Abstract
Cotton fibers are single-celled hairs that elongate to several centimeters long from the seed coat epidermis of the tetraploid species (Gossypium hirsutum and Gossypium barbadense). Thus, cotton fiber is a unique system to study the mechanisms of rapid cell expansion. Previous work has shown a transient closure of plasmodesmata during fiber elongation (Y.-L. Ruan, D.J. Llewellyn, R.T. Furbank [2001] Plant Cell 13: 47–60). To examine the importance of this closure in fiber elongation, we compared the duration of the plasmodesmata closure among different cotton genotypes differing in fiber length. Confocal imaging of the membrane-impermeant fluorescent molecule carboxyfluorescein revealed a genotypic difference in the duration of the plasmodesmata closure that positively correlates with fiber length among three tetraploid genotypes and two diploid progenitors. In all cases, the closure occurred at the rapid phase of elongation. Aniline blue staining and immunolocalization studies showed that callose deposition and degradation at the fiber base correlates with the timing of plasmodesmata closure and reopening, respectively. Northern analyses showed that the expression of a fiber-specific β-1,3-glucanase gene, GhGluc1, was undetectable when callose was deposited at the fiber base but became evident at the time of callose degradation. Genotypically, the level of GhGluc1 expression was high in the short fiber genotype and weak in the intermediate and long fiber genotypes. The data provide genotypic and developmental evidence that (1) plasmodesmata closure appears to play an important role in elongating cotton fibers, (2) callose deposition and degradation may be involved in the plasmodesmata closure and reopening, respectively, and (3) the expression of GhGluc1 could play a role in this process by degrading callose, thus opening the plasmodesmata.
Each cotton fiber is a single cell that initiates from the epidermis of the outer integument of the ovules at or just prior to anthesis (Basra and Malik, 1984). Thereafter, the fibers elongate rapidly for about 3 weeks before they switch to intensive secondary cell wall cellulose synthesis. The final length attained by the fibers could range from 1.8 to 5.0 cm in the tetraploid cotton, depending on genotype. This renders cotton fiber the longest single cell known in higher plants (Ruan et al., 2001). Another important feature of the fiber growth is that the fibers interconnect the underlying seed coat only at their basal ends, where influx of solute, water, and other molecules occurs through either plasmodesmata or plasma membrane. Thus, cotton fiber is regarded as an excellent model system in which to study the role of plasmodesmata gating in growth and development at the single-cell level in a defined direction (Pfluger and Zambryski, 2001; Ruan et al., 2001).
Although fibers elongate more than 2,000-fold after initiation, the adjacent seed coat cells expand by only 5- to 10-fold during this period. This indicates a high degree of cell autonomy of the fibers. One mechanism for such a discrepancy in cell growth and function is symplastic isolation between the respective tissue or cell types through the closure of plasmodesmata (e.g. van Bel and van Rijen, 1994; for review, see Roberts and Oparka, 2003; Zambryski, 2004). By confocal imaging of the movement of membrane-impermeant fluorescent solute carboxyfluorescein (CF), Ruan et al. (2001) observed a 5-d period of transient closure of plasmodesmata during the rapid phase of fiber elongation. The plasmodesmata closure, together with the elevated expression of plasma membrane Suc and K+ transporters, coincides with a rise of turgor in the fibers, while the reopening of the plasmodesmata appears to correlate with the termination of the elongation, possibly by releasing the high turgor. These observations indicate a direct role of plasmodesmatal regulation in plant cell development and raise several questions for further studies (Martin et al., 2001; Pfluger and Zambryski, 2001). The critical issues which remain to be determined are whether the closure of plasmodesmata is crucial for the rapid elongation of the cotton fiber cells (Ruan et al., 2001) and what is the physical mechanism enabling this closure.
The investigation of the in vivo functions of plasmodesmata regulation in plant development by using molecular genetic approaches has been proven to be a challenging task since the molecular components that control the structure and function of plasmodesmata remain virtually unknown thus far (Roberts and Oparka, 2003) and alteration in plasmodesmata function through mutational screening will likely have lethal effects on the growth of plants (Zambryski, 2004). Thus, the alternative approach chosen here is to study the role of plasmodesmatal regulation in growth and development by conducting genotypic comparisons of the gating properties of plasmodesmata in relation to the different behavior of tissues or cell types of interest (see Gisel et al., 2002).
This study aims (1) to examine the importance of plasmodesmata closure in cotton fiber elongation by comparing the gating status of the fiber plasmodesmata among five genotypes that differ in fiber length and (2) to explore the molecular basis of the plasmodesmata gating in fibers by investigating the possible involvement of callose deposition and degradation.
RESULTS
Genotypic Differences in the Duration of Plasmodesmata Closure Positively Correlate with Fiber Length
To study the gating status of fiber plasmodesmata, the phloem-mobile fluorescent probe, CF, was ester loaded into shoots through their cut ends for 24 h, unless otherwise stated (see Ruan et al., 2001). The subsequent unloading pattern of CF from the phloem of the outer seed coat to the fiber cells through plasmodesmata was monitored in situ using confocal laser scanning microscopy. The xylem discontinuity in the peduncle of developing cotton fruit (van Iersel and Oosterhuis, 1996) ensures phloem-specific transport of CF into fruits and seeds from the shoots. The movement of the CF into fibers from the phloem of seed coat indicates the opening of the plasmodesmata for CF transport at the fiber bases, while the failure of the CF entry shows the closure of the plasmodesmata.
