Abstract
Diffuse large B cell lymphoma (DLBCL) frequently harbors genetic alterations that activate the B cell receptor (BCR) and TLR pathways, which converge to activate NF-κB. While selective inhibition of BTK with ibrutinib causes clinical responses in relapsed DLBCL patients, most responses are partial and of a short duration. Here, we demonstrated that MyD88 silencing enhanced ibrutinib efficacy in DLBCL cells harboring MyD88 L265P mutations. Chemical downregulation of MyD88 expression with HDAC inhibitors also synergized with ibrutinib. We demonstrate that HDAC inhibitor regulation of MyD88 expression is mediated by STAT3. In turn, STAT3 silencing caused a decrease in MyD88 mRNA and protein levels, and enhanced the ibrutinib antilymphoma effect in MyD88 mutant DLBCL cells. Induced mutations in the STAT3 binding site in the MyD88 promotor region was associated with a decrease in MyD88 transcriptional activity. We also demonstrate that treatment with the HDAC inhibitor panobinostat decreased phosphorylated STAT3 binding to the MyD88 promotor. Accordingly, combined treatment with panobinostat and ibrutinib resulted in enhanced inhibition of NF-κB activity and caused regression of DLBCL xenografts. Our data provide a mechanistic rationale for combining HDAC inhibitors and ibrutinib for the treatment of DLBCL.
Keywords: Hematology, Oncology
Keywords: B cell receptor, Lymphomas, Protein kinases
Downregulation of MyD88 enhances ibrutinib efficacy in ABC DLBCL with L265P mutation by interrupting a newly discovered BCR signaling amplification loop involving NF-kB, STAT3, and MyD88.
Introduction
Activated B cell (ABC), a subset of diffuse large B cell lymphoma (DLBCL), is characterized by an aberrant activation of the NF-κB signaling pathway, conferring poor prognosis when treated with standard chemotherapy regimens (1–4). In ABC DLBCL, a variety of genetic alterations that aberrantly activate the B cell receptor (BCR) and TLR signaling pathways have been linked to the constitutive activation of NF-κB (5). In the BCR pathway, mutations in CD79 and CARD11 are the most frequently observed genetic alterations, whereas L265P mutation of the signaling adaptor MyD88 is the most frequent one that constitutively activates the TLR pathway (6). Knockdown of either CD79 or MyD88 has been shown to inhibit proliferation of ABC DLBCL cells, suggesting that pharmacologic targeting of these 2 pathways may have a therapeutic value for the treatment of ABC DLBCL (5, 6).
In the BCR pathway, selective targeting of the bruton tyrosine kinase (BTK) has been shown to inhibit NF-κB activation and to induce cell death of ABC DLBCL (5, 7). At the present time, there is no drug available that can directly inhibit MyD88. However, because IL-1 receptor-associated kinase 4 (IRAK4) mediates most of the biological functions of MyD88, recent investigation has focused on selective targeting of IRAK4 in diseases driven by aberrant MyD88 signaling (8). While the BCR and TLR pathways converge distally to activate NF-κB, they may also crosstalk proximally in certain lymphoma subtypes. This complex interaction resulted in conflicting reports on the role of MyD88 mutations in mediating resistance to BCR pathway–targeted agents, such as ibrutinib. In ABC DLBCL, isolated MyD88 mutations were associated with resistance to ibrutinib, whereas lymphomas with concurrent MyD88 and CD79 mutations remained sensitive to ibrutinib (9). Nevertheless, these responses where mainly partial and of a short duration, raising the question of whether a combined inhibition of MyD88 and ibrutinib would have resulted in a better treatment outcome.
In the present study, we examined the potential therapeutic value of dual inhibition of the BCR and TLR pathways in DLBC. We found that genetic silencing of MyD88 enhanced the efficacy of ibrutinib in ABC DLBCL carrying MyD88 mutations. We also report that histone deacetylase inhibitors (HDACi) transcriptionally downregulated MyD88, inhibited MyD88-driven NF-κB activation, and enhanced ibrutinib efficacy in MyD88 mutant ABC DLBCL. Furthermore, we describe a central role of STAT3 in mediating HDACi regulation of MyD88 transcription. Our data provide a mechanistic rationale for combining HDACi with ibrutinib in ABC DLBCL.
Results
MyD88 mutation sustains ABC DLBCL survival and attenuates the antiproliferative activity of ibrutinib.
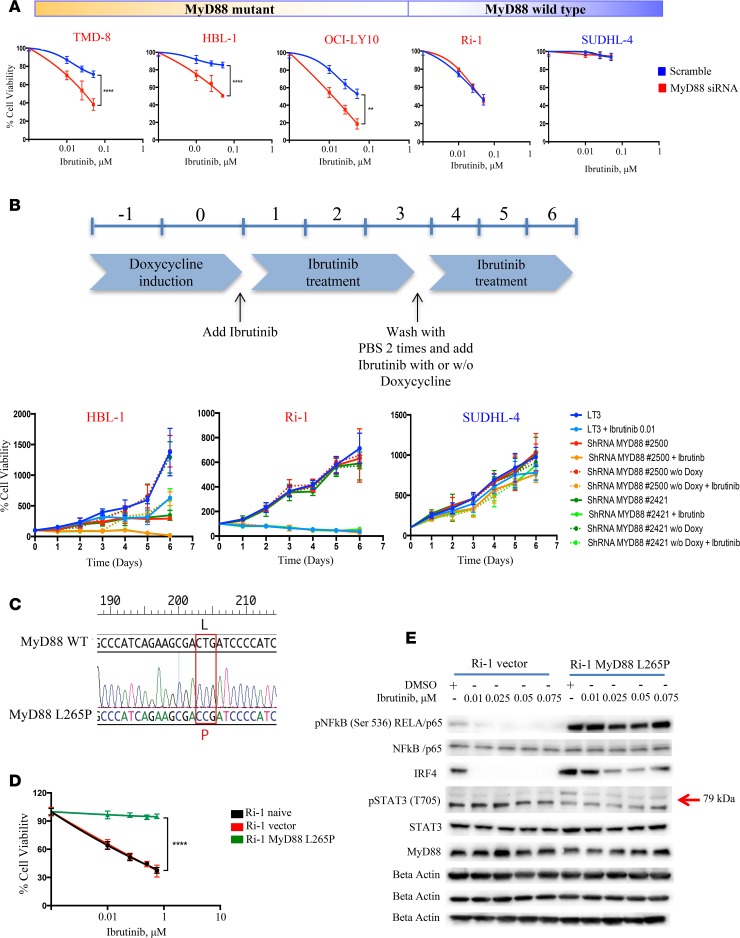
To evaluate the sensitivity of DLBCL cells to ibrutinib treatment, we treated a panel of 9 human lymphoma cell lines (6 ABC and 3 germinal center B cell-like [GCB]) with increasing drug concentrations (0.01–0.075 μM) for 24 hours. Ibrutinib showed lower efficacy in the ABC lymphoma cell lines with MyD88 L265P mutation compared with those with WT MyD88 and was not effective in the GCB cells (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.90196DS1). Knockdown of MyD88 using a small interfering RNA (siRNA) (Supplemental Figure 1B) enhanced ibrutinib efficacy in ABC cells bearing MyD88 L265P mutation, but did not enhance those with MyD88 WT or in the GCB subtype (Figure 1A). To further investigate the contribution of mutant MyD88 to ibrutinib resistance, we used tetracycline-inducible shRNA to knock down MyD88 in 2 ABC cell lines (HBL-1 and Ri-1, MyD88 mutant and WT, respectively) and 1 GCB (SUDHL-4) cell line (Supplemental Figure 1C). MyD88 knockdown enhanced ibrutinib efficacy in HBL-1 but not in Ri-1 or SUDHL-4. Notably, after the doxycyxline washout from the culture media, MyD88 expression rebounded and HBL-1 became less sensitive to ibrutinib (Figure 1B). Overexpression of the constitutively activated MyD88 mutant L265P in Ri-1 cells (Figure 1C and Supplemental Figure 2A) resulted in activation of the NF-κB pathway (Supplemental Figure 2B), enhanced proliferation and survival (Supplemental Figure 2, C and D), and decreased ibrutinib efficacy (Figure 1D). Ibrutinib treatment of the Ri-1 parent cell line was associated with profound inhibition of the NF-κB pathway, which was not observed in the Ri-1 cells that stably expressed MyD88 mutation (Figure 1E). Collectively, these data demonstrate that MyD88 L265P mutation enhances ABC DLBCL survival and may attenuate the antiproliferative activity of ibrutinib in DLBCL. Our data suggest that dual inhibition of BTK and MyD88 may provide more potent antitumor activity in ABC DLBCL harboring MyD88 mutation.
