Abstract
ClpC is a molecular chaperone of the Hsp100 family. In higher plants there are two chloroplast-localized paralogs (ClpC1 and ClpC2) that are approximately 93% similar in primary sequence. In this study, we have characterized two independent Arabidopsis (Arabidopsis thaliana) clpC1 T-DNA insertion mutants lacking on average 65% of total ClpC content. Both mutants display a retarded-growth phenotype, leaves with a homogenous chlorotic appearance throughout all developmental stages, and more perpendicular secondary influorescences. Photosynthetic performance was also impaired in both knockout lines, with relatively fewer photosystem I and photosystem II complexes, but no changes in ATPase and Rubisco content. However, despite the specific drop in photosystem I and photosystem II content, no changes in leaf cell anatomy or chloroplast ultrastructure were observed in the mutants compared to the wild type. Previously proposed functions for envelope-associated ClpC in chloroplast protein import and degradation of mistargeted precursors were examined and shown not to be significantly impaired in the clpC1 mutants. In the stroma, where the majority of ClpC protein is localized, marked increases of all ClpP paralogs were observed in the clpC1 mutants but less variation for the ClpR paralogs and a corresponding decrease in the other chloroplast-localized Hsp100 protein, ClpD. Increased amounts of other stromal molecular chaperones (Cpn60, Hsp70, and Hsp90) and several RNA-binding proteins were also observed. Our data suggest that overall ClpC as a stromal molecular chaperone plays a vital role in chloroplast function and leaf development and is likely involved in photosystem biogenesis.
In all plant cells the protein environment is a constantly changing one, and there exists several distinct mechanisms by which these intricate protein interactions are controlled. Two well-recognized mechanisms are that of molecular chaperones and proteases. These ubiquitous regulatory proteins act directly upon the structure and function of many different polypeptides. Chaperones assist in protein folding/unfolding, protein subunit assembly, and the import of many nuclear-encoded polypeptides into organelles. Chaperone activity is vital throughout the plant lifecycle, as well as under different growth conditions, especially adverse ones such as high temperatures. Many work in concert to facilitate correct protein folding, assembly, and repair, while others interact with proteolytic components to degrade terminally damaged proteins that might otherwise accumulate to potentially harmful levels (Parsell and Lindquist, 1993). Indeed, degradation of cellular proteins is a constant and ongoing process critical for continued plant growth and development. Apart from removing abnormal or otherwise damaged polypeptides, proteolysis also regulates the stability of key enzymes and regulatory proteins and facilitates the recycling of valuable amino acids (Spremulli, 2000).
Chloroplasts are particularly dynamic organelles, not only importing approximately 3,000 different nuclear-encoded proteins from the cytosol, but also producing approximately 120 proteins from its own plastome (Leister, 2003). Not surprisingly, many different chaperones and proteases function inside chloroplasts, spread throughout the various compartments. Chaperones of the Hsp70, Hsp100, and Cpn60 (Hsp60) families, for example, exist in the stroma and bind to envelope membranes, whereas peptidyl-prolyl isomerase is in the thylakoid lumen (Fulgosi et al., 1998). Similarly, proteases like the ATP-dependent Clp and FtsH exist in the stroma and stroma-exposed thylakoid membranes, respectively, whereas the ATP-independent DegP proteases occur within the thylakoid lumen and on both sides of thylakoid membranes. These types of proteases are homologous to bacterial counterparts best characterized in Escherichia coli but have many paralogs in higher plants (Adam and Clarke, 2002).
Clp proteases are a well-defined group of ATP-dependent, Ser-type proteases present in eubacteria, plants, and mammals. They are characteristically a two-component enzyme, a ClpP endopeptidase that requires the ATP-dependent unfolding activity of a Hsp100 molecular chaperone (Gottesman, 1996). Hsp100 chaperones belong to the family of AAA+ proteins (ATPases associated with various cellular activities) that often drive molecular processes such as protein unfolding, disassembly of protein complexes, and different protein-translocating activities (Dougan et al., 2002). The model Clp protease from E. coli has a central proteolytic core comprised of two opposing heptameric rings of ClpP, which are flanked at one or both ends by a single hexameric ring of either ClpA or ClpX, both of which are Hsp100 proteins (Grimaud et al., 1998). The regulatory chaperones confer substrate specificity, translocating the unfolded proteins into the catalytic chamber of ClpP (Gottesman et al., 1997; Ishikawa et al., 2001). Within this chamber, ClpP rapidly degrades the polypeptide into smaller fragments that later diffuse out.
Despite plants having by far the greatest known number and diversity of Clp proteins, with at least 23 individual proteins identified in the model species Arabidopsis (Arabidopsis thaliana), relatively little is known about their precise functions. Of those proteins identified, 10 are Hsp100/Clp chaperones of different types (four ClpB, two ClpC, one ClpD, and three ClpX), six proteolytic subunits (ClpP), and four ClpP-like proteins coined ClpR (Adam et al., 2001). Of the Hsp100 proteins, three ClpB paralogs were located in the cytosol, whereas the fourth (ClpB3) is chloroplastic (Peltier et al., 2004), as are the ClpC and ClpD proteins (Moore and Keegstra, 1993; Weaver et al., 1999). Most, if not all, ClpX proteins are in mitochondria along with ClpP2 (Halperin et al., 2001a; Peltier et al., 2004), while the four nuclear-encoded ClpP proteins (ClpP3–6) and plastomic ClpP1 are chloroplast proteins, as are the four ClpR paralogs (Nakabayashi et al., 1999; Peltier et al., 2001).
All chloroplast Clp proteins are primarily localized in the stroma (Moore and Keegstra, 1993; Shanklin et al., 1995; Halperin and Adam, 1996; Sokolenko et al., 1998; Weaver et al., 1999; Zheng et al., 2002; Peltier et al., 2004). Of the Hsp100 proteins, ClpB3 is almost certainly an HSP (Keeler et al., 2000) and does not associate with ClpP in an active protease, instead probably functioning solely as a chaperone in the solubilization of heat-induced protein aggregates (Weibezahn et al., 2004). ClpC and ClpD, however, are likely partners of ClpP, as both contain the conserved motifs (ILGF with an upstream K/R) important for complexing to ClpP (Kim et al., 2001). ClpC is constitutively more abundant than ClpD, which is inducible by stresses such as prolonged low temperatures (Zheng et al., 2002). Binding between chloroplast ClpC and ClpP has been demonstrated (Desimone et al., 1997; Sokolenko et al., 1998; Halperin et al., 2001b), inferring the existence of active ClpCP proteases in the stroma. The ClpP and ClpR paralogs apparently form a single, heterogeneous 14-subunit proteolytic core in chloroplasts, which also includes two new types of Clp protein of unknown function that are unique to higher plants (Peltier et al., 2004).
Many of the different Clp proteins perform important functions in chloroplasts. Evidence shows that plastomic ClpP1 plays a role in the degradation of the cytochrome b6f and PSII complexes in Chlamydomonas reinhardtii (Majeran et al., 2000, 2001), and disruption of its expression in tobacco (Nicotiana tabacum) retards chloroplast development and causes ablation of the shoot system (Shikanai et al., 2001; Kuroda and Maliga, 2003). Early attempts to repress clpC expression in tobacco using the antisense technique failed to produce viable cell lines with significant decreases in ClpC content (Shanklin et al., 1995), suggesting ClpC was an essential chloroplast protein. Similarly, the closely related ClpC in cyanobacteria is also necessary for cell viability and phototrophic growth (Clarke and Eriksson, 1996). The majority of ClpC in the stroma is believed to function as a housekeeping enzyme, both in its capacity as an independent molecular chaperone and as the regulatory component of the Clp protease (for review, see Adam and Clarke, 2002). ClpC has also been implicated in the stromal degradation of aberrant imported preproteins normally targeted to the thylakoid membranes (Halperin and Adam, 1996). The small proportion of ClpC associated to the envelope membrane is also thought to act as a molecular chaperone in the import process of cytosolic preproteins via the Tic-Toc pathway (Akita et al., 1997; Nielsen et al., 1997; Kouranov et al., 1998). To better understand the importance of ClpC in chloroplasts, we have examined the effect of decreased amounts of this essential protein in Arabidopsis. In this study, we demonstrate that the inactivation of the clpC1 gene by T-DNA insertion causes a significant drop in total ClpC content in chloroplasts and produces a pleiotropic phenotype that includes retardation in plant growth and development, chlorosis of leaves, impairment to the photosynthetic process, and a specific loss in PSI and PSII content.
