Significance
Phosphorylation catalyzed by protein kinases governs many aspects of cellular behavior. Apoptosis signal-regulating kinases (ASK1–3) trigger responses to stress, but the structural basis of their regulation remains unclear. Here, we show that a domain directly adjacent to the ASK1 kinase domain promotes activity of ASK1 on a key substrate and also orients an additional ASK1 domain nearby to suppress kinase activity. The structure of this regulatory domain appears to be shared by all ASK kinases and provides a versatile mechanism to control ASK activity in response to various stress stimuli.
Keywords: ASK1, MAP kinase, scaffolding, signaling, MKK6
Abstract
Apoptosis signal-regulating kinases (ASK1–3) are apical kinases of the p38 and JNK MAP kinase pathways. They are activated by diverse stress stimuli, including reactive oxygen species, cytokines, and osmotic stress; however, a molecular understanding of how ASK proteins are controlled remains obscure. Here, we report a biochemical analysis of the ASK1 kinase domain in conjunction with its N-terminal thioredoxin-binding domain, along with a central regulatory region that links the two. We show that in solution the central regulatory region mediates a compact arrangement of the kinase and thioredoxin-binding domains and the central regulatory region actively primes MKK6, a key ASK1 substrate, for phosphorylation. The crystal structure of the central regulatory region reveals an unusually compact tetratricopeptide repeat (TPR) region capped by a cryptic pleckstrin homology domain. Biochemical assays show that both a conserved surface on the pleckstrin homology domain and an intact TPR region are required for ASK1 activity. We propose a model in which the central regulatory region promotes ASK1 activity via its pleckstrin homology domain but also facilitates ASK1 autoinhibition by bringing the thioredoxin-binding and kinase domains into close proximity. Such an architecture provides a mechanism for control of ASK-type kinases by diverse activators and inhibitors and demonstrates an unexpected level of autoregulatory scaffolding in mammalian stress-activated MAP kinase signaling.
Mitogen-activated protein (MAP) kinase cascades transmit signals from membrane-associated receptors to intracellular targets to effect changes in cellular behavior. They form a hierarchical system in which activated upstream kinases (MAP3Ks) phosphorylate intermediate MAP kinase kinases (MAP2Ks), which in turn phosphorylate terminal MAP kinases, primarily ERK, p38, and JNK and their isoforms (1). Extensive studies have focused on the activation of RAS-RAF-MEK upstream in the ERK pathway and provided fertile ground for the discovery of new therapeutics (2). In contrast to the ERK pathway, which primarily promotes cellular proliferation, JNK and p38 phosphorylate a range of substrates to promote inflammation and cell death (1, 3). In addition, cross-regulation among the p38, JNK, and ERK pathways is important for the efficacy of various cancer therapies that are in use or in development (4, 5). Molecular details on the more diverse upstream regulation of the p38 and JNK pathways are currently less clear, however.
Apoptosis signal-regulating kinases (ASK1–3) are MAP3Ks that trigger cellular responses to redox stress and inflammatory cytokines (6, 7) and play vital roles in innate immunity and viral infection (8–11). When activated, ASK1–3 activate JNK and p38 via phosphorylation of MAP2Ks (MKK3/4/6/7) (12). The key initiator role of ASK1–3 in this pathway means that either too much or too little ASK activity can have pathological effects. For instance, inhibiting ASK1 is beneficial against gastric cancer (13, 14), but inactivating mutations in ASK1 contribute to the development of melanoma (15, 16). In addition, ASK1 inhibitors have shown promise for treatment in mouse models of amyotrophic lateral sclerosis, highlighting it as a critical factor modulating cellular survival (17).
Initiator signaling kinases such as MAP3Ks are often regulated by oligomerization and regulatory domains, rather than solely by phosphorylation (18, 19). This is especially true of ASK1–3, which share a conserved architecture in which the central kinase domain is flanked on either side by additional domains, and multimeric association appears to be crucial to the activity of these domains (Fig. 1A). The active signaling form of ASK1 in cells is thought to be an oligomer often referred to as the “ASK signalosome,” which also contains ASK2 and ASK3 (20-22). To the N terminus of the kinase domain lie several regions with regulatory roles but poorly understood structures. Separate regions have been proposed to bind to thioredoxin and TNF receptor-associated factors (TRAFs) (6, 7, 19, 23), which regulate the response of ASK1 to cytokines (Fig. 1A). The N-terminal region of ASK1 also has been implicated in binding CIB1 to detect Ca2+-based stress signaling, and in binding Fbxo21 to trigger innate antiviral signaling, among other protein–protein interactions (24–26). The region C terminal to the kinase is less well studied, but contains a 14-3-3 protein-binding site housed within a predicted disordered sequence (27, 28), followed by a region proposed to promote constitutive oligomerization of ASK1 (26). This C-terminal oligomerization region of ASK1 also has the capacity to directly bind to HIV Vif-1 and promote the innate antiviral response by APOBEC3G (8).
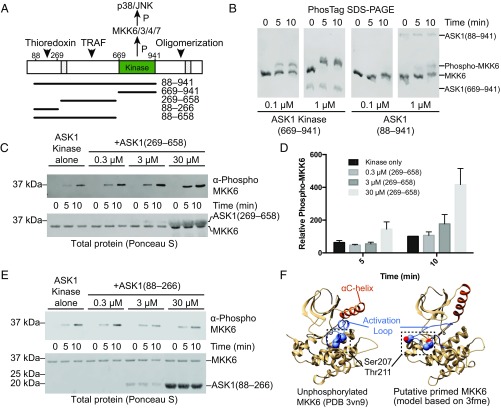
Fig. 1.
N-terminal regulatory domains of ASK1. (A) Schematic representation of ASK1 domains, with functional regions indicated, and previously proposed NCC and C-terminal coiled-coil regions hatched. Constructs used in this work are indicated below. Example purified proteins from each of these constructs are shown in Fig. S1A. (B) Phos-tag SDS/PAGE comparing phosphorylation of kinase dead MKK6 by ASK1 kinase domain (669–941), and ASK1(88–941) at matched concentrations. (C) MKK6 phosphorylation by 0.01 μM ASK1 kinase domain with increasing concentrations of ASK1(269–658) added. MKK6 was held constant (3 μM), and phosphorylation was monitored by Western blot analysis. Total protein transferred to membranes visualized by staining membrane with Ponceau S (shown below). (D) Quantitation of the independent triplicate experiments shown in C. Each band was normalized to the band intensity of the kinase-alone 10-min time point for that experiment. Mean values are plotted with error bars representing the SEM. (E) MKK6 phosphorylation by 0.01 μM ASK1 kinase domain with increasing concentrations of ASK1(88–266) added. MKK6 was held constant (3 μM), and phosphorylation was monitored by Western blot analysis. Total protein transferred to membranes visualized by staining membrane with Ponceau S (shown below). (F) Potential model of MKK6 priming. (Left) Crystal structure of unphosphorylated MKK6 (PDB ID code 3VN9), with the αC helix in orange, the activation loop in blue, and buried phosphorylation target residues shown as spheres. (Right) A model of MKK6 based on PDB 3fme, which shows a markedly different αC-helix conformation and has a disordered activation loop. In this figure, the loop was modeled using MODELER (76).
The prevailing model of ASK1 regulation is that constitutive oligomerization through the C terminus works in partnership with transient protein–protein interactions mediated by regions N terminal to the kinase domain. Regulatory factors, such as thioredoxin, associate with the N-terminal thioredoxin-binding domain to negatively regulate activity. Under conditions of redox stress, thioredoxin dissociates with and TRAF proteins associate with the region between the thioredoxin-binding domain and kinase domain, promoting ASK activation and kinase activity (19). There is some debate about the exact role of thioredoxin in negatively regulating ASK1 activity, with reports that the ASK–thioredoxin association is either dependent on or independent of thioredoxin oxidoreductase activity and disulfide bonds (6, 29–31).
Relatively little is known about the structural basis of ASK1 regulation, and thus previous studies necessarily have relied on prediction and deletion-based analysis. Although such approaches have identified various important regions for regulation of ASK proteins, the lack of atomic resolution data still confounds our understanding of how ASK proteins respond to diverse stimuli. For instance, very little is known about how thioredoxin or TRAF proteins actually might influence the recruitment of substrates to the ASK signalosome, or control the kinase activity of ASK1 on its substrate MAP2Ks. Here we present the first structure of a regulatory domain of ASK1, that of the central region that links the thioredoxin-binding and kinase domains of ASK1. This so-called “domain of unknown function” (PFAM domain DUF4071) corresponds to the region proposed to associate with TRAF proteins during ASK1 activation. The crystal structure reveals a surprisingly compact fold, with core features that are highly conserved in all human ASK proteins and in ASK orthologs throughout metazoans. Our biochemical and biophysical analyses reveal conserved residues that regulate ASK1 activity in both positive and negative manners, and show that the compact fold of the central regulatory region is crucial for bringing the thioredoxin-binding and kinase domains into close proximity and priming MAP2K substrates for phosphorylation. This model provides a structural template on which to interpret various proposed mechanisms of ASK kinase regulation by different binding partners.
Results
The ASK1 Central Regulatory Region Promotes MKK6 Phosphorylation.
The N-terminal region of ASK1 has been proposed to interact with various partners to regulate ASK kinase activity (Fig. 1A). To develop a quantitative system to analyze ASK1 activity, we used in vitro kinase assays and Phos-tag SDS/PAGE to compare the phosphorylation of kinase dead MKK6 by either the isolated ASK1 kinase domain or ASK1(88–941), which contains the N-terminal thioredoxin-binding domain, a central regulatory region, and the kinase domain (Fig. 1A). In Phos-tag SDS/PAGE, phosphorylated proteins have reduced mobility compared with unphosphorylated protein, and thus can be readily visualized (32). In this system, both the isolated ASK1 kinase domain and ASK1(88–941) were able to catalyze phosphorylation of MKK6, but activity of ASK1(88–941) was reduced by at least 10-fold (Fig. 1B). This observation supports previous reports that the N-terminal region of ASK1 suppresses kinase activity (19), and shows that a reconstituted in vitro system is a useful tool for detailed analysis of ASK1 regulation.
