Abstract
Chikungunya virus (CHIKV) is rapidly spreading across the globe, and millions are infected. Morbidity due to this virus is a serious threat to public health, but at present, there is no vaccine against this debilitating disease. We have recently developed a number of vaccine candidates, and here we have evaluated 3 of them in a nonhuman primate model. A single immunization with an attenuated strain of CHIKV (Δ5nsP3), a homologous prime-boost immunization with a DNA-launched RNA replicon encoding CHIKV envelope proteins (DREP-E), and a DREP-E prime followed by a recombinant modified vaccinia virus Ankara encoding CHIKV capsid and envelope (MVA-CE) boost all induced protection against WT CHIKV infection. The attenuated Δ5nsP3 virus proved to be safe and did not show any clinical signs typically associated with WT CHIKV infections such as fever, skin rash, lymphopenia, or joint swelling. These vaccines are based on an East/Central/South African strain of Indian Ocean lineage, but they also generated neutralizing antibodies against an isolate of the Asian genotype that now is rapidly spreading across the Americas. These results form the basis for clinical development of an efficacious CHIKV vaccine that generates both humoral and cellular immunity with long-term immunological memory.
Introduction
Chikungunya virus (CHIKV) is a mosquito-borne virus causing debilitating polyarthralgia in humans. Other signs of CHIKV infection include rapid onset of high fever, headache, skin rash, and myalgia. While most symptoms resolve within a week or two, polyarthralgia and polyarthritis can persist for months or years. CHIKV was isolated in the 1950s in Africa and was found to cause local epidemics the following decades in Africa and India (1, 2). CHIKV is typically transmitted by Aedes aegypti mosquitoes; however, coinciding with an adaptation enabling unusually efficient transmission by Aedes albopictus mosquitoes, the virus reemerged in 2004 and rapidly spread over Africa and Asia, as well as locally in Europe (3). More recently, CHIKV has spread across the Americas, with millions of people becoming infected. Morbidity due to this virus is a serious threat to global health and has now been listed as a priority pathogen by National Institutes of Allergy and Infectious Diseases (NIAID) in the US (1, 2).
CHIKV is an enveloped alphavirus of the family Togaviridae. It carries a positive single-stranded genomic RNA of 11.5 kb with 2 open reading frames that encode 4 nonstructural replicase proteins (nsP1-4) and a structural polyprotein consisting of capsid protein and the envelope proteins (C-E3-E2-6K-E1). The replicase complex serves two functions; it replicates the genomic RNA for incorporation into new virus particles and has transcriptase activity to produce an mRNA from a subgenomic promoter encoding the structural proteins (4). Mature virions bud from the plasma membrane of infected cells and carry 240 copies of E2-E1 glycoproteins arranged into 80 heterotrimeric spike complexes. The E2 protein is the receptor-binding moiety, whereas the E1 protein is involved in fusion of the virion envelope with the target cell membrane (5). Accordingly, it is the spike complex and, in particular, the E2 envelope protein that is the target for neutralizing antibodies.
At present, there is no treatment or vaccine against this CHIKV-induced disease (1, 2). Several approaches have been pursued in the quest for a CHIKV vaccine (6, 7), but in the absence of a licensed vaccine, there is an urgent need to evaluate a number of different candidates, assessing their individual merits. Moreover, given the nature of CHIKV epidemiology, a vaccine that works with only 1 or maximum 2 doses would be highly desirable. We recently developed a number of CHIKV vaccine candidates based on the La Reunion (LR-CHIKV) strain of East Central South African (ECSA) genotype. These were able to induce protective immunity against CHIKV infection in mice (8–11). Here, we have evaluated the safety, immunogenicity, and efficacy of 3 of those candidates in nonhuman primates. We used cynomolgus macaques (Macaca fascicularis) for which the immune and pathological responses to CHIKV are similar to those seen in human infections. These include persistent infection of joints, muscles, lymphoid organs, and liver, where the residual replicative virus mainly resides in macrophages (12, 13). Thus, this model lends itself well for our studies, and the results are relevant considering clinical development.
Results
Study design.
In this study ,we evaluated 3 different CHIKV vaccine candidates and regimens in nonhuman primates (Figure 1A). The first vaccine candidate, Δ5nsP3 (hereafter Δ5), is an attenuated virus in which a large portion (180 nucleotides) of the nsP3 replicase gene region has been deleted (9, 10). We have shown that this deletion is stable and does not revert (9), and results in this study have extended and corroborated these findings. The second vaccine candidate is a DNA-launched replicon vaccine, DREP-E (hereafter D) (10), a derivative of the CHIKV infectious cDNA clone from which the gene region encoding the CHIKV capsid protein (C) has been deleted. As this construct only expresses the envelope proteins (E3-E2-6K-E1) of the virus, CHIKV genomic RNA cannot be packaged and virions cannot form upon vaccination. The third candidate vaccine is a recombinant modified vaccinia Ankara virus, MVA-CE (hereafter M) that expresses the full-length CHIKV C-E3-E2-6K-E1 polyprotein (8, 10).
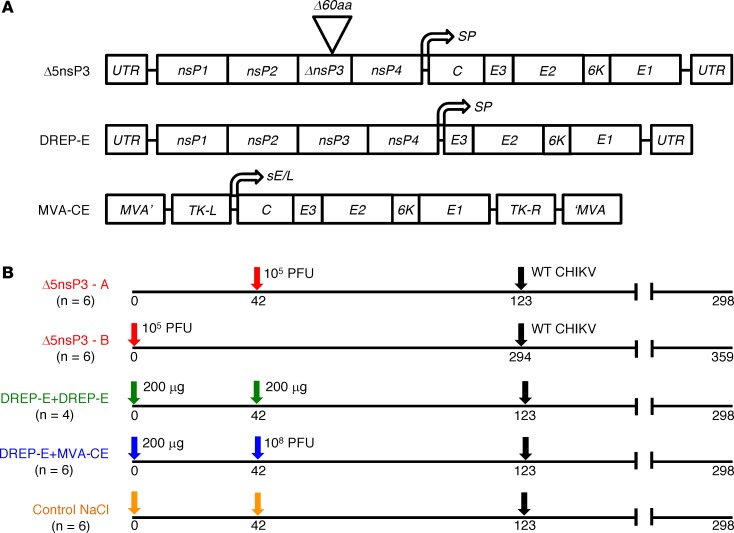
Figure 1. CHIKV vaccines and immunizations.
(A) Vaccine constructs are based on the LR-CHIKV strain LR-2006OPY1. Top, Δ5nsP3, (Δ5), an infectious attenuated virus carrying a 60 amino acid–long deletion in the nsP3 replicase protein (9). Middle, the DREP-E (D) replicon DNA vaccine (9). Bottom, the recombinant modified vaccinia virus Ankara MVA-CE (M) vaccine (8) has a cDNA copy of the CHIKV structural gene cassette (C-E3-E2-6K-E1) inserted into the MVA genome and is expressed under the control of the MVA sE/L promoter. (B) Immunization schedule of cynomolgus macaques. Group Δ5nsP3-A received one s.c. injection of 1 × 105 PFU of Δ5 virus on day 42 (red arrow). Group Δ5nsP3-B received one s.c. injection of 1 × 105 PFU of Δ5 virus on day 0 (red arrow). Group DREP-E received 200 μg of D by intradermal (i.d.) injection followed by electroporation (EP) on days 0 and 42 (green arrows). Group DREP-E+MVA-CE was primed on day 0 with 200 μg of D followed by an i.m. injection of 1 × 108 PFU of M on day 42 (blue arrows). Group 4 (controls) received 0.9% NaCl injections i.d. followed by EP on day 0 and s.c, i.m., and i.d. injections followed by EP on day 42 (orange arrows). All animals were challenged with 100 animal infectious doses 50% (AID50) WT LR-CHIKV on day 123 for groups 1A, 2, 3, and 4 and on day 294 for group 1B (black arrows). The study ended on day 298 for all groups except group 1B, which ended on day 359.
The 3 CHIKV vaccine candidates were used to immunize groups of cynomolgus macaques according to immunization schedules outlined in Figure 1B. Animals receiving the Δ5 virus were immunized only once on day 42 (group A), whereas the other animals were primed with D on day 0 and further boosted with either D (DD regimen) or M (DM regimen) on day 42. Our previous findings that a DM prime-boost gives very strong immune responses (10, 11, 14) prompted us to choose to evaluate this heterologous combination instead of testing M alone. A fourth control group of animals received saline only on days 0 and 42. On day 123 of the study (i.e., 11.5 weeks after last immunization), all animals were challenged with a high dose of WT LR-CHIKV. The immune and clinical responses were followed until day 298, the end of the study.
The CHIKV vaccines induce strong antibody responses.
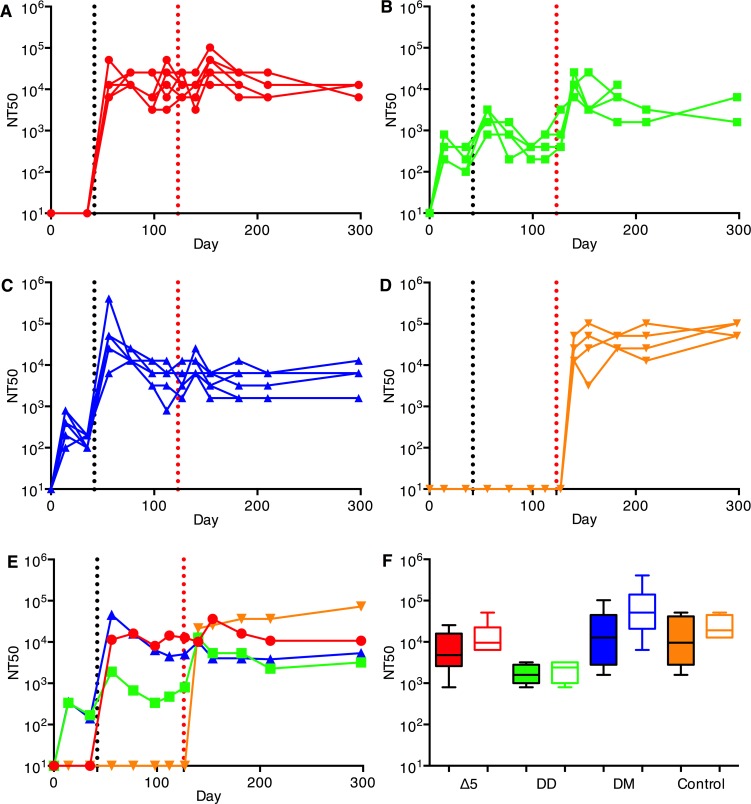
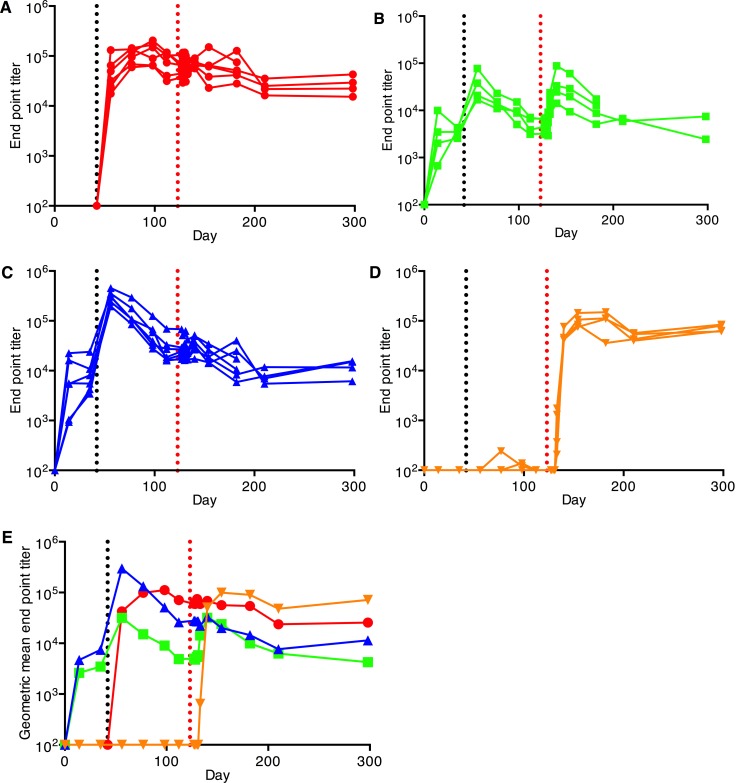
Immunization with a single dose of the attenuated Δ5 virus generated high titers of neutralizing and binding antibodies (Figure 2, A and E, and Figure 3, A and E, respectively). Antibody levels remained the same until the day of challenge (day 123). When D was used for priming, good levels of neutralizing and binding antibodies were also generated but were one order of magnitude below antibody levels obtained with Δ5 (P = 0.0001/P = 0.0005; neutralizing/binding). In addition, the antibody levels slowly declined over the weeks following the priming immunization. Antibody levels were further boosted with a second immunization of either D or M (Figure 2, B, C, and E; and Figure 3, B, C, and E, respectively). The heterologous DM combination was more potent than DD (P = 0.009/P < 0.05). At day 56, the antibody levels generated by Δ5 and DM did not differ, but those generated by DD were lower than the other 2 immunization regimens (P < 0.01/P < 0.05). For both prime-boost regimens, antibody levels declined slowly until challenge.