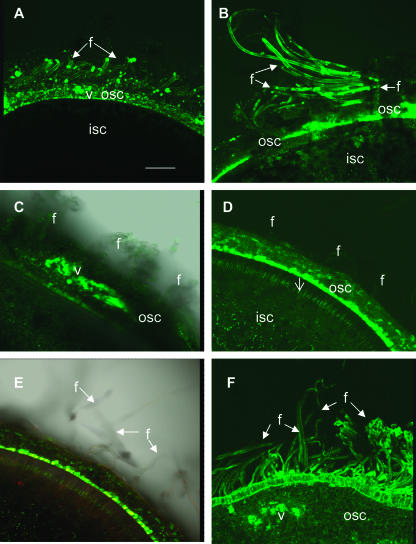
The imaging analysis of CF movement into fibers was conducted at approximately 2- to 3-d intervals over the entire elongation period initially among three tetraploid cotton genotypes (see “Materials and Methods” for more details). Together with the previously analyzed genotype Gh-c, which elongates its fibers to a length of approximately 2.8 cm (Ruan et al., 2001), two additional genotypes, Gh-r and Gb, with final fiber length of about 1.7 and 5.2 cm, respectively, were selected for comparison. Imaging analysis of the optical sections of Gh-c seed revealed a failure of dye movement into fibers from 10 to 15 d after anthesis (DAA; data not shown). This is consistent with previous observation (Ruan et al., 2001). For the short and long fiber genotypes (Gh-r and Gb, respectively), although CF moved into their fibers during the initiation and early elongation period, some distinct characteristics of fiber plasmodesmata gating were observed between the two genotypes from approximately 8 DAA onward. Some representative images from selected time points are shown in Figure 1. For the short fiber genotype Gh-r, the fluorescent CF moved into fibers from the outer seed coat at 10 (Fig. 1A) and 20 (Fig. 1B) DAA after 24-h shoot feeding. By contrast, in the long fiber genotype Gb, the fluorescent dye failed to enter the fibers from the vascular region of the seed coat at 10 DAA after the same period of feeding (Fig. 1C). Similarly, by 20 DAA, the phloem-unloaded CF still did not move into fibers, although it spread throughout the outer seed coat (Fig. 1D). The same fluorescent image was superimposed on the transmitted light to show the position of the fibers (Fig. 1E). In a separate experiment, we extended CF feeding to 48 h on shoots bearing 10- and 20-d fruit of genotype Gb, and again the CF failed to move into fibers (data not shown). However, from approximately 26 DAA onward, the fluorescent signals were readily detected in the fibers of this genotype. Figure 1F shows that CF moved extensively from the vascular region of the seed coat into epidermal cells and fibers at 30 DAA in Gb after 24-h feeding.
Figure 1.
Confocal imaging of CF transport to elongating cotton fibers in genotypes of Gh-r (A and B) and Gb (C–F) at 10 and 20 DAA. A, Optical section of seed at 10 DAA from genotype Gh-r, showing CF movement from vascular bundle of outer seed coat into fibers. Note transport and accumulation of CF from unloading area to fibers and outer seed coat but not to the inner seed coat. B, Optical section of seed at 20 DAA from genotype Gh-r, showing strong fluorescent signals of CF in fibers. Some weak signals of CF were visible in the inner seed coat at this stage. C, Blockage of CF movement into fibers at 10 DAA in genotype Gb. Note the strong and widespread CF signals in the vascular region but little or no signals in the fibers. D, Blockage of CF movement into fibers at 20 DAA in genotype Gb. Note the CF signal spread throughout the outer seed coat but was not present in fibers. Rather, the dye moved preferentially into the inner side of the outer seed coat. Some CF signals traveled to the inner seed coat through a cell layer interconnecting the outer and inner seed coats (arrow). E, The same image of D was superimposed on the transmitted light to show the position of the fibers. F, Recovery of CF movement into fibers of genotype Gb at 30 DAA. Note strong fluorescent signals of CF in fiber cells. Also note the dye appeared to move preferentially into the epidermal and fiber cells from the vascular region. f, Fiber; isc, inner seed coat; osc, outer seed coat; v, vascular bundle. Bar (in μm) = 500 in A. The scale in B to F is the same as in A.
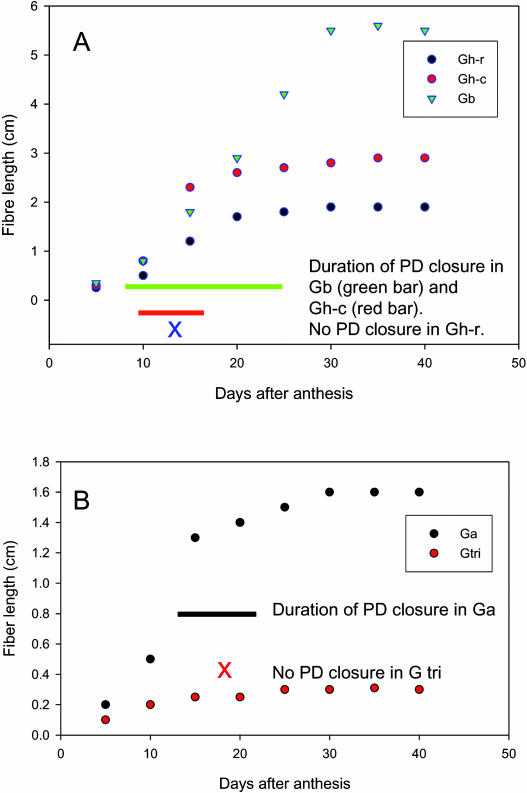
Figure 2A summarizes the data on CF movement into, hence plasmodesmatal gating of, fibers over the elongation period among the three tetraploid cotton genotypes differing in fiber length. It shows that, in the short fiber genotype Gh-r, CF moved into fibers throughout the elongation period and, thus, no plasmodesmata closure occurred at the time interval examined. For the genotype with intermediate fiber length, Gh-c, the initially open plasmodesmata closed for a 5-d period from approximately 10 DAA (Fig. 2A), as previously observed (Ruan et al., 2001). Remarkably, in the long fiber genotype Gb, fiber plasmodesmata were closed to CF import for 18 d from 8 to 26 DAA (Fig. 2A). It is also noteworthy that the closure of plasmodesmata occurred at the rapid phase of fiber elongation in both Gh-c and Gb (Fig. 2A).
Figure 2.
The kinetics of cotton fiber elongation and plasmodesmata gating among three tetraploid and two diploid genotypes. A, Elongation of cotton fiber among three tetraploid lines. The length of the bar indicates the period of plasmodesmata closure. Note the fiber plasmodesmata closed earlier and longer in the long fiber line Gb than that in Gh-c with intermediate fiber length. In both cases, the closure of plasmodesmata occurred at the rapid phase of fiber elongation. Also note no plasmodesmata closure was observed in the short fiber genotype Gh-r. B, Elongation of cotton fiber in two diploid progenitors. The black bar indicates the duration of plasmodesmata closure in an A genome line, Ga. No closure of plasmodesmata was observed in the fibers of a D genome line, Gtri, and these fibers elongate much slower than that of Ga. Each value of the fiber length in A and B is the mean of six measurements with se less than 9% of the mean.
