Abstract
The purpose of this review is to provide updated information regarding bioinformatic software for the use in the characterization of glycosylated structures since 2013. A comprehensive review by Woodin et al. Analyst 138: 2793–2803, 2013 (ref. 1) described two main approaches that are introduced for starting researchers in this area; analysis of released glycans and the identification of glycopeptide in enzymatic digests, respectively. Complementary to that report, this review focuses on mass spectrometry related bioinformatics tools for the characterization of N-linked and O-linked glycopeptides. Specifically, it also provides information regarding automated tools that can be used for glycan profiling using mass spectrometry.
Keywords: mass spectrometry, glycosylation, glycopeptide, glycan, glycoproteomics
INTRODUCTION
Overview of glycoprotein
Glycosylation, a specific enzymatic process in which glycans are attached to proteins or lipids, and is an important biological process that plays a role in cell signaling, cell adhesion, and the regulation of biochemical pathways. Of all post-translational modifications (PTMs), glycosylation is one of the most commonly observed type.2) It is believed that more than 50% proteins are glycosylated. The biological functions of glycoproteins are involved many types of biological processes. Therefore, automated tools for the identification of glycoproteins and the glycans that are attached to them, become fundamentally important. For analysis, tandem mass spectrometry (MS/MS) is a popular and efficient method in glycoproteomics because of its high sensitivity and selectivity.
Glycosylation heterogeneity
Glycoproteins generally exist as populations of glycosylated variants (glycoforms) of a single polypeptide. Although the same glycosylation machinery is available for all proteins that enter the secretory pathway in a given cell, most glycoproteins emerge with characteristic glycosylation patterns and the glycans at each glycosylation site are heterogeneous. The recognition of identical motifs in different glycans allows a heterogeneous population of glycoforms to participate in specific biological interactions. This is the most challenging factor for glycan analysis. Two major types of protein glycosylation are known: N-Linked glycans that contain asparagine-X-serine/threonine sequons (N-X-S/T) where X is any amino acid except proline. O-Linked glycans attached to the hydroxyl oxygen of either serine, threonine, tyrosine, hydroxylysine, or hydroxyproline side-chains, or oxygen atoms on lipids such as ceramide represent the second type of modification.
Glycoprotein analysis strategies for mass spectrometry
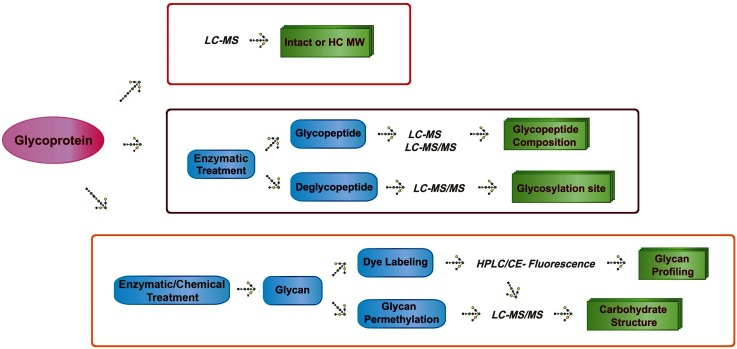
Mass spectrometry (MS) has been successfully used to determine glycan composition and their structures.3–5) Figure 1 shows MS strategies that are currently in use for glycoprotein analysis. These approaches can generally be divided into top-down and bottom-up strategies. The determination of the molecular weight of a glycoprotein represents a typical top-down analysis, which provides the most direct method for obtaining information on glycans in a glycoprotein. By calculating the molecular weight differences between the peaks, it is possible to determine the types of glycan modifications on that protein. Such an analysis, however, frequently lacks sensitivity and structure information. Because of this, glycoprotein analyses are often divided into two main strategies for collecting glycoprotein information using MS techniques. One involves an analysis of released glycans, while the other involves characterizing glycopeptides that are obtained by proteolytic digestion of the original glycoprotein.
Fig. 1. Strategies for glycoprotein analysis using mass spectrometry.
Previous reviews have dealt with software tools that permit glycosylation to be accomplished based on MS data alone, as well as software designed to speed up the interpretation of glycan and glycopeptide fragmentation from MS2 data.1) Compared to previous reviews, this review not only describes new bioinformatics tools for glycan analysis, but also focuses on software applications for glycopeptide analysis and glycan profiling.
