Abstract
Arabidopsis (Arabidopsis thaliana) and tomato (Lycopersicon esculentum) show similar physiological responses to iron deficiency, suggesting that homologous genes are involved. Essential gene functions are generally considered to be carried out by orthologs that have remained conserved in sequence and map position in evolutionarily related species. This assumption has not yet been proven for plant genomes that underwent large genome rearrangements. We addressed this question in an attempt to deduce functional gene pairs for iron reduction, iron transport, and iron regulation between Arabidopsis and tomato. Iron uptake processes are essential for plant growth. We investigated iron uptake gene pairs from tomato and Arabidopsis, namely sequence, conserved gene content of the regions containing iron uptake homologs based on conserved orthologous set marker analysis, gene expression patterns, and, in two cases, genetic data. Compared to tomato, the Arabidopsis genome revealed more and larger gene families coding for the iron uptake functions. The number of possible homologous pairs was reduced if functional expression data were taken into account in addition to sequence and map position. We predict novel homologous as well as partially redundant functions of ferric reductase-like and iron-regulated transporter-like genes in Arabidopsis and tomato. Arabidopsis nicotianamine synthase genes encode a partially redundant family. In this study, Arabidopsis gene redundancy generally reflected the presumed genome duplication structure. In some cases, statistical analysis of conserved gene regions between tomato and Arabidopsis suggested a common evolutionary origin. Although involvement of conserved genes in iron uptake was found, these essential genes seem to be of paralogous rather than orthologous origin in tomato and Arabidopsis.
Arabidopsis (Arabidopsis thaliana) serves as the reference for dicot genome analysis regarding gene sequence, gene number, and gene function. In the minimal genome concept, it is assumed that orthologous gene functions transmitted in a lineage-dependent vertical manner in related organisms should be conserved by sequence and function if they are essential (Mushegian, 1999). Using systematic gene knockout approaches and genome sequence comparisons, it was calculated that Bacillus subtilis may have about 250 to 300 essential genes (Kobayashi et al., 2003). Ninety-six percent of essential genes are conserved in bacteria and encode proteins with known functions in cell metabolism and energetics, information processing, and cell growth and division (Kobayashi et al., 2003). Yeast (Saccharomyces cerevisiae) may have about 1,000 essential genes (Winzeler et al., 1999; Giaever et al., 2002). In plants, the number of conserved and essential plant genes still remains to be experimentally determined. It is unclear whether searching for conserved sequence homologs in expressed sequence tag (EST) and genomic databases of diverse plant species may provide clues to essential gene functions. A potential obstacle for such homolog searches in plants is the fact that plant genomes underwent large evolutionary genomic rearrangements involving polyploidization, chromosome rearrangement, and partial gene loss (Gaut et al., 2000; Simillion et al., 2002). The analysis of the full genome sequence suggested that the Arabidopsis genome contained duplicated blocks corresponding to ancient and recent genome duplication events (Arabidopsis Genome Initiative, 2000; Blanc et al., 2000, 2003; Vision et al., 2000; Simillion et al., 2002; Bowers et al., 2003; Ermolaeva et al., 2003). These genome duplication events led to horizontal multiplication of homologous genes known as paralogs. The presence of paralogous genes hampers straightforward genome colinearity studies between diverged species of different families.
Full genomic sequence of the five Arabidopsis chromosomes and skeletons of genomic and EST markers mapped onto the chromosomes of other dicot species are currently available for assessing homologous gene functions (Fulton et al., 2002; Gebhardt et al., 2003; Zhu et al., 2003). Tomato (Lycopersicon esculentum) belongs to one of the plant species for which a detailed genetic map with about 1,000 mapped EST sequences is available (Tanksley et al., 1992; Fulton et al., 2002). Tomato EST markers that identify unique genes in the Arabidopsis genome were termed conserved orthologous set (COS) markers for utilization in comparative mapping (Fulton et al., 2002). Comparisons of map positions of COS markers in tomato and Arabidopsis revealed evidence of small regions with fairly conserved gene order (microsyntenies) that are often disrupted by genes that do not fit into the apparent syntenic regions (Fulton et al., 2002). Microsynteny can be observed when comparing the order of predicted genes of tomato and Arabidopsis bacterial artificial chromosome clones (Ku et al., 2000, 2001; Mao et al., 2001; Rossberg et al., 2001; Oh et al., 2002; Van der Hoeven et al., 2002). In general, more than one Arabidopsis region can be identified as potentially colinear, which may reflect ancient polyploidization events in this species.
Since iron is required for many basic enzymatic reactions and biological processes in all organisms, iron uptake is a strictly essential mechanism for growth of any organism. In plants, iron deficiency is caused when iron is not available due to low solubility, as is the case on alkaline and calcareous soils. Plants are able to cope with iron deficiency if they mobilize sufficient iron from their environment (Hell and Stephan, 2003). Lack of appropriate iron mobilization results in leaf chlorosis and severe growth retardation. Both tomato and Arabidopsis mobilize iron by reduction and increased uptake of Fe II (for review, see Bauer and Bereczky, 2003; Curie and Briat, 2003). Essential gene functions involved in iron assimilation in Arabidopsis have been demonstrated to be those of the ferric chelate reductase (FRO) gene AtFRO2 and of the root plasma membrane iron-regulated transporter (IRT) gene AtIRT1 (Robinson et al., 1999; Varotto et al., 2002; Vert et al., 2002). AtIRT1 and AtFRO2 genes belong to gene families (Eng et al., 1998; Robinson et al., 1999). Natural resistance-associated macrophage protein-like metal transporter genes (NRAMP1 and NRAMP3/NRAMP4) are up-regulated in response to iron deficiency in tomato and Arabidopsis and influence metal homeostasis in Arabidopsis (Curie et al., 2000; Thomine et al., 2000, 2003; Bereczky et al., 2003). In mammalian and yeast systems, it was shown that NRAMP homologs were essential for iron uptake (Fleming et al., 1997; Liu and Culotta, 1999). Nicotianamine synthase (NAS) encoded by the chloronerva gene (LeNAS) is required for synthesis of the metal chelator nicotianamine (Herbik et al., 1999; Higuchi et al., 1999; Ling et al., 1999). Nicotianamine is essential for iron distribution in plants (Scholz et al., 1992; Ling et al., 1999, 2002; Takahashi et al., 2003). The regulatory basic helix-loop-helix (bHLH) domain protein LeFER is essential for iron uptake and presumably acts as a transcription factor (Ling et al., 2002). LeFER controls directly or indirectly iron reduction and expression of LeIRT1, LeNRAMP1, and LeFRO1 in response to iron availability in tomato roots (Ling et al., 2002; Bereczky et al., 2003).
In this study, the term homologous genes or proteins refers to genes or proteins that have similar sequences that qualify them to share common properties, such as specific transporters or enzymes. The term homologous function specifies that the homologous genes and proteins act in a similar biological context, such as metal transporter in root iron mobilization. We made use of the essential characters of the genes FRO2, IRT1, NRAMP, NAS, and FER to analyze homology between these gene functions in Arabidopsis and tomato. By investigating sequence similarity, map position, and functional expression data, we identified gene pairs that represent the homologous functions in the two species. We discuss conservation of gene function with respect to orthologous and paralogous origin of the genes.
RESULTS
Identification and Sequence Comparison of Tomato-Arabidopsis Iron Uptake Homologs
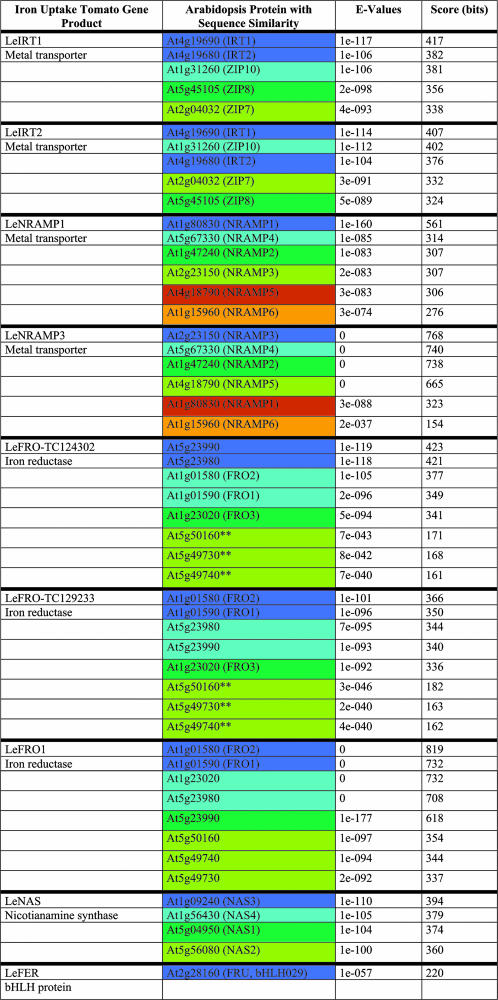
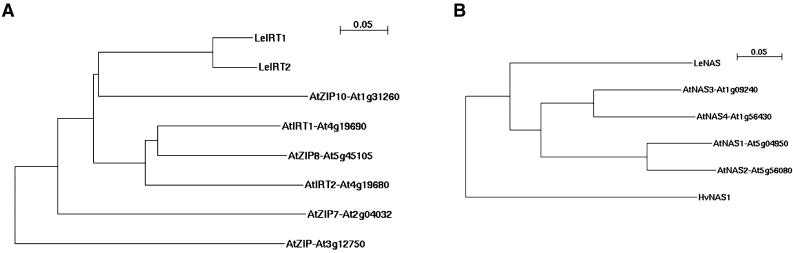
To identify homologs of iron uptake proteins from tomato and Arabidopsis, we screened the databases using the amino acid sequences of AtIRT1, LeIRT1, LeNRAMP1, LeNRAMP3, AtFRO2, LeNAS, and LeFER (“Materials and Methods;” Table I). We only retained those sequences for further analysis that were most related according to E-values (“Materials and Methods”). Partial sequences were named according to their transcript unit number in the database (e.g. LeFRO-TC129233). The available peptide sequences were aligned and represented in phylogenetic trees (Fig. 1, shown for IRT and NAS sequences).