To gain greater insight into the role of the N-terminal portions of ASK1, we designed further constructs based on secondary structure prediction. These constructs encompassed residues 88–266 (the thioredoxin-binding domain) and residues 269–658 (the central regulatory region) (Fig. 1A). Following expression in Escherichia coli and purification to homogeneity (Fig. S1A), we tested how these additional domains affected ASK1 kinase activity in trans. In these experiments, MKK6 phosphorylation was monitored using Western blot analysis, because phospho-MKK6 comigrated with the central regulatory region on Phos-tag SDS/PAGE. Surprisingly, activity of the wild-type (WT) ASK1 kinase domain was greatly enhanced by the addition of 30 μM ASK1(269–658) (Fig. 1 C and D). We did not observe any equivalent enhancement or inhibition of MKK6 phosphorylation by the thioredoxin-binding domain of ASK1 when present in similar concentrations (Fig. 1E).
Fig. S1.
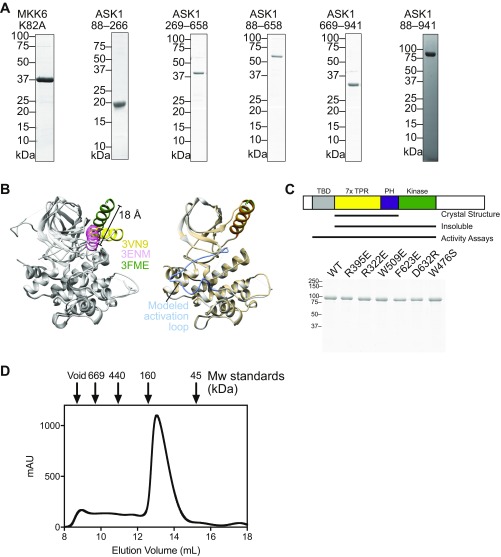
(A) Example purified sample proteins from each of the constructs used in this work. (B) Illustration of MKK6 conformational flexibility. (Left) An overlay of the three crystal structures of MKK6 currently available in the protein databank. All proteins are colored gray, with respective αC helices in yellow (2VN9), pink (3ENM), and green (3FME). The distance between the N terminus of the αC helix in 3ENM and 3FME is indicated. An illustrative model was created using Modeler to model in the disordered activation loop of structure 3FME. (Right) A comparison of 3FME with the Modeler output, with the activation loop indicated. The Modeler model is shown in Fig. 1F. (C) Purified point mutants of ASK1(88–941) used for the assays shown in Figs. 3C and 4B. A schematic summarizing the constructs is shown above, indicating that the construct containing only the central regulatory region and kinase was insoluble and so could not be tested in activity assays. (D) Preparative size-exclusion chromatography of ASK1(88–941) with molecular weight standards indicated. The majority of ASK1 protein elutes later than the 160-kDa marker, consistent with it behaving as a (∼100-kDa) monomeric species. Peak fractions elute at a concentration of ∼40 μM based on A280.
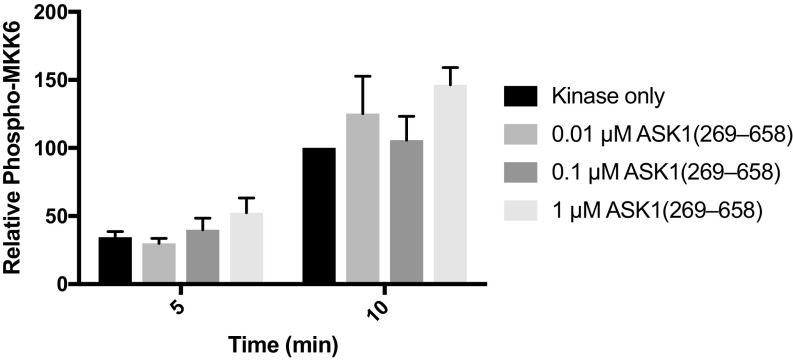
Although these results are consistent with published experiments showing that ASK1 lacking the thioredoxin-binding domain is more active than full-length ASK1 in cells (19), it has not previously been suggested that the central regulatory region is capable of stimulating ASK1 kinase activity in trans. In our simplified assay system, there are two potential mechanisms by which the central regulatory region could enhance MKK6 phosphorylation: allosterically activating the ASK1 kinase domain or priming the MKK6 substrate for phosphorylation. At concentrations of the central regulatory region up to 100-fold greater than ASK1 kinase (0.01 μM) but well below substrate MKK6 levels (3 μM), there was no significant rate enhancement (Fig. S2). Such a dose response where excess levels of the ASK1 central regulatory region relative to the substrate, rather than active kinase, are required is most consistent with the idea that the central regulatory region acts by binding to the substrate, MKK6. Attempts to investigate activity of the central regulatory region and kinase domain in one polypeptide, without the thioredoxin-binding domain, were hampered by the fact that a construct comprising residues 269–941 was completely insoluble when expressed in E. coli.
Fig. S2.
Relates to Fig. 1C. MKK6 phosphorylation by 0.01 μM ASK1 kinase domain with concentrations of ASK1(269–658) substoichiometric to the substrate (MKK6), but up to 100-fold excess to the ASK1 kinase domain. MKK6 is held constant (3 μM), and phosphorylation is monitored by Western blot analysis. Quantitation of independent triplicate experiments is shown. Each band was normalized to the band intensity of the kinase alone at the 10-min time point for that experiment. Mean values are plotted, with error bars representing the SEM.
The foregoing experiments demonstrate two interesting concepts. First, a region outside of the ASK1 kinase domain can associate with downstream substrate kinases, thereby acting as a scaffold for substrate recruitment. Second, because rate enhancement occurs when the kinase and central regulatory region are on separate polypeptides, the central regulatory region actively promotes a state of MKK6 that is primed for phosphorylation. In offering an explanation of how this might occur, we surveyed the available structures of MKK6. The three available crystal structures exhibit three different conformations of the activation loop, and movement of up to 18 Å in the N-terminal end of the αC helix, indicating that it is relatively flexible (Fig. S1B). Two of these structures contain phosphomimetic mutations (PDB ID codes 3FME and 3ENM), but the structure of unphosphorylated MKK6 (PDB ID code 3VN9) shows that the phosphorylation target residues in the activation loop (Ser207/Thr211) are buried, and that the activation loop is stabilized by the αC-helix (Fig. 1F) (33).
Structures of MKK6 bearing phosphomimetic activation loop mutations exhibit notably different positions in the αC-helix (34) (PDB ID code 3FME), which lead to different conformations of the activation loop (Fig. S1B). Thus, we propose that by maneuvering the αC-helix, it is plausible that ASK1(269–658) could manipulate access to the phosphorylation target residues and thereby “prime” MKK6 for phosphorylation (Fig. 1F). This concept provides another layer of complexity to the thoroughly investigated kinetics of MKK6 dual phosphorylation by ASK1 (35, 36), and raises the question of how the central regulatory region plays such an active scaffolding role.
Structure of the Central Regulatory Region of ASK1.
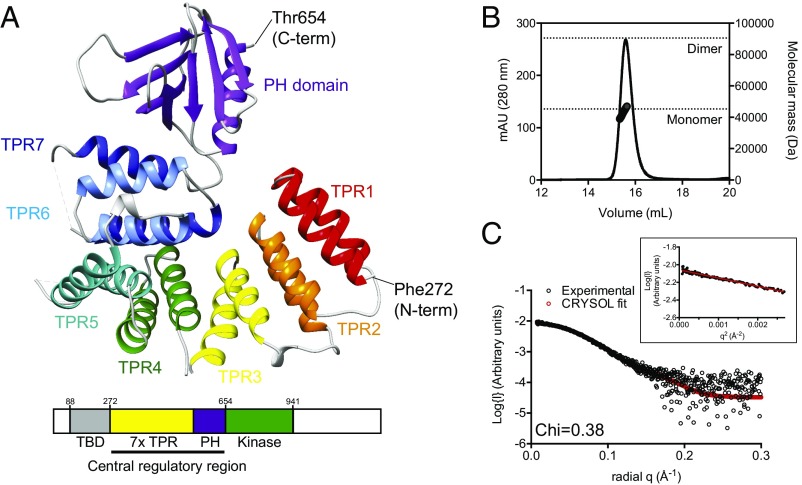
To gain insight into how the central regulatory region might prime MKK6 for phosphorylation, we expressed and purified human ASK1 residues 269–658 from E. coli, crystallized it, and solved its crystal structure to a resolution of 2.1 Å (Fig. 2A and Table 1). The final structure contains two molecules in the asymmetric unit, which are essentially identical and share an rmsd of 0.04 Å. Given the various reports of multimerization in ASK1 regulation, we also tested whether any of the crystal contacts that we observed could play a role in the formation of ASK(269–658) dimers. Neither size exclusion chromatography (SEC) coupled to both multiple-angle laser light scattering (SEC-MALLS) nor small-angle X-ray scattering (SEC-SAXS) indicated a tendency of ASK1(269–658) to form multimers in solution at the concentrations tested (Fig. 2 B and C and Table 2).
Fig. 2.
Structure of the ASK1 central regulatory region. (A) Cartoon representation of the crystal structure of ASK1(269–658). Individual TPRs are labeled, and the pleckstrin homology domain is in purple. A modified domain schematic (used hereinafter) is shown below. (B) SEC-MALLS of ASK1(269–658), with theoretical molecular weights of a putative monomer and dimer species indicated. (C) Overlay of experimental SAXS data (black circles) and scattering profile calculated using CRYSOL for the crystal structure of ASK1(269–658). Agreement between the experimental data and calculated scatter pattern is signified by χ = 0.379. (Inset) Guinier analysis.
Table 1.
Crystallographic data
| Variable | ASK1(269–658) SeMet | ASK1(269–658) |
| Beamline | AS-MX2 | AS-MX2 |
| Wavelength, Å | 0.9793 | 0.979 |
| Resolution (outer shell), Å | 47.16–2.88 (3.03–2.88) | 47.1–2.1 (2.16–2.10) |
| Space group | P1211 | P1211 |
| Unit cell parameters | a = 74.12 Å | a =74.23 Å |
| b = 56.92 Å | b = 57.12 Å | |
| c = 103.44 Å | c = 103.57 Å | |
| α = 90° | α = 90° | |
| β = 105.1° | β = 104.9 ° | |
| γ = 90° | γ = 90° | |
| Rmerge (outer shell) | 0.142 (0.367) | 0.125 (0.620) |
| Rpim (outer shell) | 0.092 (0.250) | 0.097 (0.483) |
| Mean I/σI (outer shell) | 10.7 (4.2) | 8.3 (2.3) |
| Completeness (outer shell) | 99.3 (95.5) | 99.8 (97.6) |
| Multiplicity (outer shell) | 6.0 (5.8) | 5.0 (5.0) |
| Total no. of reflections | 113052 | 244014 |
| No. of unique reflections | 18933 | 49197 |
| Mean (I) half-set correlation CC(1/2) (outer shell) | 0.989 (0.899) | 0.994 (0.788) |
| Wilson B-factor, Å2 | 22.3 | 17.7 |
| Refinement statistics | ||
| Rcryst | 0.233 (0.303) | |
| Rfree | 0.267 (0.335) | |
| rmsd for bonds, Å | 0.012 | |
| rmsd for angles, ° | 1.435 | |
| rmsd for chiral volume, Å3 | 0.086 | |
| No. of protein atoms | 6101 | |
| No. of solvent atoms | 349 | |
| Average B-factor overall, Å2 | 16.6 | |
| Average main chain B-factor, Å2 | 14.2 | |
| Average side chain and solvent B-factor, Å2 | 18.78 | |
| Ramachandran plot statistics, % | ||
| Favored regions | 98 | |
| Allowed regions | 2 | |
| Outliers | 0 | |
| PDB ID code | 5ULM |
Table 2.
