Significance
Vital physiological processes—such as the cytotoxic immune response—require the coordinated action of the atypical fusion protein Syntaxin 11 (STX11) and the Sec/Munc protein Munc18-2 for releasing effector proteins housed in membrane-enclosed secretory granules. Human mutations in STX11 and Munc18-2 genes lead to severe immunodeficiency and hemostasis disorders. However it is still unclear how STX11, a lipid-anchored SNARE, and Munc18-2 mediate membrane fusion. By using an in vitro fusion assay, we found that STX11 mainly mediates lipid mixing when combined with human lymphocyte-interacting SNAREs. Remarkably, Munc18-2 induces association among these SNAREs and facilitates the transition from a hemifusion-like state to complete fusion. Our findings support a model in which SM proteins play a direct role in membrane merging.
Keywords: SNARE, Syntaxin 11, Munc18-2, CTL, SM protein
Abstract
The atypical lipid-anchored Syntaxin 11 (STX11) and its binding partner, the Sec/Munc (SM) protein Munc18-2, facilitate cytolytic granule release by cytotoxic T lymphocytes (CTLs) and natural killer (NK) cells. Patients carrying mutations in these genes develop familial hemophagocytic lymphohistiocytosis, a primary immunodeficiency characterized by impaired lytic granule exocytosis. However, whether a SNARE such as STX11, which lacks a transmembrane domain, can support membrane fusion in vivo is uncertain, as is the precise role of Munc18-2 during lytic granule exocytosis. Here, using a reconstituted “flipped” cell–cell fusion assay, we show that lipid-anchored STX11 and its cognate SNARE proteins mainly support exchange of lipids but not cytoplasmic content between cells, resembling hemifusion. Strikingly, complete fusion is stimulated by addition of wild-type Munc18-2 to the assay, but not of Munc18-2 mutants with abnormal STX11 binding. Our data reveal that Munc18-2 is not just a chaperone of STX11 but also directly contributes to complete membrane merging by promoting SNARE complex assembly. These results further support the concept that SM proteins in general are part of the core fusion machinery. This fusion mechanism likely contributes to other cell-type–specific exocytic processes such as platelet secretion.
Cytotoxic T lymphocytes (CTLs) and natural killer (NK) cells selectively kill virally infected or cancerous target cells by releasing their lytic granule (LG) contents into the site of contact between the two cells called the immunological synapse (IS) (1, 2). Upon CTL stimulation, lytic granules undergo a series of maturation steps to become “fully armed” and subsequently are released at the IS (3). These processes require membrane fusion steps that are mediated by SNARE proteins. In particular, Syntaxin 11 (STX11) and the Sec/Munc (SM) protein Munc18-2 (STXBP2)—with which STX11 physically interacts—play critical roles in these processes (4, 5). Accordingly, patients carrying mutations in the STX11 or STXBP2 genes give rise to familial hemophagocytic lymphohistiocytosis (F-HLH, types 4 and 5, respectively), a primary immunodeficiency characterized by defective CTL and NK cell cytotoxic activity (6–8). STX11 is unusual among syntaxin family members in that it lacks a transmembrane domain and is anchored to the membrane by prenyl- and palmitoyl-lipid modifications (9–12). Cognate SNAREs that cooperate with STX11 in CTLs and NK cells to mediate fusion are still unknown. Pull-down experiments in HeLa cells and platelets suggest that STX11 interacts with SNAP23, VAMP8, and Munc18-2 and is required for fusion events during platelet granule secretion (13–15). However, in macrophages, STX11 selectively interacts with the endosomal t-SNARE, Vti1b, but not with SNAP23; this suggested that STX11 does not function as a classical fusion protein, but rather regulates fusion by sequestering Vti1b (16). Thus, whereas inactivation of STX11 or Munc18-2 impairs granule release (17), it is not yet clear whether SNAREs that lack a transmembrane domain can directly support membrane fusion in vivo or function only as inhibitors in other steps of membrane fusion. Moreover, much less is understood about how Munc18-2 participates in these processes.
The transmembrane domain (TMD) of fusion proteins, such as the hemagglutinin (HA) protein of the influenza virus or the SNARE proteins in eukaryotes, is required to drive complete merging of two lipid bilayers and fusion pore opening in vitro (18–20). When the TMD of the HA protein is replaced with glycosylphosphatidylinositol (GPI), a lipid anchor that spans only the outer leaflet of the membrane, membrane fusion ends in a hemifusion state where the outer monolayers of the membranes are fused, whereas the inner monolayers and the aqueous contents remain segregated (20–22). Similarly, replacement of the TMD of SNARE proteins by a lipid or protein anchor that spans a single leaflet of the bilayer inhibited complete fusion in vitro (23–27) and in vivo (28). Crystallographic analysis of a neuronal SNARE complex containing the TMDs revealed that SNARE motifs and the TMDs form continuous interacting α-helices (29), potentially explaining the role of the TMD in full fusion. Nonetheless, it has been reported that yeast lipid-anchored SNARE Nyv1p, which cannot support membrane fusion when it is combined with its cognate SNAREs alone, indeed mediates membrane fusion upon addition of the HOPS complex containing the SM protein VPS33 (27). Similarly, a recent study showed that the expression of lipid-anchored Syntaxin-1 or VAMP2 restored spontaneous and Ca2+-triggered exocytosis to Syntaxin-1 or Vamp2 knockout cultured neurons, respectively (30), consistent with a mechanism of membrane fusion in which SNARE-complex assembly may be sufficient to destabilize the phospholipid membrane and induce fusion. However, the contribution of additional cytosolic factors, e.g., SNARE-interacting proteins that could facilitate this process in vivo have not been investigated.
Here we tested whether a lipid-anchored version of STX11 can mediate membrane fusion and investigated how Munc18-2 functions with STX11 in cell-mediated cytotoxicity. We show that endogenous STX11 mainly interacts with SNAP23 and VAMP8 in stimulated CTLs and forms a stable SNARE complex with them in vitro. Using a reconstituted “flipped” cell–cell fusion assay we show that when coexpressed with SNAP23, STX11 bearing an artificial TMD mainly promotes complete fusion with cognate VAMP8-expressing cells but lipid-anchored STX11 primarily supports lipid mixing. Strikingly, addition of Munc18-2 substantially and selectively facilitates complete fusion mediated by lipid-anchored STX11 by promoting and stabilizing the assembly of SNARE complexes. Our data indicate that SM proteins are an integral part of the membrane fusion machinery and can promote membrane fusion events mediated by lipid-anchored syntaxins facilitating the assembly of SNARE complexes.
Results
Lipid-Anchored Flipped STX11 Mainly Promotes Incomplete Fusion in a Cell–Cell Fusion Assay.
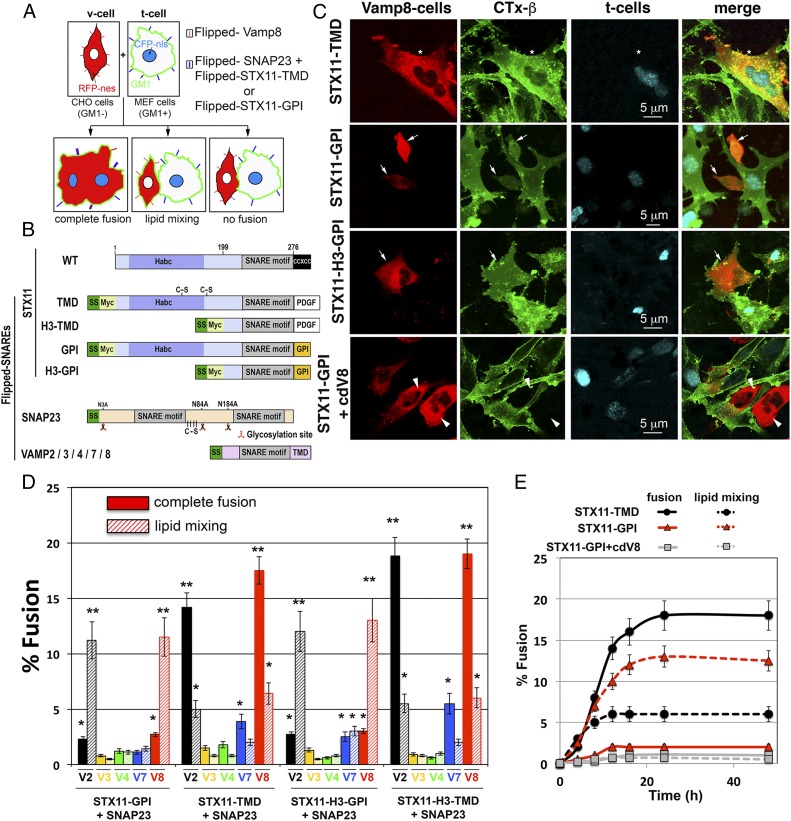
To determine whether STX11 can function as a fusogenic SNARE when combined with different partners we used the flipped SNARE cell fusion assay (25, 31). In this assay, v- and t-SNAREs are ectopically expressed in reverse topology on the surface of two different populations of cell lines expressing distinct fluorescent markers; cognate SNARE interactions mediate either complete cell–cell fusion that can be visualized by mixing of the two markers, or lipid mixing as detailed below. V cells express flipped v-SNAREs and the DsRed fluorescent protein in the cytoplasm, but do not express GM1 ganglioside on the cell surface. T cells express flipped t-SNAREs, CFP in the nucleus, and endogenous GM1 on the cell surface. Complete cell–cell fusion results in large cells with multiple CFP+ nuclei, DsRed+ cytoplasm, and GM1 ganglioside (Fig. 1 A–C, Top). If the SNAREs mediate only lipid mixing, only lipids of the outer leaflet of the plasma membrane (PM), such as GM1, are transferred between cells; this transfer is detected by FITC-cholera toxin β-subunit (FITC-CTxB) that specifically binds to GM1 (Fig. 1 A and C, Middle). We constructed full-length flipped STX11 with either a GPI-anchored motif (STX11–GPI) to mimic its physiological prenyl- and palmitoyl-lipid anchoring to the membrane or with a transmembrane domain [from platelet derived-growth factor (PDGF) receptor; STX11–TMD to resemble other Syntaxin family members] (Fig. 1B). Flipped SNAP23 and -29 were generated as previously described for flipped SNAP25 (31) in which cysteine residues were mutagenized to serine, and putative N-glycosylation sites were mutagenized to alanine, and flipped VAMP3, VAMP4, VAMP7, and VAMP8 were generated as described for flipped VAMP2 (31). Both STX11–GPI and STX11–TMD were delivered to the PM when coexpressed with flipped SNAP23 and SNAP25 but not with flipped SNAP29 or when they were expressed alone (Fig. S1), indicating that complete t-SNARE complexes were generated.
Fig. 1.
Flipped STX11–TMD drives complete fusion, whereas flipped STX11–GPI mainly promotes lipid mixing. (A) Cell fusion assay design monitoring lipid mixing and content mixing. CHO cells (GM1−) that expressed flipped VAMP2, -3, -4, -7, or -8 were labeled in the cytoplasm with RFP fused to nuclear export signal (RFP-nes). MEF 3T3 cells (GM1+) that express flipped t-SNAREs, including STX11 with the PDGF receptor transmembrane domain or a GPI anchor, were labeled in the nucleus with CFP-nls. The t cells were harvested with an EDTA buffer and overlaid on the v cells. Cells were fixed after 12 h at 37 °C. GM1 was stained with FITC-cholera toxin β-subunit (green). Complete fusion resulted in cells contained red cytoplasm, cyan nuclei, and green cell surface staining. In cells that underwent lipid mixing, GM1 transferred from t cells to the contacting CHO v cells in the absence of the mixing of the cytoplasmic markers. In the no-fusion cells, all of the markers remained within the original cells. (B) The domain structure of flipped SNAREs or GPI-anchored SNAREs. The preprolactin signal sequence (SS, dark green) was fused to the N terminus of VAMPs, SNAP23, and the STX11. The cysteine-rich domain of STX11 (CCxCC residues 277–286) was replaced by either the transmembrane domain (TMD) of the platelet-derived growth factor (PDGF) or by the GPI-anchoring sequence of decay accelerating factor. A Myc tag (light green) was engineered between the N terminus of the STX11 full-length (residues 1–276, STX11, or to the STX11–H3 domain (amino acids 199–276, STX11H3) and the signal sequence. Cysteine residues of STX11 and SNAP23 were mutated for serines and the putative N-glycosylation of SNAP23 sites were mutated from asparagine to alanine. (C) Representative confocal images of a cell fusion experiment showing complete fusion events (Top row, asterisk), lipid mixing events (Middle rows, arrows), and no fusion (Lower row, arrowheads). (Scale bar, 5 μm.) (D) CHO v cells expressing flipped VAMPs and RFP-nes were detached and overlaid onto MEF 3T3 cells expressing flipped STX11 constructs, flipped SNAP-23, and CFP-nls. The cells were fixed after 12 h at 37 °C. GM1 was stained with FITC-cholera toxin β-subunit (green). Graph shows the percentage of v and t cells in contact that underwent complete fusion (solid bars) or lipid mixing (dashed bars). (E) Time course of complete fusion and lipid mixing mediated by flipped STX11–TMD, –GPI, or STX11–GPI in presence of cytoplasmic domain of VAMP8 (cdV8). CHO v cells were mixed with MEF 3T3 stable t cells. At different time points, the percentage of v and t cells in contact that underwent complete fusion (solid lines) or lipid mixing (dashed lines) was determined using the assay described in D. Images in 50 random fields were used for calculation of each time point. Values are mean ± SE of three independent experiments. *P < 0.1; **P < 0.01.