Figure 1. MyD88 confers resistance to ibrutinib in ABC DLBCL.
(A) Five DLBCL cells (3 ABC MyD88 mutant [TMD-8, HBL-1, and OCI-LY10], 1 ABC MyD88 WT [Ri-1] and 1 GCB [SUDHL-4]) were treated with 1 μM scramble, and MyD88 siRNA were incubated with increasing concentrations of ibrutinib (0.1, 0.25, 0.5 μM) and cell viability assessed by MTS assay after 24 hours. MyD88 siRNA + ibrutinib viability data were normalized to the effect of MyD88 siRNA alone. Error bars represent SEM of triplicate experiments. Differences between groups were calculated with the Student t test. **P < 0.005; ****P < 0.0001. (B) HBL-1, Ri-1 (ABC MyD88 mutant and WT, respectively) and SUDHL-4 (GCB) cells were infected with either tet inducible control shRNA or shRNA against MyD88, selected with puromycin, and induced with doxycycline for 2 days before treating with ibrutinib and seeding for MTS assay. After 3 days from the beginning of drug treatment, doxycyxline was washed out and ibrutinib added fresh either with or without fresh doxycycline. Error bars represent SEM of triplicate experiments. (C) Analyses of DNA sequences of a MyD88 WT and MyD88 L265P mutation, showing the replacement of reference sequence CTG with CCG. (D) Ri-1 naive cells transduced with either empty vector or activated MyD88 mutant L265P were incubated with increasing concentrations of ibrutinib (0.1, 0.25, 0.5, 0.75 μM) and cell viability assessed by MTS assay after 24 hours. MyD88 mutant + ibrutinib viability data were normalized to the effect of MyD88 mutant alone. Error bars represent SEM of triplicate experiments. Differences between groups were calculated with the Student t test. ****P < 0.0001. (E) Representative Western blot demonstrating inhibition of NF-κB pathway (pNF-κB p65, IRF4, pSTAT3 (T705]) after treatment with increasing concentrations of ibrutinib (0.1, 0.25, 0.5, 0.75 μM) for 24 hours in Ri-1 MyD88 WT but not in the Ri-1 stably expressing MyD88 mutation.
HDACis transcriptionally regulate MyD88 expression.
Previous studies demonstrated that bromodomain and extraterminal domain protein inhibitors (BETis) inhibited oncogenic NF-κB activity in ABC DLBCL by reducing IκB kinase (IKK) activity (10). We investigated whether BETi-induced inhibition of NF-κB activity is mediated by regulating MyD88 expression and whether other epigenetic modifying agents, such as HDACis, have a similar effect. HDACis and BETis reduced MyD88 protein abundance in the HBL1 cells irrespective of poly (ADP-ribose) polymerase (PARP) cleavage (Figure 2A). The decrease in MyD88 protein level was due to inhibition of MyD88 transcription, which was evident as early as at 3 hours of incubation (Figure 2B). Treatment with HDACis inhibited MyD88 transcription independent of the cell of origin or MyD88 mutation status (Figure 2C). This was also observed at the protein level, as determined by an ELISA assay (Figure 2D) or Western blotting (Figure 2E). To identify which HDAC isoform is responsible for downregulating MyD88, we knocked down each individual HDAC using 4 hairpins per gene. Multiple HDACs were found to be involved in regulating MyD88 expression, with the strongest MyD88 inhibition observed with HDAC1 and HDAC7 silencing (Supplemental Figure 3).
Figure 2. HDAC inhibitors transcriptionally decrease MyD88.
(A) Western blot showing downregulation of MyD88 in HBL-1 after treatment with 8 different HDAC inhibitors (SNDX-275, mocetinostat, romidepsin, SAHA, belinostat, and panobinostat) and 2 BET inhibitors (JQ1 and CPI-203) at their IC50 dose for 12 and 24 hours. Drug activity is demonstrated by increased acetylation of histone H3. Cell death is associated to PARP cleavage. (B) Change in relative mRNA levels of MyD88 over time in HBL-1 after treatment with 3 different HDAC inhibitors (panobinostat, romidepsin, and SAHA) at their IC50 dose (0.05, 0.01, and 3 μM, respectively) for the indicated time. Error bars represent SEM of triplicate experiments. (C) Change in relative mRNA levels of MyD88 over time in B cell lymphoma cell line panel (GCB n = 3 and ABC n = 4) after treatment with romidepsin 0.01 μM for 6 and 24 hours. Error bars represent SEM of triplicate experiments. (D) Box and whiskers plot showing decrease of MyD88 in B cell lymphoma cell line panel (GCB n = 3 and ABC n = 5) after treatment with either 0.05 μM panobinostat or romidepsin 0.01 μM for 6 hours. Error bars represent SEM of triplicate experiments. (E) Representative Western blot confirming decrease in MyD88 level after treatment with 0.01 μM romidepsin for 6 and 24 hours in our B cell lymphoma panel. (F) Relative MyD88 promoter-luciferase activity in 2 representative ABC (HBL-1 and OCI-LY-10) cell lines and 1 GCB (SUDHL-4) cell line. Cells were treated for 12 hours with indicated concentration of either panobinostat, romidepsin, or DMSO. Cells were incubated with 500 ng/ml of INF-γ as positive control for TLR activation. Error bars represent SEM of triplicates. Differences between groups were calculated with 2-way ANOVA with Bonferroni’s test. ***P < 0.0005; ****P < 0.0001.
HDACi transcriptionally regulates MyD88 expression by a STAT3-dependent mechanism.