SAXS data collection and analysis statistics
| Variable | ASK1(269–658) | ASK1(88–658) | ASK1 (88-941) | W476S ASK1(88–941) |
| Data collection parameters | ||||
| Instrument | Australian Synchrotron, SAXS/WAXS beamline | |||
| Beam geometry | 120-μm point source | |||
| Wavelength, Å | 1.033 | |||
| Exposure time, s | 2 | |||
| Temperature, K | 285 | |||
| q range, Å−1* | 0.0048–0.290 | 0.0057–0.334 | 0.0057–0.334 | 0.0057–0.334 |
| Protein concentration | 50 μL of 7.7 mg/mL protein via inline gel filtration chromatography | 50 μL of 9.7 mg/mL protein via inline gel filtration chromatography | 50 μL of 9.3 mg/mL protein via inline gel filtration chromatography | 50 μL of 11.5 mg/mL protein via inline gel filtration chromatography |
| Structural parameters | ||||
| I(0), cm−1, from P(r) | 0.0087 ± 0.0001 | 0.0274 ± 0.0002 | 0.0239 ± 0.0001 | 0.0182 ± 0.0001 |
| Rg, Å, from P(r) | 25.46 ± 0.21 | 32.54 ± 0.19 | 47.01 ± 0.25 | 44.68 ± 0.27 |
| Dmax, Å | 75 | 100 | 145 | 130 |
| I(0), cm−1, from Guinier | 0.0087 ± 0.0001 | 0.0273 ± 0.0003 | 0.0243 ± 0.0002 | 0.0187 ± 0.0003 |
| Rg, Å, from Guinier | 25.10 ± 0.49 | 32.10 ± 0.53 | 47.30 ± 0.53 | 45.9 ± 1.03 |
| Software used | ||||
| Primary data reduction | Scatterbrain (Australian Synchrotron) | |||
| Data processing | PRIMUS, GNOM | |||
| Computation of model intensities | CRYSOL | |||
| Rigid body modeling 3D graphical representations | BUNCH Chimera |
q is the magnitude of the scattering vector, which is related to the scattering angle (2θ) and the wavelength (λ) as follows: q = (4π/λ)sinθ.
The overall structure of ASK1(269–658) encompasses an extended series of 14 helices, which form seven tetratricopeptide repeats (TPRs), followed by a pleckstrin homology domain that had not previously been predicted within ASK1 (Fig. 2A). To the best of our knowledge, the overall compact arrangement of TPRs capped by a pleckstrin homology domain has no close matches to previously solved structures. In contrast to the extended arrangements observed in many TPR proteins (37), the ASK1 TPR region progressively folds back on itself and forms a compact arrangement, with a close physical association between residues from helices that are significantly separated in sequence. Such a stable globular arrangement is supported by SAXS analyses showing that the ASK1(269–658) monomer crystal structure is highly representative of its solution behavior, with excellent agreement between the theoretical scatter pattern calculated from the crystal structure coordinates and the experimental scattering data of (χ = 0.379) (Fig. 2C and Table 2). Close interactions between residues that are well separated in sequence are centered around the first helix of TPR 7 (residues 508–524), which makes contact with constituent residues from helices of TPRs 2–7.
Although sequence-based predictions and experimental evidence have implicated an N-terminal coiled-coil (NCC) region as responsible for mediating interactions at the N terminus of ASK, we observed a surprisingly compact protein. Crucially, the crystallized ASK1 construct contains the predicted NCC region (residues 297–324) (19). The structure shows that residues 297–324 reside stably within TPRs 1 and 2, forming numerous interactions with surrounding TPR helices. Thus, although it is clearly important for ASK1 structure, the NCC is more accurately described as an integral part of the TPR domain and seems unlikely to directly mediate conventional coiled-coil type oligomerization.
The pleckstrin homology domain of ASK1 adopts the typical form of two antiparallel β-sheets followed by a C-terminal amphipathic helix, but lacks the tryptophan found within the terminal helix of most conventional pleckstrin homology domains (38). It is not uncommon for widely disparate sequences to produce the pleckstrin homology fold, and in ASK1 the lack of a locking tryptophan residue (which is Phe646 in ASK1) may explain why it had not been recognized previously. The interface between the base of the pleckstrin homology domain and the TPR region is highly hydrophobic and forms an extensive network of interactions with helices 12 and 14 from TPRs 6 and 7 (Fig. S3). The complementarity of these interactions, in conjunction with scattering data described above, suggests that the intimate association between the TPRs and pleckstrin homology domain of ASK1 is the stable form of ASK1 in solution. In line with this idea, attempts to express the isolated ASK1 pleckstrin homology domain in the absence of the TPR region yielded completely insoluble protein, in contrast to the solubility and stability of ASK1(269–658).
Fig. S3.
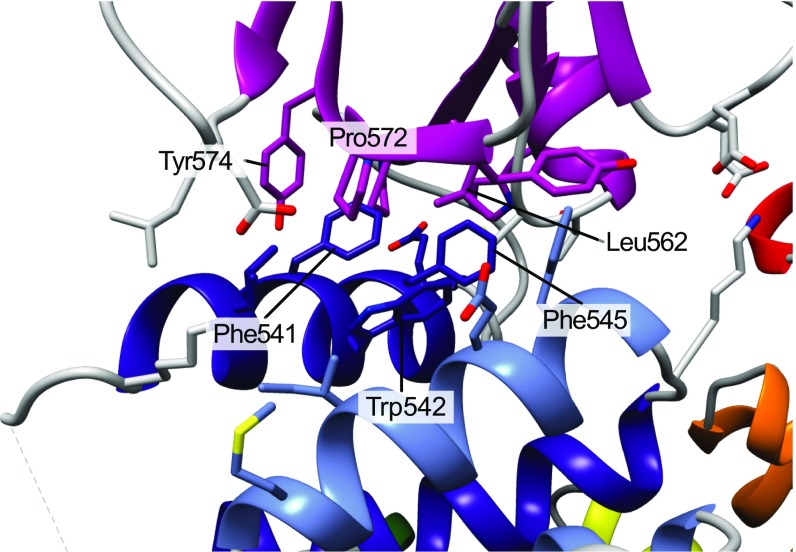
Detailed view of the interface between TPR repeats 6 and 7 and the pleckstrin homology domain. Hydrophobic residues at the core of the interface are indicated.
ASK1 Has a Cryptic Pleckstrin Homology Domain That Promotes Substrate Phosphorylation.
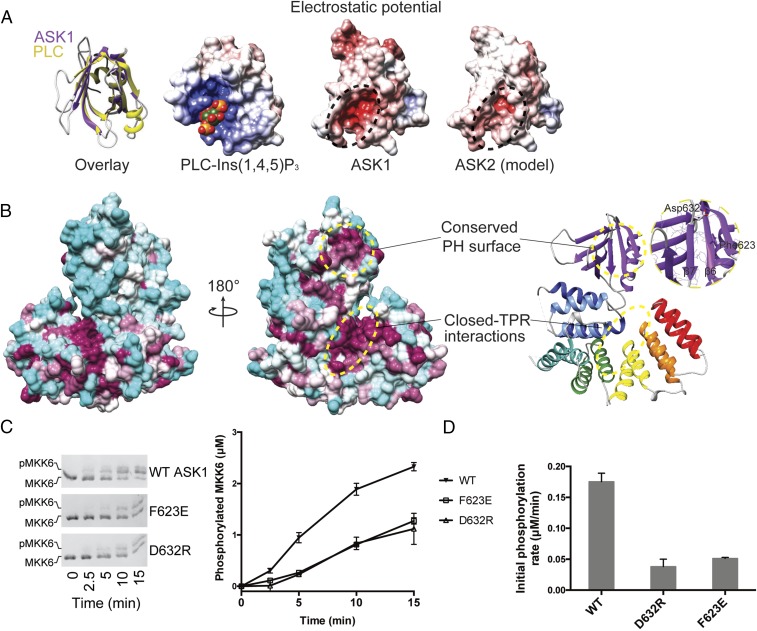
The yeast MAPK scaffolding protein Ste5 contains a pleckstrin homology domain that has been previously proposed to bind phospholipids, and is crucial for protecting the yeast MAPK pathway from inappropriate activation and localizing the activated scaffold to the plasma membrane (39, 40). Interestingly, neither the ASK1 pleckstrin homology domain nor a model of the equivalent region of ASK2 has a surface patch of increased positive charge, as is observed in bona fide phosphoinositide-binding pleckstrin homology domains, such as that from phospholipase C (Fig. 3A). In fact, the ASK1 pleckstrin homology cavity is notably negatively charged, as is the equivalent region of a homology model generated for the ASK2 pleckstrin homology domain. This is in line with genome-wide analysis in yeast showing that the majority of pleckstrin homology domains do not bind lipid head groups (41), and since their discovery, pleckstrin homology domains have been ascribed diverse roles in mediating protein–protein interactions (38).
Fig. 3.
The ASK1 pleckstrin homology domain mediates protein-protein interactions and activity. (A, Left) Superposition of the ASK1 pleckstrin homology domain (purple) with the canonical pleckstrin homology domain of phospholipase C (PLC, in yellow; PDB ID code 1MAI) (77). (A, Right) Three electrostatic surface representations calculated in APBS (78) for the pleckstrin homology domains of PLC bound to Ins(1,4,5)P3, ASK1, and a model of the same region of ASK2 generated using MODELER. (Conservation between ASK1 and ASK2 can be viewed in the alignment in Fig. S4.) (B) Surface representation of ASK1(269–658), color-coded according to the degree of conservation in the alignment in Fig. S4. The least conserved residues are in cyan; the most conserved, in maroon. Areas of high conservation are also indicated with circles. (Inset) Close-up view of the conserved pleckstrin homology surface. (C) Phos-tag SDS/PAGE monitoring MKK6 (3 μM) phosphorylation by ASK1(88–941) (1 μM) WT or indicated pleckstrin homology domain mutants. Quantitation of independent triplicate experiments is shown alongside as mean values, with error bars representing SEM. (D) Rates of MKK6 phosphorylation calculated over the first 5 min from C. Error bars represent the SD of the linear rate fit.