The tetraploid cotton is derived from polyploidization of diploid A and D genomes during evolution. Although both A and D genome progenitors initiate fiber cells from the ovular epidermis at anthesis, only the fibers from the A, but not D, genome progenitor are able to elongate to over 1.0 cm (see Applequist et al., 2001). Thus, the confocal imaging analysis of CF movement was extended to examine whether the difference in fiber plasmodesmata gating exists among the diploid progenitors of the tetraploid cotton. Figure 2B shows an A genome line, Ga, closed the fiber plasmodesmata for about 10 d during the rapid phase of elongation and produced about 1.5-cm-long fiber, while a D genome line, G tri, did not close fiber plasmodesmata, and virtually no fiber elongation occurred. These results are consistent with the genotypic correlation between the duration of plasmodesmata closure and fiber length observed in the tetraploid genotypes (Fig. 2, B versus A).
Evidence on the Involvement of Callose in Plasmodesmata Gating of Cotton Fiber
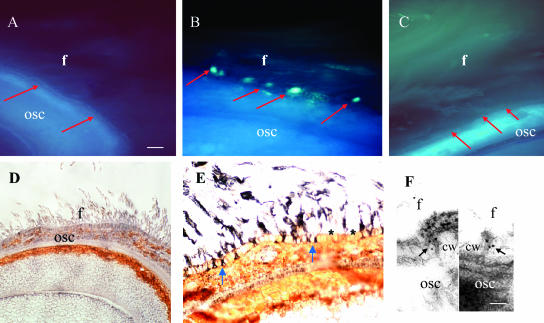
The molecular basis of plasmodesmata gating is virtually unknown. However, callose deposition at the neck region of plasmodesmata has been implicated in closing plasmodesmata (Schulz, 1999). Further experiments were therefore conducted to examine whether callose is involved in the plasmodesmata gating in cotton fiber. The timing and the duration of callose deposition in fiber base were measured in the cotton genotypes Gh-c and Gb from fluorescent signals of aniline blue, a callose-specific stain. Appearance of the fluorescence for callose at the fiber base correlated with the timing of plasmodesmata closure in both cases. Representative images from genotype Gh-c are presented in Figure 3, A to C. Fluorescent labeling with aniline blue showed that callose was undetectable in the fiber bases at 5 and 20 DAA (Fig. 3, A and C, respectively) when the plasmodesmata were open (Fig. 2A). However, strong fluorescent signals of callose were specifically detected at the fiber bases located in the seed coat epidermis at 12 DAA (Fig. 3B), when plasmodesmata were closed (Fig. 2A; Ruan et al., 2001). To verify these observations, immunolocalization experiments were further conducted by using a monoclonal antibody against callose (Meikle et al., 1991). Similar to the results from the aniline blue labeling experiments (Fig. 3, A–C), immunogold labeling on callose was undetectable in fibers at 5 DAA (Fig. 3D) but became evident at the fiber base at 12 DAA (Fig. 3E). Some immunogold signals were also seen in the upper part of the fibers (Fig. 3E). At the electron microscopic level, the immunogold-labeled callose was localized at the neck region of the plasmodesmata at the fiber base at 12 DAA (Fig. 3F). The controls were free of label (normal serum or omitting primary antibody; data not shown). The number of gold particles of callose in plasmodesmata was, however, significantly reduced at 20 DAA (Table I), when the plasmodesmata were reopened (Fig. 2A).
Figure 3.
Fluorescent and immunogold localization of callose at the fiber base of genotype Gh-c. A, Free-hand section of 5-d seed labeled with callose-specific aniline blue. Fluorescent signals of callose were undetectable at the fiber base (red arrows). B, Free-hand section of 12-d seed labeled with callose-specific aniline blue, showing strong callose signals specifically at the fiber base (red arrows) interconnecting the underlying seed coat. Note the timing of the callose localization correlates with the closure of fiber plasmodesmata (Fig. 2A). C, Free-hand section of 20-d seed labeled with callose-specific aniline blue. Note the callose at the fiber bases previously seen in B has been degraded (red arrows). Also note the plasmodesmatal permeability restored at this stage (Fig. 2A). D, Immunogold labeling with monoclonal antibody against callose in cross section of 5-d seed. Callose signals were undetectable at this stage. E, Immunogold labeling with monoclonal antibody against callose in cross section of 12-d seed, showing strong immunogold-labeled callose at the fiber bases (blue arrows). Note the labeling is specific to the fibers. No label was seen the adjacent epidermal cells (star). F, Electron microscopic immunolocalization of callose in plasmodesmata at the fiber base at 12 d. Note gold particles of callose (arrows) at the neck region of two plasmodesmata. cw, Cell wall; f, fiber cell; osc, outer seed coat. Bar = 200 μm in A and 50 nm in F. The scale in B to E is the same as A.
Table I.
Number of immunogold particles for callose at plasmodesmata of cotton fibera
| DAA | Gating Status of PDb | Gold Particles per PD | Total PD Examined |
|---|---|---|---|
| 5 | Open | 0.3 ± 0.2c | 20 |
| 12 | Closed | 3.0 ± 0.5 | 26 |
| 20 | Open | 1.1 ± 0.1 | 18 |
Genotype Gh-c.
PD, Plasmodesmata.
Each value is the mean ± se of gold particles from the total PD examined.
The Expression of a Fiber-Specific β-1,3-Glucanase Gene Correlates with Callose Degradation and Plasmodesmata Opening
The observation that the timing of callose deposition and degradation matches with the closure and opening of plasmodesmata at the fiber bases, respectively (see above), indicates the involvement of callose in plasmodesmata gating of fibers. Callose accumulation is controlled by the joint action of β-1,3-glucanase and β-1,3-glucan synthase, which degrade and synthesize callose, respectively (Bucher et al., 2001). To understand the molecular nature of the callose turnover in cotton fiber, two partial cDNAs encoding β-1,3-glucanase, designated as GhGluc1 and GhGluc2, were cloned from fiber tissue using reverse transcription-PCR. A BLAST search of the GenBank database showed that the amino acid sequences encoded by GhGluc1 and GhGluc2 shared homology exclusively to known β-1,3-glucanase in the first 20 matches. In the first match, GhGluc1 and GhGluc2 shared 63% and 60% amino acid identity with known β-1,3-glucanase from barley (Hordeum vulgare) and Arabidopsis (Arabidopsis thaliana), respectively (GenBank accession nos. M91814 and AC003058).