RESULTS
Screening of Arabidopsis clpC1 Mutant Lines
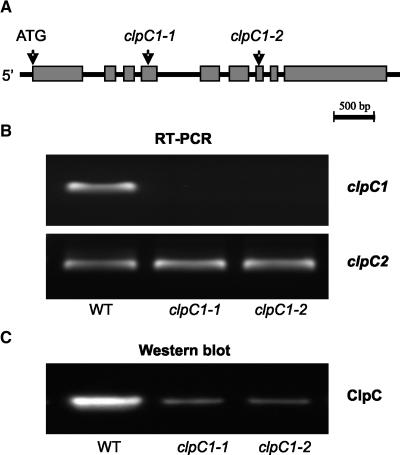
To investigate the role of ClpC in chloroplasts, searches were made of all available T-DNA insertion libraries for Arabidopsis clpC1 and clpC2 mutant lines. Although no putative clpC2 knockout was found, two independent mutant lines with a proposed T-DNA insertion in the clpC1 gene were obtained from the Salk Institute of Genomic Analysis Laboratory (SALK) and Syngenta Arabidopsis Insertion Library (SAIL). For the clpC1-SALK line, the T-DNA is inserted in the fifth exon 2,044 bp from the clpC1 start codon, whereas for the clpC1-SAIL line the insertion is in the seventh exon 2,603 bp from the start codon (Fig. 1A). Homozygote knockout lines with single T-DNA insertions were identified by seed segregation (data not shown). Inactivation of the clpC1 gene was confirmed by reverse transcription (RT)-PCR (Fig. 1B). Using gene-specific primers, no clpC1 mRNA was detected in either mutant line, in contrast to the wild type, whereas the transcript level for the clpC2 paralog remained unaffected. Using a ClpC antibody that does not distinguish between the two paralogs (Zheng et al., 2002), total ClpC content in leaves dropped approximately 65% in the mutant lines compared to the wild type (Fig. 1C). Following confirmation, the mutants were renamed to clpC1-1 for the SALK line and clpC1-2 for the SAIL line.
Figure 1.
Confirmation of the T-DNA insertion lines for clpC1. A, Schematic picture of the genomic clpC1 gene in Arabidopsis. Gray boxes and black lines represent exons and introns, respectively. Arrows indicate the ATG start codon and the proposed T-DNA insertion sites for the two independent mutant lines, clpC1-1 and clpC1-2. B, RT-PCR analysis of clpC1 and clpC2 gene expression of wild type (WT), and clpC1-1 and clpC1-2 mutant lines. Reactions were performed with equal total RNA, with the resulting RT-PCR products visualized by staining with GelStar. C, Immunoblot analysis of total ClpC protein in wild type, and clpC1-1 and clpC1-2 mutant lines. Total proteins were extracted from leaves of each plant and separated by denaturing PAGE on the basis of equal fresh weight. Total ClpC protein was detected by immunoblotting with an antibody that crossreacts with both ClpC1 and ClpC2.
Pleiotropic Phenotype for clpC1 Mutants
Both mutant lines showed a distinct slow-growing, chlorotic phenotype (Fig. 2A). Under standard growth conditions, mutant plants grew vegetatively only to a diameter of 11 to 14 cm prior to influorescence at 13 to 14 weeks, compared to 18 to 21 cm for the wild type after 8 to 9 weeks. Throughout vegetative growth, all mutant leaves including the cotyledons exhibited a homogeneous, chlorotic appearance. Measurements confirmed mutant leaves had approximately two-thirds lower chlorophyll content than the wild type (Fig. 3A) but no significant change in relative protein content (data not shown). During reproductive development, the mutant lines formed secondary influorescences more perpendicular to the primary influorescence, which was also slightly shorter than that in wild type, thereby giving the mutants a more bushy appearance (Fig. 2B). The various influorescences, cauline leaves, and siliques also showed the same chlorotic phenotype as the vegetative leaves.
Figure 2.
The clpC1 mutants exhibit visible phenotypes. A, Photograph of 55-d-old wild-type (WT), clpC1-1, and clpC1-2 plants grown side by side under the standard conditions of 23°C/18°C day/night temperatures, an 8-h photoperiod with 150 μmol white light m−2 s−1, and approximately 65% humidity. B, Photographs of fully grown flowering plants showing the shorter primary and more perpendicular secondary influorescences (magnified in the insert for clpC1-2) in the clpC1 mutant lines compared to the wild type. Scale bars = 2 cm.
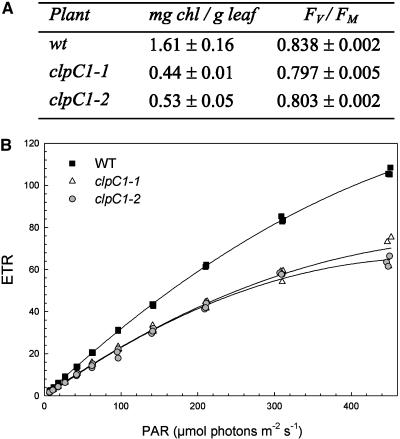
Figure 3.
Loss of ClpC1 reduces chlorophyll content and impairs photosynthesis. A, Table showing the reduced chlorophyll content and lower photochemical efficiency of PSII (Fv/Fm) in the clpC1 mutants. Values shown are the mean value from measurements done on replicate plants of each line ± se (n = 3). B, Light response curves of photosynthesis for wild type (WT) and clpC1 mutant lines. Photosynthetic ETRs as determined by chlorophyll fluorescence were measured at irradiances (photosynthetically active radiation [PAR]) from 0 to 460 μmol photons m−2 s−1. Measurements were done on fully grown leaves from three separate plants of each line grown under identical conditions and of identical age.
Because of the chlorotic, retarded growth phenotype of the mutant lines, the physiological state of the leaves was further examined by measuring different photosynthetic parameters. As determined by chlorophyll fluorescence, the photochemical efficiency of PSII (Fv/Fm) in 6-week-old plants was slightly, but significantly, lower in the mutants than in the wild type (Fig. 3A). Estimates of electron transport rates (ETRs) showed more dramatic differences, with mutant leaves having markedly reduced photosynthetic efficiency (i.e. quantum yield as determined from the initial slope of the light-response curve) and an almost halved photosynthetic capacity as measured at the highest light intensity (Fig. 3B).