Table I.
Amino acid sequence comparisons of tomato and Arabidopsis homologs and alignment results using BLASTP at http://www.arabidopsis.org/Blast
Arabidopsis iron uptake homologs were only represented and considered for further analysis if they showed E-values similar to the best hits. The only exceptions to this were three FRO proteins marked by **, which were retained due to their map position. For the purpose of highlighting tomato-Arabidopsis conserved gene clusters in Figure 3, the iron uptake genes were marked by a color code (compare Table I with Fig. 3 and Supplemental Table I). Arabidopsis iron uptake homologs located within a distance of ±400 genes (about ±2 Mb) were marked by the same color (e.g. At4g19680-IRT2 and At4g19690-IRT1). Homologous Arabidopsis iron uptake genes that were located in different regions of the genome (at a distance of more than 400 genes) were highlighted by different colors. The best hits were highlighted in dark blue, thereafter in light blue, dark green, light green, brown-orange, and light orange.
Figure 1.
Phylogenetic trees of Arabidopsis and tomato iron uptake homologs. Sequences were aligned using ClustalX. N-J Trees were generated; relative branch lengths are indicated. Arabidopsis sequences can be retrieved by their gene locus number from http://mips.gsf.de/proj/thal/db/search/search_frame.html. Tomato sequences can be retrieved using the TC numbers at http://www.tigr.org/tdb/tgi/lgi/searching/reports.html. The comparisons include homologs of IRT metal transporter (A) and NAS (B).
LeIRT1 and LeIRT2 showed highest sequence similarity with each other (Fig. 1A). Among the 15 Arabidopsis zinc and iron-regulated (ZIP) transporter sequences (Mäser et al., 2001), five of them could be grouped together with LeIRT1 and LeIRT2 (Fig. 1A). One of these, AtIRT1, was shown to be essential for iron uptake, whereas AtIRT2 was iron regulated but not essential for iron uptake (Vert et al. 2001, 2002; Varotto et al., 2002). In the sequence comparisons, AtIRT1 and AtIRT2 did not appear to be the closest homologs. In fact, AtIRT1 was most similar to AtZIP8, and LeIRT1 and LeIRT2 were most similar to AtZIP10. AtZIP7 was distantly related to IRT proteins. All other ZIP sequences from the database were even more distant to IRT than AtZIP7.
LeNRAMP1, LeNRAMP3, AtNRAMP1, AtNRAMP3, and AtNRAMP4 are encoded by iron-regulated genes (Curie et al., 2000; Thomine et al., 2000; Bereczky et al., 2003). As shown previously, LeNRAMP1 could be aligned with AtNRAMP1 and AtNRAMP6, and LeNRAMP3 could be aligned with AtNRAMP3 and AtNRAMP4 (Bereczky et al., 2003). LeNRAMP-AI778139 could be aligned with AtNRAMP2 (phylogenetic tree not shown).
AtFRO2 is required for iron reduction upon low iron supply (Robinson et al., 1999). Three LeFRO sequences and four Arabidopsis FRO sequences had significant similarity among each other and to AtFRO2. The LeFRO-TC124302 sequence was most similar to Arabidopsis FRO homologs At5g23980 and At5g23990. LeFRO1 and LeFRO-TC129233 were most related to AtFRO2 and AtFRO1. The three FRO-like proteins—At5g50160, At5g49730, and At5g49740—were not as related in sequence. Their genes were only retained because of their mapping location, as will be shown later.
NAS is essential for iron homeostasis in tomato. Four different Arabidopsis NAS sequences were in the database (see also Suzuki et al., 2001; Becher et al., 2004), which were more related to each other rather than to the tomato LeNAS (CHLORONERVA; Fig. 1B). Arabidopsis NAS sequences could be divided into two subgroups, encoded by genes on chromosomes 1 and 5, respectively.
A single Arabidopsis protein that we named FER-like regulator of iron uptake (AtFRU;At2g28160, bHLH029; Heim et al., 2003) corresponded to LeFER (see also Jakoby et al., 2004). All other predicted bHLH domain protein sequences from either Arabidopsis or tomato differed significantly outside the bHLH domain (data not shown).
In summary, it was possible to predict a unique homologous Arabidopsis-tomato gene pair in only a single case, namely that of LeFER-AtFRU. For the other four gene functions Arabidopsis had more and larger gene families encoding these functions than tomato.
Mapping of Iron Uptake Genes in Tomato
A further criterion for gene homology between two species is location in a colinear region of the two genomes. For map position comparison, it was necessary first to map the tomato genes.
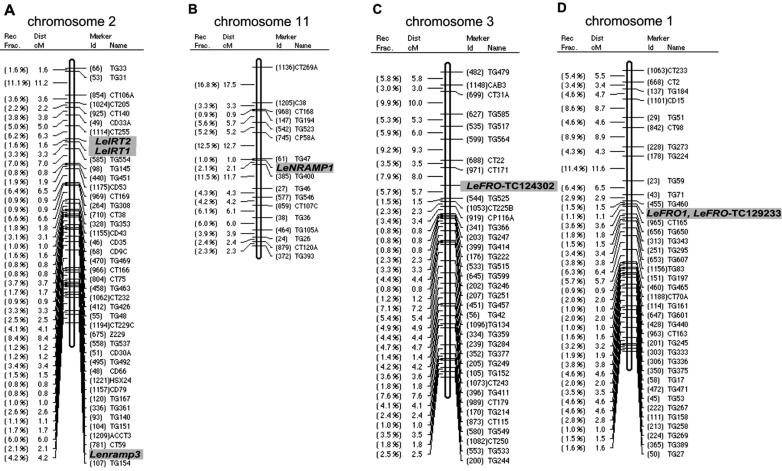
LeNAS and LeFER map positions were known to be on tomato chromosomes 1 and 6, respectively (Ling et al., 1999, 2002). Here, we mapped IRT1, FRO, and NRAMP homologs by restriction fragment length polymorphism analysis onto the tomato genome (“Materials and Methods;” Fig. 2). We found that LeIRT1 and LeIRT2 were mapped on chromosome 2 between CT255 and TG554. LeNRAMP1 was localized on chromosome 11 between TG47 and TG400. LeNRAMP3 was localized on chromosome 2 between CT59 and TG154. LeFRO-TC124302 could be mapped to chromosome 3 between CT171 and TG525. LeFRO-TC129233 and LeFRO1 were both localized in between TG460 and CT165 on chromosome 1. All genes were mapped as codominant markers with the exception of Lefro-TC124302, which was mapped as dominant marker in the L. esculentum background. Since a homolog of Lefro-TC124302 also could not be found in the potato EST database, we hypothesize that this gene is lacking from the potato and Lycopersicon pennellii genomes.
Figure 2.
Mapping of tomato iron uptake genes. A, LeIRT1, LeIRT2, and LeNRAMP3 were located on chromosome 2; B, LeNRAMP1 on chromosome 11; C, LeFRO-TC124302 on chromosome 3; D, LeFRO1 and LeFRO-TC129233 on chromosome 1. LeNAS (chloronerva) was previously fine mapped to chromosome 1 (Ling et al., 1996, 1999), and LeFER was fine mapped to chromosome 6 (Ling et al., 1996, 2002).