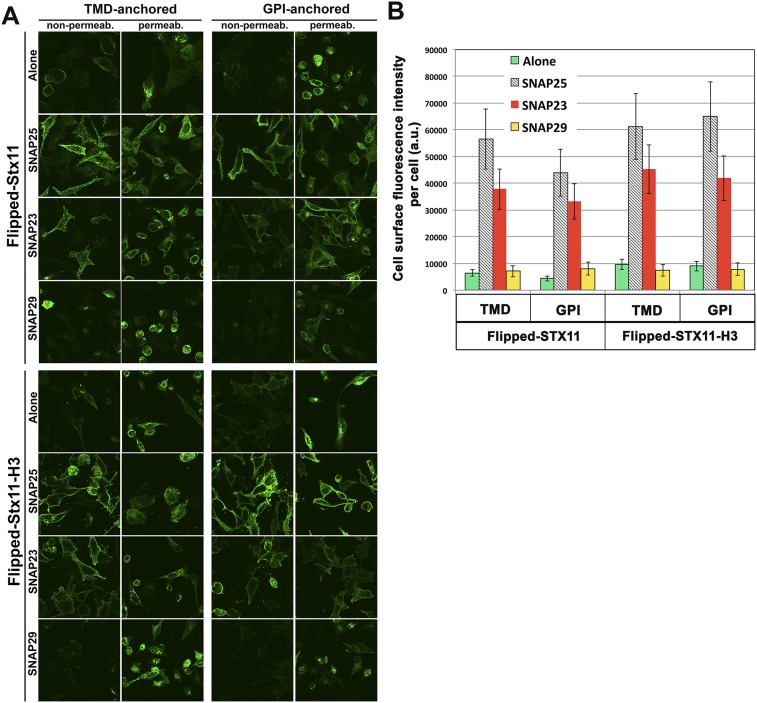
Fig. S1.
Flipped SNAP25 and SNAP23, but not SNAP29, promote cell surface expression of flipped STX11 constructs. (A) MEF 3T3 cells were transiently cotransfected with either STX11–PDGF, Syx11H3–PDGF, STX11–GPI, or STX11H3–GPI and with flipped SNAP25, -23, -29, or vector alone. Cells were fixed and incubated in the presence (permeab.) or absence (nonpermeab) of 0.1% Triton X-100. Cells were stained with anti–c-myc antibody and images were taken using confocal microscopy. (B) Cell surface quantification of different flipped STX11 constructs. Total fluorescent intensity per cell in nonpermeabilized cells stained with anti–c-myc antibody as shown in A were quantified in 75 cells for each condition. Results are mean ± SD of three independent experiments.
Because CTLs do not express SNAP25 (Fig. S2) we analyzed whether STX11–GPI and STX11–TMD expressed at the cell surface with flipped SNAP23 are sufficient to support cell fusion. To this end, different combinations of v cells expressing flipped VAMPs 2, 3, 4, 7, or 8 were tested for fusion with t cells expressing STX11 variants and SNAP23 as detailed in Materials and Methods and schematized in Fig. 1A. Among all tested combinations, the only fully fusogenic ones that showed lipid and content mixing were those in which cognate STX11–TMD/SNAP23 cells were partnered with flipped VAMP2- or VAMP8-expressing cells (Fig. 1 C and D); these combinations resulted primarily in complete fusion (about 15%), with a small fraction of the cells (about 5%) undergoing lipid mixing in which the v cells acquired CTxB, but no content mixing occurred. These results support the specificity of STX11/SNAP23 for VAMP2 and VAMP8 and show that these SNARE complexes are functional. Intriguingly, STX11–GPI/SNAP23 mainly promoted lipid mixing (about 12%) when it was partnered with flipped VAMP2 or VAMP8, but not with VAMPs 3, 4, or 7 (Fig. 1 C and D). This finding is consistent with previous studies showing that SNAREs anchored by an artificial GPI motif mainly promote lipid mixing outcomes reminiscent of hemifusion (25). To rule out the possibility that the N-terminal regulatory domain (Habc domain) of the Syntaxin family proteins interferes with the fusion activity of STX11, we generated an additional set of flipped-STX11 constructs encoding only the SNARE motif (also known as the H3 domain) fused to either a GPI-anchored motif (STX11H3–GPI) or the PDGF receptor transmembrane domain (STX11H3–TMD). The extent of fusion, ratio of full fusion to lipid mixing, and the specificity of fusion with different VAMPs obtained with these constructs highly resembled those obtained with full-length STX11 (Fig. 1D). Together, these data support the functionality of a STX11/SNAP23 SNARE complex with either VAMP2 or VAMP8 in membrane fusion.
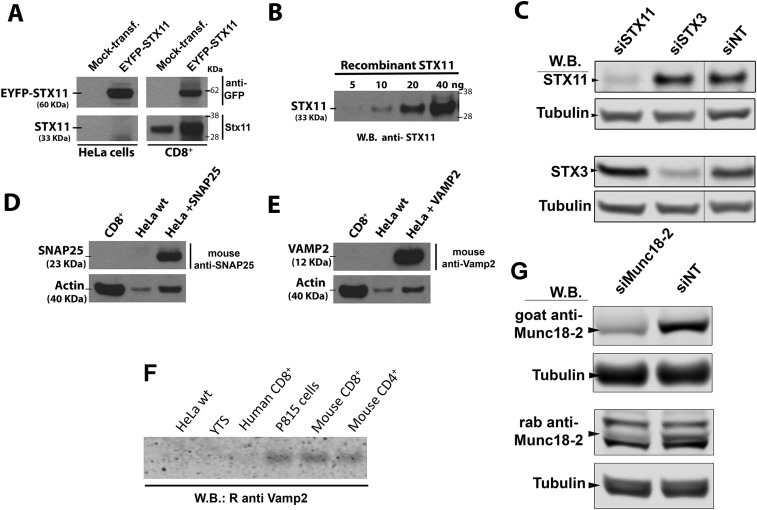
Fig. S2.
Characterization of the antibodies used in assays. (A) Anti-STX11 antibody recognizes both endogenous STX11 as well as ectopically expressed EYFP–STX11 construct in human CTL (CD8+) and HeLa cells. HeLa cells and CTLs were transiently transfected using an empty vector (mock-transf.) or EYFP–STX11 and lysates of these cells were analyzed by Western blot (W.B.) using both anti-GFP (Upper) and anti-STX11 (Lower) antibodies. (B) Anti-STX11 antibody recognizes recombinant STX11 expressed and purified from bacteria. Different amounts of recombinant STX11 were loaded in a gel and analyzed by Western blot using anti-STX11 antibody. (C) Anti-STX11 antibody specifically recognizes human STX11. Human CTLs were treated with either STX11, STX3, or nontargeting (NT) siRNAs for 48 h and cell lysates were analyzed by Western blot using the rabbit anti-STX11, anti-STX3, or mouse anti-tubulin. Data show that anti-STX11 antibody specifically recognizes STX11 but not STX3, which is the syntaxin with the highest amino acid sequence homology within the epitope recognized by the antibody. (D) Human CTLs do not express SNAP25. The expression of SNAP25 was analyzed in lysates of CTLs (CD8+), wild-type HeLa cells (HeLa-wt), or HeLa cells transiently transfected with SNAP25 construct by Western blot using a mouse monoclonal anti-SNAP25 antibody (clone 71.2). (E) CTLs cells do not express VAMP2. The expression of VAMP2 was analyzed in lysates of CTLs (CD8+), wild-type HeLa cells (HeLa-wt), or HeLa cells transiently transfected with VAMP2 construct by Western blot using a mouse monoclonal anti-VAMP2 antibody (clone 61.9). (F) Expression of endogenous VAMP2 in human CTLs, YTS (human NK cell line), and HeLa cells is below the detection level of the affinity-purified anti-VAMP2 polyclonal antibody. Equivalent amounts of cell lysates of the indicated cells were analyzed by Western blot for the expression of VAMP2 using a polyclonal antibody anti-VAMP2. Blots in the figures are representatives of three independents experiments. (G) Specificity of Munc18-2 antibodies. Human CTLs were treated with either Munc18-2 or nontargeting (NT) siRNAs for 48 h and cell lysates were analyzed by Western blot using the goat anti–Munc18-2, rabbit anti–Munc18-2, or mouse anti-tubulin. Data show a reduction of the only band detected with the goat anti–Munc18-2 antibody (Upper, arrowhead) and of one of the three bands detected by the rabbit anti–Munc18-2 antibody (Lower, arrowhead).
To ensure that the high level of lipid mixing obtained with STX11–GPI did not merely reflect a slower rate of fusion compared with the transmembrane domain-containing constructs, we measured the different fusion outcomes kinetically. The extent of both complete fusion and lipid mixing obtained with STX11–GPI plateaus at about 16 h (Fig. 1E) and no further fusion was observed beyond this period, indicating that lipid mixing does not reflect an intermediate state of a slower fusion reaction. The slow kinetics of the cell–cell fusion assay were consistent with previous studies (25, 31) and mostly is limited by the time required by the cells to attach to the substrate and to interact with each other. The inability of lipid-linked STX11 to mediate full fusion might reflect either an inability to assemble into a SNAREpin with cognate SNAREs or an inability to transduce the force of SNAREpin assembly into apposed membrane destabilization. However, the lipid mixing outcome we observed, required SNARE-complex assembly because the addition of the soluble cytoplasmic domain of VAMP8 (cdV8), a competitor inhibitor of SNARE-mediated fusion, decreased the extent of lipid mixing to less than 2% (Fig. 1E). Although we cannot rule out other mechanisms of lipid transfer between cells, such as asymmetric lipid transfer or transient membrane rupture followed by reannealing (32), the extensive lipid mixing most likely reflects a deficit of the GPI-linked SNARE complexes in destabilizing the lipid bilayers and opening a fusion pore.
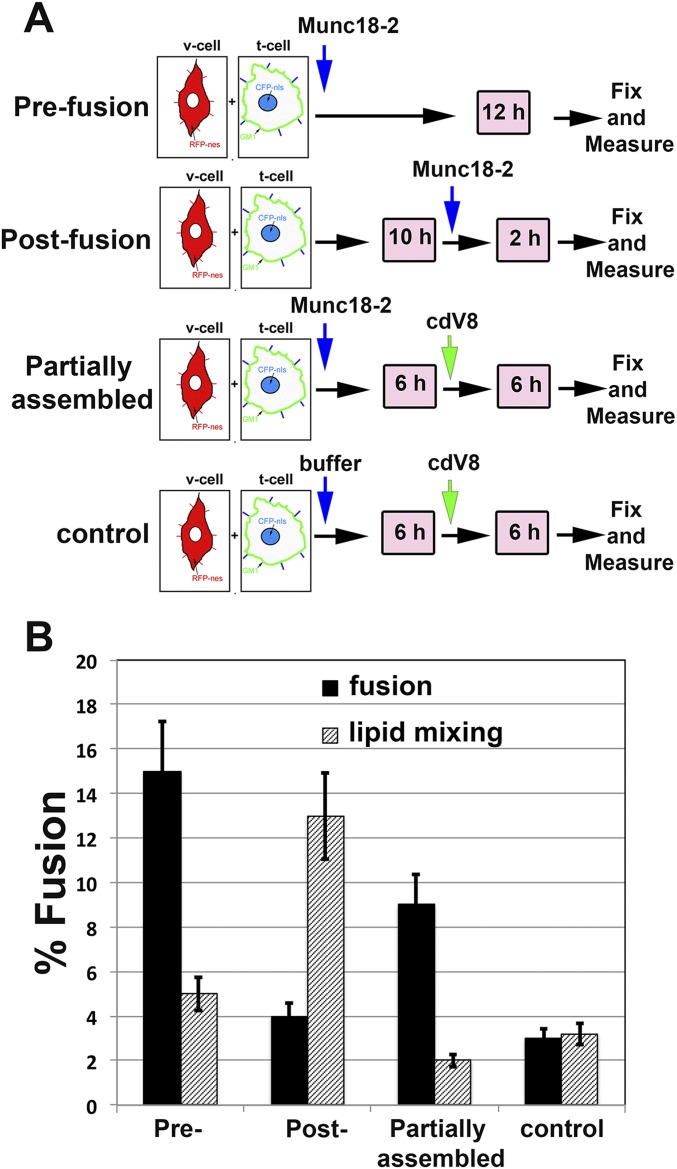
Munc18-2 Facilitates Transition from Lipid Mixing to Complete Fusion.
