and blocks their translation. Our X-ray structural analysis reveals that the cap and four additional nucleotides are encircled by IFIT1 through a central tunnel in an adaptable manner, which gives it the flexibility required to defend against many different viruses, and to deter their ability to rapidly evolve. Host mRNA, normally ribose methylated at the first and second nucleotides following the cap, avoids IFIT1 recognition through tight complementary interfaces at these positions. This study uncovers the molecular basis for how IFIT1 selectively recognizes viral mRNAs and will help guide development of viral vaccines and mRNA therapeutics.
Keywords: IFIT1 crystal structure, innate immunity, mRNA cap recognition, self vs. nonself, 2′-O methylation
Abstract
IFIT1 (IFN-induced protein with tetratricopeptide repeats-1) is an effector of the host innate immune antiviral response that prevents propagation of virus infection by selectively inhibiting translation of viral mRNA. It relies on its ability to compete with the translation initiation factor eIF4F to specifically recognize foreign capped mRNAs, while remaining inactive against host mRNAs marked by ribose 2′-O methylation at the first cap-proximal nucleotide (N1). We report here several crystal structures of RNA-bound human IFIT1, including a 1.6-Å complex with capped RNA. IFIT1 forms a water-filled, positively charged RNA-binding tunnel with a separate hydrophobic extension that unexpectedly engages the cap in multiple conformations (syn and anti) giving rise to a relatively plastic and nonspecific mode of binding, in stark contrast to eIF4E. Cap-proximal nucleotides encircled by the tunnel provide affinity to compete with eIF4F while allowing IFIT1 to select against N1 methylated mRNA. Gel-shift binding assays confirm that N1 methylation interferes with IFIT1 binding, but in an RNA-dependent manner, whereas translation assays reveal that N1 methylation alone is not sufficient to prevent mRNA recognition at high IFIT1 concentrations. Structural and functional analysis show that 2′-O methylation at N2, another abundant mRNA modification, is also detrimental for RNA binding, thus revealing a potentially synergistic role for it in self- versus nonself-mRNA discernment. Finally, structure-guided mutational analysis confirms the importance of RNA binding for IFIT1 restriction of a human coronavirus mutant lacking viral N1 methylation. Our structural and biochemical analysis sheds new light on the molecular basis for IFIT1 translational inhibition of capped viral RNA.
Infection by a virus relies on its ability to exploit the host’s translational machinery to convert its genome into protein products that can ultimately be used to assemble new viral particles. In eukaryotes, endogenous mRNA is protected by a highly conserved 5′ cap structure consisting of an N7-methylguanosine triphosphate (m7Gppp/Cap0) moiety. This is recognized by the eukaryotic translation initiation factor 4E to promote cap-dependent translation (eIF4E together with eIF4G and eIF4A comprise eIF4F) (1). In higher eukaryotes, the mRNA cap is further modified by ribose 2′-O methylation on the first and sometimes second cap-proximal nucleotides (N1 and N2, where N is any nucleotide) (Fig. S1A), resulting in Cap1- (m7GpppNmN) or Cap2- (m7GpppNmNm) mRNA (2, 3). N1 methylation was recently shown to serve as a molecular signature of “self,” which can subvert mammalian antiviral responses (4, 5). As such, many viruses also produce Cap1-mRNA, either through the action of host- or virally encoded 2′-O methyltransferases (MTases) or through viral “cap-snatching” enzymes (6, 7). Hence, Cap0-mRNAs (along with other virus-derived RNAs) are marked as “nonself” and can trigger responses, such as the type I IFN antiviral program (5, 8, 9), which culminates in the induction of hundreds of IFN-stimulated genes (ISGs) (10).
Fig. S1.
RNA binding and inhibition of in vitro translation. (A) Schematic of mRNA cap structure. Note that italics are used to distinguish m7G atoms from the cap-proximal nucleotides N1 and N2. (B) Predicted secondary structure and thermodynamic stability for each RNA determined by the Mfold web server. MHV is predicted to have a 5-nt overhang whereas HCoV is predicted to have only a 4-nt overhang. Also, MHV is predicted to have a less stable hairpin than HCoV [compare their respective minimum free energy (ΔG), more negative indicates greater stability]. (C) EMSA between IFIT1 and OH- or PPP-RNA. PPP-ss44 is a previously characterized RNA, which binds IFIT1 in its PPP- form, likely because it lacks any 5′- secondary structure. PPP-MHV also binds to IFIT1, but PPP-HCoV and PPP-GGG42 do not. (D) Translation assay with IFIT5. At 5 µM, the inhibition of FF translation is likely because of weak cap binding or nonspecific RNA binding. Data represent the mean of two independent measurements performed in duplicate ± SD.
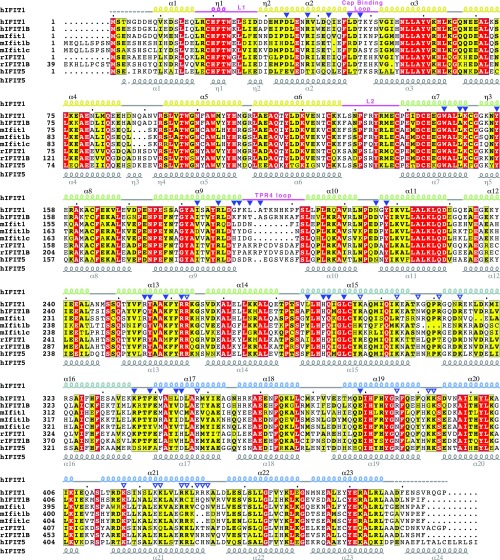
Among the most potently induced of the ISGs are the IFITs (IFN-induced proteins with tetratricopeptide repeats), a family of antiviral effectors whose expression can also be triggered downstream of IFN-independent signaling (11). IFITs are conserved throughout vertebrate evolution, with humans and most mammals encoding five paralogues—IFIT1, IFIT1B, IFIT2, IFIT3, and IFIT5—although many also possess species-specific duplications and deletions. For example, mice lack IFIT5 and were only recently discovered to have also lost IFIT1 (12). Therefore, what is currently known as mouse Ifit1 (54% sequence identity with human IFIT1) is actually an ortholog of human IFIT1B. In humans, IFIT1B (67% sequence identity with human IFIT1) is not known to be IFN-inducible (11), and recent data suggest that it may be nonfunctional (12). IFITs are structurally related and are composed of tandem copies of the tetratricopeptide repeat (TPR), a helix–turn–helix motif. Structures of several IFITs have shown that their TPRs coalesce into distinct superhelical subdomains that form clamp-shaped structures (13–16).
Recently, it was discovered that IFITs play a prominent role in impeding viral replication by directly binding the 5′ end of viral RNA (17). Thus, IFIT1 and IFIT1B can compete with eIF4F to selectively bind and sequester viral Cap0-mRNA, resulting in its translational inhibition (18–20). In this manner, mouse Ifit1 has been shown to restrict a broad spectrum of wild-type and mutant viruses lacking 2′-O MTase activity, including alphaviruses, coronaviruses, flaviviruses, and vaccinia virus (4, 18, 21–25), whereas mutating viral N1 methylation enhanced coronavirus and flavivirus sensitivity to human IFIT1 (18, 23, 24, 26). In contrast, host cellular mRNA is not targeted as it bears N1 2′-O methylation, which interferes with IFIT1 and IFIT1B binding (18–20). That many cytoplasmic virus families have adapted by acquiring 2′-O MTases to generate their own Cap1-mRNA, thereby potentially escaping IFIT1/IFIT1B restriction, underscores the importance of these proteins in this process (7). Furthermore, alphaviruses, which display only Cap0-mRNA, can still subvert mouse Ifit1 activity by encoding cap-proximal structural elements (21, 22), which has also been shown to interfere with RNA binding and enhance pathogenicity.
IFIT1, along with IFIT5, can also recognize uncapped viral triphosphate (PPP)-RNA (another nonself marker of infection) to potentially inhibit the replication of some negative-sense single-stranded (ss) RNA viruses (13, 17). The crystal structure of human IFIT5 bound to uncapped PPP-RNA revealed that the RNA sits in a narrow, positively charged tunnel at the core of the protein, with a network of electrostatic interactions specifically recognizing the PPP moiety (13). Up to four nucleotides are also stably bound within the tunnel in a sequence-nonspecific manner. The sequence identity between IFIT5 and IFIT1 (55%) and a structure of the N-terminal region of IFIT1 suggested that IFIT1 accommodates capped RNA in a similar fashion. However, IFIT5 cannot bind capped mRNA (13, 18, 19), and indeed protein residues at the base of the tunnel would block any further progression beyond the PPP moiety. Thus, how IFIT1-like proteins can accommodate Cap0-mRNA remains unclear.
We report here several crystal structures of RNA-bound human IFIT1. The structures reveal that the positively charged RNA-binding tunnel of IFIT1 is distinct from that found in IFIT5 and further extended to allow binding of both capped and uncapped RNAs. Strikingly, mRNA binding and cap recognition by IFIT1 appears to be adaptable and its mechanism is evolutionarily divergent from eIF4E and other cap-binding proteins. The shape of the tunnel in the vicinity of the 2′-hydroxyls of N1 and N2 sterically occludes RNA methylated at these positions. A comprehensive analysis of the interaction between human IFIT1 and differentially methylated capped RNAs corroborates the structural findings, revealing that either N1 or N2 methylation alone interferes with IFIT1 binding, but in an RNA-dependent manner. Combining N1 and N2 methylation resulted in an additive and potentially synergistic effect in inhibiting IFIT1 activity, particularly toward susceptible RNA sequences and at high IFIT1 concentrations. Our structural and biochemical analysis therefore sheds light on IFIT1 antiviral activity and reveals a previously uncharacterized role for N2 ribose methylation and Cap2 structures as signatures of self mRNA.
Results
RNA Binding and Inhibition of in Vitro Translation by Human IFIT1.
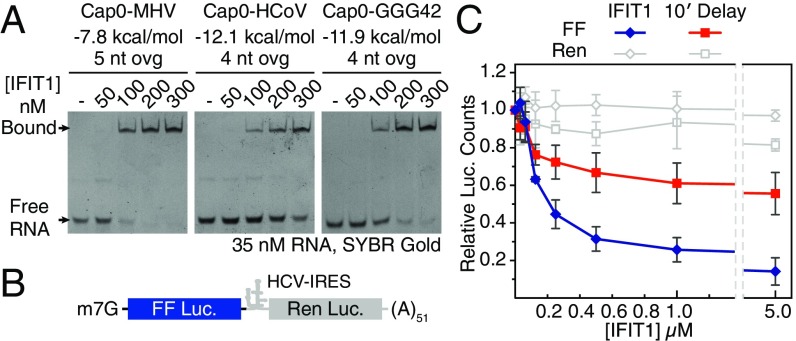
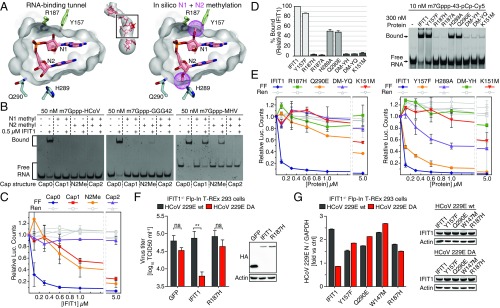
The interaction between IFIT1 and capped-RNA is well established, but the precise structural determinants of the viral RNA important for binding are as yet unclear. Thus, we began by carrying out EMSAs between human IFIT1 and two 5′ capped sequences derived from genomes of coronaviruses known to be restricted by human IFIT1 or mouse Ifit1: human coronavirus strain 229E (HCoV) and murine hepatitis virus strain A59 (MHV) (18). Human IFIT1 bound the capped-RNAs with apparent affinities of ∼250 nM and <100 nM, respectively (Fig. 1A). Binding strength decreased as the stability and proximity of RNA secondary structure to the 5′-end increased (Fig. 1A and Fig. S1B), confirming the preference for ssRNA as previously demonstrated for human IFIT1 and mouse Ifit1 (13, 21). As with IFIT5, RNA binding is generally sequence-nonspecific and replacing the first 3 nt of HCoV (sequence ACU) with GGG resulted in only a modest enhancement of binding (Fig. 1A, Right). IFIT1 also binds uncapped PPP-RNA, but this is inherently weaker and more sensitive to the presence of predicted secondary structure at the 5′-end (Fig. S1C). This finding is in contrast to IFIT5, which binds PPP-RNA but cannot accommodate capped RNA, as shown by its crystal structure and a variety of biochemical assays from several groups (13, 18, 19).
Fig. 1.
RNA binding and inhibition of in vitro translation by human IFIT1. (A) EMSAs between human IFIT1 and capped-RNA visualized by SYBR Gold staining. Cap0-MHV, first 41 nt of MHV strain A59; Cap0-HCoV, first 42 nt of HCoV strain 229E; Cap0-GGG42, ACU to GGG modification of HCoV. The RNA secondary structure minimum free energy (kcal/mol) and 5′-overhang length (ovg) are indicated (see also Fig. S1B). (B) Schematic of bicistronic mRNA reporter. (C) Translation assay with IFIT1 titrated into Krebs extracts programmed with Cap0/m7Gppp reporter, and titration following a 10-min preincubation of the reporter with extracts. FF and Ren luciferase (luc.) activities at each concentration were normalized against buffer control, which was set to 1. Data represent the mean of two independent measurements performed in duplicate ± SD.
To understand the contribution of IFIT1 binding to capped RNA in a more physiological context, we used an in vitro translation system to assess the effect on translation initiation. The system consists of Krebs extracts programmed with a bicistronic Cap0-mRNA reporter (27). The 5′-cistron expresses a Firefly luciferase (FF) reporter that is translated in a cap-dependent manner, whereas the 3′-cistron expresses a Renilla luciferase (Ren) reporter under the control of an internal ribosome entry site (IRES) from hepatitis C virus (HCV) (Fig. 1B). Ren expression serves as an internal control for nonspecific translational inhibition by IFIT1. Titrating human IFIT1 into these extracts at concentrations ranging from ∼30 nM to 5 µM showed that IFIT1 could inhibit Cap0-dependent translation with IC50 values of ∼50–200 nM (Fig. 1C, and other figures herein). Interestingly, addition of IFIT1 after the reporter was preincubated with translation extracts for 10 min resulted in its inhibitory activity being reduced by more than an order-of-magnitude (IC50 > 5 µM), presumably because of the formation of a closed-loop mRNP that facilitates ribosome reinitiation (28). This result suggests that optimal IFIT1 activity in vivo is probably only realized in cells that are already expressing the protein before infection, as might be the case for cells activated by IFN signaling in a paracrine manner.
In all cases, cap-independent translation of Ren was reduced by at most 15–20%, which is likely because of nonspecific binding of IFIT1 to either the IRES, ribosomal RNA (19), transfer RNA (19), or translation factors (e.g., eIF3e) (29). This finding is in contrast to one report showing nearly complete inhibition of HCV–IRES-mediated translation by 600 nM IFIT1 in rabbit reticulocyte lysates (30). This discrepancy may be attributed to differences in translation efficiency between Krebs extracts and rabbit reticulocyte lysates. Finally, titration of IFIT5 in these assays did not produce the same level of translational inhibition (Fig. S1D) (IC50 ≥ 5 µM), consistent with the notion that IFIT5 cannot specifically bind capped-RNA. Taken together, our data are consistent with an IFIT1 antiviral mechanism that is dependent on the recognition of mRNA cap structures to compete with eIF4F (18, 19).
Overall Structure of Full-Length Human IFIT1.