Figure 2. Neutralizing antibodies against CHIKV vaccine.
Animals were immunized on days 0 and/or 42 (y axis and black vertical dotted line). Animals were bled on days 0, 14, 34, 56, 77, 98, 112, 123, 127, 140, 154, 182, 210, and 298 (end of followup), and antibody levels in serum were determined. (A) Δ5 virus (red, 1×) corresponding to group Δ5-A (see Figure 1B), (B) DD (green), (C) DM (blue), (D) saline (orange). Animals were challenged on day 123 (red vertical dotted line). Panels A–D show antibody levels in individual animals. Panel E compares the geometric mean titers obtained for animals in panels A–D. Panel F shows NT titer of sera against Caribbean (CB, filled boxes) CHIKV isolate compared with response against East/Central/South African (ECSA) strain from day 56 (14 days after Δ5 or after D or M boost, respectively) for the vaccinated sera (panels A–C) or from day 140 (14 days after challenge with WT CHIKV) for the vaccine control animals (D). Sera from control animals were sampled after challenge with WT CHIKV. Statistical analyses were performed using the Kruskal-Wallis test followed by a 2-tailed Mann-Whitney U test to analyze differences between two.
Figure 3. Binding antibody responses to CHIKV vaccines as described in Figure 2 (A–D) determined by ELISA.
Animals were immunized on days 0 and/or 42 (y axis and black vertical dotted line). (A) Δ5 virus (red, 1×), (B) DD (green), (D) DM (blue), (D) saline (orange). Animals were challenged on day 123 (red vertical dotted line). Panels (A–D) show antibody levels in individual animals. Panel E compares the geometric mean titers obtained for animals in panels A–D.
After challenge with a high dose of WT CHIKV on day 123, animals that had been DD immunized exhibited a clear anamnestic neutralizing and binding antibody response (P = 0.028/P = 0.02, respectively) (Figure 2B and Figure 3B), whereas the antibody levels in the animals that had been immunized with the Δ5 or DM regimens showed no anamnestic response (P = 0.16/0.09 and P = 0.18/0.39, respectively) (Figure 2, A and C; and Figure 3, A and C). This indicates that the immune responses for the latter vaccine regimens were high enough to prohibit any significant replication of the challenge virus. Following a contraction period after the peak, binding and neutralizing antibody levels plateaued and remained high until the end of the study. Control unvaccinated animals that had received the CHIKV challenge developed strong neutralizing and binding antibody response that reached the same levels as those generated by the Δ5 and DM vaccine regimens (Figure 2D and Figure 3D).
The antibody responses induced by the CHIKV vaccines are cross-neutralizing.
The LR virus strain used in the vaccine constructs of this study is of ECSA genotype and belongs to the Indian Ocean lineage, while the virus now spreading in the Americas is of the Asian genotype. Thus, it was of interest to determine whether LR antibodies induced by our LR-CHIKV vaccine candidates would neutralize a Caribbean (CB) strain, which differs from LR-CHIKV by 35 amino acid residues in the envelope proteins. To perform the neutralization assays, virus replicon particles (VRPs) carrying the envelope proteins of the CB isolate were used. We found that all vaccine candidates generated neutralizing antibodies against the CB strain to levels similar to those generated against the WT LR-CHIKV virus in the challenge experiment (Figure 2F).
T cell responses induced by the CHIKV vaccines.
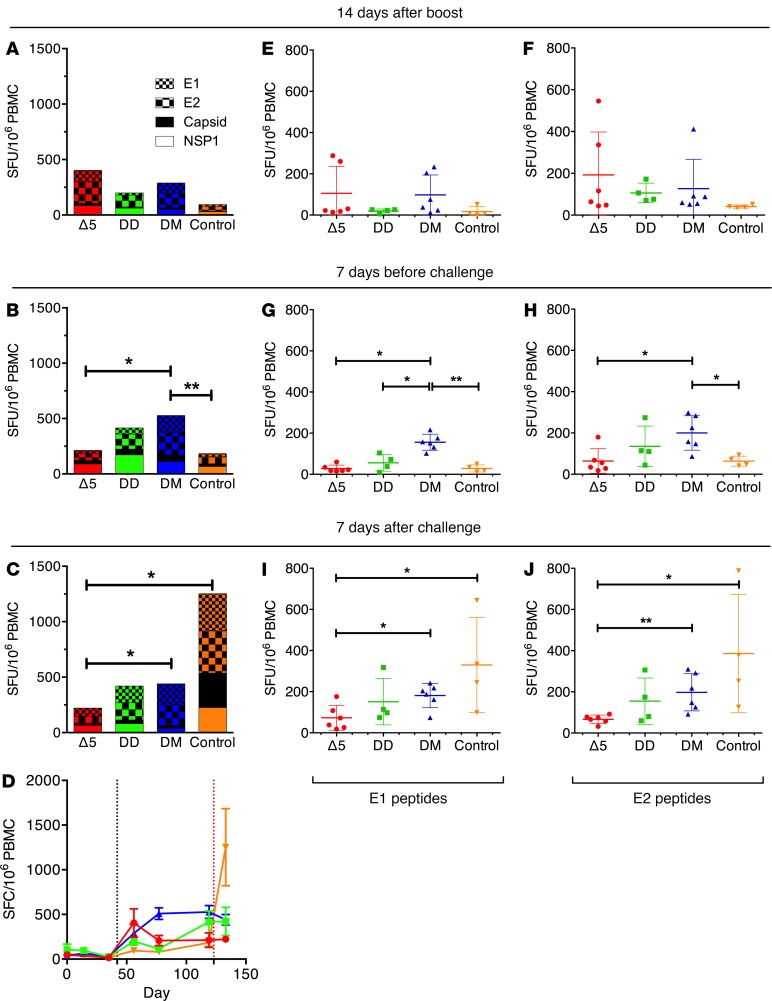
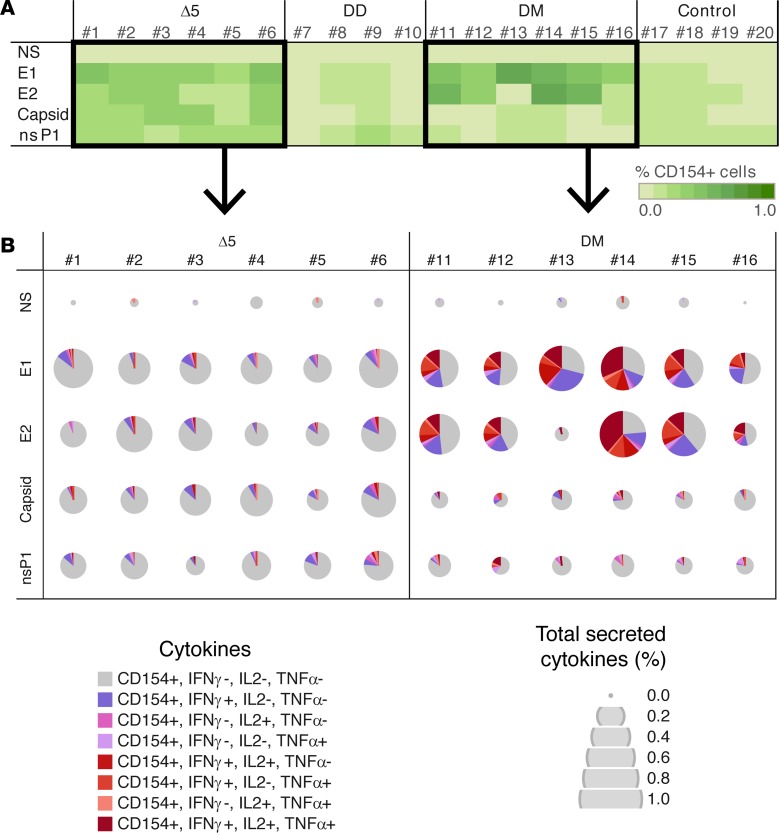
All vaccines generated significant IFN-γ T cell responses (Figure 4). The strongest responses were generated by the Δ5 and DM vaccination regimen. The T cell responses generated by the DD and DM regimens increased between booster responses and time of challenge, whereas the responses generated by the Δ5 vaccine slowly declined (Figure 4, A–C). After challenge, there was no clear expansion of T cells in any of the groups (Figure 4, C and D). Among the IFN-γ T cell responses, the envelope antigens E1 (Figure 4, E, G, and I) and E2 were mainly recognized (Figure 4, F, H, and J). Again initial envelope-specific T cell responses were highest in the Δ5 group, whereas levels were surpassed in particular by the DM group. As for the total T cell responses, we could not detect any significant anamnestic responses against E1 or E2 after challenge in any of the vaccine groups (Figure 4, I and J). T cell responses were largely CD4 restricted (data not shown), and the highest levels were obtained in the groups immunized with Δ5 and DM regimens. Activated CD154 (CD40L) T cells form a central part of the CD4+ follicular T helper cell population and are crucial for promoting B cell maturation and long-lived memory B cells. As antibody responses correlate with protection against CHIKV infection, the induction of this group of cells is of great importance. We therefore first sorted the activated CD4+-specific T cells by CD154 expression followed by detailed cytokine expression analysis (Figure 5). We found that the Δ5 and DM regimens were both potent in generating CD154+-activated T cells with strong polyfunctional responses induced by the DM regimen (Figure 5B). Overall, the observed T cell responses were consistent with the observed antibody responses.
Figure 4. CHIKV-specific T cell responses induced by CHIKV vaccine candidates.
Δ5, attenuated virus (n = 6) (red); DD, homologous prime-boost (n = 4) (green); DM, heterologous prime-boost (n = 6) (blue); Control, no vaccine but NaCl injection only (n = 4) (orange). Panels compare the sum of IFN-γ T cell responses from each group measured by FluoroSpot assays on PBMC collected at 14 days after boost (A), 7 days before challenge (B), and 7 days after challenge (C) and stimulated with envelope E1 and E2, Capsid, and nsP1 peptide pools. Statistical comparisons were performed at 14 days after boost, 7 days prechallenge, and 7 days after challenge. Panel D compares cumulated IFN-γ responses against all peptides for each group (mean ± SEM). Animals were immunized on days 0 and/or 42 (y axis and black vertical dotted line) and challenged on day 123 (red vertical dotted line). CHIKV-specific IFN-γ T cells responses against E1 (E, G, I) and E2 (F, H, J) peptide pools at 14 days after boost (E and F), 7 days before challenge (G and H), and 7 days after challenge (I and J). Panels show IFN-γ responses in individual animals with mean (± SEM). For statistical analyses, the Kruskal-Wallis test followed by the Mann-Whitney U test were used to identify differences between groups. Significance is represented by *P < 0.05 or **P < 0.01; 95% CI.