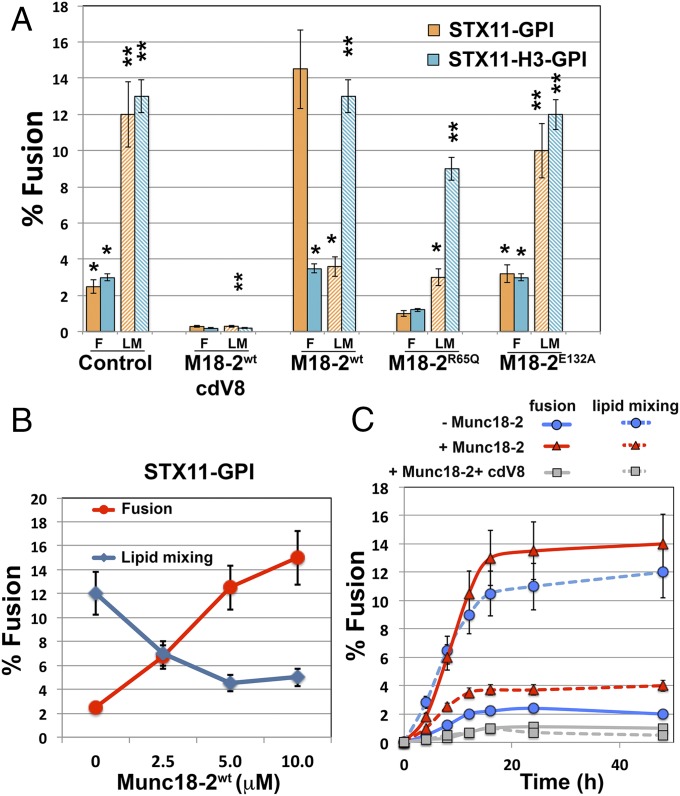
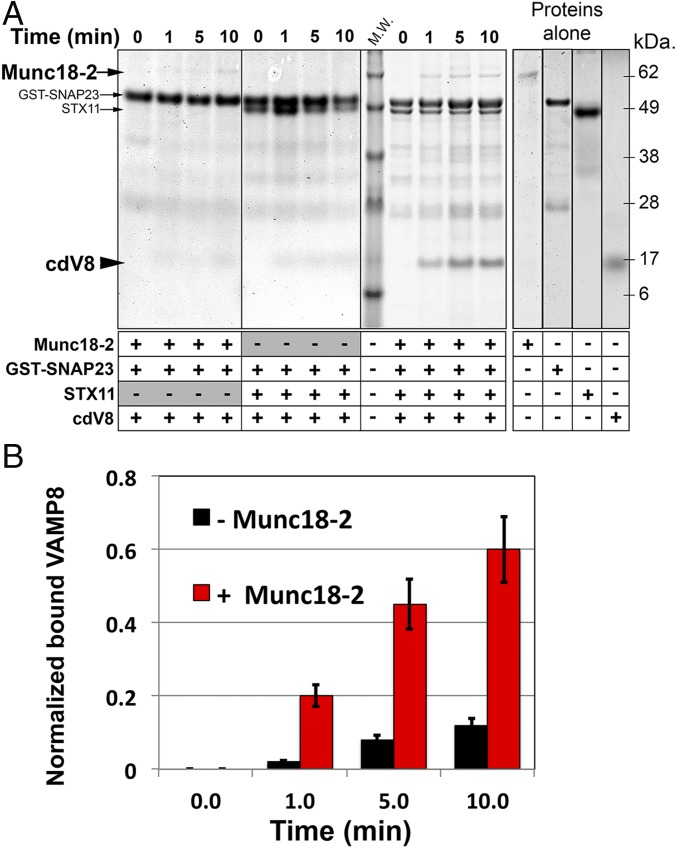
If lipid-anchored STX11 mainly promotes incomplete fusion, how can it be essential for degranulation and cell-mediated cytotoxicity in CTLs and NK cells? A recent study has shown that lipid-anchored STX1 mediates synaptic vesicle fusion when expressed in cultured neurons (30), whereas in vitro fusion assays show that it mainly promotes incomplete fusion events (23, 25, 26). This finding could indicate that additional factors may facilitate the transition from incomplete to complete fusion in vivo. One candidate that could perform this transition is Munc18-2, an SM protein that binds STX11 (13) and is expressed coordinately with STX11 (7). To test whether Munc18-2 affects the fusion activity of lipid-anchored STX11, we assayed cell fusion of t cells expressing flipped STX11–GPI and flipped SNAP23 with v cells expressing flipped VAMP8 in the presence or absence of recombinant Munc18-2 protein. Strikingly, addition of Munc18-2 to the reaction significantly increased the complete fusion events from 3.2 ± 0.6% to about 14.5 ± 1.3%, whereas the lipid mixing events inversely decreased from 12.0 ± 1.4% to 4.6 ± 0.8% (Fig. 2A). The increase in complete fusion events mediated by Munc18-2 was dose dependent (Fig. 2B) and reflected increased efficiency in full fusion but not faster kinetics than the lipid mixing events observed in the absence of Munc18-2 (Fig. 2C). Moreover, the effect of Munc18-2 required the presence of the STX11 Habc domain, because Munc18-2 had no effect on fusion mediated by STX11H3–GPI under the same conditions (Fig. 2A) and was inhibited by the addition of soluble VAMP8 cytoplasmic domain (cdV8) (Fig. 2 A and C). These results indicate that Munc18-2 acts as a positive regulator of STX11-mediated membrane fusion, and that lipid-anchored STX11 requires Munc18-2 to support complete membrane fusion when combined with SNAP23 and VAMP8.
Fig. 2.
Munc18-2 promotes the transition from incomplete to complete fusion. (A) Cell fusion experiments were performed as described in Fig. 1 using STX11–GPI (orange bars) or STX11H3–GPI (cyan bars) in the absence (control) or presence of either 5.0 μM Munc18-2WT and 5.0 μM cdV8, 5.0 μM Munc18-2WT, 5.0 μM Munc18-2R65Q, or 5.0 μM Munc18-2E132A. Graph shows the percentage of v and t cells in contact that underwent complete fusion (solid bars) or lipid mixing (dashed bars). Values are mean ± SE of three independent experiments using different preps of Munc18-2 proteins. (B) Dose-dependent effect of Munc18-2WT of cell fusion outcomes mediated by STX11–GPI. Cell fusion experiment was performed as Fig. 3C in the presence of increasing concentration (0, 2.5, 5.0, and 10 μM) of Munc18-2WT. (C) Time course of complete fusion and lipid mixing mediated by flipped STX11–GPI in the absence (blue lines) or presence of either 5.0 μM Munc18-2WT alone (red lines) or with cytoplasmic domain of VAMP8 (cdV8, gray lines). CHO v cells were mixed with MEF 3T3 stable t cells. At different time points, the percentage of v and t cells in contact that underwent complete fusion (solid lines) or lipid mixing (dashed lines) was determined using the assay described in B. Images in 50 random fields were used for calculation of each time point. Values are mean ± SD of three independent experiments. *P < 0.1; **P < 0.01.
Munc18-2R65Q is a recently described disease-associated mutant that displayed stronger binding to STX11 and inhibits membrane fusion in vitro and in vivo in a dominant-negative manner (33). Addition of Munc18-2R65Q to the flipped SNARE fusion assay with STX11–GPI drastically reduced both lipid mixing and complete fusion (Fig. 2A). However, consistent with the requirement for the Habc domain in associating with Munc18-2 (34), addition of Munc18-2R65Q had only a modest effect on fusion outcomes mediated by STX11H3–GPI (Fig. 2A). Consistently, addition of Munc18-2E132A—a Munc18-2 disease-associated mutant also located within the N lobe of Munc18-2 that reduced binding to STX11 (34, 35)—had no effect on the extent of lipid mixing or complete fusion mediated by either STX11–GPI or STX11H3–GPI (Fig. 2A). These data suggest that initial binding of Munc18-2 with the Habc domain of STX11 is critical for Munc18-2R65Q to arrest STX11-mediated membrane fusion at a step before lipid mixing.
Endogenous STX11 Mainly Interacts with SNAP23 and VAMP8 in Primary Human CTLs.
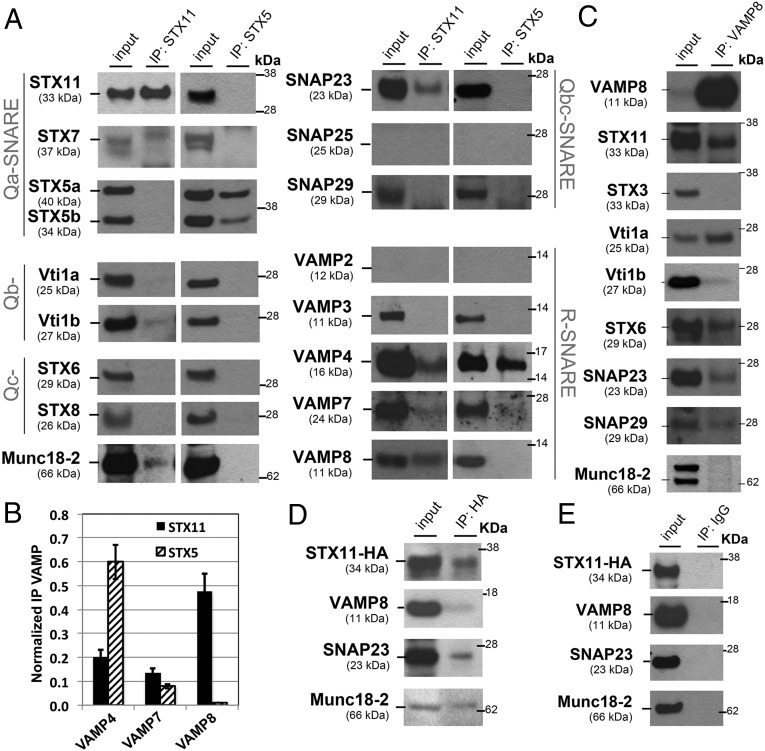
We evaluated whether cognate SNAREs that cooperate with STX11 in membrane fusion in vitro also participate with STX11 during cell-mediated cytotoxicity in primary CTLs. To this end, we used coimmunoprecipitation (co-IP) and immunoblotting to probe binding partners for endogenous STX11 (or STX5 as a negative control) in lysates of normal human CTLs that were activated with beads coated with anti-CD3 and anti-CD28 antibodies to induce lytic granule exocytosis. Lysates normalized for equal protein content were subjected to IP, and an equivalent aliquot was loaded in the input lanes for each blot. Among numerous Qb, Qc, and Qbc SNAREs tested, SNAP23 most prominently bound to STX11, but not to STX5—another Qa-SNARE expressed in CTL (Fig. 3A). Similarly, low levels of VTI1b also specifically co-IP with STX11, whereas SNAP25 was not detected in the cell lysates (Fig. 3A and Fig. S2D). As expected, Munc18-2 was only detected in the STX11 co-IP (Fig. 3A); the specificity of the Munc18-2 antibody was validated in CTLs treated with control or Munc18-2–specific small-interfering RNAs (siRNAs) (Fig. S2G). Among v-SNAREs (also known as R-SNAREs), VAMP4, VAMP7, and VAMP8 co-IP with STX11 to different degrees (Fig. 3A). In contrast, STX5 selectively co-IP VAMP4 and VAMP7, to a lesser extent, but not VAMP8. Normalization of the intensity of the v-SNARE bands in the co-IP to that of the cell homogenate (input, Fig. 3B) revealed that STX11 preferentially bound VAMP8 relative to VAMP7 or VAMP4, whereas STX5 preferentially bound VAMP4 relative to VAMP8 and -7. This indicates that SNAP23, VAMP8, and Munc18-2 specifically bind to STX11 but not STX5. The specificity of the anti-STX11 antibody was confirmed by transfection of cells lacking STX11 with control or STX11 expression constructs (Fig. S2A) and in CTLs treated with control or STX11-specific siRNAs (Fig. S2C). Similar results were obtained in co-IP experiments using a different anti-STX11 antibody that recognizes a central region of the protein. Although VAMP2 was suggested as the R-SNARE responsible for lytic granule release in mouse CTLs (36), we could not detect expression of VAMP2 in human CD8+ T cells (Fig. 3A and Fig. S2 E and F) or the NK cell line YTS (Fig. S2F). This lack of detection did not reflect lack of sensitivity of our anti-VAMP2 antibodies, as they detected endogenous VAMP2 in mouse CD4+, CD8+, and P815 cells and ectopically expressed human VAMP2 in HeLa cells (Fig. S2 E and F). These results show that endogenous STX11 physically interacts with SNAP23 and VAMP8 in human CTLs and suggest they might form a functionally active SNARE complex.
Fig. 3.