BIOINFORMATIC TOOLS FOR THE ANALYSIS OF RELEASED GLYCANS
The analysis of glycans released from a glycoprotein by enzymatic digestion is the most common method for characterizing glycoproteins. Although less site-specific when compared with glycopeptide analysis, some types of bioinformatics software that can be used in conjunction with this method are listed below.
MS approaches for released glycan analysis
Goldberg et al. described a software program called “Cartoonist” for profiling N-glycans based on MALDI TOF mass spectra. Cartoonist has some interesting features. It uses a library of biosynthetically plausible cartoons that contain about 2800 cartoons derived from 300 archetypes. In addition, it computes a confidence score for each assignment a glycan peak according match mass.6) “GlycoWorkbench” was developed by the EUROCarbDB initiative.7) In this system, MS data can be interpreted by using GlycoWorkbench annotations. According to a theoretical list of fragment masses compared with a list of peaks derived from the spectrum, GlycoWorkbench provides glycan structural constituents based on a collection of fragmentation types.8) “SysBioWare” is a web-based interface that allows raw MS data to be processed. The unique function of SysBioWare is peak isotopic grouping, charge deconvolution, and continuous wavelet analysis. Candidate structures are generated based on a database search by this algorithm.9) Yu et al. presented a software called “MultiGlycan” that packages an automated annotation of a user-defined list of of glycans based on MALDI-TOF or LC-ESI-MS data. MultiGlycan functions by matching theoretical isotopic envelopes of glycans and accounts for overlapping glycan isotope distributions. Especially for ESI, isotopic envelopes can be simultaneously calculated by using specified adducts or multiple adducts to mass data. Both label-free and labeled glycans can be process by MultiGlycan for quantifying glycans.10,11)
MS/MS approaches for released glycan analysis
“STAT,” which was designed by Gaucher et al., represents the first web-based computational program for the determination of glycan composition using MS/MS spectra. STAT has the ability to rapidly analyze sequence information from a set of MS/MS spectra for up to ten monosaccharide residues. Another function of STAT is that possible structures are listed and given a ranking system based on correct sequence when more than one candidate glycan matches the data.12) The “StrOligo” program can successfully analyze tandem mass spectra of complex N-linked oligosaccharides. StrOligo finds the most intense peak in a tandem mass spectrum and compares the potential experimental isotopic distribution to one or two theoretical isotopic distributions. Some of the optimized parameters can be used by this algorithm to remove isotopic peaks and retain monoisotopic peaks; the overall intensities of both isotopic distributions, the position of the first isotopic distribution, and the m/z separation between both distributions.13) “GlycoFragment” enables the easy generation of all theoretically possible A-, B-, C-, X-, Y- and Z-fragments of a defined glycan structure according to the definitions of Domon and Costello.14) The algorithm uses the Sweet-II15,16) program to interpret nomenclature and chooses suitable templates from a database according to linkage information. “GlycoSearchMS” imports a mass spectrum to a database search of theoretically calculated spectra and identifies the best candidate spectra. It includes most of the theoretically possible spectra of N- and O-glycans, which were extracted from SweetDB.17,18)
The Oligosaccharide Subtree Constraint Algorithm (“OSCAR”) restructures analyst-selected fragments into branching or linkage glycan structures and provides a de novo algorithm for identifying a glycan structure without being limited by presumed biosynthetic structures. In particular, OSCAR can interpret MSn data for permethylated O-linked oligosaccharides, but the disadvantage is that it cannot process LC-MS data smoothly.19,20) Goldberg et al. also described an algorithm called “CartoonistTwo” that generates all possible cartoons and ranks them by score, similar to Cartoonist, but with a more sophisticated scoring function.6) This feature of CartoonistTwo permits fragmentation ions of O-linked glycans to be automatically annotated.21) “Glyco-Peakfinder” is useful for the de novo determination of glycan compositions. Knowledge concerning for this software computation is not based on a biologcal background or fragmentation information. The fragment ions of monosaccharide cross-ring cleavage products or multiply charged ions can be annotated using Glyco-Peakfinder.22) “SimGlycan” is a commercially available software program that uses MS/MS raw data files obtained from many different types of mass spectrometers. Based on exact mass spectra, SimGlycan has a built-in database system for use in glycan database searches and special scoring techniques that provides the most likely glycan structures.23) “GlycanID” is a software program that can be used for the analysis of LC-MS/MS data for profiling and identifying glycans. A glycan profile is generated with feature detection and alignment tools developed for proteomics. The features of GlycanID is its ability to distinguish the complexity fragmentation ions caused by salt adducted ions or contaminants, multiple charge states, and possible in-source decay.24)
In conclusion, “Glyco-Peakfinder” can be easily used for released glycan analysis on both sections of MS and MS/MS approaches (http://www.glycoworkbench.org/).