To gain insight into the mechanisms of viral RNA binding by IFIT1, we initially crystallized RNA-bound, full-length, wild-type human IFIT1 (residues 1–478) in complex with short PPP- and m7Gppp-containing oligoadenosines. The IFIT1–RNA complex purified and crystallized as a dimer with two molecules in the asymmetric unit, but diffracted X-rays to only ∼2.7 Å. To improve the resolution, we mutated the dimerization interface at the C-terminal end of the protein to produce a monomeric version that crystallized in a different space group and diffracted X-rays to 1.58-Å resolution (Table S1). The overall folds of the wild-type and monomeric mutant are essentially the same (rmsd 0.35 Å). Henceforth, we describe only the high-resolution structures with respect to RNA binding, whereas all functional assays were performed with wild-type protein.
Table S1.
Data collection and refinement statistics
| Value | ||||
| Property | IFIT1 (L457E/L464E) m7Gppp-AAAA | IFIT1 (L457E/L464E) Gppp-AAAA | IFIT1 (L457E/L464E) PPP-AAAA | IFIT1 (N216A/L457E/L464E) m7Gppp-AAAA |
| Data collection | ||||
| PDB ID code | 5UDI | 5UDJ | 5UDK | 5UDL |
| Space group | P4222 | P4222 | P4222 | P4222 |
| Molecule per ASU | 1 | 1 | 1 | 1 |
| Cell dimensions | ||||
| a, b, c (Å) | 111.7, 111.7, 93.2 | 112.5, 112.5, 93.5 | 112.4, 112.4, 93.2 | 111.8, 111.8, 93.0 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50–1.58 (1.61–1.58) | 50–1.69 (1.72–1.69) | 50–1.65 (1.68–1.65) | 50–1.65 (1.68–1.65) |
| Completeness (%) | 100 (100) | 100 (100) | 99.7 (95.2) | 100 (99.9) |
| Rpim (%) | 2.4 (43.8) | 4.1 (52.6) | 2.4 (36.8) | 2.8 (39.6) |
| <I/σ(I)> | 33.6 (1.74) | 22.2 (1.9) | 30.3 (1.96) | 27.4 (1.78) |
| Redundancy | 14.5 (12.4) | 14.5 (12.2) | 14.2 (9.4) | 14.6 (12.3) |
| Refinement | ||||
| Resolution (Å) | 40.1–1.58 (1.6–1.58) | 40.3–1.69 (1.71–1.69) | 44.3–1.65 (1.67–1.65) | 40.1–1.64 (1.67–1.64) |
| No. reflections | 81,063 (7,992) | 67,665 (6,640) | 71,917 (6,888) | 71,805 (6,876) |
| Rwork/Rfree | 15.7/17.8 (25.0/27.6) | 15.9/18.1 (24.8/30.3) | 16.4/18.4 (24.2/26.8) | 16.4/18.2 (24.3/24.6) |
| No. of non-H atoms | ||||
| Protein | 3,842 | 3,852 | 3,833 | 3,816 |
| Ligand - RNA | 168 | 116 | 97 | 117 |
| Ligand - PEG | 10 | 10 | 41 | 10 |
| Ligand - ion | 2 | 2 | 2 | 2 |
| Solvent | 394 | 382 | 335 | 313 |
| Average B factors (Å2) | ||||
| All atoms | 30.65 | 27.64 | 30.61 | 30.86 |
| Protein | 30.7 | 27.6 | 30.4 | 30.9 |
| Ligand–RNA | 18.8 | 17.0 | 19.3 | 22.2 |
| Ligand–PEG | 49.2 | 48.5 | 57.4 | 62.6 |
| Ligand–ion | 32.1 | 30.8 | 26.6 | 39.5 |
| Solvent | 34.8 | 30.8 | 32.6 | 32.6 |
| Rms deviations | ||||
| Bond lengths (Å) | 0.017 | 0.014 | 0.012 | 0.008 |
| Bond angles (°) | 1.579 | 1.283 | 1.166 | 1.005 |
The highest-resolution shell is in parenthesis.
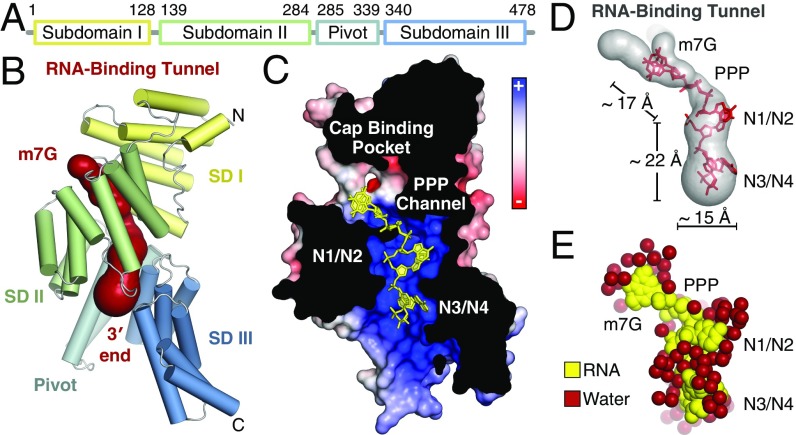
Human IFIT1 is made up of 23 α-helices, 18 of which form 9 TPR motifs that together form three distinct subdomains interrupted by non-TPR structural elements (Fig. 2 A and B and Fig. S2 A–C). The overall structure is similar to the previously determined RNA-bound structure of human IFIT5 (rmsd 1.9 Å) (Fig. S2D) and the N-terminal region of human IFIT1 (rmsd 0.8 Å) (Fig. S2E) (13). The subdomains are arranged to form a clamp-shaped structure with a central RNA-binding tunnel that is ∼30–40 Å in length and 12–19 Å in width, accommodating only ssRNA with a total of five nucleotides (cap + four RNA nucleotides) (Fig. 2 B–D). As with IFIT5, a pair of long non-TPR pivot helices connect the second and third subdomains and likely function in an analogous fashion to regulate closure of the protein around the RNA (13) (Fig. S2C). About 30–40% of the tunnel volume is occupied by bound water molecules (Fig. 2E), which appears to be an important facet for recognition of different RNA sequences and structures (discussed below). We demarcate four distinct regions of the tunnel according to their role in RNA binding: (i) the cap-binding pocket, which houses the N7-methylguanosine moiety; (ii) the triphosphate channel, which links the cap-binding pocket to the 5′-end of the RNA; (iii) the first dinucleotide (N1 and N2), where the presence of 2′-O methylation is sensed; and (iv) the second dinucleotide (N3 and N4), where the requirement for single stranded 5′-ends is reinforced.
Fig. 2.
Overall structure of monomeric, RNA-bound human IFIT1. (A) Schematic of IFIT1 subdomains. (B) Cartoon representation of human IFIT1 colored by subdomain (SD) and surface representation of the tunnel (dark red) determined by CAVER (50). (C) Cross-section of IFIT1 colored by surface electrostatic potential from negative (−10 kTe−1; red) to positive (+10 kTe−1; blue) with capped-RNA (yellow sticks). (D) Dimensions of the IFIT1 tunnel (gray surface) and capped RNA (red sticks). (E) Waters surrounding the RNA inside the tunnel.
Fig. S2.
Overall structure of monomeric, RNA-bound human IFIT1. (A) Secondary structure, TPR motif, and subdomain organization of human IFIT1, with TPR motif sequence numbers above. Note that because of an insertion between α9 and α10, TPR4 was annotated based on sequence and structure rather than sequence alone. All other TPR motifs were predicted based on sequence using TPRpred (https://toolkit.tuebingen.mpg.de/tprpred). (B) Overall structure of monomeric human IFIT1 with subdomains colored according to A. The non-TPR helices α1 and α2 are bridged by loop L1, which houses a highly conserved CHFTW motif (residues 19–23) (see Fig. S5) that mediates contacts with α7. Subdomains I and II are linked by loop L2. The two non-TPR pivot helices (α15/α16) connect subdomains II and III. (C) TPR4 (α9/α10) has an unusually long intra-TPR loop (purple) which forms a lid over the 3′ exit of the RNA-binding tunnel. Additionally, it mediates contacts between subdomains II and III (indicated by purple/blue dashes). The sequence of this loop is not conserved, but the insertion is found in almost all IFIT1- and IFIT5-like proteins. The double-headed arrow indicates putative subdomain III motions between RNA-free and RNA-bound forms of IFIT1, based on the structural analysis of human IFIT5. (D) Superposition of RNA-bound IFIT1 over RNA-bound IFIT5 (PDB ID code 4HOR). The two structures share a high degree of global similarity. The C-terminal regions appear to diverge, but that is because of intersubdomain angle differences between IFIT1 and IFIT5. (E) Superposition of RNA bound IFIT1 over the structure of RNA free, N-terminal IFIT1 (PDB ID code 4HOU). In the partial structure of RNA-free, N-terminal IFIT1, the cap-binding loop (blue) adopts a different conformation than in full-length (yellow; see also F). However, in one of the two molecules of the asymmetric unit (of 4HOU), this loop has high B-factors (loop average 61 Å2, protein average 46 Å2), whereas in the other it is disordered, indicating that this loop is mobile in the absence of RNA. The TPR4 loop (green) is also disordered in the structure of N-terminal IFIT1 (4HOU), likely because of the lack of RNA and missing subdomain contacts. (F) The cap-binding pocket is formed at the interface of subdomains I and II, with one wall formed by helix α2 and the cap-binding loop (connecting α2 and α3), and the other by subdomain II.
The IFIT1 RNA-Binding Tunnel Houses a Functionally Distinct Cap-Binding Pocket.
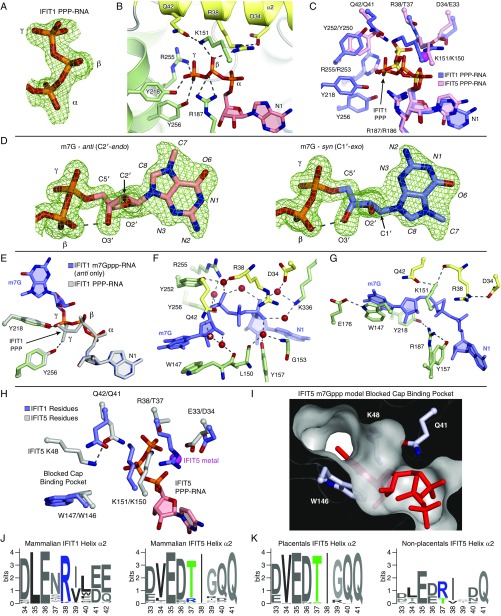
IFIT1 and IFIT5 were previously characterized as PPP-RNA binding proteins (17), although more recent evidence revealed that the primary role of IFIT1 is in binding capped-RNA. Conversely, the role of IFIT5 remains restricted to recognition of 5′-phosphorylated RNAs (13, 14, 16). The structure of IFIT1 bound to PPP-RNA revealed that, like IFIT5, the PPP moiety is ligated by numerous specific electrostatic interactions from protein side chains (Fig. S3 A–C). However, there are some key differences. IFIT5 recognition of PPP-RNA uses a positively charged metal ion bound between the α- and γ-phosphates, which stabilizes a bent conformation of the PPP facilitated by T37 at the base of the tunnel in IFIT5 (Fig. 3A). The corresponding position in IFIT1 is occupied by an arginine (R38), and an ion is no longer part of its PPP binding. This results in a more extended conformation of the PPP that allows it to reach toward the entrance of a neighboring unoccupied pocket.
Fig. S3.
IFIT1 PPP- and m7Gppp- binding mechanism. (A) Simulated annealing 2Fo–Fc omit map of the PPP moiety contoured at 1σ. (B) Cartoon/stick representation of residues making specific contacts with the triphosphate group from PPP-RNA–bound IFIT1. (C) Superposition of PPP-RNA bound IFIT1 residues (blue carbons with orange phosphates) over PPP-RNA–bound IFIT5 residues (pink carbons with yellow-orange phosphates). The metal from PPP-RNA–bound IFIT5 is represented with a magenta sphere. The PPP from IFIT1 is in a different conformation, and makes additional H-bonds with Y256, Y218, and R187 not present in IFIT5. R38/D34 from IFIT1 are replaced with T37/E33 in IFIT5. Depending on the region, IFIT5 residue positions are offset by −1 or −2 compared with IFIT1 residue positions. (D) Simulated annealing 2Fo–Fc omit map of the m7Gppp moiety contoured at 1σ. Syn- and anti-configuration carbons are colored light blue and salmon, respectively. The hydrogen bond between the β-phosphate and the 3′-OH is indicated with the dashed line. Because of the constraints imposed on the base and bridging PPP binding, the ribose sugar pucker adapts by switching from a C2′-endo conformation in the anti-configuration, to C1′-exo in the syn-configuration. The final refined occupancies are ∼0.5 for each conformation. (E) Superposition of PPP-RNA bound IFIT1 (gray) over m7Gppp-RNA bound IFIT1 (green side-chain carbons and blue RNA carbons). The extended PPP conformation is common to both PPP- and m7Gppp-RNA binding, except that the γ-phosphate of m7Gppp- is repositioned at the entrance of the cap-binding pocket, away from Y256 and toward Y218, for more optimal m7G binding inside the pocket. (F) Water-mediated hydrogen bonds between IFIT1 and the m7Gppp moiety. Starting with the water at Q42 (*) and going clockwise, the isotropic B-factors of these waters are 27.6, 18.2, 17.6, 18.0, 19.4, 24.1, 15.3, 17.8 Å2, most of which are lower than the crystal isotropic B factor average (30.65 Å2). (G) Coordination between residues involved in m7Gppp-RNA binding. R187 is coordinated by Y218 and Y157; R38 by D34; K151 by R38 and Q42; and W147 by E176. (H) Superposition of m7Gppp-RNA–bound IFIT1 over IFIT5 PPP-RNA. In IFIT5, T37 facilitates recognition of a bent/compact PPP conformation that is further stabilized by a metal bound to the α- and γ-phosphates and coordinated by E33. This draws in K150 (which is H-bonded to T37), Q41 (which interacts with the PPP directly), and K48 (which is coordinated by Q41). (I) In IFIT5, K48 sits atop the cap-binding residue to block access to the cap-binding pocket as indicated by the attempt to model m7Gppp. In IFIT1, K49 from its cap-binding loop is facing the solvent. Also, in IFIT1, Q42 is not involved in binding the PPP directly. (J) WebLogo (weblogo.berkeley.edu/) sequence consensus of helix α2 from 86 IFIT1- and IFIT1B-like sequences, and 60 IFIT5-like sequences. (K) The mammalian IFIT5 sequences were split into placentals and nonplacentals (i.e., Tazmanian devil, opossum, platypus). Only nonplacental IFIT5-like sequences have an arginine at position 37 (human IFIT5 numbering).
Fig. 3.
IFIT1 mRNA cap-binding mechanism. (A) The IFIT1 PPP (blue) adopts an extended conformation compared with the “bent” IFIT5 PPP (pink). The γ-phosphate from PPP–RNA-bound IFIT1 points toward the nearby unoccupied cap-binding pocket. (B) Protein cross-section and close-up the cap-binding pocket. This view is rotated by ∼180° compared with Fig. 2C. (C) Simulated annealing 2Fo–Fc omit map of the m7Gppp-moiety contoured at 1σ. Syn- and anti-configuration carbons are colored light blue and salmon, respectively. (D) Surface/stick representation of residues (colored by subdomain) abutting the m7G base moiety from above and below. The interplanar distance between m7G and Trp-147 is 3.4–3.7 Å. (E and F) Cartoon/stick representation of residues interacting with the m7Gppp-moiety. See also Movie S1. Shown are both conformations of the m7GpppA dinucleotide, which was modeled as a single residue during model building and refinement. (G) Cross-sections of the IFIT1 and IFIT5 RNA-binding tunnels (gray/black) with m7Gppp- or PPP-RNA (red sticks). The asterisk (*) shows where K48 and Q41 block the IFIT5 putative cap-binding pocket (Fig. S3 H–K).