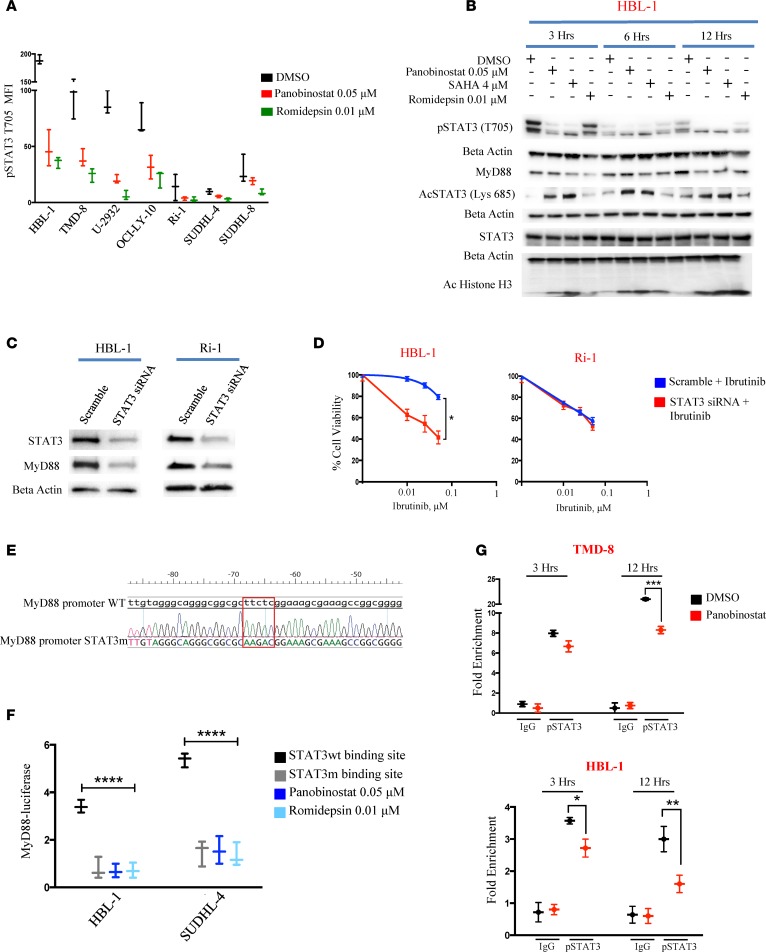
Using a luciferase-based promoter assay with the MyD88 promoter, we observed decreased luciferase activity in a time-dependent manner when ABC cell lines (HBL-1 and OCI-LY-10) and GCB (SUDHL-4) cell line were treated with HDACi (Figure 2F). MyD88 gene expression is regulated by multiple regulatory domains in the promoter region (11, 12). Accordingly, we examined the effect of HDACi on the expression of transcription factors that are known to interact with MyD88 promotor. HDACi-induced downregulation of MyD88 was associated with a significant decrease in STAT3 phosphorylation (tyrosine 705 [T705]) (Supplemental Figure 4A). ABC cell lines had a higher level of phosphorylated STAT3 at base line compared with GCB cell lines and were more prone to HDACi-induced STAT3 dephosphorylation (Figure 3A). In contrast, HDACi treatment had no effect on pSTAT1 (T701) (Supplemental Figure 4B). A concomitant decrease in pSTAT3 (T705) and MyD88 protein levels was observed in a time-dependent manner when the HBL-1 cells were treated with 3 different HDACi. On the contrary, an increase in STAT3 acetylation (lysine 685 [Lys 685]) was observed after HDACi treatment (Figure 3B and Supplemental Figure 4C). Because the acetylated Lys 685 is situated on the same SH2 domain where tyrosine residue 705 is located (13), our data indicate that HDACi-induced STAT3 lysine acetylation may interfere with T705 phosphorylation and consequently inhibit STAT3 activation (14).
Figure 3. STAT3 plays a central role in modulating MyD88 expression.
(A) Box and whiskers plot showing decrease of pSTAT3 (T705) in B cell lymphoma cell line panel (ABC n = 5 and GCB n = 2) after treatment with either 0.05 μM panobinostat or romidepsin 0.01 μM for 12 hours. Error bars represent SEM of triplicates. (B) Western blot showing the concomitant decrease of pSTAT3 (T705) and MyD88 and increase in acetylation of STAT3 (Lys 685) in HBL-1 cells after treatment with either panobinostat (0.05 μM), SAHA (4 μM), or romidepsin (0.01 μM) for 3, 6 and 12 hours. Duplicate samples run on parallel gels are shown. (C) Western blot showing the effects of STAT3 depletion by RNA interference on MyD88 levels in HBL-1 and Ri-1 cells. Both cells were treated with 2 μM scramble or STAT3 siRNA for 48 hours, and the effects on the expression levels of STAT3 and MyD88 were assessed by Western blotting. (D) HBL-1 and Ri-1 cells transfected with 2 μM scramble, and STAT3 siRNA were incubated with increasing concentrations of Ibrutinib (0.1, 0.25, 0.5 μM). Cell viability assessed by MTS assay after 48 hours. STAT3 siRNA + Ibrutinib viability data were normalized to the effect of STAT3 siRNA alone. Error bars represent SEM of triplicate experiments. Differences between groups were calculated with the Student t test. *P = 0.01. (E) Analyses of DNA sequences of a MyD88 promoter WT and MyD88 promoter with STAT3 binding site mutation, showing the replacement of reference sequence TTCTC with AAGAC. (F) Box and whiskers plot showing decrease in luciferase activity in the cells with STAT3 mutant binding site compared with the WT MyD88 promoter. No further decrease in luciferase activity is observed in the mutant cells treated with indicated concentration of either panobinostat or romidepsin for 12 hours. Error bars represent SEM of triplicates. Differences between groups were calculated with 2-way ANOVA with Bonferroni’s test. ****P < 0.0001. (G) TMD-8 and HBL-1 cells were treated with either panobinostat (0.05 μM) or DMSO, and ChIP was performed with pSTAT3 (T705) antibody or control IgG. Primers to amplify the STAT3-binding regions of the MyD88 promoter were used in qPCR to determine fold enrichment relative to a noncoding region. Error bars represent SEM of 3 independent experiments. ANOVA with Dunnett’s test was performed to compare STAT3 WT binding site versus the other conditions. *P < 0.05; **P < 0.005; ***P < 0.0005.
To confirm the role of STAT3 in regulating MyD88 expression, we silenced STAT3 gene expression using siRNA. STAT3 silencing was associated with a decrease in MyD88 protein levels (Figure 3C and Supplemental Figure 4, D and E) and mRNA levels (Supplemental Figure 4F). These changes were associated with an enhanced sensitivity of MyD88 mutant ABC cells to ibrutinib (Figure 3D and Supplemental Figure 4G). This effect was specific for MyD88 mutant cells, as knockdown of STAT3 did not increase the efficacy of ibrutinib in MyD88 WT ABC cells (Figure 3D) or in GCB cell lines (Supplemental Figure 3G). A second siRNA targeting construct for STAT3 was used in order to exclude a potential off-target effect and confirmed our data (Supplemental Figure 4, H and I). To further demonstrate the role of STAT3 in mediating an HDACi regulatory effect on MyD88 expression, we mutated the STAT3 binding site in the MyD88 promotor region (15) (Figure 3E). Cells with a STAT3 mutant binding site had a decrease in MyD88 luciferase activity compared with cells with a WT MyD88 promoter. No further decrease of luminescence was observed in the mutant cells treated with either panobinostat or romidepsin to the mutant cells (Figure 3F).
Using a ChIP qPCR assay, panobinostat treatment decreased pSTAT3 (T705) binding to the MyD88 promoter in a time-dependent manner (Figure 3G). In accordance to this, we also observed a marked reduction of the H3K4me3 binding that marks active transcriptional promoters at MyD88 promoter in response to panobinostat (Supplemental Figure 4F). Collectively, these data demonstrate a critical role for STAT3 in mediating HDACi regulation of MyD88 expression.
Panobinostat enhances ibrutinib activity in MyD88 mutant ABC DLBCL lines.