clpC1 Mutants Have Lower Photosystem Content But No Change in Leaf Anatomy and Chloroplast Ultrastructure
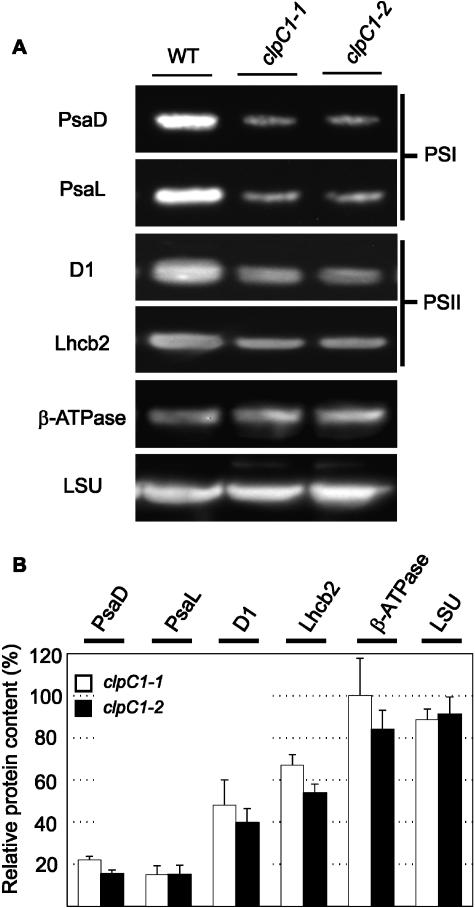
Because of the distinct morphological and physiological changes to the clpC1 mutants, an extensive examination of the leaf cell anatomy and chloroplast ultrastructure was performed. In terms of leaf anatomy, light microscopy was used to analyze chloroplast number per cell, the percent of intercellular areas within the mesophyll cross section, and the percent of chlorophyll area per cell area. For chloroplast ultrastructure, electron microscopy was used to analyze the average size of chloroplasts, thickness of granal stacks, and amount of lamellae measured from the middle of the chloroplast. All microscopy measurements were done on both palisade and spongy mesophyll cells. Surprisingly, no significant differences were observed for any of these parameters in the mutant leaves (data not shown). Because of this, the relative amount of different photosynthetic protein complexes in the mutant lines was examined by immunoblotting. Antibodies specific for the following marker proteins were used to quantify the relative amounts of each photosynthetic complex: PsaD and PsaL for PSI, D1 and Lhcb2 for PSII, CF1 β-subunit for ATPase, and large subunit for Rubisco. As based on leaf fresh weight, the level of both chlorophyll-containing PSI and PSII complexes was reduced by approximately 85% and 50%, respectively, in the mutant lines (Fig. 4). In contrast, amounts of the ATPase and Rubisco complexes showed no significant change in the mutants compared to the wild type (Fig. 4).
Figure 4.
Altered levels of PSI and PSII proteins in the clpC1 mutants. Relative amounts of different photosynthetic protein complexes were compared between the wild type (WT) and clpC1 mutant lines, clpC1-1 and clpC1-2. Total cell proteins were extracted from 55-d-old leaves from each plant and separated by denaturing PAGE on the basis of equal fresh weight (1 μg). Marker proteins for each complex were detected by immunoblotting using specific polyclonal antibodies (A) and quantified (B). Selected marker proteins were PsaD and PsaL for PSI, D1, and Lhcb2 for PSII, CF1 β-subunit for ATPase, and large subunit for Rubisco. Amounts of each marker protein in the clpC1-1 (white bar) and clpC1-2 (black bar) lines are shown as a percentage of the relative wild-type level (± se, n = 4), which was set at 100%.
Changes in the Regulation of Chloroplast Clp Proteins in the clpC1 Mutant
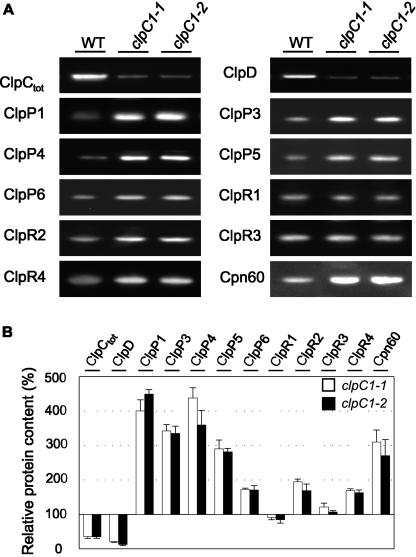
To further investigate how the other chloroplast Clp proteins were affected by the loss of ClpC1 and decrease in total ClpC content, we analyzed their relative levels in whole leaf extracts by immunoblotting using specific antibodies. As shown in Figure 5, the loss in ClpC content in the mutants produced a similar decrease (approximately 80%) in the level of ClpD, the other member of the Hsp100 chaperone family in chloroplasts. In contrast, amounts of the chloroplast paralogs for the proteolytic ClpP proteins (ClpP1, ClpP3–6) all increased between 2- and 4-fold in the mutant lines, whereas the four ClpR paralogs showed less variation, with significant rises observed only for ClpR2 and ClpR4 (Fig. 5). In addition to Clp proteins, we also investigated the amount of Cpn60, another molecular chaperone believed to interact with ClpC following import of preproteins (Jackson-Constan et al., 2001) and found a 3-fold rise in both mutant lines.
Figure 5.
Altered levels of various Clp proteins in the clpC1 mutants. Relative amounts of different chloroplast Clp proteins and the chaperone Cpn60 were compared between the wild type (WT) and clpC1 mutant lines, clpC1-1 and clpC1-2. Total cell proteins were extracted from 55-d-old leaves from each plant and separated by denaturing PAGE on the basis of equal fresh weight (1 μg). Various chloroplast Clp proteins and Cpn60 were then detected by immunoblotting, using specific polyclonal antibodies (A) and quantified (B). Proteins detected were ClpC (total), ClpD, ClpP1, ClpP3–6, ClpR1–4, and Cpn60. Amounts of each protein in the clpC1-1 (white bar) and clpC1-2 (black bar) lines are shown as a percentage of the relative wild-type level (± se, n = 4), which was set at 100%.
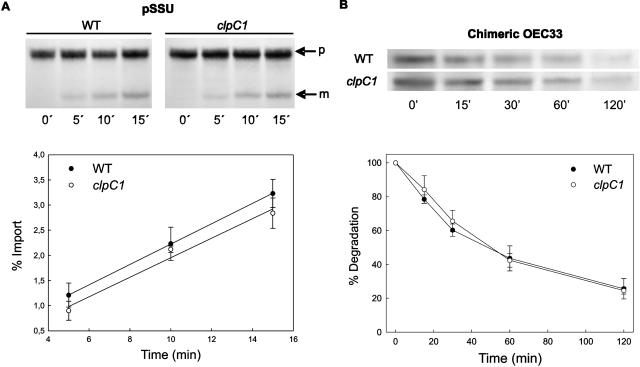
Reduced ClpC Content Does Not Impair Chloroplast Import of Preproteins in Vitro
The small proportion of ClpC associated to envelope membranes has long been thought to be involved in chloroplast protein import based on its interaction with the translocation protein Tic110 and preproteins under certain conditions (Akita et al., 1997; Nielsen et al., 1997; Kouranov et al., 1998). Because of this, we examined the import efficiency (both rate and quantity) of different preproteins into chloroplasts isolated from the clpC1 mutants and compared this to that of wild-type chloroplasts. Using in vitro translation, radiolabeled preproteins of both the small subunit of Rubisco (SSU) and the oxygen-evolving complex 33 (OEC33) of PSII were prepared and successfully imported into an equal number of intact wild-type and mutant chloroplasts. SSU was specifically chosen for this import assay since ClpC has previously been shown to interact with the SSU precursor (Nielsen et al., 1997). The rates and amount of precursor import in this study were comparable to that previously reported for Arabidopsis chloroplasts (Aronsson and Jarvis, 2002). As shown in Figure 6A, however, no significant change was observed in the import and processing efficiency of the SSU preprotein in mutant chloroplasts, despite the loss of approximately 65% of ClpC protein. A similar result was obtained for the OEC33 preprotein (data not shown).
Figure 6.