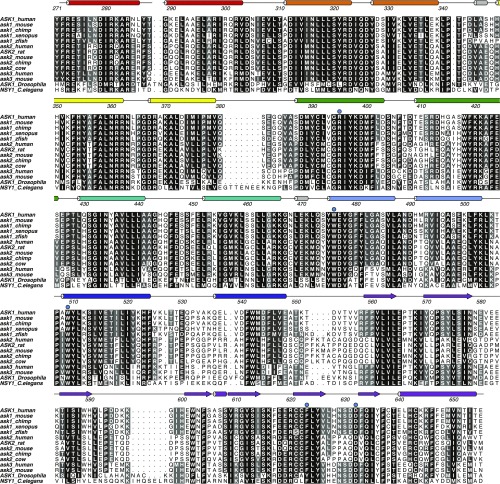
To identify a function for the ASK1 pleckstrin homology domain beyond phospholipid binding, we mapped the conservation of sequences from ASK1, ASK2, and ASK3 from diverse mammalian species, as well as sequences from dASK1 (Drosophila melanogaster) and NSY1 (Caenorhabditis elegans), onto the crystal structure of ASK1(269–658) (Fig. 3B and Fig. S4). This analysis uncovered two regions of high conservation within the central regulatory region of ASK1: residues at the core of the ASK1 closed TPR repeat region that folds back on itself, and a clear enrichment of conserved residues on the face of the ASK1 pleckstrin homology domain formed by β5–β7. The reverse side of the protein shows relatively little conservation (Fig. 3B).
Fig. S4.
Sequence alignment of ASK1–3 and orthologs from various species over the central regulatory domain, with the secondary structure indicated. Point mutants generated in this work are indicated above with a blue circle. The figure was generated using ALINE (79).
At the heart of the β5–β7 pleckstrin homology surface lie Phe623 from β6 and Asp632 from β7, which are invariant across the ASK homologs analyzed (Fig. 3B and Fig. S4). To experimentally verify the importance of these conserved residues, we created mutants in the ASK1(88–941) construct (Phe623Glu and Asp632Arg; Fig. S1C) and analyzed their ability to phosphorylate MKK6 using Phos-tag kinase assays. We hypothesized that if the conserved surface is important for autoinhibition (as observed in Fig. 1B), then the activity of mutants should be increased, whereas if the surface is important for priming MKK6 (Fig. 1C), then the Phe623Glu and Asp632Arg mutants should show decreased activity. Both mutants had markedly lower initial rates of MKK6 phosphorylation (Fig. 3 C and D), clearly supporting the hypothesis that the conserved pleckstrin homology surface plays an important positive role in facilitating MAP2K phosphorylation by ASK1. The position of the ASK1 pleckstrin homology domain directly adjacent to the kinase domain makes it an ideal location for transient docking of MAP2Ks for both localization and priming of the activation loop for phosphorylation.
Having established that the conserved pleckstrin homology surface is important for ASK1 activity on downstream substrates, we next sought to understand how the remainder of the conserved closed TPR facilitates signal control and reduced activity of ASK1(88–941) relative to the isolated kinase domain.
Closed TPR Interactions Facilitate ASK Kinase Regulation.
In the compact arrangement of the central regulatory region, its N and C termini are separated by only ∼50 Å (Fig. 2A). Based on previous reports showing that deleting the thioredoxin-binding domain leads to more active full-length ASK1, we hypothesized that a major role of the central regulatory region is to bring the thioredoxin-binding domain into close proximity to the kinase domain to inhibit its activity. Other possible effects of such an interaction may be to protect the β5–β7 pleckstrin homology surface and impede MAP2K recruitment and priming (Fig. 1).
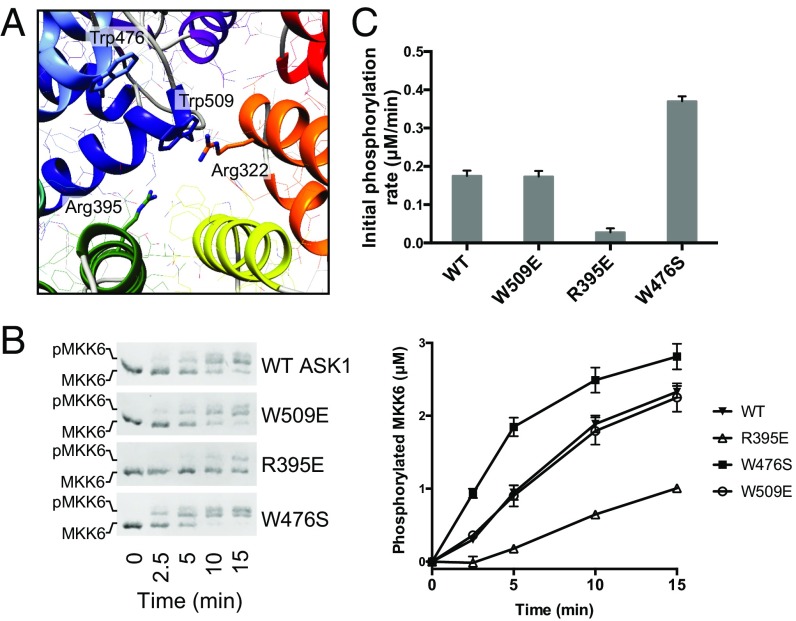
As described above, stabilizing interactions within the TPR region are centered around the first helix of TPR 7 (helix 13, residues 508–524), which makes contacts across the TPR fold. The most long-range of these contacts involve π-cation stacking between Trp509 from helix 13 and Arg322 in TPR 2 (Fig. 4A). Trp509 is also one of the most conserved residues across the sequence logo of diverse occurrences of DUF4071 in PFAM. To investigate the importance of the closed TPR interactions, we designed two mutants, one substituting Trp509 with glutamate and the other replacing Arg395 with glutamate. These mutants serve two different purposes. Trp509Glu could reasonably be expected to form a salt bridge with Arg322 and maintain the overall TPR architecture, but to disrupt ASK1 function if the closed TPR interaction is mobile and Trp509 takes on a different conformation in an active signaling form. Arg395Glu was designed as a disruptive mutant to destabilize the compact closed TPR structure. Analyzing both the time course and initial rates of MKK6 phosphorylation by these ASK1 variants showed that disruption of the closed TPR by Arg395Glu markedly reduced ASK1 activity, whereas Trp509Glu was indistinguishable from WT protein (Fig. 4 B and C). Based on these findings, we conclude that the closed TPR must remain intact to allow the full activity of ASK1(88–941).
Fig. 4.
TPR mutants activate and inhibit ASK1 activity. (A) Detailed view of the closed TPR region, with residues of interest subjected to mutagenesis shown as sticks and labeled. (B) Phos-tag SDS/PAGE monitoring MKK6 (3 μM) phosphorylation by ASK1(88–941) WT or indicated TPR mutants (1 μM). Quantitation of independent triplicate experiments is shown alongside as mean values, with error bars representing SEM. (C) Rates of MKK6 phosphorylation calculated over the initial 5 min. Error bars represent the SD of the linear rate fit.
Trp476 is another notable residue conserved within the ASK1 central domain. Trp476 is one of three invariant residues over the consensus definition of DUF4071, along with Trp509, and Trp542 (which is buried at the interface between the TPR region and the pleckstrin homology domain). In contrast to the latter two residues, which play clear roles in stabilizing the structure, Trp476 is unusually surface-exposed. It is located at the N-terminal end of TPR helix 11 (residues 475–488) and faces toward the center of the closed TPR region. To ascertain a function of Trp476, we mutated the residue to serine in the context of ASK1(88–941). Surprisingly, ASK1 W476S phosphorylated MKK6 more effectively than WT ASK1, with a roughly twofold higher initial rate (Fig. 4). This finding is consistent with Trp476 playing a role in stabilizing autoinhibitory interactions that suppress the activity of the kinase domain.
Interacting closed TPR residues (displayed in Fig. 4A) are remarkably well conserved among ASK-type kinases (Fig. S4). Namely, kinases including human ASK1, ASK2, and ASK3; Drosophila ASK1 and NSY1; and the C. elegans ASK homolog all maintain residues that mediate long-range TPR interactions, suggesting that this compact fold and function are highly conserved. Overall, these results, along with previous studies showing that deletion of residues 297–324 (the NCC) disrupts ASK1 regulation, show that the integrity of the TPR region is important for both function and regulation of ASK1 signaling. Whereas conformational changes cannot be discounted, it appears that a major role of the central closed TPR of ASK1 is to bring the kinase domain into relative proximity of the N-terminal (thioredoxin-binding) regulatory domain to mediate the regulation of kinase activity.
Architecture of the ASK Autoregulatory Scaffold in Solution.
