DNA damage is a frequent and detrimental event faced by all living organisms. Decades of research have characterized the repair pathways that counteract this threat at genetic, biochemical, and structural levels. More recently, genome sequencing has revealed patterns of mutation demonstrating that DNA repair proteins fight damage more efficiently in some regions than others (1). Techniques that monitor DNA repair directly at a genome-wide scale promise greater insight into the mechanistic basis of these repair biases and provide a stringent testbed for models derived from biochemical data (2, 3). In PNAS, Adebali et al. (4) provide the first genome-wide picture of nucleotide excision repair (NER) in bacteria.
NER is a broad-specificity pathway present in both eukaryotes and prokaryotes. Substrates include the pyrimidine dimers induced by a potent environmental mutagen, UV light. There are at least two major subpathways of NER: Damage in untranscribed DNA is recognized and repaired by “global” NER, whereas faster, transcription-coupled NER processes target lesions in genes that are being expressed (5). The two pathways differ primarily at the damage recognition step. During global NER, lesions are detected by dedicated repair proteins. In transcription-coupled repair (TCR), the damage is first detected by RNA polymerase (RNAP), which stalls at a DNA lesion and triggers the assembly of repair complexes. Because only one of the two DNA strands is used as a template for transcription, TCR is strand-specific: Most lesions only cause RNAP to stall when located in the transcribed, or template, strand (TS), and so only lesions within the TS are repaired at a faster rate. Lesions in the nontranscribed strand (NTS) do not cause RNAP to stall, and are repaired by the slower global repair pathway.
Adebali et al. (4) have analyzed patterns of NER in Escherichia coli, a bacterium in which damage recognition is undertaken by a complex of the repair proteins UvrA and UvrB (6). UvrB recruits a nuclease, UvrC, which cuts the damaged DNA strand on either side of the lesion. The resulting oligonucleotide, which contains the lesion, is released from the DNA by the helicase UvrD, leaving a single-stranded gap that is filled in by DNA polymerase and DNA ligase. During TCR, RNAP does not directly recruit repair proteins; indeed, RNAP stalled at a lesion prevents its detection by UvrA and UvrB, and so inhibits rather than enhances repair (7). A search for the missing factor that permitted enhanced repair of the TS identified the product of the mutation frequency decline (mfd) gene (8). Subsequent analyses have shown Mfd to be a DNA translocase that binds to stalled RNAP and pushes it “forward,” dissociating it from the lesion-bearing DNA (9–11). The Mfd/RNAP complex docks with UvrAB, which then initiates repair, which proceeds via dual incision, oligonucleotide release, and repair patch synthesis as for global NER (8, 12). In vivo, deletion of the mfd gene abolished TCR of UV-induced lesions in model genes, but the requirement for Mfd varied with transcription level, and even at different locations within a single gene (13, 14).
Recent results have identified an additional link between RNAP and NER. UvrD is a helicase that plays many roles in DNA metabolism, including postincision events in NER and mismatch repair as well as the resolution of conflicts between transcription complexes and DNA replication forks (15). It interacts with many partner proteins, including RNAP (16, 17). UvrD can slide RNAP “backward” along the DNA (a process called backtracking), and this activity can facilitate the access of repair proteins to lesions that would otherwise be hidden within the transcription complex (16). Although no direct stimulation of DNA repair has been shown to be associated with UvrD-mediated backtracking of RNAP, it has been suggested that the direct interaction of UvrD with UvrB, or the recruitment of UvrA to RNAP via the NusA transcription factor, might result in faster repair at sites of UvrD action (18). Backtracking by UvrD requires the high concentrations of the protein that are produced during the bacterial response to DNA damage that is termed the SOS response. It is also enhanced by a small-molecule alarmone, (p)ppGpp, which is transiently induced following exposure of cells to UV light (16, 19, 20). In addition to mapping the rates of repair across the genome in fully repair-proficient cells, a key question addressed by Adebali et al. (4) is, therefore, what are the roles of UvrD and Mfd during the response to acute UV-induced DNA damage?
Their analysis used a technique called eXcision repair sequencing (XR-seq) (2), which involves the extraction, immunoprecipitation, and sequencing of all of the short lesion-containing oligonucleotides that are produced by UvrABC (Fig. 1). This approach allowed the relative rates of excision to be determined for both DNA strands across the entire genome. The oligonucleotides produced by E. coli NER proteins in vitro are predominantly 12–13 nt long, and in agreement with this fact, the most prominent product isolated by immunoprecipitation from wild-type cells was 13 nt long. However, the abundance of these excised fragments was low due to their rapid degradation in cells. To overcome this problem, cells lacking the main single-stranded DNA nucleases were used. In addition to 13-nt oligonucleotides, an abundance of predominantly 10-nt oligonucleotides could now be observed. This shortening can be explained by the action of an unidentified 3′ to 5′ nuclease that stops 1 nt away from the lesion.