Endogenous STX11 interacts with SNAP23 and VAMP8 in activated CTLs. (A) Normal human donor CTLs purified from peripheral blood were activated using beads coated with anti-CD3 and -CD28 antibodies for 4 h. Endogenous STX11 and STX5 were immunoprecipitated from cell lysates using either rabbit anti-STX11 or rabbit anti-STX5 antibody and the indicated coimmunoprecipitated SNAREs or Munc18-2 were analyzed by Western blotting. (B) Bands in the Western blot that corresponded to the fraction of VAMP4/7/8 that coprecipitated with either STX11 or STX5 were quantified by densitometry and normalized to the total amount of STX11 or STX5, respectively, and immunoprecipitated in the same lane. Results represent mean ± SD of three independent experiments. (C) Endogenous VAMP8 was immunoprecipitated from activated CTL lysates as in A, and the indicated coimmunoprecipitated SNAREs or Munc18-2 was analyzed by Western blotting. Note that a goat anti–Munc18-2 antibody was used for Western blot in A, D, and E, giving a single band, whereas a rabbit anti–Munc18-2 antibody was used in C, giving a doublet. (D and E) HeLa cells were transiently transfected with HA–STX11. Immunoprecipitation from cell lysates was performed using either anti-HA (D) or IgG control antibody (E), and coimmunoprecipitated VAMP8, SNAP23, and Munc18-2 were analyzed by Western blotting. Blots are representative of three experiments.
To confirm these results, we performed complementary co-IP experiments in which endogenous VAMP8 was immunoprecipitated from CD3/CD28-bead activated CTLs. The results confirm that both STX11 and SNAP23 co-IP with VAMP8 (Fig. 3C), but additional interacting SNAREs were also detected, including VTI1a, VTI1b, STX6, and SNAP29. These data suggest that VAMP8 is involved in multiple trafficking steps in CTL as previously described (37, 38). To further validate the interaction among STX11, SNAP23, and VAMP8 we ectopically expressed HA–STX11 in HeLa cells and compared the complement of SNAREs that co-IP with anti-HA or an isotype control antibody. The data showed that endogenous VAMP8, SNAP23, and Munc18-2 specifically co-IP with HA–STX11 using anti-HA antibodies (Fig. 3 D and E). Together, these data support the view that in ex vivo CTLs, STX11 preferentially associates with VAMP8 and SNAP23.
STX11 Interacts in Vitro with SNAP23, VAMP8, and VAMP2 but Not with VAMPs 3, 4, or 7.
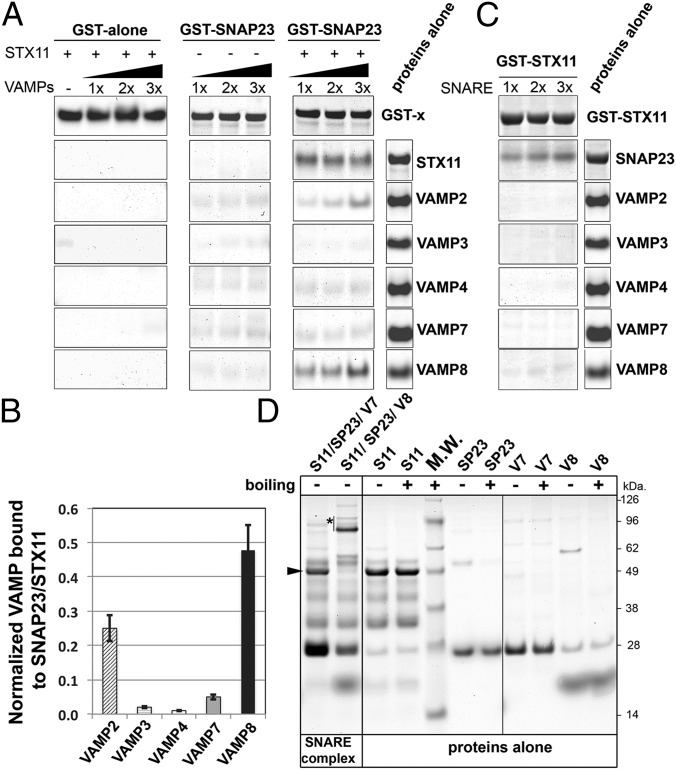
To analyze whether STX11 can bind directly to SNAP23 and VAMP8 and to evaluate the specificity of these interactions, we performed pull-down experiments using recombinant proteins expressed in bacteria. GST alone or a GST–SNAP23 fusion protein was coupled to glutathione-Sepharose (GS) beads and incubated with increased concentrations of VAMPs 2, 3, 4, 7, or 8 in the presence or absence of STX11, and bound proteins were harvested by centrifugation and analyzed by SDS/PAGE and Coomassie blue staining. STX11 bound to GST–SNAP23 but not to GST alone (Fig. 4A, Right). None of the VAMPs bound to GST alone regardless of the presence of STX11 (Fig. 4A, Left) or to GST–SNAP23 in the absence of STX11 (Fig. 4A, Middle). In the presence of STX11, however, VAMP2 and VAMP8—but not VAMPs 3, 4, or 7—bound in a dose-dependent manner to GST–SNAP23 (Fig. 4A, Right). Quantification of the bound fractions revealed that the STX11/SNAP23 complex binds more avidly to VAMP8 than to VAMP2 (Fig. 4B). Conversely, immobilized GST–STX11 bound to SNAP23 but not to any of the tested VAMPs in the absence of SNAP23 (Fig. 4C). These data suggested that VAMP8, and to a lesser extent VAMP2 (which is not present in human CTLs), binds STX11 in complex with SNAP23.
Fig. 4.
VAMP8 or VAMP2 bind to a recombinant STX11/GST–SNAP23 complex. (A) Equivalent amounts of recombinant GST (GST alone) or GST–SNAP23 were bound to glutathione-Sepharose beads, and increasing concentrations (0.5, 1.0, or 1.5 μg) of recombinant VAMP2, -3, -4, -7, and -8 were added to the beads in the absence or presence of 1.0 μg recombinant STX11 (amino acids 1–276) and incubated for 1 h at room temperature. Bound fractions were analyzed by SDS/PAGE and Coomassie blue staining. Proteins-alone lane shows representative images of the total load of each protein used in these experiments. (B) Plot showing quantification of the amount of VAMP protein bound to the beads normalized to the amount of STX11 retained in each experiment. Data represent mean ± SD of three independent experiments. (C) Recombinant GST–STX11 was bound to glutathione-Sepharose beads, and increasing concentrations (0.5, 1.0, or 1.5 μg) of recombinant SNAP23 or VAMP2, -3, -4, -7, and -8 were added to the beads. Bound fractions were analyzed by SDS/PAGE and Coomassie blue staining. Proteins-alone lane is identical to the one shown in A because both experiments were performed in parallel with the same batch of proteins. (D) Recombinant His–SUMO–STX11 was incubated with SNAP23 and either VAMP8 or VAMP7 for 1 h at room temperature, and the formation of high molecular weight complexes was analyzed by SDS/PAGE without boiling the samples. Arrowhead shows the band corresponding to His–SUMO–STX11, which largely disappeared upon SNARE complex formation with SNAP23 and VAMP8 (asterisk), but not with VAMP7. Proteins-alone lane shows the load of each protein used to generate the SNARE complexes. M.W., molecular weight markers (shown to the Right of the gel).
To test directly whether STX11 forms a stable, SDS-resistant SNARE complex, we incubated STX11 with SNAP23 and either VAMP8 or VAMP7 at room temperature and fractionated the resultant mixtures by SDS/PAGE and Coomassie blue staining. Following incubation with SNAP23 and VAMP8, but not with SNAP23 and VAMP7, the Mr 48,000 band corresponding to STX111–276 fused to a small ubiquitin-like modifier (SUMO) tag (Fig. 4D, arrowhead) largely shifts to Mr 80–96 kDa as expected for the SDS-resistant SNARE complex (Fig. 4D, asterisk). These data support a preferential association of STX11 with SNAP23 and VAMP8 as predicted from the co-IP analyses from CTL homogenates (Fig. 3) and from the in vitro fusion experiments (Fig. 1C). Moreover, these data show that neither STX11 nor SNAP23 alone can directly interact with any of the tested VAMPs, but rather that a STX11/SNAP23 t-SNARE complex specifically binds VAMP8 and VAMP2. Therefore, the STX11/SNAP23/VAMP8 complex likely mediates CTL-mediated cytotoxicity. The association of STX11 with VAMP4 and VAMP7 detected by co-IP from CTLs most likely represents either the formation of alternative SNARE complexes by a small fraction of STX11, indirect associations mediated through another protein, or a postlysis association.
Munc18-2 Binds Directly to both STX11 Alone and to the STX11/SNAP23/VAMP8 SNARE Complex.
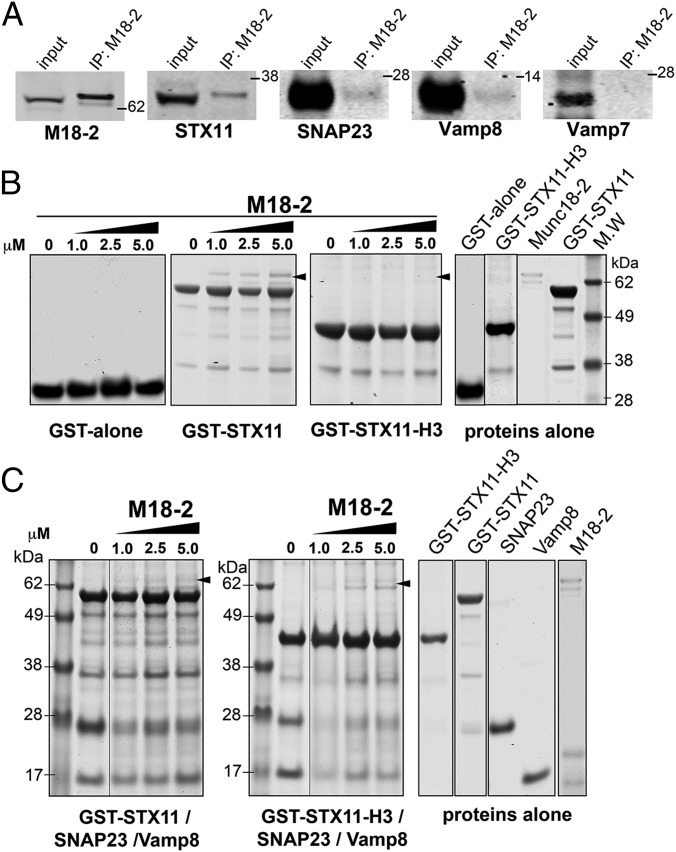
Previous studies have suggested that Munc18-2 can interact with the monomeric Syntaxins STX3 (39, 40), STX2 (41), and STX11 (13, 33–35). However, other SM proteins have distinct binding modes in which they engage either monomeric syntaxins or SNARE complexes (42–45). To determine whether endogenous Munc18-2 can directly interact with STX11 alone or with a STX11/SNAP23/VAMP8 SNARE complex and/or promotes its formation, we performed a series of experiments. First, we tested exploited co-IP experiments from lysates of activated CTLs. Consistent with the detection of Munc18-2 in immunoprecipitates of both STX11 and VAMP8 (Fig. 3 A and C), STX11, SNAP23, and VAMP8, but not VAMP7, were detected in Munc18-2 immunoprecipitates (Fig. 5A). Similar results were obtained using a different anti–Munc18-2 antibody. Because the detection of VAMP7 in lysate was significantly lower than VAMP8 and SNAP23, the results from our co-IP experiments could not completely rule out a potential association between Munc18-2 and VAMP7. We next tested whether these associations can be recapitulated in vitro by performing pull-down experiments in which we incubated immobilized GST alone, GST–STX11, or GST–STX11–H3, which lacks the N-terminal residues 1–157, with increasing concentrations of Munc18-2. Bound fractions were analyzed by SDS/PAGE followed by Coomassie blue staining. Data showed that Munc18-2 specifically associates with GST–STX11 beads in a dose-dependent manner (Fig. 5B). The Munc18-2/STX11 interaction requires of the STX11 N-terminal domain because the amount of Munc18-2 bound to GST–STX11–H3 was significantly reduced compared with full-length GST–STX11 (Fig. 5B). We then tested whether Munc18-2 can bind to STX11/SNAP23/VAMP8 complex. To this end, we first incubated either GST–STX11 or GST–STX11–H3 beads with soluble SNAP23 and VAMP8. Beads were washed to remove unbound fractions and then incubated with increasing concentrations of Munc18-2. Bound fractions were analyzed by SDS/PAGE followed by Coomassie blue staining. The results showed a dose-dependent association of Munc18-2 with both GST–STX11 and GST–STX11–H3/SNAP23/VAMP8 complexes (Fig. 5C, arrowheads), although to a lesser extent than with GST–STX11 alone (Fig. 5B). Taken together, the results show that Munc18-2 interacts with both STX11 alone and with the STX11/SNAP23/VAMP8 SNARE complex both in CTLs and in vitro. Additionally, our biochemical data indicate that the STX11 N-terminal domain is necessary for the binary association STX11/Munc18-2, but it is dispensable for the Munc18-2 association with the STX11/SNAP23/VAMP8 complex.
Fig. 5.