AUTOMATED ANALYSIS OF GLYCOPEPTIDES
Glycopeptide-based analysis is used to inform researchers the nature of glycans on one or more proteins.25) The method’s key attractive feature is its ability to link information regarding glycosylation to exact locations (glycosylation sites) on proteins. Some commonly used tools for glycopeptide identification, which are divided into MS and MS/MS analysis of N-linked or O-linked glycans are listed below.
Glycopeptide MS Data
“GlycoMod” is a software program that can help researchers find all possible compositions of a glycan structure from their experimentally data. The program can be used to annotate the composition not only underivatised reducing ends, but derivatised by methylated or acetylated monosaccharides as well. The algorithm will match the experimentally determined masses against in silico predicted enzymatic digested peptides obtained using SWISS-PROT or TrEMBL databases which have the potential to be glycosylated with either N- or O-linked glycans.26) “GlycoX” computes, not only the assignment of site-specific glycosylation without any glycan information, but also provides information on glycan heterogeneity. The GlycoX program has three main functions, 1. Isotope Filter. 2. Oligosaccharide Calculator. 3. Determination of Glycosylation Sites. It supports the interpretation of MS data obtained for fragments from a glycoprotein produced by nonspecific protease treatment.27) Desaire and colleagues presented a web-based tool, “GlycoPep DB.” The principle is similar to GlycoMod and is designed to find all possible compositions for glycopeptides by comparing experimentally measured masses to all calculated glycopeptide masses from a carbohydrate database for N-linked glycans. In comparison to GlycoMod, GlycoPep DB has the ability to process data for multiply charged ions and for making glycopeptide compositional assignment more efficient from the concept of “smart searching.”28) “GlycoSpectrumScan” was developed by Deshpande et al. It computes the masses of all possible glycopeptides, similar to GlycoMod and GlycoPep DB. Notably, the relative abundance based on signal intensities in a mass spectrum can also be calculated.29)
Here, the “GlycoPep DB” provides friendly user experience and it is available for free (http://hexose.chem.ku.edu/sugar.php).
Glycopeptide MS/MS Data
“GlycoPep ID” is a freely accessible web-based program that was specifically developed to identify peptide portions of glycopeptides. The glycopeptides may be generated by proteolytic cleavage with either a specific or a nonspecific enzyme. When the glycosylation sites are determined, the program generates a table consisting of all possible peptide sequences around these glycosylation sites. The most notable feature is that negatively charged glycopeptides can also be identified by GlycoPep ID.30) “GlycopeptideID” is a web tool developed to identify intact glycopeptides. The emphasis is on resolving a complicated peptide and a glycan that is unknown. Peptides are identified by matching the MS/MS spectra against a protein database and the glycans against a glycan database created from the GlycomeDB, and pre-loaded to the GlycopeptideID web server. The features of GlycopeptideID include a de novo glycan search, assigning the peptide and glycan modification, and uses probability based scoring for both peptide and glycan.31,32) “GlycoMiner” was developed by Ozohanics et al. It has the ability to automatically identify tandem mass spectra which correspond to N-glycopeptides by evaluating low mass oxonium ions, deduces oligosaccharide losses from the protonated molecule, and identifies the mass of the peptide residue. The disadvantage of GlycoMiner is that the glycan compositions assignments when the quality the spectra input is low.33) “Protein Prospector” combines CID and ETD search results into a single output file for glycopeptide identification by a database search. In addition, the software permits a manual comparison of the potential site assignments and the annotation of the glycosylated and de-glycosylated fragment ions in the same spectrum, which is a particularly useful feature for CID spectra of O-linked glycopeptides, where both fragment types are generally present.34)
Mayampurath et al. modified the “GlypID” algorithm,35) and developed a new software tool that implements several algorithmic approaches for utilizing MS information including accurate precursor mass and spectral patterns from both HCD and CID spectra, thus allowing for an unequivocal and accurate characterization of N-linked glycosylation sites of proteins.36) “GlycoPeptide Search (GPS)” incorporates a tool that simplifies the interpretation of N-glycopeptide CID MS/MS data and conjunction with the GlycomeDB database.37) The output results must have, not only glycopeptide oxonium ions, but also glycopeptide fragment ions that are consistent with the intact mass of the peptides.38) The “Glycolyzer” contains a full data analysis pipeline in one software package to allow for minimal user intervention. The general modules of this algorithm include data importing and exporting, FT-ICR signal preprocessing, internal calibration, noise threshold calculation, peak selection, isotope grouping and filtering, glycan annotation, intensity normalization, missing value filling, multiple spectra averaging, hypothesis testing, and multiple testing corrections. It can be applied to identifying glycan biomarkers.39)