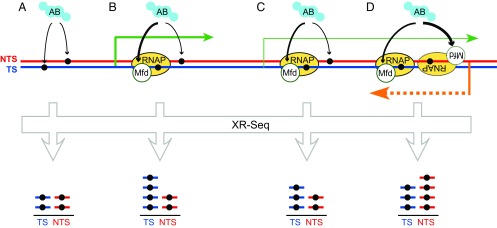
Fig. 1.
Patterns of NER across the genome. (A) In untranscribed regions, damage in both strands is detected by UvrAB. XR-seq detects equal proportions of excised oligonucleotides from both strands. (B and C) In regions with simple transcription patterns (green arrows), greater excision of the TS is detected because UvrAB binds tightly to the Mfd/RNAP complex. Transcription levels affect the degree of strand bias. (D) Antisense transcription in some genes (orange arrow) can result in preferential repair of the annotated NTS.
Fascinatingly, in cells lacking UvrD, the abundance of the lesion-containing oligonucleotides was greatly increased, and the predominant fragment length was increased to 13 nt. These changes were seen even in cells that retained the nucleases that normally rapidly degrade the oligonucleotide. How can loss of a protein known to be important for NER appear to stimulate formation of NER products? Adebali et al. (4) suggest that in the absence of UvrD, the oligonucleotide is retained in the postincision complex, and is thus protected from degradation or shortening by nucleases. This finding is consistent with the fact that UvrD is necessary in vitro both for the removal of the excised nucleotide carrying the DNA lesion and for the displacement of UvrC from the postincision complex of UvrB and DNA (21).
XR-seq analysis of fully repair-proficient cells revealed widespread preferential repair of the TS of annotated genes. In cells that lacked Mfd, this strand bias was reversed, with a tendency for the NTS of genes to be repaired more quickly. In contrast, cells that lacked UvrD retained the ability to prioritize the repair of the TS of active genes; in fact, it seemed that the bias toward repair of the TS may be slightly greater in the absence of UvrD. These results confirm that the Mfd protein plays a global role in TCR of UV-induced lesions in E. coli, and indicate that whereas UvrD is critical for postincision steps in NER, it is not required for the preferential repair of lesions in the TS of most genes, at least under the conditions of this experiment.
The observation that loss of UvrD increases the proportion of TS-derived lesion-containing oligonucleotides is interesting. A potential explanation suggested by Adebali et al. (4) is that because UvrD is required for both the displacement of the oligonucleotide and the turnover of UvrC, the oligonucleotides analyzed in cells that lack UvrD disproportionally represent “first-hit” repair events (i.e., the locations to which UvrAB first recruited UvrC). Because the cells contain Mfd, and thus an active TCR pathway, TS lesions are enriched in this first-hit sample. An alternative, although not exclusive, possibility is that this observation reflects competition between Mfd and UvrD at the stalled transcription complex. Mfd has been shown both to push RNAP forward and to recruit repair proteins, and it is this combination of activities that results in strand-specific enhancement of repair. In contrast, UvrD moves RNAP in the opposite direction to Mfd, and although models for subsequent recruitment of UvrA or UvrB have been proposed, there is as yet no direct evidence that this recruitment occurs. If UvrD backtracks RNAP without recruiting UvrA or UvrB, then events in which UvrD acts on RNAP before Mfd does will reduce the number of Mfd-dependent TCR events. Deletion of UvrD would remove competition between UvrD and Mfd, and so increase the level of Mfd-dependent TCR.
Although Mfd and UvrD clearly have differing effects on a genome-averaged level, the data reveal greater complexity when analyzed in finer detail. The effects of each of these proteins differ between genes, and within individual genes, and there are locations where the strand bias is reversed and the annotated NTS is repaired more quickly than the TS. To understand some of these complexities, the XR-seq data were compared with genome-wide transcription data. An important observation was that many annotated genes were subject to considerable levels of antisense transcription (arising from either overlapping genes or opportunistic transcription initiation events). This “pervasive transcription” (22) means that designating a strand as TS or NTS is not as clear-cut as might first appear, and partly explains why the relative strand bias measured by XR-seq is not as great as previous analysis in simpler systems had led us to expect.
This study has revealed the process of NER in bacteria in unprecedented breadth, and has identified the pathway responsible for the global TS/NTS repair bias following UV-induced DNA damage. The data point to a rich complexity of behavior at the single gene level, and further work will be needed to understand fully the nature of the repair “hot-spots” identified within some genes, and the characteristics of the genes where UvrD and Mfd appear to play atypical roles. Further questions to which this powerful approach might be fruitfully applied include the repair of alternative types of lesions, and patterns of repair during periods of prolonged genotoxic stress: Under conditions that have induced the SOS response, the concentration of repair proteins is higher, patterns of gene expression differ, and it has been reported that the TCR pathway is no longer detectable (23). Will repair be more uniform under such conditions, or will interesting new patterns emerge?