Analysis of Conserved Gene Content in Arabidopsis and Tomato Chromosomal Regions Containing Iron Uptake Genes
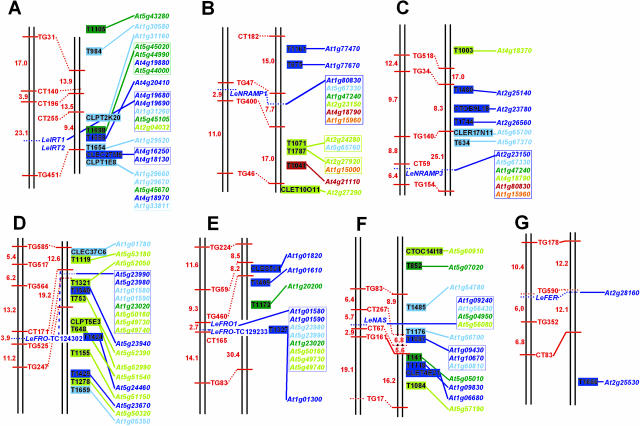
Chromosomal regions harboring iron uptake genes of Arabidopsis and tomato genomes were analyzed for their level of conserved genes. For this analysis, we selected COS markers that were mapped by Fulton et al. (2002) within an approximately 10-cM distance of tomato iron uptake genes and for which we could identify the corresponding Arabidopsis homologs by sequence similarity searches (“Materials and Methods;” Supplemental Table I, available at www.plantphysiol.org). Among the Arabidopsis COS markers, we identified those that were located up to 400 genes upstream or downstream of the respective iron uptake genes (approximately ±2 Mb) and defined them as being located in a similar region (“Materials and Methods;” Supplemental Table I; Fig. 3). For illustration, COS markers and iron uptake genes that are homologous and colocalize in the tomato and Arabidopsis genomes were highlighted by the same color (Table I; Supplemental Table I; Fig. 3).
Figure 3.
COS marker analysis of tomato and Arabidopsis chromosomal regions containing iron uptake genes. Represented were two tomato chromosome linkage maps that were modified from the high density molecular marker map (on the left side: Tanksley et al., 1992) and the COS marker map (on the right side: Fulton et al., 2002). The original maps were downloaded from http://www.sgn.cornell.edu/maps/tomato_arabidopsis/synteny_map.html. Gene locus names of the corresponding Arabidopsis iron uptake genes were indicated. Arabidopsis gene locus names reflect the chromosomal position. In most cases, multiple Arabidopsis iron uptake homologs were found corresponding to a tomato homolog. Such multiple homologs were boxed. COS markers that were located within ±10-cM distance of tomato iron uptake genes and corresponded to Arabidopsis COS markers located within a range of 400 genes upstream or downstream of the iron uptake homologs (about ±2 Mb) were indicated. Multiple Arabidopsis COS markers that were homologs of a single tomato COS marker were boxed. To better illustrate the presence of conserved gene clusters in between tomato and Arabidopsis iron uptake gene regions, a color code was used to indicate clustered Arabidopsis genes (compare Table I and Supplemental Table I). Homologous iron uptake genes within ±400 genes (about ±2 Mb) are denoted by the same color (e.g. At4g19680-IRT2 and At4g19690-IRT1). Homologous Arabidopsis iron uptake genes that were located in different regions of the genome (at a distance of more than 400 genes) were highlighted by different colors. Similarly, Arabidopsis COS markers received the colors of the neighboring iron uptake genes if they were located within ±400 genes (about ±2 Mb; e.g. COS marker At4g20410 has the same color as At4g19690-IRT1). Therefore, iron uptake genes and COS markers represented by the same color on the same map form a cluster on a chromosome. The tomato regions analyzed contain LeIRT1, LeIRT2 on chromosome 2 (A); LeNRAMP1 on chromosome 11 (B); LeNRAMP3 on chromosome 2 (C); LeFRO-TC124302 on chromosome 3 (D); LeFRO1 and LeFRO-TC129233 on chromosome 1 (E); LeNAS on chromosome 1 (F); LeFER on chromosome 6 (G). With the exception of LeFER-AtFRU, tomato-Arabidopsis regions harboring homologous iron uptake genes showed levels of conserved gene content.
We found that 8 tomato COS markers out of 24 were located in the region of LeIRT1/LeIRT2 and recognized at least one Arabidopsis region containing either AtIRT1/AtIRT2, AtZIP8, or AtZIP10 (Fig. 3A). Two of these tomato COS markers were homologous to multiple Arabidopsis COS markers that were located in two and three IRT/ZIP regions, respectively. Therefore, we could identify three Arabidopsis regions harboring IRT/ZIP genes that shared several conserved gene sequences with the LeIRT1/LeIRT2 region in tomato.
Although only 10 COS markers were available for the LeNRAMP1 region, we could identify 2 that were located near AtNRAMP1 (Fig. 3B). Single LeNRAMP1-neighboring COS markers identified regions of AtNRAMP4, AtNRAMP5, and AtNRAMP6. Three COS markers were located near AtNRAMP3. The LeNRAMP3 region showed similarity to the region of AtNRAMP3 (3 COS markers out of 26), and, to a lesser extent, to that of AtNRAMP4 (two COS markers) and AtNRAMP5 (one COS marker; Fig. 3C).
The tomato region of LeFRO-TC124302 showed similar gene sequences to the Arabidopsis regions around the FRO-like homologs At5g23990/At5g23980 (4 COS markers out of 35), AtFRO2/AtFRO1 (one COS marker), and At5g50160/At5g49730/At5g49740 (seven COS markers; Fig. 3D). The LeFRO1/LeFRO-TC129233 region showed similarity to the regions of AtFRO2/AtFRO1 (3 COS markers out of 21) as well as At5g23980/At5g23990 (one COS marker; Fig. 3E). No indication for conserved genes was found for the region of Arabidopsis FRO3.
For the LeNAS regions we identified four corresponding regions in Arabidopsis (twice 2 and twice 3 COS markers out of 29; Fig. 3F), indicating that all four Arabidopsis NAS regions were related.
Only a single COS marker out of 21 recognized the LeFER and AtFRU regions (Fig. 3G).
With the exception of the LeFER/AtFRU regions, we could thus determine that 20% to 50% of the analyzed tomato COS markers recognized Arabidopsis genes located in clusters in the vicinity of the respective iron uptake genes.
We realized that clusters of conserved COS markers could also be found between regions that did not appear related by the presence of homologous iron uptake genes. To analyze this point further, we searched for matches between COS markers of 6 unrelated iron uptake regions (42 comparisons of nonhomologous regions and the 7 comparisons of homologous regions as control; Table II). We found that the number of matching COS markers was highest when comparing Arabidopsis and tomato regions with corresponding iron uptake genes (three to five conserved genes; only exception was LeFER/AtFRU). Only in the case of LeFER was the highest level of gene conservation observed with a different region. Only 4 out of 42 comparisons of regions with nonhomologous iron uptake genes showed similar levels of conserved gene content as the regions with corresponding iron uptake genes (three and four conserved COS markers), namely Lefro-TC124302-AtIRT1/AtIRT2, LeFRO-TC124302-At1g09240 (AtNAS3), LeFER-At5g23990 (FRO-like), and LeNRAMP1-AtFRU. In 19 comparisons with nonhomologous iron uptake gene regions, no matching COS marker was detected. In 12 cases, a single matching COS marker was found. In six cases, two matching COS markers were found. These observations indicate that the level of conserved gene sequences between homologous iron uptake gene regions generally tends to be higher between homologous iron uptake gene regions than between regions with unrelated iron uptake genes.
Table II.
Number of COS marker matches between seven Arabidopsis and seven tomato regions containing iron uptake homologs, resulting in 42 comparisons of regions with nonhomologous iron uptake genes and seven control comparisons of regions with homologous iron uptake genes (in bold)
| At4g19690 AtIRT | At1g80830 AtNRAMP1 | At2g23150 AtNRAMP3 | At5g23990 AtFRO-Like | At1g01580 AtFRO2 | At1g09240 AtNAS3 | At2g28160 AtFRU | |
|---|---|---|---|---|---|---|---|
| LeIRT1/LeIRT2 | 4 + 1 | 2 | 1 | – | – | 2 | 2 |
| LeNRAMP1 | 1 | 2 + 1 | 2 | 2 | – | – | 3 |
| LeNRAMP3 | 1 | – | 3 + 1 | – | – | 1 | 2 |
| LeFRO-TC124302 | 4 | – | – | 3 + 1 | 2 | 4 | – |
| LeFRO1/LeFRO-TC129233 | 1 | 1 | – | – | 3 + 1 | 1 | – |
| LeNAS | – | 1 | 1 | 1 | – | 3 + 1 | – |
| LeFER | – | – | 1 | 3 | 1 | – | 1 + 1 |
Iron uptake homologs themselves are counted as +1 in the controls. –, No COS marker matches were found.