Munc18-2 binds to STX11 alone and to STX11/SNAP23/VAMP8 SNARE complex. (A) Coimmunoprecipitation experiments using lysates generated from normal control human CTLs activated with beads coated with anti-CD3 and CD28 antibodies. Endogenous Munc18-2 was immunoprecipitated using an anti–Munc18-2 antibody and the amount of the indicated SNAREs that coimmunoprecipitated was analyzed by Western blotting. (B) Pull-down experiments in which equivalent amounts of recombinant GST protein alone (GST alone), GST–STX11(amino acids 1–276), or GST–STX11–H3 (amino acids 158–276) were bound to GS beads and increasing concentrations (0, 1.0, 2.5, or 5.0 μM) of recombinant Munc18-2WT was added to the beads. Bound fractions were analyzed by SDS/PAGE and Coomassie blue staining. Proteins-alone lanes show representative images of the proteins used during the experiment. (C) Pull-down experiments in which equivalent amounts of recombinant GST–STX11 or GST–STX11–H3 were bound to GS beads in the presence of SNAP23 and soluble VAMP8. Beads were extensively washed to remove any unbound soluble protein and then they were incubated with increasing concentrations (0, 1.0, 2.5, or 5.0 μM) of recombinant Munc18-2WT. Bound fractions were analyzed by SDS/PAGE and Coomassie blue staining. Gels are representative of two independent experiments.
Munc18-2 Acts During SNARE Complex Assembly but Not on Fully Assembled SNARE Complexes.
To better understand the mechanism by which Munc18-2 facilitates the transition from lipid mixing to complete fusion mediated by lipid-anchored STX11, we carried out a series of “order-of-addition” experiments in the cell fusion assay with flipped STX11–GPI/SNAP23 t cells and flipped VAMP8 v cells (Fig. S3A). First, we tested whether Munc18-2WT had a similar effect when added at the time we mixed v and t cells (prefusion) or 10 h later (postfusion) when the cells have already exchanged lipids. The results showed that Munc18-2 did not significantly affect the relative ratios of the distinct fusion events when it is added postfusion (Fig. S3B), suggesting that Munc18-2 cannot act on fully assembled SNAREpins that had already undergone lipid mixing. Furthermore, these data rule out the possibility of a nonspecific effect of Munc18-2 on lipid mixing independently of SNARE complexes as described for denatured squid Munc18-1 (46). We next investigated whether Munc18-2 accelerates or stabilizes the formation of partially assembled SNARE complexes resistant to cdV8. It would be expected that cdV8 titrates down the available t-SNAREs and blocks the formation of new SNARE complexes, thus inhibiting membrane fusion. Indeed, the results showed that addition of cdV8 6 h after Munc18-2 addition modestly reduced the level of complete fusion observed, but did not impact the ratio of full fusion to lipid mixing. By contrast, in control experiments in which buffer was added instead of Munc18-2, both complete fusion and lipid mixing were inhibited. These data suggest that Munc18-2 accelerates/stabilizes the formation of partially assembled SNARE complexes, which are not accessible to cdV8, and also facilitates the progression from incomplete to complete fusion during the fusion reaction.
Fig. S3.
Munc18-2 promotes transition from incomplete to complete fusion by acting on partially assembled SNARE complex. (A) Schematic representation of order of addition experiments using the cell fusion assay as described in Fig. 1A in which Munc18-2WT was added at different times as indicated. Prefusion condition, Munc18-2WT was added at the time the v- ant t- cells were mix (Pre); Postfusion, Munc18-2WT was added after 10 h of the v ant t cells were mix (post); Munc18-2WT (partially assembled) or buffer (control) were added at the time of mixing v ant t cells, incubated for 6 h and cdV8 was added for an additional 6 h. (B) Graph showing the percentage of cells that underwent complete fusion (black bars) and lipid mixing (striped bars) in the cell fusion experiments described above in A.
Munc18-2 Promotes the Assembly of STX11-Containing SNARE Complexes.
To investigate whether Munc18-2 directly facilitates SNARE-complex assembly we set up two different assays. First, we measured the formation of STX11/SNAP23/VAMP8 complex over time in the presence or absence of Munc18-2 by pull-down experiments using recombinant proteins. We immobilized GST–SNAP23 on GS beads, added an equimolar amount of cdVAMP8 with or without STX11 and Munc18-2, and incubated the mixtures for 0–10 min. Bound fractions were analyzed by SDS/PAGE and Coomassie blue staining. As expected, in the absence of STX11, neither VAMP8 nor Munc18-2 bound to GST–SNAP23 over a 10-min period (Fig. 6A, Left). Similarly, a small amount of VAMP8 bound to GST–SNAP23 in the presence of STX11 and absence of Munc18-2 after these short incubations (Fig. 6A, Middle). Strikingly, addition of Munc18-2 to the reaction increased by over fivefold the amount of VAMP8 retained with GST–SNAP23 and STX11 (Fig. 6A, Right, and B). These data support a direct role for Munc18-2 in assisting the formation of STX11/SNAP23/VAMP8 SNARE complexes in vitro. Interestingly, the results also indicate that the formation of the STX11/SNAP23 t-SNARE complex is not influenced by Munc18-2.
Fig. 6.
Munc18-2 induces STX11/SNAP23/VAMP8 complex formation. (A) Pull-down experiments in which equivalent amount of recombinant GST–SNAP23 was bound to GS beads and incubated for a short period (0, 1, 5, and 10 min) with either 5.0 μM cdV8 and 5.0 μM Munc18-2 in the absence of His–SUMO–STX11 (Left), or with of 5.0 μM cdV8 and 5.0 μM His–SUMO–STX11 in the absence (Middle) or in the presence of 5.0 μM Munc18-2 (Right). Bound fractions were analyzed by SDS/PAGE and Coomassie blue staining. Proteins-alone lane shows representative images of the proteins used during the experiment. (B) Plot shows the quantification of the amount of cdV8 protein bound to the beads normalized by the amount of STX11 retained on each condition in the absence (black) or in the presence (red) of Munc18-2. Gel and plots are mean ± SD of three independent experiments.
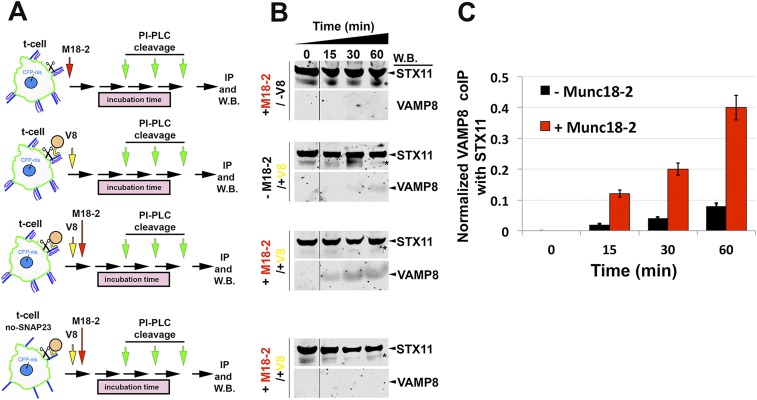
In a second approach, we developed an assay to test whether Munc18-2 facilitates the formation of trans-SNARE complexes—i.e., association of v- and t-SNAREs inserted in apposing membranes—while preserving the physical and topological barrier of the lipid layers. We incubated flipped t cells expressing STX11–GPI with or without SNAP23 together with liposomes containing VAMP8 in the presence or absence of Munc18-2 for the indicated time to allow for the formation of trans-SNARE complexes. Cells were then washed and STX11 was released from the membrane by the addition of phosphatidylinositol-specific phospholipase C (PI-PLC), which cleaves the GPI anchor between the glycerol backbone and the phosphate group. Released STX11 was then immunoprecipitated from the medium, and STX11 and VAMP8 within the immunoprecipitates were quantified by Western blotting (Fig. S4A). The data show that fourfold more VAMP8 was co-IPed with STX11 from samples in which Munc18-2 was added relative to those in which Munc18-2 was absent (Fig. S4 A and B, Middle, and C). No VAMP8 was co-IPed with STX11 when the t cells lack flipped SNAP23, regardless of the presence of Munc18-2, indicating that VAMP8 association requires the complete t-SNARE complex (Fig. S4 A and B, Lower). Taken together, these results strongly suggest that Munc18-2 increases the efficiency of trans-SNARE complex formation. The increased number of such complexes working in a concerted manner may provide enough energy to overcome the energetic barrier to transition from lipid mixing to complete fusion.
Fig. S4.
Munc18-2 induces STX11/SNAP23/VAMP8 complex formation. (A) Schematic representation showing the assessed conditions for the trans-SNARE complex formation assay. Cells expressing flipped STX11–GPI, with or without flipped SNAP23 (Lower), were incubated with VAMP8 protein reconstituted onto artificial liposomes in the presence or absence of recombinant Munc18-2 protein for the indicated period 0, 15, 30, or 60 min. After this time, cells were washed with cold PBS and treated with 1 unit/mL of PI-PLC for 15 min at 37 °C to cleave off STX11 from its GPI-anchor motif. Supernatants were collected and STX11 was IP using anti-STX11 antibody. (B) Bound fractions for each IP described in A were analyzed by Western blot using anti-STX11 and anti-VAMP8 antibodies. Asterisks mark an unspecified band obtained with anti-STX11 antibody. (C) Plot shows the quantification of the amount of co-IPed VAMP8 protein normalized by the amount of STX11 retained on each condition in the absence (black) or in the presence (red) of Munc18-2. Gel and plots are mean ± SD of three independent experiments.
Munc18-2 and STX11 Are Required for CTL-Mediated Cytotoxicity.
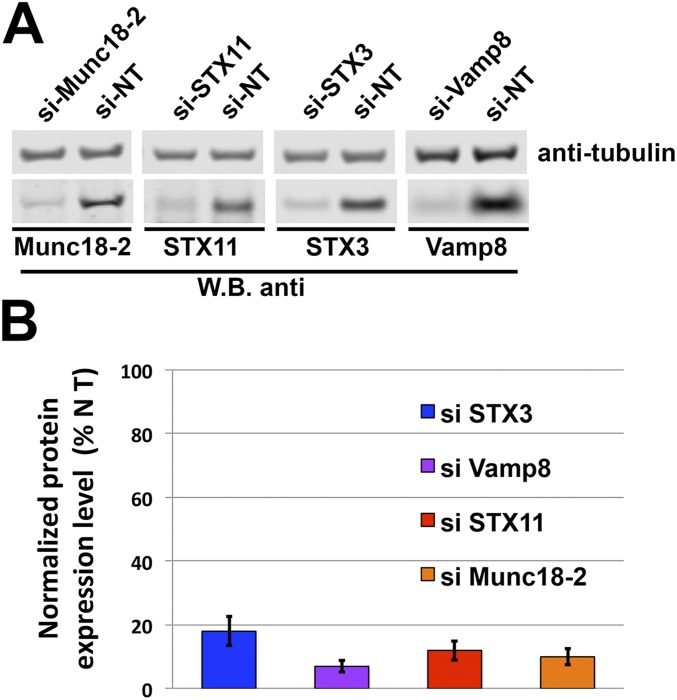
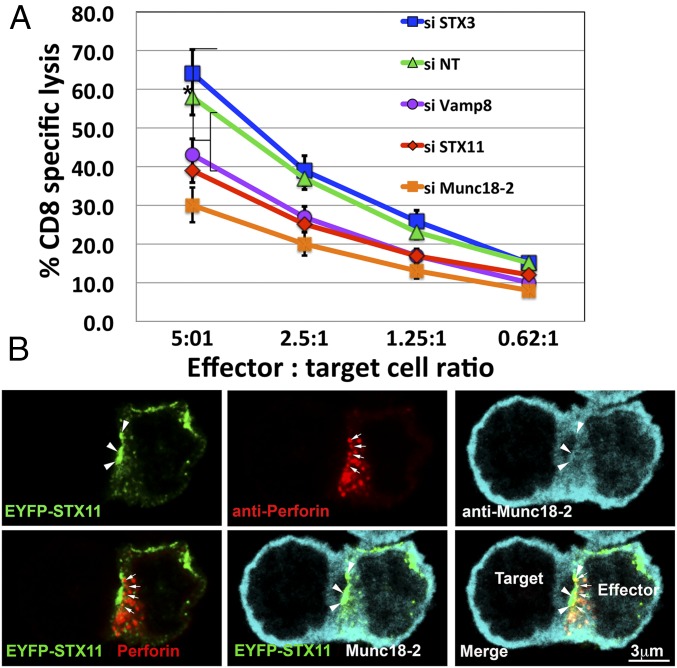
CTLs from STX11 knockout mice (47, 48) and of patients with F-HLH-4 and -5, which have mutations in STX11 and Munc18-2, respectively (7, 17, 49), display reduced cytotoxic activity toward sensitive target cells. In particular, the defective cytotoxic activity associated with F-HLH–associated missense mutations STX11–L58P (50), Munc18-2–E132A (34) and Munc18-2–R65Q (33), which interfere with the STX11/Munc18-2 association, reflect a direct role of this interaction for mediating lytic granule content release. To directly evaluate the requirement of the Munc18-2/STX11/SNAP23/VAMP8 complex for CTL-mediated cytotoxicity, we knocked down their expression in human CTLs and measured the effect on killing activity toward sensitive target cells. To do this, we treated human CTLs with siRNAs targeted to Munc18-2, STX11, or VAMP8, or to STX3—another Munc18-2 interacting syntaxin—as a negative control, and compared their cytotoxic activity with that of cells treated with nontargeting (NT) siRNA. All of the siRNAs were effective and resulted in more than 80% knockdown efficiency when assessed by Western blotting (Fig. S5 A and B). Data from cell-killing assays showed that CTLs treated with siRNAs targeted to Munc18-2, STX11, and VAMP8, but not to STX3, displayed a significantly reduced cytotoxic activity at different effector/target cell ratios (Fig. 7A). Similar results were obtained using different siRNAs for each candidate gene, thus diminishing the possibility of off-target effects. These results are consistent with previous findings in F-HLH patient cells and mouse models and with a direct role of Munc18-2, STX11, and VAMP8 on CTL-mediated cytotoxicity.