The crystal structure of IFIT1 bound to m7Gppp-RNA revealed that this adjacent pocket harbors the cap moiety. Whereas most of the RNA-binding tunnel is positively charged, the cap-binding pocket is generally more hydrophobic and interactions with the cap occur predominantly through nonspecific van der Waals contacts (Figs. 2C and 3B). Surprisingly, we found that the m7G base adopts both syn- and anti-conformations with approximately equal occupancies (Fig. 3C and Fig. S3D) (discussed in detail below). In either conformation, m7G sits atop a tryptophan residue (W147) making π–π stacking interactions, reminiscent of other cap-binding proteins, such as eIF4E (31) (Fig. 3 D and E). Additionally, the base is abutted by I183 from the same side as W147, and on the other side by L46 and T48 emanating from a flexible loop that forms the outer wall of the pocket, which we term the “cap-binding loop” (Fig. 3 D and E and Fig. S2F). The ribose of m7G is similar in the syn- and anti-modes of binding, adopting an S-type conformation that is stabilized by an intramolecular hydrogen bond between the ribose 3′-OH and the bridging β-phosphate (Fig. S3D). It sits in a pocket formed by Q42, L46, R187, Y218, I183, and L150 (Fig. 3E), and two ordered water molecules: the first coordinated by Q42 and the second bridging the ribose 3′-OH to the backbone carbonyl of W147 (Fig. S3F).
As in PPP-RNA–bound IFIT1, the bridging triphosphate in the cap-bound structure is in an extended conformation stabilized by numerous electrostatic interactions, although pulled slightly toward the cap-binding pocket (Fig. 3F and Fig. S3E). The γ-phosphate interacts with K151, R255, Y218, and R187, and the β-phosphate is coordinated by K151, R187, and R38. Additionally, six highly ordered water molecules mediate hydrogen bonds with the α- and γ-phosphates (Fig. S3F). Finally, m7Gppp binding is facilitated by a high degree of interresidue coordination: for example, R38 is held in place by D34, and W147 is coordinated by E176 (Fig. S3G).
In IFIT5, although most of these cap-binding residues are conserved, substitutions at a few key positions render it unable to bind cap productively. As described above, replacement of R38 in IFIT1 with T37 in IFIT5 causes it to recognize a more compact conformation of the PPP in a metal-dependent manner. This positions the γ-phosphate away from the putative cap-binding pocket, which draws in several residues, such as Q41 (from helix α2) and K48 (from the putative cap-binding loop), causing them to block access to its putative cap-binding pocket (Fig. 3G and Fig. S3 H and I). Therefore, the formation of a positively charged RNA-binding tunnel with an accessible and spatially separated cap-binding pocket in IFIT1 explains, at least in part, why it can bind capped-RNA, whereas IFIT5 cannot. The preference for an arginine or threonine on helix α2 is highly conserved among IFIT1/1B-like and IFIT5-like sequences, respectively (Fig. S3J), and the identity of this PPP bridging residue (Arg or Thr) appears to play a major role in determining the 5′ specificities of IFIT1/1B- or IFIT5-like proteins. Interestingly, a small group of mammalian IFIT5-like proteins retain an arginine at this position; these sequences probably have a hybrid IFIT1/IFIT5 character, and possibly resemble an ancestral IFIT1/IFIT5 precursor protein, because they all belong to nonplacental mammals (e.g., opossum and platypus) (Fig. S3K).
IFIT1 Can Nonspecifically Accommodate Multiple Forms and Conformations of the Cap.
The high-resolution structure of the monomeric IFIT1 mutant (1.58 Å) allowed us to unambiguously build two conformations for the m7G base that are consistent with the electron density, which has a relatively symmetric shape because of an ∼180° rotation about the N-glycosidic bond connecting the base to the ribose. Multiple base conformations were also evident in the lower-resolution wild-type m7Gppp-RNA cocrystals. This results in the interactions at the base periphery being quite distinct in the two conformations (Fig. 4A). Notably, there are no direct hydrogen bonds from the protein toward the base in either conformation, but there are a small number of water-mediated interactions. In the anti-orientation, N3 of the base is weakly hydrogen bonded to a water molecule that is coordinated by Q42. The N7-methyl (C7) and O6 groups make van der Waals contacts with N216, and the remainder of the base is partially oriented toward water molecules near the proximal opening of the tunnel leading to bulk solvent. In the syn-orientation, the N7-methyl and O6 are instead pointing toward the bulk solvent, whereas N2 is nestled between Y218 and N216.
Fig. 4.
IFIT1 can accommodate multiple forms and conformations of the cap. (A) m7G base interactions at its periphery in the anti- or syn-modes of binding. The water H-bonded to Q42 (*) is 3.3 Å away from N3 in the anti-mode. (B, Left) The Gppp-moiety adopts only anti, and approaches N216 to form a weak H-bond through O6; m7Gppp-RNA bound IFIT1 is superposed in gray. (Right) The water structure surrounding the G moiety changes compared with m7G, and satisfies almost all H-bond donor and acceptor groups. The arrows depict the movement of waters from the m7Gppp-bound form (gray circles) to the Gppp-bound form (red spheres). The water molecule H-bonded to Q42 and N3 becomes more ordered in the Gppp-RNA structure (same resolution and crystal form but B-factor decreases from 28 Å2 to 15.3 Å2). The water at N7 (§) appears only in the Gppp-bound form. The water at N1 appears to be in two conformations and was modeled as two water molecules.
Because of the presence of the N7-methyl group on m7G, it acquires a delocalized positive charge on its imidazole ring that could in principle enhance the stacking with W147 through additional cation–π interactions (31, 32). However, the geometry of cation–π stacking changes with the base orientation (Fig. 4A). Whereas the anti-conformation places the positive charge at an angle away from W147, the syn-conformation places it directly over the indole ring of W147. To test whether N7-methylation and associated positive charge controls cap orientation, we determined the structure of Gppp-RNA (lacking the N7-methyl group) bound to IFIT1 (1.7 Å) (Fig. 4B and Fig. S4A). Here, the base exists only in the anti-conformation, indicating that the ability of the base to adopt two conformations depends, at least in part, on N7-methylation. The presence of two base conformations is also determined by the chemical environment surrounding the base, because the structure of an N216 mutant of monomeric IFIT1 (N216A) bound to m7Gppp-RNA also resulted in the base adopting only the anti-conformation (Fig. S4B).
Fig. S4.
IFIT1 can accommodate multiple forms and conformations of the cap and mutational analysis of cap recognition. (A) Simulated annealing 2Fo–Fc omit map of the Gppp moiety contoured at 1σ. (B) Simulated annealing 2Fo–Fc omit map of m7Gppp moiety bound to monomeric IFIT1 N216A, contoured at 1σ. (C) EMSA between IFIT1 and m7Gppp- or Gppp-RNA. For HCoV, the lack of N7-methyl appears to enhance RNA binding. Whether this is a common feature of all IFIT1-like proteins remains to be validated. (D) EMSA between Gppp-GGG42 and IFIT1, IFIT1 N216D, or IFIT1 N216A. (E) In vitro translation assay with extracts programed with Cap0-, Gppp-, or Appp-capped reporter. Data represent the mean of two independent measurements performed in duplicate ± SD. (F) EMSA between IFIT1 and Appp- or PPP-GGG42 (Left), and between IFIT1 and Gppp- or Appp-GGG42 (Right). (G) Simulated annealing 2Fo–Fc omit map of the PEG molecules inside the cap-binding pocket in PPP-RNA bound IFIT1, contoured at 1σ. The m7G from m7Gppp-RNA bound IFIT1 is superposed in light blue. The PEG molecules interact nonspecifically with residues lining the cap-binding pocket. (H) At physiological pH, the m7G moiety exists in an equilibrium between a cationic Keto tautomer, and a zwitterionic enolate tautomer in which N1 (red atom) is deprotonated. (I) Schematic of residues lining the cap-binding pocket and PPP channel. Residues are colored by subdomain, and red lines indicate hydrogen bonds or salt bridges. (J) EMSA between IFIT1 mutants and PPP-ss44 or OH-ss44. Except for R38A and K151M (which target the bridging triphosphate), all mutants retain binding to PPP-ss44, confirming that the protein fold is not affected by the mutations. Dashed lines indicate lanes that were cropped out. (K) Y50 and F45 from the cap-binding loop are distal from the m7G moiety. F45 from subdomain I packs against E280 and T251 from subdomain II, whereas Y50 stacks against R93, which in turn is salt-bridged to E176. These interactions may be important for subdomain contacts, and/or preorganizing the cap-binding loop. (L) EMSA between pCp-Cy5 labeled m7Gppp-43 and IFIT1, IFIT1 N216D, or IFIT1 Q42E, similar to Fig. 5A. (M) Composite WebLogo sequence consensus from 86 IFIT1- and IFIT1B-like sequences showing residues involved in m7Gppp-RNA binding. See also Fig. S5.
Despite these observations, IFIT1 appears not to be selective for N7-methylation. In fact, gel-shift assays suggest that Gppp-RNA binding is, in some cases, more efficient than m7Gppp-RNA binding (Fig. S4C) (18, 19), in stark contrast to the case with eIF4E, whose cap binding is strongly dependent on proper methylation (31). In the absence of the methyl group, the guanine ring moves closer to N216, making a hydrogen bond with it through O6 (Fig. 4B, Left), although removing this hydrogen bond through mutation (N216A or N216D) does not weaken Gppp-RNA binding (Fig. S4D). Interestingly, the water structure surrounding Gppp-RNA changes compared with m7Gppp-RNA, such that almost all hydrogen bond donor and acceptor sites on the unmethylated base are now satisfied (Fig. 4B, Right), and this may be a contributing factor for maintaining strong Gppp-RNA binding (relative to m7Gppp-RNA).
The physiological relevance of Gppp-RNA binding by IFIT1 is unclear, but one possibility is that it may facilitate targeting of transient intermediates formed during viral mRNA capping. We therefore tested IFIT1 activity in extracts programmed with a Gppp-capped reporter (Fig. S4E). In this system, although overall translation is less efficient than in Cap0-programmed extracts, translation initiation still proceeds through binding of the cap-proximal nucleotides via eIF4G (33). IFIT1 titration resulted in translational inhibition similar to m7Gppp-capped mRNA (IC50 ∼ 200 nM), highlighting the importance of binding not only the cap, but also the proximal nucleotides to provide additional affinity to allow competition with eIF4F (19).
Because cap binding does not rely on any guanine-specific hydrogen bonds, we wondered whether IFIT1 could also recognize Appp-capped RNAs. Indeed, IFIT1 can bind Appp-RNA (Fig. S4F) and inhibit translation initiation from an Appp-capped reporter (IC50 ∼ 500 nM) (Fig. S4E). Thus, it appears that IFIT1 has evolved to recognize not only canonically capped mRNA, but rather diverse 5′-5′ linked base modifications of the mRNA through relatively nonspecific interactions in the pocket. The notion of lack of specificity is underscored by the structure of IFIT1 with PPP-RNA, where the cap-binding pocket is occupied by PEG (polyethylene glycol) molecules from the crystallization solution, which form interactions that mimic cap binding (Fig. S4G).
Mutational Analysis of Cap Recognition.
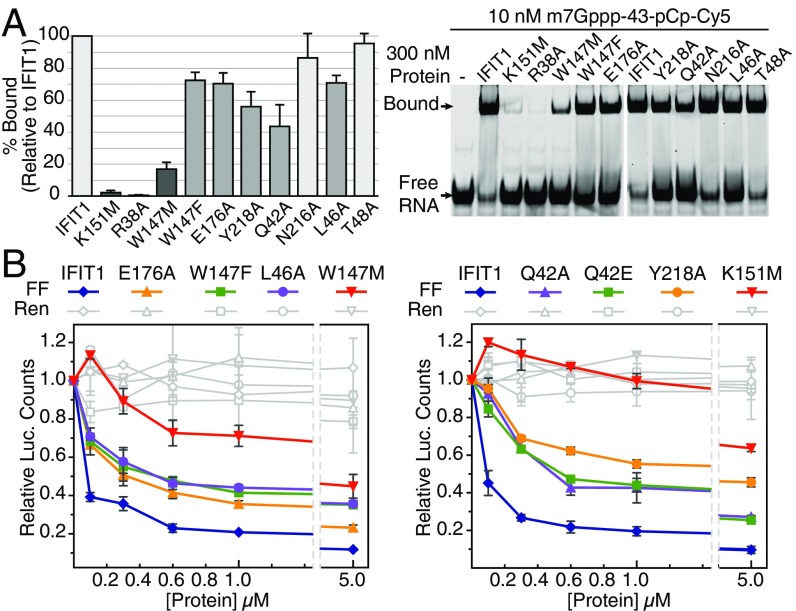
To test whether our structural findings are functionally valid, we mutated several residues involved in cap binding and assayed them in fluorescent gel-shift binding assays (with m7Gppp-43 RNA) (Table S2) and the translational inhibition assays described above (Fig. 5 and Fig. S4I). Both R38A and K151M are critical for binding, and K151M reduces IFIT1 inhibitory activity by one to two orders-of-magnitude (IC50 > 5 µM). Y218A and Q42A weakened binding to capped RNA and reduced IFIT1 inhibitory activity. N216A retains full binding to m7Gppp-RNA, indicating that the N216–cap interactions (e.g., with the N7-methyl) are dispensable. W147 is perhaps the most important residue inside the cap-binding pocket as W147M essentially abolished m7Gppp-RNA binding, whereas mutation to another aromatic residue (W147F) largely retained binding. Accordingly, W147M reduced IFIT1 translation inhibition ∼40-fold, and W147F mostly retained inhibitory activity (compared with W147M). Mutation of E176, which coordinates W147, had similar effects as W147F. From the cap-binding loop, T48 was deemed dispensable, but L46 was required for optimal binding and translational inhibition. All cap-binding pocket mutants tested here retained binding toward PPP-RNA, except for R38A and K151M (which target the PPP moiety), indicating that the protein fold was not disrupted by the mutations (Fig. S4J). Taken together, the data in the mutational analysis confirm the importance of cap binding and the role of the RNA-binding tunnel in mediating translational inhibition by IFIT1.
Table S2.
Sequences of in vitro transcribed RNA and DNA oligos used for in vitro transcription
| Name | Sequence |
| Transcribed RNA | |
| MHV | GUAUAAGAGUGAUUGGCGUCCGUACGUACCCUCUCAACUCU |
| HCoV | ACUUAAGUACCUUAUCUAUCUACAGAUAGAAAAGUUGCUUUU |
| GGG42 | GGGUAAGUACCUUAUCUAUCUACAGAUAGAAAAGUUGCUUUU |
| ss44 | GGGAGAGAGAGAGAGAGAGUAAGGGCGUCGUCGCCCCGAGAAUU |
| 43* | GGGUAAGUACCUUAUCUAUCUACAGAUAGAAAAGUUGCUAAAA |
| Reporter 5′-UTR† | GAAUACACGGAAUUCAGCAAUUCGAGCUCGCCCGGGGAUCUGCGAUCUAAGUAAGCUUGGCAUU CCGGUACUGUUGGUAAAGCCACCAUG |
| DNA Oligo templates (5′ → 3′) | |
| T7‡ | TAATACGACTCACTATAG |
| T7Φ2.5§ | TAATACGACTCACTATTA |
| MHV | AGAGTTGAGAGGGTACGTACGGACGCCAATCACTCTTATACTATAGTGAGTCGTATTA |
| HCoV | AAAAGCAACTTTTCTATCTGTAGATAGATAAGGTACTTAAGTAATAGTGAGTCGTATTA |
| GGG42 | AAAAGCAACTTTTCTATCTGTAGATAGATAAGGTACTTACCCTATAGTGAGTCGTATTA |
| ss44 | AATTCTCGGGGCGACGACGCCCTTACTCTCTCTCTCTCTCTCCCTATAGTGAGTCGTATTA |
| 43 | TTTTAGCAACTTTTCTATCTGTAGATAGATAAGGTACTTACCCTATAGTGAGTCGTATTA |
This RNA is similar to GGG42, except that it is designed to have a 5-nt overhang to allow strong IFIT1 binding, and a free 3′-end with sequence AAA to improve pCp-Cy5 labeling efficiency with T4 RNA ligase.