To analyze the significance of these findings, we determined the probability of the occurrence of tomato-Arabidopsis homolog clusters at random within 2-Mb intervals in the Arabidopsis genome. Similar results were obtained for 4-Mb (±2 Mb) windows (data not shown). We calculated P-values for the occurrence of clustering of the selected Arabidopsis gene homologs within ±1-Mb windows by taking into account the number of neighbors recognizing tomato homologs in the iron uptake region, the number of homologous genes of the same tomato region in the entire Arabidopsis genome, and the number of all genes in the Arabidopsis genome (“Materials and Methods”). Small P-values (P < 0.05) indicate that the number of homologous Arabidopsis neighbors recognized by the corresponding tomato region exceeded significantly the number of neighbors according to random clustering within the ±1-Mb window. We found that 7 out of 31 Arabidopsis iron uptake genes analyzed (22.6%) had P-values less than 0.05, and 43 out of 279 noniron uptake genes analyzed (15%) showed clustering with P-values less than 0.05 (Supplemental Table II). The cumulative distribution of P-values for iron uptake genes indicated that overall iron uptake genes were significantly more clustered than noniron uptake genes (data not shown). Three Arabidopsis homologs of LeIRT1/LeIRT2 had P-values less than 0.015, namely At1g31260, At5g45105, and At4g19690/At4g19680 (Table III). Moreover, 9 out of 11 COS homologs that we suspected to be located in these regions (compare Supplemental Table I and Fig. 3) had P-values less than 0.02 (Table III). The 2 out of 11 remaining markers had P-values of about 0.06, just slightly above the cutoff value of 0.05 below which we considered results to be statistically significant (Table III). Thus, LeIRT1 maps together with four other tomato markers from the ±10-cM region surrounding the LeIRT1 locus to a single ±1-Mb window on the Arabidopsis genome region containing At1g31260 together with 419 genes. This means that in this Arabidopsis region the homologs of tomato markers of the LeIRT1/LeIRT2 region are highly enriched, which is reflected by the average density of about one of these markers every 83 genes (5/419). We would expect only about one of these markers every 500 genes (52/26,404) if the genes were randomly distributed. The significantly similar gene content of the IRT/ZIP regions of tomato and Arabidopsis suggests a common evolutionary origin. An equally significant conservation of clustered genes was found for LeFRO-TC124302/At5g47930/At5g49740/At5g50160 (Table III). Significant clustering was obtained for LeFRO1/LeFRO-TC129233/At1g01580/At1g01590 as well as LeNAS/At1g09240 (Table III). P-values between 0.05 and 0.075 were found for the regions of LeNRAMP1/At2g23150 and LeFRO-TC124302/At1g01580/At1g01590 (Table III). All other comparisons of iron uptake regions between tomato and Arabidopsis showed no significant clustering, with P-values higher than 0.1 (Table III). However, we speculate that, for some of the NRAMP, FRO, and NAS gene regions, P-values would decrease if more COS markers were available for the analysis of these regions.
Table III.
P-values indicating the occurrence of tomato homolog clusters at random within ±1-Mb intervals in the Arabidopsis genome
| Tomato Region | Tomato Gene/Marker | Arabidopsis Gene | P-Value |
|---|---|---|---|
| LeIRT1/LeIRT2 | LeIRT1, LeIRT2 | At4g19680/At4g19690 | 0.014 |
| T1668 | At4g20410 | 0.016 | |
| T1698 | At4g19880 | 0.015 | |
| CLEC27M9 | At4g16250 | 0.607 | |
| At4g18130 | 0.002 | ||
| CLPT1E8 | At4g18970 | 0.014 | |
| LeIRT1, LeIRT2 | At1g31260 | 0.001 | |
| T1654 | At1g29520 | 0.002 | |
| CLPT2K20 | At1g31160 | 0.001 | |
| CLPT1E8 | At1g29660/At1g29670 | 0.002 | |
| At1g33811 | 1.000 | ||
| T984 | At1g30580 | 0.001 | |
| LeIRT1, LeIRT2 | At5g45105 | 0.012 | |
| T1105 | At5g43280 | 0.062 | |
| T1698 | At5g45020/At5g44990 | 0.012 | |
| At5g4400 | 0.012 | ||
| CLPT1E8 | At5g45670 | 0.064 | |
| LeNRAMP1 | LeNRAMP1 | At1g80830 | 1.000 |
| T1161 | At1g77470 | 0.353 | |
| T675 | At1g77670 | 0.353 | |
| LeNRAMP1 | At5g67330 | 0.229 | |
| T1071 | At5g65760 | 0.328 | |
| LeNRAMP1 | At2g23150 | 0.334 | |
| T1071 | At2g24280 | 0.320 | |
| T1787 | At2g27920 | 0.056 | |
| CLET10O11 | At2g27290 | 0.055 | |
| LeNRAMP1 | At4g18790 | 0.337 | |
| T1014 | At4g21110 | 0.342 | |
| LeNRAMP1 | At1g15960 | 0.377 | |
| T1787 | At1g1500 | 0.377 | |
| LeNRAMP3 | LeNRAMP3 | At2g23150 | 0.280 |
| T1480 | At2g25140 | 0.073 | |
| CTOB9L18 | At2g23780 | 0.262 | |
| T1744 | At2g26560 | 0.626 | |
| LeNRAMP3 | At5g67330 | 0.144 | |
| T634 | At5g67370 | 0.140 | |
| CLER17N11 | At5g65700 | 0.276 | |
| LeNRAMP3 | At4g18790 | 0.655 | |
| T1003 | At4g18370 | 0.659 | |
| LeFRO-TC124302 | LeFRO-TC124302 | At5g23990/At5g23980 | 0.072 |
| T1540 | At5g23940 | 0.074 | |
| T1424 | At5g24460 | 0.069 | |
| T1429 | At5g23670 | 0.075 | |
| LeFRO-TC124302 | At1g01580/At1g01590 | 0.526 | |
| CLEC37C6 | At1g01780 | 0.542 | |
| T1659 | At1g05350 | 1.000 | |
| LeFRO-TC124302 | At5g49730/At5g49740 | 0.005 | |
| At5g50160 | 0.001 | ||
| T1119 | At5g53180 | 0.030 | |
| T1321 | At5g52050 | 0.000 | |
| T1155 | At5g51150 | 0.000 | |
| T648 | At5g51540 | 0.000 | |
| T753 | At5g52390 | 0.001 | |
| T1278 | At5g50320 | 0.001 | |
| Lefro1/Lefro-129233 | Lefro1/Lefro-129233 | At1g01580/At1g01590 | 0.018 |
| CLES5J1 | At1g01820 | 0.021 | |
| T1409 | At1g01610 | 0.019 | |
| T1327 | At1g01300 | 0.015 | |
| Lefro1/Lefro-129233 | At1g23020 | 1.000 | |
| T1173 | At1g20200 | 1.000 | |
| LeNAS | LeNAS | At1g09240 | 0.015 |
| T1118 | At1g09830 | 0.069 | |
| T1687 | At1g09430 | 0.016 | |
| At1g10670 | 0.065 | ||
| CLET4E21 | At1g06680 | 0.254 | |
| LeNAS | At1g56430 | 0.190 | |
| T1176 | At1g56700 | 0.183 | |
| T1485 | At1g54780 | 0.226 | |
| T1687 | At1g60810 | 1.000 | |
| LeNAS | At5g04950 | 0.262 | |
| T141 | At5g05010 | 0.261 | |
| T852 | At5g07020 | 0.260 | |
| LeNAS | At5g56080 | 0.629 | |
| T1084 | At5g27190 | 0.643 | |
| CTOC14I18 | At5g60910 | 1.000 |
P-values were calculated by Fisher's exact test (“Materials and Methods”). P-values less than 0.05 are considered to show significant nonrandom clustering. Tandemly repeated genes were counted as one. P-values for iron uptake genes are in bold.
In summary, Arabidopsis and tomato genome regions with homologous iron uptake genes were generally characterized by the presence of multiple conserved genes. Since the Arabidopsis genome frequently contained more than one region with conserved gene content for any of the studied tomato iron uptake regions, it did not appear to be sufficient to determine a functional homology based on map position and sequence alone.
Functional Analysis of Tomato-Arabidopsis Iron Uptake Genes
Within gene families, specific biological functions of gene family members are conferred by their specific expression patterns. Iron uptake genes should be expressed in the root and/or induced by iron deficiency, indicating a function in iron mobilization of external or internal iron. Here, we investigated which of the iron uptake homologs fulfilled these expression pattern criteria in tomato and Arabidopsis. Gene expression was surveyed by analyzing EST expression data available at http://www.tigr.org for tomato genes (Supplemental Table III), as well as experimental gene expression studies in tomato and Arabidopsis (“Materials and Methods;” Figs. 4 and 5).
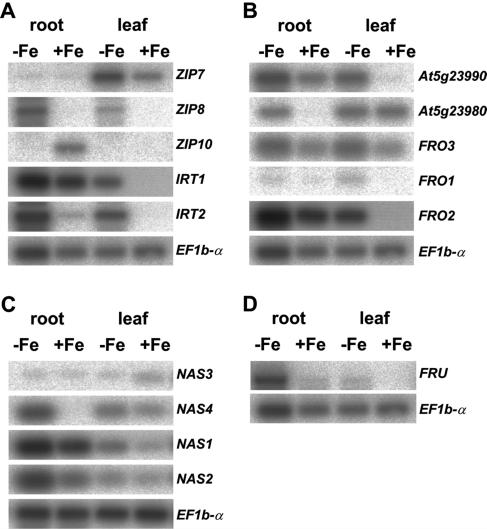
Figure 4.