Footnotes
The authors declare no conflict of interest.
See companion article on page E2116.
References
- 1.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463(7278):191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu J, Adar S, Selby CP, Lieb JD, Sancar A. Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 2015;29(9):948–960. doi: 10.1101/gad.261271.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu S, et al. Global genome nucleotide excision repair is organized into domains that promote efficient DNA repair in chromatin. Genome Res. 2016;26(10):1376–1387. doi: 10.1101/gr.209106.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adebali O, Chiou Y-Y, Hu J, Sancar A, Selby CB. Genome-wide transcription-coupled repair in Escherichia coli is mediated by the Mfd translocase. Proc Natl Acad Sci USA. 2017;114:E2116–E2125. doi: 10.1073/pnas.1700230114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9(12):958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 6.Kisker C, Kuper J, Van Houten B. Prokaryotic nucleotide excision repair. Cold Spring Harb Perspect Biol. 2013;5(3):a012591. doi: 10.1101/cshperspect.a012591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selby CP, Sancar A. Transcription preferentially inhibits nucleotide excision repair of the template DNA strand in vitro. J Biol Chem. 1990;265(34):21330–21336. [PubMed] [Google Scholar]
- 8.Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260(5104):53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 9.Park JS, Marr MT, Roberts JW. E. coli Transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109(6):757–767. doi: 10.1016/s0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 10.Deaconescu AM, et al. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006;124(3):507–520. doi: 10.1016/j.cell.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 11.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor. I. Structural domains and binding properties. J Biol Chem. 1995;270(9):4882–4889. doi: 10.1074/jbc.270.9.4882. [DOI] [PubMed] [Google Scholar]
- 12.Fan J, Leroux-Coyau M, Savery NJ, Strick TR. Reconstruction of bacterial transcription-coupled repair at single-molecule resolution. Nature. 2016;536(7615):234–237. doi: 10.1038/nature19080. [DOI] [PubMed] [Google Scholar]
- 13.Kunala S, Brash DE. Intragenic domains of strand-specific repair in Escherichia coli. J Mol Biol. 1995;246(2):264–272. doi: 10.1006/jmbi.1994.0082. [DOI] [PubMed] [Google Scholar]
- 14.Mellon I, Champe GN. Products of DNA mismatch repair genes mutS and mutL are required for transcription-coupled nucleotide-excision repair of the lactose operon in Escherichia coli. Proc Natl Acad Sci USA. 1996;93(3):1292–1297. doi: 10.1073/pnas.93.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dillingham MS. Superfamily I helicases as modular components of DNA-processing machines. Biochem Soc Trans. 2011;39(2):413–423. doi: 10.1042/BST0390413. [DOI] [PubMed] [Google Scholar]
- 16.Epshtein V, et al. UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature. 2014;505(7483):372–377. doi: 10.1038/nature12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gwynn EJ, et al. The conserved C-terminus of the PcrA/UvrD helicase interacts directly with RNA polymerase. PLoS One. 2013;8(10):e78141. doi: 10.1371/journal.pone.0078141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamarthapu V, Nudler E. Rethinking transcription coupled DNA repair. Curr Opin Microbiol. 2015;24:15–20. doi: 10.1016/j.mib.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasouly A, Pani B, Nudler E. A magic spot in genome maintenance. Trends Genet. 2017;33(1):58–67. doi: 10.1016/j.tig.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamarthapu V, et al. ppGpp couples transcription to DNA repair in E. coli. Science. 2016;352(6288):993–996. doi: 10.1126/science.aad6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orren DK, Selby CP, Hearst JE, Sancar A. Post-incision steps of nucleotide excision repair in Escherichia coli. Disassembly of the UvrBC-DNA complex by helicase II and DNA polymerase I. J Biol Chem. 1992;267(2):780–788. [PubMed] [Google Scholar]
- 22.Wade JT, Grainger DC. Pervasive transcription: Illuminating the dark matter of bacterial transcriptomes. Nat Rev Micro. 2014;12(9):647–653. doi: 10.1038/nrmicro3316. [DOI] [PubMed] [Google Scholar]
- 23.Crowley DJ, Hanawalt PC. The SOS-dependent upregulation of uvrD is not required for efficient nucleotide excision repair of ultraviolet light induced DNA photoproducts in Escherichia coli. Mutat Res. 2001;485(4):319–329. doi: 10.1016/s0921-8777(01)00068-4. [DOI] [PubMed] [Google Scholar]