Start codon underlined, this transcript was prepared with a plasmid and not a DNA oligo.
Promoter top strand for all sequences except HCoV and mRNA reporter.
Promoter top strand for HCoV.
Fig. 5.
Functional validation of cap recognition. (A) Mutational analysis of cap-binding residues investigated by fluorescent EMSA with 3′-end–labeled (pCp-Cy5) RNA. (Left) Quantification of percent bound (Upper band, Right) for each mutant normalized against IFIT1. Data represent the mean of three independent measurements ± SD. (B) In vitro translation assays with RNA binding mutants and Cap0 reporter. Data represent the mean of two measurements ± SD.
Our results are in agreement with previous mutational binding assays based on in silico modeling (19). In this model, Phe-45 and Tyr-50 from the cap-binding loop were also predicted to interact with the base; however, our structures reveal that these two residues are distal from the m7G moiety and are probably important for maintaining subdomain contacts, or helping preorganize the cap-binding loop (Fig. S4K).
The Cap-Binding Mechanism Is Conserved in IFIT1 and IFIT1B Proteins Across Mammalian Evolution.
The mode of cap binding identified here likely applies to all mammalian IFIT1- and IFIT1B-like proteins, as the residues involved in N7-methylguanosine triphosphate recognition are highly conserved (Figs. S4M and S5). Two notably prevalent differences in cap-binding residues compared with human IFIT1 include Q42 and N216, which are replaced with a glutamate and aspartate, respectively, in many of the orthologs and paralogs (including human IFIT1B). Both substitutions are conservative, because neither would disrupt hydrogen-bonding patterns nor interfere with the van der Waals interactions with the cap. We tested this by carrying out an EMSA between m7Gppp-RNA and IFIT1 N216D or IFIT1 Q42E (Fig. S4L). Whereas N216D had no impact on RNA binding, Q42E weakened the interaction and in translation assays, Q42E reduced IFIT1 activity similarly to Q42A (Fig. 5B, Right). In other IFIT1-like proteins, such as rabbit IFIT1 and rabbit IFIT1B [both of which bind m7Gppp-RNA with ∼20 and 10 nM affinity, respectively (19)], the natural Q42E variation is likely overcome by compensatory interactions.
Fig. S5.
ClustalO sequence alignment of select IFIT sequences from humans, mice, and rabbits. Filled triangles indicate residues directly in contact with the RNA. Open triangles are positively charged residues lining the protein surface outside of the tunnel (in the groove). Secondary structure elements are indicated for human IFIT1 (colored by subdomain) and human IFIT5 (PDB ID code 4HOR). This figure was prepared using ESPript (espript.ibcp.fr/ESPript/ESPript/). National Center for Biotechnology Information reference numbers for mIfit1, mIfit1b, and mIfitc are NP_032357.2, NP_444447.1, and NP_001103987.1, respectively.
Unlike IFIT1B from other species, human IFIT1B lacks an apparent function in RNA binding (12). Sequence comparison shows that, along with Q42E, human IFIT1B has acquired additional substitutions that could impact RNA recognition: L150 is replaced with an Ala, which would affect cap ribose interactions, and R255 with Gln, which would disrupt a salt-bridge with the γ-phosphate (Fig. 3 E and F). Supporting this theory, mutation of R255 in human IFIT1 (R255M) was shown to disrupt capped- and PPP-RNA binding (13, 18). On the other hand, mice lack a bona fide IFIT1 ortholog and instead encode three copies of IFIT1B-like proteins (12), currently annotated as mouse Ifit1, mouse Ifit1b, and mouse Ifit1c (Fig. S5). Mouse Ifit1b and Ifit1c harbor several substitutions that would disrupt RNA binding, such as R255G and Q42T in both, and R187H in Ifit1b (Fig. S5). Consistent with this finding, pull-downs showed that mouse Ifit1c cannot bind capped RNA directly (18).
IFIT1 Cap Binding Is Distinct from Canonical Cap-Binding Proteins.
The IFIT1 cap-binding mechanism described here is quite distinct from canonical cap-binding proteins such as eIF4E (34), cap-binding complex (CBC) (35), and VP39 (vaccinia virus N1 2′-O MTase) (36). Through convergent evolution these proteins evolved a highly specific cap-binding slot between two aromatic side-chains that engage the methylated guanine in a cation–π sandwich (31). Charge–charge interactions with the delocalized positive charge and van der Waals contacts with the N7-methyl also play a role (31). In these proteins, the absence of N7-methylation and associated positive charge on the base results in >100-fold loss in binding affinity (37). These proteins also rely on hydrogen bonds targeting groups at the m7G base periphery. The cumulative effect of these restrictions results in highly specific recognition of the cap in a single anti-conformation of m7G (Fig. 6).
Fig. 6.
Comparison with the canonical cap-binding proteins eIF4E (PDB ID code 1EJ1), CBC (PDB ID code 1H2T), and VP39 (PDB ID code 1V39). In IFIT1, some of the nearby water molecules are shown as transparent red spheres, and T48 adopts two conformations.
In contrast, IFIT1 engagement of the cap is relatively less specific, permitting both syn- and anti-base orientations, as described above. Although IFIT1 does use one aromatic residue for cap stacking, the remainder of its sandwich is formed by aliphatic side chains (Leu-46, Thr-48, and Ile-183) rather than another aromatic residue. Therefore, the lack of an electron-rich, aromatic cap-binding slot reduces the dependence on an electron-deficient, N7-methylated base. Additionally, protein contacts with the N7-methyl are dispensable (e.g., the N216A mutant), and they are altogether absent when the cap is in the syn-configuration. Importantly, IFIT1 lacks any sequence specific hydrogen bonding from protein residues, and instead uses a more plastic, water-mediated hydrogen-bonding network for base recognition.
Finally, at physiological pH m7G exists in an equilibrium between two forms: a positively charged “keto tautomer” and a zwitterionic “enolate tautomer” (in which N1 is deprotonated) (Fig. S4H) (34). The canonical cap-binding proteins are highly selective for guanine as the base and in particular, its keto form. These aspects are enforced by two elements: (i) the cation–π sandwich, which is only compatible with an electron-deficient, positively charged keto tautomer (38); and (ii) Asp or Glu residues at one end of the cap-binding slot, which hydrogen bond with a protonated N1 and the N2-amino group. Conversely, IFIT1 does not form any keto- or enolate-specific interactions with the base, suggesting that IFIT1 is not selective for the tautomerization state, reinforcing the lack of guanine specificity.
Binding of Cap-Proximal Nucleotides.
Recognition of the four RNA nucleotides following the cap is conformation-specific and can be divided into two distinct dinucleotide groups diverging between N2 and N3 (Fig. 2 C–E). The RNA backbone lies along the superhelical axis of the protein, and is recognized by specific hydrogen bonds and salt bridges from protein residues targeting the 5′-phosphates and 2′-hydroxyls of N2 and N3 (Fig. S6 A and B). In contrast, recognition of the bases is predominantly through sequence-nonspecific van der Waals and stacking interactions. The first dinucleotide (N1 and N2) adopts geometry similar to CpG dinucleotides found within Z-form RNA and UUCG tetraloops (39), and is tightly sandwiched between multiple protein residues (Fig. S6 A and C). The second dinucleotide (N3 and N4) adopts A-form helical geometry with the bases also stacked upon each other and abutted by protein residues from above and below (Fig. S6 B and D). The large water network inside the tunnel interacts with all groups of the RNA, and mediates both intermolecular protein–RNA and intramolecular RNA–RNA interactions (Fig. S6E). Interestingly, a large part of this extensive water network is involved in protein–base contacts, allowing IFIT1 to recognize a wide variety of RNA sequences that may exist at the 5′-end of viral RNA. A small degree of sequence-dependent binding affinity variation may exist because there are two adenine-specific hydrogen bonds at N2 and N4 (Fig. S6 A and B), although RNA binding assays show that adenosines are not strictly required at these positions.
Fig. S6.
RNA binding mechanism at N1-N4. (A) Interactions between IFIT1 and RNA at N1/N2 and (B) at N3/N4. The protein is oriented such that we are looking along the RNA-binding tunnel from the 3′-exit (similar orientation as Fig. S2C). Note that K336 and the backbone carbonyl of G190 are H-bonded to N2 and N4, respectively. Concerning Fig. 8 and the multiple roles of R187, Y157, H289, and Q290: R187 packs against the ribose of N1, coordinates Y218 and Y157 (Fig. S3G), and interacts with m7G and bridging PPP (Fig. 3 E and F); Y157 contacts the N1 ribose and N1 adenine through van der Waals; Q290 is H-bonded to the 3′ oxygen of N2 and the internucleotide phosphate between N2 and N3; H289 is H-bonded to the 2′-OH of N2, and contacts the ribose of N3 via van der Waals. V372 could potentially clash with methylation at the N6 position of the first adenosine (see Discussion). (C) Simulated annealing 2Fo–Fc omit map of N1/N2 contoured at 1σ. Two views rotated by 180° are shown. The dinucleotide conformation here resembles that found in CpG dinucleotides of Z-form RNA and UUCG tetraloop sequences (39). The defining feature of this rare dinucleotide motif is the antiparallel arrangement of the two riboses (with their respective O4′ atoms pointing toward each other), and a lone pair–π stack between the O4′ atom of N1 and the base of N2. (D) Simulated annealing 2Fo–Fc omit map of N3/N4 contoured at 1σ. N3/N4 adopt standard A-form helical geometry. (E) Hydrogen-bonds between the RNA bases and the waters (red spheres) inside the tunnel. N6 methylation of the first adenosine could disrupt water mediated interactions at the first nucleotide (see Discussion). (F) N1 and N2 adopt C2′-endo and C3′-endo conformations respectively. RNA nucleotides are typically in equilibrium between the two, but generally favor the C3′-endo conformation.
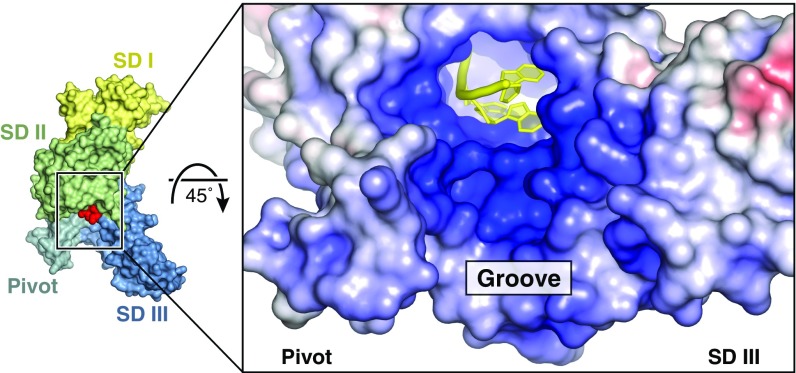
The 3′-end of the RNA (N4) emerges from the C-terminal opening of the tunnel and points toward a positively charged, solvent-exposed groove formed by the pivot helices and the third subdomain (Fig. 7). This surface is contiguous with the RNA-binding tunnel and also appears to contribute to RNA interactions because the analogous region in IFIT5 can apparently bind tRNA (14). In IFIT1, the groove does play some role in RNA binding, as primer-extension toe-printing assays suggested that IFIT1 has a 6- to 8-nt footprint at the 5′-end of mRNA, and mutational analysis of this region had an impact on mRNA binding (19). However, cocrystal structures of IFIT1 with longer oligonucleotides (6–8 nt in length) revealed extra electron density for only the 5′-phosphate of a fifth nucleotide, as was shown for IFIT5 (13), suggesting that only the first four nucleotides are stably bound by IFIT1, whereas residues in the positively charged groove probably contribute to nonspecific RNA binding.
Fig. 7.
IFIT1 forms a positively charged, solvent-exposed RNA binding groove. Fig. S5 for residues in this region.
IFIT1 Senses Ribose 2′-O Methylation at N1 and N2.
The mRNA of higher eukaryotes is normally modified by ribose 2′-O methylation at N1 and N2 (40). Whereas all cellular mRNAs are methylated at N1 in the nucleus by the endogenous Cap1-methyltransferase (CMTr1) (2), ribose methylation at N2 arises from secondary methylation in the cytoplasm through the action of CMTr2 (3, 41), and accompanies N1 methylation on up to 50% of cellular mRNAs (42, 43). N1 ribose methylation is a molecular determinant of self that can protect mRNA from IFIT1/IFIT1B recognition (18, 19), but the role of N2 methylation in this process is unknown. To gain additional insight into self- vs. nonself-mRNA discernment by IFIT1, and to explore the uncharacterized role of ribose N2 methylation in this process, we examined the interaction between human IFIT1 and differentially methylated RNA. Note that, to distinguish the naturally occurring Cap1 and Cap2 structures (m7GpppNmN- and m7GpppNmNm-) from capped RNAs that contain ribose N2 methylation only (m7GpppNNm-), we refer to the latter as Cap0N2Me-RNA.
When bound to IFIT1, N1 and N2 adopt a rare Z-RNA–like conformation that is dependent on their respective ribose conformations (Fig. S6F) (39). Whereas N2 is in the favorable C3′-endo conformation, N1 adopts a C2′-endo conformation and places its 2′-OH in close proximity to the side chains of two highly conserved residues, R187 and Y157 (Fig. 8A and Fig. S6A). Modeling of ribose 2′-O methylation on N1 to mimic Cap1-mRNA shows that the methyl group would clash with these protein residues (Fig. 8A). Rotating the methyl group away introduces a steric clash with the RNA itself and interferes with the water network. Interestingly, N2 ribose methylation is also predicted to disrupt RNA binding to IFIT1, because of hydrogen bonds with H289 and steric hindrance by Q290 (Fig. 8A and Fig. S6A). Thus, the IFIT1 tunnel is restricted to interact with RNAs not methylated at these 2′-hydroxyls.
Fig. 8.