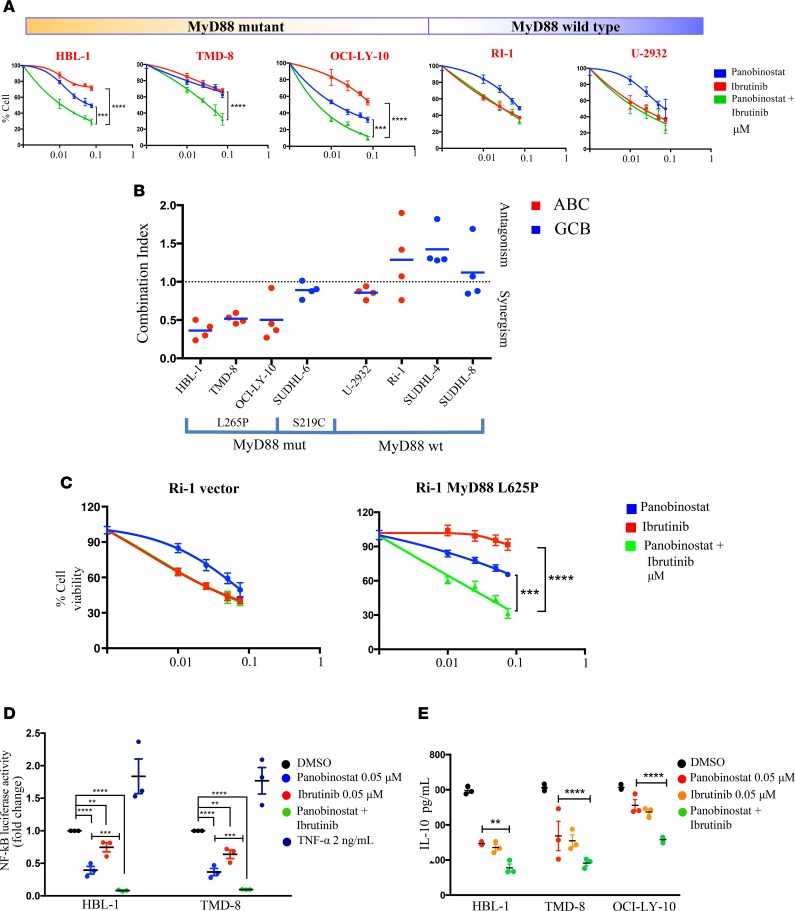
Next, we determined whether HDACi-mediated downregulation of MyD88 enhanced ibrutinib antilymphoma activity. We found that the combination of panobinostat and ibrutinib is synergistic in ABC DLBCL cell lines carrying MyD88 mutations but not in GCB cells or ABC DLBCL with MyD88 WT (Figure 4, A and B, and Supplemental Figure 5A). Moreover, this combination also demonstrated a synergistic antitumor effect in Ri-1 cells that were induced to stably express the MyD88 mutant L265P but did not demonstrate it in the WT (Figure 4C). Taken together, these data confirm the relevance of the MyD88 mutation status in predicting an attenuated response to ibrutinib therapy and support a combination strategy with HDACis.
Figure 4. Panobinostat synergizes with ibrutinib in killing ABC DLBCL harboring MyD88 mutation.
(A) MTS assay confirming enhanced antiproliferative effects of panobinostat in combination with ibrutinib in ABC cells harboring MyD88 mutation (HBL-1, TMD-8, and OCI-LY-10) but not in ABC cells with MyD88 WT (Ri-1 and U-2932). Cells were incubated with increasing concentrations of panobinostat (0.01, 0.025, 0.05, 0.075 μM) and ibrutinib (0.01, 0.025, 0.05, 0.075 μM), and viability was assessed after 24 hours. Error bars represent SEM of 3 independent experiments. Differences between groups were calculated with 2-way ANOVA with Bonferroni’s test. ***P < 0.0005; ****P < 0.0001. (B) Scatterplots summarizing the results of panobinostat + ibrutinib combinations in our cell line panel after 24 hours of incubation. Cells were incubated with increasing concentrations of panobinostat (0.01, 0.025, 0.05, and 0.075 μM) and ibrutinib (0.01, 0.025, 0.05, and 0.075 μM), and combination index was calculated according to the Chou-Talalay method. All the experiments were repeated 3 times. (C) MTS assay showing enhanced antiproliferative effects of panobinostat in combination with ibrutinib in Ri-1 stably expressing MyD88 mutation but not in Ri-1 with MyD88 WT. Cells were incubated with increasing concentrations of panobinostat (0.01, 0.025, 0.05, and 0.075 μM) and ibrutinib (0.01, 0.025, 0.05, and 0.075 μM), and viability was assessed after 24 hours. Error bars represent SEM of 3 independent experiments. Differences between groups were calculated with 2-way ANOVA with Bonferroni’s test. ***P < 0.0005; ****P < 0.0001. (D) Relative NF-κB-luciferase activity in 2 representative ABC (HBL-1 and TMD-8) cell lines. Cells were treated for 16 hours with indicated concentration of either panobinostat, ibrutinib, or the combination of the 2 drugs. Cells were incubated with 2 ng/ml of TNF-α as positive control for NF-κB activation. Error bars represent SEM of triplicates. Differences between groups were calculated with 2-way ANOVA with Bonferroni’s test. **P < 0.005; ***P < 0.0005; ****P < 0.0001. (E) IL-10 level change in 3 ABC DLBCL cell lines harboring MyD88 mutation (HBL-1, TMD-8, and OCI-LY-10). Cells were treated for 12 hours with indicated concentration of either panobinostat, ibrutinib, or the combination of the 2 drugs. Error bars represent SEM of triplicates. Differences between groups were calculated with 2-way ANOVA with Bonferroni’s test. **P < 0.005; ****P < 0.0001.
Given the convergence of BTK and MyD88 on NF-κB pathway, we investigated whether the combination of HDACis and ibrutinib is associated with an enhanced inhibition of NF-κB. As shown in Figure 4D, the combination of panobinostat and ibrutinib was more effective than either drug alone in decreasing NF-κB-luciferase activity. Similarly, the combination of panobinostat and ibrutinib was more effective than either drug alone in decreasing IKK-β and IRF4 levels in MyD88 mutant cells (Supplemental Figure 5B), which are associated with a stronger NF-κB and STAT3 activation status (Supplemental Figure 5, C and D). Because autocrine IL-6 and IL-10 production has been demonstrated to be regulated by JAK-STAT3 and MyD88 signaling in ABC DLBCL (6, 8, 16), we investigated the effect of panobinostat and ibrutinib on the level of these cytokines. The combination of the 2 drugs was more potent than either drug alone in decreasing IL-6 and IL-10 levels in the supernatants of 3 ABC DLBCL cell lines, which is consistent with an enhanced inhibitory effect on NF-κB signaling pathway (Figure 4E and Supplemental Figure 5E). Since IL-6 and IL-10 have been demonstrated to increase STAT3 phosphorylation (17), we evaluated the effect of these 2 cytokines on MyD88 transcription. HBL-1 cells were induced with 100 ng/ml of rIL-6 and rIL-10 for 30 minutes, 3 hours, and 6 hours. MyD88 mRNA expression increased in a time-dependent manner, with the maximal increase observed at 6 hours (Supplemental Figure 5F). Using a luciferase-based promoter assay with the MyD88 promoter, we confirmed the stimulatory effect of IL-6 and IL-10 on MyD88 in 2 representative ABC (HBL-1 and OCI-LY-10) cell lines and 1 GCB (SUDHL-4) cell line (Supplemental Figure 5G). Taken together, these data suggest the existence of an amplification loop that links MyD88 and JAK-STAT signaling through IL-6 and IL-10 (Figure 5).