Fig. S5.
Munc18-2 and the STX11/SNAP23/VAMP8 SNARE complex is required for CTL-mediated cytotoxicity. (A) Human CTLs were electroporated with either nontargeting (NT), Munc18-2, STX11, STX3, or VAMP8 siRNAs and cultured for 48 h. Cell lysates were analyzed by Western blot for the indicated proteins. (B) Plot showing the quantification of the knockdown efficiency of each protein compared with NT siRNA-treated cells as shown in A. Blots are representative of three independent experiments.
Fig. 7.
Munc18-2 and the STX11/SNAP23/VAMP8 SNARE complex is required for CTL-mediated cytotoxicity. (A) Cytotoxicity assay to measure CTL-mediated cell killing of human CTLs electroporated with either nontargeting (NT), Munc18-2, STX11, STX3, or VAMP8 siRNAs and cultured for 48 h. Equivalent numbers of CTLs (effectors) from each condition were incubated with anti-CD3 antibody in the presence or absence of P815 target cells (targets) at the indicated cell ratios. The killing assay was run for 4 h at 37 °C and the amount of lactate dehydrogenase (LDH) released into the supernatant was quantified using a Cytotox-96 assay. Knockdown efficiency for each siRNAs is shown in Fig. S5 A and B. Results are the mean ± SD of three independent measurements for each condition. *P < 0.01. (B) Human CTLs transfected with EYFP–STX11 using the Neon electroporation system were incubated in the presence of anti-CD3 antibody and P815 cells at a 1:1 ratio for 15 min at 37 °C and seeded onto polylysine-coated coverslips. Cells were fixed, permeabilized, and stained using first goat anti–Munc18-2 and followed by anti-perforin–Alexa 647 antibodies. (Scale bar, 3 μm.) Arrows show perforin granules close to the IS. Arrowheads show accumulation of EYFP–STX11 and Munc18-2 at the IS membrane. (Scale bar, 3 μm.)
To test whether the SNAREs and Munc18-2 function directly in cytolytic granule secretion, we next examined their subcellular localization during cytolysis using stimulated emission depletion (STED) superresolution microscopy. To this end, human CTLs expressing EYFP–STX11 were incubated with P815 target cells for 15 min to allow for the formation of a physiological IS before fixation. Analysis of cells that were labeled for Munc18-2 and perforin revealed that a fraction of STX11 accumulated at the IS (Fig. 7B, arrowheads) where the polarized perforin-containing granules make contact (Fig. 7B, arrows). Although the majority of Munc18-2 appeared diffuse throughout the cytoplasm, a cohort colocalized with STX11 at the IS membrane (Fig. 7B, arrowheads). In summary, our results provide direct evidence that STX11 and Munc18-2 are present at the IS during lytic granule exocytosis and subsequently are required for granule content release for cell-killing activity.
Discussion
The molecular fusion mechanisms underlying the release of lytic granule content by CTLs and NK cells for cell-mediated cytotoxicity have remained elusive. By using an in vitro cell–cell fusion assay, we demonstrate here for first time that STX11 is sufficient to drive membrane fusion in complex with SNAP23 and either VAMP8 or VAMP2. More generally, we show that the nature of the STX11 membrane anchor within this complex determines the final fusion outcome. Whereas STX11 that is artificially linked to a membrane spanning domain drives primarily complete fusion events, STX11 with a glycophospholipid anchor—mimicking its natural prenyl- and palmitoyl-lipid anchors—primarily mediates incomplete fusion events resulting in extensive lipid mixing reminiscent of a hemifusion state. Strikingly, we found that a transition from incomplete to complete fusion mediated by lipid anchored STX11 could be stimulated by the SM protein, Munc18-2. Because the GPI motif we appended to STX11 for our cell fusion assay intrinsically differs from its natural prenyl- and palmitoyl-lipid modification, it cannot be assumed that Munc18-2 will have a similar effect on STX11 carrying natural lipid modifications. Moreover, the presence of glycoconjugated molecules on the extracellular leaflet of the plasma membrane in our cell–cell fusion assay might be another confounding factor that may increase the energy barrier for membrane fusion and thus favoring lipid mixing. Further experiments will be necessary to confirm this. Taken together, our results reveal that Munc18-2 does not act just as an accessory or a chaperone for STX11 but rather play a direct role on membrane merging. Thus, providing a potential explanation for the physical and functional requirements of STX11 and Munc18-2 during cell-mediated cytotoxicity and the association of STX11 and STXBP2 mutations with F-HLH.
Although our findings are consistent with numerous data attributing a key role for the TMD of SNARE proteins in promoting membrane fusion and fusion pore opening (23–29, 51), they also extend our understanding about how SM proteins can assist lipid-anchored SNAREs in mediating complete membrane fusion. In these lines, these data agree with a report showing that addition of the yeast HOPS complex, which includes the SM protein VPS33, restores complete fusion in a liposome-based fusion reaction containing reconstituted lipid-anchored v-SNARE, Nyv1p (27). A general role for SM proteins in supporting fusion by lipid-anchored SNAREs could also explain how the expression of lipid-anchored neuronal SNAREs can restore neurotransmitter release in Stx1 or Vamp2 knockout cultured neurons (30).
Our data provide mechanistic insight into how Munc18-2 function is coupled to STX11-mediated fusion. We found that the ability of SM proteins to bind to SNARE complexes (42–45, 52) is conserved in Munc18-2. Moreover, we found that Munc18-2 accelerates and stabilizes the assembly of STX11/SNAP23/VAMP8 trans-SNARE complexes, which is particularly important to promote the transition to full fusion by lipid-anchored STX11, thus supporting for a direct role of SM proteins on membrane merging. This activity of Munc18-2 requires the presence of the STX11 N-terminal domain containing the conserved N peptide and the regulatory Habc region as shown by the inability of the F-HLH–associated Munc18-2E132A mutant to drive the transition from lipid mixing to complete fusion. These results are consistent with previous binding studies that show that the mutation E132A, which lies within Munc18-2 N-lobe domain, severely affects the interaction with STX11 N-terminal domain (34, 35). On the other hand, Munc18-2R65Q mutant, which binds SNARE complexes more efficiently than Munc18-2WT but acts in dominant-negative fashion most likely arresting SNAREpin formation (33), had an inhibitory effect on the extent of both lipid mixing and complete fusion mediated by either STX11–GPI or STX11H3–GPI. These results suggest that the Munc18-2R65Q mutant stabilizes the assembly of SNARE complexes at an intermediate state before lipid mixing and interferes with complete zippering of the SNARE complex. Therefore, the Munc18-2R65Q mutant seems to dissociate two distinct functions attributed to SM proteins: the stimulatory effect on membrane fusion observed in ensemble lipid mixing assays (43, 53, 54) and the inhibitory function recently described in single molecule experiments (55). Order-of-addition experiments show that Munc18-2 accelerates the formation cdV8-resistant SNARE complexes, therefore supporting a role of Munc18-2 in proof-reading trans-SNARE complexes (52, 55–57). Additionally, our data also suggest that Munc18-2 facilitates SNAREpin zippering similarly to other SM proteins (43, 54, 58–63). Thus, although the GPI anchor may be less effective than the transmembrane domain in transducing the energy of SNARE assembly to drive membrane fusion, the increased number of assembled STX11–GPI SNARE complexes generated in the presence of Munc18-2 likely cooperate, giving rise to a higher energetically favorable state that could be sufficient to destabilize the phospholipid bilayers and result in complete membrane fusion. These interpretations are supported by data on liposome fusion assays showing that the extent of lipid mixing directly correlated with the number of assembled SNARE complexes (64).
Our results, together with data from previous functional studies in F-HLH patient cells, strongly suggest that STX11/SNAP23/VAMP8 SNARE complex mediates lytic granule fusion at the immunological synapse and that Munc18-2 is necessary to drive these membrane fusion events. We found that endogenous STX11 and Munc18-2 mainly associate with SNAP23 and VAMP8 in human CTLs upon stimulation. These findings corroborate previous studies showing that STX11 interacts with SNAP23 and VAMP8 in platelets and facilitates the secretion of lysosomes, dense granules and alpha granules, which is also impaired in patients with F-HLH-4 and -5 (13, 14). Our high-resolution microscopy analyses show that STX11 and Munc18-2 localize to the IS membrane where the perforin-containing granules make contact before being released, supporting the requirement for these SNARE complexes in cytolytic granule release as previously suggested. Therefore, this fusion machinery seems to be essential for mediating regulated exocytosis in different cell types. Nonetheless, our co-IP data show a weak interaction of STX11 with VTI1b, VAMP4 and -7, consistent with the interaction of STX11 with VTI1b and inhibitory function in endolysosomal fusion in a macrophage cell line (16). We thus cannot rule out additional functions of STX11 in the endolysosomal pathway, and that such functions—such as in phagocytosis—might be more pronounced in other cell types. However, our studies suggest that these are not the primary interactions in CTLs and NK cells.
Although VAMP2 was recently proposed as the vesicular SNARE that mediates LG exocytosis in mouse CTLs (36), our data show that human CTLs do not express VAMP2 at detectable levels, and that VAMP8 knockdown substantially impairs the cytolytic activity of human CTLs. It is possible that there is substantial divergence in the machinery that controls LG exocytosis in mouse and human cells. However, other groups have observed that VAMP8 colocalizes with LG markers, and Vamp8 knockout mice displayed reduced granule exocytosis and defective cytotoxic activity (65, 66). Moreover, a recent study proposed that human CTL cytotoxicity depends on VAMP8-mediated recycling of endosomes at the IS (37), although these fusion events may involve a different set of SNAREs such as VTI1a, VTI1b, STX6, or SNAP29 that we identified as VAMP8 binding partners in Fig. 3. Therefore, the current data suggest that VAMP8 plays a key role in CTL cytotoxicity by directing multiple membrane trafficking steps during lytic granule maturation and secretion.
The LG exocytic machinery constitutes an ideal system in which to study the contribution of SM proteins in stimulating membrane fusion. Because the artificially lipid-anchored STX11 can only mediate incomplete fusion events in the absence of Munc18-2, it is easier to dissect the contribution of the SM protein for late stages of the fusion reaction than for other fusion reactions that use transmembrane domain-containing syntaxins and VAMPs. Taken together, our data support a model in which SM proteins are part of the core fusion machinery and have a direct role in inducing membrane fusion. Moreover, our results provide insights into how a key component of the exocytic machinery—the SM proteins—may promote complete fusion events by SNAREs lacking a traditional transmembrane domain in vivo by promoting SNARE complex assembly. This process may represent a critical step for exocytosis in CTLs, NK cells, and platelets in which other regulatory proteins, most likely calcium sensor proteins, tightly control to prevent the unnecessary release of harmful mediators to the extracellular space.
Materials and Methods
Human Samples.
Written consent was obtained from healthy donors for primary CD8+ T cells using a protocol approved by the Institutional Review Board at The Children's Hospital of Philadelphia.
Antibodies.
Mouse anti-GFP was from Roche. Monoclonal anti-myc (clone 9E10) and anti-HA (clone 11) from Covance. Antibodies anti-STX5 and anti-VAMP7 were produced and kindly provided by Dr. James Rothman, Yale University, New Haven, CT. Rabbit anti-VAMP3 was from Novus Biologicals. All other antibodies against SNARE proteins including rabbit anti-STX11 and rabbit anti–MUNC18-2 were purchased from Synaptic Systems. Alternatively, we also used rabbit anti-human STX11 from Sigma and goat anti-Munc18-2 from Santa Cruz Biotechnology.
Cell–Cell Fusion Assay.