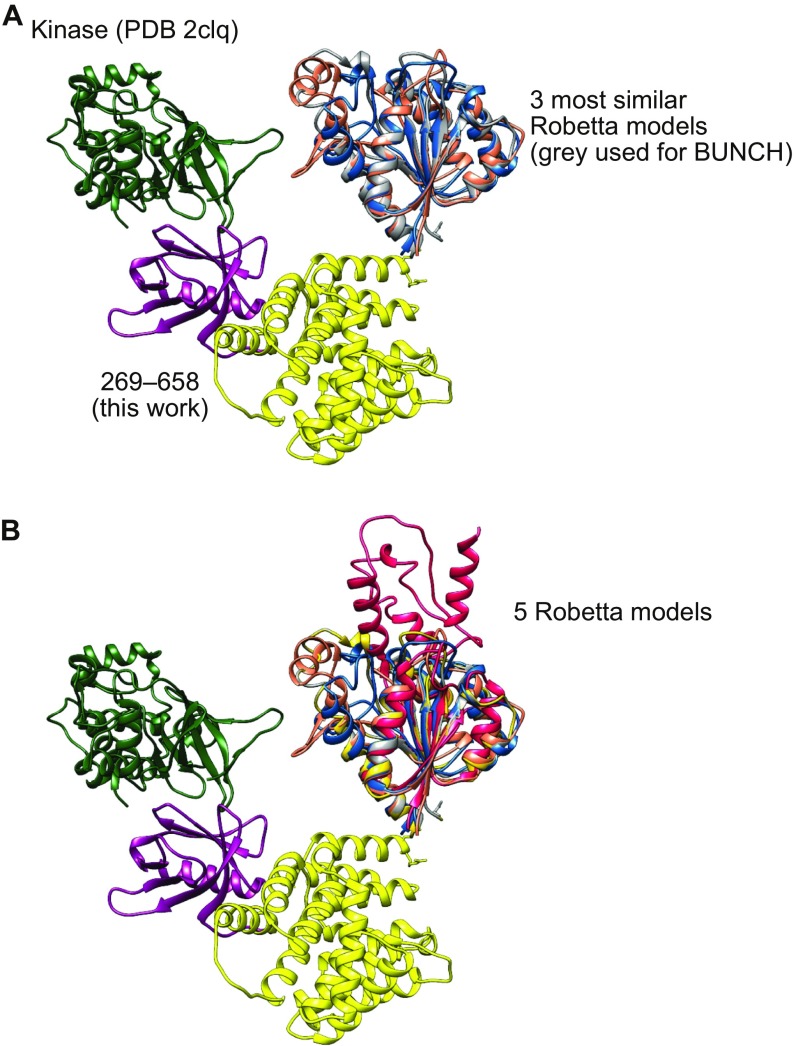
To investigate how the architecture of the thioredoxin-binding and central regions of ASK1 may facilitate signal regulation, we turned to SAXS analysis, first analyzing the ASK1 N-terminal regulatory region ASK1(88–658) alone (Table 2). Guinier analysis showed that the sample was monodispersed, and under the reducing SEC-SAXS conditions when optimal data were collected, we found no evidence of the dimerization previously shown to occur through the NCC region of ASK1. Because the structure of the N-terminal thioredoxin-binding domain has not been solved, we used the Robetta server to generate a homology model of ASK1(89–266), which predicted a globular fold based around an α-β sandwich (Fig. S5) (42).
Fig. S5.
(A) BUNCH model of ASK1(88–941) with three closely related Robetta models of the thioredoxin-binding domain overlaid. (B) As for A, but with all five models output by Robetta shown. The two additional models in B have a similar beta sheet core but differ in their decorating helices.
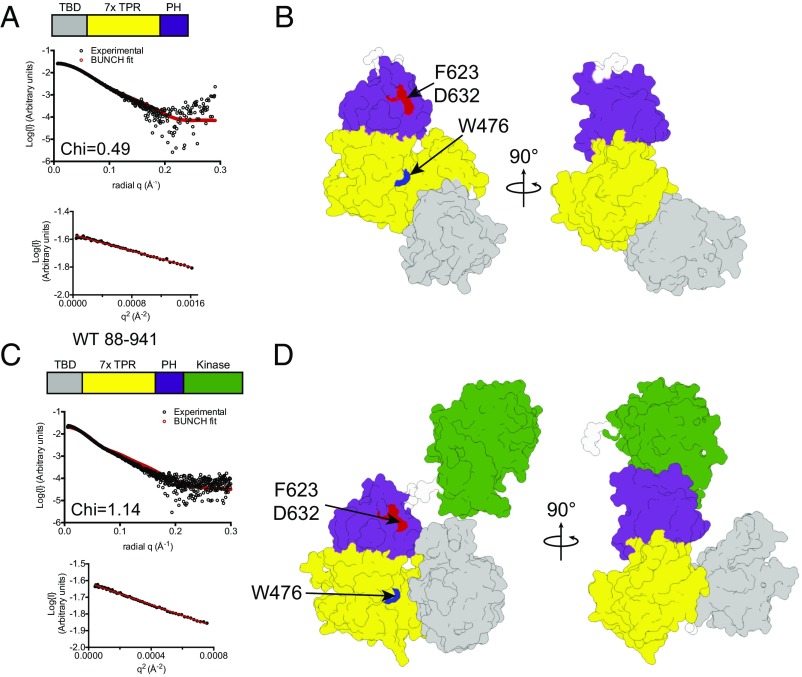
Because our earlier SAXS analysis of the central regulatory region alone showed a stable fold and limited flexibility, when analyzing scattering data, we treated residues 89–266 and 272–654 as two separate rigid bodies. Using BUNCH (43), we generated a model for ASK1(88–658) that provided an excellent fit to the scattering data (χ = 0.49) (Fig. 5 A and B, Table 2, and Fig. S6A). In this model, the ASK1 thioredoxin-binding domain sits adjacent to the N terminus of the central regulatory region, occupying a position toward the conserved face of the central regulatory region that contains both W476 and F623/D632 (Fig. 5B).
Fig. 5.
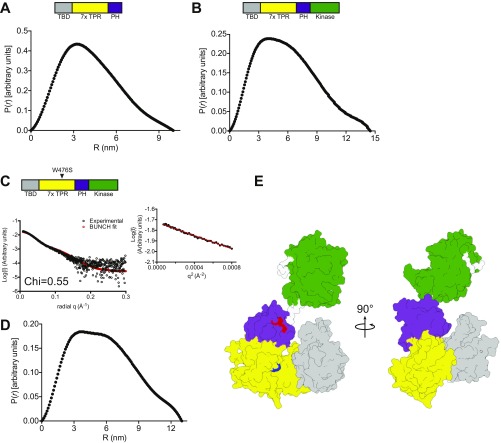
ASK1 autoregulatory scaffolding in solution. (A) Overlay of experimental scattering data (black circles) and a scattering profile calculated using BUNCH for the model of ASK1(88–658). A Guinier plot for the dataset is shown below, indicating that aggregates do not measurably contribute to the scattering profile. Agreement between the experimental data and calculated scatter pattern is indicated by χ = 0.49. (B) Surface representation of the BUNCH model of ASK1(88–658), with the thioredoxin-binding domain in gray, the TPR region in yellow, and the pleckstrin homology domain in purple. Residues that affect activity when mutated are indicated. (C and D) Experimental scattering data (C) and BUNCH model (D) for ASK1(88–941). The ASK1 kinase domain (PDB ID code 2CLQ), is colored green. Agreement between the experimental data and calculated scatter pattern is indicated by χ = 1.14.
Fig. S6.
(A and B) Interatomic distance distributions of ASK1(88–658) (A) and ASK1(88–941) (B). (C) Overlay of experimental scattering data (black circles) and scattering profile calculated using BUNCH for the model of ASK1(88–941) W476S. A Guinier plot for the dataset is shown below, indicating that aggregates do not measurably contribute to the scattering profile. (D) Interatomic distance distributions of ASK1(88–941) W476S. (E) Surface representation of the BUNCH model of ASK1(88–941) W476S, with the thioredoxin-binding domain in gray, the TPR region in yellow, and the pleckstrin homology domain in purple. Residues that affect activity when mutated are indicated in red (F623 and D632) and blue (W476). Agreement between the experimental data and the calculated scatter pattern is indicated by χ = 0.55.
We next collected scattering data for ASK1(88–941) under equivalent conditions to ASK1(88–658) (Fig. 5 C and D, Table 2, and Fig. S6B), and derived a model by incorporating the crystal structure of the isolated ASK1 kinase domain (PDB ID code 2CLQ) into rigid-body (BUNCH) analysis (44). Again, the scattering data suggested a monomeric species, consistent with the elution profile of ASK1(88–941) when loaded onto size exclusion chromatography at a high concentration (Fig. S1D). The model revealed a more compact arrangement of the thioredoxin-binding domain relative to the central regulatory region, with the thioredoxin-binding domain folding toward the conserved surface that contains W476, bringing it closer to both the pleckstrin homology and kinase domains (Fig. 5D). We also collected scattering data from the mutant ASK1(88–941) W476S construct, which displayed elevated activity (Table 2 and Fig. S6 C–E). Apart from a small reorientation of the kinase domain, the W476S model did not differ markedly from WT protein in its overall arrangement. Attempts to collect data from inhibitory mutants within the pleckstrin homology domain were hampered by protein instability at high concentrations.
The main conclusion that we draw from these solution studies is that ASK1(88–941) likely exists in dynamic continuum between an active open form and a closed conformation in which the thioredoxin-binding domain is in close proximity to the ASK kinase domain. Subtle changes (such as the W476S mutation) can alter the structural ensemble present in solution, but we are reticent to propose specific interdomain contacts, given that our modeling relies on a de novo model of the thioredoxin-binding domain. In the proposed model, the thioredoxin-binding domain is ideally placed to inhibit activity of the ASK kinase domain and impede access to the MKK6 activating surface of the ASK1 pleckstrin homology domain—in effect acting as a stimulus-responsive toggle to control ASK1 kinase activity and substrate recruitment and priming.
Discussion
Based on the results of our biochemical, biophysical, and structural experiments, we are now able to put forward a model to interpret the regulation of ASK proteins (Fig. 6). The central regulatory region spans ∼400 residues between the thioredoxin-binding domain and the kinase domain. More importantly, it provides a platform for the recruitment and priming of MAP2K substrates, as well as a link that brings the N-terminal thioredoxin-binding domain and C-terminal kinase domains of ASK1 into proximity for autoinhibition. Although our experiments have focused on ASK1, functional residues also are highly conserved in ASK2 and ASK3, and from mammals to nematodes, and so this architecture is likely to be functionally conserved throughout ASK-type kinases.
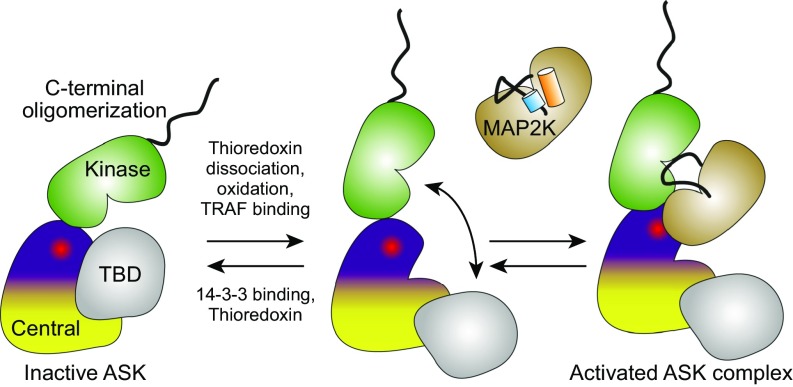
Fig. 6.
Proposed model of ASK1 autoregulatory scaffolding. An autoinhibited conformation with limited activity is shown on the left. Activity can be induced by oxidation, thioredoxin dissociation, or TRAF association (among other stimuli), at which point the activating pleckstrin homology surface becomes available for MAP2K association, activation loop priming, and phosphorylation.