Expression analysis of Arabidopsis iron uptake homologs in roots and leaves of plants grown upon low (−Fe) and sufficient iron supply (+Fe). A, IRT/ZIP genes; B, FRO genes; C, NAS genes; D, FRU gene. Expression analysis was performed by semiquantitative RT-PCR and Southern-blot analysis. Amplification of elongation factor cDNA EF1b-α (At5g19510) served as constitutive control.
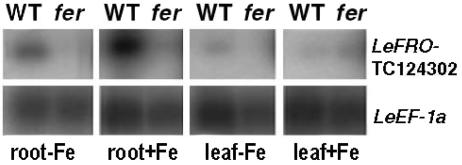
Figure 5.
Expression analysis of tomato LeFRO-TC124302 in roots and leaves of wild-type and fer mutant tomato plants, grown upon sufficient iron supply and iron deficiency. Expression analysis was performed by semiquantitative RT-PCR and Southern-blot analysis. Amplification of elongation factor cDNA LeEF1a served as constitutive control. No expression was detected for LeFRO-TC129233 (data not shown).
First, we analyzed expression of IRT/ZIP genes (Fig. 4A). LeIRT1 and LeIRT2 were previously shown to be expressed in the root, whereby LeIRT1 was iron regulated (Bereczky et al., 2003). LeIRT2 EST sequences were available in the EST database, but no data were available for LeIRT1. LeIRT2 EST sequences were all derived from root tissue (data not shown). In Arabidopsis (Fig. 4A), AtIRT1 and AtIRT2 were mainly expressed in roots and expression was induced upon iron deficiency, as expected (Eide et al., 1996; Vert et al., 2001). AtIRT2 appeared to be induced at a higher level than AtIRT1. The AtZIP8 expression pattern was similar to that of AtIRT2. AtZIP10 transcripts were detected in roots upon sufficient iron supply. AtZIP7 was mainly expressed in leaves and induced upon low iron supply.
Expression of NRAMP genes was not further analyzed here. LeNRAMP1 and AtNRAMP1 were previously found to be root specific and iron regulated (Curie et al., 2000; Bereczky et al., 2003). LeNRAMP3, AtNRAMP3, and AtNRAMP4 were iron regulated and also expressed in the leaf (Curie et al., 2000; Thomine et al., 2000; Bereczky et al., 2003). EST sequences were available for LeNRAMP3 and were derived from different tissues (data not shown).
EST sequences for LeFRO-TC124302 were found in roots and callus (data not shown). Experimental expression analysis showed that expression of LeFRO-TC124302 was not only root specific, but also slightly iron regulated and dependent on a functional LeFER gene in tomato (Fig. 5). On the other hand, transcripts for LeFRO-TC129233 were not detected experimentally in leaves and roots (data not shown). All EST sequences were derived from flower libraries, indicating that LeFRO-TC129233 was not involved in root iron uptake. As analyzed previously, LeFRO1 was expressed in roots and leaves, up-regulated by iron deficiency, and dependent on a functional LeFER gene (Li et al., 2004). Since LeFRO-TC124302 appeared to be specific to the L. esculentum genome, we tested whether LeFRO-TC124302 was essential for iron reduction in L. esculentum M82 introgression lines containing at the expected LeFRO-TC124302 locus an introgressed L. pennellii chromosomal fragment devoid of LeFRO-TC124302 (Eshed et al., 1992). We found that these introgression lines had similar levels of iron reductase activity as the control line M82 and grew in a very similar way (data not shown). LeFRO-TC124302 was therefore not essential for iron uptake and iron reduction in L. esculentum, indicating that presumably its function was performed by the redundant LeFRO1 gene. Arabidopsis FRO-like genes were all expressed in roots and leaves and tended to be induced upon iron deficiency (Fig. 4B). AtFRO1, AtFRO2, and At5g23990 shared a similar expression pattern. Expression of these genes was induced by iron deficiency in roots and leaves. Upon sufficient iron supply, AtFRO1, AtFRO2, and At5g23990 were expressed at a higher level in roots than in leaves. At5g23980 and AtFRO3 were both expressed in leaves upon sufficient iron supply. FRO3 was also expressed in roots under sufficient iron. Upon low iron supply, the two genes were induced in roots. FRO3 was also induced in leaves upon iron deficiency, whereas expression of At5g23980 was at a similar level than in leaves upon sufficient iron supply. Thus, for all Arabidopsis FRO genes, the expression patterns may suggest a function in iron deficiency responses.
We found expression of Arabidopsis NAS genes in leaves and roots upon sufficient and low iron supply (Fig. 4C), supporting the tomato data that NAS activity was indeed required constantly (Ling et al., 1999). However, we observed that AtNAS1 and AtNAS2 were expressed in roots stronger than in leaves. Upon low iron supply, AtNAS1 and AtNAS2 were induced in the root. AtNAS4 was induced by iron deficiency in roots and leaves. At sufficient iron supply, AtNAS4 was mainly expressed in leaves. AtNAS3 was mainly expressed in the leaf upon sufficient iron supply. Taking our expression data together indicates that Arabidopsis NAS genes are iron regulated. We identified Arabidopsis lines with T-DNA insertions in the coding regions of AtNAS1, AtNAS2, and AtNAS4 (data not shown). None of these lines segregated for any discernible phenotypes, suggesting that NAS genes are at least partially redundant.
LeFER is expressed in the root but not in the leaves and cotyledons (Ling et al., 2002). The tomato BHLH genes with weak sequence similarity to the tomato LeFER gene did not seem to be expressed in roots since all EST data were derived from flower tissue or callus (data not shown). AtFRU was mainly expressed in roots and hardly in leaves, whereby root expression was induced by iron deficiency (Fig. 4D). The expression pattern of AtFRU therefore suggests that the gene was indeed involved in iron deficiency responses in Arabidopsis. AtBHLH021 was only expressed in flowering tissues (data not shown). Spliced transcripts for BHLH genes AtBHLH022, AtBHLH090, AtBHLH035, and AtBHLH027 were not detected at all (data not shown).
Overall, expression data contributed to assigning homologous functions. In general, we found that gene family members from tomato differed more significantly in their expression patterns than did the homologous Arabidopsis gene family members, which tended to retain similar expression patterns.
DISCUSSION
In this study, we assigned functionally and structurally homologous gene functions involved in iron uptake between tomato and Arabidopsis. For this purpose, we based our studies not only on sequence comparisons but also took into account map position and functional expression data. Analyzing, in addition, levels of conserved gene content adjacent to the genes of interest allowed us to predict whether gene family members were redundant genes resulting from internal genome duplication events or nonredundant single-acting genes with distinct biological functions. We found that it was easier to determine the functional homologs in tomato using the Arabidopsis information than vice versa. Due to the lower gene complexity in tomato, we hypothesize that it might be generally convenient to utilize Arabidopsis genome information to predict tomato homologs.
Gene Homology and Map Position Reflect Genome Evolution in Tomato and Arabidopsis
Most comparative genome programs are based on finding orthologs by using the criterion sequence similarity (e.g. http://www.tigr.org/tdb/tgi/lgi/GO.html). In this study, sequence similarity alone was not a sufficient criterion for determining functional homologs for iron uptake between Arabidopsis and tomato. For a given gene, we frequently identified more than one related gene in the other species. The Arabidopsis genome in particular had a higher number of homologs compared to tomato. Extrapolating from our data would suggest that Arabidopsis may have more and larger gene families than tomato. Taking the number of most related FER-like, NAS, IRT, NRAMP, and FRO genes together would indicate that Arabidopsis had 21 genes (1+4+5+6+5), whereas tomato had only 10 genes for these functions (1+1+2+3+3). Van der Hoeven et al. (2002) calculated that only 50% of tomato genes were uncovered from the EST analysis. In our study, 7 of the 10 tomato genes were uncovered by EST analysis. Correcting our numbers to an estimated 14 tomato genes versus 21 Arabidopsis genes would hence suggest that Arabidopsis had about 33% more genes than tomato for the families analyzed. This result was supported by the similar findings of Van der Hoeven et al. (2002) on the comparison of multigene family copy numbers between tomato EST data and Arabidopsis genome data.