“GlycoPep Grader (GPG)” is a free software tool designed to analyze MS/MS data obtained for N-linked glycopeptides. The scoring approach relies on the identification of unique dissociation patterns shown for high mannose, hybrid, and complex N-linked glycoprotein types, including patterns that are specific to those structures that contain fucose or sialic acid residues. The useful function of this tool is the scoring algorithm that was specifically designed for dealing with low resolution CID data.40) “Byonic” is a commercially available software package that can be used for identifying peptides and proteins by tandem mass spectrometry. Byonic provides some features: Top-down and bottom-up search, Modification Fine Control™, Wildcard Search™, glycopeptide search, sequence variant search and modification site localization. One of the items, glycopeptide search, provides three ways to specify glycopeptide searches: internal tables, external tables, and fine control modification.41) “GlycoMaster DB” incorporates an N-linked glycan database that was extracted from GlycomeDB. Using the GlycoMaster DB algorithm involves three steps. The first is filtration of glycopeptide spectra, the second is glycan assignment, and the third is peptide identification. the HCD/ETD spectrum can be used for glycopeptide analysis using GlycoMaster DB that permits glycopeptides and deglycosylated glycopeptides to be analyzed simultaneously to obtain glycans and peptide sequences. It also provides a GSM score scheme, similar to the PSM score in proteomics.42)
“GPQuest” is an algorithm used for the site-specific identification of intact glycopeptides using higher-energy collisional dissociation (HCD). The application of the GPQuest algorithm requires matching the spectra of HCD-fragmented glycopeptides with the experimental spectral library (ESL). The advantage of GPQuest is that tandem mass spectra of intact glycopeptides containing oxonium ions and some of the fragment ions generated through HCD fragmentation from their deglycosylated counterparts can be analyzed.43) “MAGIC” is a mass spectrometry-based automated glycopeptide identification platform that permits peptide sequences and glycan compositions to be identified directly by means of a conventional database sequence search. MAGIC uses the Y1 (peptideY0+GlcNAc) ion for filtering out unknown glycoproteins with a novel algorithm called Trident that detects a triplet pattern from the fragmentation of the common trimannosyl core of N-linked glycopeptide. MAGIC also provides fast computing power for large-scale complex proteome data sets.44) “GlycoSeq” uses a heuristic iterated glycan sequencing algorithm that incorporates prior knowledge of the N-linked glycan synthetic pathway to achieve rapid glycan sequencing. The limiting factor for GlycoSeq is glycan and glycopeptides is that peptides containing more than one potential glycosylation site cannot be distinguished due to the lack of signature fragment ions to identify peptide backbone information.45) “pGlyco” is a novel pipeline for the identification of intact glycopeptides by integrating HCD-MS/MS, CID-MS/MS and MS3 information. Both HCD-MS/MS and CID-MS/MS could be used to optimize glycopeptide identification with a false discovery rate. Data-dependent acquisition of MS3 for the most intense peaks of HCD-MS/MS was used to provide fragments to identify the peptide backbones.46)
In this “Glycopeptide MS/MS Data” section, “GlycoPeptide Search (GPS)” can be downloaded from https://edwardslab.bmcb.georgetown.edu/trac/GlycoPeptideSearch/. It is also a free software for glycopeptide tandem mass spectrum database search.
CONCLUSION AND PERSPECTIVES
In the recent years, mass spectrometry has become a powerful tool for the identification and structural characterization of glycans. With the development of the mass spectrometer, the trend for this research has led to two main strategies for use in studies of protein glycosylation: Glycan analysis and glycopeptide analysis. Compared with glycan analysis, glycopeptide identification by tandem mass spectrometry provides for the site-specific elucidation and collection of glycan information concerning a sequence derived from an enzyme digested glycopeptide. Bioinformatic tools for glycopeptide analysis is rapidly advancing and promises to continue to expand, since it provides for the smooth and relatively rapid analysis of glycosylation end products.