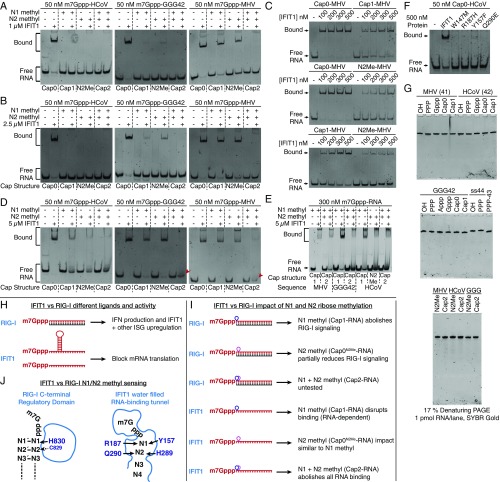
Sensing of N1 and N2 ribose 2′-O methylation by IFIT1. (A, Left) Cross-section of the IFIT1 tunnel van der Waals surface (gray) and residues predicted to clash with N1 and N2 methylations. (Right) In silico rigid body modeling of N1 and N2 methylations (purple dots). (B) SYBR Gold-stained EMSAs between 0.5 µM IFIT1 and differentially methylated m7Gppp-RNA. The dashed lines demarcate lanes with different cap structures, as indicated by the labels below the gel. Fig. S7 A–E for additional gel shifts. (C) Translation assay with differentially methylated reporter mRNA. Data represent the mean of three independent measurements performed in duplicate ± SD. (D) Mutational analysis of 2′-O methyl sensing residues investigated by fluorescent EMSA similar to Fig. 5A. (E) In vitro translation assays with 2′-O methyl sensing mutants and Cap0 reporter. Data represent the mean of two measurements ± SD. (F) Flp-In T-REx 293 IFIT1 knockout cells were cotransfected with expression plasmids for CD13 and either GFP control, wild-type IFIT1, or IFIT1 R187H and infected with HCoV 229E wild-type or HCoV 239E DA. Virus titers in supernatants were determined by TCID50 18 h postinfection. Data represent the mean of three independent experiments ± SD. **P < 0.01 as analyzed by two-way ANOVA with Bonferroni posttest. The Western blots show expression of proteins at the time of infection. (G) Similar to F, except IFIT1 knockout cells were reconstituted with the indicated IFIT1 constructs or GFP, and virus growth determined by quantitative PCR. Data represent the mean fold-change ± SD of triplicate measurements of the viral N-gene signal, relative to GFP control (ctrl). One representative experiment of three is shown. Western blots show protein expression at the end of the experiment.
Consistent with this finding, either N1 or N2 methylation of HCoV and GGG42 RNAs is sufficient to disrupt binding with up to 2.5 µM IFIT1 (Fig. 8B and Fig. S7 A and B). Surprisingly, at the same concentrations, individual N1 or N2 methylation only partially reduced the interaction between IFIT1 and MHV RNA, and combining both was required to fully abolish binding for this sequence (Fig. 8B and Fig. S7 A–C). This RNA-dependent effect is likely because of the longer overhang and decreased secondary structure stability of the MHV sequence compared with HCoV/GGG42 (Fig. 1A and Fig. S1B), allowing it to maintain relatively strong binding to IFIT1, even when modified with a single ribose methylation, and requiring the additive effect of multiple methylations (N1+N2) to avoid IFIT1 recognition. However, we cannot rule out the existence of RNA-sequence or other structural elements within MHV that intrinsically enhance its affinity for IFIT1. At higher protein concentrations (5 µM IFIT1) where nonspecific interactions may play a role, the additive effects of N1+N2 ribose methylations also became apparent for the GGG42 RNA (Fig. S7 D and E).
Fig. S7.
IFIT1 senses N1 and N2 ribose 2′-O methylation (A and B) EMSAs between 1 µM or 2.5 µM IFIT1 and differentially methylated m7Gppp-RNA (C) Comparison of IFIT1 binding to 35 nM Cap0-MHV, Cap1-MHV, or N2Me-MHV. Cap1 methylation reduces the apparent affinity to m7Gppp-MHV by ∼fourfold, as the apparent Kd for Cap1-MHV is in the 200–300 nM range, whereas the apparent Kd for Cap0-MHV is ∼75 nM (see Fig. 1A). (D) EMSAs between 5 µM IFIT1 and differentially methylated m7Gppp-RNA. Note that at these protein concentrations, a second band is more prominent at the top of the gel, which are likely nonspecific or higher-order interactions. For Cap2-GGG42 and Cap2-MHV, the red arrowhead points to the smeared band corresponding to unbound RNA. (E) Similar to D, except a higher concentration of RNA was used to improve the staining sensitivity. This gel-shift demonstrates a clear additive effect of N1+N2 for GGG42. (F) SYBR Gold-stained EMSA between IFIT1 mutants targeting 2′-O methyl-sensing residues and Cap0-HCoV RNA. (G) 1× TBE, 17% (vol/vol) 8M urea denaturing PAGE of RNA used in this study. (H–I) Comparison between IFIT1 and RIG-I self vs. nonself discernment of capped RNAs (H) In addition to recognizing base-paired, blunt-ended RNAs with a 5′-PPP, RIG-I is activated by m7Gppp/Cap0 containing RNAs (8, 9). Recognition of viral, self, or synthetic Cap0-RNAs by RIG-I leads to IFN production, and IFIT1 and other ISG up-regulation. In contrast, IFIT1 targets single-stranded mRNAs, or structured mRNAs if they have an ∼4-nt overhang, leading to inhibition of mRNA translation. (I) N1 methylation (i.e., Cap1-RNA) has been shown to abolish RIG-I activity, whereas N2 methylation alone has a partial effect; the combination of N1+N2 (i.e., Cap2-RNA) has not been tested with RIG-I. In contrast, N1 and N2 methylation alone have a similar effect on IFIT1 RNA binding, and N1+N2 methylation has an additive and potentially synergistic effect in preventing RNA binding, which may be important for protecting inherently susceptible self-mRNAs from IFIT1 recognition. (J) RIG-I is a multidomain protein that binds RNA through a central helicase domain (not drawn) and a C-terminal Regulatory domain (RD). The RIG-I RD houses a basic cleft, which recognizes the blunt-end, the bridging PPP, and the 2′-OH of N1, but makes no contacts with the m7G moiety. From the RD, H830 hydrogen-bonds with the N1 2′-OH to prevent binding of Cap1-RNA, whereas C829 makes minimal contacts with N2 but is predicted to clash with methylation at this position. Mutation of H830A does not affect Cap0-RNA binding but impairs RIG-I ability to discern self (Cap1) from nonself (Cap0) RNA. IFIT1 cap recognition differs, using a narrow tunnel that makes contacts with the m7G, the bridging PPP, and the cap-proximal nucleotides. Several conserved residues mediate 2′-O methyl-sensing and general RNA binding, and their mutation is deleterious for IFIT1 activity. These differences between IFIT1 and RIG-I reflect their complementary roles in self vs. nonself-mRNA discernment.
Consistent with the above findings, single N1 or N2 methylation of the reporter mRNA reduced IFIT1 inhibitory activity in translation assays by ∼10-fold (IC50 ∼ 1 µM) (Fig. 8C), whereas translation of Cap1- and Cap0N2Me-mRNA was still strongly inhibited by 5 µM IFIT1. This finding intriguingly suggests that N1- or N2-methyl steric hindrance can be overcome at very high IFIT1 concentrations, possibly from nonspecific RNA interactions contributed by the solvent exposed groove of IFIT1. As before, combining N1 and N2 methylations (Cap2 reporter) resulted in a striking rescue of translational inhibition, restoring FF levels to 90% even in the presence of 5 µM IFIT1 (Fig. 8C). Taken together, our combined structural and functional analysis confirms the role of N1 methylation in interfering with IFIT1 inhibitory activity, and reveals an analogous function for N2 methylation. Importantly, our data suggest that the combination of N1 and N2 methylation, as found in nearly half of endogenous mRNAs, produces an additive and potentially synergistic protective effect against IFIT1 recognition, which is particularly evident under circumstances where IFIT1 can overcome individual 2′-O methylation in an RNA-dependent or protein concentration-dependent manner.
To further confirm the importance of 2′-O methyl sensing for IFIT1 activity, we mutated the residues predicted to clash with N1 or N2 ribose methylations, and tested their impact on RNA binding and translational inhibition (Fig. 8 D and E). At N1, Y157F had only a minor effect on capped RNA binding, but both R187H and R187A abolished the interaction. At N2, mutating either H289 (H289A) or Q290 (Q290E) partially reduced binding, and combining either mutant with Y157F (DM-YH, Y157F/H289A; or DM-YQ, Y157F/Q290E) completely disrupted binding. Translation assays also showed reductions in IFIT1 inhibitory activity for all mutants, with R187H and the two double mutants having the greatest effect. It should be noted, however, that these residues are highly conserved (Fig. S4M), and as such, play an integral role in general RNA binding that extends beyond 2′-O methyl sensing (Fig. S6 A and B). Thus, capped-RNA recognition and 2′-O methyl sensing by IFIT1 are two tightly linked processes that have likely coevolved.
Functional Validation of IFIT1 Activity Against 2′-O Methyltransferase Deficient Human Coronavirus.
Human IFIT1 has been shown to inhibit replication of viruses lacking N1 ribose 2′-O methylation, such as HCoV 229E bearing a D129A (DA) mutation in its viral 2′-O MTase gene (18). Therefore, to functionally validate our results in a biological context, we tested the antiviral activity of IFIT1 RNA-binding mutants against wild-type HCoV 229E and HCoV 229E DA. First, we verified that the IFIT1 mutants used in cell-based assays (R187H, W147M, Y157F, and Q290E) disrupted the interaction with Cap0-HCoV RNA (Fig. S7F). Next, we reconstituted Flp-In T-REx 293 IFIT1 knockout cells with human IFIT1 or IFIT1 mutants, and assayed HCoV growth in these cells (Fig. 8 F and G). Although expression of a control protein (GFP) led to comparable accumulation of both wild-type and DA virus in the supernatant of infected cells, expression of IFIT1 significantly reduced growth of the DA mutant virus, but not wild-type virus (Fig. 8F). In contrast, IFIT1 R187H, which disrupts interactions with the cap ribose and bridging triphosphate (Fig. 3 E and F), was unable to impair HCoV 229E DA virus growth (Fig. 8F). Similarly, W147M, which disrupts cap recognition, or Y157F and Q290E, which impair binding to Cap0-HCoV RNA (Fig. S7F), lost their antiviral activity against HCoV 229E DA (Fig. 8G). Thus, IFIT1 binding to 2′-O unmethylated viral RNA is required for its antiviral properties.
Discussion
The ability of many viruses to cap their mRNA and mimic the host’s allows them to hijack a cell’s translational machinery and replicate new virus particles. To counteract this, host cells have evolved as part of their antiviral program, the IFIT proteins. By competing with eIF4E/eIF4F for binding to capped RNA, IFIT1 can prevent viral propagation by latching onto the ends of mRNA and preventing assembly of ribosomal initiation complexes (18, 19). Whereas recognition of the cap by eIF4E and other cap-binding proteins occurs in a highly specific manner (31, 37), we surprisingly found that recognition of the cap moiety by IFIT1 is instead nonspecific with regards to both sequence and structure. Through its highly water-filled cap-binding pocket, IFIT1 can accommodate not only bona fide cap in different orientations, but also an unmethylated cap, adenine cap, and presumably other structures too. This built-in plasticity may in part be to allow IFIT1 to maintain a broad spectrum of antiviral activity, and to thwart the ability of viral structures to rapidly evolve. Another possibility is that IFIT1 genes simply have not had enough time to evolve exquisite cap specificity, because they emerged relatively recently in evolution [jawed vertebrates (12)] compared with eIF4E and CBC, which are essential genes in all eukaryotes (34, 35). Regardless, the penalty for this plasticity is likely a reduction in affinity for the cap moiety and in this respect, the recognition of nucleotides beyond the cap provides IFIT1 the additional affinity required to compete with an otherwise very tight eIF4F–5′-cap complex.
The recognition of cap-proximal nucleotides by IFIT1 also plays a critical role in discerning self from nonself. Our structural analysis revealed that IFIT1 forms a tight interacting surface around the ribose 2′-hydroxyls of N1 and N2, thus preventing recognition of endogenous mRNAs methylated at these positions and restricting IFIT1 activity to unmethylated viral mRNAs. This finding is supported by a comprehensive gel-shift analysis that showed a preference for recognizing Cap0 structures over N1 or N2 methylated RNA, in vitro translation assays that showed a reduction in IFIT1’s ability to inhibit translation of N1 or N2 methylated mRNA reporters, and human coronavirus infectivity assays that showed enhanced IFIT1 antiviral activity when viral N1 methylation was mutated. In this way, IFIT1 effector function complements RIG-I receptor activity (summarized in Fig. S7 H–J), as RIG-I detects blunt-ended, base-paired PPP- and Cap0-RNAs to up-regulate IFIT1 and other ISGs (8, 9).
Cellular N1 methylation was generally thought to be the primary determinant of self, protecting endogenous mRNA from IFIT1 recognition, but we discovered that the ability to discern between Cap0 and Cap1 structures is diminished for one of our RNAs (MHV) (Fig. 8B), an effect that is possibly linked to its 5′-sequence or secondary structure. This finding is in accordance with a recent study by Daugherty et al., who demonstrated that human IFIT1 can target both Cap0 and Cap1 mRNAs when overexpressed in a yeast system (12). Furthermore, while this manuscript was under revision, Young et al. similarly showed that N1 methylation of a reporter gene only partially reduced its sensitivity to human IFIT1 in an in vitro translation system (44). Interestingly, using viral mRNA in the same system, Young et al. also noted potential RNA-dependent effects for N1 methylation. Taken together, these observations lead to the conclusion that N1 methylation alone may not be enough to protect all endogenous mRNAs from IFIT1, and that there are other determinants of self that govern IFIT1 activity.
Our structural and functional analysis reveals that N2 methylation by CMTr2 could fulfill this role, providing an additional safeguard against aberrant recognition of mRNAs that are otherwise susceptible to IFIT1 (Fig. 8B). However, as Cap2 structures are not as ubiquitous as Cap1 (40), other elements may prevent self-recognition. For example, actively translating mRNAs are generally found in preformed mRNP complexes and, as indicated by our order-of-addition experiment (Fig. 1C), would be protected from IFIT1 competition. Similarly, newly synthesized mRNAs undergo a pioneer round of translation directed by the CBC (45), which may also offer protection from IFIT1. Adenosine N6-methylation of the first transcribed nucleotide is another modification that accompanies ribose N1 methylation on ∼20–30% of cellular mRNAs, in the form of N6,O2′-dimethyladenosine (m6Am) (46, 47). Our structure suggests that m6Am could protect self-mRNAs by disrupting water-mediated interactions and impinging on nearby residues (Fig. S6 A and E), and thus merits further investigation. Finally, cap-proximal secondary structure elements could combine with mRNA modifications to further prevent self-recognition.
To what extent do viruses exploit these mechanisms to alter IFIT1 activity? Our observations support the model whereby viral N1 methylation evades or dampens IFIT1 activity (Fig. 8 F and G), and is consistent with previous studies showing enhanced sensitivity of coronaviruses and flaviviruses to IFIT1 when viral N1 methylation was mutated (18, 23, 24, 26). Similarly, Young et al. recently showed that parainfluenza virus type 5 (PIV5) was more sensitive to human IFIT1-mediated restriction than PIV3, partly because PIV5 mRNAs were not completely N1-methylated during infection (44). On the other hand, vesicular stomatitis virus (VSV) has been shown to uniformly N1-methylate its mRNAs in vivo (48, 49), yet it remains sensitive to IFIT1 restriction (12). One explanation is that the short and potentially unstructured 5′ UTRs of VSV mRNAs could allow IFIT1 to overcome N1 methylation (as described here for MHV RNA). Alternatively, VSV mRNAs may display another pattern specifically recognized by IFIT1 (as proposed by Daugherty et al.), such as RNA-sequence or -structural elements (12). Further work is needed to validate either notion, and to determine if this ability to overcome viral N1 methylation is an adaptation that allows IFIT1 to target other Cap1-containing viruses, or if it is restricted to VSV and related viruses.