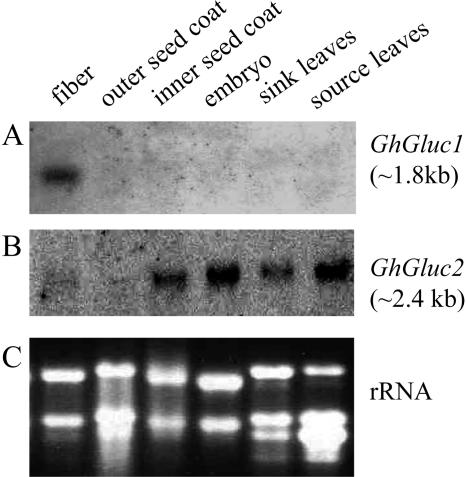
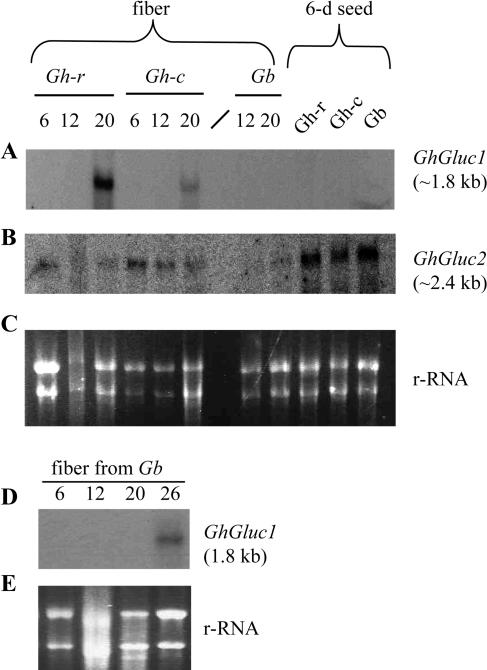
The tissue specificity of the expression of the two β-1,3-glucanase genes was examined using northern-blot analysis. Figure 4A shows that the expression of GhGluc1 was highly specific to fibers. Its mRNA (approximately 1.8 kb) was strongly detected in 20-d fibers, but neither in other parts of the seed nor in sink and source leaves (Fig. 4A). Rehybridization of the membrane with GhGluc2 cDNA probe, however, revealed a very different expression pattern (Fig. 4B). The GhGluc2 mRNA (approximately 2.4 kb) was not only weakly detected in fibers but also evidently expressed in the inner seed coat, embryo, as well as in sink and source leaves (Fig. 4B). The ethidium bromide-stained rRNA was shown as a loading control for each sample (Fig. 4C).
Figure 4.
Transcript expression of GhGluc1 and GhGluc2 in 20-d fibers and other tissues of genotype Gh-c. RNA gel blot with 20 μg of total RNA in each lane was sequentially hybridized with GhGluc1 (A) and GhGluc2 (B) cDNA probes. The equal loading of the RNA samples was indicated by the ethidium bromide-stained rRNA bands in each lane (C).
To explore whether the expression of the GhGluc1 and GhGluc2 relates to callose degradation and plasmodesmata opening in fibers, further northern analyses were performed on RNA samples isolated from representative stages of fibers among the three tetraploid genotypes differing in plasmodesmata gating and fiber length (Fig. 2A). Figure 5A shows that, in the genotype Gh-c, the expression of GhGluc1 is detected at 20 DAA, coincident with the timing of callose degradation (Fig. 3C). Importantly, the mRNA of GhGluc1 is undetectable at 12 DAA (Fig. 5A), when callose is deposited at the fiber base (Fig. 3, B, E, and F). Similarly, in the genotype Gb, GhGluc1 transcript was not detected at 12 and 20 DAA (Fig. 5A), when plasmodesmata were closed (Figs. 1 and 2A), but was detectable at 26 DAA (Fig. 5D), when the plasmodesmata were reopened (Fig. 2A).
Figure 5.
Transcript expression of GhGluc1 and GhGluc2 in developing cotton fibers and 6-d seeds among different genotypes differing in fiber length. RNA gel blot with 20 μg of total RNA in each lane was hybridized with GhGluc1 (A and D) and GhGluc2 (B) cDNA probes. The ethidium bromide-stained rRNA was shown as a loading control for each lane (C and E). The number above each lane indicates DAA.
Genotypically, the expression of GhGluc1 in 20-d fibers is strong, weak, and undetectable in Gh-r, Gh-c, and Gb (Fig. 5A), representing short, intermediate, and long fiber genotypes, respectively (Fig. 2A). Notably, the absence of GhGluc1 expression in Gb from 6 to 20 DAA (Fig. 5, A and D) correlates with the longest duration of plasmodesmata closure (Figs. 1, C and D, and 2A). In contrast with the developmental- and fiber-specific expression pattern of GhGluc1 (Figs. 4A and 5A), the transcript of GhGluc2 was detected in fibers of Gh-c at all the stages examined as well as in 6- and 20-d fibers of Gh-r and 20-d fibers of Gb (Fig. 5B). The GhGluc2 was also expressed in 6-d seeds of all the three genotypes (Fig. 5B). The experiment was repeated three times with RNA isolated from cotton plants grown in different batches. Consistent results were obtained (Fig. 5).
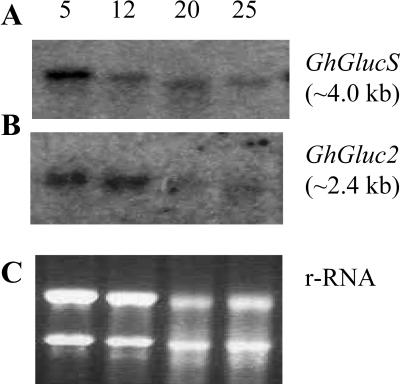
To better understand the basis of callose synthesis in fiber, a partial cDNA encoding β-1,3-glucan synthase (GhGlucS) was also cloned from fiber using reverse transcription-PCR based on sequence information in cotton fiber expressed sequence tag (EST) database (accession no. AI730469). Northern analysis revealed that expression pattern of GhGlucS was similar to that of GhGluc2 (Fig. 6) and was not fiber specific (data not shown).
Figure 6.
Transcript expression of GhGlucS and GhGluc2 in developing cotton fibers of genotype Gh-c. RNA gel blot with 20 μg of total RNA in each lane was sequentially hybridized with GhGlucS (A) and GhGluc2 (B) cDNA probes. The ethidium bromide-stained rRNA was shown as a loading control for each lane (C). The number above each lane indicates DAA.