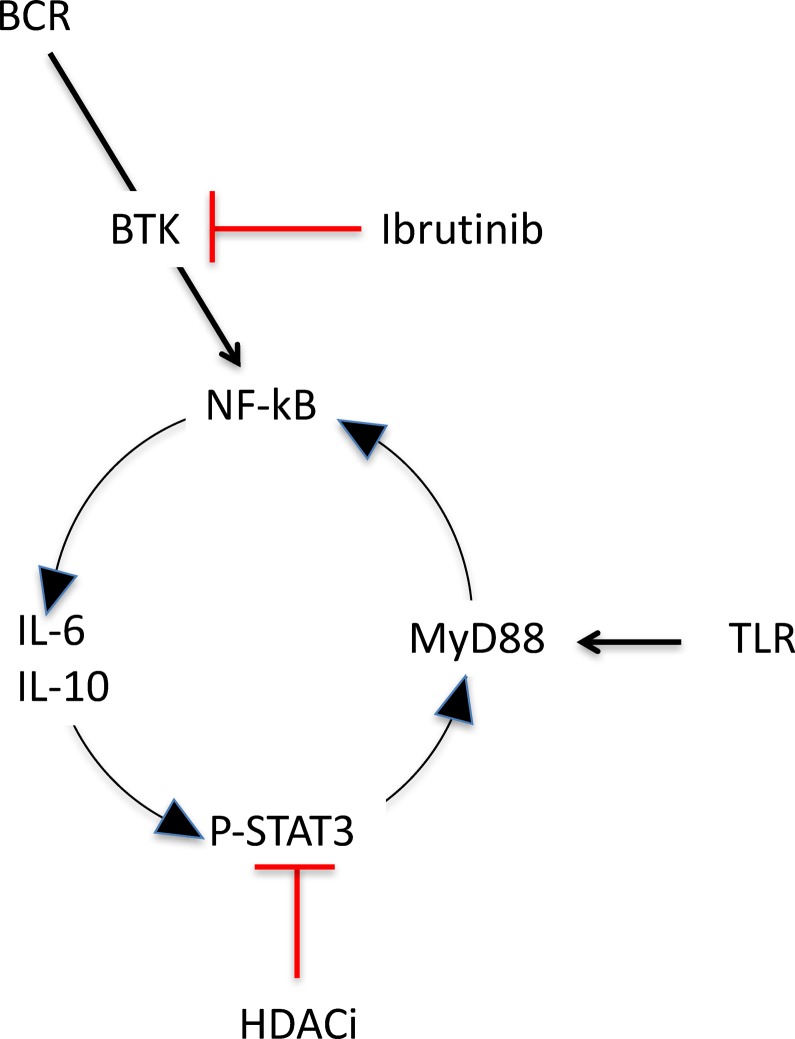
Figure 5. Model of BCR and TLR4 pathways in ABC DLBCL.
A Schematic representation of the B cell receptor (BCR) and TLR pathways. Following BCR engagement, BTK coordinates the activation of IκB kinase (IKK) and leads to NF-κB signaling activation. Similarly, during signaling, MyD88 adaptor proteins stabilize and act as initial platform for IRAK4 recruitment and subsequently IRAK2 recruitment, resulting in NF-κB activation. The downstream activation of STAT3 triggers an autocrine loop, which automantains the MyD88 transcription.
Efficacy of panobinostat plus ibrutinib in an ABC DLBCL xenograft model.
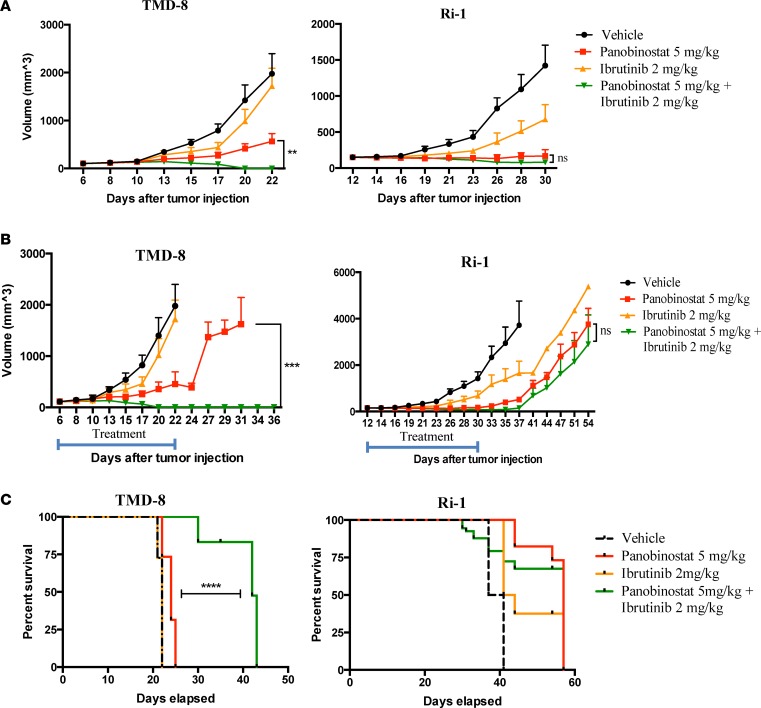
We next explored the synergy between panobinostat and ibrutinib in a xenograft model of ABC DLBCL. NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice implanted s.c. with TMD-8 (MyD88 mutant) or Ri-1 (MyD88 WT) cells were treated i.p. daily with either vehicle, panobinostat (5 mg/kg), ibrutinib (2mg/kg), or the 2 drugs together, 5 days weekly for 3 weeks. Consistent with the in vitro data, the drug combination demonstrated an enhanced therapeutic effect in ABC DLBCL with MyD88 mutation but not in MyD88 WT (Figure 6A). Both panobinostat and the combination of panobinostat and ibrutinib demonstrated antitumor efficacy. However, when the animals were observed after 3 weeks of therapy, mice with MyD88 mutant tumor that were treated with single agent panobinostat demonstrated rapid regrowth of their tumors, whereas no tumor relapse was observed in animals that were treated with the combination of panobinostat with ibrutinib (Figure 6B). Mice with WT MyD88 had a similar pattern of disease progression among different treatment groups (Figure 6B). Futhermore, the combination therapy improved survival of mice with MyD88 mutant lymphoma compared with mice with WT lymphoma (Figure 6C). Overall, panobinostat and ibrutinib were well tolerated, as therapy did not induce a significant weight loss compared with mice treated with each drug alone or vehicle (Supplemental Figure 6).
Figure 6. Panobinostat synergizes with ibrutinib in xenograft model of ABC DLBCL.
(A) NSG mice (n = 8 per treatment group) bearing TMD-8 (MyD88 mutant) and Ri-1 (MyD88 WT) tumors were treated i.p. daily with either vehicle, panobinostat (5 mg/kg), ibrutinib (2mg/kg), or the 2 drugs together, 5 times weekly. Tumor volumes were measured 3 times per week. Differences between groups were calculated with the ANOVA with Dunnett’s test. **P < 0.005. (B) NSG mice (n = 11 and 8 per treatment group, in TMD-8 and Ri-1 experiment, respectively) were treated i.p. daily with either vehicle, panobinostat (5 mg/kg), ibrutinib (2mg/kg), or the 2 drugs together, 5 times weekly for 3 weeks and observed until death after the end of the treatment. Differences between groups were calculated with ANOVA with Dunnett’s test. ***P < 0.0005. (C) Kaplan-Meier plot of the percent survival as a function of time from last drug administration. Data are from n = 11 and 8 for all groups in TMD8 and Ri-1 tumors, respectively. ****P < 0.0001.
We further confirmed these results using Ri-1 cells that were induced to stably express the MyD88 mutant L265P in a xenograft model. The combination of panobinostat and ibrutinib was more effective than either drug alone in the mutant but not in the WT tumor model (Supplemental Figure 7A). Furthermore, when treatment was stopped after 3 weeks, a sustained antitumor effect was observed in the mutant but not in the WT tumor model (Supplemental Figure 7B). Finally, the 2-drug combination also improved survival in the mutant but not in the WT tumor model (Supplemental Figure 7C).
Discussion
ABC DLBCL is a molecularly defined subgroup of DLBCL with a poor prognosis when treated with standard chemotherapy regimens (1–4). The recent discovery of recurrent genetic alterations in the BCR and TLR signaling pathways identified potential vulnerable mechanisms that can be explored therapeutically (5, 6). One such example is the use of ibrutinib, a selective BTK inhibitor, which showed promising clinical activity in patients with relapsed ABC DLBCL (9). In a trial that involved 80 patients with relapsed or refractory DLBCL, ibrutinib produced a 37% response rate of those with ABC DLBCL. However, most responses were partial and of a short duration (9). This prompted us to search for other vulnerable mechanisms that can be therapeutically explored in combination with ibrutinib. In this study, we focused our attention on the interaction between the BCR pathway and TLR pathway by exploring the potential role of targeting MyD88.