The presence of adjacent pleckstrin homology and kinase domains is reminiscent of the domain architecture of AKT proteins. This similarity is only superficial, however, given that AKT pleckstrin homology domains are bona fide binders of phosphoinositides. Structures of near full-length AKT1 have revealed that its pleckstrin homology domain and kinase antagonize each other in a reciprocal manner—the pleckstrin homology domain forces the kinase into an inactive confirmation, and the kinase domain blocks the phospholipid binding site of the pleckstrin homology domain (45, 46). Instead, it appears that the pleckstrin homology domain plays a positive role in ASK1 activity, closer to that of the pleckstrin homology domain within the yeast MAP kinase scaffold Ste5 (40). Ste5 contains a predicted pleckstrin homology domain that has been shown to bind the MAP3K Ste11 and promote activation of the mating pathway (39). In contrast to Ste5, ASK proteins already contain a MAP3K domain, and use their pleckstrin homology domain as a recruitment site for their primary substrate, MAP2Ks, thereby forming their own scaffold.
Whereas some scaffold proteins act passively by colocalizing participating active signaling proteins, other scaffolds play more active roles by activating or deactivating participating proteins to promote signal fidelity (47). For instance, in mammals, KSR1/2 act as scaffolds in the RAF-MEK-ERK MAPK pathway and promote signaling by forming RAF-KSR pseudokinase heterodimers that activate RAF kinase activity (48, 49). Directly relevant to our MKK6-ASK1 data, the yeast Ste5 scaffold contains a von Willebrand type A (VWA) domain that primes the Fus3 MAPK for phosphorylation by the MAP2K Ste7 (50). We propose that the ASK pleckstrin homology domain plays a role analogous to that of the VWA domain of Ste5, promoting a conformation of MKK6 that is primed for phosphorylation. Beyond the aforementioned Ste5 from yeast, there have been few examples of substrate priming of MAP2Ks described in metazoan MAPK pathways. The diversity of the activation loop and αC helix conformations observed for various MAP2K proteins suggests that they may be particularly sensitive to such regulation (33, 34, 51–53).
Previous work has shown the isolated ASK1 kinase domain is intrinsically active (35, 44). Cell-based studies also have shown that ASK1 lacking the N-terminal thioredoxin-binding domain is more active than full-length protein (19), suggesting that it plays an important role in suppressing ASK1 activity. Our experiments, which used recombinant proteins in the absence of other possible interactors, show the seemingly contradictory results that ASK1(88–941) is autoinhibited relative to the kinase domain alone, but that the central regulatory region promotes MKK6 phosphorylation in trans. Our point mutants surrounding the closed TPR also show an interesting dichotomy; disruption at the core of the closed TPR (R395) abrogates ASK1 activity, whereas mutation of the surface-exposed W476, which presumably retains the overall structure of the central regulatory region, appears to disrupt autoinhibition. This suggests that the overall integrity of the central regulatory domain is important for activity, but also mediates autoinhibitory interactions. The results of the SAXS analysis presented here are entirely consistent with such a role, with the thioredoxin-binding domain well positioned to restrict access of MAP2K to the pleckstrin homology docking site and to suppress ASK1 activity. Our observation that the thioredoxin-binding domain does not markedly autoinhibit kinase activity in trans (Fig. 1E) reinforces the idea that the central regulatory domain is vital, but the exact structural basis of ASK1 kinase autoinhibition remains a key outstanding question. Similarly, it remains to be determined whether ASK1 priming is specific to MKK6 or occurs across all MAP2Ks that are substrates of ASK proteins.
The ASK1 kinase domain has been shown to form a relatively tight dimer (with a dissociation constant of ∼0.2 μM) (44), and it is possible that formation of a kinase domain dimer could represent the activated state of ASK proteins. ASK1(88–941) was monomeric in solution in our SAXS experiments, in contrast with previous reports regarding the isolated kinase domain (28, 44), which may provide some insight into a potential activation mechanism. Our model do not preclude ASK kinase dimer formation, but the scattering data do suggest that it occurs less readily with longer proteins than the kinase domain. However, the presence of the C-erminal region of ASK1, which likely predisposes the protein to form oligomers, could enable kinase dimerization to predominate in the context of the full-length protein. In addition, 14-3-3 proteins bind adjacent to the kinase domain and can themselves form dimers (28). Active kinase dimers also would be analogous to RAF MAP3Ks, which have been the topic of intense study in the ERK pathways (54–57). Our functional experiments are consistent with ASK1 regulation in either a monomer form or a dimer form. In this regard, one possibility is that MKK6 primed by one pleckstrin homology domain could be phosphorylated by an ASK kinase across the kinase dimer interface. There is a wealth of data suggesting that ASK1 functions as part of an oligomeric multiprotein complex, and our observation of autoregulatory scaffolding could be amplified or regulated in the presence of ASK1–3 oligomers. No doubt many intriguing questions remain to be addressed by future biochemical and structural studies investigating how ASK1–3 oligomerization affects kinase regulation.
The precise molecular basis for manipulation of the autoregulatory scaffold from ASK-type proteins by various partners is a clear avenue for future study. For instance, thioredoxin forms both covalent and noncovalent complexes with the thioredoxin-binding domain (6, 30), either of which may be capable of interfering with the regulation of kinase activity in the context of the higher-order assembly known as the ASK signalosome. Cysteine residues that are essential for activation of ASK1 by reactive oxygen species are located very close to the linker between the central and thioredoxin-binding domains of ASK1 (31, 58). It is easy to envisage that disulfide bond formation by these cysteines could restrict ASK1 dynamics in a conformation that favors MAP2K recruitment and activity. Furthermore, the central regulatory region is also essential for binding of TRAFs to ASK proteins (7, 10, 59), which could disrupt the autoinhibitory arrangement between the ASK1 kinase and thioredoxin-binding domains.
In conclusion, the model provided here for autoregulatory scaffolding by ASK1 N-terminal regulatory domains is an enticing framework on which to interpret the various reported stimuli that control ASK1 activity. In addition, the role of substrate kinase priming in MAP kinase signaling has been underappreciated, and it will be intriguing to observe the prevalence of substrate priming for ASK-type kinases on different substrates, for other MAP3K-MAP2K phosphorylation events, or for MAP2K-MAPK activity. Further insight into each of these phenomena will allow a greater understanding of how ASK-type proteins become dysregulated in disease, as well as the fundamental regulation of kinase signaling networks.
Materials and Methods
Protein Expression and Purification.
For biochemical studies and native crystallization, all proteins were expressed in E. coli BL21(DE3). Fragments of the gene encoding ASK1 were amplified from the MegaMan Human Transcriptome Library (Agilent) and cloned into modified pET-LIC vectors (a kind gift from the Netherlands Cancer Institute Protein Facility, with funding from Grant 175.010.2007.012). ASK1(269–658) and ASK1(88–658) were expressed incorporating an N-terminal 6×His tag and 3C protease cleavage site. ASK1(88–941) was cloned with the same N-terminal 6×His tag and 3C protease cleavage site, but also with an additional StrepII tag at its C terminus, and coexpressed with human thioredoxin-1 and lambda protein phosphatase. All mutants were generated using the QuikChange Mutagenesis Kit (Agilent).
ASK1(269–658) and ASK1(88–658) were initially purified by Ni2+ affinity chromatography and then purified to homogeneity after cleavage with 3C protease using anion-exchange chromatography (Resource Q) and size exclusion chromatography (Superdex 200 Increase column; GE Healthcare). Isolated proteins and complexes were flash-frozen for storage in 10 mM Hepes pH 7.6, 300 mM NaCl, and 2 mM DTT. Selenomethionine-labeled ASK1(269–658) was produced in the methionine auxotroph E. coli 834(DE3) using SelenoMet medium (Molecular Dimensions) according to the manufacturer’s instructions. Purification proceeded as for native proteins.
ASK1(88–941) was purified by Ni2+ affinity chromatography and cleaved overnight using 3C protease while dialyzing against a buffer consisting of 50 mM Tris pH 8.0, 150 mM NaCl, 1 mM EDTA, and 2 mM DTT. Dialyzed protein was bound to Strep-Tactin high-capacity or Strep-TactinXT resin, washed with additional dialysis buffer, and eluted with 5 mM desthiobiotin (for Strep-Tactin high-capacity) or 5 mM D-Biotin (for Strep-TactinXT) diluted in dialysis buffer. Eluted proteins were then subsequently dialysed against 50 mM Tris pH 8.0, 150 mM NaCl. Samples for enzymatic assays were used directly, For SAXS analysis, eluted protein was further purified using size exclusion chromatography (Superdex 200 Increase 10/300).
Kinase dead MKK6 (MKK6 K82A) was purified as an N-terminal 6xHis tag and 3C protease cleavage and further purified by size-exclusion chromatography (Superdex 200 Increase 10/300).
Crystallization and Structure Solution.
ASK1(269–658) was initially crystallized in 0.2 M sodium fluoride and 20% (wt/vol) PEG 3350. Poor initial diffraction was improved through seeding and additive screening, with final data collected from crystals grown in 0.1 M sodium fluoride and 10% (wt/vol) PEG 3350 and frozen with the addition of 20% (vol/vol) glycerol. The structure was solved by single-wavelength anomalous dispersion, using a 2.9-Å peak dataset created by merging data from two separate selenomethionine-labeled ASK1 crystals collected at 0.9793 Å. Thirteen of 14 possible selenium sites from two ASK1 molecules in the asymmetric unit were located using PhenixAutosol (60). An initial backbone built by Buccaneer (61) was rebuilt manually. The model was further improved using ArpWarp and finally refined against native data to 2.1 Å using Refmac and the PDB_REDO web server, with cycles of manual rebuilding in Coot (62–65). Analysis of diffraction data by Phenix.Xtriage indicated a large (23% relative to origin) Patterson Peak consistent with the presence of translational pseudosymmetry, which contributed to marginally higher refinement statistics than may be expected at 2.1-Å resolution (Rcryst/Rfree, 0.233/0.267). Nevertheless, the final model has excellent geometry and, aside from several disordered loops connecting TPR helices, is clearly defined in both molecules of the asymmetric unit. Structural figures were generated using UCSF Chimera (66).
SAXS.
SAXS data collection was performed at the Australian Synchrotron SAXS/WAXS beamline using an inline gel filtration chromatography setup (67), essentially as described previously (68–71). Summary statistics for data collection and analysis are reported in Table 2. Here 50 μL of purified recombinant ASK1(269–658) at 7.7 mg/mL, ASK1(88–658) at 9.7 mg/mL, or ASK1(88–941) at 9.3 mg/mL (WT) or 11.5 mg/mL (W476S), were injected onto an inline Superdex 200 5/150 column (GE Healthcare) and eluted at a flow rate of 0.2 mL/min via a 1.5-mm glass capillary positioned in the X-ray beam in 500 mM NaCl, 10 mM Hepes (pH 7.5), 5% (vol/vol) glycerol, and 0.2 mM TCEP at 12 °C. Coflow SAXS was used to minimize sample dilution and maximize signal to noise (72).