Loss of ClpC1 has no effect on chloroplast import of preproteins or the degradation of mistargeted preproteins. A, 35S-labeled precursor of Rubisco small subunit (pSSU) was imported into chloroplasts isolated from 21-d-old wild type (WT) and clpC1-1 mutant. Samples were taken at selected times during the import assay and separated by denaturing PAGE. The top picture shows a representative import assay, indicating the pSSU (p) and processed mature SSU (m). Rate of import, as measured by the increase in mature SSU, was plotted as a percent of the total radiolabeled pSSU at the start of the assay. Values are averages ± se (n = 3) fitted to a second-order linear regression. B, 35S-labeled precursor of a chimeric OEC33 protein lacking thylakoid membrane targeting sequences was imported into isolated chloroplasts from 21-d-old wild type and clpC1-1 mutant. After preprotein import for 20 min, chloroplasts were treated with thermolysin to halt import and remove any unimported preprotein. Chloroplasts were then incubated at 25°C, with aliquots taken at different time points. Proteins were separated by denaturing PAGE. Shown is a representative assay over 120 min, with the degradation rate of the chimeric OEC33 preprotein quantified relative to the amount of chimeric OEC33 at the 0-min time point, which was set as 100%. Values shown are averages ± se (n = 3).
Reduced ClpC Content Does Not Impair Degradation of Mistargeted Preproteins in Vitro
Another function proposed for chloroplastic ClpC is in the degradation of mistargeted preproteins as part of a Clp proteolytic complex (Halperin and Adam, 1996; Halperin et al., 2001b). To test this possibility, we examined the efficiency of chloroplasts isolated from both mutant lines and wild type to degrade a chimeric form of OEC33 that is unable to be transported into the thylakoid lumen upon chloroplast import. This aberrant form of OEC33 that accumulates in the stroma has previously been shown to be degraded in a Clp protease-like manner (Halperin and Adam 1996; Halperin et al., 2001b). As for the previous import assays, the chimeric preprotein was first radiolabeled by in vitro translation and then imported into isolated chloroplasts. In wild-type chloroplasts, the imported OEC33 preprotein was rapidly degraded over a 2-h period (Fig. 6B), similar to that previously shown (Halperin and Adam, 1996). However, the degradation rate of the OEC preprotein was identical in the mutant chloroplasts (Fig. 6B), indicating the decreased amount of ClpC had no significant effect on this proteolytic activity.
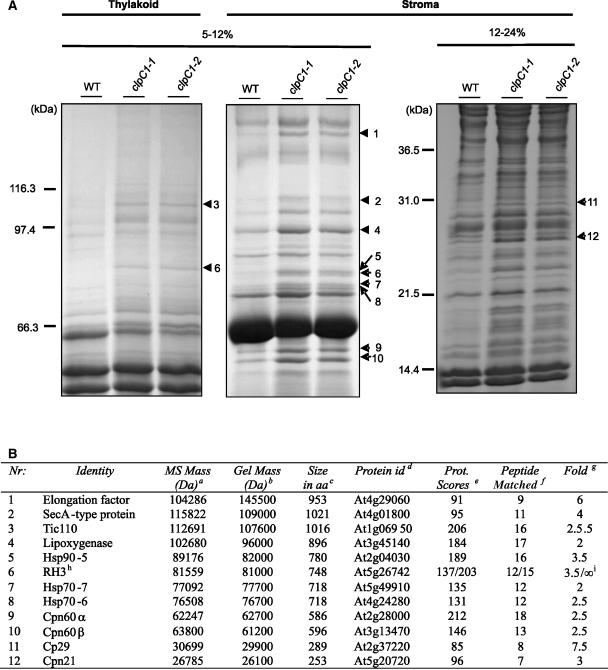
Protein Profiling of Chloroplasts from the clpC1 Mutants
Given the lack of clear effects on proposed specific ClpC functions, we profiled proteins isolated from mutant chloroplasts to detect those whose level may potentially change in response to the severe phenotypes in the mutant lines. For this study, chloroplasts from both wild-type and mutant lines were fractionated into stroma and thylakoid membranes. SDS-PAGE using long gels (20 cm) and various acrylamide gradients to separate selected protein mass ranges were chosen over two-dimensional PAGE because in our hands they better resolved a wider range of proteins. After separation of the stromal and thylakoid membrane fractions, those proteins that were most up-regulated in the mutant lines relative to the wild type were excised for identification by matrix-assisted laser-desorption ionization time of flight (MALDI-TOF) mass spectrometry (MS; Fig. 7). Of these, 12 had a significant protein identification hit (P < 0.05; Fig. 7B). Six corresponded to different types of molecular chaperone: Hsp90-5, Hsp70-6, Hsp70-7, Cpn60α, Cpn60β, and Cpn21. Four of the other proteins contained RNA-binding features and included SecA, RNA helicase 3 (RH3), cp29, and an unclassified protein containing three elongation factor Ts motifs. Of the remaining proteins identified, one was Tic110 (the subunit of the Tic protein import complex previously shown to associate with ClpC) and the other a lipoxygenase, a protein usually involved in metabolism and stress responses (Bell et al., 1995).
Figure 7.
Changes in chloroplast protein composition in clpC1 mutants. A, Stromal and thylakoid membrane proteins extracted from isolated chloroplasts of wild type (WT) and clpC1 mutants (clpC1-1 and clpC1-2) were separated by denaturing PAGE. After staining with coomassie blue, proteins showing the greatest quantitative change in the clpC1 mutants that were identifiable by MALDI-TOF MS are indicated with arrowheads. B, Table containing MALDI-TOF data from analyzed proteins. Footnote a, Molecular mass of protein determined by MS; footnote b, molecular mass calculated from PAGE size markers; footnote c, predicted size in amino acids (aa) from the MIPS Arabidopsis database (http://mips.gsf.de); footnote d, AGI gene code for identified protein; footnote e, significant protein scores >73 (P < 0.05); footnote f, peptide match at mass tolerance ± 100 ppm, allowing max one missed cleavage; footnote g, approximate up-regulation of protein based on stained gel bands; footnote h, RH3 found both in stroma and thylakoid (137/203), respectively; footnote i, no stained band could be located in wild type, therefore ∞ = interminable.
DISCUSSION
Higher plants commonly have two clpC genes encoding stromal-localized paralogs of the Hsp100 molecular chaperones termed ClpC1 and ClpC2 (Gottesman et al., 1990; Adam et al., 2001; A. Clarke, unpublished data). In this study we have characterized two Arabidopsis clpC1 mutants lacking approximately 65% of wild-type levels of ClpC protein. Interestingly, expression of the second clpC gene in the mutants was unaffected by the loss of clpC1 expression. Given that the ClpC1 and ClpC2 proteins have almost identical amino acid sequences and thus are likely to be functionally similar, an up-regulation of clpC2 gene expression should in theory compensate for the loss of clpC1 transcript. However, unaltered levels of clpC2 mRNA in the mutants suggest this gene is unable to be up-regulated in response to reduced clpC1 transcripts. It also suggests the 35% of ClpC remaining in the mutant lines is the proportion of total ClpC in wild-type chloroplasts normally derived from the clpC2 gene. Furthermore, the level of the other chloroplast Hsp100 protein, ClpD, decreased in proportion to the drop in ClpC content, inferring a possible association between these two closely related Hsp100 proteins. Although the constitutive level of ClpD is relatively low and its exact function remains a mystery, it is possible its down-regulation is linked to the phenotypic changes in the clpC1 mutants.