The comparative mapping studies presented here showed that 20% to 50% of analyzed COS markers were located in the vicinity of corresponding iron uptake genes in tomato and in Arabidopsis (except for those of the LeFER region). In contrast, only 0% to 14% of these COS markers matched with Arabidopsis regions containing nonhomologous iron uptake genes. The clustering of conserved COS marker positions in between tomato and Arabidopsis was calculated to be significant for several of them, and so it can be excluded that all clusters occurred at random. For some of the compared Arabidopsis-tomato regions with P-values greater than 0.05, we speculate that an increase of the number of analyzed markers would decrease the P-values and make the clustering statistically significant. We avoid utilizing the term colinearity for these observations since the actual gene order was not conserved in all cases. Similar gene content suggests a common evolutionary origin of the corresponding Arabidopsis and tomato genome regions. The analysis also showed internally duplicated regions in the Arabidopsis genome. It seems likely that the reason for higher multigene family copy numbers in Arabidopsis was the genome structure. Some duplications uncovered here have been found in previous studies as internally duplicated regions in the Arabidopsis genome; for example, the duplication events involving chromosome 1, chromosome 4, and chromosome 5 containing IRT/ZIP gene regions as well as the chromosome 1 duplication involving NAS genes (Blanc et al., 2003). We also detected signs of duplications that had not been previously detected and might reflect hidden duplications (Simillion et al., 2002), such as the chromosome 5 duplication involving NAS genes. Due to similar gene content and sequence similarity, we predict that NAS genes reveal an older duplication event responsible for the duplication of NAS regions on chromosomes 1 and 5 as well as one or two more recent duplication events involving chromosomes 1 and 5. The tomato genome was presumably also partially duplicated, even though to a lower degree. LeIRT1/LeIRT2 appear as recent duplicates based on high sequence similarity. Both LeFRO1/LeFRO-TC129233 and LeFRO-TC124302 regions showed levels of gene conservation with the AtFRO2 region as well as the region containing the three related FRO genes At5g50160/At5g49730/At5g49740. The latter three Arabidopsis genes were related to tomato FRO genes in terms of gene content despite being more distantly related in sequence. These observations might suggest that either the genomic region was also duplicated in tomato or that the duplication occurred early in dicot evolution. We hypothesize that LeFRO1 and LeFRO-TC129233 are present in tandem duplication in the tomato genome.
Importance of Functional Expression Data for Assignment of Functional Homology and Redundancy in Iron Uptake
This study dealt mainly with essential gene functions required in the root and/or for iron regulation. We found that expression data greatly contributed to assigning appropriate and unique homologous gene pairs. For IRT and FRO sequences, it was indispensable to consider sequence, map position, and gene expression aspects together. For example, among the 15 Arabidopsis ZIP genes (Mäser et al., 2001), AtIRT1 and AtIRT2 as well as LeIRT1 and LeIRT2 are the most similar pairs, respectively, in each genome. AtIRT1 and LeIRT1 are likely encoding the essential and iron-regulated iron transporters in both species (Varotto et al., 2002; Vert et al., 2002; Bereczky et al., 2003). AtIRT2 is iron regulated but not essential (Varotto et al., 2002). In contrast, LeIRT2 is neither iron regulated nor dependent on the regulator FER and thus not likely involved in iron mobilization (Eckhardt et al., 2001; Bereczky et al., 2003). In addition, LeIRT1 and LeIRT2 are very closely related in sequence, suggesting that the two genes are recent tandem duplications. In contrast, AtIRT1 and AtIRT2, which are also located in tandem, are much more diverged in sequence as if the tandem duplication event was much older than that in tomato. We found that AtZIP8 shared a similar expression pattern with AtIRT2. AtZIP8 was most related in sequence to AtIRT1 and AtIRT2. Hence, it is a possibility that AtIRT2 function overlaps with a redundant function provided by AtZIP8. It seems, however, unlikely that, despite their tandem location with IRT1, AtIRT2 and LeIRT2 are functioning as orthologs. Similarly, our study suggests that LeIRT2 and AtZIP10 may share a common function.
The most drastic example of why functional gene analysis data are needed for investigating homologous gene functions was provided by the FRO genes. Among the eight Arabidopsis FRO-like genes, AtFRO2 was unequivocally identified as the FRO gene in Arabidopsis due to genetic experiments (Robinson et al., 1999). From expression studies, we suggest that four other Arabidopsis FRO gene homologs could share additional functions in iron mobilization. In tomato, LeFRO1 and LeFRO-TC124302 may both be root FRO genes acting upon iron mobilization. Both genes are iron regulated in roots and dependent on FER (see also Li et al., 2004) The analysis of introgression L. esculentum lines devoid of LeFRO-TC124302 suggested that LeFRO-TC124302 was not essential and that a redundant iron reductase gene must exist in the Lycopersicon genome. This redundant function could well be provided by LeFRO1. In contrast, LeFRO-TC129233 was also similar in sequence to AtFRO2 and the genes were located in regions with conserved gene content. However, LeFRO-TC129233 was not found to be expressed in roots or leaves, nor upon iron deficiency, suggesting that LeFRO-TC129233 may be involved as putative iron reductase in a different biological process than LeFRO1 and LeFRO-TC124302.
The chloronerva (nas) tomato mutant is characterized by a distinct leaf chlorosis and root phenotype upon low iron supply (Scholz et al., 1992). A similar mutant has not been described in Arabidopsis. Our analysis of the four NAS genes in Arabidopsis indicated that NAS genes were duplicated at least twice. Interestingly, despite of a strong sequence conservation, we found differential regulation of NAS genes in iron deficiency responses, whereby three of the four NAS genes were up-regulated by low iron supply. We established previously that NAS was required for induction of LeFER-mediated LeIRT1 and LeNRAMP1 induction upon iron deficiency (Bereczky et al., 2003). In accordance with these observations, iron regulation of NAS genes in Arabidopsis might suggest that nicotianamine could have a function in iron mobilization or signaling. Due to the high sequence conservation, NAS genes form a partially redundant gene family. Absence of single functional genes did not show obvious phenotypes. However, up-regulation of individual gene family members such as observed in the metallophyte and zinc hyperaccumulator Arabidopsis halleri may lead to discrete dominant phenotypes—for instance in A. halleri zinc tolerance (Becher et al., 2004).
The Arabidopsis genome contains 162 predicted BHLH genes (Bailey et al., 2003). Sequence comparisons show that outside the conserved bHLH domain, the AtFRU protein is quite distinct from all other bHLH proteins. The genome regions of the two genes did not show apparent conservation of gene clusters. LeFER and AtFRU were both expressed in roots, whereby AtFRU was also expressed to a low level in leaves. AtFRU was induced upon iron deficiency. Recent genetic experiments suggest that LeFER and AtFRU serve a conserved biological function in iron regulation (Jakoby et al., 2004). Perhaps genes encoding regulatory components have lower evolutionary constraints on sequence conservation than genes encoding transporters and enzymes.
Arabidopsis and tomato show similar physiological responses to iron deficiency (strategy I), so that most likely iron regulation and iron uptake are conserved processes in these two species that involve conserved gene functions. If these gene functions were not performed by orthologs, at least they were expected to be compensated by paralogs. We predict from our analysis that essential homologous gene functions of iron uptake are indeed involved and conserved between Arabidopsis and tomato. However, detailed sequence and map position analysis indicated that these conserved genes are most likely of paralogous origin rather than of orthologous origin. Despite the similarities of genes and proteins involved in iron mobilization, tomato induces root morphological alterations, root hair proliferation, and transfer cell development as a response to iron deficiency (Schmidt, 1999; Schikora and Schmidt, 2001, 2002). These morphological alterations are supposed to aid iron uptake. In Arabidopsis, morphological alterations to iron deficiency are less pronounced, and it is possible that Arabidopsis relies instead on its duplicated gene functions.
MATERIALS AND METHODS
Sequence Analysis and Database Searches
Amino acid sequences of AtFRO2 (Robinson et al., 1999), AtIRT1 (Eide et al., 1996), LeIRT1 (Eckhardt et al., 2001), AtNRAMP1 (Curie et al., 2000), AtNRAMP3 (Thomine et al., 2000), LeNRAMP1 and LeNRAMP3 (Bereczky et al., 2003), LeNAS (CHLORONERVA; Ling et al., 1999), and LeFER (Ling et al., 2002) were used to search homologous sequences from tomato (Lycopersicon esculentum) and Arabidopsis (Arabidopsis thaliana) by BLASTP and BLASTX. Arabidopsis amino acid sequence homologs were identified using the BLAST program (http://www.Arabidopsis.org/Blast) and retrieved at http://mips.gsf.de/proj/thal/db/search/search_frame.html. Tomato amino acid sequences were identified and retrieved using the BLAST program (http://tigrblast.tigr.org/tgi). Sequences were retained for analysis if they were most significantly related by comparable E-values. Sequences with drastically increased E-values were not taken further into account (exception FRO). A general E-value cutoff was not applied since different protein classes showed different E-values. Multiple amino acid sequences were aligned using ClustalX and N-J Trees were generated at http://www-igbmc.u-strasbg.fr/BioInfo/ClustalX/Top.html. N-J Trees were represented by NJPLOT (Perrière and Gouy, 1996). Functional expression data were available at http://www.tigr.org for tomato genes.
Mapping of Tomato Genes and Analysis of Conserved Gene Regions
Genomic tomato DNA fragments were mapped by restriction fragment length polymorphism analysis using 43 F2 individuals of an L. esculentum/Lycopersicon pennellii mapping population according to Tanksley et al. (1992). Genetic maps were generated using the MAPMAKER program (Lander et al., 1987).
Tomato COS markers that mapped within a distance of ±10 cM of iron uptake genes were selected for analysis of conserved gene regions at http://www.sgn.cornell.edu/maps/tomato_Arabidopsis/synteny_map.html. The encoded amino acid sequences of the selected COS markers were used to BLAST for corresponding Arabidopsis amino acid sequences at http://www.Arabidopsis.org/Blast. Arabidopsis genes were used as COS markers if their encoded peptides gave expected values below e-20 in alignments with encoded peptides of tomato COS markers (Supplemental Table I). Arabidopsis COS markers located up to 400 genes from iron uptake genes (approximately ±2 Mb) were considered to be in that same chromosomal region. For example, genes in the region of At4g19690 would be expected to have gene locus numbers between At4g15690 and At4g23690.