Scattering data were collected in 2-s exposures over the course of the elution and 2D intensity plots with consistent scatter intensities from the peak of the sizing exclusion chromatography run were radially averaged, normalized to sample transmission, and background subtraction performed using Scatterbrain software (Stephen Mudie, Australian Synchrotron). Background scatter was assessed by averaging scattering profiles from earlier in the size exclusion chromatography run, before protein elution. Guinier analysis of each scatter pattern across the single elution peak showed consistent radius of gyration (Rg) values, and superimposable patterns were averaged. Four profiles for ASK1(269–658), eight profiles for ASK1(88–658), 21 profiles for ASK1(88–941), and four profiles for ASK1(88–941) W476S were averaged and background-subtracted using Scatterbrain to generate the averaged scatter patterns presented in the manuscript. Guinier data analyses were performed using PRIMUS (73). Indirect Fourier transform with GNOM (74) was used to obtain the distance distribution function, P(r), and the maximum dimension, Dmax, of the scattering particle. CRYSOL (75) was used to calculate theoretical scattering curves from crystal structure atomic coordinates and compare them with experimental scattering curves.
SEC-MALLS.
SEC-MALLS was conducted using a Wyatt Dawn 8+ detector (Wyatt Technology) coupled to a Superdex 200 10/300 column (GE Healthcare) and a refractive index detector. Samples were run in 10 mM Hepes (pH 7.6), 500 mM NaCl, 5% (vol/vol) glycerol, and 0.2 mM TCEP and loaded at 2.2 mg/mL. All data were analyzed using ASTRA V software.
Kinase Assays.
Each kinase assay was carried out at room temperature with final concentrations of 25 mM Hepes pH 7.6, 20 mM MgCl2, 2 mM DTT, 100 mM NaCl, and 3 µM kinase dead MKK6, along with 0.01–1 µM kinase and 0.01–30 µM ASK1 regulatory domains and 50 µM ATP. Assays were set up as master mixes containing all components except kinase and ATP, to ensure equal substrate addition to all reactions. For kinase assays, including separate ASK1 regulatory domains, ASK1 kinases was added to the master mix. The master mix was then aliquoted into eight-well PCR strip tubes to facilitate the use of a multichannel pipette and simultaneous addition and removal of samples. ASK1 kinases and regulatory domains were diluted in serial dilutions. The reactions were started by the addition of ATP. At each time point, an aliquot from each tube was removed in parallel, and the reaction was terminated by immediate mixing into 4× Laemmli sample buffer [240 mM Tris pH 6.8, 32% (vol/vol) glycerol, 8% (wt/vol) SDS, and 0.02% (wt/vol) bromophenol blue]. Samples were briefly spun down and stored at −20 °C until downstream analysis.
For analysis by Phos-tag gels, 15-well, 1-mm-thick Phos-tag analysis gels were hand-poured to contain final concentration of 20 µM Phos-tag, 100 µM MnCl2, and 10% (wt/vol) polyacrylamide. Gels were run as conventional SDS/PAGE gels. Total protein was visualized with Coomassie brilliant blue and imaged with an Odyssey Fc imaging system (LI-COR) in the 700 channel. Quantitation was performed by measuring the intensity of both phosphorylated and unphosphorylated MKK6 bands. The intensity of phosphorylated bands was expressed as a fraction of total intensity, and converted to an absolute concentration by multiplying the fraction of phosphorylated species by the 3 μM total concentration of MKK6 in all assays. Using mass spectrometry, we confirmed that phosphorylation of MKK6 followed the precisely ordered phosphorylation events established by Humphreys et al. (35), with the more rapidly appearing band on Phos-tag SDS/PAGE corresponding to Thr211 of MKK6 (Figs. 3 and 4).
For analysis by Western blot, samples were run in a conventional manner and transferred by a semidry method to 0.45 µm nitrocellulose (GE Healthcare). Total protein transferred to membrane was visualized by staining membrane in 0.5% (wt/vol) Ponceau S solution for 5 min at room temperature. Excess Ponceau S was removed by rinsing in distilled water. Ponceau S-stained blots were imaged using the Odyssey Fc imaging system in the 800 channel. After imaging, blots were rinsed further before blocking in 5% (wt/vol) BSA in Tris-buffered saline (TBS) for 1 h at room temperature. Blots were then incubated with rabbit polyclonal [p-MKK3/6 (Ser187), sc-7994-R; Santa Cruz Biotechnology], diluted 1/2,500 in TBST with 1% (wt/vol) BSA and allowed to bind overnight at 4 °C. Blots were then washed three times for 5 min each in TBST before incubation with secondary antibody (goat anti-rabbit IRdye 680LT; LI-COR) diluted 1/20,000 in TBST with 1% (wt/vol) BSA. The secondary antibody was allowed to bind for 1 h at room temperature before being washed another three times in TBST. Blots were developed in the Odyssey Fc imaging system. Quantification of blots and Phos-tag gels was performed using ImageStudioLite (LI-COR).
Acknowledgments
We thank the New Zealand synchrotron group for facilitating access to the MX beamlines, the staff at the MX and SAXS beamlines for their assistance with data collection, and Dr. Torsten Kleffmann for mass spectrometry analysis. P.D.M. and J.F.W. are currently supported by a Rutherford Discovery Fellowship from the New Zealand government administered by the Royal Society of New Zealand (to P.D.M.). Additional support was provided by University of Otago research grants (to P.D.M.), the Victorian State Government Operational Infrastructure Support, National Health and Medical Research Council (NHMRC) Independent Research Institute Infrastructure Support Scheme Grant 9000220, and NHMRC Fellowship 1105754 (to J.M.M.). This research was undertaken on both the macromolecular crystallography (MX1 and MX2) and small-angle X-ray scattering beamlines at the Australian Synchrotron.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5ULM).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620813114/-/DCSupplemental.
References
- 1.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26(22):3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 2.Samatar AA, Poulikakos PI. Targeting RAS-ERK signalling in cancer: Promises and challenges. Nat Rev Drug Discov. 2014;13(12):928–942. doi: 10.1038/nrd4281. [DOI] [PubMed] [Google Scholar]
- 3.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298(5600):1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 4.Ritt DA, et al. Inhibition of Ras/Raf/MEK/ERK pathway signaling by a stress-induced phospho-regulatory circuit. Mol Cell. 2016;64(5):875–887. doi: 10.1016/j.molcel.2016.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vin H, et al. BRAF inhibitors suppress apoptosis through off-target inhibition of JNK signaling. eLife. 2013;2:e00969. doi: 10.7554/eLife.00969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saitoh M, et al. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 1998;17(9):2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishitoh H, et al. ASK1 is essential for JNK/SAPK activation by TRAF2. Mol Cell. 1998;2(3):389–395. doi: 10.1016/s1097-2765(00)80283-x. [DOI] [PubMed] [Google Scholar]
- 8.Miyakawa K, et al. ASK1 restores the antiviral activity of APOBEC3G by disrupting HIV-1 Vif-mediated counteraction. Nat Commun. 2015;6:6945. doi: 10.1038/ncomms7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okazaki T, et al. The ASK family kinases differentially mediate induction of type I interferon and apoptosis during the antiviral response. Sci Signal. 2015;8(388):ra78. doi: 10.1126/scisignal.aab1883. [DOI] [PubMed] [Google Scholar]
- 10.Matsuzawa A, et al. ROS-dependent activation of the TRAF6-ASK1-p38 pathway is selectively required for TLR4-mediated innate immunity. Nat Immunol. 2005;6(6):587–592. doi: 10.1038/ni1200. [DOI] [PubMed] [Google Scholar]
- 11.Geleziunas R, Xu W, Takeda K, Ichijo H, Greene WC. HIV-1 Nef inhibits ASK1-dependent death signaling, providing a potential mechanism for protecting the infected host cell. Nature. 2001;410(6830):834–838. doi: 10.1038/35071111. [DOI] [PubMed] [Google Scholar]
- 12.Ichijo H, et al. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science. 1997;275(5296):90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- 13.Hayakawa Y, et al. Apoptosis signal-regulating kinase 1 and cyclin D1 compose a positive feedback loop contributing to tumor growth in gastric cancer. Proc Natl Acad Sci USA. 2011;108(2):780–785. doi: 10.1073/pnas.1011418108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayakawa Y, et al. Apoptosis signal-regulating kinase-1 inhibitor as a potent therapeutic drug for the treatment of gastric cancer. Cancer Sci. 2012;103(12):2181–2185. doi: 10.1111/cas.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stark MS, et al. Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing. Nat Genet. 2011;44(2):165–169. doi: 10.1038/ng.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prickett TD, et al. NISC Comparative Sequencing Program Somatic mutations in MAP3K5 attenuate its proapoptotic function in melanoma through increased binding to thioredoxin. J Invest Dermatol. 2014;134(2):452–460. doi: 10.1038/jid.2013.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujisawa T, et al. The ASK1-specific inhibitors K811 and K812 prolong survival in a mouse model of amyotrophic lateral sclerosis. Hum Mol Genet. 2016;25(2):245–253. doi: 10.1093/hmg/ddv467. [DOI] [PubMed] [Google Scholar]
- 18.Peti W, Page R. Molecular basis of MAP kinase regulation. Protein Sci. 2013;22(12):1698–1710. doi: 10.1002/pro.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujino G, et al. Thioredoxin and TRAF family proteins regulate reactive oxygen species-dependent activation of ASK1 through reciprocal modulation of the N-terminal homophilic interaction of ASK1. Mol Cell Biol. 2007;27(23):8152–8163. doi: 10.1128/MCB.00227-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iriyama T, et al. ASK1 and ASK2 differentially regulate the counteracting roles of apoptosis and inflammation in tumorigenesis. EMBO J. 2009;28(7):843–853. doi: 10.1038/emboj.2009.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeda K, et al. Apoptosis signal-regulating kinase (ASK) 2 functions as a mitogen-activated protein kinase kinase kinase in a heteromeric complex with ASK1. J Biol Chem. 2007;282(10):7522–7531. doi: 10.1074/jbc.M607177200. [DOI] [PubMed] [Google Scholar]
- 22.Federspiel JD, et al. Assembly dynamics and stoichiometry of the apoptosis signal-regulating kinase (ASK) signalosome in response to electrophile stress. Mol Cell Proteomics. 2016;15(6):1947–1961. doi: 10.1074/mcp.M115.057364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoeflich KP, Yeh WC, Yao Z, Mak TW, Woodgett JR. Mediation of TNF receptor-associated factor effector functions by apoptosis signal-regulating kinase-1 (ASK1) Oncogene. 1999;18(42):5814–5820. doi: 10.1038/sj.onc.1202975. [DOI] [PubMed] [Google Scholar]