The clpC1 mutants have a pleiotropic phenotype that suggests ClpC plays an important role throughout the lifecycle of the plant. Vegetative growth is significantly retarded, with all leaves showing a homogeneous chlorotic appearance; a chlorosis confirmed by a severalfold reduction in leaf Chl content. This type of chlorosis is in contrast to the variegated leaf appearance of mutants of the chloroplast FtsH proteases, VAR1 and VAR2 (for review, see Sakamoto, 2003). Interestingly, a recent mutant Arabidopsis line with greatly reduced ClpC2 content was shown to have no obvious phenotypic changes (Park and Rodermel, 2004) unlike the clpC1 knockout lines. The clpC2 knockdown line, however, suppressed the variegated leaf phenotype of the VAR2 (FtsH2) mutant and the requirement for FtsH protease in thylakoid biogenesis (Park and Rodermel, 2004), a result that was not conferred by antisense repression of clpC1 expression (S. Park and S. Rodermel, unpublished data), suggesting the two ClpC paralogs may not be as functionally indistinct as their primary sequence suggests. The slow growth and leaf chlorosis of the clpC1 mutants were consistent with the observed lower photosynthetic rates. The fact that PSII photochemical efficiency was less inhibited than the overall ETR infers more of an indirect down-regulation of photosynthesis and less of a direct stress-induced inhibition. The decrease in PSI and PSII complexes is also consistent with the reduced photosynthetic capacity and Chl content. Inconsistent with an overall down-regulation of photosynthesis, however, was the unaltered levels of ATPase and Rubisco complexes in the mutant leaves, as well as the lack of overt changes to chloroplast ultrastructure and number per cell. This result suggests the mutant lines have a more specific defect in PSI and PSII biogenesis, one that does not greatly influence other photosynthetic protein complexes or the overall thylakoid membrane structure.
A selective decrease in PSI and PSII similar to that in the clpC1 mutants has been seen previously in the chlorotic double mutant cpSRP43−/cpSRP54− (Hutin et al., 2002) deficient in the signal recognition particle (SRP) preprotein translocation pathway. Since one of the functions proposed for ClpC is in chloroplast preprotein import and processing (Akita et al., 1997; Nielsen et al., 1997; Kouranov et al., 1998) and that both PSI and PSII contain nuclear-encoded subunits, it is possible that the clpC1 mutant phenotypes are linked to impaired import, processing, and/or translocation of one or more photosystem subunits. ClpC supposedly functions in chloroplast import/postimport by interacting with envelope translocation components and Tic110 (Akita et al., 1997; Nielsen et al., 1997; Kouranov et al., 1998). Tic110 is thought to act as a docking site for preproteins as they emerge from the Tic translocon, conferring a unidirectional transport into the stroma by preventing the reverse translocation of preproteins (Inaba et al., 2004). Binding of ClpC to Tic110 is presumed to maintain the solubility of the associated preproteins, preventing their misfolding or aggregation prior to being transferred to Hsp70 and/or Cpn60 chaperones (Nielsen et al., 1997) and to provide the driving force for complete translocation into the stroma (Jackson-Constan et al., 2001). If ClpC indeed performs such a vital role, then a significant decrease in ClpC content would be expected to impair the import process to some extent. However, no such impairment was observed for the import and processing of both SSU of Rubisco and OEC33 of PSII in the clpC1 mutants. It is plausible that the elevated levels of Tic110 and other chaperones involved in processing and folding of imported preproteins (Cpn60, Hsp70, and Hsp90) compensated for the lower ClpC content in the mutants, maintaining efficient preprotein import and processing into chloroplasts. Nevertheless, it appears ClpC does not play a vital role in the chloroplast preprotein import/processing pathway.
Given the lack of significant inhibition in chloroplast preprotein import, it is almost certain that the phenotypic changes in the mutants are due to a reduced activity of ClpC in the stroma, which normally contains most ClpC protein (Moore and Keegstra, 1993; Zheng et al., 2002). It has been proposed that ClpC performs more of a housekeeping role within chloroplasts as either an independent chaperone or within a Clp protease (Adam and Clarke, 2002). In terms of protein turnover, the increased level of ClpP paralogs in the clpC1 mutants is almost certainly due to fewer active Clp proteases in the stroma. By analogy with the E. coli Clp protease, ClpC likely forms a hexameric ring that binds to one or both ends of a ClpP proteolytic core. Recently, a stromal 320- to 350-kD complex containing the five ClpP paralogs plus the four ClpR proteins has been identified in Arabidopsis chloroplasts (Peltier et al., 2004). Although no interaction between ClpC and this ClpPR complex has yet been shown, binding of stromal ClpP to ClpC has been demonstrated (Desimone et al., 1997; Sokolenko et al., 1998; Halperin et al., 2001b). The induction of all five chloroplastic ClpP paralogs in the clpC1 mutants is likely a compensatory response to the lower ClpC content and the inability to form sufficient active stromal Clp proteases. The induction of only two ClpR paralogs (i.e. ClpR2 and ClpR4), and that to a lesser degree than their ClpP counterparts, was surprising given that all ClpR paralogs supposedly complex with the five ClpP paralogs in a single proteolytic core complex (Peltier et al., 2004). The significance of this disparity, however, is unclear while the exact function of the ClpR proteins remains unknown.
Although specifically assaying Clp proteolytic activity in the stroma has so far proven difficult, we used the stability of a chimeric OEC33 preprotein previously suggested as being degraded by a Clp protease (Halperin and Adam, 1996; Halperin et al., 2001b) to test for possible impairment in proteolytic activity in the mutant lines. However, since the aberrant OEC33 preprotein was as readily degraded in the mutant chloroplasts as in those from wild type infers that ClpC is not primarily involved in this particular proteolytic activity. Nevertheless, an impairment of housekeeping Clp proteolytic activity remains one likely contributor to the clpC1 mutant phenotypes. Interestingly, the Clp protease has been implicated in the degradation of PSII in Chlamydomonas (Majeran et al., 2001), inferring ClpC is involved in PSII turnover. Indeed, the specific decrease in PSII and PSI content in the clpC1 mutants suggests ClpC could well participate in the biogenesis and turnover of both photosystems in Arabidopsis chloroplasts.
As a housekeeping enzyme, stromal ClpC is also likely to function as a chaperone independently. Chloroplasts have their own protein synthesis machinery and thus accordingly a range of chaperones to maintain protein quality control in terms of solubility, folding, and assembly. Hsp100 proteins are intricately involved in protein quality control systems in several different organisms (Dougan et al., 2002). The most striking outcome of the protein profiling done on the clpC1 mutants was the increased levels of most other stromal chaperones: Hsp90-5 (Hsp90), Hsp70-6, Hsp70-7 (Hsp70), Cpn60α, Cpn60β, and Cpn21 (Hsp60). These chaperone families are highly conserved in all species and perform many functions, although most are associated with general protein stability, folding/unfolding, and assembly. All three also often act in association with each other and other cochaperones. Indeed, another Hsp100 protein, ClpB, is known to rescue aggregated proteins in cooperation with Hsp70 (Weibezahn et al., 2004). As a consequence, the higher level of stromal molecular chaperones in the clpC1 mutant lines is likely in response to reduced protein solubility, folding, and/or assembly processes in the stroma that normally involve ClpC.
Other up-regulated proteins in the clpC1 mutants also implicate ClpC as a housekeeping enzyme in the chloroplast transcriptional/translational machinery. SecA is central to the Sec-pathway for translocating certain stromal preprotein intermediates into the thylakoid membranes or lumen (Mori and Cline, 2001). OEC33 is one such protein, although no significant change in its translocation rate was observed in the clpC1 mutants. SecA, however, also contains RH activity (Park et al., 1997), which is similar to another induced protein in the mutant lines, RH3. Numerous so-called DEAD-box RHs exist in Arabidopsis (Gorbalenya et al., 1989). Although the precise role of most RHs is unknown, they are required for many different cellular processes, including transcription, pre-mRNA processing, ribosome biogenesis, translation initiation, and RNA turnover (Rocak and Linder, 2004). Another putative RNA-binding protein up-regulated in the clpC1 mutants is Cp29, a protein apparently necessary for chloroplast function in Arabidopsis (Ohta et al., 1995). Another identified protein was one with unknown function but which contained three elongation factor Ts motifs, suggesting it is involved in the chloroplast protein biosynthetic machinery. Again, induction of these stromal proteins may be compensatory for the loss in ClpC activity normally involved in some aspect of chloroplast gene expression, RNA processing, or protein translation. If so, likely candidates affected by lower ClpC content would be plastid genes coding for PSI and PSII subunits given the specific decrease of these photosynthetic complexes in both clpC1 mutant lines.