We demonstrated that genetic silencing of mutant MyD88 enhanced ibrutinib efficacy in ABC DLBCL. Conversely, ectopic expression of mutant L265P MyD88 gene resulted in ibrutinib resistance. Our data indicated that dual inhibition of BTK and MyD88 is more effective as an antiproliferative strategy against ABC DLBCL and should be explored therapeutically. Interestingly, ibrutinib as a single agent demonstrated a high efficacy in Waldenström’s Macroglobulinemia (WM), in which MyD88 mutation is highly prevalent (18–20). However, most responses were partial, raising the possibility that downregulation of MyD88 may have improved the quality of responses and treatment outcome of ibrutinib in this disease.
Our data demonstrated that HDACi can inhibit MyD88 transcription via a STAT3-dependent mechanism. This observation provided an opportunity to explore the efficacy of the combination of HDACi and ibrutinib. The advantage of this strategy is the ability to test this combination immediately, using commercially available and FDA-approved HDACi. Some of these HDACs have already been tested in patients with relapsed hematologic malignancies and demonstrated a modest clinical activity (21–24). The availability of clinical data from each drug alone will facilitate the design and interpretation of clinical trials using the 2-drug combination. Alternatively, one can explore combining ibrutinib with other investigational agents targeting downstream kinases of MyD88, such as IRAK4.
Given the fact that HDACi have broad biologic activities, we attempted to identify which HDAC isoform is responsible for downregulating MyD88. Our goal was to design a combination regimen that is mechanistically more selective and, therefore, potentially less toxic. Using a loss-of-function shRNA library screening, we found several HDACs to moderately downregulate MyD88, but only HDAC1 and HDAC7 robustly downregulated MyD88 in the 2 cell lines that were examined. However, within the confinement of our study, it seems that more than one HDAC is involved in regulating MyD88. As we showed in the in vivo animal studies, we are reassured that the combination of a pan-HDACi, such as panobinostat, with ibrutinib seems to be well tolerated. Whether more chronic administration of this combination is also tolerable will need to be determined in the context of a phase-I clinical trial.
In this study, we identified STAT3 as a transcription factor involved in regulating MyD88 expression upon HDACi treatment. Nevertheless, the mechanism underlying the HDAC-mediated regulation of STAT3 remains unclear. Previous studies suggested that HDACi treatment may increase STAT3 acetylation at Lys 685 while decreasing STAT3 phosphorylation at T705. These 2 posttranslational modifications may be mutually exclusive because both are located on the same SH2 domain; therefore, STAT3 lysine acetylation might prevent its tyrosine phosphorylation (13, 14). Our data also suggest that the decrease in STAT3 phosphorylation might also be secondary to the decrease in cytokine production, such as IL-6 and IL-10 (16, 17). Treatment with IL-6 or IL-10 increased STAT3 phosphorylation (17) and MyD88 transcriptional activity, supporting the presence of an amplification loop, which links MyD88 and JAK-STAT signaling through IL-6 and IL-10. Collectively, our study provides preclinical rationale for evaluating the combination of HDACis, such as panobinostat, with ibrutinib in ABC DLBCL.
Methods
Cell lines and reagents.
The human DLBCL-derived cell lines SUDHL-4, SUDHL-6, U-2932, and Ri-1 were obtained from the German Collection of Microorganisms and Cell Cultures, Department of Human and Animal Cell Cultures (Braunschweig, Germany); SUDHL-8 was obtained from ATCC. The DLBCL-derived cell lines (HBL-1, TMD-8) were provided by Richard E. Davis (Houston, Texas, USA). The cell lines OCI-LY-10 was provided by Ethel Cesarman (New York, New York, USA). All cell lines were grown to log phase at 37 °C, in the presence of 5% CO2 and cultured in RPMI 1640 medium supplemented with 10%–20% heat-inactivated FBS (Hyclone, GE Healthcare Life Sciences), 1% L-glutamine, and penicillin-streptomycin in a humid environment of 5% CO2 at 37°C. OCI-LY-10 was cultured in Hyclone Iscove’s Modified Dulbecco’s Medium (GE Healthcare Life Sciences) with 20% heat-inactivated FBS (Hyclone, GE Healthcare Life Sciences), 1% penicillin-streptomycin, 0.1% 2-Mercaptoethanol (GIBCO). All cell lines were authenticated by MSKCC Genomic Core Facility using a targeted deep sequencing assay of 585 cancer genes (HemePACT). Barcoded pools were sequenced on Illumina HiSeq 2,500 to 500–1,000× coverage per sample. Sequencing was compared with pooled normal tissue for control. We excluded mutations present in 2 databases of inherited variants (the SNP data base [dbSNP], https://www.ncbi.nlm.nih.gov/projects/SNP/, and 1000 genomes, http://www.internationalgenome.org/) and included mutations found in COSMIC. Mutations absent in the databases of inherited variant and COSMIC but found in our human lymphoma database were included. The following compounds were purchased from Selleckchem: panobinostat, SNDX-275, mocetinostat, romidepsin, SAHA, belinostat, CPI-203, ibrutinib, and S31-201. JQ1 was purchased from BPS Bioscience. Recombinant human IL-6 and IL-10 were purchased from R&D systems.
In vitro proliferation assay.
Cells were seeded in 96-well plates at 50,000 cell/100 μl/well with either vehicle (DMSO 0.1%) or increasing concentrations of drugs for 24–72 hours. Cell viability was assessed with the nonradioactive cell proliferation MTS assay, using CellTiter 96 AQueous One Solution Reagent (Promega). The MTS reagent was added to the culture medium at 1:5 dilution, according to manufacturer’s instructions. Procedures to determine the effects of certain conditions on cell proliferation and apoptosis were performed in 3 independent experiments. The 2-tailed Student t test and Wilcoxon Rank test were used to estimate the statistical significance of differences between results from the 3 experiments.
Western blotting.
Cells were pelleted by centrifugation, washed once with ice-cold PBS, and lysed on ice for 30 minutes using the Cell Signaling lysis buffer (no. 9803) according to manufacturer’s extraction protocol. Protein quantitation was done using the Direct Detect system (Millipore). A total of 30 μg of protein was denatured in Laemli buffer at 95ºC for 5 minutes, and Western immunoblotting was performed using the Bio-Rad system (TGX 4%–15% gels). Transfer was performed using the Trans Blot turbo system (Bio-Rad) into PVDF membranes. Images were acquired by using the Bio-Rad Imaging Chemidoc MP system. Secondary anti-rabbit and anti-mouse HRP–conjugated antibodies were purchased from Bio-Rad (catalogs 170-6515, 170-6516). Proteins were detected using the following antibodies purchased from Cell Signaling Technology: Acetyl-Histone H3 Lys9 (catalog 9649), Caspase3 (catalog 9668), PARP (catalog 9542), CARD11 (catalog 4440), MYD88 (catalog 4283), HDAC1 (catalog 2062), HDAC2 (catalog 5113), HDAC3 (catalog 2632), HDAC4 (catalog 7628), HDAC6 (catalog 7558), p-cJUN (catalog 2994), p-cFOS (catalog 5348), p-cATF2 (catalog 9221), pSTAT3 (catalog 9138), STAT3 (catalog 4904), IRF1 (catalog 8478), pSTAT1 (catalog 9167), STAT1 (catalog 9172), pNFKB p65 (catalog 3033), IκBa (catalog 4812), IKKb (catalog 2370), and GFP (catalog 2956). The following antibodies were purchased from Abcam: HDAC5 (catalog 1439), HDAC7 (catalog 166911), HDAC8 (catalog 187139), HDAC9 (catalog 18970), HDAC10 (catalog 108934), and HDAC11 (catalog 166907). β-Actin was from Sigma-Aldrich (catalog A5316).