IFIT1B (which includes mouse Ifit1) is the only other IFIT family member known to specifically recognize capped RNAs to inhibit viral replication (18, 19). Our analysis supports the notion that both proteins use a similar mode of cap-recognition, and thus should display overlapping antiviral activities. However, recent evidence suggested otherwise (12). Based on our structural and functional data, we propose that both IFIT1 and IFIT1B can target Cap0-containing viruses, but they may differ in their sensitivity to cap-proximal modifications, such as methylation, RNA-sequence, or RNA-structure. These differences and underlying molecular mechanisms are not entirely clear yet, but one possibility is that the ability to overcome N1 methylation in an RNA-dependent manner could distinguish IFIT1 from IFIT1B proteins, and may explain why IFIT1 overexpression inhibited wild-type VSV replication (a Cap1-containing ssRNA virus), whereas IFIT1B overexpression did not (12). Regardless, in humans IFIT1B appears to be nonfunctional, and has been deleted or pseudogenized in several other mammals, consistent with the notion that widespread viral evasion strategies (e.g., N1 methylation) have generally defeated IFIT1B (12), whereas IFIT1 was retained possibly because of its adaptable nature.
Taken together, through a relatively nonspecific cap-binding pocket and a potentially plastic RNA binding mechanism, IFIT1 appears to have grafted adaptability onto an otherwise germ-line–encoded member of the innate immune system, to broadly defend against rapidly evolving viral pathogens. At the same time, the host evolved multiple mechanisms that combine to limit detrimental IFIT1 activity against endogenous mRNAs. Clearly, further work is needed to validate the physiological relevance of these notions, particularly with respect to understanding how IFIT1 can overcome N1 methylation in an RNA-dependent manner, the differential specificities of IFIT1 and IFIT1B proteins, and if viruses exploit CMTr2 and other enzymes to modify their mRNA and evade IFIT1. Finally, it has been established that human IFIT1 can form complexes with other IFIT family members (IFIT2 and IFIT3) and several host factors (17), which could play a role in modulating self- vs. nonself-mRNA recognition and translational inhibition. Our structural and functional analysis of IFIT1-capped RNA interactions will provide a framework for future structure-guided studies of IFIT function. Moreover, these efforts will provide important contributions to the development of mRNA therapeutics and to vaccine design, as emerging research suggests that rendering viruses more susceptible to IFIT1-like antiviral responses, by inhibiting their mRNA 2′-O methylation or modifying their 5′-secondary structure, is a strategy for the rapid development of live, attenuated vaccines (e.g., refs 21–24).
Materials and Methods
Detailed discussions of the materials and methods used in this study are provided in SI Materials and Methods.
Protein Expression and Purification.
IFIT proteins, RNMT (Human N7-MTase), and TbMTr2 (Trypanosoma brucei Cap2 MTase) were expressed in Escherichia coli, and purified by Ni-affinity, ion-exchange, and size-exclusion chromatography. CMTr2 (Human Cap2 MTase) was expressed in Sf9 insect cells and purified by Ni-affinity and size-exclusion chromatography.
IFIT1 Crystallography.
IFIT1 monomeric mutants (L457E/L464E or N216A/L457E/L464E) were purified to homogeneity and mixed with molar excess chemically synthesized oligos (SI Materials and Methods for synthesis, purification, and MS of oligos) and crystallized in 27–32% (vol/vol) PEG 200, 0.1 M Tris pH 8.1, and 200 mM CaCl2. Crystals were flash-frozen in liquid nitrogen without additional cryoprotection, and diffraction data were collected at Canadian Light Source beamline 08ID-1 (Table S1). Initial structures were determined by molecular replacement, subsequent structures by rigid body refinement.
RNA EMSA.
RNAs for EMSAs were prepared using T7 RNA polymerase and purified by denaturing PAGE. Appp-GGG42 was capped cotranscriptionally; other modifications were performed with CIP (New England Biolabs), vaccinia capping enzyme (New England Biolabs), Cap1 2′-O MTase (New England Biolabs), or home-made CMTr2 or TbMTr2. All 5′ caps and RNA sequences were confirmed by LC/MS. RNA sequences and mass spec summary are in Tables S2 and S3, respectively. The 3′-end labeling of m7Gppp-43 with pCp-Cy5 was performed with T4 RNA ligase (New England Biolabs). For EMSAs, purified proteins and RNA were mixed at the indicated concentrations in each figure, and resolved by native PAGE [1× TBE, 10% (vol/vol) gels]. Bands were visualized with SYBR Gold and UV imaging (unlabeled RNA), or with a Typhoon R3 imager (for pCp-Cy5 labeled RNA). Band densitometry was performed with ImageJ.
Table S3.
Summary of MS data
| Sequence | Expected (Da) | Detected (Da) |
| PPP-AAAA | 1,516.14 (Na adduct) | 1,516.12 (Na adduct) |
| Gppp-AAAA | 1,759.23 | 1,759.18 |
| m7Gppp-AAAA | 1,773.25 | 1,773.19 |
| PPP-MHV | 13,300.71 (Na adduct) | 13,300.05 (Na adduct) |
| m7Gppp-MHV | 13,557.99 | 13,556.99 |
| Cap1-MHV | 13,572.02 | 13,570.97 |
| N2Me-MHV | 13,572.02 | 13,571.42 |
| Cap2-MHV | 13,586.04 | 13,585.45 |
| Cap2-MHV (TbMTr2 modified) | 13,586.04 | 13,584.69 |
| PPP-HCoV | 13,569.86 (Na adduct) | 13,569.42 (Na adduct) |
| m7Gppp-HCoV | 13,827.14 | 13,825.91 |
| Cap1-HCoV | 13,841.16 | 13,840.38 |
| N2Me-HCoV | 13,841.16 | 13,840.61 |
| Cap2-HCoV (TbMTr2 modified) | 13,855.08 | 13,854.48 |
| PPP-GGG42 | 13,664.93 (Na adduct) | 13,664.47 (Na adduct) |
| Gppp-GGG42 | 13,908.18 | 13,908.09 |
| Appp-GGG42 | 13,892.18 | 13,892.22 |
| m7Gppp-GGG42 | 13,922.21 | 13,921.55 |
| Cap1-GGG42 | 13,936.23 | 13,935.91 |
| N2Me-GGG42 | 13,936.23 | 13,934.95 |
| Cap2-GGG42 (TbMTr2 modified) | 13,950.25 | 13,949.29 |
| PPP-ss44 | 14,705.73 (Na adduct) | 14,704.64 (Na adduct) |
All values are reported as molecular weight, except for PPP-AAAA, Gppp-AAAA, and m7Gppp-AAAA, which are reported as exact monoisotopic neutral mass. For all RNA except the short oligoadenosines, only the most intense peak is reported, as they all contained Na or K adducts. All PPP-RNA contain variable amounts of PP- and P-RNA, likely because of hydrolysis of the PPP under the alkaline conditions of LC-MS. For all m7Gppp-, Cap1-, N2Me-, and Cap2-RNA, expected values are calculated based on the zwitterionic enolate form of m7G (in which N1 is deprotonated; see Fig. S4H). Unless otherwise specified, N2 methylations to generate N2Me- or Cap2-RNA were performed with the human CMTr2 protein.
Reporter mRNA Preparation and in Vitro Translation Assay.
The bicistronic reporter (5′-UTR sequence is in Table S2) was in vitro transcribed with SP6 RNA polymerase in the presence of either m7GpppG, GpppG, or ApppG RNA Cap Analog (New England Biolabs). N1/N2 methylations were performed post transcriptionally using mRNA Cap1 2′-O MTase (New England Biolabs) and/or TbMTr2. In vitro translations were set up at a final volume of 10 µL with 4 ng/μL reporter mRNA (∼4 nM final) and 1 µL protein in untreated Krebs-2 extracts, and performed as described in SI Materials and Methods.
Virus Infectivity Assay.
Flp-In T-REx 293 IFIT1−/− cells were cotransfected with plasmids for IFIT1 and CD13 (HCoV receptor). Cells were treated with 20 units of IFN-β or left untreated and 24 h later infected with HCoV 229E wild-type and HCoV 229E DA with a multiplicity of infection of 1 and 1.25, respectively. Eighteen hours postinfection, virus accumulation was tested by quantitative RT-PCR or TCID50 analysis.
SI Materials and Methods
Cloning, Protein Expression, and Purification of IFIT Proteins.
IFIT1 (UniProt accession number P09914) and IFIT5 (UniProt accession number Q13325) were cloned into a pSMT3 vector (pET-28a backbone) between BamHI and NotI sites, resulting in fusion proteins encoding an N-terminal, Ulp1-cleavable 6xHis-Sumo tag (51). Point mutants were generated by site-directed mutagenesis using iProof High-Fidelity DNA polymerase (Bio-Rad). All IFITs were expressed in BL21 (DE3) or Rosetta 2 (DE3) pLysS cells using standard protocols, and purified by Ni-affinity chromatography followed by cleavage of the tag. Cleaved proteins carry over an N-terminal serine residue encoded by the BamHI site. For IFIT1 and mutants, cleaved proteins were further purified by passing over a 5-mL HiTrap SP HP column (GE Healthcare) equilibrated with 50 mM Hepes pH 7.5 and 1 mM DTT, and eluted with a salt gradient between 100 and 400 mM NaCl. For IFIT5, a second Ni column was performed followed by a Mono Q 4.6/100 PE (GE Healthcare) column in 25 mM Tris pH 8.0 and 1 mM DTT, and eluted over a shallow salt gradient between 100 and 200 mM NaCl. All proteins were further purified by gel filtration using superdex 200 10/300 or 16/60 columns (GE Healthcare) in 20 mM Tris pH 7.6, 150 mM NaCl and 1 mM DTT (gel-filtration buffer). All pure proteins had a final A260/280 ratio ∼0.5, indicating no RNA contamination.
Cloning, Protein Expression, and Purification of RNA Modifying Enzymes.
The Human N7-methyltransferase (RNMT, a gift from Stewart Shuman, Sloan Kettering Institute, New York, NY) residues 2–476 was cloned into pSMT3 between BamHI and NotI, expressed in Rosetta 2 (DE3) pLysS and the purification was adapted from ref. 52. RNMT was purified by Ni-affinity chromatography followed by cleavage of the tag as above, then passed over a 5-mL HiTrap SP HP column (GE Healthcare) equilibrated with 20 mM Tris pH 7, 10% (vol/vol) glycerol, and 5 mM β-ME, and eluted with a salt gradient between 50 and 500 mM NaCl. RNMT was further purified on a superdex 200 16/60 column in 20 mM Tris pH 7.6, 300 mM NaCl, 10% (vol/vol) glycerol, and 5 mM β-ME. RNMT protein-containing fractions were dialyzed overnight against 20 mM Tris pH 7.6, 300 mM NaCl, 50% (vol/vol) glycerol, and 5 mM β-ME, and stored at −20 °C at 0.8 mg/mL A synthetic gene encoding Trypanosoma brucei brucei Cap2 methyltransferase (TbMTr2, also called TbCom1, reference gene name Tb11.02.2500, UniProt Q385S9) was ordered as an Escherichia coli codon optimized gBlock Gene Fragment (IDT) and cloned into pProEX HTb between BamHI and NotI, resulting in a fusion protein with a tobacco etch virus (TEV) protease cleavable 6xHis N-terminal tag. TbMTr2 was expressed in BL21 (DE3) and purified by two-step Ni affinity chromatography using standard protocols. The second Ni flow-through was applied to a 5 mL HiTrap Q HP column (GE Healthcare) equilibrated with 25 mM Tris pH 8.0 and 1 mM DTT, and the column washed with the same buffer containing 50 mM NaCl. Cleaved TbMTr2 was present in the Q column flow-through and low salt wash, and was applied to a 5-mL HiTrap SP HP column equilibrated with 50 mM Hepes pH 7.5, 1 mM DTT, and eluted with a salt gradient between 50 and 500 mM NaCl. Protein-containing fractions were concentrated and injected onto a superdex 200 10/300 column equilibrated with gel-filtration buffer (see above). Fractions corresponding to TbMTr2 dimers were pooled and an aliquot stored at 3.84 mg/mL (80 µM) at –20 °C in 50 mM Tris pH 7.4, 5 mM DTT, 50% (vol/vol) glycerol, and 25 mM NaCl. Human Cap2 methyltransferase (CMTr2/FTSJD1, UniProt Q8IYT2) was ordered from DNASU and cloned into pFastBac HTb between BamHI and NotI. Recombinant baculovirus was generated according to the Bac-to-Bac Baculovirus Expression System protocol (Invitrogen) using DH10MultiBac cells (53). Virus was added to Sf9 cells grown in I-Max medium (Wisent Bioproducts), and CMTr2 was expressed at 27 °C for 64–72 h. Cells were pelleted at 1,000 × g at 4 °C for 10 min, and pellets were resuspended in 20 mM Tris pH 8, 10 mM imidazole, 500 mM NaCl, 10% (vol/vol) glycerol, and 5 mM β-ME (Ni A Buffer), supplemented with cOmplete EDTA-free Protease Inhibitor Mixture (Roche) and 1 mM PMSF. Cells were lysed by sonication on ice (10 s ON, 20 s OFF, 50% amplitude, six total pulses), and clarified by ultracentrifugation at 173,021 × g. Supernatants were applied to Ni resin, washed three times with 10 bed volumes Ni A, and eluted with 4 bed volumes Ni A with 350 mM imidazole. Eluted protein was cleaved overnight with TEV protease in Ni A with 2 mM DTT instead of β-ME. A second Ni bead purification was performed to remove TEV protease, and the flow-through was concentrated and injected onto superdex 200 10/300 equilibrated in 20 mM Tris pH 7.6, 150 mM NaCl, and 3 mM DTT. Fractions corresponding to monomeric protein were pooled, concentrated to 4 mg/mL, and stored at 4 °C.
Crystallization and Data Collection.
The monomeric mutants of IFIT1 (L457E/L464E) or IFIT1 (N216A/L457E/L464E) were mixed with molar excess chemically synthesized oligos (see below for RNA preparation) either directly before crystallization or before gel filtration. The protein buffer was 20 mM Tris pH 7.6, 150 mM NaCl and 1 mM TCEP. The cocrystals were obtained with 5–10 mg/mL protein drops mixed 1:1 with reservoir solution containing 27–32% (vol/vol) PEG 200 (Sigma), 0.1 M Tris pH 8.1, and 200 mM CaCl2 using the hanging-drop vapor diffusion method at 4 °C. Crystals were flash-frozen in liquid nitrogen without additional cryo-protection. All data were collected at 100K with 0.979 Å X-rays on beamline 08ID-1 at the Canadian Light Source, which is equipped with a Mar300CCD detector (54), and the images integrated, scaled, and merged using the HKL2000 suite (55). Data were truncated with Ctruncate (ccp4 suite) (56) and 5% of reflections were set aside for the Rfree set. Only the highest-resolution dataset was used as a master file to generate all Rfree sets. Data collection statistics from HKL2000 are in Table S1.
Structure Determination, Model Building, and Refinement.
The initial crystal structure was determined by molecular replacement using the PHASER program in Phenix (57). The structure of wild-type human IFIT1 was used as a search model. Subsequent structures were determined with rigid body refinement with the protein only. Restraints for m7GpppA and GpppA, as well as bound PEG molecules, were calculated using Phenix eLBOW (58). The structure was refined iteratively using Phenix (59) with manual model building in Coot (60). The refinement strategy included all-isotropic B-factor refinement, occupancy refinement, and TLS. All final models contained protein residues 8–467. Structure validation was performed with MOLPROBITY in Phenix (61). Ramachandran statistics are as follows: IFIT1 L457E/L464E with m7Gppp-RNA, 98.5% favored, 0% outliers, 0.2% rotamer outliers; IFIT1 L457E/L464E with Gppp-RNA: 98.9% favored, 0% outliers, 0% rotamer outliers; IFIT1 L457E/L464E with PPP-RNA: 98.9% favored, 0.2% outliers, 0.2% rotamer outliers; IFIT1 N216A/L457E/L464E with m7Gppp-RNA: 98.7% favored, 0% outliers, 0.2% rotamer outliers. The MOLPROBITY overall score for each was 1.00, 0.99, 0.93, and 0.92 respectively. Refinement statistics are in Table S1.