DISCUSSION
Genotypic and Developmental Evidence for the Role of Plasmodesmata Regulation in Cotton Fiber Elongation
In this study, we present data to show that duration of plasmodesmata closure in cotton fiber cells positively correlates with the fiber length attained among three tetraploid lines and two diploid progenitors examined (Figs. 1 and 2). Consistently, a tetraploid lintless mutant (fls), which produces fuzz-like fiber of less than 0.5 cm, does not close its fiber plasmodesmata (data not shown). Furthermore, in all cases, the closure of plasmodesmata assayed by CF import takes place at the onset of rapid fiber elongation (Figs. 1 and 2). These observations indicate that the closure of plasmodesmata plays a key role in controlling fiber length.
Developmentally programmed symplastic isolation has been observed in both vegetative and reproductive tissue or cell types. This typically includes developing leaves during sink-source transition (Turgeon, 1996; Oparka et al., 1999), sieve element-companion cell complex of stem (van Bel and van Rijen, 1994), shoot apex at the onset of flower induction (Gisel et al., 1999), and embryos of Arabidopsis at the torpedo stage of development (Kim et al., 2002). Although many of the symplastic isolation processes are permanent, some are indeed reversible. For example, the symplastic pathway shut down in the shoot apex of birch tree at the end of the growing season (Rinne and van der Schoot, 1998). However, the plasmodesmatal connection is reestablished at the breakage of bud dormancy after a chilling winter season (Rinne et al., 2001). The developmentally controlled down-regulation of plasmodesmatal permeability is generally believed to be required for respective tissues or cells to establish their identities or to perform distinct functions (Roberts and Oparka, 2003). However, the assessment of the role of the symplastic isolation in plant development is experimentally difficult, owing to the multicellular complexity of plant tissues and the lack of knowledge on the molecular components of plasmodesmata (Roberts and Oparka, 2003; Zambryski, 2004). In this regard, the cotton fiber is a unique system to address the in vivo function of plasmodesmata gating for a number of reasons: its single-celled nature, yet with significant size for manipulation and observation; the occurrence of plasmodesmatal closure during elongation; and the wealth of existing genotypes with different fiber length for comparison (Pfluger and Zambryski, 2001; Ruan et al., 2001). The genotypic and developmental evidence presented here (Figs. 1 and 2) provide a set of strong correlative evidence on the role of plasmodesmatal closure in rapid expansion of plant cell at a well-defined single-cell level. Pertinently, a recent study shows that the transitory restriction of symplastic communication in shoot apex of Arabidopsis correlates with the timing of floral induction among five different flowering-timing mutants and wild-type plants (Gisel et al., 2002). However, it is not possible to locate the cellular site at which the restriction and the restoration of symplastic tracer movement occurs due to the complex multicellular differentiation processes in the shoot apical meristem (Gisel et al., 2002).
The rapid elongation of cotton fiber is a specialized single-cell expansion, which must be achieved by concerted action of pushing against the cell wall exerted by turgor and loosening of the cell wall (see Cosgrove, 1997; Martin et al., 2001). Previously, we observed that the closure of fiber plasmodesmata follows a significant rise of turgor and rapid phase of fiber elongation in the intermediate line, Gh-c (Ruan et al., 2001). Coincident with the closure of plasmodesmata are the increased expression of phosphoenolpyruvate carboxylase, plasma membrane H+-ATPase (Smart et al., 1998), and transporters for uptake of Suc and K+ (Ruan et al., 2001). The elevated expression of these genes would result in the observed accumulation of malate, sugars, and K+, the major osmoticum, hence the influx of water and the generation of high turgor in the fiber cells (Dhinsda et al., 1975; Ruan et al., 1997; Ruan et al., 2003). Given the enormous discrepancy in the expansion rate of fibers versus adjacent seed coat epidermal cells, the closure of plasmodesmata at the fiber bases may well provide a cellular basis to maintain the high turgor (Ruan et al., 2001), analogous to stomatal guard cells (Palevitz and Hepler, 1985) and Arabidopsis root hairs (Rigas et al., 2001). This notion is supported by the observation that the reopening of plasmodesmata correlates with the release of turgor (Ruan et al., 2001) and termination of the elongation process (Fig. 2). Based on these analyses, we envisage that the extended period of plasmodesmata closure in the long fiber genotype Gb (Figs. 1 and 2), may allow the fibers to sustain a high turgor for a longer period, leading to the much longer fiber than the intermediate line, Gh-c (Fig. 2A).
It is noteworthy that during the early elongation period, the fiber plasmodesmata are open among all the genotypes examined (Fig. 2). In addition, the short fiber lines, the tetraploid Gh-r and the diploid progenitor, Gtri, do not close their plasmodesmata over the entire elongation period at the interval examined. These data indicate that, although closure of plasmodesmata appears to play an important role in elongating fibers to extended length (see above), it is not required for early or moderate elongation. Consistent with the opening of the plasmodesmata in early elongation, plasmolysis studies revealed similar osmotic, hence turgor, potential between the fiber initials and adjacent seed coat epidermal cells (Ruan et al., 2000). Therefore, the achievement of the relatively slow elongation without the closure of plasmodesmata may be accomplished through the relaxation of the cell wall mediated by the expression of an expansin gene (Ruan et al., 2000). Indeed, high-level expression of an expansin gene has been observed in fibers early in elongation (Shimizu et al., 1997; Orford and Timmis, 1998; Ruan et al., 2001) when the plasmodesmata are open (Fig. 1A).
Callose Deposition May Be Required for the Closure of Plasmodesmata in Cotton Fiber
Although the molecular nature of plasmodesmata gating remains elusive, callose deposition around the plasmodesmata neck has been considered a potentially important mechanism for the regulation of plasmodesmatal permeability (e.g. Lucas et al., 1993). The possibility of localized callose turnover in closing and opening of fiber plasmodesmata was therefore explored here in parallel with the plasmodesmata gating analysis. Callose-specific labeling with aniline blue and a monoclonal antibody revealed strong signals of callose at the basal region of fibers when plasmodesmata were closed but not at the early stage when they were open (Fig. 3). At the electron microscopic level, the gold particles of callose were indeed localized in or around the plasmodesmata of fibers (Fig. 3F). Significantly, the callose was degraded from the plasmodesmata when the fiber plasmodesmata were reopened (Table I). For the short fiber line Gh-r that shows no closure of PD (Figs. 1 and 2), no callose signals were detected in the fiber base during the elongation period (data not shown). Collectively, these results show that callose deposition and degradation correlates with the closure and opening of fiber plasmodesmata, respectively. The analyses indicate that the deposition of callose may directly or indirectly contribute to the closure of plasmodesmata in cotton fiber. The plugging of plasmodesmata by callose could result in firm osmotic isolation and restriction of water movement (e.g. Rinne and van der Schoot, 1998). This may help the fiber cells to maintain the high turgor for sustained elongation (see “Discussion” above).