In conclusion, our results clearly demonstrate that ClpC plays an important role in chloroplasts of Arabidopsis. No evidence was found for ClpC being an essential factor in either chloroplast import of nuclear-encoded preproteins or the degradation of aberrant polypeptides in the stroma. Up-regulation of proteins involved in both processes (e.g. Tic110 and ClpP) certainly supports the involvement of ClpC, but it is almost certain ClpC also performs additional roles of equal or greater importance. To elucidate the details of such functions, the clpC1 mutants will undoubtedly remain an invaluable tool for future studies.
MATERIALS AND METHODS
Plant Growth Conditions
Wild-type seeds from Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 and clpC1 mutant seeds clpC1-1 (accession no. SALK_014058), clpC1-2 (Garlic_873_G11.b.b.1a.Lb3Fa) were sown on soil containing 20% perlite. All seeds were vernilized at 4°C for 48 h to break dormancy. All plants were grown under a standardized condition: 8-h photoperiod with white light (150 μmol m−2 s−1), 23°C/18°C day-night temperatures, and 65% relative air humidity.
Identification of Putative clpC1 T-DNA Insertion Mutant Lines
Putative mutants were screened by electronic BLAST searches of available populations of sequence-indexed Arabidopsis T-DNA-insertion mutants using the Arabidopsis genomic clpC1 (At5g50920) and clpC2 (At3g48870) gene sequences. A single putative clpC1 knockout was found in both the SALK (Alonso et al., 2003) and SAIL (Sessions et al., 2002) populations. Seeds for the clpC1-1 (SALK) and clpC1-2 (SAIL) lines were obtained and then screened using either kanamycin or BASTA as selective marker. For clpC1-1, seeds were sterilized by shaking for 4 min in 70% ethanol with 1% Triton X-100, rinsing them thrice in 100% ethanol, and then air drying them on a piece of filter paper before sowing on Murashige and Skoog media plates containing 20 μg mL−1 kanamycin. For clpC1-2, seeds were grown on soil for 10 d in thin lawns and then sprayed with BASTA as previously described (Altman et al., 1992). Seed batches that contained heterozygotes with single T-DNA insertion, showing a one-quarter death ratio on selection plates/pots, were identified and used for screening homozygote clpC1 mutant lines in the next generation.
RT-PCR
For analysis of the clpC1 mutants, total RNA was isolated from leaves of 6-week-old wild-type and mutant plants grown under standard conditions. RNA was extracted using the Concert Plant RNA Reagent (Invitrogen, Carlsbad, CA) and treated with RNase-free DNase I (Amersham Pharmacia, Uppsala). RT-PCR was performed using SuperScript One-Step RT-PCR with Platinum Taq (Invitrogen), with the following concentrations of total RNA for each gene analyzed: clpC1, 2 μg mL−1; clpC2, 20 μg mL−1; and tic110, 24 μg mL−1. Primers used in the amplifications were as described previously (Zheng et al., 2002) for the clpC genes, while for tic110 primers used were forward 5′-CCTGCTCTACTGTGTAACTGGAG and reverse 5′-CTCTGAGCGAGATCATGGACAAC. RT-PCR products size separated by agarose electrophoresis were visualized by GelStar staining (Biowhittaker Molecular Applications, Rockland, ME) and quantified using the ChemiGenius2 Imaging system and associated software (Syngene, Frederick, MD).
Chlorophyll Quantification and Photosynthetic Measurements
Chlorophyll was extracted from 11-mm diameter leaf discs of 55-d-old wild-type Arabidopsis and clpC1 mutants according to Porra et al. (1998) in 80% buffered acetone. Absorbance at 663.6 and 646.6 nm was measured by a UV-2401PC spectrophotometer (Shimadzu, Columbia, MD), and the Chl content was calculated as described previously (Porra et al., 1989). The photochemical efficiency of PSII (Fv/Fm) and the photosynthetic ETR of the same 55-d-old leaves were measured by chlorophyll fluorescence using a pulse-amplitude modulated fluorimeter (PAM 2000; Heinz-Walz, Effeltrich, Germany) according to the manufacturer's recommendation. ETR values obtained from the PAM measurements were adjusted according to Evans (1996) to take into account the variable chlorophyll contents between the mutant lines and wild type.
Light and Electron Microscopy
Leaves from 55-d-old wild-type, clpC1-1, and clpC1-2 plants were fixed in 1.5% glutaraldehyde, 1.5% paraformaldehyde, 0.15 m Suc, 2 mm CaCl2 in 0.05 m cacodylate buffer, pH 7.0, at 4°C, with additional fixation done according to Soikkeli (1980). Three leaves per sample tube were prepared for light microscopic analyses as described in Sutinen (1987). The whole-leaf cross section (magnification in the microscope 40×), and palisade and spongy cells (magnification in the microscope 100×) were digitally photographed. Digital images were analyzed with Adobe Photoshop (version 6.0; Adobe Systems, Mountain View, CA). Thin sections for transmission electron microscopy were prepared from the same leaf samples as used for the light microscopy. Palisade and spongy cells of one leaf per treatment were photographed with transmission electron microscope (JEOL JEM EX; Tokyo) at 5,000× magnification. The negatives were scanned and digital images were analyzed using Adobe Photoshop 6.0.
Production of ClpR-Specific Antibodies
Polyclonal antibodies specific to Arabidopsis ClpR1, -R2, -R3, and -R4 were generated using synthetic peptides corresponding to unique amino acid sequences at the C-terminal end of the proteins: ClpR1, ADSQDSSFEKRDYDGTLAQR; ClpR2, VRPPRIKEDAPRQDESAGLG; ClpR3, KIIADVVPSEEFDKNAGIKS; and ClpR4, NERGSQDRGVVSDLKKAQLI. The peptides were conjugated to bovine serum albumin and then injected into rabbits intramuscularly and subcutaneously (AgriSera, Hällnäs, Sweden).
Leaf Protein Extracts and Immunoblotting
Total cell proteins were extracted from approximately 200 mg of leaves ground in liquid N2 using a precooled mortar and pestle. The frozen powder was transferred to a precooled microfuge tube and weighed. NuPAGE lithium dodecyl sulfate sample buffer (Invitrogen) was then added to a final concentration of 0.3 mg mL−1. Samples were mixed and heated to 75°C for 5 min, cooled to 4°C, and centrifuged at 20,000g for 10 min at 4°C. Supernatants containing solubilized proteins were kept at 4°C prior to SDS-PAGE and immunoblot analysis.
Because of the chlorotic appearance of clpC1 mutant leaves, protein samples were compared on the basis of equal leaf fresh weight (i.e. 1 μg), which was also equivalent to equal protein content. Proteins were separated using different types of gels depending on the protein size range being analyzed: precast 3% to 8% Tris-Acetate gels (Invitrogen) for ClpC and ClpD detection, or 12% linear Bis-Tris gels (Invitrogen) using MES buffer for ClpP1, ClpP3-6, ClpR1-4, PsaD, PsaL, D1, Lhcb2, and MOPS buffer for Cpn60, large subunit of Rubisco, and the CF1 β-subunit of ATPase. Separated proteins were electrophoretically transferred to Immun-Blot polyvinylidene difluoride Membrane (Bio-Rad Laboratories, Hercules, CA) using an XCell blotting apparatus (Invitrogen). Arabidopsis Clp proteins were detected using specific polyclonal antibodies as previously described (Zheng et al., 2002) or as detailed above. Antibodies specific for other chloroplast proteins were obtained as gifts: Lhcb2, PsaL, and β-ATPase (S. Jansson, Umeå University, Sweden), D1 and Rubisco large subunit (G. Öquist, Umeå University, Sweden) and PsaD (H. Scheller, University of Copenhagen), except for Cpn60 that was bought commercially (StressGen Biotechnologies, Victoria, Canada). Primary antibodies were detected using horseradish peroxidase-linked, antirabbit secondary antibody from donkey (Amersham Pharmacia) and visualized by enhanced chemiluminescence using the SuperSignal West Dura Extended Duration Substrate (Pierce Chemical, Rockford, IL). Chemiluminescent signals were detected and quantified using the ChemiGenius2 Imaging system (Syngene) and associated software.