Figure 5. Activation of CD154+ CD4 T cells and CHIKV-specific cytokine secretion induced by different CHIKV vaccine candidates using ICS assay.
Δ5, attenuated virus (n = 6) (red); DD, homologous prime-boost (n = 4) (green); DM, heterologous prime-boost (n = 6) (blue); Control, no vaccine but NaCl injection only (n = 4) (orange). Antigens used for stimulation of T cells: medium only (NS), envelope E1, envelope E2, Capsid, and nsP1. (A) The percentage of CD154+ cells among the CD4 T cells (green scale with 10 steps from 0.0%–1.0%) from each animal in each group at 14 days after boost. (B) The quality and the quantity of the cytokine response (IFN-γ, IL-2, and TNF-α) by the CD154+ CD4 T cells in Δ5 and DM groups at 14 days after boost. Secreted cytokines are represented with color: no cytokine (gray), one secreted cytokine (range of pink), 2 or more secreted cytokines (range of red). Quantity of secreted cytokines is presented as percentage among the CD4 T cells and represented by a 5-size scale of pies (0.0%–1.0%).
All CHIKV vaccines generated protective immune responses.
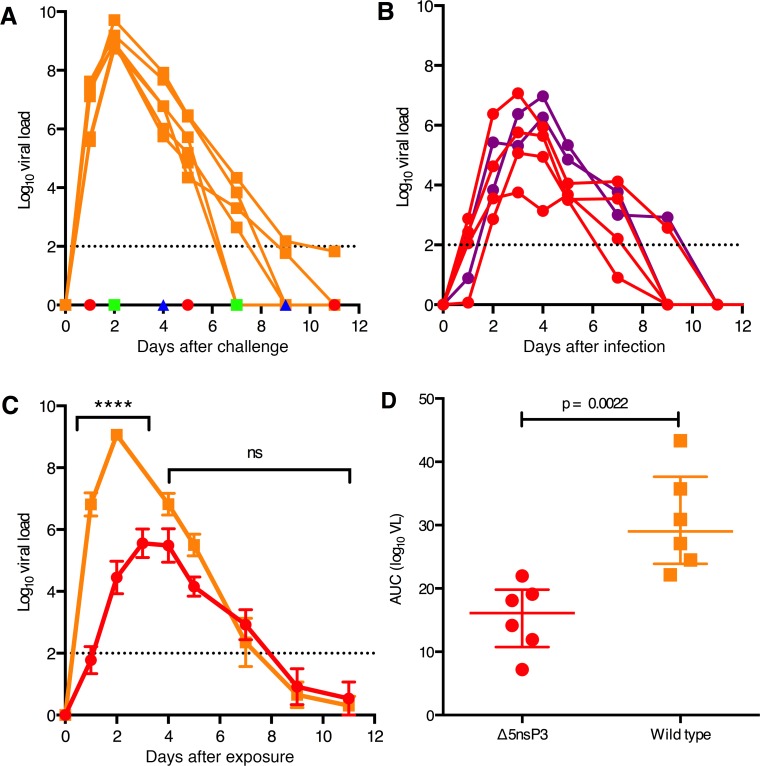
To evaluate whether the vaccinated animals in this study were protected from CHIKV infection, we determined the plasma viral load immediately after challenge with a dose of CHIKV close to 100 times the 50% Animal Infectious Dose (AID50). LR-CHIKV infection of cynomolgus macaques typically results in plasma viremia that peaks at day 2–3 after challenge (13, 15). Consistent with this, we found that all control animals that had received only saline displayed high peak viremia that declined, reaching undetectable levels by day 10 after challenge (Figure 6A). In contrast, despite the high virus dose used, none of the vaccinated animals showed any sign of viremia at any time point.
Figure 6. Viremia in macaques following challenge with WT LR-CHIKV (vaccine study) or infection with Δ5 virus (safety study).
(A) Plasma viremia of vaccinated animals after challenge. Vaccinated animals (groups Δ5, DD, DM) or control animals that had received saline only (group 4) were challenged with 100 50% animal infections dose (AID50) (about 7,000–10,000 PFU) of WT LR-CHIKV. Only the control group (NaCl, orange squares) displayed any viremia. The groups Δ5 (red circle), DD (green square), and DM (blue triangle) did not display any viremia. (B) Plasma viremia of naive animals infected with 1 × 105 PFU of the Δ5 virus (group Δ5-B in Figure 1). Animals shown in red (n = 4) were infected by inoculating the virus s.c. in the back, whereas animals inoculated s.c. in the wrist (SCw) (n = 2) are indicated in purple. (C) Comparison of mean viremia + SEM in LR-CHIKV (WT CHIKV) challenged control animals (from A) with mean viremia + SEM in Δ5 infected animals (from B). The viremia from WT and Δ5 differ significantly at the time point indicated (****P < 0.0001; 2-way ANOVA + Bonferroni’s multiple comparisons test). (D) Total viral production in LR-CHIKV challenged control animals and Δ5 infected animals. AUC were computed using trapezoid rules taking in account a baseline setup at the limit of quantitation of the qPCR we used (100 copies/ml = 2 in Log10). Statistics was done using Mann & Whitney U test.
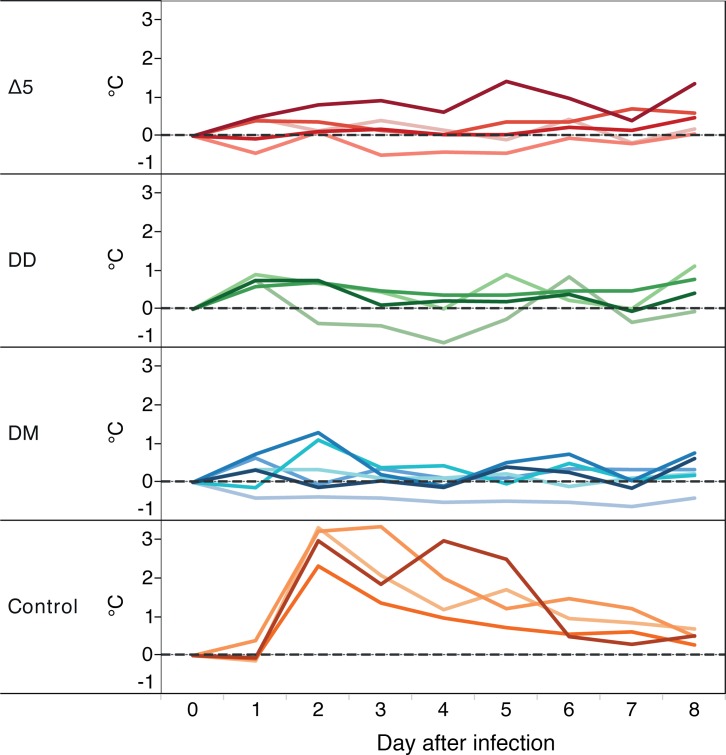
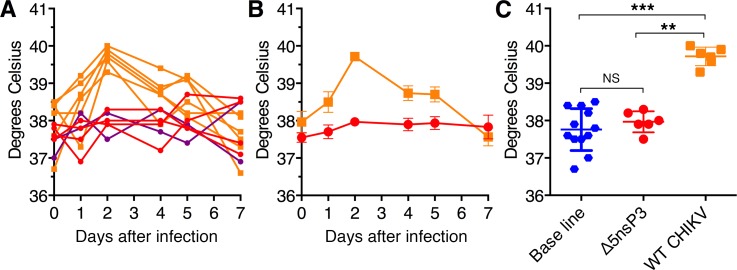
Typical for CHIKV infection of humans (16) and macaques (13) is an onset of high fever by day 2 after infection, which resolves usually in a week’s time. This was also clearly the case in the control animals that had received the challenge WT virus (Figure 7). In contrast, none of the vaccinated animals displayed any sign of fever at any time.
Figure 7. Vaccines protect against Chikungunya-associated fever.
Variation of body temperature compared with baseline (gray horizontal dotted line) from cynomolgus macaques immunized with the different CHIKV vaccine candidates and challenged at day 0. Animals were immunized with Δ5 virus (red), DD (green), or DM (blue) or injected with saline (i.v. route; orange). Temperature measured at midnight each day is plotted during the first 8 days following the challenge. Temperature at day 0 is the reference temperature and calculated with the mean of day –6 to day 0 (7 measures) from temperature of each animal at midnight (black horizontal dotted line).
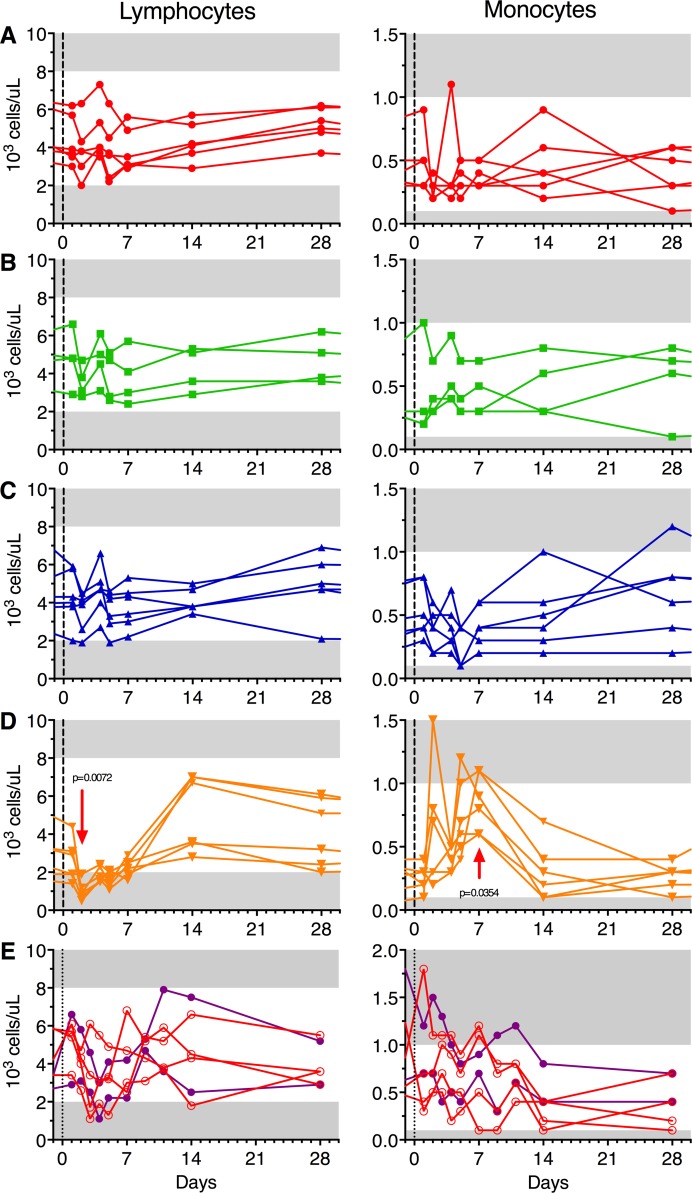
However, another way to characterize CHIKV infection is by hematological parameters (13). The control animals that received the challenge virus displayed transient lymphopenia and increased levels of monocytes, while the vaccinated animals had no such manifestations (Figure 8, A–D). Collectively, these criteria underscore that all vaccine candidates protected the animals from CHIKV infection and disease.
Figure 8. Vaccines protect against WT Chikungunya–induced lymphopenia and monocytosis.