The molecular basis of callose deposition and degradation was further examined by expression analysis of two β-1,3-glucanase cDNAs, GhGluc1 and GhGluc2, and one β-1,3-glucan synthase (GhGlucS), cloned from cotton fiber. Although the expression of GhGluc2 and GhGlucS does not correlate to callose turnover and plasmodesmata gating at the fiber bases (Figs. 3, 5, and 6), it is possible that GhGlucS may play a role in the synthesis of callose observed in the plasmodesmata (Fig. 3) as well as in other parts of the fiber cells (see Pillonel et al., 1980; Okuda et al., 1993). Once callose is synthesized in the fibers, it may be subject to, spatially and temporally, differential degradation by GhGluc1 and GhGluc2. While GhGluc2 appears to be expressed in all the tissues examined (Fig. 4B), the expression of GhGluc1 was found to be highly fiber specific (Figs. 4A and 5A). The timing of its expression in fibers (Fig. 5, A and D) correlates to that of callose degradation and plasmodesmata opening in the genotypes Gh-c and Gb (Figs. 1–3). By contrast, the mRNA of this β-1,3-glucanase gene was undetectable at 12 DAA (Fig. 5A), when callose was deposited in Gh-c (Fig. 3; Table I) and plasmodesmata were closed (Fig. 2A). These data suggest GhGluc1 may be responsible for the degradation of callose in the plasmodesmata, which either prevents plasmodesmata closure or allows reopening, hence shortening the elongation period. Consistent with this view is the observation that the mRNA levels of this gene in 20-d fibers are strongest in Gh-r, weak in Gh-c, and hardly detectable in Gb, representing short, intermediate, and long fiber genotypes, respectively (Fig. 5A). These expression patterns of GhGluc1 (Figs. 4 and 5), together with the callose localization analyses (Fig. 3; Table I), point to callose deposition and degradation as a likely basis for plasmodesmata closure and opening in cotton fiber.
The involvement of callose turnover in reversible gating of plasmodesmata is not unique to cotton fiber. In the shoot apical meristem of birch, for example, callose was undetectable in plasmodesmata when they were open for transport of the small molecule Lucifer Yellow CH, but became evident when the plasmodesmata were closed, induced by short photoperiod (Rinne and van der Schoot, 1998). Remarkably, the symplastic organization is restored at the breakage of bud dormancy, coincident with the degradation of callose, mediated by β-1,3-glucanase (Rinne et al., 2001). To determine the role of this enzyme in callose degradation and plasmodesmata function, Iglesias and Meins (2000) analyzed transgenic tobacco (Nicotiana tabacum) with reduced expression of β-1,3-glucanase. As expected, the transgenic plants showed enhanced callose deposition and reduced plasmodesmatal size exclusion limit, leading to delayed intercellular trafficking of virus via plasmodesmata. Conversely, hydrolyzing callose by overexpression of β-1,3-glucanase increases cell-to-cell movement of virus in tobacco leaves (Bucher et al., 2001). These results, together with the observation made from cotton fiber here, show that, at least in some plant species or tissues/cell types, callose deposition and degradation could play an important role in down- and up-regulation of plasmodesmatal permeability, respectively.
In conclusion, the data obtained in this study represent genotypic and developmental evidence that (1) plasmodesmata closure appears to play an important role in elongating cotton fibers, (2) callose deposition and degradation correlates with plasmodesmata closure and reopening, respectively, and (3) the expression of GhGluc1 could play a role in this process by degrading callose, thus opening the plasmodesmata.
MATERIALS AND METHODS
Plant Material
The cotton genotypes selected for comparative studies were three tetraploid lines, Gossypium hirsutum L. var. Coker 315 (Gh-c) and var. Riverina Poplar (Gh-r) and Gossypium barbadense L. var. S7 (Gb), and two diploid progenitors, an A genome line, Gossypium arboretum (Ga), and a D genome line, Gossypium trilobum (Gtri; see Percival et al. [1999] for detailed information about the taxonomical and morphological characteristics of these genotypes). All the cotton plants were grown in soil mixture under controlled conditions as described previously (Ruan et al., 1997). Cotton fruit age was determined by tagging the flowering truss when the flower was fully opened. Shoots, each bearing two to three fruits and developed leaves, were excised from the plant for loading of 5(6)-CF. For RNA extraction, samples were frozen in liquid N2 and stored at −70°C until analysis. Fresh samples were used for fixation for immunolocalization analysis.
Loading of Carboxyfluorescein and Confocal Laser Scanning Microscopy
The loading of CF and confocal imaging of the movement of CF into fibers were carried out as described previously (Ruan et al. 2001) at an interval of 2 to 3 d after the initiation of the fibers. For each time point, the imaging analysis was performed on 3 to 4 seeds with a total of at least 12 optical sections. The results were reproducible and are summarized in Figure 2. Some representative images were shown in Figure 1.
Aniline Blue Fluorescent Labeling of Callose
Fresh cotton seeds with fiber attached were boiled in ethanol for 10 min immediately after harvesting from plants to minimize the wound-induced callose production (Radford and White, 2001). Thereafter, free-hand sections were cut and briefly rinsed in water to remove cell debris. The sections were then incubated in 0.05% aniline blue in 67 mm phosphate buffer, pH 8.5, for 5 min following two 0.5-min washes with the buffer. The yellow-green fluorescent signals of aniline blue were viewed under UV, and digital images were taken from a Leica microscope (Leica Microsystems, Wetzlar, Germany).
Light Microscopic Immunolocalization of Callose
For immunolocalization of callose at the light microscopic level, cotton seeds at specified developmental stages were fixed in formalin-acetic acid, dehydrated in ethanol, and embedded in paraffin, immediately after harvesting from plants. Immunogold silver staining was then conducted as described previously (Ruan and Chourey, 1998) using the HISTOGOLD kit (ZYMED HISTOGOLD SYSTEM for Immuno-Histological Staining; ZYMED Laboratories, San Francisco). The deparaffined sections (10 μm each in thickness) were incubated with 1:100 diluted monoclonal antibody to (1→3)-β-glucan (Biosupplies Australia, Parkville, Australia) for 1 h. After washing in phosphate-buffered saline, the sections were incubated for 45 min in 1:300 diluted goat anti-mouse IgG linked to colloidal gold (ICN Biomedicals, Aurora, OH). Slides were then washed thoroughly with phosphate-buffered saline (four times, 3 min each), incubated for 4 min with freshly prepared silver enhancement reagents, and washed with excess distilled water. Thereafter, slides were dehydrated in an ethanol series and permanently mounted in Permount (ProSciTech, Thuringowa, Australia) for microscopic examination. Pairs of immuno- and preimmunostained sections were treated on the same slide for better comparison.