- 24.Yoon KW, et al. CIB1 functions as a Ca(2+)-sensitive modulator of stress-induced signaling by targeting ASK1. Proc Natl Acad Sci USA. 2009;106(41):17389–17394. doi: 10.1073/pnas.0812259106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu Z, et al. Lys29-linkage of ASK1 by Skp1-Cullin 1-Fbxo21 ubiquitin ligase complex is required for antiviral innate response. eLife. 2016;5:249. doi: 10.7554/eLife.14087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawarazaki Y, Ichijo H, Naguro I. Apoptosis signal-regulating kinase 1 as a therapeutic target. Expert Opin Ther Targets. 2014;18(6):651–664. doi: 10.1517/14728222.2014.896903. [DOI] [PubMed] [Google Scholar]
- 27.Cockrell LM, Puckett MC, Goldman EH, Khuri FR, Fu H. Dual engagement of 14-3-3 proteins controls signal relay from ASK2 to the ASK1 signalosome. Oncogene. 2010;29(6):822–830. doi: 10.1038/onc.2009.382. [DOI] [PubMed] [Google Scholar]
- 28.Petrvalska O, et al. Structural Insight into the 14-3-3 protein-dependent inhibition of protein kinase ASK1 (apoptosis signal-regulating kinase 1) J Biol Chem. 2016;291(39):20753–20765. doi: 10.1074/jbc.M116.724310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kosek D, et al. Biophysical and structural characterization of the thioredoxin-binding domain of protein kinase ASK1 and its interaction with reduced thioredoxin. J Biol Chem. 2014;289(35):24463–24474. doi: 10.1074/jbc.M114.583807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nadeau PJ, Charette SJ, Toledano MB, Landry J. Disulfide bond-mediated multimerization of Ask1 and its reduction by thioredoxin-1 regulate H(2)O(2)-induced c-Jun NH(2)-terminal kinase activation and apoptosis. Mol Biol Cell. 2007;18(10):3903–3913. doi: 10.1091/mbc.E07-05-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nadeau PJ, Charette SJ, Landry J. REDOX reaction at ASK1-Cys250 is essential for activation of JNK and induction of apoptosis. Mol Biol Cell. 2009;20(16):3628–3637. doi: 10.1091/mbc.E09-03-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kinoshita E, Kinoshita-Kikuta E, Koike T. Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nat Protoc. 2009;4(10):1513–1521. doi: 10.1038/nprot.2009.154. [DOI] [PubMed] [Google Scholar]
- 33.Matsumoto T, et al. Crystal structure of non-phosphorylated MAP2K6 in a putative auto-inhibition state. J Biochem. 2012;151(5):541–549. doi: 10.1093/jb/mvs023. [DOI] [PubMed] [Google Scholar]
- 34.Min X, et al. The structure of the MAP2K MEK6 reveals an autoinhibitory dimer. Structure. 2009;17(1):96–104. doi: 10.1016/j.str.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humphreys JM, Piala AT, Akella R, He H, Goldsmith EJ. Precisely ordered phosphorylation reactions in the p38 mitogen-activated protein (MAP) kinase cascade. J Biol Chem. 2013;288(32):23322–23330. doi: 10.1074/jbc.M113.462101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piala AT, Humphreys JM, Goldsmith EJ. MAP kinase modules: The excursion model and the steps that count. Biophys J. 2014;107(9):2006–2015. doi: 10.1016/j.bpj.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeytuni N, Zarivach R. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure. 2012;20(3):397–405. doi: 10.1016/j.str.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 38.Scheffzek K, Welti S. Pleckstrin homology (PH)-like domains: Versatile modules in protein–protein interaction platforms. FEBS Lett. 2012;586(17):2662–2673. doi: 10.1016/j.febslet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 39.Garrenton LS, Young SL, Thorner J. Function of the MAPK scaffold protein, Ste5, requires a cryptic PH domain. Genes Dev. 2006;20(14):1946–1958. doi: 10.1101/gad.1413706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zalatan JG, Coyle SM, Rajan S, Sidhu SS, Lim WA. Conformational control of the Ste5 scaffold protein insulates against MAP kinase misactivation. Science. 2012;337(6099):1218–1222. doi: 10.1126/science.1220683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu JW, et al. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell. 2004;13(5):677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- 42.Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32(Web Server issue):W526–31. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J. 2005;89(2):1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bunkoczi G, et al. Structural and functional characterization of the human protein kinase ASK1. Structure. 2007;15(10):1215–1226. doi: 10.1016/j.str.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu W-I, et al. Crystal structure of human AKT1 with an allosteric inhibitor reveals a new mode of kinase inhibition. PLoS One. 2010;5(9):e12913. doi: 10.1371/journal.pone.0012913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashwell MA, et al. Discovery and optimization of a series of 3-(3-phenyl-3H-imidazo[4,5-b]pyridin-2-yl)pyridin-2-amines: Orally bioavailable, selective, and potent ATP-independent Akt inhibitors. J Med Chem. 2012;55(11):5291–5310. doi: 10.1021/jm300276x. [DOI] [PubMed] [Google Scholar]
- 47.Good MC, Zalatan JG, Lim WA. Scaffold proteins: Hubs for controlling the flow of cellular information. Science. 2011;332(6030):680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rajakulendran T, Sahmi M, Lefrançois M, Sicheri F, Therrien M. A dimerization-dependent mechanism drives RAF catalytic activation. Nature. 2009;461(7263):542–545. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 49.McKay MM, Ritt DA, Morrison DK. Signaling dynamics of the KSR1 scaffold complex. Proc Natl Acad Sci USA. 2009;106(27):11022–11027. doi: 10.1073/pnas.0901590106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Good M, Tang G, Singleton J, Reményi A, Lim WA. The Ste5 scaffold directs mating signaling by catalytically unlocking the Fus3 MAP kinase for activation. Cell. 2009;136(6):1085–1097. doi: 10.1016/j.cell.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsumoto T, Kinoshita T, Kirii Y, Tada T, Yamano A. Crystal and solution structures disclose a putative transient state of mitogen-activated protein kinase kinase 4. Biochem Biophys Res Commun. 2012;425(2):195–200. doi: 10.1016/j.bbrc.2012.07.066. [DOI] [PubMed] [Google Scholar]
- 52.Ohren JF, et al. Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat Struct Mol Biol. 2004;11(12):1192–1197. doi: 10.1038/nsmb859. [DOI] [PubMed] [Google Scholar]
- 53.Fischmann TO, et al. Crystal structures of MEK1 binary and ternary complexes with nucleotides and inhibitors. Biochemistry. 2009;48(12):2661–2674. doi: 10.1021/bi801898e. [DOI] [PubMed] [Google Scholar]
- 54.Thevakumaran N, et al. Crystal structure of a BRAF kinase domain monomer explains basis for allosteric regulation. Nat Struct Mol Biol. 2015;22(1):37–43. doi: 10.1038/nsmb.2924. [DOI] [PubMed] [Google Scholar]
- 55.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464(7287):427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hatzivassiliou G, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464(7287):431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 57.Hu J, et al. Allosteric activation of functionally asymmetric RAF kinase dimers. Cell. 2013;154(5):1036–1046. doi: 10.1016/j.cell.2013.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kylarova S, et al. Cysteine residues mediate high-affinity binding of thioredoxin to ASK1. FEBS J. 2016;283(20):3821–3838. doi: 10.1111/febs.13893. [DOI] [PubMed] [Google Scholar]
- 59.Lu Y-Y, et al. TRAF1 is a critical regulator of cerebral ischaemia-reperfusion injury and neuronal death. Nat Commun. 2013;4:2852. doi: 10.1038/ncomms3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adams PD, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55(1):94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cowtan K. The Buccaneer software for automated model building, 1: Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 9):1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 62.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3(7):1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Joosten RP, Long F, Murshudov GN, Perrakis A. The PDB_REDO server for macromolecular structure model optimization. IUCrJ. 2014;1(Pt 4):213–220. doi: 10.1107/S2052252514009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 66.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 67.Kirby NM, et al. A low-background-intensity focusing small-angle X-ray scattering undulator beamline. J Appl Cryst. 2013;46(6):1670–1680. [Google Scholar]
- 68.Murphy JM, et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity. 2013;39(3):443–453. doi: 10.1016/j.immuni.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 69.Murphy JM, et al. Insights into the evolution of divergent nucleotide-binding mechanisms among pseudokinases revealed by crystal structures of human and mouse MLKL. Biochem J. 2014;457(3):369–377. doi: 10.1042/BJ20131270. [DOI] [PubMed] [Google Scholar]
- 70.Carr PD, et al. Crystal structure of the mouse interleukin-3 β-receptor: Insights into interleukin-3 binding and receptor activation. Biochem J. 2014;463(3):393–403. doi: 10.1042/BJ20140863. [DOI] [PubMed] [Google Scholar]
- 71.Murphy JM, et al. Molecular mechanism of CCAAT-enhancer binding protein recruitment by the TRIB1 pseudokinase. Structure. 2015;23(11):2111–2121. doi: 10.1016/j.str.2015.08.017. [DOI] [PubMed] [Google Scholar]
- 72.Kirby N, et al. Improved radiation dose efficiency in solution SAXS using a sheath flow sample environment. Acta Crystallogr D Struct Biol. 2016;72(Pt 12):1254–1266. doi: 10.1107/S2059798316017174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J Appl Cryst. 2003;36(5):1277–1282. [Google Scholar]
- 74.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Cryst. 1992;25(4):495–503. [Google Scholar]
- 75.Svergun D, Barberato C, Koch MHJ. CRYSOL: A program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Cryst. 1995;28(6):768–773. [Google Scholar]
- 76.Webb B, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci. 2016;86:2.9.1–2.9.37. doi: 10.1002/cpps.20. [DOI] [PubMed] [Google Scholar]
- 77.Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB. Structure of the high- affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell. 1995;83(6):1037–1046. doi: 10.1016/0092-8674(95)90219-8. [DOI] [PubMed] [Google Scholar]
- 78.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bond CS, Schüttelkopf AW. ALINE: A WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 5):510–512. doi: 10.1107/S0907444909007835. [DOI] [PubMed] [Google Scholar]