Statistical analysis and calculation of P-values for random clustering of homologous genes between Arabidopsis and tomato were performed as follows: For each of the Arabidopsis genes of interest, an interval of ±1 Mb was considered in which the gene of interest was in the center. If K was the total number of Arabidopsis genes in the ±1-Mb interval, L was the number of Arabidopsis genes located in this interval and homologous to genes from the corresponding ±10-cM tomato region minus tandem duplicates, M was the total number of genes in the Arabidopsis genome, and N was the number of genes in the Arabidopsis genome homologous to genes from the ±10-cM tomato region minus tandem duplicates, then the probability pL of finding L genes by chance given K, M, and N was computed by Fisher's exact test as the right tail of the hypergeometric distribution:
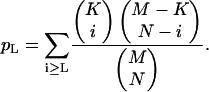 |
When determining K, L, M, and N, the gene in the center of the ±1-Mb interval was not counted. The statistical significance was computed as the probability that L or more than L genes could occur by chance in the ±1-Mb region if the numbers K, M, and N were fixed. The P-values were assigned to the genes in the center of the ±1-Mb intervals, and P-values less than 0.05 were considered significant for nonrandom clustering. For example, At1g31260 maps at position 11,175,540 bp on Arabidopsis chromosome 1. The ±1-Mb interval centered at position 11,175,540 contains 420 genes, including At1g31260 (K = 419). Out of those 419 genes, five are homologs of COS markers from the ±10-cM tomato region containing LeIRT1 (L = 5). In total, there are 26,405 Arabidopsis genes, including At1g31260 (M = 26,404). Out of those 26,404 genes, 51 are homologs of COS markers from the ±10-cM region of LeIRT1 (n = 51). The density of 5 genes out of 419 is 6-fold higher than the density of 51 genes out of 26,404, and the P-value is 0.001.
Plant Growth and Plant Material
Tomato plants used in RNA expression analysis were derived from the lines L. esculentum T3238fer (fer mutant phenotype) and L. esculentum Moneymaker (wild type). Homozygous plants of the first-generation introgression lines from L. pennellii (LA 716) in the genetic background of the processing tomato variety M82 were propagated and utilized in this study (Eshed et al., 1992).
For tomato iron uptake studies, 12-d-old plants were grown in a hydroponic Hoagland medium containing 0.1 (low iron) or 10 μm (sufficient iron) FeNaEDTA for 1 week according to Bereczky et al. (2003). Iron reductase assays were performed on roots as described by Stephan and Prochazka (1989).
For expression studies in Arabidopsis, 2-week-old Arabidopsis Columbia plants grown on solid Hoagland medium in the presence of 10 μm FeNaEDTA were transferred to Hoagland medium containing 10 μm FeNaEDTA (sufficient iron) or no iron and 200 μm bathophenanthroline disulfonic acid (low iron) for 5 d.
nas1, nas2, and nas4 T-DNA insertion lines were identified by database searches at the SALK Institute Web site (http://signal.salk.edu) and ordered from the Arabidopsis Biological Resource Center.
Gene Expression Analysis
Semiquantitative reverse transcription (RT)-PCR analysis was performed according to Bereczky et al. (2003). Total RNA was extracted from tomato using the Purescript RNA Isolation kit (Gentra Systems, Minneapolis) and from Arabidopsis using the Invisorb Spin Plant RNA Mini kit (Invitek, Berlin). From 0.1 to 2 μg RNA were reverse transcribed into cDNA using oligo(dT) primer and the RevertAid First Strand cDNA synthesis kit (Fermentas GmbH, St. Leon-Rot, Germany). cDNA was amplified using specific primers according to standard procedures (Table IV). The number of amplification cycles was determined experimentally so that the reaction was analyzed in the exponential phase. PCR fragments were separated by agarose gel electrophoresis, blotted, and hybridized according to standard procedures. Amplification of elongation factor cDNA was used as a constitutive control to normalize all samples.
Table IV.
Oligonucleotide primers used in RT-PCR experiments
| Gene Name | Number | 5′ Primer 5′–3′ | 3′ Primer 5′–3′ |
|---|---|---|---|
| LeFRO-TC129233 | TC129233 | atgggggttatgggtgcatcagag | tctatgcctgcattatgcttcctggt |
| LeFRO-TC124302 | TC124302 | tgtgaacgtgccaagtgtatcca | ggcccacaaacaacaactcca |
| LeEF-1a | TC123773 | cctcttgggctcgttaatctggct | ctggtggttttgaagctggtatct |
| AtZIP7 | At2g04032 | gtgcgtatgcgaggagaact | atcaaagtgaaggacagaagtaaaga |
| AtZIP8 | At5g45105 | atgcgagaccgattcaacag | tttcataaaagtcgagaggataatgt |
| AtZIP10 | At1g31260 | tacagcttctgcggtatcgtat | catctatttgtaagctccgctct |
| AtIRT1 | At4g19690 | gcatgggtcttggcggttgt | atccacatgatttcaatcccgcaat |
| AtIRT2 | At4g19680 | ggtaagaactcagtcggaccagt | gtcgccttgaataataaatcaaataa |
| AtFRO-like | At5g23990 | gataaggactccaagaagcaggta | caaacatatatagttagaacatggaataga |
| AtFRO-like | At5g23980 | ggataaggagttcaagaatcaggta | aacacacatagttagaacatggaataca |
| AtFRO1 | At1g01590 | cgacaacttatctccggtgatt | ttgtaacccaaacatctatgataaaa |
| AtFRO2 | At1g01580 | tctccaacatcttctcctacctcatcat | caacacatagtgaaaacagagttatatacgcaa |
| AtNAS3 | At1g09240 | caactccgtggtgatctctaaa | atagagaattaggaaacaagaaacg |
| AtNAS4 | At1g56430 | aggtgaagatgctaatggtgtt | acacagcatttttctaggtaagtt |
| AtNAS1 | At5g04950 | ggggttaatggtactcgtgg | agacatgaaatgaaaagagcagtt |
| AtNAS2 | At5g56080 | gccagatcggacggtgt | ctcgatcaaattcttctccatac |
| AtFRU BHLH029 | At2g28160 | atggaaggaagagtcaacgc | tcaatatagtgcagaaccgg |
| AtBHLH 090 | At1g10610 | atgatgatgatgagaggtggtgagagagtg | ttacgttactaccttgacattgagaacggtcat |
| AtBHLH 035 | At5g57150 | atggaggatatcgtcgaccaagaattaagc | ttagagagacaagagagatagagagagaaagatgctga |
| AtBHLH 027 | At4g29930 | atggaagatctcgaccatgagtacaagaattac | tcaaaccaaaacaagacacgtacagtattctct |
| AtBHLH 022 | At4g21330 | atgggtggaggaagcagatttcaag | ttatggattgcttctcataacttccaaaaga |
| AtBHLH 021 | At2g16910 | atgagcctgaacgggacagtggtc | ttattggttgtggtaatggttgatgtgttgg |
| AtEF1b-a | At5g19510 | aggagagggaggctgctaag | aatcttgttgaaagcgacaatg |
Supplementary Material
Acknowledgments
Tomato mapping filters were provided by Dr. M. Ganal. Help with the MAPMAKER program by Dr. X. Huang and Dr. M. Röder is greatly acknowledged.
This work was supported by Deutsche Forschungsgemeinschaft grants in the Emmy Noether program (Ba1610/3–1 to P.B.) and in the Arabidopsis Functional Genomics Network program (Ba1610/4–1 to P.B. and R.H.), and by the Bundesministerium für Bildung und Forschung to the Bioinformatics Center Gatersleben-Halle (grant to I.G.).