Mass Spectrom (Tokyo) 2017; 6(2): S0064
References
- 1) C. L. Woodin, M. Maxon, H. Desaire. Software for automated interpretation of mass spectrometry data from glycans and glycopeptides. Analyst (Lond.) 138: 2793–2803, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2) R. Apweiler. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Bio. Biophys. Acta (BBA)-General Subjects 1473: 4–8, 1999. [DOI] [PubMed] [Google Scholar]
- 3) W. Morelle, K. Canis, F. Chirat, V. Faid, J. C. Michalski. The use of mass spectrometry for the proteomic analysis of glycosylation. Proteomics 6: 3993–4015, 2006. [DOI] [PubMed] [Google Scholar]
- 4) D. S. Dalpathado, H. Desaire. Glycopeptide analysis by mass spectrometry. Analyst (Lond.) 133: 731–738, 2008. [DOI] [PubMed] [Google Scholar]
- 5) S. J. North, P. G. Hitchen, S. M. Haslam, A. Dell. Mass spectrometry in the analysis of N-linked and O-linked glycans. Curr. Opin. Struct. Biol. 19: 498–506, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6) D. Goldberg, M. Sutton-Smith, J. Paulson, A. Dell. Automatic annotation of matrix-assisted laser desorption/ionization N-glycan spectra. Proteomics 5: 865–875, 2005. [DOI] [PubMed] [Google Scholar]
- 7) C. W. von der Lieth, A. A. Freire, D. Blank, M. P. Campbell, A. Ceroni, D. R. Damerell, A. Dell, R. A. Dwek, B. Ernst, R. Fogh, M. Frank, H. Geyer, R. Geyer, M. J. Harrison, K. Henrick, S. Herget, W. E. Hull, J. Ionides, H. J. Joshi, J. P. Kamerling, B. R. Leeflang, T. Lutteke, M. Lundborg, K. Maass, A. Merry, R. Ranzinger, J. Rosen, L. Royle, P. M. Rudd, S. Schloissnig, R. Stenutz, W. F. Vranken, G. Widmalm, S. M. Haslam. EUROCarbDB: An open-access platform for glycoinformatics. Glycobiology 21: 493–502, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8) A. Ceroni, K. Maass, H. Geyer, R. Geyer, A. Dell, S. M. Haslam. GlycoWorkbench: A tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome Res. 7: 1650–1659, 2008. [DOI] [PubMed] [Google Scholar]
- 9) S. Y. Vakhrushev, D. Dadimov, J. Peter-Katalinic. Software platform for high-throughput glycomics. Anal. Chem. 81: 3252–3260, 2009. [DOI] [PubMed] [Google Scholar]
- 10) C. Y. Yu, A. Mayampurath, Y. Hu, S. Zhou, Y. Mechref, H. Tang. Automated annotation and quantification of glycans using liquid chromatography-mass spectrometry. Bioinformatics 29: 1706–1707, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11) Y. Hu, S. Zhou, C. Y. Yu, H. Tang, Y. Mechref. Automated annotation and quantitation of glycans by liquid chromatography/electrospray ionization mass spectrometric analysis using the MultiGlycan-ESI computational tool. Rapid Commun. Mass Spectrom: RCM 29: 135–142, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12) S. P. Gaucher, J. Morrow, J. A. Leary. STAT: A Saccharide Topology Analysis Tool used in combination with tandem mass spectrometry. Anal. Chem. 72: 2331–2336, 2000. [DOI] [PubMed] [Google Scholar]
- 13) M. Ethier, J. A. Saba, W. Ens, K. G. Standing, H. Perreault. Automated structural assignment of derivatized complex N-linked oligosaccharides from tandem mass spectra. Rapid Commun. Mass Spectrom: RCM 16: 1743–1754, 2002. [DOI] [PubMed] [Google Scholar]
- 14) B. Domon, C. E. Costello. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj. J. 5: 397–409, 1988. [Google Scholar]
- 15) A. Bohne, E. Lang, C.-W. von der Lieth. W3-SWEET: Carbohydrate modeling by internet. J. Mol. Model. 4: 33–43, 1998. [Google Scholar]
- 16) A. Bohne, E. Lang, C. W. von der Lieth. SWEET—WWW-based rapid 3D construction of oligo- and polysaccharides. Bioinformatics 15: 767–768, 1999. [DOI] [PubMed] [Google Scholar]