White blood cell subset absolute counts at each bleeding point after challenge. Animals were immunized with Δ5 virus (A; red), DD (B; green), DM (C; blue), or saline (D; orange) and challenged at day 0 (black vertical dotted line corresponding to day 123 in the Figure 1 schedule). Panel E shows animals that were infected with Δ5 virus at day 0 either s.c. (clear red circles; n = 4) or s.c. in the wrist (SCw, filled purple dots; n = 2). Horizontal gray area on panels represent the values out of normal range (lymphocytes normal range: 2 × 103 to 8 × 103 cells/μl; monocytes normal range: 0.1 × 103 to 1 × 103 cells/μl). Statistics by Friedman + Dunn’s post test for multiple comparison of time point values to baseline.
Characterization of the Δ5 CHIKV vaccine candidate.
Given the promising results with the Δ5 vaccine candidate, it was important to further assess its level of attenuation. We have shown that the deletion mutation in nsP3 causing the attenuation is stable and does not revert. However, it was important to assess to what extent the virus is attenuated. Accordingly, we infected a separate group of 6 naive cynomolgus macaques (Figure 1; Δ5nsP3-B) with a dose of the Δ5 virus corresponding to that used for vaccination (1 × 105 plaque-forming units, PFU). The s.c. route of injection in the upper back (s.c., n = 4) was chosen to mimic the route used for vaccination and challenge, while the route of inoculation s.c. at the back of the wrist (i.w., n = 2) was used to allow monitoring of possible localized joint swelling as it was done in mouse models (8–10). Within the first days after infection, viremia could readily be detected in all animals (Figure 6B). However, plasma virus titers were significantly lower (approx. 4 orders of magnitude) as compared with those obtained after WT LR-CHIKV infections (P = 0.009) (Figure 6, A–D). Furthermore, the peak titers were delayed by 1–2 days for the Δ5 virus. There was no fever in the group infected by the Δ5 virus, whereas the group infected by WT virus typically had a fever peak around day 2 (Figure 9, A–C). Furthermore, the Δ5-infected group did not demonstrate any lymphopenia or increase in monocyte counts, whereas the group infected with WT virus showed clear and typical hematological effects (Figure 8, D and E).
Figure 9. Attenuated virus (Δ5) vaccine does not induce fever.
Rectal body temperature were recorded each morning at time of bleeding and clinical examination. (A and B) Day-to-day followup of macaques infected with the Δ5 virus (red and purple lines, dots) and of control animals infected with LR-CHIKV WT strain (orange lines, squares). Temperature at day 0 is the reference temperature and calculated with the mean of day –6 to day 0 (2 measures) from temperature of each animal. (C) Comparison of rectal temperature increase between day 0 and day 2 after infection. (**P < 0.01; ***P < 0.001; Kruskal-Wallis followed by Mann & Whitney U rank test).
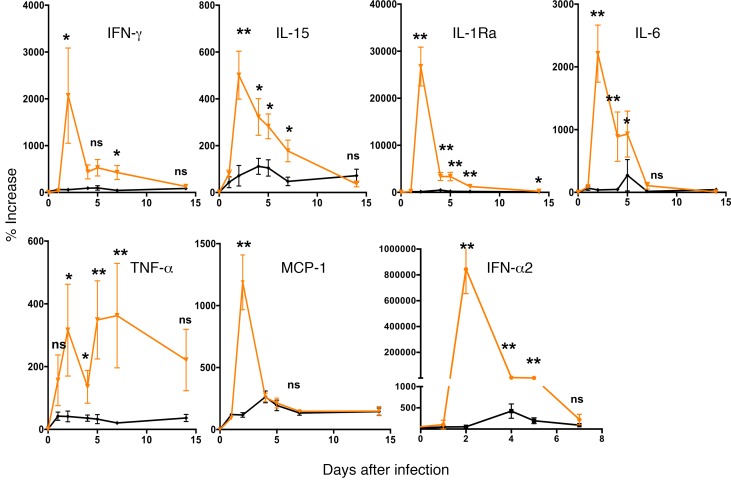
In CHIKV infection of both humans or macaques, viral load and severity of disease are strongly correlated to the upregulation of a number of cytokines such as IFNs, IL-6, IL-1Ra, TNF-α, and MCP-1 (12, 13, 17–24). We therefore analyzed cytokine upregulation during infection with WT CHIKV or with Δ5 virus. We scored for a large number of cytokines and found 7 cytokines that were particularly upregulated in the WT CHIKV-infected animals (Figure 10). In contrast, these cytokines were not upregulated when macaques were infected with the Δ5 virus, further underscoring the differences between these two CHIKV variants. We also represented the cytokine profiles of animals that had been vaccinated with the three different vaccine candidates and subsequently challenged with WT CHIKV (Figure 11). This analysis showed that indeed the vaccines resulted in protection from CHIKV infections, as the cytokines typically upregulated by WT CHIKV, such as IFN-α2, IL-1Ra, IL-6, and MCP-1, were almost exclusively expressed by animals infected by the WT virus (G4) (Figure 11). In cases where upregulation of cytokines was observed in vaccinated animals that had received a challenge with WT CHIKV (G1/Δ5, G2/DD, or G3/DM), the cytokines were partly different, such as IL-2 and IL-4, and can be attributed to typical memory immune responses.
Figure 10. Profile of cytokines associated with infection and inflammation induced after infection of naive animals with WT CHIKV (orange curves) or with attenuated Δ5 virus (black curves).
Animals were inoculated with virus on day 0. Statistical analysis using Kruskall-Wallis test (P < 0.0001) for all except IFN-γ P < 0.01, then Mann & Whitney U test to compare each days between WT and attenuated virus groups. **P < 0.01; *P < 0.05.
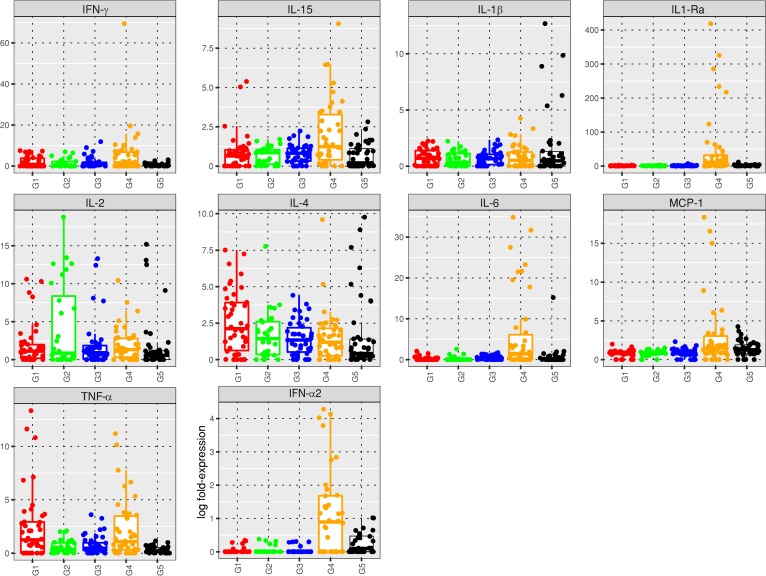
Figure 11. Box plot and plot-jittering representations of the cytokine levels reached in plasma of macaques within 1 week after exposure to WT CHIKV after vaccination (G1 to G4) or Δ5 attenuated virus (G5, safety study).
Animals were inoculated with virus on day 0, which corresponded to day 123 of immunization schedule for groups G1 to G4. G1 = 1× Δ5 virus (red), G2 = DD (green), G3 = DM (blue), G4 = control saline (orange), G5 = safety study (black). For each cytokine, the box plots show the first and third quartiles, together with the medians of the cytokine expressions.
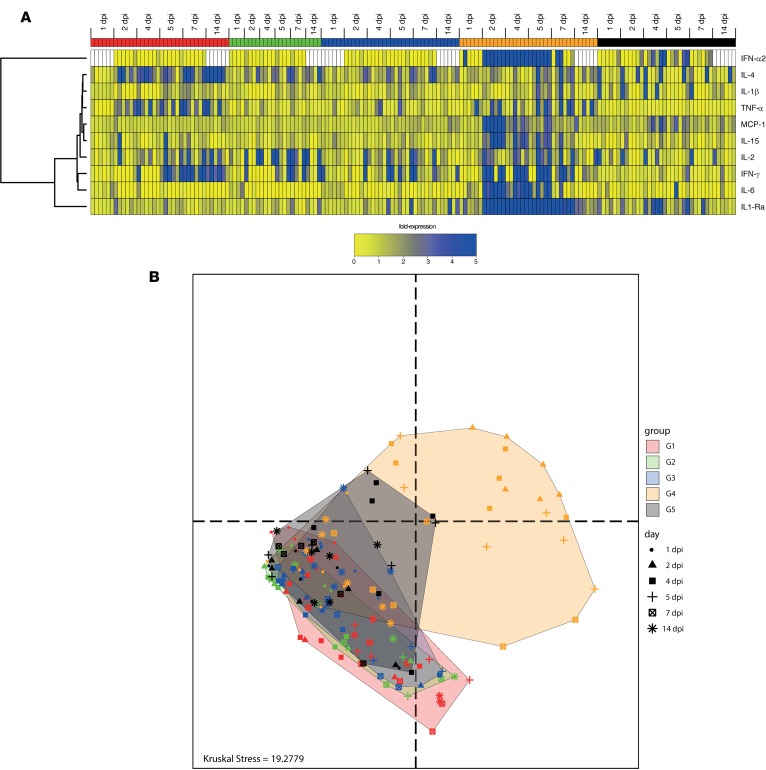
In order to have a kinetic resolution of the cytokine expressions in the whole dataset, we generated a heatmap restricted to 10 of the most relevant cytokines (Figure 12A). A hierarchical clustering was performed in order to classify the cytokines based on their expression profiles. WT infected animals (G4, orange) showed an earlier, higher, and persistent expression of cytokines compared with those infected by the attenuated virus (G5, black). As expected IFN-α2 was highly induced (66 ng/ml) in the animals infected with the WT virus but within a very short window from 2 days to 5 days postinfection (dpi). IL-1RA was the most abundant cytokine and was detected at all time points. IL-6, IFN-γ, IL-15, and MCP-1 were also highly expressed in animals infected by the WT virus, mainly at 2 dpi with a decreasing expression from 4–14 dpi. IL-4 and IL-1β were expressed in WT virus–infected animals starting 5 dpi. In contrast, only scarce expression of IFN-α2 was detected in animals inoculated with the attenuated Δ5 virus vaccine (G5) at 4 dpi or 5 dpi, at levels (0.13 ng/ml) that can be seen in some animals at baseline (data not shown). Animals exposed to the attenuated virus (G5) showed a remarkable delayed and shorter expression of IL-1RA between 4 and 7 dpi. Animals vaccinated with the Δ5 attenuated virus (G1), DD (G2), and DM (G3) showed distinct cytokine patterns compared with G4 and G5, with no expression of inflammatory cytokines, such as IL-1RA, IL-6, MCP-1, and IL-15 (Figure 12A). Animals in all of these groups (G1–G3) expressed cytokines associated with recall immune response (IL-4, IL-2) and antiviral responses (IFN-γ). This suggests a concomitant activation of humoral and cellular responses against the viral infection after challenge. Interestingly, animals vaccinated with attenuated virus (G5) also showed high expression of TNF-α, suggesting macrophage activation. This macrophage activation is independent of MCP-1 activation, which is upregulated in naive animals infected by the WT virus (G4) but not in any of the vaccine groups (G1-3).
Figure 12. Cytokine levels and profiles in macaques.
(A) Heatmap of cytokine levels in plasma of macaque within 1 week after exposition to WT CHIKV after vaccination (G1 to G4) or infection of naive animals with Δ5 virus (G5, safety study). Animals were inoculated with virus on day 0, which corresponded to day 123 in Figure 1 for groups 1–4. Δ5 virus (1×, red), DD (green), DM (blue), control saline (orange), Δ5 virus in the safety study in (black). Missing values are indicated by white boxes. Hierarchical clustering of cytokines and samples were created based on the Euclidean distance and using the complete linkage method. (B) Multidimensional scaling (MDS) representation of samples based on their cytokine profiles from day 1 to day 14 after exposition to the virus (data from A). Each dot represents the cytokine profile of a sample, plotted in the space of cytokine expressions. The distances between the dots are proportional the Euclidian distances computed between the samples, restricted to the cytokines as in A. Each group is delineated by a convex hull (i.e., the smallest convex set containing the points). The Kruskal Stress indicated at the bottom of the representation quantifies the quality of the representation as the percentage of information lost in the dimensionality reduction process.