PCR pathway arrays and qPCR.
Total RNA was extracted with the Qiagen RNeasy mini kit protocol. A total of 1 μg of RNA was converted to cDNA using iScript cDNA synthesis kit (Bio-Rad). Real-time PCR was performed using the model CFX96 (Bio-Rad). Primers for MyD88 and GAPDH were purchased from Bio-Rad: MyD88 (qHsaCED0046947), GAPDH (qHsaCED0038674).
SiRNA experiments.
siRNA transfections were performed by using the Amaxa 4D–Nucleofector Unit (Lonza). Briefly, 3 × 106 cells per condition were transfected with 1–2 μM siRNA or scramble, and resuspended in RPMI 10% FBS with no antibiotics. siRNAs were purchased from Invitrogen: STAT3 (catalog 4390824, clones s743 and s744), MyD88 (catalog 4390824, clone s9136), Negative Control 1 (catalog 4390643). HBL-1, TMD8, Ri-1, SUDHL-4, and OCI-LY-10 cells were transfected using the EN-150, EN-138, EN-150, DS-150, and DN-100 electroporation programs, respectively, using Solution F. Optimization was done using the Amaxa Cell Line Optimization 4D-Nucleofector X kit (catalog V4XC-9064). Transfection was performed using the 4D- Nucleofector X kit L (catalog V4XC-2024).
ELISA.
Standard ELISA was performed using the human myeloid differentiation factor 88 (Myd88) ELISA kit from MyBiosource (catalog MBS086853), STAT3 pT705 kit from Abcam (catalog ab126458), human IL-6 (catalog 550799) and human IL-10 kit (catalog 550613) from BD Biosciences, according to manufacturer’s instructions.
NF-κB Reporter Assays.
To generate the NF-κB transcriptional reporters, HBL and TMD8 cells were first transduced with a CMV Renilla luciferase lentivirus, to be used as internal control for normalization (QIAGEN). After hygromycin selection, cells were transduced with a NF-κB reporter lentivirus, where the expression of firefly luciferase is under control of a synthetic minimal cytomegalovirus promoter (mCMV) promoter and tandem repeats of the NFκB transcriptional response element (TRE) (QIAGEN). Puromycin was used for this second step of selection. For drug experiments, cells were counted and seeded in fresh media at 1 × 106 cells per ml before treatment for the indicated times. For shRNA experiments, cells were counted and seeded in fresh media at 1 × 106 cells per ml before induction for the indicated times. Luciferase activity was measured in 96-well plates by using the Dual-Luciferase Reporter Assay System (Promega) according to manufacturer’s instruction. Readings were performed on a Microtiter Plate Luminometer (PerkinElmer).
MyD88 Promoter Assays.
Gaussia Luciferase (GLuc) luciferase promoter was obtained from GeneCoepia with the MyD88 promoter cloned upstream of the GLuc enzyme. Normalization was done to secreted alkaline phosphatase (SEAP) (under the constitutively active SV40 promoter).
Cells were seeded at 1 × 106 cells per ml and treated with indicated drugs for the indicated times. Luminescence quantitation was assayed using the Secrete-Pair Dual Luminescence Assay Kit (GeneCoepia).
Mutation of the Stat binding site in the MyD88 promoter region were obtained based on previous publication (15). In particular, for the mutation of the Stat site, the reference sequence TTCTC was replaced by AAGAC. The mutant promoter construct was generated by site-directed mutagenesis using the Q5 Site Directed mutagenesis kit (New England Biolabs).
Mutation of MyD88.
The lentivirus vector encoding MyD88 harboring L265P mutation was used to stably transduce Ri-1 cell lines using HEK293T/Amphotericin cells, and then the cells were selected in 2 μg/ml puromycin for 7 days. Based on previous publication (25), the reference sequence CTG was replaced by CCG. The mutant promoter construct was generated by site-directed mutagenesis using the Q5 Site Directed mutagenesis kit (New England Biolabs)
shRNA library screening.
shRNA was expressed from a Tet-regulated lentivirus using the miR-E Vector LT3GEPIR, provided by Johannes Zuber (Research Institute of Molecular Pathology, Vienna, Austria) (26). In brief, this vector includes a Tet3G promoter, driving expression of EGFP with the 3′ UTR modified with an shRNA target sequence embedded in a miR-30 backbone. In addition, this vector constitutively expresses a puromycin resistance gene and the third-generation Tet-transactivator from the PGK promoter, coupled with an IRES sequence. miR-E shRNAs for HDAC1, -2, -3, -4, -5, -6, -7, and -9 and Myd88 (see sequences in Table 1) were designed by DSIR-Sensor rule (26) and cloned into the lentiviral expression vector LT3GEPIR. This method failed to generate effective shRNA knockdown of HDAC8, -10, and -11. For these genes, we used SPLASH (http://splashrna.mskcc.org), a machine learning approach to shRNA sequence prediction (Table 2).
Table 1. The RNAi sequences of shRNA generated using the DSIR-Sensor rule are as follows:
Table 2. The RNAi sequences of shRNA generated using the SPLASH design are as follows:
Lentiviral particles were generated using a second-generation system in HEK293FT cells in 6-well plates. The LT3GEPIR vector expressing the desired shRNA sequence was cotransfected with psPAX2, MD2.G, and pSuper expressing shRNA to DGCR8 at a ratio of 7:7:2:1, with a total of 19 ml DNA per well. A calcium phosphate transfection system was used to express the plasmids. Media was changed on the 6-well plates the morning following transfection, and media was collected from the cells at 48 and 72 hours following transfection. This media containing lentiviral particles was centrifuged at 3,000 g for 5 minutes to remove cell debris and filtered through a 0.45 µM filter. This filtered media was used without freezing to infect the target cell population using a spinfection protocol. Viral supernatant was added to 1 million cells in a 50 ml conical tube with polybrene (8 µg/ml) and centrifuged at 2,000 g for 2 hours. Media was then changed and the cells returned to normal growth conditions. Puromycin (1 µg/ml) was added 48 hours after infection. Doxycycline (2 µg/ml) was added following selection, and protein knockdown was measured at third day.
ChIP.
HBL-1 and TMD-8 cells were treated with 1% formaldehyde for 15 minutes at room temperature and quenched with ice-cold 125 nM glycine for 5 minutes at room temperature. Cells were lysed and sonicated on ice to yield 200–800 bp DNA fragments. Chromatin was incubated overnight at 4°C with 2 μg of specific antibodies (pSTAT3 [Y 705] and histone H3 [tri methyl K4], Abcam) or nonspecific IgG (Santa Cruz Biotechnology Inc.). Immunocomplexes were precipitated by incubation overnight with protein G-conjugated beads. Immunoprecipitates were washed and crosslinks were reversed by heating to 65°C for 6 hours and then treated with proteinase K for 1 hour at 55°C. Chromatin was purified using QiaQuick PCR clean-up columns (Qiagen). ChIP-qPCR was performing using the ChIP primers: Actin control, forward, 5′-AGACCTTCAACACCCCAGCC-3′, reverse, 5′- GTCACGCACGATTTCCCGCT-3′. MyD88 primers were purchased from QIAGEN: binding sequence GGCTTCCC, binding positions: chr3 38174114-38174121 (GPH1009200(-)06A); chr3 38169918-38169925 (GPH1009200(-)11A); chr3 38160234-38160241 (GPH1009200(-)20A).
Xenograft studies.