The on-line version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.047233.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bailey PC, Martin C, Toledo-Ortiz G, Quail PH, Huq E, Heim MA, Jakoby M, Weisshaar B (2003) Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 15: 2497–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer P, Bereczky Z (2003) Gene networks involved in iron acquisition strategies in plants. Agronomie 23: 1–8 [Google Scholar]
- Becher M, Talke IN, Krall L, Kramer U (2004) Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J 37: 251–268 [DOI] [PubMed] [Google Scholar]
- Bereczky Z, Wang HY, Schubert V, Ganal M, Bauer P (2003) Differential regulation of nramp and irt metal transporter genes in wild type and iron uptake mutants of tomato. J Biol Chem 278: 24697–24704 [DOI] [PubMed] [Google Scholar]
- Blanc G, Barakat A, Guyot R, Cooke R, Delseny M (2000) Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12: 1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Hokamp K, Wolfe KH (2003) A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res 13: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 383–384 [DOI] [PubMed] [Google Scholar]
- Curie C, Alonso JM, Le Jean M, Ecker JR, Briat JF (2000) Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochem J 347: 749–755 [PMC free article] [PubMed] [Google Scholar]
- Curie C, Briat J-F (2003) Iron transport and signaling in plants. Annu Rev Plant Biol 54: 183–206 [DOI] [PubMed] [Google Scholar]
- Eckhardt U, Marques AM, Buckhout TJ (2001) Two iron-regulated cation transporters from tomato complement metal uptake-deficient yeast mutants. Plant Mol Biol 45: 437–448 [DOI] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng BH, Guerinot ML, Eide D, Saier MH (1998) Sequence analyses and phylogenetic characterization of the ZIP family of metal ion transport proteins. J Membr Biol 166: 1–7 [DOI] [PubMed] [Google Scholar]
- Ermolaeva MD, Wu M, Eisen JA, Salzberg SL (2003) The age of the Arabidopsis thaliana genome duplication. Plant Mol Biol 51: 859–866 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Abu-Abied M, Saranga Y, Zamir D (1992) Lycopersicon esculentum lines containing small overlapping introgressions from L. pennellii. Theor Appl Genet 83: 1027–1034 [DOI] [PubMed] [Google Scholar]
- Fleming MD, Trenor CC, Su MA, Foernzler D, Beier DR, Dietrich WF, Andrews NC (1997) Microcytic anaemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nat Genet 16: 383–386 [DOI] [PubMed] [Google Scholar]
- Fulton TM, Van der Hoeven R, Eannetta NT, Tanksley SD (2002) Identification, analysis, and utilization of conserved ortholog set markers for comparative genomics in higher plants. Plant Cell 14: 1457–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Le Thierry d'Ennequin M, Peek AS, Sawkins MC (2000) Maize as a model for the evolution of plant nuclear genomes. Proc Natl Acad Sci USA 97: 7008–7015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt C, Walkemeier B, Henselewski H, Barakat A, Delseny M, Stuber K (2003) Comparative mapping between potato (Solanum tuberosum) and Arabidopsis thaliana reveals structurally conserved domains and ancient duplications in the potato genome. Plant J 34: 529–541 [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol Biol Evol 20: 735–747 [DOI] [PubMed] [Google Scholar]
- Hell R, Stephan UW (2003) Iron uptake, trafficking and homeostasis in plants. Planta 216: 541–551 [DOI] [PubMed] [Google Scholar]
- Herbik A, Koch G, Mock HP, Dushkov D, Czihal A, Thielmann J, Stephan UW, Baumlein H (1999) Isolation, characterization and cDNA cloning of nicotianamine synthase from barley—a key enzyme for iron homeostasis in plants. Eur J Biochem 265: 231–239 [DOI] [PubMed] [Google Scholar]
- Higuchi K, Suzuki K, Nakanishi H, Yamaguchi H, Nishizawa NK, Mori S (1999) Cloning of nicotianamine synthase genes, novel genes involved in the biosynthesis of phytosiderophores. Plant Physiol 119: 471–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Wang HY, Reidt W, Weisshaar B, Bauer P (2004) FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett (in press) [DOI] [PubMed]
- Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, et al (2003) Essential Bacillus subtilis genes. Proc Natl Acad Sci USA 100: 4678–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku HM, Liu J, Doganlar S, Tanksley SD (2001) Exploitation of Arabidopsis-tomato synteny to construct a high-resolution map of the ovate-containing region in tomato chromosome 2. Genome 44: 470–475 [DOI] [PubMed] [Google Scholar]
- Ku HM, Vision TJ, Liu J, Tanksley SD (2000) Comparing sequenced segments of the tomato and Arabidopsis genomes: large-scale duplication followed by selective gene loss creates a network of synteny. Proc Natl Acad Sci USA 97: 9121–9126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181 [DOI] [PubMed] [Google Scholar]
- Li L, Cheng X, Ling HQ (2004) Isolation and characterization of Fe(III)-chelate reductase gene LeFRO1 in tomato. Plant Mol Biol 54: 125–136 [DOI] [PubMed] [Google Scholar]
- Ling HQ, Bauer P, Bereczky Z, Keller B, Ganal M (2002) The tomato fer gene encoding a bHLH protein controls iron- uptake responses in roots. Proc Natl Acad Sci USA 99: 13938–13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Koch G, Baumlein H, Ganal MW (1999) Map-based cloning of chloronerva, a gene involved in iron uptake of higher plants encoding nicotianamine synthase. Proc Natl Acad Sci USA 96: 7098–7103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Pich A, Scholz G, Ganal MW (1996) Genetic analysis of two tomato mutants affected in the regulation of iron metabolism. Mol Gen Genet 252: 87–92 [DOI] [PubMed] [Google Scholar]
- Liu XF, Culotta VC (1999) Mutational analysis of Saccharomyces cerevisiae Smf1p, a member of the Nramp family of metal transporters. J Mol Biol 289: 885–891 [DOI] [PubMed] [Google Scholar]
- Mao L, Begum D, Goff SA, Wing RA (2001) Sequence and analysis of the tomato JOINTLESS locus. Plant Physiol 126: 1331–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJ, Sanders D, et al (2001) Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol 126: 1646–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushegian A (1999) The minimal genome concept. Curr Opin Genet Dev 9: 709–714 [DOI] [PubMed] [Google Scholar]
- Oh K, Hardeman K, Ivanchenko MG, Ellard-Ivey M, Nebenfuhr A, White TJ, Lomax TL (2002) Fine mapping in tomato using microsynteny with the Arabidopsis genome: the Diageotropica (Dgt) locus. Genome Biol 3: 0049.0041 [DOI] [PMC free article] [PubMed]
- Perrière G, Gouy M (1996) WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78: 364–369 [DOI] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Rossberg M, Theres K, Acarkan A, Herrero R, Schmitt T, Schumacher K, Schmitz G, Schmidt R (2001) Comparative sequence analysis reveals extensive microcolinearity in the lateral suppressor regions of the tomato, Arabidopsis, and Capsella genomes. Plant Cell 13: 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schikora A, Schmidt W (2001) Iron stress-induced changes in root epidermal cell fate are regulated independently from physiological responses to low iron availability. Plant Physiol 125: 1679–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schikora A, Schmidt W (2002) Formation of transfer cells and H(+)-ATPase expression in tomato roots under P and Fe deficiency. Planta 215: 304–311 [DOI] [PubMed] [Google Scholar]
- Schmidt W (1999) Mechanisms and regulation of reduction-based iron uptake in plants. New Phytol 141: 1–26 [Google Scholar]
- Scholz G, Becker R, Pich A, Stephan UW (1992) Nicotianamine—a common constituent of strategy-I and strategy-II of iron acquisition by plants—a review. J Plant Nutr 15: 1647–1665 [Google Scholar]
- Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y (2002) The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA 99: 13627–13632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan UW, Prochazka Z (1989) Physiological disorders of the nicotianamine-auxotroph tomato mutant chloronerva at different levels of iron nutrition. I. Growth characteristics and physiological abnormalities related to iron and nicotianamine supply. Acta Bot Neerl 38: 147–153 [Google Scholar]
- Suzuki K, Nakanishi H, Nishizawa NK, Mori S (2001) Analysis of upstream region of nicotianamine synthase gene from Arabidopsis thaliana: presence of putative ERE-like sequence. Biosci Biotechnol Biochem 65: 2794–2797 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Terada Y, Nakai I, Nakanishi H, Yoshimura E, Mori S, Nishizawa NK (2003) Role of nicotianamine in the intracellular delivery of metals and plant reproductive development. Plant Cell 15: 1263–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD, Ganal MW, Prince JP, de Vicente MC, Bonierbale MW, Broun P, Fulton TM, Giovannoni JJ, Grandillo S, Martin GB, et al (1992) High density molecular linkage maps of the tomato and potato genomes. Genetics 132: 1141–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomine S, Lelievre F, Debarbieux E, Schroeder JI, Barbier-Brygoo H (2003) AtNRAMP3, a multispecific vacuolar metal transporter involved in plant responses to iron deficiency. Plant J 34: 685–695 [DOI] [PubMed] [Google Scholar]
- Thomine S, Wang RC, Ward JM, Crawford NM, Schroeder JI (2000) Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc Natl Acad Sci USA 97: 4991–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoeven R, Ronning CM, Giovannoni JJ, Martin GB, Tanksley S (2002) Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. Plant Cell 14: 1441–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varotto C, Maiwald D, Pesaresi P, Jahns P, Salamini F, Leister D (2002) The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. Plant J 31: 589–599 [DOI] [PubMed] [Google Scholar]
- Vert G, Briat JF, Curie C (2001) Arabidopsis IRT2 gene encodes a root-periphery iron transporter. Plant J 26: 181–189 [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD (2000) The origins of genomic duplications in Arabidopsis. Science 290: 2114–2117 [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Zhu H, Kim DJ, Baek JM, Choi HK, Ellis LC, Kuester H, McCombie WR, Peng HM, Cook DR (2003) Syntenic relationships between Medicago truncatula and Arabidopsis reveal extensive divergence of genome organization. Plant Physiol 131: 1018–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.