In Vitro Import Assays
Template plasmid DNA was amplified using Z-competent Escherichia coli cells DH5α (Zymo Research, Orange, CA) and SNAP MidiPrep kit (Invitrogen). Radiolabeled protein was translated using the TNT coupled wheat germ extract system (Promega, Madison, WI) using T7 polymerase for SSU (Wong et al., 1992) and SP6 polymerase for PSII OEC33 (Halperin and Adam, 1996). Import reactions were performed on chloroplasts isolated from 21-d-old plants in HMS buffer according to Aronsson and Javis (2002). An equal number of intact chloroplasts, as determined by phase-contrast microscopy, were used for each import reaction to enable both import rates and quantities to be calculated (Aronsson and Jarvis, 2002). Samples were resolved on 12% BisTris precast NuPAGE gels (Invitrogen), blotted to nitrocellulose membrane (Transblot; Bio-Rad Laboratories), and exposed to x-ray film (Hyperfilm; Amersham Pharmacia). X-ray films were developed using Kodak GBX developer and fixer (Eastman Kodak, Rochester, NY), then imaged and quantified using the ChemiGenius2 imaging system and associated software (Syngene).
Protein Degradation Assay
Radiolabeled chimeric OEC33 was translated using the TNT coupled wheat germ extract system (Promega) and imported into isolated chloroplasts for 20 min as described by Aronsson and Jarvis (2002). Following import, chloroplasts were treated with thermolysin (100 μg mL−1 final concentration) on ice for 20 min. The mixture was carefully pelleted by centrifugation at 1,000g for 2 min and resuspended in HMS buffer (Aronsson and Jarvis, 2002) containing 20 mm gluconic acid, 10 mm NaHCO3, 0.2% (w/v) bovine serum albumin, 10 mm Met, and 5 mm ATP. The suspension was incubated under 100 μmol m−2 s−1 white light at 25°C. Aliquots were taken at 0, 15, 30, 60, and 120 min, then pelleted and frozen in liquid nitrogen to stop all protein degradation. Pelleted samples were dissolved in NuPAGE sample buffer (Invitrogen), heated for 5 min at 75°C, and then separated by SDS-PAGE. Radiolabeled proteins were detected and analyzed as for the import assays described above.
Profiling of Stromal and Thylakoid Membrane Proteins
Stromal and thylakoid membrane proteins were isolated from fully expanded leaves of wild type and clpC1 mutants just prior to influorescence as previously described by Zheng et al. (2002). Protein concentration of the stromal fraction was determined using the bicinchoninic acid protein assay kit according to manufacturers protocol (Pierce Chemical), while the Chl concentration of the thylakoid membrane fraction was done as described above. Samples (75 μg protein for stroma, 15 μg Chl for thylakoid membrane) were separated on 20-cm long, gradient SDS-PAGE gels, with an acrylamide gradient of either 5% to 12% or 12% to 28%, depending on the optimal size range. After electrophoresis, proteins were stained with Coomassie Blue (Simply Blue Safestain; Invitrogen) according to manufacturer's recommendation. Proteins up-regulated in the clpC1 mutant lines were excised from each gel and sent for identification by MALDI-TOF MS (Swegene Proteomics Center, Göteborg University, Sweden).
Acknowledgments
We thank Henrik Aronsson for the pSSU construct; Zach Adam for the chimeric pOEC33 and OEC33 constructs; and Stefan Jansson, Henrik Scheller, and Gunnar Öquist for providing different antibodies. We also thank Doug Campbell for help with calculating the photosynthetic ETR values.
This work was supported by the Swedish Research Council (VR), the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (Formas), the Swedish Foundation for International Cooperation in Research and Education (STINT), and an overseas postgraduate scholarship from the Natural Sciences and Engineering Research Council of Canada (to T.M.M.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.053835.
References
- Adam Z, Adamska I, Nakabayashi K, Ostersetzer O, Haussuhl K, Manuell A, Zheng B, Vallon O, Rodermel SR, Shinozaki K, et al (2001) Chloroplast and mitochondrial proteases in Arabidopsis: a proposed nomenclature. Plant Physiol 125: 1912–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam Z, Clarke AK (2002) Cutting edge of chloroplast proteolysis. Trends Plant Sci 7: 451–456 [DOI] [PubMed] [Google Scholar]
- Akita M, Nielsen E, Keegstra K (1997) Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J Cell Biol 136: 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Altman T, Damm B, Halfter U, Willmitzer L, Morris P-C (1992) Protoplast transformation and methods to create specific mutants in Arabidopsis thaliana. In C Koncz, N-H Chua, J Schell, eds, Methods in Arabidopsis Research. World Scientific, London, pp 310–330
- Aronsson H, Jarvis P (2002) A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett 529: 215–220 [DOI] [PubMed] [Google Scholar]
- Bell E, Creelman RA, Mullet JE (1995) A chloroplast lipoxygenase is required for wound-induced jasmonic accumulation in Arabidopsis. Proc Natl Acad Sci USA 92: 8675–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AK, Eriksson M-J (1996) The cyanobacterium Synechococcus sp. PCC 7942 possesses a close homologue to the chloroplast ClpC protein of higher plants. Plant Mol Biol 31: 721–730 [DOI] [PubMed] [Google Scholar]
- Desimone M, Weiss-Wichert C, Wagner E, Altenfeld U, Johanningmeier U (1997) Immunochemical studies on the Clp-protease in chloroplasts: evidence for the formation of a ClpC/P complex. Bot Acta 110: 234–239 [Google Scholar]
- Dougan DA, Mogk A, Zeth K, Turgay K, Bukau B (2002) AAA+ proteins and substrate recognition, it all depends on their partner in crime. FEBS Lett 529: 6–10 [DOI] [PubMed] [Google Scholar]
- Evans JR (1996) Developmental constraint on photosynthesis: effects of light and nutrition. In NR Baker, ed, Photosynthesis and the Environment. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 281–304
- Fulgosi H, Vener AV, Altschmied L, Herrmann RG, Andersson B (1998) A novel multi-functional chloroplast protein: identification of a 40 kDa immunophilin-like protein located in the thylakoid lumen. EMBO J 17: 1577–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM (1989) Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res 17: 4713–4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S (1996) Proteases and their targets in Escherichia coli. Annu Rev Genet 30: 465–506 [DOI] [PubMed] [Google Scholar]
- Gottesman S, Maurizi MR, Wickner S (1997) Regulatory subunits of energy-dependent proteases. Cell 91: 435–438 [DOI] [PubMed] [Google Scholar]
- Gottesman S, Squires C, Pichersky E, Carrington M, Hobbs M, Mattick JS, Darlymple B, Kuramitsu H, Shiroza T, Foster T, et al (1990) Conservation of the regulatory subunit for the Clp ATP-dependent protease in prokaryotes and eukaryotes. Proc Natl Acad Sci USA 87: 3513–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud R, Kessel M, Beuron F, Stevens AC (1998) Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J Biol Chem 273: 12476–12481 [DOI] [PubMed] [Google Scholar]
- Halperin T, Adam Z (1996) Degradation of mistargeted OEE33 in the chloroplast stroma. Plant Mol Biol 30: 925–933 [DOI] [PubMed] [Google Scholar]