NSG mice (The Jackson Laboratory) were used for in vivo studies. Six-week-old female mice were injected s.c. with either 10 million TMD8 cells or 10 million Ri-1 cells together with matrigel. Once tumors reached an average volume of 100 mm3, mice were randomized to receive either vehicle control, Ibrutinib, panobinostat, or ibrutinib + panobinostat. Ibrutinib was dosed at 2 mg/kg, i.p. daily, 5 times weekly, while panobinostat was dosed at 5 mg/kg i.p. daily, 5 times weekly for up to 3 weeks. Mice were observed daily throughout the treatment period for signs of morbidity/mortality. Tumors were measured twice weekly using calipers, and volume was calculated using the formula: length × width2 × 0.52. Body weight was also assessed twice weekly. Upon treatment end, mice were monitored twice weekly for tumor regrowth and clinical signs.
Statistics.
The 2-tailed Student t test, the ANOVA test, and Wilcoxon Rank test were used to estimate the statistical significance of differences between results from the 3 experiments. The PRISM software was used for the statistical analyses. P < 0.05 were considered significant (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). Survival was estimated with the Kaplan-Meier survival curve method, and differences in survival were calculated by long-rank test (Graph Pad Prism 6.0).
Study approval.
The present xenograft studies were reviewed and approved by the Memorial Sloan-Kettering Cancer Center Institutional Animal Care and Use Committee.
Author Contributions
PM designed and performed the experiments and wrote the paper. AY designed the experiments and wrote the paper. EJB assisted in designing viral experiments. EDS performed in vivo studies. ET helped in performing ChIP experiments. AJC assisted in performing the mutational animal model. MF and RG designed the shRNA library. SV assisted in performing the statistical analysis. HGW, JB, and DAS assisted with data interpretation.
Supplementary Material
Acknowledgments
AY and this work were supported in part by the Vogelstein Fund for Lymphoma Research. EDS was supported by the Memorial Sloan Kettering Cancer Center Core grant U54 OD02035501. DAS was supported by NCI R01 CA 55349.
Version 1. 03/23/2017
Electronic publication
Version 2. 11/20/2018
The title was update.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2017;2(6):e90196. https://doi.org/10.1172/jci.insight.90196.
Contributor Information
Patrizia Mondello, Email: patriziamondello@hotmail.it.
Elisa De Stanchina, Email: destance@mskcc.org.
Eneda Toska, Email: toskae@mskcc.org.
Myles Fennell, Email: fennellm@mskcc.org.
Venkatraman Seshan, Email: seshanv@mskcc.org.
Ralph Garippa, Email: garippar@mskcc.org.
David A. Scheinberg, Email: d-scheinberg@ski.mskcc.org.
Hans-Guido Wendel, Email: wendelh@mskcc.org.
Anas Younes, Email: younesa@mskcc.org.
References
- 1.Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2001;194(12):1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lenz G, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci USA. 2008;105(36):13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenwald A, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(25):1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 4.Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci USA. 2003;100(17):9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis RE, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ngo VN, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470(7332):115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y, et al. Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer Cell. 2012;21(6):723–737. doi: 10.1016/j.ccr.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelly PN, et al. Selective interleukin-1 receptor-associated kinase 4 inhibitors for the treatment of autoimmune disorders and lymphoid malignancy. J Exp Med. 2015;212(13):2189–2201. doi: 10.1084/jem.20151074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson WH, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med. 2015;21(8):922–926. doi: 10.1038/nm.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ceribelli M, et al. Blockade of oncogenic IκB kinase activity in diffuse large B-cell lymphoma by bromodomain and extraterminal domain protein inhibitors. Proc Natl Acad Sci USA. 2014;111(31):11365–11370. doi: 10.1073/pnas.1411701111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Juilland M, et al. CARMA1- and MyD88-dependent activation of Jun/ATF-type AP-1 complexes is a hallmark of ABC diffuse large B-cell lymphomas. Blood. 2016;127(14):1780–1789. doi: 10.1182/blood-2015-07-655647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamawaki Y, Kimura H, Hosoi T, Ozawa K. MyD88 plays a key role in LPS-induced Stat3 activation in the hypothalamus. Am J Physiol Regul Integr Comp Physiol. 2010;298(2):R403–R410. doi: 10.1152/ajpregu.00395.2009. [DOI] [PubMed] [Google Scholar]
- 13.O’Shea JJ, Kanno Y, Chen X, Levy DE. Cell signaling. Stat acetylation--a key facet of cytokine signaling? Science. 2005;307(5707):217–218. doi: 10.1126/science.1108164. [DOI] [PubMed] [Google Scholar]
- 14.Gupta M, Han JJ, Stenson M, Wellik L, Witzig TE. Regulation of STAT3 by histone deacetylase-3 in diffuse large B-cell lymphoma: implications for therapy. Leukemia. 2012;26(6):1356–1364. doi: 10.1038/leu.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu M, Xu Y, Lin S, Zhang X, Xiang L, Yuan Z. Hepatitis B virus polymerase inhibits the interferon-inducible MyD88 promoter by blocking nuclear translocation of Stat1. J Gen Virol. 2007;88(Pt 12):3260–3269. doi: 10.1099/vir.0.82959-0. [DOI] [PubMed] [Google Scholar]
- 16.Lam LT, et al. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-{kappa}B pathways in subtypes of diffuse large B-cell lymphoma. Blood. 2008;111(7):3701–3713. doi: 10.1182/blood-2007-09-111948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta M, et al. Elevated serum IL-10 levels in diffuse large B-cell lymphoma: a mechanism of aberrant JAK2 activation. Blood. 2012;119(12):2844–2853. doi: 10.1182/blood-2011-10-388538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Treon SP, et al. MYD88 L265P somatic mutation in Waldenström’s macroglobulinemia. N Engl J Med. 2012;367(9):826–833. doi: 10.1056/NEJMoa1200710. [DOI] [PubMed] [Google Scholar]
- 19.Hunter ZR, et al. The genomic landscape of Waldenstrom macroglobulinemia is characterized by highly recurring MYD88 and WHIM-like CXCR4 mutations, and small somatic deletions associated with B-cell lymphomagenesis. Blood. 2014;123(11):1637–1646. doi: 10.1182/blood-2013-09-525808. [DOI] [PubMed] [Google Scholar]
- 20.Treon SP, et al. Ibrutinib in previously treated Waldenström’s macroglobulinemia. N Engl J Med. 2015;372(15):1430–1440. doi: 10.1056/NEJMoa1501548. [DOI] [PubMed] [Google Scholar]
- 21.Campbell P, Thomas CM. Belinostat for the treatment of relapsed or refractory peripheral T-cell lymphoma. J Oncol Pharm Pract. 2017;23(2):143–147. doi: 10.1177/1078155216634178. [DOI] [PubMed] [Google Scholar]
- 22.Olsen EA, et al. Phase IIb multicenter trial of vorinostat in patients with persistent, progressive, or treatment refractory cutaneous T-cell lymphoma. J Clin Oncol. 2007;25(21):3109–3115. doi: 10.1200/JCO.2006.10.2434. [DOI] [PubMed] [Google Scholar]
- 23.Piekarz RL, et al. Phase II multi-institutional trial of the histone deacetylase inhibitor romidepsin as monotherapy for patients with cutaneous T-cell lymphoma. J Clin Oncol. 2009;27(32):5410–5417. doi: 10.1200/JCO.2008.21.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.San-Miguel JF, et al. Panobinostat plus bortezomib and dexamethasone versus placebo plus bortezomib and dexamethasone in patients with relapsed or relapsed and refractory multiple myeloma: a multicentre, randomised, double-blind phase 3 trial. Lancet Oncol. 2014;15(11):1195–1206. doi: 10.1016/S1470-2045(14)70440-1. [DOI] [PubMed] [Google Scholar]
- 25.Poulain S, et al. MYD88 L265P mutation in Waldenstrom macroglobulinemia. Blood. 2013;121(22):4504–4511. doi: 10.1182/blood-2012-06-436329. [DOI] [PubMed] [Google Scholar]
- 26.Fellmann C, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5(6):1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.