We also performed a multidimensional scaling (MDS) representation of animals based on their cytokines profiles (Figure 12B). This dimensional reduction method allows visualization of the similarities (and dissimilarities) between high-dimensional samples in a 2-dimensional space. In such a representation, the distance between the dots are proportional to the Euclidian distances computed between the animals based on their cytokine profiles. All animals at 1 dpi were grouped in the same location. From this location, the cytokine burst of the animals from G4 (i.e., infected with the WT virus) was visible, especially from 2–5 dpi. Animals vaccinated and challenged, but protected from infection (i.e., G1, G2, and G3), moved shorter distances compared with G4 and in a different direction within the MDS space. Finally, animals from G5 (i.e., infected with the Δ5 attenuated virus) occupied a space more similar to protected animals compared with infected animals. These findings underscore that the WT and Δ5 virus have very different infection and inflammation properties.
Clinical assessment in the safety study.
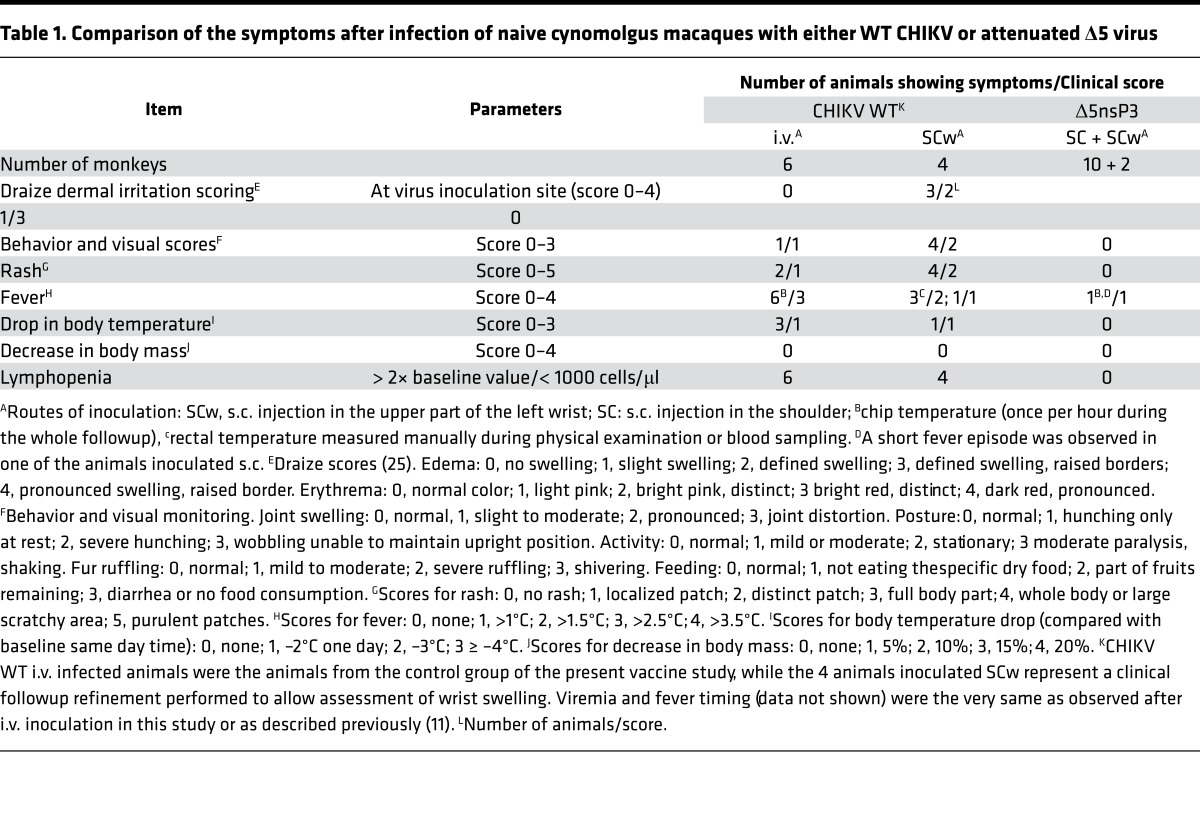
To assess local effect of infection/vaccination that was performed, we added a group of macaques that were infected with WT CHIKV by s.c. inoculation in the wrist (SCw). This allowed us to observe regularly local clinical signs occurring in the very same area used for the injection. Previous work (ref. 13 and data not shown) indicated that the viral replication kinetics is the same, whatever route of WT inoculation was used. After inoculation with the WT and Δ5, CHIKV animals were examined for the first 6 hours for behavioral and clinical signs and further during 2 weeks at times of sampling (once a day the first 5 days, then every 2 days). Following the Draize dermal irritation score (25), the SCw animals of the WT virus could be classified as level 2–3 immediately after vaccine inoculation due to edema with defined swelling with distinct or raised boarders and by distinct bright pin or red erythema (Table 1). In contrast, the Δ5 virus did not result in any such clinical signs. For behavioral and visual scores, the WT virus clearly induced level 1–2 signs including hunching, decreased activity, fur ruffling, decreased food intake, and joint swelling (day 7–10 after inoculation). Again, the Δ5 virus did not result in these clinical signs. A similar picture was seen for rash, fever, and drop in body temperature where the WT virus, irrespective of inoculation route, resulted in clinical abnormalities, whereas the Δ5 virus did not. Lymphopenia (Figure 8) was also evident in the WT-infected animals but not in Δ5-infected animals.
Table 1. Comparison of the symptoms after infection of naive cynomolgus macaques with either WT CHIKV or attenuated Δ5 virus.
Antibody responses in the safety study.
The safety study differed from the vaccine study in that the time between infection and challenge was much longer than in the vaccine study. Thus, we were able to assess whether neutralizing and binding antibody responses against Δ5 waned over time after initial induction, an important aspect to evaluate for the attenuated vaccine candidate. Both neutralizing and binding antibody levels were induced to the same levels as found in the vaccine study (Figure 13). In neither case had the antibody responses decreased by day 294, when the animals were challenged with WT CHIKV. Upon challenge, there was no anamnestic response indicating that the initial Δ5 virus infection had resulted in protective immune responses. Antibody levels in the control animals reached the same level as in the control group in the vaccine study (Figure 2D and Figure 3D) and reached the same levels as those induced by the Δ5 virus (Figure 10).
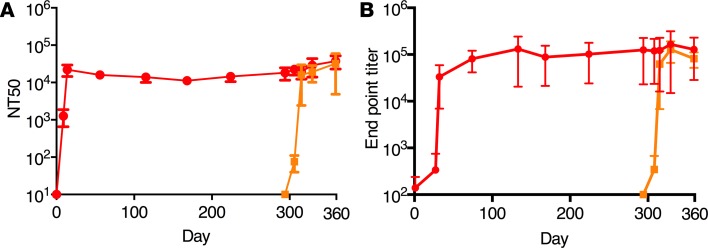
Figure 13. Antibody levels in the safety study.
CHIKV specific neutralizing (A) and binding (B) antibodies in sera of cynomolgus macaques infected with CHIKV. Animals were infected on day 0 with Δ5 virus (n = 6, red curve) or challenged with WT CHIKV on day 294 (n = 2, orange curve).
Discussion
In this study, we evaluated 3 very different CHIV vaccine candidates in nonhuman primates. Utilizing a truly validated animal model mimicking CHIKV infection in humans, we were able to address how these three different vaccine modalities compare head-to-head in terms of immunogenicity and protective efficacy. Previous studies of CHIKV vaccine candidates in smaller animals (mice) have shown that a large number of CHIKV vaccine candidates are able to provide protective immunity (6, 26–28). Thus, it was of importance to assess the individual merits of new CHIKV vaccine platforms in a more stringent animal model that more closely resembles a clinical setting. Antibody responses are known to correlate with protection against CHIKV (29–36), but CD4 T cell responses may also be important (37–39). Thus, it was of importance to determine the potential immunological differences that our vaccine candidate might display.
Our first vaccine candidate, Δ5, is an attenuated strain of CHIKV carrying a stable large deletion in the nsP3 replicase–encoding region. This vaccine candidate generated very strong, durable, and protective antibody responses upon a single immunization. These antibody levels did not decline over the following months after immunization (81 and 294 days in the vaccine and safety studies, respectively), suggesting a very robust initial response with a good memory potential. The fact that this vaccine also generated significant CD154-specific CD4 T cell responses required for promoting B cell maturation and development of long-lived memory B cells underscores the potency of this vaccine candidate. Upon challenge, vaccinated animals did not display any anamnestic response, suggesting that even 10 months after immunization, the antibody levels were high enough to prohibit CHIKV replication. Individuals that have suffered from CHIKV infection and disease tend to be protected for many years, perhaps even for life. It is therefore noteworthy that the attenuated Δ5 vaccine virus generated similar antibody levels as were induced after infections with WT virus.
An attenuated CHIKV strain, TSI-GSD-218 (181/clone 25), was clinically evaluated years ago and showed good potency with generation of long-term memory. However, its further development was abandoned due to safety concerns, assumingly because the vaccine could cause arthralgia (40, 41). However, the number of volunteers was low; thus, it is difficult to determine causality. If this attenuated vaccine was unsafe, it could be attributed to reversions, as the attenuation was based on only 2-point mutations (42). In the case of the Δ5 virus, we have shown that reversions do not occur, probably due to the large deletion. Collectively, the results obtained with both of these CHIKV vaccine candidates suggest that an attenuated virus vaccine strategy, should it prove safe, would make a very potent vaccine (43, 44).
The D vaccine, a DNA-launched RNA replicon deficient in virion formation, was less immunogenic than the two other regimens, although it also provided full protection from CHIKV infection. We have previously shown that a single dose of D protects mice from a stringent challenge (10), but in this study, we only were able to evaluate protection following two doses (DD) of the vaccine. Thus, it remains to be demonstrated whether this vaccine platform is able to protect after a single immunization. We did observe an anamnestic response upon challenge with WT LR-CHIKV, but this could be the result of a very limited virus replication or due to a reaction against the incoming virus protein antigen inoculum. The complete absence of viremia or any other pathological signs upon challenge suggests that, if there was any replication of the incoming challenge virus, it must have been very restricted. This conclusion is further supported by the lack of anamnestic T cell responses and the absence of any upregulated cytokines normally associated with WT CHIKV infection. The successful results with the DREP platform show that a DNA launched replicon approach mimicking an infection by an RNA virus could be an alternative to conventional DNA vaccines, as significantly lower amounts of DNA (and number of doses) would be needed to achieve strong protective immune responses (4, 14, 45). A conventional DNA vaccine expressing the envelope antigens of CHIKV, a construct very similar to our D in terms of insert, has been previously evaluated in nonhuman primates (33). In that particular study, 5 immunizations, each with 1,000 μg of the CHIKV-DNA vaccine, were required to achieve significant neutralizing antibody titers compared with one immunization of 200 μg in the data presented here. This interpretation should be considered in the light of the differences in animals (rhesus vs. cynomolgus macaques) and neutralization assays used in the two studies. However, similar results from others do suggest that a replicon derivative of a DNA vaccine may be much more potent (4).