- 17) A. Loß, P. Bunsmann, A. Bohne, A. Loß, E. Schwarzer, E. Lang, C. W. von der Lieth. SWEET-DB: An attempt to create annotated data collections for carbohydrates. Nucleic Acids Res. 30: 405–408, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18) K. K. Lohmann, C. W. von der Lieth. GlycoFragment and GlycoSearchMS: Web tools to support the interpretation of mass spectra of complex carbohydrates. Nucleic Acids Res. 32(Web Server): W261–W266, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19) A. J. Lapadula, P. J. Hatcher, A. J. Hanneman, D. J. Ashline, H. Zhang, V. N. Reinhold. Congruent strategies for carbohydrate sequencing. 3. OSCAR: An algorithm for assigning oligosaccharide topology from MSn data. Anal. Chem. 77: 6271–6279, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20) D. J. Ashline, A. J. Lapadula, Y. H. Liu, M. Lin, M. Grace, B. Pramanik, V. N. Reinhold. Carbohydrate structural isomers analyzed by sequential mass spectrometry. Anal. Chem. 79: 3830–3842, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21) D. Goldberg, M. Bern, B. Li, C. B. Lebrilla. Automatic determination of O-glycan structure from fragmentation spectra. J. Proteome Res. 5: 1429–1434, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22) K. Maass, R. Ranzinger, H. Geyer, C. W. von der Lieth, R. Geyer. “Glyco-peakfinder”–de novo composition analysis of glycoconjugates. Proteomics 7: 4435–4444, 2007. [DOI] [PubMed] [Google Scholar]
- 23) A. Apte, N. S. Meitei. Bioinformatics in glycomics: Glycan characterization with mass spectrometric data using SimGlycan. Methods Mol. Biol. 600: 269–281, 2010. [DOI] [PubMed] [Google Scholar]
- 24) H. Peltoniemi, S. Natunen, I. Ritamo, L. Valmu, J. Rabina. Novel data analysis tool for semiquantitative LC-MS-MS2 profiling of N-glycans. Glycoconj. J. 30: 159–170, 2013. [DOI] [PubMed] [Google Scholar]
- 25) C. C. Nwosu, R. R. Seipert, J. S. Strum, S. S. Hua, H. J. An, A. M. Zivkovic, B. J. German, C. B. Lebrilla. Simultaneous and extensive site-specific N- and O-glycosylation analysis in protein mixtures. J. Proteome Res. 10: 2612–2624, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26) C. A. Cooper, E. Gasteiger, N. H. Packer. GlycoMod—A software tool for determining glycosylation compositions from mass spectrometric data. Proteomics 1: 340–349, 2001. [DOI] [PubMed] [Google Scholar]
- 27) H. J. An, J. S. Tillinghast, D. L. Woodruff, D. M. Rocke, C. B. Lebrilla. A new computer program (GlycoX) to determine simultaneously the glycosylation sites and oligosaccharide heterogeneity of glycoproteins. J. Proteome Res. 5: 2800–2808, 2006. [DOI] [PubMed] [Google Scholar]
- 28) E. P. Go, K. R. Rebecchi, D. S. Dalpathado, M. L. Bandu, Y. Zhang, H. Desaire. GlycoPep DB: A tool for glycopeptide analysis using a “Smart Search.” Anal. Chem. 79: 1708–1713, 2007. [DOI] [PubMed] [Google Scholar]
- 29) N. Deshpande, P. H. Jensen, N. H. Packer, D. Kolarich. GlycoSpectrumScan: Fishing glycopeptides from MS spectra of protease digests of human colostrum sIgA. J. Proteome Res. 9: 1063–1075, 2010. [DOI] [PubMed] [Google Scholar]
- 30) J. Irungu, E. P. Go, D. S. Dalpathado, H. Desaire. Simplification of mass spectral analysis of acidic glycopeptides using GlycoPep ID. Anal. Chem. 79: 3065–3074, 2007. [DOI] [PubMed] [Google Scholar]
- 31) S. Joenväärä, I. Ritamo, H. Peltoniemi, R. Renkonen. N-glycoproteomics—An automated workflow approach. Glycobiology 18: 339–349, 2008. [DOI] [PubMed] [Google Scholar]
- 32) H. Peltoniemi, S. Joenvaara, R. Renkonen. De novo glycan structure search with the CID MS/MS spectra of native N-glycopeptides. Glycobiology 19: 707–714, 2009. [DOI] [PubMed] [Google Scholar]
- 33) O. Ozohanics, J. Krenyacz, K. Ludanyi, F. Pollreisz, K. Vekey, L. Drahos. GlycoMiner: A new software tool to elucidate glycopeptide composition. Rapid Commun. Mass Spectrom: RCM 22: 3245–3254, 2008. [DOI] [PubMed] [Google Scholar]
- 34) Z. Darula, R. J. Chalkley, P. Baker, A. L. Burlingame, K. F. Medzihradszky. Mass spectrometric analysis, automated identification and complete annotation of O-linked glycopeptides. Eur. J. Mass Spectrom. (Chichester, Eng.) 16: 421–428, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35) Y. Wu, Y. Mechref, I. Klouckova, M. V. Novotny, H. Tang. in Systems Biology and Computational Proteomics (Ed.: T. Ideker, V. Bafna), Springer, Berlin/Heidelberg, 2007, pp. 96–107.