Cell–cell fusion assay was performed as previously described (25). Briefly, 24 h before performing transfection, 7 × 104 MEF 3T3 cells were seeded on sterile 12-mm glass coverslips contained in 24-well plates and 8 × 105 CHO cells were seeded on 6-well plates. MEF 3T3 cells were transiently cotransfected with pCFP fused to nuclear localization signal (pCFP-nls), flipped STX11 construct, and flipped SNAP23/25 or -29 constructs using Lipofectamine 2000. CHO cells were cotransfected with pCH19 and the indicated flipped VAMP constructs to generate v cells. Twenty-four hours posttransfection, CHO v cells were detached from the dishes with EDTA [Cell Dissociation Solution (Sigma)] and 1 × 105 cells were added to each coverslip already containing the t cells. After various times at 37 °C in 5% (vol/vol) CO2, the coverslips were gently washed once with PBS supplemented with 0.1 g/L CaCl2 and 0.1 g/L MgCl2 (PBS++), then fixed with 4% (wt/vol) paraformaldehyde for 30 min, washed three times with PBS++, and incubated for 15 min with 0.5 μg/mL FITC-cholera toxin β-subunit (Sigma). After three washes with PBS++ the coverslips were mounted with Prolong Antifade Gold mounting medium (Molecular Probes). Confocal images were collected as indicated in Image Acquisition. At each time point the total number of fused cells (f) or hemifused-like cells (LM) and the total number of v cells (V) and t cells (T) in contact with each other (but not yet fused or hemifused) in random fields were determined. Images were analyzed using FIJI software (67). Objects were identified using the “analyze particle” function in each channel (red for v cells, CFP for t cells, and green for lipid marker) and setting up the minimum threshold as 20 fluorescence intensity arbitrary units (AU). Then we quantified the total number of each cell population and identified red cells with either blue nucleus and/or green staining to determine fused (f) vs. lipid mixing (LM) cells. The efficiency of fusion (F) or lipid mixing (LM) as percentage were calculated as follows: F = 2f/( V + T + 2f) × 100; LM = 2LM/(V + T + 2LM) × 100.
Image Acquisition.
Images were acquired on a Leica SP5-STED confocal microscope, equipped with Leica LAF software and usually using a HCX PL APO 20×, 1.25 N.A. objective. For higher magnification images a HCX PL APO 63×, 1.4 N.A. oil immersion objective was used. The images were analyzed using FIJI software and processed with Adobe Photoshop software.
Additional materials and methods are described in SI Materials and Methods.
SI Materials and Methods
Cells and Cell Lines.
Written consent was obtained from healthy donors for primary CD8+ T cells using a protocol approved by the Institutional Review Board at the Children’s Hospital of Philadelphia. Samples were collected in EDTA tubes and processed within 24 h of venipuncture. PBMC cells were obtained by density gradient centrifugation (Lymphoprep, Axis-Shield). CD8+ T cells were isolated from PBMC fraction using Dynabeads untouched human CD8+ T cell kit from Invitrogen (Thermo Fisher Scientific) and resuspended in complete medium [RPMI 1640 supplemented with 10% (vol/vol) FBS, l-glutamine, penicillin, and streptomycin, all from Invitrogen/Life Technologies]. CTLs were activated and expanded using Dynabeads (Human T-Expander CD3/CD28, Life Technologies) for 5 d in complete medium supplemented with 100 units/mL recombinant IL2 (Proleukin-Novartis). After this time, beads were removed using a magnet and cells were used for the experiments. For cell–cell fusion assay, CHO cells (v cells) and MEF 3T3 cells (t cells) were cultured in complete medium [DMEM supplemented with 10% (vol/vol) FBS, l-glutamine, penicillin, and streptomycin, all from Invitrogen/Life Technologies].
Constructs.
To generate flipped SNARE constructs and the fluorescent markers used in the cell fusion assay—DsRed fused to nuclear export signal (DsRed-nes, pCH19) and CFP fused to nuclear localization signal (CFP-nls)—we followed the same procedure as previously described (25, 31). Briefly, full-length human VAMP3, -4, -7, and -8 were amplified by PCR and swapped with mouse VAMP2 cDNA that was cloned in frame with the bovine preprolactin signal sequence (SS) of the pCH1 construct. Flipped human STX11 constructs encoding for amino acids 1–276 (STX11) or amino acids 199–276 (STX11H3) were generated by amplifying by PCR the indicated regions of STX11 cDNA. PCR products were digested with EcoRI and ApaI and subcloned into plasmids encoding for the preprolactin SS, a myc epitope tag, and either the GPI-anchor motif of the decay-acceleranting factor (pCH53) to generate flipped STX11–GPI or the transmembrane domain of the platelet-derived growth factor receptor (pCH71) to generate flipped STX11–TMD. Flipped SNAP23 and SNAP29 were generated by PCR amplification of the human SNAP23/29 cDNAs. PCR products were used to replace SNAP25 cDNA from a plasmid encoding for the preprolactin SS fuse to SNAP25 (pCH9). Cysteine residues on flipped STX11 and on SNAP23/29 were mutagenized to serine residues as well as asparagines of putative N-glycosylation sites were mutagenized to alanine. Full-length human STX11 was subcloned in frame into the pEYFP–C1 or pCMV–HA vector (Clontech) to generate EYFP–STX11 or HA–STX11. The bacterial expression construct pET28a–Hisx6–SUMO–hMunc18-2, pET28a–Hisx6–SUMO–hSTX11–C102S/C157/S were generated by PCR amplification of full-length human Munc18-2 or residues 1–276 of STX11 followed by subcloning into pET28a–SUMO–rat Munc18-1 WT (33, 43). Munc18-2 R65Q and E132A constructs were generated by mutagenizing pET28a–Hisx6–SUMO–hMunc18-2 using the Quick-Change Site-Directed mutagenesis kit (Agilent Technologies). GST–SNAP23, GST–STX11 (amino acids 1–276), and GST–STX11–H3 (amino acids 158–276) constructs were produced by PCR amplification of the human full-length SNAP23 or STX111–276–C102S/C157S cDNAs and subcloned into the pDEST-60 vector (Thermo Scientific) using a gateway recombination technology following manufacturer’s instruction. The cytoplasmic domain (CD) of mouse VAMP2 (1–94), human VAMP3 (1–77), human VAMP4 (1–116), human VAMP7 (1–187), and human VAMP8 (1–77) were amplified by PCR and subcloned into pET28a–Hisx6 plasmid.
Protein Expression and Purification.
Full-length Hisx6–SUMO–Munc18-2 WT, R65Q, and E132A; Hisx6–SUMO–STX11, Hisx6–CD–VAMP proteins; and Hisx6–SNAP23 were expressed in BL21(DE3) Escherichia coli bacteria and purified by nickel affinity chromatography as previously described (33, 43). In brief, his-tagged proteins were expressed and purified according to The QIA Expressionist handbook (Qiagen). For Sumo-his-tagged protein, on-column cleavage was performed using SUMO protease at 30 °C for 90 min. GST protein alone, GST–SNAP23, GST–STX11, and GST–STX11–H3 were expressed in BL21(DE3) E. coli bacteria and purified using glutathione–Sepharose beads (GE Healthcare) following manufacturer specifications.
Pull-Down Assay.
Purified GST-alone or GST-tagged SNARE were bound to GS beads, washed three times with PBS buffer, and then incubated with the indicated purified recombinant protein combination for 2 h at room temperature. After three washes with PBS buffer, the protein samples were analyzed by SDS/PAGE.
siRNA Knockdown.
Small-interference RNA (siRNA) knockdown experiments were performed by incubating 5.0 × 106 human CTLs with 1.0 nmol of human siGenome siRNAs (GE-Dharmacon) followed by electroporation using the Gene Pulser-II system (Bio-Rad) following manufacturer’s instructions. Nontargeting (NT) siRNA consist of a pool of siRNAs catalog no. d-001206–13-20 sequences: 5′-uagcgacuaaacacaucaa-3′; 5′-uaaggcuaugaagagauac-3′; 5′-auguauuggccuguauuag-3′; and 5′-augaacgugaauugcucaa-3′. Cells were cultured for 48 h in complete medium and then used for different experiments.
Coimmunoprecipitation Assays.
HA–STX11 was transfected into HeLa cells using Lipofectamine 2000 (Life Technologies) or into human CD8+ using the Neon Microporation system (Thermo Scientific) following manufacturer’s protocol for PBMCs. Twenty-four hours later, HeLa or primary human CD8+ cells previously activated with anti-CD3– and anti-CD28–coated beads for 4 h were lysed [25 mM Tris, 150 mM NaCl, 1 mM EDTA, 5% (vol/vol) glycerol, 1% Nonidet P-40; pH 7.4] and centrifuged at 20,000 × g for 15 min. Coimmunoprecipitation (co-IP) experiments were carried out using a co-IP kit from Pierce (Thermo Scientific). Briefly, after lysis and centrifugation, supernatants were collected and incubated with agarose beads coupled with the corresponding antibody for 20 h at 4 °C. Complexes bound to the beads were pelleted by centrifugation, followed by five washes with cold lysis buffer. Proteins bound to the antibodies were eluted and examined by Western blotting.
Immunostaining and STED Microscopy.
CD3/CD28-activated CTLs were mixed with anti-CD3–coated P815 target cells at a 1:1 ratio, pelleted by centrifugation, and incubated for 15 min to generate conjugates. Cells were then seeded in 24-well plates containing polylysine-coated glass coverslips for 10 min at 37 °C. Cells were fixed for 10 min with PBS containing 4% (wt/vol) paraformaldehyde. Cells were permeabilized with 0.1% Triton X-100 in PBS buffer and then blocked with 3% (wt/vol) BSA in PBS for 30 min at room temperature. Immunostaining was performed using the indicated primary antibodies with secondary Atto-425 goat anti-mouse and Dylight-488 goat anti-rabbit antibodies. Coverslips were extensively washed with PBS and mounted with Prolong Gold anti-fade reagent (Thermo Fisher Scientific). Images were collected using a Leica SP5-STED-CW microscope (Leica Microsystems). To obtain maximum resolution, the depletion laser (592 nm) was used at maximum power.
Cytotoxicity Assays.
Cytotoxicity was evaluated using a nonradioactive assay (Cytotox-96, Promega). To evaluate CTL killing, expanded PBMCs were supplemented with 0.5 μg/mL anti-CD3 mAb, mixed with 2 × 104 target P815 cells and incubated in quadruplicate for 4 h at 37 °C. Effector-to-target cell ratios ranged from 10 to 0.65 in 100 μL medium in 96-well V-bottom plates. After 4 h, plates were centrifuged at 250 × g for 4 min and 50 μL of supernatant was transferred to a new flat bottom 96-well plate. Fifty microliters of substrate was added to each well, followed by incubation for 30 min at room temperature. The reaction was stopped using 50 μL of stop solution per well. Lactate dehydrogenase (LDH) release was measured at 490 nm using a 96-well spectrophotometer (SpectraMax, Molecular Devices). Percent cytotoxicity was calculated as: cytotoxicity (%) = (experiment − effector spontaneous − target spontaneous/target maximum − target spontaneous) × 100.
Acknowledgments
We thank Drs. Michael Marks and Thomas J. Melia for helpful discussion and comments on the manuscript. This work was supported by the PEW Biomedical Scholar Award, NIH National Institute of Allergy and Infectious Diseases Grants R01AI123538-01A1 and R01GM123020, the Histiocytosis Association Award, the W. W. Smith Charitable Trust Award, and the American Association for Immunologists Career Award (all to C.G.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617981114/-/DCSupplemental.