Sequence, Structure, and RNA Analysis.
APBS was used to calculate the surface electrostatic potential (62), and PyMol to general all molecular figures (https://www.pymol.org/). For surface electrostatic analysis, all surfaces are colored by electrostatic potential from negative (−10 kTe−1; red) to positive (+10 kTe−1; blue). CAVER (50) was used to draw the protein tunnel in Fig. 2. ESPript was used to generate the sequence alignment in Fig. S5 (63). RNA secondary structure prediction was performed with Mfold (64). In silico N1/N2 methylation for Fig. 8 was performed by rigid body docking. For sequence conservation analysis, all mammalian IFIT1-like and IFIT5-like annotated sequences were retrieved from the nonredundant protein sequences database using human IFIT1 as a BLAST query. The results were manually curated to remove highly related isoforms, and to remove sequences with large N- and C-terminal deletions (for example, those missing R38/T37). The final dataset contained 86 IFIT1- or IFIT1B-like sequences, and 60 IFIT5-like sequences. Sequences were aligned with ClustalOmega, and edited in Jalview (www.jalview.org/) to remove columns arising from insertions in nonhuman IFIT1 or IFIT5 sequences. This was done to maintain human IFIT1 or human IFIT5 residue numbering. The aligned and edited sequences were used to generate the sequence logos with WebLogo (65), and only blocks related to m7Gppp- binding residues are shown.
Enzymatic Preparation and Purification of RNA for EMSAs.
All PPP-RNAs used for EMSAs were prepared enzymatically by run-off transcription using T7 RNA polymerase. RNA sequences are listed in Table S2. Single-stranded oligo templates (also in Table S2) were mixed 1:1 with the appropriate promoter sequence at a final concentration of 10 µM (each), heated to 95 °C for 2–3 min, and slow-cooled on the bench. Transcription was carried out at 37 °C with 1 µM template (bottom strand) in 1× buffer (New England Biolabs) supplemented with 10 mM DTT, 2 mM ATP, 2 mM UTP, 2 mM CTP, 2 mM GTP, 2 U/mL pyrophosphatase (E. coli; New England Biolabs), RNasin (Promega), and 2.5 U/µL T7 RNA polymerase (New England Biolabs), in 0.5- to 1-mL final volume. MgCl2 concentrations between 5 and 40 mM were used and optimized for each template. Appp-capped GGG42 was prepared by replacing the GTP with 8:1 A(5′)ppp(5′)G:GTP. All RNAs were purified by 10% (vol/vol) or 12% (vol/vol) denaturing PAGE. The ss44-, MHV-, HCoV-, and GGG42-transcribed RNA were validated by MS to confirm the purity and sequence of the final product, all of which contained minor amounts of n+1 contamination carried over from the transcription and purification, and to confirm the presence of an intact triphosphate or Appp- cap (details below). The 3′-end labeling of m7Gppp-43 with pCp-Cy5 (Jena Biosciences) was performed with T4 RNA ligase (New England Biolabs), and the labeled product purified by gel extraction on 8% (vol/vol) denaturing PAGE. OH-RNA was prepared by treating the PPP-RNA with CIP (New England Biolabs). m7Gppp- and Gppp-RNA were prepared by posttranscriptional capping of PPP-RNA with the vaccinia capping system (New England Biolabs) with or without S-adenosylmethionine (SAM). Cap1-RNA was prepared by including the Cap1 2′-O-Methyltransferase (New England Biolabs) in the capping reactions. New England Biolabs reactions were performed according to the manufacturer’s instructions. Ribose N2 methylations were performed with purified CMTr2 or TbMTr2 as indicated in Table S3. For N2 methylation with TbMTr2, the protocol was adapted from ref. 66. Next, 200 pmol of Cap0- or Cap1-RNAs were incubated with 800 pmol TbMTr2 in 200 µL reactions containing 50 mM Tris pH 7.5, 5 mM DTT, 2 mM SAM and RNasin. Reactions were carried out for 2 h at 27 °C. For CMTr2 N2 methylation, the protocol was adapted from ref. 3. Then 150–200 pmol of Cap0- or Cap1-RNAs were incubated with 500 pmol CMTr2 in 100-μL reactions containing 30 mM Tris pH 7.4, 50 mM KCl, 10 mM EDTA, 2.5 mM SAM and RNasin. Reactions were carried out for 2 h at 37 °C. Modified RNAs were purified by phenol/chloroform extraction and ethanol precipitation. The purity of RNA used in this study was also confirmed by 17% (vol/vol) denaturing PAGE (Fig. S7G). All short RNA used for Cap0/Cap1/Cap02/Cap2 binding analyses were validated using LC-MS to confirm > 90–95% homogeneity of 5′ modification (details below).
RNA EMSA.
Stocks (10×) of purified protein were prepared in 25 mM Tris pH 7.6, 150 mM NaCl, 50% (vol/vol) glycerol, 5 mM β-ME, 0.5 mM PMSF, and 1 mM EDTA, and stored at −20 °C. RNAs were diluted in MilliQ water at a final concentration of 1 µM and heat refolded before use. For capped-RNA, the RNA was heated at 95 °C for 2–3 min and slow-cooled on the bench. For PPP-RNA, heating was at 65 °C for 10 min with slow-cooling on the bench. Protein and RNA were mixed with 10× buffer (500 mM Tris pH 7.4, 1.25 M NaCl, 10 mM EDTA, and 0.1 mg/mL Heparin) at a final volume of 20 µL and the binding proceeded on the bench for at least 15 min. For Fig. S7E, only 12 µL was loaded in each well. For EMSAs with pCp-Cy5 labeled RNA, the final volume was 10 µL. The final reactions all contained 5% (vol/vol) glycerol (carried over from the protein storage buffer) and no loading dyes were used. The final RNA and protein concentrations are indicated in each figure. Bound and unbound RNA were resolved on 1× TBE, 10% (vol/vol) 19:1 acrylamide:bisacrylamide native gels supplemented with 25 mM NaCl. The gels were prerun in a cold room in 0.5× TBE running buffer for at least 30 min, loaded with samples, and run at 120–180 V for approximately 45–60 min. The temperature of the system remained 8–14 °C during the run. For EMSAs with fluorescent RNA, the gel apparatus was covered with aluminum foil to minimize light exposure. Gels were stained with SYBR Gold (Invitrogen) and visualized with a UV transilluminator equipped with a SYBR Green filter. For Figs. 5A and 8D and Fig. S4L, pCp-Cy5 labeled RNA was visualized with a Typhoon R3 imager (GE Healthcare). Band densitometry was performed with ImageJ (https://imagej.nih.gov/ij/).
Preparation of Reporter mRNA.
The bicistronic reporter, pSP-(CAGless)/FF/HCV/Ren was derived from pSP(CAG)33/FF/HCV/Ren·pA51 by exchanging the EcoRI/SacI fragment with an oligonucleotide (5′GAATTCACAATTCGAGCTC3′, restriction sites are underlined) (67). The 5′ UTR of (CAGless)-FF/HCV/Ren is described in Table S2. For in vitro transcription, pSP-(CAGless)/FF/HCV/Ren was linearized with BamHI. In vitro transcriptions were performed with SP6 RNA polymerase in the presence of m7G(5′)ppp(5′)G, G(5′)ppp(5′)G or A(5′)ppp(5′)G RNA Cap Analog (New England Biolabs). N1 and N2 methylations were performed post transcriptionally using mRNA Cap1 2′-O-Methyltransferase (New England Biolabs) or purified TbMTr2. Reporter mRNA was heated at 65 °C for 5 min, then placed on ice for 5 min before reactions were set up. To generate Cap1 reporter, N1 methylation was performed according to New England Biolabs protocols, except the reaction was performed at 30 °C for 2 h. N2 methylation of Cap0- and Cap1-reporter were performed as described above for the short RNA. Modified mRNA was purified by phenol/chloroform extraction, ethanol precipitation, and desalting columns.
In Vitro Translation Assay with Krebs Extracts.
Preparation of the mouse-derived Krebs-2 extracts for cell-free translation is described in ref. 68. In vitro translations were set up with a final volume of 10 µL with 4 ng/μL reporter mRNA (∼4 nM final) and 1 µL of purified protein (in gel-filtration buffer) in untreated Krebs-2 extracts, as described previously (27). The reactions were set up on ice and translation was allowed to proceed for 1 h at 30 °C. The reactions were stopped on ice, and FF and Ren luciferase activities (RLU) were measured on a Berthold Lumat LB 9507 luminometer. Values obtained were normalized against buffer control, which was set at 1. In Fig. 1C and Figs. S1D and S4E, the following final protein concentrations were used: 0.031, 0.062, 0.125, 0.250, 0.5, 1, and 5 µM. For Figs. 5B and 8 C and E, 0.1, 0.3, 0.6, 1, and 5 µM final protein concentration was used. Except for Figs. 5B and 8E, all data are represented by the mean of two to three independent measurements performed in duplicate (as indicated in each figure legend) ± SD.
Mammalian Constructs, Cells, Reagents, and Viruses.
Mammalian IFIT1 expression constructs were generated by targeted mutagenesis PCR and cloned into pTO-SII-HA-GW (17) using gateway cloning (Invitrogen). Sequences were verified by Sanger sequencing. Huh7 cells were a gift from Georg Kochs, University of Freiburg, Freiburg im Breisgau, Germany, Flp-In T-REx 293 IFIT1−/− cells were a gift from Giulio Superti-Furga, CeMM, Vienna, Austria. Cells were maintained in DMEM (PAA Laboratories) containing 10% (vol/vol) FCS (GE Healthcare) and antibiotics (100 U/mL penicillin, 100 µg/mL streptomycin). Antibodies used to test expression levels were actin-HRP (Santa Cruz: sc-47778), and HA-HRP (Sigma: H6533). IFN-β was a gift from Peter Stäheli, University of Freiburg, Freiburg im Breisgau, Germany. Wild-type and 2′-O-methyltransferase–deficient recombinant coronaviruses 229E (HCoV-229E) have been described previously (18).
Virus Infections and Determination of Virus Titers.
To determine the impact of IFIT1 mutants on virus growth, Flp-In T-REx 293 IFIT1−/− cells were cotransfected with plasmids for IFIT1 and CD13 (HCoV receptor). Cells were treated with 20 units of IFN-β or left untreated and 24 h later infected with HCoV 229E wild-type and HCoV 229E DA with a multiplicity of infection of 1 and 1.25, respectively. Eighteen hours postinfection, cells were harvested for quantitative RT-PCR analysis and supernatant was harvested to test for virus accumulation by TCID50 on Huh7 cells.
Real-Time RT-PCR.
Total RNA was isolated using the NucleoSpin RNA Plus kit (Machery-Nagel). Next, 200–500 ng of RNA was reverse-transcribed with PrimeScript RT Master Mix (TAKARA) and quantified by real-time PCR using the QuantiFast SYBR Green RT-PCR Kit (Qiagen) and a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Each cycle consisted of 10 s at 95 °C and 30 s at 60 °C, followed by melting curve analysis. Primer sequences are in Table S2.
Chemical Synthesis of PPP-AAAA.
Phosphoramidite solid phase synthesis conditions were used for the synthesis of RNA in a twist oligonucleotide synthesis column (Glen Research) and adapted from refs. 69 and 70. The 5′pppAAAA3′ strand was synthesized at the 1-μmol scale, using a chain alkylamine controlled-pore glass as a solid support derivatized with N-phenoxyacetyl adenosine (ChemGenes). (5′-ODMTr-2′-TBDMS-APac)-3′-O-(2-cyanoethyl-N,N-diisopropyl)phosphoramidite was prepared as a 0.15 M solution in acetonitrile. 5-Ethylthiotetrazole was used as an activator, and 3% (wt/vol) trichloroacetic acid in dichloromethane was used to detritylate. Capping was carried out using phenoxyacetic anhydride in THF and 16% (vol/vol) N-methylimidazole in THF. Next, 0.1 M I2 in 1:2:10 pyridine/water/THF was used for oxidation. The RNA phosphoramidite was coupled for 600 s. Installation of the triphosphate moiety (or cap structure) at the 5′-end of the RNA was performed in the same twist columns in a fully automated procedure integrated within the synthesis program of the ABI-394 (69, 70).
For 5′-H-phosphitylation of solid-supported RNA, a solution of 3 mL diphenyl phosphite in 12 mL dry pyridine was prepared and applied to the column three times (delivery time 5 s, wait time 300 s each). The support was washed with ACN for 3 min. A 0.1 M solution of triethylammonium buffer was delivered six times to the column (delivery time 5 s, wait times 60 × 2 + 450 × 4 s).
To prepare solid-supported 5′-phosphoroimidazolide RNA, the oxidation solution was prepared as follows. To a solution of imidazole (900 mg) in N,O-bistrimethylsilylacetamide (3 mL), ACN (6 mL), BrCl3C (6 mL), and triethylamine (1.2 mL) were added. Activated 3-Å molecular sieves were added and the solution was stored for 24 h at 4 °C before use. The mixture was delivered to the column five times (delivery time 5 s, waiting time 1,800 s each). The solid support was then washed with ACN for 3 min.
For substitution of imidazole by pyrophosphate moiety, tributhylammonium pyrophosphate salt required for phosphorylation reactions was first prepared from tetrasodium pyrophosphate. Tetrasodium phyrosphophate decahydrate (2.3 g) was dissolved in 20 mL of sterile nuclease-free water and eluted through a glass column filled with 40 mL of wet DOWEX-50WX8 resin, H+ form. The eluted product was collected in a flask containing 40 mL of ethanol and 2.5 mL of tributylamine while stirring at 0 °C. The resin was washed with additional 20 mL of water. Solvents were evaporated and the residue coevaporated four times with dry dioxane to yield 2.7 g of tributhylammonium pyrophosphate that was dissolved in a mixture of 3.3 mL dry DMF and 3.3 mL of anhydrous ACN. This solution was delivered five times to the column (delivery time 3 s, wait time 300 × 4 + 18,000 s). The solid support was then washed with ACN and dried under argon.
For deprotection and release from the support, the twist column was removed from the synthesizer and the CPG was transferred to a screw cap (Eppendorf). Deprotection and cleavage from the solid support was achieved with 3:1 aqueous NH4OH/EtOH for 3 h at room temperature. After decanting to remove the CPG, the deprotection solution was removed under vacuum in a speedvac lyophilizer. Desilylation was achieved by adding 150 μL of 0.1 M solution of tetrabutylammonium fluoride in THF and shaking at room temperature for 24 h. After addition of 850 μL of autoclaved water, the crude sample was desalted using NAP-25 Sephadex size-exclusion columns according to manufacturer’s protocol. Purification of PPP-AAAA is described below.
Chemical Synthesis of Gppp-AAAA.
The synthesis of Gppp-AAAA was performed following a similar procedure to PPP-AAAA except that the substitution reaction on the solid-supported 5′-phosphoroimidazolide was carried out by reaction with GDP instead of pyrophosphate (52).