- Halperin T, Ostersetzer O, Adam Z (2001. b) ATP-dependent association between subunits of Clp protease in pea chloroplasts. Planta 213: 614–619 [DOI] [PubMed] [Google Scholar]
- Halperin T, Zheng B, Itzhaki H, Clarke AK, Adam Z (2001. a) Plant mitochondria contain proteolytic and regulatory subunits of the ATP-dependent Clp protease. Plant Mol Biol 45: 461–468 [DOI] [PubMed] [Google Scholar]
- Hutin C, Havaux M, Carde J, Kloppstech K, Meiherhoff K, Hoffman N, Nussaume L (2002) Double mutation cpSRP43-/cpSRP54- is necessary to abolish the cpSRP pathway required for thylakoid targeting of the light-harvesting chlorophyll proteins. Plant J 29: 531–543 [DOI] [PubMed] [Google Scholar]
- Inaba T, Li M, Alvarez-Huerta M, Kessler F, Schnell DJ (2004) atTic110 functions as a scaffold for coordinating the stromal events of protein import into chloroplasts. J Biol Chem 278: 38617–38627 [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Beuron F, Kessel M, Wickner S, Maurizi MR, Steven AC (2001) Translocation pathway of protein substratesin ClpAP protease. Proc Natl Acad Sci USA 98: 4328–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Constan D, Akita M, Keegstra K (2001) Molecular chaperones involved in chloroplast protein import. Biochim Biophys Acta 1541: 102–113 [DOI] [PubMed] [Google Scholar]
- Keeler SJ, Boettger CM, Haynes JG, Kuches KA, Johnson MM, Thureen DL, Keeler CL Jr, Kitto SL (2000) Acquired thermotolerance and expression of the HSP100/ClpB genes of lima bean. Plant Physiol 123: 1121–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YI, Levchenko I, Fraczkowska K, Woodruff RV, Sauer RT, Baker TA (2001) Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat Struct Biol 8: 230–233 [DOI] [PubMed] [Google Scholar]
- Kouranov A, Chen X, Fuks B, Schnell DJ (1998) Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J Cell Biol 143: 991–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Maliga P (2003) The plastid clpP1 protease gene is essential for plant development. Nature 425: 86–89 [DOI] [PubMed] [Google Scholar]
- Leister D (2003) Chloroplast research in the genomic age. Trends Genet 19: 47–56 [DOI] [PubMed] [Google Scholar]
- Majeran W, Olive J, Drapier D, Vallon O, Wollman F-A (2001) The light sensitivity of ATP synthase mutants of Chlamydomonas reinhardtii. Plant Physiol 126: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Wollman FA, Vallon O (2000) Evidence for a role of ClpP in the degradation of the chloroplast cytochrome b(6)f complex. Plant Cell 12: 137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T, Keegstra K (1993) Characterization of a cDNA clone encoding a chloroplast-targeted Clp homologue. Plant Mol Biol 21: 525–537 [DOI] [PubMed] [Google Scholar]
- Mori H, Cline K (2001) Post-translational protein translocation into thylakoids by the Sec and ΔpH-dependent pathway. Biochim Biophys Acta 1541: 80–90 [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Ito M, Kiosue T, Shinozaki K, Watanabe A (1999) Identification of clp genes expressed in senescing Arabidopsis leaves. Plant Cell Physiol 40: 504–514 [DOI] [PubMed] [Google Scholar]
- Nielsen E, Akita M, Davila-Aponte J, Keegstra K (1997) Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J 16: 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Sugita M, Sugiura M (1995) Three types of nuclear genes encoding chloroplast RNA-binding proteins (cp29, cp31 and cp33) are present in Arabidopsis thaliana: presence of cp31 in chloroplasts and its homologue in nuclei/cytoplasms. Plant Mol Biol 27: 529–539 [DOI] [PubMed] [Google Scholar]
- Park S, Rodermel SR (2004) Mutations in ClpC2/Hsp100 suppress the requirement for FtsH in thylakoid membrane biogenesis. Proc Natl Acad Sci USA 101: 12765–12770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Kim DW, Choe J, Kim K (1997) RNA helicase activity of Escherichia coli SecA protein. Biochem Biophys Res Commun 235: 593–594 [DOI] [PubMed] [Google Scholar]
- Parsell DA, Lindquist S (1993) The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet 27: 437–496 [DOI] [PubMed] [Google Scholar]
- Peltier J, Ripoll DR, Friso G, Rudella A, Cai Y, Ytterberg J, Giacomelli L, Pillardy P, van Wijk KJ (2004) Clp protease complexes from photosynthetic and non-photosynthetic plastids and mitochondria of plants, their predicted three-dimensional structures, and functional implications. J Biol Chem 279: 4768–4781 [DOI] [PubMed] [Google Scholar]
- Peltier J, Ytterberg J, Liberles DA, Roepstorff P, van Wijk KJ (2001) Identification of a 350-kDa ClpP protease complex with 10 different Clp isoforms in chloroplast of Arabidopsis thaliana. J Biol Chem 276: 16318–16327 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Rocak S, Linder L (2004) DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol 5: 232–241 [DOI] [PubMed] [Google Scholar]
- Sakamoto W (2003) Leaf-variegated mutations and their responsible genes in Arabidopsis thaliana. Genes Genet Syst 78: 1–9 [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanklin J, Dewitt ND, Flanagan JM (1995) The stroma of higher plant plastids contain ClpP and ClpC, functional homologs of Escherichia coli ClpP and ClpA: an archetypal two-component ATP-dependent protease. Plant Cell 7: 1713–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T, Shimizu K, Ueda K, Nishimura Y, Kuroiwa T, Hashimoto T (2001) The chloroplast clpP gene encoding a proteolytic subunit of ATP-dependent protease and is indispensable for chloroplast development in tobacco. Plant Cell Physiol 42: 264–273 [DOI] [PubMed] [Google Scholar]
- Soikkeli S (1980) Ultrastructure of the mesophyll in Scots pine and Norway spruce: seasonal variation and molarity of fixative buffer. Protoplasma 103: 241–252 [Google Scholar]
- Sokolenko A, Lerbs-Machr S, Altschmied L, Herrmann RG (1998) Clp protease complexes and their diversity in chloroplasts. Planta 207: 286–295 [DOI] [PubMed] [Google Scholar]
- Spremulli L (2000) Protein synthesis, assembly, and degradation. In B Buchanan, W Gruissem, R Jones, eds, Biochemistry & Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 412–456
- Sutinen S (1987) Cytology of Norway spruce needles: changes during ageing. Eur J For Pathol 17: 65–73 [Google Scholar]
- Weaver LM, Froehlich JE, Amasino RM (1999) Chloroplast-targeted ERD1 protein declines but its mRNA increases during senescence in Arabidopsis. Plant Physiol 119: 1209–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibezahn J, Bukau B, Mogk A (2004) Unscrambling an egg: protein disaggregation by AAA+ proteins. Microb Cell Fact 3: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong EY, Hironaka CM, Fischhoff DA (1992) Arabidopsis thaliana small subunit leader and transit peptide enhance the expression of Bacillus thuringiensis proteins in transgenic plants. Plant Mol Biol 20: 81–93 [DOI] [PubMed] [Google Scholar]
- Zheng B, Halperin T, Hruskova-Heidingsfeldova O, Adam Z, Clarke AK (2002) Characterization of chloroplast Clp proteins in Arabidopsis: localization, tissue specificity and stress responses. Physiol Plant 114: 92–101 [DOI] [PubMed] [Google Scholar]