- 36) A. M. Mayampurath, Y. Wu, Z. M. Segu, Y. Mechref, H. Tang. Improving confidence in detection and characterization of protein N-glycosylation sites and microheterogeneity. Rapid Commun. Mass Spectrom: RCM 25: 2007–2019, 2011. [DOI] [PubMed] [Google Scholar]
- 37) R. Ranzinger, S. Herget, T. Wetter, C. W. von der Lieth. GlycomeDB—Integration of open-access carbohydrate structure databases. BMC Bioinformatics 9: 384, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38) P. Pompach, K. B. Chandler, R. Lan, N. Edwards, R. Goldman. Semi-automated identification of N-glycopeptides by hydrophilic interaction chromatography, nano-reverse-phase LC-MS/MS, and glycan database search. J. Proteome Res. 11: 1728–1740, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39) S. R. Kronewitter, M. L. De Leoz, J. S. Strum, H. J. An, L. M. Dimapasoc, A. Guerrero, S. Miyamoto, C. B. Lebrilla, G. S. Leiserowitz. The glycolyzer: Automated glycan annotation software for high performance mass spectrometry and its application to ovarian cancer glycan biomarker discovery. Proteomics 12: 2523–2538, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40) C. L. Woodin, D. Hua, M. Maxon, K. R. Rebecchi, E. P. Go, H. Desaire. GlycoPep grader: A web-based utility for assigning the composition of N-linked glycopeptides. Anal. Chem. 84: 4821–4829, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41) M. Bern, Y. J. Kil, C. Becker. Byonic: Advanced peptide and protein identification software. Curr. Protoc. Bioinformatics 40: 13.20.1–13.20.14, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42) L. He, L. Xin, B. Shan, G. A. Lajoie, B. Ma. GlycoMaster DB: Software to assist the automated identification of N-linked glycopeptides by tandem mass spectrometry. J. Proteome Res. 13: 3881–3895, 2014. [DOI] [PubMed] [Google Scholar]
- 43) S. Toghi Eshghi, P. Shah, W. Yang, X. Li, H. Zhang. GPQuest: A spectral library matching algorithm for site-specific assignment of tandem mass spectra to intact N-glycopeptides. Anal. Chem. 87: 5181–5188, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44) K. S. Lynn, C. C. Chen, T. M. Lih, C. W. Cheng, W. C. Su, C. H. Chang, C. Y. Cheng, W. L. Hsu, Y. J. Chen, T. Y. Sung. MAGIC: An automated N-linked glycoprotein identification tool using a Y1-ion pattern matching algorithm and in silico MS2 approach. Anal. Chem. 87: 2466–2473, 2015. [DOI] [PubMed] [Google Scholar]
- 45) C. Y. Yu, A. Mayampurath, R. Zhu, L. Zacharias, E. Song, L. Wang, Y. Mechref, H. Tang. Automated glycan sequencing from tandem mass spectra of N-linked glycopeptides. Anal. Chem. 88: 5725–5732, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46) W. F. Zeng, M. Q. Liu, Y. Zhang, J. Q. Wu, P. Fang, C. Peng, A. Nie, G. Yan, W. Cao, C. Liu, H. Chi, R. X. Sun, C. C. Wong, S. M. He, P. Yang. pGlyco: A pipeline for the identification of intact N-glycopeptides by using HCD- and CID-MS/MS and MS3. Sci. Rep. 6: 25102, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]