Electron Microscopic Immunolocalization of Callose
For immunolocalization of callose at the electron microscopic level, cotton ovules were dissected out in a drop of the fixative under a dissecting microscope. Material was fixed in 2% (v/v) paraformaldehyde and 0.1% (v/v) glutaraldehyde in 25 mm phosphate buffer, pH 7.2, for 2 h at room temperature. After washing in buffer, the ovules were dehydrated in an ethanol series and embedded in LR White resin (medium grade; Alltech, Deerfield, IL) and polymerized at 70°C for approximately 90 min under nitrogen gas.
Ultrathin sections were cut and collected on parlodion-coated nickel grids. Sections were preincubated in TBST (20 mm Tris, 154 mm NaCl, 0.1% Tween 20, pH 7.5) containing 1% bovine serum albumin (Sigma, St. Louis) for 30 min, then incubated in a 1:500 dilution in TBST of a primary monoclonal antibody against β-1,3-glucans (Biosupplies Australia) for 7 h at room temperature then overnight at 4°C. Grids were rinsed in buffer at room temperature for 30 min, then incubated in 10 nm gold-labeled goat anti-mouse monoclonal secondary antibody (at 1:500 dilution) for 45 min before rinsing in buffer, staining in uranyl acetate and lead citrate, and observation with a JEOL 100CX TEM (Brookvale, Australia) at 80 kV.
Control treatments included omitting the primary antibody or replacing the primary antibody with nonimmune mouse IgG or normal mouse serum.
Reverse Transcription-PCR and Cloning
Total RNA was isolated from fibers of Gh-c at 18 DAA according to Ruan et al. (1997). First-strand cDNA was obtained by reverse transcription of 2 μg of RNA with an oligo(dT)20 primer. For cloning a partial β-1,3-glucanase cDNA, GhGluc1, a pair of gene-specific primers, forward 5′-ATATGGGTTTTTAATCTCAGCAATGG-3′ and reverse 5′-AGGGACGATGTTGGTGTTAACCC-3′, was synthesized based on the sequence information in the GenBank (accession no. D88416; also see Shimizu et al., 1997). A putative β-1,3-glucanase DNA fragment (GhGluc1) at the expected size of 303 bp was amplified with the following conditions: 45 cycles of denaturation at 94°C for 1.0 min, annealing at 58°C for 0.5 min, and elongation at 72°C for 2 min. Using the same conditions, a PCR product corresponding to a 547 bp of a second β-1,3-glucanase gene GhGluc2 was amplified with the following primers: forward 5′-ATCAAAGCTGGGATTGGCAGC-3′ and reverse 5′-CAAAATTCCCTGGATCAATGC-3′. This set of primers was based on a cDNA EST sequence of a putative β-1,3-glucanase from cotton fiber (accession no. AI728205).
Similarly, a PCR product of 603 bp of a β-1,3-glucan synthase gene GhGlucS was amplified with primers forward 5′-TGGAAGGGCTTATTTGGCAC-3′ and reverse 5′-GTCCGCCTCTGTAAAATAGC-3′. This set of primers was based on a cDNA EST sequence of a putative β-1,3-glucan synthase from cotton fiber (accession no. AI730469).
The three PCR products, GhGluc1, GhGluc2, and GhGlucS, were purified with Wizard PCR Preps DNA purification system (Promega, Madison, WI), cloned into pGEM-T Easy vector (Promega), and sequenced using big dye terminator (Applied Biosystems 373A DNA sequencing system; Applied Biosystems, Foster City, CA).
RNA Gel-Blot Analysis
After denaturing at 65°C for 5 min in MOPS buffer, pH 7.0, with 50% (v/v) formamide and 18% (v/v) formaldehyde, 20 μg of total RNA from each sample was loaded on a 1.4% agarose gel containing 5% formaldehyde (v/v) for electrophoresis in the MOPS buffer. The fractionated RNA samples were then transferred to nylon membrane, hybridized with P32-labeled probe at 60°C, and washed in 6× SSC and then 0.2× SSC at the same temperature as described previously (Ruan et al., 1997). The entire lengths of GhGluc1, GhGluc2, and GhGlucS were released for probe making by digestion with EcoR1.
Measuring Fiber Length
The fiber length from seed at various developmental stages was measured according to Schubert et al. (1973). In all cases, the measurement was done from the chalazal end of the seed.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers D88416, AI728205, and AI730469.
Acknowledgments
We thank Drs. Greg Constable and Danny Llewellyn for providing the genotypes Gb and Gh-r, and Curt Brubaker for supplying the diploid progenitor lines Ga and Gtri. We appreciate discussions and comments from Drs. Bill Taylor and Tony Arioli during the course of this study.
The work was supported in part by Australian Cotton Research Development Corporation.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.051540.