MVA in itself represents a strong vaccine platform (46, 47). However, our previous findings that a DM prime-boost gives unprecedented strong immune responses (10, 11, 14) prompted us to choose to evaluate the heterologous DM combination in particular, since we were restricted in the number of animals available. Indeed, this regimen resulted in peak antibody responses comparable to those obtained with Δ5 and with superior T cell responses. Hence, the DM is promising as a combined CHIKV vaccine candidate protocol. Polyfunctional T cells may be important to combat long-term persistence of the CHIKV in macrophages (13, 38). It will be important to study in what way the immune responses differ between vaccine regimens. In nonhuman primates (this study) and in mice (10), the DM regimen generated approximately 10-fold stronger antibody and T cell responses compared with the homologous reciprocals. From a qualitative viewpoint, a single B cell epitope in the E2 envelope protein is mutually targeted by humans, macaque, and mice (amino acid 3,033–3,066), while other epitopes are species specific. Specifically, in nonhuman primates, WT CHIKV induces responses to E2 epitopes 2,961–2,978; 2,985–3,002; and 3,025–3,066 (48). In contrast, in mice, WT CHIKV and Δ5 target the E2 3,033–3,066; 3,113–3,138; and 3,185–3,210 epitopes (10). However, the DD, MM, and DM regimens target the 3,033–3,066 and 3,113–3,138 epitopes (10). These epitopes are located in three different domains of the E2 protein (5).
The Δ5 virus vaccine was clearly our top candidate in terms of immunogenicity, but use of a live attenuated virus will be dependent on its safety profile. For this reason, we conducted a separate safety study in which the Δ5 and WT CHIKV viruses were compared on the basis of a number of criteria and the following key observations were made: (i) the Δ5 virus infection resulted in viral loads that were several orders of magnitude lower than for WT virus, and the onset of viremia was delayed; (ii) Δ5 virus did not induce any fever in contrast to WT virus; (iii) Δ5 virus did not result in lymphopenia or increased levels of monocytes so typical for WT CHIKV infection; (iv) Δ5 virus did not display an increase in cytokine production commonly associated with CHIKV infection and inflammation; and (v) Δ5 infected animals did not show any clinical signs of infection such as edema, erythema, joint swelling, hunching, fur ruffling, or rash that were observed in WT-infected animals.
A potential concern with certain virus vaccines is the possibility that vaccination could exacerbate disease upon reinfection later. In our study, we did not have any indication of such exacerbation. Vaccination neither had any effect on viral replication capacity upon challenge nor on cytokine upregulation, fever, lymphopenia, monocytosis, or other clinical signs.
In the present study, it was not possible to determine mononuclear cell infiltration as a possible manifestation of degree of arthralgia, because joint swelling is very faint and does not normally occur when animals are challenged by i.v. route. Swelling may be reproducible and quantified in the wrist of animals inoculated by SCw route. Thus, despite the low number of animals we inoculated by this way (see Table 1), we clearly showed that the attenuated Δ5 virus we use as a vaccine does not induce wrist swelling compared with the negative control (PBS + inactivated virus) or to the positive control (WT virus). Furthermore, arthralgia manifests itself as pain of the joints, but it is hard to assess the degree of pain in animals we manipulated after anesthesia, taking into account that, in humans, its degree of severity is more dependent on self-perception than external scaling.
Recently, two clinical trials were conducted evaluating two CHIKV vaccine platforms, one based on virus-like particles (VLPs) (29, 49) and the other on a recombinant measles virus (50, 51). Both were immunogenic and safe but required at least two immunizations to achieve substantial antibody responses. The attenuated Δ5 virus and the previously evaluated TSI-GSD-218 attenuated vaccine candidates both generated antibody responses that have persisted for about a year. It remains to be determined whether the immune responses generated by any of the CHIKV vaccine candidates tested to date are sufficient to be protective against a CHIKV infection that could occur after several years after vaccination. For any practical purposes, it certainly appears that WT CHIKV infection results in life-long protection; thus, an attenuated vaccine approach seems most promising. Our results regarding the safety profile of the attenuated Δ5 virus vaccine candidate provide a good basis for moving forward.
Methods
CHIKV and vaccine candidates.
The Δ5, D, and M vaccine candidates have been described (8–10). The stability of the 180 nucleotides long deletion in Δ5 was originally determined after 5 passages in Vero cells (9), and herein, the analysis was extended to include 10 passages. The D replicon DNA vaccine (10) is derived from the WT CHIKV infectious clone after deletion of the capsid encoding region. Upon immunization, virions will not form, as the capsid protein needed for packaging of replicon RNAs is absent. M (previously named MVA-CHIKV, ref. 8) was constructed by inserting the cDNA encoding the structural polyprotein of CHIKV into MVA (8).
Nonhuman primate model.
To evaluate the CHIKV vaccines, we used cynomolgus macaques, Macaca fascicularis (13). No randomization was used; however, the groups were balanced for MHC haplotypes to avoid the susceptible and resistant bias (52, 53). Sample size was chosen knowing that 100% infection was expected in the control group based on previous titration of the challenge stock. In CHIKV-infected animal fever, joint swelling and rash were monitored at each sampling date. Adult cynomolgus macaques (3–4 years old, body weights ranging between 3 kg and 5 kg) were imported from international accredited breeding facilities on Mauritius Island and housed in the Atomic Energy Commission (CEA) facilities in Fontenay-aux-Roses, France. Animals were sedated with ketamine (10 mg/kg, Rhône-Mérieux) before handling. Clinical examinations were performed, and rectal temperature and weight were recorded 10 minutes after sedation, before immunization, challenge or bleeding.
Immunization and challenge.
The animals were inoculated as follows: (i) The first group received 1 × 105 PFU of Δ5 virus by a single s.c. injection of 0.4 ml in the right upper back side on day 42. (ii) The second group of animals received 200 μg of the D vaccine by intradermal (i.d.) injection followed by electroporation (EP) on days 0 (4 inoculations of 0.1 ml in right back side) and day 42 (4 inoculations of 0.1 ml in left back side). The i.d. injections were performed by the Mantoux method (54) using syringes for i.d. skin test. EP using a CU21 EDIT Nepagen were performed after the 4 i.d. injections (55). (iii) The third group of animals was primed on day 0 with 200 μg of D followed by an i.m. injection of 1 × 108 PFU of M on day 42. (iv) The fourth group of animals received 0.9% NaCl injections i.d. followed by EP on day 0 and then 0.9% NaCl s.c., i.m., and i.d. injections followed by EP on day 42. For challenge, the CHIKV strain LR2006-OPY1 stock was used as described (13). The AID50 was estimated at 7.07 × 103 ± 3.15 × 103 viral RNA (vRNA) copies using the method of Spouge (56). Macaques were challenged i.v. (saphenous vein) with 100 AID50 corresponding to 7,000 – 10,000 PFU (16). The in vitro titer of the used stock of CHIKV strain LR2006-OPY1 was 1.8 × 108 ± 0.5 × 108 PFU/ml in Vero cells, which corresponded to 1.8 × 1010 ± 0.9 × 1010 vRNA equivalents per ml.
Safety study.
All animals in the immunogenicity study receiving either WT or ΔnsP3 virus were also used to assess safety of the ΔnsP3 vaccine candidate. This includes the clinical assessment (Table 1) and levels of cytokine expression. Local effects of virus injection was studied in macaques that were infected with WT or ΔnsP3 CHIKV by SCw. This allowed us to regularly observe local clinical signs occurring in the same area as used for the injection.
Determination of plasma viremia.
qPCR and viral titration were performed as described (16). For measurement of viral load, viral RNA was prepared from 100 μl of EDTA anticoagulated, cell-free plasma, using the Nucleospin 96 Virus Kit (Macherey Nagel) following the manufacturer’s instructions. CHIKV RNA diluted in an EDTA plasma sample from CHIKV-noninfected macaques was used to generate a standard curve by serial 10-fold dilutions. Extracted RNAs (10 μl) were subjected to reverse transcription using the Superscript One-Step RT-PCR kit (Invitrogen) following the manufacturer’s recommendations as previously described (13). qPCR was performed in an iCycler real-time thermocycler (Bio-Rad) with the following cycling conditions: 30 minutes at 56°C, then 5 minutes at 95°C, followed by 50 cycles at 95°C for 15 seconds and 60°C for 1 minute. The standard RNA template dilution showed a correlation coefficient of 98-99% over 7 orders of magnitude, with a limit of detection (LOD) of 300 copies of vRNA/ml or 3 copies per tested samples in 100 μl. For negative samples at day 3 and 4 after challenge of the protected animals, a second PCR was done on another serum sample of 500 μl of plasma, followed by elution in 50 μl of the RNA to reduce the LOD to 30 copies of vRNA/ml.
Detection of antibodies.
ELISA plates (Maxisorp, Nunc) were coated with 2 μg/ml recombinant soluble CHIKV envelope (p62-E1) protein ectodomain (5, 9) diluted in PBS and stored at least overnight at 4ºC. Plates were first rinsed 3 times with PBS-Tween (0.05%) and blocked with PBS containing 5% dry milk for 1 hour at 37°C. Sera were inactivated for 45 minutes at 56°C and diluted in PBS containing 2.5% milk. Then, sera were further diluted in 3-fold serial dilutions before the plates were incubated overnight at 4ºC. The plates were then washed 3 times with PBS-Tween 0.05% before HRP-conjugated mouse anti-monkey IgG (Southern Biotechnology) diluted 1/5,000 in PBS containing 1.25% milk (Gibco/Invitrogen) was added to each well. Plates were incubated at room temperature for 1.5 hours and washed 5 times with PBS-Tween before o-phenylenediamine dihydrochloride (OPD) (Sigma-Aldrich) was added. The reaction was stopped by addition of 1 M HCl after 15 minutes, and the absorbance was read at 490 nm. CHIKV neutralization assays were performed essentially as described using CHIKV VRPs expressing Gaussia luciferase (Gluc) (57, 58). Neutralization potency was determined as percentage of measured Gluc activity compared with Gluc readout after VRP application without serum.
Helper vectors encoding for capsid and envelope proteins of CHIKV from the recent CB outbreak were based on a sequence of the structural region of virus isolated in St. Martin, Antilles, France in December 2013. The amino acid sequence of the structural polyprotein of this isolate is highly similar to that of other viruses isolated from the South Pacific/CB outbreak (59). The appropriate fragments of the synthetic cDNA were used to replace the regions encoding for the capsid protein in LR the helper vectors (57).
T cell assays.
T cell assays were performed on peripheral blood mononuclear cells (PBMC) from peripheral blood using Li-Heparin CPT vacutainer (BD Biosciences). After centrifugation (500 g, 30 minutes), recovered cells were washed with complete Roswell Park Memorial Institute medium (RPMI 1640 [Sigma-Aldrich]) supplemented with 5 % FCS (Laboratoires Eurobio), 2 mM L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (all from Gibco, Invitrogen) and then treated with red blood cell lysis buffer (Sigma-Aldrich) for 2 minutes, washed, and resuspended in complete RPMI medium. Viable cells were counted using a ViCell counter (Beckman-Coulter), and parts of them were frozen at 10 million cells/ml in heat-inactivated FCS, 10% DMSO and stored at -135°C. A bank of peptides (404 oligomers, 15 amino acids long with 9 amino acid overlap [MIMOTOPE]) covering the sequence of nonstructural protein nsP1 (2 pools) and structural proteins E1, E2, and Capsid (1 pool each) were used to assess the specific response of T cells.
ELISpot assay.