For the capping reaction with GDP, the bis(trin-butylammonium) salt of GDP was prepared from the commercially available sodium salt purchased from Sigma-Aldrich. GDP sodium salt (0.5 g) was dissolved in 25 mL of sterile nuclease-free water and eluted through a glass column filled with 20 mL of wet DOWEX-50WX8 resin, H+ form. The eluted product was collected in a flask containing 40 mL of ethanol and 2.5 mL of tributylamine while stirring at 0 °C. The resin was rinsed with addition of 20 mL of water. The solvents were evaporated and the residue was coevaporated two times with absolute ethanol and three times with dry DMF to yield a hygroscopic powder (0.90 g). Then, 103 mg of this solid was transferred to a glass vial and dissolved in 0.5 mL of dry DMF. Next, 28 mg of ZnCl2 were also added to the mixture. The resulting solution was applied to the column containing the solid-supported 5′-phosphoroimidazolide RNA and was allowed to react for 24 h at room temperature. The solution was removed, and the support was washed with MeCN and DMF. The column was dried for 1 min by blowing argon. The deprotection and purification steps were similar to those described above for PPP-AAAA. Note that this procedure differs from the one described in ref. 52. The work by Debart and colleagues (52) reports the synthesis of capRNA using 2′-PivOM protected amidites meanwhile herein we use the classical 2′-O-TBDMS chemistry instead of only TBDMS. Similar results were obtained. Purification of Gppp-AAAA is described below.
Purification of Chemically Synthesized PPP-AAAA and Gppp-AAAA.
Purification was performed on an HPLC using a Protein Pak DEA 5PW analytical anion exchange HPLC column. A stationary phase of Milli-Q water and a mobile phase of 1 M LiClO4 in water was used for analysis and purification using a gradient of 0–50% over 46 min. Following purification, excess LiClO4 salts were removed using NAP-25 Sephadex size-exclusion columns. The oligos were characterized by LC-MS (details below).
Preparation of m7Gppp-AAAA.
m7Gppp-AAAA was prepared by enzymatically modifying Gppp-AAAA according to ref (52). Reaction conditions contained 40 mM Tris pH 8.0, 5 mM DTT, and 4 mM SAM. For every 3.3 nmol of Gppp-AAAA, 20 pmol RNMT in an 80-µL reaction was used. The reactions were scaled up according to crystallization needs. The modified oligo was mixed with purified protein and the complex cleaned up by gel filtration before crystallization. LC-MS was used to verify that the modification was quantitative (see below).
Characterization of RNA by LC-MS.
RNA were analyzed by LC-MS using a Dionex Ultimate 3000 coupled to a Bruker Maxis Impact QTOF in negative ESI mode. Samples were run through an Acclaim RSLC 120 C18 column (2.2 μM 120 Å 2.1 × 50 mm) using a gradient of 98% mobile phase A (100 mM HFIP and 5 mM TEA in H2O) and 2% mobile phase B (MeOH) to 40% mobile phase A and 60% mobile phase B in 8 min. The data were processed and deconvoluted using the Bruker DataAnalysis software v4.1. MS data are summarized in Table S3.
Supplementary Material
Acknowledgments
We thank the Canadian Macromolecular Crystallography Facility staff for X-ray data collection (Canadian Light Source); Dr. A. Wahba for LC-MS (McGill University); A. Gorelik, Dr. N. Siddiqui, and Dr. M. W. Górna for feedback and reading of the manuscript; A. Gorelik and K. Illes for Sf9 expression; and P. Sénéchal for assistance with translation assays. Support was provided by the Canadian Institutes of Health Research Strategic Training Initiative in Chemical Biology and the Natural Sciences and Engineering Research Council CREATE Training Program in Bionanomachines (Y.M.A.); a Canada Research Chair (B.N.); Canadian Institutes of Health Research Grants MOP-133535 (to B.N.) and MOP-115126 (to J.P.); a Discovery grant from the Natural Sciences and Engineering Research Council of Canada (to M.J.D.); the McGill Canadian Institutes of Health Research Drug Development Training Program (S.M.-M.); and the Max-Planck Free Floater program, the European Research Council (StG iVIP, 311339), InfectERA (ERASE), and the German Federal Ministry of Education and Research (A.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org/pdb/home/home.do (PDB ID codes 5UDI, 5UDJ, 5UDK, and 5UDL).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612444114/-/DCSupplemental.
References
- 1.Pelletier J, Graff J, Ruggero D, Sonenberg N. Targeting the eIF4F translation initiation complex: A critical nexus for cancer development. Cancer Res. 2015;75(2):250–263. doi: 10.1158/0008-5472.CAN-14-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bélanger F, Stepinski J, Darzynkiewicz E, Pelletier J. Characterization of hMTr1, a human Cap1 2'-O-ribose methyltransferase. J Biol Chem. 2010;285(43):33037–33044. doi: 10.1074/jbc.M110.155283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Werner M, et al. 2′-O-ribose methylation of cap2 in human: Function and evolution in a horizontally mobile family. Nucleic Acids Res. 2011;39(11):4756–4768. doi: 10.1093/nar/gkr038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daffis S, et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468(7322):452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Züst R, et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12(2):137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Decroly E, Ferron F, Lescar J, Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol. 2011;10(1):51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hyde JL, Diamond MS. Innate immune restriction and antagonism of viral RNA lacking2׳′-O methylation. Virology. 2015;479-480:66–74. doi: 10.1016/j.virol.2015.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuberth-Wagner C, et al. A conserved histidine in the RNA sensor RIG-I controls immune tolerance to N1-2'O-methylated self RNA. Immunity. 2015;43(1):41–51. doi: 10.1016/j.immuni.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devarkar SC, et al. Structural basis for m7G recognition and 2′-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc Natl Acad Sci USA. 2016;113(3):596–601. doi: 10.1073/pnas.1515152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fensterl V, Chattopadhyay S, Sen GC. No love lost between viruses and interferons. Annu Rev Virol. 2015;2(1):549–572. doi: 10.1146/annurev-virology-100114-055249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fensterl V, Sen GC. Interferon-induced Ifit proteins: Their role in viral pathogenesis. J Virol. 2015;89(5):2462–2468. doi: 10.1128/JVI.02744-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daugherty MD, Schaller AM, Geballe AP, Malik HS. Evolution-guided functional analyses reveal diverse antiviral specificities encoded by IFIT1 genes in mammals. elife. 2016;5:60. doi: 10.7554/eLife.14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abbas YM, Pichlmair A, Górna MW, Superti-Furga G, Nagar B. Structural basis for viral 5′-PPP-RNA recognition by human IFIT proteins. Nature. 2013;494(7435):60–64. doi: 10.1038/nature11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katibah GE, et al. tRNA binding, structure, and localization of the human interferon-induced protein IFIT5. Mol Cell. 2013;49(4):743–750. doi: 10.1016/j.molcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z, et al. Crystal structure of ISG54 reveals a novel RNA binding structure and potential functional mechanisms. Cell Res. 2012;22(9):1328–1338. doi: 10.1038/cr.2012.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng F, et al. Crystal structure and nucleotide selectivity of human IFIT5/ISG58. Cell Res. 2013;23(8):1055–1058. doi: 10.1038/cr.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pichlmair A, et al. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat Immunol. 2011;12(7):624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- 18.Habjan M, et al. Sequestration by IFIT1 impairs translation of 2′O-unmethylated capped RNA. PLoS Pathog. 2013;9(10):e1003663. doi: 10.1371/journal.ppat.1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar P, et al. Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5′-terminal regions of cap0-, cap1- and 5'ppp- mRNAs. Nucleic Acids Res. 2014;42(5):3228–3245. doi: 10.1093/nar/gkt1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimura T, et al. Ifit1 inhibits Japanese encephalitis virus replication through binding to 5′ capped 2′-O unmethylated RNA. J Virol. 2013;87(18):9997–10003. doi: 10.1128/JVI.00883-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyde JL, et al. A viral RNA structural element alters host recognition of nonself RNA. Science. 2014;343(6172):783–787. doi: 10.1126/science.1248465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reynaud JM, et al. IFIT1 differentially interferes with translation and replication of alphavirus genomes and promotes induction of type I interferon. PLoS Pathog. 2015;11(4):e1004863. doi: 10.1371/journal.ppat.1004863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menachery VD, et al. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2′-o-methyltransferase activity. J Virol. 2014;88(8):4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Züst R, et al. Rational design of a live attenuated dengue vaccine: 2′-o-methyltransferase mutants are highly attenuated and immunogenic in mice and macaques. PLoS Pathog. 2013;9(8):e1003521. doi: 10.1371/journal.ppat.1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szretter KJ, et al. 2′-O methylation of the viral mRNA cap by West Nile virus evades ifit1-dependent and -independent mechanisms of host restriction in vivo. PLoS Pathog. 2012;8(5):e1002698. doi: 10.1371/journal.ppat.1002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinto AK, et al. Human and murine IFIT1 proteins do not restrict infection of negative-sense RNA viruses of the orthomyxoviridae, bunyaviridae, and filoviridae families. J Virol. 2015;89(18):9465–9476. doi: 10.1128/JVI.00996-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novac O, Guenier AS, Pelletier J. Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res. 2004;32(3):902–915. doi: 10.1093/nar/gkh235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amrani N, Ghosh S, Mangus DA, Jacobson A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature. 2008;453(7199):1276–1280. doi: 10.1038/nature06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo J, Hui DJ, Merrick WC, Sen GC. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 2000;19(24):6891–6899. doi: 10.1093/emboj/19.24.6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C, et al. Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J Virol. 2003;77(7):3898–3912. doi: 10.1128/JVI.77.7.3898-3912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quiocho FA, Hu G, Gershon PD. Structural basis of mRNA cap recognition by proteins. Curr Opin Struct Biol. 2000;10(1):78–86. doi: 10.1016/s0959-440x(99)00053-6. [DOI] [PubMed] [Google Scholar]
- 32.Dougherty DA. The cation-π interaction. Acc Chem Res. 2013;46(4):885–893. doi: 10.1021/ar300265y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Gregorio E, Preiss T, Hentze MW. Translational activation of uncapped mRNAs by the central part of human eIF4G is 5′ end-dependent. RNA. 1998;4(7):828–836. doi: 10.1017/s1355838298980372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89(6):951–961. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 35.Mazza C, Segref A, Mattaj IW, Cusack S. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 2002;21(20):5548–5557. doi: 10.1093/emboj/cdf538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hodel AE, Gershon PD, Shi X, Wang SM, Quiocho FA. Specific protein recognition of an mRNA cap through its alkylated base. Nat Struct Biol. 1997;4(5):350–354. doi: 10.1038/nsb0597-350. [DOI] [PubMed] [Google Scholar]
- 37.Fechter P, Brownlee GG. Recognition of mRNA cap structures by viral and cellular proteins. J Gen Virol. 2005;86(Pt 5):1239–1249. doi: 10.1099/vir.0.80755-0. [DOI] [PubMed] [Google Scholar]
- 38.Hu G, Tsai AL, Quiocho FA. Insertion of an N7-methylguanine mRNA cap between two coplanar aromatic residues of a cap-binding protein is fast and selective for a positively charged cap. J Biol Chem. 2003;278(51):51515–51520. doi: 10.1074/jbc.M307654200. [DOI] [PubMed] [Google Scholar]
- 39.D’Ascenzo L, Leonarski F, Vicens Q, Auffinger P. ‘Z-DNA like’ fragments in RNA: A recurring structural motif with implications for folding, RNA/protein recognition and immune response. Nucleic Acids Res. 2016;44(12):5944–5956. doi: 10.1093/nar/gkw388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banerjee AK. 5′-Terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol Rev. 1980;44(2):175–205. doi: 10.1128/mr.44.2.175-205.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perry RP, Kelley DE. Kinetics of formation of 5′ terminal caps in mRNA. Cell. 1976;8(3):433–442. doi: 10.1016/0092-8674(76)90156-2. [DOI] [PubMed] [Google Scholar]
- 42.Wei CM, Moss B. Methylated nucleotides block 5′-terminus of vaccinia virus messenger RNA. Proc Natl Acad Sci USA. 1975;72(1):318–322. doi: 10.1073/pnas.72.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cleaves GR, Dubin DT. Methylation status of intracellular dengue type 2 40 S RNA. Virology. 1979;96(1):159–165. doi: 10.1016/0042-6822(79)90181-8. [DOI] [PubMed] [Google Scholar]
- 44.Young DF, et al. Human IFIT1 inhibits mRNA translation of rubulaviruses but not other members of the paramyxoviridae family. J Virol. 2016;90(20):9446–9456. doi: 10.1128/JVI.01056-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maquat LE, Tarn W-Y, Isken O. The pioneer round of translation: Features and functions. Cell. 2010;142(3):368–374. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei C, Gershowitz A, Moss B. N6, O2′-dimethyladenosine a novel methylated ribonucleoside next to the 5′ terminal of animal cell and virus mRNAs. Nature. 1975;257(5523):251–253. doi: 10.1038/257251a0. [DOI] [PubMed] [Google Scholar]
- 47.Wei CM, Gershowitz A, Moss B. 5′-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA. Biochemistry. 1976;15(2):397–401. doi: 10.1021/bi00647a024. [DOI] [PubMed] [Google Scholar]
- 48.Rose JK. Heterogneeous 5′-terminal structures occur on vesicular stomatitis virus mRNAs. J Biol Chem. 1975;250(20):8098–8104. [PubMed] [Google Scholar]
- 49.Liang B, et al. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell. 2015;162(2):314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pavelka A, et al. CAVER: Algorithms for analyzing dynamics of tunnels in macromolecules. IEEE/ACM Trans Comput Biol Bioinformatics. 2016;13(3):505–517. doi: 10.1109/TCBB.2015.2459680. [DOI] [PubMed] [Google Scholar]
- 51.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5(5):865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 52.Thillier Y, et al. Synthesis of 5′ cap-0 and cap-1 RNAs using solid-phase chemistry coupled with enzymatic methylation by human (guanine-N7)-methyl transferase. RNA. 2012;18(4):856–868. doi: 10.1261/rna.030932.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berger I, Fitzgerald DJ, Richmond TJ. Baculovirus expression system for heterologous multiprotein complexes. Nat Biotechnol. 2004;22(12):1583–1587. doi: 10.1038/nbt1036. [DOI] [PubMed] [Google Scholar]
- 54.Grochulski P, Fodje MN, Gorin J, Labiuk SL, Berg R. Beamline 08ID-1, the prime beamline of the Canadian Macromolecular Crystallography Facility. J Synchrotron Radiat. 2011;18(Pt 4):681–684. doi: 10.1107/S0909049511019431. [DOI] [PubMed] [Google Scholar]
- 55.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 56.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moriarty NW, Grosse-Kunstleve RW, Adams PD. Electronic Ligand Builder and Optimization Workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 10):1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42(Web Server issue):W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schneider TD, Stephens RM. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18(20):6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hall MP, Ho CK. Functional characterization of a 48 kDa Trypanosoma brucei cap 2 RNA methyltransferase. Nucleic Acids Res. 2006;34(19):5594–5602. doi: 10.1093/nar/gkl573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robert F, et al. Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2.GTP.Met-tRNA(i)(Met) ternary complex availability. Mol Biol Cell. 2006;17(11):4632–4644. doi: 10.1091/mbc.E06-06-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Svitkin YV, Sonenberg N. 2004. An efficient system for cap- and poly(A)-dependent translation in vitro. mRNA Processing and Metabolism, Methods in Molecular Biology, ed Schoenberg DR (Humana, Totowa, NJ), Vol 257, pp 155–170.
- 69.Zlatev I, et al. Efficient solid-phase chemical synthesis of 5′-triphosphates of DNA, RNA, and their analogues. Org Lett. 2010;12(10):2190–2193. doi: 10.1021/ol1004214. [DOI] [PubMed] [Google Scholar]
- 70.Zlatev I, Manoharan M, Vasseur J-J, Morvan F. 2012. Solid-phase chemical synthesis of 5′-triphosphate DNA, RNA, and chemically modified oligonucleotides. Curr Protoc Nucleic Acid Chem Chapter 1:Unit1.28. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.