References
- 1.Dustin ML, Long EO. Cytotoxic immunological synapses. Immunol Rev. 2010;235(1):24–34. doi: 10.1111/j.0105-2896.2010.00904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stinchcombe JC, Griffiths GM. Secretory mechanisms in cell-mediated cytotoxicity. Annu Rev Cell Dev Biol. 2007;23:495–517. doi: 10.1146/annurev.cellbio.23.090506.123521. [DOI] [PubMed] [Google Scholar]
- 3.Ménager MM, et al. Secretory cytotoxic granule maturation and exocytosis require the effector protein hMunc13-4. Nat Immunol. 2007;8(3):257–267. doi: 10.1038/ni1431. [DOI] [PubMed] [Google Scholar]
- 4.de Saint Basile G, Ménasché G, Fischer A. Molecular mechanisms of biogenesis and exocytosis of cytotoxic granules. Nat Rev Immunol. 2010;10(8):568–579. doi: 10.1038/nri2803. [DOI] [PubMed] [Google Scholar]
- 5.Luzio JP, Hackmann Y, Dieckmann NM, Griffiths GM. The biogenesis of lysosomes and lysosome-related organelles. Cold Spring Harb Perspect Biol. 2014;6(9):a016840. doi: 10.1101/cshperspect.a016840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.zur Stadt U, et al. Linkage of familial hemophagocytic lymphohistiocytosis (FHL) type-4 to chromosome 6q24 and identification of mutations in syntaxin 11. Hum Mol Genet. 2005;14(6):827–834. doi: 10.1093/hmg/ddi076. [DOI] [PubMed] [Google Scholar]
- 7.zur Stadt U, et al. Familial hemophagocytic lymphohistiocytosis type 5 (FHL-5) is caused by mutations in Munc18-2 and impaired binding to syntaxin 11. Am J Hum Genet. 2009;85(4):482–492. doi: 10.1016/j.ajhg.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Côte M, et al. Munc18-2 deficiency causes familial hemophagocytic lymphohistiocytosis type 5 and impairs cytotoxic granule exocytosis in patient NK cells. J Clin Invest. 2009;119(12):3765–3773. doi: 10.1172/JCI40732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hellewell AL, Foresti O, Gover N, Porter MY, Hewitt EW. Analysis of familial hemophagocytic lymphohistiocytosis type 4 (FHL-4) mutant proteins reveals that S-acylation is required for the function of syntaxin 11 in natural killer cells. PLoS One. 2014;9(6):e98900. doi: 10.1371/journal.pone.0098900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang BL, Low DY, Hong W. Syntaxin 11: A member of the syntaxin family without a carboxyl terminal transmembrane domain. Biochem Biophys Res Commun. 1998;245(2):627–632. doi: 10.1006/bbrc.1998.8490. [DOI] [PubMed] [Google Scholar]
- 11.Prekeris R, Klumperman J, Scheller RH. Syntaxin 11 is an atypical SNARE abundant in the immune system. Eur J Cell Biol. 2000;79(11):771–780. doi: 10.1078/0171-9335-00109. [DOI] [PubMed] [Google Scholar]
- 12.Advani RJ, et al. Seven novel mammalian SNARE proteins localize to distinct membrane compartments. J Biol Chem. 1998;273(17):10317–10324. doi: 10.1074/jbc.273.17.10317. [DOI] [PubMed] [Google Scholar]
- 13.Al Hawas R, et al. Munc18b/STXBP2 is required for platelet secretion. Blood. 2012;120(12):2493–2500. doi: 10.1182/blood-2012-05-430629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye S, et al. Syntaxin-11, but not syntaxin-2 or syntaxin-4, is required for platelet secretion. Blood. 2012;120(12):2484–2492. doi: 10.1182/blood-2012-05-430603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valdez AC, Cabaniols JP, Brown MJ, Roche PA. Syntaxin 11 is associated with SNAP-23 on late endosomes and the trans-Golgi network. J Cell Sci. 1999;112(Pt 6):845–854. doi: 10.1242/jcs.112.6.845. [DOI] [PubMed] [Google Scholar]
- 16.Offenhäuser C, et al. Syntaxin 11 binds Vti1b and regulates late endosome to lysosome fusion in macrophages. Traffic. 2011;12(6):762–773. doi: 10.1111/j.1600-0854.2011.01189.x. [DOI] [PubMed] [Google Scholar]
- 17.Bryceson YT, et al. Defective cytotoxic lymphocyte degranulation in syntaxin-11 deficient familial hemophagocytic lymphohistiocytosis 4 (FHL4) patients. Blood. 2007;110(6):1906–1915. doi: 10.1182/blood-2007-02-074468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han X, Wang CT, Bai J, Chapman ER, Jackson MB. Transmembrane segments of syntaxin line the fusion pore of Ca2+-triggered exocytosis. Science. 2004;304(5668):289–292. doi: 10.1126/science.1095801. [DOI] [PubMed] [Google Scholar]
- 19.Deák F, Shin OH, Kavalali ET, Südhof TC. Structural determinants of synaptobrevin 2 function in synaptic vesicle fusion. J Neurosci. 2006;26(25):6668–6676. doi: 10.1523/JNEUROSCI.5272-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melikyan GB, Markosyan RM, Roth MG, Cohen FS. A point mutation in the transmembrane domain of the hemagglutinin of influenza virus stabilizes a hemifusion intermediate that can transit to fusion. Mol Biol Cell. 2000;11(11):3765–3775. doi: 10.1091/mbc.11.11.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kemble GW, Danieli T, White JM. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 1994;76(2):383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 22.Melikyan GB, White JM, Cohen FS. GPI-anchored influenza hemagglutinin induces hemifusion to both red blood cell and planar bilayer membranes. J Cell Biol. 1995;131(3):679–691. doi: 10.1083/jcb.131.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNew JA, et al. Close is not enough: SNARE-dependent membrane fusion requires an active mechanism that transduces force to membrane anchors. J Cell Biol. 2000;150(1):105–117. doi: 10.1083/jcb.150.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rohde J, Dietrich L, Langosch D, Ungermann C. The transmembrane domain of Vam3 affects the composition of cis- and trans-SNARE complexes to promote homotypic vacuole fusion. J Biol Chem. 2003;278(3):1656–1662. doi: 10.1074/jbc.M209522200. [DOI] [PubMed] [Google Scholar]
- 25.Giraudo CG, et al. SNAREs can promote complete fusion and hemifusion as alternative outcomes. J Cell Biol. 2005;170(2):249–260. doi: 10.1083/jcb.200501093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y, Zhang F, Su Z, McNew JA, Shin YK. Hemifusion in SNARE-mediated membrane fusion. Nat Struct Mol Biol. 2005;12(5):417–422. doi: 10.1038/nsmb921. [DOI] [PubMed] [Google Scholar]
- 27.Xu H, Zick M, Wickner WT, Jun Y. A lipid-anchored SNARE supports membrane fusion. Proc Natl Acad Sci USA. 2011;108(42):17325–17330. doi: 10.1073/pnas.1113888108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grote E, Baba M, Ohsumi Y, Novick PJ. Geranylgeranylated SNAREs are dominant inhibitors of membrane fusion. J Cell Biol. 2000;151(2):453–466. doi: 10.1083/jcb.151.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460(7254):525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou P, Bacaj T, Yang X, Pang ZP, Südhof TC. Lipid-anchored SNAREs lacking transmembrane regions fully support membrane fusion during neurotransmitter release. Neuron. 2013;80(2):470–483. doi: 10.1016/j.neuron.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu C, et al. Fusion of cells by flipped SNAREs. Science. 2003;300(5626):1745–1749. doi: 10.1126/science.1084909. [DOI] [PubMed] [Google Scholar]
- 32.Zick M, Stroupe C, Orr A, Douville D, Wickner WT. Membranes linked by trans-SNARE complexes require lipids prone to non-bilayer structure for progression to fusion. eLife. 2014;3:e01879. doi: 10.7554/eLife.01879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spessott WA, et al. Hemophagocytic lymphohistiocytosis caused by dominant-negative mutations in STXBP2 that inhibit SNARE-mediated membrane fusion. Blood. 2015;125(10):1566–1577. doi: 10.1182/blood-2014-11-610816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hackmann Y, et al. Syntaxin binding mechanism and disease-causing mutations in Munc18-2. Proc Natl Acad Sci USA. 2013;110(47):E4482–E4491. doi: 10.1073/pnas.1313474110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bin NR, Jung CH, Piggott C, Sugita S. Crucial role of the hydrophobic pocket region of Munc18 protein in mast cell degranulation. Proc Natl Acad Sci USA. 2013;110(12):4610–4615. doi: 10.1073/pnas.1214887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matti U, et al. Synaptobrevin2 is the v-SNARE required for cytotoxic T-lymphocyte lytic granule fusion. Nat Commun. 2013;4:1439. doi: 10.1038/ncomms2467. [DOI] [PubMed] [Google Scholar]
- 37.Marshall MR, et al. VAMP8-dependent fusion of recycling endosomes with the plasma membrane facilitates T lymphocyte cytotoxicity. J Cell Biol. 2015;210(1):135–151. doi: 10.1083/jcb.201411093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pryor PR, et al. Combinatorial SNARE complexes with VAMP7 or VAMP8 define different late endocytic fusion events. EMBO Rep. 2004;5(6):590–595. doi: 10.1038/sj.embor.7400150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riento K, Kauppi M, Keranen S, Olkkonen VM. Munc18-2, a functional partner of syntaxin 3, controls apical membrane trafficking in epithelial cells. J Biol Chem. 2000;275(18):13476–13483. doi: 10.1074/jbc.275.18.13476. [DOI] [PubMed] [Google Scholar]
- 40.Peng RW, Guetg C, Abellan E, Fussenegger M. Munc18b regulates core SNARE complex assembly and constitutive exocytosis by interacting with the N-peptide and the closed-conformation C-terminus of syntaxin 3. Biochem J. 2010;431(3):353–361. doi: 10.1042/BJ20100145. [DOI] [PubMed] [Google Scholar]
- 41.Fukuda M, Imai A, Nashida T, Shimomura H. Slp4-a/granuphilin-a interacts with syntaxin-2/3 in a Munc18-2-dependent manner. J Biol Chem. 2005;280(47):39175–39184. doi: 10.1074/jbc.M505759200. [DOI] [PubMed] [Google Scholar]
- 42.Dulubova I, et al. Munc18-1 binds directly to the neuronal SNARE complex. Proc Natl Acad Sci USA. 2007;104(8):2697–2702. doi: 10.1073/pnas.0611318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen J, Tareste DC, Paumet F, Rothman JE, Melia TJ. Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell. 2007;128(1):183–195. doi: 10.1016/j.cell.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 44.Carr CM, Grote E, Munson M, Hughson FM, Novick PJ. Sec1p binds to SNARE complexes and concentrates at sites of secretion. J Cell Biol. 1999;146(2):333–344. doi: 10.1083/jcb.146.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burkhardt P, Hattendorf DA, Weis WI, Fasshauer D. Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide. EMBO J. 2008;27(7):923–933. doi: 10.1038/emboj.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Y, Seven AB, Su L, Jiang QX, Rizo J. Membrane bridging and hemifusion by denaturated Munc18. PLoS One. 2011;6(7):e22012. doi: 10.1371/journal.pone.0022012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sepulveda FE, et al. Distinct severity of HLH in both human and murine mutants with complete loss of cytotoxic effector PRF1, RAB27A, and STX11. Blood. 2013;121(4):595–603. doi: 10.1182/blood-2012-07-440339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kögl T, et al. Hemophagocytic lymphohistiocytosis in syntaxin-11-deficient mice: T-cell exhaustion limits fatal disease. BlCood. 2013;121(4):604–613. doi: 10.1182/blood-2012-07-441139. [DOI] [PubMed] [Google Scholar]
- 49.Cetica V, et al. STXBP2 mutations in children with familial haemophagocytic lymphohistiocytosis type 5. J Med Genet. 2010;47(9):595–600. doi: 10.1136/jmg.2009.075341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Müller ML, et al. An N-terminal missense mutation in STX11 causative of FHL4 abrogates Syntaxin-11 binding to Munc18-2. Front Immunol. 2014;4:515. doi: 10.3389/fimmu.2013.00515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi L, et al. SNARE proteins: One to fuse and three to keep the nascent fusion pore open. Science. 2012;335(6074):1355–1359. doi: 10.1126/science.1214984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baker RW, et al. A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science. 2015;349(6252):1111–1114. doi: 10.1126/science.aac7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parisotto D, Malsam J, Scheutzow A, Krause JM, Söllner TH. SNAREpin assembly by Munc18-1 requires previous vesicle docking by synaptotagmin 1. J Biol Chem. 2012;287(37):31041–31049. doi: 10.1074/jbc.M112.386805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lou X, Shin J, Yang Y, Kim J, Shin YK. Synaptotagmin-1 is an antagonist for Munc18-1 in SNARE zippering. J Biol Chem. 2015;290(16):10535–10543. doi: 10.1074/jbc.M114.631341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, et al. Munc18a does not alter fusion rates mediated by neuronal SNAREs, synaptotagmin, and complexin. J Biol Chem. 2015;290(16):10518–10534. doi: 10.1074/jbc.M114.630772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma L, et al. Munc18-1-regulated stage-wise SNARE assembly underlying synaptic exocytosis. eLife. 2015;4:4. doi: 10.7554/eLife.09580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baker RW, Hughson FM. Chaperoning SNARE assembly and disassembly. Nat Rev Mol Cell Biol. 2016;17(8):465–479. doi: 10.1038/nrm.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hashizume K, Cheng YS, Hutton JL, Chiu CH, Carr CM. Yeast Sec1p functions before and after vesicle docking. Mol Biol Cell. 2009;20(22):4673–4685. doi: 10.1091/mbc.E09-02-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scott BL, et al. Sec1p directly stimulates SNARE-mediated membrane fusion in vitro. J Cell Biol. 2004;167(1):75–85. doi: 10.1083/jcb.200405018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Diao J, et al. Single-vesicle fusion assay reveals Munc18-1 binding to the SNARE core is sufficient for stimulating membrane fusion. ACS Chem Neurosci. 2010;1(3):168–174. doi: 10.1021/cn900034p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma C, Su L, Seven AB, Xu Y, Rizo J. Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science. 2013;339(6118):421–425. doi: 10.1126/science.1230473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen C, et al. The trans-SNARE-regulating function of Munc18-1 is essential to synaptic exocytosis. Nat Commun. 2015;6:8852. doi: 10.1038/ncomms9852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lobingier BT, Merz AJ. Sec1/Munc18 protein Vps33 binds to SNARE domains and the quaternary SNARE complex. Mol Biol Cell. 2012;23(23):4611–4622. doi: 10.1091/mbc.E12-05-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ji H, et al. Protein determinants of SNARE-mediated lipid mixing. Biophys J. 2010;99(2):553–560. doi: 10.1016/j.bpj.2010.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loo LS, et al. A role for endobrevin/VAMP8 in CTL lytic granule exocytosis. Eur J Immunol. 2009;39(12):3520–3528. doi: 10.1002/eji.200939378. [DOI] [PubMed] [Google Scholar]
- 66.Dressel R, Elsner L, Novota P, Kanwar N, Fischer von Mollard G. The exocytosis of lytic granules is impaired in Vti1b- or Vamp8-deficient CTL leading to a reduced cytotoxic activity following antigen-specific activation. J Immunol. 2010;185(2):1005–1014. doi: 10.4049/jimmunol.1000770. [DOI] [PubMed] [Google Scholar]
- 67.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]