T cell responses were determined by IFN-γ IL-2 responses FluoroSpot (FS-2122-10 Monkey IFN-γ/IL-2 FluoroSpot kit from Mabtech) on freshly isolated PBMC. Briefly, FluoroSpot plates were saturated with 35% ethanol, washed, and coated overnight by incubation with monoclonal antibodies against monkey IFN-γ (clone GZ-4, Mabtech) and against monkey IL-2 (clones IL2M-I/249, Mabtech) at a concentration of 15 μg/ml in sterile phosphate-buffered saline (PBS, Laboratories Eurobio) at 4°C. Plates were washed 5 times with PBS and then blocked by incubation for 1 hour at 37°C with RPMI 1640 medium with glutamax-1 (Gibco, Paisley, UK) supplemented with 10% FCS. PBMC (2.5 × 105) were added to each well with 0.1 μg/ml of monoclonal antibody CD28-A (Mabtech). Peptide pools (described in the T cell assay paragraph) were added in duplicate to a final concentration of each peptide in the culture medium of 2 μM. PMA/ionomycin was used as positive control. Plates were incubated for 24 hours at 37°C in an atmosphere containing 5% CO2. After incubation, plates were washed 5 times with PBS. FITC anti–IFN-γ antibody (clone 7-B6-1-FS-FITC, Mabtech) and biotinylated anti–IL-2 antibody (IL2-II-biotin) were added at a concentration of 1:200 and 1 μg/ml, respectively, in 0.5% FCS in PBS; the plates were incubated 2 hours at 37°C. After 5 washes with PBS, incubation was performed with anti-FITC-Green (1:200, Mabtech) and SA-Red (1:200, Mabtech) diluted in 0.5% FCS in PBS for 1 hour at room temperature and washed 5 times with PBS. Spots were developed by adding fluorescence enhancer. Spots were counted with the FluoroSpot Reader ELRIFL04 (Autoimmun Diagnostika). Responses of at least 25 SFU/1 × 106 PBMCs and at least 4 times the background were regarded as positive.
Intracellular cytokine staining.
T cell responses measured the frequency of PBMCs expressing IL-2, IFN-γ, TNF-α (Becton Dickinson), and CD154 (BD Pharmingen) upon stimulation with nsP1, E1, and E2 and C peptide pools. Briefly, 1 × 106 to 2 × 106 thawed viable PBMCs were resuspended in 150 μl of complete medium containing 0.2 μg of each costimulatory antibody CD28 and CD49b (Becton Dickinson). Stimulation was performed in 96-well plates using 2 μM of each peptide pool or 800 ng of staphylococcal enterotoxin B (SEB, Sigma-Aldrich) (positive control) or medium alone (negative control). Brefeldin A (Sigma-Aldrich) was added to each well at a final concentration of 10 μg/ml, and the plates were incubated at 37°C, 5% CO2 overnight. Cells were then washed, stained with a viability dye (violet fluorescent reactive dye, Invitrogen), and fixed and permeabilized with the BD Cytofix/Cytoperm reagent. Permeabilized cell samples were stored at –80°C before staining (60). Antibody staining was performed in a single step following permeabilization. After 30 minutes of incubation on ice in the dark, cells were washed in BD Perm/Wash buffer. Cells were analyzed with Canto II (BD Biosciences) or LSR-II immediately after staining. FlowJo software was used for sample analysis. CD4+ T cells were first sorted by CD154 (CD40L, clone TRAP1, Becton Dickinson) expression.
Cytokine determination.
Plasma levels of selected cytokines and growth factors were measured using the Milliplex Non-Human Primate Cytokine Magnetic Bead Panel for 14 soluble markers: IFN-γ, IL-2, IL-10, Il12/23(p40), IL-15, IL-17, IL-1Rα, IL-1β, IL-4, IL-6, IL-8, MCP-1 (CCL2), TNF-α, and IL-18 (Millipore) according to the manufacturer’s instructions. Results were acquired with a Bio-Plex Instrument 200 and analyzed with Bio-Plex Manager Software version 6.1 (Bio-Rad). IFN-α2 was measured with IFN-α Cynomolgus/Rhesus ELISA kit (VeriKine, PBL-Assay Science). Cytokine data were statistically analyzed using R/Bioconductor. Cytokine expression levels are expressed as percentage of increase relative to baseline obtained at week 12 after infection. The sample and cytokine hierarchical clusterings were generated based on the Euclidean metric and based on the complete linkage method. The box plot and plot-jittering representations were generated using the ggplot2 library available on R/Bioconductor. The MDS representation was generated using the SVD-MDS algorithm (61). MDS methods aim to represent the similarities and differences among high-dimensionality objects into a space of lower dimensions, generally in 2 or 3 dimensions for visualization purposes.
Measurements of body temperature.
Body temperature was monitored using STAR ODDI chips (www.star-oddi.com) implanted s.c. in the upper back, between scapulae. Temperature was recorded each hour from 2 weeks before immunization and during all the following experiments after immunization/infection. Data were extracted from the chip using Mercury software. Rectal temperature was also manually assessed at time of each blood sampling as described (13)
Hematology.
Hematology (CBC) was performed using a HMX A/L (Beckman Coulter) on 185 μl of blood samples with EDTA vacutainer tubes (BD Biosciences). Measured parameters were count of white blood cells (WBC), red blood cells (RBC), platelets (PLT), hemoglobin (HGB), and hematocrit (HCT). Percentage and absolute counts of lymphocytes (LY), monocytes (MO), polynuclear cells (Neutrophile [NE]; basophils [BA], eosinophils [EO]. Red blood cell parameters (HGB, HCT, MCV, mean corpuscular hemoglobin [MCH], MCH concentration [MCHC], RBC distribution width [RDW]) and platelets (MPV, reticulocytes [RETC], immature reticulocyte fraction [IRF], and MRV) were also measured.
Statistics.
Statistical analysis was performed using the GraphPad Prism 6 software (GraphPad Software). A Kruskal-Wallis test was used for multiple comparisons, followed by a 2-tailed Mann-Whitney U test to analyze differences between two groups, and for comparisons between multiple groups, Dunn’s test for multiple comparisons was used. Wilcoxon rank test was used to analyze immune response kinetics. Significant differences were obtained for P < 0.05 (*), P < 0.01 (**) and P < 0.001 (***). All tests were performed with a 95% CI. For statistical analyses of T cell responses, the Kruskal-Wallis test followed by the Mann-Whitney U test were used to identify differences between groups. To compare the viremia over time, a 2-way ANOVA followed by Bonferroni’s multiple comparisons test were used for each individual time point.
Study approval.
Monkey studies were approved by the regional animal care and use committee (“Comite Regional d’Ethique sur l’experimentation animale Ile de France Sud”, Fontenay-aux-Roses, France) with approval numbers A10-024, A12-008, and A13-028. Nonhuman primates are used at the CEA in accordance with French national regulation, and CEA facilities are fully authorized under the number B-92-032-02 for animal use and under the number 2005-69 for nonhuman primate breeding. National veterinarian inspectors regularly inspect these facilities. The CEA is in compliance with ETS123 recommendations, European Union Directive 2010/63/CE, 2010 September 22nd, for animal breeding and with Standards for Human Care and Use of Laboratory Animals (Animal Welfare Assurance, OLAW number A5826-01). The monkeys were housed individually in stainless-steel squeeze-back cages (cage size following the directive 2010/63/CE and French law “décret 2013-118 from 2013 February 1st”) in a climate-controlled room (temperature 21°C ± 1°C and relative humidity 55% ± 5%) with a 12-hour light/dark cycle. In CHIKV-infected aniNonhuman primate inspected for any behavior by animal caretakers who report directly to the veterinarians in charge of the animal facilities and animal welfare. The head of the veterinarian staff is empowered to interrupt the protocol in case of animal pathology or suffering. Animals were daily feed ad libitum with specific food and fruits. Enrichments such as toys and special “hidden” foods were provided by staff that was not part of the study team. The studies were performed as a continuation of experiments performed at Karolinska Institutet that were approved by the local ethics committee (Stockholm’s Norra Djurförsöksetiska Nämnd), permit number N74/11.
Author contributions
PR, ME, A Merits, RLG, and PL conceived and designed the experiments. PR, KL, BMK, and DH performed the experiments. PR, KL, BMK, LG, NDB, NT, and PL analyzed the data. JGA, A Merits, ME, and A. Meinke contributed materials/reagents. PR, KL, NT, and PL wrote the manuscript with review and comments of all the authors.
Acknowledgments
This work was supported by the European Union FP7 project grant agreement no. 261202 (ICRES) and by the Swedish Research Council grant no. K2013-56X-09494-23-3. The project was hosted by the infectious disease models for innovative therapies (IDMIT) infrastructure supported by the ANR (Agence Nationale pour la Recherche, Paris, France) grant “Programme d’investissements d’avenir” N° ARN11-INBS-0008. We thank the technical expertise of staff at IDMIT.
Footnotes
DH’s present address is: Affibody AB, Solna, Sweden.
Conflict of interest: A. Meinke is an employee of Valneva Austria GmbH.
Reference information:JCI Insight. 2017;2(6):e83527. https://doi.org/10.1172/jci.insight.83527.
Contributor Information
Pierre Roques, Email: pierre.roques@cea.fr.
Karl Ljungberg, Email: Karl.Ljungberg@ki.se.
Leslie Gosse, Email: Leslie.Gosse@gmail.com.
Nathalie Dereuddre-Bosquet, Email: nathalie.bosquet@cea.fr.
Nicolas Tchitchek, Email: nicolas.tchitchek@cea.fr.
David Hallengärd, Email: david.hallengard@affibody.com.
Juan García-Arriaza, Email: jfgarcia@cnb.csic.es.
Andreas Meinke, Email: Andreas.MEINKE@valneva.com.
Mariano Esteban, Email: mesteban@cnb.csic.es.
Andres Merits, Email: andres.merits@ut.ee.
Roger Le Grand, Email: roger.le-grand@cea.fr.
Peter Liljeström, Email: Peter.Liljestrom@ki.se.