References
- Applequist WL, Cronn R, Wendel JF (2001) Comparative development of fiber in wild and cultivated cotton. Evol Dev 3: 3–17 [DOI] [PubMed] [Google Scholar]
- Basra A, Malik CP (1984) Development of the cotton fiber. Int Rev Cytol 89: 65–113 [Google Scholar]
- Bucher GL, Tarina C, Heinlein M, Serio FD, Meins F Jr, Iglesias VA (2001) Local expression of β-1,3-glucanase enhances symptoms of TMV infection in tobacco. Plant J 28: 361–369 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (1997) Relaxation in a high-stress environment: the molecular bases of extensible cell walls and cell enlargement. Plant Cell 9: 1031–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhindsa RS, Beasley CA, Ting IP (1975) Osmoregulation in cotton fiber. Plant Physiol 56: 394–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisel A, Barella S, Hempel FD, Zambryski PC (1999) Temporal and spatial regulation of symplastic trafficking during development in Arabidopsis thaliana apices. Development 126: 1879–1889 [DOI] [PubMed] [Google Scholar]
- Gisel A, Hempel FD, Barella S, Zambryski PC (2002) Leaf-to-shoot apex movement of symplastic tracer is restricted coincident with flowering in Arabidopsis. Proc Natl Acad Sci USA 99: 1713–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias VA, Meins F Jr (2000) Movement of plant virus is delayed in a β-1,3-glucanase-deficient mutant showing a reduced plasmodesmatal size exclusion limit and enhanced callose deposition. Plant J 21: 157–166 [DOI] [PubMed] [Google Scholar]
- Kim I, Hempel FD, Sha K, Pfluger J, Zambryski PC (2002) Identification of a developmental transition in plasmodesmatal function during embryogenesis in Arabidopsis thaliana. Development 129: 1261–1272 [DOI] [PubMed] [Google Scholar]
- Lucas WJ, Ding B, Van der Schoot C (1993) Plasmodesmata and the supracellular nature of plants. New Phytol 125: 435–476 [DOI] [PubMed] [Google Scholar]
- Martin C, Bhatt K, Baumann K (2001) Shaping in plant cells. Curr Opin Plant Biol 4: 540–549 [DOI] [PubMed] [Google Scholar]
- Meikle PJ, Bonig I, Hoogenraad NJ, Clarke AE, Stone BA (1991) The location of (1→3)-β-glucans in the walls of pollen tubes of Nicotiana alata using a (1→3)-β-glucan-specific monoclonal antibody. Planta 185: 1–8 [DOI] [PubMed] [Google Scholar]
- Okuda K, Li L, Kudlicka K, Kuga S, Brown RM Jr (1993) β-Glucan synthesis in the cotton fiber. I. Identification of β-1,4- and β-1,3-glucans synthesized in vitro. Plant Physiol 101: 1131–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka KJ, Roberts AG, Boevink P, Cruz SS, Roberts I, Pradel KS, Imlau A, Kotlizky G, Sauer N, Epel B (1999) Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell 97: 743–754 [DOI] [PubMed] [Google Scholar]
- Orford SJ, Timmis JN (1998) Specific expression of an expansin gene during elongation of cotton fibers. Biochim Biophys Acta 1398: 342–346 [DOI] [PubMed] [Google Scholar]
- Palevitz BA, Hepler PK (1985) Changes in dye coupling of stomatal cells of Allium and Commelina demonstrated by microinjection of Lucifer yellow. Planta 164: 473–479 [DOI] [PubMed] [Google Scholar]
- Percival AE, Wendel JF, Stewart JM (1999) Taxonomy and germplasm resources. In CW Smith, JT Cothren, eds, Cotton: Origin, History, Technology and Production. John Wiley & Sons, New York, pp 33–65
- Pfluger J, Zambryski PC (2001) Cell growth: the power of symplastic isolation. Curr Biol 11: R436–R439 [DOI] [PubMed] [Google Scholar]
- Pillonel C, Buchala AJ, Meier H (1980) Glucan synthesis by intact cotton fibers fed with different precursors at the stages of primary and secondary cell wall formation. Planta 149: 306–312 [DOI] [PubMed] [Google Scholar]
- Radford JE, White RG (2001) Effect of tissue-preparation-induced callose synthesis on estimates of plasmodesmata size exclusion limits. Protoplasma 216: 47–55 [DOI] [PubMed] [Google Scholar]
- Rigas S, Debrosses G, Haralampidis K, Vicente-Agullo F, Feldmann KA, Grabov A, Dolan L, Hatzopoulos P (2001) TRH1 encodes a potassium transporter required for tip growth in Arabidopsis root hairs. Plant Cell 13: 139–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinne PL, van der Schoot C (1998) Symplastic fields in the tunica of the shoot apical meristem coordinate morphogenetic events. Development 125: 1477–1485 [DOI] [PubMed] [Google Scholar]
- Rinne PL, Kaikuranta PM, van der Schoot C (2001) The shoot apical meristem restores its symplasmic organization during chilling-induced release from dormancy. Plant J 26: 249–264 [DOI] [PubMed] [Google Scholar]
- Roberts AG, Oparka KJ (2003) Plasmodesmata and the control of symplastic transport. Plant Cell Environ 26: 103–124 [Google Scholar]
- Ruan Y-L, Chourey PS (1998) A fiberless seed mutation in cotton is associated with lack of fiber cell initiation in ovule epidermis and alterations in sucrose synthase expression and carbon partitioning in developing seeds. Plant Physiol 118: 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y-L, Chourey PS, Delmer PD, Perez-Grau L (1997) The differential expression of sucrose synthase in relation to diverse patterns of carbon partitioning in developing cotton seed. Plant Physiol 115: 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y-L, Llewellyn DJ, Furbank RT (2000) Pathway and control of sucrose import into initiating cotton fibers. Aust J Plant Physiol 27: 795–800 [Google Scholar]
- Ruan Y-L, Llewellyn DJ, Furbank RT (2001) The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. Plant Cell 13: 47–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y-L, Llewellyn DJ, Furbank RT (2003) Suppression of sucrose synthase expression represses cotton fiber cell initiation, elongation and seed development. Plant Cell 15: 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert AM, Benedict CR, Berlin JD, Kohel RJ (1973) Cotton fiber development kinetics of cell elongation and secondary wall thickening. Crop Sci 13: 704–709 [Google Scholar]
- Schulz A (1999) Physiological control of plasmodesmatal gating. In AJE Van Bel, WJP van Kesteren, eds, Plasmodesmata—Structure, Function, Role in Cell Communication. Springer-Verlag, Berlin, pp 173–204
- Shimizu Y, Satoshi S, Hasegawa O, Kawada T, Sakuno T, Sakai F, Hayashi T (1997) Changes in levels of mRNAs for cell wall-related enzymes in growing cotton fiber cells. Plant Cell Physiol 38: 375–378 [DOI] [PubMed] [Google Scholar]
- Smart LB, Vojdani F, Maeshima M, Wilkins TA (1998) Genes involved in osmoregulation during turgor-driven cell expansion of developing cotton fibers are differentially regulated. Plant Physiol 116: 1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon R (1996) Phloem loading and plasmodesmata. Trends Plant Sci 1: 418–423 [Google Scholar]
- van Bel AJE, van Rijen HVM (1994) Microelectrode-recorded development of the symplastic autonomy of the sieve element/companion cell complex in the stem phloem of Lupinus luteus L. Planta 192: 165–175 [Google Scholar]
- van Iersel MW, Oosterhuis DM (1996) Drought effects on the water relations of cotton fruits, bracts, and leaves during ontogeny. Environ Exp Bot 36: 51–59 [Google Scholar]
- Zambryski PC (2004) Cell-to-cell transport of proteins and fluorescent tracers via plasmodesmata during plant development. J Cell Biol 162: 165–168 [DOI] [PMC free article] [PubMed] [Google Scholar]