References
- 1.Rougeron V, Sam IC, Caron M, Nkoghe D, Leroy E, Roques P. Chikungunya, a paradigm of neglected tropical disease that emerged to be a new health global risk. J Clin Virol. 2015;64:144–152. doi: 10.1016/j.jcv.2014.08.032. [DOI] [PubMed] [Google Scholar]
- 2.Weaver SC, Lecuit M. Chikungunya virus and the global spread of a mosquito-borne disease. N Engl J Med. 2015;372(13):1231–1239. doi: 10.1056/NEJMra1406035. [DOI] [PubMed] [Google Scholar]
- 3.Delisle E, et al. Chikungunya outbreak in Montpellier, France, September to October 2014. Euro Surveill. 2015;20(17):17. doi: 10.2807/1560-7917.es2015.20.17.21108. [DOI] [PubMed] [Google Scholar]
- 4.Ljungberg K, Liljeström P. Self-replicating alphavirus RNA vaccines. Expert Rev Vaccines. 2015;14(2):177–194. doi: 10.1586/14760584.2015.965690. [DOI] [PubMed] [Google Scholar]
- 5.Voss JE, et al. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature. 2010;468(7324):709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- 6.Ahola T, et al. Therapeutics and vaccines against chikungunya virus. Vector Borne Zoonotic Dis. 2015;15(4):250–257. doi: 10.1089/vbz.2014.1681. [DOI] [PubMed] [Google Scholar]
- 7.Erasmus JH, Rossi SL, Weaver SC. Development of Vaccines for Chikungunya Fever. J Infect Dis. 2016;214(suppl 5):S488–S496. doi: 10.1093/infdis/jiw271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.García-Arriaza J, et al. A novel poxvirus-based vaccine, MVA-CHIKV, is highly immunogenic and protects mice against chikungunya infection. J Virol. 2014;88(6):3527–3547. doi: 10.1128/JVI.03418-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hallengärd D, et al. Novel attenuated Chikungunya vaccine candidates elicit protective immunity in C57BL/6 mice. J Virol. 2014;88(5):2858–2866. doi: 10.1128/JVI.03453-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hallengärd D, et al. Prime-boost immunization strategies against Chikungunya virus. J Virol. 2014;88(22):13333–13343. doi: 10.1128/JVI.01926-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knudsen ML, et al. Kinetic and phenotypic analysis of CD8+ T cell responses after priming with alphavirus replicons and homologous or heterologous booster immunizations. J Virol. 2014;88(21):12438–12451. doi: 10.1128/JVI.02223-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoarau JJ, et al. Persistent chronic inflammation and infection by Chikungunya arthritogenic alphavirus in spite of a robust host immune response. J Immunol. 2010;184(10):5914–5927. doi: 10.4049/jimmunol.0900255. [DOI] [PubMed] [Google Scholar]
- 13.Labadie K, et al. Chikungunya disease in nonhuman primates involves long-term viral persistence in macrophages. J Clin Invest. 2010;120(3):894–906. doi: 10.1172/JCI40104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knudsen ML, et al. Superior induction of T cell responses to conserved HIV-1 regions by electroporated alphavirus replicon DNA compared to that with conventional plasmid DNA vaccine. J Virol. 2012;86(8):4082–4090. doi: 10.1128/JVI.06535-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roy CJ, et al. Chikungunya vaccine candidate is highly attenuated and protects nonhuman primates against telemetrically monitored disease following a single dose. J Infect Dis. 2014;209(12):1891–1899. doi: 10.1093/infdis/jiu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borgherini G, et al. Outbreak of chikungunya on Reunion Island: early clinical and laboratory features in 157 adult patients. Clin Infect Dis. 2007;44(11):1401–1407. doi: 10.1086/517537. [DOI] [PubMed] [Google Scholar]
- 17.Chow A, et al. Persistent arthralgia induced by Chikungunya virus infection is associated with interleukin-6 and granulocyte macrophage colony-stimulating factor. J Infect Dis. 2011;203(2):149–157. doi: 10.1093/infdis/jiq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gardner J, et al. Chikungunya virus arthritis in adult wild-type mice. J Virol. 2010;84(16):8021–8032. doi: 10.1128/JVI.02603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelvin AA, et al. Inflammatory cytokine expression is associated with chikungunya virus resolution and symptom severity. PLoS Negl Trop Dis. 2011;5(8):e1279. doi: 10.1371/journal.pntd.0001279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng LF, et al. IL-1beta, IL-6, and RANTES as biomarkers of Chikungunya severity. PLoS One. 2009;4(1):e4261. doi: 10.1371/journal.pone.0004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schilte C, et al. Type I IFN controls chikungunya virus via its action on nonhematopoietic cells. J Exp Med. 2010;207(2):429–442. doi: 10.1084/jem.20090851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teng TS, et al. A Systematic Meta-analysis of Immune Signatures in Patients With Acute Chikungunya Virus Infection. J Infect Dis. 2015;211(12):1925–1935. doi: 10.1093/infdis/jiv049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wauquier N, Becquart P, Nkoghe D, Padilla C, Ndjoyi-Mbiguino A, Leroy EM. The acute phase of Chikungunya virus infection in humans is associated with strong innate immunity and T CD8 cell activation. J Infect Dis. 2011;204(1):115–123. doi: 10.1093/infdis/jiq006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venugopalan A, Ghorpade RP, Chopra A. Cytokines in acute chikungunya. PLoS One. 2014;9(10):e111305. doi: 10.1371/journal.pone.0111305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Draize JH, Woodard G, Calvery HO. Methods for the study of irritation and toxicity of substances applied topically to the skin and mucous membranes. J Pharmacol Exp Ther. 1944;2:377–390. [Google Scholar]
- 26.García-Sastre A, Mena I. Novel vaccine strategies against emerging viruses. Curr Opin Virol. 2013;3(2):210–216. doi: 10.1016/j.coviro.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Powers AM. Chikungunya virus control: is a vaccine on the horizon? Lancet. 2014;384(9959):2008–2009. doi: 10.1016/S0140-6736(14)61290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weaver SC, Osorio JE, Livengood JA, Chen R, Stinchcomb DT. Chikungunya virus and prospects for a vaccine. Expert Rev Vaccines. 2012;11(9):1087–1101. doi: 10.1586/erv.12.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akahata W, et al. A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nat Med. 2010;16(3):334–338. doi: 10.1038/nm.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu H, et al. Deciphering the protective role of adaptive immunity to CHIKV/IRES a novel candidate vaccine against Chikungunya in the A129 mouse model. Vaccine. 2013;31(33):3353–3360. doi: 10.1016/j.vaccine.2013.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fric J, Bertin-Maghit S, Wang CI, Nardin A, Warter L. Use of human monoclonal antibodies to treat Chikungunya virus infection. J Infect Dis. 2013;207(2):319–322. doi: 10.1093/infdis/jis674. [DOI] [PubMed] [Google Scholar]
- 32.Goh LY, et al. Neutralizing monoclonal antibodies to the E2 protein of chikungunya virus protects against disease in a mouse model. Clin Immunol. 2013;149(3):487–497. doi: 10.1016/j.clim.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 33.Mallilankaraman K, et al. A DNA vaccine against chikungunya virus is protective in mice and induces neutralizing antibodies in mice and nonhuman primates. PLoS Negl Trop Dis. 2011;5(1):e928. doi: 10.1371/journal.pntd.0000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pal P, et al. Development of a highly protective combination monoclonal antibody therapy against Chikungunya virus. PLoS Pathog. 2013;9(4):e1003312. doi: 10.1371/journal.ppat.1003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plante K, et al. Novel chikungunya vaccine candidate with an IRES-based attenuation and host range alteration mechanism. PLoS Pathog. 2011;7(7):e1002142. doi: 10.1371/journal.ppat.1002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Selvarajah S, et al. A neutralizing monoclonal antibody targeting the acid-sensitive region in chikungunya virus E2 protects from disease. PLoS Negl Trop Dis. 2013;7(9):e2423. doi: 10.1371/journal.pntd.0002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakaya HI, Gardner J, Poo YS, Major L, Pulendran B, Suhrbier A. Gene profiling of Chikungunya virus arthritis in a mouse model reveals significant overlap with rheumatoid arthritis. Arthritis Rheum. 2012;64(11):3553–3563. doi: 10.1002/art.34631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poo YS, et al. Multiple immune factors are involved in controlling acute and chronic chikungunya virus infection. PLoS Negl Trop Dis. 2014;8(12):e3354. doi: 10.1371/journal.pntd.0003354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teo TH, et al. A pathogenic role for CD4+ T cells during Chikungunya virus infection in mice. J Immunol. 2013;190(1):259–269. doi: 10.4049/jimmunol.1202177. [DOI] [PubMed] [Google Scholar]
- 40.Edelman R, Tacket CO, Wasserman SS, Bodison SA, Perry JG, Mangiafico JA. Phase II safety and immunogenicity study of live chikungunya virus vaccine TSI-GSD-218. Am J Trop Med Hyg. 2000;62(6):681–685. doi: 10.4269/ajtmh.2000.62.681. [DOI] [PubMed] [Google Scholar]
- 41.Hoke CH, et al. US Military contributions to the global response to pandemic chikungunya. Vaccine. 2012;30(47):6713–6720. doi: 10.1016/j.vaccine.2012.08.025. [DOI] [PubMed] [Google Scholar]
- 42.Gorchakov R, et al. Attenuation of Chikungunya virus vaccine strain 181/clone 25 is determined by two amino acid substitutions in the E2 envelope glycoprotein. J Virol. 2012;86(11):6084–6096. doi: 10.1128/JVI.06449-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Plotkin SA, Orenstein WA, Offit PA. Vaccines. Philadelphia, PA: Saunders; 2013. [Google Scholar]
- 44.Plotkin SA, Plotkin SL. The development of vaccines: how the past led to the future. Nat Rev Microbiol. 2011;9(12):889–893. doi: 10.1038/nrmicro2668. [DOI] [PubMed] [Google Scholar]
- 45.Knudsen ML, Ljungberg K, Tatoud R, Weber J, Esteban M, Liljeström P. Alphavirus replicon DNA expressing HIV antigens is an excellent prime for boosting with recombinant modified vaccinia Ankara (MVA) or with HIV gp140 protein antigen. PLoS One. 2015;10(2):e0117042. doi: 10.1371/journal.pone.0117042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gómez CE, Perdiguero B, Garcia-Arriaza J, Esteban M. Poxvirus vectors as HIV/AIDS vaccines in humans. Hum Vaccin Immunother. 2012;8(9):1192–1207. doi: 10.4161/hv.20778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gómez CE, Nájera JL, Krupa M, Perdiguero B, Esteban M. MVA and NYVAC as vaccines against emergent infectious diseases and cancer. Curr Gene Ther. 2011;11(3):189–217. doi: 10.2174/156652311795684731. [DOI] [PubMed] [Google Scholar]
- 48.Kam YW, et al. Unique epitopes recognized by antibodies induced in Chikungunya virus-infected non-human primates: implications for the study of immunopathology and vaccine development. PLoS One. 2014;9(4):e95647. doi: 10.1371/journal.pone.0095647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang LJ, et al. Safety and tolerability of chikungunya virus-like particle vaccine in healthy adults: a phase 1 dose-escalation trial. Lancet. 2014;384(9959):2046–2052. doi: 10.1016/S0140-6736(14)61185-5. [DOI] [PubMed] [Google Scholar]
- 50.Brandler S, et al. A recombinant measles vaccine expressing chikungunya virus-like particles is strongly immunogenic and protects mice from lethal challenge with chikungunya virus. Vaccine. 2013;31(36):3718–3725. doi: 10.1016/j.vaccine.2013.05.086. [DOI] [PubMed] [Google Scholar]
- 51.Ramsauer K, et al. Immunogenicity, safety, and tolerability of a recombinant measles-virus-based chikungunya vaccine: a randomised, double-blind, placebo-controlled, active-comparator, first-in-man trial. Lancet Infect Dis. 2015;15(5):519–527. doi: 10.1016/S1473-3099(15)70043-5. [DOI] [PubMed] [Google Scholar]
- 52.Aarnink A, et al. Influence of the MHC genotype on the progression of experimental SIV infection in the Mauritian cynomolgus macaque. Immunogenetics. 2011;63(5):267–274. doi: 10.1007/s00251-010-0504-6. [DOI] [PubMed] [Google Scholar]
- 53.Blancher A, et al. Study of cynomolgus monkey (Macaca fascicularis) Mhc DRB gene polymorphism in four populations. Immunogenetics. 2012;64(8):605–614. doi: 10.1007/s00251-012-0613-5. [DOI] [PubMed] [Google Scholar]
- 54.Mantoux C. Intradermo-réaction de la tuberculine. Comp Rend Acad Sci. 1908;147:355–357. [Google Scholar]
- 55.Adam L, Le Grand R, Martinon F. Electroporation-mediated intradermal delivery of DNA vaccines in nonhuman primates. Methods Mol Biol. 2014;1121:309–313. doi: 10.1007/978-1-4614-9632-8_27. [DOI] [PubMed] [Google Scholar]
- 56.Spouge JL. Statistical analysis of sparse infection data and its implications for retroviral treatment trials in primates. Proc Natl Acad Sci USA. 1992;89(16):7581–7585. doi: 10.1073/pnas.89.16.7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gläsker S, et al. Virus replicon particle based Chikungunya virus neutralization assay using Gaussia luciferase as readout. Virol J. 2013;10:235. doi: 10.1186/1743-422X-10-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smerdou C, Liljeström P. Two-helper RNA system for production of recombinant Semliki forest virus particles. J Virol. 1999;73(2):1092–1098. doi: 10.1128/jvi.73.2.1092-1098.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lanciotti RS, Valadere AM. Transcontinental movement of Asian genotype chikungunya virus. Emerging Infect Dis. 2014;20(8):1400–1402. doi: 10.3201/eid2008.140268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guenounou S, Bosquet N, Dembek CJ, Le Grand R, Cosma A. OMIP-016: Characterization of antigen-responsive macaque and human T-cells. Cytometry A. 2013;83(2):182–184. doi: 10.1002/cyto.a.22233. [DOI] [PubMed] [Google Scholar]
- 61.Bécavin C, Tchitchek N, Mintsa-Eya C, Lesne A, Benecke A. Improving the efficiency of multidimensional scaling in the analysis of high-dimensional data using singular value decomposition. Bioinformatics. 2011;27(10):1413–1421. doi: 10.1093/bioinformatics/btr143. [DOI] [PubMed] [Google Scholar]